- Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX, USA
The post-translational modification AMPylation is emerging as a significant regulatory mechanism in both prokaryotic and eukaryotic biology. This process involves the covalent addition of an adenosine monophosphate to a protein resulting in a modified protein with altered activity. Proteins capable of catalyzing AMPylation, termed AMPylators, are comparable to kinases in that they both hydrolyze ATP and reversibly transfer a part of this primary metabolite to a hydroxyl side chain of the protein substrate. To date, only four AMPylators have been characterized, though many more potential candidates have been identified through amino acid sequence analysis and preliminary in vitro studies. This modification was first discovered over 40 years ago by Earl Stadtman and colleagues through the modification of glutamine synthetase by adenylyl transferase; however research into this mechanism has only just been reenergized by the studies on bacterial effectors. New AMPylators were revealed due to the discovery that a bacterial effector having a conserved Fic domain transfers an AMP group to protein substrates. Current research focuses on identifying and characterizing various types of AMPylators homologous to Fic domains and adenylyl transferase domains and their respective substrates. While all AMPylators characterized thus far are bacterial proteins, the conservation of the Fic domain in eukaryotic organisms suggests that AMPylation is omnipresent in various forms of life and has significant impact on a wide range of regulatory processes.
Introduction
The post-translational modification AMPylation, previously referred to as adenylylation, was first discovered by Earl Stadtman et al. in the 1960s when he observed that a tyrosine residue of Escherichia coli glutamine synthetase was modified with AMP (Brown et al., 1971). This modification is defined as the stable and reversible covalent addition of an adenosine mono phosphate group to a hydroxyl side chain of a protein (Figures 1A,B). While AMPylation has only been observed to modify threonine and tyrosine residues, it is likely that serine can function as a target as well. AMPylation is distinct from transient adenylylation events that involve the addition of AMP to the protein targets and use the energy of ATP to drive an enzymatic reaction (e.g., processes like ubiquitin activation (Worby et al., 2009; Yarbrough and Orth, 2009; Luong et al., 2010) or prokaryotic thiamine and molybdenum biosynthesis (Lake et al., 2001; Duda et al., 2005).
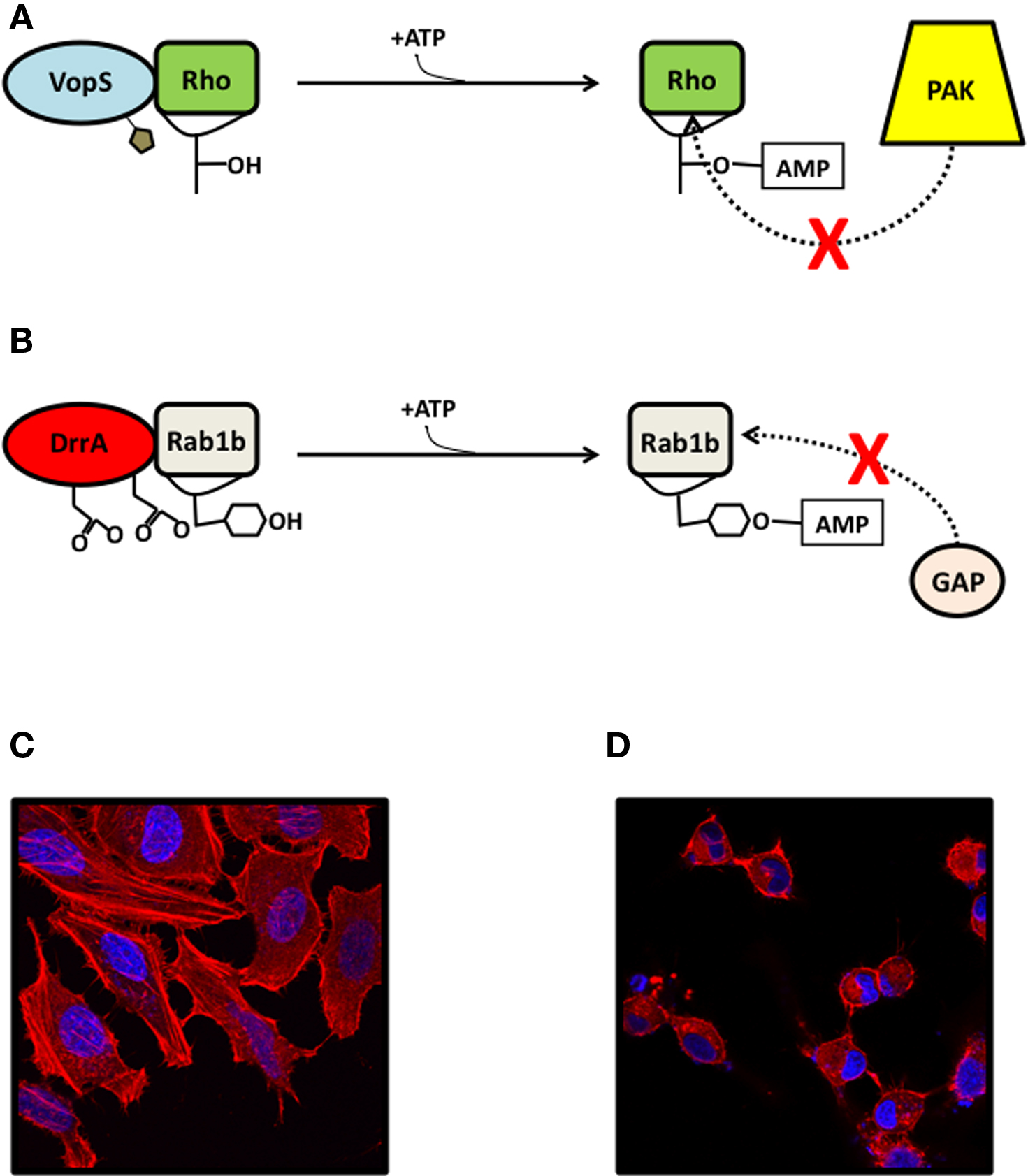
Figure 1. AMPylation in pathogenicity. (A) The Fic effector AMPylators VopS (shown) and IbpA are secreted into eukaryotic cells and modify a threonine or tyrosine residue on the Switch 1 loop of Rho family GTPases, sterically blocking their association with downstream substrates like PAK and rhotekin. (B) The adenylyl transferase effector AMPylator DrrA catalyzes the exchange of GDP for GTP in the GTPase Rab1b through its GEF domain (not shown) and AMPylates a tyrosine in the Switch 2 loop, preventing its association with GAPs. (C, D) HeLa cells transfected with mock vector (C) or with VopS (D), visualized with confocal microscopy for rhodamine phalloidin stain (actin, red) and Hoechst stain (DNA, blue).
In pathogenicity, AMPylators act as bacterial effectors that are translocated into eukaryotic host cells by Type III or IV Secretion Systems (T3SS or T4SS) (Broberg and Orth, 2010) or a two-partner secretion system (Jacob-Dubuisson et al., 2001). They typically disable the cell by AMPylating components of essential signaling pathways, such as the regulation of the actin cytoskeleton by Rho GTPases (Yarbrough et al., 2009), and alter their function. In metabolism, the regulation of glutamine synthetase through the addition and removal of AMP by glutamine synthetase adenylyl transferase (GS-ATase) is a well characterized part of the complex regulation of nitrogen levels in the bacterial cell, and represents an important metabolic function for AMPylation (Brown et al., 1971; Jiang et al., 1998). The Fic and adenylyl transferase domains comprise the current known AMPylators and each have distinct primary sequence and structural features. AMPylation by these domains has been demonstrated to have roles in both the pathogenicity of bacterial species and in endogenous metabolic regulation. A significant amount of structural and kinetic data about the mechanism of AMPylation by both domains has been elucidated. Herein are described the mechanisms for known AMPylators, including their substrates, structural features, and enzyme kinetics.
The Fic and Adenylyl Transferase Domains: Two of a Kind
The Fic domain is a member of the Fido (Fic/doc) protein superfamily and is widely conserved across most non-plant species. Similarities in catalytic motif and structural organization led to the merging of the Fic and doc families (Kinch et al., 2009). The Fic and doc domains share the conserved HPFx[D/E]GN[G/K]R motif, in which the invariant histidine residue has proven to be essential for AMPylation activity in Fic proteins (Luong et al., 2010) and cytotoxicity in doc (Garcia-Pino et al., 2008). Fic and doc also share similar structural features, but doc is not known to catalyze AMPylation. Doc has been shown to catalyze its toxic phenotype in Escherichia coli by associating with the 30S ribosomal subunit and inhibiting translational elongation (Lehnherr et al., 1993; Liu et al., 2008). In terms of distribution, the comprehensive protein family database Pfam currently identifies the Fic domain in over 2,000 bacterial proteins that are cataloged among 984 bacterial species, including human pathogens and commensals in addition to environmental bacteria. Proteobacteria and firmicutes constitute approximately one-half and one-quarter of this number, respectively, and maintain a roughly 2:1 overall ratio of Fic proteins per species. In contrast, only 59 Fic proteins from 43 eukaryotic species have been identified, and metazoans Drosophila menanogaster, Caenorhabditis elegans, mice, and humans contain only one copy each (Finn et al., 2010). Additionally, the Fic domain has been cataloged in 49 archaeal proteins and in seven viral proteins. It is very rare for one protein to contain more than one Fic domain, though the Histophilus somni protein IbpA is one of the few exceptions with two.
Characterization of the Fic domain has thus far progressed mostly with bacterial effector proteins, though in vitro auto-AMPylation activity has been observed for both the human protein HYPE (Worby et al., 2009) and the Drosophila protein CG9523 (Kinch et al., 2009). Speculation on the function of the Fic domain in eukaryotic proteins has centered around their domain organization, which is well covered in Kinch et al. (2009). Briefly, bioinformatic analysis of Fic proteins has revealed its association with DNA binding domains, transmembrane regions, a variety of protein–protein interaction and enzymatic domains, and perhaps most interestingly, a low-density lipoprotein receptor domain class A (LDL) (Kinch et al., 2009). This association with the LDL receptor domain could imply a role in cholesterol metabolism in eukaryotes. Despite these insights, the only two Fic proteins that have been characterized are the secreted bacterial effectors VopS and IbpA (Worby et al., 2009; Yarbrough et al., 2009).
The adenylyl transferase domain (GlnE family) is part of the larger nucleotidyl transferase protein family and in addition to the Fic domain has been identified as capable of catalyzing AMPylation. It is characterized by a conserved G-X11-D-X-D motif, of which the aspartate residues have been shown to be essential for the AMPylation activity (Figure 1B) (Jiang et al., 2007a; Muller et al., 2010). This domain has been identified in more than 1,400 bacterial proteins among 685 bacterial species, of which the large majority are proteobacteria (Finn et al., 2010). Two members of this family have been identified as AMPylators: initially with GS-ATase and more recently with the characterization of the Legionella pneumophila effector DrrA/SidM (Muller et al., 2010).
AMPylators in Pathogenicity
Secretion systems are often used by Gram negative bacterial species to translocate virulence factors, also called effectors, into eukaryotic cells. These proteins typically subvert cellular processes during infection by locking the target into an active or inactive state, often by mimicking a normal cellular signaling mechanism. While Gram negative bacteria have been shown to use a broad arsenal of effectors with different mechanisms, effectors from three different species have been shown to use AMPylation (Table 1). The proteins VopS, IbpA, and DrrA are secreted into eukaryotic cells from Vibrio parahaemolyticus (V. para), H. somni, and L. pneumophila, respectively, and have been demonstrated to AMPylate host GTPases, resulting in changes in the actin cytoskeleton (Worby et al., 2009; Yarbrough et al., 2009; Muller et al., 2010).
GTPases: A Common Target of AMPylators
The families of Rho, Rab, and Arf GTPases have fundamental roles in actin cytoskeleton dynamics and vesicular trafficking and control cellular processes such as phagocytosis in the host cell. During infection, the pathogen must control the inhibition or induction of host cell phagocytosis to prevent or promote its internalization. Thus, small G proteins involved in these processes are often the target of numerous bacterial virulence factors such as bacterial surface proteins, effectors, and toxins (Boquet and Lemichez, 2003). The GTPase activating protein (GAP) and GTPase exchange factor (GEF) activity of YopE from Yersinia spp. and SopE from Salmonella enterica target Rho GTPases, while the aforementioned DrrA has a modular domain with GEF activity targeted toward Rab GTPases (Hardt et al., 1998; Nagai et al., 2002). A large number of post-translational mechanisms are employed by bacterial effectors to modify and inhibit GTPases, including GAP, GEF, ADP-ribosylation, and proteolytic cleavage (Boquet and Lemichez, 2003).
Protein AMPylation by effectors VopS, IpbA, and DrrA have provided a novel biochemical mechanism used during bacterial pathogenesis that is distinct from those previously described. Protein AMPylation by these effectors occurs in the conserved switch 1 or switch 2 regions of GTPases and sterically hinders downstream binding of Rho GTPase substrates. Similarly, clostridial toxins such as ToxA and ToxB from C. difficile have glucosyltransferase activity that mediates the covalent modification of a threonine residue in the switch 1 region of Rho GTPases with glucose (Richard et al., 1999). This is the same threonine residue AMPylated by VopS (Yarbrough et al., 2009). Modification by glucosylation or AMPylation sterically hinders downstream substrate binding, inhibiting the function of Rho GTPases and leading to breakdown of actin cytoskeleton signaling.
VopS
Vibrio parahaemolyticus is an extracellular Gram negative bacterium that causes gastroenteritis from eating undercooked seafood (Daniels et al., 2000a,b). An essential virulence factor for many Gram negative pathogens, including V. para, is the T3SS, a needle-like structure that extends from the bacterium and penetrates a host cell to inject effectors (Makino et al., 2003). V. para secretes a variety of effectors, each of which individually are responsible for a broad range of phenotypes in the infected cell. VopS was the first example of a Fic domain functioning as an AMPylator. VopS modifies threonine 35 on the switch 1 region of the Rho family GTPases resulting in GTPases that are unable to bind to downstream effectors like p21 activated kinase 1 protein (PAK) (Yarbrough et al., 2009) (Figure 1A). The loss of this interaction disables the host cell’s control of the actin cytoskeleton, which leads to cell rounding (Figure 1C,D).
IbpA
IbpA is secreted from H. somni, an obligate and opportunistic pathogen in cattle (Martin et al., 1998). H. somni is a Gram negative bacterium that commonly infects the respiratory epithelium of cattle and is often responsible for bovine respiratory disease complex, an economically important cause of bovine mortality. H. somni secretes immunoglobulin binding proteins through a two-partner secretion system, which have been shown to be an important virulence factor and are likely to mediate bacterial adhesion to the host epithelium (Corbeil et al., 1997; Jacob-Dubuisson et al., 2001). One of these is IbpA, which contains putative adhesin domains at the N-terminal region that are predicted to mediate its binding and internalization into host eukaryotic cells after secretion from the bacterium (Zekarias et al., 2010). IbpA also has two Fic domains at the C-terminal region that are responsible for its cytotoxicity (Worby et al., 2009). This cytotoxicity is a result of the IbpA Fic domains’ AMPylation of Rho family GTPases, which blocks their binding to downstream substrates like rhotekin and PAK. Cells transfected with the Fic domains of IbpA have a rounded phenotype similar to those transfected with VopS (Figure 1C,D). However, IbpA’s AMPylation of the switch 1 region in GTPases occurs on a tyrosine, rather than a threonine.
DrrA
The most recently characterized AMPylator, DrrA, is yet another bacterial effector, but contains an adenylyl transferase domain rather than a Fic domain Muller et al. (2010). This virulence factor is secreted from L. pneumophila, an intracellular pathogen that survives in Legionella containing vacuoles (LCV) in the cell. Legionella uses a T4SS to secrete proteins from the LCV into the host cell. DrrA is composed of three domains: an N-terminal adenylyl transferase domain, a central guanine nucleotide exchange factor (GEF) domain, and a C-terminal phosphatidylinositol-4 phosphate- binding (P4M) domain (Murata et al., 2006; Brombacher et al., 2009). The first domain of DrrA very closely resembles the C-terminal adenylyl transferase domain of GS-ATase (Figure 2) and contains the conserved G-X11-D-X-D motif (Muller et al., 2010). The GEF domain is capable of catalyzing the exchange of GDP for GTP in the GTPase Rab1b, which plays a role in the regulation of vesicular transport from the endoplasmic reticulum (Murata et al., 2006). The P4M domain anchors the effector to the cytoplasmic side of the LCV membrane (Brombacher et al., 2009). DrrA has been shown to hijack the function of Rab1b by locking it into its GTP-bound active state through the function of the DrrA GEF and adenylyl transferase domains. The GEF domain of DrrA binds Rab1b and activates it by exchanging GDP for GTP. The adenylyl transferase domain then AMPylates the Y77 residue of the Switch 2 region of Rab1b, which blocks its interaction with GAPs and prevents the hydrolysis of GTP. Due to DrrA’s localization to the LCV membrane through its P4M domain, it is thought that the permanently activated, AMPylated and prenylated GTP-Rab1b will be localized with DrrA at the LCV and dominantly target ER vacuoles to the LCV (Muller et al., 2010). Transfection of DrrA into eukaryotic cells causes a rounding phenotype similar to VopS and IbpA, albeit by a distinct mechanism (Figure 1C,D).
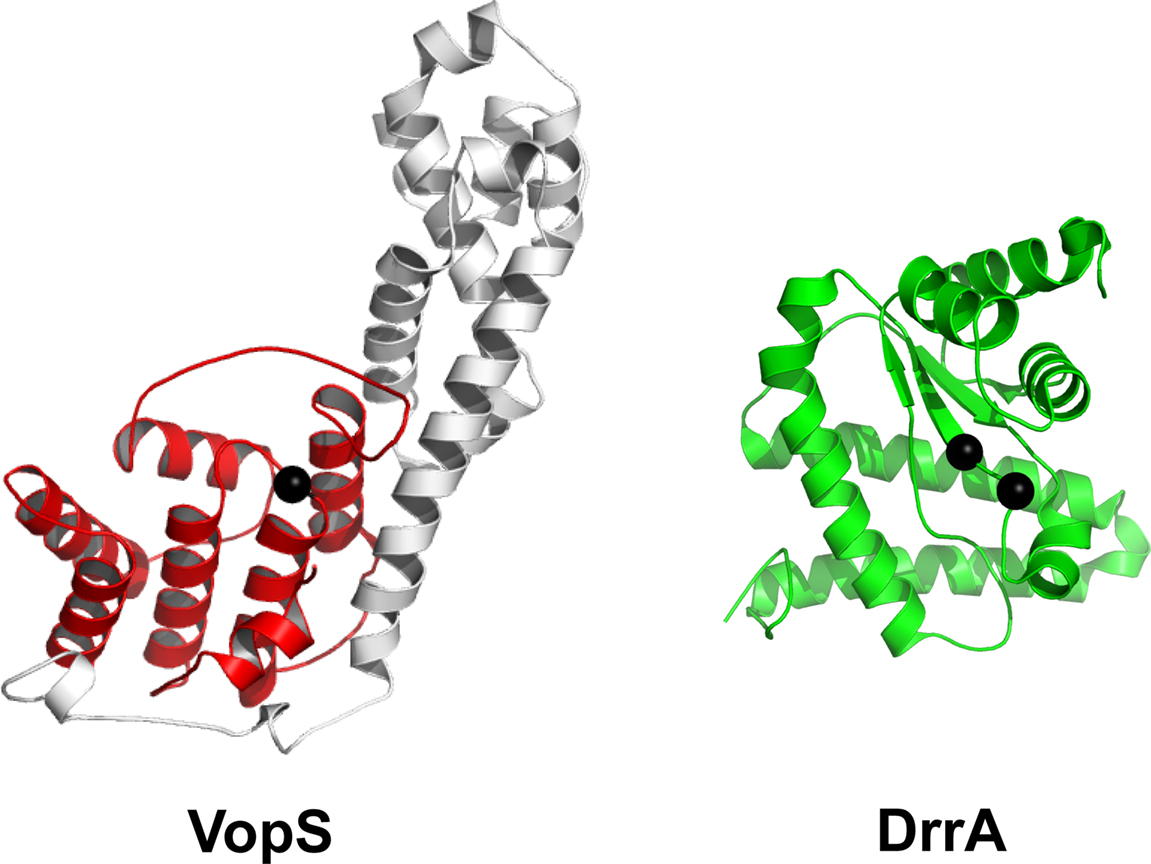
Figure 2. Domain architecture of the Fic-containing protein VopS and the adenylyl transferase region of DrrA. The Fic domain is highlighted in red with the non-conserved subdomain in white. Black spheres in VopS display the conserved catalytic histidine of the Fic motif. The DrrA adenylyl transferase region is colored green and black spheres correspond to the two conserved aspartates in adenylyl transferases.
AnkX
Legionella pneumophila also contains another likely AMPylator, the Fic protein effector AnkX. It is one of a group of effectors containing ankyrin repeat homology domains (ARHDs) that are translocated into a host cell via the T4SS. Though its molecular target remains unknown, it has been demonstrated to cause fragmentation of the Golgi apparatus and inhibit secretory transport after transfection in mammalian cells (Pan et al., 2008; Roy and Mukherjee, 2009). AMPylation by AnkX has not been fully demonstrated, but the induction of Golgi fragmentation is absent during transfections with truncations lacking the Fic domain or a point mutant of the catalytically essential histidine residue of the Fic domain, suggesting that AMPylation is responsible.
AMPylators in Regulatory Pathways
GS-ATase (GlnE)
GS-ATase is involved in metabolic regulation and was the first enzyme characterized to regulate another protein by modification with AMP. While it was first characterized in E. coli, the protein is conserved across many bacterial species and solved structures exist for both the E. coli (PDB ID IV4A and 3K7D) (Xu et al., 2004, 2009) and Mycobacterium tuberculosis homologues (PDB ID 2WHI and 2WGS) (Nilsson et al., 2009). The E. coli GS-ATase contains two adenylyl transferase domains that catalyze the removal and addition of AMP to glutamine synthetase at its N- and C-terminus, respectively. GS-ATase plays a role in nitrogen metabolism by controlling the AMP modified state of glutamine synthetase and thus its ability to synthesize glutamine. This modification is controlled by a complex regulatory system, in which the PII protein controls the activity of GS-ATase, which in turn controls the activity of glutamine synthetase via the addition or removal of AMP. PII activity is dependent on the intracellular concentrations of the metabolites α-ketoglutarate and glutamine, which reflect the current state of nitrogen metabolism in the cell. When levels of intracellular glutamine are low during nitrogen starvation, the PII protein exists primarily as PII-UMP. In the presence of PII-UMP and high α-ketoglutarate, GS-ATase is stimulated to remove AMP from glutamine synthetase, allowing the synthesis of glutamine (Jiang et al., 1998). PII is mostly unmodified when glutamine levels are high, during which GS-ATase transfers AMP to Y397 of glutamine synthetase, inhibiting its activity (Jiang et al., 1998). It has become clear, more than 40 years after its discovery, that GS-ATase catalyzes the same reaction that has since been termed AMPylation. Thus, the GS-ATase acts as the only example of an endogenous signaling system that is regulated by AMPylation, a reversible post-translational modification.
Kinetic and Structural Features of AMPylators
Enzyme Kinetics
Enzymatic studies with VopS enzyme and mutants at conserved residues in the Fic domain provided insight into the catalytic mechanism used by the Fic domain to modify Rho GTPases with AMP (Luong et al., 2010). Steady-state studies revealed the apparent affinity for ATP binding and the catalytic contribution of conserved Fic residues toward AMPylation (Luong et al., 2010). The histidine of the Fic motif was shown to be the most critical residue, acting as a general base during catalysis (Luong et al., 2010). Based on these kinetic studies predictions were made about the coordinate binding of substrates where Fic motif residues and the beta/hairpin loop element are important for ATP and protein substrate binding, respectively. Kinetic studies that supported a direct transfer mechanism were validated by the crystal structures of the Fic region from IbpA alone and in complex with Cdc42 Rho GTPase (see below) (Xu et al., 2009).
Both DrrA and GS-ATase contain the catalytic motif G-X11-D-X-D, with each aspartate coordinating a magnesium ion. Mutations of the conserved D110A and D112A in DrrA was demonstrated to abrogate AMPylation activity (Muller et al., 2010). The conserved aspartate residues D701 and D703 of GS-ATase were also shown to be critical for activity in steady-state kinetic studies (Jiang et al., 2007a). Further mechanistic studies with GS-ATase supported a sequential reaction mechanism where enzyme, substrate, and ATP form a ternary complex for catalysis, which matches the findings of similar studies with VopS and GTPases (Jiang et al., 2007b; Luong et al., 2010).
Structural Features
Structures of Fic-containing proteins have been determined from the structural genomic consortium and include Helicobacter pylori [PDB ID 2F6S], Bartonella henselae [PDB ID 2JK8], Shewanella oneidensis [PDB ID 3EQX], Bacteroides thetaiotaomicron [PDB ID 3CUC], and Neisseria meningitidis [PDB ID 2GO3] (Kinch et al., 2009; Luong et al., 2010). However, the endogenous substrates of these proteins have remained elusive. Protein structures of VopS [PDB ID 3LET] and IbpA [PDB ID 3N3U and 3N3V] with their Fic domain have been elucidated and these enzymes have been shown to AMPylate the Rho GTPase family on a threonine and tyrosine residue, respectively (Luong et al., 2010; Xiao et al., 2010). The Fic domain family displays a conserved topology, with a central two-helix bundle encircled peripherally by up to 6 helices. The Fic motif HPFx[D/E]GN[G/K]R lies within the loop connecting the two central helices. A conserved beta hairpin/loop structural element positioned near the Fic motif is also present in all solved structures (Kinch et al., 2009). In addition to their Fic domain most of the proteins contain topologically diverse subdomains, which mediates their unique biological function.
The initial crystal structure of the Fic region from VopS and the subsequent structures from native IbpA and in complex with Cdc42 Rho GTPase have revealed insights into the structural mechanism of protein substrate and ATP binding (Figure 2) (Luong et al., 2010; Xiao et al., 2010). The structure of IbpA–Cdc42 is an end-product complex with its tyrosine AMPylated and mimics the GDI-bound state of Rho GTPases. Extensive contacts are observed between the enzyme and substrate, which includes the conserved switch 1 and switch 2 regions of Cdc42. As predicted by the bioinformatics studies by Kinch et al., the beta hairpin/loop of Fic domains coordinates binding of substrates. The beta/hairpin loop of IbpA interacts in strand-to-strand fashion with the switch 1 region of Cdc42. The beta/hairpin loop is disordered in the native structure of IbpA but, as predicted based on previously solved Fic domains (Kinch et al., 2009; Luong et al., 2010), transition to ordered state upon protein substrate binding. Structural rearrangements also take place within the non-conserved subdomain of IbpA and support in protein substrate binding (Xiao et al., 2010). The VopS non-conserved subdomain was further demonstrated to have a role in protein substrate binding (Xiao et al., 2010). The Fic structure of BepA [PDB ID 2JK8] from B. henselae shows residues Asp/Glu and Asn of the Fic motif have roles in coordination of magnesium pyrophosphate (Luong et al., 2010).
The structure of the AMPylation domain of GS-ATase from E. coli was revealed to be structurally homologous to the nucleotidyl transferase family (Xu et al., 2009). This family possesses a core alpha/beta-fold comprised of a three-stranded mixed beta sheet flanked by four alpha helices (Kuchta et al., 2009). The N-terminal region of L. pneumophilia DrrA (residues 9-218) was determined by X-ray crystallography and is structurally similar to GS-ATase (Figure 2) (Muller et al., 2010). Concurrently a structure of AMPylated Rab1b modified at switch 2 tyrosine-77 GppNHp bound was also solved (Muller et al., 2010). DrrA was demonstrated to disrupt vesicular trafficking through AMPylation of Rab1b GTPase. There is no DrrA Rab1b GTPase complex structure available yet, but structure comparisons of modified Rab1b with native structure of closely related Rab3A implies AMP is not inducing structural rearrangements but inhibits downstream signaling by sterically hindering the binding of native substrates (Muller et al., 2010).
De-AMPylation
GS-ATase is a single enzyme that catalyzes two reactions, the de-AMPylation and AMPylation of glutamine synthetase (Jiang et al., 2007a). The domains for each activity are located at its N- and C-terminus, respectively. The de-AMPylation activity of GS-ATase removes AMP from modified glutamine synthetase and forms ADP and unmodified glutamine synthetase. Similar to the AMPylation domain, the de-AMPylase region of GS-ATPase is structurally homologous to the nucleotidyl transferase family and contains the G-X11-D-X-D motif (Xu et al., 2004). Structural comparisons revealed the N-terminal de-AMPylating domain and the C-terminal AMPylating region of GS-ATase are very similar with a root mean square deviation (RMSD) of 2.4 Å with both regions sharing only 24% sequence identity. Positions of active site residues involved in ATP binding and catalysis are conserved in both structures (Xu et al., 2004). Differences in secondary structural elements and conformations at the active site help explain differences in enzymatic activity between the two structurally homologous domains. No other bona fide de-AMPylases have been discovered.
A phosphodiester bond covalently links AMP and a hydroxyl chain residue of a protein, and it is possible that the phosphodiesterase (PDE) family of enzymes could potentially remove AMP from AMPylated proteins. Supporting this notion, the promiscuous snake venom PDE was demonstrated to remove AMP from a modified tyrosine in Rac1 GTPase (Worby et al., 2009).
Concluding Remarks
The role of AMPylation is in its infancy and many questions about the nature and diversity of this post-translational modification remain to be answered. First and foremost is the role of AMPylation in endogenous eukaryotic signaling. Bacterial AMPylators have shown a preference for GTPase substrates, but no biologically relevant data supports GTPases as substrates for eukaryotic Fic domain-containing proteins (Worby et al., 2009). As shown previously, the domain organization of several eukaryotic Fic proteins suggests membrane localization and a role in metabolism or stress response. Secondly, bioinformatic studies show the existence of Fic domains in bacteria as housekeeping genes. In fact the name “Fic” is derived from genetic observation (filamentation induced by cAMP) whereby the cell morphology of E. coli lacking a Fic protein was altered in the presence of cAMP. Understanding the role of Fic domains in the bacterial host will provide valuable insight for many Fic domain-containing proteins. A third important aspect of this field of research should include understanding the mechanism of de-AMPylation. Undoubtedly, the role of de-AMPylases will be important for our understanding the regulatory cycle of this post-translational modification. This is clearly the case with the GS-ATase, in which the N-terminal adenylyl transferase domain is a de-AMPylase and is intimately involved in the regulation of a metabolic system. It remains to be determined if de-AMPylation is catalyzed by other adenylyl transferases, phosphodiesterases, or another yet-to-be-discovered enzyme.
Further research into the crosstalk between the vast numbers of post-translational modifications on proteins is essential for understanding their impact on cellular signaling. Although the reductionist approach to cellular signaling has been incredibly productive in increasing our understanding of cellular signaling, it has also made clear how little we understand about the complexity of most, if not all signaling pathways. The manner in which different post-translational modifications compete for the same and proximal residues is poorly understood but likely represents an important aspect of signaling. This aspect of signaling becomes ever more daunting as more reversible post-translational modifications are discovered. When one considers that AMPylation can modify the same residues as phosphorylation, it becomes clear that the significance of AMPylation in signaling will continue to grow. Tackling the regulatory and molecular mechanisms of AMPylation, as is being done with other modifications such as acetylation, methylation, glycosylation, and lipidation, is imperative for understanding cellular signaling.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Boquet, P., and Lemichez, E. (2003). Bacterial virulence factors targeting Rho GTPases: parasitism or symbiosis? Trends Cell Biol. 13, 238–246.
Broberg, C. A., and Orth, K. (2010). Tipping the balance by manipulating post-translational modifications. Curr. Opin. Microbiol. 13, 34–40.
Brombacher, E., Urwyler, S., Ragaz, C., Weber, S. S., Kami, K., Overduin, M., and Hilbi, H. (2009). Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J. Biol. Chem. 284, 4846–4856.
Brown, M. S., Segal, A., and Stadtman, E. R. (1971). Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the P II-regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 68, 2949–2953.
Corbeil, L. B., Bastida-Corcuera, F. D., and Beveridge, T. J. (1997). Haemophilus somnus immunoglobulin binding proteins and surface fibrils. Infect. Immun. 65, 4250–4257.
Daniels, N. A., MacKinnon, L., Bishop, R., Altekruse, S., Ray, B., Hammond, R. M., Thompson, S., Wilson, S., Bean, N. H., and Griffin, P. M. (2000a). Emergence of a new Vibrio parahaemolyticus serotype in raw oysters: a prevention quandary. JAMA 284, 1541–1545.
Daniels, N. A., Ray, B., Easton, A., Marano, N., Kahn, E., McShan, A. L., 2nd, Del Rosario, L., Baldwin, T., Kingsley, M. A., and Puhr, N. D. (2000b). Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181, 1661–1666.
Duda, D. M., Walden, H., Sfondouris, J., and Schulman, B. A. (2005). Structural analysis of Escherichia coli ThiF. J. Mol. Biol. 349, 774–786.
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., Gavin, O. L., Gunasekaran, P., Ceric, G., and Forslund, K. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222.
Garcia-Pino, A., Christensen-Dalsgaard, M., Wyns, L., Yarmolinsky, M., Magnuson, R. D., Gerdes, K., and Loris, R. (2008). Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J. Biol. Chem. 283, 30821–30827.
Hardt, W. D., Chen, L. M., Schuebel, K. E., Bustelo, X. R., and Galan, J. E. (1998). S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93, 815–826.
Jacob-Dubuisson, F., Locht, C., and Antoine, R. (2001). Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40, 306–313.
Jiang, P., Mayo, A. E., and Ninfa, A. J. (2007a). Escherichia coli glutamine synthetase adenylyltransferase (ATase, EC 2.7.7.49): kinetic characterization of regulation by PII, PII-UMP, glutamine, and alpha-ketoglutarate. Biochemistry 46, 4133–4146.
Jiang, P., Pioszak, A. A., and Ninfa, A. J. (2007b). Structure-function analysis of glutamine synthetase adenylyltransferase (ATase, EC 2.7.7.49) of Escherichia coli. Biochemistry 46, 4117–4132.
Jiang, P., Peliska, J. A., and Ninfa, A. J. (1998). The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry 37, 12802–12810.
Kinch, L. N., Yarbrough, M. L., Orth, K., and Grishin, N. V. (2009). Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One 4(6): p. e5818. doi: 10.1371/journal.pone.0005818.
Kuchta, K., Knizewski, L., Wyrwicz, L. S., Rychlewski, L., and Ginalski, K. (2009). Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 37, 7701–7714.
Lake, M. W., Wuebbens, M.M., Rajagopalan, K.V., and Schindelin, H. (2001). Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 414, 325–329.
Lehnherr, H., Maguin, E., Jafri, S., and Yarmolinsky, M. B. (1993). Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233, 414–428.
Liu, M., Zhang, Y., Inouye, M., and Woychik, N. A. (2008). Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc. Natl. Acad. Sci. U.S.A. 105, 5885–5890.
Luong, P., Kinch, L. N., Brautigam, C. A., Grishin, N. V., Tomchick, D. R., and Orth, K. (2010). Kinetic and structural insights into the mechanism of AMPylation by VopS Fic domain. J. Biol. Chem. 285, 20155–20163.
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., Iijima, Y., Najima, M., Nakano, M., and Yamashita, A. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749.
Mangum, J. H., Magni, G., and Stadtman, E. R. (1973). Regulation of glutamine synthetase adenylylation and deadenylylation by the enzymatic uridylylation and deuridylylation of the PII regulatory protein. Arch. Biochem. Biophys. 158, 514–525.
Martin, S. W., Harland, R. J., Bateman, K. G., and Nagy, E. (1998). The association of titers to Haemophilus somnus, and other putative pathogens, with the occurrence of bovine respiratory disease and weight gain in feedlot calves. Can. J. Vet. Res. 62, 262–267.
Muller, M. P., Peters, H., Blumer, J., Blankenfeldt, W., Goody, R. S., and Itzen, A. (2010). The legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949.
Murata, T., Delprato, A., Ingmundson, A., Toomre, D. K., Lambright, D. G., and Roy, C. R. (2006). The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8, 971–977.
Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A., and Roy, C. R. (2002). A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679–682.
Nilsson, M. T., Krajewski, W. W., Yellagunda, S., Prabhumurthy, S., Chamarahally, G. N., Siddamadappa, C., Srinivasa, B. R., Yahiaoui, S., Larhed, M., Karlen, A. (2009). Structural basis for the inhibition of Mycobacterium tuberculosis glutamine synthetase by novel ATP-competitive inhibitors. J. Mol. Biol. 393, 504–513.
Pan, X., Luhrmann, A., Satoh, A., Laskowski-Arce, M. A., and Roy, C. R. (2008). Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320, 1651–1654.
Richard, J. F., Petit, L., Gibert, M., Marvaud, J. C., Bouchaud, C., and Popoff, M. R. (1999). Bacterial toxins modifying the actin cytoskeleton. Int. Microbiol. 2, 185–194.
Yarbrough, M. L., and Orth, K. (2009). AMPylation is a new post-translational modiFICation. Nat. Chem. Biol. 5, 378–379.
Yarbrough, M. L., Li, Y., Kinch, L. N., Grishin, N. V., Ball, H. L., and Orth, K. (2009). AMPylation of rho GTPases by vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272.
Worby, C. A., Mattoo, S., Kruger, R. P., Corbeil, L. B., Koller, A., Mendez, J. C., Zekarias, B., Lazar, C., and Dixon, J. E. (2009). The fic domain: regulation of cell signaling by adenylylation. Mol. Cell. 34, 93–103.
Xiao, J., Worby, C. A., Mattoo, S., Sankaran, B., and Dixon, J. E. (2010). Structural basis of Fic-mediated adenylylation. Nat. Struct. Mol. Biol. 17, 1004–1010.
Xu, Y., Zhang, R., Joachimiak, A., Carr, P. D., Huber, T., Vasudevan, S. G., and Ollis, D. L. (2004). Structure of the N-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase. Structure 12, 861–869.
Xu, Y., Carr, P. D., Vasudevan, S. G., and Ollis, D. L. (2009). Structure of the adenylylation domain of E. coli glutamine synthetase adenylyl transferase: evidence for gene duplication and evolution of a new active site. J. Mol. Biol. 396, 773–784.
Keywords: AMPylation, adenylylation, GTPase, effector, Fic, Type III secretion system, adenylyl transferase
Citation: Woolery AR, Luong P, Broberg CA and Orth K (2010) AMPylation: something old is new again. Front. Microbio. 1:113. doi: 10.3389/fmicb.2010.00113
Received: 04 September 2010;
Paper pending published: 15 September 2010;
Accepted: 22 September 2010;
Published online: 19 October 2010
Edited by:
Rey Carabeo, Imperial College London, UKCopyright: © 2010 Woolery, Luong, Broberg and Orth. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Kim Orth, Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX 75390-9148, USA. e-mail:a2ltLm9ydGhAdXRzb3V0aHdlc3Rlcm4uZWR1