- Shengjing Hospital of China Medical University, Shenyang, China
Purpose: Retinopathy of prematurity (ROP) is a common retinal vascular disease in premature neonates. In recent years, there is increasing evidence that the long non-coding RNA taurine upregulated gene 1 (TUG1) plays a regulatory role in vascular diseases, suggesting a potential role for TUG1 in vascular endothelial cells. We hypothesized that TUG1 may be associated with ROP. Our aim, therefore, was to explore the biological functions of TUG1 in aberrant retinal development.
Methods: We used the mouse oxygen-induced retinopathy (OIR) model to simulate the pathological changes of retinal in ROP. Quantitative real-time polymerase chain reaction was used to detect the expression of TUG1, miR-145-5p and cellular communication network factor 1 (CCN1). Human retinal endothelial cells (HRECs) were treated with CoCl2 to mimic hypoxia conditions. Cellular functional changes were observed after transfection with RNA interference (RNAi)-TUG1 and miR-145-5p mimics. The apoptosis of HRECs was detected by flow cytometry, the migration ability was detected by wound healing and transwell migration assays, and the ability of angiogenesis was detected by tube formation assay. The potential binding sites between TUG1, miR-145-5p, and CCN1 were verified by dual-luciferase reporter assays. The degree of retinopathy was evaluated by staining retinal sections with hematoxylin and eosin, and the expression of CCN1, HIF-1α, VEGF, caspase-3, Bcl-2, IL-1β, and TNF-α protein was analyzed by Western blotting and immunohistochemistry.
Results: In the retina tissue of OIR mice, TUG1, miR-145-5p, and CCN1 were differentially expressed. Knocking down TUG1 attenuated apoptosis, migration, and angiogenesis induced by hypoxia on HRECs, as did miR-145-5p overexpression. Results from reporter assays indicate direct interactions between TUG1, miR-145-5p, and CCN1. Intravitreal injection of miR-145-5p mimics reduced the degree of retinopathy.
Conclusion: TUG1 acts as a molecular sponge of miR-145-5p to regulate CCN1 expression and thus regulate the development of retinal neovascularization. This regulatory mechanism may provide a new theoretical basis for the prevention and treatment of ROP.
Introduction
Retinopathy of prematurity (ROP) is a retinal vascular disease that occurs in preterm and low birth weight infants. While the details of the pathogenic process are not clear, it is believed that the formation of retinal non-perfusion areas and pathological retinal neovascularization (RNV) lead to ROP (1, 2). At present, retinal laser photocoagulation and vitreous injection of anti-vascular endothelial growth factor (VEGF) drugs are commonly used to control the development of ROP. However, laser photocoagulation may damage the retina, and drugs injected into the vitreous cavity may enter the blood circulation, adversely affecting the underdeveloped organ systems of premature infants. Even if drugs are combined with laser therapy, there are serious adverse reactions (3–5). Therefore, at present, anti-VEGF drugs are still used cautiously in the treatment of ROP. In light of these drawbacks, the prevention and treatment of ROP is still an unsolved problem, and there is an urgent need to identify new strategies to combat this disease.
Long non-coding RNAs (lncRNAs) are a class of RNA transcripts that are greater than 200 bp in length and have almost no potential to encode proteins (6). The lncRNA taurine upregulated gene 1 (TUG1) was discovered relatively early in the history of lncRNA investigations, and it is indispensable in the development of the retina (7). It has also been implicated in various disease-linked cellular processes, including cell proliferation, cell differentiation, cell invasion, radiation resistance, angiogenesis, and inflammation (8, 9). In particular, it plays an important role in neovascularization and the malignant degree and prognosis of many tumor types (10). It plays an important role in biological regulation by acting as a competitive endogenous RNA (or sponge) for microRNAs (miRNAs), thereby regulating downstream mRNAs (11). The miRNAs that bind to TUG1 include miR-1299, miR-186-5p, miR-212-3p, miR-145, miR-26a, and so on (12–14). MiRNAs are endogenous non-coding small RNAs that are 21–23 nucleotides in length and highly conserved in evolution (15, 16). LncRNAs can compete with miRNAs to regulate the proliferation and migration of human retinal endothelial cells (HRECs) and the formation of RNV in diabetic retinopathy (17). Based on the fact that the molecular mechanism of RNV in ROP is similar to that of diabetic retinopathy, we speculated that lncRNAs and miRNAs may also be involved in the regulation of ROP. Cellular communication network factor 1 (CCN1) is a stromal cell protein involved in vascular development and tissue repair. In our previous study, we found that CCN1 is highly expressed during RNV in the formation of ROP (18), and a study that analyzed human clinical specimens also showed that the level of CCN1 increased in some ocular vascular complications (19). Results from bioinformatics analyses (via the starBase database 2.0.1) (20) predict that miR-145-5p is a target of TUG1 and CCN1 and suggest that TUG1 may affect the expression of CCN1 by regulating miR-145-5p. Therefore, this study aimed to investigate the expression and function of the lncRNA TUG1 and miR-145-5p in a mouse model of oxygen-induced retinopathy (OIR) and HRECs. Our results provide new insights for clinical investigations focused on improving treatments for ROP.
Materials and Methods
Animals and Oxygen-Induced Retinopathy Model
All animal experiments were performed according to the ARRIVE guidelines and in accordance with the EU Directive 2010/63/EU for animal experiments. These experiments also complied with the Chinese Guidelines for Ethical Review of Laboratory Animal Welfare (GB/T 35892-20181). The Medical Ethics Committee of Shengjing Hospital of China Medical University approved the use of animals for the experiments (ethics code: 2018PS239K) on 1 March 2018. C57BL/6J mice were purchased from Shenyang Changsheng Biological Technology Co., Ltd. [Shenyang, China; license No.: SCXK (Liao) 2015-0001]. Mice were reared in the standard pathogen-free facility for experimental animals at the Benxi base of Shengjing Hospital under temperature conditions of 23 ± 2°C, with a 12-h light/dark cycle. The pups were randomly divided into the control, OIR, OIR-negative control (NC), and OIR-mimics groups, with 60 pups per group. An OIR model was established using the classical Smith method (21). The pups in the control groups were placed in normoxia up to postnatal day (PD) 17. On PD 7, pups in OIR, OIR-NC, OIR-mimics groups were placed in a hyperoxia chamber (75 ± 2%) for 5 days and subsequently removed to normoxic condition for a further 5 days up to PD 17. OIR-NC and OIR-mimics groups received intravitreal injection under isoflurane inhalation anesthesia on PD12. The pups in OIR-NC group received intravitreal injection of 0.5 μL of miR-145-5p mimics negative control reagent (miR-145-5p NC mimics) + 0.5 μL of animal infection reagent (IVG2101) (InvivoGene, China) + 0.5 μL of PBS, and the OIR-mimics group was treated with 0.5 μL of miR-145-5p mimics reagent (miR-145-5p mimics) + 0.5 μL of animal infection reagent + 0.5 μL of PBS. The sequences of miR-145-5p NC mimics were as follows: sense: 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense: 5′-ACGUGACACGUUCGGAGAATT-3′. The miR-145-5p mimics sequences were as follows: sense: 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′; antisense: 5′-GGAUUCCUGGGAAAACUGGACUU-3′. The retinal tissue for polymerase chain reaction (PCR) was collected on PD14, and on PD17 for Western blotting and other follow-up experiments.
Cell Lines and Cell Culture
The HREC and human renal epithelial (HEK293T) cell lines were purchased from American Type Culture Collection (ATCC, Gaithersburg, MD, United States). The cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS) (Gibco, NY, United States) in a humidified incubator (Thermo Scientific, MA, United States) at 37°C, with 5% CO2. To simulate hypoxic conditions, cells were treated with 200 μmol/L CoCl2 (Sigma-Aldrich, St. Louis, MO, United States) (22). When knocking down TUG1, cell culture experiments comprised the following groups: HRECs cultured under normal conditions (control group), HRECs treated with CoCl2 (CoCl2 group), HRECs transfected with negative control RNA interference (RNAi) and treated with CoCl2 (CoCl2 + NC group), and HRECs transfected with TUG1-RNAi and treated with CoCl2 (CoCl2 + RNAi group). When overexpressing miR-145-5p, cell culture experiments comprised the following groups: control group, CoCl2 group, HRECs transfected with miR-145-5p NC mimics and treated with CoCl2 (CoCl2 + NC group), and HRECs transfected with miR-145-5p mimics and treated with CoCl2 (CoCl2 + mimics group).
Transient Transfections
Human retinal endothelial cells were evenly seeded into a six-well plate and transfected at 60–70% confluency. TUG1 RNAi (TUG1-RNAi), negative control TUG1-RNAi (RNAi-NC) (Genechen, Shanghai, China), miR-145-5p mimics (miR-145-5p mimics), and miR-145-5p NC mimics (NC mimics) (GenePharma, Shanghai, China) were transfected into the cells using the liposome transfection reagent Lipofectamine 3000 (Invitrogen, Waltham, MA, United States), according to the manufacturer’s instructions. Before the experiment, we examined the transfection efficiency of the plasmids and mimics by quantitative real-time PCR (qRT-PCR). The sequences of TUG1-RNAi, RNAi-NC, miR-145-5p mimics, and NC mimics are as follows:
TUG1-RNAi 1: 5′-CGAGATGATTCCTACCACCTT-3′; TUG1-RNAi 2: 5′-GCCAAATTCAAATACTGGCAA-3′; TUG1-RNAi 3: 5′-CCTCCATGAATACCTGAATTA-3′; RNAi-NC: 5′-TTCTCCGAACGTGTCACGT-3′; miR-145-5p mimics: sense: 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′; miR-145-5p mimics: antisense: 5′-GGAUUCCUGGGAAAACUGGACUU-3′; NC mimics: sense: 5′-UUCUCCGAACGUGUCACGUTT-3′; NC mimics: antisense: 5′-ACGUGACACGUUCGGAGAATT-3′.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from cultured cells and retina tissue using TRIzol (Takara Biotechnology, Otsu, Japan), and the expression of TUG1, miRNA-145-5p, and CCN1 mRNA was detected by qRT-PCR. cDNA was synthesized using the Mir-X miRNA First-Strand Synthesis kit (Cat. No. 638313, Takara Biotechnology, Otsu, Japan) and the PrimeScript RT reagent kit with gDNA Eraser (Code No. RR047Q, Takara Biotechnology). PCR was then performed on a 7500 Fast Real-Time PCR machine (Applied Biosystems, Warrington, United Kingdom) using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus, code no. RR820A, Takara Biotechnology). The reaction conditions were as follows: 40 cycles of denaturation for 30 s at 95°C, amplification for 3 s at 95°C and 30 s at 60°C. β-Actin and U6 were used as internal reference genes for mRNA and miRNA analyses, respectively. For quantitative analysis of differential expression, data were processed using the 2–ΔΔCt method, with the mRNA expression presented as the relative fold change compared to β-actin, and miRNA expression presented as the relative fold change compared to U6. The primers (Shenggong, Shanghai, China) used are shown in Table 1.
Flow Cytometry for Apoptosis
Twenty-four hours after transfection, HRECs were treated with 200 μmol/L CoCl2 for 24 h. Then, the cells were collected by treating with trypsin without EDTA and subjected to AnnexinV-PE/7-AAD labeling using an apoptosis detection kit (Vazyme, Nanjing, China). The cells were incubated at room temperature and protected from light for 10 min according to the manufacturer’s instructions and analyzed by flow cytometry (BD, FACSCalibur, NJ, United States). Cell Quest Pro software (Becton-Dickinson, CA, United States) was used for data analysis.
Wound Healing Assay
Human retinal endothelial cells were uniformly seeded into a six-well culture plate, and three horizontal lines were marked on the bottom of each well. The cells were transfected and treated with CoCl2 as described earlier. When the cells reached a confluency of approximately 90%, the tip of a 10 μL micropipette was used to make a uniform scratch through the cell monolayer, perpendicular to the horizontal line. The disrupted cell mass was gently washed off with PBS, and the remaining adherent cells were incubated in a serum-free culture medium. After 48 h, wound healing was examined and photographed.
Transwell Migration Assay
The cells were transfected and treated with CoCl2 as described earlier. Next, 3 × 104 cells were suspended in 200 μL of serum-free RPMI1640 medium and transferred to the upper chamber of the transwell insert in a 24-well plate (Corning, NY, United States), and 500 μL of medium containing 10% FBS was added to the lower chamber. The cells were incubated at 37°C for 24 h, after which the remaining cells in the upper chamber were gently scraped off with cotton swabs. The transwell membrane was then fixed with 4% paraformaldehyde for 20 min, washed with PBS, stained with 0.5% hematoxylin solution for 20 min, and washed again with PBS. Finally, we analyzed the number of cells on the bottom of the membrane using an inverted microscope (ECLIPSE Ni-U, Nikon, Tokyo, Japan). For each well, the average number of cells was calculated from five randomly selected fields of view.
Tube Formation Assay
The bottom of a 96-well plate was evenly coated with 50 μL of Matrigel (Corning, NY, United States) diluted 2:1 in a basic medium and incubated at 37°C for 30 min to form a gel. The cells were suspended in a medium containing 10% FBS, and 1 × 104 cells were seeded per well in the Matrigel-coated 96-well plate. After 6 h of incubation, each well was examined and photographed using an inverted microscope (ECLIPSE Ni-U, Nikon). Each treatment group comprised three wells, and 1–2 fields of view were captured for each well. The overall experiment was repeated five times. In each field of view, we counted the number of nodes and meshes and measured the total branching length of the tubes.
Dual-Luciferase Reporter Assays
Dual luciferase reporter assays were used to verify binding of miR-145-5p with TUG1 and CCN1. The sites in miR-145-5p that facilitate binding with TUG1 and CCN1 were predicted via the starBase database. An expression vector was designed and constructed. First, 5 × 103 HEK293T cells were seeded into each well in a 96-well plate. After 12 h, 3 pmol miR-145-5p mimics or miR-145-5p NC mimics were co-transfected with 0.1 μg TUG1-WT or TUG1-MUT using Lipofectamine 3000, according to the manufacturer’s instructions. After a further 36 h, the cells were lysed with 1 × passive lysis buffer from the Dual-Luciferase Reporter Assay System (Promega, WI, United States), and luciferase activity was measured on a luminometer (Synergy H1 microplate reader, BioTek, VT, United States). Firefly luciferase activity was normalized to Renilla luciferase activity. Each transfection reaction was performed in triplicate. The same procedure was used for the co-transfection of miR-145-5p mimics or miR-145-5p NC mimics with CCN1-WT or CCN1-MUT.
Retinal Vascular Staining
Under isoflurane inhalation anesthesia, eyeballs from 17-day-old mice were removed and fixed in 4% paraformaldehyde overnight. The limbus cornea was incised and the anterior segment and vitreous body were removed. Retinal tissue was separated and cleaned with PBS, the Isolectin IB4 Alexa Fluor® dye conjugate (Invitrogen, United States) was dissolved in PBS solution containing 1% TritonX-100 (Solarbio, Beijing, China), and incubated with the retinal tissue at 4°C overnight. The retinal tissue was taken out the next day, cleaned with PBS, placed on glass slides, and cut into four sections under the microscope. The slices were sealed with an anti-quenching agent and photographed with a fluorescence microscope (ECLIPSE Ni-U, Nikon). ImageJ software 1.51 (NIH, United States) was used to calculate the retinal avascular area.
Detailed steps are as follows:
1. Open the picture, click Image → Type → 8-bit.
2. Select the heart-shaped circle, the Freehand Selections are displayed.
3. Circle the entire area of the retina in the picture with the mouse, click Analyze → Measurement, to get the total area of the retina.
4. Re-select the non-perfusion area of the retina and click Analyze → Measurement, to obtain the non-perfusion area of the retina.
5. Enlarge the picture and circle the neovascularization part of the retina as much as possible to get the neovascularization area of the retina.
6. Non-perfusion area/Total retinal area = percentage of areas without vascular perfusion; Neovascularization area/Total retinal area = percentage area of neovascularization.
Hematoxylin and Eosin Staining
The eyeballs of 17-day-old mice were fixed in 4% paraformaldehyde overnight, embedded in paraffin, and cut into 4 μm thick sections. The slices were dried at 60°C for 6, dewaxed, stained with hematoxylin and eosin (HE), dehydrated, and sealed with neutral gum. The number of nuclei penetrating the inner limiting membrane of the retina was directly counted by the double-blind method.
Immunohistochemistry
Immunohistochemistry was performed with the Streptavidin-Biotin Complex (SABC) kit (Boster Bio, Pleasanton, CA, United States). The paraffin sections were first dewaxed and thermally repaired with a 3% citric acid repair solution. Then, 3% H2O2 was added dropwise and incubated at room temperature for 10 min. The sections were washed three times with PBS and blocked with goat serum at 37°C for 30 min. Then, the sections were incubated with diluted primary antibody [CCN1, hypoxia-inducible factor-1 α (HIF-1α), VEGF, caspase-3, Bcl-2, IL-1β, and TNF-α, 1:200, ImmunoWay, Plano, TX, United States] at 4°C overnight. On the second day, the sections were washed with PBS three times. Biotinylated goat anti-rabbit IgG was dropped and incubated at 37°C for 30 min, then washed with PBS three times. The SBC-POD reagent was added dropwise to the samples and incubated at 37°C for 30 min. For negative control groups, the primary antibody was replaced with PBS. Under the microscope, 3, 3′-diaminobenzidine was applied for 50 s and counterstained with hematoxylin, sealed with neutral gum, and then observed and photographed under the microscope. Brown cells were positive for the respective antibody, and ImageJ was used to calculate the cumulative optical density of each immunohistochemical section by the double-blind method.
Western Blotting
The treated cells and retinal tissue were lysed in RIPA lysis buffer (Solarbio) containing 1% PMSF (Solarbio), and the protein concentration was determined using BCA assay (BCA Protein Quantification Kit, Vazyme, Nanjing, China). Then, protein samples were separated with a 10% fast electrophoresis gel preparation kit (EpiZyme, Shanghai, China) and transferred to a PVDF membrane (Merck Millipore, Burlington, MA, United States). After blocking with 5% bovine serum albumin for 1 h, membranes were incubated in primary antibody overnight at 4°C. The primary antibodies used were rabbit-derived anti-CCN1 (1:1,000) and rabbit-derived anti-tubulin (1:1,000) (both Immunoway). The secondary antibody was horseradish peroxidase-labeled goat anti-rabbit IgG (H + L) (1:1,000, Immunoway). After washing with TBST (0.05 M Tris, 0.15 M NaCl, pH 7.4 with 0.1% Tween-20), the PVDF membrane was developed on an Amersham Imager 680 with chemiluminescent reagents (BeyoECL Plus, Biyuntian, Shanghai, China). ImageJ software 1.51 (NIH) was used to normalize the intensity of the target protein signal to that of tubulin.
Statistical Analysis
All analyses were performed in GraphPad Prism 8.0 software (GraphPad Software Inc., CA, United States). All values are presented as the mean ± standard deviation (SD) of three or more independent experiments. Statistical analysis was performed using Student’s t-test or one-way ANOVA. *P < 0.05, **P < 0.01, and ***P < 0.001 indicate significant differences.
Results
Tug1 Expression Was Upregulated and MiR-145-5p Was Down-Regulated in Oxygen-Induced Retinopathy Mice
We analyzed the expression of Tug1 and miR-145-5p in the control group and the OIR group by using qRT-PCR. The results showed that the expression of Tug1 in the OIR group was significantly higher than that in the control group (Figure 1A, ***P < 0.001), while the expression of miR-145-5p significantly lower (Figure 1B, **P < 0.01).
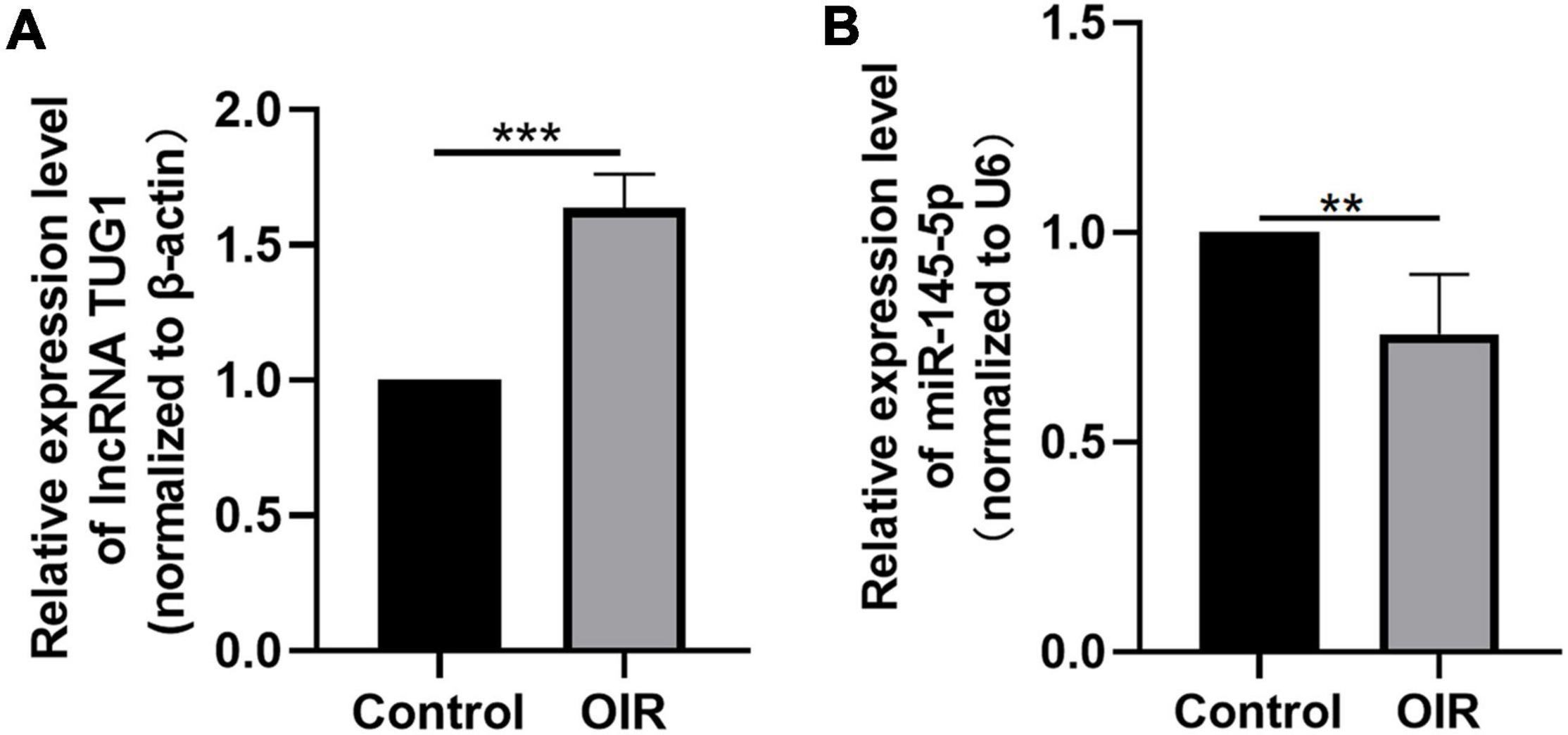
Figure 1. Tug1 expression was upregulated and miR-145-5p was down-regulated in OIR mice. (A) Tug1 expression level (mean ± SD, n = 5). ***P < 0.001. (B) MiR-145-5p expression level (mean ± SD, n = 5). **P < 0.01.
Knocking Down Taurine Upregulated Gene 1 Inhibits Apoptosis, Migration, and Angiogenesis in Human Retinal Endothelial Cells Induced by Hypoxia
As shown in Figure 2A, TUG1 expression was successfully knocked down in normal HRECs when transfected with TUG1-RNAi, and TUG1-RNAi 1 was selected for subsequent experiments. Next, we determined the relative expression of TUG1 in the control, CoCl2, CoCl2 + NC, and CoCl2 + RNAi groups (Figure 2B). TUG1 expression was significantly higher in the CoCl2 group compared to the control (***P < 0.001), and expression of the TUG1 was significantly lower in the CoCl2 + RNAi group compared to the CoCl2 + NC (***P < 0.001). These results suggest that TUG1-RNAi successfully inhibited the increase in TUG1 induced by hypoxia. Flow cytometry analysis indicated that the apoptosis was increased in the CoCl2 group compared with the control (Figures 2C,D, ***P < 0.001) and was significantly inhibited in the CoCl2 + RNAi group compared with the CoCl2 + NC (***P < 0.001). The wound healing and transwell migration assays showed that treatment with CoCl2 promoted HREC migration compared with that in the control (Figures 2E–H, wound healing assay***P < 0.001, transwell assay***P < 0.001), the migratory ability of HRECs in the CoCl2 + RNAi group was lower than that in the CoCl2 + NC group (wound healing assay***P < 0.001, transwell migration assay***P < 0.001). Together, these data indicate that hypoxia induced by CoCl2 promoted HREC migration and knocking down TUG1 inhibited the migratory ability of these cells. The formation of RNV is related to the tube-forming ability of endothelial cells, and therefore, we examined the angiogenic activity of HRECs using a tube formation assay. The results demonstrated that, compared with the control group (Figures 2I,J), the CoCl2 group exhibited a greater number of meshes (***P < 0.001), which promoted tube formation in the HRECs. Compared with the CoCl2 + NC group, the CoCl2 + RNAi group exhibited fewer meshes (***P < 0.001), indicating that loss of TUG1 reduces the angiogenic capacity of HRECs.
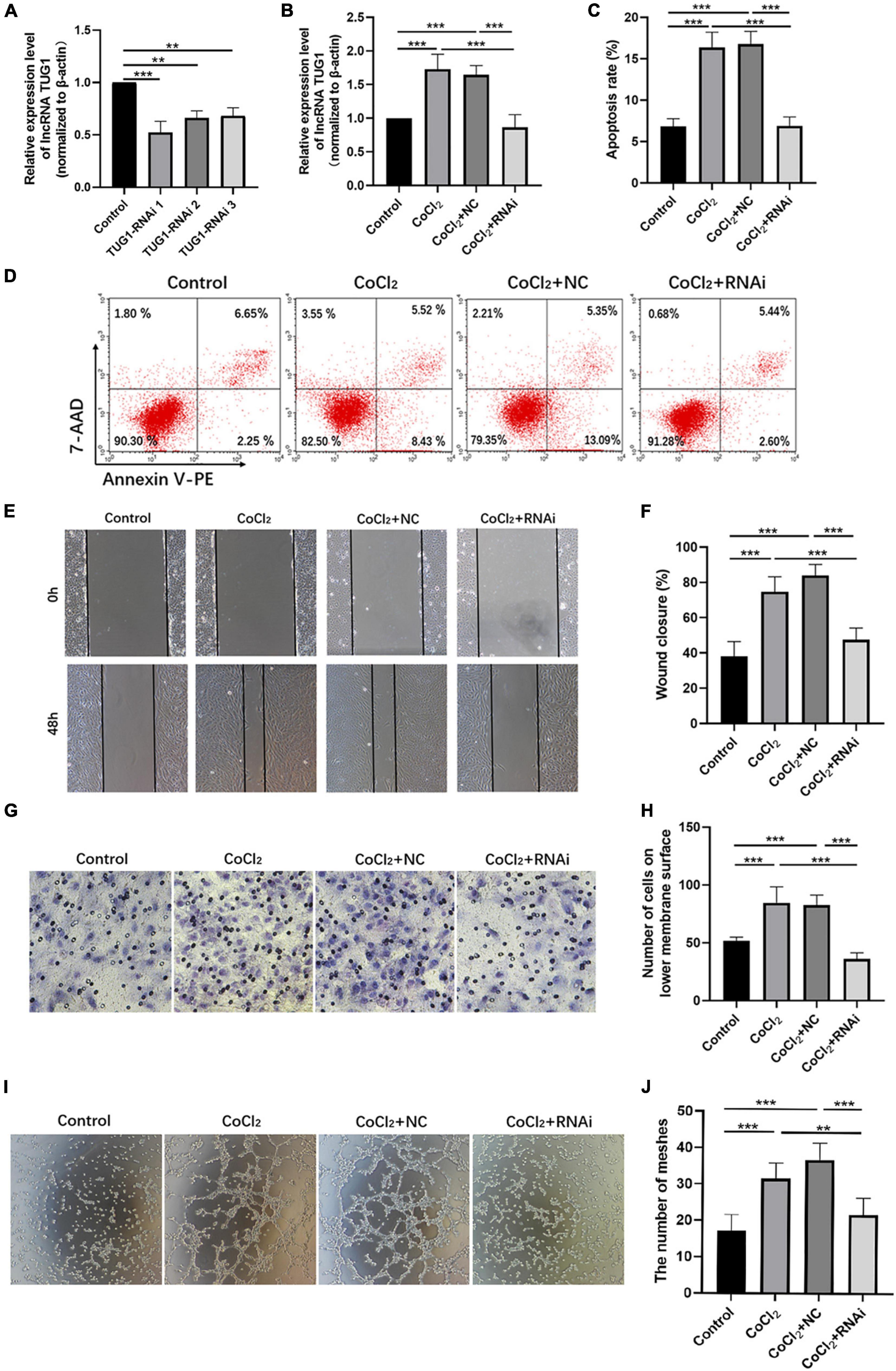
Figure 2. Knocking down Tug1 inhibits apoptosis, migration, and angiogenesis in HRECs induced by hypoxia. (A) The transfection efficiency of TUG1-RNAi (mean ± SD, n = 3). ***P < 0.001, **P < 0.01. (B) TUG1 expression level (mean ± SD, n = 5). ***P < 0.001. (C) Flow cytometry results showing the percentage of apoptotic cells (mean ± SD, n = 5). ***P < 0.001. (D) Results of flow cytometry with cells labeled with Annexin-PE and 7-Aminoactinomycin D (7-AAD). (E) Results of wound healing assay, 100× magnification. (F) Quantitation of results from the wound healing assay in (E) (mean ± SD, n = 5). ***P < 0.001. (G) Results of transwell migration assay, 200× magnification. (H) Quantitation of results from the transwell migration assay in (G) (mean ± SD, n = 5). ***P < 0.001. (I) Results of tube formation assay, 100× magnification. (J) Comparison of the number of meshes in different treatment groups (mean ± SD, n = 5), ***P < 0.001, *P < 0.05.
MiR-145-5p Interacts With Taurine Upregulated Gene 1 and Cellular Communication Network Factor 1
Bioinformatic analyses using the starBase interface indicated potential binding sites between TUG1 and miR-145-5p (Figure 3A). The transfection efficiency of miR-145-5p mimics was confirmed by qRT-PCR (Figure 3B, ***P < 0.001). Results from luciferase reporter assays showed that in the TUG1-WT group, transfection with miR-145-5p mimics significantly reduced relative luciferase activity compared with that of the NC mimics (Figure 3C, ***P < 0.001), suggesting an interaction between them.
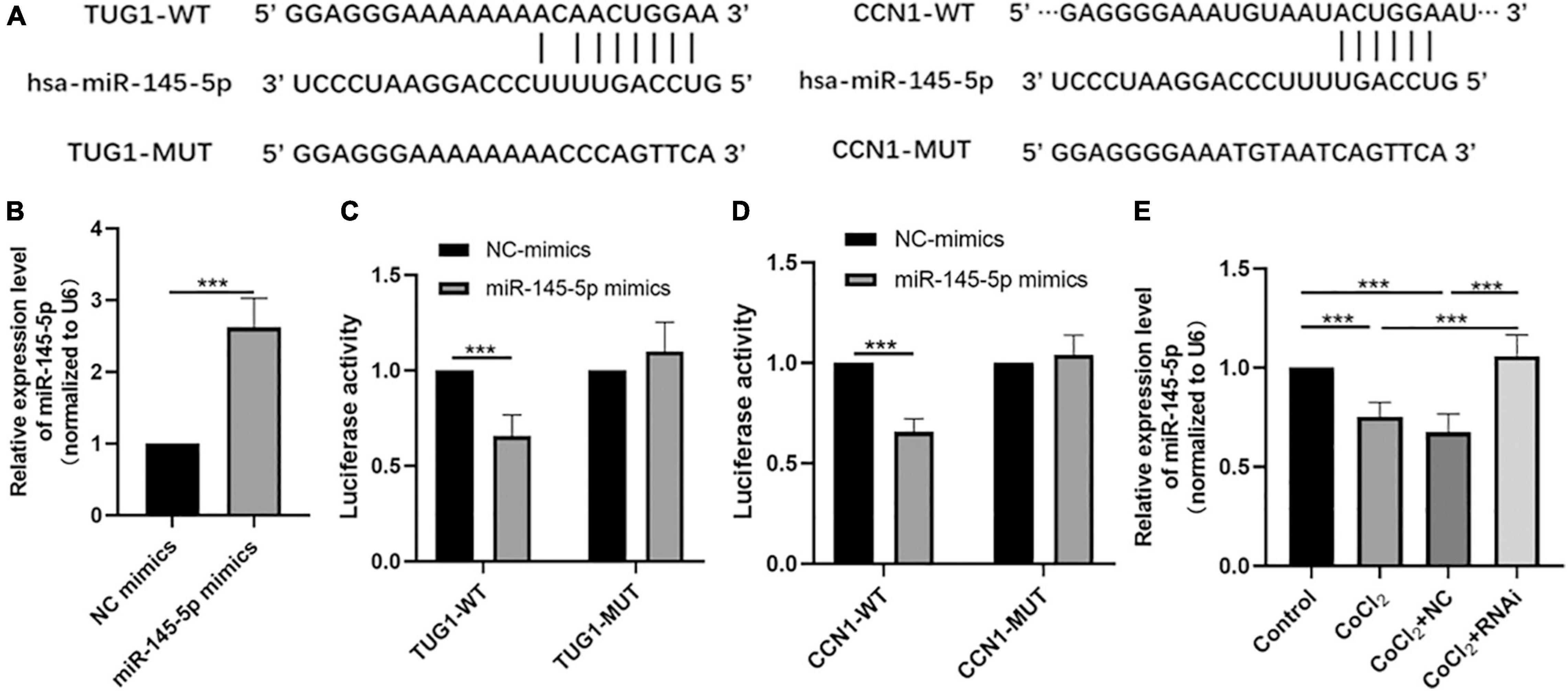
Figure 3. MiR-145-5p interacts with TUG1 and CCN1. (A) The predicted binding sites between miR-145-5p and TUG1, and those between miR-145-5p and CCN1 from bioinformatic analysis using starBase interface. (B) The transfection efficiency and expression of miR-145-5p (mean ± SD, n = 5). ***P < 0.001. (C) Luciferase reporter assays of TUG1 and miR-145-5p (mean ± SD, n = 5). ***P < 0.001. (D) Luciferase reporter assays of miR-145-5p and CCN1 (mean ± SD, n = 5). ***P < 0.001. (E) MiR-145-5p expression level (mean ± SD, n = 5). ***P < 0.001.
Our bioinformatic analyses also predicted the presence of potential binding sites between miR-145-5p and CCN1 (Figure 3A). The results of luciferase reporter assays also indicated a direct binding interaction between CCN1 and miR-145-5p (Figure 3D, ***P < 0.001). We next examined the expression of miR-145-5p in HRECs in the control, CoCl2, CoCl2 + NC, and CoCl2 + RNAi groups using qRT-PCR (Figure 3E). As expected, the expression of miR-145-5p in the CoCl2 group was lower than that in the control group (***P < 0.001), while miR-145-5p expression was higher in the CoCl2 + RNAi compared to that in the CoCl2 + NC group (***P < 0.001). These results suggest that changes in TUG1 expression can produce corresponding changes in miR-145-5p levels.
Overexpressing MiR-145-5p Inhibits Apoptosis, Migration, and Angiogenesis in Human Retinal Endothelial Cells Induced by Hypoxia
We further explored the influence of miR-145-5p on the biological functions of HRECs. The relative expression of miR-145-5p in the control, CoCl2, CoCl2 + NC, and CoCl2 + mimics groups was determined using qRT-PCR analysis (Figure 4A). The results showed that miR-145-5p expression decreased in the CoCl2 group compared to the control group (*P < 0.05) but increased in the CoCl2 + mimics group compared to the CoCl2 + NC group (***P < 0.001). The percentage of apoptotic cells was reduced in the CoCl2 + mimics group compared with the CoCl2 + NC group (Figures 4B,C, ***P < 0.001), suggesting that the overexpression of miR-145-5p inhibits apoptosis. The results from the wound healing and transwell migration assays revealed that the migration rate of the CoCl2 + mimics group was lower than that of the CoCl2 + NC group (Figures 4D–G, wound healing assay ***P < 0.001, transwell migration assay***P < 0.001), supporting the role of overexpression of miR-145-5p in inhibiting cell migration. The results from the tube formation assay revealed a similar trend (Figures 4H,I). Compared with the CoCl2 + NC group, the CoCl2 + mimics group exhibited fewer meshes (***P < 0.001).
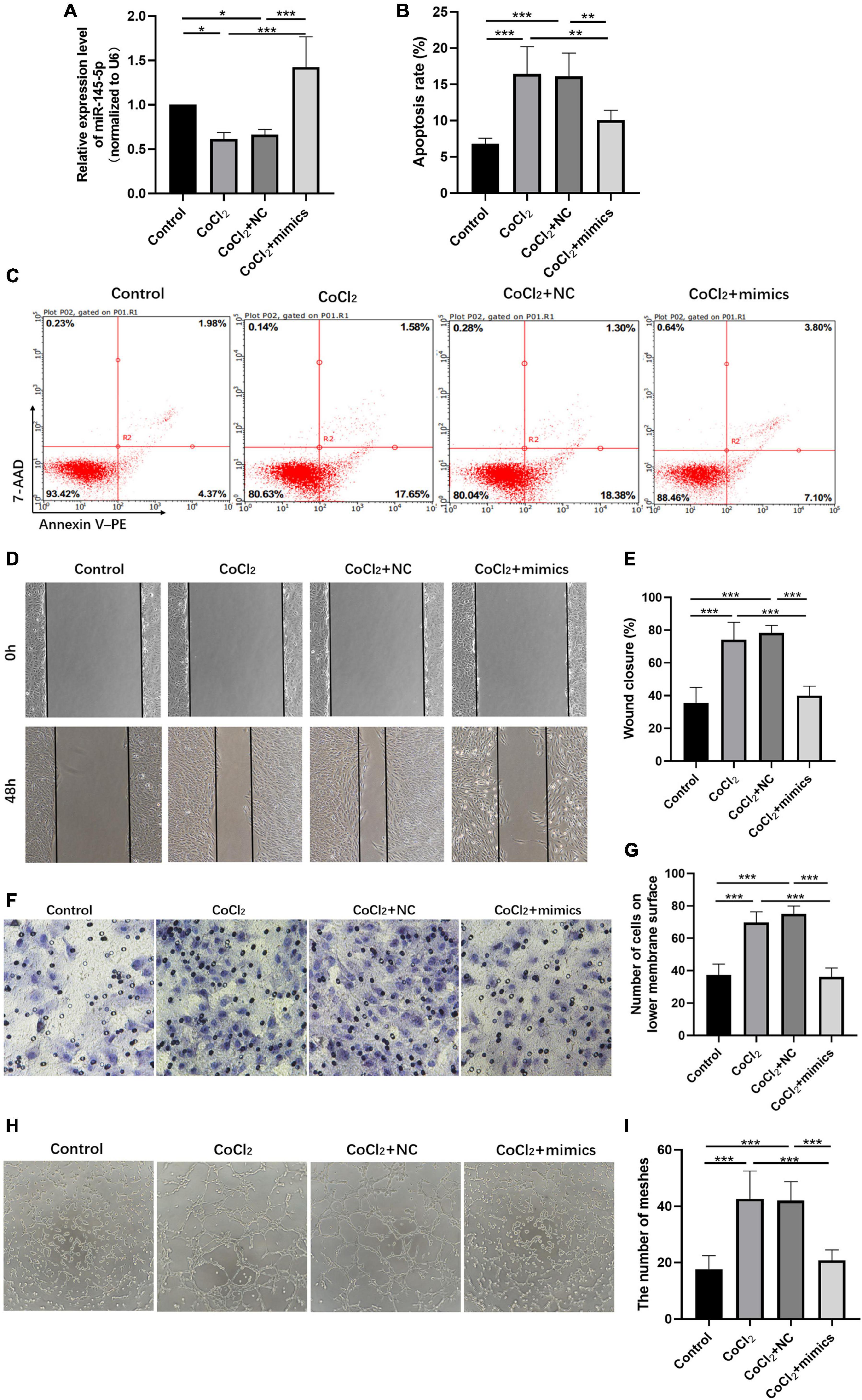
Figure 4. Overexpressing miR-145-5p inhibits apoptosis, migration, and angiogenesis in HRECs induced by hypoxia. (A) MiR-145-5p expression level (mean ± SD, n = 5). *P < 0.05. (B) Flow cytometry results showing the percentage of apoptotic cells (mean ± SD, n = 5). ***P < 0.001. (C) Results of flow cytometry with cells labeled with Annexin-PE and 7-Aminoactinomycin D (7-AAD). (D) Results of the wound healing assay, 100× magnification. (E) Quantitation of results from the wound healing assay in (D) (mean ± SD, n = 5). ***P < 0.001. (F) Results of transwell migration assay, 200× magnification. (G) Quantitation of results from the transwell migration assay in (F) (mean ± SD, n = 5). ***P < 0.001. (H) Results of tube formation assay, 100× magnification. (I) Comparison of the number of meshes in different treatment groups (mean ± SD, n = 5), ***P < 0.001, **P < 0.01.
Knocking Down Taurine Upregulated Gene 1 and Overexpressing MiR-145-5p Inhibit Cellular Communication Network Factor 1 Expression
The qRT-PCR results showed that the expression of CCN1 mRNA in the CoCl2 + RNAi group was significantly lower than that in the CoCl2 + NC group (Figure 5A, **P < 0.01), the Western blot results showed decreased expression of CCN1 in the CoCl2 + RNAi compared to the CoCl2 + NC group (Figures 5B,C, ***P < 0.001). Similarly, overexpression of miR-145-5p effectively reduced CCN1 mRNA and protein levels (Figures 5D–F, qRT-PCR ***P < 0.001, **P < 0.01, Western blot ***P < 0.001).
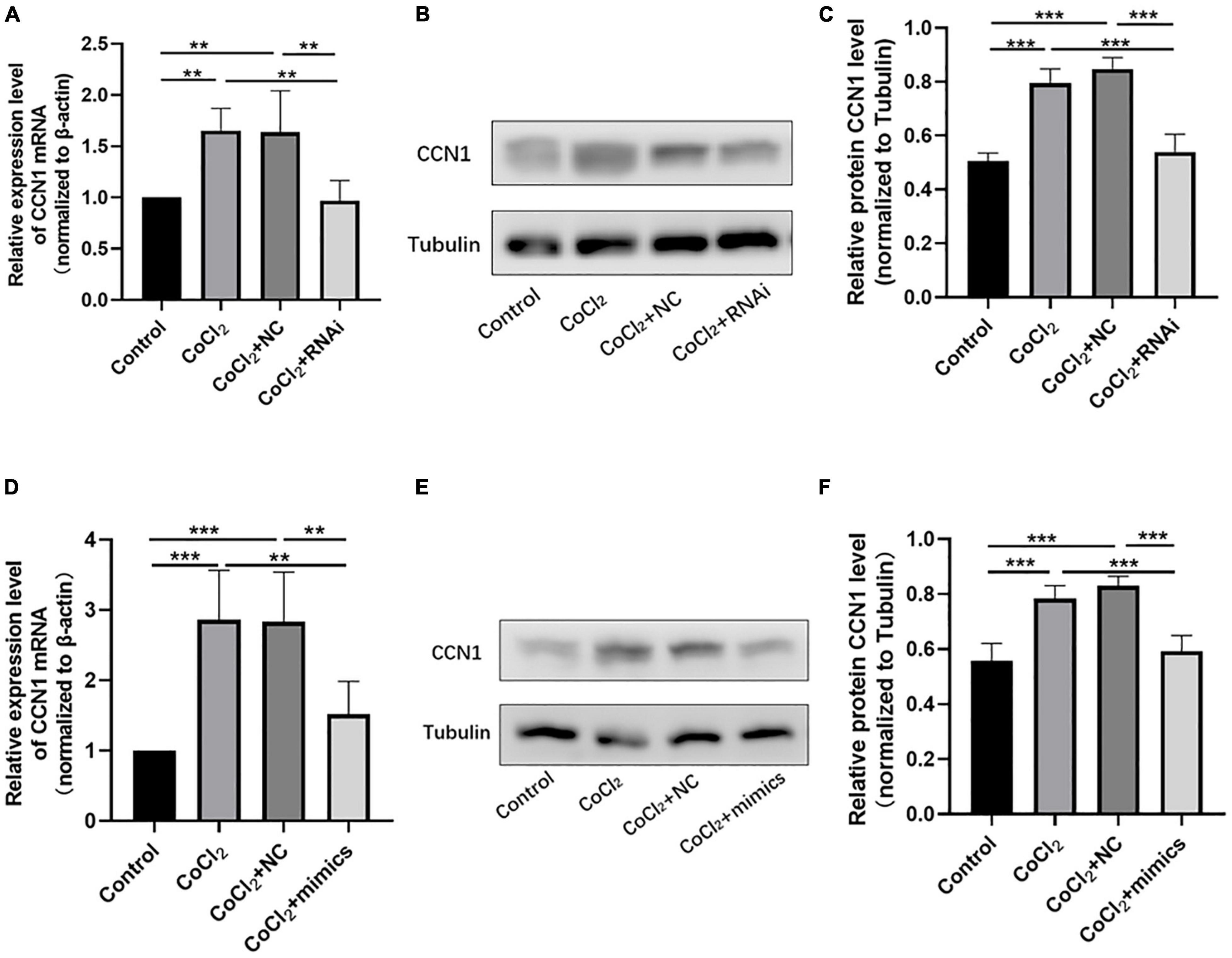
Figure 5. Knocking down TUG1 and overexpressing miR-145-5p inhibits CCN1 expression. (A) CCN1 mRNA expression level (mean ± SD, n = 5). **P < 0.01. (B) Western blot results of CCN1 expression. (C) Quantitative analysis of band intensities from Western blots shown in (B) (mean ± SD, n = 5). ***P < 0.001. (D) CCN1 mRNA expression level (mean ± SD, n = 5). ***P < 0.001, **P < 0.01. (E) Western blot results of CCN1 expression. (F) Quantitative analysis of band intensities from Western blots shown in (E) (mean ± SD, n = 5). ***P < 0.001.
Overexpression of MiR-145-5p Can Reduce the Degree of Oxygen-Induced Retinopathy Retinal Lesions
Retinal images of 17-day-old mice are shown in Figure 6A. Compared with the control group, the OIR group had a larger non-perfusion area and more neovascularization, while the OIR-mimics group injected with miR-145-5p mimics had a significantly lower non-perfusion area and neovascularization than the OIR-NC group (Figures 6A–C, ***P < 0.001).
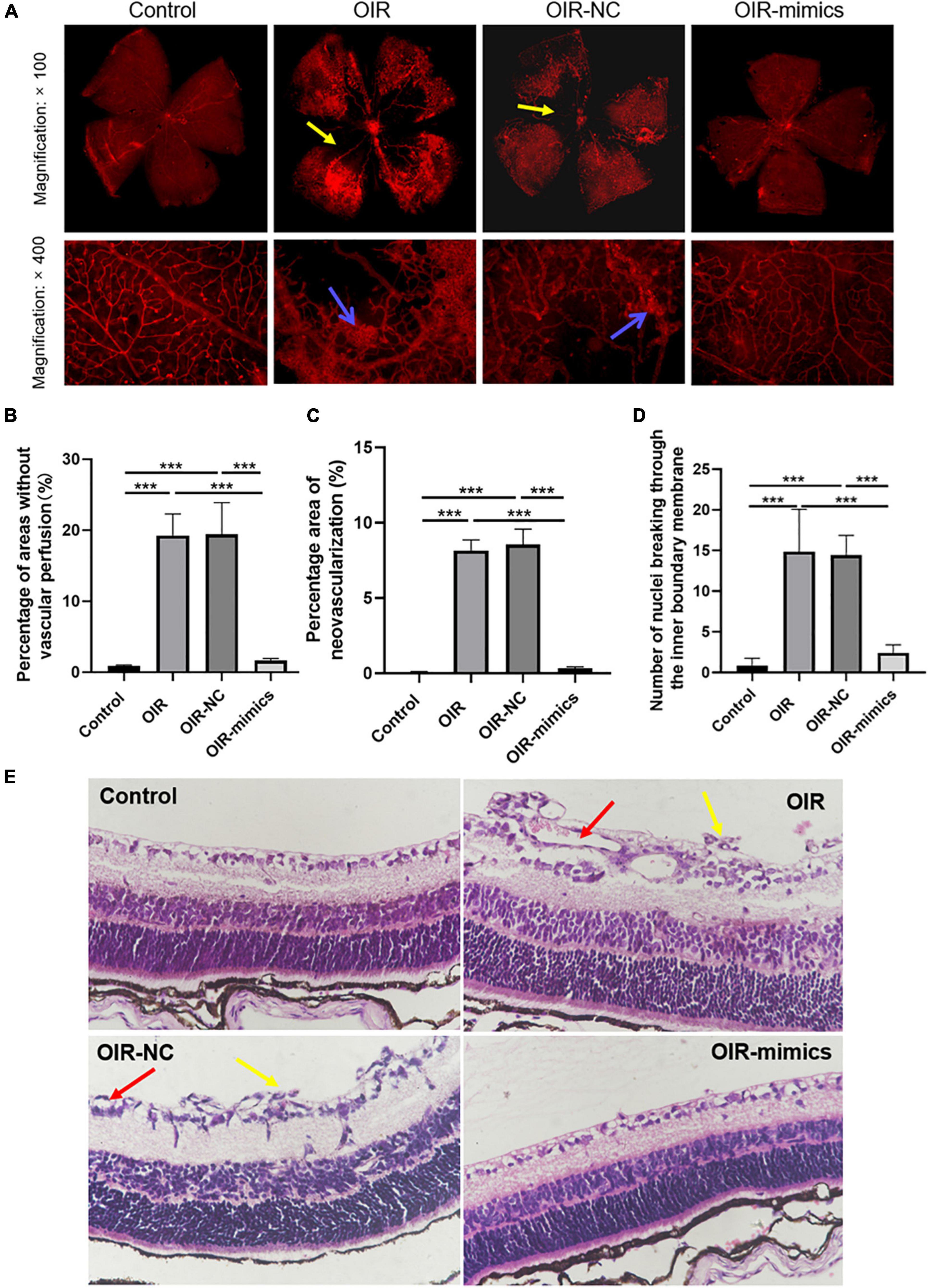
Figure 6. Overexpression of miR-145-5p can reduce the degree of OIR retinal lesions. (A) Retinal vascular morphology of mice in different treatment groups, yellow arrows indicate non-perfusion area, blue arrows indicate neovascularization. (B) Percentage of areas without vascular perfusion (mean ± SD, n = 7). ***P < 0.001. (C) Percentage area of neovascularization (mean ± SD, n = 7), ***P < 0.001. (D) Number of nuclei breaking through the inner limiting membrane (mean ± SD, n = 7), ***P < 0.001. (E) HE staining images in different treatment groups, yellow arrows indicate nuclei breaking through the inner limiting membrane, red arrows indicate neovascularization.
Hematoxylin and eosin staining revealed that the retinal cells in the control group were arranged neatly, while those in the OIR group were disorderly, and the number of endothelial nuclei breaking through the inner limiting membrane into the vitreous lumen was significantly increased (Figures 6D,E, ***P < 0.001). Compared with the OIR-NC group, the endothelial cells in the OIR-mimics group were arranged neatly and the number of endothelial nuclei breaking through the inner boundary membrane decreased significantly (***P < 0.001). These results are consistent with our observations of HREC overexpressing miR-145-5p that exhibited reduced migration and angiogenesis capacity. From outside to inside, the retina is as follows: pigment epithelium, optic rod cone layer, outer limiting membrane, outer nuclear layer, outer reticular layer, inner nuclear layer, inner reticular layer, ganglion cell layer, nerve fiber layer, and inner limiting membrane. Further inside the inner limiting membrane is the vitreous body.
Overexpression of MiR-145-5p Inhibits Inflammation and Apoptosis in Oxygen-Induced Retinopathy Retinal Tissue
The expression of Tug1 and miR-145-5p in retinal tissue was detected by qRT-PCR analysis. The expression of Tug1 in the OIR-mimics group was significantly lower than that in the OIR-NC group (Figure 7A, ***P < 0.001), while the expression of miR-145-5p significantly higher (Figure 7B, ***P < 0.001). The results of Western blot showed that compared with the control group, the expression levels of IL-1β, TNF-α, and caspase-3 increased significantly in the OIR group, whereas Bcl-2 expression level decreased (Figures 7C–F, ***P < 0.001). Compared with the OIR-NC group, IL-1β, TNF-α, and caspase-3 decreased and Bcl-2 increased in the OIR-mimics group (***P < 0.001). Immunohistochemistry indicated the same trend (Figures 7G,H, ***P < 0.001).
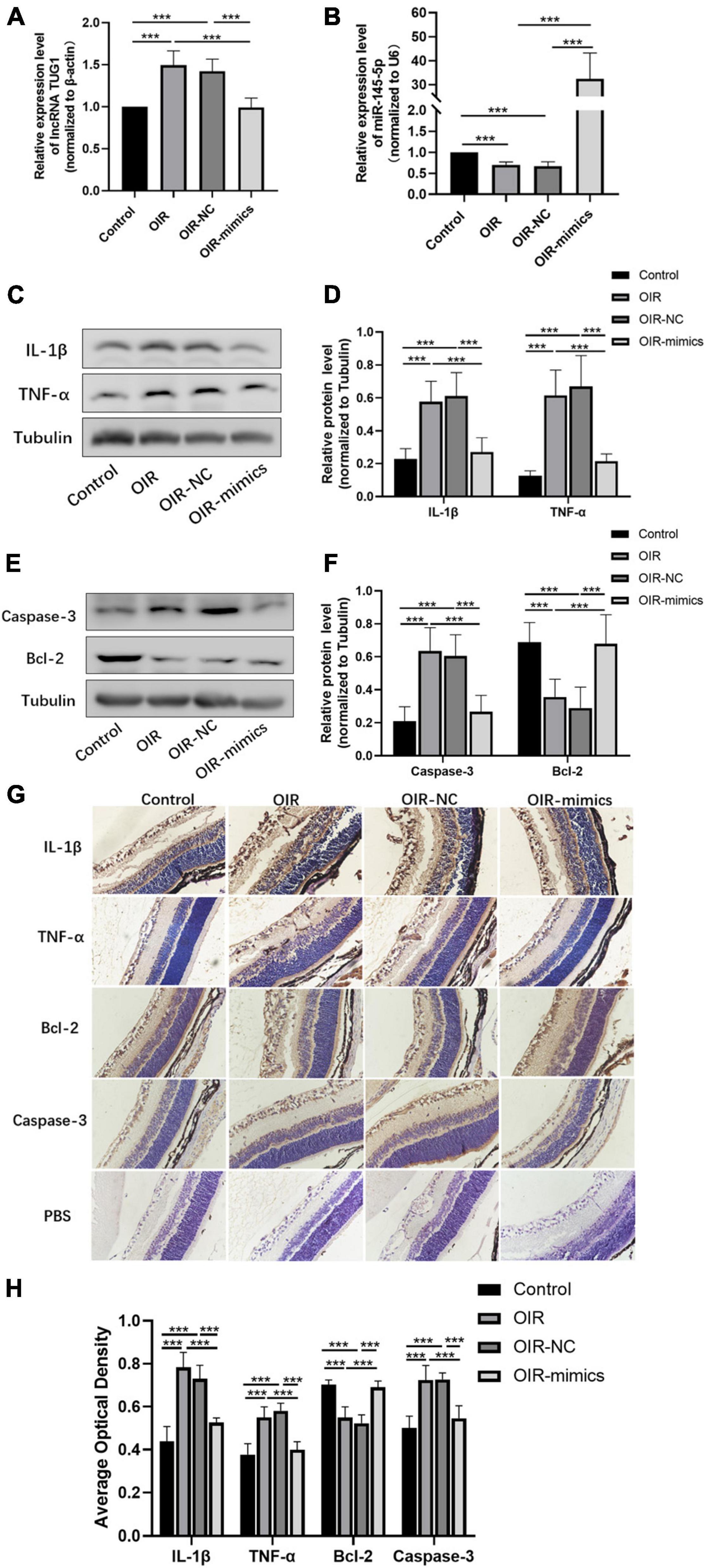
Figure 7. Overexpression of miR-145-5p inhibits inflammation and apoptosis in OIR retinal tissue. (A) Tug1 expression level (mean ± SD, n = 6), ***P < 0.001. (B) MiR-145-5p expression level (mean ± SD, n = 6), ***P < 0.001. (C) Western blot results of IL-1β, and TNF-α expression. (D) Quantitative analysis of band intensities in (C) (mean ± SD, n = 7). ***P < 0.001. (E) Western blot results of caspase-3, and Bcl-2 expression. (F) Quantitative analysis of band intensities in (E) (mean ± SD, n = 7). ***P < 0.001. (G) Immunohistochemical map of IL-1β, TNF-α, Bcl-2, and caspase-3 in the retinal tissues of each group. (H) The average optical density of the immunohistochemical map in different treatment groups (mean ± SD, n = 7). ***P < 0.001.
Over Expression of MiR-145-5p Inhibits the Expression of Cellular Communication Network Factor 1, Hypoxia-Inducible Factor-1 α, and Vascular Endothelial Growth Factor
The qRT-PCR results showed that CCN1 mRNA expression was increased in OIR and miR-145-5p decreased in the OIR-mimics group (Figure 8A, ***P < 0.001). Western blot results showed that the expression of CCN1, HIF-α, and VEGF were significantly increased in the OIR group compared with the control group, while in the OIR-mimics group the same were significantly decreased compared with the OIR-NC group (Figures 8B,C,E,F, ***P < 0.001). Immunohistochemistry also showed the same trend (Figures 8D,G, ***P < 0.001).
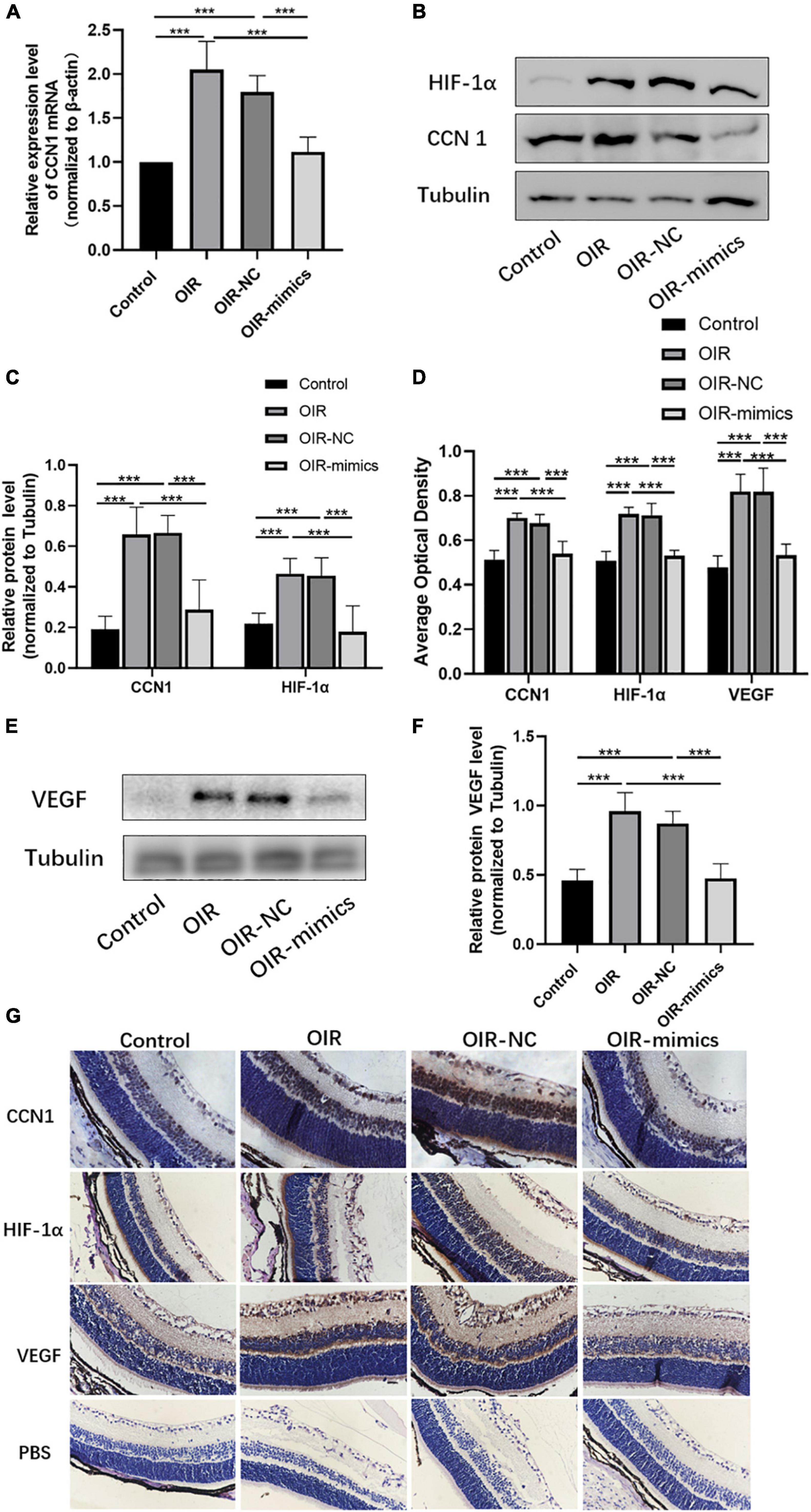
Figure 8. Overexpression of miR-145-5p inhibits the expression of CCN1, HIF-α, and VEGF expression. (A) CCN1 mRNA expression level (mean ± SD, n = 6), ***P < 0.001. (B) Western blot results of CCN1 and HIF-α. (C) Quantitative analysis of band intensities from Western blots shown in (B) (mean ± SD, n = 6). ***P < 0.001. (D) The average optical density of the immunohistochemical map in different treatment groups (mean ± SD, n = 7). ***P < 0.001. (E) Western blot results of VEGF. (F) Quantitative analysis of band intensities from Western blots shown in (E) (mean ± SD, n = 6). ***P < 0.001. (G) Immunohistochemical map of CCN1, HIF-α, and VEGF in the retinal tissues.
Discussion
The increase in the mean age of mothers and global advances in perinatal medicine have resulted in a high number of premature births and survival, leading to an increased rate of incidence of ROP. The development of ROP is generally believed to be related to the relative hypoxia of the retina. When premature neonates are exposed to supplemental oxygen in incubation chambers, returning them to a normoxic environment can cause vasoconstriction and occlusion, which is mainly seen in the developing retinal vessels (23). Exposure to high levels of oxygen may lead to an increase in potentially destructive reactive oxygen species (ROS), and the return to normal oxygen environment reduces antioxidants to neutralize ROS in the retina of preterm infants, leading to endothelial cell damage, apoptosis, inflammation, and capillary constriction (24, 25).
The OIR model is commonly used to study retinal vascular diseases, including ROP because it simulates the relative hypoxia stage of ROP created by hyperoxia followed by normoxia (26, 27). Many previous reports have described miR-145-5p as a target gene of TUG1, and that knockdown of TUG1 can inhibit cell migration and damage, which may have a protective effect on cells via targeting miR-145-5p (28–30). In our study, we found that the expression of the lncRNA Tug1 was upregulated and the expression of miR-145-5p was down-regulated in the OIR mouse model. In our in vitro experiments, we induced hypoxia in HREC with CoCl2 to simulate the relative hypoxia state of OIR, and this is method has been used previously by Du et al. to study OIR (22). Our results also showed that when TUG1 was knocked down or miR-145-5p was overexpressed, apoptosis, migration, and angiogenesis were inhibited, which have a protective effect on cells. Furthermore, the expression of CCN1 mRNA and protein levels were both affected, suggesting that CCN1 is regulated prior to CCN1 mRNA translation. Therefore, we propose that TUG1 acts as a molecular sponge of miR-145-5p to regulate CCN1 expression, the dual-luciferase reporter assays also confirmed the binding relationship among them.
The changes in inflammatory and apoptotic factors are observed in many retinal diseases, including ROP (27, 31). We also found the expression of apoptotic and inflammatory factors increased and anti-apoptotic factors decreased in OIR mouse models. When we overexpressed miR-145-5p in vivo, retinal apoptosis and inflammation were reduced. Furthermore, it is generally believed that hypoxia leads to the increase of hypoxia-inducible factor-1 α (HIF-1α) and VEGF, which leads to the apoptosis and migration of endothelial cells resulting in RNV (32). In our study, HIF-1α and VEGF were also significantly altered in OIR mice, and overexpressing of miR-145-5p in vivo, decreased retinal apoptosis and neovascularization significantly. These results indicate that miR-145-5p overexpression has a protective effect on RNV/OIR. Moreover, we have previously demonstrated that knocking down TUG1 can reduce retinal apoptosis, inflammation, and damage in OIR mice (33). In conclusion, TUG1 acts as a molecular sponge of miR-145-5p to regulate CCN1 expression and TUG1/miR-145-5p/CCN1 together are involved in the occurrence and development of RNV in OIR mice.
The current clinical treatment for ROP is mainly vitreous injection. However, the effects of vitreous injections are short-lived and most patients may require multiple injections to achieve the effect of treatment, which increases the risk of infection. Therefore, it is essential to study alternative target molecules. One of the advantages of gene therapy is that it can shorten the treatment time and work continuously (34, 35). Combined with our previous and current studies, it can be seen that a single knockdown of TUG1 or overexpression of miR-145-5p in vivo can achieve good therapeutic effects. Our findings reveal gene therapy as a potential long-term treatment for ROP and provide some new insights for gene therapy of neovascularization diseases. However, gene regulation is complex, and gene therapy technology as clinical treatment is not very mature, therefore, the safety of injection into the eye and the feasibility of gene therapy as a clinical application needs further research. However, we believe that with more research, gene therapy will solve not only ROP but also many other medical conditions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University. This committee approved the use of animals for the experiments (ethics code: 2018PS239K) on 1 March 2018.
Author Contributions
YxW: conceptualization, methodology, formal analysis, and writing—original draft preparation. YeW: data curation and visualization. XW: investigation and software. YM: resources and validation. ZL: writing—reviewing and editing. YD: funding acquisition and supervision. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (81600747), Liaoning province Department of Education fund (QNZR2020010), and 345 Talent Project of Shengjing Hospital of China Medical University (40B).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to extend our thanks to the R&D Center of Shengjing Hospital Affiliated to China Medical University for research environment, to the tutor YD for guidance with respect to experimental ideas, to the co-authors for the experimental help, language help and manuscript proofreading.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.803214/full#supplementary-material
Footnotes
References
1. Hellstrom A, Smith LE, Dammann O. Retinopathy of prematurity. Lancet. (2013) 382:1445–57. doi: 10.1016/S0140-6736(13)60178-6
2. Dai C, Webster KA, Bhatt A, Tian H, Su G, Li W. Concurrent physiological and pathological angiogenesis in retinopathy of prematurity and emerging therapies. Int J Mol Sci. (2021) 22:4809. doi: 10.3390/ijms22094809
3. Choi AY, Cho H, Kim YC. Effect of two different doses of intravitreal bevacizumab with temporal retina-sparing laser photocoagulation for retinopathy of prematurity. Int J Ophthalmol. (2018) 11:166–9. doi: 10.18240/ijo.2018.01.26
4. Heiduschka P, Plagemann T, Li L, Alex AF, Eter N. Different effects of various anti-angiogenic treatments in an experimental mouse model of retinopathy of prematurity. Clin Exp Ophthalmol. (2019) 47:79–87. doi: 10.1111/ceo.13368
5. Jalali S, Balakrishnan D, Zeynalova Z, Padhi TR, Rani PK. Serious adverse events and visual outcomes of rescue therapy using adjunct bevacizumab to laser and surgery for retinopathy of prematurity. The Indian Twin Cities Retinopathy of Prematurity Screening database Report number 5. Arch Dis Child Fetal Neonatal Ed. (2013) 98:F327–33. doi: 10.1136/archdischild-2012-302365
6. Da M, Zhuang J, Zhou Y, Qi Q, Han S. Role of long noncoding RNA taurine-upregulated gene 1 in cancers. Mol Med. (2021) 27:51. doi: 10.1186/s10020-021-00312-4
7. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. (2005) 15:501–12. doi: 10.1016/j.cub.2005.02.027
8. Li Z, Shen J, Chan MT, Wu WK. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. (2016) 49:471–5. doi: 10.1111/cpr.12269
9. Zhang Z, Xiong R, Li C, Xu M, Guo M. LncRNA TUG1 promotes cisplatin resistance in esophageal squamous cell carcinoma cells by regulating Nrf2. Acta Biochim Biophys Sin (Shanghai). (2019) 51:826–33. doi: 10.1093/abbs/gmz069
10. Wang L, Zhao Z, Feng W, Ye Z, Dai W, Zhang C, et al. Long non-coding RNA TUG1 promotes colorectal cancer metastasis via EMT pathway. Oncotarget. (2016) 7:51713–9. doi: 10.18632/oncotarget.10563
11. Yu G, Li S, Liu P, Shi Y, Liu Y, Yang Z, et al. LncRNA TUG1 functions as a ceRNA for miR-6321 to promote endothelial progenitor cell migration and differentiation. Exp Cell Res. (2020) 388:111839. doi: 10.1016/j.yexcr.2020.111839
12. Zhou H, Gao Z, Wan F. Taurine-upregulated gene 1 contributes to cancers through sponging microRNA. Acta Biochim Biophys Sin (Shanghai). (2019) 51:123–30. doi: 10.1093/abbs/gmy156
13. Pei Y, Li K, Lou X, Wu Y, Dong X, Wang W, et al. miR1299/NOTCH3/TUG1 feedback loop contributes to the malignant proliferation of ovarian cancer. Oncol Rep. (2020) 44:438–48. doi: 10.3892/or.2020.7623
14. Zhan FL, Chen CF, Yao MZ. LncRNA TUG1 facilitates proliferation, invasion and stemness of ovarian cancer cell via miR-186-5p/ZEB1 axis. Cell Biochem Funct. (2020) 38:1069–78. doi: 10.1002/cbf.3544
15. Zeinali T, Mansoori B, Mohammadi A, Baradaran B. Regulatory mechanisms of miR-145 expression and the importance of its function in cancer metastasis. Biomed Pharmacother. (2019) 109:195–207. doi: 10.1016/j.biopha.2018.10.037
16. Mansoori B, Mohammadi A, Shirjang S, Baradaran B. Micro-RNAs: the new potential biomarkers in cancer diagnosis, prognosis and cancer therapy. Cell Mol Biol (Noisy-Le-Grand). (2015) 61:1–10.
17. Fan G, Gu Y, Zhang J, Xin Y, Shao J, Giampieri F, et al. Transthyretin upregulates long Non-coding RNA MEG3 by affectingPABPC1 in diabetic retinopathy. Int J Mol Sci. (2019) 20. doi: 10.3390/ijms20246313
18. Di Y, Zhang Y, Yang H, Wang A, Chen X. The mechanism of CCN1-enhanced retinal neovascularization in oxygen-induced retinopathy through PI3K/Akt-VEGF signaling pathway. Drug Des Devel Ther. (2015) 9:2463–73. doi: 10.2147/DDDT.S79782
19. Choi J, Lin A, Shrier E, Lau LF, Grant MB, Chaqour B. Degradome products of the matricellular protein CCN1 as modulators of pathological angiogenesis in the retina. J Biol Chem. (2013) 288:23075–89. doi: 10.1074/jbc.M113.475418
20. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. (2014) 42:D92–7. doi: 10.1093/nar/gkt1248
21. Smith LE, Wesolowski E, McLellan A, Kostyk SK, D’Amato R, Sullivan R, et al. Oxygen-induced retinopathy in the mouse. Invest Ophthalmol Vis Sci. (1994) 35:101–11.
22. Du W, Yu W, Huang L, Zhao M, Li X. Ephrin-a4 is involved in retinal neovascularization by regulating the VEGF signaling pathway. Invest Ophthalmol Vis Sci. (2012) 53:1990–8. doi: 10.1167/iovs.11-8788
23. Shah PK, Prabhu V, Karandikar SS, Ranjan R, Narendran V, Kalpana N. Retinopathy of prematurity: past, present and future. World J Clin Pediatr. (2016) 5:35–46. doi: 10.5409/wjcp.v5.i1.35
24. Auten RL, Davis JM. Oxygen toxicity and reactive oxygen species: the devil is in the details. Pediatr Res. (2009) 66:121–7. doi: 10.1203/PDR.0b013e3181a9eafb
25. Lu F, Liu Y, Guo Y, Gao Y, Piao Y, Tan S, et al. Metabolomic changes of blood plasma associated with two phases of rat OIR. Exp Eye Res. (2020) 190:107855. doi: 10.1016/j.exer.2019.107855
26. Guo Y, Du F, Tan YL, Luo J, Xiong D, Song WT. VEGF-mediated angiogenesis in retinopathy of prematurity is co-regulated by miR-17-5p and miR-20a-5p. Biochem Cell Biol. (2021) 99:414–23. doi: 10.1139/bcb-2020-0357
27. Yao Y, Cai Y, Sui A, Yao Y, Su T, Zhu Y, et al. Etanercept as a TNF-alpha inhibitor depresses experimental retinal neovascularization. Graefes Arch Clin Exp Ophthalmol. (2021) 259:661–71. doi: 10.1007/s00417-020-04956-6
28. Shi L, Tian C, Sun L, Cao F, Meng Z. The lncRNA TUG1/miR-145-5p/FGF10 regulates proliferation and migration in VSMCs of hypertension. Biochem Biophys Res Commun. (2018) 501:688–95. doi: 10.1016/j.bbrc.2018.05.049
29. Wu Z, Zhao S, Li C, Liu C. LncRNA TUG1 serves an important role in hypoxia-induced myocardial cell injury by regulating the miR1455pBinp3 axis. Mol Med Rep. (2018) 17:2422–30. doi: 10.3892/mmr.2017.8116
30. Shan W, Chen W, Zhao X, Pei A, Chen M, Yu Y, et al. Long noncoding RNA TUG1 contributes to cerebral ischaemia/reperfusion injury by sponging mir-145 to up-regulate AQP4 expression. J Cell Mol Med. (2020) 24:250–9. doi: 10.1111/jcmm.14712
31. Lim RR, Hainsworth DP, Mohan RR, Chaurasia SS. Characterization of a functionally active primary microglial cell culture from the pig retina. Exp Eye Res. (2019) 185:107670. doi: 10.1016/j.exer.2019.05.010
32. Campochiaro PA, Akhlaq A. Sustained suppression of VEGF for treatment of retinal/choroidal vascular diseases. Prog Retin Eye Res. (2021) 83:100921. doi: 10.1016/j.preteyeres.2020.100921
33. Wang Y, Wang X, Wang YX, Ma Y, Di Y. The long-noncoding RNA TUG1 regulates oxygen-induced retinal neo-vascularization in mice via MiR-299. Invest Ophthalmol Vis Sci. (2022) 63:37. doi: 10.1167/iovs.63.1.37
34. Nishida K, Suzuki T, Kakutani K, Yurube T, Maeno K, Kurosaka M, et al. Gene therapy approach for disc degeneration andassociated spinal disorders. Eur Spine J. (2008) 17(Suppl 4):459–66. doi: 10.1007/s00586-008-0751-5
Keywords: long non-coding RNA taurine up-regulated gene 1 (TUG1), miR-145-5p, cellular communication network factor 1 (CCN1), human retinal endothelial cells (HRECs), retinopathy of prematurity (ROP), retinal neovascularization (RNV)
Citation: Wang Y, Wang Y, Wang X, Ma Y, Li Z and Di Y (2022) LncRNA TUG1 Promotes Apoptosis, Invasion, and Angiogenesis of Retinal Endothelial Cells in Retinopathy of Prematurity via MiR-145-5p. Front. Med. 9:803214. doi: 10.3389/fmed.2022.803214
Received: 27 October 2021; Accepted: 11 March 2022;
Published: 04 April 2022.
Edited by:
Leah Owen, The University of Utah, United StatesReviewed by:
Folami Powell, Augusta University, United StatesPeiquan Zhao, Shanghai Jiao Tong University, China
Halesha Dhurvigere Basavarajappa, Beckman Research Institute of City of Hope, United States
Copyright © 2022 Wang, Wang, Wang, Ma, Li and Di. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Di, ZGl5dWpzZHh0QDE2My5jb20=
 Yuexia Wang
Yuexia Wang Yue Wang
Yue Wang Yu Di
Yu Di