- State Key Laboratory of Mariculture Breeding, Fisheries College, Jimei University, Xiamen, Fujian, China
Oysters are notably susceptible to accumulating substantial amounts of cadmium (Cd), leading to food safety concerns, yet the molecular mechanisms are not fully understood. Previous study identified a significant correlation between polymorphisms in the Zip1 and Zip3 of Fujian oyster (Crassostrea angulata) and the levels of Cd accumulation. Zip1 and Zip3, belonging to the Zip family, are primarily responsible for the cellular transport of zinc and other metal ions. To delve deeper into their roles in Cd transport within oysters, this research cloned and analyzed the sequences of Fujian oyster Zip1 and Zip3. The open reading frames for Zip1 and Zip3 were 960 and 978 bp, respectively, encoding proteins of 319 and 325 amino acids. Following Cd exposure, the expression levels of these genes in the mantle, gills, and other tissues initially increased significantly before decreasing, demonstrating a dose-dependent response. The lowest expression levels were observed in the adductor muscle, with Zip1 reaching its highest expression in the mantle and Zip3 in the visceral mass. In vitro RNAi experiments revealed that silencing Zip1 led to a 1.26-fold decrease in new 113Cd accumulation in the mantle, whereas silencing Zip3 resulted in a 1.29-fold increase. Immunohistochemistry showed weak expression of Zip1 and Zip3 proteins in the connective tissue of the mantle. In summary, Zip1 and Zip3 in Fujian oyster can transport Cd as a substrate, and played different roles in the absorption and efflux of Cd.
1 Introduction
Oyster represents a significant aquaculture specie cultivated widely across the globe. According to 2022 FAO statistics, oyster cultivation reached a production of 6.06 million tons, accounting for 34.2% of the global mollusk farming volume (FAO, 2022). In recent years, rapid urbanization and industrialization in coastal areas have led to metal pollution, posing substantial challenges to oyster farming. Oysters possess a remarkable capacity to accumulate metals, particularly cadmium (Cd), zinc (Zn), and copper (Cu) (Ke and Wang, 2001; Pan and Wang, 2009). The bioaccumulation factor of Cd can exceed 48,000 (Wu et al., 2013). After consumption, humans can absorb 78.3–87.8% of Cd from oysters, with the frequency and amount of consumption significantly correlating with Cd levels in human blood (Copes et al., 2008; He and Wang, 2013). Cd, a non-essential element for humans, can accumulate in the body through various pathways. Excessive intake may lead to acute poisoning and irreversible damage to the kidneys, liver, bones, and reproductive systems (Kumar and Sharma, 2019; Wang et al., 2021). Consequently, countries have established safety limits for Cd content in oysters (Satapathy et al., 2019). However, excessive safety limits of Cd are not uncommon. An investigation in India found an average Cd concentration of 10.6 µg/g dry weight in oyster soft tissues, 19 times higher than that of the surrounding water (Patra et al., 2021). In some areas of the South China Sea, the Cd concentration in oyster tissues ranged from 5.87 to 17.62 µg/g dry weight (Luo et al., 2018). The health risks posed by consuming oysters with excessive Cd content significantly limit the sustainable development of the oyster industry. Several scholars have made considerable progress in reducing metal content through genetic improvements by elucidating the molecular mechanisms of metal hyperaccumulation plant (Delhaize et al., 2004; Sasaki et al., 2004; Tang et al., 2017). Referring to these studies, exploring the molecular mechanisms controlling Cd accumulation in oysters and attenuating this capability through genetic improvements is of paramount importance.
Oysters, as typical Cd-accumulating organisms, have been studied for Cd accumulation kinetics and detoxification methods through isotope tracing and gel chromatography (Engel, 1999; Boisson et al., 2003; Mark et al., 2005). Most current researches focus on toxicodynamics, the impact of Cd exposure on gene or protein expression, and the estimation of the heritability of Cd accumulation. Toxicodynamic studies have partially explained the phenomenon of oyster Cd accumulation: oysters absorb Cd at a rate higher than other bivalves but excrete it more slowly (Ke and Wang, 2001; Pan and Wang, 2012). Although some scholars have investigated the molecular mechanisms behind oyster Cd accumulation, such as the relationship between Cd exposure and oyster gene polymorphisms (Meng et al., 2021), the specific roles of particular genes in Cd accumulation remain unknown. Thus, further exploration of the molecular genetic mechanisms is necessary.
Previous genome-wide association analyses in Fujian oyster identified a strong linkage between Cd accumulation traits and gene polymorphism on chromosome 3, where Zip1 and Zip3 were located (Wu et al., 2023). Prior research indicates Zip1 is ubiquitously expressed in human tissues, serving as the primary Zn uptake transporter in K562 leukemia cells and prostate cells, transporting Zn from outside to inside the cytoplasm (Franklin et al., 2003; Zhao and Eide, 1996; Gaither and Eide, 2001). Besides Zn, Zip1 has also been found to transport other metal ions, such as Cd, manganese, and iron, in yeast and Arabidopsis (Gitan et al., 1998; Korshunova et al., 1999; Connolly et al., 2002). However, in oysters and scallops, only the expression correlation between Zip1 and Cd exposure duration has been confirmed (Engel, 1999; Zhao et al., 2020). Zip3 is related to Zn uptake in mammalian cells and is widely expressed in many tissue types (Kelleher and Lönnerdal, 2005; Eide, 2006). Studies suggest that in cabbage varieties with high Cd levels, Zip3 expression is higher than in those with low levels, hypothesizing that it may transport Cd from the roots to the shoots (Yu et al., 2017). Therefore, further exploration of Zip1 and Zip3’s functions in Cd transport of oyster is warranted.
Fujian oyster is the most cultivated oyster specie in southern China. This study selected Fujian oyster as the research subject, cloning cDNA encoding Zip1 and Zip3 and analyzing their expression levels to explore their correlation with Cd accumulation. We then identified the Cd accumulation levels after Zip1 and Zip3 RNA interference (RNAi). Additionally, we attempted to explore the tissue and cellular localization of Zip1 and Zip3 in response to Cd stress. The results of this study provide a basis for researching the differential molecular mechanisms of oyster Cd accumulation and for breeding oyster strains with low Cd content.
2 Materials and methods
2.1 Oysters and Cd exposure
The oysters (C. angulata) used in the experiment were one-year-old, with an average weight of 74 ± 16 g, and were sourced from an oyster farm near Xiaocheng Town, Lianjiang City, Fujian Province. Approximately 400 similarly sized and well-formed oysters were collected, brought back to the laboratory, cleaned, removed from their attachment, and domesticated for one week. The oysters were cultured in twelve 30 L polyethylene containers, with daily complete water changes. They were fed commercial Chlorella spp. powder at a rate of approximately 2% of their soft tissue dry weight per day.
After a 7-day acclimation, oysters were randomly divided into three groups. Each group was subjected to different exposure conditions: 0ppb, 2ppb, and 5ppb Cd2+. Stock solutions of 10 ppm Cd (CdCl2; MW:183.32 g/mol; Sigma-Aldrich) were prepared with Milli-Q-filtered water. After exposure, eight individuals per group were randomly sampled at 0 d, 1 d, 3 d, 7 d, and 15 d. Their soft tissues were rinsed several times with distilled water before dissecting for mantle, gills, adductor muscle, and visceral mass. Samples were partly designated for Cd concentration determination and partly in liquid nitrogen and stored at -80°C for gene expression pattern analysis.
2.2 Zip1 and Zip3 ORF clone
Total RNA from the oyster mantle was extracted using TRIzol® Reagent (Invitrogen, USA) (Ni et al., 2013). First-strand cDNA was synthesized using M-MuLV reverse transcriptase (Thermo Scientific, USA). Zip1 and Zip3 were amplified by PCR using primers Zip1-F and Zip1-R listed in Table 1, based on mRNA sequences for encoding Zip1 and Zip3 selected from our lab’s mantle transcriptome database. The PCR protocol was as follows: 1 cycle of predenaturation at 94°C for 5 min, 35 cycles of 94°C for 30 s, Zip1 is 55.3°C for 30 s; Zip3 is 52.5°C for 30 s, and 72°C for 1 min, followed by a 10 min extension at 72°C. All primer sequences are provided in Table 1. Nucleotide and protein sequences were characterized with DNAMAN software. Multiple sequence alignment was performed using Clustal W (University College Dublin, Dublin, Ireland) and MEGA 7.0’s neighbor-joining (NJ) method, generating a phylogenetic tree.
2.3 Gene expression pattern analysis
2.3.1 Quantitative real-time PCR analysis
The expression levels of Zip1 and Zip3 in different tissues under Cd stress were detected using qRT-PCR. RNA extraction from Cd-exposed samples followed the method described in Section 2.2. Each 20 μL PCR reaction contained 10 μL 2×ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China), 4 μL of fivefold diluted cDNA, and 10 µM of gene-specific primers (Table 1). The amplification conditions were: 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s, with a final extension at 95°C for 15 s and 60°C for 60 s, ending with 95°C for 15 s. EF1α from Fujian oyster served as the reference gene. Each sample was analyzed in six biological replicates, with relative expression levels of Zip1 and Zip3 calculated according to the 2−ΔΔCT method (Rao et al., 2013).
2.3.2 Cd content determination in oyster tissues
Cd content in oyster tissues was determined using Inductively Coupled Plasma Mass Spectrometry (ICP-MS-7700, Agilent) (Luo et al., 2014; Weng and Wang, 2014). Oyster samples were dried at 80°C for 24 hours until constant weight, digested with nitric acid at room temperature for 12 hours, followed by hot digestion at 80°C for 12 hours until complete dissolution. After appropriate dilution, 114Cd concentration in oyster samples was measured by ICP-MS. A multi-element standard solution (Agilent) was used for ICP-MS calibration, with 103Rh as an internal standard to correct for instrumental drift and sensitivity changes. Quality control involved re-measuring a previously tested sample every twenty samples.
2.4 Immunohistochemistry assays
The localization of Zip1 and Zip3 within Fujian oyster tissues was determined via immunohistochemistry (Kingtong et al., 2007). Oysters subjected to 5ppb Cd stress were dissected on the third day of the experiment to collect mantle tissue, which was then fixed in 4% paraformaldehyde for 24 hours. Subsequently, the tissues underwent hydration in graded ethanol and dewaxing in xylene. The samples were incubated overnight at 4°C with polyclonal antibodies against Zip1 and Zip3 (1:20) (GL Biochem, China). The control group used pre-immune serum at the same concentration, followed by three 5-minute washes in 0.01M phosphate-buffered saline (PBS). Goat anti-rabbit IgG conjugated with horseradish peroxidase (HRP) (GL Biochem, China) was used as the secondary antibody (1:50) and incubated at room temperature for 1 hour before developing with DAB. A separate set of identical samples was stained with HE (Liu, 2022) and observed under a light microscope for comparison of tissue structures with those subjected to immunohistochemistry.
2.5 RNA interference
2.5.1 dsRNA synthesis
PCR was employed to amplify cDNA fragments of Zip1 and Zip3, along with a GFP cDNA (612 bp) fragment from the pEGFP-N1 plasmid for in vitro transcription and dsRNA synthesis. Primers designed using siDirect 2.1 (University of Tokyo, Japan) included a T7 promoter sequence (5’-TAATACGACTCACTATAGGG-3’) at the 5’ end for dsRNA synthesis. Linear templates were amplified using these primers, followed by DNA precipitation with ethanol. T7 RNA Polymerase (Thermo Scientific, USA) was used for in vitro transcription to synthesize dsRNA, which was then stored at -80°C after electrophoresis verification and concentration measurement.
2.5.2 Ex vivo tissue culture and dsRNA interference
Following dsRNA preparation, tissue culture and RNAi were conduted according to the methods of in vitro culture of the mud crab (Scylla paramamosain) tissue and the calm (Meretrix meretrix) primary cells with slight modifications (You et al., 2012; Liu, 2022). Furthermore, based on qPCR results and existing studies on metal accumulation, the mantle is identified as the primary tissue for metal accumulation and target gene expression (Shi et al., 2019a). Hence, we have selected the mantle as the tissue material for our in vitro RNA interference experiments. Mantle tissue was excised under sterile conditions, washed thrice in sterile seawater containing 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B (filtered through 0.22 μm), with each wash lasting 20, 60, and 100 minutes, respectively. Tissues were then cut into 1 mm3 pieces and pre-cultured in modified sterile L15 medium (Gibco, USA) supplemented with 1 g/L glucose, 2.4 g/L HEPES, 12 g/L NaCl, 1.2 g/L NaHCO3, and 5 g/L yeast extract, along with the aforementioned antibiotics, in a 48-well cell culture plate (LABSELECT, Beijing) at 26°C for 60 minutes. After confirming no significant change in medium color, the medium was replaced.
The tissues were divided into three groups: Zip1 interference, Zip3 interference, and Zip1+Zip3 interference, with each group including an EGFP dsRNA as a control. L15 medium containing the respective dsRNA (final concentration 1 μg/µL) was gently mixed into the culture plates. After 24 hours of RNAi, samples from the experimental and control groups were collected for Cd content determination. Additional corresponding dsRNA was then added to the remaining tissues to maintain RNAi efficiency, along with 113Cd to a final concentration of 5 μg/L. After a further 24 hours of RNAi and 113Cd exposure, mantle tissues were collected for qRT-PCR analysis and 113Cd content determination. Each group had three replicates.
2.5.3 113Cd concentration measurement
To assess the changes in 113Cd accumulation in the mantle tissues following Zip1 and Zip3 interference, the concentrations of 113Cd and 111Cd were measured using the method described in section 2.3.2, and calculated following the approach by Zhong et al. (2020). The newly accumulated Cd in oyster tissues was calculated using the formula:
Here, [total Cd]111 and [total Cd]113 are the total Cd concentrations measured by ICP-MS when selecting 111Cd and 113Cd for analysis, respectively. 12.22% represents the natural abundance of 113Cd. Thus, this formula allows for the calculation of newly accumulated Cd in oyster tissues.
2.6 Statistical analysis
All data are presented as mean ± SE (n=8 or 3). Statistical significance was determined through two-way ANOVA and paired sample T-tests. Graphs were created using GraphPad Prism 9.0 (San Diego, USA), with statistical significance set at p< 0.05.
3 Results
3.1 Sequence and phylogenetic analysis of Zip1 and Zip3
The ORF of Fujian oyster Zip1 was 960 bp (GenBank accession number: OR759776), encoding a protein of 319 amino acids with a predicted molecular weight of 35.7 kDa and an isoelectric point (pI) of 5.78. The predicted Zip1 protein lacks a signal peptide but contains eight transmembrane helices located at amino acids 10–31, 43–65, 80–102, 160–182, 192–214, 223–245, 260–279, and 292–310 (Figure 1A). The ORF of Zip3 was 978 bp long (GenBank accession number: OR759777), predicted to encode 325 amino acids, with a molecular weight of 35.9 kDa and a pI of 6.27, featuring eight α-helical transmembrane domains located at amino acids 10–31, 44–66, 81–103, 176–198, 203–225, 237–259, 269–289, and 302–321 (Figure 1B).
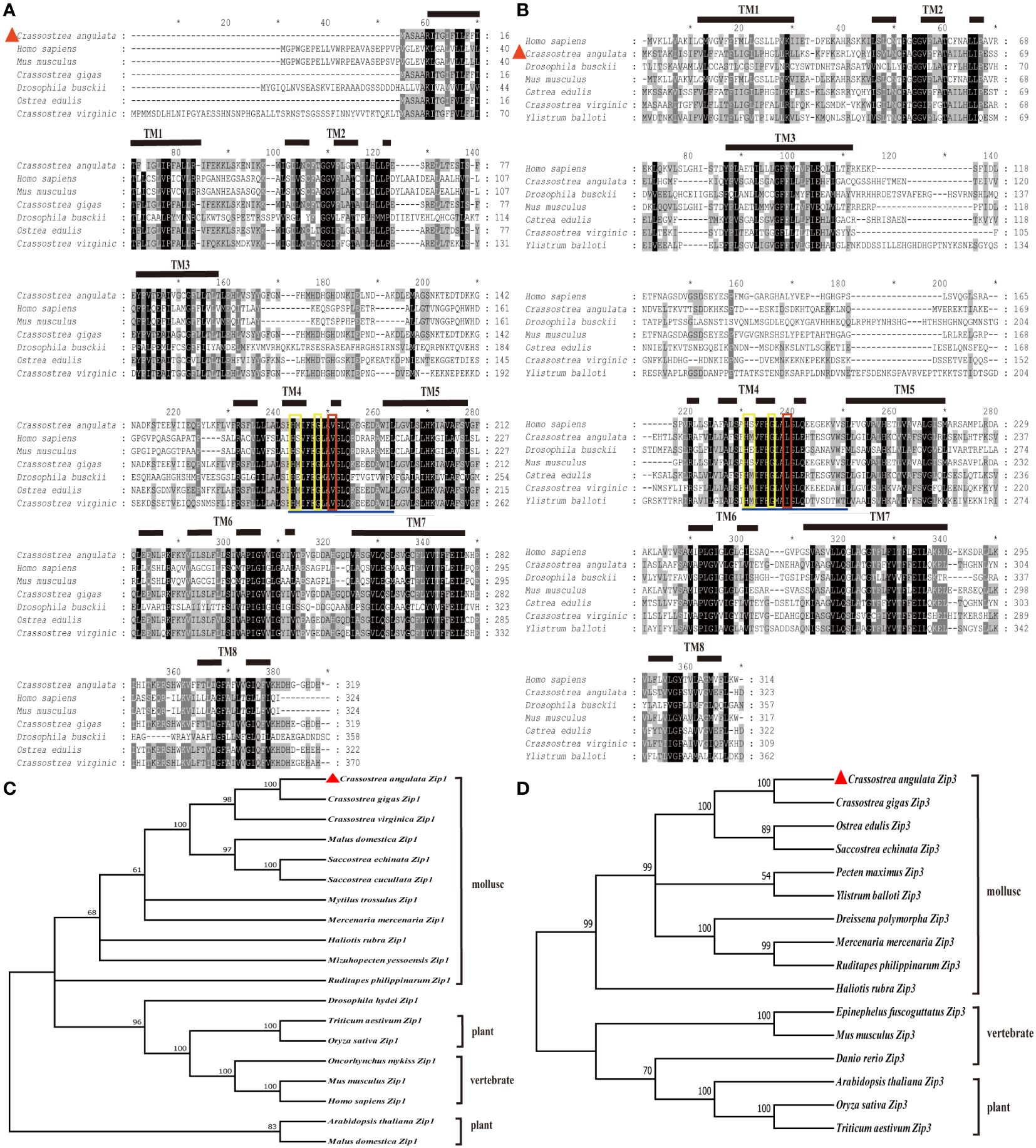
Figure 1 Illustrates the multiple sequence alignment and phylogenetic tree analysis of Zip1 and Zip3 with amino acid sequences from other species. Similar amino acids are highlighted, and the eight presumed transmembrane domains (TM) are denoted by solid and dashed lines. The fourth presumed transmembrane domain includes conserved histidine, serine or methionine, and glycine residues (highlighted in yellow boxes). (A) Multiple sequence alignment of Zip1 with amino acid sequences from various species. (B) Multiple sequence alignment of Zip3 with amino acid sequences from different species. (C) Phylogenetic tree analysis of Zip1. (D) Phylogenetic tree analysis of Zip3. Zip1 and Zip3 proteins were submitted to the GenBank database, with other GenBank accession numbers as follows: Homo sapiens (Zip1, NP_001258888.1), Mus musculus (Zip1, NP_038929.2), Crassostrea gigas (Zip1, XP_011453296.2), Drosophila busckii (Zip1, ALC42177.1), Crassostrea virginica (Zip1, XP_022330862.1), Ostrea edulis (Zip1, XP_048736341.1), Crassostrea gigas (Zip1, XM_011454994.3), Crassostrea virginica (Zip1, XM_022475156.1), Malus domestica (Zip1, NM_001294116.1), Saccostrea echinata (Zip1, XM_061318195.1), Saccostrea cucullata (Zip1, XM_062749567.1), Mytilus trossulus (Zip1, XM_063579755.1), Mercenaria mercenaria (Zip1, XM_045356306.2), Haliotis rubra (Zip1, XM_046712745.1), Mizuhopecten yessoensis (Zip1, XM_021487195.1), Ruditapes philippinarum (Zip1, XM_060749981.1), Drosophila hydei (Zip1, XM_030225087.1), Triticum aestivum (Zip1, XM_044496491.1), Oryza sativa (Zip1, NM_001397949.1), Oncorhynchus mykiss (Zip1, NM_001124481.1), Mus musculus (Zip1, NM_001411505.1), Homo sapiens (Zip1, NM_001271957.2), Arabidopsis thaliana (Zip1, NM_112111.3), Malus domestica (Zip1, NM_001294116.1), Homo sapiens (Zip3, NP_653165.2), Drosophila busckii (Zip3, ALC45898.1), Mus musculus (Zip3, NP_001345827.2), Ostrea edulis (Zip3, XP_048735000.2), Crassostrea virginica (Zip3, XP_022330863.1), Ylistrum balloti (Zip3, XP_060079049.1), Crassostrea gigas (Zip3, XM_034467256.1), Ostrea edulis (Zip3, XM_048879043.2), Saccostrea echinate (Zip3, XM_061316256.1), Pecten maximus (Zip3, XM_061316256.1), Ylistrum balloti (Zip3, XM_060223066.1), Dreissena polymorpha (Zip3, XM_052384496.1), Mercenaria mercenaria (Zip3, XM_053532146.1), Ruditapes philippinarum (Zip3, XM_060744456.1), Haliotis rubra (Zip3, XM_046728897.1), Epinephelus fuscoguttatus (Zip3, XM_049588280.1), Mus musculus (Zip3, NM_001358898.2), Danio rerio (Zip3, NM_001080619.1), Arabidopsis thaliana (Zip3, NM_128786.4), Oryza sativa (Zip3, NM_001419857.1), Triticum aestivum (Zip3, XM_044469838.1). Multiple alignments were performed using Clustal W with default parameters. Identical and similar residues are displayed against black and grey backgrounds, respectively. α-helices are represented by horizontal lines and dashed lines in the figures and red triangle symbol indicate the location of Zip1 and Zip3.
Phylogenetic trees constructed by comparing the amino acid sequences of Zip1 and Zip3 from a variety of published organisms revealed that Fujian oyster, like other animals, plants, and fungi, have eight transmembrane helices. All possess an extended sequence of 12 amino acids (underlined in blue) within the fourth transmembrane domain, characteristic of the Zip family, containing conserved histidine, serine, and glycine residues (highlighted in yellow). Notably, the serine has been replaced by methionine in mollusks, differentiating them from mammals. Additionally, Zip3 features a conserved amino acid difference (marked in red) compared to Zip1, whose function and significance remain unclear. The evolutionary tree indicates that the Fujian oyster is separately clustered with other bivalves, showing the highest similarity with the Pacific oyster (Figures 1C, D).
3.2 Gene expression pattern
3.2.1 Cd accumulation in oyster tissues
Cd content of Fujian oyster tissues (gills, mantle, adductor muscle, and visceral mass) was detected using ICP-MS before and after Cd stress (Figure 2). Before exposure, the average Cd content was in the order: visceral mass> gills> mantle> adductor muscle; after exposure, the average Cd content followed: gills> mantle> visceral mass> adductor muscle. As the exposure time and Cd dose increased, the accumulation of Cd in all tissues also increased, reaching the highest on the 15th day after exposure. In the gills of the oysters exposed to 5ppb, the Cd content on the 15th day was 138 times that of the control group.
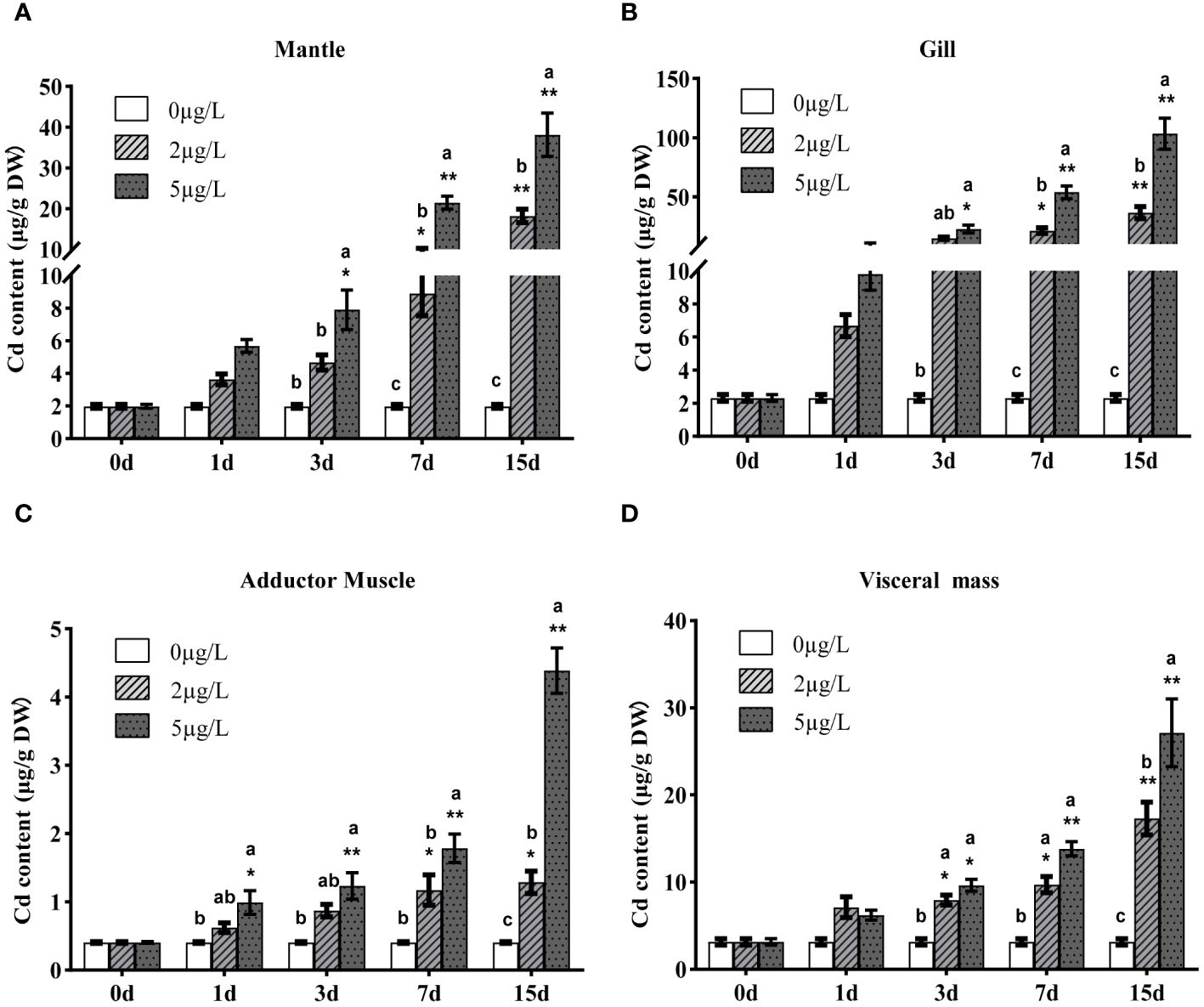
Figure 2 Accumulations of 114Cd in mantle (A), gills (B), adductor muscle (C) and visceral mass (D) of Fujian oyster after Cd exposure. Data are presented as mean ± SE (n=8). Different lowercase letters indicate significant differences between different Cd concentrations at the same exposure time (p<0.05), while * indicates significant differences between different exposure times at the same Cd level. (*p<0.05, **p<0.01).
3.2.2 Expression spectrum of Zip1 and Zip3 after Cd exposure
To analyze the expression patterns of Zip1 and Zip3 in oysters during Cd exposure, oysters were exposed to different concentrations of Cd. As shown in Figures 3A–D and 4A–D, at the same Cd concentration, the expression levels of Zip1 and Zip3 in all tissues generally first increased and then decreased over exposure time. Before Cd exposure, the highest expression of Zip1 was in the gills, with the lowest in the adductor muscle. Zip3 was most highly expressed in the visceral mass and least in the gills. After Cd exposure, the highest expression of Zip1 was in the mantle, while Zip3 was most highly expressed in the visceral mass, with both showing the lowest expression in the adductor muscle. Within the same exposure time, the expression levels of Zip1 and Zip3 overall showed dose-dependency, increasing with the concentration of Cd exposure. In the 5ppb group, the expression of Zip1 reached its peak on the 3rd day in the adductor muscle and visceral mass, while in the mantle and gill, it peaked on the 7th and 1st days, respectively, before gradually decreasing to control levels. In the gills, the expression of Zip3 also peaked on the 3rd day, while in the mantle, it was highest on the 1st day, and in other tissues, it peaked on the 7th day.
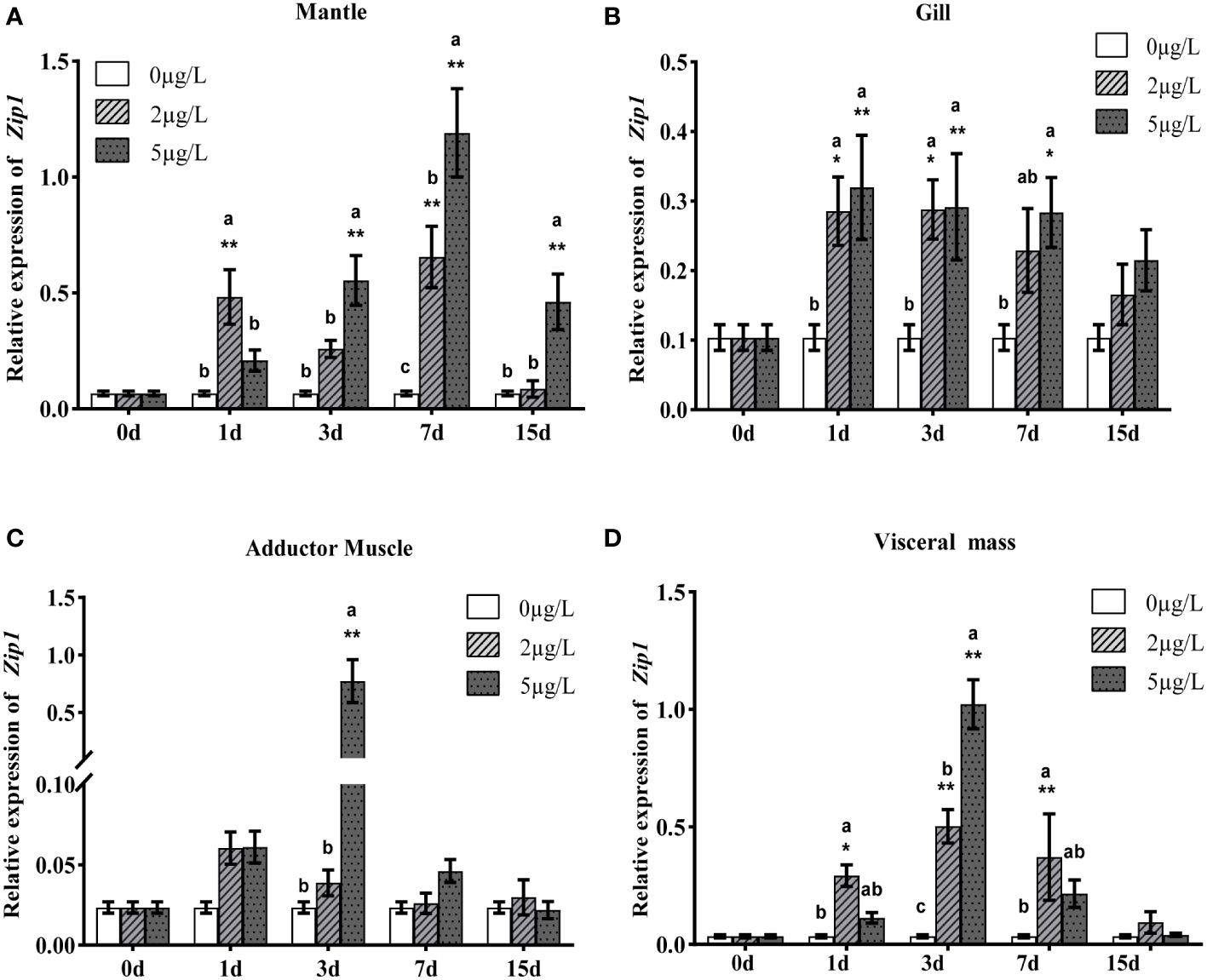
Figure 3 Gene expression analysis of Zip1 in mantle (A), gills (B), adductor muscle (C) and visceral mass (D) of Fujian oyster. qRT-PCR data are represented as mean ± SE (n=8). EF1α was used as an internal reference to normalize Zip1 expression levels. Different lowercase letters indicate significant differences between different Cd concentrations at the same exposure time (p<0.05), while * indicates significant differences between different exposure times at the same Cd level. (*p<0.05, **p<0.01).
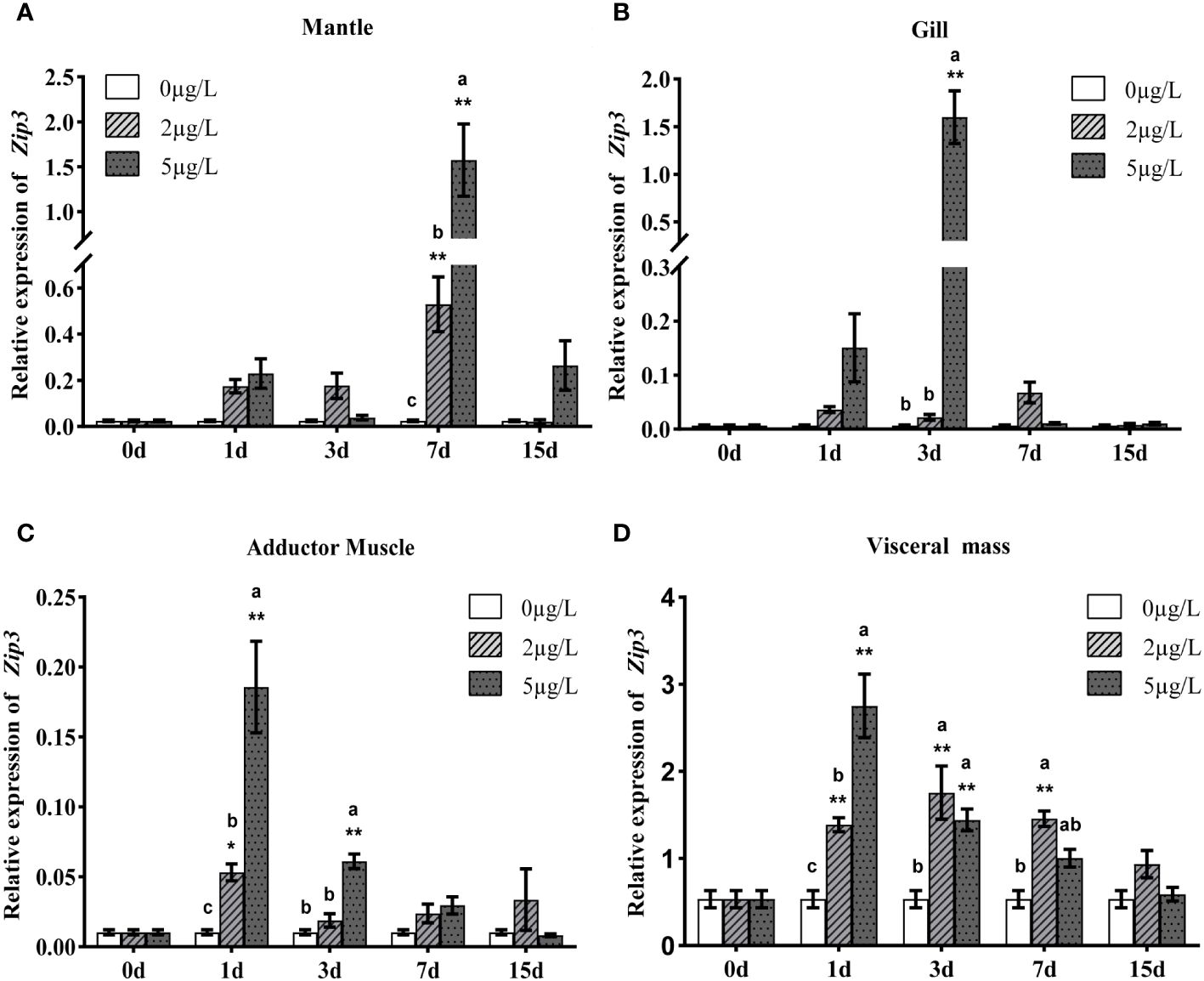
Figure 4 Analysis of Zip3 gene expression in mantle (A), gills (B), adductor muscle (C) and visceral mass (D) of Fujian oyster, following the same presentation and statistical notation as Zip1. Different lowercase letters indicate significant differences between different Cd concentrations at the same exposure time (p<0.05), while * indicates significant differences between different exposure times at the same Cd level. (*p<0.05, **p<0.01).
3.3 Histological structure and tissue-cell localization of Zip1 and Zip3
Further investigation into the localization of Zip1 and Zip3 within Fujian oyster tissues was conducted through immunohistochemistry. Location in the connective tissue was confirmed through comparison with a hematoxylin and eosin–stained mantle tissue section, which provided a more refined image of the target mantle structures (Figures 5A, B). No signal was detected in mantle tissue of control sections treated with preimmune sera (Figures 5C, D). Zip1 (Figures 5E, F) and Zip3 (Figures 5G, H) showed weak expression in the connective tissue of the mantle.
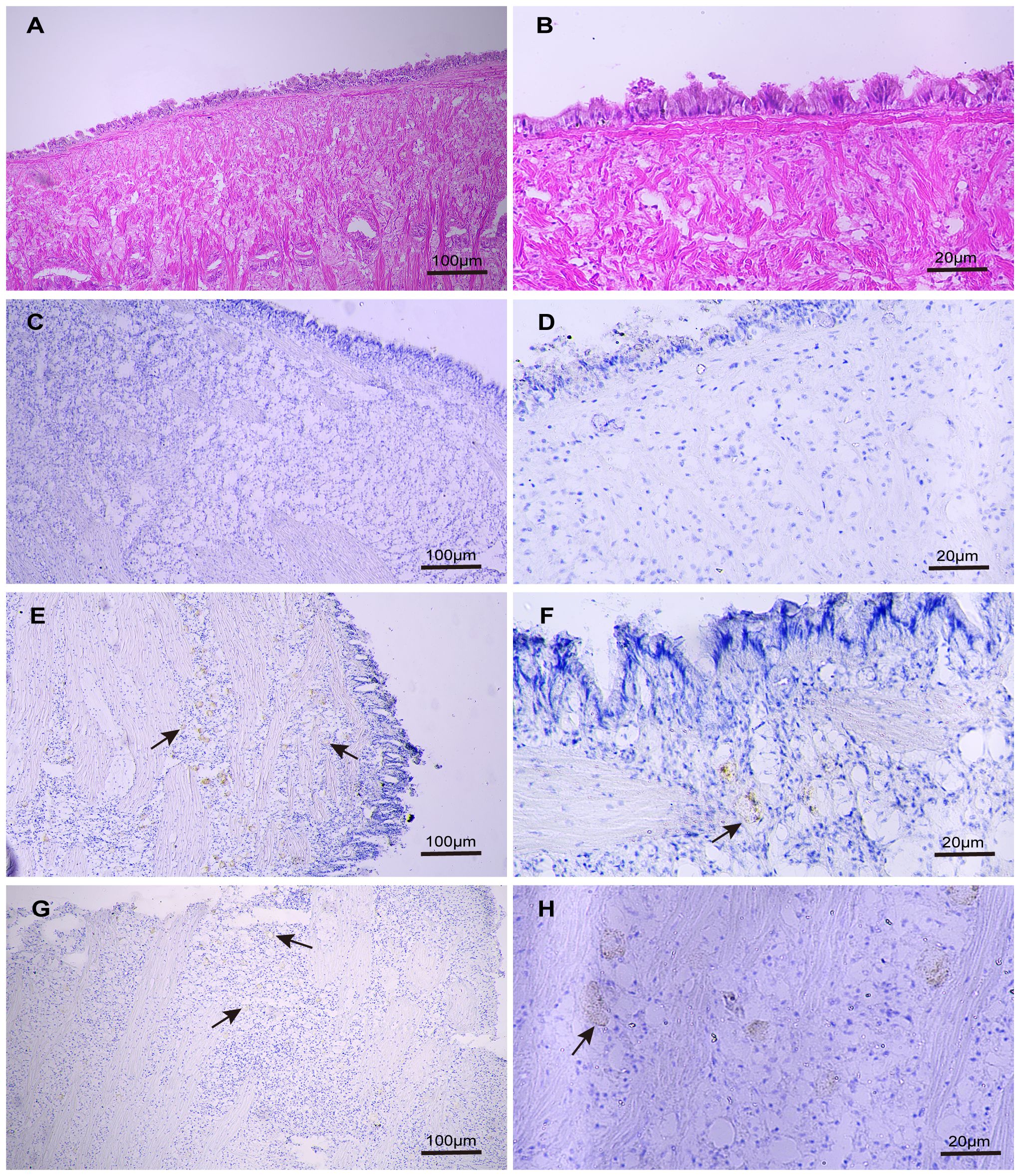
Figure 5 The distribution of Zip1 and Zip3 expression in the mantle was examined using optical microscopy images of immunoperoxidase-stained sections from paraffin-embedded Fujian oyster material exposed to 5ppb Cd for 3 days, counterstained with hematoxylin and eosin (H&E). Panels (A, B) display H&E-stained mantle sections from the same tissue not exposed to Cd. Panels (C, D) serve as negative controls for the mantle structure not exposed to Cd. Both Zip1 (E, F) and Zip3 (G, H) are weakly expressed in the connective tissue of the mantle.
3.4 The impact of dsRNA-induced silencing of Zip1 and Zip3 on Cd accumulation
3.4.1 mRNA expression levels after silencing Zip1 and Zip3
We conducted individual as well as combined interference experiments on Zip1 and Zip3. After 48 hours of interference, the expression levels of both Zip1 and Zip3 significantly decreased, as shown in Figure 6. Compared to the control group, silencing Zip1 and Zip3 for 48 hours resulted in a reduction of expression by 84% and 52%, respectively. Simultaneous silencing of Zip1+Zip3 for 48 hours led to a reduction in their expression by 64% and 62%, respectively, demonstrating that the addition of dsRNA in vitro significantly reduces the expression levels of Zip1 and Zip3 in mantle cells.
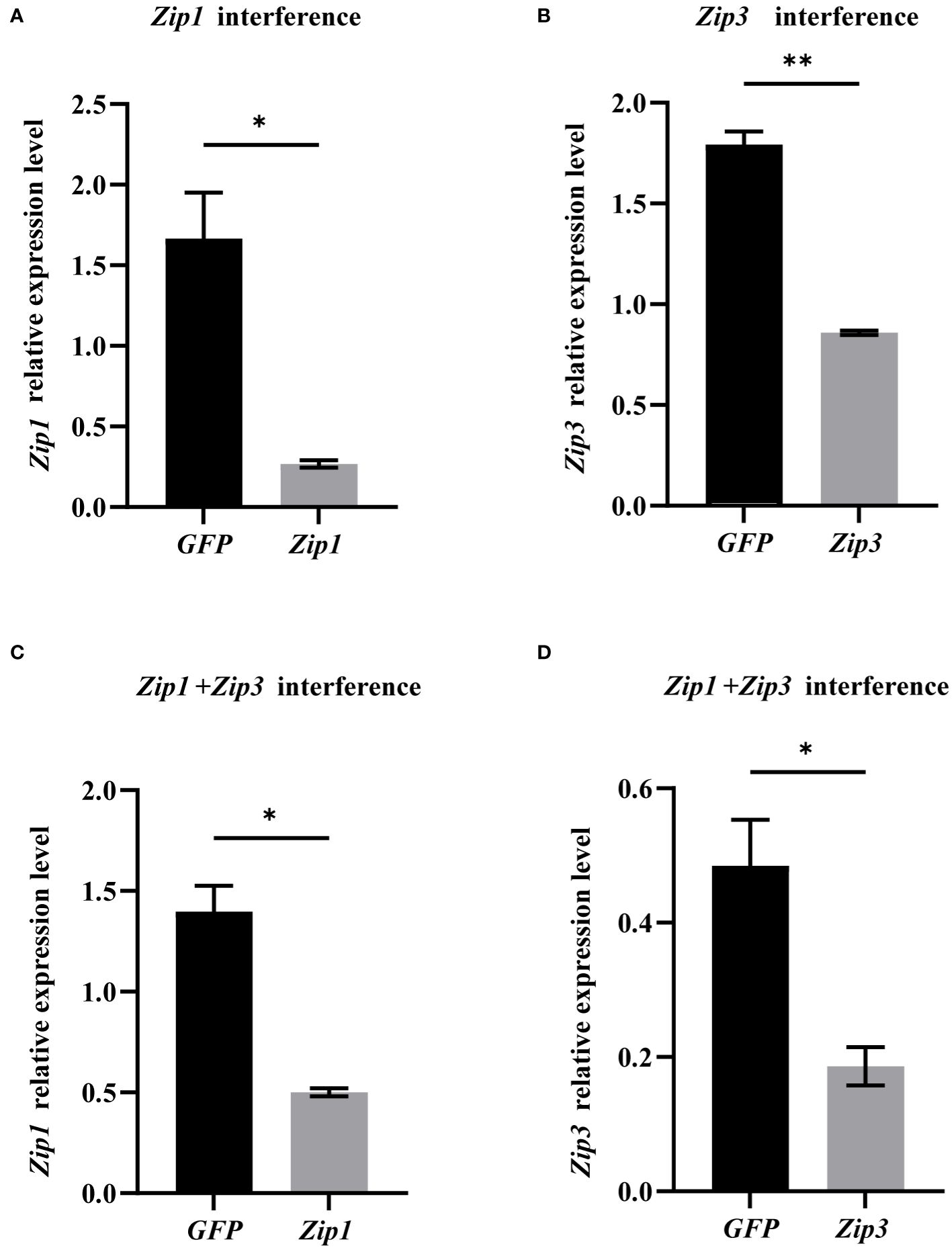
Figure 6 mRNA expression levels of Zip1 and Zip3 after gene silencing. (A) Silencing efficiency of Zip1 after 48 hours. (B) Silencing efficiency of Zip3 after 48 hours. (C) Silencing efficiency of Zip1 after dual silencing of Zip1 and Zip3 for 48 hours. (D) Silencing efficiency of Zip3 after dual silencing of Zip1 and Zip3 for 48 hours. The significant differences compared to controls are denoted with *. (*p<0.05,**p<0.01).
3.4.2 Cd content in mantle tissue after dsRNA-induced silencing of Zip1 and Zip3
The data in Figure 7 revealed that, although Cd content significantly increased in both dsGFP and dsZip1 groups after silencing Zip1 for 48 hours, but the Cd content in the dsGFP group was 1.26 times that of the dsZip1 group. Thus, Zip1 silencing significantly reduced the accumulation of Cd in the mantle. After silencing Zip3, the Cd content in the dsZip3 group was 1.29 times that of the dsGFP group. The Cd content in the group with both Zip1 and Zip3 silenced was 1.15 times that of the control group, slightly lower than the individual interference group.
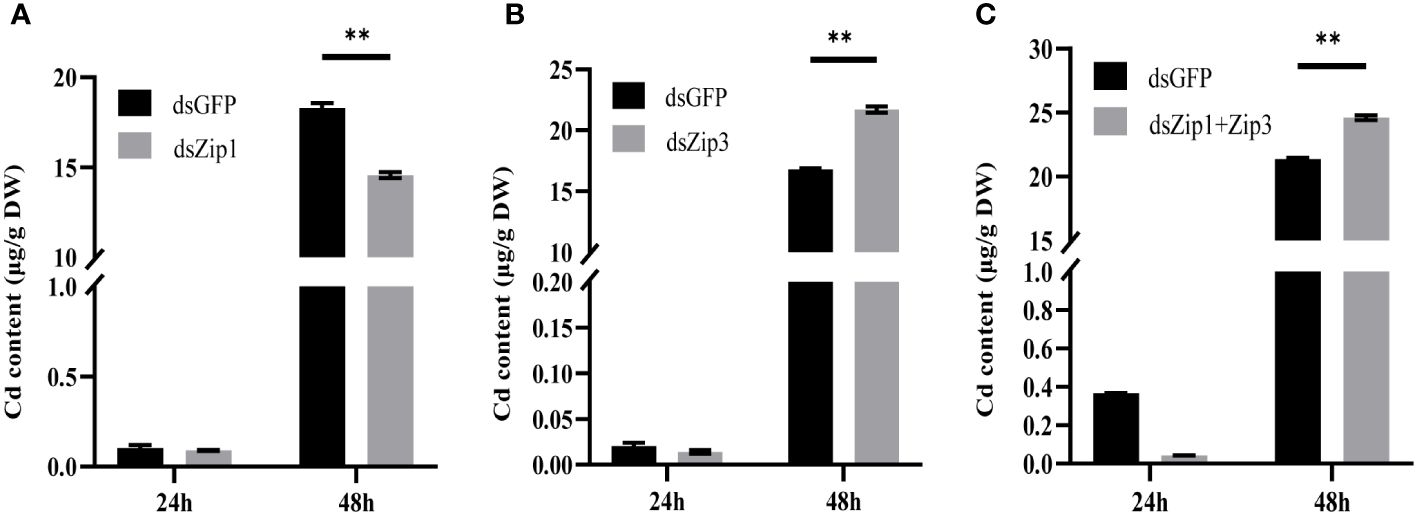
Figure 7 Cd content in oyster mantle tissue after gene silencing. Cd new accumulation in the mantle 24 hours after adding dsRNA (or dsGFP for the control group), followed by exposure to 5ppb 113Cd for 24 hours. (A) After adding dsZip1, (B) after adding dsZip3, (C) after adding both dsZip1 and dsZip3. The significant differences compared to controls are denoted with **, p<0.01.
4 Discussion
Our preliminary genome-wide association analysis of Cd accumulation traits in Fujian oyster revealed significant correlations between Cd accumulation and the polymorphisms of Zip1 and Zip3, suggesting their potential involvement in the regulation of Cd accumulation in oysters. To date, research on Zip1 and Zip3 in aquatic animals has primarily focused on their role in Zn transport regulation in fish (Chen et al., 2020, 2020; Puar et al., 2020), crabs (Liu et al., 2023), and bivalves (Zhao et al., 2020; Meng et al., 2021). However, in bivalves, only the expression changes of Zip1 and Zip3 following Cd stress have been observed, with their involvement in the cellular transport of Cd in oysters remaining unclear. To further elucidate the function of Zip1 and Zip3 in bivalves and uncover their role in Cd transport, we cloned the cDNA of Zip1 and Zip3 from Fujian oyster. Multiple sequence alignments and phylogenetic analysis show that Zip1 and Zip3 are highly conserved and closely related to orthologues in other bivalve and gastropod species. Like other members of the Zip family, they contain eight α-helical transmembrane domains and similar topologies, demonstrating strong conservation across animals, plants, and fungi (Eide, 2006). Between their TM3 and TM4 domains lies a His-rich loop region associated with metal-binding capacity. Studies have shown that, besides Zn, this region can also bind Cd and cobalt (Eide, 2006), indicating the potential of Fujian oyster Zip1 and Zip3 to bind Cd. Moreover, a conserved amino acid difference in TM4 between Zip3 and Zip1 suggests functional divergence.
In this study, as Cd exposure concentration and duration increased, oyster tissues exhibited similar Cd accumulation patterns, showing dose and time-dependent increases, peaking on the 15th day. Another study also reported significant increases in oyster Cd content after continuous exposure to various Cd concentrations for 96 hours (Moncaleano-Nino et al., 2017). Additionally, before Cd exposure, the highest Cd accumulation was in the visceral mass, followed by gills and least in the adductor muscle. After exposure, the highest accumulation shifted to the gills, then the mantle, with the visceral mass third. Previous research has shown that oyster tissues acquire Cd from both water and food, with increased waterborne Cd concentrations leading to rapid internal accumulation through the mantle and gills (Cao et al., 2018). The expression patterns under Cd stress indicated that Zip1 and Zip3 in the mantle, gills, adductor muscle, and visceral mass are rapidly upregulated by Cd stress, significantly increasing before gradually returning to control levels over time, showing dose and time-dependent changes. Similar Cd-induced expression of Zip1 and Zip3 has been observed in Pacific oyster, suggesting their potential role in regulating oyster Cd accumulation (Meng et al., 2021). Existing studies confirm that Zip1 and Zip3 expression in different mouse cells is associated with Zn homeostasis and tightly regulated by Zn (Bogdanovic et al., 2022). In humans, high Zn levels activate the transcription factor RREB1, which downregulates Zip1 (Zou et al., 2011) and upregulates Zip3 (Franklin et al., 2014) to regulate cellular Zn homeostasis. Thus, Cd stress may activate transcription factors for oyster Zip1 and Zip3, causing gene expression changes. Additionally, competition for binding sites between Zn and Cd in oyster cells may occur (Kandhol et al., 2023; Liu et al., 2023). During exposure, Cd may compete with Zn for ion channels, leading to changes in the homeostasis of Zn and thereby enhancing the expression of Zn transporters (Fontanili et al., 2016). Moreover, the significant expression of Zip1 and Zip3 in the mantle and gills, the primary Cd accumulation tissues, compared to lower accumulating tissues like the adductor muscle, and higher expression in individuals with greater accumulation (Wu et al., 2023) suggests their involvement in the transport and accumulation of Cd in oyster cells.
Zip1 and Zip3 show weak expression in the blood or nerve cells within the connective tissue of the oyster mantle. Studies in humans and mice have found Zip1 expressed in almost all tissues, localized on the plasma membrane, whereas Zip3’s localization on the plasma membrane or intracellular compartments is influenced by Zn ion concentration (Franklin et al., 2003; Jeong and Eide, 2013; Bowers and Srai, 2018). The mantle, composed of inner and outer epithelia, connective tissue, muscle fibers, and secretory gland cells, acts as a filter-feeding tissue directly in contact with the external environment and is well-supplied with blood vessels, participating in oyster respiration and secretion, indicating the mantle serves as a primary barrier between the environment, hemolymph, and internal organs. Previous studies have shown that Cd stress in oysters activates the “hemocyte-neuron” neuroendocrine-immune axis in connective tissue and induces oyster hemocyte apoptosis to mediate neuroimmune regulation (Sokolova et al., 2004; Liu et al., 2017). Furthermore, hemocytes in connective tissue can store large amounts of metals like Zn, Cu, and Cd (Shi et al., 2019b). Based on the localization of Zip1 and Zip3 in the mantle’s connective tissue, it is speculated that ZIP transport proteins are involved in Cd absorption and accumulation in the mantle.
Cd is a non-essential element for biological systems, and its absorption and excretion processes require the utilization of channels for other essential metals. Studies in animals, plants, and fungi have shown that Cd can enter cells via transport channels for Ca (Washko and Cousins, 1976; Min et al., 2008, 2015), Fe (Ragan, 1977; Flanagan et al., 1978), Zn (Waalkes and Perantoni, 1988; Dalton et al., 2005; Girijashanker et al., 2008) and Mn (Sun et al., 2019). The mantle is an important organ for oysters to accumulate metals (Shi et al., 2019a). In our study, disruption of Zip1 led to a significant reduction in Cd accumulation in the oyster mantle compared to controls, indicating that Zip1 transcription levels affect Cd accumulation in oyster mantles. Zip1 has been confirmed to transport Zn from the extracellular environment into cells in humans, mice, and yeast (Zhao and Eide, 1996; Dufner-Beattie et al., 2003; Wang et al., 2004). Furthermore, research has indicated that in Arabidopsis thaliana, genome-wide bioinformatics screening revealed a significant upregulation of the expression of Zinc Transporter Protein 1 (Zip1) upon exposure to Cd. It is hypothesized that this may play a certain role in the uptake of Cd in plants (Zheng et al., 2018). Thus, we propose that Zip1 in oyster cells is a crucial transport protein for Cd absorption. Silencing of Zip1 reduced Cd absorption, leading to decreased Cd accumulation in the mantle. However, Cd accumulation still occurred in the mantle after Zip1 silencing, suggesting that the interference was not completely blocking (efficiency of 84%) or that Cd might enter cells through other metal transport channels.
After disrupting Zip3, we observed a significant increase in Cd accumulation in the mantle. Zip3 has been confirmed to have multiple Zn transport functions across the plasma membrane and intracellular compartments (Costello et al., 2011). Studies have shown that in Zn-deficient yeast, Zrt3 can transport Zn stored in vacuoles to the cytoplasm (Zhao and Eide, 1996). Additionally, in Zn-deficient mice, Zip3 is localized on the surface of mammary epithelial cells; after Zn treatment, the protein relocates to intracellular compartments of secretory-type mammary epithelial cells, facilitating Zn transport from the cytoplasm into these compartments, which helps transfer Zn from maternal circulation to milk (Kelleher and Lönnerdal, 2005). When oysters face metal stress, they can mitigate metal toxicity through enhanced excretion, metal chelation, and compartmentalization (Engel, 1983; Liu and Wang, 2016). Some studies on human and mouse Cd exposure have shown that absorbed Cd enters blood cells or lysosomes, forming Cd-metallothionein (MT) complexes, where Cd is released from MT and appears in the cytoplasm, altering the electrochemical gradient on membranes to facilitate Cd excretion through renal tubules with urine (Nordberg, 2009). In addition, numerous studies have investigated transport proteins involved in Cd ion transportation in plants. A. thaliana AtABCG36/AtPDR8, a member of the ABC transporter family, is localized to the cell membrane. It functions by transporting Cd from the cell interior to the exterior, reducing Cd toxicity in plants and correlating with lead tolerance (Kim et al., 2007; Park et al., 2012). Additionally, the three genes of the ABCC family, AtABCC1, AtABCC2 and AtABCC3, sequester Cd by transporting it from the cytoplasm into vacuoles, thereby mitigating Cd’s harmful effects on plants (Park et al., 2012; Brunetti et al., 2015). Based on the reported mechanisms of Cd efflux and the results of this experiment, we hypothesized that Cd efflux was blocked after Zip3 interference, resulting in a significant increase in Cd content in the mantle. Therefore, based on the expression patterns, interference, and expression localization analysis of Zip1 and Zip3, we hypothesize that they play pivotal roles in the accumulation of Cd in oysters. Further research is needed to evaluate the roles of Zip1 and Zip3 in intracellular and intercellular Cd transport in oysters.
5 Conclusion
Genetic improvements are highly beneficial for the sustainable development of the oyster aquaculture industry. This study has identified tissue-specific expressions of Zip1 and Zip3 in Fujian oyster. Results from RNAi and immunohistochemistry further confirm the critical roles of Zip1 and Zip3 in Cd accumulation in Fujian oyster. Altogether, these findings provide a basis for future research into the molecular mechanisms of Cd accumulation in Fujian oyster and the cultivation of low-Cd oyster strains.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, accession numbers: OR759776 and OR759777.
Ethics statement
The animal study was approved by the Animal Care and Use Committee of the School of Aquatic Sciences, Jimei University (Animal Ethics Number 1067). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HL: Writing – original draft, Writing – review & editing. ZL: Data curation, Software, Writing – review & editing. LW: Data curation, Software, Writing – review & editing. YK: Funding acquisition, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – review & editing. HQ: Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Visualization, Writing – review & editing. BS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the National Natural Science Foundation of China (grant number 32172949), the Natural Science Foundation of Fujian Province (grant number 2020J05135) and the Creation and Demonstration Application of Excellent New Germplasm of Fujian oyster (grant number FJHYF-L-2023-2).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bogdanovic M., Asraf H., Gottesman N., Sekler I., Aizenman E., Hershfinkel M. (2022). The ZIP3 zinc transporter is localized to mossy fiber terminals and is required for kainate-induced degeneration of CA3 neurons. J. Neurosci. 42, 2824–2834. doi: 10.1523/JNEUROSCI.0908-21.2022
Boisson F., Goudard F., Durand J. P., Barbot C., Pieri J., Amiard J. C., et al. (2003). Comparative radiotracer study of cadmium uptake, storage, detoxification and depuration in the oyster Crassostrea gigas: potential adaptive mechanisms. Mar. Ecol. Prog. Ser. 254, 177–186. doi: 10.3354/meps254177
Bowers K., Srai S. K. S. (2018). The trafficking of metal ion transporters of the Zrt- and Irt-like protein family. Traffic 19, 813–822. doi: 10.1111/tra.12602
Brunetti P., Zanella L., De Paolis A., Di Litta D., Cecchetti V., Falasca G., et al. (2015). Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 66, 3815–3829. doi: 10.1093/jxb/erv185
Cao R., Liu Y., Wang Q., Dong Z., Yang D., Liu H., et al. (2018). Seawater acidification aggravated cadmium toxicity in the oyster Crassostrea gigas: Metal bioaccumulation, subcellular distribution and multiple physiological responses. Sci. Total Environ. 642, 809–823. doi: 10.1016/j.scitotenv.2018.06.126
Chen S., Wu K., Lv W., Song C., Luo Z. (2020). Molecular characterization of ten zinc (Zn) transporter genes and their regulation to Zn metabolism in freshwater teleost yellow catfish Pelteobagrus fulvidraco. J. Trace Elem. Med. Biol. 59, 126433. doi: 10.1016/j.jtemb.2019.126433
Connolly E. L., Fett J. P., Guerinot M. L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell. 14, 1347–1357. doi: 10.1105/tpc.001263
Copes R., Clark N. A., Rideout K., Palaty J., Teschke K. (2008). Uptake of cadmium from Pacific oysters (Crassostrea gigas) in British Columbia oyster growers. Environ. Res. 107, 160–169. doi: 10.1016/j.envres.2008.01.014
Costello L. C., Levy B. A., Desouki M. M., Zou J., Bagasra O., Johnson L. A., et al. (2011). Decreased zinc and downregulation of ZIP3 zinc uptake transporter in the development of pancreatic adenocarcinoma. Cancer Biol. Ther. 12, 297–303. doi: 10.4161/cbt.12.4.16356
Dalton T. P., He L., Wang B., Miller M. L., Jin L., Stringer K. F., et al. (2005). Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis. Proc. Natl. Acad. Sci. U. S. A. 102, 3401–3406. doi: 10.1073/pnas.0406085102
Delhaize E., Ryan P. R., Hebb D. M., Yamamoto Y., Sasaki T., Matsumoto H. (2004). Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc. Natl. Acad. Sci. U. S. A. 101, 15249–15254. doi: 10.1073/pnas.0406258101
Dufner-Beattie J., Langmade S. J., Wang F. D., Eide D., Andrews G. K. (2003). Structure, function, and regulation of a subfamily of mouse zinc transporter genes. J. Biol. Chem. 278, 50142–50150. doi: 10.1074/jbc.M304163200
Eide D. J. (2006). Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta-Mol. Cell Res. 1763, 711–722. doi: 10.1016/j.bbamcr.2006.03.005
Engel D. W. (1983). The intracellular partitioning of trace-metals in marine shellfish. Sci. Total Environ. 28, 129–140. doi: 10.1016/S0048-9697(83)80013-8
Engel D. W. (1999). Accumulation and cytosolic partitioning of metals in the American oyster Crassostrea virginica. Mar. Environ. Res. 47, 89–102. doi: 10.1016/S0141-1136(97)00100-1
FAO (2022). The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation (Rome: Food & Agriculture Organization of the United). doi: 10.4060/cc0461zh
Flanagan P. R., McLellan J. S., Haist J., Cherian G., Chamberlain M. J., Valberg L. S. (1978). Increased dietary cadmium absorption in mice and human subjects with iron deficiency. Gastroenterology 74, 841–846. doi: 10.1016/0016-5085(78)90138-5
Fontanili L., Lancilli C., Suzui N., Dendena B., Yin Y., Ferri A., et al. (2016). Kinetic analysis of zinc/cadmium reciprocal competitions suggests a possible Zn-insensitive pathway for root-to-shoot cadmium translocation in rice. Rice 9, 1–13. doi: 10.1186/s12284-016-0088-3
Franklin R. B., Ma J., Zou J., Guan Z., Kukoyi B. I., Feng P., et al. (2003). Human ZIP1 is a major zinc uptake transporter for the accumulation of zinc in prostate cells. J. Inorg. Biochem. 96, 435–442. doi: 10.1016/S0162-0134(03)00249-6
Franklin R. B., Zou J., Costello L. C. (2014). The cytotoxic role of RREB1, ZIP3 zinc transporter, and zinc in human pancreatic adenocarcinoma. Cancer Biol. Ther. 15, 1431–1437. doi: 10.4161/cbt.29927
Gaither L. A., Eide D. J. (2001). The human ZIP1 transporter mediates zinc uptake in human K562 erythroleukemia cells. J. Biol. Chem. 276, 22258–22264. doi: 10.1074/jbc.M101772200
Girijashanker K., He L., Soleimani M., Reed J. M., Li H., Liu Z., et al. (2008). Slc39a14 gene encodes ZIP14, a metal/bicarbonate symporter: similarities to the ZIP8 transporter. Mol. Pharmacol. 73, 1413–1423. doi: 10.1124/mol.107.043588
Gitan R. S., Luo H., Rodgers J., Broderius M., Eide D. (1998). Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273, 28617–28624. doi: 10.1074/jbc.273.44.28617
He M., Wang W. X. (2013). Bioaccessibility of 12 trace elements in marine molluscs. Food. Chem. Toxicol. 55, 627–636. doi: 10.1016/j.fct.2013.01.046
Jeong J., Eide D. J. (2013). The SLC39 family of zinc transporters. Mol. Aspects. Med. 34, 612–619. doi: 10.1016/j.mam.2012.05.011
Kandhol N., Rai P., Pandey S., Singh S., Sharma S., Corpas F. J., et al. (2023). Zinc induced regulation of PCR1 gene for cadmium stress resistance in rice roots. Plant Sci. 337, 111783. doi: 10.1016/j.plantsci.2023.111783
Ke C., Wang W. X. (2001). Bioaccumulation of Cd, Se, and Zn in an estuarine oyster (Crassostrea rivularis) and a coastal oyster (Saccostrea glomerata). Aquat. Toxicol. 56, 33–51. doi: 10.1016/S0166-445X(01)00185-0
Kelleher S. L., Lönnerdal B. (2005). Zip3 plays a major role in zinc uptake into mammary epithelial cells and is regulated by prolactin. Am. J. Physiol. Cell Physiol. 288, C1042–C1047. doi: 10.1152/ajpcell.00471.2004
Kim D. Y., Bovet L., Maeshima M., Martinoia E., Lee Y. (2007). The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218. doi: 10.1111/j.1365-313X.2007.03044.x
Kingtong S., Chitramvong Y., Janvilisri T. (2007). ATP-binding cassette multidrug transporters in Indian-rock oyster Saccostrea forskali and their role in the export of an environmental organic pollutant tributyltin. Aquat. Toxicol. 85, 124–132. doi: 10.1016/j.aquatox.2007.08.006
Korshunova Y. O., Eide D., Clark W. G., Guerinot M. L., Pakrasi H. B. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 40, 37–44. doi: 10.1023/A:1026438615520
Kumar S., Sharma A. (2019). Cadmium toxicity: effects on human reproduction and fertility. Rev. Environ. Health 34, 327–338. doi: 10.1515/reveh-2019-0016
Liu A. (2022). Roles and mechanism of allatostain in reproductive endocrine regulation of the mud crab, Scylla paramamosain. [dissertation/doctor's thesis]. Xiamen, Fujian: Xiamen University (in Chinese).
Liu J., Wang E., Cheng Z., Gao Y., Chen C., Jia R., et al. (2023). Zinc alleviates cadmium-induced reproductive toxicity via regulating ion homeostasis, metallothionein expression, and inhibiting mitochondria-mediated apoptosis in the freshwater crab Sinopotamon henanense. Ecotoxicol. Environ. Saf. 262, 115188. doi: 10.1016/j.ecoenv.2023.115188
Liu X., Wang W. (2016). Antioxidant and detoxification responses of oysters Crassostrea hongkongensis in a multimetal-contaminated estuary. Environ. Toxicol. Chem. 35, 2798–2805. doi: 10.1002/etc.3455
Liu Z., Zhou Z., Jiang Q., Wang L., Yi Q., Qiu L., et al. (2017). The neuroendocrine immunomodulatory axis-like pathway mediated by circulating haemocytes in pacific oyster Crassostrea gigas. Open Biol. 7, 160289. doi: 10.1098/rsob.160289
Luo L., Ke C., Guo X., Shi B., Huang M. (2014). Metal accumulation and differentially expressed proteins in gill of oyster (Crassostrea hongkongensis) exposed to long-term heavy metal-contaminated estuary. Fish Shellfish Immunol. 38, 318–329. doi: 10.1016/j.fsi.2014.03.029
Luo H., Wang Q., Nie X., Ren H., Shen Z., Xie X., et al. (2018). Heavy metal contamination in the cultivated oyster Crassostrea rivularis and associated health risks from a typical mariculture zone in the South China Sea. Bull. Environ. Contam. Toxicol. 101, 33–41. doi: 10.1007/s00128-018-2360-2
Mark D. C., Stephen M. G., Sanford E. (2005). Can selective breeding reduce the heavy metals content of Pacific oysters (Crassostrea gigas), and are there trade-offs with growth or survival? J. Shellfish Res. 24, 979–986. doi: 10.2983/0730–8000(2005)24[979:CSBRTH]2.0.CO;2
Meng J., Wang W., Li L., Zhang G. (2021). Accumulation of different metals in oyster Crassostrea gigas: Significance and specificity of SLC39A (ZIP) and SLC30A (ZnT) gene families and polymorphism variation. Environ. pollut. 276, 116706. doi: 10.1016/j.envpol.2021.116706
Min K. S., Sano E., Ueda H., Sakazaki F., Yamada K., Takano M., et al. (2015). Dietary deficiency of calcium and/or iron, an age-related risk factor for renal accumulation of cadmium in mice. Biol. Pharm. Bull. 38, 1557–1563. doi: 10.1248/bpb.b15-00341
Min K. S., Ueda H., Tanaka K. (2008). Involvement of intestinal calcium transporter 1 and metallothionein in cadmium accumulation in the liver and kidney of mice fed a low-calcium diet. Toxicol. Lett. 176, 85–92. doi: 10.1016/j.toxlet.2007.10.011
Moncaleano-Nino A. M., Barrios-Latorre S. A., Poloche-Hernandez J. F., Becquet V., Huet V., Villamil L., et al. (2017). Alterations of tissue metallothionein and vitellogenin concentrations in tropical cup oysters (Saccostrea sp.) following short-term (96 h) exposure to cadmium. Aquat. Toxicol. 185, 160–170. doi: 10.1016/j.aquatox.2017.02.011
Ni J., Zeng Z., Ke C. (2013). Sex steroid levels and expression patterns of estrogen receptor gene in the oyster Crassostrea angulata during reproductive cycle. Aquaculture 376, 105–116. doi: 10.1016/j.aquaculture.2012.11.023
Nordberg G. F. (2009). Historical perspectives on cadmium toxicology. Toxicol. Appl. Pharmacol. 238, 192–200. doi: 10.1016/j.taap.2009.03.015
Pan K., Wang W. (2009). Biodynamics to explain the difference of copper body concentrations in five marine bivalve species. Environ. Sci. Technol. 43, 2137–2143. doi: 10.1021/es802888u
Pan K., Wang W. X. (2012). Reconstructing the biokinetic processes of oysters to counteract the metal challenges: physiological acclimation. Environ. Sci. Technol. 46, 10765–10771. doi: 10.1021/es302040g
Park J., Song W. Y., Ko D., Eom Y., Hansen T. H., Schiller M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288. doi: 10.1111/j.1365-313X.2011.04789.x
Patra P., Mohandass C., Chakraborty P., Seleyi S. C. (2021). Biochemical responses of oysters in evaluating environmental quality of tropical Indian estuarine systems. Chemosphere 278, 130338. doi: 10.1016/j.chemosphere.2021.130338
Puar P., Niyogi S., Kwong R. W. M. (2020). Regulation of metal homeostasis and zinc transporters in early-life stage zebrafish following sublethal waterborne zinc exposure. Aquat. Toxicol. 225, 105524. doi: 10.1016/j.aquatox.2020.105524
Ragan H. A. (1977). Effects of iron deficiency on the absorption and distribution of lead and cadmium in rats. J. Lab. Clin. Med. 90, 700–706.
Rao X., Huang X., Zhou Z., Lin X. (2013). An improvement of the 2^ΔΔCT method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma Biomath. 3, 71–85. doi: 10.1016/S0920-5489(99)92176-1
Sasaki T., Yamamoto Y., Ezaki B., Katsuhara M., Ahn S. J., Ryan P. R., et al. (2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653. doi: 10.1111/j.1365-313X.2003.01991.x
Satapathy S., Panda C. R., Jena B. S. (2019). Risk-based prediction of metal toxicity in sediment and impact on human health due to consumption of seafood (Saccostrea cucullata) found in two highly industrialised coastal estuarine regions of Eastern India: a food safety issue. Environ. Geochem. Health 41, 1967–1985. doi: 10.1007/s10653-019-00251-4
Shi B., Wu Y. D., Zhou L., You W. W., Ke C. H. (2019a). Influence of gonadal development on metal accumulation in the Portuguese oyster, Crassostrea angulata, in subtropical areas. Aquac. Res. 50, 1142–1152. doi: 10.1111/are.13988
Shi B., Wang T., Zeng Z., Zhou L., You W., Ke C. (2019b). The role of copper and zinc accumulation in defense against bacterial pathogen in the Fujian oyster (Crassostrea angulata). Fish Shellfish Immunol. 92, 72–82. doi: 10.1016/j.fsi.2019.05.049
Sokolova I. M., Evans S., Hughes F. M. (2004). Cadmium-induced apoptosis in oyster hemocytes involves disturbance of cellular energy balance but no mitochondrial permeability transition. J. Exp. Biol. 207, 3369–3380. doi: 10.1242/jeb.01152
Sun Q., Cui P., Zhu M., Fan T., Ata-Ul-Karim S. T., Gu J., et al. (2019). Cd (II) retention and remobilization on δ-MnO2 and Mn (III)-rich δ-MnO2 affected by Mn (II). Environ. Int. 130, 104932. doi: 10.1016/j.envint.2019.104932
Tang L., Mao B., Li Y., Lv Q., Zhang L., Chen C., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 7, 14438. doi: 10.1038/s41598–017-14832–9
Waalkes M. P., Perantoni A. (1988). In vitro assessment of target cell specificity in cadmium carcinogenesis: interactions of cadmium and zinc with isolated interstitial cells of the rat testes. In Vitro Cell Dev. Biol. 24, 558–565. doi: 10.1007/BF02629091
Wang M., Chen Z., Song W., Hong D., Huang L., Li Y. (2021). A review on cadmium exposure in the population and intervention strategies against cadmium toxicity. Bull. Environ. Contam. Toxicol. 106, 65–74. doi: 10.1007/s00128-020-03088-1
Wang F. D., Dufner-Beattie J., Kim B. E., Petris M. J., Andrews G., Eide D. J. (2004). Zinc-stimulated endocytosis controls activity of the mouse ZIP1 and ZIP3 zinc uptake transporters. J. Biol. Chem. 279, 24631–24639. doi: 10.1074/jbc.M400680200
Washko P. W., Cousins R. J. (1976). Metabolism of 109Cd in rats fed normal and low-calcium diets. J. Toxicol. Environ. Health 1, 1055–1066. doi: 10.1080/15287397609529408
Weng N., Wang W. (2014). Variations of trace metals in two estuarine environments with contrasting pollution histories. Sci. Total Environ. 485, 604–614. doi: 10.1016/j.scitotenv.2014.03.110
Wu L., Gong S., Li H., Ke C., Shi B. (2023). Genome - wide association study and candidate gene analysis of cadmium accumulation in Fujian oysters (Crasosstrea angulata). Aquaculture 572, 739546. doi: 10.1016/j.aquaculture.2023.739546
Wu Z., Wang Y., Li X., Liu T., Sun Y., Li H., et al. (2013). Dynamics studies on accumulation and elimination of cadmium by two economic shellfishes. Period. Ocean Univ. China. 43, 56–61. doi: 10.16441/j.cnki.hdxb.2013.12.009
You Y., Huan P., Liu B. (2012). RNAi assay in primary cells: a new method for gene function analysis in marine bivalve. Mol. Biol. Rep. 39, 8209–8216. doi: 10.1007/s11033-012-1668-y
Yu R., Li D., Du X., Xia S., Liu C., Shi G. (2017). Comparative transcriptome analysis reveals key cadmium transport-related genes in roots of two pak choi (Brassica rapa L. ssp chinensis) cultivars. BMC Genomics 18, 587. doi: 10.1186/s12864-017-3973-2
Zhao H., Eide D. (1996). The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. U. S. A. 93, 2454–2458. doi: 10.1073/pnas.93.6.2454
Zhao Y., Kang X., Shang D., Ning J., Ding H., Zhai Y., et al. (2020). Hyperaccumulation of cadmium by scallop Chlamys farreri revealed by comparative transcriptome analysis. Biometals 33, 397–413. doi: 10.1007/s10534-020-00257-x
Zheng X., Chen L., Li X. (2018). Arabidopsis and rice showed a distinct pattern in ZIPs genes expression profile in response to Cd stress. Bot. Stud. 59, 43 (2024). doi: 10.1186/s40529-018-0238-6
Zhong G., Lu S., Chen R., Chen N., Tan Q. (2020). Predicting risks of cadmium toxicity in salinity-fluctuating estuarine waters using the toxicokinetic-toxicodynamic model. Environ. Sci. Technol. 54, 13899–13907. doi: 10.1021/acs.est.0c06644
Keywords: Fujian oyster, cadmium accumulation, Zip1, Zip3, expression analysis, RNAi, immunohistochemistry
Citation: Li H, Liang Z, Wu L, Ke Y, Que H and Shi B (2024) The role of Zip1 and Zip3 in cadmium accumulation in Fujian oyster (Crassostrea angulata). Front. Mar. Sci. 11:1412127. doi: 10.3389/fmars.2024.1412127
Received: 04 April 2024; Accepted: 09 May 2024;
Published: 24 May 2024.
Edited by:
Jinhyun Kim, Korea Polar Research Institute, Republic of KoreaReviewed by:
Shaharior Hossen, Chonnam National University, Republic of KoreaMaocang Yan, Zhejiang Mariculture Research Institute, China
Copyright © 2024 Li, Liang, Wu, Ke, Que and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Shi, c2hpYm9Aam11LmVkdS5jbg==
 Huanhuan Li
Huanhuan Li Huayong Que
Huayong Que Bo Shi
Bo Shi