- Department of Microbiology, Institute of Health Biosciences, The University of Tokushima Graduate School, Tokushima, Japan
Human immunodeficiency virus type 1 (HIV-1) is the causative virus of human acquired immunodeficiency syndrome (AIDS). Due to the lack of appropriate animal models, basic studies on HIV-1 replication, pathogenesis, and evolution, have been limited to cellular and molecular levels (Nomaguchi et al., 2008). Moreover, applied clinical studies in vivo also have been forced to use simian immunodeficiency virus isolated from macaques (SIVmac) and/or chimeric virus between SIVmac and HIV-1 (SHIV) based on our pioneering work 20 years ago (Shibata et al., 1991; Sakuragi et al., 1992). This experimental hindrance is mainly originated from the extremely narrow host range of HIV-1. To overcome the difficulty, we and others have made every effort to develop and establish suitable virus/animal systems. Because HIV-1 has adapted itself from the ancestral SIVs in Africa to replicate in humans in a marvelously strict way (Kirchhoff, 2009), the most promising one would be the use of macaque-tropic HIV-1 and macaques (Ambrose et al., 2007; Nomaguchi et al., 2008). As input viruses for infection of macaques, simian-tropic (st)HIV-1, and macaque-tropic HIV-1 (HIV-1mt) were designed, generated and characterized in vitro (Hatziioannou et al., 2006; Kamada et al., 2006; Thippeshappa et al., 2011) and in vivo (Igarashi et al., 2007; Hatziioannou et al., 2009; Saito et al., 2011; Thippeshappa et al., 2011). However, any of these HIV-1 derivatives neither replicate similarly robustly with the SIVmac standard clone in macaque cells nor are pathogenic for macaques so far. We, therefore, started improving the prototype HIV-1mt clone designated NL-DT5R by viral adaptation in macaque cells and genome manipulation based on bioinformatics (Figure 1A). NL-DT5R contains a 21-nucleotide SIVmac Gag-capsid (CA) element that lacks cyclophilin A (CypA) binding activity of the corresponding HIV-1 Gag-CA loop [CypA binding loop (−) in Figure 1A] and the entire SIVmac vif gene (Kamada et al., 2006). NL-DT5RS contains a loop sequence between helices 6 and 7 (h6/7L in Figure 1A) of SIVmac Gag-CA relative to NL-DT5R. MN4–8 carries three adaptive mutations in the pol-integrase and env-gp120 regions. MN4–8S contains the h6/7L of SIVmac Gag-CA and three adaptive mutations. MN4Rh-3 has an additional mutation in Gag-CA relative to MN4–8S. We now have a newest clone MN4Rh-3V (CXCR4-tropic), which was generated by the introduction of several mutations deduced from structural analysis into Gag-CA. Because CCR5-tropic viruses are thought to be clinically more important than CXCR4-tropic viruses, we similarly have generated a CCR5-tropic clone designated MN5Rh-3V.
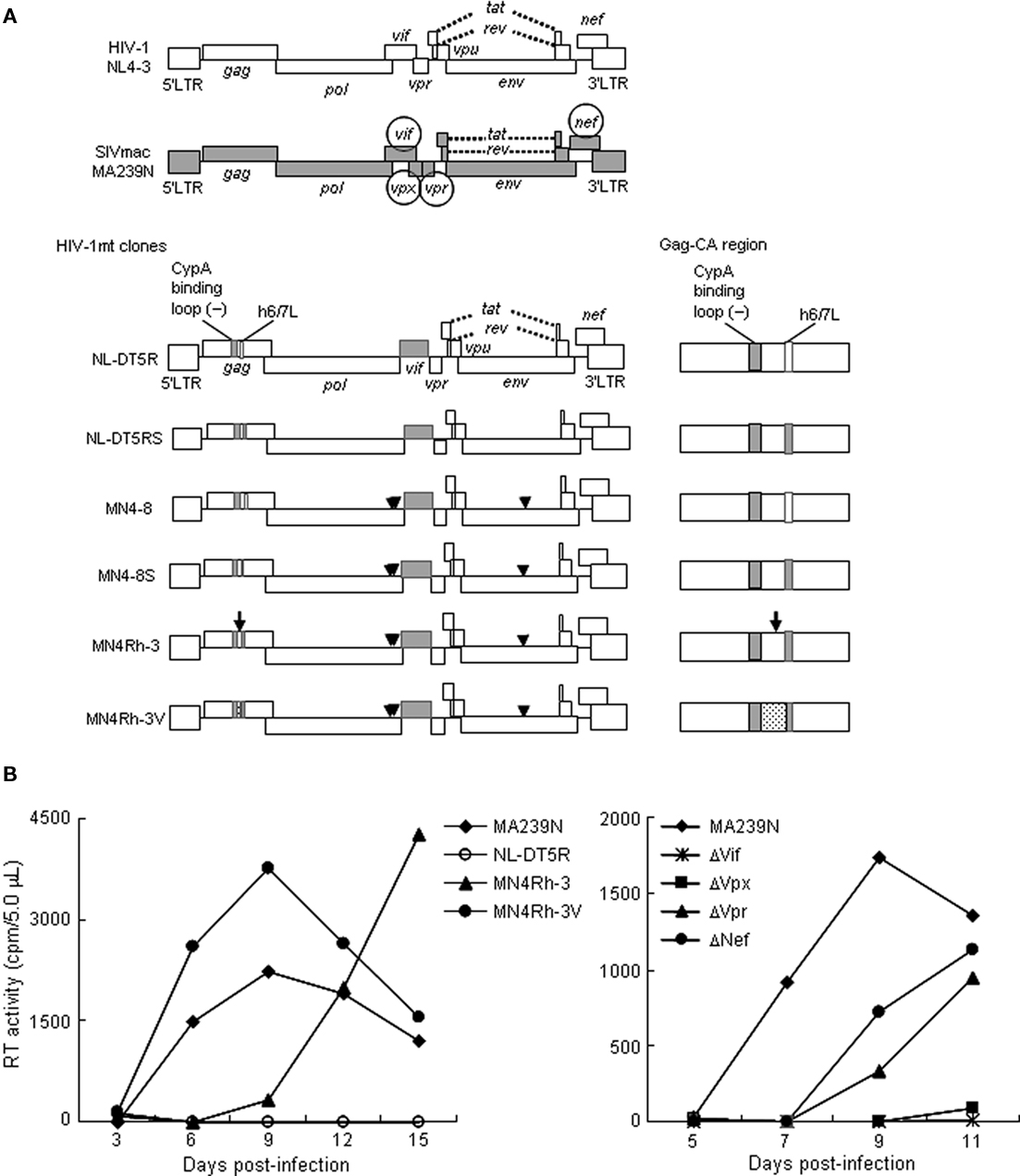
Figure 1. Virological characteristics of HIV-1/SIVmac clones. (A) Proviral genome structure of various viruses used for infection of M1.3S. Genomes of various HIV-1mt clones generated in our laboratory are schematically illustrated. Parental clones HIV-1 NL4-3 (Adachi et al., 1986) and SIVmac MA239N (Doi et al., 2010) are shown at the top. Accessory gene-inactivated mutants of SIVmac MA239N (circled) were constructed by substituting an internal stop codon in the nef gene of MA239 mutants (Shibata et al., 1991) with a Glu codon as reported for construction of MA239N (Doi et al., 2010). White and gray areas indicate the genomic regions of NL4-3 and MA239N, respectively. An arrow and arrowheads indicate the site of the four mutations in gag, pol, and env genes, respectively. The dotted region contains additional single-nucleotide mutations relative to the other clones. Enlarged Gag-CA region is shown on the right. For details, see the text. (B) Replication kinetics of the viruses in M1.3S cells. Cell-free viruses were prepared from 293T cells transfected with proviral clones indicated, and inoculated into 1 × 106 cells of M1.3S. Viral replication was monitored at intervals by reverse transcriptase (RT) activity in the culture supernatants. The experiments were done as described previously (Kamada et al., 2006; Doi et al., 2010). For infection, 6.3 × 104 and 2.4 × 106 RT units of MA239N and HIV-1mt clones (left), respectively, and 5.0 × 104 RT units of MA239N clones (right) were used. The representative data in two independent experiments are shown. Replication of NL-DT5RS, MN4–8, and MN4–8S was not detected as observed for NL-DT5R.
To achieve our purpose to obtain HIV-1mt clones potentially pathogenic for macaques, as a prerequisite, it is critical to have macaque cell lines suitable and easily usable to characterize many of the virus clones generated. However, macaque cell lines that are sensitive to various SIV and HIV-1mt clones and maintain the characteristics of natural target cells such as peripheral blood mononuclear cells were not readily available to us. We therefore searched for the appropriate cell lines and finally paid attention to the macaque cell lines immortalized by Herpesvirus saimiri (Akari et al., 1996; Fujita et al., 2003; Doi et al., 2010). Of various cynomolgus and rhesus macaque cell lines in these reports, we were particularly interested in the rhesus MT-IL2I cell line (Doi et al., 2010). It is the most refractory cell line among those examined by us to infection of SIVmac and HIV-1mt clones, but is CD4-positive, CXCR4-positive, and CCR5-positive. Therefore, it was predicted that this cell line expresses potent intracellular restriction factors against viruses. In fact, the TRIM5 allele of this cell line was found to be Mamu 1/Mamu 3 (Doi et al., 2010), a genotype that phenotypically shows a strong resistance to SIVmac infection (Lim et al., 2010). A subline of the MT-IL2I, after CD4-positive cells had been enriched by sorting, was designated M1.3S and used for infection experiments thereafter. Representative results obtained in M1.3S are shown in Figure 1B. Viruses of the early generation (from the prototype NL-DT5R up to MN4–8S) did not replicate at all. However, in cynomolgus HSC-F cells that are very sensitive to virus infection, MN4–8S replicated better than NL-DT5R, NL-DT5RS, and MN4–8. Interestingly, MN4Rh-3 acquired replication ability in M1.3S cells, and MN4Rh-3V exhibited further improved replication potential (Figure 1B left). On the other hand, the mutational effects of SIVmac accessory proteins on viral replication are also readily observed in M1.3S cells (Figure 1B right). Generally, these effects cannot be easily recognized in the established cell lines. As clearly observed in Figure 1B, each mutation to knock out the expression of individual accessory protein has greatly affected SIVmac replication. In particular, the mutations in vif and vpx genes have demonstrated a drastic negative effect. Therefore, it is quite evident that restriction factors against virus replication (counteracted by viral accessory proteins) are amply present in M1.3S cells.
In conclusion, the M1.3S cell line is exquisitely suitable to characterize macaque-tropic viruses and to investigate anti-viral cellular factors. Although primary cells are indispensable for basic studies on HIV/SIV that have highly adapted themselves to replicate in these cells, M1.3S cells can alternatively be used to address important issues to understand the biology and molecular biology of HIV/SIV.
References
Adachi, A., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A., and Martin, M. A. (1986). Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59, 284–291.
Akari, H., Mori, K., Terao, K., Otani, I., Fukasawa, M., Mukai, R., and Yoshikawa, Y. (1996). In vitro immortalization of Old World monkey T lymphocytes with Herpesvirus saimiri: its susceptibility to infection with simian immunodeficiency viruses. Virology 218, 382–388.
Ambrose, Z., KewalRamani, V. N., Bieniasz, P. D., and Hatziioannou, T. (2007). HIV/AIDS: in search of an animal model. Trends Biotechnol. 25, 333–337.
Doi, N., Fujiwara, T., Adachi, A., and Nomaguchi, M. (2010). Growth ability in various macaque cell lines of HIV-1 with simian cell-tropism. J. Med. Invest. 57, 284–292.
Fujita, M., Yoshida, A., Sakurai, A., Tatsuki, J., Ueno, F., Akari, H., and Adachi, A. (2003). Susceptibility of HVS-immortalized lymphocytic HSC-F cells to various strains and mutants of HIV/SIV. Int. J. Mol. Med. 11, 641–644.
Hatziioannou, T., Ambrose, Z., Chung, N. P. Y., Piatak, M. Jr., Yuan, F., Trubey, C. M., Coalter, V., Kiser, R., Schneider, D., Smedley, J., Pung, R., Gathuka, M., Estes, J. D., Veazey, R. S., KewalRamani, V. N., Lifson, J. D., and Bieniasz, P. D. (2009). A macaque model of HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A. 106, 4425–4429.
Hatziioannou, T., Princiotta, M., Piatak, M. Jr., Yuan, F., Zhang, F., Lifson, J. D., and Bieniasz, P. D. (2006). Generation of simian-tropic HIV-1 by restriction factor evasion. Science 314, 95.
Igarashi, T., Iyengar, R., Byrum, R. A., Buckler-White, A., Dewar, R. L., Buckler, C. E., Lane, H. C., Kamada, K., Adachi, A., and Martin, M. A. (2007). Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J. Virol. 81, 11549–11552.
Kamada, K., Igarashi, T., Martin, M. A., Khamsri, B., Hatcho, K., Yamashita, T., Fujita, M., Uchiyama, T., and Adachi, A. (2006). Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U.S.A. 103, 16959–16964.
Kirchhoff, F. (2009). Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat. Rev. Microbiol. 7, 467–476.
Lim, S.-Y., Rogers, T., Chan, T., Whitney, J. B., Kim, J., Sodroski, J., and Letvin, N. L. (2010). TRIM5α modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 6, e1000738. doi: 10.1371/journal.ppat.1000738
Nomaguchi, M., Doi, N., Kamada, K., and Adachi, A. (2008). Species barrier of HIV-1 and its jumping by virus engineering. Rev. Med. Virol. 18, 261–275.
Saito, A., Nomaguchi, M., Iijima, S., Kuroishi, A., Yoshida, T., Lee, Y. J., Hayakawa, T., Kono, K., Nakayama, E. E., Shioda, T., Yasutomo, Y., Adachi, A., Matano, T., and Akari, H. (2011). Improved capacity of a monkey-tropic HIV-1 derivative to replicate in cynomolgus monkeys with minimal modifications. Microbes Infect. 13, 58–64.
Sakuragi, S., Shibata, R., Mukai, R., Komatsu, T., Fukasawa, M., Sakai, H., Sakuragi, J., Kawamura, M., Ibuki, K., Hayami, M., and Adachi, A. (1992). Infection of macaque monkeys with a chimeric human and simian immunodeficiency virus. J. Gen. Virol. 73, 2983–2987.
Shibata, R., Kawamura, M., Sakai, H., Hayami, M., Ishimoto, A., and Adachi, A. (1991). Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65, 3514–3520.
Thippeshappa, R., Polacino, P., Kimata, M. T. Y., Siwak, E. B., Anderson, D., Wang, W., Sherwood, L., Arora, R., Wen, M., Zhou, P., Hu, S.-L., and Kimata, J. T. (2011). Vif substitution enables persistent infection of pig-tailed macaques by human immunodeficiency virus type 1. J. Virol. 85, 3767–3779.
Citation: Doi N, Fujiwara S, Adachi A and Nomaguchi M (2011) Rhesus M1.3S cells suitable for biological evaluation of macaque-tropic HIV/SIV clones. Front. Microbio. 2:115. doi: 10.3389/fmicb.2011.00115
Received: 30 April 2011; Accepted: 11 May 2011;
Published online: 23 May 2011.
Copyright: © 2011 Doi, Fujiwara, Adachi and Nomaguchi. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: adachi@basic.med.tokushima-u.ac.jp; nomaguchi@basic.med.tokushima-u.ac.jp
