- 1State Key Laboratory of Chinese Medicine Modernization, Tianjin University of Traditional Chinese Medicine, Tianjin, China
- 2Guangzhou Laboratory, Guangzhou, China
- 3Tianjin Pharmaceutical Da Ren Tang Group Corp., Ltd. Traditional Chinese Medicine Research Institute, Tianjin, China
- 4Haihe Laboratory of Modern Chinese Medicine, Tianjin, China
Background: The global outbreak of coronavirus disease (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has raised significant public health concerns. Qingyan Dropping Pills (QDP), as a recommended drug, is issued by the National Health Commission of the People’s Republic of China for the treatment of COVID-19. However, its bioactive compounds and their mechanisms of action remain largely unidentified. In this study, the integration of computational and experimental approaches was performed to identify the bioactive compounds in QDP and elucidate its mechanisms against COVID-19.
Methods: Utilizing UPLC-Q/TOF-MS, the chemical compounds of QDP were delineated, followed by network pharmacology analysis and molecular docking targeting SARS-CoV-2 spike protein (Spro), main protease (Mpro), and papain-like protease (PLpro). To validate the inhibitory activity of these compounds, fluorescence resonance energy transfer (FRET) and surface plasmon resonance (SPR) assays were employed. The antivival efficacy was tested in Vero E6 cells infected with SARS-CoV-2 Omicron BA.5 variant. Moreover, anti-inflammatory potential was evaluated via the measurement of inflammatory markers, including nitric oxide (NO), interleukin-6 (IL-6), interleukin-1 beta (IL-1β), and tumor necrosis factor-alpha (TNF-α).
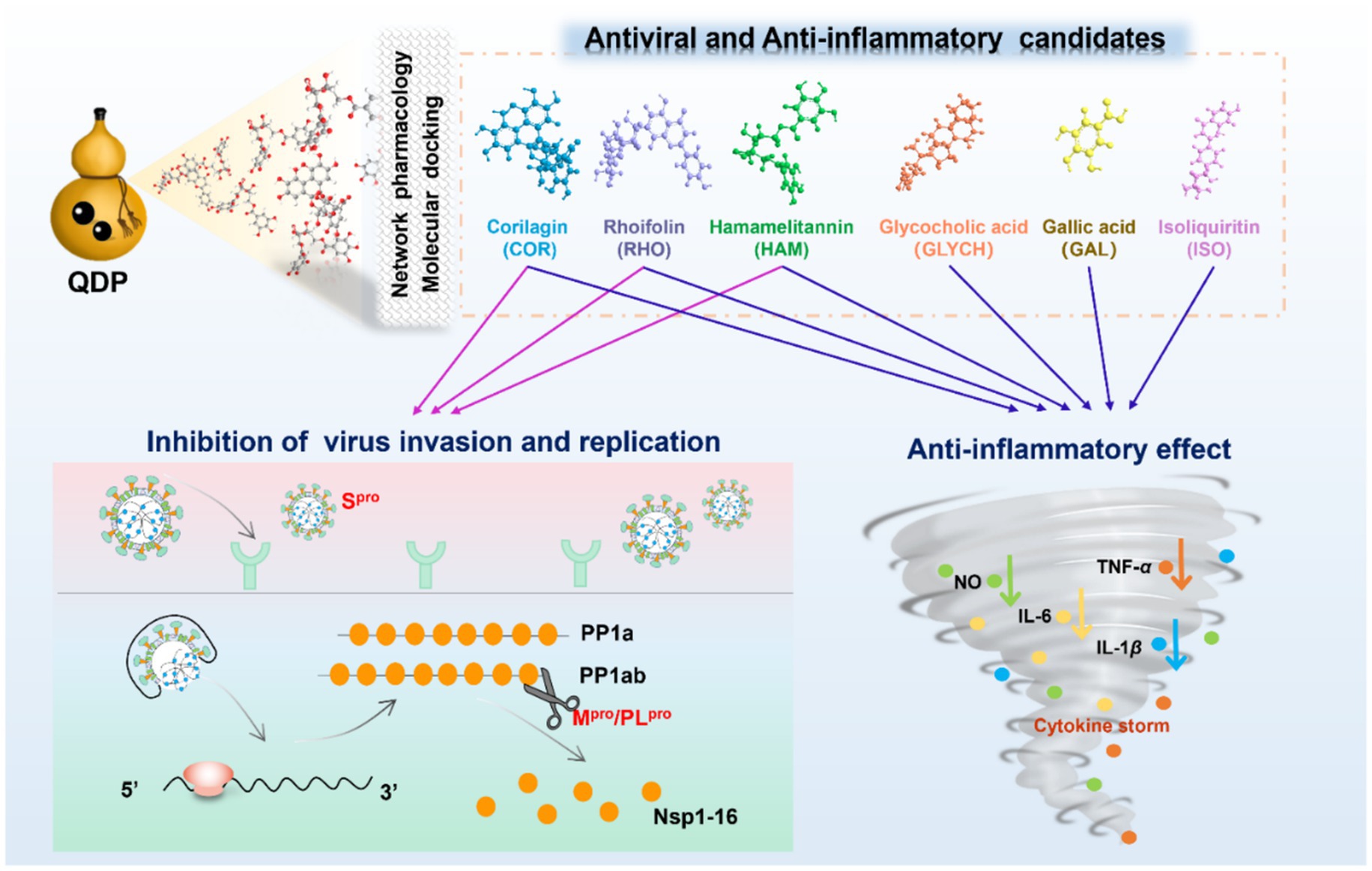
Results: Among the 48 identified compounds, 33 demonstrated potential antiviral activity against COVID-19. Notably, Hamamelitannin (HAM), corilagin (COR), and rhoifolin (RHO) effectively interacted with Spro, Mpro and PLproin silico. In SPR assays, the equilibrium dissociation constant (KD) for COR and RHO ranged from 4.515 × 10−8 M to 7.718 × 10−6 M, while HAM showed strong binding affinity to Spro (KD = 9.33 × 10−8 M) but weaker affinity for Mpro and PLpro. In FRET assays, COR and RHO inhibited Mpro with IC50 valuse of 0.73 μM and 21.61 μM, respectively. Additionally, COR proved effective against the Omicron BA.5 variant. The compounds COR, HAM, RHO, isoliquiritin (ISO), glycocholic acid (GLYCH), and gallic acid (GAL) displayed significant anti-inflammatory activity by inhibiting the crucial inflammatory factors, indicating their dual therapeutic potential in managing COVID-19.
Conclusion: Our study focused on Chinese patent medicine QDP to highlight the anti-SARS-CoV-2 and anti-inflammatory bioactives, providing evidence and insights into its clinical practice in the treatment of COVID-19.
1 Introduction
The outbreak of COVID-19 has posed a severe threat to global public health (1). SARS-CoV-2 is the primary pathogen responsible for this pandemic. Although remarkable progress has been made in vaccine development and antiviral drug discovery-including BNT162b2, mRNA-1273, ChAdOx1, and BBIBP-CorV, as well as in therapeutic options such as remdesivir and corticosteroids, the ongoing mutation of SARS-CoV-2 and the emergence of novel variants have sustained the pandemic’s worldwide impact (1). The development of effective inhibitors against SARS-CoV-2 remains crucial in controlling viral infection. In addition, the coexistence of COVID-19 with chronic diseases, such as diabetes mellitus, has increased disease severity and mortality, highlighting the need for comprehensive prevention and treatment strategies. Traditional Chinese medicine (TCM) has played a significant role in the fight against COVID-19, owing to its holistic regulatory effects, multi-target therapeutic mechanisms, and personalized treatment approaches (2–4). Among the various treatments used in clinical practice, three drugs-Jinhua Qinggan granule, Lianhua Qingwen capsule, and Xuebijing injection-and three prescriptions-Xuanfei Baidu formula, Huashi Baidu formula, and Qingfei Paidu decoction-have demonstrated effectiveness in reducing mortality rates, alleviating symptoms, preventing the progression of mild to severe disease and promoting recovery (5, 6). These findings provide valuable insights and complementary approaches to combating emerging infectious diseases. Therefore, elucidating the bioactive compounds and underlying mechanisms is imperative for optimizing the use of TCM in clinical practice.
QDP is a multi-herbal Chinese prescription, mainly used for treating acute or chronic throat inflammation in the clinic. Based on its demonstrated efficacy, QDP has been endorsed by the National Health Commission of the People’s Republic of China as a recommended treatment for COVID-19 patients (7). The formulation comprises Terminalia chebula Retz. (Chebulae Fructus, CF), Bovis Calculus Artificialis (Artificial Calculus Bovis, ACB), Indigo Naturalis (Natural Indigo, NI), Glycyrrhizae Radix et Rhizoma (Licorice root, LR), Borneolum Syntheticum (borneol, BO), and Mentholum (L-menthol, LM). Previous studies have identified QDP as a common remedy for treating respiratory diseases, demonstrating inhibitory effects against pathogens such as Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) (8). Therefore, investigating the bioactive compounds and mechanisms of action that underlie its antiviral and anti-inflammatory activities is both urgent and meaningful for clinical application in managing COVID-19.
SARS-CoV-2, a single-stranded RNA virus belonging to the β-coronavirus family, encodes 29 proteins, including four structural proteins, sixteen non-structural proteins, and nine accessory proteins (9, 10). The life cycle of SARS-CoV-2 contains four main stages, invasion, replication, assembly, and release. Spro serves as the primary target as it binds to angiotensin-converting enzyme 2 (ACE2) receptors, facilitating membrane fusion and mediating viral entry into host cells (11). Upon release of the viral genome into the host cell, viral replication is activated. Mpro and PLpro play pivotal roles by cleaving the PP1a and PP1ab polyproteins, producing non-structural proteins essential for SARS-CoV-2 transcription and replication. Following the assembly of mature viral particles, these are released into the extracellular space via exocytosis, triggering an inflammatory response that can lead to a cytokine storm (12). Given their critical functions in the viral life cycle, Spro, Mpro, and PLpro are targeted for screening potential antiviral compounds (13, 14).
Accordingly, in this study, we first clarified the chemical profile of QDP using UPLC-Q/TOF-MS analysis. Subsequently, we constructed pharmacological networks to identify potential active compounds, which were filtered through molecular docking techniques. Finally, focusing on the identified compounds, SPR assay, FRET screening, antiviral experiments, and cell models were performed to validate the active compounds and elucidate their mechanisms of the antivirus and anti-inflammation. This study aims to contribute valuable insights and robust scientific foundation into the clinical application of QDP for treating COVID-19 patients.
2 Materials and methods
2.1 Reagents and materials
The QDP (Lot. 620,054) were generously provided by Tianjin pharmaceutical DA REN TANG group corporation limited NO.6 traditional Chinese medicine factory (Tianjin, China). High-quality MS grade methanol and acetonitrile were purchased from Thermo Fisher Scientific Co., Ltd. (Fair Lawn, NJ, USA), as well as formic acid and dimethyl-sulfoxide (DMSO) from Shanghai Aladdin Biochemical Technology Co., Ltd. (Shanghai, China). Mpro and PLpro were purified in our lab and Spro was purchased from Sino Biological Inc. (Beijing, China). CM5 sensor chips, amine-coupling kit, and running buffer were purchased from General Electric Company (Boston, USA). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum were purchased from Gibco Invitrogen Corp. (New York, USA). Nitric oxide (NO) assay kit was purchased from the Beyotime Institute of Biotechnology (Shanghai, China). Enzyme-linked immunosorbent assays (ELISA) kits of IL-6, TNF-α, and IL-1β were supplied by ZCIBIO Technology Co., Ltd. (Shanghai, China).
The 24 standard compounds (HPLC purity ≥ 98%) were used as the reference standards. 24 compounds are derived from the three institutions, National Institutes for Food and Drug Control, Sichuan Weiqi Technology Co., Ltd. and Shanghai Yuanye Bio-Technology Co., Ltd. Glycyrrhizic acid (GLYCZ), glycyrrhetinic acid (GLYCT), liquiritigenin (LIQN), liquiritin (LIQ), isoliquiritin (ISO), gallic acid (GAL), glycocholic acid (GLYCH), indirubin (IND), cholic acid (CHO), and hyodeoxycholic acid (HYO) were obtained from National Institutes for Food and Drug Control (Beijing, China) and Sichuan Weiqi Technology Co., Ltd. (Sichuan, China). Rhoifolin (RHO), corilagin (COR), hamamelitannin (HAM), chebulic acid (CHE), tauroursodeoxycholic acid (TAUD), erucamide (ERU), licoricesaponin G2 (LIC), ononin (ONO), glycoursodeoxycholic acid (GLYCA), piperine (PIP), glycycoumarin (GLYC), 16-hydroxyhexadecanoic acid (HYDRA), chenodeoxycholic acid (CHEA) and oleamide (OLE) were purchased from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China). Ritonavir (RIT) as a positive drug was obtained from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China).
2.2 Preparation of reference and sample solutions for chemical analysis
QDP (80 mg) was extracted with 3 mL 70% methanol (v/v) by ultrasound extraction for 30 min at 35 °C, filtered through a 0.45 μm nylon syringe filter. The filtrate was collected for further analysis.
The reference compounds were dissolved in methanol with the concentration of 1 mg/mL as the standard stock solutions, respectively. Then, the reference stock solutions were mixed and diluted with methanol to obtain the standard working solution. The final concentration for each compound was 20 μg/mL.
2.3 UPLC-Q/TOF-MS analysis
Analysis was performed on an Agilent 1,290 coupled to an Agilent 6,520 Q/TOF MS spectrometer with an electrospray ionization (ESI) source (CA, USA), which was operated by a MassHunter workstation. Gradient elution was undertaken on the ACQUITY UPLC® BEH C18 (2.1 × 100 mm, 1.7 μm) at 35 °C. The mobile phase consisted of acetonitrile (A) and water with 0.1% formic acid (B), which was applied as follows: 0–4 min, 5% − 9% A; 4–10 min, 9% − 44.5% A; 10–12 min, 44.5% − 51% A; 12–14 min, 51% − 58% A; 14–16 min, 58% − 62% A; and 16–25 min, 62% − 95% A. The flow rate was set at 0.3 mL/min, and the sample injection volume was 3 μL.
Mass spectra were analyzed in both positive ion and negative ion modes with a mass range of m/z 100–1,500 Da. Optimal MS parameters were as follows: capillary voltage, −2.5 kV in negative ion mode and +3.0 kV in positive ion mode, capilary temperature at 120 °C, desolvation temperature at 350 °C, flow rate of collision gas at 0.20 mL/min, nebulizer gas pressure at 30 psig, and collision energy at 30 eV. Data acquisition was performed using MassHunter workstation.
2.4 Network pharmacology analysis
Potential targets associated with the identified compounds in QDP were retrieved from TCMSP database1 and the Swiss Target Prediction database2 (15). We selected “COVID-19” and “coronavirus pneumonia” as the keywords to acquire the disease-related targets from the GeneCards database3 and OMIM database.4 Then, the shared targets were displayed in Venn diagram (16). Based on the shared targets, the herbs-preparation-compounds-targets network and protein–protein interaction (PPI) network were, respectively, constructed to screen the potential active compounds and the core targets. Homo sapiens was used to restrict the organism, and the minimum interaction threshold was set at the highest confidence (0.900). In the network, the degree value of the target was calculated to assess the significance (17).
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using the DAVID database5 to illustrate the biological process (BP), cellular component (CC), molecular function (MF), and signaling pathways (18). The top 10 GO terms and 30 KEGG pathways ordered by p-value were exhibited in the bar graphs.
2.5 Molecular docking
Molecular docking is a useful approach in structure-based drug discovery in silico (19). To further discover the bioactive compounds in QDP against SARS-CoV-2, the compounds that were filtered by network pharmacology were docked with the key targets of SARS-CoV-2, including Mpro, PLpro, and Spro (PDB ID: 6 LU7, 7CJM, and 7T9L). Structures of the tested compounds and protein-ligand complexes were obtained from the PubChem database6 and the protein data bank (PDB) database7 (20).
The preparation of ligands and proteins was performed using Discovery Studio 2020 software. Proteins were prepared via removing extra water molecules, adding hydrogens, and repairing missing residues. The location of the original ligand was defined as the active site, which was deleted from the prepared protein. The cdocker mode was employed for docking. Molecular docking was carried out using the cdocker protocol as implemented in Discovery Studio 2020, adhering to its validated default settings. The CHARMm-based scoring function accounted for van der Waals and electrostatic interactions via the Lennard-Jones potential and a distance-dependent dielectric constant (cut-off: 14 Å). The docking procedure involved random rigid-body rotations of the ligands, simulated annealing, and a final grid-based refinement. Pose ranking was based on the CDOCKER Interaction Energy, which comprises the ligand’s internal strain energy and its interaction energy with the protein. The results were visualized by Pymol 2.6 software. Ritonavir (RIT) as a positive compound that was reported to inhibit SARS-CoV-2 (21), N3 as the original ligand of Mpro, and GRL0617 as the original ligand of PLpro, were docked to the corresponding targets as the positive references (22, 23).
2.6 Cloning, protein expression, and purification of SARS-CoV-2 Mpro and PLpro
The plasmid containing full-length gene of SARS-CoV-2 Mpro and PLpro were constructed and transferred into E. coli BL21 (DE3), respectively, which were separately cultured in Luria broth medium with 100 μg/mL ampicillin for expressing Mpro and 50 μg/mL kanamycin for expressing PLpro at 37 °C for 6 ~ 8 h. Then, the proteins were induced for expression with 500 μM isopropyl β-D-thiogalactoside (IPTG) at 16 °C for 16 ~ 20 h. A centrifugated deposit were collected by 4,000 rpm for 20 min at 4 °C, which were crushed through the high-pressure homogenizer. Ni-NTA affinity column was used for purification with the buffer at pH 8.0 containing 20 mM Tris–HCl, 150 mM NaCl, 5% Glycerol, and 300 mM imidazole. The C-terminal 6 x His tag of Mpro was removed by human rhinovirus 3C protease, and the small ubiquitin-like modifier (SUMO) linking to PLpro was removed by SUMO enzyme. The Mpro and PLpro were further subjected to purification by size-exclusion chromatography (Superdex 200 Increase 10/300 GL) and ion exchange chromatography (HiTrapTM Q HP 5 mL). SDS-PAGE is employed to verify the purification of the tested proteins above 95%.
2.7 SPR assay
Surface plasmon resonance (SPR) assay was performed by Biacore T200 to validate the binding affinity of the focused compounds to the targeted proteins. The Spro, Mpro, and PLpro as the target proteins were, respectively, immobilized on the activated CM5 sensor via an amine coupling reaction. Firstly, the focused compounds were dissolved with DMSO at 2.5 mM as the standard stock solution, which was diluted for 20 times with the buffer (1.05 × PBS-P solution) to obtain the initial concentration samples (125 μM). Then, the samples were diluted step by step in the running buffer (1.05 × PBS-P, 5% DMSO, pH = 7.4) with the concentration ranging from 0.030 to 125 μM. The flow rate was set at 30 μL/min, and the association time and dissociation time was separately set at 120 s and 180 s. The entire experiment was conducted at a temperature of 25 °C. Data were analyzed by Biaevaluation software 2.0 in kinetics mode.
2.8 Determination of the enzymatic inhibition activities on Mpro and PLpro
Fluorescence resonance energy transfer assay (FRET) was employed to screen enzymatic inhibitors according to the properties of Mpro and PLpro as the drug targets. The substrates can specifically recognize the proteins to change the distance between the fluorescent receptor and the donor, which has been used to investigate intermolecular interactions. The substrate of Mpro is designed and synthesized as MCA-AVLQSGFR-Lys (Dnp)-Lys-NH2 and the substrate sequence of PLpro is Dabcyl-FTLKGGYAPTKVTE-Edans by GL biochem Co., Ltd. (Shanghai, China). The enzymatic activity evaluating system of Mpro contains 0.2 μM protein, the different concentrations of compounds (0.3125–160 μM), and 20 μM of substrate. And the enzymatic activity evaluating system of PLpro contains 1 μM protein, the different concentrations of compounds (0.049–100 μM), and 10 μM of substrate. The inhibitions (%) of the tested compounds were evaluated by Graphpad prism 8.0 (24).
2.9 Antiviral activity assay
The antiviral activity of the tested compound was evaluated by cytopathic effects (CPE) inhibition assay in Vero E6 cells in vitro, which was performed in a biosafety level 3 (BSL-3) laboratory. Vero E6 cells were seeded in a 96-well plate with 1 × 105 cells/well overnight, which were cultured in DMEM supplemented with 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 U/mL penicillin, at 37 °C with 5% CO2 and 95% air. The mixed sample with Omicron BA.5 (MOI = 0.01) and the tested compound (concentration from 0.0137 μM to 30.00 μM) was added into each well to incubate for 48 h at 37 °C. Then, CPE rate was scored by the Celigo Image Cytometer, which was employed to calculate the value of half maximal effective concentration (EC50).
2.10 Anti-inflammatory activity assay
RAW 264.7 cells were cultured in DMEM with 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 U/mL penicillin at 37 °C and supplemented with 5% CO2 and 95% air. After seeded and cultured in a 96-well plate (2 × 105 cells/well, 100 μL medium/well) for overnight, RAW 264.7 were treated with the tested compounds for 24 h, respectively. Then, 50 μL MTT (2.5 mg/mL) was added into each well and incubated at 37 °C for 4 h. Finally, the absorbance was recorded at 490 nm by a microplate reader to assess the cell viability.
NO is an essential pro-inflammatory factor, which can be not only highly expressed in cells but also promoted the expression of other inflammatory cytokines. TNF-α, IL-6, and IL-1β were considered as the main inflammatory cytokines, which were screened as the key targets in network pharmacology. So, the cytokines, including NO, TNF-α, IL-6, and IL-1β, were selected as indicators to evaluate the anti-inflammatory effects. After cultured in a 96-well plate overnight for 1 × 105 cells/well, RAW 264.7 cells were simultaneously treated with the tested compounds (concentration from 5 μM to 100 μM) and lipopolysaccharide (LPS) (concentration with 1 μg/mL) for 24 h. Then, the supernatant was collected to measure the cytokines levels with the corresponding assay kits, respectively.
2.11 Statistical analysis
All statistical analyses were performed using GraphPad Prism 8. Statistical significance for each endpoint was assessed with an unpaired two-tailed Student’s t-test. Data are presented as the mean ± standard deviation (SD). A significant level of p < 0.05 was considered statistically significant for all analyses.
3 Results
3.1 Characterization of the chemical compounds in QDP
To qualitative analysis the chemical compounds in QDP, the chromatographic analysis was performed by UPLC-Q/TOF-MS and the base peak chromatogram is shown in Figures 1A,B, from which 48 compounds (Supplementary Table S1) were tentatively identified and 24 compounds were unambiguously characterized by matching with reference compounds (GAL, LIQ, ISO, LIQN, GLYCZ, CHO, HYO, IND, GLYCT, CHE, RHO, COR, GLYCH, HAM, TAUD, ERU, LIC, PIP, OLE, GLYCA, GLYC, HYDRA, HYO, and CHEA). The radar map summarizes the cross-herb distribution of major chemical classes. Each axis denotes a chemical class, and the radial value represents its normalized relative abundance within each herb. This visualization enables rapid comparison of chemical fingerprints across herbs, highlights dominant compound families and overall chemical diversity, and provides a rationale for prioritizing compound classes for subsequent bioassays and mechanistic studies. Among the 24 compounds, as shown in the radar map (Figure 1C), 15 compounds were from glycyrrhizae radix et rhizome (LR), including 5 flavones, 5 fatty acids, 4 terpenoids, and 1 coumarin. 13 compounds were from chebulae fructus (CF), including 8 phenolic acids, 4 fatty acids, and 1 flavone. 11 compounds were from bovis calculus artifactus (ACB), including 9 steroids and 2 alkaloids. One alkaloid was identified from Indigo Naturalis (NI).
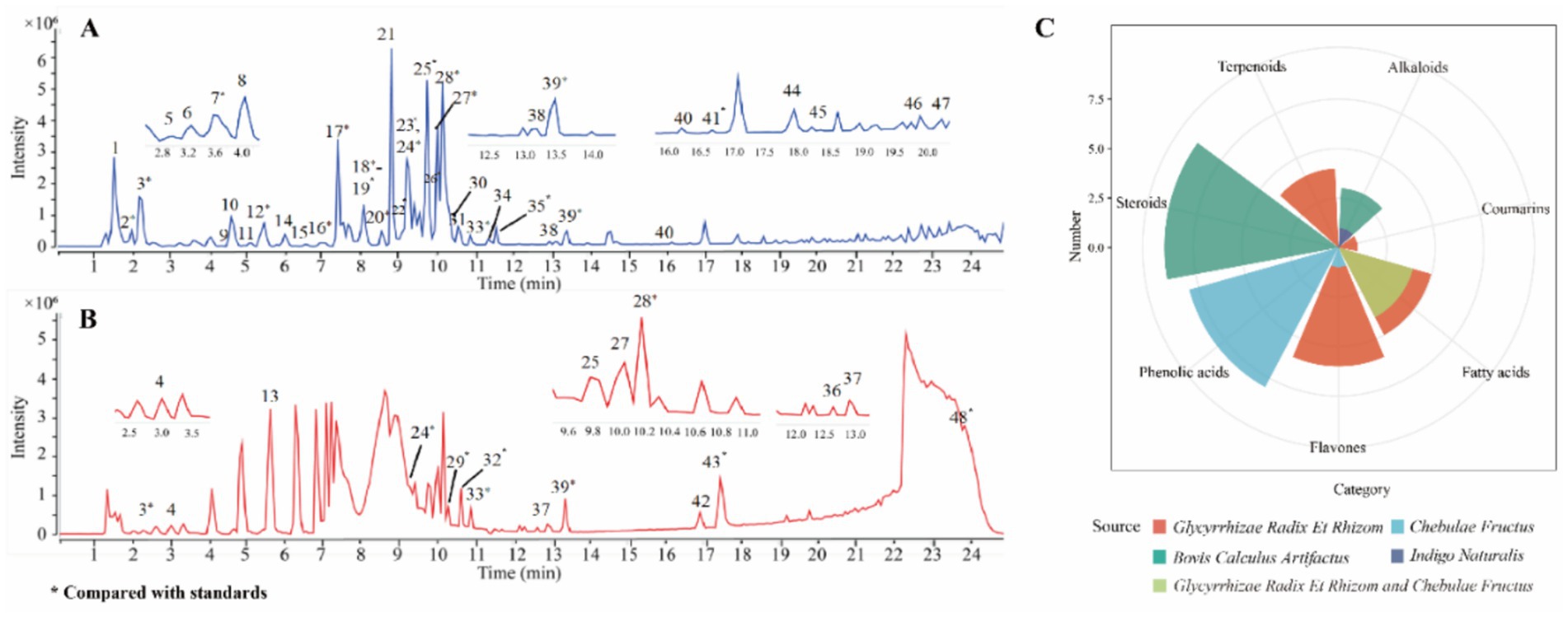
Figure 1. Base peak intensity chromatograms and compound source analysis of QDP extracts. (A) Negative ion mode. (B) Positive ion mode. (C) Radar map of compound sources.
Chebulic acid (peak 2) was taken as a case to introduce identification of compounds. In the negative ion mode, a quasi-molecular ion was detected at m/z 355.0315 [M − H]−, which, respectively, produced the fragment ions at m/z 337.0210 [M-H-H2O] − and 311.0409 [M-H-CO2] − by loss of H2O and CO2. The compound was confirmed as chebulic acid (25).
3.2 Identification of active compounds and illumination of core targets and pathways associated with COVID-19 by network pharmacology analysis
Focusing on the identified 48 compounds, we employed network pharmacology to preliminarily confirm active compounds in QDP against COVID-19. 468 targets associated with the characterized compounds and 1,958 targets related with COVID-19 were overlapped to obtain the 102 shared targets, which were displayed in the Venn diagram in Supplementary Figures S1A,B. In Figure 2A, the network of herbs-preparation-compounds-targets showed that interactions happened between the complicated compounds and multiple targets to perform antiviral and anti-inflammatory effects, from which 33 compounds were unveiled as the potential active candidates (Supplementary Table S2).
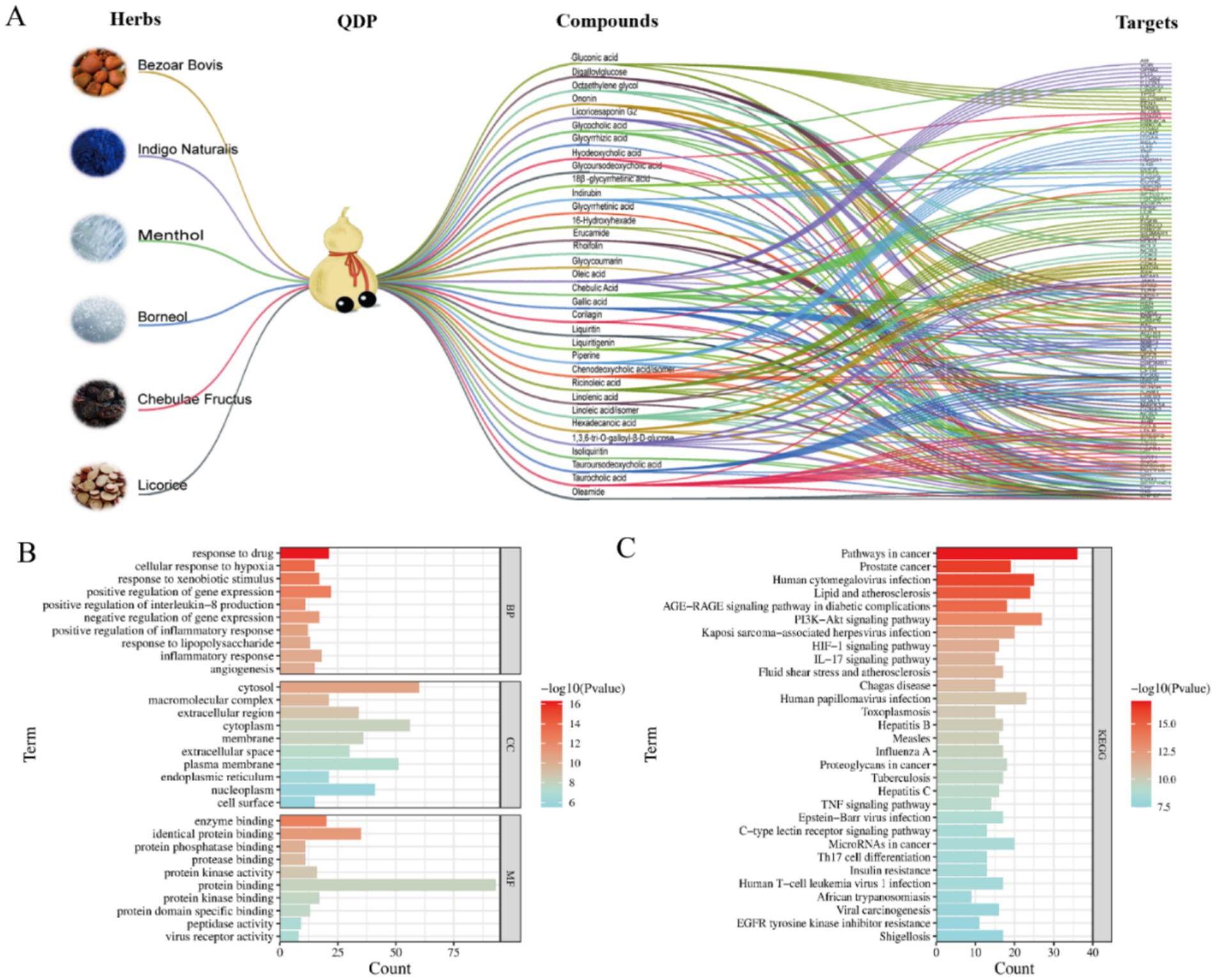
Figure 2. Network pharmacology analysis of herbs-preparation-compounds-targets and functional enrichment. (A) Herb-preparation-compound-target network. (B) GO enrichment analysis. (C) KEGG enrichment analysis.
In order to obtain the core targets, the PPI network was constructed according to the shared 102 targets, and the degree value of each target was calculated and ranked to identify core targets. As shown in Supplementary Figure S2C, the higher the degree value was, the more important the node was in the PPI network. The key nodes in the network were considered as the core targets. Accordingly, β-actin (ACTB), tumor necrosis factor (TNF), tumor protein p53 (TP 53), vascular endothelial growth factor A (VEGFA), and interleukin-6 (IL-6) were suggested as the core targets against COVID-19.
To further explore the biological functions and pathways, GO and KEGG enrichment analyses were undertaken. As shown in Figure 2B, the top 10 terms were, respectively, ranked by p-value and displayed in BP, CC, and MF analyses, from which the important biological functions were enriched, including the response to drug, cellular response to hypoxia, enzyme binding, identical protein binding, and so on. Notably, several immune-related biological processes including IL-8 production, inflammatory response, and response to LPS were significantly enriched (p < 0.01). KEGG pathway enrichment analysis indicated that the core targets were enriched in viral infection-related pathways, including human cytomegalovirus infection, Kaposi sarcoma-associated herpesvirus infection, human papillomavirus infection, and so on (Figure 2C). Moreover, as for inflammation regulation, phosphatidylinositol 3-kinase-AKT (PI3K-Akt), interleukin-17 (IL-17), and TNF signaling pathways were suggested as the key pathways.
3.3 Exploration in the binding mode of potential active compounds to Spro, Mpro, and PLpro via molecular docking
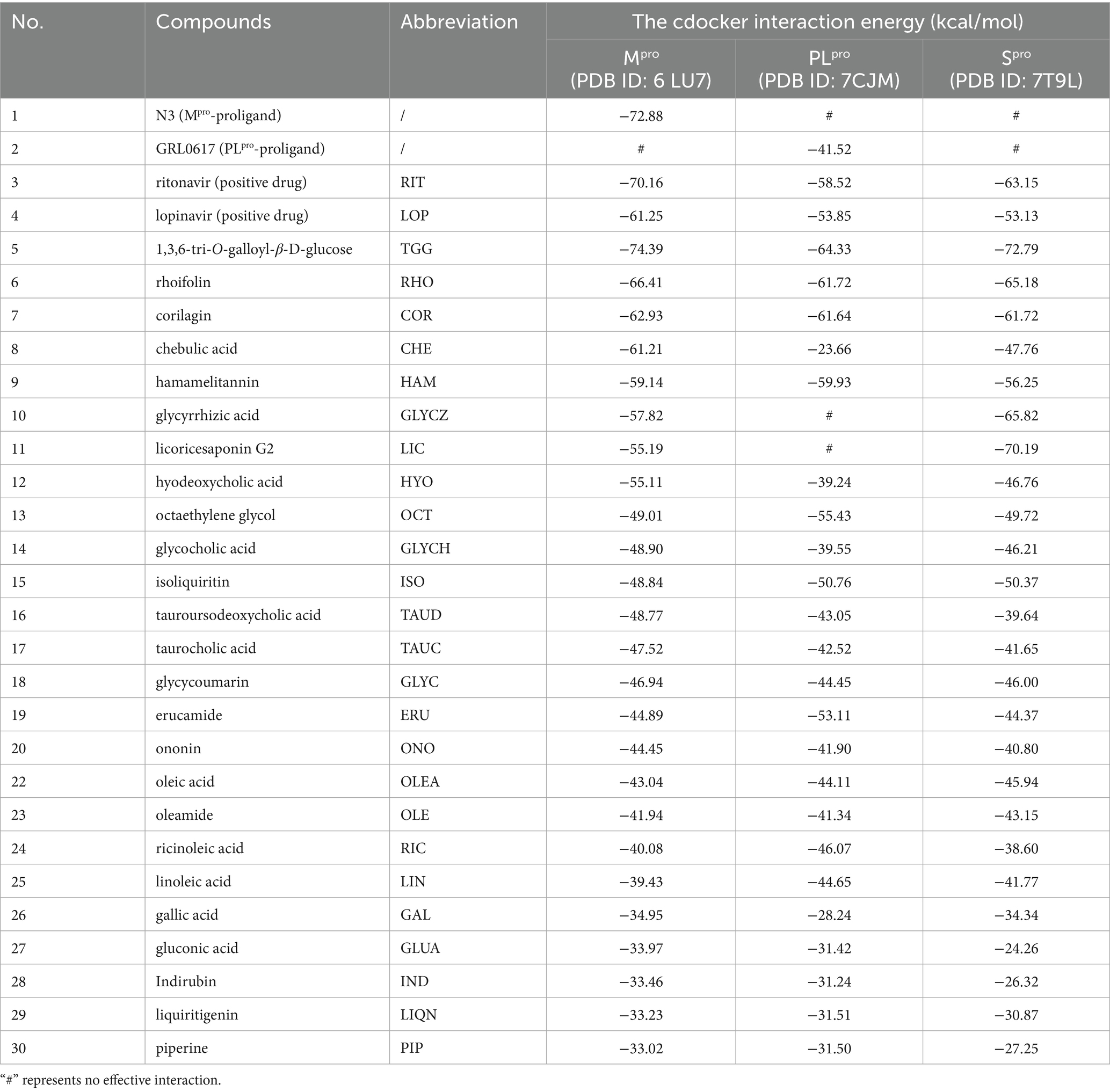
As the key targets for invasion and replication of SARS-CoV-2, Spro, Mpro, and PLpro were employed to dock with the 33 potential active compounds, respectively. Lower cdocker interaction energy indicates the more stable conformation of the target-ligand complex as shown in Table 1. As the original ligand of Mpro (PDB:6 LU7), N3 was docked with Mpro at a binding energy of −72.88 kcal/mol, while GRL0617, as an original ligand of PLpro (PDB:7CJM), was docked with PLpro at a binding energy of −41.52 kcal/mol (Supplementary Figure S2), resulting in validation of the feasibility of docking model. RIT and LOP, which were reported as the inhibitors of SARS-CoV-2, were proved to have perfect binding with Spro, Mpro, and PLpro as positive drugs. Twenty-six compounds from thirty-three potential bioactive compounds were successfully docked to thees three targets, respectively. As displayed in Figure 3A, the heatmap of binding energy has shown that COR, RHO, and HAM exhibited satisfactory binding with all three targets, whose binding energy was as low as below the score of the original ligand.
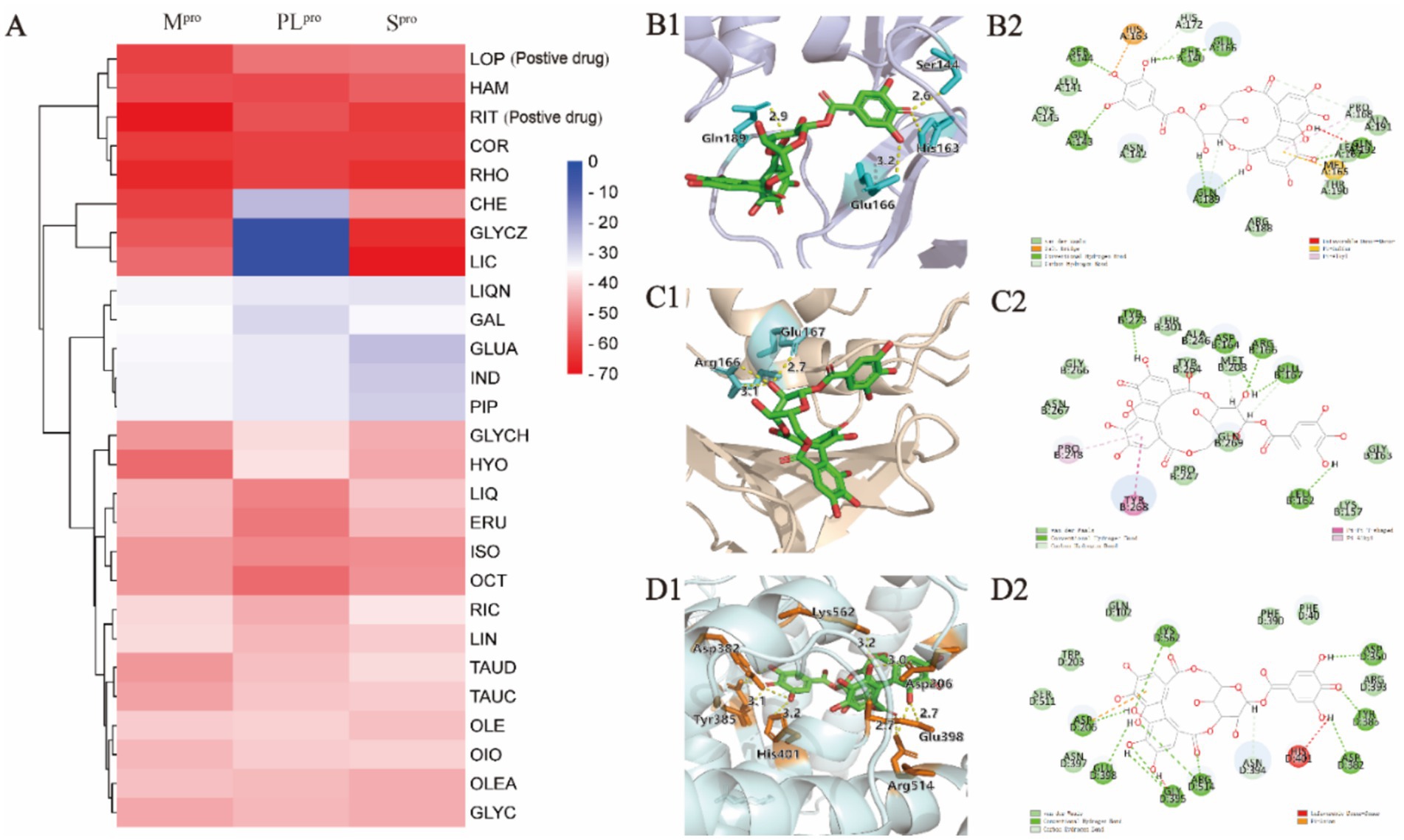
Figure 3. Molecular docking analysis of compounds against viral proteases. (A) Heatmap of CDOCKER interaction energies for compounds-Mpro, compounds-PLpro, and compounds-Spro complexes. (B1,B2) Binding conformations of COR-Mpro complex. (C1,C2) Binding conformations of COR-PLpro complex. (D1,D2) Binding conformations of COR-Spro complex.
COR was taken as an example to display the docking mode with the different targets in Figures 3B1,B2-D1,D2, from which hydrogen bonds and π-π stacking were observed. For example, the COR-Mpro complex obviously showed the formation of hydrogen bonds, which were formed with the residues of Ser144 (2.6 Å), Glu166 (3.2 Å), and Gln189 (2.9 Å) in the binding site. The interactions between COR and PLpro were witnessed at residues of Glu167 (2.7 Å) and Arg166 (2.8 Å) to form hydrogen bonds. The hydrogen bonds were exhibited in the COR-Spro complex at residues of Asp350 (2.8 Å), Asp382 (3.1 Å), Tyr385 (3.0 Å), Lys562 (3.2 Å), Arg514 (2.7 Å), Glu398 (2.7 Å), and Asp206 (3.0 Å). Moreover, the aromatic ring of COR formed π-π stacking with the side chain residue of Asp206.
For RHO and HAM, we have placed the result graph of the supplementary molecular docking in Supplementary Figure S3. The RHO-Mpro complex obviously showed the formation of hydrogen bonds, which were formed with the residues of Glu166 (2.4 Å), Glu166 (2.9 Å), Phe140 (2.0 Å), Phe140 (1.9 Å), Asn142 (2.3 Å), Asn142 (2.8 Å) and Thr26 (1.9 Å) in the binding site. The interactions between RHO and PLpro were witnessed at residues of Thr301 (1.9 Å), Gly163 (2.8 Å) and Asp164 (1.9 Å) to form hydrogen bonds. The hydrogen bonds were exhibited in the RHO-Spro complex at residues of Asp206 (2.0 Å), Asp350 (1.9 Å), Ala348 (2.0 Å) and Ala348 (2.0 Å).
The HAM-Mpro complex obviously showed the formation of hydrogen bonds, which were formed with the residues of Thr190 (2.3 Å), Asn142 (2.4 Å) and Cys145 (2.0 Å) in the binding site. The interactions between HAM and PLpro were witnessed at residues of Glu167 (1.9 Å), Tyr268 (3.2 Å), Arg166 (3.3 Å) and Asp164 (2.4 Å) to form hydrogen bonds. The hydrogen bonds were exhibited in the HAM-Spro complex at residues of Asn349 (2.1 Å), Lys562 (1.9 Å) and Lys562 (1.8 Å).
3.4 SPR assay for validation of binding affinity with Mpro, PLpro, and Spro
Mpro and PLpro were purified with a purify exceeding 95% in our lab as shown in Supplementary Figures S4. By taking the availability of compounds in the market into account, as the representative compounds with the most satisfactory interaction with the focused targets, COR, RHO, and HAM were employed to perform validation of binding affinity by determining the equilibrium dissociation constant (KD). The lower KD value of compounds indicates a stronger binding affinity to the targets. In Figures 4A1-A3, targeting Spro, the KD values for COR, RHO, and HAM were determined at 1.920 × 10−7 M, 7.068 × 10−6 M, and 9.330 × 10−8 M, respectively. In comparison, other natural compounds, such as epigallocatechin gallate (KD = 1.15 × 10−5 M), isobavachalcone (KD = 5.70 × 10−6 M) and isochlorogenic acid A (KD = 1.83 × 10−5 M) (26), also demonstrated binding affinity to the Spro RBD and were verified as promising SARS-CoV-2 inhibitors. Therefore, the evaluation of KD values proves to be an effective strategy for screening potential inhibitors. For PLpro and Mpro, as illustrated in Figures 4B1-B3,C1-C3, using RIT as a positive drug, the KD values were evaluated at 6.565 × 10−5 M for PLpro and 7.035 × 10−5 M for Mpro. The KD values for RHO and COR were appraised to fall within the ranges of 4.515 × 10−8 M and 7.718 × 10−6 M, respectively. Conversely, HAM exhibited negligible affinity toward both PLpro and Mpro.
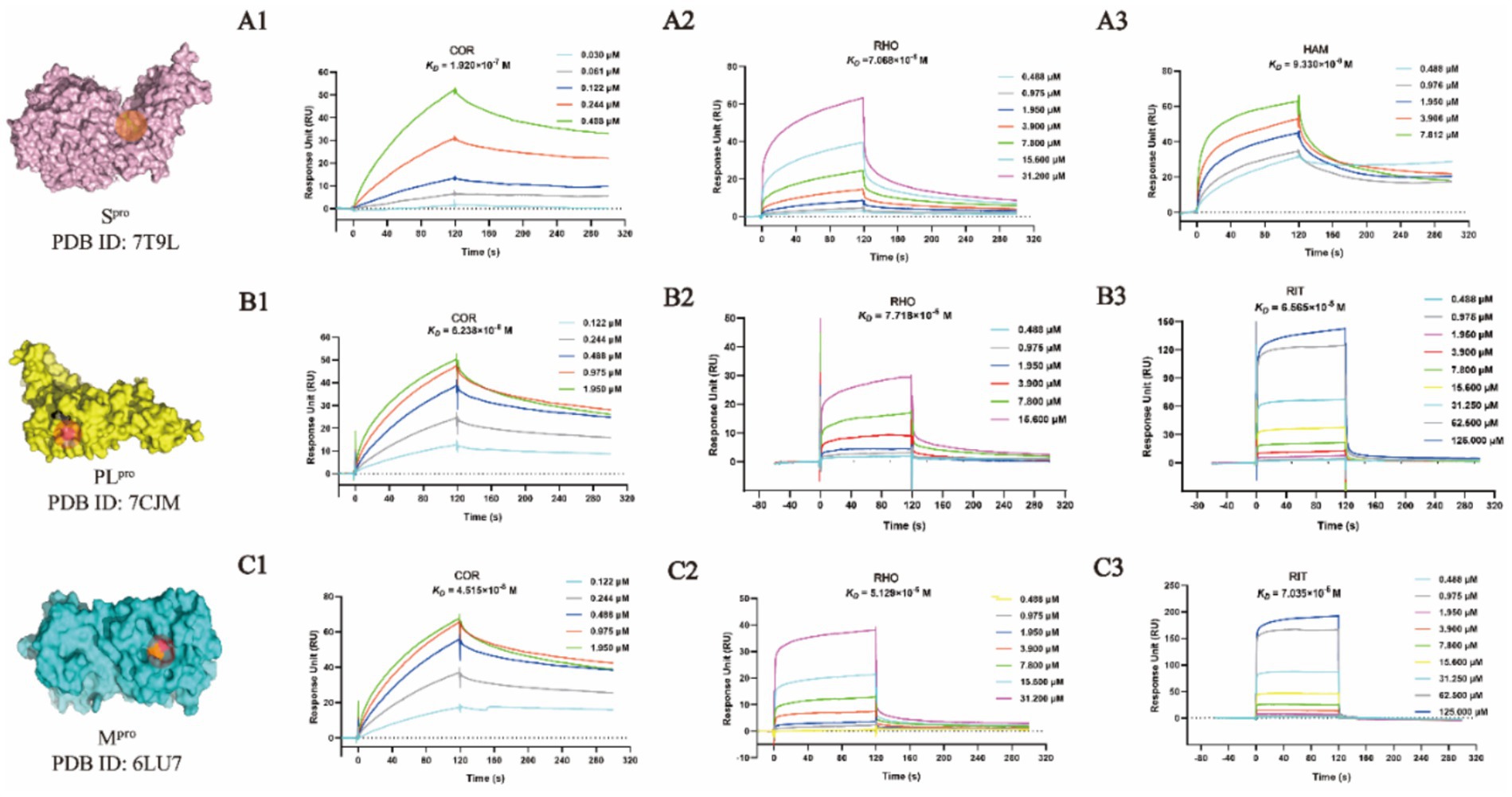
Figure 4. SPR binding affinity analysis of compounds against viral proteases. (A1-A3) Spro-compound interactions. (B1-B3) PLpro-compound interactions. (C1-C3) Mpro-compound interactions.
Despite the docking results, HAM, RHO and COR showed different binding affinities with Mpro, PLpro, and Spro. The virtual screenings, one of the most widely practiced strategies for discovering potential active compounds, presents some design shortcomings. Typically, a scoring function is used to evaluate the strength of the interaction between receptors and ligands. However, this approach often suffers from issues of accuracy and applicability. Furthermore, in virtual screening, the active site is predefined, which may not accurately reflect its true nature. Therefore, while virtual screening could rapidly focus on the potential active compounds and predict their mechanisms, it is imperative to corroborate these predictions with a series of experimental validations.
3.5 High-throughput screening inhibitors of Mpro and PLpro by FRET
FRET has been widely employed in screening the inhibitors of Mpro and PLpro because of its high sensitivity and specificity (27). Owing to the excellent binding affinity with Mpro and PLpro, RHO and COR were further used to evaluate the inhibitory activity on Mpro and PLpro. As showed in Figure 5, ebselen, as the Mpro inhibitor, showed the strongest inhibition on Mpro with IC50 of 0.10 ± 0.05 μM (Figure 5B). RHO and COR exhibited satisfactory inhibitory activities with IC50 of 21.61 ± 8.78 μM (Figure 5C) and 0.73 ± 0.05 μM (Figure 5D). Furthermore, COR demonstrated potent inhibition of the Mpro with an IC50 value of 0.73 μM, outperforming 1,2,3,4,6-penta-O-galloyl-β-D-glucose and 1,2,3,6-tetra-O-galloyl-β-D-glucose with IC50 values ranging from 1.33 to 27.37 μM (9). GRL0617, as the PLpro inhibitor, showed inhibitory effects with IC50 of 2.69 ± 0.43 μM (Supplementary Figure S5A). Unlike Mpro, COR and RHO showed poor inhibition on PLpro at 40 μM and IC50 beyond 100 μM (Supplementary Figure S5B). The results suggested that COR and RHO exhibited preferentially inhibition activity on Mpro by comparing with PLpro.
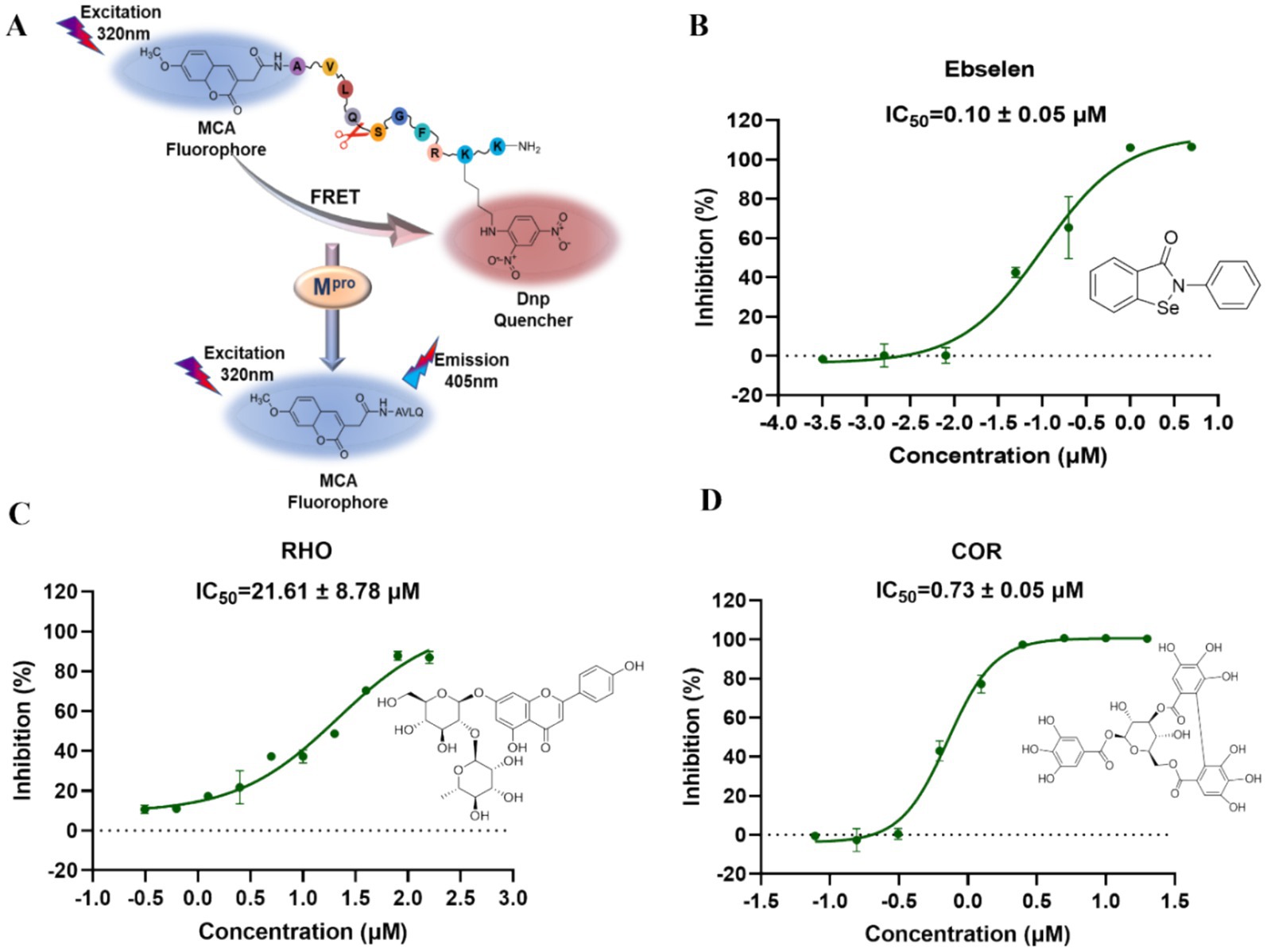
Figure 5. Inhibition activity of the focused compounds on SARS-CoV-2 Mpro. (A) The experimental principle of Mpro inhibitory activity. (B) The IC50 inhibition curve of ebselsen. (C) The IC50 inhibition curve of RHO. (D) The IC50 inhibition curve of COR.
Natural compounds were a rich source to discover the antiviral agents. Due to the safety in clinical practice, the natural products receive more attention. As the natural products, COR and RHO showed significantly inhibitory activity on Mpro at a comparable level with the chemical molecules and exhibited specifical inhibitory activity on Mpro. This difference is related to the structural characteristics of these proteins. In QDP, COR mainly comes from the herb CF, and RHO comes from the herb LR, which were suggested as the main active compounds for antiviral activity by inhibiting Mpro.
3.6 Evaluation of antiviral activity on SARS-CoV-2 omicron BA.5 in vitro
COR exhibited the strongest affinity and inhibitory activity on Mpro, while KD of COR with Spro displayed excellent interaction, which was considered as the most potential antiviral compound and employed to perform antiviral validation. In the MTT assay, COR was witnessed to be no cytotoxicity after incubating with Vero E6 cells for 48 h with CC50 values more than 100 μM (Figure 6A).
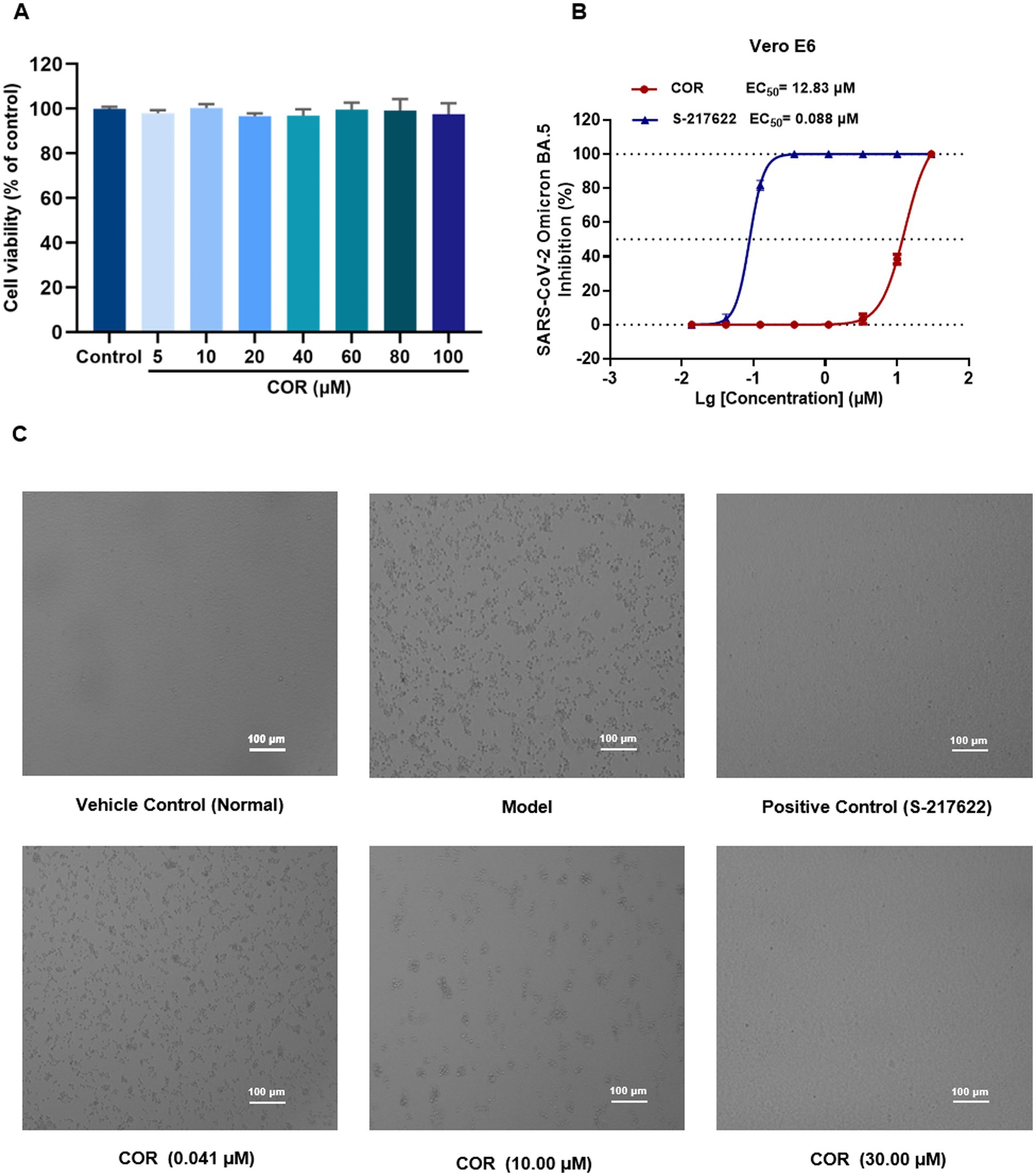
Figure 6. The cytotoxicity and antiviral activity evaluation of COR against SARS-CoV-2 Omicron BA.5. (A) Cytotoxicity assessment in Vero E6 cells by MTT assay. (B) Inhibition of SARS-CoV-2 Omicron BA.5 replication (mean ± SD, n = 3). (C) Representative cytopathic effect (CPE) images in Vero E6 cells.
S-217622, the first oral non-covalent and non-peptidic SARS-CoV-2 Mpro inhibitor, is used as a positive control from the clinical candidate (28). S-217622 and COR was diluted into the different concentrations and incubated with SARS-CoV-2 Omicron BA.5 variant in Vero E6 cells, respectively. As shown in Figure 6B, a dose-dependent manner of COR was observed by protecting the cells damaged by SARS-CoV-2 Omicron BA.5. The EC50 of COR was obtained at 12.83 μM and that of S-217622 was 0.088 μM, showing the excellent antiviral activity. Representative images of CPE formation were displayed in Figure 6C. We speculated that COR possibly exerts antiviral activity through Spro, Mpro, and PLpro to interfere with the viral invasion and replication (29).
As a phenolic acid, COR derives from CF in QDP, which has been proved to inhibit the activity of reverse transcriptase of RNA tumor viruses and show negligible toxicity on normal cells and tissues (30). In this study, COR was screened from the identified compounds library in QDP by an integrated strategy of dry and wet method, which displays specifically inhibitory activity on Mpro. Additionally, COR has a good solubility and stability, which is considered as a promising scaffold for developing inhibitors against coronavirus (31).
3.7 Evaluation of anti-inflammatory activity in vitro
As the main lethal factors of COVID-19, the fatal pneumonia was caused by the cytokine storm. Anti-inflammatory therapy is considered as an effective strategy for reducing the damage on the important organs (32). In order to screen the active compounds in QDP with anti-inflammatory activity, 24 compounds from 33 potential active compounds, which accessibly were deserved from market, were used to verify the anti-inflammatory effects in vitro by evaluating the NO, IL-6, IL-1β, and TNF-α level in LPS-induced RAW 264.7 cells. To establish the LPS stimulation condition, a preliminary dose–response experiment was conducted in RAW 264.7 cells. Cells were treated with LPS at 1, 2, 5, or 10 μg/mL, and NO levels in the culture supernatants were measured by the Griess method using a kit-derived standard curve (Supplementary Figure S6A). All LPS concentrations significantly increased NO compared with the blank control, with no significant differences among the LPS groups (Supplementary Figure S6B), indicating a response plateau from 1 to 10 μg/mL. Therefore, 1 μg/mL-the lowest dose achieving a significant response-was chosen for subsequent experiments to ensure consistent activation while minimizing potential nonspecific effects. As shown in Supplementary Figure S7, RHO, COR, GAL, and ISO exhibited no cytotoxicity up to 100 μΜ in RAW264.7 cells (CC50 > 100 μΜ), while for HAM and CLYCH, cell viability significantly decreased (p < 0.05) above 40 μΜ. When the RAW264.7 cells were exposed to LPS (1 μg/mL), NO level increased significantly. As shown in Figures 7A–F, comparing model group, the production of NO can be inhibited by RHO, COR, HAM, GAL, ISO, and GLYCH within the tested concentration in the dose-dependent manners.
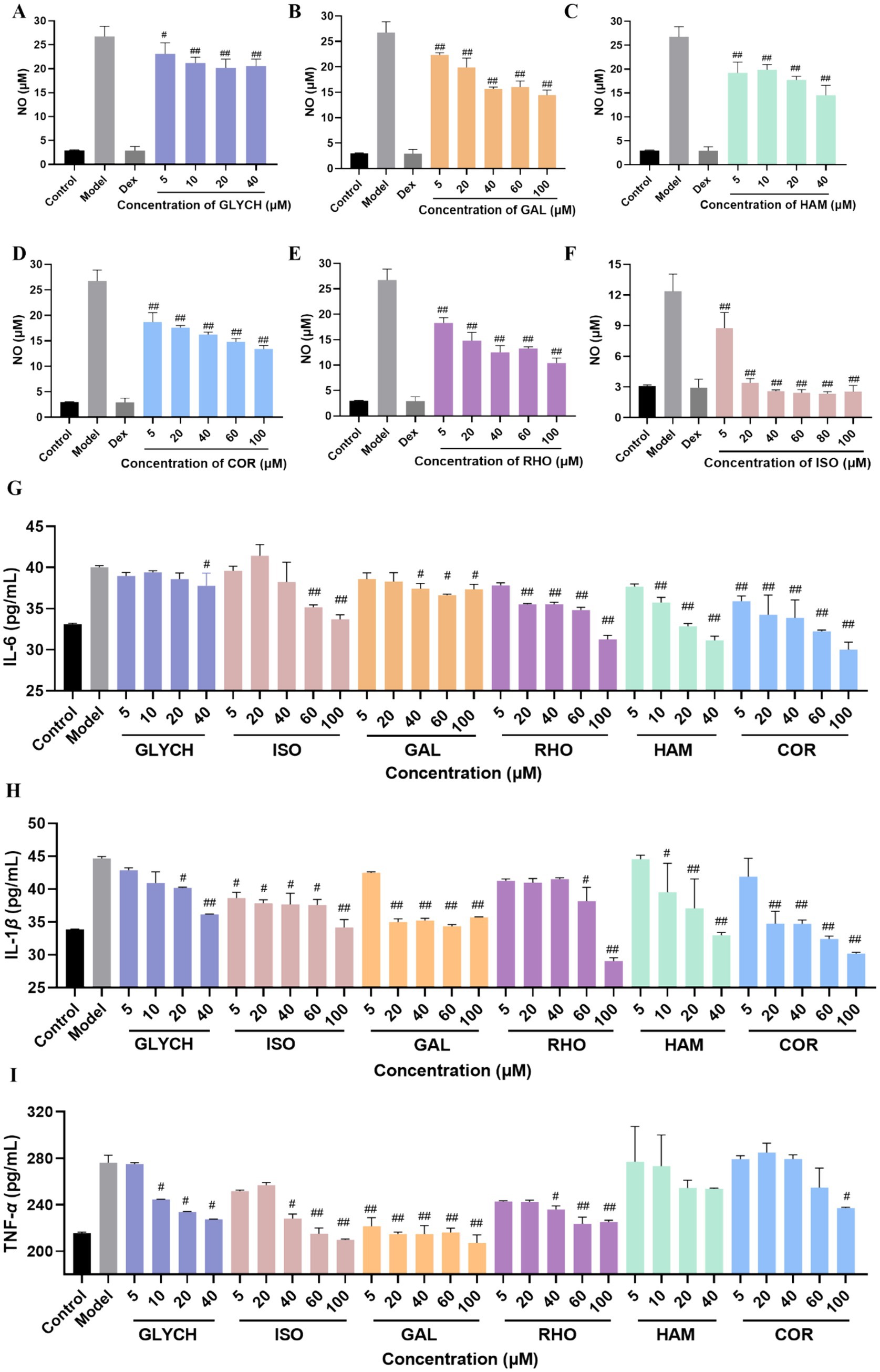
Figure 7. Anti-inflammatory activity evaluation of the tested compounds by the level of NO (A–F), IL-6 (G), IL-1 β (H), and TNF-α (I). (mean ± SD, n = 3). p# < 0.05 and p## < 0.01 vs. model group.
Furthermore, the main inflammatory factors were tested at different concentrations. As displayed in Figures 7G–I, RHO, COR, HAM, GAL, ISO, and GLYCH can significantly reduce the level of IL-6 and IL-1β. Except for HAM, RHO, COR, GAL, ISO, and GLYCH can also decrease the level of TNF-α.
Generally, through this study, by integrating of in silico screening and bioactivity validation, the features of QDP were unveiled to combat COVID-19 via the multi-compounds, multi-targets, and multi-pathways. Also, the focused compounds can interfere with viral invasion, viral replication, and cytokine storm, showing satisfactory performance in multi-stage of COVID-19.
4 Discussion
QDP is officially recommended in the “Home Care Guideline for COVID-19 Patients” issued by the Joint Prevention and Control Mechanism of the State Council of China. Its real-world clinical evidence supports the effectiveness of QDP in pharyngitis and related inflammatory conditions. Clinical studies report significant improvements in sore throat, pharyngeal dryness, and swelling in patients with acute or chronic pharyngitis, with a rapid onset of action and favorable patient adherence (33, 34). Moreover, pharmacokinetic studies in rats show that key compounds such as borneol and menthol are rapidly absorbed after oral administration (Tmax: 20–25 min), which is consistent with the prompt symptom relief observed in clinical settings and may support QDP’s therapeutic effects (8).
The COVID-19 pandemic has exerted unparalleled impacts on global health and economic stability. The onging emergence of SARS-CoV-2 variants continues to pose a significant public health risk, manifesting with severe respiratory symptoms, serious complications like viral neumonia, long-term ramifications known as “Long COVID” (32) and elevated mortality rates especially among vulnerable populations. Notably, recent genomic surveillance data (GISAID, 2024) highlight that variants like JN.1 carry mutations in PLpro such as R168H, which experimentally confer resistance to clinical protease inhibitors by distorting the conformation of S3/S4 subpocket (35). This underscores the urgent need for broad-spectrum therapeutics targeting conserved viral elements.
As crucial targets in the viral lifecycle, Spro, Mpro, and PLpro play key roles in viral entry and replication. Spro facilitates the identification of ACE2 receptors, while PLpro and Mpro are involved in cleaving of polyproteins. Although both Mpro and PLpro are enssential for viral replication within the host, their catalytic Mechanisms are distinct. Mpro operates through a catalytic dyad (Cys145-His41), cleaving polyproteins at glutamine (P1) residues within a conserved substrated-binding groove comprising four subpockets (S1’, S1, S2, S3/S4). Structural studies confirm that the S2 subpocket displays pronounced hydrophobicity, accommodating bulky P2 residues such as leucine (36). In contrast, PLpro relies on a catalytic triad (Cys111-His272-Asp286) and a zinc-binding domain essential for structural integrity. PLpro exhibits dual deubiquitinating and deISGylating activities through extended substrate-recognition grooves, with allosteric modulation via C270 critically regulating catalytic efficiency (37).
These mechanistic distinctions necessitate tailored therapeutic strategies, particularly given PLpro’s higher mutational flexibility compared to the evolutionarily constrained Mpro active site (38, 39). Critically, PLpro cleaves both ubiquitin (Ub) and interferon-stimulated gene 15 (ISG15) from host proteins, disrupting innate immune signaling pathways. This immune evasion occurs via the direct cleavage of STING, which abolishes TBK1 phosphorylation and IRF3 nuclear translocation—a mechanism absent in Mpro, which exclusively processes viral polyproteins (40). This functional distinction categorizes PLpro as a dual-function viral protease and immune modulator.
Cryo-EM structures demonstrate that PLpro’s BL2 loop undergoes hinge-like movements to accommodate bulky Ub/ISG15 substrates, while its zinc finger domain maintains structural integrity via Zn2+ chelation (40). Distinct from Mpro’s compact active site, PLpro features extended substrate-binding grooves (including BL2 loop and Ub-binding region) and dual enzymatic activities create pharmacological barriers insurmountable by Mpro-targeted compounds. Moreover, divergent electrostatic potentials govern substrate recognition: Mpro’s S1 pocket is characterized by electronegativity (mediated by His163/Glu166), whereas PLpro’s Ub-binding region features cationic residues (Lys157/Arg166) (41). Therefore, developing inhibitors for PLpro is more challenging.
In the quest for effective COVID-19 treatments, TCM has demonstrated considerable efficacy in managing symptoms and enhancing recovery, leveraging its diverse array of bioactive compounds. These compounds are noted for their direct antiviral, anti-inflammatory, and immunomodulatory properties. The structural diversity of phytochemicals present in Chinese herbs highlights their promise for the development of innovative antiviral therapies, which are capable of both neutralizing the virus and modulating excessive inflammatory reactions (42). Importantly, the TCM formulation known as QDP, traditionally employed in treating pharyngitis, has been endorsed by the Joint Prevention and Control Mechanism of the State Council as an effective remedy for at-home treatment of COVID-19, as outlined in the guidelines published in December 2022. This recommendation aligns with the WHO’s 2023 guidelines on integrating traditional medicines into pandemic preparedness, though it calls for standardized quality control of herbal preparations.
Currently available anti-coronavirus TCMs, such as Lianhua Qingwen capsules (43) and Huashi Baidu formula (44), rely heavily on the antiviral and anti-inflammatory effects of compounds. The volatile compounds borneol and isoborneol in QDP provide immediate mucosal permeability. The non-volatile compounds, berberine, alkaloids and so on, sustained the anti-inflammatory and antiviral activities (45, 46). The integration of volatile and non-volatile compounds forms a unique fast-acting and long-lasting synergy, highlighting the chemical uniqueness of QDP. This chemical composition may potentially translate into multiple pharmacological advantages. The unique troche formulation and volatile compounds enable direct action at the throat and mouth, possibly offering faster local immune regulation and physical barrier reinforcement. Studies have shown that both chebulagic acid and punicalagin, at noncytotoxic concentrations, can reduce virus-induced plaque formation in Vero E6 monolayers. These compounds appear to function as allosteric regulators, targeting SARS-CoV-2 Mpro (43). Corilagin binds directly to RdRp, robustly inhibiting its polymerase activity, as evidenced by both cell-free and cell-based assays (47). Tannic acid may contribute to the treatment of inflammation by decreasing MPO enzyme activity (48).
Our investigation employed a dual strategy, integrating in silico screening and bioactivity assays, to discover and validate the therapeutic potential of QDP against COVID-19. Notably, compared with the single-step screening methods, this “dry-wet combined” strategy not only saves the use of standards and solvents, but also significantly accelerates the identification speed of bioactive candidates. In the face of emerging and sudden diseases, it can quickly screen out potential effective components. This research represents the first isolation and characterization of chemical compounds in QDP. Among the 48 identified compounds, network pharmacology analysis revealed likely active compounds and elucidated their mechanisms of action against COVID-19. We targeted key proteins, including Spro, Mpro, and PLpro, performing exhaustive virtual screenings of QDP to assess its inhibitory capabilities against SARS-CoV-2. Subsequently, we verified the inhibitory interactions of three principal phytocompounds—COR, RHO, and HAM—through molecular docking studies. Moreover, these compounds demonstrated consistent inhibitory effects on Mpro and exhibited anti-inflammatory properties.
Among the compounds examined, COR, a polyphenolic gallotannin recognized for its anti-inflammatory and antioxidant properties, emerged as a notably potent agent (49). It displayed a strong binding affinity to Spro, Mpro, and PLpro, evidenced by a low KD. Functional assays further confirmed its efficacy, with COR significantly inhibiting Mpro in FRET assay (IC50 = 0.73 ± 0.05 μM) and demonstrating antiviral efficacy in Vero E6 cells infected with the SARS-CoV-2 Omicron BA.5 variant (EC50 = 12.83 μM). Additionally, COR reduced inflammatory cytokine production, indicating a dual role in both viral inhibition and immune modulation. Given its broad-spectrum activity against other pathogens, including influenza (50) and herpes simplex viruses, COR represents a promising candidate for further antiviral therapeutic exploration.
RHO, another potential agent, is a flavonoid glycoside known for its anti-inflammatory, antioxidant, and anticancer properties. However, it exhibited a limited activity in our assays (51). While RHO was effective against Mpro, it failed to inhibit the SARS-CoV-2 Omicron BA.5 variant. This specificity highlights the necessity for deeper molecular investigations to fully ascertain RHO’s mechanistic roles and its potential therapeutic utility against SARS-CoV-2.
From a therapeutic standpoint, these findings are encouraging, offering a robust scientific foundation for the clinical application of QDP in treating COVID-19. A thorough examination of the interactions and the required inhibitory concentrations can inform dosage and formulation strategies in clinical contexts. Nonetheless, translating these results into clinical practice necessitates comprehensive clinical trials to ascertain efficacy and safety thoroughly.
In addition to the virus invasion inhibition investigated in this work, we plan to further explore the effects of QDP on other key stages of the viral life cycle, including replication, assembly, and release. Moreover, more complex models such as organoids will be employed to systematically evaluate the antiviral immune regulatory effects of the drug in a simulated human respiratory microenvironment.
Overall, this study not only underscores the therapeutic potential of TCM addressing COVID-19 but also contributes to the global quest for antiviral drug discovery by identifying novel bioactive compounds. Future research should focus on validating these findings in clinical settings and elucidating the mechanistic pathways of these compounds to maximize their therapeutic potential.
5 Conclusion
As endorsed by the National Health Commission of the People’s Republic of China for COVID-19 treatment, QDP was comprehensively investigated through multiple scientific approaches. The study involved the clarification of chemical compounds, network pharmacology analysis, molecular docking simulations, binding affinity assessments by SPR, enzymatic activity measurements via FRET, and evaluations of antiviral and anti-inflammatory activities. This integrated virtual and experimental strategy enabled the rapid screening of components, identifying COR as exceptionally potent in antiviral capacity, and RHO, COR, HAM, GAL, ISO, and GLYCH as significant anti-inflammatory agents. In general, this study provides a strategy and procedures for rapid and effective identification of active compounds and elucidation of their mechanism, conducing to the scientific application of Chinese patent medicine in treating COVID-19 patients. Furthermore, this study underscores the critical role of integrating traditional medicine with contemporary scientific techniques to explore potential therapies for emerging infectious diseases.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
LL: Data curation, Investigation, Validation, Writing – original draft. XL: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. XW: Investigation, Validation, Writing – review & editing. PZ: Writing – review & editing. QY: Validation, Writing – review & editing. TG: Investigation, Writing – review & editing. YW: Writing – review & editing. JZ: Writing – review & editing. CW: Writing – review & editing. JY: Writing – review & editing. MZ: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Science and Technology Project of Haihe Laboratory of Modern Chinese Medicine (Enhancement of Quality Standards for Proprietary Chinese Medicines in the Tianjin Pharmaceutical Sector and 22HHZYJC00007), the National Natural Science Foundation of China (Grant No. 82104372).
Acknowledgments
We are grateful to the staff at Research Centre of Modern Analytical Technology, Tianjin University of Science and Technology to supply SPR assay technical support.
Conflict of interest
TG was employed by Tianjin Pharmaceutical Da Ren Tang Group Corp., Ltd. Traditional Chinese Medicine Research Institute.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1684713/full#supplementary-material
Footnotes
Edited by:
Yu Wang, Shanghai Shidong Hospital of Yangpu District, ChinaReferences
1. Deshmukh, R, Harwansh, RK, Garg, A, Mishra, S, Agrawal, R, and Jangde, R. COVID-19: recent insight in genomic feature, pathogenesis, immunological biomarkers, treatment options and clinical updates on SARS-CoV-2. Curr Genomics. (2024) 25:69–87. doi: 10.2174/0113892029291098240129113500
2. Tian, M, Liu, H, Peng, S, Yang, Z, Tao, W, Che, H, et al. Report on the 4th board meeting of the international human phenome consortium. Phenomics. (2024) 4:254–6. doi: 10.1007/s43657-023-00139-5
3. Al-Kuraishy, HM, Al-Gareeb, AI, Mostafa-Hedeab, G, Dubey, R, Prabhakar, PK, and Batiha, GE. COVID-19 and diabetes: will novel drugs for diabetes help in COVID-19? Curr Mol Pharmacol. (2023) 16:494–506. doi: 10.2174/1874467215666220908091604
4. Su, YY, Wang, WP, Wang, Y, Wang, C, Sun, S, Zhu, XH, et al. Application and development of targeted fishing technology in natural product screening - a simple minireview. Curr Pharm Anal. (2024) 20:231–40. doi: 10.2174/0115734129301241240429114323
5. Huang, M, Liu, YY, Xiong, K, Yang, FW, Jin, XY, Wang, ZQ, et al. The role and advantage of traditional Chinese medicine in the prevention and treatment of COVID-19. J Integr Med. (2023) 21:407–12. doi: 10.1016/j.joim.2023.08.003
6. Lyu, M, Xiao, G, Wang, S, Wang, R, Tan, L, Ma, S, et al. “Three medicines and three formulas” in COVID-19: from bench to bedside. Acupunct Herbal Med. (2023) 3:309–22. doi: 10.1097/HM9.0000000000000082
7. National Health Commission of the People’s Republic of China. Guidelines for home treatment of people infected with the novel coronavirus [Internet]. (2022). Available online at: http://www.nhc.gov.cn/xcs/zhengcwj/202212/2b6c16cc176b4806b399ea5588353b3c.shtml (Accessed: 29 September 2023).
8. Xu, X, Li, Y, Hou, J, Zhang, S, Xu, Y, Wang, Y, et al. Pharmacokinetic study of borneol and menthol in rats after oral administration of qingyan drop pills. Planta Med. (2011) 77:1600–4. doi: 10.1055/s-0030-1270998
9. Wang, C, Cao, Y, Yang, Q, Zhang, H, Wang, X, Liu, G, et al. High-throughput screening of dual-target inhibitors for SARS-CoV-2 main protease and papain-like protease from Chebulae fructus: in silico prediction and experimental verification. Front Microbiol. (2024) 15:1510665. doi: 10.3389/fmicb.2024.1510665
10. Wang, CJ, Yang, ZY, Chai, X, Wang, YF, Wang, WL, Zhang, M, et al. Tea as a natural gift for discovering antiviral candidates. Acupunct Herb Med. (2022) 4:211–20. doi: 10.1097/HM9.0000000000000048
11. Chen, W, Yuan, P, Yang, M, Yan, Z, Kong, S, Yan, J, et al. SARS-CoV-2 entry factors: ACE2 and TMPRSS2 are expressed in peri-implantation embryos and the maternal-fetal interface. Engineering. (2020) 6:1162–9. doi: 10.1016/j.eng.2020.07.013
12. Chen, LYC, and Quach, TTT. COVID-19 cytokine storm syndrome: a threshold concept. Lancet Microbe. (2021) 2:e49–50. doi: 10.1016/S2666-5247(20)30223-8
13. Jin, Z, Du, X, Xu, Y, Deng, Y, Liu, M, Zhao, Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. (2020) 582:289–93. doi: 10.1038/s41586-020-2223-y
14. Shin, D, Mukherjee, R, Grewe, D, Bojkova, D, Baek, K, Bhattacharya, A, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. (2020) 587:657–62. doi: 10.1038/s41586-020-2601-5
15. Ru, J, Li, P, Wang, J, Zhou, W, Li, B, Guo, Z, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. (2014) 6:13. doi: 10.1186/1758-2946-6-13
16. Zhu, Y, Yu, J, Zhang, K, Feng, Y, Guo, K, Sun, L, et al. Network pharmacology analysis to explore the pharmacological mechanism of effective Chinese medicines in treating metastatic colorectal cancer using meta-analysis approach. Am J Chin Med. (2021) 49:1839–70. doi: 10.1142/S0192415X21500877
17. Tao, Q, Du, J, Li, X, Zeng, J, Tan, B, Xu, J, et al. Network pharmacology and molecular docking analysis on molecular targets and mechanisms of Huashi Baidu formula in the treatment of COVID-19. Drug Dev Ind Pharm. (2020) 46:1345–53. doi: 10.1080/03639045.2020.1788070
18. Chen, L, Zhang, YH, Wang, S, Zhang, Y, Huang, T, Cai, YD, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One. (2017) 12:e0184129. doi: 10.1371/journal.pone.0184129
19. Meng, XY, Zhang, HX, Mezei, M, and Cui, M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. (2011) 7:146–57. doi: 10.2174/157340911795677602
20. Burley, SK, Berman, HM, Kleywegt, GJ, Markley, JL, Nakamura, H, Velankar, S, et al. Protein data Bank (PDB): the single global macromolecular structure archive. Methods Mol Biol. (2017) 1607:627–41. doi: 10.1007/978-1-4939-7000-1_26
21. Reina, J, and Iglesias, C. Nirmatrelvir más ritonavir (Paxlovid) una potente combinación inhibidora de la proteasa 3CLpro del SARS-CoV-2 [Nirmatrelvir plus ritonavir (Paxlovid) a potent SARS-CoV-2 3CLpro protease inhibitor combination]. Rev Esp Quimioter. (2022) 35:236–40. doi: 10.37201/req/002.2022
22. Rehman, SU, and Yoo, HH. COVID-19 challenges and its therapeutics. Biomed Pharmacother. (2021) 142:112015. doi: 10.1016/j.biopha.2021.112015
23. Wang, C, Zhang, H, Wang, X, Yang, Q, Liu, G, Cao, Y, et al. Comprehensive review on fruit of Terminalia chebula: traditional uses, phytochemistry, pharmacology, toxicity, and pharmacokinetics. Molecules. (2024) 29:5547. doi: 10.3390/molecules29235547
24. Zhang, M, Liu, L, Zhao, Y, Cao, Y, Zhu, Y, Han, L, et al. Discovery and evaluation of active compounds from Xuanfei Baidu formula against COVID-19 via SARS-CoV-2 Mpro. Chin Med. (2023) 18:94. doi: 10.1186/s13020-023-00790-0
25. Xu, J, Wang, X, Yu, H, Zhang, LH, Qiu, F, Wang, Y, et al. Study on quality characteristic of Chebulae fructus and its adulterants and degradation pathway of hydrolyzable tannins. Molecules. (2024) 29:2399. doi: 10.3390/molecules29102399
26. Zhang, D, Hamdoun, S, Chen, R, Yang, L, Ip, CK, Qu, Y, et al. Identification of natural compounds as SARS-CoV-2 entry inhibitors by molecular docking-based virtual screening with bio-layer interferometry. Pharmacol Res. (2021) 172:105820. doi: 10.1016/j.phrs.2021.105820
27. Zhang, X, Hu, Y, Yang, X, Tang, Y, Han, S, Kang, A, et al. Förster resonance energy transfer (FRET)-based biosensors for biological applications. Biosens Bioelectron. (2019) 138:111314. doi: 10.1016/j.bios.2019.05.019
28. Unoh, Y, Uehara, S, Nakahara, K, Nobori, H, Yamatsu, Y, Yamamoto, S, et al. Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J Med Chem. (2022) 65:6499–512. doi: 10.1021/acs.jmedchem.2c00117
29. Yang, LJ, Chen, RH, Hamdoun, S, Coghi, P, Ng, JPL, Zhang, DW, et al. Corilagon prevents SARS-CoV-2 infection by targeting RBD-ACE2 binding. Phytomedicine. (2021) 87:153591. doi: 10.1016/j.phymed.2021.153591
30. Xu, J, Zhang, G, Tong, Y, Yuan, J, Li, Y, Song, G, et al. Corilagin induces apoptosis, autophagy and ROS generation in gastric cancer cells in vitro. Int J Mol Med. (2019) 43:967–79. doi: 10.3892/ijmm.2018.4031
31. Binette, V, Côté, S, Haddad, M, Nguyen, PT, Bélanger, S, Bourgault, S, et al. Corilagin and 1,3,6-tri-O-galloyl-β-D-glucose: potential inhibitors of SARS-CoV-2 variants. Phys Chem Chem Phys. (2021) 23:14873–88. doi: 10.1039/D1CP01790J
32. GISAID. Genomic epidemiology of SARS-CoV-2 variants [Internet]. (2024). Available online at: https://www.gisaid.org/ (Accessed: 15 May 2024).
33. Zhan, KS, Wei, W, and Yu, NS. Clinical research on the treatment of chronic pharyngitis with qingyandiwan. Chin J Prim Med Pharm. (2003) 10:41–2. Available at: https://www.cnki.net/
34. Shu, L, Qiu, H, He, Y, Zhang, S, Qian, J, Liu, S, et al. Chemical composition analysis of Qingyan dropping pills using ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. (2024) 38:e9833. doi: 10.1002/rcm.9833
35. Shao, Q, Xiong, M, Li, J, Hu, H, Su, H, and Xu, Y. Unraveling the catalytic mechanism of SARS-CoV-2 papain-like protease with allosteric modulation of C270 mutation using multiscale computational approaches. Chem Sci. (2023) 14:4681–96. doi: 10.1039/d3sc00166k
36. Breidenbach, J, Voget, R, Si, Y, Hingst, A, Claff, T, Sylvester, K, et al. Macrocyclic Azapeptide nitriles: structure-based discovery of potent SARS-CoV-2 Main protease inhibitors as antiviral drugs. J Med Chem. (2024) 67:8757–90. doi: 10.1021/acs.jmedchem.4c00053
37. Wang, X, Xiong, L, Zhu, Y, Liu, S, Zhao, W, Wu, X, et al. Covalent DNA-encoded library workflow drives discovery of SARS-CoV-2 nonstructural protein inhibitors. J Am Chem Soc. (2024) 146:33983–96. doi: 10.1021/jacs.4c12992
38. Lin, C, Zhu, Z, Jiang, H, Zou, X, Zeng, X, Wang, J, et al. Structural basis for coronaviral main proteases inhibition by the 3CLpro inhibitor GC376. J Mol Biol. (2024) 436:168474. doi: 10.1016/j.jmb.2024.168474
39. Rhodin, MHJ, Reyes, AC, Balakrishnan, A, Bisht, N, Kelly, NM, Gibbons, JS, et al. The small molecule inhibitor of SARS-CoV-2 3CLpro EDP-235 prevents viral replication and transmission in vivo. Nat Commun. (2024) 15:6503. doi: 10.1038/s41467-024-50931-8
40. Tan, B, Zhang, X, Ansari, A, Jadhav, P, Tan, H, Li, K, et al. Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model. Science. (2024) 383:1434–40. doi: 10.1126/science.adm9724
41. Yu, W, Zhao, Y, Ye, H, Wu, N, Liao, Y, Chen, N, et al. Structure-based Design of a Dual-Targeted Covalent Inhibitor against Papain-like and Main Proteases of SARS-CoV-2. J Med Chem. (2022) 65:16252–67. doi: 10.1021/acs.jmedchem.2c00954
42. Wang, X, Xu, J, Zhang, LH, Qiu, F, Wang, Y, Liu, G, et al. Global profiling of the antioxidant constituents in Chebulae fructus based on an integrative strategy of UHPLC/IM-QTOF-MS, MS/MS molecular networking, and spectrum-effect correlation. Antioxidants. (2023) 12:2093. doi: 10.3390/antiox12122093
43. Chen, Y, Zhang, C, Wang, N, and Feng, Y. Deciphering suppressive effects of Lianhua Qingwen capsule on COVID-19 and synergistic effects of its major botanical drug pairs. Chin J Nat Med. (2023) 21:383–400. doi: 10.1016/S1875-5364(23)60455-8
44. Xu, H, Li, S, Liu, J, Cheng, J, Kang, L, Li, W, et al. Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19. Proc Natl Acad Sci USA. (2023) 120:e2301775120. doi: 10.1073/pnas.2301775120
45. Xu, XF, Wang, Y, Chen, ZJ, Haji, J, Zhang, YJ, and He, X. Study on the rule of buccal absorption and the release discipline of borneol and menthol in qingyan drop pills. Chin J Clin Pharm Ther. (2009) 14:974–8. Available at: https://www.cnki.net/
46. Du, R, Cooper, L, Chen, Z, Lee, H, Rong, L, and Cui, Q. Discovery of chebulagic acid and punicalagin as novel allosteric inhibitors of SARS-CoV-2 3CLpro. Antivir Res. (2021) 190:105075. doi: 10.1016/j.antiviral.2021.105075
47. Li, Q, Yi, D, Lei, X, Zhao, J, Zhang, Y, Cui, X, et al. Corilagin inhibits SARS-CoV-2 replication by targeting viral RNA-dependent RNA polymerase. Acta Pharm Sin B. (2021) 11:1555–67. doi: 10.1016/j.apsb.2021.02.011
48. Soyocak, A, Kurt, H, Cosan, DT, Saydam, F, Calis, IU, Kolac, UK, et al. Tannic acid exhibits anti-inflammatory effects on formalin-induced paw edema model of inflammation in rats. Hum Exp Toxicol. (2019) 38:1296–301. doi: 10.1177/0960327119864154
49. Lv, HL, Hong, L, Tian, Y, Yin, C, Zhu, C, Feng, H, et al. Corilagin alleviates acetaminophen-induced hepatotoxicity via enhancing the AMPK/GSK3β-Nrf2 signaling pathway. Cell Commun Signal. (2019) 17:2. doi: 10.1186/s12964-018-0314-2
50. Zu, M, Li, C, Fang, JS, Lian, WW, Liu, AL, Zheng, LS, et al. Drug discovery of host CLK1 inhibitors for influenza treatment. Molecules. (2015) 20:19735–47. doi: 10.3390/molecules201119653
Keywords: COVID-19, SARS-CoV-2, Qingyan Dropping Pills (QDP), antivirus, anti-inflammation
Citation: Liu L, Li X, Wang X, Zhang P, Yang Q, Geng T, Wang Y, Zhang J, Wang C, Yang J and Zhang M (2025) Elucidation of anti-SARS-CoV-2 and anti-inflammatory bioactives in Qingyan Dropping Pills via integrated in silico screening and bioactivity validation. Front. Med. 12:1684713. doi: 10.3389/fmed.2025.1684713
Copyright © 2025 Liu, Li, Wang, Zhang, Yang, Geng, Wang, Zhang, Wang, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changjian Wang, d2FuZ2NoYW5namlhbjIzQDE2My5jb20=; Jing Yang, eWFuZ2ppbmdvZmZpY2VAMTYzLmNvbQ==; Min Zhang, emhhbmdtMDM2QHRqdXRjbS5lZHUuY24=
†These authors have contributed equally to this work
 Liting Liu1†
Liting Liu1† Yuefei Wang
Yuefei Wang Junhua Zhang
Junhua Zhang Min Zhang
Min Zhang