- 1Department of Medical Research, E-DA Hospital, I-Shou University, Kaohsiung, Taiwan
- 2Department of Statistics, National Taipei University, New Taipei City, Taiwan
- 3Department of Obstetrics and Gynecology, National Cheng Kung University Hospital, National Cheng Kung University, Tainan, Taiwan
- 4Department of Obstetrics and Gynecology, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Kaohsiung, Taiwan
- 5Department of Obstetrics and Gynecology, Jen-Ai Hospital, Taichung, Taiwan
Background: Biological barriers are essential for maintaining integrity and function and preventing microbial invasion. Maternal barrier dysfunction may play a role in preterm birth (PTB). However, the link between maternal barrier function and PTB is still unknown. This study aims to identify genetic evidence supporting the role of maternal barrier genes in PTB risk.
Methods: We examined 201 barrier-related genes to assess their association with PTB susceptibility. We utilized the FinnGen study, published literature's whole-genome sequencing (WGS) summary statistics and Early Growth Genetics (EGG) meta-analysis to identify the maternal barrier gene associated with PTB.
Results: Findings from the analysis of the maternal genome highlighted several barrier genes (NOTCH1, LAMA4, F11R, MAGI1, MAGI2, TJP1, PARD3, CLDN10, CLDN14, CLDN15, GRHL3, CGNL1, LAMB2, RHOA, and LRP5) associated with PTB. Notably, NOTCH1 was supported by at least two independent genomic datasets.
Conclusion: The established roles of NOTCH1 in vascular barrier function, angiogenesis, decidualization, intestinal epithelial barrier, and inflammation support its mechanistic involvement. Our research enhances our understanding of maternal barrier genes linked to PTB, providing valuable insights for future prevention and intervention strategies.
1 Introduction
Preterm birth (PTB) refers to the birth of a baby before completing 37 weeks of gestation (1). Epidemiological evidence indicates that PTB occurred in ~9.9% of all live births worldwide in 2020 (1). Unfortunately, ~70% of PTBs are spontaneous PTBs (sPTBs; including preterm prelabor rupture of membranes and idiopathic PTB), and remain poorly understood, with limited tools available for early identification or prevention (2). The degree of prematurity is directly proportional to the risks of mortality and morbidity (1). Moreover, PTB is associated with increased risks of long-term health and neurodevelopmental problems (3). The etiology of PTB is intricate and remains to be further explored. Cumulative evidence indicates that maternal medical disorders, antenatal risk factors, inflammatory diseases, genetic predispositions, socioeconomic factors, and environmental factors are associated with the risk of PTB (2, 4).
Biological barriers are crucial for maintaining their integrity and function, as well as preventing microbial invasion (5). Various organs possess different biological barriers, such as the skin, the intestine, the reproductive system, the lung, the central nervous system, the placental villi, and the cervix (5, 6). Several inflammatory disorders are linked to barrier dysfunction, including inflammatory bowel diseases (IBD) (7), allergic diseases (8), atopic dermatitis (9), central nervous system disorders (10), and infections. Inflammatory processes are hypothesized to play an important role in PTB, and the origin of inflammation may be due to infection or sterile inflammation (7). Approximately 60% of PTBs could be ascribed to sterile inflammation (7). Emerging evidence has linked maternal IBD (8), allergic diseases (asthma, allergic rhinitis, allergic conjunctivitis, food allergy, drug allergy, and contact dermatitis) (9), systemic maternal infections, and bacterial vaginosis (10) with risks for PTB. Barrier-related genes are those that regulate the structural and functional integrity of biological barriers. Most of these genes encode junctional or structural proteins (e.g., CLDN and TJP1), while others are involved in signaling pathways (e.g., the Wnt signaling pathway) or tissue-specific functions (e.g., mucins in the intestine and galectins in the reproductive tract). However, whether barrier dysfunction plays a role in preterm is still unknown. Thus, investigating the correlation between maternal barrier function and PTB is particularly interesting in light of these findings.
It is crucial to note that women with a history of PTB in the past are at high risk for recurrent PTB (11). These findings provide evidence of genetic predisposition to PTB. Based on epidemiological research, sPTB is influenced by both maternal and fetal genomes, but predominantly by the maternal genome (12–14). Genome-wide association studies (GWAS) (15–19), whole-exome sequencing (20), and whole-genome sequencing (WGS) (21) studies have indicated that genetic variants in maternal genomes contribute to the risk of PTB. Previous studies have identified over 750 single-nucleotide polymorphisms (SNPs) in more than 240 genes in the maternal and fetal genomes that may be associated with PTB or gestational duration at birth (2, 22). These genes involved in tissue remodeling, vascular, endothelial, metabolic, inflammatory, and immune processes are implicated (2, 22). Together, these genetic approaches can be used to confirm known associations of genetic variants and/or discover novel genetic variants.
In the present study, we hypothesized that impaired maternal barrier function may contribute to PTB. To this end, we utilized available data sources, including the FinnGen study, WGS summary statistics from published literature, and the EGG meta-analysis, to identify evidence for the involvement of maternal barrier genes in susceptibility to PTB.
2 Materials and methods
2.1 The study design
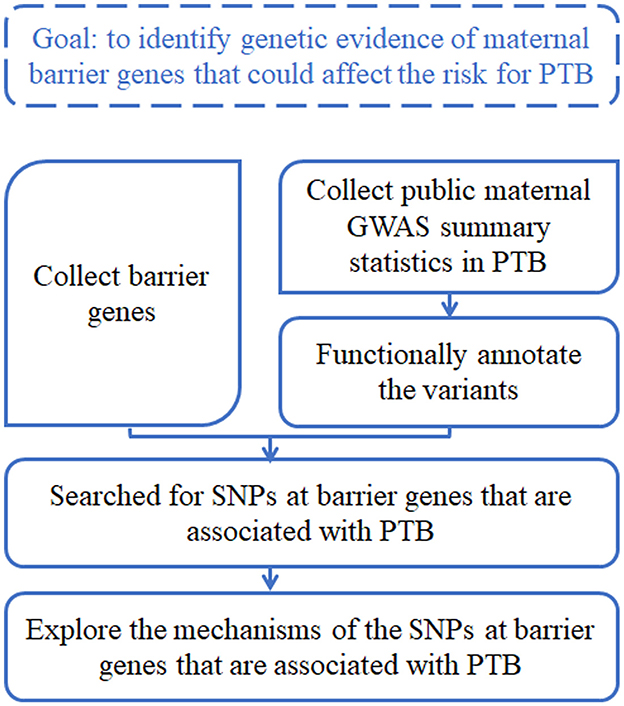
The goal of this study was to identify genetic evidence of maternal barrier genes that could affect the risk for PTB. The analysis workflow was briefly described as follows (Figure 1), and the details were shown in respective sections. First, we collected a list of barrier genes from the literature search. Second, we surveyed and collected public maternal GWAS summary statistics in PTB, and functionally annotated the variants using computational tools. Third, we searched for SNPs at barrier genes that were associated with PTB. Fourth, we explored the mechanisms of those SNPs at barrier genes that are associated with PTB.
2.2 Barrier genes
We collected 201 barrier-related genes from the literature (Supplementary Table S1). Of these, 22 genes associated with epidermal malignancy were categorized into structural components, microenvironmental factors, and differentiation-related groups (23). A total of 146 genes related to intestinal epithelial barrier dysfunction were identified, including those involved in the mucus layer, tight junctions, adherens junctions, desmosomes, hemidesmosomes, cytoskeleton, extracellular matrix, and regulatory proteins (24). A total of 12 genes associated with the blood–brain barrier were classified as central nervous system endothelial cell genes involved in angiogenesis and barriergenesis (25). In total, 19 genes encoding galectins were included based on their role in the vaginal microenvironment as a defense barrier (26), along with two additional barrier-related genes (27, 28).
2.3 Datasets
We surveyed three existing maternal GWAS studies in PTB. The first study was the PTB GWAS results in FinnGen (29). FinnGen is a research project that combines genotype data from Finnish biobanks and digital health record data from Finnish health registries (https://www.finngen.fi/en) to provide new insights into disease genetics, and it has conducted a GWAS of 1,932 diseases in its 224,737 participants. We downloaded the PTB GWAS results from FinnGen release 8, which included 20,153,666 variants and a sample size of 7,678 cases and 148,153 controls. It was a maternal GWAS study, where the cases were females with a history of PTB, and the controls were females without a history of PTB. The second study aimed to identify molecular characteristics of PTB using multi-omic data (21). It used a cohort of 791 family trios from various ancestries, of which 270 had PTB. The study integrated WGS data from the fathers, mothers, and newborns in these family trios and RNA-seq gene expression and DNA methylation data from maternal blood samples and gathered comprehensive clinical information concerning pregnancy, delivery, and newborn health. We downloaded their released maternal GWAS results, which included the variants showing FDR < 0.1. The third study was a meta-analysis of results from multiple PTB GWASs (19), with a total sample size of 15,419 cases and 217,871 controls. The summary statistics were derived from maternal GWASs, where the cases consisted of females with a history of PTB, and the controls were females without a history of PTB. The data were downloaded from the website of the Early Growth Genetics (EGG) Consortium (http://egg-consortium.org).
2.4 Variant functional annotation of GWAS summary statistics
We searched for SNPs at barrier genes that are associated with PTB. For studies that released full GWAS summary statistics (i.e., the FinnGen and EGG datasets), the variant annotation was applied by the Functional Mapping and Annotation (FUMA) of the GWASs web application (30), where the SNPs showing p-value < 1 × 10−5 could be lead SNPs for further analysis. The obtained data of the published literature's WGS summary statistics were not full GWAS summary statistics and included only the variants showing FDR < 0.1, so we used the threshold of FDR-adjusted p-value < 0.1 for this dataset. We used a less stringent threshold of p-values, often termed “suggestive threshold,” to increase discovery power (31, 32) for understanding what possible mechanisms behind the associations between the variants and PTB. Existing studies have used the suggestive threshold in the GWAS and found valuable insights (33–35). A SNP was mapped to a gene when at least one of the following conditions was met. First, the SNP was located on the gene body or up to 10 kb apart from the gene. Second, the SNP was an eQTL of a gene in at least one available tissue type of GTEx v8. Third, the SNP was located at least one of the known chromatin interaction regions in FUMA. A detailed setup for FUMA annotations is shown in Table 1. The regional association plot was generated by R.
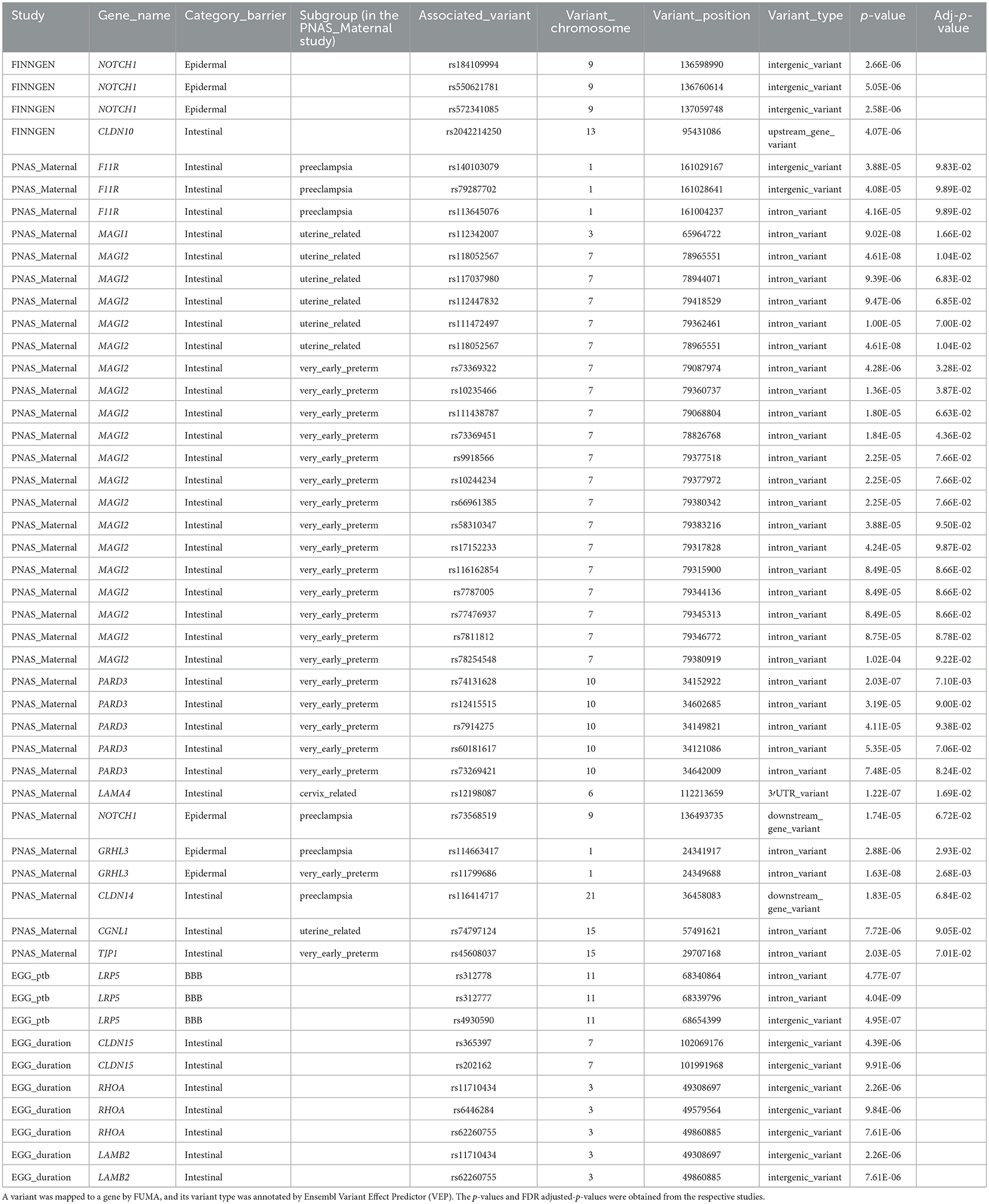
Table 1. The barrier genes that overlapped with significantly suggestive variants of GWAS results in PTB.
2.5 Ethical statement
The Institutional Review Board of the E-DA Hospital approved the study (EMRP-113-099).
3 Results
3.1 NOTCH1 and other barrier genes exhibit suggestive associations with PTB in maternal GWAS datasets
There are several types of barrier genes, such as the epidermal barrier, intestinal barrier, blood–brain barrier, and galectin genes. We thoroughly investigated all 201 barrier genes to explore their role in susceptibility to PTB (Supplementary Table S1). A comprehensive screening of various PTB datasets, such as GWAS and WGS, was conducted to identify associations with barrier genes. First, the FinnGen study was used to conduct maternal GWAS analysis on 7,678 PTB and 148,153 term cases. Genetic variants at 4 loci were associated with PTB at suggestive significance (Figure 2, upper panel, and Table 1). Two barrier genes (NOTCH1 and CLDN10) showed potential links to PTB in the FinnGen dataset among the maternal genome. Second, a maternal GWAS analysis was performed on 270 PTB and 521 term cases using the published literature's WGS summary statistics of maternal genomes (21). Genetic variants at 35 loci were associated with PTB at suggestive significance (Figure 2, middle panel, and Table 1). Ten barrier genes (NOTCH1, LAMA4, F11R, MAGI1, MAGI2, TJP1, PARD3, CLDN14, GRHL3, and CGNL1) showed potential links to PTB in the published literature's dataset among the maternal genome. Notably, the F11R, NOTCH1, GRHL3, and CLDN14 genes were associated with preeclampsia-associated PTB. Third, the EGG meta-analysis was used to conduct maternal GWAS analysis on 15,419 PTB and 217,871 term cases. Genetic variants at 10 loci were associated with PTB or gestational duration at birth at suggestive significance (Supplementary Figure S1 and Table 1). Four barrier genes (CLDN10, LAMB2, RHOA, and LRP5) showed potential links to PTB in the EGG meta-analysis among the maternal genome (Table 1). Maternal NOTCH1 was associated with PTB in the FinnGen dataset, which overlapped with findings in the published literature's WGS data. Unfortunately, we found no significant association for NOTCH1 in the EGG meta-analysis.
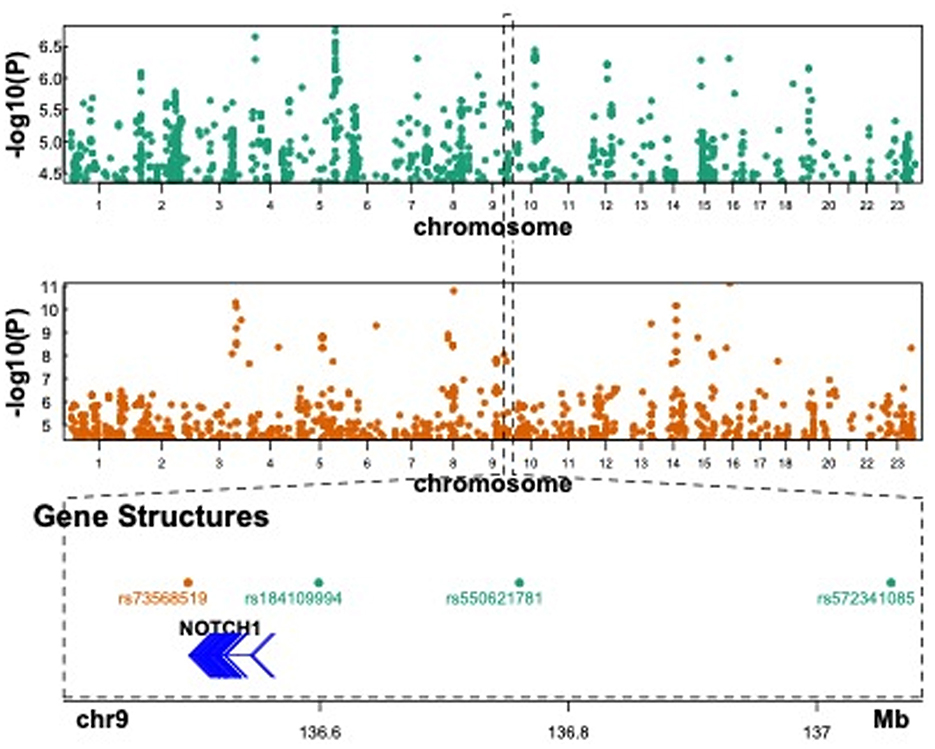
Figure 2. Published GWAS results in PTB and suggestive association at NOTCH1. The upper panel showed the Manhattan plot of Finngen GWAS in PTB when the p-value < 1 × 10−5. The middle panel showed the Manhattan plot of the maternal GWAS results in PTB with preeclampsia when FDR < 0.1. The lower panel showed a focal region in chromosome 9 where significant variants were found in both GWAS results.
3.2 Functional annotation of the NOTCH1 SNPs
Functional annotation of the significant SNPs from 15 imputed genes was displayed in Table 1. Four maternal SNPs (rs184109994, rs550621781, rs572341085, and rs73568519) at the NOTCH1 locus were associated with PTB. We highlight a genomic region spanning 136–137 Mb (rs184109994, rs550621781, rs572341085, and rs73568519) on chromosome 9 in Figure 2, lower panel. The alleles linked to PTB have not been previously reported. We mapped the associated variants through 3-D chromatin interaction. The circos plot clearly illustrates numerous chromatin interactions between the genomic risk locus and NOTCH1 (Figure 3). Maternal SNPs (rs184109994) lie within the enhancer region of NOTCH1. The mapped placenta is functionally involved in providing nutrients to the fetus, and therefore has implications for its association with the etiology of PTB (Supplementary Table S3). However, according to the GTEx database, whether the four SNPs on NOTCH1 can impact the messenger RNA expression is uncertain.
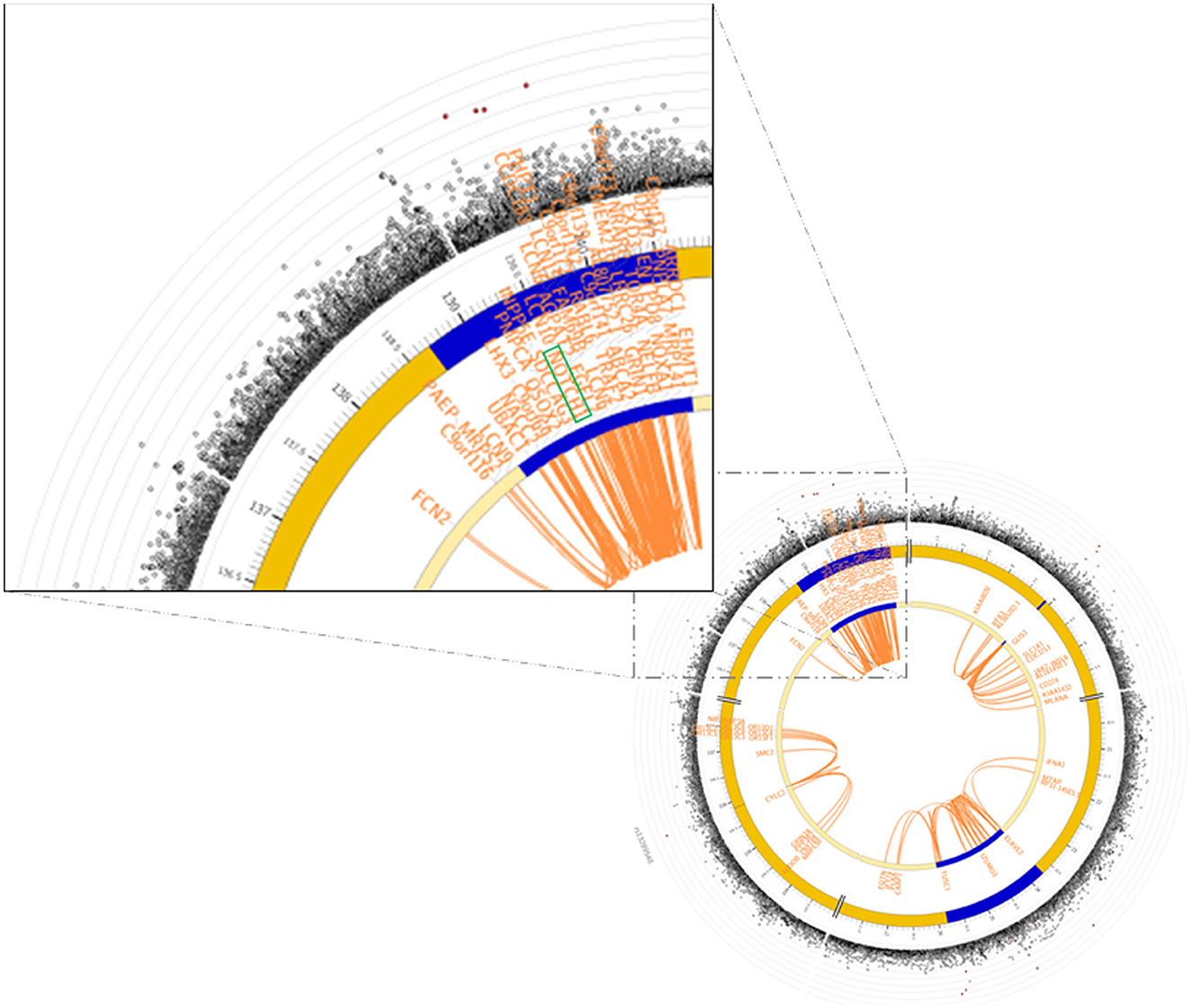
Figure 3. GWAS significant SNPs at NOTCH1 in Finngen overlapped a known chromatin interaction region. The circos plot on the lower-right side showed the chromatin interaction mapping results of chromosome 9, and the zoom-in on the NOTCH1 region was on the upper-left side. In the circos plot, the Manhattan plot on the most outer layer showed the significance of the SNPs with p-value < 0.05, and the SNPs colored in red were in strong LD (i.e., r2 > 0.8) to one of the independent significant SNPs in the locus. The second and third layers showed the chromosomal locations, and the genomic risk loci were highlighted in blue. The gene(s) being mapped by chromatin interaction were shown within the two layers, and were colored in orange. The links colored orange in the innermost region were chromatin interactions. Further details of the plot were shown in FUMA (https://fuma.ctglab.nl).
4 Discussion
GWAS has provided valuable insights into genetic risk factors and associated genomic regions for PTB. This is the first GWAS report on identifying maternal barrier genes associated with PTB. Analysis of the maternal genome's GWAS and EGG meta-analysis revealed several barrier genes (NOTCH1, LAMA4, F11R, MAGI1, MAGI2, TJP1, PARD3, CLDN10, CLDN14, CLDN15, GRHL3, CGNL1, LAMB2, RHOA, and LRP5) associations with PTB. At least two genomic datasets revealed associations of NOTCH1.
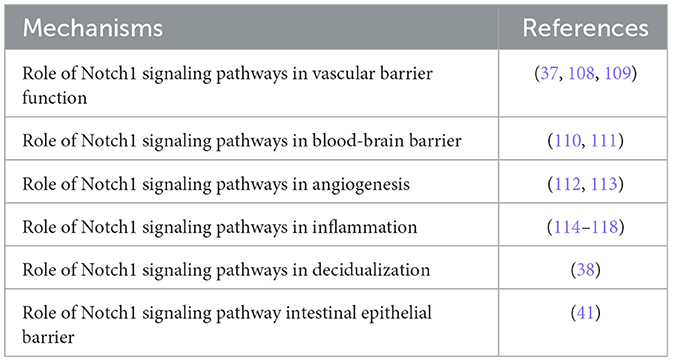
NOTCH1 was found to be expressed widely but with different tissue distributions (36). High expression was detected in the intermediate suprabasal layers, whereas low to intermediate expression was detected in lymphocytes in peripheral lymphoid tissues (36). The NOTCH1 signaling pathway in preterm has potential molecular functions, including vascular barrier function, angiogenesis, blood–brain barrier, decidualization, intestinal epithelial barrier, and inflammation (Table 2). First, NOTCH1 signaling is crucial in maintaining vascular stability (Supplementary Figure S2). Upon binding to Delta-like ligand 4, NOTCH1 releases the Notch intracellular domain (NICD), which translocates to the nucleus to induce anti-inflammation and pro-angiogenesis while suppressing endothelial cell proliferation. The non-canonical pathway involves the activation of NOTCH1 to release the transmembrane domain, which forms a complex with VE-cadherin to promote endothelial junction formation (37). Second, NOTCH1 signaling is crucial for decidualization progression (38). The decidua acts as a barrier during pregnancy by regulating trophoblast invasion and the immune response. A previous study indicated that PTB-associated genes RPBJ interact with NOTCH1 according to the STRING tool (21). The NICD regulates the expression of target genes with the DNA-binding protein RBPJ (39). NOTCH1 signaling via RBPJ regulates the expression of ovarian steroid receptor PGR and glucose transporter SLC2A1 during decidualization (40). Decidualization defects result in recurrent pregnancy loss, preeclampsia, preterm labor, and intrauterine growth restriction (40). Third, a previous study indicated that NOTCH1 regulates intestinal epithelial barrier function via balanced tight junction protein complexes and plays a vital role in the mucosal immune response (41). NOTCH1 is essential in early pregnancy, particularly during implantation and placentation. It enables interactions between the endometrium and trophectoderm, regulates extravillous trophoblast invasion, and aids spiral artery remodeling (38, 42). Additionally, it plays a role in placental angiogenesis by guiding vascular branching and maturation (38, 42). Disruption of NOTCH1 signaling has been linked to complications such as preeclampsia, intrauterine growth restriction, polycystic ovary syndrome, endometriosis, adenomyosis, infertility, and endometrial cancer (38, 42). Together, NOTCH1 is crucial for regulating vascular barrier function, angiogenesis, decidualization, intestinal epithelial barrier function, and inflammation during pregnancy, and is critical in preterm delivery (43–45).
The tight junction genes, which include F11R, MAGI1, MAGI2, and TJP1, encode proteins that interact with each other according to the STRING tool (46). Tight junction barrier disruption can increase paracellular permeability, allowing luminal pro-inflammatory molecules to activate the mucosal immune system, causing inflammation and tissue damage (47). F11R, which encodes the F11 receptor, is a tight junction protein that connects neighboring epithelial or endothelial cells (48). F11R is associated with microscopic colitis (49). In addition, F11R, E-cadherin, occludin, claudin-1, and ZO-1 are abundant in the human endocervix (50). A lack of tight junctions in the lower female reproductive tract allows pathogens and immune cells to move between epithelial cells (51). Furthermore, previous findings indicated that F11R is one of the candidate genes for preeclampsia (48, 52). TJP1 encodes tight junction protein 1, also known as Zonula occludens-1 (ZO-1). TJP1 is a tight junction protein that connects neighboring epithelial cells and provides cellular integrity (53). TJP1 downregulation in IBD impairs mucosal repair and promotes progression (54). In human placental development, TJP1 also plays a crucial role in trophoblast cell-cell fusion and differentiation (55). In addition, a previous study has shown that the downregulation of TJP1 due to inflammation may be a critical factor in the development of PROM (56). MAGI1 (MAGUK with inverted domain structure-1) is a tight junction protein that connects neighboring epithelial cells or vascular endothelial cells (57, 58). MAGI-1 and its interacting proteins localize to the tight junctions of epithelial cells, resulting in enhanced epithelial integrity (57). MAGI-1 is associated with Crohn's disease (CD) and microscopic colitis (49, 59). In addition, MAGI1 is crucial for adherens junction maturation and cell-cell adhesion mediated by VE-cadherin (58). It regulates vascular functions like permeability, NO production, and angiogenesis (57). MAGI2 (MAGUK with inverted domain structure-2) plays a crucial role in maintaining the barrier function of the kidney (60). In addition, previous studies indicated that significant associations were found between MAGI2 and celiac disease, IBD, CD, as well as ulcerative colitis (UC) (46, 61–63).
The FinnGen, EGG, and WGS data from published literature indicate that the maternal CLDN gene family (CLDN10, CLDN14, and CLDN15) was associated with PTB. Claudins in paracellular channels have three types of selectivity: anion, cation, and water (64). CLDN10, CLDN14, and CLDN15 are members of the claudin family associated with tight junctions (65). CLDN proteins are associated with regulating the differentiation of the intestinal epithelium (66). CLDN10 is associated with HELIX syndrome (67). CLDN14 acts as a barrier to cations in epithelial cells and is associated with non-syndromic hearing loss and hypercalciuric nephrolithiasis (68–71). The formation of CLDN15-based tight junctions plays a pivotal role in regulating the microenvironment of the small intestine, particularly in controlling ion conductance and ensuring normal-sized morphogenesis (72).
Our data indicate that the maternal laminin family (LAMA4 and LAMB2) was associated with PTB. LAMA4, which encodes the laminin subunit alpha 4, is vital in promoting cell migration, proliferation, apoptosis, angiogenesis in trophoblast cells, microvessel maturation, and maintaining endothelial cell tightness (73–75). LAMA4 is one of the isoforms of laminin that regulates the maturation and function of the blood-brain barrier (76). Previous studies have shown that LAMA4 is implicated in regulating the onset and progression of preeclampsia (73, 77, 78). LAMA4 is a critical factor in the differentiation and invasion of trophoblasts (78). Laminin β2 (LAMB2) is a crucial component present in the intestine, glomerular basement membrane, neuromuscular junctions, and various ocular structures, and it is associated with Pierson syndrome (79).
Additionally, some genes related to cell junctions (PARD3 and CGNL1) have been identified, and some genes associated with barrier functions (RHOA, GRHL3, and LRP5) have also been linked to PTB. PARD3 (Par-3 Family Cell Polarity Regulator, also known as PAR-3) is a regulator of cell polarity in tight junctions of epithelial cells. Previous studies suggest that PARD3 is linked to IBD, celiac disease, CD, and UC (63). CGNL1 (cingulin-like 1) co-localizes with actin filament bundles, suggesting it could be a key modulator linking intercellular junction assembly to actin cytoskeleton-regulated morphogenesis in angiogenesis (80). RHOA is essential for endothelial barrier function (81). RhoA regulates signal transduction, actomyosin dynamics, cell shape, adhesion, division, migration, trafficking, and proliferation (81). A previous study revealed a significant increase in GTP-bound RHOA in the myometrium of women undergoing spontaneous preterm labor (82). Grainyhead-like 3 (GRHL3) is essential for maintaining skin barrier function and epidermal proliferation (83). GRHL3 is linked to Van der Woude Syndrome and Neural tube defects (84–87). Low-density lipoprotein-related receptors 5 (LRP5) is a co-receptor of Wnt/β-catenin signaling and plays a significant role in retinal vasculature development (88). A previous study indicated that intronic variants of the LRP5 gene may be associated with obesity due to their impact on the WNT signaling pathway or lipid metabolism (89).
4.1 Clinical and research implications
The microbiome is a multifaceted characteristic influenced by various factors, such as host genetics and the environment (90). Given the mechanistic link between barrier defects, dysbiosis, and inflammation, it is tempting to speculate that barrier dysfunction leads to microbiota dysbiosis, with resultant inflammation and PTB. For instance, dysregulation of the barrier results in microbiota dysbiosis. The current gut–placenta axis hypothesis indicates that microbiota-derived metabolites or pathogenic microorganisms may pass from mother to fetus through the placenta and harm the fetus (91). Additionally, intrauterine infection leading to PTB is a result of pathogens ascending from the vagina (92). Finally, vaginal dysbiosis is linked to bacterial vaginosis, PTB, premature membrane rupture, and chorioamnionitis (93, 94). Although our current study does not include microbiome analysis, given these facts, it is worthwhile to investigate the relationship between tight junction genes (F11R, MAGI1, MAGI2, and TJP1), dysbiosis, and PTB in future studies.
Ambient air pollution, including PM2.5, nitrogen dioxide (NO2), and O3, is associated with adverse perinatal outcomes, including PTB (95–97), which may be attributed to inflammation (98), placental inflammation, and reduced blood flow (99, 100). Additionally, air pollutants can disrupt the epithelial barrier, contributing to respiratory diseases such as asthma and chronic obstructive pulmonary disease (101). These findings parallel our observations of barrier-gene dysregulation in PTB, suggesting that genetic susceptibility in barrier-related pathways may exacerbate the inflammatory effects of environmental exposures. Such dysfunction of the barriers could make the maternal–fetal interface more susceptible to inflammation caused by pollutants, thereby increasing the risk of PTB. This underscores a potential interaction between genetic factors and environmental influences in PTB.
Preeclampsia and PTB may be linked to maternal barrier defects, indicating that they may share similar mechanisms (21). For example, endothelial dysfunction is prevalent in preeclampsia, characterized by barrier disruption and reduced vasodilatory capacity, which can lead to PTB (102). VE-cadherin is a key protein in endothelial cells that regulates vascular permeability and cell–cell contacts. If it is disrupted, the endothelial barrier function may be compromised, causing inflammation and other cellular dysfunctions (103, 104). Our findings indicate a correlation between these genes and PE, as well as VE-cadherin, such as NOTCH1, LAMA4, MAGI1, and F11R. Unfortunately, we could not find variants at the NOTCH1, LAMA4, MAGI1, and F11R loci associated with PE in the FinnGen database (Supplementary Table S2).
4.2 Strengths and limitations
The major strength of the study is its examination of the potential association between maternal barrier genes and PTB. We identified several maternal barrier genes in different datasets. Our study has provided new insights into the dysfunction of the barrier, which can disrupt microbial homeostasis, leading to inflammation and PTB.
We have identified certain limitations in our study. First, our data were unable to differentiate between medically indicated PTB and spontaneous PTB (preterm pre-labor rupture of membranes and idiopathic PTB). Second, we did not observe a significant association for NOTCH1 in the EGG meta-analysis. This could be explained by the large sample sizes, which have resulted in modest discoveries for PTB due to small effect sizes (2, 105). In addition, concerns have been raised about the suitability of meta-analysis methodologies in GWAS due to preterm heterogeneity observed among studies investigating the same trait (2, 105). The factors contributing to variability among studies can differ significantly due to variations in measurement techniques and research methodologies, the incorporation of diverse ethnic populations, exposure to varied environmental influences, and the use of different genotyping platforms (2, 105, 106). Third, the three significant NOTCH1 variants are located in UTR regions and have no direct effects attributed to these SNPs.
5 Conclusion
Early detection of the risk of PTB can reduce the global burden of adverse neonatal outcomes (107). This study confirms that genomic constitutions may contribute to the risk of PTB in women before or during pregnancy. Our findings, based on GWAS, provide novel insights into maternal barrier function and PTB. Further investigations are warranted to replicate the association between barrier genes and PTB and to explore the mechanisms of barrier defects in the pathogenesis of PTB.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the Institutional Review Board of the E-DA Hospital approved the study (EMRP-113-099). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
K-RC: Funding acquisition, Investigation, Methodology, Writing – review & editing, Writing – original draft. S-KC: Investigation, Methodology, Writing – review & editing, Conceptualization. P-LK: Conceptualization, Investigation, Methodology, Writing – review & editing, Funding acquisition, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the Cheng Chen Foundation, National Science and Technology Council (MOST 110-2314-B-006-039-MY2 and NSTC 112-2314-B-650-003-MY3 to P-LK and NSTC 113-2320-B-650-002-MY3 to K-RC) and the E-DA Hospital (EDAHJ113007 and EDAHS113016 to K-RC), Taiwan.
Acknowledgments
The authors thank all the participants and investigators in the FinnGen and EGG studies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1580877/full#supplementary-material
References
1. Ohuma EO, Moller AB, Bradley E, Chakwera S, Hussain-Alkhateeb L, Lewin A, et al. National, regional, and global estimates of preterm birth in 2020, with trends from 2010: a systematic analysis. Lancet. (2023) 402:1261–71. doi: 10.1016/S0140-6736(23)00878-4
2. Mead EC, Wang CA, Phung J, Fu JY, Williams SM, Merialdi M, et al. The role of genetics in preterm birth. Reprod Sci. (2023) 30:3410–27. doi: 10.1007/s43032-023-01287-9
3. Cheong JLY, Burnett AC, Treyvaud K, Spittle AJ. Early environment and long-term outcomes of preterm infants. J Neural Transm. (2020) 127:1–8. doi: 10.1007/s00702-019-02121-w
4. Couceiro J, Matos I, Mendes JJ, Baptista PV, Fernandes AR, Quintas A. Inflammatory factors, genetic variants, and predisposition for preterm birth. Clin Genet. (2021) 100:357–67. doi: 10.1111/cge.14001
5. Archie SR, Al Shoyaib A, Cucullo L. Blood-brain barrier dysfunction in CNS disorders and putative therapeutic targets: an overview. Pharmaceutics. (2021) 13:1779. doi: 10.3390/pharmaceutics13111779
6. Vidal MS Jr, Lintao RCV, Severino MEL, Tantengco OAG, Menon R. Spontaneous preterm birth: involvement of multiple feto-maternal tissues and organ systems, differing mechanisms, and pathways. Front Endocrinol. (2022) 13:1015622. doi: 10.3389/fendo.2022.1015622
7. Keelan JA. Intrauterine inflammatory activation, functional progesterone withdrawal, and the timing of term and preterm birth. J Reprod Immunol. (2018) 125:89–99. doi: 10.1016/j.jri.2017.12.004
8. Ali MF, He H, Friedel D. Inflammatory bowel disease and pregnancy: fertility, complications and treatment. Ann Gastroenterol. (2020) 33:579–90. doi: 10.20524/aog.2020.0536
9. Saito-Abe M, Yamamoto-Hanada K, Pak K, Sato M, Irahara M, Mezawa H, et al. Association of maternal history of allergic features with preterm pregnancy outcomes in the Japan environment and children's study. Int Arch Allergy Immunol. (2021) 182:650–62. doi: 10.1159/000513749
10. Cobo T, Kacerovsky M, Jacobsson B. Risk factors for spontaneous preterm delivery. Int J Gynaecol Obstet. (2020) 150:17–23. doi: 10.1002/ijgo.13184
11. Prediction Prediction and prevention of spontaneous preterm birth: ACOG practice bulletin, number 234. Obstet Gynecol. (2021) 138, e65-90. doi: 10.1097/AOG.0000000000004479
12. Svensson AC, Sandin S, Cnattingius S, Reilly M, Pawitan Y, Hultman CM, et al. Maternal effects for preterm birth: a genetic epidemiologic study of 630,000 families. Am J Epidemiol. (2009) 170:1365–72. doi: 10.1093/aje/kwp328
13. Plunkett J, Feitosa MF, Trusgnich M, Wangler MF, Palomar L, Kistka ZA, et al. Mother's genome or maternally-inherited genes acting in the fetus influence gestational age in familial preterm birth. Hum Hered. (2009) 68:209–19. doi: 10.1159/000224641
14. York TP, Eaves LJ, Lichtenstein P, Neale MC, Svensson A, Latendresse S, et al. Fetal and maternal genes' influence on gestational age in a quantitative genetic analysis of 244,000 Swedish births. Am J Epidemiol. (2013) 178:543–50. doi: 10.1093/aje/kwt005
15. Zhang H, Baldwin DA, Bukowski RK, Parry S, Xu Y, Song C, et al. A genome-wide association study of early spontaneous preterm delivery. Genet Epidemiol. (2015) 39:217–26. doi: 10.1002/gepi.21887
16. Zhang G, Feenstra B, Bacelis J, Liu X, Muglia LM, Juodakis J, et al. Genetic associations with gestational duration and spontaneous preterm birth. N Engl J Med. (2017) 377:1156–67. doi: 10.1056/NEJMoa1612665
17. Bhattacharjee E, Thiruvengadam R, Ayushi Das C, Wadhwa N, Natchu UCM, Kshetrapal P, et al. Genetic variants associated with spontaneous preterm birth in women from India: a prospective cohort study. Lancet Reg Health Southeast Asia. (2023) 14:100190. doi: 10.1016/j.lansea.2023.100190
18. Pasanen A, Karjalainen MK, Zhang G, Tiensuu H, Haapalainen AM, Ojaniemi M, et al. Meta-analysis of genome-wide association studies of gestational duration and spontaneous preterm birth identifies new maternal risk loci. PLoS Genet. (2023) 19:e1010982. doi: 10.1371/journal.pgen.1010982
19. Solé-Navais P, Flatley C, Steinthorsdottir V, Vaudel M, Juodakis J, Chen J, et al. Genetic effects on the timing of parturition and links to fetal birth weight. Nat Genet. (2023) 55:559–67. doi: 10.1530/ey.20.12.3
20. Huusko JM, Karjalainen MK, Graham BE, Zhang G, Farrow EG, Miller NA, et al. Whole exome sequencing reveals HSPA1L as a genetic risk factor for spontaneous preterm birth. PLoS Genet. (2018) 14:e1007394. doi: 10.1371/journal.pgen.1007394
21. Knijnenburg TA, Vockley JG, Chambwe N, Gibbs DL, Humphries C, Huddleston KC, et al. Genomic and molecular characterization of preterm birth. Proc Natl Acad Sci USA. (2019) 116:5819–27. doi: 10.1073/pnas.1716314116
22. Mladenić T, Barišić A, Pereza N, Ostojić S, Peterlin B, Dević Pavlić S. Maternal genetic risk factors for spontaneous preterm birth: a systematic review and meta-analysis. Int J Gynaecol Obstet. (2025) 169:458–73. doi: 10.1002/ijgo.16056
23. Darido C, Georgy SR, Jane SM. The role of barrier genes in epidermal malignancy. Oncogene. (2016) 35:5705–12. doi: 10.1038/onc.2016.84
24. Vancamelbeke M, Vanuytsel T, Farré R, Verstockt S, Ferrante M, Van Assche G, et al. Genetic and transcriptomic bases of intestinal epithelial barrier dysfunction in inflammatory bowel disease. Inflamm Bowel Dis. (2017) 23:1718–29. doi: 10.1097/MIB.0000000000001246
25. Chow BW, Gu C. The molecular constituents of the blood-brain barrier. Trends Neurosci. (2015) 38:598–608. doi: 10.1016/j.tins.2015.08.003
26. Lujan AL, Croci DO, Rabinovich GA, Damiani MT. Galectins as potential therapeutic targets in STIs in the female genital tract. Nat Rev Urol. (2022) 19:240–52. doi: 10.1038/s41585-021-00562-1
27. Ioghen O, Chi, t oiu L, Gherghiceanu M, Ceafalan LC, Hinescu ME. CD36 - a novel molecular target in the neurovascular unit. Eur J Neurosci. (2021) 53:2500–10. doi: 10.1111/ejn.15147
28. Duchatelet S, Pruvost S, de Veer S, Fraitag S, Nitschké P, Bole-Feysot C, et al. A new TRPV3 missense mutation in a patient with Olmsted syndrome and erythromelalgia. JAMA Dermatol. (2014) 150:303–6. doi: 10.1001/jamadermatol.2013.8709
29. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. (2023) 613:508–18. doi: 10.1038/s41586-022-05473-8
30. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. (2017) 8:1826. doi: 10.1038/s41467-017-01261-5
31. Chen Z, Boehnke M, Wen X, Mukherjee B. Revisiting the genome-wide significance threshold for common variant GWAS. G3. (2021) 11:jkaa056. doi: 10.1093/g3journal/jkaa056
32. Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. (1995) 11:241–7. doi: 10.1038/ng1195-241
33. Xiao X, Li R, Wu C, Yan Y, Yuan M, Cui B, et al. A genome-wide association study identifies a novel association between SDC3 and apparent treatment-resistant hypertension. BMC Med. (2022) 20:463. doi: 10.1186/s12916-022-02665-x
34. Li S, Gui J, Passarelli MN, Andrew AS, Sullivan KM, Cornell KA, et al. Genome-wide and transcriptome-wide association studies on Northern New England and Ohio amyotrophic lateral sclerosis cohorts. Neurol Genet. (2024) 10:e200188. doi: 10.1212/NXG.0000000000200188
35. Marigorta UM, Rodríguez JA, Gibson G, Navarro A. Replicability and prediction: lessons and challenges from GWAS. Trends Genet. (2018) 34:504–17. doi: 10.1016/j.tig.2018.03.005
36. Baldi A, De Falco M, De Luca L, Cottone G, Paggi MG, Nickoloff BJ, et al. Characterization of tissue specific expression of Notch-1 in human tissues. Biol Cell. (2004) 96:303–11. doi: 10.1111/j.1768-322X.2004.tb01418.x
37. Polacheck WJ, Kutys ML, Yang J, Eyckmans J, Wu Y, Vasavada H, et al. A non-canonical Notch complex regulates adherens junctions and vascular barrier function. Nature. (2017) 552:258–62. doi: 10.1038/nature24998
38. Cuman C, Menkhorst E, Winship A, Van Sinderen M, Osianlis T, Rombauts LJ, et al. Fetal-maternal communication: the role of Notch signalling in embryo implantation. Reproduction. (2014) 147:R75–86. doi: 10.1530/REP-13-0474
39. Castel D, Mourikis P, Bartels SJ, Brinkman AB, Tajbakhsh S, Stunnenberg HG. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. (2013) 27:1059–71. doi: 10.1101/gad.211912.112
40. Strug MR, Su RW, Kim TH, Jeong JW, Fazleabas A. The Notch family transcription factor, RBPJκ, modulates glucose transporter and ovarian steroid hormone receptor expression during decidualization. Reprod Sci. (2019) 26:774–84. doi: 10.1177/1933719118799209
41. Mathern DR, Laitman LE, Hovhannisyan Z, Dunkin D, Farsio S, Malik TJ, et al. Mouse and human Notch-1 regulate mucosal immune responses. Mucosal Immunol. (2014) 7:995–1005. doi: 10.1038/mi.2013.118
42. Moldovan GE, Miele L, Fazleabas AT. Notch signaling in reproduction. Trends Endocrinol Metab. (2021) 32:1044–57. doi: 10.1016/j.tem.2021.08.002
43. Tiwari D, Choudhury SS, Nath T, Bose S. An investigation into the role of Notch signaling, altered angiogenesis, and inflammatory-induced preterm delivery and related complications in Northeast Indian patients. Placenta. (2023) 139:172–80. doi: 10.1016/j.placenta.2023.07.002
44. Jaiswal MK, Agrawal V, Pamarthy S, Katara GK, Kulshrestha A, Gilman-Sachs A, et al. Notch signaling in inflammation-induced preterm labor. Sci Rep. (2015) 5:15221. doi: 10.1038/srep15221
45. Agrawal V, Jaiswal MK, Pamarthy S, Katara GK, Kulshrestha A, Gilman-Sachs A, et al. Role of Notch signaling during lipopolysaccharide-induced preterm labor. J Leukoc Biol. (2016) 100:261–74. doi: 10.1189/jlb.3HI0515-200RR
46. Norén E, Almer S, Söderman J. Genetic variation and expression levels of tight junction genes identifies association between MAGI3 and inflammatory bowel disease. BMC Gastroenterol. (2017) 17:68. doi: 10.1186/s12876-017-0620-y
47. Lee SH. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intest Res. (2015) 13:11–8. doi: 10.5217/ir.2015.13.1.11
48. Founds SA, Shi H, Conley YP, Jeyabalan A, Roberts JM, Lyons-Weiler J. Variations in discovery-based preeclampsia candidate genes. Clin Transl Sci. (2012) 5:333–9. doi: 10.1111/j.1752-8062.2012.00413.x
49. Norén E, Mellander MR, Almer S, Söderman J. Genetic variation and gene expression levels of tight junction genes indicates relationships between PTEN as well as MAGI1 and microscopic colitis. Dig Dis Sci. (2018) 63:105–12. doi: 10.1007/s10620-017-4857-7
50. Blaskewicz CD, Pudney J, Anderson DJ. Structure and function of intercellular junctions in human cervical and vaginal mucosal epithelia. Biol Reprod. (2011) 85:97–104. doi: 10.1095/biolreprod.110.090423
51. Poholek AC. Tissue-specific contributions to control of T cell immunity. Immunohorizons. (2021) 5:410–23. doi: 10.4049/immunohorizons.2000103
52. Founds SA, Tsigas E, Ren D, Barmada MM. Associating symptom phenotype and genotype in preeclampsia. Biol Res Nurs. (2018) 20:126–36. doi: 10.1177/1099800417754140
53. Imafuku K, Iwata H, Natsuga K, Okumura M, Kobayashi Y, Kitahata H, et al. Zonula occludens-1 distribution and barrier functions are affected by epithelial proliferation and turnover rates. Cell Prolif. (2023) 56:e13441. doi: 10.1111/cpr.13441
54. Kuo WT, Zuo L, Odenwald MA, Madha S, Singh G, Gurniak CB, et al. The tight junction protein ZO-1 Is dispensable for barrier function but critical for effective mucosal repair. Gastroenterology. (2021) 161:1924–39. doi: 10.1053/j.gastro.2021.08.047
55. Pidoux G, Gerbaud P, Gnidehou S, Grynberg M, Geneau G, Guibourdenche J, et al. ZO-1 is involved in trophoblastic cell differentiation in human placenta. Am J Physiol Cell Physiol. (2010) 298:C1517–1526. doi: 10.1152/ajpcell.00484.2008
56. Li J, Liu Y, Xue R, Shen H, Wu Y, Quinn M, et al. Inflammation-related downregulation of zonula Occludens-1 in fetal membrane contributes to development of prelabor rupture of membranes. Placenta. (2020) 99:173–9. doi: 10.1016/j.placenta.2020.07.029
57. Wörthmüller J, Rüegg C. MAGI1, a scaffold protein with tumor suppressive and vascular functions. Cells. (2021) 10 :1494. doi: 10.3390/cells10061494
58. Sakurai A, Fukuhara S, Yamagishi A, Sako K, Kamioka Y, Masuda M, et al. MAGI-1 is required for Rap1 activation upon cell-cell contact and for enhancement of vascular endothelial cadherin-mediated cell adhesion. Mol Biol Cell. (2006) 17:966–76. doi: 10.1091/mbc.e05-07-0647
59. Alonso A, Domènech E, Julià A, Panés J, García-Sánchez V, Mateu PN, et al. Identification of risk loci for Crohn's disease phenotypes using a genome-wide association study. Gastroenterology. (2015) 148:794–805. doi: 10.1053/j.gastro.2014.12.030
60. Balbas MD, Burgess MR, Murali R, Wongvipat J, Skaggs BJ, Mundel P, et al. MAGI-2 scaffold protein is critical for kidney barrier function. Proc Natl Acad Sci USA. (2014) 111:14876–81. doi: 10.1073/pnas.1417297111
61. Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. (2008) 57:463–7. doi: 10.1136/gut.2007.133132
62. McGovern DP, Taylor KD, Landers C, Derkowski C, Dutridge D, Dubinsky M, et al. MAGI2 genetic variation and inflammatory bowel disease. Inflamm Bowel Dis. (2009) 15:75–83. doi: 10.1002/ibd.20611
63. McCole DF. IBD candidate genes and intestinal barrier regulation. Inflamm Bowel Dis. (2014) 20:1829–49. doi: 10.1097/MIB.0000000000000090
64. Barmeyer C, Schulzke JD, Fromm M. Claudin-related intestinal diseases. Semin Cell Dev Biol. (2015) 42:30–8. doi: 10.1016/j.semcdb.2015.05.006
65. Gupta IR, Ryan AK. Claudins: unlocking the code to tight junction function during embryogenesis and in disease. Clin Genet. (2010) 77:314–25. doi: 10.1111/j.1399-0004.2010.01397.x
66. Zeisel MB, Dhawan P, Baumert TF. Tight junction proteins in gastrointestinal and liver disease. Gut. (2019) 68:547–61. doi: 10.1136/gutjnl-2018-316906
67. Hadj-Rabia S, Brideau G, Al-Sarraj Y, Maroun RC, Figueres ML, Leclerc-Mercier S, et al. Multiplex epithelium dysfunction due to CLDN10 mutation: the HELIX syndrome. Genet Med. (2018) 20:190–201. doi: 10.1038/gim.2017.71
68. Lee K, Ansar M, Andrade PB, Khan B, Santos-Cortez RL, Ahmad W, et al. Novel CLDN14 mutations in Pakistani families with autosomal recessive non-syndromic hearing loss. Am J Med Genet A. (2012) 158a:315–21. doi: 10.1002/ajmg.a.34407
69. Lu Y, Yao J, Wei Q, Xu J, Xing G, Cao X. Genetic analysis of CLDN14 in the Chinese population affected with non-syndromic hearing loss. Int J Pediatr Otorhinolaryngol. (2018) 105:6–11. doi: 10.1016/j.ijporl.2017.11.016
70. Thorleifsson G, Holm H, Edvardsson V, Walters GB, Styrkarsdottir U, Gudbjartsson DF, et al. Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat Genet. (2009) 41:926–30. doi: 10.1038/ng.404
71. Hou J. The yin and yang of claudin-14 function in human diseases. Ann N Y Acad Sci. (2012) 1258:185–90. doi: 10.1111/j.1749-6632.2012.06529.x
72. Tamura A, Kitano Y, Hata M, Katsuno T, Moriwaki K, Sasaki H, et al. Megaintestine in claudin-15-deficient mice. Gastroenterology. (2008) 134:523–34. doi: 10.1053/j.gastro.2007.11.040
73. Shan N, Zhang X, Xiao X, Zhang H, Tong C, Luo X, et al. Laminin α4 (LAMA4) expression promotes trophoblast cell invasion, migration, and angiogenesis, and is lowered in preeclamptic placentas. Placenta. (2015) 36:809–20. doi: 10.1016/j.placenta.2015.04.008
74. Li L, Song J, Chuquisana O, Hannocks MJ, Loismann S, Vogl T, et al. Endothelial basement membrane laminins as an environmental cue in monocyte differentiation to macrophages. Front Immunol. (2020) 11:584229. doi: 10.3389/fimmu.2020.584229
75. Thyboll J, Kortesmaa J, Cao R, Soininen R, Wang L, Iivanainen A, et al. Deletion of the laminin alpha4 chain leads to impaired microvessel maturation. Mol Cell Biol. (2002) 22:1194–202. doi: 10.1128/MCB.22.4.1194-1202.2002
76. Kumarasamy M, Sosnik A. Heterocellular spheroids of the neurovascular blood-brain barrier as a platform for personalized nanoneuromedicine. iScience. (2021) 24:102183. doi: 10.1016/j.isci.2021.102183
77. Ji Y, Zhou L, Wang G, Qiao Y, Tian Y, Feng Y. Role of LAMA4 gene in regulating extravillous trophoblasts in pathogenesis of preeclampsia. Med Sci Monit. (2019) 25:9630–6. doi: 10.12659/MSM.917402
78. Shan N, Zhang X, Xiao X, Zhang H, Chen Y, Luo X, et al. The role of laminin α4 in human umbilical vein endothelial cells and pathological mechanism of preeclampsia. Reprod Sci. (2015) 22:969–79. doi: 10.1177/1933719115570913
79. Nishiyama K, Kurokawa M, Torio M, Sakai Y, Arima M, Tsukamoto S, et al. Gastrointestinal symptoms as an extended clinical feature of Pierson syndrome: a case report and review of the literature. BMC Med Genet. (2020) 21:80. doi: 10.1186/s12881-020-01019-9
80. Chrifi I, Hermkens D, Brandt MM, van Dijk CGM, Bürgisser PE, Haasdijk R, et al. Cgnl1, an endothelial junction complex protein, regulates GTPase mediated angiogenesis. Cardiovasc Res. (2017) 113:1776–88. doi: 10.1093/cvr/cvx175
81. Eckenstaler R, Hauke M, Benndorf RA. A current overview of RhoA, RhoB, and RhoC functions in vascular biology and pathology. Biochem Pharmacol. (2022) 206:115321. doi: 10.1016/j.bcp.2022.115321
82. Lartey J, Smith M, Pawade J, Strachan B, Mellor H, López Bernal A. Up-regulation of myometrial RHO effector proteins (PKN1 and DIAPH1) and CPI-17 (PPP1R14A) phosphorylation in human pregnancy is associated with increased GTP-RHOA in spontaneous preterm labor. Biol Reprod. (2007) 76:971–82. doi: 10.1095/biolreprod.106.058982
83. Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. (2006) 299:122–36. doi: 10.1016/j.ydbio.2006.07.015
84. Wang Y, Sun Y, Huang Y, Pan Y, Jia Z, Ma L, et al. Association study between Van der Woude Syndrome causative gene GRHL3 and nonsyndromic cleft lip with or without cleft palate in a Chinese cohort. Gene. (2016) 588:69–73. doi: 10.1016/j.gene.2016.04.045
85. Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, et al. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am J Hum Genet. (2014) 94:23–32. doi: 10.1016/j.ajhg.2013.11.009
86. Tian T, Wang L, Shen Y, Zhang B, Finnell RH, Ren A. Hypomethylation of GRHL3 gene is associated with the occurrence of neural tube defects. Epigenomics. (2018) 10:891–901. doi: 10.2217/epi-2018-0016
87. Kimura-Yoshida C, Mochida K, Ellwanger K, Niehrs C, Matsuo I. Fate specification of neural plate border by canonical Wnt signaling and Grhl3 is crucial for neural tube closure. EBioMedicine. (2015) 2:513–27. doi: 10.1016/j.ebiom.2015.04.012
88. Xia CH, Yablonka-Reuveni Z, Gong X. LRP5 is required for vascular development in deeper layers of the retina. PLoS ONE. (2010) 5:e11676. doi: 10.1371/journal.pone.0011676
89. Guo YF, Xiong DH, Shen H, Zhao LJ, Xiao P, Guo Y, et al. Polymorphisms of the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with obesity phenotypes in a large family-based association study. J Med Genet. (2006) 43:798–803. doi: 10.1136/jmg.2006.041715
90. Goodrich JK, Davenport ER, Clark AG, Ley RE. The relationship between the human genome and microbiome comes into view. Annu Rev Genet. (2017) 51:413–33. doi: 10.1146/annurev-genet-110711-155532
91. Ruiz-Triviño J, Álvarez D, Cadavid JÁ, Alvarez AM. From gut to placenta: understanding how the maternal microbiome models life-long conditions. Front Endocrinol. (2023) 14:1304727. doi: 10.3389/fendo.2023.1304727
92. Bayar E, Bennett PR, Chan D, Sykes L, MacIntyre DA. The pregnancy microbiome and preterm birth. Semin Immunopathol. (2020) 42:487–99. doi: 10.1007/s00281-020-00817-w
93. Dos Anjos Borges LG, Pastuschek J, Heimann Y, Dawczynski K, Schleußner E, Pieper DH, et al. Vaginal and neonatal microbiota in pregnant women with preterm premature rupture of membranes and consecutive early onset neonatal sepsis. BMC Med. (2023) 21:92. doi: 10.1186/s12916-023-02805-x
94. Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The vaginal microbiome and preterm birth. Nat Med. (2019) 25:1012–21. doi: 10.1038/s41591-019-0450-2
95. Bekkar B, Pacheco S, Basu R, DeNicola N. Association of air pollution and heat exposure with preterm birth, low birth weight, and stillbirth in the US: a systematic review. JAMA Netw Open. (2020) 3:e208243. doi: 10.1001/jamanetworkopen.2020.8243
96. Jones SI, Pruszynski JE, Spong CY, Nelson DB. Traffic-related air pollution is associated with spontaneous extremely preterm birth and other adverse perinatal outcomes. Am J Obstet Gynecol. (2023) 229:455.e1–455.e7. doi: 10.1016/j.ajog.2023.07.040
97. Cocchi E, Bellisario V, Cresi F, Plazzotta C, Cassardo C, Siniscalco C, et al. Air pollution and aeroallergens as possible triggers in preterm birth delivery. Int J Environ Res Public Health. (2023) 20:1610. doi: 10.3390/ijerph20021610
98. Li J, Gu J, Liu L, Cao M, Wang Z, Tian X, et al. The relationship between air pollutants and preterm birth and blood routine changes in typical river valley city. BMC Public Health. (2024) 24:1677. doi: 10.1186/s12889-024-19140-2
99. Becker S, Dailey LA, Soukup JM, Grambow SC, Devlin RB, Huang YC. Seasonal variations in air pollution particle-induced inflammatory mediator release and oxidative stress. Environ Health Perspect. (2005) 113:1032–8. doi: 10.1289/ehp.7996
100. Saenen ND, Martens DS, Neven KY, Alfano R, Bové H, Janssen BG, et al. Air pollution-induced placental alterations: an interplay of oxidative stress, epigenetics, and the aging phenotype? Clin Epigenetics. (2019) 11:124. doi: 10.1186/s13148-019-0688-z
101. Lee PH, Park S, Lee YG, Choi SM, An MH, Jang AS. The impact of environmental pollutants on barrier dysfunction in respiratory disease. Allergy Asthma Immunol Res. (2021) 13:850–62. doi: 10.4168/aair.2021.13.6.850
102. Opichka MA, Rappelt MW, Gutterman DD, Grobe JL, McIntosh JJ. Vascular dysfunction in preeclampsia. Cells. (2021) 10:3055. doi: 10.3390/cells10113055
103. Vestweber D. VE-cadherin: the major endothelial adhesion molecule controlling cellular junctions and blood vessel formation. Arterioscler Thromb Vasc Biol. (2008) 28:223–32. doi: 10.1161/ATVBAHA.107.158014
104. Kaur S, Ewing HT, Warrington JP. Blood-brain barrier dysfunction in hypertensive disorders of pregnancy. Curr Hypertens Rep. (2023) 25:463–70. doi: 10.1007/s11906-023-01288-8
105. Begum F, Ghosh D, Tseng GC, Feingold E. Comprehensive literature review and statistical considerations for GWAS meta-analysis. Nucleic Acids Res. (2012) 40:3777–84. doi: 10.1093/nar/gkr1255
106. Bhattacharjee E, Maitra A. Spontaneous preterm birth: the underpinnings in the maternal and fetal genomes. NPJ Genom Med. (2021) 6:43. doi: 10.1038/s41525-021-00209-5
107. Jain VG, Monangi N, Zhang G, Muglia LJ. Genetics, epigenetics, and transcriptomics of preterm birth. Am J Reprod Immunol. (2022) 88:e13600. doi: 10.1111/aji.13600
108. Moll M, Reichel K, Nurjadi D, Förmer S, Krall LJ, Heeg K, et al. Notch ligand delta-like 1 is associated with loss of vascular endothelial barrier function. Front Physiol. (2021) 12:766713. doi: 10.3389/fphys.2021.766713
109. Liu T, Zhang C, Ying J, Wang Y, Yan G, Zhou Y, et al. Inhibition of the intracellular domain of Notch1 results in vascular endothelial cell dysfunction in sepsis. Front Immunol. (2023) 14:1134556. doi: 10.3389/fimmu.2023.1134556
110. Yu L, Lu Z, Burchell S, Nowrangi D, Manaenko A, Li X, et al. Adropin preserves the blood-brain barrier through a Notch1/Hes1 pathway after intracerebral hemorrhage in mice. J Neurochem. (2017) 143:750–60. doi: 10.1111/jnc.14238
111. Lee MJ, Zhu J, An JH, Lee SE, Kim TY, Oh E, et al. A transcriptomic analysis of cerebral microvessels reveals the involvement of Notch1 signaling in endothelial mitochondrial-dysfunction-dependent BBB disruption. Fluids Barriers CNS. (2022) 19:64. doi: 10.1186/s12987-022-00363-7
112. Li M, Zhang Z, Joynauth J, Zhan X, Du L. Intrauterine growth restriction neonates present with increased angiogenesis through the Notch1 signaling pathway. Microvasc Res. (2022) 140:104308. doi: 10.1016/j.mvr.2021.104308
113. Limbourg FP, Takeshita K, Radtke F, Bronson RT, Chin MT, Liao JK. Essential role of endothelial Notch1 in angiogenesis. Circulation. (2005) 111:1826–32. doi: 10.1161/01.CIR.0000160870.93058.DD
114. Fazio C, Ricciardiello L. Inflammation and Notch signaling: a crosstalk with opposite effects on tumorigenesis. Cell Death Dis. (2016) 7:e2515. doi: 10.1038/cddis.2016.408
115. Šućur A, Filipović M, Flegar D, Kelava T, Šisl D, Lukač N, et al. Notch receptors and ligands in inflammatory arthritis - a systematic review. Immunol Lett. (2020) 223:106–14. doi: 10.1016/j.imlet.2020.04.010
116. Gallenstein N, Tichy L, Weigand MA, Schenz J. Notch signaling in acute inflammation and sepsis. Int J Mol Sci. (2023) 24:3458. doi: 10.3390/ijms24043458
117. Christopoulos PF, Gjølberg TT, Krüger S, Haraldsen G, Andersen JT, Sundlisæter E. Targeting the Notch signaling pathway in chronic inflammatory diseases. Front Immunol. (2021) 12:668207. doi: 10.3389/fimmu.2021.668207
Keywords: preterm birth, NOTCH1, maternal barrier genes, genome-wide association studies, genetic variant
Citation: Chen K-R, Chu S-K and Kuo P-L (2025) Barrier genes are associated with preterm birth. Front. Med. 12:1580877. doi: 10.3389/fmed.2025.1580877
Received: 21 February 2025; Accepted: 03 June 2025;
Published: 23 June 2025.
Edited by:
Enrico Cocchi, University of Bologna, ItalyReviewed by:
Wenbo Zhou, Changzhou Maternal and Child Health Care Hospital, ChinaTea Mladenić, University of Rijeka, Croatia
Copyright © 2025 Chen, Chu and Kuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pao-Lin Kuo, cGFvbGlua3VvQGdtYWlsLmNvbQ==
 Kuan-Ru Chen
Kuan-Ru Chen Shih-Kai Chu
Shih-Kai Chu Pao-Lin Kuo
Pao-Lin Kuo