- 1Department of Respiratory Medicine, The Second Hospital of Jilin University, Changchun, China
- 2Shandong First Medical University Affiliated Provincial Hospital, Jinan, Shandong, China
- 3Department of Pulmonary and Critical Care Medicine of Jiangbei Campus, The First Affiliated Hospital of Army Medical University, Chongqing, China
- 4Department of Internal Medicine, Hospital of Jilin University, Changchun, China
Background: Lung cancer is among the malignancies most vulnerable to coronavirus disease 2019 (COVID-19). Eosinophils have anti-tumor and antiviral effects. Since November 2021, the omicron variant of COVID-19 has become a topic of concern; however, the impact of eosinophils on the severity and outcomes of patients with lung cancer with omicron remains uncertain. This study aimed to utilize eosinophils to predict patient outcomes and guide the prevention and monitoring of omicron.
Methods: This study performed an analysis of 284 patients with lung cancer who were hospitalized in the second hospital of Jilin University, of whom 83 patients were confirmed to have omicron infection. Depending on the eosinophil counts, patients were divided into two groups: low and high eosinophil counts. The relationship between eosinophil counts and severity and outcomes was then analyzed.
Results: We found that omicron, especially severe-to-critical omicron, decreased survival in patients with lung cancer. Patients with omicron had a lower eosinophil count. Patients with eosinopenia (< 0.015 × 109/L) were more likely to have an eastern cooperative oncology group performance status ≥ 2; be undergoing anti-cancer treatment; have comorbidities; and exhibit lower disease control rates, reduced 30-day survival, and shorter overall survival (median 75 days vs. not reached). Multivariate regression analysis revealed that the eosinophil count was an independent predictor of disease severity and survival in patients with lung cancer with omicron.
Conclusion: Eosinopenia correlates with poor outcomes in patients with lung cancer with omicron, and the eosinophil count is an independent indicator for predicting the severity and outcomes in these patients.
Introduction
The coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has had a remarkable influence on the healthcare system (1). With the emergence of the second and third waves of SARS-CoV-2 (2), multiple variants, including omicron, have emerged, making the management of COVID-19 even more complicated (3).
Cancer and anti-cancer therapy can cause immunosuppression, which may make patients with cancer, compared with those without cancer, more prone to developing infections (4, 5). COVID-19 can damage multiple systems, including the vasculature and respiratory system, and leads to a series of complications, such as pneumonia and thromboembolism (6, 7). Furthermore, among patients with COVID-19, those with cancer are more prone to suffer from infection-related adverse events, such as intensive care unit (ICU) admission, mechanical ventilation, and death, than those without (8, 9). According to the 2020 Global Cancer Assessment report, lung cancer is the leading cause of cancer deaths and the most frequent new cancer in men in China (10). Lung cancer is the second most common cancer type leading to death, second only to patients with hematological malignancies (11).
The risk factors influencing prognosis have received increasing attention. Alongside demographic and oncological characteristics such as age, pneumonia, cancer stage, and eastern cooperative oncology group (ECOG) performance status, immune-related indicators also play a significant role in the survival and prognosis of patients with lung cancer with COVID-19. These indicators include neutrophil counts, procalcitonin levels, C-reactive protein concentrations, and the neutrophil-to-lymphocyte ratio (NLR) (1, 12–14).
Eosinophils are innate immune cells primarily derived from granulocytes in hematopoietic progenitor cells of the bone marrow. Previous eosinophil studies mainly focused on parasitic infections and allergic diseases such as asthma (15, 16). The impact of eosinophils on cancer has also attracted the attention of clinicians. Tumor-associated tissue eosinophilia (TATE) and peripheral blood eosinophilia have been shown to improve the prognosis of various solid tumors, including colon, breast, and lung cancer (17–19). However, Lee et al. (20) indicated that TATE-enriched head and neck squamous cell carcinoma correlates with aggressive pathological features and poor prognosis, underscoring the functional plasticity and heterogeneity of TATE across different tumor types. Additionally, the role of eosinophils in antiviral defense cannot be ignored, as seen in the case of the human influenza virus (21), respiratory syncytial virus (22), and other respiratory viruses. Our previous research found that eosinopenia was associated with increased mortality in patients with COVID-19; however, that study included only non-cancer patients (23). From December 2022 to early 2023, China experienced an epidemic of the COVID-19 omicron variant (24). Thus, this study aimed to investigate the impact of eosinopenia on disease severity and clinical outcomes in lung cancer patients infected with the omicron variant.
Materials and methods
Study approval
The studies involving human participants were reviewed and approved by the Ethics Committee of the Second Hospital of Jilin University (Number: 2023-217). Written informed consent for participation was not required from the participants or their legal guardians/next of kin in accordance with national legislation and institutional requirements. The project adhered to Helsinki principles outlined in its declaration.
Study procedures and participants
This observational study included patients hospitalized in the Second Hospital of Jilin University from December 1, 2022, to February 28, 2023. The inclusion criterion was the pathological diagnosis was lung cancer and age ≥ 18 years old. All patients were tested for COVID-19 (RT-PCR or antigen test of nasopharyngeal swabs) within 0–7 days prior to enrollment (25). Exclusion criteria included lung cancer patients with primary liver or kidney diseases (e.g., cirrhosis, hepatitis, chronic kidney disease), excluded to avoid confounding by preexisting eosinophil dysregulation or treatment-related organ toxicity. Other exclusion criteria were: patients with concurrent other malignant tumors, a history of COVID-19 infection that had resolved by enrollment, missing data on demographics, clinical symptoms, complications, comorbidities, oncological parameters, or laboratory values, and those lost to follow-up.
All variables considered were collected at enrollment or COVID-19 diagnosis and mainly included: (1) demographic characteristics: age, sex, and smoking status; (2) oncological characteristics: ECOG performance status, histological type of lung cancer, cancer stage at enrollment, anti-cancer therapy received within 3 months before enrollment, the number of lines and type of anti-cancer therapy; (3) fever (> 37.5 °C) and respiratory symptoms: cough, expectoration, dyspnea, asthma and chest pain; (4) comorbidities: hypertension, diabetes mellitus, ischemic heart disease and chronic obstructive pulmonary disease (COPD); (5) complications: pneumonia/pneumonitis and venous thromboembolism; (6) laboratory parameters: white blood cell count (WBC), absolute lymphocyte count (ALC), absolute neutrophil count (ANC), eosinophil count, platelet to lymphocyte ratio (PLR), NLR, serum albumin (ALB), lactate dehydrogenase (LDH), hypersensitive C-reactive protein (hs-CRP), β2-microglobulin (β2-MG), and D-dimer; and (7) chest computed tomography scans or chest radiographic images at enrollment.
To initially explore the long-term prognosis of patients with lung cancer with COVID-19, the follow-up period was set at ≥ 6 months after enrollment (26), with a follow-up deadline of September 1, 2023. The primary endpoint was overall survival during the follow-up period, and the secondary endpoints were disease control rate (DCR) and survival rate within 30 days. COVID-19 was diagnosed and classified according to the diagnostic and classification criteria of the Diagnosis and Treatment Plan for Novel Coronavirus Pneumonia (10th edition) (25), and all the enrolled patients with omicron infection were divided into mild cases (with mild clinical symptoms and no signs of pneumonia on imaging); moderate cases (with fever and respiratory symptoms and evidence of pneumonia on imaging); severe cases (patients meeting any of the following criteria: (1) respiratory distress (≥ 30 beats/minute); (2) oxygen saturation at rest ≤ 93%; (3) arterial partial pressure of oxygen (PaO2)/inhaled partial pressure of oxygen (FiO2) ≤ 300 mmHg; (4) cases with progressive aggravation of clinical symptoms, where lung imaging shows a significant 50% progression of the lesion within 24–48 h); and critical cases (patients meeting any of the following criteria: (1) respiratory failure requiring mechanical ventilation; (2) shock; (3) combined with other organ failure requiring ICU monitoring and treatment). DCR was defined as the percentage of patients who achieved complete response, partial response, or stable disease out of the number of evaluable cases after treatment. Progressive disease was defined as the sum of the maximum diameters of the tumor target lesions increasing by at least 20%, or new lesions appearing (27). Patients with lung cancer were followed up according to the European Society for Medical Oncology guidelines (28–30).
Statistical analysis
SPSS 26.0 and GraphPad Prism 8.0.2 software were used for statistical analyses. The Kolmogorov-Smirnov method was used to analyze the distribution of continuous variables. If the distribution was normal, mean ± standard deviation was used to describe the continuous variables. If the distribution was not normal, the median and interquartile range (IQR) were used for the description, and the Mann-Whitney U test was used for comparison between the two groups. Categorical variables were described using the number of cases and percentages, and the Chi-square tests or Fisher’s exact tests were used for the comparison between groups. The binary logistic regression model was used for the univariate and multivariate analyses (forward likelihood ratio model) of severity. Kaplan-Meier analysis and log-rank test were used to compare the cumulative survival rates. The Cox regression model was used to calculate the hazard ratio (HR) of survival and its associated 95% confidence interval (CI) and perform the univariate and multivariate survival analyses (forward likelihood ratio model). All statistical analyses were based on two-sided hypothesis tests; with α = 0.05 as the test level, p < 0.05 was considered statistically significant.
Results
Baseline characteristics
A total of 447 patients with lung cancer who were hospitalized were enrolled, of which 163 patients were excluded based on the exclusion criteria (Figure 1).
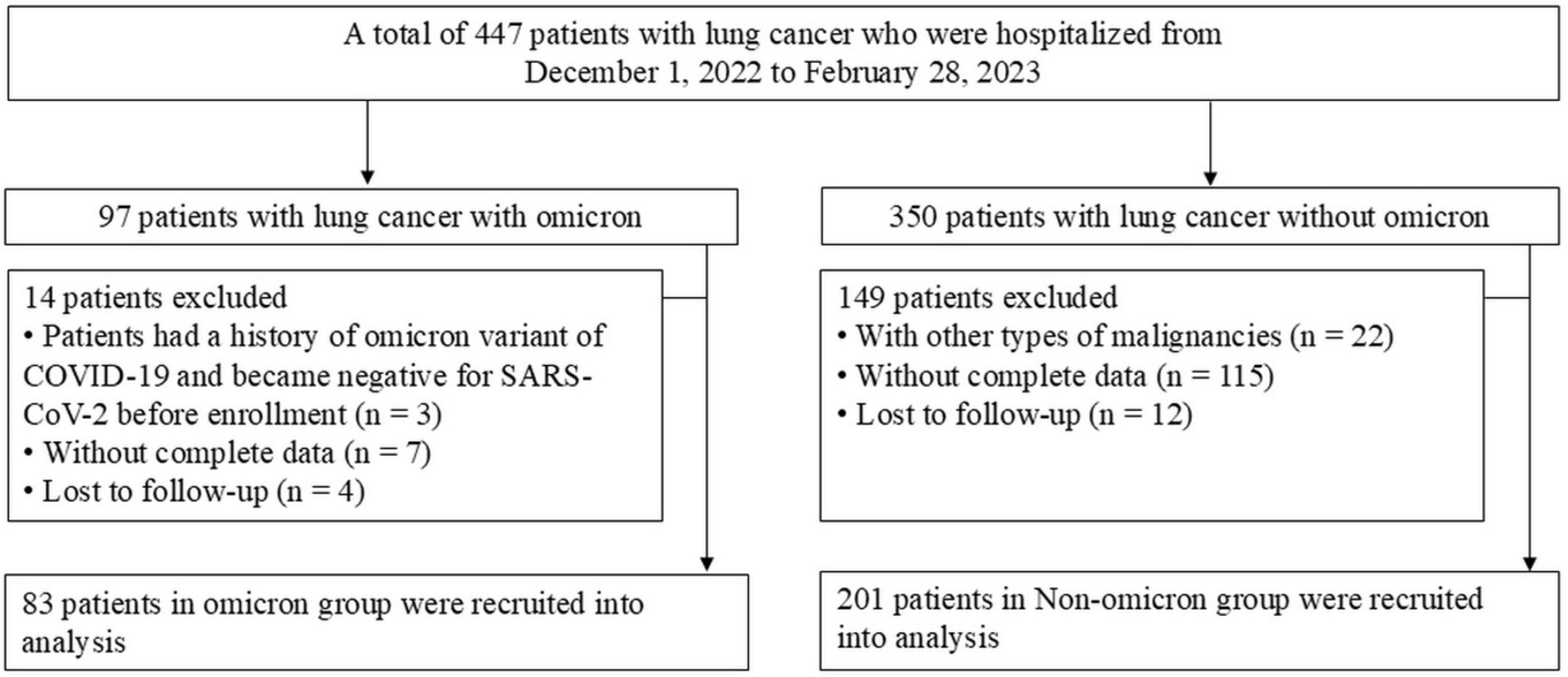
Figure 1. Flow chart for the enrolled patients. COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Our study enrolled 284 patients, including 83 with and 201 without omicron. The median age was 63 years in both groups. Significant differences in ECOG performance status and histological types were observed in the two groups (Table 1). Patients with omicron were more likely to develop pneumonia, fever, and respiratory symptoms, such as cough, expectoration, wheezing, and chest pain, than those without omicron (Supplementary Table S1). Compared with patients without omicron, those with omicron had higher levels of WBC, ANC, PLR, NLR, LDH, hs-CRP, β2-MG, and D-dimer and lower levels of ALC, eosinophil count, and ALB (Figure 2).
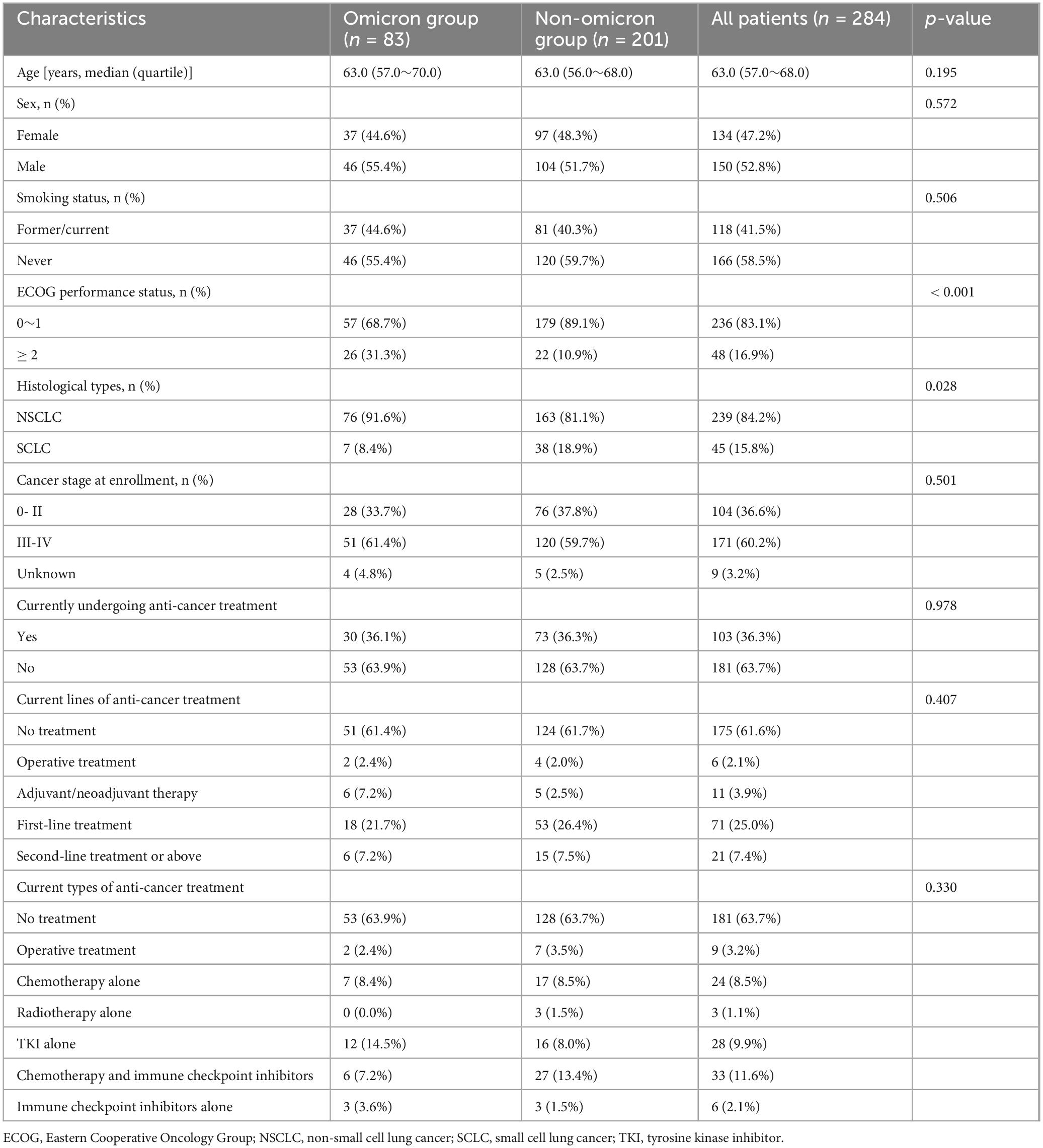
Table 1. Baseline demographics and oncological characteristics of enrolled patients with lung cancer.
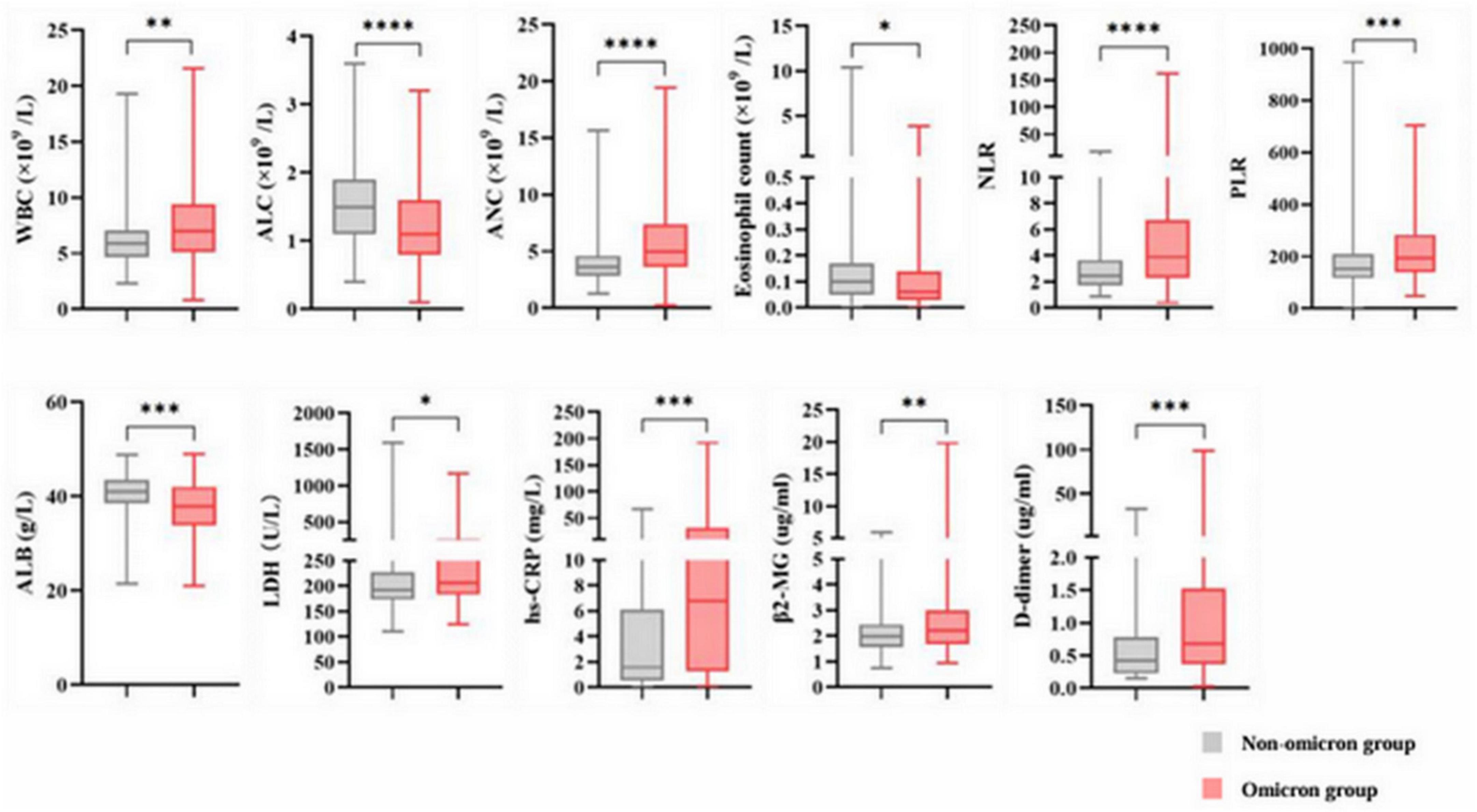
Figure 2. Abnormal baseline laboratory parameters in the non-omicron and omicron groups. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns, not significant. WBC, white blood cell count; ALC, absolute lymphocyte count; ANC, absolute neutrophil count; PLR, platelet to lymphocyte ratio; NLR, neutrophil to lymphocyte ratio; ALB, serum albumin; LDH, lactate dehydrogenase; hs-CRP, hypersensitive C-reactive protein; β2-MG, β2-microglobulin.
ROC curve analysis for eosinophil count to predict severity and mortality
According to the above results, eosinophils were significantly different in patients with and without omicron. Furthermore, the patients with lung cancer with omicron were divided into a mild-to-moderate group and a severe-to-critical group. Receiver operator characteristic (ROC) analysis found that the cut-off value of eosinophil counts for predicting disease severity and death in patients with lung cancer with omicron was 0.015 × 109/L, and the area under the ROC curve (AUC) was 0.754 (95% CI: 0.608–0.899, p = 0.001; Figure 3A) and 0.759 (95% CI: 0.634–0.884, p = 0.002; Figure 3B), respectively. Depending on whether the eosinophil count was < 0.015 × 109/L, patients with lung cancer with omicron were divided into low and high eosinophil count groups.
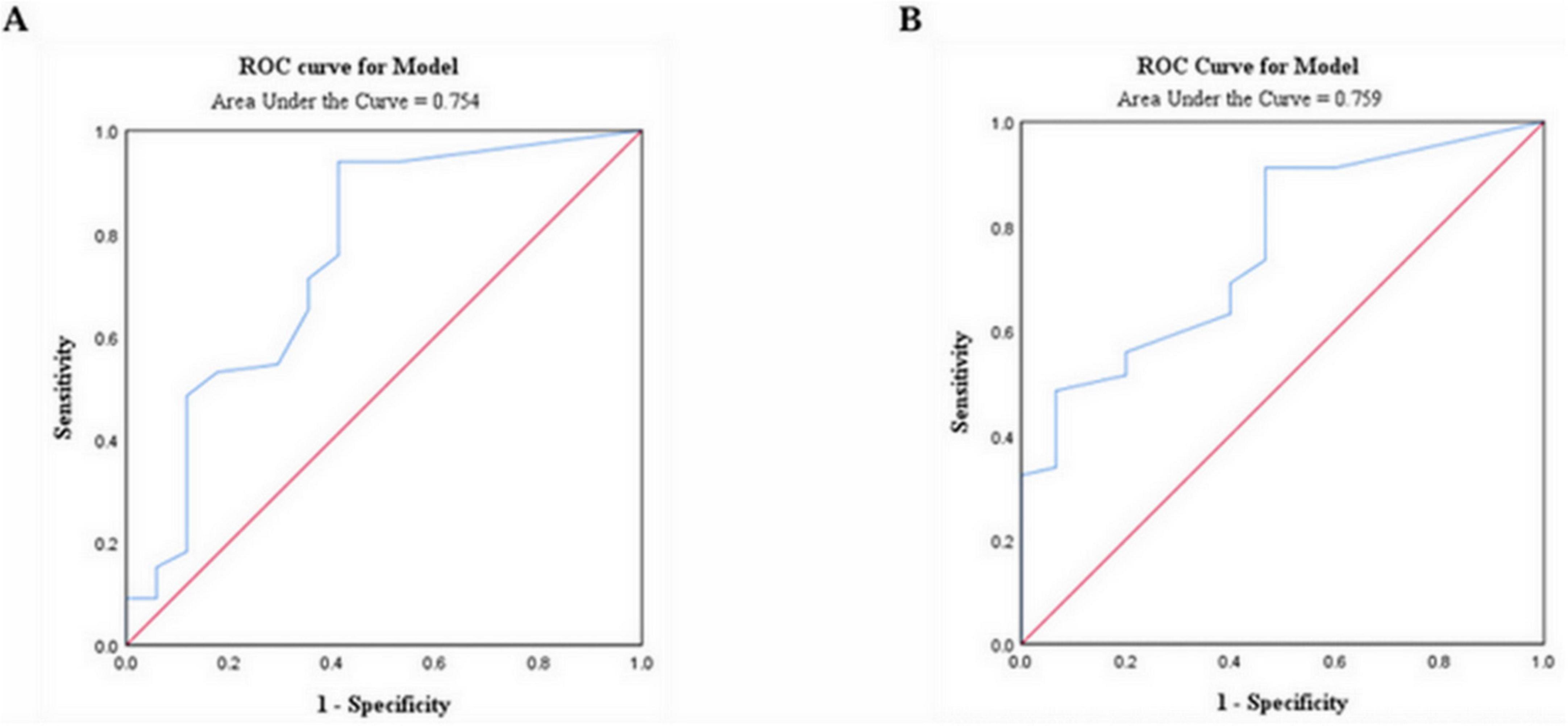
Figure 3. ROC analysis revealed a cut-off value of 0.015 × 109/L for eosinophil count to predict disease severity (A) and mortality (B) in patients with lung cancer infected with the omicron variant; ROC, receiver operating characteristic.
Associations of eosinophils with demographics, oncology characteristics, and laboratory parameters
We further explored the effects of eosinophils on demographics, oncologic characteristics, and laboratory measures. The results showed that low eosinophil count was more likely to occur in patients with ECOG performance status ≥ 2, Currently undergoing anti-cancer treatment, and with accompanying comorbidities. The low eosinophil count group exhibited higher levels of WBC, ANC, PLR, NLR, LDH, hs-CRP, β2-MG, and D-dimer, while exhibiting lower levels of ALC and ALB compared to the high eosinophil count group (Table 2).
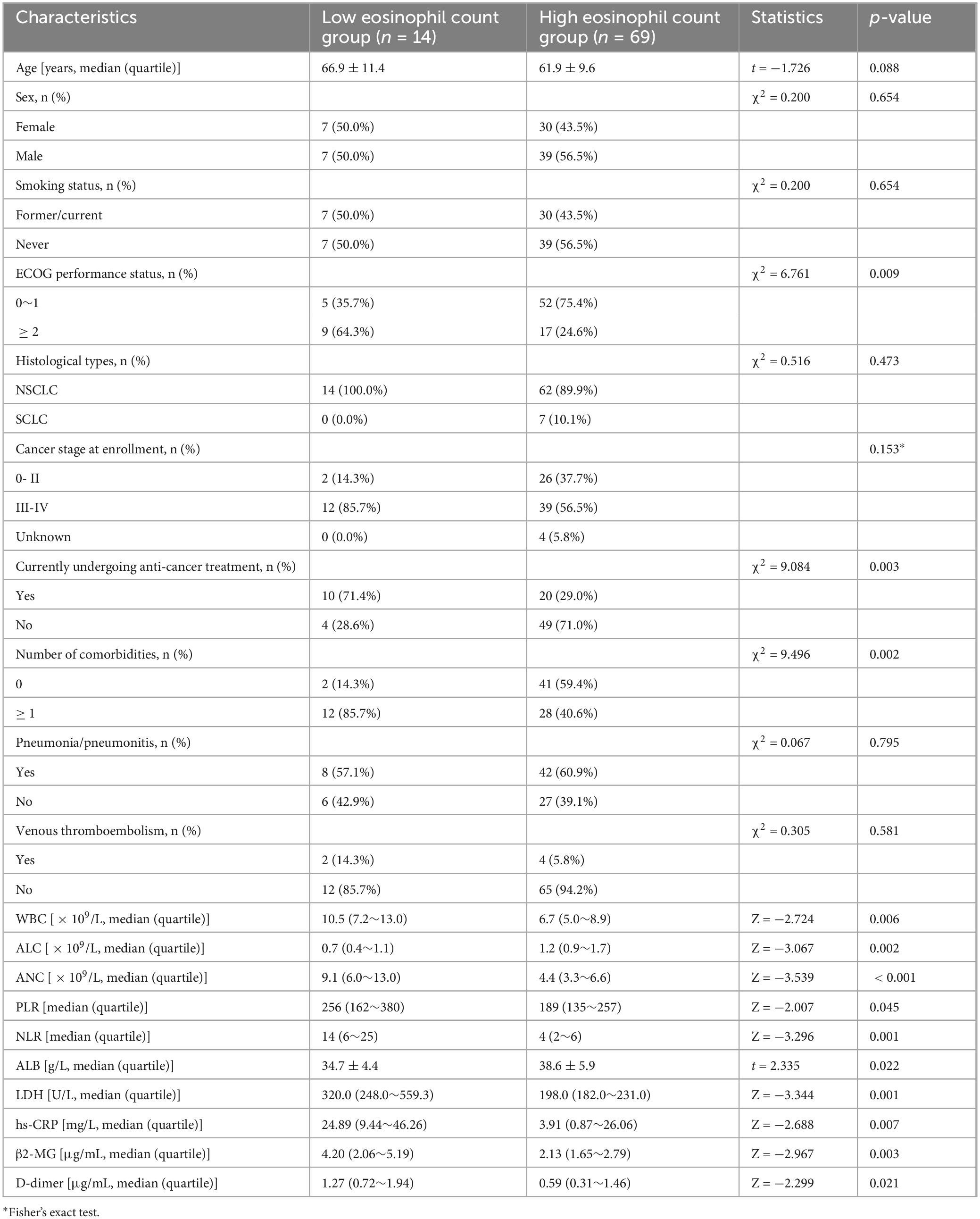
Table 2. Comparison between the low and high eosinophil count groups in patients with lung cancer with omicron.
Disease control and survival rate analysis
Based on the analysis of the impact of eosinophils on the clinical characteristics of patients with lung cancer with omicron, the effect of eosinophils on disease control and survival was further analyzed. The median follow-up time was 251 days (IQR, 229–260) for the high eosinophil count group and 121 days (IQR, 24–257) for the low eosinophil count group.
Eosinophils between the disease control and the progressive disease groups showed no meaningful difference (0.055 vs. 0.070 × 109/L, p = 0.1319) (Figure 4A). To investigate the impact of eosinophils on short-term survival, we examined the relationship between eosinophils and 30-day survival. Our findings revealed that patients who died within 30 days had lower eosinophil counts compared to those who survived during the same period (0.000 vs. 0.070 × 109/L, p = 0.0029) (Figure 4B). The eosinophil counts observed during follow-up were significantly lower among patients who died compared to those who survived (0.010 vs. 0.080 × 109/L, p = 0.0013) (Figure 4C).
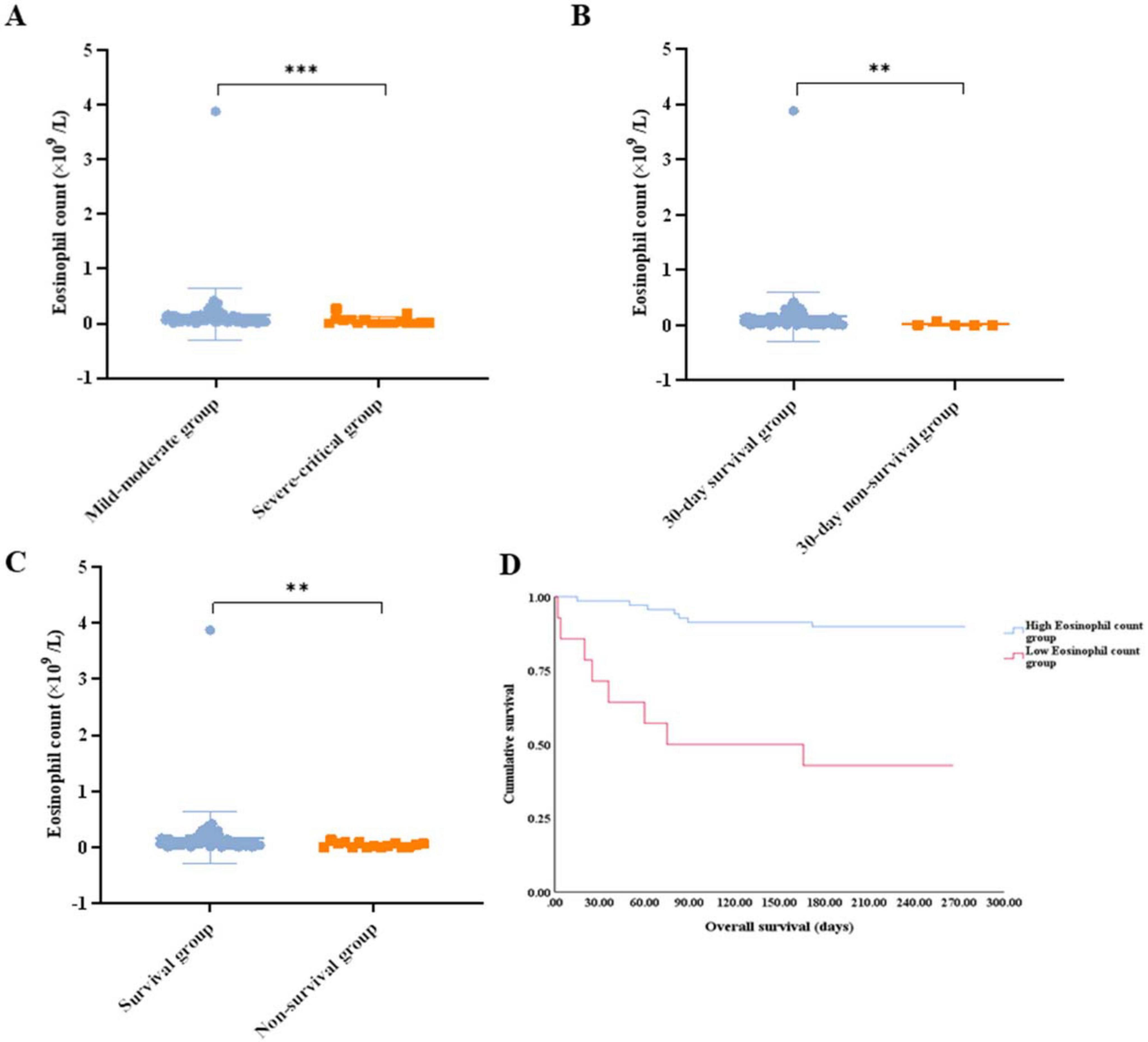
Figure 4. (A) Comparison of the eosinophil count between the disease control group and the progressive disease group. (B) Comparison of the eosinophil count between patients that survived for 30 days and those that did not. (C) Comparison of the eosinophil count between patients that survived and those that did not. (D) Survival analysis of the patients with lung cancer with omicron in the high eosinophil count group vs. low eosinophil count group. ** p < 0.01; ns, not significant.
Patients with a low eosinophil count displayed a lower 30-day survival rate (71.4% vs. 98.6%, p = 0.002), worse survival (42.9% vs. 89.9%, p < 0.001), and shorter median OS (75 days vs. not reached, p < 0.001) (Figure 4D). Patients with eosinopenia tended to have lower DCR, although no statistical difference was reached (57.1% for low eosinophil counts vs. 82.6% for high eosinophil counts, p = 0.064).
Eosinopenia was associated with poor outcomes
Compared to the non-omicron group, the omicron group had a higher mortality rate (18.1% vs. 7.5%, p = 0.008), especially in the severe-to-critical group (58.8% for the severe-to-critical group vs. 7.6% for the mild-to-moderate group, p < 0.001). Therefore, we further investigated the risk factors associated with disease severity and mortality. ROC analysis identified cut-off values of 220.63 and 4.86 for PLR and NLR, respectively.
Univariate logistic regression analysis showed that patients with ECOG performance status ≥ 2, Cancer stage III-IV at enrollment, currently undergoing anti-cancer treatment, with accompanying comorbidities, increased WBC, ANC, NLR, LDH, hs-CRP, β2-MG, and D-dimer levels, and decreased eosinophil count and ALB levels were more serious (Table 3). Further multivariate analysis revealed that ECOG performance status (OR = 6.341, 95% CI: 1.289–31.189; p = 0.023), ANC (OR = 13.559, 95% CI: 2.709–67.879; p = 0.002), and eosinophil count (OR = 7.365, 95% CI: 1.531–35.433; p = 0.013) determinants of the severity (Table 4).
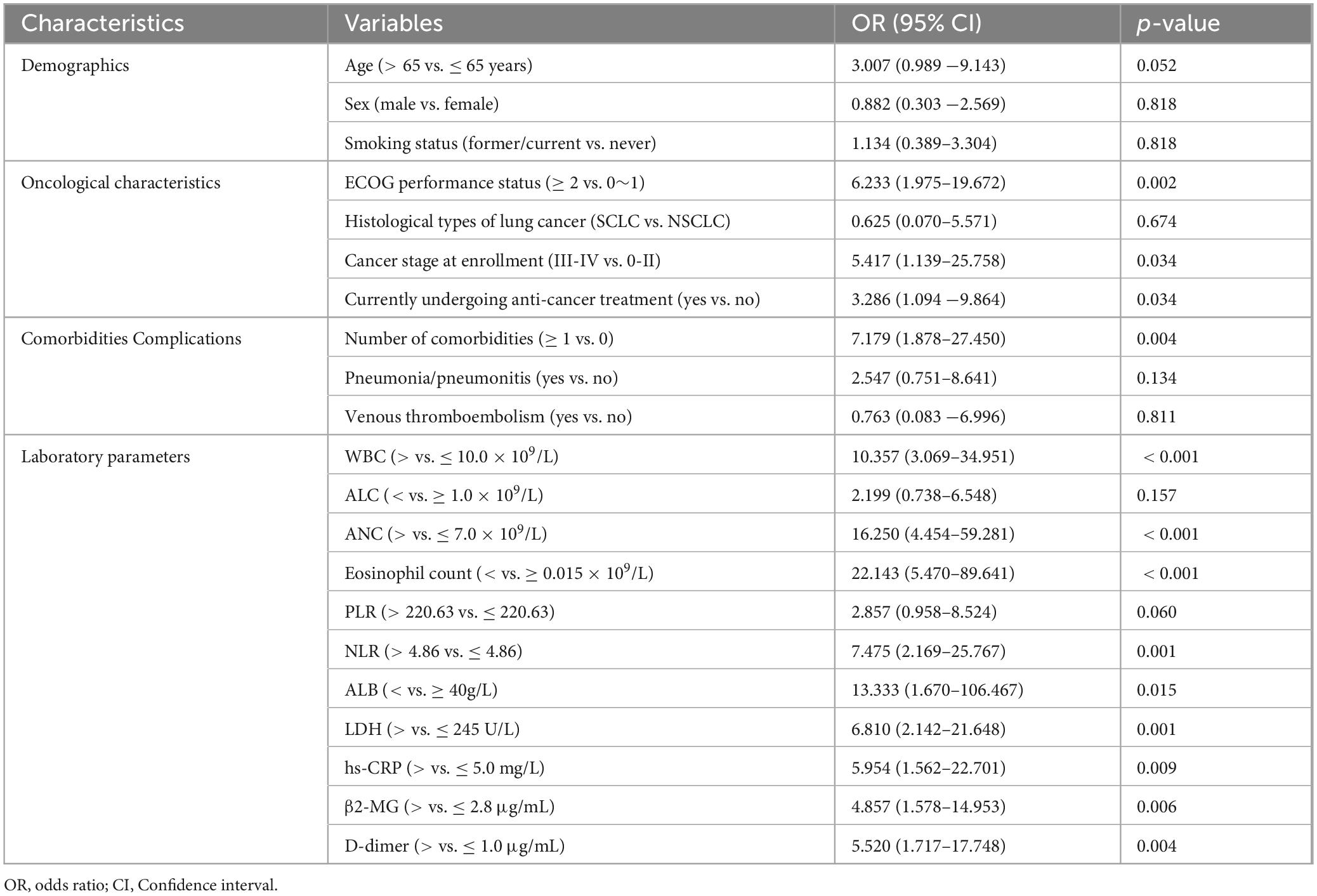
Table 3. Univariate logistic regression analysis for the severity of patients with lung cancer with omicron.
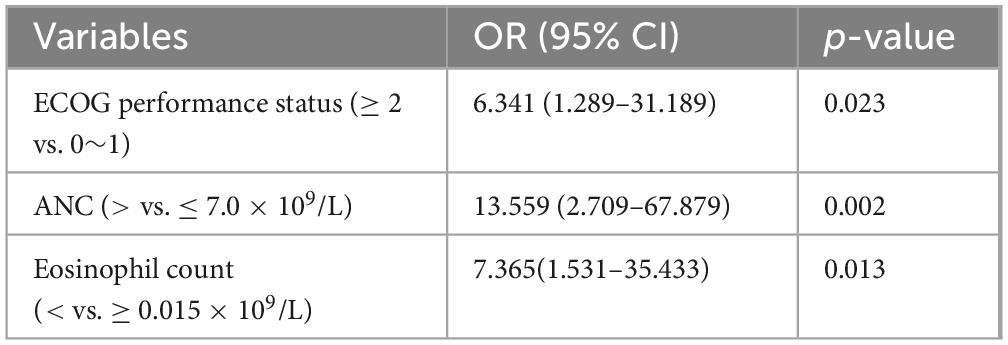
Table 4. Multivariate logistic regression model for the severity of patients with lung cancer with omicron.
Univariate survival analysis indicated that Cancer stage III-IV at enrollment, comorbidities, increased WBC, ANC, NLR, LDH, and β2-MG, and decreased eosinophil count were associated with an increased risk of death (Figure 5). Further multivariate analysis revealed that histological types of lung cancer (HR = 7.283, 95% CI: 1.626–32.618; p = 0.009), ANC (HR = 4.119, 95% CI: 1.289–13.165; p = 0.017), and eosinophil count (HR = 7.660, 95% CI: 2.157–27.195; p = 0.002) independently correlated with the survival (Table 5). Thus, the eosinophil count was an independent prognostic factor for the severity and survival of patients with omicron, and eosinopenia was obviously associated with poor outcomes.
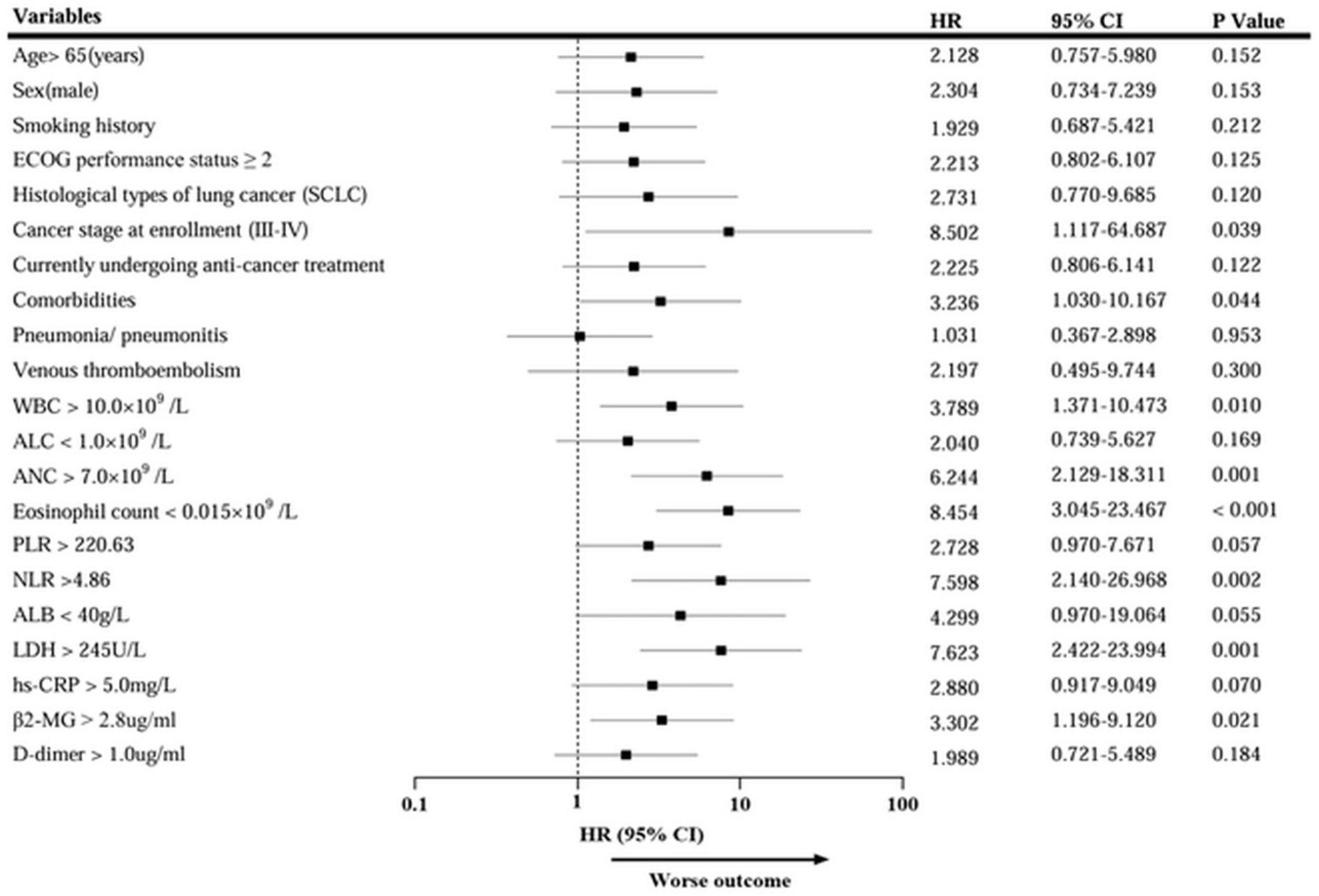
Figure 5. Univariate Cox regression analysis of mortality in patients with lung cancer with omicron. HR, hazard ratio; CI, Confidence interval.
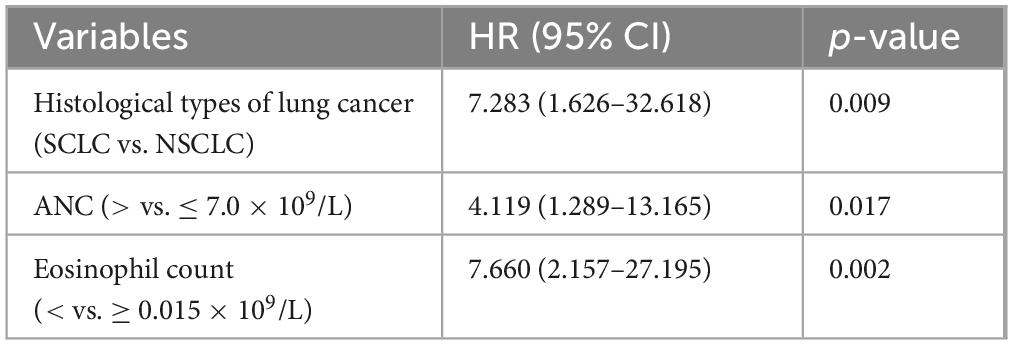
Table 5. Multivariate Cox regression model for the mortality of patients with lung cancer with omicron.
Discussion
This study revealed that patients with lung cancer with omicron had a higher mortality and a lower eosinophil count. In patients with lung cancer with omicron, the cut-off value of eosinophil count for predicting disease severity and death was 0.015 × 109/L, consistent with another study by our team (31). The AUC values were 0.754 for severity and 0.759 for death, respectively, indicating good predictive performance. Further analysis revealed that patients with eosinopenia (< 0.015 × 109/L) had worse 30-day survival and OS. Patients with a low eosinophil count tended to have lower DCR. After multivariate analysis, the eosinophil count was an independent predictor of disease severity and survival, and eosinopenia was significantly associated with poor outcomes.
Since March 2020, we have experienced the COVID-19 pandemic (32). Among patients with lung cancer, the mortality rate of those who developed COVID-19 was as high as 24–39% (1, 12, 33, 34). In this study, the mortality rate of patients with the omicron variant of COVID-19 was 18.1%, which is lower than the mortality rates reported in the aforementioned studies; the difference may be attributed to the variation in COVID-19 variants. Since November 2021, omicron has become the fifth variant of concern, apart from the Alpha, Beta, Gamma, and Delta variants (35). Multiple studies also have revealed that the omicron variant poses a lower risk of mortality compared with other variants (36, 37).
Since the emergence of the omicron variant (B.1.1.529), various sublineages have emerged, such as BA.1, BA.2, BA.4/BA.5 (38), as well as the subsequent omicron XBB sublineage (39), JN.1 sublineage (40), and BF.7 sublineage (41). To date, few studies have explored the role of eosinophils in the infection and disease progression of these sublineages. Zhu et al. (42) analyzed eosinophil counts in 1,157 patients infected with the SARS-CoV-2 omicron/BA.2 variant and found that eosinopenia was an independent risk factor for disease severity (OR 1.34, p = 0.006). Another study showed that patients infected with the SARS-CoV-2 omicron/BF.7 variant may also exhibit eosinopenia, although its association with disease severity and clinical outcomes remains unclear (41). Furthermore, some studies analyzing clinical characteristics of SARS-CoV-2-infected patients included those with delta variant but did not specifically examine the relationship between eosinophils and disease severity or outcomes in delta-infected patients (43, 44). Previous research has demonstrated that compared with the parental omicron lineage, each subvariant differs in transmissibility, virulence, and immune evasion ability (38, 40, 45). Omicron variants not only exhibit reduced virulence compared to other SARS-CoV-2 variants but also harbor more mutations and demonstrate stronger immune evasion capabilities (46–48). This study focused on patients with lung cancer who were infected with the omicron variant. Given the dynamic evolution of SARS-CoV-2 variants and the resulting differences in transmissibility, virulence, and immune evasion, the applicability of these findings to other omicron sublineages and earlier variants of SARS-CoV-2 requires cautious interpretation. Continuous monitoring of SARS-CoV-2 variants is warranted to clarify the impact of eosinophils across different strains.
Further analysis in this study revealed that the severe-to-critical omicron increased the mortality of patients with lung cancer (58.8% vs. 7.6%, p < 0.001), which was consistent with the results of GRAVID’s study (13). Severe COVID-19 leads to increased severe lung tissue damage (49), a higher proportion of mechanical ventilation, a higher chance of admission to the ICU (13), and a higher likelihood of post-COVID-19 syndrome (14, 50), which eventually leads to irreversible damage to lung function, delayed and reduced tolerance of anti-cancer treatment, and decreased survival.
The prognostic risk factors affecting patients with lung cancer who have COVID-19 have been the subject of numerous studies (1, 12, 13). Several studies have revealed that advanced cancer stages or metastasis at the time of COVID-19 diagnosis (1, 13), accompanying comorbidities (12, 34), and elevated WBC, neutrophil (1, 13), NLR (13, 51), and LDH (13) suggest poor prognosis. Beyond these established factors, our study identified eosinopenia as an independent predictor of disease severity and poor survival in lung cancer patients with omicron infection.
Eosinophils are important innate immune cells that have anti-cancer properties. They can be activated by costimulatory ligands, such as major histocompatibility complex class I (MHC-I), MHC-II, CD80, and CD86. This activation enhances eosinophil-dependent antigen presentation to CD4 + and CD8 + T cells, which promotes T cell proliferation and the release of associated cytokines. Moreover, eosinophils aid in the recruitment of CD8 + T cells by secreting chemokines, such as C-C motif chemokine ligand 5 (CCL5), C-X-C motif chemokine ligand (CXCL) 9, and CXCL10, which enhance the cytotoxic activity of T cells (52). Additionally, eosinophils stimulate the proliferation of natural killer (NK) cells and facilitate the CCL5-dependent recruitment of NK cells to the lungs in mice, contributing to anti-cancer effects (53). Other studies have found that the products of eosinophil lysis may also be involved in tumor destruction (54).
Wang et al. (55) followed 443,542 adults for an average of 5.8 years and found that the eosinophil count in peripheral blood was inversely associated with the occurrence of eight non-hematologic cancers, including lung cancer (aHR range: 0.65–0.95). In addition to being associated with lung cancer development, eosinophils are related to the efficacy of anti-cancer therapies. A study involving 189 patients with NSCLC found that a higher eosinophil proportion after treatment was associated with a better major pathological response (18). In patients with NSCLC treated with immune checkpoint inhibitors, although elevated eosinophils will increase the incidence of checkpoint inhibitor pneumonitis, high eosinophil levels can improve objective response rate and progression-free survival (PFS) (56). An increase in the eosinophil ratio > 1.43 was associated with an improved 5-year PFS and OS (10% vs. 8% and 21% vs. 10%, respectively) in patients with advanced NSCLC who received radiotherapy (57). Moreover, in patients with pleural effusion due to lung cancer, eosinophilia in pleural effusion contributes to improved survival (58). However, the present study revealed that patients with lung cancer with omicron who received current anti-cancer treatment were more prone to develop eosinopenia, which may be connected to the type of treatment and the effect of omicron.
Additionally, eosinophils exhibit antiviral effects. Eosinophils express Toll-like receptors (TLR), including TLR7, to recognize single-stranded RNA viruses (59). Meanwhile, eosinophils can release eosinophil cationic protein (ECP) and eosinophil-derived neurotoxin (EDN) to degrade the viral RNA genome. Eosinophils can also produce cytokines, such as interleukin-12 and interferon-γ, which can express MHC-I and MHC-II, promoting antigen presentation and recruiting virus-specific CD8 + T cells to the lungs to play an antiviral role (60). Besides, Li et al. (61) unexpectedly indicated that Patients with asthma are less susceptible to COVID-19 than the general population. Patients with COVID-19 with asthma had a lower mortality rate than those without asthma (62). Camiolo et al. (63) found a negative correlation between peripheral blood eosinophil count and the expression of angiotensin-converting enzyme 2 (ACE2) in the bronchial epithelium of patients with asthma. ACE2 has a high affinity to the C-terminal domain of SARS-CoV-2 and can promote the entry of SARS-CoV-2 into the cells (64). Therefore, eosinophils may also contribute to the prevention of SARS-CoV-2 infection by downregulating the expression of ACE2.
Numerous studies have shown that patients with severe COVID-19 are more prone to develop eosinopenia (65, 66). Eosinopenia was associated with aggravated chest CT findings (67), the elevated proportion of receiving drug therapy and oxygen support therapy (68), prolonged hospital stay (69), increased risk of admission to ICU (70), increased mortality (68, 71) and the occurrence of Long-COVID (72). Furthermore, the vaccination status of COVID-19 may also affect the level of eosinophils in patients infected with SARS-CoV-2. Li et al. (73) found that compared with vaccinated COVID-19 patients, eosinophil levels in the peripheral blood of unvaccinated COVID-19 patients were lower. Among patients infected with omicron, the third booster of the COVID-19 vaccine showed a more effective and sustained promoting effect on eosinophils, and this effect persisted 5 months after the last vaccination (42). Among patients infected with BF.7, vaccination with two and three doses of the COVID-19 vaccine can reduce the abnormal rate of eosinophils (41). In this study, the influence of vaccination status could not be evaluated due to incomplete vaccination information in the study population. Prospective studies are needed to incorporate patients’ vaccination status and clarify how it influences the role of eosinophils in COVID-19 outcomes.
At present, the reason for the increased susceptibility of patients with severe COVID-19 to eosinopenia is still not apparent. The lung histology of patients who died from severe COVID-19 and developed severe eosinopenia (74) and cytokine profile analysis related to severe COVID-19 (75) have found that depletion and suppression of eosinophil production may play a crucial role. In summary, SARS-CoV-2, especially severe-to-critical cases of SARS-CoV-2, may induce depletion of eosinophils and suppression of their production, leading to eosinopenia in peripheral blood. This will further weaken the antiviral and anti-cancer ability of patients with lung cancer, ultimately leading to cancer progression and reduced survival, which is consistent with our findings. As previously reported in the literature, eosinophilia is positively correlated with the prognosis of lung cancer patients receiving immunotherapy (56, 76). This appears to contradict the above conclusions and the research findings of Lindsley et al. (60). However, this may be related to the fact that immune checkpoint inhibitors (such as PD-1 inhibitors) can relieve immunosuppression, leading to increased secretion of IL-5 by CD4 + T cells. This in turn promotes the recruitment and activation of eosinophils, as well as enhancing eosinophil-mediated cytotoxicity and anti-tumor immunity (77). Another study has shown that increased secretion of granulocyte-macrophage colony-stimulating factor by type II innate lymphoid cells may also be involved (52). The divergent roles of eosinophils in the immunotherapy of COVID-19 and lung cancer reflect the functional plasticity of these cells in different immune microenvironments. However, randomization and large sample clinical studies as well as mechanistic studies in vivo and in vitro are still needed to explore the impacts of eosinopenia on patients with lung cancer with omicron.
Our study has the following limitations. First, it is a single-institution study with a small sample size, which is not ideal for meeting the per-variable event requirements of multivariate regression. Therefore, we need multi-center, large-sample studies in the future. Second, we have not conducted detailed studies to elucidate the fundamental role and mechanism of eosinopenia in patients with lung cancer with omicron, and we will carry out basic studies to further explore this area. Third, while eosinophils can help predict a patient’s severity and risk of death, incorporating additional features and indicators can enhance the robustness and accuracy of predictive models. Finally, because influenza virus and mycoplasma infections began to occur in China after our follow-up period, we had a short follow-up period for enrolled patients. After that, we will continue to follow the patients for a longer period and further analyze the difference in the impact of omicron and other pathogens on the outcomes of patients with lung cancer.
Conclusion
This study is the first to systematically analyze the effect of eosinophils on the severity and outcomes of patients with lung cancer with omicron. We found that omicron, especially the severe-to-critical omicron, reduced the survival of patients with lung cancer. Patients with omicron had a lower eosinophil count. Those with eosinopenia tended to have lower DCR, reduced 30-day survival, and decreased OS. The eosinophil count was an independent predictor for disease severity and survival in patients with lung cancer infected with omicron. Eosinopenia was significantly associated with poor outcomes. These insights not only elucidate the pathophysiological role of eosinophils in lung cancer and omicron but also provide valuable guidance for the prevention and surveillance of omicron during the epidemic. Multi-center and large sample clinical studies and in vitro and in vivo pathogenesis studies are necessary to reveal the mechanism of eosinophils in lung cancer and omicron.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the Second Hospital of Jilin University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
XH: Conceptualization, Data curation, Investigation, Methodology, Software, Writing – original draft. JT: Conceptualization, Data curation, Investigation, Writing – review & editing. RL: Formal Analysis, Software, Writing – review & editing. DD: Methodology, Project administration, Writing – review & editing. MY: Project administration, Supervision, Writing – review & editing. JY: Conceptualization, Funding acquisition, Methodology, Project administration, Writing – review & editing. QX: Data curation, Formal Analysis, Methodology, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Jilin Province of China (Grant No. YDZJ202401406ZYTS) and Jilin Province Medical and Health Talent (Grant No. 2024WSZX-B11).
Acknowledgments
We thank the Natural Science Foundation of Jilin Province of China for supporting our project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1583843/full#supplementary-material
References
1. Whisenant J, Baena J, Cortellini A, Huang L, Lo Russo G, Porcu L, et al. A definitive prognostication system for patients with thoracic malignancies diagnosed with coronavirus disease 2019: An update from the TERAVOLT registry. J Thorac Oncol. (2022) 17:661–74. doi: 10.1016/j.jtho.2021.12.015
2. Bowe B, Xie Y, Al-Aly Z. Acute and postacute sequelae associated with SARS-CoV-2 reinfection. Nat Med. (2022) 28:2398–405. doi: 10.1038/s41591-022-02051-3
3. Zhou Z, Zhu Y, Chu M. Role of COVID-19 vaccines in SARS-CoV-2 variants. Front Immunol. (2022) 13:898192. doi: 10.3389/fimmu.2022.898192
4. Kamboj M, Sepkowitz K. Nosocomial infections in patients with cancer. Lancet Oncol. (2009) 10:589–97. doi: 10.1016/S1470-204570069-5
5. Li J, Duan X, Wang L, Xu Y, Huang L, Zhang T, et al. Selective depletion of regulatory T cell subsets by docetaxel treatment in patients with nonsmall cell lung cancer. J Immunol Res. (2014) 2014:286170. doi: 10.1155/2014/286170
6. Davis H, Assaf G, McCorkell L, Wei H, Low R, Re’em Y, et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine. (2021) 38:101019. doi: 10.1016/j.eclinm.2021.101019
7. Xie Y, Xu E, Bowe B, Al-Aly Z. Long-term cardiovascular outcomes of COVID-19. Nat Med. (2022) 28:583–90. doi: 10.1038/s41591-022-01689-3
8. Khoury E, Nevitt S, Madsen W, Turtle L, Davies G, Palmieri C. Differences in outcomes and factors associated with mortality among patients with SARS-CoV-2 infection and cancer compared with those without cancer: A systematic review and meta-analysis. JAMA Netw Open. (2022) 5:e2210880. doi: 10.1001/jamanetworkopen.2022.10880
9. Zhang L, Zhu F, Xie L, Wang C, Wang J, Chen R, et al. Clinical characteristics of COVID-19-infected cancer patients: A retrospective case study in three hospitals within Wuhan. China. Ann Oncol. (2020) 31:894–901. doi: 10.1016/j.annonc.2020.03.296
10. Sung H, Ferlay J, Siegel R, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
11. Venkatesulu B, Chandrasekar V, Girdhar P, Advani P, Sharma A, Elumalai T, et al. A systematic review and meta-analysis of cancer patients affected by a novel coronavirus. JNCI Cancer Spectr. (2021) 5:kaa102. doi: 10.1093/jncics/pkaa102
12. Garassino M, Whisenant J, Huang L, Trama A, Torri V, Agustoni F, et al. COVID-19 in patients with thoracic malignancies (TERAVOLT): First results of an international, registry-based, cohort study. Lancet Oncol. (2020) 21:914–22. doi: 10.1016/S1470-204530314-4
13. Provencio M, Mazarico Gallego J, Calles A, Antoñanzas M, Pangua C, Mielgo Rubio X, et al. Lung cancer patients with COVID-19 in Spain: GRAVID study. Lung Cancer. (2021) 157:109–15. doi: 10.1016/j.lungcan.2021.05.014
14. Yong S. Long COVID or post-COVID-19 syndrome: Putative pathophysiology, risk factors, and treatments. Infect Dis (Lond). (2021) 53:737–54. doi: 10.1080/23744235.2021.1924397
15. Jacobsen E, Jackson D, Heffler E, Mathur S, Bredenoord A, Pavord I, et al. Eosinophil knockout humans: Uncovering the role of eosinophils through eosinophil-directed biological therapies. Annu Rev Immunol. (2021) 39:719–57. doi: 10.1146/annurev-immunol-093019-125918
16. Rosenberg H, Dyer K, Foster P. Eosinophils: Changing perspectives in health and disease. Nat Rev Immunol. (2013) 13:9–22. doi: 10.1038/nri3341
17. Fernández-Aceñero M, Galindo-Gallego M, Sanz J, Aljama A. Prognostic influence of tumor-associated eosinophilic infiltrate in colorectal carcinoma. Cancer. (2000) 88:1544–8. doi: 10.1002/(SICI)1097-0142(20000401)88:73.0.CO;2-S
18. Huai Q, Luo C, Song P, Bie F, Bai G, Li Y, et al. Peripheral blood inflammatory biomarkers dynamics reflect treatment response and predict prognosis in non-small cell lung cancer patients with neoadjuvant immunotherapy. Cancer Sci. (2023) 114:4484–98. doi: 10.1111/cas.15964
19. Artham S, Juras P, Goyal A, Chakraborty P, Byemerwa J, Liu S, et al. Estrogen signaling suppresses tumor-associated tissue eosinophilia to promote breast tumor growth. Sci Adv. (2024) 10:ead2442. doi: 10.1126/sciadv.adp2442
20. Lee T, Chen T, Kuo Y, Lan H, Yang M, Chu P. Tumor-associated tissue eosinophilia promotes angiogenesis and metastasis in head and neck squamous cell carcinoma. Neoplasia. (2023) 35:100855. doi: 10.1016/j.neo.2022.100855
21. Samarasinghe A, Woolard S, Boyd K, Hoselton S, Schuh J, McCullers J. The immune profile associated with acute allergic asthma accelerates clearance of influenza virus. Immunol Cell Biol. (2014) 92:449–59. doi: 10.1038/icb.2013.113
22. Phipps S, Lam C, Mahalingam S, Newhouse M, Ramirez R, Rosenberg H, et al. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood. (2007) 110:1578–86. doi: 10.1182/blood-2007-01-071340
23. Yan B, Yang J, Xie Y, Tang X. Relationship between blood eosinophil levels and COVID-19 mortality. World Allergy Organ J. (2021) 14:100521. doi: 10.1016/j.waojou.2021.100521
24. Gong S, Ma R, Zhu T, Ge X, Xie R, Tao Q, et al. Elevated serum beta-2 microglobulin level predicts short-term poor prognosis of patients with de novo acute omicron variant COVID-19 infection. Front Cell Infect Microbiol. (2023) 13:1204326. doi: 10.3389/fcimb.2023.1204326
25. Epstein Shochet G, Brook E, Bardenstein-Wald B, Shitrit D. TGF-β pathway activation by idiopathic pulmonary fibrosis (IPF) fibroblast derived soluble factors is mediated by IL-6 trans-signaling. Respir Res. (2020) 21:56. doi: 10.1186/s12931-020-1319-0
26. Pinato D, Scotti L, Gennari A, Colomba-Blameble E, Dolly S, Loizidou A, et al. Determinants of enhanced vulnerability to coronavirus disease 2019 in UK patients with cancer: A European study. Eur J Cancer. (2021) 150:190–202. doi: 10.1016/j.ejca.2021.03.035
27. Eisenhauer E, Therasse P, Bogaerts J, Schwartz L, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer. (2009) 45:228–47. doi: 10.1016/j.ejca.2008.10.026
28. Planchard D, Popat S, Kerr K, Novello S, Smit E, Faivre-Finn C, et al. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2018) 29:iv192–237. doi: 10.1093/annonc/mdy275
29. Postmus P, Kerr K, Oudkerk M, Senan S, Waller D, Vansteenkiste J, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2017) 28:iv1–21. doi: 10.1093/annonc/mdx222
30. Dingemans A, Früh M, Ardizzoni A, Besse B, Faivre-Finn C, Hendriks L, et al. Small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2021) 32:839–53. doi: 10.1016/j.annonc.2021.03.207
31. Tan J, Fang H, Hu X, Yue M, Yang J. Serum beta2-microglobulin and peripheral blood eosinophils for the assessment of severity and prognosis with omicron variant COVID-19 infection. Front Mol Biosci. (2024) 11:1476080. doi: 10.3389/fmolb.2024.1476080
32. Gonfiotti A, Gatteschi L, Salvicchi A, Bongiolatti S, Lavorini F, Voltolini L. Clinical courses and outcomes of five patients with primary lung cancer surgically treated while affected by severe acute respiratory syndrome coronavirus 2. Eur J Cardiothorac Surg. (2020) 58:598–604. doi: 10.1093/ejcts/ezaa233
33. Lee L, Cazier J, Starkey T, Briggs S, Arnold R, Bisht V, et al. COVID-19 prevalence and mortality in patients with cancer and the effect of primary tumour subtype and patient demographics: A prospective cohort study. Lancet Oncol. (2020) 21:1309–16. doi: 10.1016/S1470-204530442-3
34. Luo J, Rizvi H, Preeshagul I, Egger J, Hoyos D, Bandlamudi C, et al. COVID-19 in patients with lung cancer. Ann Oncol. (2020) 31:1386–96. doi: 10.1016/j.annonc.2020.06.007
35. Chen Z, Zhang Y, Wang M, Islam M, Liao P, Hu Y, et al. Humoral and cellular immune responses of COVID-19 vaccines against SARS-Cov-2 Omicron variant: A systemic review. Int J Biol Sci. (2022) 18:4629–41. doi: 10.7150/ijbs.73583
36. Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 omicron wave compared with previous waves. JAMA. (2022) 327:583–4. doi: 10.1001/jama.2021.24868
37. Nyberg T, Ferguson N, Nash S, Webster H, Flaxman S, Andrews N, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: A cohort study. Lancet. (2022) 399:1303–12. doi: 10.1016/S0140-673600462-7
38. Xia S, Wang L, Zhu Y, Lu L, Jiang S. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct Target Ther. (2022) 7:241. doi: 10.1038/s41392-022-01105-9
39. Davis-Gardner M, Lai L, Wali B, Samaha H, Solis D, Lee M, et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA bivalent booster. N Engl J Med. (2023) 388:183–5. doi: 10.1056/NEJMc2214293
40. Planas D, Staropoli I, Michel V, Lemoine F, Donati F, Prot M, et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat Commun. (2024) 15:2254. doi: 10.1038/s41467-024-46490-7
41. Gao X, Wang F, Liu H, Chai J, Tian G, Yao L, et al. BF.7: A new Omicron subvariant characterized by rapid transmission. Clin Microbiol Infect. (2024) 30:137–41. doi: 10.1016/j.cmi.2023.09.018
42. Zhu Z, Cai J, Tang Q, Mo Y, Deng T, Zhang X, et al. Circulating eosinophils associated with responsiveness to COVID-19 vaccine and the disease severity in patients with SARS-CoV-2 omicron variant infection. BMC Pulm Med. (2023) 23:177. doi: 10.1186/s12890-023-02473-w
43. Bie F, Yuan W, Chen Y, Gao Q. Clinical characteristics of the delta variant of COVID-19 in Jingmen, China. Medicine (Baltimore). (2022) 101:e30812. doi: 10.1097/MD.0000000000030812
44. Yan Y, Lyu C, Davgadorj C, Wang X, Sha M. [Analyze the clinical characteristics of mild and severe SARS-CoV-2 infected patients based on the local Omicron variant epidemic]. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. (2023) 35:32–6. doi: 10.3760/cma.j.cn121430-20220406-00339
45. Kimura I, Yamasoba D, Tamura T, Nao N, Suzuki T, Oda Y, et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell. (2022) 185:3992–4007.e16. doi: 10.1016/j.cell.2022.09.018
46. Meng B, Abdullahi A, Ferreira I, Goonawardane N, Saito A, Kimura I, et al. Altered TMPRSS2 usage by SARS-CoV-2 omicron impacts infectivity and fusogenicity. Nature. (2022) 603:706–14. doi: 10.1038/s41586-022-04474-x
47. Ai J, Zhang H, Zhang Y, Lin K, Zhang Y, Wu J, et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg Microbes Infect. (2022) 11:337–43. doi: 10.1080/22221751.2021.2022440
48. Zhang L, Li Q, Liang Z, Li T, Liu S, Cui Q, et al. The significant immune escape of pseudotyped SARS-CoV-2 variant omicron. Emerg Microbes Infect. (2022) 11:1–5. doi: 10.1080/22221751.2021.2017757
49. Filbin M, Mehta A, Schneider A, Kays K, Guess J, Gentili M, et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep Med. (2021) 2:100287. doi: 10.1016/j.xcrm.2021.100287
50. Karuna S, Gallardo-Cartagena J, Theodore D, Hunidzarira P, Montenegro-Idrogo J, Hu J, et al. Post-COVID symptom profiles and duration in a global convalescent COVID-19 observational cohort: Correlations with demographics, medical history, acute COVID-19 severity and global region. J Glob Health. (2023) 13:06020. doi: 10.7189/jogh.13.06020
51. Sutandyo N, Jayusman A, Widjaja L, Dwijayanti F, Imelda P, Hanafi A. Neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio as mortality predictor of advanced stage non-small cell lung cancer (NSCLC) with COVID-19 in Indonesia. Eur Rev Med Pharmacol Sci. (2021) 25:3868–78. doi: 10.26355/eurrev_202105_25954
52. Grisaru-Tal S, Rothenberg M, Munitz A. Eosinophil-lymphocyte interactions in the tumor microenvironment and cancer immunotherapy. Nat Immunol. (2022) 23:1309–16. doi: 10.1038/s41590-022-01291-2
53. Schuijs M, Png S, Richard A, Tsyben A, Hamm G, Stockis J, et al. ILC2-driven innate immune checkpoint mechanism antagonizes NK cell antimetastatic function in the lung. Nat Immunol. (2020) 21:998–1009. doi: 10.1038/s41590-020-0745-y
54. Mattes J, Hulett M, Xie W, Hogan S, Rothenberg M, Foster P, et al. Immunotherapy of cytotoxic T cell-resistant tumors by T helper 2 cells: An eotaxin and STAT6-dependent process. J Exp Med. (2003) 197:387–93. doi: 10.1084/jem.20021683
55. Wang J, Rabkin C, Engels E, Song M. Associations between eosinophils and cancer risk in the UK Biobank. Int J Cancer. (2024) 155:486–92. doi: 10.1002/ijc.34986
56. Chu X, Zhao J, Zhou J, Zhou F, Jiang T, Jiang S, et al. Association of baseline peripheral-blood eosinophil count with immune checkpoint inhibitor-related pneumonitis and clinical outcomes in patients with non-small cell lung cancer receiving immune checkpoint inhibitors. Lung Cancer. (2020) 150:76–82. doi: 10.1016/j.lungcan.2020.08.015
57. Wang N, Zhang X, Sui J, Wang Y, Wu Y, Lei Q, et al. Radiation-induced eosinophil increase ratio predicts patient outcomes in non-small celllung cancer. Front Oncol. (2022) 12:999555. doi: 10.3389/fonc.2022.999555
58. Takeuchi E, Okano Y, Machida H, Atagi K, Kondou Y, Kadota N, et al. Eosinophilic pleural effusion due to lung cancer has a better prognosis than non-eosinophilic malignant pleural effusion. Cancer Immunol Immunother. (2022) 71:365–72. doi: 10.1007/s00262-021-02994-5
59. Nagase H, Okugawa S, Ota Y, Yamaguchi M, Tomizawa H, Matsushima K, et al. Expression and function of toll-like receptors in eosinophils: Activation by toll-like receptor 7 ligand. J Immunol. (2003) 171:3977–82. doi: 10.4049/jimmunol.171.8.3977
60. Lindsley A, Schwartz J, Rothenberg M. Eosinophil responses during COVID-19 infections and coronavirus vaccination. J Allergy Clin Immunol. (2020) 146:1–7. doi: 10.1016/j.jaci.2020.04.021
61. Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. (2020) 146:110–8. doi: 10.1016/j.jaci.2020.04.006
62. Liu S, Cao Y, Du T, Zhi Y. Prevalence of comorbid asthma and related outcomes in COVID-19: A systematic review and meta-analysis. J Allergy Clin Immunol Pract. (2021) 9:693–701. doi: 10.1016/j.jaip.2020.11.054
63. Camiolo M, Gauthier M, Kaminski N, Ray A, Wenzel S. Expression of SARS-CoV-2 receptor ACE2 and coincident host response signature varies by asthma inflammatory phenotype. J Allergy Clin Immunol. (2020) 146:315–324.e7. doi: 10.1016/j.jaci.2020.05.051
64. Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. (2020) 181:894–904.e9. doi: 10.1016/j.cell.2020.03.045
65. Bao J, Li C, Zhang K, Kang H, Chen W, Gu B. Comparative analysis of laboratory indexes of severe and non-severe patients infected with COVID-19. Clin Chim Acta. (2020) 509:180–94. doi: 10.1016/j.cca.2020.06.009
66. Zhao L, Zhang Y, Yang X, Liu X. Eosinopenia is associated with greater severity in patients with coronavirus disease 2019. Allergy. (2021) 76:562–4. doi: 10.1111/all.14455
67. Xie G, Ding F, Han L, Yin D, Lu H, Zhang M. The role of peripheral blood eosinophil counts in COVID-19 patients. Allergy. (2021) 76:471–82. doi: 10.1111/all.14465
68. Wang X, Dian Y, Zhou Q, Deng G, Wei R, Zeng F. Association of eosinopenia with worsening prognosis in hospitalized Azvudine-treated COVID-19 patients: A retrospective cohort study. Front Immunol. (2023) 14:1320973. doi: 10.3389/fimmu.2023.1320973
69. Tan Y, Zhou J, Zhou Q, Hu L, Long Y. Role of eosinophils in the diagnosis and prognostic evaluation of COVID-19. J Med Virol. (2021) 93:1105–10. doi: 10.1002/jmv.26506
70. Valverde-Monge M, Cañas J, Barroso B, Betancor D, Ortega-Martin L, Gómez-López A, et al. Eosinophils and chronic respiratory diseases in hospitalized COVID-19 patients. Front Immunol. (2021) 12:668074. doi: 10.3389/fimmu.2021.668074
71. Cortés-Vieyra R, Gutiérrez-Castellanos S, Álvarez-Aguilar C, Baizabal-Aguirre V, Nuñez-Anita R, Rocha-López A, et al. Behavior of eosinophil counts in recovered and deceased COVID-19 patients over the course of the disease. Viruses. (2021) 13:1675. doi: 10.3390/v13091675
72. Proal A, VanElzakker M. Long COVID or post-acute sequelae of COVID-19 (PASC): An overview of biological factors that may contribute to persistent symptoms. Front Microbiol. (2021) 12:698169. doi: 10.3389/fmicb.2021.698169
73. Li X, Xu Y, Li X, Liu W, Yao D, Chen W, et al. Real-world effectiveness and protection of SARS-CoV-2 vaccine among patients hospitalized for COVID-19 in Xi’an, China, December 8, 2021, to January 20, 2022: A retrospective study. Front Immunol. (2022) 13:978977. doi: 10.3389/fimmu.2022.978977
74. Cauchois R, Pietri L, Dalmas J, Koubi M, Capron T, Cassir N, et al. Eosinopenia as predictor of poor outcome in hospitalized COVID-19 Adult patients from waves 1 and 2 of 2020 pandemic. Microorganisms. (2022) 10:2423. doi: 10.3390/microorganisms10122423
75. Jesenak M, Banovcin P, Diamant Z. COVID-19, chronic inflammatory respiratory diseases and eosinophils-Observations from reported clinical case series. Allergy. (2020) 75:1819–22. doi: 10.1111/all.14353
76. Alves A, Dias M, Campainha S, Barroso A. Peripheral blood eosinophilia may be a prognostic biomarker in non-small cell lung cancer patients treated with immunotherapy. J Thorac Dis. (2021) 13:2716–27. doi: 10.21037/jtd-20-3525
Keywords: omicron, COVID-19, lung cancer, eosinopenia, outcomes
Citation: Hu X, Tan J, Luan R, Ding D, Yue M, Yang J and Xue Q (2025) Eosinopenia predicts poor outcomes in patients with lung cancer with the omicron variant of COVID-19. Front. Med. 12:1583843. doi: 10.3389/fmed.2025.1583843
Received: 11 March 2025; Accepted: 20 June 2025;
Published: 09 July 2025.
Edited by:
Haider Abdul-Lateef Mousa, University of Basrah, IraqReviewed by:
Matteo Stravalaci, IRCCS Ca ’Granda Foundation Maggiore Policlinico Hospital, ItalyRicardo H. Pineda, University of Pittsburgh, United States
Copyright © 2025 Hu, Tan, Luan, Ding, Yue, Yang and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junling Yang, anVubGluZ0BqbHUuZWR1LmNu; Qianfei Xue, NDk3NDQ5MTI4QHFxLmNvbQ==
 Xiao Hu
Xiao Hu Jie Tan1
Jie Tan1 Junling Yang
Junling Yang