- 1Miocrobiology Laboratory, Department of Genetic Engineering and Biotechnology, University of Rajshahi, Rajshahi, Bangladesh
- 2Department of Genetic Engineering and Biotechnology, University of Rajshahi, Rajshahi, Bangladesh
- 3Department of Pathology, College of Korean Medicine, Kyung Hee University, Seoul, South Korea
- 4Ferdows School of Paramedical and Health, Birjand University of Medical Sciences, Birjand, Iran
- 5Drug Exploration and Development Chair (DEDC), Department of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 6Nutrition and Bromatology Group, Department of Analytical Chemistry and Food Science, Faculty of Science, University of Vigo, Ourense, Spain
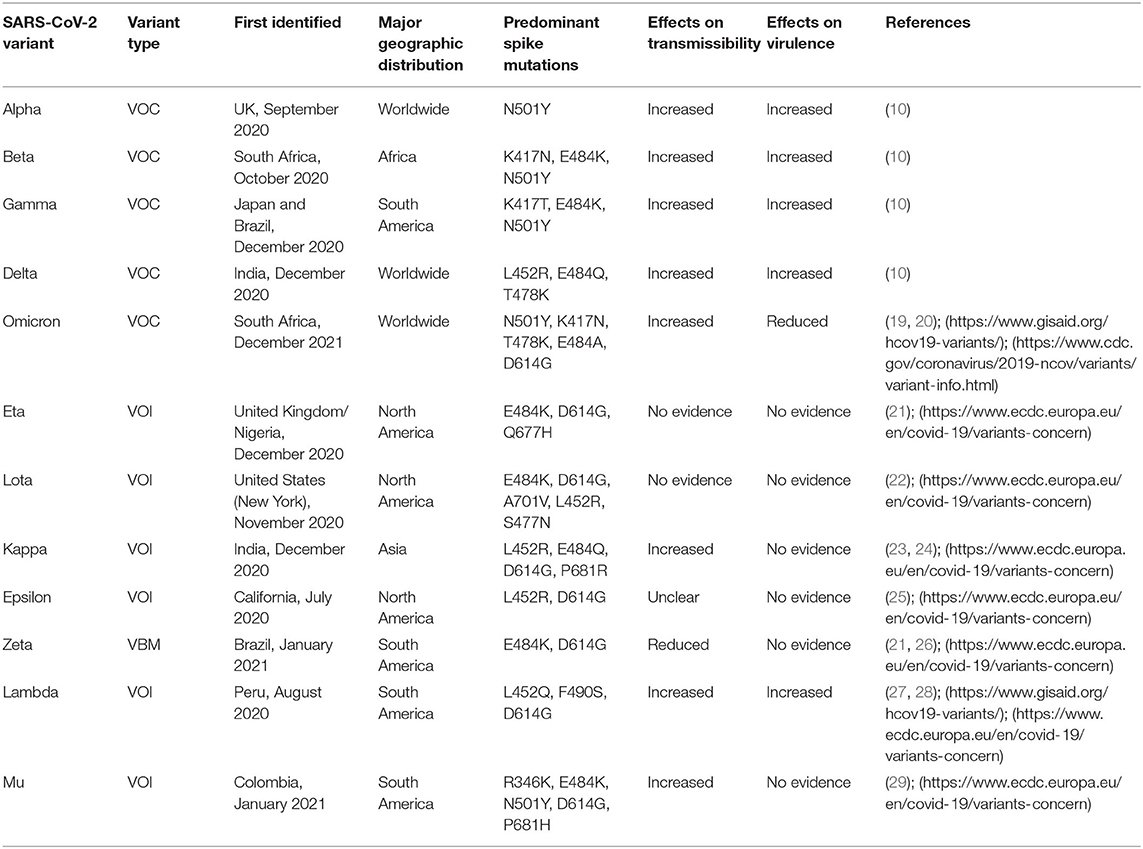
The emergence of several novel SARS-CoV-2 variants regarded as variants of concern (VOCs) has exacerbated pathogenic and immunologic prominences, as well as reduced diagnostic sensitivity due to phenotype modification-capable mutations. Furthermore, latent and more virulent strains that have arisen as a result of unique mutations with increased evolutionary potential represent a threat to vaccine effectiveness in terms of incoming and existing variants. As a result, resisting natural immunity, which leads to higher reinfection rates, and avoiding vaccination-induced immunization, which leads to a lack of vaccine effectiveness, has become a crucial problem for public health around the world. This study attempts to review the genomic variation and pandemic impact of emerging variations of concern based on clinical characteristics management and immunization effectiveness. The goal of this study is to gain a better understanding of the link between genome level polymorphism, clinical symptom manifestation, and current vaccination in the instance of VOCs.
Highlights
- SARS-CoV-2 has evolved many variants as a result of genome-level mutations, worsening the current pandemic situation.
- SARS-CoV-2 variants increase transmissibility, viral virulence, and reduce diagnostic sensitivity.
- The vaccine's efficacy has been brought into question due to the emergence of variants containing a new mutation.
- Vaccine effectiveness and clinical management vary among variants.
- Natural immunity hedging and vaccine-induced immunization evasion have become major public health concerns.
SARS-CoV-2 GENOME AND MUTATIONS
The largest (amid 26 kb and 32 kb) single-stranded positive-sense RNA genome of SARS-CoV-2 shows low genome stability, with about 1,516 nucleotide-level variations in genome-wide annotations and over 9.8 × 10−4 substitutions/site yearly (1–4). The genome or viral transcript of SARS-CoV-2 contains two open reading frames (ORFs) expressing non-structural proteins (NSPs) and four genes encoding structural proteins, namely N (nucleocapsid), M (membrane), E (envelop), and S (spike). ORF1a encodes 11 non-structural proteins (NSP1–11), whereas ORF1b encodes five non-structural proteins (NSP12–16), and ORF8, ORF7b, ORF7a, ORF6, and ORF3a genes encode six accessory proteins, the non-structural proteins being primarily functional proteins (enzymes) that act as a prerequisite for viral replication in tandem with methylation to provoke host responses during infection (5–11).
The viral transcript is notable for a large number of recurrent mutations (>15 occurrences) in the Orf1ab region, namely in three sites (Nsp6, Nsp11, and Nsp13 encoding sites) and one in the spike (S) protein (4, 5, 12, 13). In comparison to the original viral genome, a variant is a virus strain with a considerable phenotypic alteration that exhibits unusual characteristics in terms of virulence, transmissibility, and antigenicity (10, 14). It results from either a complicated combinatorial aberration or an abnormal mutation caused by the combination of three factors, including viral replication errors, recombination between two different viral lineages during coinfection, and the stimulation of host RNA-editing mechanisms. Furthermore, each genetic mutation is incapable of causing significant changes in the essential protein for viral replication and infectivity modification (3, 4, 15–18). The Global Initiative for Sharing Avian Influenza Data (GISAID) managed the global SARS-CoV-2 sequence database by submitting over 1.4 million sequences by May 2021, with a total of 3,913 major representative variants genomes being identified. Additionally, variants of concern (VOCs), which include the Alpha (B.1.1.7 and Q lineages) variant, Beta (B.1.351+B.1.351.2+B.1.351.3) variant, Gamma (P.1 and descendent lineages) variant, Delta (B.1.617.1, B.1.617.2 and AY lineages) variant, the and Omicron (B.1.1529 and BA lineages) variant, predominately emerge from the mutation of the spike gene, where ORF1a region of the genome operates as a pre-eminent NSP mutations site (https://www.gisaid.org/hcov19-variants/). There are two VOIs abbreviated as variants of interest: Lambda (C.37+ C.37.1) and Mu (B.1.621+B.1.621.1), as well as one variant under monitor or VUM, which includes an unidentified (B.1.640 and descendent lineages) variant. However, according to CDC, only omicron and delta variants are considered as the VOCs whereas Alpha, Beta, Gamma, and Mu in conjunction with Eta (B.1.525) variant, Lota (B.1.526) variant, Kappa (B.1.617.1) variant, Epsilon (B.1.427 and B.1.429) variant, Zeta (P.2), and an unknown (B.1.617.3) variant are listed as variants being monitored or VBM (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html). Of these, kappa is the most recent significant variant evolved from the second COVID-19 wave. (https://www.axios.com/variants-tracker). Our primary emphasis will be the VOCs, VOIs, VUM, or VBMs that play significant roles in SARS-CoV-2-related public health issues (Table 1).
Spike Mutations
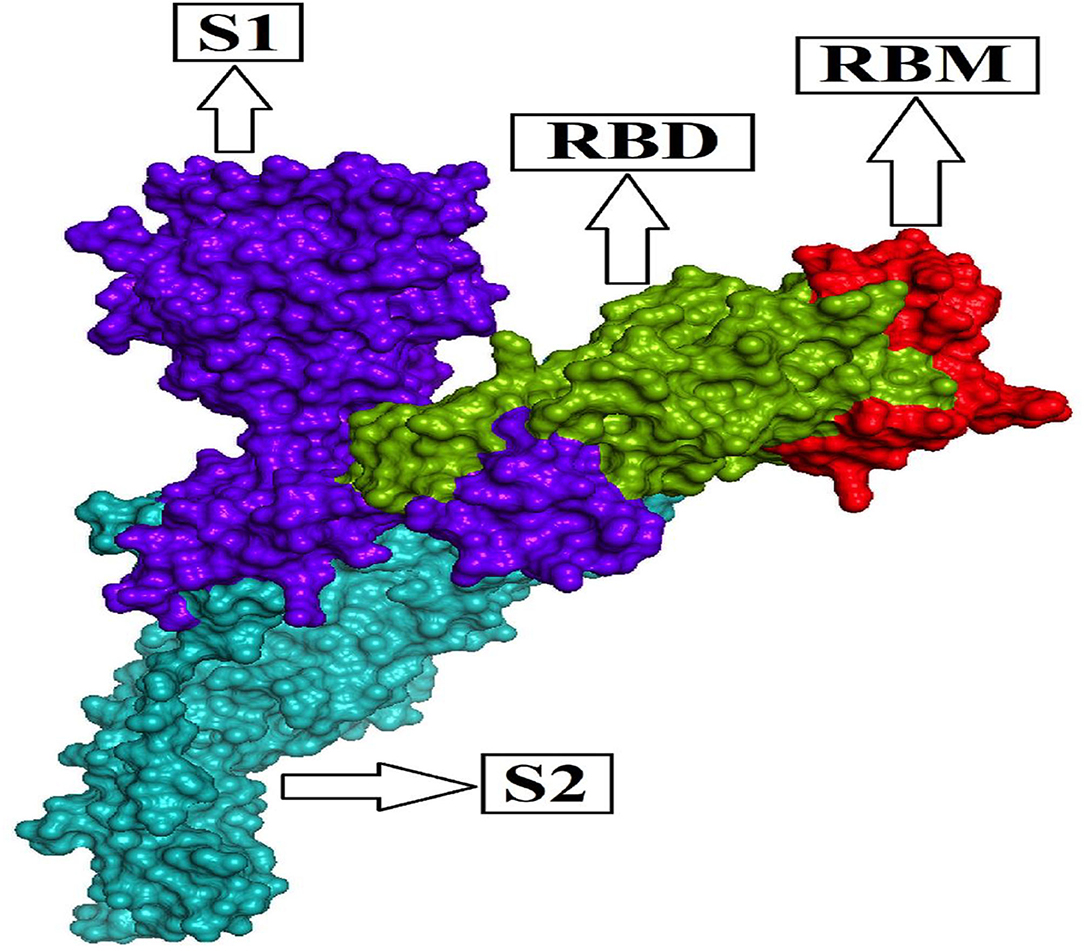
The homo-trimeric spike protein (S) of SARS-CoV-2 is similar to an obligatory protein that conducts viral entry for virus attachment during infection by recognizing receptors [angiotensin-converting enzyme (ACE2)] in conjunction with cell membrane fusion form. The spike protein is divided into two subunits: S1 subunit comprises the receptor-binding domain (RBD), which can bind to the PD (peptidase domain) of ACE2, and S2 subunit conducts cell membrane fusion via the explicit two-heptad repeat region using the six-helical bundle generation (9, 30–33). The RBD's most important operative motif, known as the receptor-binding motif (RBM; Figure 1) evolves the interface between hACE2 and the S protein while maintaining RBD structural stability. As a result, the S1 subunit, which is considered a mutation hotspot with significant clinical relevance, including host immune evasion, transmissibility, and virulence, provides a common key for antibody (Ab) neutralization, as well as future cross-reactive antibody recognition (34–37). The Alpha variant identified in the UK in September 2020 has numerous spike glycoprotein alterations, including K1191N, D1118H, S982A, T716I, P681H, D614G, A570D, N501Y, S494P, E484K, 144del, 70del, 69del, T478I, F490S, E484Q, T478K, T478A, S477N, L455F, Y449S, Y449H, and K417T (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html); (https://www.gisaid.org/hcov19-mutation-dashboard/).
The N501Y mutation of the alpha variant denotes the substitution of asparagine (N) for tyrosine (Y) at amino acid residue 501; similarly, K417N mutations denote the substitution of lysine (K) for asparagine (N) at amino acid residue 417. However, an Alpha (B.1.1.7) descending evolving variation occupies the E484K mutation, which results in the glutamic acid E being replaced by lysine K at the 484 residues. The Beta variation has the E484K mutation, but the Gamma variant has the K417T mutation in tandem with the E484K mutation, indicating that Beta has numerous substitutions in combination with N501Y, as discovered in South Africa in October 2020 and Brazil/Japan in December 2020 (https://www.gisaid.org/hcov19-variants/). Spike protein substitutions such as A701V, D614G, N501Y, E484K, K417N, 243del, 242del, 241del, D215G, and D80A are provided by the Beta variants, whereas the Gamma variants provide T1027I, H655Y, D614G, N501Y, E484K, K417T, R190S, D138Y, P26S, T20N, and L18F spike protein substitutions (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html). Delta and Kappa variants, which were first identified in India in December 2020 and are notable as recent influential variants with constant mutations, include E484Q, which refers to the substitution of E (glutamic acid) by Q (glutamine) at the 484 residues, and L452R, which refers to the substitution of L (leucine) by R (arginine) at the 452 residues (https://www.gisaid.org/hcov19-variants/). Additionally, The Delta variant occupies diverse spike glycoprotein substitutions including D950N, P681R, D614G, T478K, L452R, K417N, W258L, A222V, R158G, G142D, T95I, V70F, T19R, G504D, V503F, N501Y, N501T, P499L, S494P, S494L, Q493E, Q493L, F490W, F490L, Y489L, N487T, F486Y, E484Q, E484K, S477C, S477N, S477I, A475T, K458N, L455F, G446V, V445I, and K417T (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html); (https://www.gisaid.org/hcov19-mutation-dashboard/). The T478K mutation, which refers to the substitution of T (threonine) for K (lysine) at amino acid position 478, is a strange Delta variant mutation.
In addition, numerous spike protein substitutions are notable concerning the Eta (F888L, Q677H, D614G, E484K, 144del, 70del, 69del, and A67V), Lota (Q957R, D950H, T859N, A701V, D614G, E484K, S477N, L452R, D253G, F157S, T95I, D80G, and L5F), and Kappa (Q1071H, T95I, P681R, D614G, E484Q, L452R, E154K, and G142D) variants according to the CDC (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html). T76I, L452Q, G75V, F490S, D614G, and T859N substitution for the lambda (C.37) variant (37) and S13I, W152C, D614G, and L452R substitution for the epsilon (B.1.427/B.1.429) variant were recognized in the spike gene titled S gene (https://covdb.stanford.edu/page/mutation-viewer/#sec_epsilon). The recent variant named “Omicron” first identified in South Africa has several spike protein substitutions such as del142-144, Y145D, del211, A67V, del69-70, T95I, L212I, ins214EPE, G339D, K417N, N440K, G446S, S371L, S373P, S375F, S477N, T478K, E484A, Q493R, Y505H, T547K, D614G, G496S, Q498R, N501Y, H655Y, N679K, P681H, Q954H, N969K, L981F, N764K, D796Y, and N856K. The omicron mutation N501Y is identical to the mutations mentioned in alpha, beta, gamma, and delta variants. K417N mimics the substitution of alpha and beta variants that differ from gamma and delta variants, and it has the T478K strange delta variant substitution in tandem with E484A substitution that is not observed in any of the above-mentioned-variants, and notable D614G substitution is also present. (https://www.gisaid.org/hcov19-variants/);(https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html). However, on 26 November 2021, the World Health Organization (WHO) designated this “Omicron (B.1.1.529)” as a Variant of Concern due to its high transmissibility and danger of immunological deficiency (38). Table 1 shows the most common spike mutations for SARS-CoV-2 variants.
However, the alterations in the S1 subunit result in a significant increase in S RBD binding affinity for the ACE2 receptor, as well as a decreased affinity for antibody (Ab) neutralization. For example, the B.1 lineage of Beta variants with the D614G mutation in the spike protein shows a 4.3-fold antibody reduction and a 3.5-fold antibody neutralization rebate on average. However, a new Beta variation (501Y.V2), capable of reinfection in COVID-19 convalescent patients, resides in the E484K spike protein mutation, demonstrating the ability to evade first-wave anti-SARS-CoV-2 neutralizing antibodies.
Furthermore, the presence of E484K or N501Y mutations in the S1 subunits causes greater virulence and transmissibility, as well as a higher fatality rate and morbidity (39–42). The indicated D614G mutation at non-RBD sites alters the spike protein structure, leading to monoclonal antibody neutralization and increased SARS-CoV-2 replication via virion infectivity enhancement is notable as a prominent spreading mutation extant in more than 99% prevalent variants. Despite not being capable of boosting binding affinity for ACE2 or neutralization sensitivity, the D614G improves infectivity by increasing the amount of functional S protein, as well as improved spike density due to S1 shedding escape and spike integrity shielding. As a result, the presence of the D614G mutation is associated with more agile transmission in vivo and increased replication during an in vitro investigation (43–47). The L452R mutation, which was discovered in the spike RBM of SARS-CoV-2, allows the virus to escape HLA-A24-restricted cellular immunity, resulting in increased fusogenicity and viral infectivity, as well as increased viral replication (48). Recurrent deletion areas (RDRs) encompassing (90%) four separate regions in the NTD show an Ab-recognizing neutralization domain with increasing deletions remaining in the S1 subunit (N-terminus). Deletions occurring in RDRs is notable in maximum in Alpha-originated variants (e.g., S: ΔHV 69–70, S: ΔY144 in ΔRDR1, and ΔRDR2 respectively), in tandem with Beta-stemmed variants (e.g., S: ΔLAL 242–244, ΔRDR4) and B.1.36 (e.g., S: ΔI210, ΔRDR3), which ends in the resistance for antibody neutralization, wiped epitopes, support in host's immune evasion together with vaccines, or Abs neutralizing declination (10, 49). Furthermore, the top 10 RBD region mutations comprise S494P, T478K, E484K, N501Y, K417N, L452R, K417T, N439K, F490S, and S477N substitutions, observed in 2385, 83587, 19505, 96499, 1129, 83717, 8646, 1930, 499, and 6102 SARs-CoV-2 isolated sequence among 298,376 sample sequences, respectively. Notably, the most prevailing mutations N501Y were detected majorly in the Unites States during April 2021, while L452R and T478K were identified in the United Kingdom during June 2021 (https://www.cbrc.kaust.edu.sa/covmt/index.php?p=top-rbd-variants-heatmap).
Among the rapidly disseminating arising variants that include alpha, beta, gamma, delta, kappa, eta, lota, epsilon, lambda, mu, and omicron variants, the most well-known D614G mutation (44, 46, 50) provides a reasonable benefit in terms of infectivity (47, 51, 52) and improves transmissibility (53), implying a higher fatality and infectivity rate (54–56). Similarly, the N501Y alteration observed in alpha, beta, gamma, delta, and omicron imparts better ACE2 binding, demonstrating (57–59) the massive increase in ACE2 affinity with a single RBD mutation (57). Furthermore, the E484K mutation in alpha, beta, gamma, delta, kappa, eta, zeta, and lota stimulates escape from multiple mAbs [monoclonal antibodies; (60–62)], as well as antibodies against convalescent plasma (61–63). Again, the K417N/T mutations in the RBD show immune evasion from antibodies and vaccines produced by natural infection (50, 64, 65). Furthermore, both the omicron, delta, and beta variants of K417N and the delta, gamma, and alpha variants of K417T are anticipated to have a lower ACE2-binding affinity (57). Furthermore, the L18F mutation of gamma is responsible for the escape of certain NTD-binding mAbs, resulting in reduced antibody neutralization (66). Similarly, the appearance of the S477N mutation in alpha, delta, lota, and omicron is responsible for resistance to RBD-targeting mAbs-derived neutralization, as well as improved affinity to a lesser extent for the ACE2 receptor. Among the top 10 RBD region mutations, the N439K change improves affinity, but to a lower level in the case of the ACE2 receptor (57, 66). However, an antigenic consequence of the Y144 mutation, which is found in the alpha, eta, and omicron variants, has been observed to prevent neutralization by a number of neutralizing antibodies (66). Furthermore, delta variants with p. 475 (Ala to Val), delta, lota, kappa, and epsilon variants with p. 452 (Leu to Arg), and delta variants with p. 490 (Phe to Leu) increase resistance to a variety of neutralizing antibodies (55). Figures 2–4 show the prominent residues of spike glycoprotein where various mutations occur, resulting in the evolution of different SARS-CoV-2 variants.
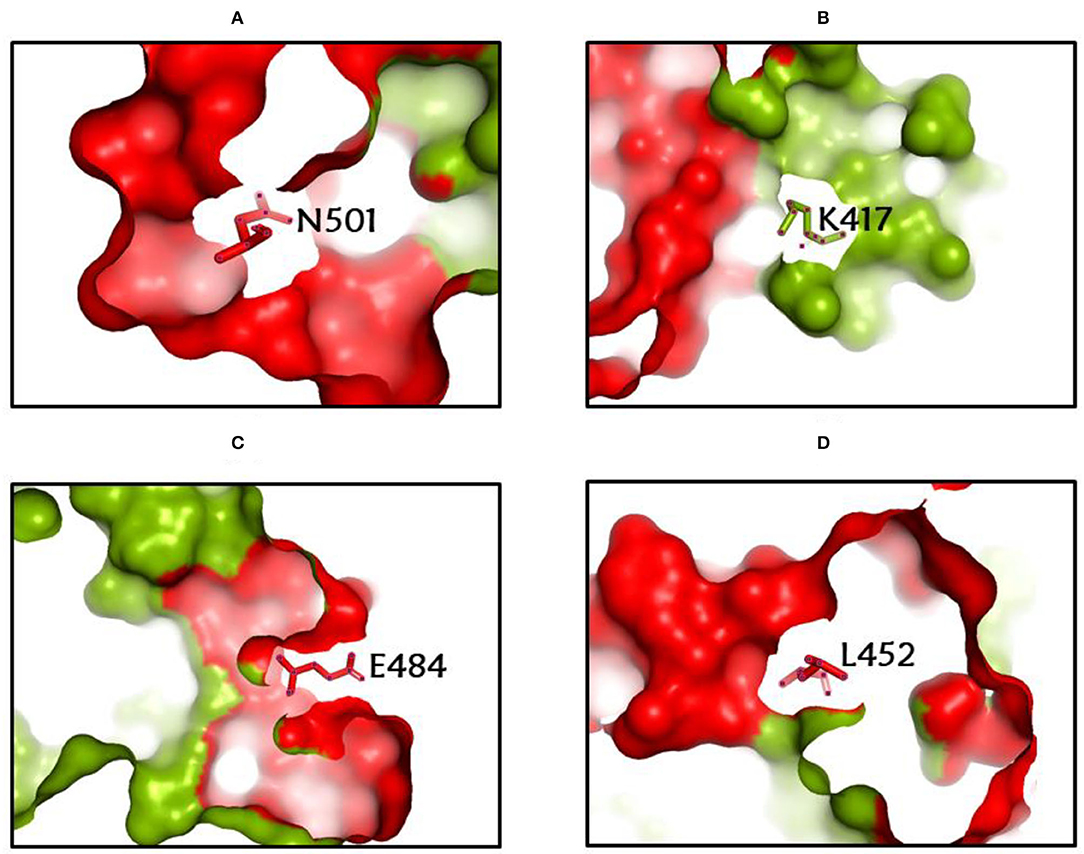
Figure 2. (A-D) The most common spike glycoprotein residues (N501, K417, E484, L452), where diverse mutations occur, resulting in different SARS-CoV-2 VOCs, VOIs, and VBMs.
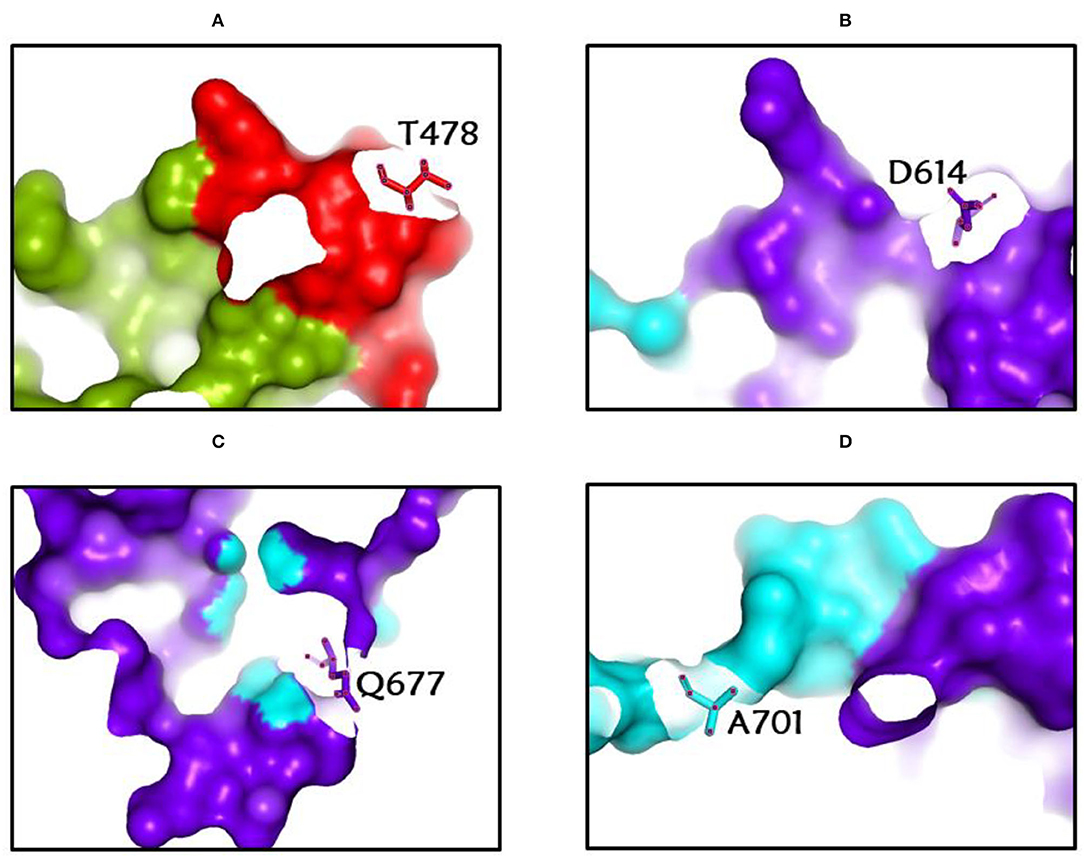
Figure 3. (A-D) The most common spike glycoprotein residues (T478, D614, Q677, A701) where diverse mutations occur, resulting in different SARS-CoV-2 VOCs, VOIs, and VBMs.
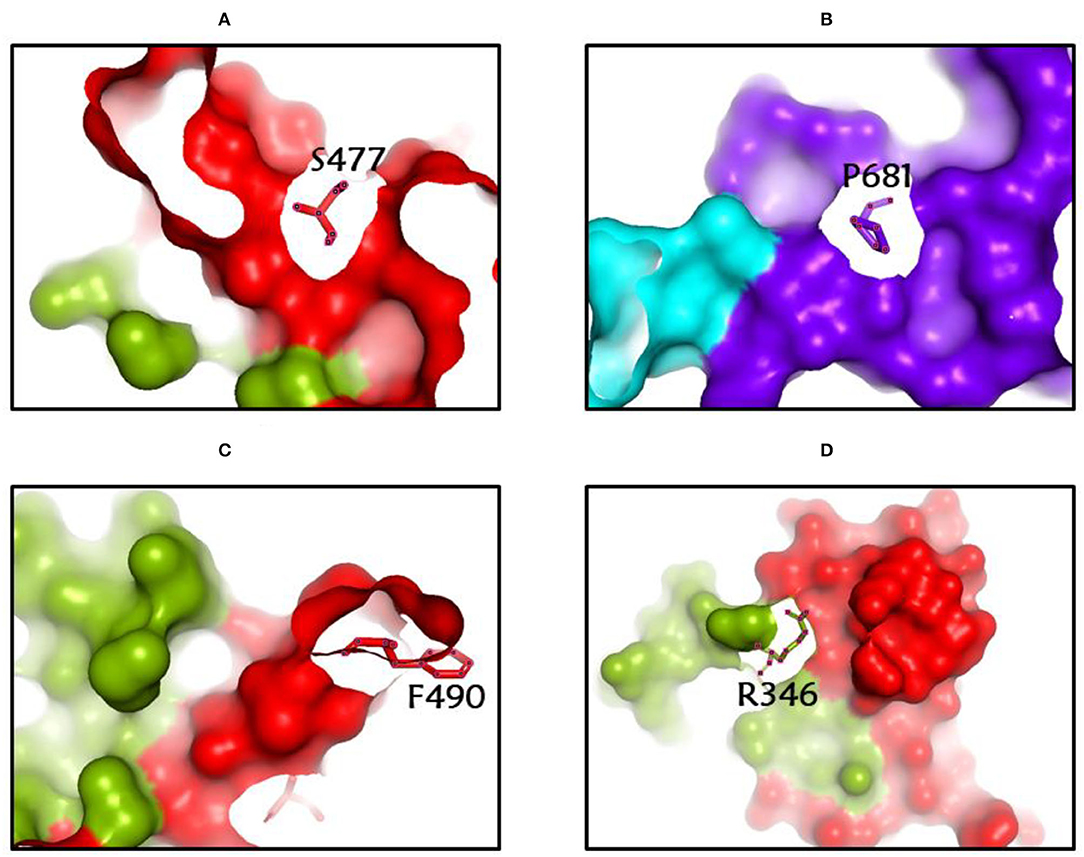
Figure 4. (A-D) The most common spike glycoprotein residues (S477, P681, F490, R346) where diverse mutations occur, resulting in different SARS-CoV-2 VOCs, VOIs, and VBMs.
NSP Mutations
ORF8 downregulates host cell major histocompatibility complex class I (MHC-I) in addition to NSP1 positioned in ORF1a/ORF1ab antagonizing activation of type I interferon in host cells, which is associated with SARS-CoV-2 transmissibility and virulence. It is established that the potent suppression of IFN-I signaling causes antiviral activity by combating viral reproduction via nsp 1 and nsp 6 in concert with antagonization of the IFN-I response via nsp 6, nsp 13, and (ORF6), resulting in the avoidance of the host immune response. The D500-532 mutation in Nsp1 reduces IFN-I response in SARS-CoV-2-infected host cells, as well as during transcription and protein translation in transfected cell lines [HEK293T and A549; (63, 67)]. Additionally, an ill-timed stop codon identified in the Alpha version at position 27 is found in the immune-evasive ORF8 protein, which conducts evasion functions and unique immunological suppression.
However, the most recent major African variants, ORF8: 382 variant and NSP1: 500–532 variant, both of which affect around 5% of global infections and were found in Singapore and China, respectively, contain both ORF8 and NSP1 partial deletions, resulting in lower SARs infection with the CoV-2 virus [https://www.gisaid.org/hcov19-variants/; (68–70)]. Furthermore, 382 SARS-CoV-2 moderate infection variants shorten ORF7b and abolish ORF8 transcription, reducing severe COVID-19-associated proinflammatory cytokines, chemokines, and growth factors (70–72). Important mutations in nsp 2 (T265I), nsp 12 (P4715L), and nsp 13 (P5828L and Y5865C), which serve as helicase or replicase, have also been identified in the United States (73). However, V121D destabilizing NSP-1, G1691C reducing NSP-3 flexibility in combination with V843F, A889V substitution, and V843F dominating substitution in combination with A889V in PLPro has indicated the possibility of an attenuated vaccine in combination with PLPro inhibitors (74, 75).
PROBABLE CLINICAL IMPACTS OF SARS-CoV-2 VARIANTS
Increased Transmissibility and Viral Virulence
The Delta variation was linked to a high viral load, high transmission rates, and reinfection (PANGO lineage: B.1.617.2). Unlike the Alpha variant and monoclonal antibodies used in SARS-COV-2 medicines, this version increases rather than decreases susceptibility to the virus (76, 77). In comparison to the Alpha VOC, the Delta (B.1.617.2) VOC was predominantly observed in the younger age group, putting patients at risk of a second hospitalization if they had more than five comorbidities. In vitro neutralization experiments using convalescent serum and monoclonal antibodies, and a subject testing of immunized serum show that the Delta variation increases vaccination resistance, especially in individuals with one dosage (76–78).
The Delta SARS-CoV-2 mutant was shown to be 60% more infectious than the wild type and able to evade adaptive immunity in half the time. The S-protein mutation D614G of the Delta variant has been shown to affect virulence and virus transmissibility by preserving a stronger affinity for olfactory epithelium and enhanced virion stability (10, 79). On the other hand, because of their increasing transmissibility and massive mutations in the spike gene, both the alpha variant B.1.1.7 and the beta variant B.1.351 are gaining popularity. It is turbulent for monoclonal antibodies to neutralize the N-terminal region of the spike protein in the B.1.1.7 variant. Evidence suggests that the transmission rate of Alpha and Beta variants of concerns is growing by about 50% in children and younger people (10, 65, 80).
When comparing clinical outcome records, it appears that B.1.1.7 infection has a 30% higher fatality rate than other SARS-CoV-2 variants, which could be attributable to alterations in the receptor-binding domain that make them immune to neutralizing antibodies (10, 81, 82). Furthermore, the B.1.351 variant improves the neutralization of many monoclonal antibodies against the RBD at receptor-binding sites, resulting in an E484K substitution mutation and a 9.4-fold increase in resistance through plasma convalescence (65, 83). Several studies show that the B.1.1.7 variation is around 35–45% more transmissible across the country and gains frequency at a double pace every one and a half weeks (82).
The Epsilon variant, CAL.20C (B.1.427/B.1.429), is characterized by three mutations: L452R mutation in the RBD, and W152C and S13I mutations in the N-terminal domain (NTD). In conjunction with higher viral shedding, these variants increase transmissibility by up to 24% (10, 84, 85). In response to the currently circulating strains, various genome sequences of B.1.427/B.1.429 variants arrayed a rapid increase in viral prevalence, with 50% exceeding the transmissibility rate (85, 86). The Gamma variant P.1 (originating in Brazil) indicated a 20.0% increase in hospitalizations compared with non-VOC patients, and spike mutations may have increased virulence, raising ACE2 rapport, although significant information about viral pathogenicity is currently unavailable in these genotypes (10, 83, 87).
Decreased Diagnostic Sensitivity
Several studies (13, 88, 89) have described a transitory genetic evolution as a result of the geographical viral transmission of SARS-CoV-2 since the genomic sequence of SARS-CoV-2 became easily accessible. Newly arising variants of concerns can impair the sensitivity of RT-PCR-based identification if a mutation arises in a region where both primers and probes may bind. In the RT-PCR experiment, 79% of the primer binding sites are utilized, but the genome has altered in the meanwhile, with the most important GGG AAC substitution (10, 23). During the expanding SARS-CoV-2 outbreak, novel genetic variants may reduce the specificity and sensitivity of RT-PCR-based detection. Active viral recombination and mutation rates, in particular, might undoubtedly disrupt oligonucleotide annealing and impact sensitivity or inclusiveness. An analysis of genetic variants in the widely distributed SARS-CoV-2 genomes reveals a total of 27 probe- or primer-binding sites with a variant frequency of <1% (89–91).
Diagnostic failures are implicated in the Alpha (B.1.1.7) lineage as considerably higher false-negative results by RTPCRs that target the spike (S) gene. Diagnostic performance was unaffected by the Berlin–Charité technique, with nearly 98% of the sequences being detected using contemporary primers/probe sets because the S protein-producing gene was never used as a target in this procedure. For detection tests, expensive qPCR equipment that relies on signal absences rather than the positive indication for a variant presence is required (92, 93). Sequencing the entire variant's genome for both alpha (B.1.1.7) and beta (B.1.351) variant identification in the next generation sequencing (NGS) approach may result in incorrect or inconclusive detection. Several tests on the Delta B.1.1.7 variant reveal that in the three-target gene of RT-PCR diagnostic assay, where positive cases of SARS-CoV-2 infection are on the rise, this variant exhibits an increase in S-gene target failure rather than positive ORF1ab, N target genes (94–96). An exploratory study of the B.1.1.7. lineage revealed the presence of polymorphisms in the amplified sequences, indicating the discovery of new haplotypes. These haplotypes have a low frequency of single nucleotide polymorphisms (SNPs) that affect oligobinding site areas, hindering accurate identification and resulting in false-negative test findings (93, 97).
In France, a novel variant of concern 202012/01 (VOC) with the deletion of the spike (S) at H69–V70 (H69/V70) location was detected, which is also 43–82% transmissible compared with other SARS-CoV-2 variants. This deletion process is linked to an S-gene target failure in (ORF) 1ab, S, and nucleocapsid (N) gene targets, according to an RT-PCR assay (TaqPath kit) (98). The findings from the TaqPath RT-PCR kit uncovers 0.6% of overall prevalence, indicating a limited variations circulation with H69/V70 during the second wave. Three RBD mutations, Y453F, N501Y, and N439K, have been linked to the H69/V70 gene, which reduces SARS-CoV-2 antibody sensitivity (98, 99). Several laboratories experimented with diagnostic primers or probes alignment with a short viral sequence exhibiting mismatches that led to false-negative results due to a worldwide pandemic emergency (100).
Mutant viruses or genetic diversity were shown to have potential mismatches in the primer or probe binding region of the viral genome, resulting in false-negative results. While a single mismatch has little impact, two or three mismatches reduce technique sensitivity, and having more than three mismatches can result in a complete reaction failure (100, 101). SARS-CoV-2 has newly emerged variants of concern, as well as likely mismatches, indicating the importance of molecular surveillance and providing fresh diagnostic tools for future prevalence. On the other hand, it also provides an identification scheme with high sensitivity and specificity, allowing the CRISPR-based diagnostic tools to be developed (10, 102).
Potential Influence on Vaccination
Spike protein plays a key role in the pathogenicity of SARS-CoV-2, and vaccines based on targeting this spike protein are being developed (10). Meanwhile, B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.526 (Delta) forms of SARS-CoV-2 are propagating over the world (103, 104). In this part, we will explore the effectiveness of numerous vaccines against these variants.
Genetic Vaccines
Pfizer-BioNTech and Moderna developed two anti-SARS-CoV-2 vaccines, “BNT162b2” and “mRNA-1273”, both mRNA-based vaccines that were previously licensed (10). According to several studies, the BNT162b2 vaccination was projected to be 89.5% effective against the Alpha variant and 75.0% effective against the Beta form (105). BNT162b2 was found to be 84% effective against the Gamma version (103), and 88% effective against the Delta variant (106). The mRNA-1273 vaccination was shown to be 94.1 % (107–109) effective for the Alpha variant and 96.4 % effective for the Beta form (110). mRNA-1273, on the other hand, had a lower neutralization rate against Gamma and Delta variants (111). According to multiple studies, the effectiveness of BNT162b2 and mRNA-1273 vaccines against Omicron was only 36% after the second dose but increased to 61% after the third dose (38).
Another study looked into whether using various vaccines as booster doses could increase the immunological response to Omicron. Two doses of BNT162b2 vaccination plus one booster dose of BNT162b2 vaccine, as well as two doses of CoronaVac vaccine plus one booster dose of BNT162b2 vaccine provided protection against Omicron. The vaccine effectiveness of these two groups increases by 95% after a booster dose against this variation (112).
Adenovirus-Based Vaccines
Adenovirus-based vaccines have been approved for both emergency and routine use (10). Among them, the University of Oxford and AstraZeneca's ChAdOx1 nCoV-19 (chimpanzee adenovirus type Y25 vector) or the AZD1222 vaccine was 74.5% effective against the Alpha variant (106, 113, 114) and 10.4% effective against the Beta variant (115). Though the ChAdOx1 nCoV-19 vaccine has yet to be proved to be effective against the Gamma version, it has shown to be 59.8% effective against the Delta form (116). Another Ad26.COV2.S vaccine which is a recombinant, replication-incompetent human adenovirus type 26 vector encoding full-length and stagnates SARS-CoV-2 spike protein (JANSSEN) was found to have about 86% decreased efficiency against Alpha variant (117, 118) as well as a 64% protection against the Beta variant (119). Furthermore, this vaccination appears to be quite practical against the Gamma variation, although no information on its efficacy against the Delta variant has been released. (https://www.covid19immunitytaskforce.ca/literature-review-effectiveness-of-the-covid-19-vaccines-approved-for-use-in-canada-against-circulating-variants-of-concern/). The Gamaleya Research Institute's Sputnik V vaccine has a high virus-neutralizing efficiency against B.1.351, B.1.617.2, and P.1, as well as other variations (115).
Subunit Vaccines
The recombinant NVX-CoV2373 (Novavax) vaccine contains prefusion, full-length spike protein with 85.6 % and 51% effectiveness against Alpha and Beta variants, respectively (10, 106, 111). None of the protein-based vaccines, on the other hand, have been approved for widespread use.
Inactivated Virus-Based Vaccines
BBIBP-CorV, BBV152, and CoronaVac are three inactivated virus-based vaccines that have been approved and are widely used in China, India, and Brazil, respectively (10). Among them, BBIBP-CorV is a vaccine manufactured by Sinopharm (Beijing, China), producing vaccine antisera that are compatible for neutralizing the Beta variant (120). BBV152 (Bharat Biotech, India) is a vaccine that showed efficacy against Alpha and Beta variants and was 652% effective against Delta variant (121, 122), whereas CoronaVac (Sinovac Biotech) is 42% efficient against the Gamma variant (123).
To put an end to this discussion about vaccines, it appears that none of them are effective against all SARS-CoV-2 variants, but the majority of the licensed vaccines are partially effective against the Alpha and Beta types.
Limitations
According to CDC (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html), previously called VOCs (epsilon, alpha, beta, and gamma) are now designated as the VBMs in the USA, while alpha, beta, and gamma are still mentioned as VOCs according to GISAID (https://www.gisaid.org/hcov19-variants/) because variants are classified based on their potential impact on critical SARS-CoV-2 countermeasures, including vaccines, treatments, and diagnostics, as they are important for public health. As a result, the viewpoint of variant monitoring is at a conflict with time and research data. Furthermore, new spike protein mutations, in combination with genetic and host predisposition, impact the current vaccination efficacy (124). Furthermore, as new strains appear, the need to reexamine vaccination efficiency by experimenting with in vivo reduction of viral infection, as well as antibody quantification obtained from in vitro exposure reveals a dearth of understanding about vaccine efficacy (125). Furthermore, as SARS-CoV-2 is constantly mutating, accumulating around one new mutation every 2 weeks in the genome (126), Delta Plus is observed with several new mutations concerning ORF1a (A1146T, A3209V, P1604L, T3750I, and V3718S) evolving from delta variant (127), and RBD-ACE2 system analysis of newly emergent variants such as Omicron revealed 32 mutations in S protein, raising significant concern for its transmissibility. As a result, the shifting variations identified through dynamic research in mutation findings necessitate more investigation for vaccination efficacy and variant tracking (128). As a result, it may be inferred that our understanding of variation mutation and the impact of a variant in conjunction with vaccination efficacy data is evolving, and that, while our analysis depicts the current landscape of variant tracking perfectly, it may alter over time.
Conclusion
When spike protein mutations are combined with non-structural protein mutations reported in emerging VOCs, VOIs, and VBMs, the clinical relevance of each variance changes. As a result, a potentially devastating global health catastrophe occurs from either a novel variant of concern or a variant of interest that has the potential to worsen the infected individual's clinical status. Furthermore, changes in either the spike protein or the NSP protein has a conceivable impact on vaccination, which is an important problem in vaccine efficacy. As a result of this review, it appears that more vaccine development research is needed to ensure that vaccines are effective against all SARS-CoV-2 variants.
Author Contributions
SB, SM, MM, SA, MH, JS-G and MSa: study concept and design. SB, SM, MM, SA, MH, GP, AO, and MSa: acquisition of data, analyses, and interpretation of data. SB, SM, MM, SA, MH, AO, and MSh: drafting the manuscript. SZ, MU, MSa, MP, AS, BK, AJO, and JS-G: critical revision of the manuscript for important intellectual content. SZ, MU, BK and MSa: technical or material support. MSa, BK, and JS-G: study supervision. BK: fund acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1I1A2066868), and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1A5A2019413). Funding for open access charge: University of Vigo/CISUG.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Islam MR, Hoque MN, Rahman MS, Alam ASMRU, Akther M, Puspo JA, et al. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci Rep. (2020) 10:1–9. doi: 10.1038/s41598-020-70812-6
2. Cao C, Cai Z, Xiao X, Rao J, Chen J, Hu N, et al. The architecture of the SARS-CoV-2 RNA genome inside virion. Nat Commun. (2021) 12:1–14. doi: 10.1038/s41467-021-22785-x
3. van Dorp L, Richard D, Tan CCS, Shaw LP, Acman M, Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat Commun. (2020) 11:5986. doi: 10.1038/s41467-020-19818-2
4. van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol. (2020) 83:104351. doi: 10.1016/j.meegid.2020.104351
5. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. (2020) 181:914–21.e10. doi: 10.1016/j.cell.2020.04.011
6. Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. (2016) 96:59–126. doi: 10.1016/bs.aivir.2016.08.008
7. Sola I, Almazán F, Zúñiga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. (2015) 2:265–88. doi: 10.1146/annurev-virology-100114-055218
8. Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Reports. (2020) 19:100682. doi: 10.1016/j.genrep.2020.100682
9. Li F, Li W, Farzan M, Harrison SC. Structural biology: structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. (2005) 309:1864–8. doi: 10.1126/science.1116480
10. Khateeb J, Li Y, Zhang H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit Care. (2021) 25:1–8. doi: 10.1186/s13054-021-03662-x
11. Panigrahi GK, Sahoo SK, Sahoo A, Behera S, Sahu S, Dash A, et al. Bioactive molecules from plants: a prospective approach to combat SARS-CoV-2. Adv Tradit Med. (2021) 1–14. doi: 10.1007/s13596-021-00599-y
12. Huston NC, Wan H, Strine MS, de Cesaris Araujo Tavares R, Wilen CB, Pyle AM. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol Cell. (2021) 81:584–598.e5. doi: 10.1016/j.molcel.2020.12.041
13. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. (2020) 579:265–9. doi: 10.1038/s41586-020-2008-3
14. Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2-what do they mean? J Am Med Assoc. (2021) 325:529–31. doi: 10.1001/jama.2020.27124
15. Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LLM, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. (2003) 331:991–1004. doi: 10.1016/S0022-2836(03)00865-9
16. Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, et al. Discovery of an RNA virus 3′ → 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A. (2006) 103:5108–13. doi: 10.1073/pnas.0508200103
17. Fraser C, Crook D, Peto T, Andersson M, Jeffries K, Eyre D, et al. Shared SARS-CoV-2 diversity suggests localised transmission of minority variants. bioRxiv. (2020) 372:eabg0821. doi: 10.1126/SCIENCE.ABG0821
18. Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. (2003) 424:99–103. doi: 10.1038/nature01709
19. Lupala CS, Ye Y, Chen H, Su XD, Liu H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun. (2022) 590:34–41. doi: 10.1016/j.bbrc.2021.12.079
20. Ulloa AC, Buchan SA, Daneman N, Brown KA. Early estimates of SARS-CoV-2 Omicron variant severity based on a matched cohort study, Ontario, Canada. medRxiv. (2022). doi: 10.1101/2021.12.24.21268382 [Preprint].
21. Chakraborty C, Bhattacharya M, Sharma AR. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev Med Virol. (2021) 32:e2270. doi: 10.1002/rmv.2270
22. Dong Y, Dai T, Wang B, Zhang L, Zeng-hui L, Huang J, et al. The way of SARS-CoV-2 vaccine development: success and challenges. Signal Transduct Target Ther. (2021) 6:1–14. doi: 10.1038/s41392-021-00796-w
23. Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. (2017) 22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494
24. Kumar V, Singh J, Hasnain SE, Sundar D. Possible link between higher transmissibility of alpha, kappa and delta variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. Int J Mol Sci. (2021) 22:9131. doi: 10.3390/ijms22179131
25. Johnson BA, Xie X, Kalveram B, Lokugamage KG, Muruato A, Zou J, et al. Furin cleavage site is key to SARS-CoV-2 pathogenesis. bioRxiv Prepr Serv Biol. (2020). doi: 10.1101/2020.08.26.268854 [Preprint].
26. Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet. (2021) 22:757–73. doi: 10.1038/s41576-021-00408-x
27. Acevedo ML, Alonso-Palomares L, Bustamante A, Gaggero A, Paredes F, Cortés CP, et al. Infectivity and immune escape of the new SARS-CoV-2 variant of interest Lambda. medRxiv. (2021). doi: 10.1101/2021.06.28.21259673 [Preprint].
28. Kimura I, Kosugi Y, Wu J, Yamasoba D, Butlertanaka EP, Tanaka YL, et al. SARS-CoV-2 Lambda variant exhibits higher infectivity and immune resistance. bioRxiv. (2021) 7:2021.07.28.454085. doi: 10.1101/2021.07.28.454085
29. Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. (2021) 372:eabg3055. doi: 10.1126/science.abg3055
30. Song W, Gui M, Wang X, Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. (2018) 14:1–19. doi: 10.1371/journal.ppat.1007236
31. Zhou D, Duyvesteyn HME, Chen CP, Huang CG, Chen TH, Shih SR, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. (2020) 581:215–20. doi: 10.1038/s41586-020-2180-5
32. Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci U S A. (2004) 101:4240–5. doi: 10.1073/pnas.0306446101
33. Yan R, Zhang Y, Guo Y, Xia L, Zhou Q. Structural basis for the recognition of the 2019-nCoV by human ACE2. (2020) 2762:1–10. doi: 10.1101/2020.02.19.956946
34. Cagliani R, Forni D, Clerici M, Sironi M. Computational inference of selection underlying the evolution of the novel coronavirus, severe acute respiratory syndrome coronavirus 2. J Virol. (2020) 94:e00411–20. doi: 10.1128/JVI.00411-20
35. Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. (2020) 21:730–8. doi: 10.1002/cbic.202000047
36. Yi C, Sun X, Ye J, Ding L, Liu M, Yang Z, et al. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell Mol Immunol. (2020) 17:621–30. doi: 10.1038/s41423-020-0458-z
37. Huang Y, Yang C, Xu-feng X, Xu W, Liu-wen S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. (2020) 41:1141–9. doi: 10.1038/s41401-020-0485-4
38. Nasreen S, Chung H, He S, Brown KA, Gubbay JB, Buchan SA, et al. Effectiveness of COVID-19 vaccines against symptomatic SARS-CoV-2 infection and severe outcomes with variants of concern in Ontario. Nat Microbiol. (2022) 7:379–85. doi: 10.1038/s41564-021-01053-0
39. Nelson G, Buzko O, Patricia S, Niazi K, Rabizadeh S, Soon-Shiong P. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the 1 combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an esc. bioRxiv. (2021). doi: 10.1101/2021.01.13.426558 [Preprint].
40. Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. (2021) 29:44–57.e9. doi: 10.1016/j.chom.2020.11.007
41. Edara VV, Norwood C, Floyd K, Lai L, Davis-Gardner ME, Hudson WH, et al. Reduced binding and neutralization of infection- and vaccine-induced antibodies to the B.1.351 (South African) SARS-CoV-2 variant. bioRxiv Prepr Serv Biol. (2021). doi: 10.1101/2021.02.20.432046 [Preprint].
42. Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. (2020) 183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037
43. Weissman D, Alameh MG, de Silva T, Collini P, Hornsby H, Brown R, et al. D614G Spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. (2021) 29:23–31.e4. doi: 10.1016/j.chom.2020.11.012
44. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 Virus. Cell. (2020) 182:812–27.e19. doi: 10.1016/j.cell.2020.06.043
45. Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. (2020) 11:1–9. doi: 10.1038/s41467-020-19808-4
46. Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. (2021) 592:116–21. doi: 10.1038/s41586-020-2895-3
47. Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. (2020) 370:1464–8. doi: 10.1126/science.abe8499
48. Motozono C, Toyoda M, Zahradnik J, Saito A, Nasser H, Tan TS, et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. (2021) 29:1124–36.e11. doi: 10.1016/j.chom.2021.06.006
49. McCarthy KR, Rennick LJ, Nambulli S, Robinson-McCarthy LR, Bain WG, Haidar G, et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. (2021) 371:1139–42. doi: 10.1126/science.abf6950
50. Gong SY, Chatterjee D, Richard J, Prévost J, Tauzin A, Gasser R, et al. Contribution of single mutations to selected SARS-CoV-2 emerging variants spike antigenicity. Virology. (2021) 563:134–45. doi: 10.1016/j.virol.2021.09.001
51. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. (2021) 19:409–24. doi: 10.1038/s41579-021-00573-0
52. Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile T, Wang Y, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. SSRN Electron J. (2020) 183:739–51. doi: 10.2139/ssrn.3657338
53. Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole Á, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. (2021) 184:64–75.e11. doi: 10.1101/2020.07.31.20166082
54. Toyoshima Y, Nemoto K, Matsumoto S, Nakamura Y, Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J Hum Genet. (2020) 65:1075–82. doi: 10.1038/s10038-020-0808-9
55. Li L, Shuai L, Sun J, Li C, Yi P, Zhou Z, et al. The role of 1-methylcyclopropene in the regulation of ethylene biosynthesis and ethylene receptor gene expression in mangifera indica L. (Mango Fruit). Food Sci Nutr. (2020) 8:1284–94. doi: 10.1002/fsn3.1417
56. Bakhshandeh B, Jahanafrooz Z, Abbasi A, Goli MB, Sadeghi M, Mottaqi MS, et al. Mutations in SARS-CoV-2; consequences in structure, function, and pathogenicity of the virus. Microb Pathog. (2021) 154:104831. doi: 10.1016/j.micpath.2021.104831
57. Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. (2020) 182:1295–310.e20. doi: 10.1016/j.cell.2020.08.012
58. Prévost J, Richard J, Gasser R, Ding S, Fage C, Anand SP, et al. Impact of temperature on the affinity of SARS-CoV-2 Spike glycoprotein for host ACE2. J Biol Chem. (2021) 297:101151. doi: 10.1016/j.jbc.2021.101151
59. Zhu X, Mannar D, Srivastava SS, Berezuk AM, Demers JP, Saville JW, et al. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. (2021) 19:1–17. doi: 10.1371/journal.pbio.3001237
60. Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, et al. Individual antibodies. Science. (2020) 369:1014–8. doi: 10.1126/science.abd0831
61. Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JCC, et al. Escape from neutralizing antibodies 1 by SARS-CoV-2 spike protein variants. Elife. (2020) 9:1. doi: 10.7554/eLife.61312
62. Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. (2021) 29:477–88.e4. doi: 10.1016/j.chom.2021.01.014
63. Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, et al. SARS-CoV-2 escape from a highly neutralizing COVID-19 convalescent plasma. Proc Natl Acad Sci USA. (2021) 118:e2103154118. doi: 10.1073/pnas.2103154118
64. Amanat F, Thapa M, Lei T, Ahmed SMS, Adelsberg DC, Carreño JM, et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell. (2021) 184:3936–48.e10. doi: 10.1016/j.cell.2021.06.005
65. Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B1351 and B117. Nature. (2021) 593:130–5. doi: 10.1038/s41586-021-03398-2
66. McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. (2021) 184:2332–47.e16. doi: 10.1016/j.cell.2021.03.028
67. Zhang Y, Zhang J, Chen Y, Luo B, Yuan Y, Huang F, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv. (2021) 118:e2024202118. doi: 10.1073/pnas.2024202118
68. Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. Structure of SARS-cov-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci U S A. (2021) 118:1–6. doi: 10.1073/pnas.2021785118
69. Lin J-W, Tang C, Wei H-C, Du B, Chen C, Wang M, et al. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe. (2021) 29:489–502.e8. doi: 10.1016/j.chom.2021.01.015
70. Young BE, Fong SW, Chan YH, Mak TM, Ang LW, Anderson DE, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. (2020) 396:603–11. doi: 10.1016/S0140-6736(20)31757-8
71. Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. (2020) 55:102763. doi: 10.1016/j.ebiom.2020.102763
72. Huang C, Wang Y, Li X, Ren L, Zhao L, Zang Li, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:470–3. doi: 10.1016/S0140-6736(20)30183-5
73. Banerjee S, Seal S, Dey R, Mondal KK, Bhattacharjee P. Mutational spectra of SARS-CoV-2 orf1ab polyprotein and signature mutations in the United States of America. J Med Virol. (2021) 93:1428–35. doi: 10.1002/jmv.26417
74. Hossain MU, Bhattacharjee A, Emon MTH, Chowdhury ZM, Ahammad I, Mosaib MG, et al. Novel mutations in NSP-1 and PLPro of SARS-CoV-2 NIB-1 genome mount for effective therapeutics. J Genet Eng Biotechnol. (2021) 19::1–10. doi: 10.1186/s43141-021-00152-z
75. Moniruzzaman M, Hossain MU, Islam MN, Rahman MH, Ahmed I, Rahman TA, et al. Coding-complete genome sequence of SARS-CoV-2 isolate from Bangladesh by sanger sequencing. Microbiol Resour Announc. (2020) 9::e00626–20. doi: 10.1128/MRA.00626-20
76. Sheikh A, McMenamin J, Taylor B, Robertson C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. (2021) 397:2461–2. doi: 10.1016/S0140-6736(21)01358-1
77. Abdelnabi M, Mora B, Eshak N, Nugent K. SARS-CoV-2 Delta variant: What do we know so far? Southwest Respir Crit Care Chronicles. (2021) 9:1–2. doi: 10.12746/swrccc.v9i40.905
78. Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. (2021) 596:276–80. doi: 10.1038/s41586-021-03777-9
79. Yang W, Shaman J. COVID-19 pandemic dynamics in India and impact of the SARS-CoV-2 Delta (B.1.617.2) variant. MedRxiv. (2021). [Preprint]. doi: 10.1101/2021.06.21.21259268
80. Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B117 in England. Nature. (2021) 593:266–9. doi: 10.1038/s41586-021-03470-x
81. Iacobucci G. Covid-19: New UK variant may be linked to increased death rate, early data indicate. BMJ. (2021) 372:n230. doi: 10.1136/bmj.n230
82. Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor Barrett KM, et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. (2021) 184:2587–94.e7. doi: 10.1016/j.cell.2021.03.052
83. Faria NR, Claro IM, Candido D, Franco LAM, Andrade PS, Thais M, et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological Org. (2021) 372:815–21.
84. Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. (2020) 5:1403–7. doi: 10.1038/s41564-020-0770-5
85. McCallum M, Bassi J, De Marco A, Chen A, Walls AC, Di Iulio J, et al. SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv Prepr Serv Biol. (2021) 373:648–54. doi: 10.1126/science.abi7994
86. Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N, et al. Increased transmissibility and global spread of SARSCoV- 2 variants of concern as at June 2021. Eurosurveillance. (2021) 26:1–6. doi: 10.2807/1560-7917.ES.2021.26.24.2100509
87. Funk T, Pharris A, Spiteri G, Bundle N, Melidou A, Carr M, et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Eurosurveillance. (2021) 26:2100348. doi: 10.2807/1560-7917.ES.2021.26.16.2100348
88. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. (2020) 26:450–2. doi: 10.1038/s41591-020-0820-9
89. Peñarrubia L, Ruiz M, Porco R, Rao SN, Juanola-Falgarona M, Manissero D, et al. Multiple assays in a real-time RT-PCR SARS-CoV-2 panel can mitigate the risk of loss of sensitivity by new genomic variants during the COVID-19 outbreak. Int J Infect Dis. (2020) 97:225–9. doi: 10.1016/j.ijid.2020.06.027
90. Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med. (2020) 58:1070–6. doi: 10.1515/cclm-2020-0285
91. Jain A, Rophina M, Mahajan S, Krishnan BB, Sharma M, Mandal S, et al. Analysis of the potential impact of genomic variants in global SARS-CoV-2 genomes on molecular diagnostic assays. Int J Infect Dis. (2021) 102:460–2. doi: 10.1016/j.ijid.2020.10.086
92. Sherrill-Mix S, Van Duyne GD, Bushman FD. Molecular beacons allow specific RT-LAMP detection of B.1.1.7 variant SARS-CoV-2. medRxiv. (2021) 32:98–101. doi: 10.7171/jbt.21-3203-004
93. Ramírez JD, Muñoz M, Patiño LH, Ballesteros N, Paniz-Mondolfi A. Will the emergent SARS-CoV2 B117 lineage affect molecular diagnosis of COVID-19? J Med Virol. (2021) 93:2566–8. doi: 10.1002/jmv.26823
94. Yaniv K, Ozer E, Plotkin N, Bhandarkar NS, Kushmaro A. RT-qPCR assay for detection of British (B . 1 . 1 . 7) and South Africa (B . 1 . 351) variants of SARS-CoV-2. (2021). doi: 10.1101/2021.02.25.21252454 [Preprint].
95. Andrés C, Garcia-Cehic D, Gregori J, Piñana M, Rodriguez-Frias F, Guerrero-Murillo M, et al. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg Microbes Infect. (2020) 9:1900–11. doi: 10.1080/22221751.2020.1806735
96. Lopez-Rincon A, Perez-Romero CA, Tonda A, Mendoza-Maldonado L, Claassen E, Garssen J, et al. Design of specific primer sets for the detection of B.1.1.7, B.1.351 and P.1 SARS-CoV-2 variants using deep learning. bioRxiv. (2021) 70:2021.01.20.427043. doi: 10.1101/2020.12.29.424715
97. Ramírez JD, Muñoz M, Hernández C, Flórez C, Gomez S, Rico A, et al. Genetic diversity among sars-cov2 strains in South America may impact performance of molecular detection. Pathogens. (2020) 9:1–14. doi: 10.3390/pathogens9070580
98. Bal A, Destras G, Gaymard A, Stefic K, Marlet J, Eymieux S, et al. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69–V70, France, August to December 2020. Eurosurveillance. (2021) 26:1–5. doi: 10.2807/1560-7917.ES.2021.26.3.2100008
99. Thomson EC, Rosen LE, Shepherd JG, Spreafico R, da Silva Filipe A, Wojcechowskyj JA, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. (2021) 184:1171–87.e20. doi: 10.1016/j.cell.2021.01.037
100. Khan KA, Cheung P. Presence of mismatches between diagnostic PCR assays and coronavirus SARS-CoV-2 genome: Sequence mismatches in SARS-CoV-2 PCR. R Soc Open Sci. (2020) 7:200636. doi: 10.1098/rsos.200636
101. Gand M, Vanneste K, Thomas I, Van Gucht S, Capron A, Herman P, et al. Use of whole genome sequencing data for a first in silico specificity evaluation of the rt-qpcr assays used for sars-cov-2 detection. Int J Mol Sci. (2020) 21:1–25. doi: 10.3390/ijms21155585
102. Kuchinski KS, Jassem AN, Prystajecky NA. Assessing oligonucleotide designs from early lab developed PCR diagnostic tests for SARS-CoV-2 using the PCR_strainer pipeline. J Clin Virol. (2020) 131:104581. doi: 10.1016/j.jcv.2020.104581
103. Nasreen S, He S, Chung H, Brown KA, Gubbay JB, Buchan SA, et al. Effectiveness of COVID-19 vaccines against variants of concern, Canada. medRxiv. (2021). [Preprint]. doi: 10.1101/2021.06.28.21259420
104. Chen Y, Shen H, Huang R, Tong X, Wu C. Serum neutralising activity against SARS-CoV-2 variants elicited by CoronaVac. Lancet Infect Dis. (2021) 21:1071–2. doi: 10.1016/S1473-3099(21)00287-5
105. Abu-Raddad LJ, Chemaitelly H, Butt AA. Effectiveness of the BNT162b2 Covid-19 vaccine against the B117 and B1351 variants. N Engl J Med. (2021) 385:187–9. doi: 10.1056/NEJMc2104974
106. Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. (2021) 385:585–94. doi: 10.1101/2021.05.22.21257658
107. Heath PT, Galiza EP, Baxter DN, Boffito M, Browne D, Burns F, et al. Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine. N Engl J Med. (2021) 385:1172–83. doi: 10.1056/NEJMoa2107659
108. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
109. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
110. Chemaitelly H, Yassine HM, Benslimane FM, Al Khatib HA, Tang P, Hasan MR, et al. mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar. Nat Med. (2021) 27:1614–21. doi: 10.1038/s41591-021-01446-y
111. Li Q, Wang J, Tang Y, Lu H. Next-generation COVID-19 vaccines: Opportunities for vaccine development and challenges in tackling COVID-19. Drug Discov Ther. (2021) 15:118–23. doi: 10.5582/ddt.2021.01058
112. Khong KW, Liu D, Leung KY, Lu L, Lam HY, Chen L, et al. Antibody response of combination of BNT162b2 and coronavac platforms of COVID-19 vaccines against omicron variant. Vaccines. (2022) 10:1–10. doi: 10.3390/vaccines10020160
113. Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. (2020) 396:467–78. doi: 10.1016/S0140-6736(20)31604-4
114. Xu K, An Y, Li Q, Huang W, Han Y, Zheng T, et al. Recombinant chimpanzee adenovirus AdC7 expressing dimeric tandem-repeat spike protein RBD protects mice against COVID-19. Emerg Microbes Infect. (2021) 10:1574–88. doi: 10.1080/22221751.2021.1959270
115. Gushchin VA, Dolzhikova IV, Shchetinin AM, Odintsova AS, Siniavin AE, Nikiforova MA, et al. Neutralizing activity of sera from sputnik V-vaccinated people against variants of concern (VOC: B117, B1351, P1, B16172, B16173) and Moscow endemic SARS-CoV-2 variants. Vaccines. (2021) 9:779. doi: 10.3390/vaccines9070779
116. Williams S V, Vusirikala A, Ladhani SN, Fernandez Ruiz De Olano E, Iyanger N, Aiano F, et al. An outbreak caused by the SARS-CoV-2 Delta (B.1.617.2) variant in a care home after partial vaccination with a single dose of the COVID-19 vaccine Vaxzevria, London, England, April 2021. Eurosurveillance. (2021) 26:1–5. doi: 10.2807/1560-7917.ES.2021.26.27.2100626
117. Jahangir MA. A Review on the Contemporary Status of Mutating Coronavirus and Comparative Literature Study of Current COVID-19 Vaccines. Int J Pharm Pharmacol. (2021) 5:1–19. doi: 10.31531/2581-3080.1000153
118. Sadoff J, Gray G, Vandebosch A, Cárdenas V, Shukarev G, Grinsztejn B, et al. Safety and efficacy of single-dose Ad26COV2S vaccine against Covid-19. N Engl J Med. (2021) 384:2187–201. doi: 10.1056/NEJMoa2101544
119. Gupta RK. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat Rev Immunol. (2021) 21:340–1. doi: 10.1038/s41577-021-00556-5
120. Huang B, Dai L, Wang H, Hu Z, Tan W, Gao GF, et al. Neutralization of SARS-CoV-2 VOC 501Y.V2 by human antisera elicited by both inactivated BBIBP-CorV and recombinant dimeric RBD ZF2001 vaccines. bioRxiv. (2021). doi: 10.1101/2021.02.01.429069 [Preprint].
121. Yadav PD, Sapkal GN, Ella R, Sahay RR, Nyayanit DA, Patil DY, et al. Neutralization of Beta and Delta variant with sera of COVID-19 recovered cases and vaccinees of inactivated COVID-19 vaccine BBV152/Covaxin. J Travel Med. (2021) 1:1–3. doi: 10.1101/2021.06.05.447177
122. Basu A. Review of: “Efficacy, safety, and lot to lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): a double-blind, randomised, controlled phase 3 trial.” Qeios. (2021). doi: 10.32388/R013IB [Preprint].
123. Ranzani OT, Hitchings M, Dorion Nieto M, Lang TD, Cardoso de Paula R, Ferreira Pereira de Paula O, et al. Effectiveness of the CoronaVac vaccine in the elderly population during a P.1 variant-associated epidemic of COVID-19 in Brazil: a test-negative case-control study. nedRxiv. (2021) 374:n2015. doi: 10.1101/2021.05.19.21257472
124. Darbeheshti F, Rezaei N. Genetic predisposition models to COVID-19 infection. Med Hypotheses. (2020) 142:109818. doi: 10.1016/j.mehy.2020.109818
125. Luchsinger LL, Hillyer CD. Vaccine efficacy probable against COVID-19 variants. Science (80-). (2021) 371:1116. doi: 10.1126/science.abg9461
126. Erol A. Are the emerging SARS-COV-2 mutations friend or foe? Immunol Lett. (2021) 230:63–4. doi: 10.1016/j.imlet.2020.12.014
127. Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, Quinn TP, et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun. (2021) 124:102715. doi: 10.1016/j.jaut.2021.102715
Keywords: SARS-CoV-2, variant of concern, antibody, genomic variation, clinical perspective
Citation: Biswas S, Mahmud S, Mita MA, Afrose S, Hasan MR, Paul GK, Shimu MSS, Uddin MS, Zaman S, Park MN, Siyadatpanah A, Obaidullah AJ, Saleh MA, Simal-Gandara J and Kim B (2022) The Emergence of SARS-CoV-2 Variants With a Lower Antibody Response: A Genomic and Clinical Perspective. Front. Med. 9:825245. doi: 10.3389/fmed.2022.825245
Received: 30 November 2021; Accepted: 21 March 2022;
Published: 06 May 2022.
Edited by:
Sanjay Kumar, Armed Forces Medical College, Pune, IndiaReviewed by:
Sajjad Ahmad, Foundation University, Islamabad, PakistanCopyright © 2022 Biswas, Mahmud, Mita, Afrose, Hasan, Paul, Shimu, Uddin, Zaman, Park, Siyadatpanah, Obaidullah, Saleh, Simal-Gandara and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Md. Abu Saleh, c2FsZWhAcnUuYWMuYmQ=; Jesus Simal-Gandara, anNpbWFsQHV2aWdvLmVz; Bonglee Kim, Ym9uZ2xlZWtpbUBraHUuYWMua3I=
†These authors have contributed equally to this work
 Suvro Biswas
Suvro Biswas Shafi Mahmud
Shafi Mahmud Mohasana Akter Mita
Mohasana Akter Mita Shamima Afrose
Shamima Afrose Md. Robiul Hasan
Md. Robiul Hasan Gobindo Kumar Paul
Gobindo Kumar Paul Mst. Sharmin Sultana Shimu2
Mst. Sharmin Sultana Shimu2 Md. Salah Uddin
Md. Salah Uddin Md. Abu Saleh
Md. Abu Saleh Jesus Simal-Gandara
Jesus Simal-Gandara