- 1Division of Cancer Therapeutics, The Institute of Cancer Research, London, UK
- 2Cancer Center, Humanitas Clinical and Research Center, Milano, Italy
- 3The Royal Marsden NHS Foundation Trust, London, UK
Hepatocellular carcinoma (HCC) is a primary malignancy of the liver with poor prognosis and limited therapeutic options. Over the past few years, many studies have evaluated the role of non-coding RNAs (ncRNAs) in hepatocarcinogenesis and tumor progression. ncRNAs were shown to have diagnostic, prognostic, and therapeutic potential in HCC. In this manuscript, we review the latest major discoveries concerning microRNAs and long ncRNAs in HCC pathogenesis, and discuss the potentials and the limitations for their use in clinical practice.
Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is a primary tumor of the liver and represents the third cause of cancer deaths worldwide (1). Only few patients are eligible for curative treatments, while majority of cases are diagnosed at later stages (1). HCCs often arise on a background of liver cirrhosis and, as such, early diagnosis is frequently missed. Local ablative therapies, such as transarterial chemoembolization (TACE) and radiofrequency ablation (RFA), are used when the tumor is localized within the liver, while the multikinase-inhibitor sorafenib is the only approved systemic therapy for advanced HCC (2, 3). However, overall survival (OS) of patients affected by HCC remains poor. A growing effort has been addressed toward the study of genomics and molecular biology in order to unravel the mechanisms of liver carcinogenesis and therefore identifying novel targets of therapies as well as early diagnostic and prognostic markers to improve the clinical management of HCC patients. Along with an extensive characterization of the protein-coding genome of liver tumors (4–7), there has been a great interest in the study of non-coding RNAs (ncRNA). Hereby, we will review the role of ncRNA in liver carcinogenesis and their clinical implication.
Non-Coding RNAs in HCC
Most of the eukaryotic genome is transcribed into RNA transcripts that do not translate into proteins. These RNA transcripts can be generally divided into two classes according to their size (with 200 nt as cut-off): short ncRNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). Although they are not translated into protein products, they exert essential functions within the cell by modulating the expression of protein-coding mRNAs, interacting with proteins to affect their function, and interacting one with each other to finely tune their expression (8).
microRNAs
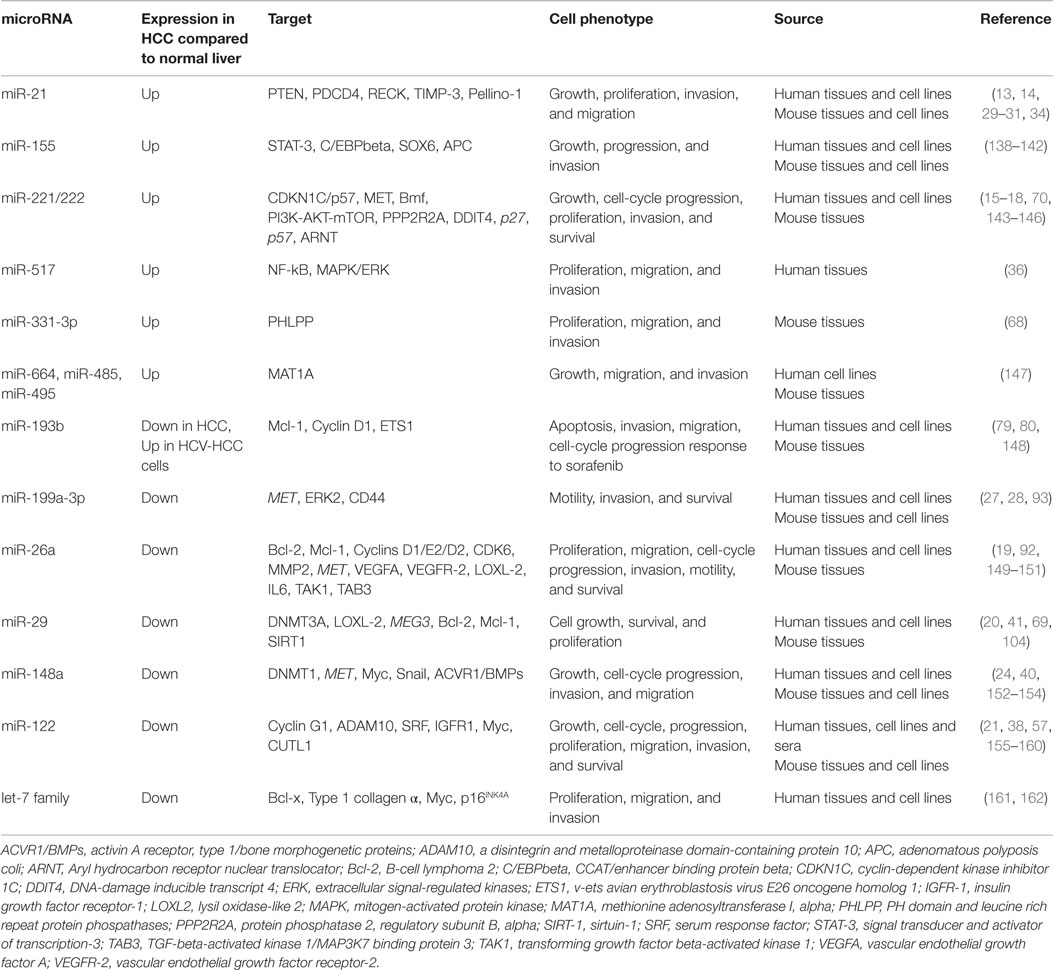
microRNAs are 18–24 nt long and represent key actors in the processes of tumor development, progression, and resistance to anti-tumor agents (9). Many studies have identified alterations in miRNAs in HCC (10–12). Some miRNAs, such as miR-21 and miR-221/222, were found increased in expression, and acting as oncogenes by affecting several cancer-related pathways (13–18). On the other hand, selected miRNAs, such as miR-26, miR-29, miR-122, miR-148a, and miR-199a, were reported to be reduced in HCC and to promote cancer by the lack of their oncosuppressive activity (19–24).
Data on the expression of miRNAs have mainly come from real time PCR-based and microarray-based profiling. More recently, the introduction of novel technologies, such as RNA next generation sequence (NGS) (RNA-seq), has led to the identification of novel miRNAs. NGS analysis produces miRNA expression profile that is reproducible and comparable to that produced by microarray, but has the advantage of discovering novel miRNAs (as for the case of miR-9986 for HCC), and of providing a detailed profile of expression of miRNA isoforms (25, 26). If down-regulation of miR-199a-3p in HCC was a solid data achieved through old technologies (27, 28), Wojcicka et al. showed through NGS that there are nine different isoforms of miR-199a-3p, which include three different seed regions. All the miR-199a-3p isoforms are lower in HCC tumors, and apparently the gene targeting is conserved across the isoforms (26). These findings add interesting insights into the understanding of miRNAs, because they imply that the message delivered by each miRNA may be amplified according to the expression of their isoforms, and probe designing should take into account the variability among these.
Several reports have confirmed that the aberrant expression of miRNAs in HCC cells is associated with the derangement of a number of pathways and processes, which all together initiate and maintain cancer, promote cell growth, mediate apoptosis escape, and induce migration and invasion (Table 1). An example among all is miR-21, whose over-expression results into the silencing of several targets including phosphatase and tensin homolog (PTEN), programed cell death protein 4 (PDCD4), reversion-inducing-cysteine-rich protein with kazal motifs (RECK), metalloproteinase inhibitor 3 (TIMP-3), and Pellino-1 (13, 14, 29–31), but is concomitantly finely tuned by a plethora of factors whose source can either be the tumor cell itself [nuclear factor kinase B (NF-KB), hepatitis B virus X protein (HBV x)] (32, 33), or microenvironment cells [interleukin-6 (IL-6) or monocyte chemotactic protein-1 (MCP-1)] (33, 34). It is interesting to note that normal liver tissue seems to express a limited number of miRNAs, including miR-199a and miR-122 (35). Thus, it is not surprising that HCC tissues exhibit loss of these miRNAs along with over-expression of others, such as miR-21, miR-221/222, and miR-517 (13, 18, 36).
The mechanisms through which miRNA expression is modulated during liver carcinogenesis are variable, and include chromosomal rearrangements, promoter methylation, and transcriptional induction through direct control of transcription factors such as Myc (9, 37–40). Recent evidence suggests that alpha-feto-protein (AFP) can alter miR-29 expression and can induce changes in the methylome of liver cancer cells that are responsible for the more aggressive behavior of AFP + HCC (41). Growing evidence show that nanovesicles-mediated delivery of miRNAs is another way of miRNA regulation and intercellular communication. Tumor cells were shown to actively secrete miRNAs through exosomes, which can then be internalized by other cells altering miRNA profiling, and modulating gene expression in donor cells (42, 43). Vacuolar protein sorting-associated protein 4A (Vps4A) has been identified as one of the regulators of exosome bioactivity, as it facilitates the release of exosome containing onco-miRs and alters the accumulation of those containing oncosuppressor miRNAs (44). miRNAs are highly conserved genes, and therefore, alterations of their sequence have been studied as potential causes of aberrant miRNA expression. It was observed that selected polymorphisms in miRNA genes might result in increased production of mature miRNA forms and therefore induce miRNA-dependent liver carcinogenesis. For instance, a G > C polymorphism located in the stem region of miR-146a is associated with increased predisposition to develop HCC (45). To date, polymorphisms in a number of miRNAs have been associated to increased risk of HCC (46–51). However, these studies have been carried out mainly in Chinese or Turkish populations, and their extrapolation to other ethnicities is not clear.
Clinical Implications of miRNAs in HCC
Diagnostic role. As previously discussed early diagnosis is one of the main challenges in the clinical management of HCC. miRNAs have shown to distinguish HCC from adjacent normal or cirrhotic tissues (18, 22, 36, 52). However, the main advances have been achieved through the detection of circulating miRNAs. Tissutal analyses are indeed a limitation in this field given HCC diagnosis is made on a combination of blood tests and imaging with no recommendation for mandatory biopsy in most of cases (1). Conversely, miRNAs can be detected in sera and plasma (53). They can circulate as free RNAs, which are bound to Argonaut RISC catalytic component 2 (Ago2) or included in exosomes; in either case, they are protected by RNases and remain stable after harsh conditions (54, 55). There are now a number of reports that showed miRNAs are detectable in the plasma and sera of HCC patients and their expression profile is different between patients with HCC and patients with cirrhosis, as reviewed by Roderburg et al. (56). The larger study includes a series of 934 participants among healthy controls, and patients with chronic hepatitis B, cirrhosis, and hepatitis B virus (HBV)-related HCC (57). A panel of seven plasma miRNAs (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801) was shown to have a high-diagnostic accuracy of HCC. The diagnostic performance persisted regardless of disease stage and was able to differentiate HCC from healthy controls, chronic hepatitis B, and cirrhosis (57), suggesting a potential use of this panel in the early diagnosis of HBV-related HCC. However, it looks like circulating miRNA profiles differ according to the etiology, and therefore, these data cannot be extrapolated to all cases of HCC. Indeed, circulating miRNAs not only can distinguish patients who have developed HCC among all the HBV carriers but can also distinguish between patients with HBV and those with hepatitis C virus (HCV) (58), suggesting that specific miRNAs may be identified in each subtype of HCC. These data are not surprising given that (1) miRNA expression profiles were shown to be different in tissues from HBV-related versus HCV-related HCC (59), (2) miRNAs can facilitate replication of hepatitis viruses (60, 61), and (3) circulating miRNAs can reflect the status of liver injury in inflammatory diseases (62). Nonetheless, we need to remember that circulating miRNAs may also originate from blood cells, and therefore, an appropriate sample collection and processing, which excludes contamination by leukocytes and erythrocytes, is mandatory in this kind of analysis (63, 64). To date, lack of standardization of sample collection and data normalization, along with limited sample size in most of the reports impair the reproducibility and comparability across the studies. Thus, despite the interesting potential of circulating miRNAs as biomarker in HCC, further investigations are still warranted before they can be taken into clinical practice.
Prognostic role. microRNAs induce malignant phenotypic changes in liver cancer cells and contribute to the acquisition of invasive and metastatic properties (65, 66). Therefore, it comes with no surprise that their prognostic value has been widely investigated in HCC patients undergoing either curative treatments or local ablative therapies. Despite, Jiang et al. have initially postulated that a global loss of miRNA expression is associated with poorer clinical outcome (67), subsequent analyses in larger cohorts of patients identified single miRNAs as potential prognostic markers. Over-expression of oncogenic miRNAs, such as miR-331-3p (68) or down-regulation of oncosuppressive miRNAs, such as miR-29 (20, 69) has been associated to poor prognosis in unselected cohorts of human HCC tissues. High miR-221 levels were associated with tumor multifocality (15) and reduced time to recurrence after surgery (70). In addition, miR-221 was shown to have a prognostic value, with a significantly lower OS in patients with high-serum miRNA expression (71). Sato et al. looked at miRNA expression in 73 human resected HCCs that had not received any pre-operative therapy and observed that miRNAs expression recapitulates the risk of early or late recurrence in analogy to mRNA profiles (6, 72). The miRNA profile of tumoral tissue could predict early recurrence, while the miRNA profile of non-tumoral tissue was predictive of late recurrence and of de novo carcinogenesis. Ji et al. studied miRNA expression in a large (>400 cases) cohort of resected human HCC and observed a remarkable down-regulation of miR-26, which correlated with poor prognosis (19). Patients with high miR-26a expression had longer time to recurrence and longer OS. Despite having worse OS, patients with low miR-26 expressing tumors had an increased benefit from adjuvant therapy with interferon (IFN)-alpha compared to those with tumors with high miR-26a. This was due to the parallel predominant activation of the signaling pathway NF-KB and IL-6 in this cohort of patients, with enhanced oncogenic potential. Thus, miR-26 expression acts not only as a prognostic factor but also as an independent predictor of the response to IFN-alpha (19). On the bases of these findings, a clinical trial is undergoing to look at the effect of adjuvant IFN in patients with HBV-related HCC and low miR-26 expression, and to our knowledge, this is the only clinical trial including miRNA expression as inclusion criteria or stratifying factor, to date. The possibility to detect circulating miRNAs in the sera/plasma of liver cancer patients has recently increased the potential of studying the prognostic values of miRNAs in the clinical setting, resulting in a growing number of reports on the role of circulating miRNAs in predicting relapse after curative treatment. Sugimachi et al. observed that low expression of circulating exosome-related miR-718 was associated to poor histological differentiation, high incidence of tumors beyond the Milan criteria, and a trend to increased recurrence after liver transplantation (73). Some authors have suggested that serum miRNAs levels can be monitored after radical resection to assess disease relapse (74, 75), while others have noticed a correlation between serum levels of miR-200 and response to TACE (76). In addition, some studies have shown that miRNAs can affect survival by modulating invasiveness and metastasis, i.e., miR-135a was found over-expressed in portal vein thrombus tissues and was related to poor clinical outcome (77). Luk et al. (78) found that all miRNAs included in the DLK1–DIO3 cluster at 14q32.2 are coordinately up-regulated in a subset of HCC patients with stem-like features, vascular invasion, and shorter survival. However, the limited sample size of these studies warrants further confirmations before these findings can be incorporated into clinical management.
Predictive role. Some miRNAs were found to be predictors of response to anticancer therapy in HCC both in vitro and in vivo. We have shown that miR-193b can facilitate sorafenib-induced apoptosis through modulation of myeloid leukemia cell differentiation protein (Mcl-1) in human HCC cell lines (79). Similarly, restoration of miR-193b was shown to sensitize HBV+ HCC cells to sorafenib (80). However, expression of this miRNA in patient samples has not been investigated yet. Loss of miR-122 was associated to lower sensitivity of HCC cells to sorafenib in in vitro experiments (81). A retrospective analysis showed that high levels of miR-425-3p in HCC bioptic tissues were associated with longer time to progression and OS in patients treated with sorafenib (82). However, further prospective evaluation is needed before miRNAs can be used as stratifying factors for first line treatment. Moreover, given the lack of liver biopsies in the routine clinical setting studies on the role of circulating miRNAs on the prediction of response to sorafenib are likely to represent more useful tools to customize treatment.
Therapeutic role. Several approaches to normalize ncRNA expression have been described to date. Therapeutic inhibition of oncogenic miRNAs can be managed through different technologies. Antisense oligonucleotides (ASO) are single-strand DNA molecules that pair to complementary RNA. They may be delivered intravenously but have poor stability (83). On the contrary, locked nucleic acid antimiRs (LNA-antimiRs) seem to be more stable and specific than ASO as they are composed of DNA and a phosphorothioate backbone (84). miRNA sponges contain binding site for several miRNAs and act as competitive inhibitors. They are potentially useful to inhibit a plethora of miRNAs that finally act on the same pathways, or a number of isoforms of the same miRNA (85). The best example of effective anti-miRNA therapy comes from the Miraversen studies. Miravirsen is a LNA-antimiR against miR-122, which is known to promote HCV RNA accumulation within the cells. Miraversen was tested in 36 patients with HCV genotype 1 infection within a phase II clinical trial. Treatment resulted in a dose dependent and prolonged decrease of HCV RNA that lasted beyond the end of active therapy and was not associated with viral resistance and dose-limiting adverse events (86). Given the association noticed between reduced levels of miR-122 and development of liver cancer, the safety of anti-miR-122 therapy has been carefully evaluated, and none of the patients treated with Miravirsen were reported to have developed HCC or other liver-related complications (87).
With regards to HCC treatment, preclinical studies have been successful in achieving tumor growth inhibition through silencing of miR-221/222. MiR-221 and -222 are encoded in tandem from a gene cluster on the chromosome X (Xp11.3) and share the same 5′ region. They regulate and promote cell-cycle progression through down-regulation of cyclin-dependent kinase inhibitors (p27Kip1 and CDKN1C/p57) (88), and pro-apoptotic proteins Bcl-2-modifying factor (Bmf) (15), and can also modulate cell survival through controlling the phosphoinositide 3-kinase–protein kinase B–mammalian target of rapamycin (PI3K–AKT–mTOR) pathway (18). miR-221/222 is frequently up-regulated in human HCC and it was found to be associated with aggressive clinical features. Some reports suggest that MET induces miR-221/222 transcription through the activator protein-1 (AP-1) transcription factor, and that miR-221/222 can account for the aggressive biology of the “high MET” liver cancers (89). Intravenous delivery of cholesterol- and 2′-O-methyl phosphorothioate-modified anti-miR-221 oligonucleotide (anti-miR oligonucleotide, AMO) led to reduced tumor cell proliferation, increased apoptosis, cell-cycle arrest, and increased survival in an orthotopic mouse model of HCC (90). Delivery of AMO anti-miR-221 confirmed anti-tumor effects in a transgenic mouse model over-expressing miR-221 in the liver, which spontaneously develops HCC and accelerates diethylnitrosamine-induced HCC growth (17). Interestingly, three injections over a period of 30 days were sufficient to inhibit miR-221 expression and to cause HCC growth inhibition in this model (17). More recently, Callegari et al. have provided evidence that adeno-associated viruses (AAV) genetically modified to drive the expression of multiple binding sites for miR-221 can act as sponges that sequester miR-221 cellular molecules and exhibit anti-tumor activity in HCC cells and may be tested for in vivo miRNA inhibition (91).
miRNA-based therapeutics have also been developed to restore the expression of miRNAs, which are down-regulated in liver cancer. Delivery of miR-26 was attempted trough viral delivery in a mouse model of HCC and proved to be successful in inhibiting cancer cell proliferation, inducing of tumor-specific apoptosis, and blocking disease progression without liver toxicities (92). Intrahepatic delivery of an adenovirus expressing miR-199a in newborn mice led to virus replication and fast removal of implanted HepG2 liver cancer cells, as well as reduced tumor growth in different HCC mouse models (93). Selected evidences showed miR34a is lost in HCC and its expression has been linked to the status of p53 (94, 95). Thus, attempts to over-express miR-34 have been pursued for the treatment of liver cancer. On one hand, a small molecular modulator termed Rubone, was shown to induce miR34a expression specifically in HCC cells by enhancing the occupancy of p53 on the miR34a promoter, and showed anti-tumor activity in a xenograft HCC mouse model (96). In p53 non-deleted HCC, Rubone exhibited a preclinical anti-HCC potency comparable to sorafenib without showing any additional toxicity (96). Therapeutics based on the restoration of miR-34 expression has also been pursued through liposome-mediated miRNA delivery technologies. Indeed, one ongoing phase I study is evaluating the safety of MRX34, a liposomal formulation of miR-34, in patients with primary HCC or those with liver metastases from other cancers (97). This is the first and only example of clinical trial, which assess miRNA-based therapeutics in humans, and results are highly expected from the scientific community. Therapies aiming at blocking or restoring miRNAs are promising and could become a new cornerstone in the treatment of HCC, either in monotherapy or in combination with sorafenib. However, additional investigations are needed to establish the real therapeutic benefit of these approaches, leading eventually to an improvement of survival in HCC.
Long Non-Coding RNAs
Long non-coding RNAs can vary in length from 200 nt to 100 kb. Although the majority of lncRNAs have yet to be characterized thoroughly, they have been shown to exhibit cell type-specific expression, localization to subcellular compartments, and association with cancer. lncRNAs can be intronic or intergenic and can be transcribed either in sense or antisense. Their sequence is characterized by a paucity of introns and low-cytosine–guanine (CG) content, which may account for the low level of expression of these transcripts (98). In vitro analyses have shown that antisense and intergenic lncRNAs are more stable than others (99). Growing evidence is supporting the involvement of lncRNAs in carcinogenesis. They may modulate cancer initiation and progression by affecting several biological pathways (100). However, their actual mechanism of function is not yet clear. Some evidence suggests that lncRNAs can modulate gene activity and affect the expression of other protein-coding genes. For instance, X-inactive specific transcript (XIST) was shown to modulate gene transcription by “coating Chromosome X” and creating a nuclear compartment that excludes RNA Polymerase-II (RNAPol-II) (101). Interaction with proteins has also been postulated, i.e., lncRNAs interact with the histone modification proteins and act as scaffold molecules for chromatin remodeling complexes (102). Growing evidence is now suggesting that lncRNAs can modulate the microRNome by binding one or multiple miRNAs and act like miRNA sponges (103).
Long Non-Coding RNAs in HCC
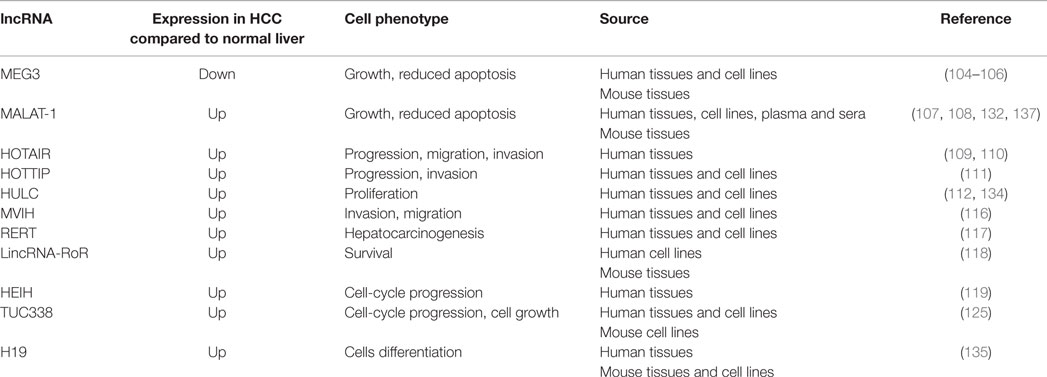
Long non-coding RNAs have been found to be aberrantly expressed in HCC and to play a role in modulating malignant phenotypes (Table 2). Maternally expressed gene 3 (MEG3) is an imprinted ncRNA located on chromosome 14q32.3 within the DLK-1 locus. We observed down-regulation of MEG3 in human HCC tissues in comparison to cirrhotic tissues secondary to hypermethylation of its promoter. In vitro experiments confirmed its oncosuppressive properties as over-expression of MEG3 reduced tumor cell growth and induced apoptosis (104). A recent report confirmed down-regulation of MEG3 in more than 70 cases of human HCC and found an association between low levels of MEG3 and poor clinical outcome with reduced overall and relapse-free survival (105). MEG3 was found to be the most frequently deregulated ncRNA in primary HCC when a set of 16 expression profiles from Oncomine comprising altogether 953 primary human HCC specimens was analyzed (106). Indeed, it was down-regulated in 60% of cases and there was correlation between increased DNA methylation and reduced MEG3 expression. Interestingly, no alterations in DNA methylation at the DLK1-MEG3 imprinting locus were found in hepatocellular adenomas or focal nodular hyperplasia. The metastatic lung adenocarcinoma transcript 1 (MALAT-1) is a lncRNA located on chromosome 11q13.1 that was initially identified in metastatic lung cancer and was then found increased in a mouse model of HCC as well as in human HCC cell lines and tissues (107, 108). MALAT-1 seems to mediate carcinogenesis by modulating apoptosis and cell growth in HCC cell lines and its expression correlates with risk of recurrence after liver transplantation (107). HOX transcript antisense RNA (HOTAIR) is expressed from the developmental HOX-C locus located on chromosome 12q13.13. HOTAIR expression was found increased in patients with large primary HCC and those with nodal involvement (109, 110). It acts as an oncogene, which may positively regulate the expression levels of multiple genes involved in the promotion of metastatic process, such as vascular endothelial growth factor (VEGF) and matrix metallopeptidase 9 (MMP9) (109). HOTAIR acts as a scaffolding molecule that binds polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1 (LSD1) and increases recruitment of enzymes involved in the epigenetic modification, with subsequent repression of tumor suppressor genes (102). HOXA transcript at the distal tip (HOTTIP) is another lncRNA located in physical contiguity with a HOX locus. HOTTIP is located at the distal tip of the HOXA13 gene, which encodes transcription factors regulating embryonic development. HOTTIP directly controls HOXA locus gene expression, but is also controlled by HOXA13 showing that a fine regulatory feedback loop is necessary for its activation (111). HOTTIP is remarkably increased in human HCC tissues from liver biopsies and its expression predicts clinical outcome in patients who had not received any HCC treatments, suggesting that liver biopsy may be an important source of information for the understanding of molecular biology of inoperable HCC and for the definition of molecular prognostic markers. Highly up-regulated in liver cancer (HULC) is a <500 nt lncRNA, which show high conservation across species (112). It is present in the cytoplasm of liver cancer cells, where it localizes within the ribosomes, suggesting it may modulate translational activity. Indeed, silencing of HULC in HCC cells-induced global mRNA changes in genes involved in hepatocarcinogenesis (112). Interestingly, not only HULC increased in liver cancer tissues but was also found to be increased in cells from peripheral bloods of HCC patients, suggesting that its involvement in liver cancer may be exerted also through a modulation of the immune system. HULC expression positively correlates with that of HBV x. HBV x was shown to upregulate HULC, which in turn promotes proliferation of hepatoma cells by suppressing the oncosuppressor p18 (113). In other studies, HULC was shown to promote hepatoma cells proliferation by modulation of lipid metabolism (114). Moreover, a particular variant genotype (rs 7763881) in HULC has been found to contribute to decreased HCC development in HBV+ patients (115). Microvascular invasion in HCC (MVIH) is a long ncRNA independently transcribed in human HBV-related HCC tissues compared to adjacent normal counterparts (116). Over-expression of MVIH correlated to microvessel invasion, advanced stage, and poorer OS in a large and unselected cohort of human HBV–HCC patients. Interestingly, MVIH could also significantly predict relapse in patients with early HCC who underwent radical treatment. In vitro experiments have shown that MVIH can physically interact with the protein phosphoglycerate kinase 1 (PGK1), an anti-angiogenic protein. It looks like the interaction results in reduced secretion of PGK1, and enhancement of angiogenesis and tumor growth. Indeed, MVIH expression in the primary tumor inversely correlated with PGK1 levels in serum of HCC patients (116). RERT is an lncRNA whose sequence overlaps with that of Prolyl-hydroxylase 1 (EGLN2) (117). EGLN2 is one of the three enzymes able to determine degradation of hypoxia inducible factor (HIF) by poly-ubiquitylation and proteasomal degradation. It was shown that a 4-bp deletion polymorphism (rs10680577) within RERT significantly correlated with higher expression of RERT and subsequent up-regulation of EGLN2 in human HCC (117). Finally, EGLN2 over-expression made cells more sensitive to hypoxia stress, leading to less HIF-alpha stabilization and HIF activation, which were detrimental for hepatic cell survival. These findings provided an example of how up-regulation of lncRNA can promote hepatocarcinogenesis through regulation of transcription of close genes and modulation of cell response to stress (117). Long intergenic ncRNA regulator of reprograming (lincRNA-RoR) is another hypoxia-responsive lncRNA, which is increased in malignant human liver cancer cells, and in the hypoxic regions of tumor cell xenografts in vivo (118). Interestingly, linc-ROR was detected in extracellular vesicles released by tumor cells during hypoxia, suggesting that this lncRNA may contribute to the intercellular signaling promoting cell survival in hypoxic stress (118). High expression in HCC (HEIH) is an lncRNA identified in human liver tissues and was named after its over-expression in human HBV–HCC compared to cirrhotic samples (119). It was shown to act as an oncogene in vitro and in vivo models and was found to interact with the enhancer of zeste homolog 2, an essential subunit of PRC2 complex. HEIH over-expression was found significantly associated with higher recurrence in HBV–HCC patients and was an independent prognostic factor for OS (119). lincRNA–UFC1 was found over-expressed in HCC tissues and associated with advanced stages and poor clinical outcome (120). lincRNA–UFC1 seems to control expression of beta-catenin not by direct interaction but through the binding to HuR, a RNA-binding proteins that can in turn interact with beta-catenin mRNA. Interestingly, lincRNA–UFC1 was also found to be a direct target of miR-34, whose loss in HCC was postulated to be the driver of lincRNA–UFC1 over-expression (120).
Transcribed Ultra-Conserved Regions
Ultra-conserved regions (UCRs) comprise 481 genomic sequences longer than 200 bp, which are totally conserved among mouse, rat, and human genomes (121). SNPs and mutations are normally under-represented in UCR genes (122). Many UCRs are transcribed (T-UCRs) in normal human tissue (123). Some of them have a ubiquitous expression, while others are tissue-specific (124). T-UCRs were shown to have distinct genome-wide expression profiles in different human cancers and this evidence supports their role in human carcinogenesis (123–127). The function of T-UCRs is still partly unknown, but they seem to modify the microRNome of the cell (128). Several mechanisms may be responsible for the deregulation of T-UCRs, including promoter methylation (127) and transcriptional activation (123). T-UCRs have been investigated in HCC and their expression profile was found to be deregulated in malignant compared to normal hepatocytes (125). The ultra-conserved element 338 (uc.338) is partly overlapping a protein-coding gene but was found to be transcribed as part of an independent lncRNA (TUC338). TUC338 expression is increased in human and murine malignant hepatocytes as well as in human HCC tissues in comparison to normal liver. Interestingly, not only the sequence but also the functional activity in promoting cellular growth is conserved across the species, suggesting that TUC338 is essential for the normal homeostasis of liver cells and that its aberrant over-expression may be responsible for driving carcinogenesis (125). Kogure et al. have also demonstrated that T-UCRs can be found in extracellular vesicles secreted by cancer cells, strengthening the hypothesis that they exert an important role in modulating tumor cell growth and they are part of the intercellular signaling through which HCC may grow and spread (129). Growing evidence is supporting the role of T-UCRs also in other types of cancer (124, 126, 130), and given their conservation it is likely that they play an essential part in driving carcinogenesis and may therefore represent valuable targets for novel therapeutics.
Interactions between lncRNAs and miRNAs
Recent evidence suggests that miRNAs and lncRNAs are likely to modulate each other by acting in a complex network. miRNAs can directly or indirectly regulate the expression of lncRNAs. Calin et al. showed that miRNAs can bind to the sequence of lncRNAs and negatively regulate their expression (123). MALAT-1 was found to be a target of miR-125b, which can directly bind the lncRNA and control its expression in bladder cancer (131). Over-expression of MALAT-1 (107) and down-regulation of miR-125 (132) have been separately reported in human HCC, suggesting that this mechanism may account for up-regulation of MALAT-1 in liver cancer as well. miRNAs were shown to control lncRNA expression also through indirect mechanisms. For instance, the miRNA-dependent control of the methylation machinery may have implications on the oncogenic role of selected lncRNAs in HCC. This is the case of MEG3 and miR-29. MiR-29 can modulate de novo methyltranferase (DMNT) 1 and 3. In case of low expression of miR-29, methylation-dependent tissue-specific regulation of MEG3 does not occur, and the lncRNA is suppressed (104).
If miRNAs can regulate lncRNAs, there is also evidence that lncRNAs may, in turn, regulate miRNAs by acting as competing endogenous RNAs or by being processed into small RNAs (133). HULC was shown to have binding sites for miR-372 and therefore to act as an endogenous “sponge.” HULC-mediated inhibition of miR-372 leads to reduction in repression of the target gene PRKACB and subsequent phosphorylation of the proto-oncogene c-AMP response element-binding protein (CREB) in HCC cells (134). Linc-RoR, a hypoxia-induced lncRNA, was shown to deplete HCC cells from miR-145 in hypoxic conditions and to mediate its biological effect through the interaction with miRNAs (118). Liz et al. have recently provided solid evidence that T-UCR can affect miRNA processing by preventing the release of mature forms. Indeed, uc.283 was shown to interact with the lower stem region of pri-miR-195 transcript preventing the miRNA cleavage by Drosha (128). Some authors have reported that the lncRNA H19, observed to be up-regulated in HCC (135), can mediate muscle differentiation by releasing miR-675, which is encoded within its sequence (136). Ren et al. have speculated that MALAT-1 can be fragmented in several small RNAs that can then be released from cancer cells and found in the plasma of HCC patients (137). Despite growing evidence is reported on the interplay between different classes of ncRNAs, further investigation is warranted in order to better understand the function of ncRNA and exploit their potential as therapeutic targets.
Conclusion
Non-coding RNAs participate in genomic regulation from transcription, post-transcription, and epigenetic modification. They can interact with each other and create networks of signal transduction that have a crucial role in hepatocarcinogenesis and disease progression. Moreover, they may modulate cell response to anticancer agents and have an intrinsic antineoplastic effect complementary to that of systemic therapies. In recent years, a large number of ncRNAs have been identified and several studies have investigated the role of ncRNA in HCC providing valuable knowledge for the understanding of liver carcinogenesis. However, despite the increasing knowledge on ncRNAs in liver cancer, none of them has entered clinical practice, and only few phase I and II trials have been conducted up to now. The process of translation of preclinical results into clinic seems challenging and hard to pursue. Indeed, the safety of ncRNAs as clinical therapeutic targets needs to be established with certainty. For example, the use of viral delivery systems may activate the innate immune responses with subsequent serious adverse events, while targeting ncRNAs involved in regulation of gene expression could bring to unexpected off target gene effects. Moreover, modulation of miRNA expression and competition with endogenous miRNAs may have unexpected effects on cellular physiology.
Hepatocellular carcinoma has still poor prognosis and limited therapeutic options. To date, only one drug, sorafenib, has been approved for first line treatment, and several trials investigating novel drugs have failed. Several ncRNAs directly involved in HCC promotion and progression have been effectively targeted in preclinical studies. It is hoped that these efforts will be soon translated into clinical practice.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CB is currently funded by an Institute of Cancer Research Clinician-Scientist Fellowship, a Marie Curie Career Integration Grant (European Union), and a Research Innovation Fund by Pancreatic Cancer UK.
References
1. Llovet JM, Lok A. Hepatitis B virus genotype and mutants: risk factors for hepatocellular carcinoma. J Natl Cancer Inst (2008) 100(16):1121–3. doi: 10.1093/jnci/djn261
2. Bruix J, Llovet JM, Castells A, Montana X, Bru C, Ayuso MC, et al. Transarterial embolization versus symptomatic treatment in patients with advanced hepatocellular carcinoma: results of a randomized, controlled trial in a single institution. Hepatology (1998) 27(6):1578–83. doi:10.1002/hep.510270617
3. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med (2008) 359(4):378–90. doi:10.1056/NEJMoa0708857
4. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet (2012) 44(6):694–8. doi:10.1038/ng.2256
5. Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Méndez-González J, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology (2015) 61(6):1945–56. doi:10.1002/hep.27732
6. Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med (2008) 359(19):1995–2004. doi:10.1056/NEJMoa0804525
7. Marquardt JU, Seo D, Andersen JB, Gillen MC, Kim MS, Conner EA, et al. Sequential transcriptome analysis of human liver cancer indicates late stage acquisition of malignant traits. J Hepatol (2014) 60(2):346–53. doi:10.1016/j.jhep.2013.10.014
8. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov (2013) 12(11):847–65. doi:10.1038/nrd4140
9. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet (2009) 10(10):704–14. doi:10.1038/nrg2634
10. Braconi C, Henry JC, Kogure T, Schmittgen T, Patel T. The role of microRNAs in human liver cancers. Semin Oncol (2011) 38(6):752–63. doi:10.1053/j.seminoncol.2011.08.001
11. Braconi C, Patel T. MicroRNA expression profiling: a molecular tool for defining the phenotype of hepatocellular tumors. Hepatology (2008) 47(6):1807–9. doi:10.1002/hep.22326
12. Braconi C, Patel T. Non-coding RNAs as therapeutic targets in hepatocellular cancer. Curr Cancer Drug Targets (2012) 12(9):1073–80. doi:10.2174/156800912803987904
13. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology (2007) 133(2):647–58. doi:10.1053/j.gastro.2007.05.022
14. Zhou L, Yang ZX, Song WJ, Li QJ, Yang F, Wang DS, et al. MicroRNA-21 regulates the migration and invasion of a stem-like population in hepatocellular carcinoma. Int J Oncol (2013) 43(2):661–9. doi:10.3892/ijo.2013.1965
15. Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res (2009) 15(16):5073–81. doi:10.1158/1078-0432.CCR-09-0092
16. Wong QW, Ching AK, Chan AW, Choy KW, To KF, Lai PB, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res (2010) 16(3):867–75. doi:10.1158/1078-0432.CCR-09-1840
17. Callegari E, Elamin BK, Giannone F, Milazzo M, Altavilla G, Fornari F, et al. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology (2012) 56(3):1025–33. doi:10.1002/hep.25747
18. Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A (2010) 107(1):264–9. doi:10.1073/pnas.0907904107
19. Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med (2009) 361(15):1437–47. doi:10.1056/NEJMoa0901282
20. Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology (2010) 51(3):836–45. doi:10.1002/hep.23380
21. Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene (2009) 28(40):3526–36. doi:10.1038/onc.2009.211
22. Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene (2006) 25(17):2537–45. doi:10.1038/sj.onc.1209283
23. Yang X, Zhang XF, Lu X, Jia HL, Liang L, Dong QZ, et al. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology (2014) 59(5):1874–85. doi:10.1002/hep.26941
24. Gailhouste L, Gomez-Santos L, Hagiwara K, Hatada I, Kitagawa N, Kawaharada K, et al. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology (2013) 58(3):1153–65. doi:10.1002/hep.26422
25. Murakami Y, Tanahashi T, Okada R, Toyoda H, Kumada T, Enomoto M, et al. Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PLoS One (2014) 9(9):e106314. doi:10.1371/journal.pone.0106314
26. Wojcicka A, Swierniak M, Kornasiewicz O, Gierlikowski W, Maciag M, Kolanowska M, et al. Next generation sequencing reveals microRNA isoforms in liver cirrhosis and hepatocellular carcinoma. Int J Biochem Cell Biol (2014) 53:208–17. doi:10.1016/j.biocel.2014.05.020
27. Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol (2009) 50(3):453–60. doi:10.1016/j.jhep.2008.06.010
28. Henry JC, Park JK, Jiang J, Kim JH, Nagorney DM, Roberts LR, et al. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun (2010) 403(1):120–5. doi:10.1016/j.bbrc.2010.10.130
29. Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao Y, et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene (2013) 32(27):3296–305. doi:10.1038/onc.2013.150
30. Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol (2010) 298(4):G535–41. doi:10.1152/ajpgi.00338.2009
31. Chen M, Liu Y, Varley P, Chang Y, He XX, Huang H, et al. High mobility group box-1 promotes hepatocellular carcinoma progression through miR-21-mediated matrix metalloproteinase activity. Cancer Res (2015) 75(8):1645–56. doi:10.1158/0008-5472.CAN-14-2147
32. Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM, et al. Hepatocyte nuclear factor 4alpha-nuclear factor-kappaB feedback circuit modulates liver cancer progression. Hepatology (2014) 60(5):1607–19. doi:10.1002/hep.27177
33. Li CH, Xu F, Chow S, Feng L, Yin D, Ng TB, et al. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. Eur J Cancer (2014) 50(15):2560–9. doi:10.1016/j.ejca.2014.07.008
34. Shih YT, Wang MC, Zhou J, Peng HH, Lee DY, Chiu JJ. Endothelial progenitors promote hepatocarcinoma intrahepatic metastasis through monocyte chemotactic protein-1 induction of microRNA-21. Gut (2014). doi:10.1136/gutjnl-2013-306302
35. Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell (2011) 19(2):232–43. doi:10.1016/j.ccr.2011.01.001
36. Toffanin S, Hoshida Y, Lachenmayer A, Villanueva A, Cabellos L, Minguez B, et al. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology (2011) 140(5):e–28. doi:10.1053/j.gastro.2011.02.009
37. Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene (2012) 31(13):1609–22. doi:10.1038/onc.2011.354
38. Wang B, Hsu SH, Wang X, Kutay H, Bid HK, Yu J, et al. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology (2014) 59(2):555–66. doi:10.1002/hep.26712
39. Calin GA, Croce CM. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J Clin Invest (2007) 117(8):2059–66. doi:10.1172/JCI32577
40. Han H, Sun D, Li W, Shen H, Zhu Y, Li C, et al. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology (2013) 57(6):2378–89. doi:10.1002/hep.26302
41. Parpart S, Roessler S, Dong F, Rao V, Takai A, Ji J, et al. Modulation of miR-29 expression by alpha-fetoprotein is linked to the hepatocellular carcinoma epigenome. Hepatology (2014) 60(3):872–83. doi:10.1002/hep.27200
42. Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology (2011) 54(4):1237–48. doi:10.1002/hep.24504
43. Patel T. Extracellular vesicle noncoding RNA: new players in the diagnosis and pathogenesis of cholangiocarcinoma. Hepatology (2014) 60(3):782–4. doi:10.1002/hep.27185
44. Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology (2014) 61(4):1284–94. doi:10.1002/hep.27660
45. Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis (2008) 29(11):2126–31. doi:10.1093/carcin/bgn195
46. Xu Y, Liu L, Liu J, Zhang Y, Zhu J, Chen J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer (2011) 128(2):412–7. doi:10.1002/ijc.25342
47. Qi P, Dou TH, Geng L, Zhou FG, Gu X, Wang H, et al. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol (2010) 71(6):621–6. doi:10.1016/j.humimm.2010.02.017
48. Li XD, Li ZG, Song XX, Liu CF. A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. Pathology (2010) 42(7):669–73. doi:10.3109/00313025.2010.522175
49. Liu YM, Xia Y, Dai W, Han HY, Dong YX, Cai J, et al. Cholesterol-conjugated let-7a mimics: antitumor efficacy on hepatocellular carcinoma in vitro and in a preclinical orthotopic xenograft model of systemic therapy. BMC Cancer (2014) 14:889. doi:10.1186/1471-2407-14-889
50. Liang TJ, Liu HJ, Zhao XQ, Yu CH, Li CS. Lack of association of MiR-34b/c polymorphism (rs4938723) with hepatocellular carcinoma: a meta-analysis. PLoS One (2013) 8(7):e68588. doi:10.1371/journal.pone.0068588
51. Akkiz H, Bayram S, Bekar A, Akgollu E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepat (2011) 18(7):e399–407. doi:10.1111/j.1365-2893.2010.01414.x
52. Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res (2007) 67(13):6092–9. doi:10.1158/0008-5472.CAN-06-4607
53. Mitchell AP. A VAST staging area for regulatory proteins. Proc Natl Acad Sci U S A (2008) 105(20):7111–2. doi:10.1073/pnas.0803384105
54. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A (2011) 108(12):5003–8. doi:10.1073/pnas.1019055108
55. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol (2008) 110(1):13–21. doi:10.1016/j.ygyno.2008.04.033
56. Roderburg C, Luedde T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J Hepatol (2014) 61(6):1434–7. doi:10.1016/j.jhep.2014.07.017
57. Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol (2011) 29(36):4781–8. doi:10.1200/JCO.2011.38.2697
58. Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res (2010) 70(23):9798–807. doi:10.1158/0008-5472.CAN-10-1001
59. Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology (2009) 49(4):1098–112. doi:10.1002/hep.22749
60. Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A (2007) 104(31):12884–9. doi:10.1073/pnas.0704894104
61. Song K, Han C, Zhang J, Lu D, Dash S, Feitelson M, et al. Epigenetic regulation of MicroRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology (2013) 58(5):1681–92. doi:10.1002/hep.26514
62. Roderburg C, Benz F, Vargas Cardenas D, Koch A, Janssen J, Vucur M, et al. Elevated miR-122 serum levels are an independent marker of liver injury in inflammatory diseases. Liver Int (2014) 35(4):1172–84. doi:10.1111/liv.12627
63. Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (2012) 5(3):492–7. doi:10.1158/1940-6207.CAPR-11-0370
64. Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA (2014) 311(4):392–404. doi:10.1001/jama.2013.284664
65. Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell (2012) 22(3):291–303. doi:10.1016/j.ccr.2012.07.023
66. Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology (2008) 47(3):897–907. doi:10.1002/hep.22160
67. Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res (2008) 14(2):419–27. doi:10.1158/1078-0432.CCR-07-0523
68. Chang RM, Yang H, Fang F, Xu JF, Yang LY. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology (2014) 60(4):1251–63. doi:10.1002/hep.27221
69. Bae HJ, Noh JH, Kim JK, Eun JW, Jung KH, Kim MG, et al. MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene (2014) 33(20):2557–67. doi:10.1038/onc.2013.216
70. Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer (2013) 13:21. doi:10.1186/1471-2407-13-21
71. Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun (2011) 406(1):70–3. doi:10.1016/j.bbrc.2011.01.111
72. Sato F, Hatano E, Kitamura K, Myomoto A, Fujiwara T, Takizawa S, et al. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One (2011) 6(1):e16435. doi:10.1371/journal.pone.0016435
73. Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer (2015) 112(3):532–8. doi:10.1038/bjc.2014.621
74. Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One (2011) 6(12):e28486. doi:10.1371/journal.pone.0028486
75. Liu Y, Hei Y, Shu Q, Dong J, Gao Y, Fu H, et al. VCP/p97, down-regulated by microRNA-129-5p, could regulate the progression of hepatocellular carcinoma. PLoS One (2012) 7(4):e35800. doi:10.1371/journal.pone.0035800
76. Liu M, Liu J, Wang L, Wu H, Zhou C, Zhu H, et al. Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS One (2014) 9(10):e109347. doi:10.1371/journal.pone.0109347
77. Liu S, Guo W, Shi J, Li N, Yu X, Xue J, et al. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol (2012) 56(2):389–96. doi:10.1016/j.jhep.2011.08.008
78. Luk JM, Burchard J, Zhang C, Liu AM, Wong KF, Shek FH, et al. DLK1-DIO3 genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem (2011) 286(35):30706–13. doi:10.1074/jbc.M111.229831
79. Braconi C, Valeri N, Gasparini P, Huang N, Taccioli C, Nuovo G, et al. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res (2010) 16(3):957–66. doi:10.1158/1078-0432.CCR-09-2123
80. Mao K, Zhang J, He C, Xu K, Liu J, Sun J, et al. Restoration of miR-193b sensitizes hepatitis B virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett (2014) 352(2):245–52. doi:10.1016/j.canlet.2014.07.004
81. Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem (2009) 284(46):32015–27. doi:10.1074/jbc.M109.016774
82. Vaira V, Roncalli M, Carnaghi C, Faversani A, Maggioni M, Augello C, et al. MicroRNA-425-3p predicts response to sorafenib therapy in patients with hepatocellular carcinoma. Liver Int (2014) 35(3):1077–86. doi:10.1111/liv.12636
83. Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci U S A (1978) 75(1):285–8. doi:10.1073/pnas.75.1.285
84. Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature (2008) 452(7189):896–9. doi:10.1038/nature06783
85. Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods (2007) 4(9):721–6. doi:10.1038/nmeth1079
86. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med (2013) 368(18):1685–94. doi:10.1056/NEJMoa1209026
87. van der Ree MH, van der Meer AJ, de Bruijne J, Maan R, van Vliet A, Welzel TM, et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res (2014) 111:53–9. doi:10.1016/j.antiviral.2014.08.015
88. Le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J (2007) 26(15):3699–708. doi:10.1038/sj.emboj.7601790
89. Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell (2009) 16(6):498–509. doi:10.1016/j.ccr.2009.10.014
90. Park JK, Kogure T, Nuovo GJ, Jiang J, He L, Kim JH, et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res (2011) 71(24):7608–16. doi:10.1158/0008-5472.CAN-11-1144
91. Moshiri F, Callegari E, D’Abundo L, Corra F, Lupini L, Sabbioni S, et al. Inhibiting the oncogenic mir-221 by microRNA sponge: toward microRNA-based therapeutics for hepatocellular carcinoma. Gastroenterol Hepatol Bed Bench (2014) 7(1):43–54.
92. Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell (2009) 137(6):1005–17. doi:10.1016/j.cell.2009.04.021
93. Callegari E, Elamin BK, D’Abundo L, Falzoni S, Donvito G, Moshiri F, et al. Anti-tumor activity of a miR-199-dependent oncolytic adenovirus. PLoS One (2013) 8(9):e73964. doi:10.1371/journal.pone.0073964
94. Xie K, Liu J, Chen J, Dong J, Ma H, Liu Y, et al. Methylation-associated silencing of microRNA-34b in hepatocellular carcinoma cancer. Gene (2014) 543(1):101–7. doi:10.1016/j.gene.2014.03.059
95. He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature (2007) 447(7148):1130–4. doi:10.1038/nature05939
96. Xiao Z, Li CH, Chan SL, Xu F, Feng L, Wang Y, et al. A small-molecule modulator of the tumor-suppressor miR34a inhibits the growth of hepatocellular carcinoma. Cancer Res (2014) 74(21):6236–47. doi:10.1158/0008-5472.CAN-14-0855
97. Clinicaltrials.gov. A Multicenter Phase I Study of MRX34, MicroRNA miR-RX34 Liposomal Injection. Available from: https://clinicaltrials.gov/ct2/show/NCT01829971?term=miR34&rank=1.
98. Niazi F, Valadkhan S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3’ UTRs. RNA (2012) 18(4):825–43. doi:10.1261/rna.029520.111
99. Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res (2012) 22(5):885–98. doi:10.1101/gr.131037.111
100. He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett (2014) 344(1):20–7. doi:10.1016/j.canlet.2013.10.021
101. Muscatelli F, Lena D, Mettei MG, Fontes M. A male with two contiguous inactivation centers on a single X chromosome: study of X inactivation and XIST expression. Hum Mol Genet (1992) 1(2):115–9. doi:10.1093/hmg/1.2.115
102. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res (2011) 71(20):6320–6. doi:10.1158/0008-5472.CAN-11-1021
103. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature (2010) 465(7301):1033–8. doi:10.1038/nature09144
104. Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene (2011) 30(47):4750–6. doi:10.1038/onc.2011.193
105. Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, et al. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol Carcinog (2015). doi:10.1002/mc.22270
106. Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, et al. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One (2012) 7(11):e49462. doi:10.1371/journal.pone.0049462
107. Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol (2012) 29(3):1810–6. doi:10.1007/s12032-011-0004-z
108. Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene (2007) 26(6):851–8. doi:10.1038/sj.onc.1209846
109. Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep (2013) 29(3):946–50. doi:10.3892/or.2012.2219
110. Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol (2011) 18(5):1243–50. doi:10.1245/s10434-011-1581-y
111. Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology (2014) 59(3):911–23. doi:10.1002/hep.26740
112. Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology (2007) 132(1):330–42. doi:10.1053/j.gastro.2006.08.026
113. Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem (2012) 287(31):26302–11. doi:10.1074/jbc.M112.342113
114. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res (2015) 75(5):846–57. doi:10.1158/0008-5472.CAN-14-1192
115. Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One (2012) 7(4):e35145. doi:10.1371/journal.pone.0035145
116. Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology (2012) 56(6):2231–41. doi:10.1002/hep.25895
117. Zhu Z, Gao X, He Y, Zhao H, Yu Q, Jiang D, et al. An insertion/deletion polymorphism within RERT-lncRNA modulates hepatocellular carcinoma risk. Cancer Res (2012) 72(23):6163–72. doi:10.1158/0008-5472.CAN-12-0010
118. Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci (2014) 127(Pt 7):1585–94. doi:10.1242/jcs.141069
119. Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology (2011) 54(5):1679–89. doi:10.1002/hep.24563
120. Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology (2015) 148(2):e–26. doi:10.1053/j.gastro.2014.10.012
121. Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science (2004) 304(5675):1321–5. doi:10.1126/science.1098119
122. Wojcik SE, Rossi S, Shimizu M, Nicoloso MS, Cimmino A, Alder H, et al. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis (2010) 31(2):208–15. doi:10.1093/carcin/bgp209
123. Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell (2007) 12(3):215–29. doi:10.1016/j.ccr.2007.07.027
124. Fassan M, Dall’Olmo L, Galasso M, Braconi C, Pizzi M, Realdon S, et al. Transcribed ultraconserved noncoding RNAs (T-UCR) are involved in Barrett’s esophagus carcinogenesis. Oncotarget (2014) 5(16):7162–71.
125. Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci U S A (2011) 108(2):786–91. doi:10.1073/pnas.1011098108
126. Hudson RS, Yi M, Volfovsky N, Prueitt RL, Esposito D, Volinia S, et al. Transcription signatures encoded by ultraconserved genomic regions in human prostate cancer. Mol Cancer (2013) 12:13. doi:10.1186/1476-4598-12-13
127. Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene (2010) 29(48):6390–401. doi:10.1038/onc.2010.361
128. Liz J, Portela A, Soler M, Gomez A, Ling H, Michlewski G, et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell (2014) 55(1):138–47. doi:10.1016/j.molcel.2014.05.005
129. Kogure T, Yan IK, Lin WL, Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer (2013) 4(7–8):261–72. doi:10.1177/1947601913499020
130. Ferdin J, Nishida N, Wu X, Nicoloso MS, Shah MY, Devlin C, et al. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ (2013) 20(12):1675–87. doi:10.1038/cdd.2013.119
131. Han Y, Liu Y, Zhang H, Wang T, Diao R, Jiang Z, et al. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long noncoding RNA MALAT1. FEBS Lett (2013) 587(23):3875–82. doi:10.1016/j.febslet.2013.10.023
132. Jiang JX, Gao S, Pan YZ, Yu C, Sun CY. Overexpression of microRNA-125b sensitizes human hepatocellular carcinoma cells to 5-fluorouracil through inhibition of glycolysis by targeting hexokinase II. Mol Med Rep (2014) 10(2):995–1002. doi:10.3892/mmr.2014.2271
133. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature (2014) 505(7483):344–52. doi:10.1038/nature12986
134. Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res (2010) 38(16):5366–83. doi:10.1093/nar/gkq285
135. Li X, Nong Z, Ekstrom C, Larsson E, Nordlinder H, Hofmann WJ, et al. Disrupted IGF2 promoter control by silencing of promoter P1 in human hepatocellular carcinoma. Cancer Res (1997) 57(10):2048–54.
136. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev (2014) 28(5):491–501. doi:10.1101/gad.234419.113
137. Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer (2013) 49(13):2949–59. doi:10.1016/j.ejca.2013.04.026
138. Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology (2009) 50(4):1152–61. doi:10.1002/hep.23100
139. Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN, Fu CJ, et al. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology (2013) 57(6):2274–86. doi:10.1002/hep.26257
140. Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang Y, et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer (2012) 118(9):2431–42. doi:10.1002/cncr.26566
141. Han ZB, Chen HY, Fan JW, Wu JY, Tang HM, Peng ZH. Up-regulation of microRNA-155 promotes cancer cell invasion and predicts poor survival of hepatocellular carcinoma following liver transplantation. J Cancer Res Clin Oncol (2012) 138(1):153–61. doi:10.1007/s00432-011-1076-z
142. Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, et al. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology (2012) 56(5):1631–40. doi:10.1002/hep.25849
143. Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene (2008) 27(43):5651–61. doi:10.1038/onc.2008.178
144. Fornari F, Milazzo M, Galassi M, Callegari E, Veronese A, Miyaaki H, et al. p53/mdm2 feedback loop sustains miR-221 expression and dictates the response to anticancer treatments in hepatocellular carcinoma. Mol Cancer Res (2014) 12(2):203–16. doi:10.1158/1541-7786.MCR-13-0312-T
145. Yuan Q, Loya K, Rani B, Mobus S, Balakrishnan A, Lamle J, et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology (2013) 57(1):299–310. doi:10.1002/hep.25984
146. Santhekadur PK, Das SK, Gredler R, Chen D, Srivastava J, Robertson C, et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. J Biol Chem (2012) 287(17):13952–8. doi:10.1074/jbc.M111.321646
147. Yang H, Cho ME, Li TW, Peng H, Ko KS, Mato JM, et al. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest (2013) 123(1):285–98. doi:10.1172/JCI63861
148. Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, et al. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer (2010) 46(15):2828–36. doi:10.1016/j.ejca.2010.06.127
149. Yang X, Liang L, Zhang XF, Jia HL, Qin Y, Zhu XC, et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology (2013) 58(1):158–70. doi:10.1002/hep.26305
150. Zhao N, Wang R, Zhou L, Zhu Y, Gong J, Zhuang SM. MicroRNA-26b suppresses the NF-kappaB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol Cancer (2014) 13:35. doi:10.1186/1476-4598-13-35
151. Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, et al. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res (2012) 40(10):4615–25. doi:10.1093/nar/gkr1278
152. Li L, Liu Y, Guo Y, Liu B, Zhao Y, Li P, et al. Regulatory MiR-148a-ACVR1/BMP circuit defines a cancer stem cell-like aggressive subtype of hepatocellular carcinoma. Hepatology (2015) 61(2):574–84. doi:10.1002/hep.27543
153. Zhang JP, Zeng C, Xu L, Gong J, Fang JH, Zhuang SM. MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene (2014) 33(31):4069–76. doi:10.1038/onc.2013.369
154. Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest (2013) 123(2):630–45. doi:10.1172/JCI64265
155. Koberle V, Kronenberger B, Pleli T, Trojan J, Imelmann E, Peveling-Oberhag J, et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer (2013) 49(16):3442–9. doi:10.1016/j.ejca.2013.06.002
156. Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest (2012) 122(8):2884–97. doi:10.1172/JCI63455
157. Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest (2012) 122(8):2871–83. doi:10.1172/JCI63539
158. Zeng C, Wang R, Li D, Lin XJ, Wei QK, Yuan Y, et al. A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology (2010) 52(5):1702–12. doi:10.1002/hep.23875
159. Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology (2010) 52(4):1431–42. doi:10.1002/hep.23818
160. Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res (2009) 69(14):5761–7. doi:10.1158/0008-5472.CAN-08-4797
161. Lan FF, Wang H, Chen YC, Chan CY, Ng SS, Li K, et al. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int J Cancer (2011) 128(2):319–31. doi:10.1002/ijc.25336
Keywords: non-coding RNA, microRNA, long non-coding RNA, HCC, liver cancer
Citation: Ghidini M and Braconi C (2015) Non-coding RNAs in primary liver cancer. Front. Med. 2:36. doi: 10.3389/fmed.2015.00036
Received: 17 March 2015; Accepted: 19 May 2015;
Published: 04 June 2015
Edited by:
Alfredo Fusco, Consiglio Nazionale delle Ricerche, ItalyReviewed by:
Rosa Marina Melillo, University of Naples Federico II, ItalyFrancesco Trapasso, University “Magna Graecia” of Catanzaro, Italy
Luigi Tornillo, University of Basel, Switzerland
Copyright: © 2015 Ghidini and Braconi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiara Braconi, Division of Cancer Therapeutics, The Institute of Cancer Research, 15 Cotswold Road, Sutton, Surrey SM25NG, UK,Y2hpYXJhLmJyYWNvbmlAaWNyLmFjLnVr
 Michele Ghidini
Michele Ghidini Chiara Braconi
Chiara Braconi