- 1Leprosy Laboratory, Oswaldo Cruz Institute, Fiocruz, Rio de Janeiro, Brazil
- 2Clinical Research Laboratory in Chagas Disease, Evandro Chagas National Institute of Infectious Diseases, Fiocruz, Rio de Janeiro, Brazil
- 3Laboratory Animal House, Lauro de Souza Lima Institute, Sao-Paulo, Brazil
- 4Laboratory of Cell Biology, Institute of Biological Sciences, Federal University of Juiz de Fora, Juiz de Fora, Brazil
Peripheral neuropathy is the main cause of physical disability in leprosy patients. Importantly, the extension and pattern of peripheral damage has been linked to how the host cell will respond against Mycobacterium leprae (M. leprae) infection, in particular, how the pathogen will establish infection in Schwann cells. Interestingly, viable and dead M. leprae have been linked to neuropathology of leprosy by distinct mechanisms. While viable M. leprae promotes transcriptional modifications that allow the bacteria to survive through the use of the host cell's internal machinery and the subvert of host metabolites, components of the dead bacteria are associated with the generation of a harmful nerve microenvironment. Therefore, understanding the pathognomonic characteristics mediated by viable and dead M. leprae are essential for elucidating leprosy disease and its associated reactional episodes. Moreover, the impact of the viable and dead bacteria in Schwann cells is largely unknown and their gene signature profiling has, as yet, been poorly explored. In this study, we analyzed the early differences in the expression profile of genes involved in peripheral neuropathy, dedifferentiation and plasticity, neural regeneration, and inflammation in human Schwann cells challenged with viable and dead M. leprae. We substantiated our findings by analyzing this genetic profiling in human nerve biopsies of leprosy and non-leprosy patients, with accompanied histopathological analysis. We observed that viable and dead bacteria distinctly modulate Schwann cell genes, with emphasis to viable bacilli upregulating transcripts related to glial cell plasticity, dedifferentiation and anti-inflammatory profile, while dead bacteria affected genes involved in neuropathy and pro-inflammatory response. In addition, dead bacteria also upregulated genes associated with nerve support, which expression profile was similar to those obtained from leprosy nerve biopsies. These findings suggest that early exposure to viable and dead bacteria may provoke Schwann cells to behave differentially, with far-reaching implications for the ongoing neuropathy seen in leprosy patients, where a mixture of active and non-active bacteria are found in the nerve microenvironment.
Introduction
Leprosy neuropathy is a chronic neurological condition, caused by the infection of the nerve by its etiologic agents, Mycobacterium leprae and Mycobacterium lepromatosis (1–5). M. leprae infection provokes early pathological changes in the host cell that are, to some extent, associated with the late degenerative appearance of the infected nerves (6, 7). Schwann cells, the glial cells of the peripheral nervous system, are the preferable host for M. leprae entry, persistence, and replication within the nerve (8–11). Therefore, much attention has been given to the molecular and cellular alterations driven by leprosy bacilli once inside Schwann cells in order to identify the underlying reasons that culminate in the severe neuropathy seen in patients.
The immune response elicited in the nerve microenvironment against the bacilli is a key component that may lead to the distinct clinical manifestations (4). Infected Schwann cells produce a broad panel of inflammatory chemokines and cytokines, that accompanied with cell-mediated immune response, have been linked to the manifestation of neural pain and leprosy neuritis in patients (12–15). Of importance, this immunomodulation profile seen in infected Schwann cells was reported to occur before the reprogramming of the Schwann cells to the immature phenotype, highlighting the crucial role of the immune signaling network in the context of the early stages of M. leprae infection (16, 17). Additionally, M. leprae infection of Schwann cells has been associated with alterations in the glucose/lactate metabolic pathway (18, 19), lipid/cholesterol accumulation (20, 21), mitochondrial dysfunction (11), and myelin dismantling (22–24). Some of these changes were also confirmed in leprosy patients, and are suggested to cause the ongoing neuropathy and the observed tissue fibrosis and loss of nerve function experienced by leprosy individuals (14, 25). However, while these events may arise from modifications of the host Schwann cell's supportive function of the nerve, the accompanying changes in gene expression profile are largely unknown.
Studies have shown that the phenolic glycolipid 1 (PGL-1), a major M. leprae cell wall pathogenic component, is essential for M. leprae internalization into Schwann cells and has also been extensively attributed to induce pathology in vitro and in experimental infectious models (10, 23, 26). The understanding of the immunopathogenic mechanisms related to nerve damage in leprosy patients is pivotal for the development of new therapeutic strategies to control leprosy neuropathy. The treatment of nerve damage with steroids is effective but at least 40% of patients relapse and require a further course of steroids (27, 28).
Because leprosy neuropathy is an intricate complex disease, in which both viable and dead M. leprae may have a detrimental role for disease progression, it is necessary to fully understand and decipher the contribution of viable and dead bacteria in altering Schwann cell biology. For example, it was suggested that dead bacilli, unlike viable M. leprae, make Schwann cells susceptible to attack by killer cells (29). Moreover, dead bacteria and its components, such as lipoarabinomannan (LAM), were also reported to cause neural damage via modulation of the autophagic flux (30) and the complement attack of the nerve (31).
Despite these observations, the early effects of viable and dead M. leprae on the global Schwann cell gene expression profile that may be linked to primary neural leprosy are still largely unknown. Therefore, in the present study, we analyzed the expression profile of transcripts involved in neuropathy, glial cell plasticity, nerve repair, and the inflammatory network in leprosy and non-leprosy nerve biopsies and after challenging Schwann cells with viable and dead M. leprae independently. Our utmost goal was to provide novel evidence of how viable and dead bacteria modulate Schwann cell gene expression responses along with a detailed statistical correspondence to several histopathological findings commonly observed in nerve biopsies from leprosy and non-leprosy patients.
Materials and Methods
Human Nerve Biopsy
Nerve biopsy specimens from eight patients diagnosed with pure neural leprosy (PNL) were obtained from volunteers recruited at the Souza Araujo Outpatient Unit (Leprosy Laboratory, Oswaldo Cruz Institute, Oswaldo Cruz Foundation) (Table 1). Nerve biopsy fragments, as well their nerve sections, were available for histopathological staining and PCR analysis. For the present study, patients with PNL were selected who did not present any sign of nerve endoneurial fibrosis to ensure the chosen nerve specimens were in the early stages of leprosy neuropathy progression. This selection was made after analyzing the nerve section stained with the hematoxylin and eosin and Gomori trichrome stains under a light microscope following previously published protocol (6). Exclusion criteria were patients with coinfection, metabolic comorbidities such as diabetes, and signs of endoneurial fibrosis, pregnant women and patients under 18 years. For the control group, nerve biopsy specimens from three individuals who underwent brachial plexus surgery were kindly donated by the University Hospital Clementino Fraga Filho (HUCFF-UFRJ). This study was approved by the Oswaldo Cruz Foundation Ethics Committee (number of purports: 2.227.887).
Schwann Cell Culture
The human Schwann cell line, ST88-14, was used in the present study for the in vitro assays. Prior to the assays, cells were cultured in RPMI media (Gibco BRL, Grand Island, NY, USA) supplemented with 1% penicillin, 1% streptomycin, 2 mM L-glutamine, and 10% fetal bovine serum. The cells were maintained in a controlled environment at 37°C and 5% CO2. For the assays, ST88-14 cells were suspended in culture medium without penicillin-streptomycin and cultured at a density of 5 × 105 cells/well on six-well culture plates. The cell culture was infected with viable M. leprae, gently donated by Lauro de Souza Lima Institute (Sao-Paulo, Brazil) or stimulated with dead (gamma-irradiated) M. leprae, obtained through BEI Resources (#NR-19326), at a multiplicity of infection (MOI) of 50 bacilli/cell (50:1). After 24 h of incubation, supernatants were harvested and kept frozen at −20°C until quantification of inflammatory chemokines and cytokines. Additionally, Schwann cell cultures were subjected to total RNA extraction procedures.
RNA Extraction and RT-qPCR Array
Schwann cell cultures and nerve biopsy fragments were mechanically grinded and resuspended in 1 mL TRIzol (Gibco BRL) and RNA was obtained following the manufacturer's orientations and stored at −70°C until use. After, 10 ng of total RNA was reverse-transcribed to cDNAs using the Superscript III kit (Invitrogen, Carlsbad, CA, USA) and then amplified using the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) or TaqMan assays (ThermoScientific). The GeneQuery™ Human Schwann cell PCR Primer library array kit (Realtime Primers, Elkins Park, PA, USA #GK096) was used to profile total gene expression in Schwann cells and human nerve biopsies. The full list of genes is available on https://www.sciencellonline.com/genequerytm-human-schwann-cell-biology-qpcr-array-kit.html” (accessed on 27th Jan 2022). The TaqMan Fast Universal PCR Master Mix and Human TaqMAn MGB-Probe assays (ThermoScientific), were used to determine mRNA expression of TNF (HS-99999043_m1), IL-23A (HS-0037334324_m1), CCL2 (HS-00234140_m1) and CXCL10 (HS-0017042_m1). The RT-qPCR array was performed in triplicate, and the amplifications were carried out in the ViiA7 Real-Time PCR System (Thermo Fisher Scientific Inc., Waltham, MA, USA). The ΔΔCT method (32) was used to analyze the obtained data after normalization using the endogenous control of the housekeeping gene RPL13, for SYBR Green analysis, or normalized using the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH; HS-02758991_g1), for TaqMan assays.
Enzyme-Linked Immunosorbent Assay
For cytokine/chemokine release evaluation, the supernatants from control ST88-14 cultures and M. leprae (viable or dead) infected ST88-14 cultures were harvested after 24 h and stored at −20°C until use. The following inflammatory mediators (TNF, TGF-β, IL-6, IL-8, IL-12, IL-10, MCP-1/CCL2, and IP-10/CXCL10) were quantified by ELISA technique following the manufacturer's orientations (R&D Systems, Minneapolis, MN, USA).
Statistical Analysis
Analyses of the experiments were performed by unpaired t-test, Kruskal-Wallis test or one-way ANOVA. For all statistical analyses the value of p ≤ 0.05 was considered significant. Statistical analyses were performed using the GraphPad Prism version 8.0 software (GraphPad Software, San Diego, CA, USA). Alternatively, a correlogram graph was generated to evaluate the correlation between histopathological characteristics in nerve fragments and the pattern of gene expression. A Pearson's correlation was applied to each pair of variables present in the data. The correlation value varies between −1 and 1, with negative values implying the existence of negative correlation and positive values implying positive correlation. The strength of the correlation is measured by the proximity of the value to 1 or −1, with values closer to these suggesting stronger correlation. Each regression was performed using the gene expression as the response variable and initially both PCR and Acid Fast Bacilli (AFB) as covariates. Each coefficient had its statistical significance tested by the t-test for regression coefficients. In the cases where one of the variables was not statistically significant, the model was fitted again using only the statistically significant covariate. Finally, the model's goodness of fit was evaluated using R2, a statistical measure that evaluates how much of the variation on the response variable is explained by the covariates.
Results
Gene Profiling Analysis of Leprosy and Non-leprosy Nerve Biopsies
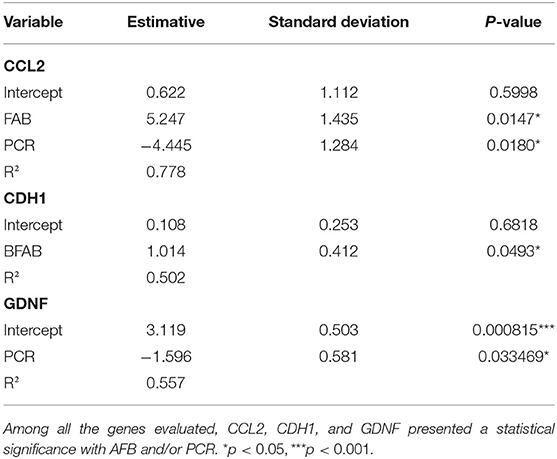
In order to identify the molecular pathways related to primary neural leprosy (PNL), a Schwann cell biology PCR array was performed comparing gene expression in nerve fragments from PNL patients and non-leprosy controls. As illustrated in Figure 1A, differentially expressed patterns of genes related to peripheral neuropathy, Schwann cell plasticity/reprogramming, and nerve support could be observed. Regarding neuropathy-related genes, some targets were statistically elevated in nerves from PNL patients, such as HLA-DRB1, APOB, and WNK, while others were downregulated, including HLA-DQB1 and PLP1. Interestingly, HLA-DRB1, previously reported to influence leprosy susceptibility (33), was upregulated 60-times more in leprosy nerves when compared to non-leprosy nerves. APOB and WNK were also augmented in leprosy nerves, by 10- and 4-times more, respectively.
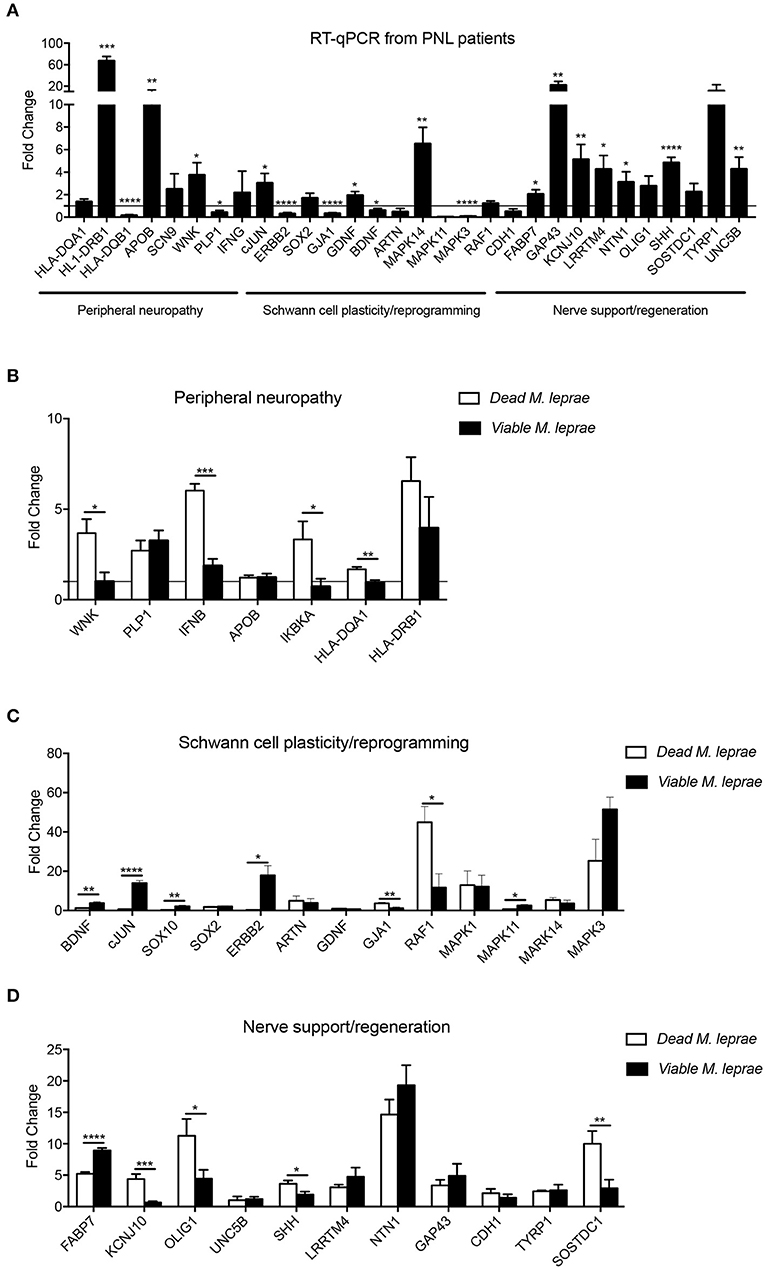
Figure 1. Expression pattern of genes involved in Schwann cell biology. Gene expression profile analysis of upregulated and downregulated mRNA transcripts from signaling pathways related to peripheral neuropathy, Schwann cell plasticity and regeneration support. Analysis were performed in human nerve biopsies from leprosy (A, black bars) and non-leprosy individuals (A, horizontal line) and in human Schwann cells (B-D) infected with dead M. leprae (white bars) and viable M. leprae (black bars); results are presented as mean ± SD from three to eight normalized independent biological replicates; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
By examining the mRNA profile involved in Schwann cell dedifferentiation, we observed that cJUN, GDNF, and MAPK14 were statistically upregulated, while ERBB2, GJA1, BDNF, and MAPK3 were significantly decreased in nerves from PNL patients. Although neurotrophins, such as BDNF, are required for axon regeneration, here we found that BDNF was downregulated in PNL patients, which is in agreement with a previous report that investigated neurotrophin expression in leprosy infection (34).
In summary, our PCR array analysis remarkably showed that most genes involved in nerve regeneration were significantly increased in leprosy nerve biopsies, possibly suggesting a continuous balance of nerve degeneration and an attempt to regrow during the course of infection in vivo.
Viable and Dead M. leprae Distinctly Modulate Schwann Cell Functional Genes
Neural damage in leprosy has been associated with the role played by viable and dead bacteria after being in contact with Schwann cells (10, 35). Therefore, we were interested in exploring the early effect of M. leprae infection on Schwann cell biology genes. For that, we infected human Schwann cells with viable and dead M. leprae and screened changes in the mRNA levels after 24 h of infection using the Schwann cell biology PCR array. As observed in Figures 1B-D, dead M. leprae increased the expression of WNK, IFNB, IKBKA, and HLA-DQA1 (genes related to neuropathy), in addition to GJA1 and RAF1 (for Schwann cell reprogramming) and KCNJ10, OLIG1, SHH, and SOSTDC1 (for neural regeneration). Interestingly, viable M. leprae appeared to modulate genes related to Schwann cell plasticity and dedifferentiation, such as BDNF, cJUN, SOX10, ERBB2, and MAPK11. In summary, this first set of analysis points to the notion that dead M. leprae induces greater expression of peripheral neuropathy and nerve regeneration support genes whereas viable M. leprae acts by modulating genes related to Schwann cell plasticity and dedifferentiation. We have summarized this gene intersection in a Venn diagram (Figure 2).
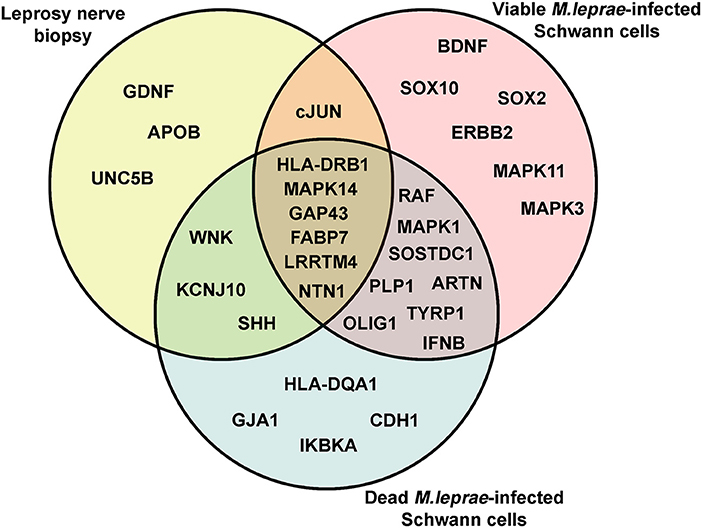
Figure 2. The Venn diagram of differentially upregulated genes related to Schwann cell biology genetic signature in leprosy. The Venn diagram was used to identify clusters of overlapping and non-overlapping mRNA transcripts, upregulated in leprosy nerve biopsies and in Schwann cells infected with viable and dead M. leprae.
Inflammatory Network Analysis Suggests That Dead and Viable M. leprae Increase CCL2 Expression
We next aimed to analyze the inflammatory network profile in leprosy and non-leprosy nerve biopsies, as well as in vitro, using Schwann cells challenged with viable and dead bacteria. The human nerve analysis indicated that IL23, TNF, CXCL10, and CCL2 were increased in PNL patients when compared with control biopsies (Figure 3A). In addition, when we analyzed changes in Schwann cells in vitro, we observed that dead M. leprae induced a higher expression of TNF, CXCL10, and IL6 in Schwann cells when compared with those infected with viable bacilli (Figure 3B). Furthermore, cells infected with viable M. leprae increased IL23 and CCL2 expression when compared to non-stimulated cultures. These results support the notion that dead bacteria are likely involved in the induction of a pro-inflammatory profile, suggesting that such pro-inflammatory mediators, in the context of neural involvement shown by the biopsies, are induced by dead bacteria present at the site of infection.
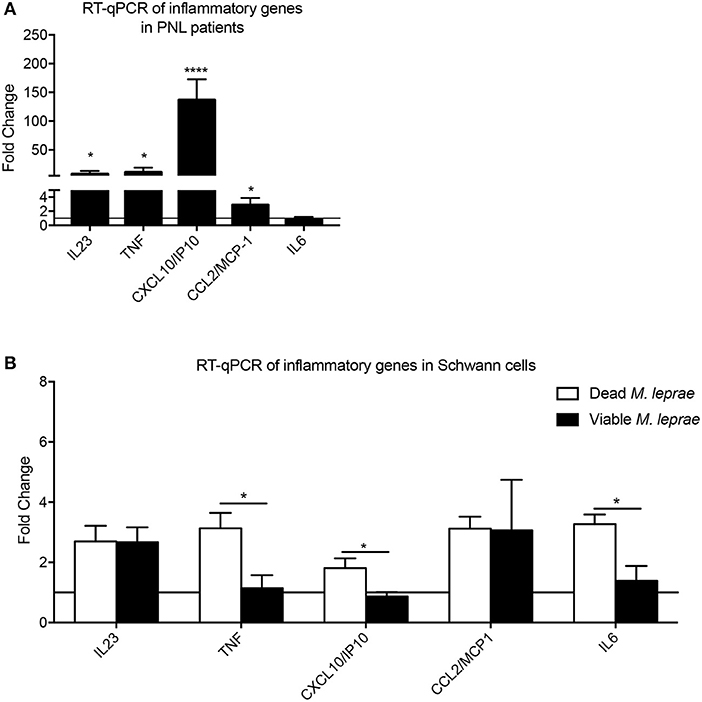
Figure 3. Analysis of inflammatory network in leprosy infection. Fold-change measurement of inflammatory cytokines/chemokines mRNA transcripts in human nerve biopsies from leprosy (A, black bars) and non-leprosy patients (A, horizontal line) and in human Schwann cells (B) infected with dead M. leprae (white bars) and viable M. leprae (black bars); results are presented as mean ± SD from three to eight normalized independent biological replicates; *p < 0.05, ****p < 0.0001.
Viable and Dead M. leprae Promote a Distinct Inflammatory Response in Schwann Cells
We substantiated our transcriptional analysis by evaluating the cytokine network production in Schwann cells challenged with viable and dead bacilli. For that, we performed analysis of cytokine levels in 24-h supernatants, and found that dead M. leprae caused an increase in the TNF, IL-8, MCP-1/CCL-2, and CXCL-10 levels in comparison to non-stimulated control cultures (Figure 4). Conversely, viable M. leprae infection led to increased TGF-β, IL-8, IL-6, IL-10, MCP-1/CCL2, and CXCL-10 in comparison to non-stimulated controls. We also observed a statistical reduction in TNF levels after viable M. leprae infection when compared to dead stimulated cultures and controls. Together, these results suggest that dead M. leprae induces pro-inflammatory mediators in human Schwann cells, whereas viable M. leprae preferably promote anti-inflammatory cytokines like IL-10 and TGF-β and reduced TNF production (Figure 4).
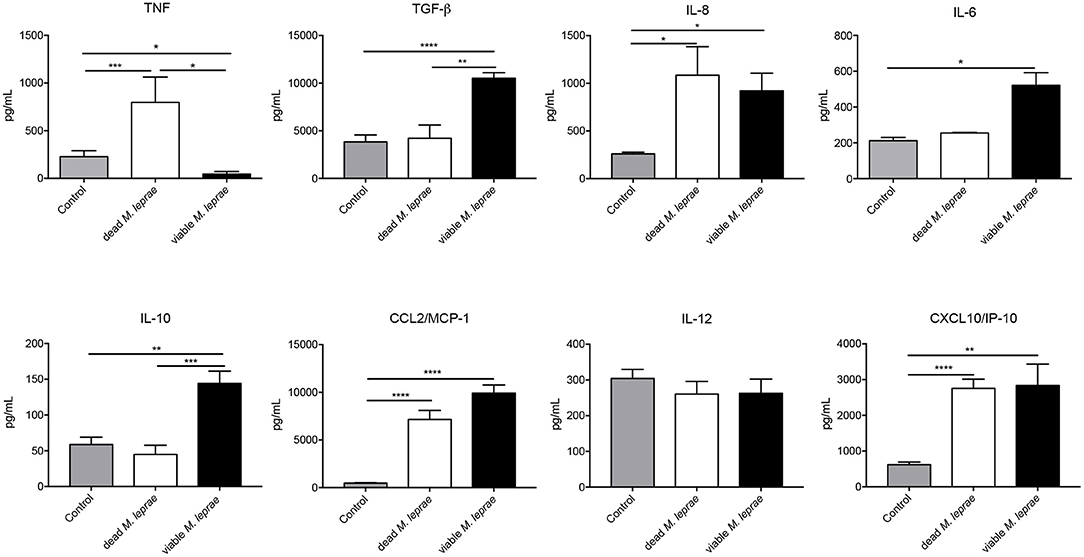
Figure 4. Differentially pattern of cytokine/chemokine release after viable and dead M. leprae infection in human Schwann cells. Two-side sandwich ELISA showing the pattern secretion of pro- and anti-inflammatory cytokines and chemokines, in human Schwann cells control (gray bars) and after infection with dead M. leprae (white bars) and viable M. leprae (black bars), at 24 h after stimuli in vitro, results are shown as mean ± SD from three to eight normalized independent biological replicates; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
CCL2 Correlates With Decreased Fiber in Histopathology, AFB and PCR From Nerve Fragments
We next applied a correlation matrix to analyze a potential relationship of histopathological findings and changes in gene profiling of Schwann cells in order to evaluate top molecular signature candidates during the early stages of nerve damage in leprosy patients. The overall nerve pathological findings are listed on the y-axis of Figure 5. Using this correlogram, we observed that, onion bulb axons, axonal regeneration, demyelination, and Schwann cell proliferation were positively correlated with SHH, TYRP1, ERBB2, and MAPK14 genes. With regards to remyelination appearance, this was positively correlated with NTN1, OLIG1, and UNC5B genes. Foamy macrophages correlated with IL6 and granulomas were positively correlated with GDNF, GJA1, and SHH (Figure 5). CCL2 was negatively correlated with lymphocytic, perineurial, and epineurial infiltrate and positively correlated with decreased fibers and Schwann cell proliferation (Figure 5).
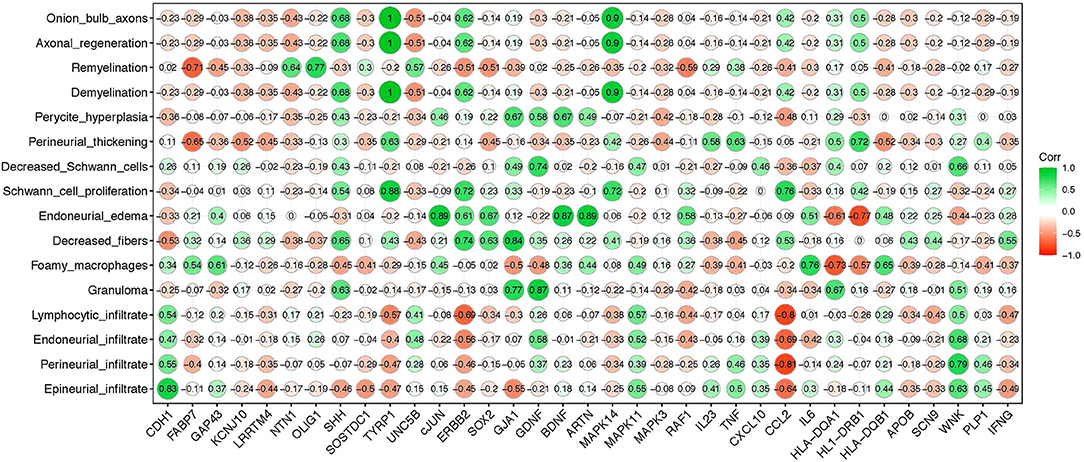
Figure 5. Correlation matrix between leprosy histopathological alterations and gene expression profile in PNL patients. The y-axis indicates major nerve histopathological alterations observed in leprosy patients; the x-axis shows Schwann cell genes evaluated in PNL patients. The green color (closer to 1 value) means stronger correlation while the red color (closer to −1 value) means weaker correlation between the gene expression and the specific histopathological alteration, respectively.
Acid Fast Bacilli (AFB) staining in nerve lesions and positive PCR are two commonly used tools for diagnosing PNL at clinics. Thus, we next searched for a potential association between the presence of the bacilli in nerve and changes in gene expression. It was statistically relevant that the expression of CCL2 and CDH1 positively correlated with AFB, while, CCL2 and GDNF expression negatively correlated with PCR (Table 2).
Discussion
The comprehension of Schwann cell-M. leprae pathological interactions is vital to fully understand disease progression in experimental models as well as in leprosy patients. Moreover, how viable and dead bacteria affect Schwann cell biology is largely unknown and poorly explored in the field. Therefore, in the present work, we used the in vitro host-pathogen interaction model to determine how viable and dead bacteria modify the global gene expression profile as soon as the bacteria comes in contact with the Schwann cells. Furthermore, we extended these early changes in the Schwann cell gene profile in response to infection to the analysis of patient nerve biopsies, establishing a potential link to the early response pattern during the disease progression in leprosy individuals.
We initially performed an exploratory, broad gene expression analysis of Schwann cells challenged with viable or dead bacteria, along with data obtained from nerve biopsies. The following families of genes involved in the nerve response to injury were investigated: (i) peripheral neuropathy, (ii) inflammatory network, (iii) Schwann cell plasticity/dedifferentiation, and (iv) nerve regeneration support.
Among the global gene expression, HLA-DRB1, MAPK14, GAP43, FABP7, NTN1, and LRRTM4 were upregulated in nerve biopsies from PNL patients and in viable and dead M. leprae-infected Schwann cells, in comparison with the respective control groups (Figure 2). It has been described that human leukocyte antigen (HLA) alleles affect the host immune response against M. leprae (36, 37). HLA-DRB1 is one of the most upregulated genes in multiple sclerosis, having a special role in inducing demyelination (38). Moreover, this transcript is consistently related to the tuberculoid leprosy (T-Lep) clinical form, while HLA-DQB1 has a strong relation with the L-Lep clinical form (39). In the present study, HLA-DQB1 was downregulated in PNL patients, in comparison with the uninfected nerve specimens (Figure 1A). Additionally, receptors for the axon guidance molecule, netrin-1 (NTN1), are expressed by Schwann cells and play a role in peripheral regeneration and induce the regeneration phenotype (40).
When evaluating the set of genes related to Schwann cell plasticity and dedifferentiation, some serine/threonine kinases, such as mitogen-activated protein kinase 14 (MAPK14), widely known for its role at the inflammasome pathways in the neuroglia (41), were found to be upregulated in PNL patients. However, there is a lack of information regarding MAPK14 in leprosy. GAP43, the major protein of the axon growth-cone that plays a role in axon growth (42) and regeneration was also upregulated, which suggests a tentative role for axonal regrowth in the injured infected nerve. SHH, WNK, and KCNJ10 were upregulated in leprosy nerve biopsies and in dead M. leprae-infected Schwann cells (Figure 2). The sonic hedgehog gene (SHH), which has been previously reported in nerve damage (43, 44), may also trigger regeneration and induce Schwann cell proliferation, as an attempt to counteract the harm caused by the bacilli (44). KCNJ10 encodes the inward-rectifying potassium channel (Kir4.1). Alteration of KCNJ10 gene expression is related to neuropathies, such as Huntington's disease East/SeSAME syndrome, by elevating the extracellular K(+), which consequently leads to abnormal neuron excitability (45, 46). It was demonstrated in mice that inflammation can silence (Kir4.1) channels, leading to hyperalgesia and trigeminal pain (47). Despite the absence of studies relating the KCNJ10 gene and M. leprae infection, it seems this may be a potential pathway to be explored for leprosy neuritis in the future. cJUN was the only gene statistically upregulated in PNL biopsies and viable M. leprae-infected Schwann cells in comparison to the respective controls (Figure 2). cJUN is a master Schwann cell regulator involved in the transition of a differentiated phenotype toward a progenitor/stem-cell like stage (48) and has also previously been reported to be upregulated, among other developmental and neural crest genes, during M. leprae infection in mouse Schwann cells (35).
PLP1, SOSTDC1, RAF1, ARTN, OLIG1, MAPK1, IFNB, and TYRP1 were upregulated in viable and dead M. leprae-infected Schwann cells, but not in the PNL biopsies. The tyrosinase-related protein 1 (TYRP1) plays a role in the melanin biosynthetic pathway, being mostly expressed by melanocytes (49). It has been documented that leprosy patients may present hyper or de-pigmented lesions due to a defective transfer of melanin (50). The melanocytes have been recently associated with the innate immune response, by producing inflammatory cytokines, such as IL-1β (50). But the role of the TYRP1 pathway in the peripheral nerve requires further investigation. It was documented that interferon beta (IFNB) is increased during cell stress conditions, but the lack of this protein leads to neuroinflammation (51, 52). IFNB is essential to start the cell fate pathway driven by NUPR1, a gene signature that contributes to a progressive infection in human cells (53). Oligodendrocyte transcription factor 1 (OLIG1) is an important protein of the remyelination phenotype, usually upregulated after a disturbance in the cell microenvironment (54). It is well described that during diabetic neuropathy, the lack of insulin leads to the demyelination phenotype due to a downregulation of OLIG1 (54). On the other hand, in this study, some genes like MAPK3 (ERK1), an important extracellular signal-regulated kinase and Schwann cell migration and proliferation molecule (55) was downregulated in PNL patients. Likewise, ErbB2, a well-known demyelination inducer and Connexin43—GJA1—were also significantly reduced in PNL patients. Despite the lack of information about GJA1 in leprosy, this has been described in neural impairment, like in the pathogenesis of Alzheimer's disease, where GJA1 downregulation leads to reduced levels of ApoE (56, 57).
While we know that the polarity of leprosy can result from the host's inflammatory response profile, multiple features of neurological involvement may also occur. For example, polymorphisms in certain genes such as IL10, ninjurin 1 and TNF have been associated with leprosy susceptibility (58–60).
In the present work, we were able to correlate the pattern of histopathological impairments with alteration in the gene expression profile induced by M. leprae infection. The correlogram analysis demonstrated, for the first time, a statistically significant correlation between the gene signature from the early stages—before fibrosis—and the histopathological alterations of the nerve damage in leprosy neuropathy. Regarding the histopathological features, TYRP1, SHH, and MAPK14 expression were strongly correlated to onion bulb axons, axon regeneration, demyelination, and Schwann cell proliferation, while MCP1/CCL2 was strongly negatively correlated with nerve inflammatory infiltrates: epineurial, endoneurial, perineurial, and lymphocytic infiltrates. It is important to realize that the weak correlation with endoneurial, perineurial, and epineurial fibrosis was due to the inclusion criteria for this study, which selected patients with no signs of fibrosis.
The biserial correlation demonstrated that the monocyte chemoattractant protein 1 (MCP1/CCL2) was positively correlated with AFB and negatively correlated with PCR, which strongly indicates the correlation of increased expression of this gene by viable bacteria. This finding corroborates a previous study by our group, which showed activation of the ESX-1 mycobacterial system by the viable M. leprae, leading to the activation of the OASL gene and the induction of CCL2, impairing the host bactericidal response, which was not observed with dead M. leprae stimuli in THP-1 cells (61). More than that, independently, Schwann cells are responsible for the triggering response of nerve damage through initiating the clearance of the debris by myelinophagy, followed by macrophage recruitment, which is especially regulated by CCL2 (62). In this sense, we see the relevance of CCL2 gene activation not only as a protective mechanism in the maintenance of mycobacterial viability, but also as a biological marker indicative of positive AFB. CCL2 has been described as one of the innate immunity genes immediately activated in the context of infection in vitro (35). Thus, despite the relevant findings, there is still a way to go in terms of understanding the role of this chemokine in infection and nerve damage during leprosy.
To strengthen the data obtained from the gene signature of M. leprae infection, we evaluated the immunomodulatory profile secreted by Schwann cells without infection and challenged with viable and dead M. leprae. We observed a pro-inflammatory profile elicited by the dead bacilli, while an anti-inflammatory microenvironment appeared to be promoted by infection with the viable bacteria. Studies have shown that macrophage infection by viable M. leprae leads to a regulatory T cell response rather than a cytotoxic T cell response, which contributes to the persistence of the infection in the host (63, 64). This characteristic is already well demonstrated in patients with L-lep, which leads to a high bacillary load. In the present work, we demonstrated that Schwann cells also behave in a way to induce the anti- or pro-inflammatory phenotype according to the bacilli viability. Since in reactional episodes there is a mixture of viable and dead bacilli, it is interesting to point out the role of the Schwann cells, and not only the macrophages, as modulators of the reaction process in the nerve, which can even lead to leprosy neuritis (14).
Conclusion
In the present study, we identified the early impact of viable and dead bacteria, independently, on modifying the gene expression profile of human Schwann cells in vitro. We also described a molecular signature associated with neural damage in early stages of pure neural leprosy from patients (i.e., HLA-DRB1, MAPK14, GAP43, FABP7, NTN1, and LRRTM4). Leprosy is a complex intricate disease and the identification of this genetic profiling may contribute for the fully understanding of leprosy neuropathy pathogenesis with the long-term goal of identifying these pathways as targets for the development of effective therapeutic strategies. We acknowledge the relatively limited number of human nerve samples, as in our experimental design we narrowed our analysis to nerve biopsies of leprosy patients with no signs of neural fibrosis and at the initial stages of neural damage. Therefore, future work with additional nerve samples from leprosy individuals, at distinct clinical stages, are important to advance this host-pathogen interactions and associated genetic analysis in larger cohorts. In summary, these results open new perspectives for the understanding of the genetic signature of neural commitment in leprosy disease.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Statement
The studies involving human participants were reviewed and approved by Oswaldo Cruz Foundation Ethics Committee (number of purports: 2.227.887). The Ethics Committee waived the requirement of written informed consent for participation.
Author Contributions
BS, RP, and BM designed and carried out experiments, collected and interpreted data, and prepared illustrations. BS, GS, RP, and BM analyzed the data. BS, MM, GS, PS-R, MOM, MJ, ES, RP, and BM reviewed and edited the manuscript. ES, RP, and BM conceptualized and designed experiments, supervised the study, interpreted data, and obtained funding. BS, RP, and BM wrote the manuscript. PS-R provided viable M. leprae. All authors read and approved the final version of the manuscript.
Funding
This study was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Fundação Oswaldo Cruz (Fiocruz) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). Dead M. leprae was obtained through BEI Resources, NIAID, NIH: Mycobacterium leprae, Strain NHDP, Gamma-Irradiated Whole Cells (lyophilized), NR-19326.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Save MP, Shetty VP, Shetty KT, Antia NH. Alterations in neurofilament protein(s) in human leprous nerves: morphology, immunohistochemistry and Western immunoblot correlative study. Neuropathol Appl Neurobiol. (2004) 30:635–50. doi: 10.1111/j.1365-2990.2004.00578.x
2. World Health Organization. Global Strategy for Further Reducing the Leprosy Burden and Sustaining Leprosy Control Activities (Plan Period: 2006-2010). Geneva: WHO (2005).
3. Save MP, Shetty VP, Shetty KT. Hypophosphorylation of NF-H and NF-M subunits of neurofilaments and the associated decrease in KSPXK kinase activity in the sciatic nerves of swiss white mice inoculated in the foot pad with Mycobacterium leprae. Lepr Rev. (2009) 80:388–401. doi: 10.47276/lr.80.4.388
4. Pinheiro RO, Schmitz V, Silva BJA, Dias AA, de Souza BJ, de Mattos Barbosa MG, et al. Innate immune responses in leprosy. Front Immunol. (2018) 9:518. doi: 10.3389/fimmu.2018.00518
5. Deps P, Collin SM. Mycobacterium lepromatosis as a second agent of Hansen's disease. Front Microbiol. (2021) 12:698588. doi: 10.3389/fmicb.2021.698588
6. Antunes SL, Chimelli L, Jardim MR, Vital RT, Nery JA, Corte-Real S, et al. Histopathological examination of nerve samples from pure neural leprosy patients: obtaining maximum information to improve diagnostic efficiency. Mem Inst Oswaldo Cruz. (2012) 107:246–53. doi: 10.1590/S0074-02762012000200015
7. Jardim M, Vital RT, Illarramendi X, Hacker M, Junqueira B, Pitta IJR, et al. The red flags of ulnar neuropathy in leprosy. PLoS One. (2021) 16:e0259804. doi: 10.1371/journal.pone.0259804
8. Scollard DM, Dacso MM, Abad-Venida ML. Tuberculosis and leprosy: classical granulomatous diseases in the twenty-first century. Dermatol Clin. (2015) 33:541–62. doi: 10.1016/j.det.2015.03.016
9. Serrano-Coll H, Salazar-Peláez L, Acevedo-Saenz L, Cardona-Castro N. Mycobacterium leprae-induced nerve damage: direct and indirect mechanisms. Pathog Dis. (2018) 76:fty062. doi: 10.1093/femspd/fty062
10. Díaz Acosta CC, Dias AA, Rosa TLSA, Batista-Silva LR, Rosa PS, Toledo-Pinto TG, et al. PGL I expression in live bacteria allows activation of a CD206/PPARγ cross-talk that may contribute to successful Mycobacterium leprae colonization of peripheral nerves. PLoS Pathog. (2018) 14:e1007151. doi: 10.1371/journal.ppat.1007151
11. Oliveira MF, Medeiros RCA, Mietto BS, Calvo TL, Mendonca APM, et al. Reduction of host cell mitochondrial activity as Mycobacterium leprea's strategy to evade host innate immunity. Immunol Rev. (2021) 301:193-208. doi: 10.1111/imr.12962
12. Spierings E, De Boer T, Zulianello L, Ottenhoff TH. Novel mechanisms in the immunopathogenesis of leprosy nerve damage: the role of Schwann cells, T cells and Mycobacterium leprae. Immunol Cell Biol. (2000) 78:349–55. doi: 10.1046/j.1440-1711.2000.00939.x
13. Spierings E, De Boer T, Zulianello L, Ottenhoff TH. The role of Schwann cells, T cells and Mycobacterium leprae in the immunopathogenesis of nerve damage in leprosy. Lepr Rev. (2000) 71(Suppl):S121-9. doi: 10.5935/0305-7518.20000083
14. Andrade PR, Jardim MR, da Silva AC, Manhaes PS, Antunes SL, Vital R, et al. Inflammatory cytokines are involved in focal demyelination in leprosy neuritis. J Neuropathol Exp Neurol. (2016) 75:272-83. doi: 10.1093/jnen/nlv027
15. Angst DBM, Pinheiro RO, Vieira JSDS, Cobas RA, Hacker MAV, Pitta IJR, et al. Cytokine levels in neural pain in leprosy. Front Immunol. (2020) 11:23. doi: 10.3389/fimmu.2020.00023
16. Masaki T, McGlinchey A, Cholewa-Waclaw J, Qu J, Tomlinson SR, Rambukkana A. Innate immune response precedes Mycobacterium leprae-induced reprogramming of adult Schwann cells. Cell Reprogram. (2014) 16:9–17. doi: 10.1089/cell.2013.0064
17. van Hooij A, Geluk A. In search of biomarkers for leprosy by unraveling the host immune response to Mycobacterium leprae. Immunol Rev. (2021) 301:175-92. doi: 10.1111/imr.12966
18. Medeiros RCA, Girardi KD, Cardoso FK, Mietto BS, Pinto TG, et al. Subversion of Schwann cell glucose metabolism by Mycobacterium leprae. J Biol Chem. (2016) 291:21375–87. doi: 10.1074/jbc.M116.725283
19. Borah K, Girardi KDCV, Mendum TA, Lery LMS, Beste DJV, Lara FA, et al. Intracellular Mycobacterium leprae utilizes host glucose as a carbon source in schwann cells. mBio. (2019) 10:e02351–19. doi: 10.1128/mBio.02351-19
20. Mattos KA, Oliveira VGC, D'Avila H, Rodrigues LS, Pinheiro RO, Sarno EN, et al. TLR6-driven lipid droplets in Mycobacterium leprae-infected schwann cells: immunoinflammatory platforms associated with bacterial persistence. J Immunol. (2011) 187:2548–58. doi: 10.4049/jimmunol.1101344
21. Rosa TLSA, Marques MAM, DeBoard Z, Hutchins K, Silva CAA, Montague CR, et al. Reductive power generated by Mycobacterium leprae through cholesterol oxidation contributes to lipid and ATP synthesis. Front Cell Infect Microbiol. (2021) 11:709972. doi: 10.3389/fcimb.2021.709972
22. Mietto BS, Andrade PR, Jardim MR, Antunes SL, Sarno EN. Demyelination in peripheral nerves: much to know from leprosy neuropathy. J Mult Scler. (2016) 3:2. doi: 10.4172/2376-0389.1000174
23. Madigan CA, Cambier CJ, Kelly-Scumpia KM, Scumpia PO, Cheng TY, Zailaa J, et al. A macrophage response to Mycobacterium leprae phenolic glycolipid initiates nerve damage in leprosy. Cell. (2017) 170:973–85.e10. doi: 10.1016/j.cell.2017.07.030
24. Mietto BS, de Souza BJ, Rosa PS, Pessolani MCV, Lara FA, Sarno EN. Myelin breakdown favours Mycobacterium leprae survival in Schwann cells. Cell Microbiol. (2020) 22:e13128. doi: 10.1111/cmi.13128
25. Petito RB, Amadeu TP, Pascarelli BM, Jardim MR, Vital RT, Antunes SL, et al. Transforming growth factor-β1 may be a key mediator of the fibrogenic properties of neural cells in leprosy. J Neuropathol Exp Neurol. (2013) 72:351–66. doi: 10.1097/NEN.0b013e31828bfc60
26. Ng V, Zanazzi G, Timpl R, Talts JF, Salzer JL, Brennan PJ, et al. Role of the cell wall phenolic glycolipid-1 in the peripheral nerve predilection of Mycobacterium leprae. Cell. (2000) 103:511–24. doi: 10.1016/S0092-8674(00)00142-2
27. Walker SL, Nicholls PG, Dhakal S, Hawksworth RA, Macdonald M, Mahat K, et al. A phase two randomised controlled double blind trial of high dose intravenous Methylprednisolone and oral prednisolone versus intravenous normal saline and oral prednisolone in individuals with leprosy type 1 reactions and/or nerve function impairment. PLoS Negl Trop Dis. (2011) 5:e1041. doi: 10.1371/journal.pntd.0001041
28. Lockwood DN, Darlong J, Govindharaj P, Kurian R, Sundarrao P, John AS. AZALEP a randomized controlled trial of azathioprine to treat leprosy nerve damage and Type 1 reactions in India: main findings. PLoS Negl Trop Dis. (2017) 11:e0005348. doi: 10.1371/journal.pntd.0005348
29. Steinhoff U, Wand-Württenberger A, Bremerich A, Kaufmann SH. Mycobacterium leprae renders Schwann cells and mononuclear phagocytes susceptible or resistant to killer cells. Infect Immun. (1991) 59:684–8. doi: 10.1128/iai.59.2.684-688.1991
30. Silva BJ, Barbosa MG, Andrade PR, Ferreira H, Nery JA, Côrte-Real S, et al. Autophagy is an innate mechanism associated with leprosy polarization. PLoS Pathog. (2017) 13:e1006103. doi: 10.1371/journal.ppat.1006103
31. Bahia El Idrissi N, Das PK, Fluiter K, Rosa PS, Vreijling J, Troost D, et al. M. leprae components induce nerve damage by complement activation: identification of lipoarabinomannan as the dominant complement activator. Acta Neuropathol. (2015) 129:653-67. doi: 10.1007/s00401-015-1404-5
32. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
33. da Silva SA, Mazini PS, Reis PG, Sell AM, Tsuneto LT, Peixoto PR, et al. HLA-DR and HLA-DQ alleles in patients from the south of Brazil: markers for leprosy susceptibility and resistance. BMC Infect Dis. (2009) 9:134. doi: 10.1186/1471-2334-9-134
34. Nogueira MRS, Amôr NG, Michellin LB, Filho NC, Rosa PS, Latini ACP, et al. Effect of Mycobacterium leprae on neurotrophins expression in human Schwann cells and mouse sciatic nerves. Mem Inst Oswaldo Cruz. (2020) 115:e200075. doi: 10.1590/0074-02760200075
35. Masaki T, Qu J, Cholewa-Waclaw J, Burr K, Raaum R, Rambukkana A. Reprogramming adult Schwann cells to stem cell-like cells by leprosy bacilli promotes dissemination of infection. Cell Vol. (2013) 152:51–67. doi: 10.1016/j.cell.2012.12.014
36. de Souza-Santana FC, Marcos EV, Nogueira ME, Ura S, Tomimori J. Human leukocyte antigen class I and class II alleles are associated with susceptibility and resistance in borderline leprosy patients from Southeast Brazil. BMC Infect Dis. (2015) 15:22. doi: 10.1186/s12879-015-0751-0
37. Dallmann-Sauer M, Fava VM, Gzara C, Orlova M, Van Thuc N, Thai VH, et al. The complex pattern of genetic associations of leprosy with HLA class I and class II alleles can be reduced to four amino acid positions. PLoS Pathog. (2020) 16:e1008818. doi: 10.1371/journal.ppat.1008818
38. Simon Enz L, Zeis T, Schmid D, Geier F, van der Meer F, Steiner G, et al. Increased HLA-DR expression and cortical demyelination in MS links with HLA-DR15. Neurol Neuroimmunol Neuroinflamm. (2019) 7:e656. doi: 10.1212/NXI.0000000000000656
39. Marcos EVC, de Souza FC, Ura S, Opromolla DVA. Study of the association of HLA antigens and ulcerated type 1 leprotic reaction. An Bras Dermatol. (2000) 75:282–90. https://dx.doi.org/10.1186%2Fs12879-015-0751-010.1186/s12879-015-0751-0
40. Dun XP, Parkinson DB. Role of Netrin-1 signaling in nerve regeneration. Int J Mol Sci. (2017) 8:491. doi: 10.3390/ijms18030491
41. She H, He Y, Zhao Y, Mao Z. Release the autophage brake on inflammation: the MAPK14/p38α-ULK1 pedal. Autophagy. (2018) 14:1097-8. doi: 10.1080/15548627.2018.1446626
42. Kawasaki A, Okada M, Tamada A, Okuda S, Nozumi M, Ito Y, et al. Growth cone phosphoproteomics reveals that GAP-43 phosphorylated by JNK is a marker of axon growth and regeneration. iScience. (2018) 4:190–203. doi: 10.1016/j.isci.2018.05.019
43. Benito C, Davis CM, Gomez-Sanchez JA, Turmaine M, Meijer D, Poli V, et al. STAT3 controls the long-term survival and phenotype of repair schwann cells during nerve regeneration. J Neurosci. (2017) 37:4255-69. doi: 10.1523/JNEUROSCI.3481-16.2017
44. Wagstaff LJ, Gomez-Sanchez JA, Fazal SV, Otto GW, Kilpatrick AM, Michael K, et al. Failures of nerve regeneration caused by aging or chronic denervation are rescued by restoring Schwann cell c-Jun. Elife. (2021) 10:e62232. doi: 10.7554/eLife.62232
45. Tong X, Ao Y, Faas GC, Nwaobi SE, Xu J, Haustein MD. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington's disease model mice. Nat Neurosci. (2014) 17:694–703. doi: 10.1038/nn.3691
46. Morin M, Forst AL, Pérez-Torre P, Jiménez-Escrig A, Barca-Tierno V, García-Galloway E, et al. Novel mutations in the KCNJ10 gene associated to a distinctive ataxia, sensorineural hearing loss and spasticity clinical phenotype. Neurogenetics. (2020) 21:135–43. doi: 10.1007/s10048-020-00605-6
47. Takeda M, Takahashi M, Nasu M, Matsumoto S. Peripheral inflammation suppresses inward rectifying potassium currents of satellite glial cells in the trigeminal ganglia. Pain. (2011) 152:2147–56. doi: 10.1016/j.pain.2011.05.023
48. Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. (2005) 6:671–82. doi: 10.1038/nrn1746
49. Dennis MK, Mantegazza AR, Snir OL, Tenza D, Acosta-Ruiz A, Delevoye C, et al. BLOC-2 targets recycling endosomal tubules to melanosomes for cargo delivery. J Cell Biol. (2015) 209:563-77. doi: 10.1083/jcb.201410026
50. Gasque P, Jaffar-Bandjee MC. The immunology and inflammatory responses of human melanocytes in infectious diseases. J Infect. (2015) 71:413–21. doi: 10.1016/j.jinf.2015.06.006
51. Ejlerskov P, Hultberg JG, Wang J, Carlsson R, Ambjørn M, Kuss M, et al. Lack of neuronal IFN-β-IFNAR causes Lewy body- and Parkinson's disease-like dementia. Cell. (2015) 163:324–39. doi: 10.1016/j.cell.2015.08.069
52. Inácio AR, Liu Y, Clausen BH, Svensson M, Kucharz K, Yang Y, et al. Endogenous IFN-β signaling exerts anti-inflammatory actions in experimentally induced focal cerebral ischemia. J Neuroinflammation. (2015) 12:211. doi: 10.1186/s12974-015-0427-0
53. Andrade PR, Mehta M, Lu J, Teles RMB., Montoya D, Scumpia PO, et al. The cell fate regulator NUPR1 is induced by Mycobacterium leprae via type I interferon in human leprosy. PLoS Negl Trop Dis. (2019) 13:e0007589. doi: 10.1371/journal.pntd.0007589
54. Schmid D, Zeis T, Schaeren-Wiemers N. Transcriptional regulation induced by cAMP elevation in mouse Schwann cells. ASN Neuro. (2014) 6:137–57. doi: 10.1042/AN20130031
55. Mantuano E, Lam MS, Shibayama M, Campana WM, Gonias SL. The NMDA receptor functions independently and as an LRP1 co-receptor to promote Schwann cell survival and migration. J Cell Sci. (2015) 128:3478-88. doi: 10.1242/jcs.173765
56. Kajiwara Y, Wang E, Wang M, Sin WC, Brennand K, Schadt E, et al. GJA1 (connexin43) is a key regulator of Alzheimer's disease pathogenesis. Acta Neuropathol Commun. (2018) 6:144. doi: 10.1186/s40478-018-0642-x
57. Mentis AA, Dardiotis E, Chrousos GP. Apolipoprotein E4 and meningeal lymphatics in Alzheimer disease: a conceptual framework. Mol Psychiatry. (2021) 26:1075-97. doi: 10.1038/s41380-020-0731-7
58. Cardoso CC, Martinez AN, Guimarães PEM, Mendes CT, Pacheco AG, et al. Ninjurin 1 asp110ala single nucleotide polymorphism is associated with protection in leprosy nerve damage. J Neuroimmunol. (2007) 190:131–8. doi: 10.1016/j.jneuroim.2007.07.015
59. Tarique M, Naqvi RA, Santosh KV, Kamal VK, Khanna N, Rao DN. Association of TNF-α-308: IL-10–819: IL-10–1082(GG) and IL-1R1+1970(CC) genotypes with the susceptibility and progression of leprosy in North Indian population. Cytokine. (2015) 73:61–5. doi: 10.1016/j.cyto.2015.01.014
60. Tarique M, Naz H, Saini C, Suhail M, Shankar H, Khanna N, et al. Association of IL-10 gene polymorphism with IL-10 secretion by CD4 and T regulatory cells in human leprosy. Front Immunol. (2020) 11:1974. doi: 10.3389/fimmu.2020.01974
61. de Toledo-Pinto TG, Ferreira AB, Ribeiro-Alves M, Rodrigues LS, Batista-Silva LR, Silva BJ, et al. STING-dependent 2'-5' oligoadenylate synthetase-like production is required for intracellular Mycobacterium leprae survival. J Infect Dis. (2016) 214:311–20. doi: 10.1093/infdis/jiw144
62. Medeiros MF, Rodrigues MM, Vital RT, da Costa Nery JA, Sales AM, de Andrea Hacker M, et al. CXCL10, MCP-1, and other immunologic markers involved in neural leprosy. Appl Immunohistochem Mol Morphol. (2015) 23:220–9. doi: 10.1097/PAI.0000000000000074
63. Montoya D, Cruz D, Teles RM, Lee DJ, Ochoa MT, Krutzik SR, et al. Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe. (2009) 6:343-53. doi: 10.1016/j.chom.2009.09.002
Keywords: leprosy, Mycobacterium leprae, peripheral nervous system, Schwann cell, host-pathogen interaction
Citation: Souza BJd, Mendes MA, Sperandio da Silva GM, Sammarco-Rosa P, Moraes MOd, Jardim MR, Sarno EN, Pinheiro RO and Mietto BS (2022) Gene Expression Profile of Mycobacterium leprae Contribution in the Pathology of Leprosy Neuropathy. Front. Med. 9:861586. doi: 10.3389/fmed.2022.861586
Received: 24 January 2022; Accepted: 14 March 2022;
Published: 15 April 2022.
Edited by:
Jessica L. Jones, United States Food and Drug Administration, United StatesReviewed by:
Héctor Serrano, CES University, ColombiaJuliana De Moura, Federal University of Paraná, Brazil
Huma Naz, University of Missouri, United States
Tarun Narang, Postgraduate Institute of Medical Education and Research, India
Copyright © 2022 Souza, Mendes, Sperandio da Silva, Sammarco-Rosa, Moraes, Jardim, Sarno, Pinheiro and Mietto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bruno Siqueira Mietto, YnJ1bm8ubWlldHRvQHVmamYuZWR1LmJy
†These authors have contributed equally to this work and share senior authorship
 Beatriz Junqueira de Souza
Beatriz Junqueira de Souza Mayara Abud Mendes
Mayara Abud Mendes Gilberto Marcelo Sperandio da Silva
Gilberto Marcelo Sperandio da Silva Patrícia Sammarco-Rosa
Patrícia Sammarco-Rosa Milton Ozorio de Moraes
Milton Ozorio de Moraes Marcia Rodrigues Jardim
Marcia Rodrigues Jardim Euzenir Nunes Sarno1
Euzenir Nunes Sarno1 Roberto Olmo Pinheiro
Roberto Olmo Pinheiro Bruno Siqueira Mietto
Bruno Siqueira Mietto