- 1Key Laboratory of Gastrointestinal Cancer (Fujian Medical University), Ministry of Education, Fuzhou, China
- 2Fujian Key Laboratory of Tumor Microbiology, Department of Medical Microbiology, Fujian Medical University, Fuzhou, China
Helicobacter pylori infection is a significant risk factor for various gastrointestinal diseases, while the standard triple therapy for its eradication is increasingly compromised by antibiotic resistance. This study investigates the role of the CrdAB-CzcBA efflux pump and its regulation by copper in tetracycline resistance in H. pylori. Using minimum inhibitory concentration (MIC) determination and growth curve analysis, we found that the deletion of crdA or czcA significantly reduced tetracycline resistance, while overexpression of CrdAB-CzcBA under the urease promoter enhanced bacterial resistance by reducing intracellular tetracycline accumulation. Ethidium bromide and tetracycline accumulation assays confirmed that CrdAB-CzcBA mediates active efflux of tetracycline, contributing to reduced intracellular drug levels. Furthermore, copper supplementation upregulated the expression of CrdAB-CzcBA via the CrdRS two-component system, thereby promoting bacterial growth under tetracycline stress. Notably, copper-induced resistance was abrogated in ΔcrdR mutants, demonstrating the dependence of this mechanism on CrdRS. These findings highlight CrdAB-CzcBA as a critical efflux system in tetracycline resistance and emphasize the role of environmental factors, such as copper, in modulating bacterial antibiotic resistance, underscoring the need for strategies that account for metal ion influences in managing H. pylori infections.
1 Introduction
Gastric cancer causes more than 720,000 deaths per year worldwide and has become a global health threat (1). Infection with Helicobacter pylori, a Gram-negative human pathogen that infects approximately 50% of the world’s population, is a major risk factor for gastric cancer. H. pylori is closely related to the development of gastrointestinal diseases, including gastritis, peptic ulcers, gastric cancer, and mucosa-associated lymphoid tissue lymphoma (MALT) (2). To eradicate H. pylori, a standard triple therapy is recommended using a proton pump inhibitor and two of five antibiotics, including amoxicillin, tetracycline, clarithromycin, metronidazole, and levofloxacin (3–5). Currently, the commonly recommended first-line regimen is bismuth quadruple therapy, which includes tetracycline. Thus, the effectiveness of this regimen may be affected by tetracycline efflux mechanisms such as CrdAB-CzcBA. However, the eradication rate has been challenged owing to the increasing drug resistance rate worldwide (6).
Tetracycline acts as a protein synthesis inhibitor of both gram-positive and gram-negative bacteria by binding to the 30S subunit of ribosomes, thus inhibiting aminoacyl-tRNA binding (7). Tetracycline resistance is mainly caused by alterations in 16S rRNA. For clinically isolated tetracycline-resistant H. pylori strains, an AGA to TTC (926–928) triple-base-pair mutation preferentially occurs, whereas single or two-base-pair substitution induces only moderate levels of resistance (8). A study also showed that AGA to TTC (965–967) mutations also caused high levels of tetracycline resistance in H. pylori clinical isolates (9). However, some clinical isolates containing no mutations in 16S rRNA genes also showed tetracycline resistance, suggesting that other mechanisms might also be involved in tetracycline resistance in H. pylori (10).
In some gram-negative bacteria, tetracycline resistance is induced through TetA, an efflux protein (11, 12). TetA expression is induced by tetracycline through the transcriptional regulator TetR, which acts by directly binding to the promoter of the tetA gene (13). In H. pylori, the protein sequence of HP1165 showed similarity to the TetA of Clostridium perfringens, and is involved in intermediate resistance and inducible tetracycline resistance (14). However, if other efflux pumps are involved in tetracycline resistance, further investigation is required.
Drug resistance of pathogens can be either acquired or intrinsic. There are various intrinsic drug-resistant strategies, and efflux pumps play an important role in the drug resistance of gram-negative bacteria (15). There are five categories of efflux pumps: the major facilitator superfamily (MFS), the ATP-binding cassette (ABC) superfamily, the multidrug and toxic compound extrusion (MATE) family, the small multidrug resistance (SMR) family, and the resistance-nodulation-division (RND) superfamily (16, 17). The RND family has been shown to be involved in multidrug resistance of gram-negative bacteria (18, 19). The RND efflux pump is a protein complex consisting of three proteins: bacterial plasma membrane active transporters, membrane fusion proteins, and outer membrane factors. RND efflux pumps, such as MexAB-Oprm in Pseudomonas aeruginosa and AcrAB-TolC in Escherichia coli have been thoroughly studied previously (20–22). It has been reported that there are only four TolC homologous genes, including HP0605, HP0971, HP1327, and HP1489 in the H. pylori 26,695 genome, comprising four RND efflux systems: HP0605-0607, HP0971-0969, HP1326-1329 and HP1487-1489 (23, 24). The relationship between these efflux systems and drug resistance has been studied in different groups (23, 25–27).
CrdAB-CzcBA (HP1326-1329) comprises an RND efflux pump in H. pylori and is involved in copper extrusion. It consists of four main components: HP1326 (CrdA), a secreted protein crucial for maintaining cytoplasmic copper homeostasis; HP1327 (CrdB), a putative outer membrane protein believed to function as the efflux channel for metal ions; HP1328 (CzcB homolog), an inner membrane protein likely involved in substrate recognition and transport across the inner membrane; and HP1329 (CzcA homolog), another inner membrane protein thought to be responsible for the active transport of copper, possibly using the proton motive force. CrdAB-CzcBA is required for resistance to high concentrations of copper (28). The expression of CrdAB-CzcBA is induced by copper through a two-component CrdRS system (29). Copper ions play an important role in bacterial metabolism; for example, copper acts as a cofactor of enzymes involved in superoxide dismutase and cytochrome c oxidase (30, 31). However, copper is required at a low concentration, while excess copper is toxic to bacteria by generating reactive oxygen species, thus causing cellular damage (30). Copper plays an important role in the pathogenesis of H. pylori, and studies have shown that it promotes H. pylori colonization of the gastric mucosa, while copper toxicity is also employed by macrophages to eliminate bacteria through phagosomes (32, 33). In this study, we have shown here that CrdAB-CzcBA is involved in the efflux activity of extruding tetracycline in H. pylori and, consequently, the resistance to tetracycline. We have also found that copper enhances the resistance of H. pylori to tetracycline through activation of CrdAB-CzcBA.
2 Materials and methods
2.1 Strains and growth conditions
H. pylori strains, including Hp26695, and the clinical isolates HpFZ068 and HpFZ169, were used in this study. H. pylori strains were cultured under a microaerobic environment (5% O2, 10% CO2, 85% N2) at 37°C. Bacteria were cultured on Columbia blood agar plates (OXOID, Thermo Fisher Scientific, UK) containing 5% sheep blood or in Brucella broth (Becton Dickinson, Sparks, MD, USA) supplemented with 10% fetal bovine serum (FBS, PAN-Biotech, Aidenbach, Germany) (BB + FBS) with agitation at a speed of 120 rpm. Kanamycin (MP Biomedicals, LLC, USA) (10 μg/mL) and chloramphenicol (MP Biomedicals, LLC, USA) (20 μg/mL) were supplied as needed. E. coli DH5α was grown in Luria-Bertani medium at 37°C.
2.2 Construction of ΔczcA, ΔcrdA and ΔcrdR isogenic mutant strains of H. pylori
To construct a ΔczcA isogenic mutant of H. pylori 26,695, primers CzcA-upF and CzcA-upR were used to amplify the upstream sequence of czcA from H. pylori 26,695 genomic DNA. Primers CzcA-downF and CzcA-DownR were used to amplify the downstream sequence of czcA. The kanamycin-resistant genes were amplified using the primers AphA-F and AphA-R. PCR reactions were performed using PrimeSTAR HS DNA Polymerase (Takara, Beijing, China). These DNA fragments were cloned into the pBluescript SK II (−) vector (Stratagene, La Jolla, CA, USA) using the ClonExpress® Ultra One Step Cloning Kit (Vazyme, Nanjing, China), generating pBlue-CzcAKO, which was then transformed into E. coli DH5α. After confirmation by colony PCR and DNA sequencing, pBlue-CzcAKO was transformed into H. pylori 26,695 by electroporation. Bacteria were selected using Columbia blood agar plates containing kanamycin (3 μg/mL), and ΔczcA was confirmed by colony PCR and DNA sequencing. CzcA-upF, CzcA-upR, CzcA-downF, and CzcA-downR were used for the construction of HpFZ068ΔczcA and HpFZ169ΔczcA, and the experiments were performed as described above. To construct ΔcrdR, primers CrdR-upF, CrdR-upR, CrdR-downF, and CrdR-downR were used. To construct ΔcrdA, primers CrdA-upF, CrdA-upR, CrdA-downF, and CrdA-downR were used. Primers used in this study were listed in Supplementary Table 1.
2.3 Construction of crdAB-czcBAhe, crdAB-ΔczcAhe and Hp26695chl
To facilitate the substitution of the crdAB-czcBA promoter with the ureAB promoter, the urease promoter fragment was obtained via amplification with primers PureAB-F and PureAB-R. Concurrently, the chloramphenicol resistance cassette (CAT) was amplified using primer pairs ChlR-F and ChlR-R. DNA segments harboring the upstream region of the crdA promoter were amplified with the primer pairs CrdAhe-upF and CrdAhe-upR, whereas downstream sequences were amplified using CrdAhe-downF and CrdAhe-downR. These fragments were subsequently ligated into the pBluescript SK II (−) vector, resulting in the construction of pBlue-CrdAhe. This construct was then introduced into H. pylori 26,695 and ΔczcA strains and selected on Columbia blood agar plates containing chloramphenicol (4 μg/mL), yielding the crdAB-czcBAhe and crdAB-ΔczcAhe strains, respectively. For the generation of chloramphenicol-resistant Hp26695 (Hp26695chl), the upstream and downstream sequences flanking the crdA promoter were amplified using the primer sets CrdAhe-upF, CrdAhe-upR1, CrdAhe-downF1, and CrdAhe-downR, and the CAT was amplified with ChlR-F and ChlR-R. These amplified products were cloned into pBluescript SK II (−), creating pBlue-ChlR, which was subsequently transformed into H. pylori 26,695. Primers utilized in this study are itemized in Supplementary Table 1.
2.4 Determination of antibiotic susceptibility
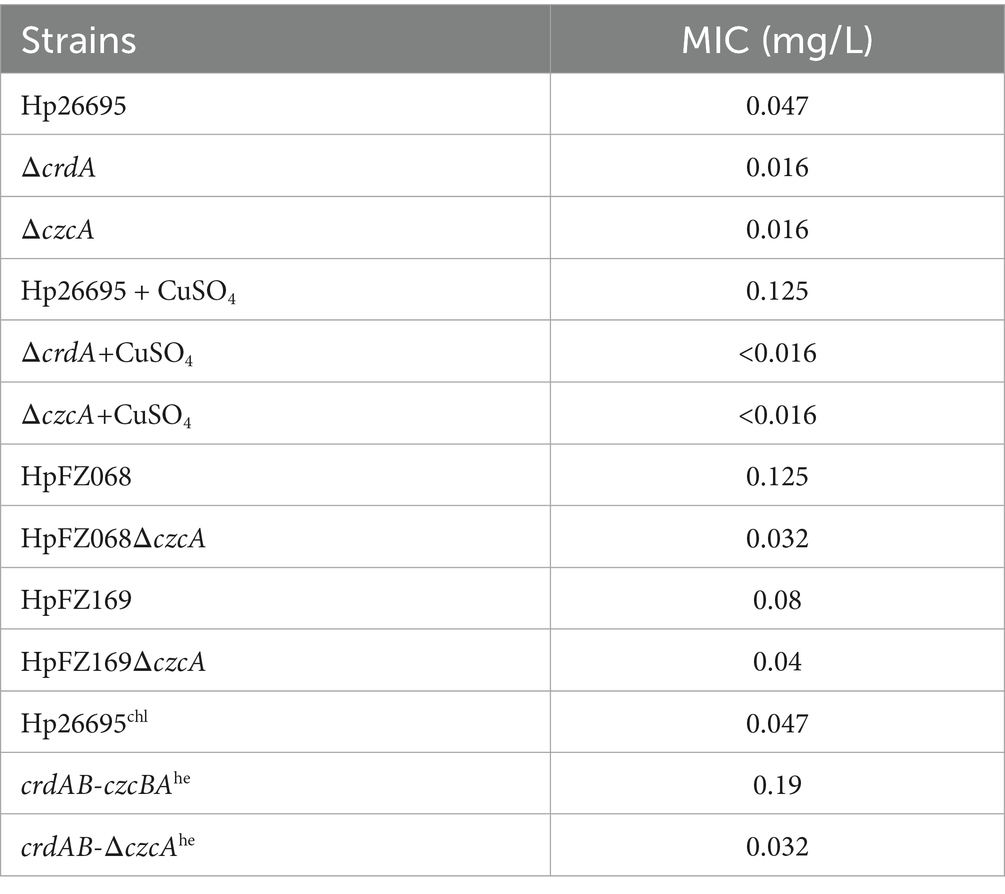
H. pylori wild type strain and clinical isolates were cultivated on Columbia Blood agar plates containing 5% sheep blood for 3 days. Then the bacterial cells were collected and resuspended in Brucella broth, and 100 μL of cell suspension with 2 McFarland standard were spread onto Mueller-Hinton agar plates (OXOID, Thermo Fisher Scientific, UK) containing 5% sheep blood. The minimum inhibitory concentration (MIC) of each strain to amoxicillin, tetracycline, clarithromycin, metronidazole, and levofloxacin were determined by MIC test strips (MTS, Liofilchem, Italy) which were placed onto the inoculated plates. The plates were incubated at 37°C under microaerobic conditions, and the MIC values were determined after 72 h. Each MIC value represents the average from three independent experiments.
2.5 RNA extraction, reverse transcription and qPCR assay
To quantify the expression of efflux pumps stimulated by copper, an overnight culture of H. pylori 26,695 was resuspended in BB + FBS with an initial OD600 of 0.2, with or without 50 μM CuSO4. Bacterial culture was maintained for 4 h and RNA was extracted as described above. All samples were analyzed in at least three biological replicates. Bacterial RNA was extracted using the TRIzol® Reagent (Life Technologies, Grand Island, NY, USA) according to the manufacturer’s protocol, and reverse transcription was performed using the HiScript®II Q RT SuperMix for qPCR kit (Vazyme, Nanjing, China) with 0.5 μg of RNA in a 20 μL reaction system. For quantitative real-time PCR (qPCR), SYBR qPCR Master Mix (Vazyme, Nanjing, China) was used, and the primers for each gene are listed in Supplementary Table 1. The 16S rRNA amplicon was used as an endogenous control, and relative mRNA levels were determined using the 2−ΔΔCt method.
2.6 Monitoring the growth of H. pylori influenced by etracycline and copper
To monitor the growth curves of H. pylori, bacteria were first cultured for 24 h in BB + FBS. Then, bacterial cells were resuspended in fresh BB + FBS at an initial OD600 of 0.1, with or without supplementation of antibiotics or CuSO4. Tetracycline (0.023 μg/mL) was supplemented at the subinhibitory concentration (0.5 × MIC), and 50 μΜ CuSO4 was supplemented as indicated. Bacterial culture was maintained for 48–72 h, and the OD600 of the culture was monitored at the indicated time points. Each experiment was independently performed at least three times.
2.7 Ethidium bromide accumulation assay and tetracycline accumulation assay
Ethidium bromide (EB) accumulation assay was performed as previously described (25). In brief, H. pylori cells were resuspended in fresh BB + FBS with an initial OD600 of 0.1 and cultured in BB + FBS until the exponential phase (OD600 = 0.6). The cells were subsequently washed twice with PBS (pH = 7.0), and the bacterial cells were harvested and resuspended in 100 μL PBS (OD600 = 0.4) in a 96-well plate (Corning3603, Carlsbad, CA, USA). Then, 100 μL of EB was added at a final concentration of 10 μg/mL. Fluorescence was measured using an EnSight™ Multimode Plate Reader (PerkinElmer, Waltham, MA, USA) under room temperature, with an excitation wavelength of 530 nm and an emission wavelength of 590 nm (34). Luminescence was recorded every 60 s for 30 min. Each experiment was performed in triplicate, and values are shown as the averages of three independent experiments.
For the tetracycline accumulation assay, the experiments were performed as described previously (35). Briefly, H. pylori strains were cultured overnight in BB + FBS until the exponential growth phase. Then, 0.8 × 109 cells/sample were washed twice with 2 mL of Mg2+ buffer (50% methanol, 10 mM Tris–HCl, 0.1 mM MgCl2, and 0.2% glucose, pH 8.0), and the bacterial cells were resuspended in 2 mL Mg2+ buffer containing tetracycline (100 μg/mL) and incubated for 15 min (36). After centrifugation, the cells were collected and resuspended in 2 mL Mg2+ buffer. For measurement of tetracycline accumulation, 100 μL of each sample was added to a 96-well black plate (Corning, Carlsbad, CA, USA), and fluorescence was measured using an EnSight™ Multimode Plate Reader (PerkinElmer, Waltham, MA, USA) under room temperature, with excitation and emission wavelengths of 400 and 520 nm, respectively (37).
2.8 Statistical analysis
Data are presented as the mean ± standard error. To assess data significance, an unpaired t-test was used to study the degree of statistical analysis of the two groups. Statistical significance was set at p < 0.05. GraphPad Prism software (version 8.0) was used to analyze the results.
3 Results
3.1 CrdAB-CzcBA contributes to the tetracycline resistance of H. pylori
To verify whether the RND efflux pump CrdAB-CzcBA is involved in drug resistance, we constructed isogenic ΔcrdA and ΔczcA mutants. We investigated the MICs of antibiotics in the H. pylori wild type, ΔcrdA, ΔczcA (Table 1; Supplementary Table S2). We found that MIC values of levofloxacin, metronidazole, clarithromycin and amoxicillin showed no significant difference between the wild type strain and mutant strains. However, the MIC of tetracycline in H. pylori ΔcrdA and ΔczcA was 0.016 mg/L, compared to 0.047 mg/L in the wild type strain Hp26695. This reduction was also observed in clinical isolates: the MIC of HpFZ068ΔczcA was 0.032 mg/L versus 0.125 mg/L in the parental strain HpFZ068; and for HpFZ169ΔczcA, the MIC was 0.04 mg/L compared to 0.08 mg/L in the parental strain HpFZ169. To confirm this result, we measured the growth curves of H. pylori 26,695 wild type, ΔcrdA, and ΔczcA mutants cultivated in Brucella broth with or without tetracycline (0.023 mg/L, 0.5 × MIC). Knockout of crdA or czcA had no effect on the growth of the wild type H. pylori in the absence of antibiotics (Figure 1A). In the presence of tetracycline, the growth of H. pylori strains was attenuated, however, ΔcrdA and ΔczcA exhibited a higher sensitivity to tetracycline compared to the wild type, with a more retarded growth (Figure 1B). These results certificated that CrdAB-CzcBA is involved in tetracycline resistance in H. pylori.
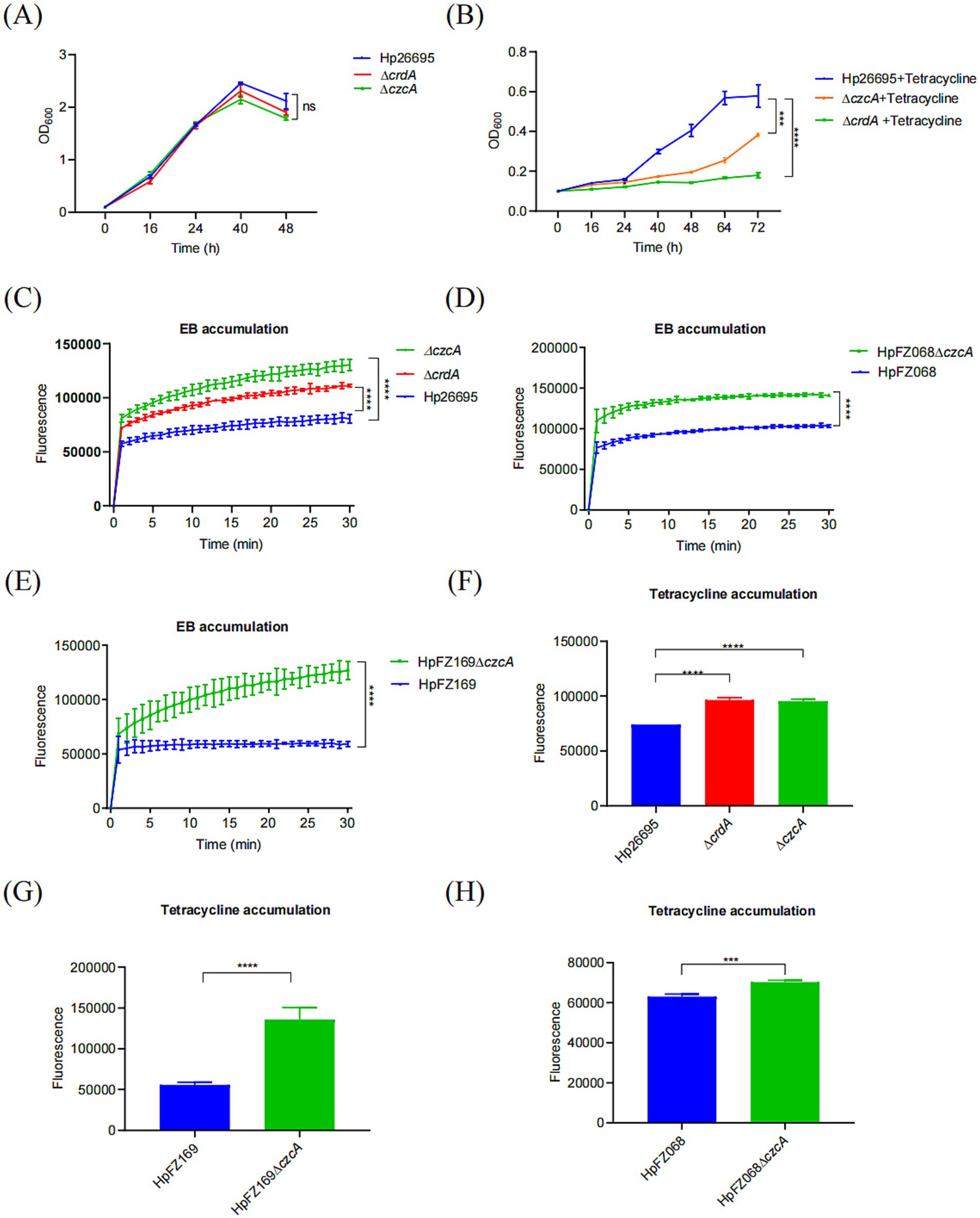
Figure 1. CrdAB-CzcBA contributes to the tetracycline resistance of H. pylori. (A,B) Growth curves for Hp26695, ΔcrdA, ΔczcA in the presence (A) or with (B) 0.023 μg/mL tetracycline. (C–E) EB accumulation measured by the fluorescence intensity, indicating active efflux in Hp26695, ΔcrdA, ΔczcA. Clinical isolates HpFZ068 and HpFZ169, along with their respective ΔczcA mutants, were included for comparison. Fluorescence intensity was recorded at 30-s intervals over 30 min. (F–H) Tetracycline accumulation assays performed for Hp26695, ΔcrdA, ΔczcA, clinical isolates HpFZ068 and HpFZ169, and their ΔczcA mutants. Each value represents the mean ± standard deviation from three independent experiments, and error bars represent the standard deviation. Statistical significance is indicated as ***p < 0.001; ****p < 0.0001; ns, non-significance.
To further investigate the role of CrdAB-CzcBA in tetracycline resistance within H. pylori, efflux activity was assessed through ethidium bromide (EB) accumulation assays. These assays revealed increased EB accumulation in the ΔczcA and ΔcrdA mutants compared to the wild type, indicating a deficit in efflux activity (Figure 1C). This pattern was also observed in two additional clinical isolates, HpFZ068 and HpFZ169, where the ΔczcA mutants accumulated more EB than their respective parent strains (Figures 1D,E). To more directly assess tetracycline efflux activity, we performed tetracycline accumulation assays. Similarly, the ΔcrdA and ΔczcA strains accumulated greater amounts of tetracycline than the wild type, suggesting CrdAB-CzcBA is involved in tetracycline efflux (Figure 1F). These findings were consistent with the clinical isolate strains, which demonstrated increased tetracycline accumulation in the ΔczcA mutants compared to their parental strains (Figures 1G,H), supporting the role of CrdAB-CzcBA in mediating tetracycline efflux and contributing to resistance by diminishing intracellular antibiotic concentrations.
3.2 Elevated expression of CrdAB-CzcBA significantly enhanced tetracycline resistance
A previous study comparing the MIC difference between ΔczcA and wild type suggested that the efflux pump CrdAB-CzcBA was not involved in antibiotic resistance in H. pylori 1,061 strain (23). However, we suspected this might be due to its low expression level under normal laboratory culture condition. So we replaced its native promoter with the robust urease (coded by ureAB) promoter in both Hp26695 wild type and ΔczcA strains, hypothesizing that augmented expression would correlate with increased resistance, aiming to provide a direct link between the overexpression of CrdAB-CzcBA efflux pump genes and enhanced antibiotic resistance phenotypes (Figure 2A). The increased expression levels of CrdA and CzcA were verified, with marked elevation under the ureAB promoter compared to the Hp26695 strain containing only the chloramphenicol resistance gene (Figure 2B). Furthermore, the overexpression of CrdAB-CzcBA did not significantly affect bacterial growth under normal cultivation conditions (Figure 2C). When challenged with tetracycline, CrdAB-CzcBA overexpression substantially improved bacterial growth, an effect not observed in the ΔczcA background (Figure 2D). We have also confirmed this result by comparing the MICs of Hp26695chl, crdAB-czcBAhe and crdAB-ΔczcAhe strains (Table 1). The results showed that MIC of tetracycline in H. pylori crdAB-czcBAhe, but not crdAB-ΔczcAhe, was 4-fold higher than that in Hp26695 wild type strain containing chloramphenicol resistance cassette (Hp26695chl). Further analysis of EB accumulation and tetracycline accumulation showed that strains overexpressing crdAB-czcBA (crdAB-czcBAhe) showed reduced EB and tetracycline accumulation levels compared to the control Hp26695chl strain, yet there was no significant difference between the crdAB-ΔczcBAhe strain and the ΔczcA strain (Figures 2E,F), certified that induction of CrdAB-CzcBA resulted in the significant increase in its efflux capacity. Collectively, these data certified that when expression of CrdAB-CzcBA was activated, H. pylori showed enhanced efflux capacity and resistance to tetracycline.
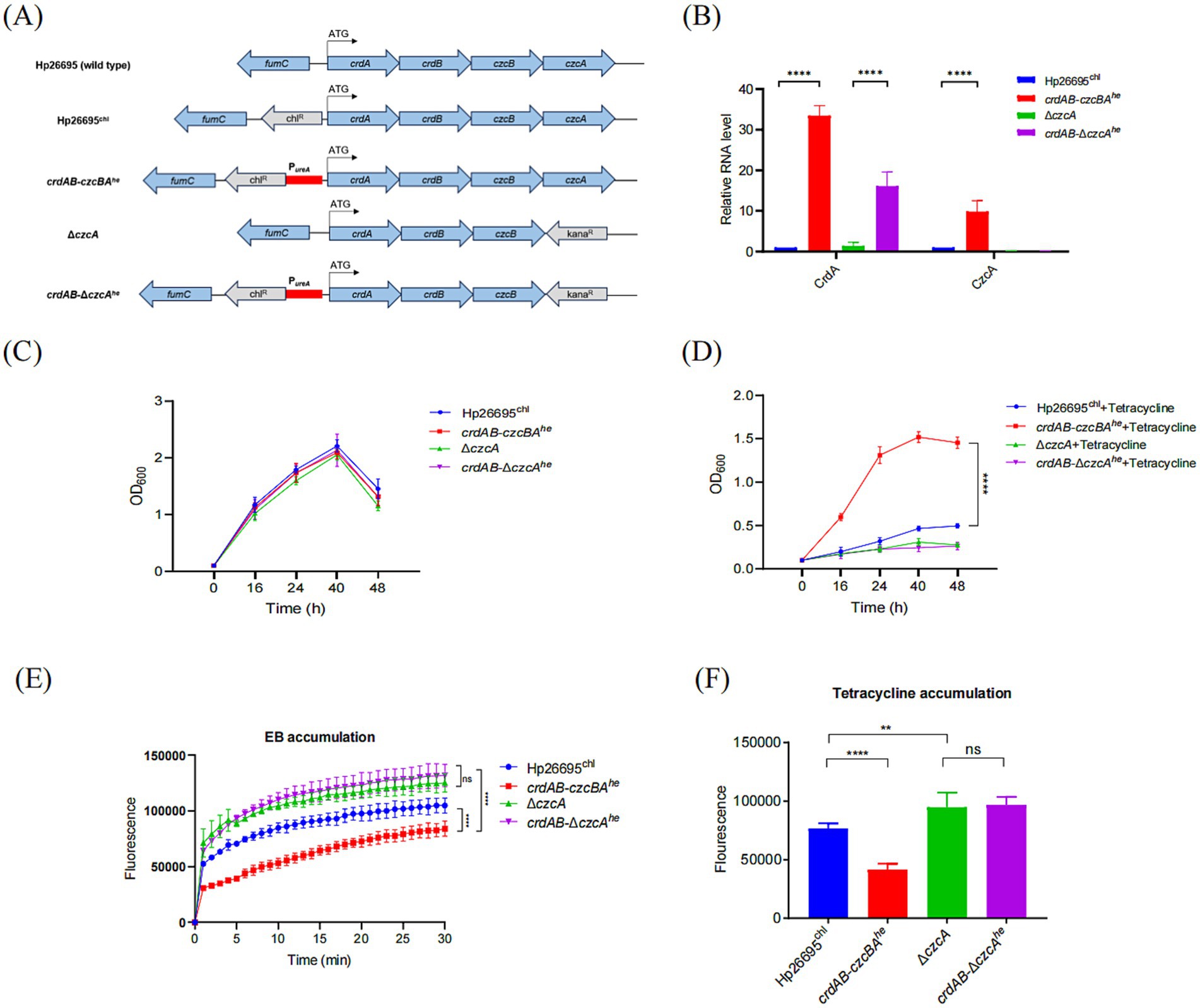
Figure 2. Overexpression of CrdAB-CzcBA enhances resistance to tetracycline. (A) Schematic representation of the constructs for high-expression strains derived from Hp26695, utilizing the urease promoter region (red bar), antibiotic resistance genes (gray), and H. pylori genes (blue). Start codon is indicated by arrows. (B) qPCR quantification of CrdA and CzcA mRNA in Hp26695chl, crdAB-czcBAhe, ΔczcA, and crdAB-ΔczcAhe, normalized to Hp26695chl. (C,D) Growth profiles in the absence (C) and presence (D) of tetracycline. (E,F) EB accumulation assays (E) and tetracycline accumulation assays (F) performed for Hp26695chl, ΔczcA, overexpression strains crdAB-czcBAhe and crdAB-ΔczcAhe. Each value represents the mean ± standard deviation of three independent experiments. Error bars represent standard deviation. Statistical significance is indicated as **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, non-significance.
3.3 Copper enhances bacterial growth under tetracycline in H. pylori
The expression of CrdAB-CzcBA is induced by copper, we then confirmed this result, showing that the expression of CzcA was greatly enhanced by copper with expression upregulated more than 14-folds (Figure 3A). We have also investigated the expression of the nine genes representing all the other efflux pumps reported in H. pylori, including ABC transporter family proteins (HP1206, HP1082, and HP0600) (38), MFS family proteins (HP1181and HP1174) (39, 40), MATE family protein (HP1184) (23), and RND family proteins (HP0607, HP0969, HP1487). The expression of HP1082, HP1206, HP1174, HP0607, HP0969, and HP1487 was not influenced by copper, while the expression of HP0600, HP1181, and HP1184 was 78, 95, and 71% higher, respectively, in the presence of copper. These results suggest that only CrdAB-CzcBA is significantly activated by copper. To verify whether copper enhances tetracycline resistance through activating expression of CrdAB-CzcBA, we cultivated H. pylori 26,695, ΔcrdA, and ΔczcA mutants in the presence or absence of 50 μM CuSO4 and tetracycline. As expected, supplementation with 50 μM CuSO4 inhibited the growth of the ΔcrdA and ΔczcA strain but had no effect on growth of the wild type strain (Figure 3B). In the presence of tetracycline, the addition of copper promoted the growth of wild type strain, but had no effect on the growth of ΔcrdA. Compared to ΔcrdA, H. pylori mutant ΔczcA showed a similar phenotype, i.e., ΔczcA was more sensitive to tetracycline than the wild type strain, and copper inhibited the growth of ΔczcA in the presence or absence of tetracycline (Figure 3C). These results suggest that copper induced tetracycline resistance through activation of CrdAB-CzcBA. CrdRS is a two-component system responsible for sensing copper and the activation of CrdAB-CzcBA (29). To prove that copper-induced tetracycline resistance was dependent on CrdRS, we constructed a ΔcrdR strain and evaluated its resistance to tetracycline in the presence or absence of copper. As previously reported, copper inhibited the growth of ΔcrdR (Figure 3B). Growth of ΔcrdR is significantly inhibited by tetracycline compared to the H. pylori wild type strain, suggesting that CrdR contributes to tetracycline resistance. Tetracycline resistance was not promoted by copper in ΔcrdR, suggesting that copper-induced tetracycline resistance is dependent on CrdR (Figure 3C). These results suggest that copper induces the expression of CrdAB-CzcBA through CrdR, enhancing bacterial resistance to tetracycline.
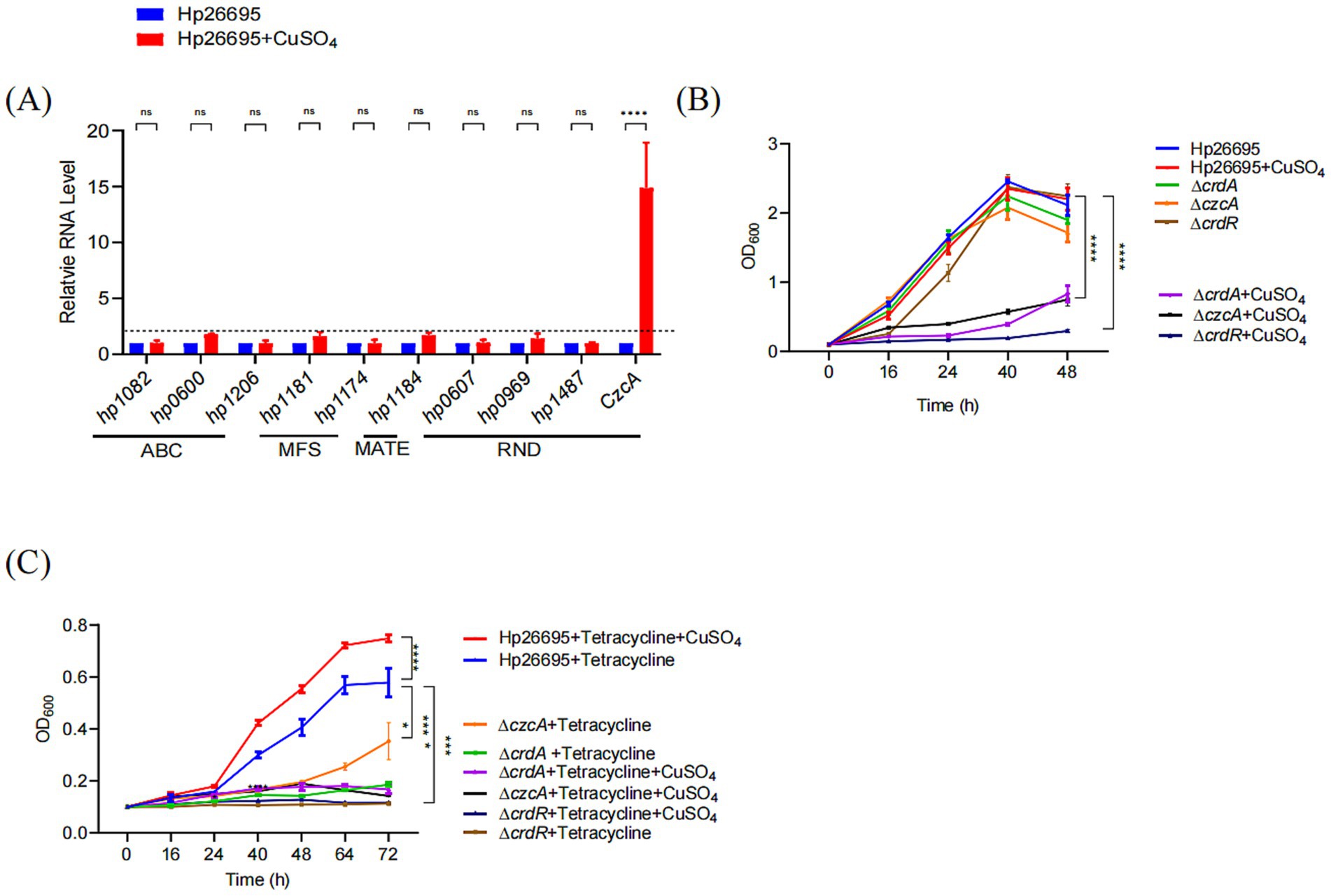
Figure 3. Copper enhances the bacterial growth under tetracycline in H. pylori. (A) Copper modulates expression levels of H. pylori efflux pump-related genes. The expression of efflux pumps was quantified by qPCR with mRNA levels normalized to those in the untreated Hp26695 control. The dashed line represents a one-fold increase in relative expression due to copper supplementation. (B) Growth curves for Hp26695, ΔcrdA, ΔczcA and ΔcrdR with or without supplementation of 50 μM CuSO4. Data points represent the mean OD600 values from three independent experiments with standard deviations indicated. Statistical significance: ****p < 0.0001. (C) Growth curves for Hp26695, ΔcrdA, ΔczcA and ΔcrdR under 0.023 μg/mL tetracycline, with or without supplementation of 50 μM CuSO4. Data points reflect the mean OD600 values from three independent experiments, with standard deviations indicated. Statistical significance: ****p < 0.0001.
4 Discussion
Efflux pumps provide intrinsic antibiotic resistance to bacteria and are thus considered therapeutic targets for the mediation of antibiotic resistance. Several methods have been proposed to inhibit the function of efflux pumps, including downregulation of their expression by interfering with the regulator system, directly inhibiting the assembly or action of these pumps, or modification of antibiotics so that they can no longer act as substrates of efflux pumps (41, 42). However, efflux pumps are present in both drug-sensitive and drug-resistant strains (15). Environmental cues that stimulate the expression of efflux pumps may lead to higher resistance to the corresponding antibiotics. In this study, we found that the copper resistance determinants CrdAB-CzcBA are involved in tetracycline resistance. Unlike some efflux pumps such as MexAB-OprM and NorA, which are involved in the efflux of distinct classes of drugs, and substrates, CrdAB-CzcBA showed no significant effect on resistance to antibiotics, including levofloxacin, metronidazole, clarithromycin, and amoxicillin (data not shown) (15, 42). This is also supported by the finding that copper showed no cross-protection of H. pylori to these antibiotics, which also failed to stimulate the expression of the CrdAB-CzcBA operon (data not shown). This suggests that the CrdAB-CzcBA efflux pump only extrudes specific antibiotics. This is also the case for efflux systems such as AbaF, which provide resistance to Fosfomycin (43). Efflux in enteric rods can also promote bile resistance, suggesting a complex role of these pumping systems (44). If CrdAB-CzcBA is involved in the resistance of other substrates, further investigation is required.
Tetracyclines inhibit protein translation by interfering with bacterial ribosomes and are widely used in both human medicine and livestock production worldwide. Approximately 11 classes, including more than 40 genes, have been characterized as tetracycline-resistant genes. Among these, approximately 60 percent are involved in efflux pumps by extruding tetracycline extracellularly with substrate specificity (12, 45, 46). All of these genes belong to the MFS family, which are single polypeptides, and are proton motive force-dependent (47, 48). In H. pylori, only HP1165 was shown to be involved in induced tetracycline resistance (14). Several studies have shown that knockout of efflux pumps in H. pylori does not alter tetracycline resistance (23). We suspect that this might be due to a relatively low level of expression of these genes in vitro. A higher expression level of these efflux pump genes in vivo might play a significant role in antibiotic resistance. One in silico study also found that there are 27 genes in H. pylori that encoding putative translocases belonging to the ABC transporter, MAT, MFS, and RND families (23). More genes involved in antibiotic resistance require further investigation.
Copper enhances the resistance of tetracycline by enhancing the expression of CrdAB-CzcBA through the two-component CrdRS system (Figure 3) (28, 29). This finding is significant as it reveals a previously unrecognized link between metal ion homeostasis and antibiotic resistance in H. pylori. Furthermore, while similar copper-induced efflux systems have been described in other bacteria, such as the CzcCBA system in P. aeruginosa, these systems are primarily associated with resistance to different antibiotics and heavy metals (49, 50). To our knowledge, this is the first report of copper-induced tetracycline resistance via an RND efflux pump in H. pylori. Besides, other factors that regulate CrdRS activity might also result in the alteration of CrdAB-CzcBA expression. Studies have shown that CrdRS-CrdA is important for survival under nitrosative stress, and the expression of CrdA is activated by CrdRS in response to nitric oxide (51). This suggests that nitrosative stress, such as that occurring during inflammation of the stomach, may alter the resistance to copper and tetracycline.
Contrary to prior observations in H. pylori strain 1,061 that discounted the role of CrdAB-CzcBA in antibiotic resistance (23), our data indicate a substantial increase in tetracycline resistance upon overexpression of CrdAB-CzcBA. This discrepancy could be attributed to the relatively low expression of CrdAB-CzcBA under standard laboratory conditions, which may mask its role in resistance. The concept that efflux pumps with low baseline expression can exhibit a pronounced resistance phenotype upon activation is supported by findings in E. coli, where the overexpression of typically lowly expressed RND family efflux pumps, such as yhiUV (52), has been linked to increased resistance against a range of antibiotics including fluoroquinolones, linezolid, and tetracycline. Hence, the functional impact of efflux pumps expressed at low levels under basal conditions may become more apparent upon induction, underlining the potential for adaptive resistance mechanisms.
H. pylori survives in the stomach environment of humans and needs to respond to environmental signals, such as pH changes, nutrient limitation, and reactive oxygen species. Transition metals participate in various processes, including acting as nutrients for living organisms by incorporation into metalloproteins (53, 54). The host limits the availability of these metals to bacteria through nutritional immunity (55). Copper has been utilized by many bacteria as a cofactor for enzymes, including superoxide dismutase and NADH dehydrogenase (30, 31). However, excess copper can cause the generation of reactive oxygen species, including superoxide radicals, through Fenton reactions, and can thus damage cellular macromolecules and cellular structures (56, 57). Our study suggests that copper homeostasis is closely related to the survival and drug resistance of the bacterium.
Environmental factors can drastically alter the expression of specific genotypes of bacteria, conferring antibiotic resistance. One study showed that Salmonella Typhimurium was found to be significantly more resistant to antibiotics when grown in an environment mimicking conditions under low pH, magnesium, and phosphate compared to grown in standard media (58). Other studies have also shown that environmental conditions that reduce the growth rate activate the drug resistance gene through a stringent response (59). The expression of CrdAB-CzcBA is silenced under normal laboratory conditions, whereas copper significantly activates its expression, suggesting that environmental signals are strongly correlated with the drug resistance of the bacterium. However, using a standard medium might fail to elucidate the role of some silenced genes involved in antibiotic resistance (28, 29). More host environmental factors involved in modulating the bacterial resistance to antibiotics deserve further investigation. Copper concentration in serum is up to 1.5 mg/L (23.6 μM) in healthy individuals (60). However, copper is important in the inflammatory response for its bactericidal effect against pathogen (61). Significant copper accumulation was found both in the serum and tissue during inflammation, this suggests that H. pylori infection which causes gastritis might lead to the activation of CrdAB-CzcBA expression (62, 63). High expression of efflux pumps plays an important role in clinical drug-resistant isolates, and this might be due to the mutations in the promoter region or in the regulatory proteins (15). We speculate that mutation in promoter of crdAB-czcBA or in CrdRS that resulted in activation of CrdAB-CzcBA might lead to a significant resistance to tetracycline in clinical isolates of H. pylori.
5 Conclusion
Taken together, our results showed that CrdAB-CzcBA comprises an efflux pump, with tetracycline and EB efflux activity, and is involved in tetracycline resistance. Copper activated CrdAB-CzcBA expression by acting on CrdRS, increasing bacterial resistance to tetracycline in H. pylori. Our study suggests that copper is an important nutrient for bacteria and plays a role in the cross-protection of tetracycline resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
FG: Methodology, Writing – review & editing, Investigation. WX: Investigation, Writing – review & editing. XZ: Methodology, Writing – review & editing. XH: Data curation, Methodology, Writing – review & editing. FS: Project administration, Writing – review & editing. YW: Data curation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (grant no. 82472286), Natural Science Foundation of Fujian Province, China (grant no. 2020Y9003, 2021Y9101, 2024J01495).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1552537/full#supplementary-material
References
1. Bray, F, Jemal, A, Grey, N, Ferlay, J, and Forman, D. Global cancer transitions according to the human development index (2008-2030): a population-based study. Lancet Oncol. (2012) 13:790–801. doi: 10.1016/s1470-2045(12)70211-5
2. Kuipers, EJ. Helicobacter pylori and the risk and management of associated diseases: gastritis, ulcer disease, atrophic gastritis and gastric cancer. Aliment Pharmacol Ther. (1997) 11:71–88. doi: 10.1046/j.1365-2036.11.s1.5.x
3. Malfertheiner, P, Megraud, F, O'Morain, CA, Gisbert, JP, Kuipers, EJ, Axon, AT, et al. Management of Helicobacter pylori infection-the Maastricht V/Florence consensus report. Gut. (2017) 66:6–30. doi: 10.1136/gutjnl-2016-312288
4. Papastergiou, V, Georgopoulos, SD, and Karatapanis, S. Treatment of Helicobacter pylori infection: past, present and future. World J Gastrointest Pathophysiol. (2014) 5:392–9. doi: 10.4291/wjgp.v5.i4.392
5. Papastergiou, V, Georgopoulos, SD, and Karatapanis, S. Treatment of Helicobacter pylori infection: meeting the challenge of antimicrobial resistance. World J Gastroenterol. (2014) 20:9898–911. doi: 10.3748/wjg.v20.i29.9898
6. Graham, DY. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. (1998) 115:1272–7. doi: 10.1016/s0016-5085(98)70100-3
7. Chopra, I, and Roberts, M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. (2001) 65:232–60. doi: 10.1128/mmbr.65.2.232-260.2001
8. Gerrits, MM, Berning, M, Van Vliet, AH, Kuipers, EJ, and Kusters, JG. Effects of 16S rRNA gene mutations on tetracycline resistance in Helicobacter pylori. Antimicrob Agents Chemother. (2003) 47:2984–6. doi: 10.1128/aac.47.9.2984-2986.2003
9. Trieber, CA, and Taylor, DE. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J Bacteriol. (2002) 184:2131–40. doi: 10.1128/jb.184.8.2131-2140.2002
10. Dailidiene, D, Bertoli, MT, Miciuleviciene, J, Mukhopadhyay, AK, Dailide, G, Pascasio, MA, et al. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob Agents Chemother. (2002) 46:3940–6. doi: 10.1128/aac.46.12.3940-3946.2002
11. Møller, TS, Overgaard, M, Nielsen, SS, Bortolaia, V, Sommer, MO, Guardabassi, L, et al. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol. (2016) 16:39. doi: 10.1186/s12866-016-0649-z
12. Schnappinger, D, and Hillen, W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. (1996) 165:359–69. doi: 10.1007/s002030050339
13. Luo, ZQ, and Farrand, SK. Cloning and characterization of a tetracycline resistance determinant present in Agrobacterium tumefaciens C58. J Bacteriol. (1999) 181:618–26. doi: 10.1128/jb.181.2.618-626.1999
14. Li, Y, and Dannelly, HK. Inactivation of the putative tetracycline resistance gene HP1165 in Helicobacter pylori led to loss of inducible tetracycline resistance. Arch Microbiol. (2006) 185:255–62. doi: 10.1007/s00203-006-0093-9
15. Webber, MA, and Piddock, LJ. The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother. (2003) 51:9–11. doi: 10.1093/jac/dkg050
16. Paulsen, IT. Multidrug efflux pumps and resistance: regulation and evolution. Curr Opin Microbiol. (2003) 6:446–51. doi: 10.1016/j.mib.2003.08.005
17. Fernando, DM, and Kumar, A. Resistance-nodulation-division multidrug efflux pumps in gram-negative Bacteria: role in virulence. Antibiotics (Basel, Switzerland). (2013) 2:163–81. doi: 10.3390/antibiotics2010163
18. Putman, M, van Veen, HW, and Konings, WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. (2000) 64:672–93. doi: 10.1128/mmbr.64.4.672-693.2000
19. Zgurskaya, HI, Krishnamoorthy, G, Tikhonova, EB, Lau, SY, and Stratton, KL. Mechanism of antibiotic efflux in gram-negative bacteria. Front Biosci. (2003) 8:s862–73. doi: 10.2741/1134
20. Nikaido, H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. (1996) 178:5853–9. doi: 10.1128/jb.178.20.5853-5859.1996
21. Tsutsumi, K, Yonehara, R, Ishizaka-Ikeda, E, Miyazaki, N, Maeda, S, Iwasaki, K, et al. Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism. Nat Commun. (2019) 10:1520. doi: 10.1038/s41467-019-09463-9
22. Shi, X, Chen, M, Yu, Z, Bell, JM, Wang, H, Forrester, I, et al. In situ structure and assembly of the multidrug efflux pump AcrAB-TolC. Nat Commun. (2019) 10:2635. doi: 10.1038/s41467-019-10512-6
23. van Amsterdam, K, Bart, A, and van der Ende, A. A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob Agents Chemother. (2005) 49:1477–82. doi: 10.1128/aac.49.4.1477-1482.2005
24. Johnson, JM, and Church, GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. (1999) 287:695–715. doi: 10.1006/jmbi.1999.2630
25. Kutschke, A, and de Jonge, BL. Compound efflux in Helicobacter pylori. Antimicrob Agents Chemother. (2005) 49:3009–10. doi: 10.1128/aac.49.7.3009-3010.2005
26. Liu, ZQ, Zheng, PY, and Yang, PC. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastroenterol. (2008) 14:5217–22. doi: 10.3748/wjg.14.5217
27. Bina, JE, Alm, RA, Uria-Nickelsen, M, Thomas, SR, Trust, TJ, and Hancock, RE. Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob Agents Chemother. (2000) 44:248–54. doi: 10.1128/aac.44.2.248-254.2000
28. Waidner, B, Melchers, K, Ivanov, I, Loferer, H, Bensch, KW, Kist, M, et al. Identification by RNA profiling and mutational analysis of the novel copper resistance determinants CrdA (HP1326), CrdB (HP1327), and CzcB (HP1328) in Helicobacter pylori. J Bacteriol. (2002) 184:6700–8. doi: 10.1128/jb.184.23.6700-6708.2002
29. Waidner, B, Melchers, K, Stähler, FN, Kist, M, and Bereswill, S. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J Bacteriol. (2005) 187:4683–8. doi: 10.1128/jb.187.13.4683-4688.2005
30. Samanovic, MI, Ding, C, Thiele, DJ, and Darwin, KH. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe. (2012) 11:106–15. doi: 10.1016/j.chom.2012.01.009
31. Andreini, C, Banci, L, Bertini, I, and Rosato, A. Occurrence of copper proteins through the three domains of life: a bioinformatic approach. J Proteome Res. (2008) 7:209–16. doi: 10.1021/pr070480u
32. Neyrolles, O, Wolschendorf, F, Mitra, A, and Niederweis, M. Mycobacteria, metals, and the macrophage. Immunol Rev. (2015) 264:249–63. doi: 10.1111/imr.12265
33. Montefusco, S, Esposito, R, D'Andrea, L, Monti, MC, Dunne, C, Dolan, B, et al. Copper promotes TFF1-mediated Helicobacter pylori colonization. PLoS One. (2013) 8:e79455. doi: 10.1371/journal.pone.0079455
34. Lin, J, Zhang, X, Wen, Y, Chen, H, and She, F. A newly discovered drug resistance gene rfaF in Helicobacter pylori. Infect Drug Resist. (2019) 12:3507–14. doi: 10.2147/idr.S231152
35. Feng, Z, Liu, D, Wang, L, Wang, Y, Zang, Z, Liu, Z, et al. A putative efflux transporter of the ABC family, YbhFSR, in Escherichia coli functions in tetracycline efflux and Na(+)(Li(+))/H(+) transport. Front Microbiol. (2020) 11:556. doi: 10.3389/fmicb.2020.00556
36. Sudano Roccaro, A, Blanco, AR, Giuliano, F, Rusciano, D, and Enea, V. Epigallocatechin-gallate enhances the activity of tetracycline in staphylococci by inhibiting its efflux from bacterial cells. Antimicrob Agents Chemother. (2004) 48:1968–73. doi: 10.1128/aac.48.6.1968-1973.2004
37. de Cristóbal, RE, Vincent, PA, and Salomón, RA. Multidrug resistance pump AcrAB-TolC is required for high-level, Tet(a)-mediated tetracycline resistance in Escherichia coli. J Antimicrob Chemother. (2006) 58:31–6. doi: 10.1093/jac/dkl172
38. Chiu, HC, Lin, TL, Yang, JC, and Wang, JT. Synergistic effect of imp/ostA and msbA in hydrophobic drug resistance of Helicobacter pylori. BMC Microbiol. (2009) 9:136. doi: 10.1186/1471-2180-9-136
39. Falsafi, T, Ehsani, A, Attaran, B, and Niknam, V. Association of hp1181 and hp1184 genes with the active efflux phenotype in multidrug-resistant isolates of Helicobacter pylori. Jundishapur J Microbiol. (2016) 9:e30726. doi: 10.5812/jjm.30726
40. Ge, X, Cai, Y, Chen, Z, Gao, S, Geng, X, Li, Y, et al. Bifunctional enzyme SpoT is involved in biofilm formation of Helicobacter pylori with multidrug resistance by upregulating efflux pump Hp1174 (gluP). Antimicrob Agents Chemother. (2018) 62:e00957-18. doi: 10.1128/aac.00957-18
41. Bhardwaj, AK, and Mohanty, P. Bacterial efflux pumps involved in multidrug resistance and their inhibitors: rejuvinating the antimicrobial chemotherapy. Recent Pat Antiinfect Drug Discov. (2012) 7:73–89. doi: 10.2174/157489112799829710
42. Sharma, A, Gupta, VK, and Pathania, R. Efflux pump inhibitors for bacterial pathogens: from bench to bedside. Indian J Med Res. (2019) 149:129–45. doi: 10.4103/ijmr.IJMR_2079_17
43. Sharma, A, Sharma, R, Bhattacharyya, T, Bhando, T, and Pathania, R. Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter-AbaF. J Antimicrob Chemother. (2017) 72:68–74. doi: 10.1093/jac/dkw382
44. Piddock, LJV. Multidrug-resistance efflux pumps? Not just for resistance. Nat Rev Microbiol. (2006) 4:629–36. doi: 10.1038/nrmicro1464
45. Zhang, T, Wang, CG, Lv, JC, Wang, RS, and Zhong, XH. Survey on tetracycline resistance and antibiotic-resistant genotype of avian Escherichia coli in North China. Poult Sci. (2012) 91:2774–7. doi: 10.3382/ps.2012-02453
46. Adelowo, OO, and Fagade, OE. The tetracycline resistance gene tet39 is present in both gram-negative and gram-positive bacteria from a polluted river, southwestern Nigeria. Lett Appl Microbiol. (2009) 48:167–72. doi: 10.1111/j.1472-765X.2008.02523.x
47. Li, XZ, and Nikaido, H. Efflux-mediated drug resistance in bacteria. Drugs. (2004) 64:159–204. doi: 10.2165/00003495-200464020-00004
48. Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. (2007) 39:162–76. doi: 10.1080/07853890701195262
49. Caille, O, Rossier, C, and Perron, K. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J Bacteriol. (2007) 189:4561–8. doi: 10.1128/JB.00095-07
50. Nguyen, THT, Nguyen, HD, Le, MH, Nguyen, TTH, Nguyen, TD, Nguyen, DL, et al. Efflux pump inhibitors in controlling antibiotic resistance: outlook under a heavy metal contamination context. Molecules. (2023) 28:2912. doi: 10.3390/molecules28072912
51. Hung, CL, Cheng, HH, Hsieh, WC, Tsai, ZT, Tsai, HK, Chu, CH, et al. The CrdRS two-component system in Helicobacter pylori responds to nitrosative stress. Mol Microbiol. (2015) 97:1128–41. doi: 10.1111/mmi.13089
52. Bohnert, JA, Schuster, S, Fähnrich, E, Trittler, R, and Kern, WV. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J Antimicrob Chemother. (2007) 59:1216–22. doi: 10.1093/jac/dkl426
53. Haley, KP, and Gaddy, JA. Metalloregulation of Helicobacter pylori physiology and pathogenesis. Front Microbiol. (2015) 6:911. doi: 10.3389/fmicb.2015.00911
54. Foster, AW, Osman, D, and Robinson, NJ. Metal preferences and metallation. J Biol Chem. (2014) 289:28095–103. doi: 10.1074/jbc.R114.588145
55. Hood, MI, and Skaar, EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. (2012) 10:525–37. doi: 10.1038/nrmicro2836
56. Rademacher, C, and Masepohl, B. Copper-responsive gene regulation in bacteria. Microbiology. (2012) 158:2451–64. doi: 10.1099/mic.0.058487-0
57. Dupont, CL, Grass, G, and Rensing, C. Copper toxicity and the origin of bacterial resistance--new insights and applications. Metallomics. (2011) 3:1109–18. doi: 10.1039/c1mt00107h
58. Kubicek-Sutherland, JZ, Heithoff, DM, Ersoy, SC, Shimp, WR, House, JK, Marth, JD, et al. Host-dependent induction of transient antibiotic resistance: a prelude to treatment failure. EBioMedicine. (2015) 2:1169–78. doi: 10.1016/j.ebiom.2015.08.012
59. Koskiniemi, S, Pränting, M, Gullberg, E, Näsvall, J, and Andersson, DI. Activation of cryptic aminoglycoside resistance in Salmonella enterica. Mol Microbiol. (2011) 80:1464–78. doi: 10.1111/j.1365-2958.2011.07657.x
61. White, C, Lee, J, Kambe, T, Fritsche, K, and Petris, MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. (2009) 284:33949–56. doi: 10.1074/jbc.M109.070201
62. Milanino, R, Marrella, M, Moretti, U, Concari, E, and Velo, GP. Copper and zinc status in rats with acute inflammation: focus on the inflamed area. Agents Actions. (1988) 24:356–64. doi: 10.1007/bf02028294
Keywords: H. pylori , copper, efflux transporter, CrdAB-CzcBA, tetracycline resistance
Citation: Gao F, Xiang W, Zhang X, Huang X, She F and Wen Y (2025) Copper enhances tetracycline resistance via the efflux transporter CrdAB-CzcBA in Helicobacter pylori. Front. Med. 12:1552537. doi: 10.3389/fmed.2025.1552537
Edited by:
Ilana L. B. C. Camargo, University of São Paulo, BrazilReviewed by:
Paweł Krzyżek, Wroclaw Medical University, PolandSasikala Muthusamy, Brigham and Women’s Hospital and Harvard Medical School, United States
Yongkang Lai, Second Military Medical University, China
Copyright © 2025 Gao, Xiang, Zhang, Huang, She and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yancheng Wen, aGl0d3ljQHFxLmNvbQ==; Feifei She, c2hlZmVpZmVpQHllYWgubmV0
†These authors have contributed equally to this work
 Fanglin Gao1,2†
Fanglin Gao1,2† Wanquan Xiang
Wanquan Xiang Yancheng Wen
Yancheng Wen