- Department of Microbiology, SRM Medical College Hospital and Research Centre, SRM Institute of Science and Technology (SRMIST), Kattankulathur, Chengalpattu, Tamil Nadu, India
Background: Carbapenem-resistant Gram-negative bacteria (CR-GNB) pose a serious global health threat, especially in low- and middle-income countries. Local surveillance is crucial for informing antimicrobial stewardship and infection control strategies. This study aimed to evaluate the prevalence, demographic distribution, and temporal fluctuations of carbapenem resistance among key Gram-negative pathogens in a South Indian tertiary care center over a two-year period.
Methods: A retrospective study was conducted on 8,359 non-duplicate Gram-negative isolates obtained from clinical specimens between July 2022 and July 2024. Organisms were identified, and antimicrobial susceptibility was determined using the VITEK® 2 Compact system (BioMérieux). Resistance to imipenem (IPM) and meropenem (MEM) was assessed. Data were stratified by age, sex, ward type, specimen source, and quarterly distribution. A subset of resistant isolates underwent molecular screening for carbapenemase genes using real time PCR.
Results: Carbapenem resistance was observed in 24% (2007) of Gram-negative isolates. Acinetobacter baumannii (48.0%) and Klebsiella pneumoniae (38.6%) accounted for the majority of resistant cases. Resistance was significantly higher in males (64.3%) and in patients aged 61–80 years (p < 0.001). Surgical wards showed greater resistance rates compared to medical departments. A peak in resistance was identified during January–March 2023, particularly for A. baumannii (76.3%). IPM-MEM resistance discrepancies were found in Citrobacter and Proteus species. Gene profiling of resistant strains revealed the predominance of blaNDM, blaVIM in all organism.
Conclusion: The findings reveal a high and fluctuating burden of carbapenem resistance, especially in elderly males and surgical settings. Continuous surveillance and targeted interventions are vital to curbing the spread of CR-GNB in high-risk healthcare environments.
1 Introduction
Carbapenem-resistant organisms (CROs) represent a significant and growing challenge in healthcare settings worldwide (1). These organisms are often associated with high morbidity, mortality, and healthcare costs due to limited treatment options and the potential for widespread outbreaks. Carbapenems, a class of β-lactam antibiotics with a broad spectrum of activity, have historically been considered drugs of last resort for treating severe infections caused by multi-drug-resistant GNB (2). However, the emergence and spread of carbapenem resistance have severely compromised their effectiveness (3).
The mechanisms of carbapenem resistance are diverse, including the production of carbapenemase enzymes (e.g., KPC, NDM, VIM, IMP, and OXA types), efflux pumps, and porin mutations that reduce antibiotic uptake (4). The spread of these resistant organisms is facilitated by plasmids and other mobile genetic elements that can be transferred between bacteria, accelerating the dissemination of resistance genes across different species and geographical regions (5, 6).
In India, the prevalence of carbapenem-resistant organisms is particularly concerning. Studies have reported high rates of carbapenem resistance among key pathogens such as K. pneumoniae, A. baumannii, and Pseudomonas aeruginosa (7). The National Antimicrobial Resistance Surveillance Network (NARS-Net) in India has highlighted alarming levels of resistance, with reports indicating that over 50% of K. pneumoniae isolates and 30% of P. aeruginosa isolates are resistant to Carbapenems. These figures underscore the critical need for enhanced surveillance, stringent infection control measures, and the development of new therapeutic strategies.
Tertiary care centre, which provide specialized medical care and serve as referral hospitals, are particularly vulnerable to the spread of CROs (8). Patients in these settings often have complex medical conditions, require invasive procedures, and are exposed to broad-spectrum antibiotics, all of which increase the risk of acquiring resistant infections. Monitoring the prevalence of carbapenem resistance in such settings is crucial for developing effective infection control strategies and guiding empirical therapy (9).
In this study, we conducted a retrospective analysis of carbapenem resistance in a tertiary care centre over a two-year period. Our objectives were to determine the prevalence of CROs, identify trends over time, and evaluate the distribution of resistance among different bacterial species. In addition to phenotypic analysis, we also performed molecular characterization to identify the specific resistance genes responsible for carbapenem resistance. By analysing these data, we aim to provide insights that can inform clinical practice and policy decisions to better manage and prevent the spread of carbapenem-resistant infections.
2 Materials and methods
2.1 Study design, setting, and population
This retrospective study was conducted to evaluate the prevalence and trends of CROs over a two-year period, from July 2022 to July 2024, at SRM Medical College Hospital and Research Centre, SRMIST, Kattankulathur, Chennai, Tamil Nadu. The study included all patients from whom clinical specimens were submitted for microbiological analysis during the study duration. Various specimen types—including blood, urine, respiratory secretions, and exudates—were processed for bacterial culture and antimicrobial susceptibility testing. All patients, irrespective of age or gender, were considered eligible, provided their samples yielded growth of GNB. Specimens that showed no bacterial growth, yielded Gram-positive organisms, or represented duplicate isolates from the same patient were excluded to avoid duplication and ensure the accuracy of the prevalence data.
2.2 Antimicrobial susceptibility testing and data collection
AST was performed using the VITEK system (BioMérieux), an advanced automated method for bacterial identification and susceptibility pattern. The panel of antibiotics tested against the CROs included AN, Amikacin; ATM, Aztreonam; CAZ, Ceftazidime; CIP, Ciprofloxacin; CS, Colistin; FEP, Cefepime; FOS, Fosfomycin; GM, Gentamicin; LEV, Levofloxacin; MNO, Minocycline; SXT, Trimethoprim/Sulfamethoxazole; TZP, Piperacillin/Tazobactam along with carbapenems such as Imipenem (IPM) and Meropenem (MEM). These antibiotics were selected based on CLSI guidelines to cover a broad spectrum of resistance mechanisms found in Enterobacterales, Pseudomonas, and Acinetobacter (10). The resistance profiles for IPM and MEM were determined separately using the same VITEK AST-N405 and VITEK AST-N406 cards that were used to test all antibiotics for GNB.
An isolate was classified as carbapenem-resistant if it exhibited resistance to either or both drugs, regardless of its susceptibility to ertapenem, in accordance with CLSI recommendations. For Enterobacteriaceae, resistance was defined as a MIC value of ≥4 μg/mL for IPM or MEM. For P. aeruginosa and A. baumannii, resistance was defined as a MIC value of ≥8 μg/mL for these antibiotics.
2.3 Molecular characterization
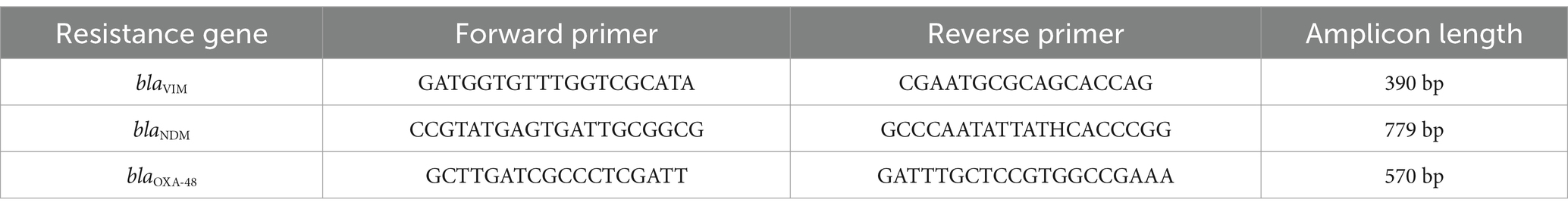
Genomic DNA was extracted from pure cultures of carbapenem-resistant isolates grown overnight on nutrient agar plates. Bacterial cells were transferred into centrifuge tubes containing sterile double-distilled water, boiled at 95°C for 15 min, centrifuged at 15000 rpm for 10 min and stored at −72°C. Plasmid DNA was isolated using Truescreen magnetic bead-based extraction kit (developed by TranScience innovative Technologies Pvt. Ltd.) after lysis with Proteinase K. DNA binding to magnetic beads was facilitated by Truescreen solution, and the bound plasmid DNA was separated from cellular debris through magnetic separation and washing. Primers targeting carbapenemase genes blaVIM, blaNDM, and blaOXA-48 were designed based on ICMR guidelines and synthesized by Eurofins Genomics (Table 1). A SYBR Green-based master mix containing AmpliTaq Gold DNA polymerase and optimized components was prepared with primers and template DNA. Real-time PCR (ABI PRISM 7900HT) was conducted for absolute quantification, with an amplification protocol consisting of initial denaturation at 95°C, followed by cycles of denaturation, annealing, and extension. Genotypically confirmed positive ATCC control strains, obtained from HiMedia (India), were included in each PCR run to validate the assay. These strains were not sequenced, as their resistance genotypes are well characterized. This confirmed the presence of the blaNDM, blaOXA-48, and blaVIM genes, identifying the molecular basis of carbapenem resistance in these isolates (5).
2.4 Statistical analysis
Data were analysed using IBM SPSS Statistics version 27 to determine the prevalence and trends of carbapenem resistance. Descriptive statistics were employed to summarize patient demographics, specimen types, bacterial species, and resistance profiles. The prevalence of CROs was calculated as the proportion of resistant isolates relative to the total number of GNB isolates. Categorical data were described using frequency and percentages and analysed using Chi square test. A p < 0.05 was considered statistically significant.
2.5 Ethical considerations
Ethical approval was obtained from the Institutional Ethics Committee of SRMMCH&RC. Given the retrospective nature of the study, individual patient consent was not required. However, patient confidentiality was strictly maintained, and data were anonymized prior to analysis to protect patient privacy.
3 Results
3.1 Prevalence of carbapenem-resistance in GNB
During the study period from July 2022 to July 2024, a total of 8,359 clinical specimens yielded growth of GNB and were included in the analysis. The overall prevalence of carbapenem-resistant GNB among the isolates was significant. A. baumannii exhibited the highest level of resistance at 48% (388 out of 808 isolates), followed by K. pneumoniae at 38.6% (733 out of 1,897 isolates). P. aeruginosa showed a resistance rate of 24.18% (290 out of 1,199 isolates), while Proteus spp. had a resistance rate of 24.1% (126 out of 522 isolates). The prevalence of carbapenem resistance in E. coli was 12.1% (433 out of 3,577 isolates), and Citrobacter spp. showed the lowest resistance rate at 10.39% (37 out of 356 isolates). These findings (Figure 1) highlight the significant burden of carbapenem resistance among GNB in the tertiary care centre.

Figure 1. Prevalence of carbapenem resistance in GNB, showing highest resistance in A. baumannii (48%) and lowest in Citrobacter spp. (10.39%).
To further evaluate the association between organism type and carbapenem resistance, a Chi-square test was performed. The test revealed a statistically significant association (p = 0.019*), with A. baumannii and K. pneumoniae emerging as the organisms most strongly associated with carbapenem resistance.
3.2 Carbapenem resistance across clinical specimens and hospital alliances
The study examined carbapenem resistance patterns in key GNB—E. coli, K. pneumoniae, P. aeruginosa, A. baumannii, Proteus spp., and Citrobacter spp.—across various clinical specimens and hospital alliances. Overall, K. pneumoniae and A. baumannii exhibited the highest resistance to both IPM and MEM, particularly in blood, respiratory and exudate samples. Among K. pneumoniae isolates, those from blood samples exhibited the highest carbapenem resistance (51.4%), whereas no Citrobacter spp. were isolated from blood. In E. coli, isolates from exudates showed the highest resistance (>18%), while Proteus spp. and Citrobacter spp. displayed greater resistance in respiratory samples (Figure 2).
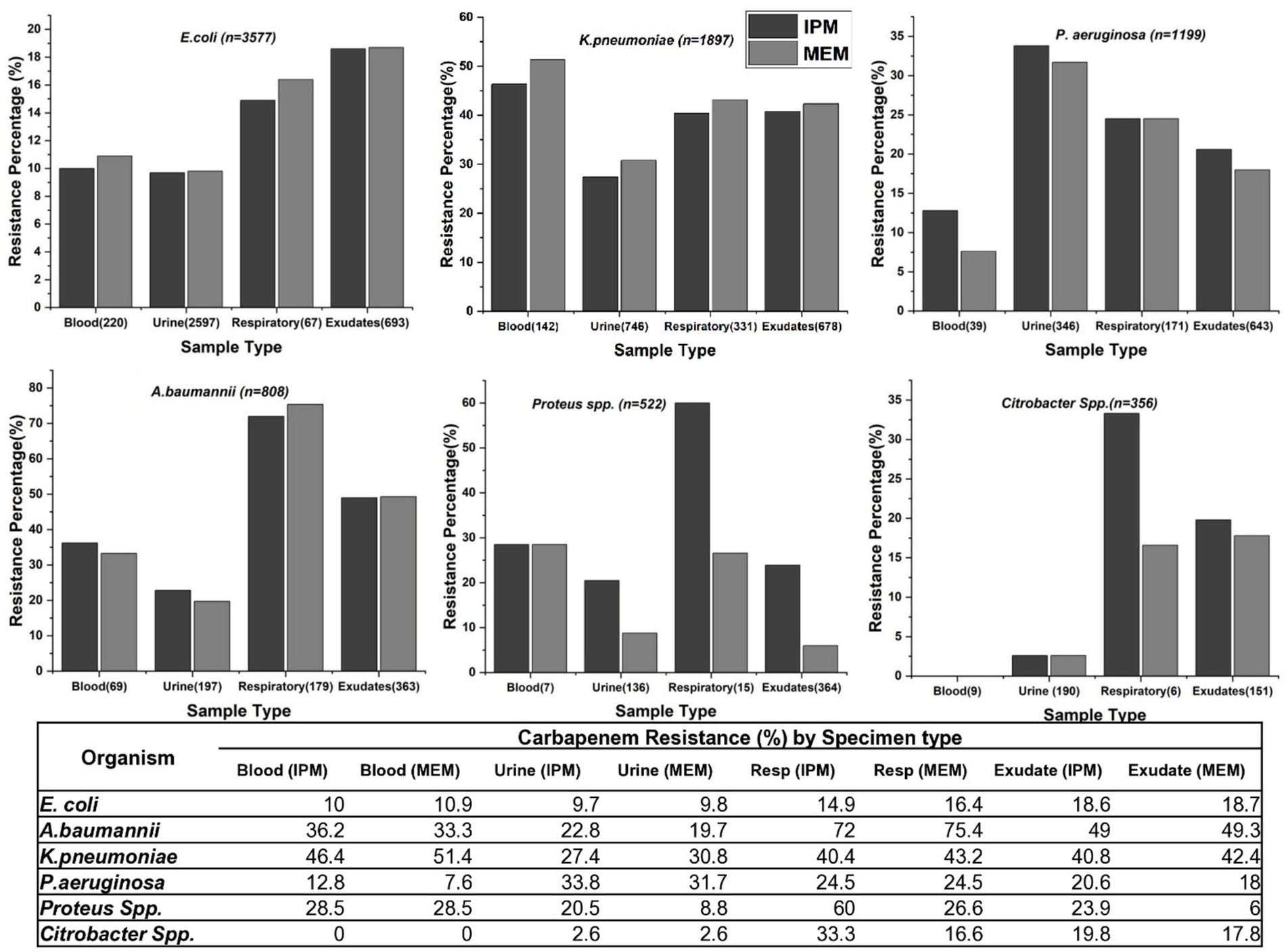
Figure 2. Carbapenem resistance rates in various clinical specimens for different bacterial species. The graphs display IPM and MEM resistance rates across blood, urine, respiratory samples, and exudates for E. coli, K. pneumoniae, P. aeruginosa, A. baumannii, Citrobacter spp. and Proteus spp.
Across hospital alliances (Figure 3), the Surgery Alliance consistently showed the highest resistance rates for all six pathogens. For instance, carbapenem resistance in P. aeruginosa, A. baumannii, and Proteus spp. exceeded 60% in this alliance. In contrast, the Paediatric Alliance demonstrated minimal resistance across all organisms (Table 2). These trends emphasize the importance of implementing targeted antimicrobial stewardship, particularly within surgical departments where resistance is most pronounced.
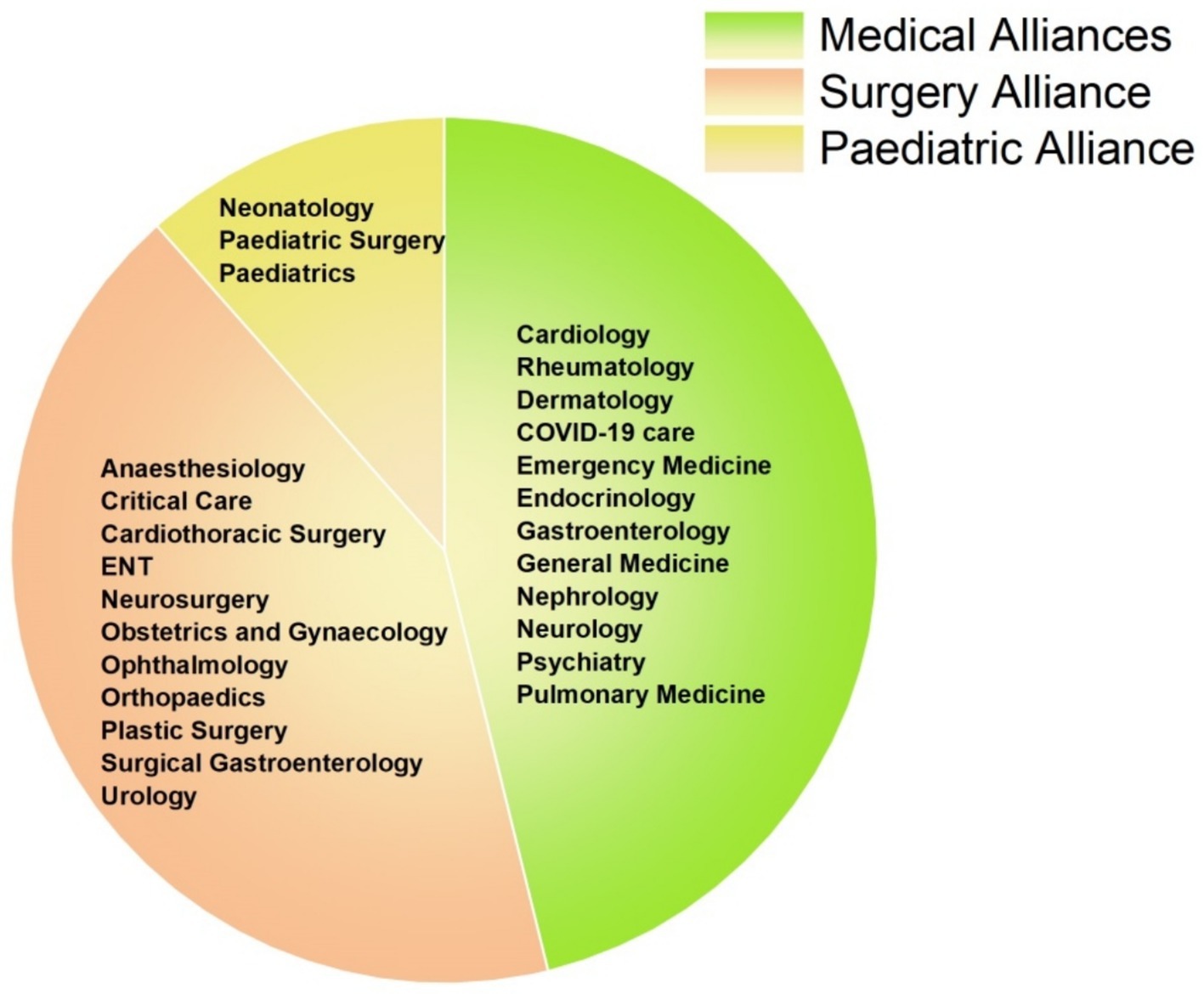
Figure 3. Distribution of hospital departments across clinical alliances. The pie chart categorizes hospital departments into three clinical alliances: Medical (green), Surgical (orange), and Paediatric (yellow). Each segment represents the departments contributing to their respective alliance for the analysis of antimicrobial resistance patterns.
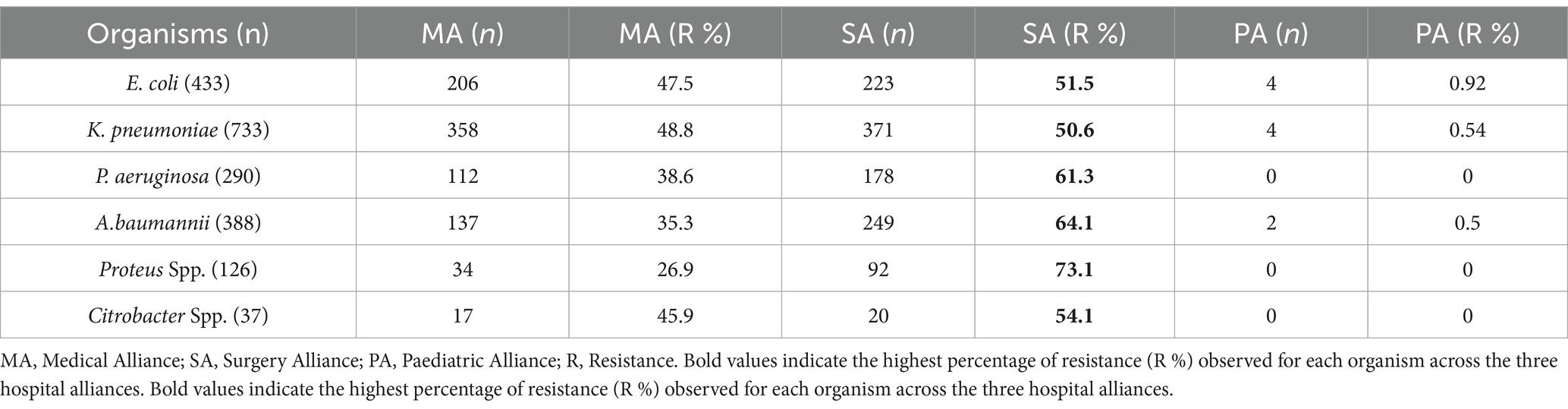
Table 2. Carbapenem resistance rates for various GNB across medical, surgery, and paediatric alliances, with the surgery alliance showing higher resistance.
Interestingly, when comparing resistance patterns between IPM and MEM, specific differences were observed. In Proteus spp., 33.4% of respiratory isolates were resistant to IPM but remained sensitive to MEM. A similar pattern was noted in Citrobacter spp., where 16.6% of respiratory isolates showed resistance exclusively to IPM. In contrast, no substantial differences between the two drugs were observed in E. coli, K. pneumoniae, or A. baumannii.
3.3 Gender-wise distribution of carbapenem-resistant organisms
The gender distribution of CROs was analysed among the isolates (Table 3). The data showed that out of 2,007 total isolates, 716 (35.7%) were from females and 1,291 (64.3%) were from males. The highest number of carbapenem-resistant isolates were found in K. pneumoniae with 733 total isolates, comprising 238 females and 495 males, showing a significant gender difference [χ2(df) = 32.74, p < 0.001**]. A. baumannii also showed a high resistance rate with 388 total isolates, where 137 were from females and 251 from males. Other organisms such as P. aeruginosa had 290 total isolates (81 females, 209 males), E. coli had 433 total isolates (201 females, 232 males), Proteus spp. had 126 total isolates (45 females, 81 males), and Citrobacter spp. had 37 total isolates (14 females, 23 males). This data highlights the higher prevalence of carbapenem resistance in male patients compared to female patients.
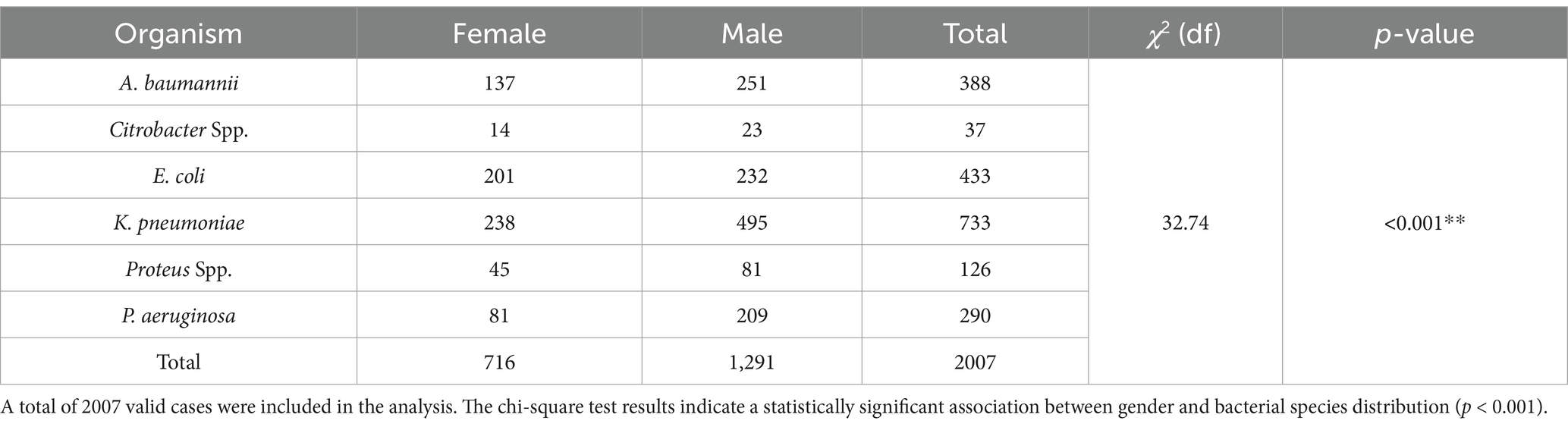
Table 3. Gender-wise distribution of CROs among isolates, showing a higher prevalence in male patients compared to female patients.
3.4 Age-wise distribution of carbapenem-resistant organisms
The age distribution of CROs shows a significant age-related difference in prevalence [χ2(df) = 55.020, p < 0.001**]. The highest prevalence of carbapenem-resistant E. coli and K. pneumoniae is in individuals over 60 years, accounting for 43.1 and 47.9%, respectively, (Figure 4). Similarly, P. aeruginosa and A. baumannii are most common in the 40–60 and over 60 age groups. Proteus shows a markedly high prevalence in the 40–60 age group (59.5%), while Citrobacter is predominantly found in the 40–60 age group (72.9%). This data underscores the higher prevalence of CROs in older age groups, particularly those over 60 years old.
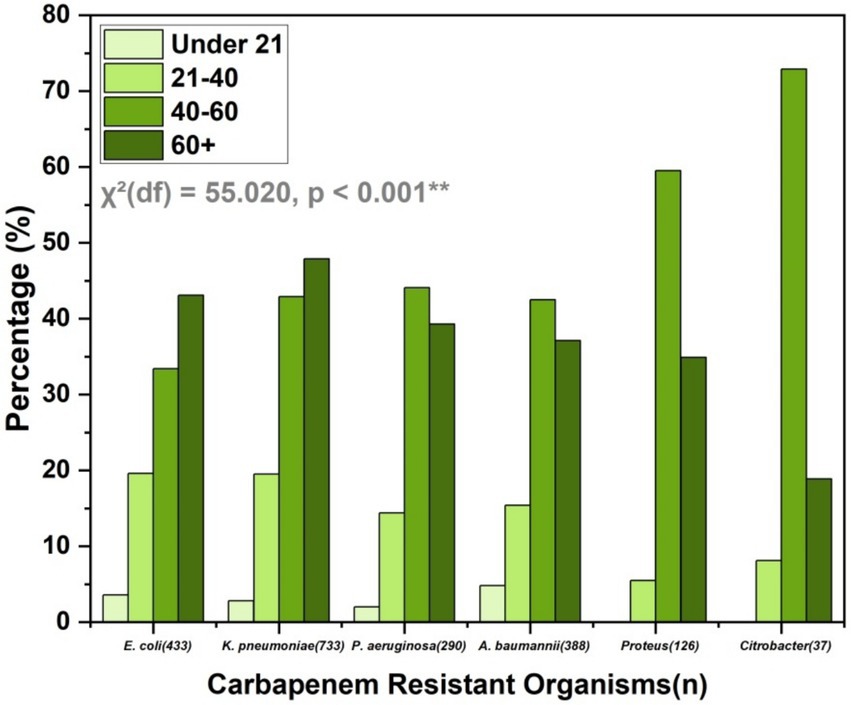
Figure 4. Age distribution of CROs, highlighting higher prevalence in individuals over 60 years, with significant age-related differences in prevalence. The chi-square test results indicate a statistically significant association between age groups and bacterial species distribution (p < 0.001).
3.5 Quarterly trends in resistance patterns
The quarterly analysis of carbapenem resistance from July 2022 to June 2024 reveals distinct temporal and seasonal variations among key Gram-negative organisms (Table 4). The periods were categorized based on regional seasonal patterns in Tamil Nadu: Southwest Monsoon (July–September), Northeast Monsoon (October–December), Dry Season (January–March), and summer (April–June). In the initial quarter (Southwest Monsoon, July–September 2022), A. baumannii exhibited the highest resistance rate at 46%, while Citrobacter spp. had the lowest at 1.5%, with a significant chi-square value [χ2(df) = 55.95, p < 0.001] (Supplementary Table 1).
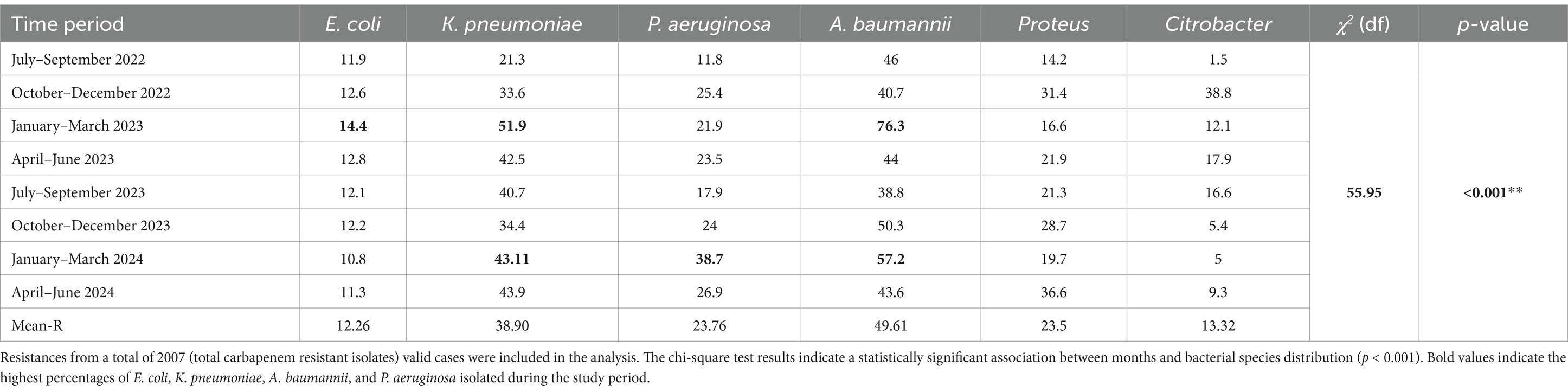
Table 4. Quarterly prevalence of carbapenem resistance in Gram-negative organisms from July 2022 to June 2024.
Throughout the study, A. baumannii consistently displayed high resistance, peaking at 76.3% during the Dry Season (January–March 2023) and averaging 49.61% across the 2 years. K. pneumoniae also showed notable resistance trends, with a mean resistance of 38.92%, peaking at 51.9% in January–March 2023. In contrast, E. coli maintained lower resistance rates, generally around 12%, with a mild increase to 14.4% in the same Winter/Dry period (January–March 2023). P. aeruginosa demonstrated considerable variability, ranging from 11.8% (July–September 2022) to a peak of 38.7% (January–March 2024), averaging 23.76% overall.
Proteus spp. showed fluctuating resistance, with an overall average of 23.8%, reaching a high of 36.6% during summer (April–June 2024). Citrobacter spp., despite exhibiting the lowest overall resistance, had sporadic surges, notably 38.8% during the Northeast Monsoon (October–December 2022), and an average of 13.32% across the study period. These findings highlight the dynamic and seasonal nature of carbapenem resistance, with notable peaks during the Dry Season, particularly in A. baumannii, E. coli, P. aeruginosa and K. pneumoniae. The highly significant chi-square result supports substantial inter-organism and temporal variability, underscoring the critical need for continuous antimicrobial surveillance and seasonally tailored infection control strategies in healthcare settings.
3.6 Resistance patterns to other antibacterial agents
The resistance patterns of CROs to various antibacterial agents reveal crucial insights (Figure 5). Notably, high resistance was observed in E. coli to Ceftazidime and Piperacillin/Tazobactam (both 100%), Ciprofloxacin (98.3%), and Cefepime (98.3%). K. pneumoniae also exhibited significant resistance to Ceftazidime (96%), Ciprofloxacin (94.4%), and Cefepime (95.2%). P. aeruginosa showed high resistance to Ciprofloxacin (94%) and Piperacillin/Tazobactam (88%), while A. baumannii exhibited considerable resistance to Ceftazidime (96.2%), Piperacillin/Tazobactam (96.2%), and Ciprofloxacin (94.9%).
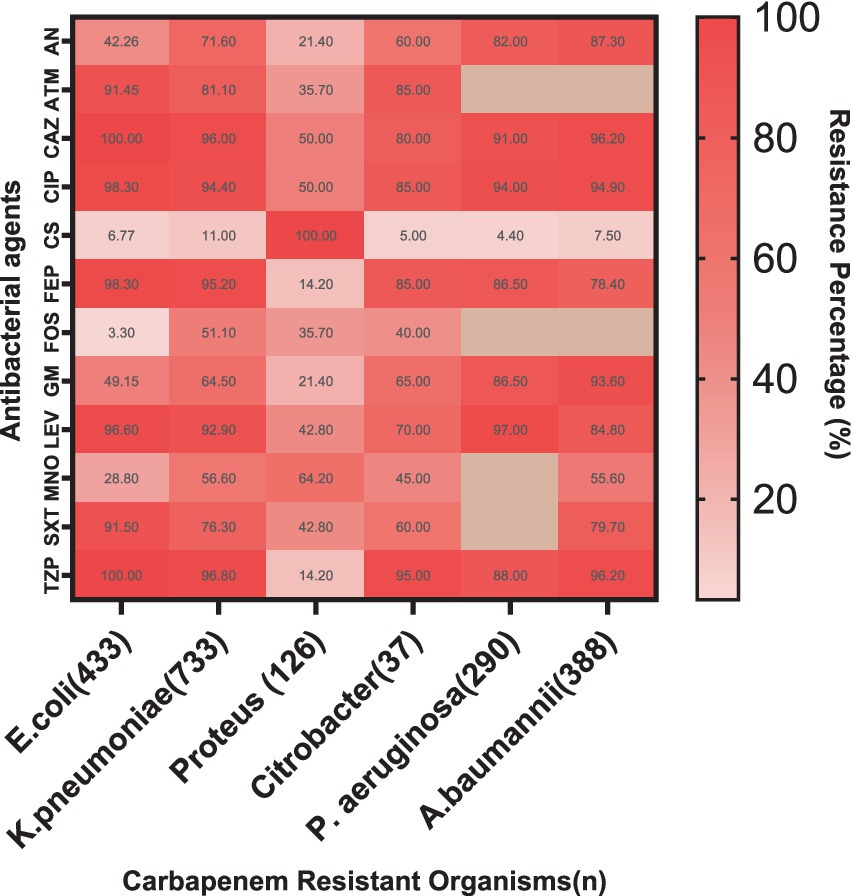
Figure 5. Resistance patterns of CROs to various antibacterial agents. The heat map illustrates high resistance levels in E. coli, K. pneumoniae, P. aeruginosa, and A. baumannii to multiple antibiotics, with lower resistance observed for Colistin, Minocycline and Fosfomycin. AN, Amikacin; ATM, Aztreonam; CAZ, Ceftazidime; CIP, Ciprofloxacin; CS, Colistin; FEP, Cefepime; FOS, Fosfomycin; GM, Gentamicin; LEV, Levofloxacin; MNO, Minocycline; SXT, Trimethoprim/Sulfamethoxazole; TZP, Piperacillin/Tazobactam.
In contrast, Citrobacter showed variable resistance patterns, with moderate resistance levels to Ceftazidime and Ciprofloxacin. Proteus also exhibited notable resistance, particularly to Ciprofloxacin and Piperacillin/Tazobactam (Supplementary Table 2).
Colistin remains effective against most strains, with notably low resistance in E. coli (6.77%), although A. baumannii showed some resistance (7.5%). Gentamicin and Levofloxacin showed varying levels of resistance across different organisms, with E. coli showing moderate resistance to Gentamicin (49.15%) and K. pneumoniae showing substantial resistance to Levofloxacin (92.9%). This data underscores the critical need for ongoing monitoring and judicious use of these antibiotics to manage and treat infections caused by CROs effectively.
3.7 Genotypic distribution
BlaNDM, blaOXA-48, and blaVIM were highly prevalent among CR-GNB, especially in E. coli, K. pneumoniae, and A. baumannii, with lower rates in Proteus and Citrobacter spp. (Figure 6).
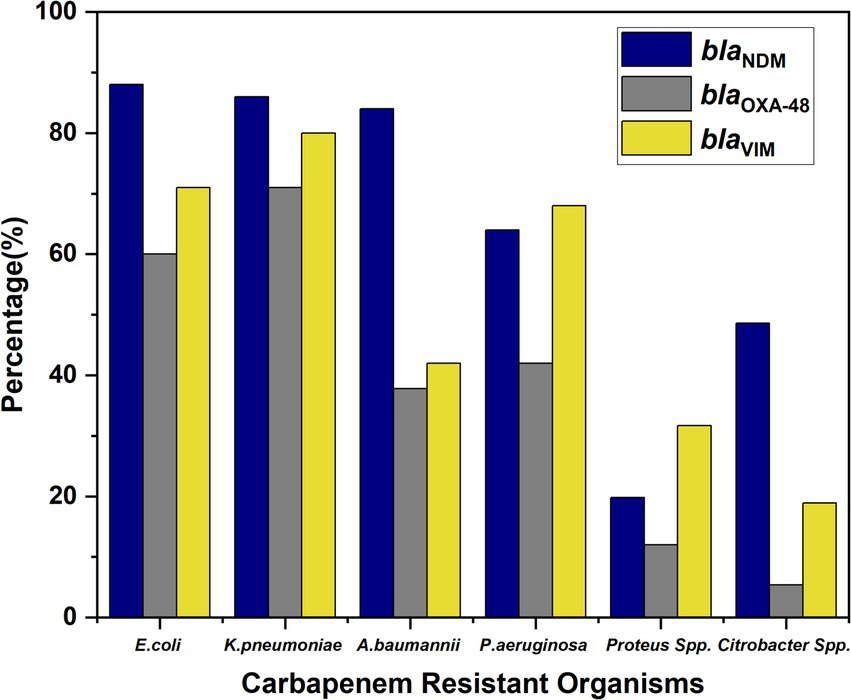
Figure 6. Distribution of carbapenemase genes (blaNDM, blaOXA-48, and blaVIM) across different bacterial isolates.
Importantly, co-carriage of multiple resistance genes was observed among several isolates. A total of 46% of isolates harbored all three genes (blaNDM + blaOXA-48 + blaVIM), indicating a high level of genetic resistance complexity. Additionally, 9% of the isolates co-harbored blaNDM + blaVIM, while 12% carried blaNDM + blaOXA-48. The presence of these combinations underscores the evolving nature of carbapenem resistance and the potential for horizontal gene transfer among MDR organisms.
Organisms isolated from respiratory samples (medical alliance) showed the highest prevalence of all three resistance genes across all six bacterial species, highlighting the respiratory tract as a key reservoir for MDR strains. In K. pneumoniae and A. baumannii, blood from surgery alliance was the second most common source, while Citrobacter spp. showed higher gene prevalence in exudates. Notably, P. aeruginosa exhibited significant resistance gene carriage in urine samples, whereas for other organisms, urine was the least common source of resistance genes. No statistically significant association was observed with gender, age, and MIC values, highlighting that the distribution of these resistance genes is independent of patient demographics.
4 Discussion
Our investigation into the 2-year prevalence of carbapenem resistance among E. coli, K. pneumoniae, Citrobacter, A. baumannii, Proteus, and P. aeruginosa isolates in a tertiary care hospital in South India reveals critical insights into the ongoing challenge of antibiotic resistance in healthcare settings. Consistent with findings from other regions in India and globally, our study underscores the high prevalence of carbapenem resistance, particularly among Klebsiella and Acinetobacter species, which are notorious for their ability to acquire and disseminate resistance mechanisms (11). These resistance patterns can be compared with similar studies conducted globally, including the recent study by Abu Hammour et al. (12) in Jordan. A study conducted by Nieto-Saucedo also indicates a similar trend of resistance in A. baumannii and K. pneumoniae (13).
The resistance patterns observed in our study mirror those reported in other Indian regions, including North India, where similar high resistance rates have been documented (14). This nationwide issue highlights the urgent need for a coordinated and comprehensive approach to antimicrobial stewardship across the country. On a global scale, our data align with reports from regions with intense antibiotic pressure, such as Southeast Asia (15) and parts of Europe, further emphasizing the widespread nature of this public health threat (16).
Our study identified that carbapenem-resistant E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, Citrobacter, and Proteus were isolated from various clinical specimens, including exudates, blood, respiratory specimens, and urine. Notably, E. coli and K. pneumoniae were predominantly found in urine samples, while A. baumannii and P. aeruginosa were more frequently isolated from respiratory specimens. Citrobacter and Proteus were also prevalent in urine samples. Changes in sample-wise distribution were observed when compared to findings from other studies (17, 18). These variations may be attributed to differences in patient demographics, hospital settings, sample collection practices, and regional prevalence of specific pathogens. Additionally, the selection pressure exerted by antibiotic usage patterns in different regions could also contribute to these differences in sample-wise distribution.
One of the most compelling aspects of this study is the observation of quarterly fluctuations in carbapenem resistance rates. These temporal variations offer a novel perspective on resistance epidemiology, suggesting that resistance trends are dynamic rather than static. Such patterns indicate that multiple external and internal factors may influence the rise and fall of resistance over time. Seasonal variations in antibiotic prescribing practices—particularly during periods such as the monsoon season when infectious diseases tend to spike—could contribute to increased antibiotic usage, thereby exerting selective pressure that promotes resistance. Additionally, modifications in hospital admission rates, changes in infection control protocols, and shifts in patient demographics may all play a role in shaping these fluctuations. For instance, an influx of critically ill patients requiring broad-spectrum antibiotics or the implementation of new antibiotic stewardship measures during certain quarters could significantly alter resistance patterns. Similar temporal dynamics have been reported in regional studies, such as that by Modi C. in Gujarat (19), underscoring the need for continuous, time-sensitive surveillance to better understand and respond to emerging resistance trends (19, 20).
In our study, we observed a significant difference in resistance patterns between IPM and MEM among the CROs. These findings suggest varying efficacy of these two carbapenems against the resistant strains, which may be attributed to differences in their molecular structure, permeability, or affinity for penicillin-binding proteins (PBPs). This disparity highlights the need for tailored antibiotic stewardship strategies when selecting carbapenem agents for empirical therapy, considering the specific resistance profiles observed. Results of a recent study conducted by Ikenoue et al. (21) positively correlate with this difference.
Our analysis revealed that the male-to-female ratio in the resistant isolates was skewed toward males, which aligns with other studies suggesting gender-based differences in susceptibility to infections or healthcare-seeking behavior. Similar results have been observed in studies conducted by Wang et al. (22). Furthermore, the age-wise distribution of resistance in our study showed a higher prevalence in older adults, particularly those over 60. This age group is often more vulnerable to infections due to comorbidities and frequent hospitalizations, making them more likely to be exposed to resistant pathogens. This observation is consistent with Zhang et al. (23) (China), who found the highest rates among individuals aged 65–79 years, and is supported by the CRACKLE study, which reported a median age of 70 years for carbapenem-resistant K. pneumoniae infections. However, a different study identified a significant proportion (48.30%) of carbapenem-resistant infections in the 36–65 age group, with lower prevalence in those aged 66–95 years and 0–33 years (24). This discrepancy highlights regional or methodological variations in age-related resistance patterns and underscores the importance of context-specific analyses in understanding resistance dynamics.
In our study, the ward-wise distribution of CROs showed a higher prevalence in ICUs and surgical wards, reflecting the increased use of broad-spectrum antibiotics and the higher risk of nosocomial infections in these settings. This finding aligns with (25), where the majority of carbapenem-resistant isolates were obtained from general wards (41.5%) and ICUs (33.2%). It is also consistent with a study conducted by Kumari N, which found a significant proportion of carbapenemase-producing isolates in the medicine ICU (47.0%) and surgery ward (35.3%) (18). However, Nair and Vaz (26) reported that the majority of CRE isolates were found in hospitalized patients (42%), followed by OPD (32%) and ICU (26%). While our study and others consistently highlight ICUs as significant hotspots for resistance, the variation in distribution across different healthcare settings may be attributed to differences in antimicrobial stewardship practices, infection control protocols, patient case-mix, and institutional diagnostic approaches.
In addition to carbapenem resistance, our study also documented the resistance profiles of these isolates to other commonly used antibiotics. The data revealed a high degree of MDR, with many CROs also showing resistance to cephalosporins, aminoglycosides, and fluoroquinolones. Our findings align with previous studies (27, 28), which reported high levels of MDR among carbapenem-resistant isolates. Similar to these studies, we observed resistance to cephalosporins, aminoglycosides, and fluoroquinolones. While specific resistance rates varied, the consistent trend across studies highlights the urgent need for alternative therapeutic strategies to combat these extensively drug-resistant pathogens.
In our study, the distribution of carbapenemase genes among GNB revealed a concerning trend, with a notably high prevalence of blaNDM, blaOXA-48, and blaVIM genes across several species. E. coli and K. pneumoniae exhibited particularly high rates of these genes, with E. coli showing 88% blaNDM, 60% blaOXA-48, and 71% blaVIM, while K. pneumoniae displayed 86, 71, and 80%, respectively. This highlights the growing threat posed by MDR Enterobacteriaceae, which are known to be efficient in acquiring and disseminating resistance determinants (5). The co-existence of these genes significantly compromises therapeutic efficacy, often leaving limited treatment options such as polymyxins or tigecycline, which themselves are associated with toxicity and emerging resistance.
Among non-fermenters, A. baumannii showed an 84% prevalence of blaNDM and moderate levels of blaOXA-48 (37.8%) and blaVIM (42%), suggesting its expanding role in healthcare-associated infections with a multidrug-resistant phenotype. P. aeruginosa also carried all three genes at appreciable levels, further compounding its known intrinsic resistance mechanisms. Though Proteus spp. and Citrobacter spp. had comparatively lower frequencies, the presence of blaNDM in 48.6% of Citrobacter spp. and the detection of all three genes even in Proteus spp. point toward the silent spread of these genes among lesser-monitored species (29, 30).
Of particular concern is the co-occurrence of multiple resistance genes within the same isolate, as seen in 46% of cases (NDM + VIM + OXA-48), 9% (NDM + VIM), and 12% (NDM + OXA-48). These combinations enhance the spectrum and level of resistance, potentially leading to complete therapeutic failure. Such MDR not only limits the choice of antimicrobials but also increases morbidity, mortality, and healthcare costs. The presence of multiple carbapenemase genes in a single isolate also raises the possibility of horizontal gene transfer through plasmids, accelerating the spread of resistance within hospital environments (5). This underlines the urgent need for robust molecular surveillance, strict antimicrobial stewardship, and effective infection control measures to mitigate the clinical and epidemiological impact of these formidable pathogens.
5 Strengthening hospital infection control in response to CRO trends
As an impact of this study, several targeted interventions were implemented within the hospital to strengthen infection prevention and control (IPC) practices and mitigate the spread of CROs. Active surveillance cultures were initiated in high-risk units such as ICUs and surgical wards to facilitate early detection of CRO colonization. Isolation precautions were reinforced for affected patients, with dedicated staff and cohorting strategies to minimize cross-transmission. Periodic audits and feedback sessions were introduced to monitor antibiotic prescribing patterns, ensuring stricter adherence to antimicrobial stewardship protocols. Hand hygiene practices were re-emphasized through staff training, compliance monitoring, and real-time feedback systems. Environmental disinfection procedures were also upgraded, incorporating enhanced terminal cleaning with sporicidal agents and UV-based disinfection in critical care areas. Additionally, laboratory reports for isolates carrying multiple carbapenemase genes were flagged with clinical alerts, enabling timely and appropriate infection management. Collectively, these measures contributed to a more vigilant and proactive IPC environment within the healthcare facility.
6 Conclusion
This 2-year surveillance study highlights the alarming burden of carbapenem resistance among GNB, particularly in K. pneumoniae, A. baumannii, and E. coli, in a tertiary care hospital setting in South India. The high prevalence of resistance to multiple antibiotic classes, along with the co-existence of major carbapenemase genes (blaNDM, blaOXA-48, and blaVIM), reflects the growing challenge posed by MDR organisms in clinical practice. The study also revealed significant variations in resistance trends based on sample type, patient age, gender, and hospital wards. Notably, the quarterly fluctuations in resistance patterns suggest a dynamic and evolving resistance profile, underscoring the importance of continuous, time-based monitoring. The widespread distribution of carbapenemase genes across various species, including lesser-monitored organisms like Citrobacter and Proteus, calls for enhanced molecular diagnostics and routine surveillance.
In conclusion, the findings reaffirm the critical need for ongoing surveillance, rational antibiotic use, and targeted research to address the threat of antimicrobial resistance. These insights contribute meaningfully to the broader understanding of carbapenem resistance patterns in healthcare settings and provide a foundation for future investigations aimed at developing effective containment strategies.
7 Limitations and future directions
This single-center study limits generalizability due to localized antimicrobial practices. Molecular analysis was restricted to gene detection without assessing expression levels. Environmental or healthcare worker surveillance was not performed. Additionally, data on prior antibiotic exposure and patient comorbidities were incomplete, limiting correlation analyses. Lack of data on regional antibiotic policies has also been acknowledged as a limitation in this study, as it restricts broader interpretation of resistance trends. Future studies should include multi-center surveillance, whole-genome sequencing, longitudinal monitoring, environmental sampling, and evaluation of targeted antimicrobial stewardship interventions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by SRM Medical College Hospital and Research Centre. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
RL: Conceptualization, Formal analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. LV: Conceptualization, Formal analysis, Supervision, Validation, Writing – review & editing. JT: Project administration, Resources, Validation, Writing – review & editing. DN: Data curation, Methodology, Software, Writing – review & editing. PV: Formal analysis, Investigation, Methodology, Writing – review & editing. VP: Data curation, Methodology, Validation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to thank Rooban Sivakumar from the Department of Biochemistry, SRM Medical College Hospital and Research Centre, for his invaluable assistance in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1571231/full#supplementary-material
Abbreviations
GNB, Gram-negative Bacilli; IPM, Imipenem; MEM, Meropenem; CRO, Carbapenem Resistant Organisms; MDR, Multi-Drug Resistance; NARS-Net, National Antimicrobial Resistance Surveillance Network; MA, Medical Alliance; SA, Surgery Alliance; PA, Paediatric Alliance; MIC, Minimum Inhibitory Concentration; DD, Disc Diffusion; SPSS, Statistical Package for the Social Sciences; IPC, Infection Prevention and Control.
References
1. Mancuso, G, De Gaetano, S, Midiri, A, Zummo, S, and Biondo, C. The challenge of overcoming antibiotic resistance in Carbapenem-resistant gram-negative Bacteria: ‘attack on titan’. Microorganisms. (2023) 11:1912. doi: 10.3390/microorganisms11081912
2. Elshamy, AA, and Aboshanab, KM. A review on bacterial resistance to Carbapenems: epidemiology, detection and treatment options. Future Sci OA. (2020) 6:FSO438. doi: 10.2144/fsoa-2019-0098
3. Jin, H-W, and Eom, Y-B. Antibacterial and anti-biofilm effects of Thymoquinone against Carbapenem-resistant Uropathogenic Escherichia coli. Indian J Microbiol. (2024) 64:1747–56. doi: 10.1007/s12088-024-01231-8
4. Aurilio, C, Sansone, P, Barbarisi, M, Pota, V, Giaccari, LG, Coppolino, F, et al. Mechanisms of action of Carbapenem resistance. Antibiotics. (2022) 11:421. doi: 10.3390/antibiotics11030421
5. Abhishek, S, Naik, SS, Leela, KV, and Maheswary, D. Genotypic distribution and antimicrobial susceptibilities of Carbapenemase-producing Enterobacteriaceae isolated in tertiary Care Hospital in South India. J Pure Appl Microbiol. (2022) 16:2488–95. doi: 10.22207/JPAM.16.4.08
6. Wang, X, Zhang, H, Yu, S, Li, D, Gillings, MR, Ren, H, et al. Inter-plasmid transfer of antibiotic resistance genes accelerates antibiotic resistance in bacterial pathogens. ISME J. (2024) 18:dwrad032. doi: 10.1093/ismejo/wrad032
7. Balkhair, A, Al Saadi, K, and Al Adawi, B. Epidemiology and mortality outcome of carbapenem- and colistin-resistant Klebsiella pneumoniae, Escherichia coli, Acinetobacter baumannii, and Pseudomonas aeruginosa bloodstream infections. IJID Regions. (2023) 7:1–5. doi: 10.1016/j.ijregi.2023.01.002
8. Shibabaw, A, Sahle, Z, Metaferia, Y, Atlaw, A, Adenew, B, Gedefie, A, et al. Epidemiology and prevention of hospital-acquired carbapenem-resistant Enterobacterales infection in hospitalized patients, Northeast Ethiopia. IJID Regions. (2023) 7:77–83. doi: 10.1016/j.ijregi.2023.02.008
9. Verwaest, C. Meropenem versus imipenem/cilastatin as empirical monotherapy for serious bacterial infections in the intensive care unit. Clin Microbiol Infect. (2000) 6:294–302. doi: 10.1046/j.1469-0691.2000.00082.x
10. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 34th ed. CLSI supplement M100. Wayne, PA: CLSI (2025) Available at: www.clsi.org
11. Sharma, K, Tak, V, Nag, VL, Bhatia, PK, and Kothari, N. An observational study on carbapenem-resistant Enterobacterales (CRE) colonisation and subsequent risk of infection in an adult intensive care unit (ICU) at a tertiary care hospital in India. Inf Prev Prac. (2023) 5:100312. doi: 10.1016/j.infpip.2023.100312
12. Hammour, KA, Abu-Farha, R, Itani, R, Karout, S, Allan, A, Manaseer, Q, et al. The prevalence of Carbapenem resistance gram negative pathogens in a tertiary teaching Hospital in Jordan. BMC Infect Dis. (2023) 23:634. doi: 10.1186/s12879-023-08610-4
13. Nieto-Saucedo, JR, López-Jacome, LE, Franco-Cendejas, R, Colín-Castro, CA, Hernández-Duran, M, Rivera-Garay, LR, et al. Carbapenem-resistant gram-negative Bacilli characterization in a tertiary care center from El Bajio, Mexico. Antibiotics. (2023) 12:1295. doi: 10.3390/antibiotics12081295
14. Chatterjee, N, Nirwan, PK, Srivastava, S, Rati, R, Sharma, L, Sharma, P, et al. Trends in carbapenem resistance in pre-COVID and COVID times in a tertiary care hospital in North India. Ann Clin Microbiol Antimicrob. (2023) 22:1. doi: 10.1186/s12941-022-00549-9
15. Handa, VL, Patel, BN, Bhattacharya, DA, Kothari, RK, Kavathia, DG, and Vyas, BRM. A study of antibiotic resistance pattern of clinical bacterial pathogens isolated from patients in a tertiary care hospital. Front Microbiol. (2024) 15:1383989. doi: 10.3389/fmicb.2024.1383989
16. Karlijn, VL, Voor In ‘t Holt Anne, F, and Vos Margreet, C. A systematic review and Meta-analyses of the clinical epidemiology of Carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother. (2017) 62:01730-17. doi: 10.1128/aac.01730-17
17. Ranganathan, SS, Wanigatunge, C, Senadheera, GPSG, and Beneragama, BVSH. A national survey of antibacterial consumption in Sri Lanka. PLoS One. 16:e0257424. doi: 10.1371/journal.pone.0257424
18. Kumari, N, Kumar, M, Katiyar, A, Kumar, A, Priya, P, Kumar, B, et al. Genome-wide identification of carbapenem-resistant gram-negative bacterial (CR-GNB) isolates retrieved from hospitalized patients in Bihar, India. Sci Rep. (2022) 12:8477. doi: 10.1038/s41598-022-12471-3
19. Modi, CM, Singh, SP, Pandya, YG, Patel, CP, and Patel, RM. Prevalence of Carbapenem resistant Enterobacteriaceae in a tertiary Care Hospital of Gujarat, India. J Clin Diagn Res. (2021) 273:19. doi: 10.7860/jcdr/2021/47332.14627
20. Moghnieh, R, Abdallah, D, Jadayel, M, Zorkot, W, el Masri, H, Dib, MJ, et al. Epidemiology, risk factors, and prediction score of carbapenem resistance among inpatients colonized or infected with 3rd generation cephalosporin resistant Enterobacterales. Sci Rep. (2021) 11:14757. doi: 10.1038/s41598-021-94295-1
21. Ikenoue, C, Matsui, M, Inamine, Y, Yoneoka, D, Sugai, M, Suzuki, S, et al. The importance of meropenem resistance, rather than imipenem resistance, in defining carbapenem-resistant Enterobacterales for public health surveillance: an analysis of national population-based surveillance. BMC Infect Dis. (2024) 24:209. doi: 10.1186/s12879-024-09107-4
22. Wang, N, Zhan, M, Wang, T, Liu, J, Li, C, Li, B, et al. Long term characteristics of clinical Distributioand resistance trends of Carbapenem-resistant and extended-Spectrum β-lactamase Klebsiella pneumoniae infections: 2014–2022. Infect Drug Resist. (2023) 16:1279–95. doi: 10.2147/IDR.S401807
23. Zhang, Y, Wang, Q, Yin, Y, Chen, H, Jin, L, Gu, B, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE network. Antimicrobial Agents Chemother. (2018) 62:10–1128. doi: 10.1128/aac.01882-17
24. Verma, G, Nayak, SR, Jena, S, Panda, SS, Pattnaik, D, Praharaj, AK, et al. Prevalence of Carbapenem-resistant Enterobacterales, Acinetobacter baumannii, and Pseudomonas aeruginosa in a tertiary Care Hospital in Eastern India: a pilot study. J Pure Appl Microbiol. (2023) 17:2243–9. doi: 10.22207/JPAM.17.4.21
25. Thomas, N, and Sarwat, T. Prevalence of Carbapenem resistant Enterobacteriaceae in a tertiary care hospital. Int J Curr Microbiol Appl Sci. (2019) 8:1418–24. doi: 10.20546/ijcmas.2019.811.166
26. Nair, PK, and Vaz, MS. Study of the prevalence of metallo-β-lactamase-producing enterobacteriaceae from a tertiary care hospital in Mumbai. J Acad Clin Microbiol. (2014) 16:53. doi: 10.4103/0972-1282.144705
27. Su, Y, Xin, L, Zhang, F, Peng, C, Li, Z, Liu, C, et al. Drug resistance analysis of three types of avian-origin carbapenem-resistant Enterobacteriaceae in Shandong Province, China. Poult Sci. (2023) 102:102483. doi: 10.1016/j.psj.2023.102483
28. Lathakumari, RH, Ravi, S, Trisal, S, Vajravelu, LK, Vishnudasan, D, Thulukanam, J, et al. Efficacy of green synthesised Iron oxide nanoparticles against various Uropathogens: a cross-sectional study. J Clin Diagn Res. (2022) 16:10. doi: 10.7860/jcdr/2022/58018.17065
29. Zhao, Q, Guo, L, Ye, K, Wang, L, Yang, J, and Ye, L. Epidemiology, phylogeny and genetic characterization of carbapenem-resistant Citrobacter spp. from 5 hospitals in China. J Glob Antimicrob Resist. (2025) 42:207–13. doi: 10.1016/j.jgar.2025.03.003
30. Wang, B, Zhang, W, Zhang, H, Li, M, Zhang, Z, Peng, X, et al. Comparative analysis of clinical characteristics and outcomes between carbapenem-resistant and carbapenem-sensitive Klebsiella pneumoniae infections: insights from a tertiary hospital in northern China. Front Med. (2025) 12:1499057. doi: 10.3389/fmed.2025.1499057
Keywords: carbapenem resistance, molecular characterization, hospital acquired infections, imipenem, meropenem
Citation: Lathakumari RH, Vajravelu LK, Thulukanam J, Nair DM, Vimala PB and Panneerselvam V (2025) Prevalence and molecular insights into carbapenem resistance: a 2-year retrospective analysis of superbugs in South India. Front. Med. 12:1571231. doi: 10.3389/fmed.2025.1571231
Edited by:
Chaitra Shankar, University of Michigan, United StatesReviewed by:
Mostafa Javanian, Babol University of Medical Sciences, IranVarun Shamanna, Kempegowda Institute of Medical Sciences, India
Copyright © 2025 Lathakumari, Vajravelu, Thulukanam, Nair, Vimala and Panneerselvam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rahul Harikumar Lathakumari, cmFodWxobDE5OTdAZ21haWwuY29t; Leela Kakithakara Vajravelu, bGVlbGF2QHNybWlzdC5lZHUuaW4=
†ORCID: Rahul Harikumar Lathakumari, orcid.org/0000-0002-7222-7090
 Rahul Harikumar Lathakumari
Rahul Harikumar Lathakumari Leela Kakithakara Vajravelu
Leela Kakithakara Vajravelu Jayaprakash Thulukanam
Jayaprakash Thulukanam Dakshina M. Nair
Dakshina M. Nair Poornima Baskar Vimala
Poornima Baskar Vimala Vishnupriya Panneerselvam
Vishnupriya Panneerselvam