- 1Clinical Laboratory, HwaMei Hospital, University of Chinese Academy of Sciences, Ningbo, China
- 2Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo, China
- 3Department of Hepatology, HwaMei Hospital, University of Chinese Academy of Sciences, Ningbo, China
- 4Department of Blood Transfusion, HwaMei Hospital, University of Chinese Academy of Sciences, Ningbo, China
Background: Polyethylene glycol interferon alpha (PEG-IFN-α) is the most frequently used pharmacotherapeutic approach in patients infected with hepatitis B virus (HBV). Numerous studies have reported that interleukin-28B (IL-28B) genetic polymorphisms are related to the therapeutic efficacy of PEG-IFN-α, but the results are inconsistent. The present meta-analysis aimed to analyze the association between IL-28B genetic polymorphisms and the prognosis of patients with chronic hepatitis B (CHB) treated with PEG-IFN-α to inform clinical practice.
Methods: PubMed, EBSCO, and Scopus databases were searched for relevant literature published before February 30, 2021. We calculated the crude odds ratios (ORs) with 95% confidence intervals (CIs) of the cited articles. A total of 2510 patients with CHB treated with PEG-IFN-α in 13 clinical cohort studies were analyzed.
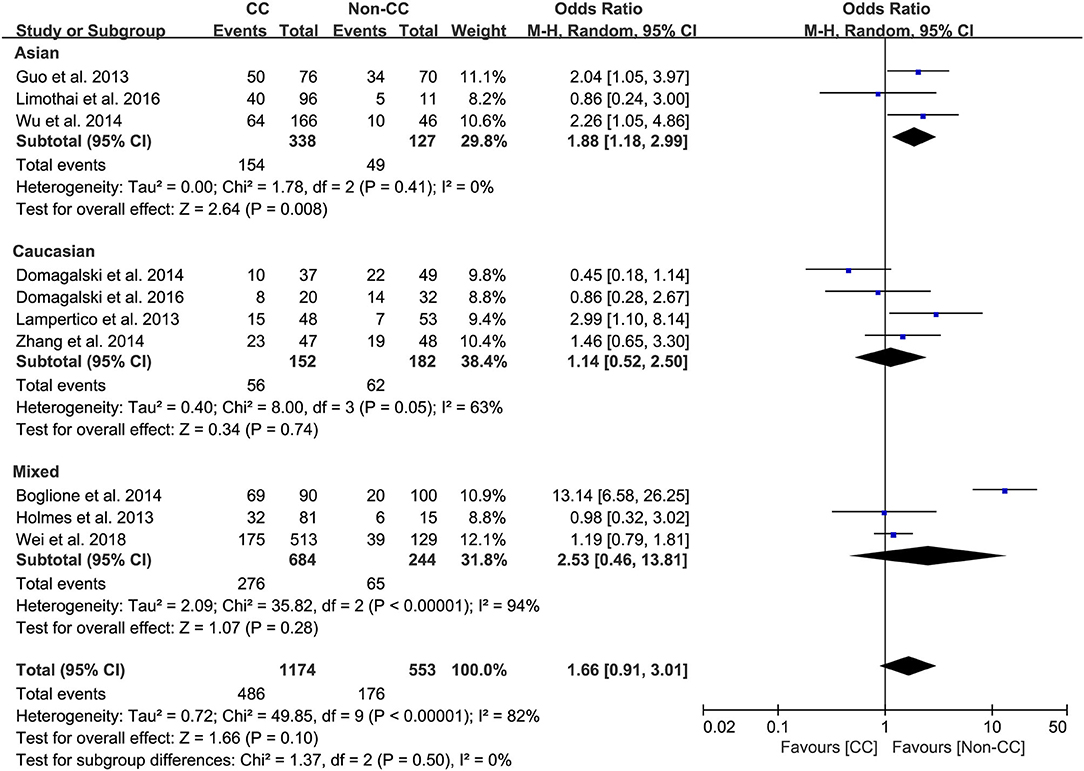
Results: The overall analysis demonstrated a potential association between IL-28B genetic polymorphisms and response to PEG-IFN-α; however, the association was not statistically significant. Furthermore, the subgroup analysis revealed that among patients with HBeAg-negative CHB, the rs12979860 CC genotype and rs8099917 TT genotype were associated with more significant treatment response to PEG-IFN-α (CC vs. non-CC: OR 2.78, 95% CI 1.00–7.76, I2 = 83%; TT vs. non-TT: OR 2.16, 95% CI 1.35–3.48, I2 = 0%). Among Asian patients with CHB, the rs12979860 CC genotype was associated with a more significant treatment response to PEG-IFN (CC vs. non-CC: OR 1.88, 95% CI 1.18–2.99, I2 = 0%).
Conclusion: This meta-analysis revealed that the IL-28B rs12979860 CC genotype and rs8099917 TT genotype indicated a better treatment response than non-CC and non-TT genotypes for PEG-IFN-α in patients with CHB.
Introduction
Hepatitis B vaccine programs have been implemented gradually. Hepatitis B virus (HBV) infection affects more than 350 million people worldwide and remains the most common cause of liver cancer and has a very low five-year survival rate (1, 2). More than 2000 million patients infected with HBV die every year due to HBV-related diseases, such as hepatocellular carcinoma (HCC) and liver cirrhosis (1). Currently, the prevention of HBV injection remains dominant in the treatment of chronic hepatitis B (CHB) and other HBV-related diseases. However, HBV complete eradication is difficult to achieve once a person is infected by HBV without effective prevention. Thus, the current therapeutic strategy for patients infected with HBV is to prevent CHB-related complications, which can be achieved by suppressing HBV replication. PEG-IFN-α is a current therapeutic strategy for patients with CHB with a remarkable effect in HBeAg-negative adults (3–5). Presently, treatment with IFN for more than 1 year is the primary therapy for adult patients with CHB, achieving complete HBsAg clearance (6). Furthermore, IFN-associated therapies also achieve a sustainable therapeutic response in most patients with CHB after 48-weeks of treatment (7, 8). However, previous studies found that the therapeutic results of PEG-IFN-α treatment in patients with CHB with HBeAg seroconversion or loss are poor (9, 10). Thus, it is essential to select patients with CHB who are sensitive to PEG-IFN-α so that PEG-IFN-α-based treatment achieves a better curative effect.
It is well-known that cytokines and regulatory molecules play critical roles in the immune response and pathogenesis of HBV infection. Interferon lambda 3 (IFNL3) is a cytokine encoded by interleukin 28B (IL-28B) and exerts anti-viral effects on HBV replication (11, 12). Based on the anti-viral and immune actions of IL-28B, researchers predicted that IL-28B genotypes might be associated with HBV infection and the therapeutic efficacy of interferons in patients with CHB. However, another study found three single-nucleotide polymorphisms (SNPs), rs12979860 C/T, rs12980275 A/G, and rs8099917 T/G, of IL-28B are not associated with the outcomes of HBV infection. Furthermore, Zhao et al. performed a meta-analysis to evaluate the role of IL-28B SNPs (rs12979860, rs12980275, and rs8099917) in the progression of HBV infection and the results showed that IL28B polymorphisms had no association with the outcome of HBV infection (13). However, the association between IL28B polymorphisms and the efficacy of PEG-IFN-α in patients with CHB was not analyzed. It is encouraging that Wu et al. found that IL-28B polymorphisms could predict the clinical outcomes of PEG-IFN-α in Chinese patients with CHB (14). However, another study performed in Asian populations found that the IL-28B genotype is not accurate in predicting outcomes in patients infected with HBVs treated with PEG-IFN-α (15). Thus, it is imperative to further analyze the association between IL-28B polymorphisms and the prognosis of patients with CHB treated with PEG-IFN-α. We then designed and performed this meta-analysis to elaborate the association between IL-28B genetic polymorphisms and the prognosis of patients with CHB treated with PEG-IFN-α to inform clinical practice.
Methods
Search Strategy and Selection Criteria
This meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (15). Two authors independently searched PubMed, Embase, and Scopus until February 30, 2021, for relevant articles using the following keywords: peginterferon alpha, IL-28B, and hepatitis B. There were no language or data restrictions.
The inclusion criteria were as follows: (1) studies of patients with HBV infection; (2) patients receiving peginterferon alpha (PEG-IFN-α) therapy; (3) studies reporting precise IL28B genotypes (CC vs. CT + TT for rs12979860; AA vs. AG + GG for rs12980275; TT vs. GT + GG for rs8099917) for included patients; (4) the primary outcome was the treatment response, including virological response (HBV DNA < 2,000 IU/mL), serological response (HBeAg seroconversion), biochemical response (ALT or AST < 40 IU/L), or combined response; and (5) study design including randomized controlled trials, non-randomized controlled trials, observational study. In addition, case reports, non-human studies, studies without adequate information, or concerning outcomes were excluded from the meta-analysis.
Data Extraction and Quality Assessment
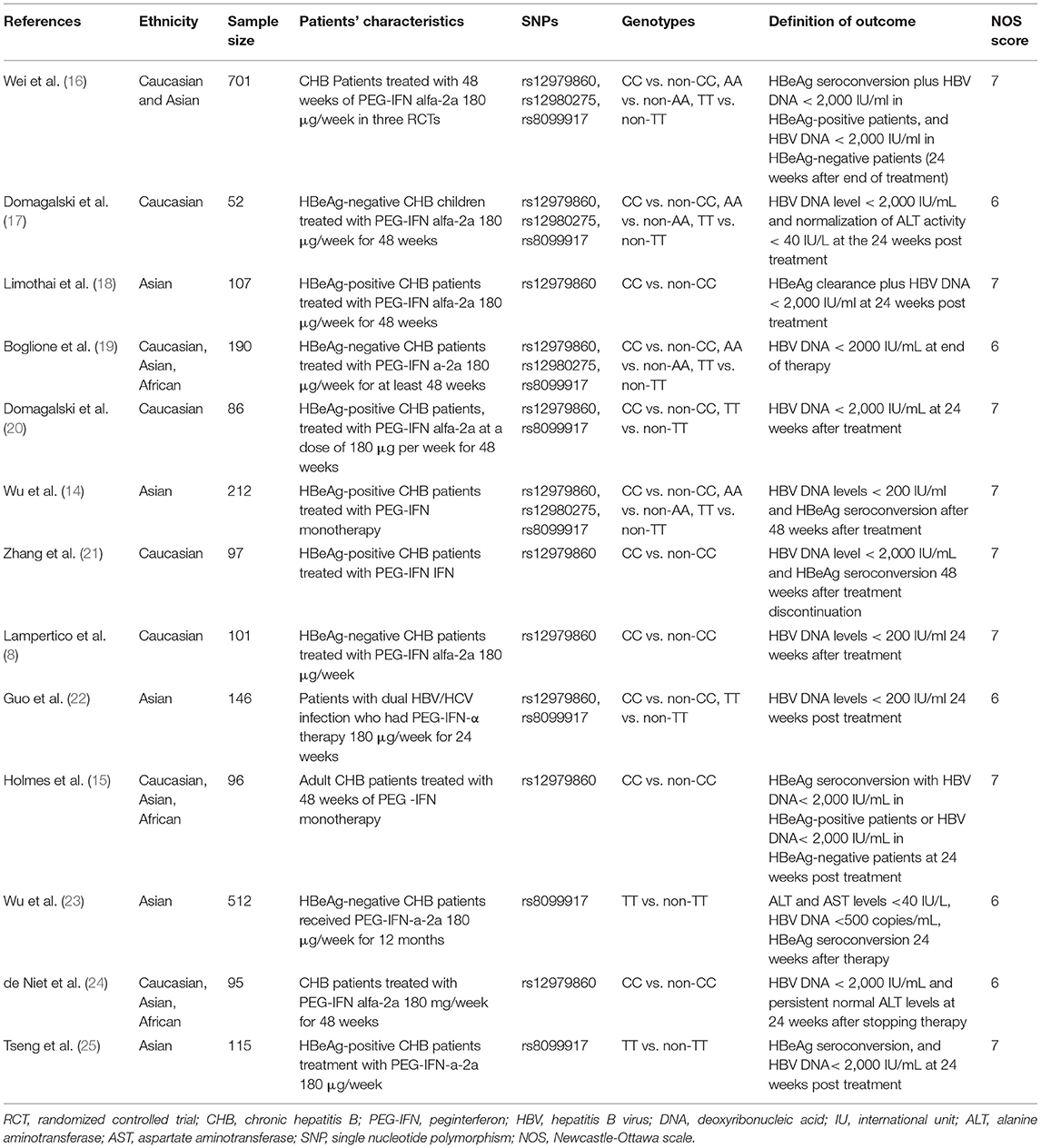
Two authors independently retrieved and extracted relevant studies. The basic characteristics of the studies (first author, year of publication, ethnicity, sample size, population characteristics, genotypes, and definition of outcomes) were recorded in Table 1. Any discrepancies in all phases were resolved through a team consensus. If relevant information was not reported in the article, we contacted the corresponding authors for further information.
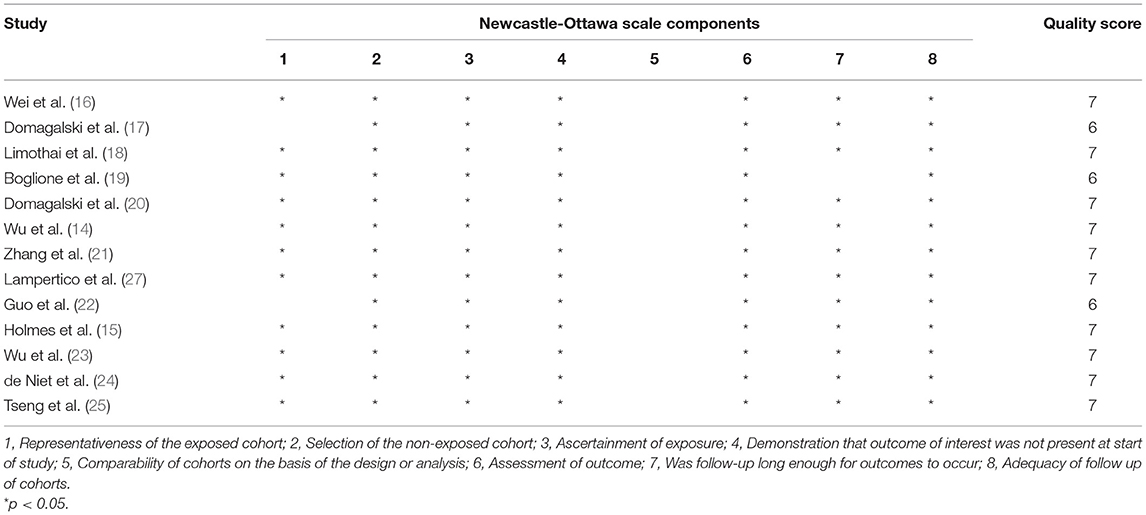
Two reviewers independently used the Newcastle-Ottawa Scale (NOS) to assess the risk of bias in the included studies. Publication bias was evaluated using Egger's regression test. Any discrepancies in all phases were resolved through a team consensus.
Statistical Synthesis and Analysis
Pooled analysis was performed to calculate the odds ratio (OR) with a 95% confidence interval (95% CI) between genotypic variations in IL-28B and treatment response of patients with HBV infection receiving PEG-IFN-α therapy. We calculated the I2 statistic to measure the proportion of total variation in the study estimates attributed to heterogeneity. I2 values of < 25, 25–75, and >75% indicate low, moderate, and high heterogeneity, respectively (26). If significant heterogeneity existed, we adopted a random-effects model to perform the analysis. Subgroup analysis was performed according to HBeAg (HBeAg negative vs. positive) and race (Asian vs. Caucasian). All analyses were performed using RevMan 5.3 and R 3.6. Statistical significance was set at P < 0.05.
Results
Study Characteristics
The search and selection processes are presented in Figure 1. A total of 171 studies were initially identified, and 47 duplicates were excluded. After screening the titles and abstracts, 40 full-text articles were assessed for eligibility. Finally, 13 studies involving 2,510 patients were included in the meta-analysis (14–25, 27). These included studies were published between 2011 and 2020, with sample sizes ranging from 52 to 701. There were four studies in the Caucasian populations, five in Asian populations, and four in admixture populations, including Caucasian, Asian, and African populations. All studies included patients with persistent HBV infection with PEG-IFN-α treatment, and three SNPs (rs12979860, rs12980275, and rs8099917) were genotyped using polymerase chain reaction. All studies reported the treatment response defined as HBV DNA < 2,000 IU/ mL for different genotype groups and nine studies combined the virological with serological or biochemical response as the primary outcome.
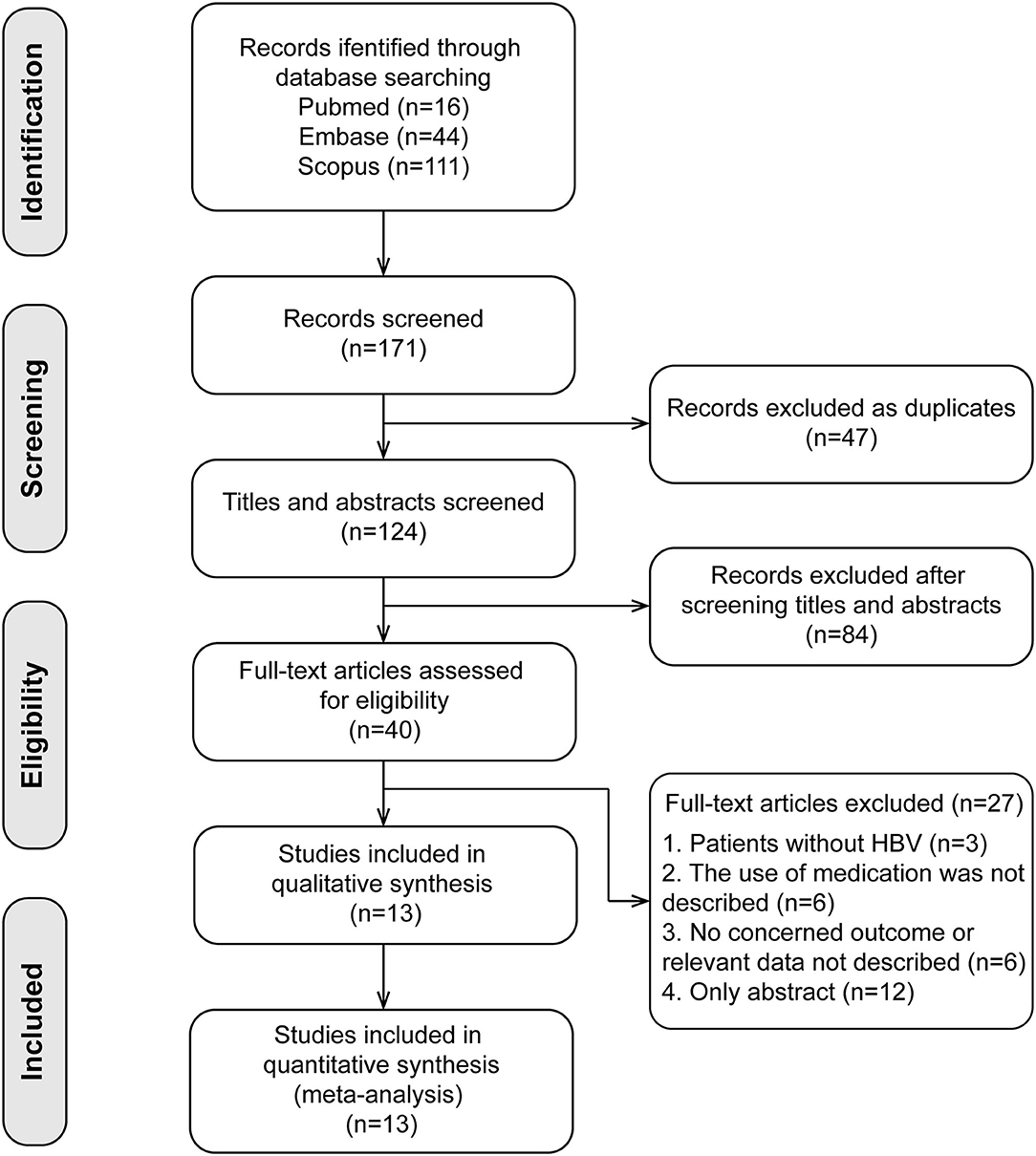
Figure 1. Flow chart of research selection process. Thirteen cohort studies were included in the present meta-analysis.
Quality assessment using the NOS score is shown in Table 2. Ten studies (14–16, 18, 20, 21, 23–25, 27) were of high quality with a total score of >6. The comparability of cohorts is a major concern. Since none of the studies included in this meta-analysis were randomized trials, patients' characteristics such as age, sex, race, baseline HBV DNA, and medication history in different groups could not be fully controlled. In addition, Domagalski et al. (17) enrolled children with HBV, and Guo et al. (22) enrolled patients with dual HBV and HCV infection. The follow-up period in the trial of Boglione et al. (19) was not long enough for the outcome to occur. Visual inspection of funnel plots and Egger's test showed no significant publication bias (Figure 2).
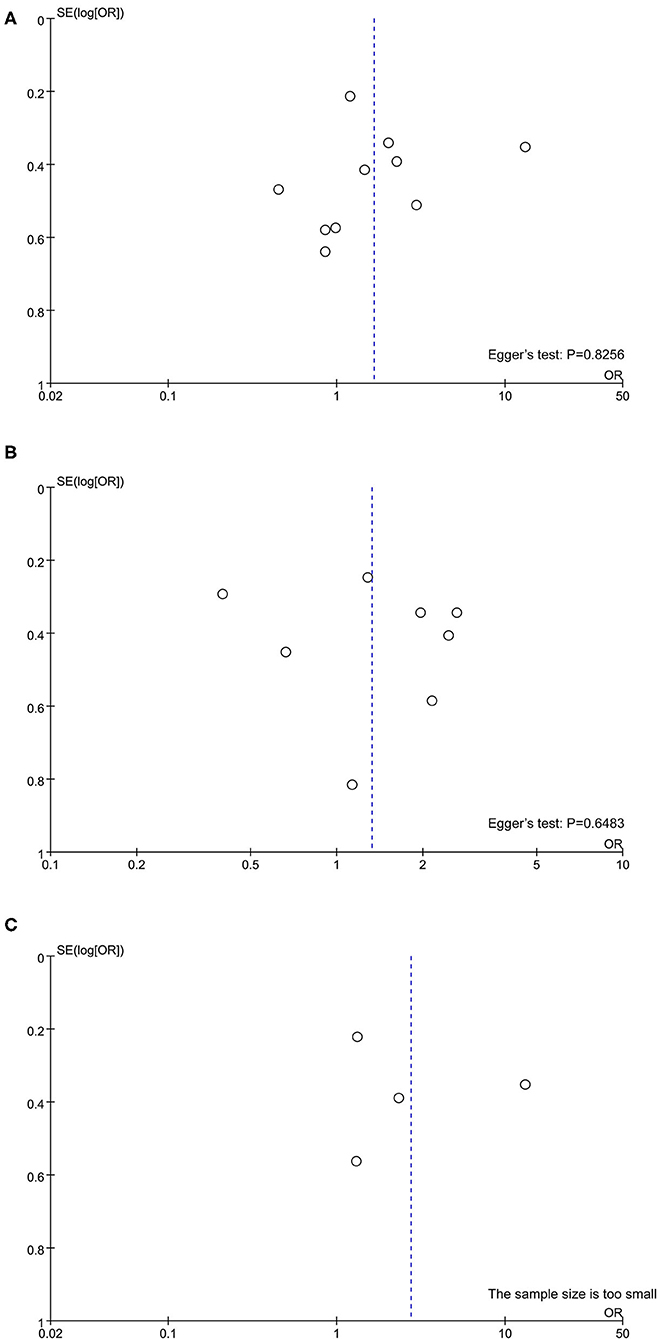
Figure 2. Publication bias assessed by funnel plot and Egger's test. (A) rs12979860; (B) rs8099917; (C) rs12980275.
Primary Analysis
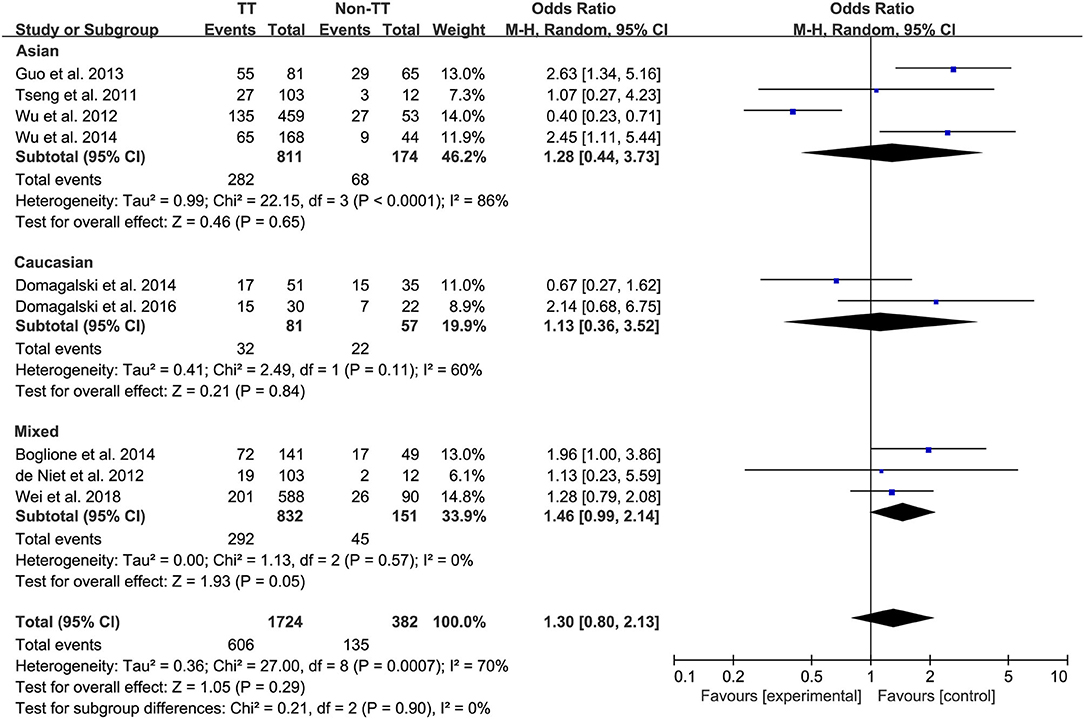
Ten studies comprising 1,174 patients with CC genotype vs. 553 patients with non-CC genotype (including CT+TT) that considered rs12979860; 9 studies comprising 1724 patients with TT genotype vs. 382 patients with non-TT genotype (including TG+GG) for rs8099917; 4 studies comprising 838 patients with AA genotype vs. 294 patients with non-CC genotype (including AG+GG) for rs12980275. A random-effects model was employed to estimate the SNP polymorphism's association with the treatment response among patients with CHB with PEG-IFN-α treatment (Figure 3). There was no significant association between rs12979860 and treatment response to PEG-IFN-α in all patients with HBV infection (CC vs. non-CC: OR 1.66, 95% CI 0.91–3.01, I2 = 82%). Similarly, no significant association was observed between rs8099917 (TT vs. non-TT: OR 1.30, 95% CI 0.80–2.13, I2 = 70%) and rs12980275 (AA vs. non-AA: OR 2.74, 95% CI 0.88–8.51, I2 = 90%).
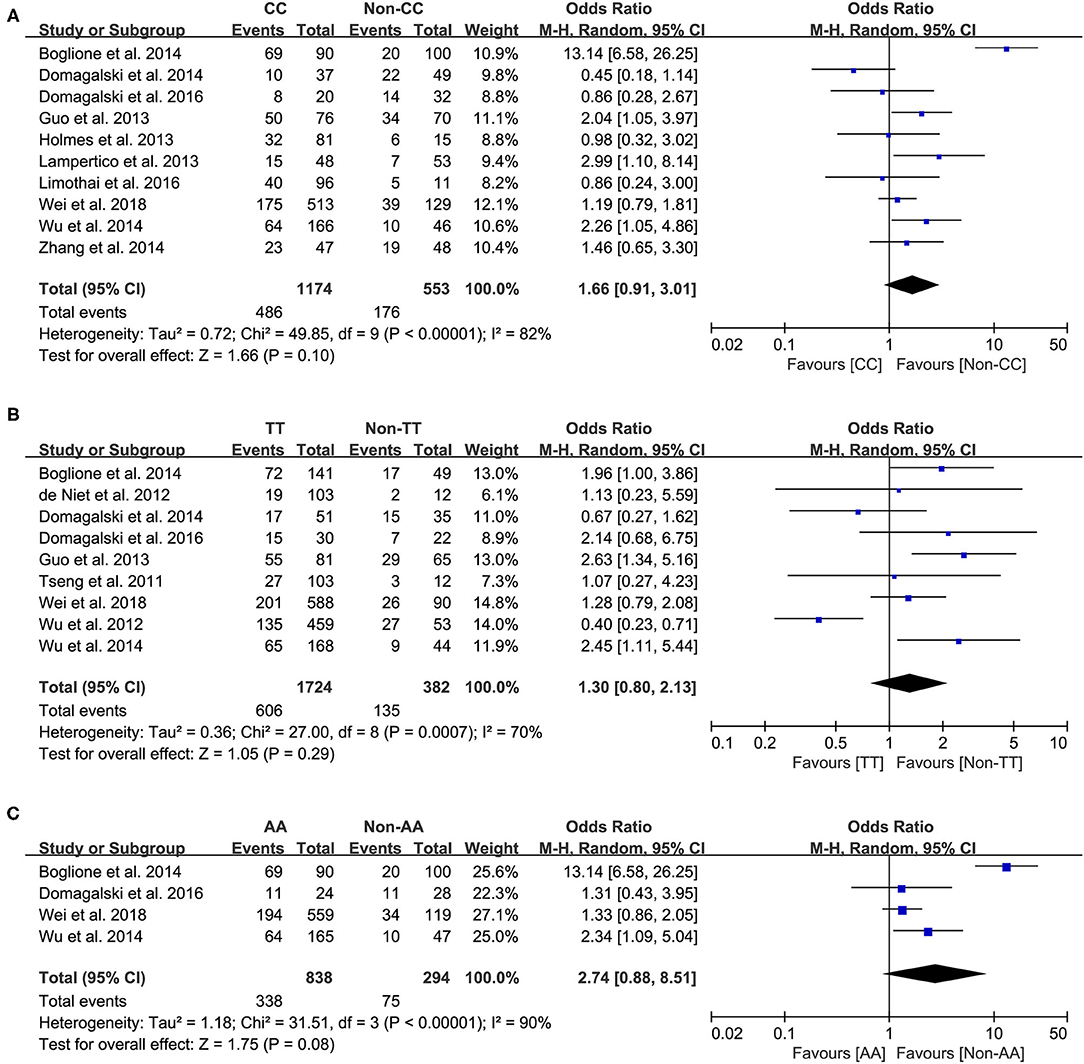
Figure 3. Forest plots for the association between IL-28B polymorphisms and treatment response of HBV infected patients treated with PEG-IFN. (A) overall analysis of rs12979860; (B) overall analysis of rs8099917; (C) overall analysis of rs12980275. HBV, hepatitis B virus; PEG-IFN, pegylated interferon.
Subgroup Analysis
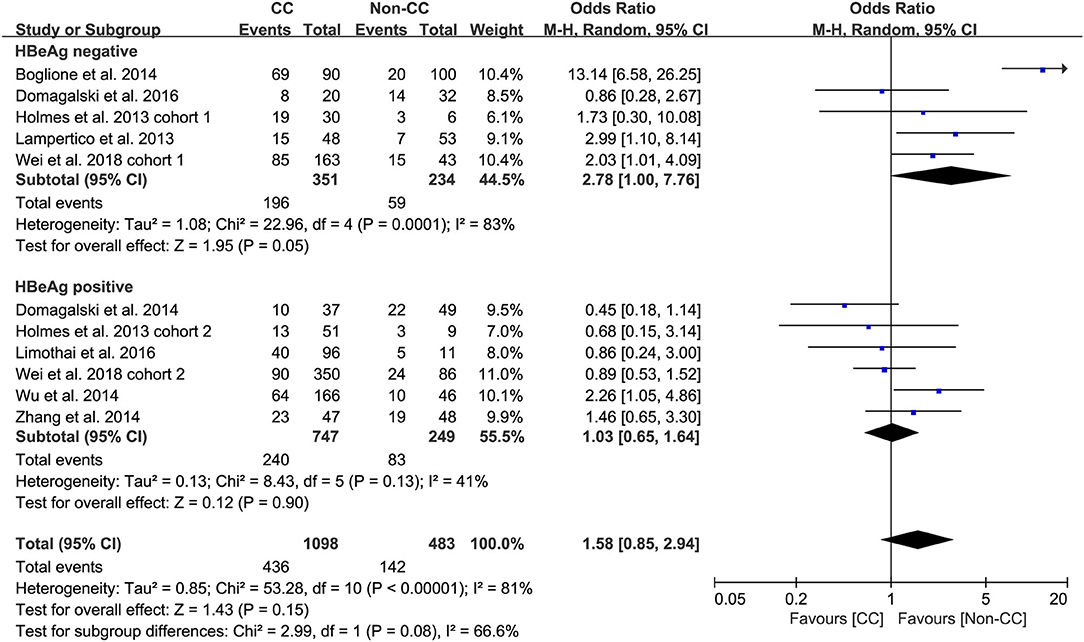
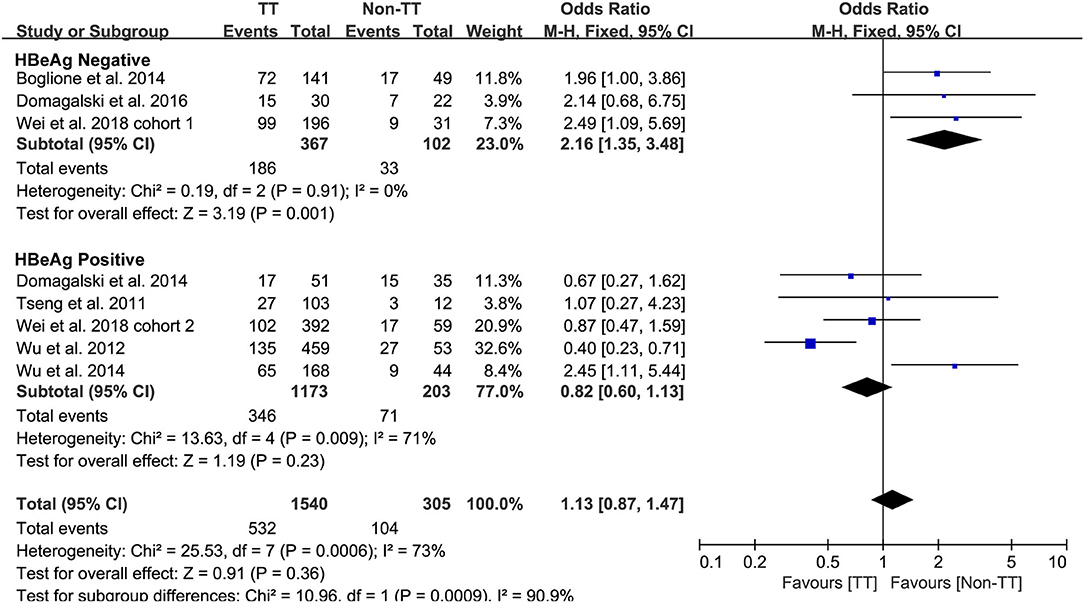
Predefined subgroup analyses were stratified by HBeAg and race to explore the discrepant treatment effect of different subgroups and heterogeneity. Among HBeAg-negative patients with CHB, the rs12979860 CC genotype and rs8099917 TT genotype were associated with a more significant treatment response to PEG-IFN-α (CC vs. non-CC: OR 2.78, 95% CI 1.00–7.76, I2 = 83%, Figure 4; TT vs. non-TT: OR 2.16, 95% CI 1.35–3.48, I2 = 0%, Figure 5). However, no differences were observed among patients with HBeAg-positive CHB.
Among Asian patients with CHB, the IL-28B rs12979860 CC genotype was associated with a more significant treatment response to PEG-IFN-α (CC vs. non-CC: OR 1.88, 95% CI 1.18–2.99, I2 = 0%, Figure 6). However, no significant differences were observed among the Caucasian CHB population. In addition, there was no association between the SNP rs8099917 with the incidence of treatment response among Asian or Caucasian populations with CHB (Figure 7).
Discussion
To date, there are three identified restriction fragment length polymorphisms (rs12980275, rs12979860, and rs8099917) of IL-28B, which have been studied in relation to the therapeutic efficacy of PEG-IFN-α in patients with CHB. Over the last few years, the association between IL-28B polymorphisms and the efficacy of PEG-IFN-α treatment in patients with CHB remains controversial. Many confounding factors can influence the results, such as sample sizes, different genotyping methods, ethnic differences, lifestyle, and environment.
In the present meta-analysis, we evaluated the association between IL-28B polymorphisms and treatment response in patients with CHB treated with PEG-IFN. We performed a meta-analysis of the correlation of IL-28B rs12979860 C/T, rs12980275 A/G, and rs8099917 polymorphisms and therapeutic outcomes, including virological response, serological response, biochemical response, or combined response, of PEG- IFN in patients infected with HBV. In this study, we found that there was no significant association between rs12979860 (CC vs. non-CC), rs8099917 (TT vs. non-TT) or rs12980275 (AA vs. non-AA) and treatment response to PEG-IFN-α in all patients with HBV infection. However, in predefined subgroup analysis, we found that among patients with HBeAg-negative CHB, the rs12979860 CC genotype and rs8099917 TT genotype were associated with significant treatment response to PEG-IFN-α. Among Asian patients with CHB, the IL-28B rs12979860 CC was significantly associated with promising treatment response to PEG-IFN-α.
Taken together, the controversy regarding IL-28B polymorphisms and the efficacy of PEG-IFN-α treatment in patients with CHB is especially apparent in the Asian population, as some studies have reported contradictory results (14, 15). Comparing 338 patients with rs12979860 CC genotype and 127 rs12979860 non-CC, the results showed that patients with rs12979860 CC genotype tended to achieve a better treatment response to PEG-IFN-α (OR 1.88, 95% CI 1.18–2.99, I2 = 0%, Figure 6). No significant differences were observed among the other populations with CHB and IL-28B polymorphisms. Thus, IL-28B rs12979860 CC can act as a predictor for the therapeutic efficacy of PEG-IFN-α in Asian patients with CHB. Previous studies have found that the IL-28B polymorphism is an additional predictor of PEG-IFN-α therapy in patients with HBeAg-negative CHB (27). Our results revealed IL-28B rs12979860 (OR 2.78, 95% CI 1.00–7.76, I2 = 83%, Figure 4) and rs8099917 (OR 2.16, 95% CI 1.35–3.48, I2 = 0%, Figure 5) genotypes could be used to predict curative effects, which is consistent with previous reports.
This study is the first to evaluate the value of three SNPs of IL-28B to predict the therapeutic outcomes of PEG-IFN-based treatment in patients with CHB. Nonetheless, our study has some limitations, and the study findings need to be interpreted cautiously. First, since none of the studies included in this meta-analysis were randomized trials, potential bias and confounding factors (e.g., age, sex, baseline HBV DNA, and medication history) could not be well-controlled. Notably, Boglione et al. (19) and Sonneveld et al. (28) performed multivariate analysis, and the adjusted OR indicated that the SNPs were associated with prognostic significance in assessing treatment response in patients with CHB. Boglione et al. (19) found that CC and TT genotypes were related to higher rates of virological response (CC vs. non-CC: adjusted OR 4.29, 95% CI 1.59-11.58; TT vs. non-TT: adjusted OR 3.75, 95% CI 1.24–11.36). Similarly, Sonneveld et al. (28) indicated that more patients with CHB with CC or AA genotype achieved HBeAg seroconversion (CC vs. non-CC: adjusted OR 2.89, 95% CI 1.15–7.80; AA vs. non-AA: adjusted OR 3.16, 95% CI 1.26–8.25). Thus, our results should be further validated in more well-designed trials that fully control potential confounders to rule out alternative explanations.
Another important limitation was the significant heterogeneity observed in the analyses. The existence of clinical heterogeneity is expected to lead to a degree of statistical heterogeneity in the results. The definitions of outcomes were inconsistent among the included studies. Four studies employed the single virological response as the primary outcome, while other studies used the virological response combined with serological or biochemical response as the primary outcome. In addition, the baseline characteristics of patients were inconsistent among included studies as well. Subgroup analysis of studies that only included patients with HBeAg-negative CHB found that patients with CC and/or TT genotype had a high incidence of treatment response to PEG-IFN-α. However, no difference was observed among the populations with positive HBeAg.
Conclusion
These results indicate that IL-28B polymorphism may be related to higher treatment response in patients infected with HBV treated with PEG-IFN. Therefore, detection of the IL-28B polymorphism may be an effective biomarker for predicting the treatment response of PEG-IFN-based therapy of patients with CHB. Regarding the limitations mentioned above, further trials warranted to better supplement and confirm our findings.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
S-YY, Y-RH, G-SG, K-HL, and ZH provided substantial contribution to the conception, drafting, editing, and final approval of this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by Ningbo Natural Science Foundation (2013A610239).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
References
1. Chan HL, Sung JJ. Hepatocellular carcinoma and hepatitis B virus. Semin Liver Dis. (2006) 26:153–61. doi: 10.1055/s-2006-939753
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
3. Papatheodoridis GV, Lampertico P, Manolakopoulos S, Lok A. Incidence of hepatocellular carcinoma in chronic hepatitis B patients receiving nucleos(t)ide therapy: a systematic review. J Hepatol. (2010) 53:348–56. doi: 10.1016/j.jhep.2010.02.035
4. Sung JJ, Tsoi KK, Wong VW, Li KC, Chan HL. Meta-analysis: Treatment of hepatitis B infection reduces risk of hepatocellular carcinoma. Aliment Pharmacol Ther. (2008) 28:1067–77. doi: 10.1111/j.1365-2036.2008.03816.x
5. Marcellin P, Lau GK, Bonino F, Farci P, Hadziyannis S, Jin R, et al. Peginterferon alfa-2a alone, lamivudine alone, and the two in combination in patients with HBeAg-negative chronic hepatitis B. N Engl J Med. (2004) 351:1206–17. doi: 10.1056/NEJMoa040431
7. Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. (2009) 50:661–2. doi: 10.1002/hep.23190
8. Lampertico P, Vigano M, Di Costanzo GG, Sagnelli E, Fasano M, Di Marco V, et al. Randomised study comparing 48 and 96 weeks peginterferon alpha-2a therapy in genotype D HBeAg-negative chronic hepatitis B. Gut. (2013) 62:290–8. doi: 10.1136/gutjnl-2011-301430
9. Lau GK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, et al. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med. (2005) 352:2682–95. doi: 10.1056/NEJMoa043470
10. Manesis EK, Hadziyannis SJ. Interferon alpha treatment and retreatment of hepatitis B e antigen-negative chronic hepatitis B. Gastroenterology. (2001) 121:101–9. doi: 10.1053/gast.2001.25524
11. Grzegorzewska AE, Jodlowska E, Mostowska A, Jagodzinski P. Effect of interferon lambda3 gene polymorphisms, rs8099917 and rs12979860, on response to hepatitis B virus vaccination and hepatitis B or C virus infections among hemodialysis patients. Pol Arch Med Wewn. (2015) 125:894–902. doi: 10.20452/pamw.3205
12. Grzegorzewska AE, Swiderska MK, Winnicka H, Niepolski L, Bura M, Lagiedo-Zelazowska M, et al. Circulating interferon-lambda3 and post-vaccination antibodies against the surface antigen of hepatitis B virus in hemodialysis patients exposed to hepatitis E virus. Cytokine. (2019) 123:154766. doi: 10.1016/j.cyto.2019.154766
13. Zhao J, Zhang X, Fang L, Pan H, Shi J. Association between IL28B polymorphisms and outcomes of hepatitis B virus infection: a meta-analysis. BMC Med Genet. (2020) 21:88. doi: 10.1186/s12881-020-01026-w
14. Wu H, Zhao G, Qian F, Liu K, Xie J, Zhou H, et al. Association of IL28B polymorphisms with peginterferon treatment response in Chinese Han patients with HBeAg-positive chronic hepatitis B. Liver Int. (2015) 35:473–81. doi: 10.1111/liv.12491
15. Holmes JA, Nguyen T, Ratnam D, Heerasing NM, Tehan JV, Bonanzinga S, et al. IL28B genotype is not useful for predicting treatment outcome in Asian chronic hepatitis B patients treated with pegylated interferon-alpha. J Gastroenterol Hepatol. (2013) 28:861–6. doi: 10.1111/jgh.12110
16. Wei L, Wedemeyer H, Liaw YF, Chan HL, Piratvisuth T, Marcellin P, et al. No association between IFNL3 (IL28B) genotype and response to peginterferon alfa-2a in HBeAg-positive or -negative chronic hepatitis B. PLoS ONE. (2018) 13:e0199198. doi: 10.1371/journal.pone.0199198
17. Domagalski K, Pawlowska M, Zalesna A, Pilarczyk M, Rajewski P, Halota W, et al. Impact of IL28B and OAS gene family polymorphisms on interferon treatment response in Caucasian children chronically infected with hepatitis B virus. World J Gastroenterol. (2016) 22:9186–95. doi: 10.3748/wjg.v22.i41.9186
18. Limothai U, Chuaypen N, Khlaiphuengsin A, Posuwan N, Wasitthankasem R, Poovorawan Y, et al. Association of interferon-gamma inducible protein 10 polymorphism with treatment response to pegylated interferon in HBeAg-positive chronic hepatitis B. Antivir Ther. (2016) 21:97–106. doi: 10.3851/IMP2992
19. Boglione L, Cusato J, Allegra S, Esposito I, Patti F, Cariti G, et al. Role of IL28-B polymorphisms in the treatment of chronic hepatitis B HBeAg-negative patients with peginterferon. Antiviral Res. (2014) 102:35–43. doi: 10.1016/j.antiviral.2013.11.014
20. Domagalski K, Pawlowska M, Zalesna A, Tyczyno M, Skorupa-Klaput M, Tretyn A, et al. The relationship between IL-28B polymorphisms and the response to peginterferon alfa-2a monotherapy in anti-HBe-positive patients with chronic HBV infection. Eur J Clin Microbiol Infect Dis. (2014) 33:2025–33. doi: 10.1007/s10096-014-2172-1
21. Zhang Q, Lapalus M, Asselah T, Laouenan C, Moucari R, Martinot-Peignoux M, et al. IFNL3 (IL28B) polymorphism does not predict long-term response to interferon therapy in HBeAg-positive chronic hepatitis B patients. J Viral Hepat. (2014) 21:525–32. doi: 10.1111/jvh.12177
22. Guo X, Yang G, Yuan J, Ruan P, Zhang M, Chen X, et al. Genetic variation in interleukin 28B and response to antiviral therapy in patients with dual chronic infection with hepatitis B and C viruses. PLoS ONE. (2013) 8:e77911. doi: 10.1371/journal.pone.0077911
23. Wu X, Xin Z, Zhu X, Pan L, Li Z, Li H, et al. Evaluation of susceptibility locus for response to interferon-alpha based therapy in chronic hepatitis B patients in Chinese. Antiviral Res. (2012) 93:297–300. doi: 10.1016/j.antiviral.2011.12.009
24. de Niet A, Takkenberg RB, Benayed R, Riley-Gillis B, Weegink CJ, Zaaijer HL, et al. Genetic variation in IL28B and treatment outcome in HBeAg-positive and -negative chronic hepatitis B patients treated with Peg interferon alfa-2a and adefovir. Scand J Gastroenterol. (2012) 47:475–81. doi: 10.3109/00365521.2011.648952
25. Tseng T-C, Yu M-L, Liu C-J, Lin C-L, Huang Y-W, Hsu C-S, et al. Effect of host and viral factors on hepatitis B e antigen-positive chronic hepatitis B patients receiving pegylated interferon-α-2a therapy. Antivir Ther. (2011) 16:629–37. doi: 10.3851/IMP1841
26. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. (2002) 21:1539–58. doi: 10.1002/sim.1186
27. Lampertico P, Viganò M, Cheroni C, Facchetti F, Invernizzi F, Valveri V, et al. IL28B polymorphisms predict interferon-related hepatitis B surface antigen seroclearance in genotype D hepatitis B e antigen-negative patients with chronic hepatitis B. Hepatology. (2013) 57:890–6. doi: 10.1002/hep.25749
Keywords: IL-28B, polymorphism, hepatitis B virus, polyethylene glycol, interferon alpha, meta-analysis
Citation: Ying S-Y, Hu Y-R, Gao G-S, Lou K-H and Huang Z (2021) Interleukin-28B Polymorphisms Predict the Efficacy of Peginterferon Alpha in Patients With Chronic Hepatitis B: A Meta-Analysis. Front. Med. 8:691365. doi: 10.3389/fmed.2021.691365
Received: 16 April 2021; Accepted: 11 June 2021;
Published: 09 July 2021.
Edited by:
Jessica Cusato, University of Turin, ItalyReviewed by:
Amedeo De Nicolò, University of Turin, ItalyAntonio D'Avolio, University of Turin, Italy
Copyright © 2021 Ying, Hu, Gao, Lou and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Huang, aHVhbmd6aGVuMDAxQHpqdS5lZHUuY24=
 Sang-Yu Ying
Sang-Yu Ying Yao-Ren Hu2,3
Yao-Ren Hu2,3