- 1Simon F.S. Li Marine Science Laboratory, School of Life Sciences, Chinese University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 2Institute for Advance Study, Shenzhen University, Shenzhen, China
- 3Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
- 4Department of Biology, Baptist University, Hong Kong, Hong Kong SAR, China
- 5Department of Ocean Science, The Hong Kong University of Science and Technology, Kowloon, Hong Kong SAR, China
- 6Southern Marine Science and Engineering Guangdong Lab (Guangzhou), Guangzhou, China
- 7Institute of Space and Earth Information Science, The Chinese University of Hong Kong, Shatin, Hong Kong SAR, China
- 8Hong Kong Branch of the Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), The Hong Kong University of Science and Technology, Kowloon, Hong Kong SAR, China
Introduction: Climate change has resulted in elevated sea surface temperature as well as increased frequency of extreme weather events, e.g. cyclones and rainstorms, which could lead to reduced seawater salinity. While temperature effects on corals have been widely examined, the combined effects of both temperature and salinity on corals, especially their early stages, remain poorly known. This study aimed to examine how the larvae of Acropora pruinosa in a marginal coral habitat, Hong Kong, respond to high temperature (+5°C ambient, HT), low salinity (26 psu, LS), and the combined effects of both stressors (HTLS).
Methods: We recorded larval survival and settlement success under different experimental treatments, and used RNA-Seq technique to compare the gene expression patterns of these larvae to understand the underlying molecular mechanism of stress responses.
Results: Our results showed that the survivorship of coral larvae was not affected in all experimental treatments, with all larvae surviving through the 72-hour period of the experiment. Yet, larval settlement was compromised under all stress treatments. The settlement rates were 39.3%, 12%, and 0% for the elevated temperature, reduced salinity, and the combined treatment, respectively, which were all significantly lower than that under the control treatment (78%). We demonstrated that low salinity (LS) triggered responsive gene sets with functions in ATP production, protein translation, and receptor for neuroactive ligands. In addition, high temperature (HT) treatment also triggered MAPK and NF-kB signaling and apoptosis in these coral larvae. The combined stressor treatment (HTLS) acted synergistically, resulting in the up-regulation of intracellular transducers that could trigger the intrinsic apoptosis pathway. This may explain the total failure in larval settlement under HTLS that could further increase larval vulnerability in the natural environment.
Discussion: Our results provide new insights into the molecular responses of coral larvae and represent an essential first step in expanding ourunderstanding of the mechanisms of tolerance that may be exhibited by coral larvae exposed to multiple stressors.
1 Introduction
The maintenance and persistence of coral reefs require a continuous supply of larvae that would disperse, settle and recruit successfully into the population and eventually become reproductive individuals. Larval tolerance to multiple stressors in every step of this recruitment process may therefore become a bottleneck for the sustainability of the coral population (Ritson-Williams et al., 2010; Byrne, 2011). Majority of spawning coral species produce aposymbiotic larvae (Baird et al., 2009) that are strongly dependent on the endogenous energy reserves within the spawned eggs provided by the parent generation (Harii et al., 2007). Due to this limited energetic reserve, larval responses to stressors are expected to involve trade-offs, i.e., tolerance to one stressor might affect sensitivity to another (Hofmann and Todgham, 2010), and effective responses to stressors might eventually decrease larval fitness to settle as energy stores are depleted. Therefore, it is important to investigate not only the responses of coral larvae to projected multiple stressors in the future oceans, but also to understand the underlying mechanisms behind these responses.
Mean temperature of global surface has risen over the past 150 years and the trend is expected to continue by a further 2.6-4.8°C in the 21st century (IPCC, 2013; Heron et al., 2016). As global warming could also lead to an increased frequency of intense cyclones and rainfall (Emanuel, 2005; Webster et al., 2005; Lee et al., 2011; Knutson et al., 2015), additional stressors like lowering of salinity could become a particular issue of concern. Under this scenario, co-occurrence of elevated temperature and lowered salinity is expected in the near future as a result of global climate change.
Studies have explored the effects of elevated temperature on physiological performance of the early stages of coral. High temperature has been shown to alter coral fertilization success (Negri et al., 2007; Puisay et al., 2018), accelerate cell cleavage and abnormal development during embryogenesis (Negri et al., 2007; Randall and Szmant, 2009a; Chui and Ang, 2015; Chui et al., 2016; Puisay et al., 2018), reduce survivorship (Randall and Szmant, 2009b; Heyward and Negri, 2010; Weeriyanun et al., 2022), and alter larval settlement rate (Nozawa and Harrison, 2007; Randall and Szmant, 2009a; Randall and Szmant, 2009b; Heyward and Negri, 2010; Weeriyanun et al., 2022).
While the effects of temperature stress on early stages of coral have been relatively well explored, there has been comparatively little focus on examining the salinity effect (Mayfield and Gates, 2007; Berkelmans et al., 2012). Reduction in salinity has been shown to reduce fertilization success (Richmond, 1993; Humphrey et al., 2008; Chui and Ang, 2015; Hédouin et al., 2015), increase developmental abnormality (Humphrey et al., 2008; Chui and Ang, 2015; Hédouin et al., 2015; Chui et al., 2016), and increase pre- and post-settlement mortality (Vermeij et al., 2006; Scott et al., 2013; Chui and Ang, 2017). Corals are known as osmoconformers (Hoegh-Guldberg and Smith, 1989). However, it is suggested that cell-mediated responses of corals to salinity fluctuation are achieved through the regulation of compatible organic osmolytes such as glycerol, free amino acids, and a polyol (Mayfield and Gates, 2007). Previous study on organic osmolytes in the coral Fungia scutaria larvae has revealed that glycine betaine accounted for more than 90% of the identified organic osmolytes-type solutes, followed by other amino acids (taurine and glycine) and myo-inositol (Hagedorn et al., 2010).
Although studies have tested the effects of temperature or salinity on early life stages of coral, the potential combined impact of these multiple stressors on early developmental stages of corals has only starting to attract increasing attention in the recent decade (Chui and Ang, 2015; Chui et al., 2016; Chui and Ang, 2017; Dias et al., 2019; Ding et al., 2022). Responses to multiple stressors are more complex than those elicited under single stressor scenario, because the extent of effective response to single stressor may change or be modified at the presence of other stressors, which often resulted in an entirely new perspective on the capacity of organisms to respond (Hofmann and Todgham, 2010). For instance, lowered temperature together with reduced salinity (i.e., -3°C ambient, 28 psu) could synergistically increase the chances of abnormal development in Acropora valida (Chui et al., 2016) whereas elevated temperature reversed it (antagonistic effect, i.e. +3°C ambient, 26 psu) in Platygyra acuta (Chui and Ang, 2015). At the molecular level, it has been reported that sea urchin larval development at low pH resulted in a compromised ability to express genes involved in cellular defense under heat stress, indicating synergistic effects of multiple stressors (O’Donnell et al., 2009). Interestingly, warming could reverse the down-regulation of proteins under low pH, resulting in an improved metamorphosis in oyster larvae (Dineshram et al., 2016). Such complex interplay of tolerance often makes consequences of environmental changes unpredictable. In this context, RNA-Seq analyses have shed some light on the overview of the transcriptomic changes of coral embryos/larvae under thermal stress, bringing to light more information about the underlying mechanisms of the organisms’ response. For example, genes involved in processes such as metabolism, translation, protein synthesis, protein degradation, Ca2+ ion transport, intracellular signal transduction, cytoskeleton organization, apoptosis, heat shock, oxidative stress are shown to be affected (Rodriguez-Lanetty et al., 2009; Voolstra et al., 2009; Portune et al., 2010; Polato et al., 2013; Xiao et al., 2018). As such, there is a pressing need to understand the capacity of coral larvae to respond to multiple-stressors, as well as to obtain detailed insights into the mechanism behind such responses at molecular levels.
In the present study, we explored the responses of larvae from a cosmopolitan coral species Acropora pruinosa from Hong Kong to multi-stressors. Hong Kong is located in southern China at the northern edge of the South China Sea. The large annual fluctuation in seawater temperature from 13°C in winter to 30°C in summer makes Hong Kong a marginal environmental for coral growth (Ang et al., 2005). Furthermore, Hong Kong corals are also subjected to large fluctuation in salinity because of the presence of large outflow from the Pearl River to the west of its territorial water as well as the monsoonal nature of its climate that brings in heavy seasonal rainfall (Yin, 2002). Fluctuation of seawater salinity from ambient 33 psu to 26 psu that persisted for more than a day in coral communities has been recorded after heavy rainfall during spawning season (Chui and Ang, 2015). Hong Kong corals are therefore periodically exposed to temperature and salinity stresses and their responses and tolerances, especially those of their larvae, could be quite unique when compared to those from more tropical areas (Chui and Ang, 2015). The aim of this study was therefore to identify changes in gene expression patterns in competent larvae of A. pruinosa exposed to normal (control) and stressful conditions, including (i) lowered salinity, (ii) elevated temperature, and (iii) combined temperature and salinity stresses, using RNA-Seq technique. Our results provide new insights into the transcriptional profiles of coral larvae and represent an essential first step in expanding our understanding of the mechanisms of tolerance that may be exhibited by coral larvae exposed to multiple stressors.
2 Materials and methods
2.1 Coral spawning, collection of gametes, and larval rearing
Experiments were conducted on competent larvae of hermaphroditic broadcasting A. pruinosa that spawned in the summer of 2014. Expected spawning dates and timing were determined according to its previously observed spawning pattern (Chui et al., 2014). Egg-sperm bundles were collected from multiple colonies from a depth of -1 to -2 m Chart Datum in A Ye Wan (AYW), Tung Ping Chau Marine Park (TPCMP), Hong Kong (22°32’N, 114°25’E), and transferred to a make-shift laboratory on shore. Subsequent embryonic and larval rearing procedures followed those described in Chui et al. (2014) and Omori and Iwao (2014). Briefly, the collected egg-sperm bundles from different colonies were mixed in a fertilization tank and left for an hour to allow for fertilization. After sperm wash, embryos were reared in culture tanks filled with freshly prepared 40-μm filtered seawater. Seawater was changed every day in the culture tanks to keep the larvae healthy until being transferred to the Marine Science Laboratory, Chinese University of Hong Kong four days after spawning. Larvae brought back were maintained in culture tanks filled with freshly prepared 40-μm filtered seawater until the experiment.
2.2 Stress experiment
A 72-h exposure period that tested the effects of temperature and salinity stress on larvae of A. pruinosa was conducted 6.5 days after spawning. Two levels of temperature (ambient 27 and high 32°C) and salinity (ambient 33 and low 26 psu) treatments were selected and arranged in the following factorial experimental designs: (i) ambient temperature + ambient salinity (Control); (ii) ambient temperature + low salinity (LS); (iii) high temperature + ambient salinity (HT); and (iv) high temperature + low salinity (HTLS). The temperature range used encompasses current and future projected temperature conditions. Temperature of 27°C is the mean ambient temperature of the spawning months in Hong Kong, whereas the elevated temperature of 32°C represented the highest projected increase (i.e. +5°C ambient) in sea surface temperature (SST) under one of the worst-case scenarios by 2100 (i.e. RCP8.5 range 2.6 - 4.8°C, IPCC, 2013). Persistence of 26 psu seawater for more than a day throughout the coastal water column in the study area was recorded in May 2014 due to the continuous discharge of underground seepage after a heavy downpour (Chui and Ang, 2015). The frequency of such incident is likely to increase given the projected increase in the occurrence of heavy precipitation due to global climate change (IPCC, 2013). Increasing frequency of coral larvae exposure to variation in salinity can therefore be anticipated and examining coral larval exposure to salinity stress is ecologically relevant.
The desired salinity level was prepared by diluting 0.22-µm filtered natural seawater (FSW) with a volume of milli-Q water. For control and treatments (each with three replicates), approximately 250 competent larvae were randomly allocated into Duran bottles each containing 500 ml of seawater with specific salinity. Duran bottles were placed in water baths equipped with thermostats to maintain the respective stable temperatures throughout the experiment. Seawater in each container was changed every 24-h with the seawater being pre-conditioned to the specific temperature for each treatment accordingly. No noticeable change in salinity was detected within each 24-h period. Much care was taken not to disturb the larvae during replacement of fresh seawater. After 72-h exposure, a sub-sample of 50 larvae was removed from the replicate containers of each treatment to be used for settlement assay, while the remaining larvae were flash frozen with liquid nitrogen and stored at -80°C for later gene expression analysis.
2.3 Survival
A set of experiments that tested the effects of temperature and salinity stress on larval survival was conducted in parallel with the 72-h exposure experiment. For control and treatments (each with four replicates) in this survival experiment, 20 competent larvae were randomly allocated into glass vials each containing 15 ml of seawater of specific salinity. These glass vials were placed in the same water baths with different temperatures as the stress exposure experiment. The number of live larvae was counted daily for three days thereafter. Seawater in each vial was refreshed after each counting with new seawater that was pre-adjusted to respective specific temperature and salinity.
2.4 Settlement assay
After 72-h of stress exposure period, beakers containing 50 ml of seawater with specific salinity, each housing a ceramic tile (pre-conditioned for approximately 1.5 months in situ), were deployed at the same water baths as the stress exposure experiment. The sub-samples of 50 competent A. pruinosa larvae that were removed from stress exposure culture were then added to each of the beakers for settlement assay. The number of settlers was examined after 24-h of settlement induction under dissecting microscope. A larva was quantified as a settler only when it had formed flattened disks of tissue or single polyps that were attached to the settlement tiles provided.
Statistical analyses were conducted with IBM SPSS version 19 for Windows (SPSS Inc., USA). Settlement results were given as mean ± standard error. Data were arcsine transformed and tested for normality using One-Sample Kolmogorov-Smirnov Test and for homogeneity of variances using Levene’s test. A two-factor analysis of variance (ANOVA) was then used to evaluate significant differences (p-value < 0.05) of the combined effects of seawater temperature and salinity on percent settlement. Pairwise comparisons were used to compare the treatment effects of either temperature or salinity alone at each level of the other factor. Bonferroni correction to the p-value was used to adjust the probability of type I error in multiple comparisons (Dunne, 2010).
2.5 Total RNA preparation and Illumina library preparation
Total RNA was extracted from approximately 200 flash frozen larvae from each treatment. These larvae were pulverized with a clean pre-chilled glass tissue grinder in 1 ml TRIzol® Reagent (Invitrogen, USA). Total RNAs were extracted following the manufacturer’s protocol, with modification in the RNA precipitation step where ½ volume of high salt solution (1.2M sodium chloride and 0.8M sodium citrate) and ½ volume of pre-chilled isopropanol were added to the retrieved aqueous phase solution. Following isolation, RNA pellets were re-suspended in diethylpyrocarbonate (DEPC)-treated water and stored in -80°C until cDNA synthesis. RNA concentration (ng/µL) and purity (A260/A280 ratio) were estimated using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, USA), and its integrity was confirmed through agarose gel electrophoresis.
In cDNA library preparation for Illumina sequencing, a total of 12 separated libraries were constructed using total RNA (5 µg) from individual replicates of the four treatment samples. RNAs were re-precipitated by adding 1/5 volume of 10M lithium chloride at -20°C for an hour. RNA pellets were then re-suspended in DEPC-treated water. cDNA libraries were prepared using Illumina Truseq RNA Sample Preparation kit v2 (Illumina, USA). Briefly, poly(A) mRNA was isolated with Oligo (dT) attached magnetic beads and heat-fragmented into small pieces to avoid priming bias. Using these short fragments as templates, double-stranded cDNA was synthesized using random hexamer primers with the SuperScript® Double-Stranded cDNA Synthesis Kit (Invitrogen, USA). The synthesized cDNA fragments were then subjected to an end-repair process, addition of single ‘A’ base, and ligation of the sequencing adapters according to Truseq RNA Sample Preparation kit v2 protocol.
2.6 Illumina sequencing, de novo assembly, and functional annotation
Illumina sequencing, transcriptome de novo assemblies and functional annotations were performed by the Novogene. Illumina sequencing was carried out using a HiSeq™ 2000 (Illumina, USA) in paired-end mode with a read length of 125 bp. cDNA libraries from the 12 samples were sequenced separately and 2.5-3 Gbp clean data (~20 million clean reads) was targeted for each individual library. Prior to assembly, sequence data (raw reads) that contain adaptor sequences, more than 5% unknown nucleotides, or more than 20% low quality bases (base quality value ≤ 10) were discarded using the NGS QC toolkit package (version 2.3) (Patel and Jain, 2012). The reference transcriptome assembly was generated from de novo assembling of all the clean reads from each sample with the short reads assembling program Trinity (release-20130225) (Grabherr et al., 2011). All the resulting transcript sequences from all 12 sample transcriptomes were then further processed, considering sequence splicing and redundancy removing with sequence clustering software to acquire non-redundant transcripts as long as possible. Transcripts with similarities exceeding 70% were assigned into one gene family cluster, with the assigned prefix “CL” followed by cluster id. Other transcripts that remain to be singletons were assigned with the prefix “Unigene”.
Gene annotations of unigenes were performed with NCBI non-redundant (nr) protein database (NR), Swiss-Prot, KEGG, and COG protein databases using Blastx alignment, with the minimum e-value score set to 0.00001. The best alignment results were used to decide the coding region sequences and to further decide the frame direction of the unigenes. ESTScan 2 (Iseli et al., 1999; Lottaz et al., 2003) was applied to unigenes that could not be oriented by Blastx alignments. cDNA sequences with reading frame that translated to < 20 amino acids were discarded. To check the completeness of the translated protein database, BUSCO v. 5.2.2. (Simão et al., 2015) was performed. The database “metazoan_odb10” (as of 2020-09-10) was selected as the reference base. With NR annotations, the Blast2GO program (Conesa et al., 2005) was applied to assign Gene Ontology (GO) of the annotated transcripts. Taxonomical assignment of annotated transcripts was performed using the LCA assignment algorithm in MEGAN v5.10.5 (Huson et al., 2011) based on the top 20 hits for each transcript in the nr database.
2.7 Mapping of sequence reads
Clean sequence reads from each sample specific read library were mapped (aligned) against the reference transcriptome (the annotated assembly of transcriptome derived from all samples) with the alignment program Bowtie2 (Langmead and Salzberg, 2012) to produce the raw gene expression count data for further analysis. The number of aligned reads per transcript was counted using SAMtools (v2.0.1) (Quinlan and Hall, 2010). The gene expression levels in each sample are presented as FPKM (fragments per kilobase of transcript per million mapped reads) (Mortazavi et al., 2008). The FPKM method normalized the raw reads of each transcript to eliminate biases of different transcript length and sequencing level on the calculation of gene expression.
2.8 Differential expression gene (DEG) analysis
To test for DEGs, all calculations and statistical tests were conducted using the R statistical software. As quantification accuracy is positively correlated with sequence coverage (Garber et al., 2011), only the transcripts that have at least five mapped reads in at least one sample were selected for differential expression analysis (Sun et al., 2013). Differential expression analysis was conducted using edgeR (Robinson et al., 2010). The three replicate samples from each treatment were used to generate mean expression levels with associated variances. Expression differences in three pairwise comparisons between larval cultured under control and three treatments were performed (using student’s t-test): (i) Control vs. LS; (ii) Control vs. HT, and (iii) Control vs. HTLS. In each comparison, the mean and standard deviation of the log2-fold change of expression of each transcript (TPM ratio) was calculated. To ensure a high accuracy in identification of DEGs, false discovery rate (FDR) was controlled at 0.01% (FDR ≤ 0.001) (Benjamini and Yekutieli, 2001).
2.9 Enrichment analyses and protein-protein interaction network reconstruction
The quality of sequence annotation is limited by the accuracy of information provided in any database used. KEGG database, a collection of proteins from well-characterized and ubiquitous biochemical pathways, is used as the most accurate database for functional annotation (Dunlap et al., 2013). To investigate the dominant gene function involved in the responses, enrichment of particular KEGG pathways among the DEGs was tested. To avoid any biological contaminants affecting the analysis, only DEGs with blast annotation under Metazoa were selected for enrichment analysis. Cumulative hypergeometric distribution tests were conducted in BLAST2GO to extract significantly over-represented KEGG terms in DEGs shortlisted in each treatment compared to the reference transcriptome dataset. Enrichment for KEGG pathway was determined based on the p-value ≤ 0.005 to evaluate whether any processes were more highly represented than expected by chance. Enrichment analyses were carried out for the set of DEGs shortlisted (significantly up and down-regulated transcripts) in each treatment.
Annotated DEGs were selected for reconstruction of protein-protein interaction network. Human annotation (gene symbol) of the DEG was submitted to STRING (Szklarczyk et al., 2015) and the human PPI database was used as the reference for PPI network reconstruction. The reconstructed PPI networks were exported and downloaded as a tab separated values (tsv) file. Network visualization and graph optimization was conducted in cytoscape v.3.9.0 (Shannon et al., 2003).
3 Results
3.1 Larval survival and settlement
No larval mortality was observed in the control and treatment sets at either the larval survival experiment or during the larval settlement experiment. Highest settlement rate was achieved under control (78.0 ± 3.5%), followed by 39.3 ± 7.9% under HT, 12.0 ± 4.2% under LS, and total failure (0%) under HTLS (Figure 1). Results of two-way ANOVA (Table 1) demonstrated a statistically significant main effect of temperature [F(1,8) = 41.00, p < 0.001], a main effect of salinity [F(1,8) = 145.91, p < 0.001], but the interactive effect between temperature and salinity was not significant [F(1,8) = 0.35, p = 0.57].
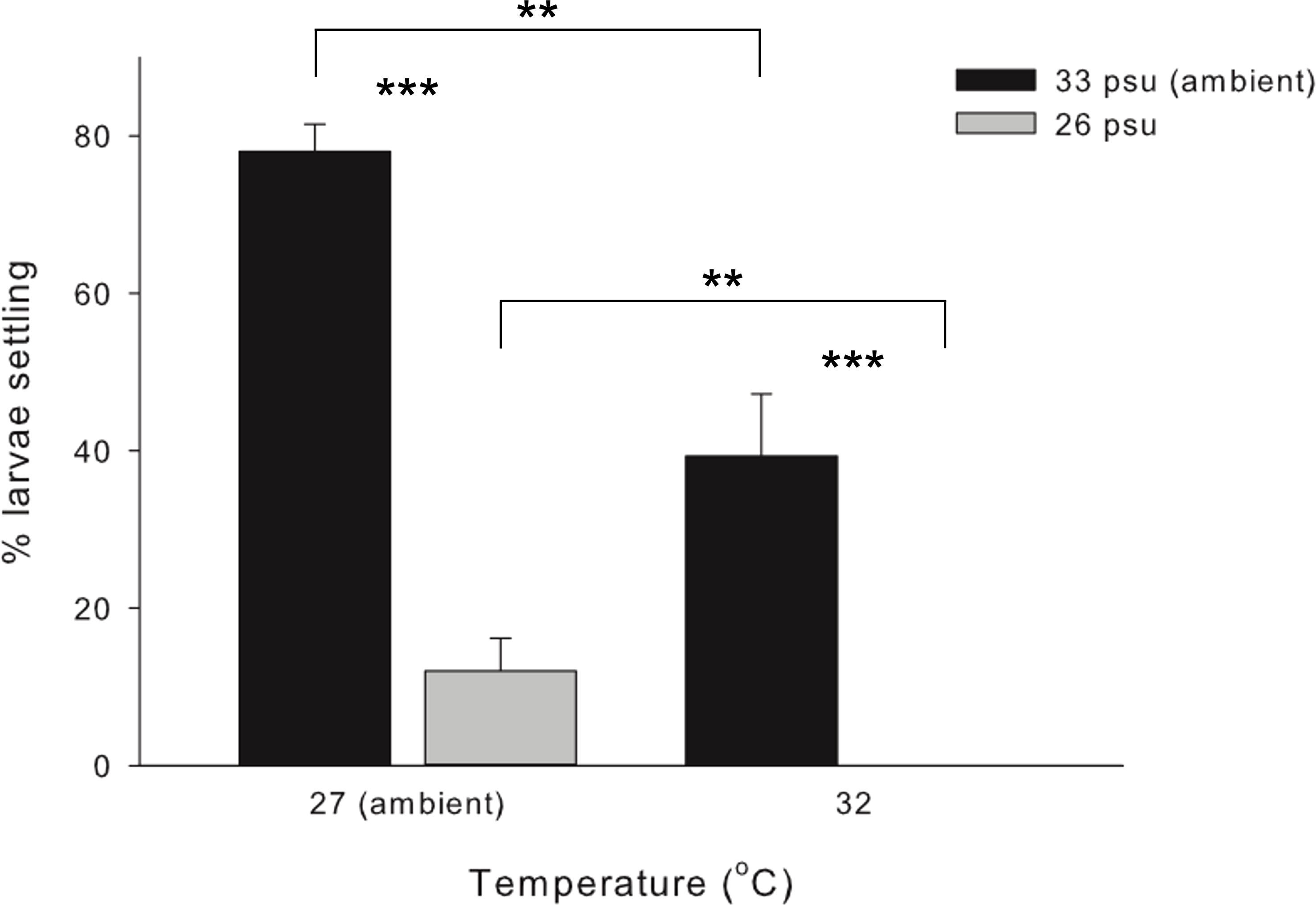
Figure 1 Mean (+ SE) percentage (%) of settlers of the coral Acropora pruinosa in response to different temperature (27 and 32°C) and salinity (33 and 26 psu) treatments (n = 3 replicates per treatment). Results from Two-way ANOVA demonstrate a significant effect of temperature (p < 0.001) and salinity (p < 0.001), with no significant interaction among stressors (p = 0.569). Pairwise comparisons with Bonferroni corrections showed significant differences between temperature and salinity treatments (**p < 0.05, ***p < 0.001).
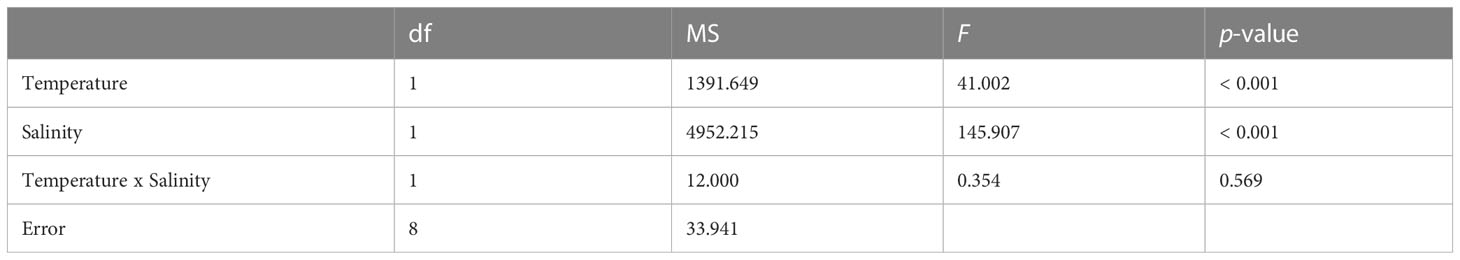
Table 1 Results of two-way ANOVA on differences in mean settlement (%) of Acropora pruinosa larvae under different temperature (27 and 32°C) and salinity (33 and 26 psu) treatments.
3.2 De novo assembly and functional annotation of Acropora pruinosa transcriptome
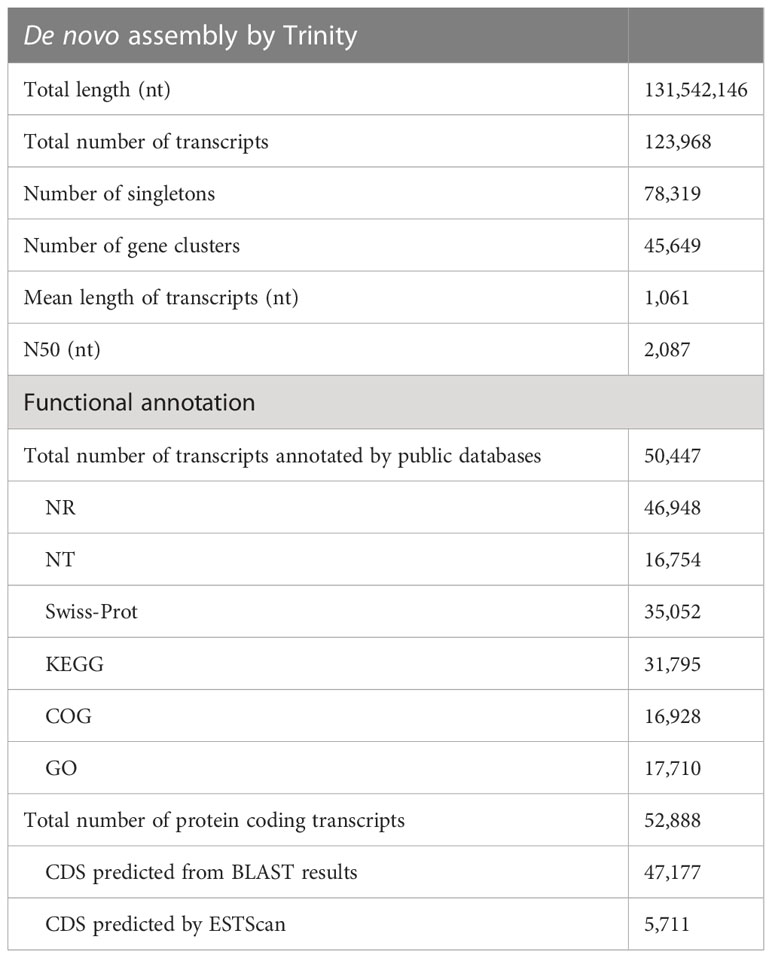
Illumina HiSeq sequencing yielded an average of 73 million raw reads from the triplicated samples from each of the control and treatment conditions (see Supplementary Table 1 for summary of Illumina sequencing). Over 91% (average of 67 million) of which remained after filtering. The reference transcriptome assembly consisted of 123,968 transcripts, which included a total of 45,649 clusters (group of transcripts representing the same gene or homologous gene) and 78,319 singletons (single gene). The average length of transcripts is 1061 bp and N50 value is 2087 bp. The length distribution of all transcripts is shown in Supplementary Figure 1. In total, 50,447 (40.69%) transcripts had at least one significant hit in the NR, Swiss-Prot, KEGG or COG protein database. The functional annotation results are shown in Table 2. The full annotation information is provided in Supplementary File 1. There were 17,710 NR-annotated transcripts assigned to major Gene Ontology (GO) categories, which included three categories: molecular function, cellular component, and biological process. The COG and GO functional categories for the annotated transcripts are shown in Supplementary Figure 2. BLAST and ESTscan search of all the aforementioned protein databases resulted in the prediction of 52,888 protein coding transcripts (Table 2). The translated protein database has a completeness of 90.4%, according to assessment by BUSCO (see full BUSCO report in Supplementary Table 2).
3.3 Differentially expressed gene analysis
We compared gene expression profiles between the control and the three treatment conditions (LS, HT and HTLS). Of the 123,968 transcript sequences detected, 89,786 (72.42%) transcripts have at least five clean reads mapped to at least one of the samples and were considered in the differential expression analysis. Applying such stringent filter can ensure high quantification accuracy among the abundantly expressed genes. By applying a 1-fold (or two times) differential expression and a q-value cutoff at 0.001, 3,997, 4,371, and 9,306 DEGs were identified in LS, HT and HTLS treatments, respectively. The majority of these DEGs were not annotated by nr. In the LS treatment stressor group, 2,670 up-regulated and 1,327 down-regulated DEGs were identified; in the HT single stressor treatment group, 3,430 up-regulated and 941 down-regulated DEGs were identified; in the HTLS double stressor treatment group, 7,050 up-regulated and 2,256 down-regulated DEGs were identified (Figure 2A). A complete list of up- and down-regulated DEGs of each treatment is provided in Files 2, 3, 4 of the Supplementary Information.
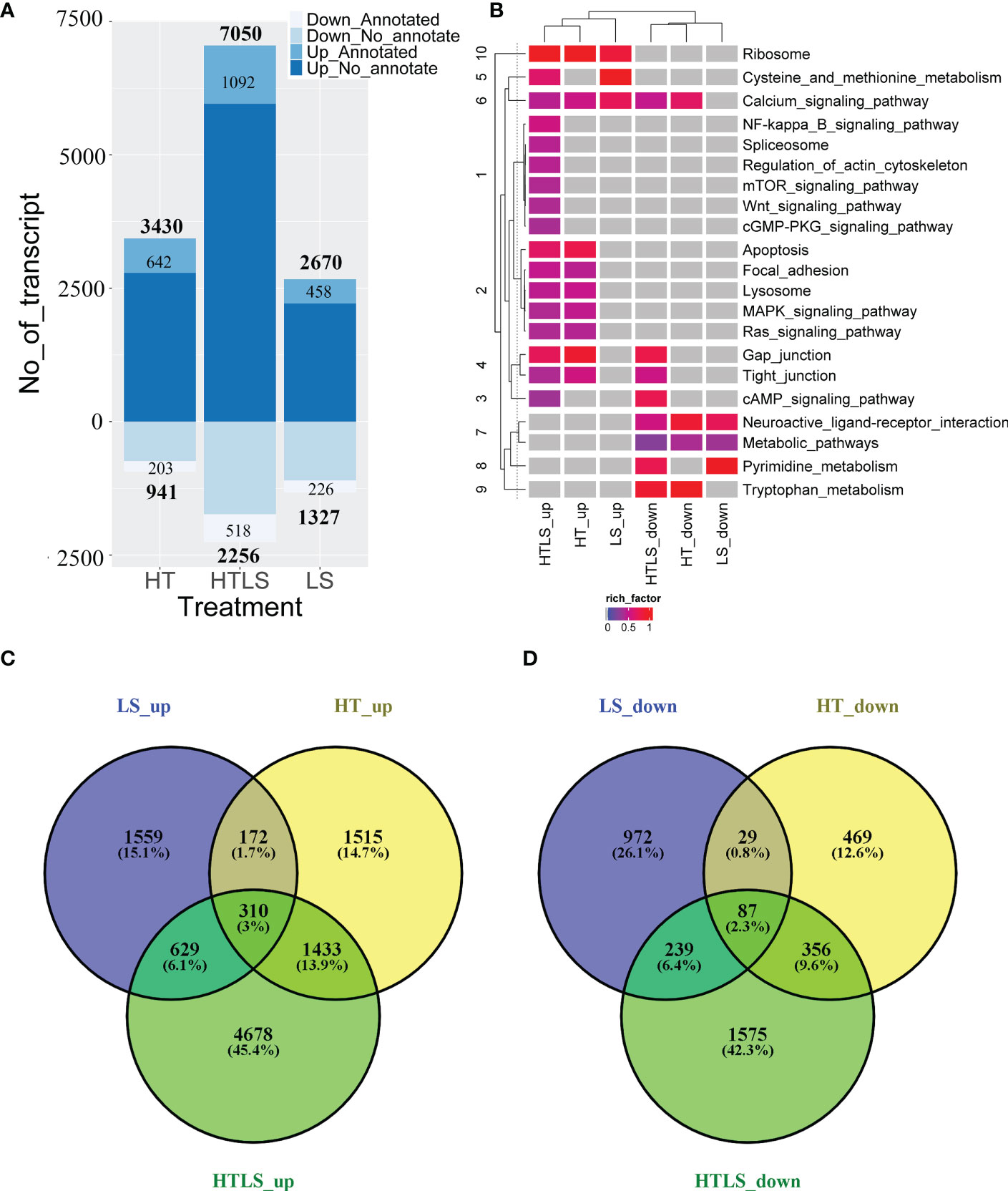
Figure 2 A summary of the results of differentially expressed gene (DEG) analysis. Bar chart in (A) summarizes the number of up- and down-regulated differentially expressed genes in each treatment, with labels specifying the number of NCBI nr-annotated and non-annotated transcript. The Venn diagrams in (B, C) compare the list of up- and down-regulated DEGs from each treatment. The heatmap in (D) summarizes the results of functional (KEGG terms) enrichment analysis results for up- and down- regulated DEGs, with the color specifying enrichment q-value of each enriched term, grey denoted no enrichment of the functional KEGG term.
The overall up- and down-regulated DEGs between treatments are presented in Venn diagrams (Figures 2B, C). When DEGs in HT and LS treatments were compared, 1,559 differentially expressed transcripts were detected only in LS but not in HT nor in HTLS treatments (Figure 2B). Among these 1,559 DEGs, 972 were nr-annotated (Figure 2C). On the other hand, 1,515 differentially expressed transcripts were detected only in HT but not in LS nor in HTLS treatments (Figure 2B); and among these 1,515 DEGs, 469 were nr-annotated (Figure 2C). Over ca. 45% or 4,678 DEGs were detected only in HTLS treatment (Figure 2B), among which 1,575 DEGs were nr-annotated (Figure 2C). Only 310 or around 3% of the total DEGs were shared by LS, HT and HTLS treatments; 629 or slightly more than 6% of the total DEGs were shared by LS and HTLS treatments; but 1,433 or almost 14% of the total DEGs were shared by HT and HTLS treatments. KEGG annotation based functional enrichment analysis of DEGs in LS, HT, and HTLS treatments revealed similarity and distinctiveness of different single and double stressor effects on A. pruinosa larvae (Figure 2D). Ribosomal protein genes (KEGG pathway “Ribosome”) were enriched in the up regulated DEGs in all three treatments; “Calcium signaling pathway” genes were enriched in up-regulated DEGs in all three treatments and down-regulated DEGs in HT and HTLS treatments; “Apoptosis”, “MAPK signaling pathway” and “Ras signaling pathway” genes were enriched in the DEGs in both HT and HTLS treatments. Structural component genes such as those involved in “Regulation of actin cytoskeleton”, “Focal adhesion”, “Gap junction” were enriched in up-regulated DEGs in HTLS treatment, or in both HT and HTLS treatments. Interestingly, genes involved in “Neuroactive ligand interaction” and “Metabolic pathways” were enriched in down-regulated DEGs in all three treatments.
3.4 The impact of single and double stressors treatments on the coral larval metabolic genes
KEGG functional enrichment analysis revealed “Ribosome” genes, genes involved in “Neuroactive ligand interaction” and “Metabolic pathway” genes were differentially expressed exclusively in both single stressors and the double stressor treatments. We anticipated that the most crucial influence of the stressor on coral larvae was perhaps on metabolism. To visualize the influence of the stressor on the coral larval metabolism, we firstly mapped DEGs from LS, HT and HTLS treatment to the KEGG “Metabolic pathway” map and compared the metabolic pathway affected by the single and double stressor treatments. Based on the KEGG map, HTLS double stressors treatment resulted in differential expression of genes involved in oxidative phosphorylation, fatty acid metabolism, lipid metabolism, nucleotide metabolism, carbohydrate metabolism (Figure 3A); LS and HT single stressor treatments resulted in differential expression of genes involved in abovementioned metabolic reactions with subtle variations (Figures 3B, C). Overall, HTLS double stressor treatment resulted in 213 differentially expressed metabolic genes and hence, according to the KEGG map, the highest number of metabolic reactions were affected. HT treatment resulted in mainly up- and down- regulation of 82 and 48 metabolic genes while 68 and 79 up- and down-regulated metabolic genes were observed in LS treatment (Figure 3D).
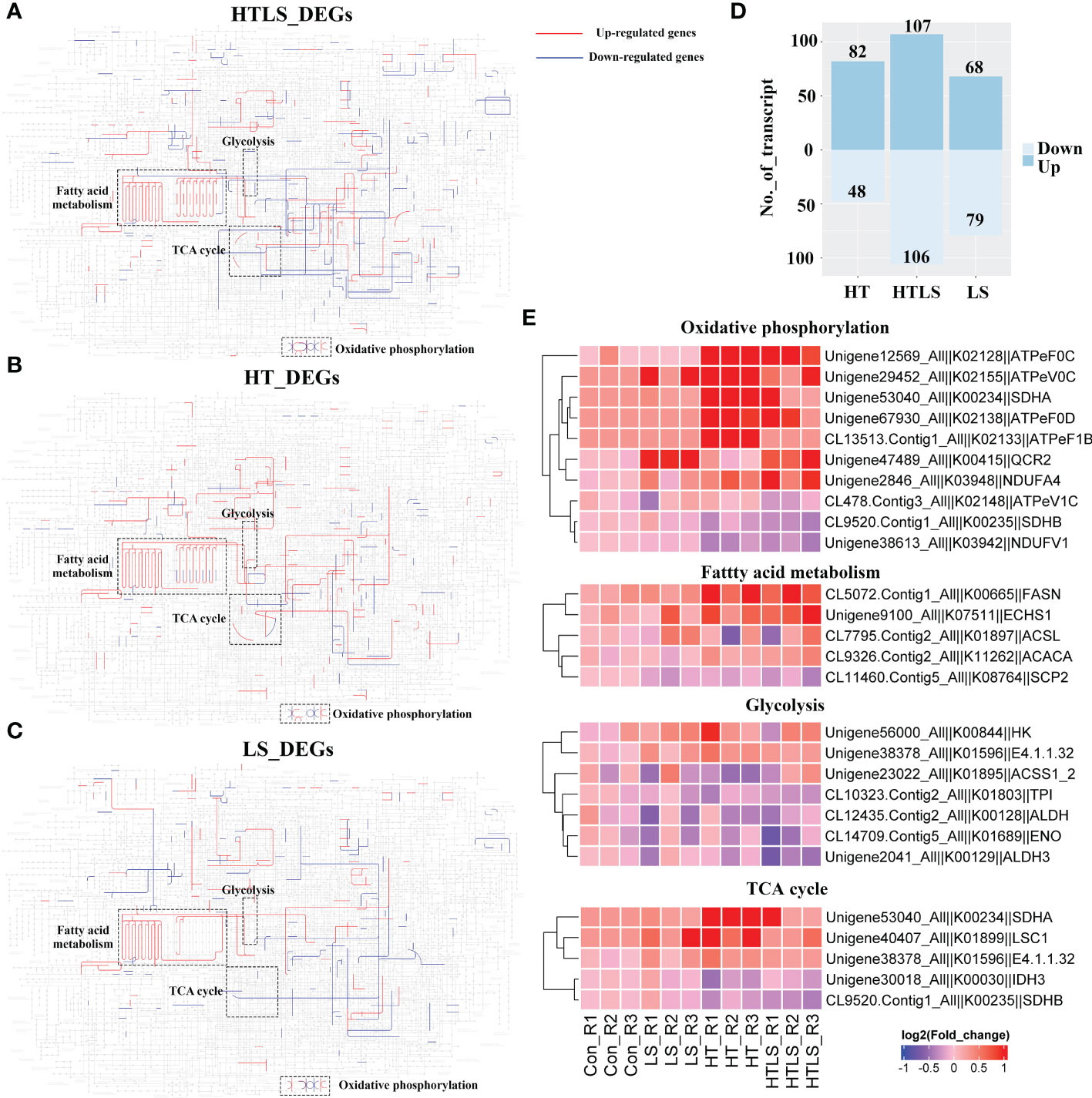
Figure 3 Differential expression of metabolic genes in coral larvae under HT, LS and HTLS treatments. The KEGG map of DEGs detected in (A) HT, (B) LS, and (C) HTLS treatment and mapped to KEGG “Metabolic pathway” are shown, with highlight of mapped up- and down-regulated DEGs in red and blue respectively, and components involved in “oxidative phosphorylation”, “glycolysis”, “TCA cycle” and “pentose phosphate pathway”. The total number of DEGs mapped to “Metabolic pathway” is summarized in the bar chart in (D) The heatmap in (E) provides expression pattern of DEGs mapped to “Metabolic pathway” in each replicate of each treatment.
We focused on the expression of genes involved in oxidative phosphorylation, fatty acid metabolism, glycolysis as genes involved in these essential metabolic processes were affected differently by HT, LS and HTLS double stressor treatments (Figure 3E). Several genes, including fatty acid synthase (fasn) involved in the elongation of fatty acid carbon chain and multiple F-type H+-transporting ATPase subunits (atpef0c, atpev0c, atpef0d, atpef1b) involved in oxidative phosphorylation were up-regulated in HT and HTLS treatments but not in LS treatment. On the other hand, ubiquinol-cytochrome c reductase core subunit 2 (qcr2) was up-regulated only in LS and HTLS treatments but not in HT treatment. Genes involved in glycolysis include aldehyde dehydrogenase (aldh), triosephosphate isomerase (tpi), enolase (eno) and aldehyde dehydrogenase 3 (aldh3). For TCA cycle, genes were differentially expressed in HT and HTLS but not in LS treatment. Succinate dehydrogenase (ubiquinone) iron-sulfur subunit (sdhb) and isocitrate dehydrogenase (NAD+) (idh) were down-regulated; phosphoenolpyruvate carboxykinase (GTP) (E4.1.1.32), succinate dehydrogenase (ubiquinone) flavoprotein subunit (sdha) and succinyl-CoA synthetase alpha subunit (lsc1) were up-regulated (Supplementary File 5 and Figure 3E).
3.5 Gene response exclusively to both single and double stressors
Ribosomal proteins are important protein translation machineries in the de novo synthesis system. There were in total 255 ribosomal protein transcripts differentially expressed in at least one of the stressor treatments (Figure 4A and Supplementary File 5). These ribosomal transcripts encoded either small or large ribosomal protein subunits (Figure 4B). It is worth noting that a remarkable number of these transcripts were not expressed in the normal larval samples (Supplementary File 5).
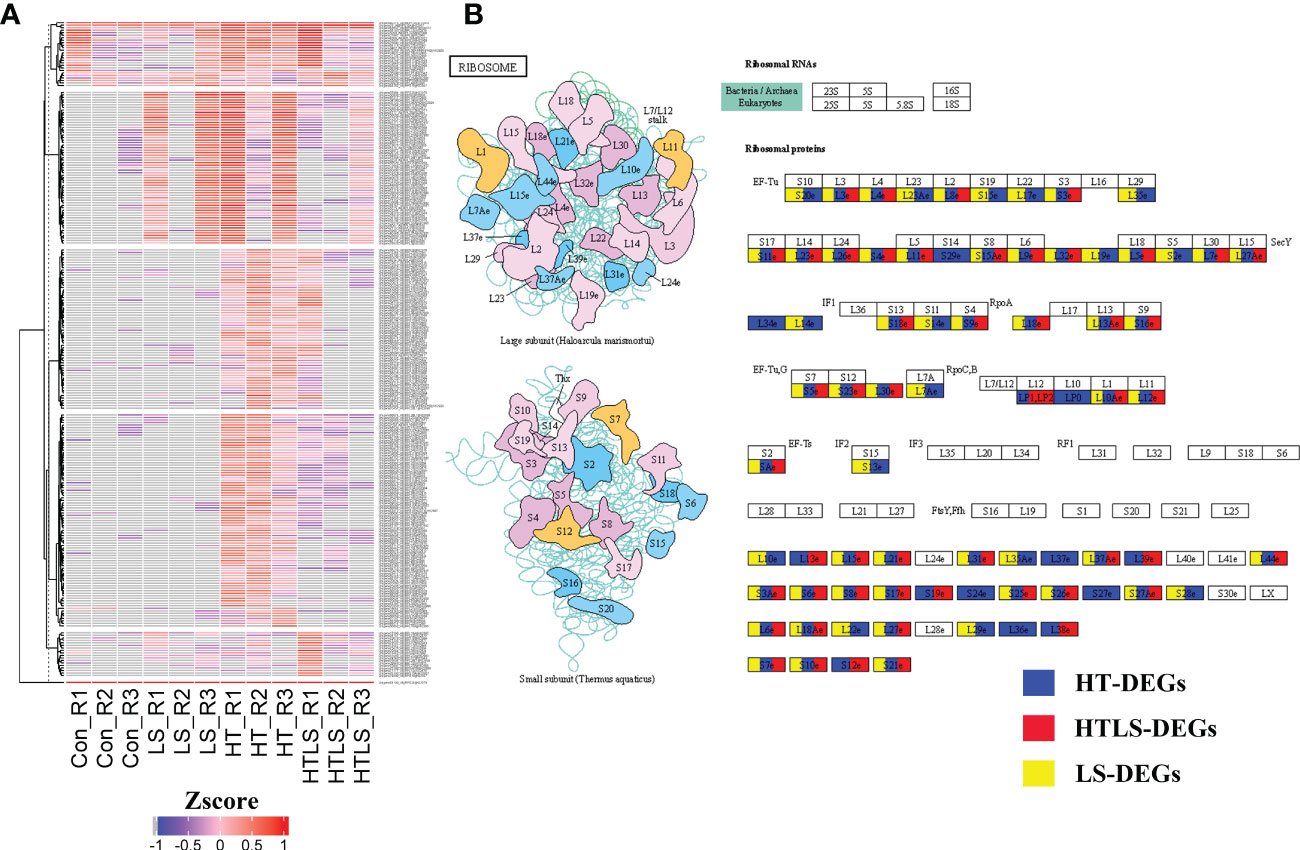
Figure 4 Differential expression of ribosomal protein genes in HT, LS and HTLS treatments. (A) Heatmap showing the expression pattern of DEGs mapped to “Metabolic pathway” in each replicate of each treatment. (B) The KEGG map of DEGs mapped to KEGG “Metabolic pathway” are shown. DEGs detected in HT, LS, and HTLS treatment are highlighted in blue, yellow, and red respectively.
The KEGG pathway “Neuroactive ligand interaction” is mainly composed of G-protein coupled receptors (GPCRs) and channel-gate receptors, which are important cell surface receptors for perceiving cellular signals and mediating downstream signal transduction. According to the KEGG annotation, many of the differentially expressed receptors genes (Supplementary File 5) are either GPCRs or channel receptors which could bind to neurotransmitters such as dopamine, serotonin, gamma-aminobutyric acid (GABA) and different amino acids such as beta-alanine, glycine (Supplementary Figure 3). Some of them were annotated as neuropeptide receptor family. These results suggested coral larvae were highly sensitive to both low salinity and high temperature when these stressors were applied individually or concomitantly.
3.6 Impact of single (high temperature) and double (high temperature+low salinity) stressors on MAPK, NF-κB signaling
MAPK signaling pathway is a highly conserved intracellular signaling pathway that is well implicated in stress response in animal cells (Gehart et al., 2010). MAPK signaling interact with signaling of NF-κB (nuclear factor kappa light chain enhancer of activated B cells), which is a family of highly conserved transcription factors that regulate many important cellular processes (Oeckinghaus et al., 2011). As shown in Figure 2D, both MAPK and NF-κB signaling were enriched among DEGs from HT and HTLS treatments but not LS treatment. KEGG pathway mapping of DEGs showed that while most of the mapped components in MAPK and NF-κB signaling pathway were differentially expressed in both HT and HTLS treatments, certain transcription factors were differentially expressed only in HT or HTLS treatment (Figures 5A, B).
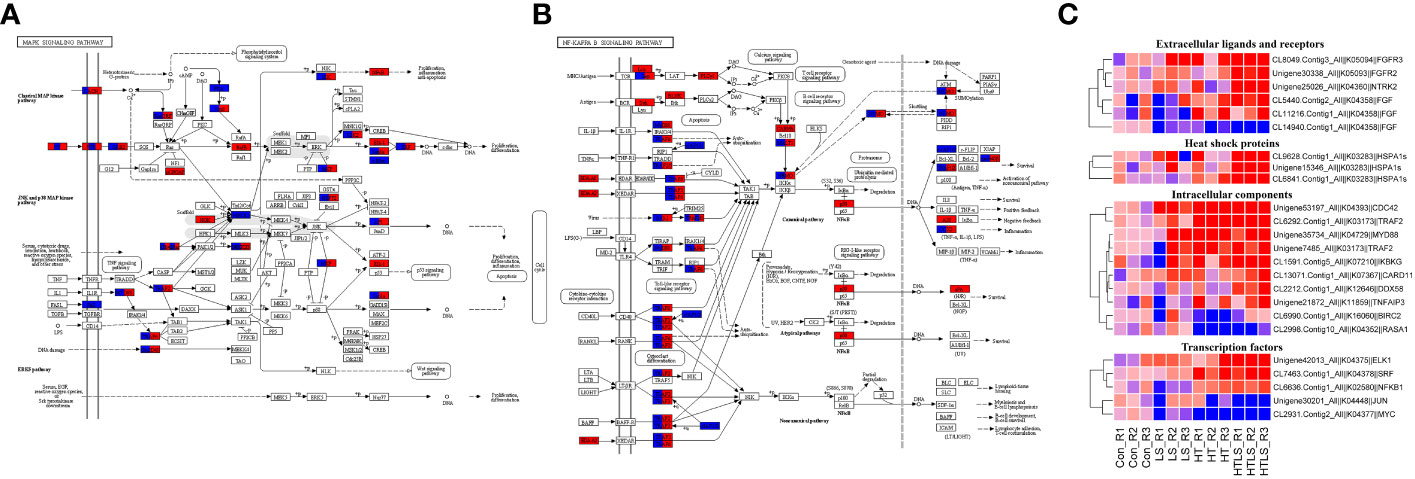
Figure 5 Differential expression of components involved in MAPK and NF-κB signaling pathways in HT and HTLS treatments. The KEGG map of DEGs mapped to KEGG (A) “MAPK signaling pathway” and (B) “NF kappa B signaling pathway” are shown. DEGs detected in HT and HTLS treatment are highlighted in blue and red respectively. The heatmap in (C) provides the expression patterns of DEGs mapped to MAPK and NF-κB signaling pathways in each replicate of each treatment. Note that the expression patterns of DEGs annotated as the molecular chaperone protein HSP70, which is also a component of MAPK pathway, are also shown in the heatmap.
As shown in the expression heatmap (Figure 5C), the majority of the mapped components in MAPK and NF-κB signaling pathway were up-regulated in both HT and HTLS treatments (see expression data in Supplementary File 5), including transmembrane receptors fibroblast growth factor receptors (fgfr4), key intracellular signal transducers TNF receptor-associated factors (traf2/3/6), mitogen-activated protein kinase kinase kinase (MAP3K-1/2/3), myeloid differentiation primary response protein (myd88) and down-stream transcription factors AP-1 (ap-1/jun) and IKBKG (inhibitor of nuclear factor kappa-B kinase subunit gamma, denoted as “IKK” in Figure 5A and “NEMO” in Figure 5B), the regulator of the key transcription factor in NF-κB signaling. However, transcription factors, including nuclear factor NF-kappa-B p105 subunit (nfkb1, denoted as “NF-κB” in Figure 5A and “p50” in Figure 5B) and ETS domain-containing protein Elk-1 (elk1) were up-regulated only in the HTLS treatment. MYC was down-regulated only in the HT treatment.
3.7 Single and double stressors treatment and apoptosis pathways
In the functional enrichment analysis, apoptosis pathway was enriched in the DEGs from HT and HTLS treatments (Figure 2D). Detail analysis of the gene expression pattern of the apoptosis pathway components revealed that most of the mapped components were up-regulated in both HT and HTLS treatments (Figure 5B and Supplementary Figure 5), including the death receptor associated signaling cascade, including FADD, Fas, TRAIL (denoted as TNFSF10 in Figure 6), were all up-regulated in both HT and HTLS treatments (Figure 6A). Yet, caspase 8, which could be triggered by the extrinsic FAS receptor mediated pathway and plays a central role in the execution-phase of cell apoptosis (Kruidering and Evan, 2000), was down-regulated in both HT and HTLS treatments. However, the pro-apoptotic gene Bcl-2 homologous antagonist/killer (Bak), which is a key apoptotic regulator of the intrinsic pathway and could induce apoptosis by promoting permeabilization of the mitochondria outer membrane (Peña-Blanco and García-Sáez, 2018), was found to be up-regulated only in the double stressor HTLS treatment. The intrinsic pathway of apoptosis could be triggered by a number of intracellular changes such as DNA damage. We found that p53 (denoted as TP53 in Figure 6), which could be triggered by DNA damage and could serve as an upstream regulator of Bak, was also up-regulated only in the HTLS treatment. Interestingly, in the reconstructed PPI network of differentially expressed apoptotic genes, components involved in the intrinsic pathway of apoptosis were differentially expressed only in the HTLS treatment, while the components involved in the extrinsic pathway were differentially expressed only in the HT treatment (Figure 6B).
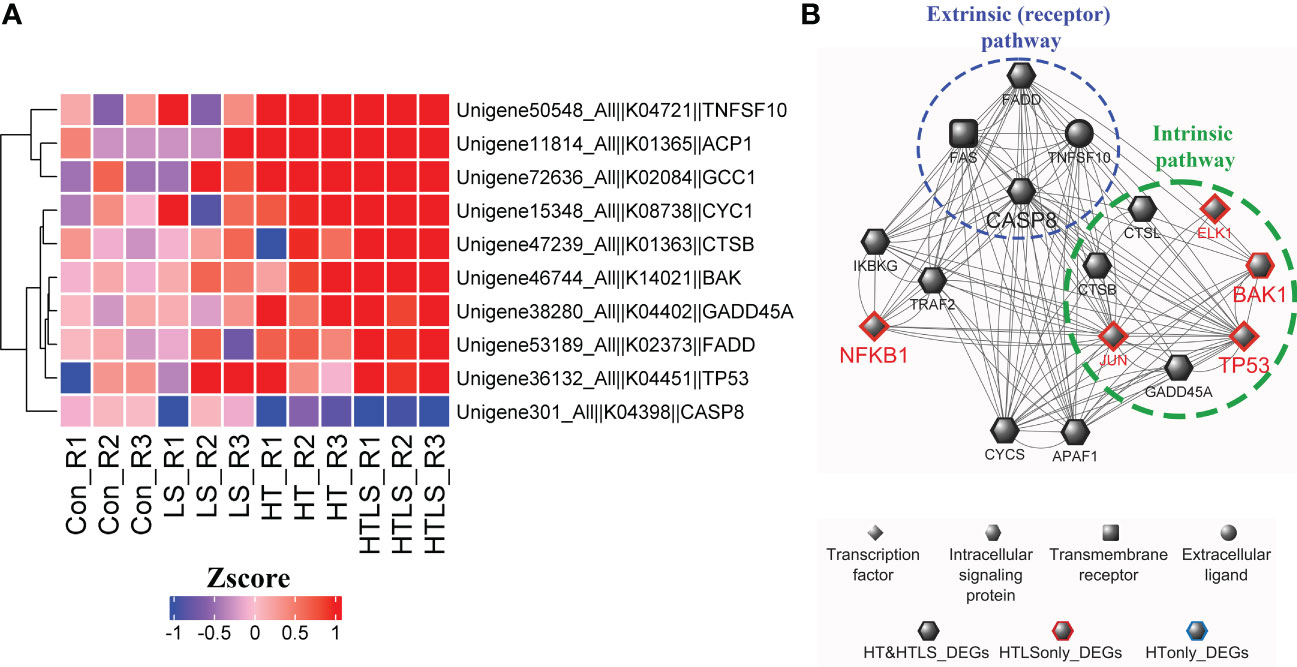
Figure 6 Differential expression of components involved in apoptosis pathways in HT and HTLS treatments. The heatmap in (A) provides the expression patterns of DEGs mapped to the KEGG apoptosis map. (B) Reconstructed based on STRING human PPI database, a protein-protein interaction network of the DEGs involved in apoptosis is illustrated. Components involved in the extrinsic (receptor-mediated) and intrinsic (caspase-independent) pathways of apoptosis were also highlighted.
4 Discussion
We investigated the interactive effects of changes in temperature and salinity on early life history processes of corals, including percent larval survival and settlement success. Our results showed that all competent A. pruinosa larvae survived well after 72-h of exposure to the different salinity and temperature treatments. However, the subsequent settlement success was significantly affected, posing a major problem to the recruitment success of this coral species. This has a significant implication suggesting how environmental stressors could jeopardize the persistence of corals in a multi-stressor environment. Given that the period of annual synchronous coral spawning event in Hong Kong usually coincides with the start of monsoon season (Chui and Ang, 2015), failure in the recruitment process could be one of the reasons why A. pruinosa is not more common in Hong Kong waters (Ang and Chui, 2022). From organismal to molecular levels, our transcriptomic data of coral larval response revealed the potential mechanisms that led to the survival of the larvae but the subsequent compromise of larval settlement ability under environmental stress. By working with aposymbiotic larvae, we ensured that expression differences detected in transcriptomic analysis were directly related only to the host-specific stress responses.
We summarized our results in Figure 7 that provides an overview on the impact of single and double stressor treatments on the transcriptomic landscape of A. pruinosa larvae. Changes in the expression of genes involved in carbohydrate metabolism and the oxidative phosphorylation (ATP metabolism), ribosomal proteins, molecular chaperone proteins (HSP70), GPCRs, components of stress-induced signaling pathways such as MAPK and NF-κB signaling pathway, and apoptosis are rather typical, as these functional proteins have been implicated in stress responses in different animal systems. For instance, changes in oxidative phosphorylation components is in line with the thermal stress response in Acropora palmata after 48-h exposure (Polato et al., 2013). Up-regulation of ribosomal proteins (RPs) was commonly observed as molecular response of corals to heat stresses, bleaching, virus infection, or disease conditions (Seneca et al., 2010; Libro and Vollmer, 2016; Wong et al., 2021); In the Pacific oyster Crassostrea gigas, distinctive gene expression patterns of GPCRs were observed in response to high temperature, salinity, air exposure, heavy metal, marine pathogens challenge (Fu et al., 2022). HSPs gene expression in the coral Acropora millepora were subjected to progressive changes in response to elevation of temperatures (Rosic et al., 2011). In another coral Pocillopora damicornis, MAPK expression increased in response to reduced osmotic pressure at night, as the coral experienced reduced level of light-dependent fluxes of dinoflagellate derived photosynthetic compounds (Mayfield et al., 2010). Heat stress suppressed the NF-κB signaling pathway component NLRC3-like protein in Acropora aculeus (Zhou et al., 2017). All of these are well characterized mechanisms for cell protection in corals under stressful conditions and might be a typical stress response among metazoans. However, continuous expression of these cell protection proteins may lead to detrimental effects, as energy is needed for the proteins to function properly. In addition, gene expression for routine metabolic processes such as carbohydrate metabolisms and amino acid metabolisms could be compromised in order to temporarily increase its tolerance limits to environmental stress.
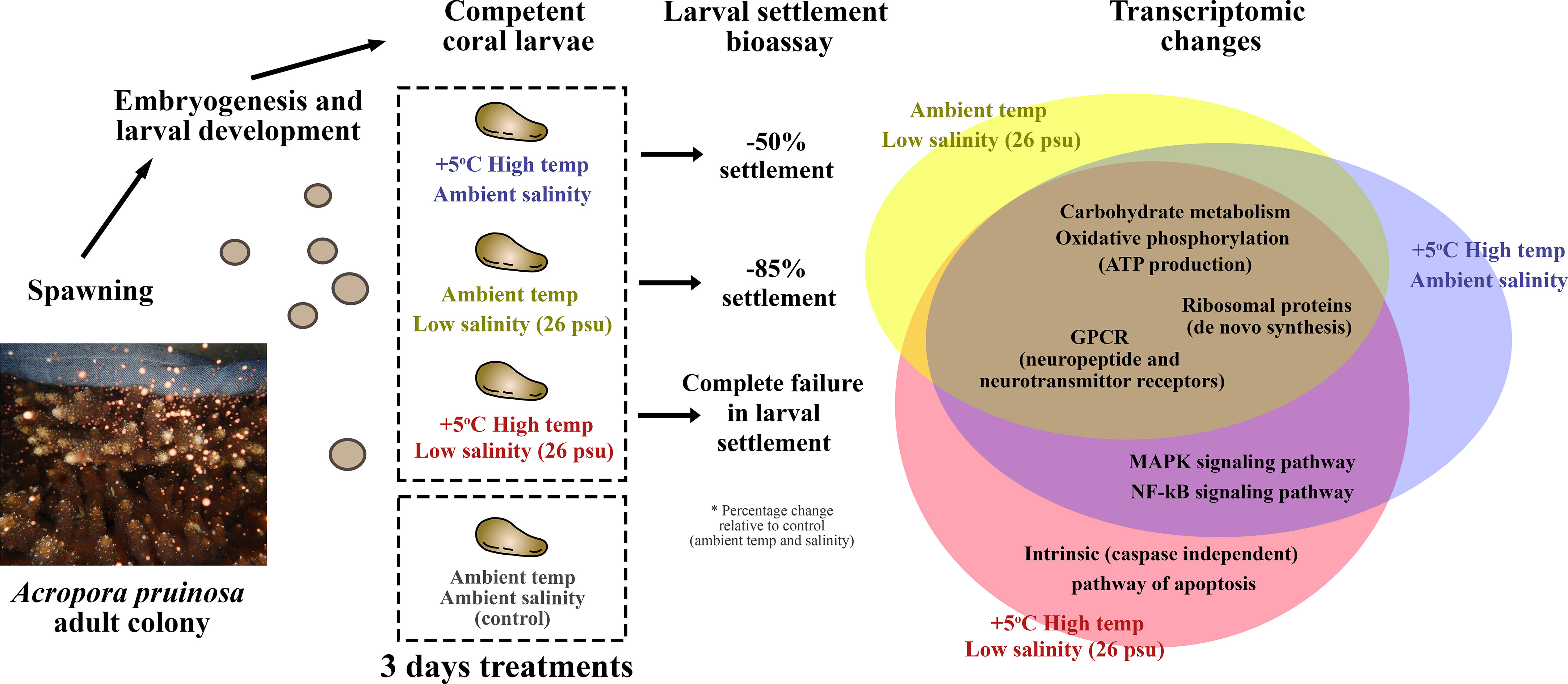
Figure 7 Summary of the major findings. HT, LS and HTLS treatments led to 39%, 12% and 0% settlement of A. pruinosa competent larvae, or 50, 85 and 100% reduction in settlement success respectively, compared to 78% settlement rate of coral larvae in the control group. The Venn diagram on the right-hand side summarizes the similarity and differences in DEGs expression in coral larvae under low salinity or high temperature single stressor treatments, and low salinity + high temperature double stressor treatment.
An important objective of this study is to determine if concomitant heat and salinity stresses will result in synergistic effect in coral larval internal environment, as larval settlement bioassay demonstrated combined double stressor treatment resulted in complete failure in larval settlement when compared with single stressor treatment. There were substantially different responses between coral larvae exposed to LS or HT alone and to combined HTLS treatment, with over two times of DEGs involved in HTLS and less than half of the DEGs overlapped between the single stressor and combined stressor treatments. The DEG sets that were involved in LS and HT alone but not in the combined HTLS treatment might suggest a reduced ability of larvae to express genes in the presence of another stressor. This result suggests that the two stressors may act synergistically to reinforce the intensity of stress and thereby the intensity of transcriptomic response, resulted in failure in larval settlement.
To understand the molecular mechanism underlying the synergistic effect of double stressor treatment, we focused on MAPK and NF-κB signaling pathways, as these two stress-induced pathways were highlighted by functional enrichment analysis and are essential for controlling various physiological processes, including programmed cell death (apoptosis), cellular proliferation, immune responses, and stress responses (Seger and Krebs, 1995; Perkins, 2007; Patterson et al., 2009; Son et al., 2011). Although activation of these two pathways requires modification at the protein level, we anticipate heat stress and concomitant heat and salinity stress to have triggered the signaling cascade, as the up-regulation of intracellular signaling molecules TNF receptor-associated factors (TRAF2, TRAF3), mitogen-activated protein kinase 1 (MEKK1) and transcription factor AP-1(c-JUN) observed in our study were reportedly linked to the activation of MAPK and NF-κB signal transduction pathways (Karin et al., 1997; Dhanasekaran et al., 2007; Dhanasekaran and Reddy, 2008; Häcker et al., 2011; Bironaite et al., 2013). In addition, differential expression of these key signaling genes is consistent with the heat stress response in adult corals observed in previous studies, which also showed up-regulation of TRAF2, TRAF3 in A. palmata (DeSalvo et al., 2010; Barshis et al., 2013) and c-JUN in A. aspera (van de Water et al., 2015).
MAPK and NF-κB signaling pathways are considered up-stream regulatory mechanisms, as activation of the signaling cascade results in increased expression of effector genes. These include some important transcription factors such as c-JUN, ELK-1 and p53 that trigger expression of genes responsible for different cellular events such as cell differentiation, proliferation and apoptosis (Townsend et al., 1999; Hickman et al., 2002; Leu et al., 2004). Of these transcription factors, the unique up-regulation of p53 in the double stressor treatment should be noted. Activation of p53 in response to DNA damage is a highly conserved mechanism (Lakin and Jackson, 1999). The expression of p53 could rapidly increase, leading to transcriptional activation of downstream effector genes (Lakin and Jackson, 1999), one of which is the pro-apoptotic gene Bcl-2 homologous antagonist/killer (Bak) (Leu et al., 2004). Bak mediates the caspase-independent intrinsic pathway of apoptosis (Leu et al., 2004). Since both p53 and Bak were up-regulated only in the double stressor (HT+LS) treatment but not in the single stressor (HT/LS) treatments, suggesting that apoptosis via caspase-independent intrinsic pathway could be one of the major reasons for the abrupt reduction of larval settlement success.
Yet, components involved in the extrinsic (receptor-mediated) apoptosis including FADD, FAS receptor and TRAIL the death ligand were also up-regulated in HT (single stressor) treatment. Why HT treatment did not result in a similar reduction in larval settlement rate as in double stressor treatment? The answer may lie in the down-regulation of caspase 8, a member of the cysteine-aspartic acid protease (caspase) family that executes the major degradation role during cellular apoptosis (Kruidering and Evan, 2000). Our data suggest that although the major components, including the receptor and the death ligand, were up-regulated in both single and double stressor treatments, the extrinsic pathway was suppressed via an unexplored mechanism.
A very crucial role of MAPK and NF-κB signaling pathways is their regulatory role on cellular apoptosis. To date, very little is known about the expression dynamics of signal transduction pathway genes in corals (DeSalvo et al., 2010; van de Water et al., 2015). For example, dual specificity protein phosphatase (MKP), which acts as a negative regulator of extracellular signal-regulated kinase (ERK) and Jun N-terminal kinase (JNK) (Patterson et al., 2009; Son et al., 2011), was found to be upregulated under HT stress. These data suggest that while there is a general transcriptional up-regulation of MAPK pathway components and downstream transcription factor c-Jun, the pathway itself may also be highly regulated. It has been suggested that the roles of reactive oxygen species in MKP expression is concentration dependent (Son et al., 2011). Activation of MAPK pathway is also cell-type dependent and involves multiple layers of control, from transcriptional control to post-translational modification (Dhanasekaran and Reddy, 2008). Further investigation into gene expressions and validation of expression profiles will be required to gain more insight into these dynamics.
4.1 Limitations and future perspective
In the present study, we performed an exposure experiment on larvae of mixed parentage. It is possible that the diverse genetic backgrounds of these larvae led to high variance between replicates, which was indeed observable in the current dataset as illustrated by the 2D PCA plot based on the expression data of the 12 samples (Supplementary Figure 4). Altogether, this high variance may mask part of the DEGs detection and enriched KEGG pathways in the experiments (Granados-Cifuentes et al., 2013). Nonetheless, more specific experiments may be carried out in the future with larvae of known parentages to address this possible complication.
While our analyses suggested activation of apoptosis via the intrinsic pathway could be the reason behind the complete failure in coral larval settlement under concomitant heat and salinity stresses, there are still much questions to answer before we could understand the full mechanism of such cellular molecular event. For instance, what was the trigger of the apoptosis intrinsic pathway? Up-regulation of p53 suggested DNA damage could be one of the triggers, but what caused DNA damage? On the other hand, components in NF-κB signaling pathway, especially NF-κB which is commonly known as a pro-survival factor, were up-regulated in larvae under double stressor treatment. How did NF-κB, MAPK and apoptosis pathway interact in different coral larval cells? We anticipate that there are both pro-survival signals and pro-cell death signals triggering differential responses in different coral larval cells, and the outcome of which is that coral larvae could not settle but could still survive the 3-day double stressor treatment period. Yet, which cells received pro-survival signal and which, the pro-cell death signals? As abovementioned, activation of MAPK pathway is cell-type dependent. Cell signals received by different cell types and their subsequent cellular dynamics could therefore be physiologically important for the fate of the larva. These questions could only be answered by a high-resolution single cell transcriptomic study, which will allow us to acquire the spatial expression information and thereby the expression levels of intracellular pathway genes in different coral larval cells.
Furthermore, as we have shown in Figure 2, there were typically more than 30% of the differentially expressed genes that were not annotated. There is still much to be learned about the comprehensive responses of coral larvae to heat, salinity or their combined stresses. Our annotation-based results only revealed mapped functions based on existing knowledge derived largely from vertebrate model systems. Our results suggest that there are many coral specific molecular responses that are lost during evolution. From this perspective, our dataset provides a list of genes of unknown functions that are sensitive to respective heat or salinity stress, or heat and salinity double stress. Further studies using gene manipulation experiments, such as gene knockdown or over-expression, are essential in elucidating the roles of these stress sensitive coral genes in coupling with environmental stresses in adult and larval stages of corals.
4.2 Summary
Overall, the present study provided novel insights into the molecular mechanisms behind coral larval responses to single and combined stresses. Acropora pruinosa larvae were capable of surviving a 3-day single or combined temperature and salinity stress condition but their settlement was compromised. RNAseq analyses revealed remarkable molecular responses relating to stress induced signaling pathways including MAPK and NF-κB. It is also evident that these signaling genes as well as apoptotic genes were more severely affected under double stress condition, in comparison to that under single stresses. Such information is extremely important to understand how coral larvae respond and cope with the highly dynamic natural field conditions. Interestingly, previous study on the larvae of another coral species in Hong Kong, Platygyra acuta, has shown high tolerance to elevated temperatures (+3 °C and +5 °C above ambient) that did not cause any negative effects on larval settlement success and post-settlement survival for up to 56 days of prolonged exposure (Chui and Ang, 2017). Further research is necessary to compare their molecular responses which might reveal the possible underlying mechanisms contributing to the difference in thermal tolerance among species. In the face of climate change, these data are particularly valuable to help understand the potential for success in coral recruitment processes and hence, the potential for coral reef recovery under stress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA901236.
Author contributions
AC and PA designed and carried out the experiments. AC, YW, and PA analyzed the experimental results and wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted final version.
Funding
This study was supported by the PEW Fellowship in Marine Conservation and the Direct Grant of the Chinese University of Hong Kong (No. 4053488) to AC, the Innovation Team Project of Universities in Guangdong Province (No. 2020KCXTD023) and the Natural Science Foundation of China (No. 42276104) to YW, and the University Grant Council (UGC) of Hong Kong General Research Fund (No. 14122215) to PA.
Acknowledgments
We thank the Marine Parks Division of the Agriculture, Fisheries and Conservation Department of the Hong Kong SAR Government for permission to work in the marine park.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1096407/full#supplementary-material
References
Ang P. O., Choi L. S., Choi M. M., Cornish A., Fung H. L., Lee M. W., et al. (2005). Status of coral reefs of the East Asian seas region: 2004 (Ministry of the Environment, Government of Japan: Japan Wildlife Research Centre), 121–152.
Ang P. O., Chui A. P. Y. (2022). “Status and trends of East Asian coral reefs: 1983-2019,” in Global coral reef monitoring network, East Asia region (Ministry of the Environment, Government of Japan: Japan Wildlife Research Centre), 62–81.
Baird A. H., Guest J. R., Willis B. L. (2009). Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu. Rev. Ecol. Evol. Syst. 40, 551–571. doi: 10.1146/annurev.ecolsys.110308.120220
Barshis D. J., Ladner J. T., Oliver T. A., Seneca F. O., Traylor-Knowles N., Palumbi S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat 29, 1165–1188. doi: 10.1214/aos/1013699998
Berkelmans R., Jones A. M., Schaffelke B. (2012). Salinity thresholds of Acropora spp. on the great barrier reef. Coral Reefs 31, 1103–1110. doi: 10.1007/s00338-012-0930-z
Bironaite D., Brunk U., Venalis A. (2013). Protective induction of Hsp70 in heat-stressed primary myoblasts: Involvement of MAPKs. J. Cell. Biochem. 114, 2024–2031. doi: 10.1002/jcb.24550
Byrne M. (2011). Impact of ocean warming and ocean acidication on marine invertebrate life history stages: Vulnerabilities and potential for persistence in a changing ocean. Oceanogr. Mar. Biol. Annual Rev. 49, 1–42.
Chui A. P. Y., Ang P. O. (2015). Elevated temperature enhances normal early embryonic development in the coral Platygyra acuta under low salinity conditions. Coral Reefs 34, 461–469. doi: 10.1007/s00338-014-1247-x
Chui A. P. Y., Ang P. O. (2017). High tolerance to temperature and salinity change should enable scleractinian coral Platygyra acuta from marginal environments to persist under future climate change. PloS One 12, e0179423. doi: 10.1371/journal.pone.0179423
Chui A. P. Y., Wong M. C., Liu S. H., Lee G. W., Chan S. W., Lau P. L., et al. (2014). Gametogenesis, embryogenesis, and fertilization ecology of Platygyra acuta in marginal nonreefal coral communities in Hong Kong. J. Mar. Biol. 2014, 1–9. doi: 10.1155/2014/953587
Chui A. P. Y., Yeung C. W., Tsang R. H. L., Leung Y. H., Ng T. Y., Chai K. H., et al. (2016). Lowered temperature and reduced salinity retarded development of early life history stages of Acropora valida from the marginal environment. Regional Stud. Mar. Sci. 8, 430–438. doi: 10.1016/j.rsma.2016.04.004
Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
DeSalvo M. K., Sunagawa S., Voolstra C. R., Medina M. (2010). Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser. 402, 97–113. doi: 10.3354/meps08372
Dhanasekaran D. N., Kashef K., Lee C. M., Xu H., Reddy E. P. (2007). Scaffold proteins of MAP-kinase modules. Oncogene 26, 3185–3202. doi: 10.1038/sj.onc.1210411
Dhanasekaran D. N., Reddy E. P. (2008). JNK signaling in apoptosis. Oncogene 27, 6245–6251. doi: 10.1038/onc.2008.301
Dias M., Ferreira A., Gouveia R., Vinagre C. (2019). Synergistic effects of warming and lower salinity on the asexual reproduction of reef-forming corals. Ecol. Indic. 98, 334–348. doi: 10.1016/j.ecolind.2018.11.011
Dineshram R., Chandramouli K., Ko G. W. K., Zhang H., Qian P. Y., Ravasi T., et al. (2016). Quantitative analysis of oyster larval proteome provides new insights into the effects of multiple climate change stressors. Global Change Biol. 22, 2054–2068. doi: 10.1111/gcb.13249
Ding D. S., Patel A. K., Singhania R. R., Chen C. W., Dong C. D. (2022). Effects of temperature and salinity on growth, metabolism and digestive enzymes synthesis of Goniopora columna. Biology 11, 436. doi: 10.3390/biology11030436
Dunlap W. C., Starcevic A., Baranasic D., Diminic J., Zucko J., Gacesa R., et al. (2013). KEGG orthology-based annotation of the predicted proteome of Acropora digitifera: ZoophyteBase - an open access and searchable database of a coral genome. BMC Genomics 14, 509. doi: 10.1186/1471-2164-14-509
Dunne R. P. (2010). Synergy or antagonism–interactions between stressors on coral reefs. Coral Reefs 29, 145–152. doi: 10.1007/s00338-009-0569-6
Emanuel K. (2005). Increasing destructiveness of tropical cyclones over the past 30 years. Nature 436, 686–688. doi: 10.1038/nature03906
Fu H., Tian J., Shi C., Li Q., Liu S. (2022). Ecological significance of G protein-coupled receptors in the pacific oyster (Crassostrea gigas): Pervasive gene duplication and distinct transcriptional response to marine environmental stresses. Mar. pollut. Bull. 185, 114269. doi: 10.1016/j.marpolbul.2022.114269
Garber M., Grabherr M. G., Guttman M., Trapnell C. (2011). Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8, 469–477. doi: 10.1038/nmeth.1613
Gehart H., Kumpf S., Ittner A., Ricci R. (2010). MAPK signalling in cellular metabolism: Stress or wellness? EMBO Rep. 11, 834–840. doi: 10.1038/embor.2010.160
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Trinity: Reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Granados-Cifuentes C., Bellantuono A. J., Ridgway T., Hoegh-Guldberg O., Rodriguez-Lanetty M. (2013). High natural gene expression variation in the reef-building coral Acropora millepora: Potential for acclimative and adaptive plasticity. BMC Genomics 14, 228. doi: 10.1186/1471-2164-14-228
Häcker H., Tseng P. H., Karin M. (2011). Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 11, 457–468. doi: 10.1038/nri2998
Hagedorn M., Carter V. L., Ly S., Andrell R. M., Yancey P. H., Leong J. C., et al. (2010). Analysis of internal osmolality in developing coral larvae, Fungia scutaria. Physiol. Biochem. Zoology 83, 157–166. doi: 10.1086/648484
Harii S., Nadaoka K., Yamamoto M., Iwao K. (2007). Temporal changes in settlement, lipid content and lipid composition of larvae of the spawning hermatypic coral Acropora tenuis. Mar. Ecol. Prog. Ser. 346, 89–96. doi: 10.3354/meps07114
Hédouin L., Pilon R., Puisay A. (2015). Hyposalinity stress compromises the fertilization of gametes more than the survival of coral larvae. Mar. Environ. Res. 104, 1–9. doi: 10.1016/j.marenvres.2014.12.001
Heron S. F., Maynard J. A., van Hooidonk R., Eakin C. M. (2016). Warming trends and bleaching stress of the world’s coral reefs 1985–2012. Sci. Rep. 6, 38402. doi: 10.1038/srep38402
Heyward A. J., Negri A. P. (2010). Plasticity of larval pre-competency in response to temperature: Observations on multiple broadcast spawning coral species. Coral Reefs 29, 631–636. doi: 10.1007/s00338-009-0578-5
Hickman E. S., Moroni M. C., Helin K. (2002). The role of p53 and pRB in apoptosis and cancer. Curr. Opin. Genet. Dev. 12, 60–66. doi: 10.1016/s0959-437x(01)00265-9
Hoegh-Guldberg O., Smith G. J. (1989). The effect of sudden changes in temperature, light and salinity on the population density and export of zooxanthellae from the reef corals Stylophora pistillata esper and Seriatopora hystrix Dana. J. Exp. Mar. Biol. Ecol. 129, 279–303. doi: 10.1016/0022-0981(89)90109-3
Hofmann G. E., Todgham A. E. (2010). Living in the now: Physiological mechanisms to tolerate a rapidly changing environment. Annu. Rev. Physiol. 72, 127–145. doi: 10.1146/annurev-physiol-021909-135900
Humphrey C., Weber M., Lott C., Cooper T., Fabricius K. (2008). Effects of suspended sediments, dissolved inorganic nutrients and salinity on fertilisation and embryo development in the coral Acropora millepora (Ehrenberg 1834). Coral Reefs 27, 837–850. doi: 10.1007/s00338-008-0408-1
Huson D. H., Mitra S., Ruscheweyh H. J., Weber N., Schuster S. C. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21, 1552–1560. doi: 10.1101/gr.120618.111
International Panel on Climate Change (2013). IClimate change 2013: The Physical Science Basis. In Stocker T. F., Qin D., Plattner G. K., Tignor M., Allen S. K., Boschung J., et al Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (UK and New York, NY, USA.: Cambridge University Press). doi: 10.1101/gr.120618.111
Iseli C., Jongeneel C. V., Bucher P. (1999). ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 11, 138–148.
Karin M., Liu Z., Zandi E. (1997). AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246. doi: 10.1016/S0955-0674(97)80068-3
Knutson T. R., Sirutis J. J., Zhao M., Tuleya R. E., Bender M., Vecchi G. A., et al. (2015). Global projections of intense tropical cyclone activity for the late twenty-first century from dynamical downscaling of CMIP5/RCP4.5 scenarios. J. Climate 28, 7203–7224. doi: 10.1175/JCLI-D-15-0129.1
Kruidering M., Evan G. I. (2000). Caspase-8 in apoptosis: The beginning of “the end”? IUBMB Life 50, 85–90. doi: 10.1080/713803693
Lakin N. D., Jackson S. P. (1999). Regulation of p53 in response to DNA damage. Oncogene 18, 7644–7655. doi: 10.1038/sj.onc.1203015
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee T., Chan K., Chan H., Kok M. (2011). Projections of extreme rainfall in Hong Kong in the 21st century. Acta Meteorol. Sin. 25, 691–709. doi: 10.1007/s13351-011-0601-y
Leu J. I.-J., Dumont P., Hafey M., Murphy M. E., George D. L. (2004). Mitochondrial p53 activates bak and causes disruption of a bak-Mcl1 complex. Nat. Cell Biol. 6, 443–450. doi: 10.1038/ncb1123
Libro S., Vollmer S. V. (2016). Genetic signature of resistance to white band disease in the Caribbean staghorn coral Acropora cervicornis. PloS One 11, e0146636. doi: 10.1371/journal.pone.0146636
Lottaz C., Iseli C., Jongeneel C. V., Bucher P. (2003). Modeling sequencing errors by combining hidden Markov models. Bioinformatics 19, ii103–ii112. doi: 10.1093/bioinformatics/btg1067
Mayfield A. B., Gates R. D. (2007). Osmoregulation in anthozoan–dinoflagellate symbiosis. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 147, 1–10. doi: 10.1016/j.cbpa.2006.12.042
Mayfield A. B., Hsiao Y. Y., Fan T. Y., Chen C. S., Gates R. D. (2010). Evaluating the temporal stability of stress-activated protein kinase and cytoskeleton gene expression in the pacific reef corals Pocillopora damicornis and Seriatopora hystrix. J. Exp. Mar. Biol. Ecol. 395, 215–222. doi: 10.1016/j.jembe.2010.09.007
Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Negri A. P., Marshall P. A., Heyward A. J. (2007). Differing effects of thermal stress on coral fertilization and early embryogenesis in four indo pacific species. Coral Reefs 26, 759–763. doi: 10.1007/s00338-007-0258-2
Nozawa Y., Harrison P. L. (2007). Effects of elevated temperature on larval settlement and post-settlement survival in scleractinian corals, Acropora solitaryensis and Favites chinensis. Mar. Biol. 152, 1181–1185. doi: 10.1007/s00227-007-0765-2
O’Donnell M. J., Hammond L. M., Hofmann G. E. (2009). Predicted impact of ocean acidification on a marine invertebrate: Elevated CO2 alters response to thermal stress in sea urchin larvae. Mar. Biol. 156, 439–446. doi: 10.1007/s00227-008-1097-6
Oeckinghaus A., Hayden M. S., Ghosh S. (2011). Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708. doi: 10.1038/ni.2065
Omori M., Iwao K. (2014). Methods of farming sexually propagated corals and outplanting for coral reef rehabilitation; With list of references for coral reef rehabilitation through active restoration measure. Okinawa: Akajima Mar. Sci. Lab. 22, 6–7.
Patel R. K., Jain M. (2012). NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PloS One 7, e30619. doi: 10.1371/journal.pone.0030619
Patterson K. I., Brummer T., O’brien P. M., Daly R. J. (2009). Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem. J. 418, 475–489. doi: 10.1042/BJ20082234
Peña-Blanco A., García-Sáez A. J. (2018). Bax, bak and beyond - mitochondrial performance in apoptosis. FEBS J. 285, 416–431. doi: 10.1111/febs.14186
Perkins N. D. (2007). Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62. doi: 10.1038/nrm2083
Polato N. R., Altman N. S., Baums I. B. (2013). Variation in the transcriptional response of threatened coral larvae to elevated temperatures. Mol. Ecol. 22, 1366–1382. doi: 10.1111/mec.12163
Portune K. J., Voolstra C. R., Medina M., Szmant A. M. (2010). Development and heat stress-induced transcriptomic changes during embryogenesis of the scleractinian coral Acropora palmata. Mar. Genomics 3, 51–62. doi: 10.1016/j.margen.2010.03.002
Puisay A., Pilon R., Goiran C., Hédouin L. (2018). Thermal resistances and acclimation potential during coral larval ontogeny in Acropora pulchra. Mar. Environ. Res. 135, 1–10. doi: 10.1016/j.marenvres.2018.01.005
Quinlan A. R., Hall I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Randall C. J., Szmant A. M. (2009a). Elevated temperature affects development, survivorship, and settlement of the elkhorn coral, Acropora palmata (Lamarck 1816). Biol. Bull. 217, 269–282. doi: 10.1086/BBLv217n3p269
Randall C., Szmant A. (2009b). Elevated temperature reduces survivorship and settlement of the larvae of the Caribbean scleractinian coral, Favia fragum (Esper). Coral Reefs 28, 537–545. doi: 10.1007/s00338-009-0482-z
Richmond R. H. (1993). Coral reefs: Present problems and future concerns resulting from anthropogenic disturbance. Am. Zool. 33, 524–536. doi: 10.1093/icb/33.6.524
Ritson-Williams R., Arnold S. V., Fogarty N., Steneck R., Vermeij M., Paul V. (2010). New perspectives on ecological mechanisms affecting coral recruitment on reefs. Smithsonian Contributions to Mar. Sci. 38, 457. doi: 10.5479/si.01960768.38.437
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodriguez-Lanetty M., Harii S., Hoegh-Guldberg O. (2009). Early molecular responses of coral larvae to hyperthermal stress: Coral molecular responses to heat stress. Mol. Ecol. 18, 5101–5114. doi: 10.1111/j.1365-294X.2009.04419.x
Rosic N. N., Pernice M., Dove S., Dunn S., Hoegh-Guldberg O. (2011). Gene expression profiles of cytosolic heat shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: Possible implications for coral bleaching. Cell Stress Chaperones 16, 69–80. doi: 10.1007/s12192-010-0222-x
Scott A., Harrison P. L., Brooks L. O. (2013). Reduced salinity decreases the fertilization success and larval survival of two scleractinian coral species. Mar. Environ. Res. 92, 10–14. doi: 10.1016/j.marenvres.2013.08.001
Seger R., Krebs E. G. (1995). The MAPK signaling cascade. FASEB J. 9, 726–735. doi: 10.1096/fasebj.9.9.7601337
Seneca F. O., Forêt S., Ball E. E., Smith-Keune C., Miller D. J., van Oppen M. J. H. (2010). Patterns of gene expression in a scleractinian coral undergoing natural bleaching. Mar. Biotechnol. 12, 594–604. doi: 10.1007/s10126-009-9247-5
Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: A software nnvironment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Son Y., Cheong Y. K., Kim N. H., Chung H. T., Kang D. G., Pae H. O. (2011). Mitogen-activated protein kinases and reactive oxygen species: How can ROS activate MAPK pathways? J. Signal Transduction 2011, 1–6. doi: 10.1155/2011/792639
Sun J., Chen Q., Lun J. C., Xu J., Qiu J. W. (2013). PcarnBase: Development of a transcriptomic database for the brain coral Platygyra carnosus. Mar. Biotechnol. 15, 244–251. doi: 10.1007/s10126-012-9482-z
Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., et al. (2015). STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi: 10.1093/nar/gku1003
Townsend K. J., Zhou P., Qian L., Bieszczad C. K., Lowrey C. H., Yen A., et al. (1999). Regulation of MCL1 through a serum response factor/Elk-1-mediated mechanism links expression of a viability-promoting member of the BCL2 family to the induction of hematopoietic cell differentiation. J. Biol. Chem. 274, 1801–1813. doi: 10.1074/jbc.274.3.1801
van de Water J. A. J. M., Leggat W., Bourne D. G., van Oppen M. J. H., Willis B. L., Ainsworth T. D. (2015). Elevated seawater temperatures have a limited impact on the coral immune response following physical damage. Hydrobiologia 759, 201–214. doi: 10.1007/s10750-015-2243-z
Vermeij M. J. A., Fogarty N. D., Miller M. W. (2006). Pelagic conditions affect larval behavior, survival, and settlement patterns in the Caribbean coral Montastraea faveolata. Mar. Ecol. Prog. Ser. 310, 119–128. doi: 10.3354/meps310119
Voolstra C. R., Schnetzer J., Peshkin L., Randall C. J., Szmant A. M., Medina M. (2009). Effects of temperature on gene expression in embryos of the coral Montastraea faveolata. BMC Genomics 10, 627. doi: 10.1186/1471-2164-10-627
Webster P. J., Holland G. J., Curry J. A., Chang H.-R. (2005). Changes in tropical cyclone number, duration, and intensity in a warming environment. Science 309, 1844–1846. doi: 10.1126/science.1116448
Weeriyanun P., Collins R. B., Macadam A., Kiff H., Randle J. L., Quigley K. M. (2022). Predicting selection–response gradients of heat tolerance in a widespread reef-building coral. J. Exp. Biol. 225, jeb243344. doi: 10.1242/jeb.243344
Wong Y. H., Zhang Y., Lun J. C. Y., Qiu J. W. (2021). A proteomic analysis of skeletal tissue anomaly in the brain coral Platygyra carnosa. Mar. pollut. Bull. 164, 111982. doi: 10.1016/j.marpolbul.2021.111982
Xiao R., Zhou H., Chen C. M., Cheng H., Li H., Xie J., et al. (2018). Transcriptional responses of Acropora hyacinthus embryo under the benzo(a)pyrene stress by deep sequencing. Chemosphere 206, 387–397. doi: 10.1016/j.chemosphere.2018.04.149
Yin K.. (2002). Monsoonal influence on seasonal variations in nutrients and phytoplankton biomass in coastal waters of Hong Kong in the vicinity of the Pearl River estuary. Mar. Ecol. Prog. Ser. 245, 111–122. doi: 10.3354/meps245111
Keywords: Acropora pruinosa, climate change, coral larvae, elevated seawater temperature, reduced salinity, transcriptomic (RNA-seq)
Citation: Chui APY, Wong YH, Sun J, Chang TKT, Qiu J-W, Qian P-Y and Ang P Jr (2023) Effects of heat and hyposalinity on the gene expression in Acropora pruinosa larvae. Front. Mar. Sci. 10:1096407. doi: 10.3389/fmars.2023.1096407
Received: 12 November 2022; Accepted: 10 March 2023;
Published: 06 April 2023.
Edited by:
Fei Xu, Institute of Oceanology (CAS), ChinaReviewed by:
Ao Li, Institute of Oceanology (CAS), ChinaBiao Chen, Guangxi University, China
Octavio R. Salazar, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2023 Chui, Wong, Sun, Chang, Qiu, Qian and Ang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Apple Pui Yi Chui, YXBwbGVweWNodWlAY3Voay5lZHUuaGs=; Put Ang Jr., YXB1dDk2MjI5QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Apple Pui Yi Chui
Apple Pui Yi Chui Yue Him Wong
Yue Him Wong Jin Sun
Jin Sun Taison Ka Tai Chang
Taison Ka Tai Chang Jian-Wen Qiu
Jian-Wen Qiu Pei-Yuan Qian
Pei-Yuan Qian Put Ang Jr.
Put Ang Jr.