- 1State Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Molecular Microbiology and Technology of the Ministry of Education, Department of Microbiology, College of Life Sciences, Nankai University, Tianjin, China
- 2Department of Molecular Genetics and Microbiology, College of Medicine, University of Florida, Gainesville, FL, USA
Post-transcriptional regulation enables bacteria to quickly response to environmental stresses. Polynucleotide phosphorylase (PNPase), which contains an N-terminal catalytic core and C-terminal RNA binding KH-S1 domains, is involved in RNA processing. Here we demonstrate that in Pseudomonas aeruginosa the KH-S1 domains of PNPase are required for the type III secretion system (T3SS) and bacterial virulence. Transcriptome analysis revealed a pleiotropic role of PNPase in gene regulation. Particularly, the RNA level of exsA was decreased in the ΔKH-S1 mutant, which was responsible for the reduced T3SS expression. Meanwhile, the pilus biosynthesis genes were down regulated and the type VI secretion system (T6SS) genes were up regulated in the ΔKH-S1 mutant, which were caused by increased levels of small RNAs, RsmY, and RsmZ. Further studies revealed that deletion of the KH-S1 domains did not affect the transcription of RsmY/Z, but increased their stabilities. An in vivo pull-down and in vitro electrophoretic mobility shift assay (EMSA) demonstrated a direct interaction between RsmY/Z and the KH-S1 fragment. Overall, this study reveals the roles of PNPase in the regulation of virulence factors and stabilities of small RNAs in P. aeruginosa.
Introduction
PNPase is a phosphate-dependent 3–5′ exonuclease with homologs identified in bacteria and eukaryotes (Guarneros and Portier, 1991; Symmons et al., 2000). PNPase consists of two RNase PH (pnp1 and pnp2) domains at the N-terminus and two RNA binding domains, KH and S1 on the C-terminus (Bermúdez-Cruz et al., 2005; Briani et al., 2007; Fernández-Ramírez et al., 2010). In bacteria, a small proportion of PNPase forms a large multi-protein complex with ribonuclease E (RNase E), a RNA helicase RhlB and a glycolytic enzyme enolase, named the RNA degradosome, which plays an important role in RNA processing (Miczak et al., 1996; Vanzo et al., 1998; Khemici et al., 2004; AJ, 2007; Nurmohamed et al., 2009).
PNPase is involved in bacterial response to environmental stresses (Rosenzweig and Chopra, 2013). For example, PNPase is required for the growth at low temperatures of E. coli, Yersinia pestis, and Yersinia pseudotuberculosis, Salmonella enterica, Campylobacter jejuni, Bacillus subtilis, and Staphylococcus aureus (Jones et al., 1987; Wang and Bechhofer, 1996; Goverde et al., 1998; Clements et al., 2002; Rosenzweig et al., 2005, 2007; Anderson and Dunman, 2009; Haddad et al., 2009). In addition, PNPase is involved in the regulation of virulence factors in several pathogenic bacteria, including Y. pestis, Y. pseudotuberculosis, S. enterica, and Dichelobacter nodosus (Rosenzweig and Chopra, 2013). In Yersinia, PNPase is required for the T3SS (Rosenzweig and Schesser, 2007), which is a highly conserved protein delivery system in various Gram-negative animal and plant pathogens (Sheahan and Isberg, 2015). The T3SS injects effector proteins into host cell cytosol, interfering with host cell signaling and other cellular processes, which facilitates bacterial pathogenesis (Hueck, 1998). In Yersinia, the effect of PNPase on the T3SS requires its S1 domain but is independent of its ribonuclease activity (Rosenzweig et al., 2005). However, the molecular mechanism remains elusive. In S. typhimurium, the wild type strain causes acute systemic infection in an intraperitoneal challenge assay, whereas the pnp mutant establishes a persistent infection in mice, suggesting a role of PNPase in the regulation of different sets of virulence factors (Clements et al., 2002).
So far, PNPase has been found to control gene expression mainly through three mechanisms: degradation of mRNA, affecting translation, and modulating sRNA stability. For example, PNPase autoregulates its own expression through RNase III dependent and independent mechanisms in E. coli (Wong et al., 2013; Carzaniga et al., 2015). In the RNase III dependent pathway, the pnp mRNA is processed by RNase III, followed by degradation in a PNPase dependent mechanism (Robert-Le Meur and Portier, 1994; Jarrige et al., 2001). In the RNase III independent pathway, PNPase binds to the 5′ untranslated region (5′UTR) of its own mRNA through its KH-S1 domains, which excludes the binding of ribosomal protein S1 and inhibits the translation (Carzaniga et al., 2015). In the cold shock response, the role of PNPase is to degrade unnecessary cold shock protein transcripts and resume growth after cold shock in both E. coli and Yersinia enterocolitica (Neuhaus et al., 2000; Polissi et al., 2003). Other than mRNAs, PNPase is involved in the degradation of small RNAs (sRNAs) that do not associate with RNA chaperone Hfq in E. coli (Andrade et al., 2012). However, PNPase was also found to be required for the stability of several sRNAs including RyhB, SgrS, and CyaR in E. coli through an unknown mechanism (De Lay and Gottesman, 2011).
Previously, we found that a pnp::Tn mutant was defective in the secretion of a T3SS effector protein ExoS in Pseudomonas aeruginosa (Li et al., 2013). P. aeruginosa is a versatile Gram-negative bacterium, which causes acute and chronic infections in humans (Stover et al., 2000; Driscoll et al., 2007). Virulence factors, including T3SS and motility play important roles in acute infections (Sadikot et al., 2005). During chronic infections, P. aeruginosa forms biofilm, in which bacteria grow inside an extracellular matrix mainly composed of polysaccharide, DNA and protein (Deretic et al., 1995; Sadikot et al., 2005). High level expression of type VI secretion system (T6SS) HSI-I is often associated with biofilm formation during chronic infection (Aubert et al., 2008; Khajanchi et al., 2009). It has been demonstrated that the T6SS plays a major role in killing target bacterial cells through translocation of toxic effector proteins in a cell–cell contact-dependent process (MacIntyre et al., 2010; Russell et al., 2011).
In P. aeruginosa, two small RNAs, RsmY, and RsmZ (RsmY/Z), reciprocally regulate acute and chronic infection associated virulence factors. RsmY/Z directly bind to RsmA and suppress its function (Brencic et al., 2009; Bordi et al., 2010). RsmA is a global post-transcriptional regulatory protein, which binds to a GGA motif in untranslated region of target mRNAs and represses their translation (Mulcahy et al., 2008). The direct target of RsmA includes mRNAs of T6SS genes fha1 and tssA1, as well as pslA, a gene involved in biofilm matrix exopolysaccharide synthesis (Brencic and Lory, 2009; Irie et al., 2010). Meanwhile, RsmA positively regulates the T3SS and the type VI pili through an unknown mechanism (Brencic and Lory, 2009).
The levels of RsmY/Z are controlled at the transcriptional and posttranscriptional level. The two-component regulatory system GacS-GacA directly controls the transcription of RsmY/Z (Brencic et al., 2009). Interaction between RsmY and Hfq protects RsmY from degradation by RNase E (Sonnleitner et al., 2006; Sorger-Domenigg et al., 2007).
In this study, we demonstrate that PNPase is essential for the T3SS and pathogenesis in P. aeruginosa. Transcriptome analysis reveals that PNPase controls multiple virulence factors, including T6SS and pilus biosynthesis genes. And we demonstrate a direct interaction between PNPase and RsmY/Z whereby PNPase controls the stability of these sRNAs and subsequently the expression of T6SS and pilus biosynthesis genes. These results provide a new insight into the regulatory mechanism of PNPase in P. aeruginosa.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The strains and plasmids used in this study are listed in Table S1. All strains were cultured in LB broth or L-agar (LA) at 37°C. Antibiotics were used at the following concentrations: for E. coli, ampicillin at 100 μg/ml, kanamycin at 50 μg/ml, gentamicin at 15 μg/ml; for P. aeruginosa, carbenicillin at 150 μg/ml, gentamicin at 50 μg/ml, tetracycline at 50 μg/ml.
Murine Acute Pneumonia Model
All animal experiments complied with Nankai University and Chinese national guidelines regarding the use of animals in research. The protocol was approved by the institutional animal care and use committee of the college of life sciences of Nankai University with permit number: NK-04-2012. Overnight bacterial culture was diluted in fresh LB and grown to an optical density at 600 nm (OD600) of 1.0. The bacterial cells were collected and resuspended in phosphate-buffered saline (PBS). Female BALB/c mice (6–8 weeks old) were anesthetized with 0.1 ml of 7.5% chloral hydrate injected intraperitoneally. Then the mice were inoculated intranasally with 2 × 107 CFU bacteria. Twelve hours post infection, the mice were sacrificed. The lungs were isolated and homogenized in 1% proteose peptone. The bacterial loads were determined by serial dilutions and plating. Statistical analysis was performed with the GraphPad Prism software.
Cytotoxicity Assay
Bacterial cytotoxicity was determined by measuring detachment of mammalian cells after bacterial infection as described previously (Li et al., 2013). 1.2 × 105 HeLa cells were seeded into each well of a 24-well plate and cultured in Dulbecco's modified Eagle medium (DMEM) with 10% (vol/vol) heat-inactivated fetal bovine serum (hiFBS), penicillin G (100U/ml), and streptomycin (100 μg/ml) at 37°C with 5% CO2 for 18 h. The medium was replaced with antibiotic free DMEM with 10% hiFBS 1 h before bacterial infection. Overnight bacterial culture was diluted 50-fold in fresh LB and grown to an OD600 of 1.0. Bacteria were washed twice and resuspended in PBS. HeLa cells were infected with indicated strains at a multiplicity of infection (MOI) of 20. After 3 h, the medium in each well was removed. Cells remaining attached were washed twice with PBS and stained with 0.1% crystal violet for 15 min. Then, each well was washed twice with water. For quantification, the stained crystal violet was dissolved in 95% ethanol and measured at the wavelength of 590 nm.
Western Blotting
Over night bacterial culture was diluted 1:100 in LB or 1:30 in LB supplemented with 5 mM EGTA, and cultured for 3.5 h. The supernatant and pellet were separated by centrifugation. Samples from equal numbers of bacteria were loaded onto a 12% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE). The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane and probed with a rabbit polyclonal antibody against ExoS or a mouse monoclonal antibody against FLAG (Sigma). Signals were detected with the ECL-plus kit (Millipore).
RNA Extraction and Real Time PCR
Bacteria were grown in LB medium to indicated growth phases. Total RNA was isolated with an RNA prep Pure cell/Bacteria Kit (Tiangen Biotec). cDNA was synthesized with a PrimeScript Reverse Transcriptase and random primers (Takara). The cDNA was mixed with indicated primers and FastStart Essential DNA Green Master (Roche). The pyrroline-5-carboxylate reductase coding gene proC was used as an internal control.
Transcriptome Sequencing and Analysis
The Transcriptome sequencing and analysis were performed by GENEWIZ (Suzhou, China). Briefly, total RNA of each sample was quantified and qualified by an Agilent 2100 Bioanalyzer (Agilent Technologies). One microgram total RNA with RIN value above seven was used for library preparation. Large ribosomal RNA was depleted from bacterial total RNA using RiboMinus Bacteria Module (Invitrogen) and the rRNA-depleted mRNA was then fragmented, and primed with random primers. Pair-end index libraries were constructed according to the manufacturer's protocol (NEBNext. Ultra. Directional RNA Library Prep Kit for Illumina). The RNA expression analysis was based on the annotations of PAO1 (www.pseudomonas.com). The RSEM software (V 1.2.15) was used to align the input reads against the reference gene with Bowtie2 and expression values were calculated using the FPKM (fragments per kilobase of transcript per million reads) method. The software edger (V3.4.2) (Bioconductor) was used to calculate p-values.
RNA Stability Analysis
Bacteria were grown to an OD600 of 1.0 and treated with 100 μg/ml rifampicin. At each indicated time point, same volume of bacteria was taken and the bacterial concentration was determined by serial dilution and plating. Next, each sample was mixed with equal numbers of gfp expressing E. coli cells. Total RNA was purified and the levels of RsmY/Z were analyzed with real time PCR. The gfp RNA level in each sample was used as an internal control for normalization.
Twitching Motility
The twitching motility was assayed on 1% LB agar. Each strain was inoculated in the agar by stabbing with a sharp toothpick. The plates were incubated at 37°C for 18 h. The twitching zones were visualized by staining with 0.1% crystal violet.
Purification of Protein and Detection of Associated RNA
The C-terminus His-tagged full length PNPase or KH-S1 fragment was constructed in pMMB67EH and transferred into wild type PAK constitutively expressing a gfp gene. Bacteria containing either one of the plasmids were grown to an OD600 of 0.8 and expression of the His-tagged protein was induced by the addition of 1 mM IPTG for 3 h. Bacteria from 100 mL culture were collected by centrifugation and resuspended in 1.5 ml lysis buffer (500 mM NaCl, 50 mM Tris, 20 mM imidazole, pH 7.9) with recombinant RNase inhibitor (RRI, Takara) and lysed by sonication. After centrifugation, the supernatant was incubated with 30 μl Ni-NTA agarose beads (Qiagen) at 4°C for 1 h. Then the beads were washed 5 times with 1 ml lysis buffer, and incubated with 30 μl elution buffer (500 mM NaCl, 50 mM Tris, 250 mM imidazole, pH 7.9) for 10 min. After centrifugation, protein and RNAs in the supernatant were collected and subjected to RNA purification with an RNA prep Pure cell/Bacteria Kit (Tiangen Biotec). The purified RNA was analyzed by real time PCR. The gfp RNA was used as the reference to determine the relative level of each sRNA.
In vitro Transcription and RNA Gel Mobility Shift Assay
The sRNA transcripts were synthesized from PCR products using the Riboprobe System-T7 (Promega) according to the manufacturer's instructions. The primers used in PCR are shown in supporting information (Table S2). The RNAs were purified with an RNA prep Pure cell/ Bacteria Kit (Tiangen Biotec) and refolded by heating at 90°C for 10 min followed by natural cooling at room temperature for 30 min. One nano mole sRNA was incubated with indicated amount of purified KH-S1-His protein in 20 μl binding buffer [10 mM Tris-HCl, pH 7.5; 50 mM KCl, 5 mM MgCl2, 10% glycerol, 1 U recombinant RNase inhibitor (Takara)] for 30 min at room temperature. Fifteen microliters of each sample was loaded onto a non-denaturing 7% polyacrylamide gel. Electrophoresis was performed at 100 V for 150 min with 0.5 × TBE buffer (Tris-borate-ethylenediaminetetraacetic acid) at 4°C. The RNA bands were observed by staining with ethidium bromide (Takara) for 15 min.
Results
PNPase is Required for ExoS Expression and Cytotoxicity
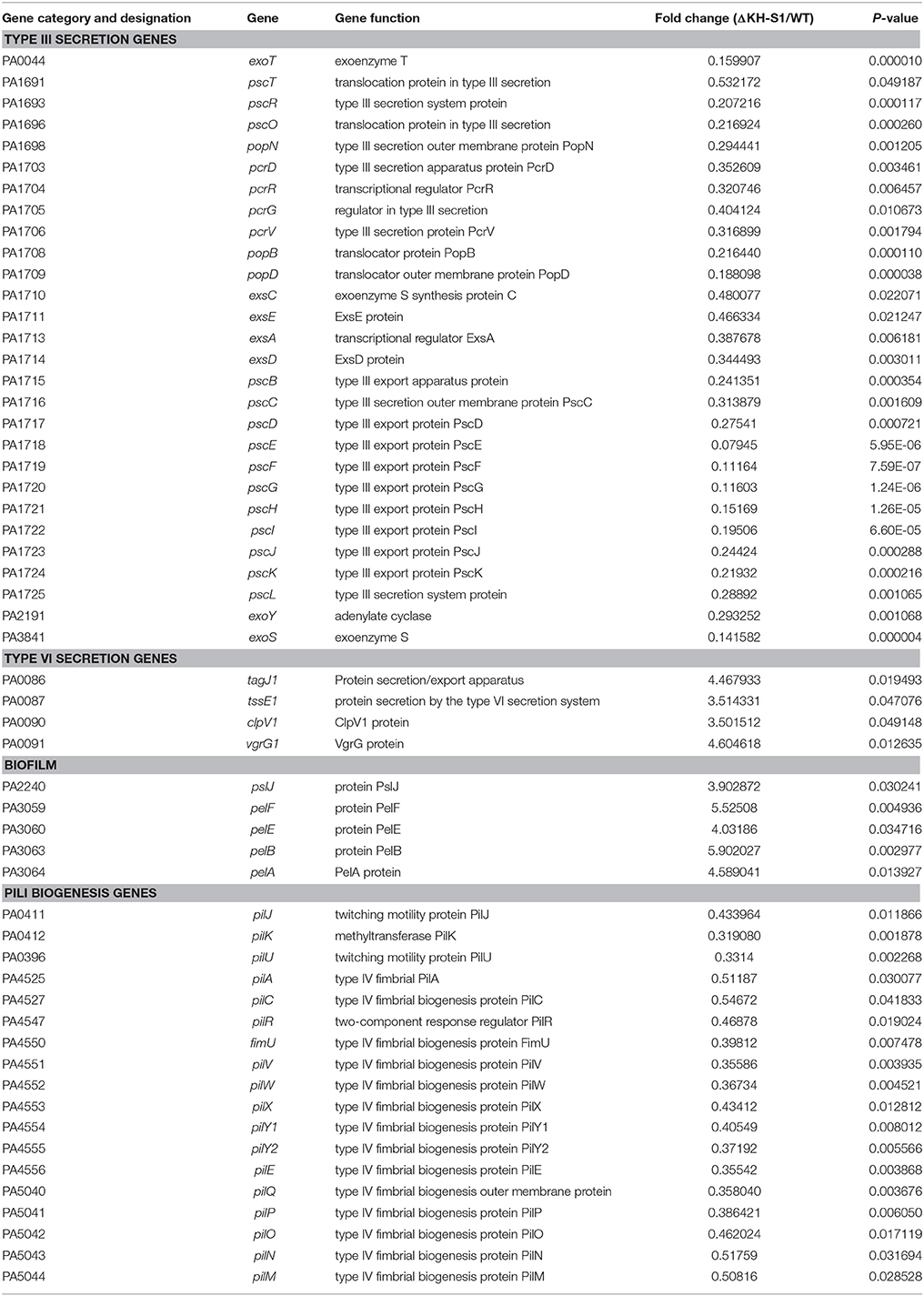
In the P. aeruginosa genome, the PNPase coding gene pnp is 2106 base pairs (bp) long. Our previous study demonstrated that transposon (Tn) insertion at the 1655 bp of the pnp gene in a wild type strain PAK resulted in reduced ExoS secretion (Li et al., 2013). In order to investigate the role of PNPase in T3SS, we firstly tried to delete the entire pnp gene in wild type PAK. However, we were unable to obtain the mutant after multiple attempts. Then, we constructed a mutant with the deletion of nucleotides 1655–2106, which includes the coding region of the KH and S1 domains of PNPase (Figure 1A), and designated ΔKH-S1. As shown in Figure 1B, the expression of ExoS was lower in this mutant under T3SS inducing condition (in the presence of EGTA). For complementation, the full length pnp gene driven by its native promoter was inserted into the chromosome by a mini-Tn7 vector (Choi and Schweizer, 2006), which restored the expression and secretion of ExoS (Figure 1B).
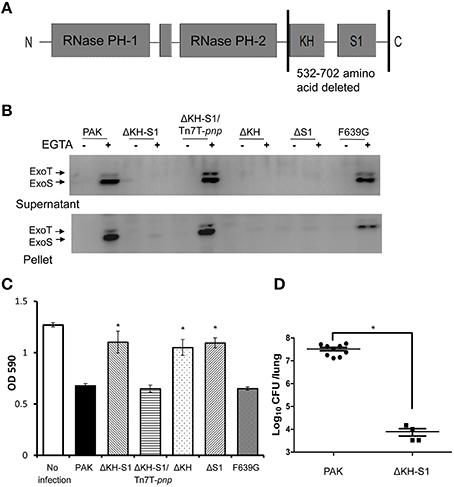
Figure 1. Role of PNPase in the expression of T3SS and pathogenesis. (A) Structure of the PNPase protein in P. aeruginosa. (B) Secretion and expression of ExoS in indicated strains. Bacteria were grown to an OD600 of 1.0 in LB with or without 5 mM EGTA. Lysates from equivalent bacterial cells were loaded onto SDS-PAGE gels and probed with the ExoS antibody. The ExoS antibody cross-recognizes another T3SS effector ExoT due to a high sequence homology between the two proteins. (C) Cytotoxicity of the indicated strains. HeLa cells were infected with the indicated strains at a MOI of 20. After 3 h, cells attached to the 24-well plate were quantified by crystal violet staining. Each assay was done in triplicates, and the error bars indicate standard deviations. *p < 0.05 compared to PAK by Student's t-test. (D) Bacterial load in the lungs of mice infected with P. aeruginosa. 6–8 weeks old female BALB/c mice were inoculated with 2 × 107 CFU wild-type PAK or the ΔKH-S1 mutant. Twelve hours post infection, the mice were sacrificed, and the lungs were isolated and homogenized. The bacteria loads were determined by serial dilution and plating. Long lines represent medians and shot lines represent standard errors of the means (SEM). *p < 0.05, by Mann Whitney test.
In P. aeruginosa, the T3SS plays a major role in killing host cells. To examine the role of PNPase in cytotoxicity, HeLa cells were infected with PAK, the ΔKH-S1 mutant and the complemented strain at a MOI of 20. When infected with PAK, majority of the cells were rounded and detached 3 h post infection. Deletion of the KH and S1 domains reduced the cytotoxicity, which was restored by the complementation with a full length pnp gene (Figure 1C). These results suggest that loss of the KH-S1 domains of PNPase results in defective T3SS in P. aeruginosa.
PNPase is Essential for the Bacterial Virulence in a Mouse Acute Pneumonia Model
The T3SS is activated during infection and plays an important role in the pathogenesis in acute infections (Roy-Burman et al., 2001; Hauser, 2009; Howell et al., 2013). To evaluate the role of PNPase in virulence, BALB/c mice were infected intranasally with 2 × 107 CFU of PAK or the ΔKH-S1 mutant. 12 h post infection, the mice were sacrificed, and the bacterial load in each lung was enumerated. Compared to the wild type PAK, significantly less ΔKH-S1 mutant bacteria were recovered from the lungs (Figure 1D), suggesting an essential role of PNPase in the colonization by P. aeruginosa.
Role of the KH and S1 Domains in the Regulation of T3SS
In E. coli, PNPase negatively regulates its own mRNA stability and translation (Wong et al., 2013; Carzaniga et al., 2015). Therefore, deletion of the KH-S1 domains in P. aeruginosa might result in an up regulation of the remaining N-terminal catalytic core domains in the chromosome and lead to defective T3SS. To test the expression level of the N-terminal domains, we cloned the coding region with its native promoter and fused it with a His tag on its C-terminus, and designated it as NTD-His (Figure S1A). The construct was inserted into the chromosomes of PAK and the ΔKH-S1 mutant. As shown in Figure S1B, the expression levels of the NTD-His were similar between the wild type PAK and the ΔKH-S1 mutant, suggesting that the defective T3SS is not due to an increase of the N-terminal catalytic core domains.
In Yersinia, the S1 domain but not the PNPase ribonuclease activity is required for the T3SS (Rosenzweig et al., 2005). Of the S1 domain, the F639 residue is conserved and plays an integral role in RNA binding. Replacement of the F639 residue with glycine reduces the PNPase catalytic activity and T3SS expression in Yersinia (Jarrige et al., 2002; Schubert et al., 2004; Rosenzweig et al., 2007). To test the role of these two domains, we complemented the ΔKH-S1 mutant with a pnp gene without either the KH or the S1 domain, or with a F639G mutation. As shown in Figures 1B,C, PNPase without the KH or S1 domain was unable to restore the T3SS and cytotoxicity, suggesting an essential role of both the KH and S1 domains. However, we cannot exclude the possibility that deletion of the KH domain affects the correct folding of the S1 domain. The F639G mutation had no influence on the T3SS expression and cytotoxicity (Figures 1B,C). In addition, complementation with a fragment containing the KH-S1 domains did not restore the T3SS expression in theΔKH-S1 mutant (Figure S2). These results indicate a difference in the PNPases in Yersinia and P. aeruginosa.
Identification of Genes Affected by PNPase
To further understand the role of PNPase in T3SS and possibly other genes' expression, we performed transcriptome analyses (RNA-seq) on wild type PAK and the ΔKH-S1 mutant. The two strains were grown to log phase and total RNAs were purified. Compared to wild type PAK, expression of 309 genes were altered, with 182 genes down regulated, and 127 genes up regulated in the ΔKH-S1 mutant (Table S3). Consistent with aforementioned results, mRNA levels of the T3SS genes were reduced in the ΔKH-S1 mutant (Table 1). Besides, the type IV pilus biosynthesis genes were down regulated (Table 1) while the T6SS and biofilm matrix exopolysaccharide genes were up regulated. Additionally, the RNA levels of ribosome proteins were increased in the absence of PNPase (Table S3). The full data set has been deposited in the NCBI SRA, with the Study accession number SRP069795.). These results revealed that the PNPase has a global impact on mRNA levels in P. aeruginosa.
Over Expression of exsA Restores the T3SS Expression in the ΔKH-S1 Mutant
ExsA directly controls the expression of T3SS genes through binding to their promoters (Hauser, 2009). Our transcriptome analysis reveals a 61% decrease of the exsA mRNA in the ΔKH-S1 mutant (Table 1), which was confirmed by real time PCR analysis (Figure 2A). To test if the defective T3SS is due to reduced expression of ExsA, we inserted a lac promoter driven exsA gene into the chromosome by the mini-Tn7 vector (Choi and Schweizer, 2006; Sun et al., 2014). As shown in Figure 2B, over expression of ExsA restored the expression and secretion of ExoS in the ΔKH-S1 mutant. These results suggest that PNPase affects the T3SS through ExsA.
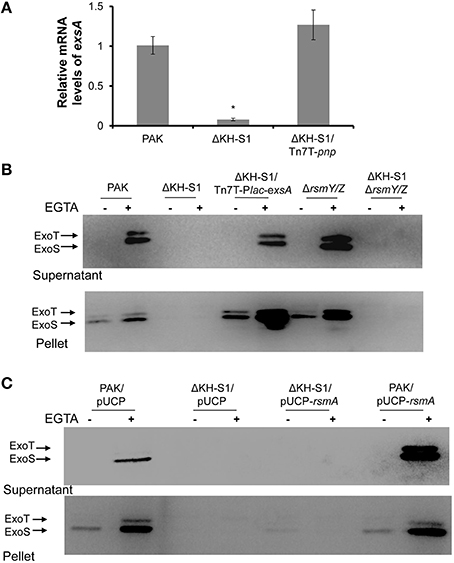
Figure 2. RsmY/Z and RsmA are not involved in PNPase-mediated regulation of T3SS. (A) Relative mRNA levels of exsA in indicated strains. Bacteria were grown to an OD600 of 1.0. Total RNA was isolated and the exsA mRNA levels were determined by real time PCR with proC as an internal control. Each assay was done in triplicates, and the error bars indicate standard deviations. *p < 0.05 compared to PAK by Student's t-test. (B,C) Secretion and expression of ExoS in indicated strains. Bacteria were grown to an OD600 around 1.0 in LB with or without 5 mM EGTA. Intracellular and secreted ExoS were separated by centrifugation. Samples from equivalent bacterial cells were loaded into SDS-PAGE gels and probed with antibody against ExoS.
Twitching Motility is Affected by the PNP Mutation
As shown in Table 1, the mRNA levels of type IV pilus biosynthesis genes were decreased in the ΔKH-S1 mutant. Real time PCR results confirmed the down regulation of type IV pilus regulatory genes pilR and structural gene pilA in the ΔKH-S1 mutant. Complementation with a pnp gene restored the expression of these two genes (Figure 3A). Consistent with the gene expression pattern, the ΔKH-S1 mutant displayed defective twitching motility, which was restored in the complementation strain (Figures 3B,C). These results suggest a role of PNPase in the regulation of twitching motility.
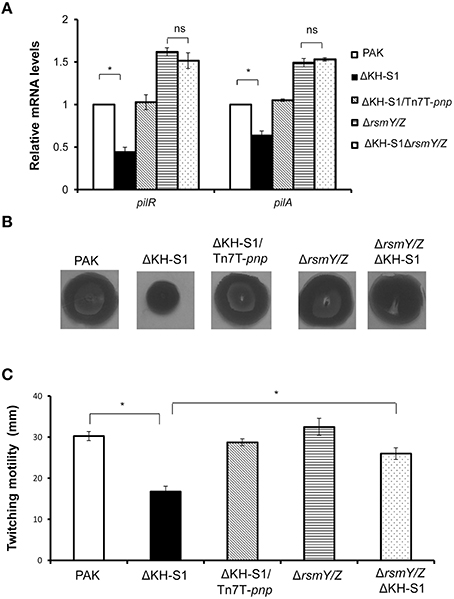
Figure 3. PNPase regulates the type IV pili through RsmY/Z. (A) Bacteria were grown to an OD600 of 1.0. Total RNAs were collected and the relative mRNA levels of pilR and pilA were determined by real time PCR. Each assay was done in triplicates, and the error bars indicate standard deviations. *p < 0.05 compared to wild type PAK by Student's t-test. (B) Twitching motilities of the indicated strains were assayed on 1% LB agar. The twitching zones were visualized by staining with 0.1% crystal violet. (C) Diameters of the twitching zones of indicated strains. The values represent the average diameters from three independent experiments. The error bars indicate standard deviations. *p < 0.05 compared to wild type PAK by Student's t-test.
T6SS is Up Regulated in the ΔKH-S1 Mutant
In our transcriptome analysis, the mRNA levels of four T6SS HSI-I genes were significantly increased in the ΔKH-S1 mutant (Table 1). Real time PCR verified the up regulation of vgrG and hcp-1 (Figure 4A). To further confirm the expression level, a hcp1 gene driven by its native promoter was tagged with a FLAG and inserted into the chromosomes of indicated strains. Consistent with the mRNA level, the Hcp-1 protein level was increased in the ΔKH-S1 mutant (Figure 4B).
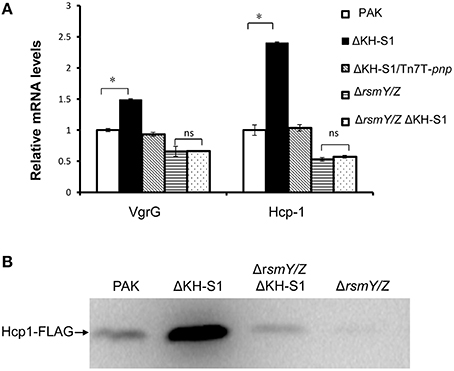
Figure 4. PNPase regulates the T6SS expression through RsmY/Z. (A) Relative mRNA levels of the T6SS genes in indicated strains. Each assay was done in triplicates, and the error bars indicate standard deviations. *p < 0.05 compared to wild type PAK by Student's t-test; ns, not significant. (B) Levels of Hcp-1 protein in indicated strains. Strains with an hcp1-FLAG in their chromosomes were grown for 7 h in LB. Samples from equivalent numbers of bacterial cells were loaded onto a SDS-PAGE gel and probed with an anti-FLAG antibody.
Increased Levels of RsmY and RsmZ are Responsible for the Altered Expression of Pili, T6SS, But Not T3SS in the PNP Mutant Strain
The down regulation of T3SS and pilus as well as up regulation of T6SS genes resemble the phenotypes of a rsmA mutant (Brencic and Lory, 2009). However, the RsmA mRNA levels were similar between wild type PAK and the ΔKH-S1 mutant (Figure S3). Since the function of RsmA is antagonized by small RNAs RsmY and RsmZ, we examined the levels of these sRNAs. Indeed, the RsmY/Z levels were higher in the ΔKH-S1 mutant at both late exponential and stationary phases, while complementation with a pnp gene restored the RNA levels (Figure 5A).
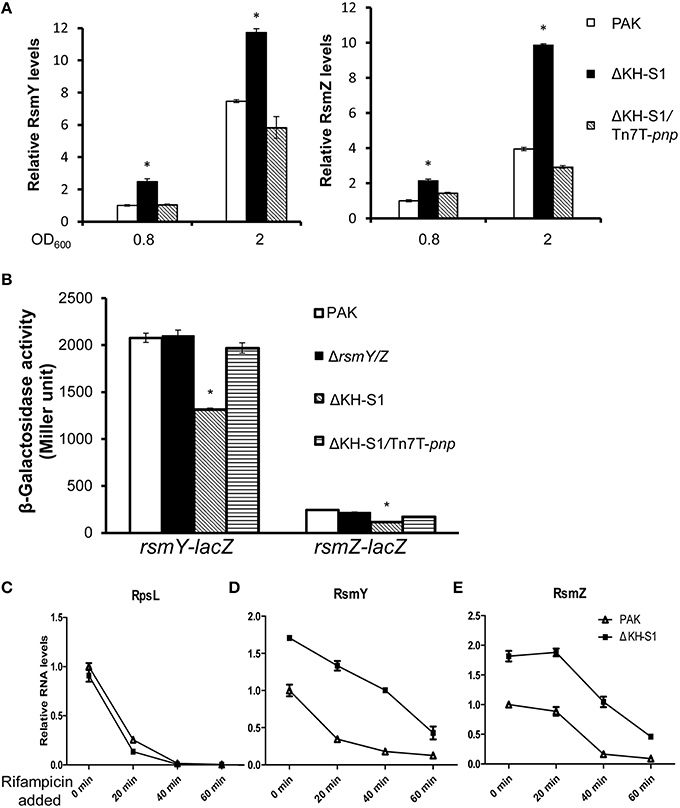
Figure 5. PNPase controls the stabilities of RsmY/Z. (A) Levels of the RsmY and RsmZ in indicated strains. Bacteria were grown to an OD600 = 0.8 or 2 in LB. Total RNAs were collected and the relative levels of RsmY and RsmZ were determined with real time PCR. The RNA levels of proC were used as internal controls. *p < 0.05 compared to wild type PAK by Student's t-test. (B) Expression of RsmY and RsmZ in wild type PAK and the ΔKH-S1 mutant. Bacteria containing rsmY-lacZ or rsmZ-lacZ transcriptional fusion were grown to the similar OD600 of 2.0 and subjected to β-galactosidase assays. Each assay was done in triplicates, and the error bars indicate standard deviations. *p < 0.05 compared to wild type PAK by Student's t-test. Degradation of RpsL (C), RsmY (D), and RsmZ (E) in wild type PAK and the ΔKH-S1 mutant. Bacterial cells with or without rifampicin treatment were spiked with equal numbers of gfp expressing E. coli cells. Total RNA was purified and the relative RNA levels were determined by real time PCR. The gfp RNA level in each sample was used as an internal control for normalization.
To test the role of RsmY/Z in the ΔKH-S1 mutant, we constructed a ΔKH-S1ΔrsmYΔrsmZ triple mutant. Expression and secretion of ExoS were similar between the triple mutant and the ΔKH-S1 mutant (Figure 2B). And over expression of RsmA had no effect on the expression of ExoS in the ΔKH-S1 mutant (Figure 2C). These results suggest that RsmA and RsmY/Z are not involved in the altered expression of T3SS in the ΔKH-S1 mutant.
However, deletion of rsmY/Z restored the expression of pilus biosynthesis genes and twitching motility (Figures 3A–C), as well as the expression of vgrG and hcp-1 in the ΔKH-S1 mutant (Figures 4A,B). Therefore, increased levels of RsmY/Z are responsible for the suppression of twitching motility and up regulation of T6SS genes.
PNPase Controls the Stabilities of RsmY/Z
The higher levels of RsmY/Z in the ΔKH-S1 mutant might be due to increased transcription or stability. To test these possibilities, we firstly constructed rsmY-lacZ and rsmZ-lacZ transcriptional fusions. Compared to wild type PAK, the LacZ levels were slightly lower in the ΔKH-S1 mutant (Figure 5B). In addition, the mRNA levels of LadS, GacS and GacA, which are known to positively regulate RsmY/Z (Brencic et al., 2009), were slightly decreased in the pnp mutant (Figure S3). These results suggest that the higher levels of RsmY/Z are not due to increased transcription.
Next, we compared the stabilities of RsmY/Z in wild type PAK and the ΔKH-S1 mutant. Bacteria were grown to an OD600 of 1.0, and 100 μg/ml rifampicin was added to block RNA transcription. After 20, 40, or 60 min, total RNA was extracted and examined with real time PCR. The ribosomal RNA (rRNA) RpsL levels were similar between PAK and the ΔKH-S1 mutant with or without rifampicin treatment, suggesting a similar degradation rate (Figure 5C). The RsmY level in the ΔKH-S1 mutant was 1.76–fold of that in PAK without rifampicin treatment, whereas 20 min after rifampicin treatment, the differences rose to 3.5-fold, and then dropped to around 3-folds after 40 or 60 min (Figure 5D). As for the RsmZ levels, the difference between the ΔKH-S1 mutant and PAK rose from 1.8-fold without rifampicin treatment to 2-, 4-, and 5-fold after 20, 40, and 60 min treatment, respectively (Figure 5E). These results suggest increased stability of the sRNAs in the ΔKH-S1 mutant. Furthermore, over expression of the PNPase in wild type PAK reduced the levels of RsmY/Z (Figures 6A,B), suggesting a role of PNPase in the degradation of RsmY/Z.
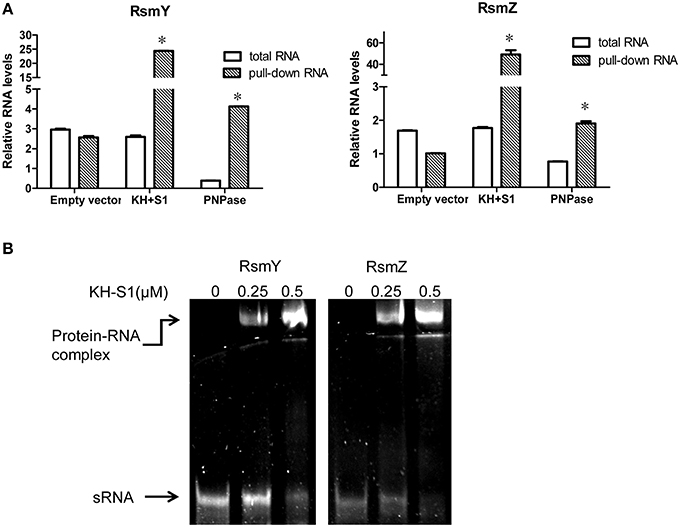
Figure 6. PNPase directly binds to RsmY/Z through the KH-S1 domain. (A) Levels of RsmY/Z in bacterial cells or co-purified with His-tagged PNPase or KH-S1 fragment. Expression of a His-tagged full length PNPase or KH-S1 fragment on plasmid pMMB67EH was induced by IPTG in wild type PAK carrying a gfp gene. The cellular levels of RsmY/Z were determined with real time PCR. The His-tagged proteins were purified by nickel affinity chromatography and associated RNAs were purified and analyzed with real time PCR. The gfp RNA level in each sample was used as an internal control. Each assay was done in triplicates, and the error bars indicate standard deviations. *p < 0.05 compared to total RNA by Student's t-test. (B) Formation of RNA-protein complexes in the presence of RsmY/Z and KH-S1-6His fragments. The RsmY, RsmZ RNAs were generated by in vitro transcription. RNA gel mobility shift assay was performed in a non-denaturing polyacrylamide gel. Positions of free RNA and RNA-protein complexes are indicated by arrows.
It has been reported that the RNA chaperone Hfq binds to and stabilizes RsmY (Sonnleitner et al., 2006; Sorger-Domenigg et al., 2007). To test whether the increased stability of RsmY is due to up regulation of Hfq, we examined the Hfq level by real time PCR. As shown in Figure S4, the mRNA levels of Hfq were the same between the wild type strain and the ΔKH-S1 mutant. Therefore, the increased stabilization of RsmY/Z in the ΔKH-S1 mutant is not due to higher level of Hfq.
PNPase Directly Binds to RsmY/Z
The RNA binding domains carried by PNPase and its role in controlling RsmY/Z suggest a possible interaction between PNPase and RsmY/Z. To test this possibility, we carried out a PNPase-RNA or KH-S1-RNA co-purification experiment. Expression of the C-terminus His-tagged full length PNPase or KH-S1 fragment on plasmid pMMB67EH was induced by IPTG in wild type PAK carrying a gfp gene. The His-tagged proteins were purified by nickel affinity chromatography and the associated RNAs were further isolated and analyzed by real time PCR. The levels of gfp RNA were used as reference to calculate the relative levels of RsmY/Z. If the relative level of RsmY or RsmZ is higher in the nickel affinity chromatography purified sample than that purified from bacterial cells, we consider there is enrichment, which indicates interaction between the protein and sRNA. As shown in Figure 6A, RsmY/Z were enriched with the purified His-tagged PNPase or KH-S1 fragment, although induction of full length PNPase reduced the cellular levels of RsmY/Z. Meanwhile, no enrichment of sRNA was observed in the empty vector control, suggesting a specific interaction between the KH-S1 fragment and RsmY/Z.
To test whether the co-purification of RsmY/Z and the KH-S1 domain is due to direct interaction, we performed a RNA gel mobility shift assay. In vitro transcribed RsmY/Z were incubated with purified His tagged KH-S1 fragment. As shown in Figure 6B, the KH-S1 fragment was able to retard both sRNAs, suggesting a direct interaction between the KH-S1 fragment and RsmY/Z. In combination, our results suggest that PNPase regulates the stability of RsmY/Z via direct PNPase–RNA binding.
Discussion
In this study, we studied the relationship between the KH-S1 domains of PNPase and the virulence factors as well as sRNAs in P. aeruginosa. In our experiments, we were unable to delete the full length pnp gene in wild type PAK even after multiple attempts. Ectopic expression of a pnp gene driven by a regulatable lac promoter enabled us to delete the pnp gene on the chromosome. In the presence of IPTG, the mutant grew as fast as the wild type strain. However, in the absence of IPTG, the strain grew more slowly (Figure S5). We suspect that the leaky expression of the pnp gene driven by the lac promoter might support the growth of the mutant. Therefore, it is likely that the RNase PH domains play an important role for the growth of P. aeruginosa under normal growth condition.
RNA metabolism and processing are essential for bacterial survival and response to environmental stresses. In E. coli, the expression of PNPase is up regulated upon cold shock (Jones et al., 1987; Cairrão et al., 2003). And PNPase is required for the growth at low temperature for a number of bacteria (Jones et al., 1987; Wang and Bechhofer, 1996; Goverde et al., 1998; Clements et al., 2002; Rosenzweig et al., 2007; Anderson and Dunman, 2009). We found that the P. aeruginosa ΔKH-S1 mutant was unable to grow at 16°C in LB medium (data not shown), suggesting a similar role of PNPase in P. aeruginosa.
Besides cold shock, PNPase is involved in the regulation of virulence factors in several pathogenic bacteria (Rosenzweig and Chopra, 2013). In Y. pseudotuberculosis and Y. pestis, it was shown that the Δpnp strain was less virulent (Rosenzweig et al., 2005, 2007). In S. enterica, PNPase promotes acute infection (Clements et al., 2002). Here in this study, we found that the KH-S1 domains of PNPase are required for the expression of acute infection associated virulence factors in P. aeruginosa, including T3SS and pili. Similarly, PNPase is required for the T3SS in Y. pseudotuberculosis and the S1 RNA binding domain of PNPase restored the T3SS activity (Rosenzweig et al., 2005, 2007). However, PNPase is required for the secretion rather than expression of the T3SS in Yersinia (Rosenzweig et al., 2005, 2007). In fact, the transcription of T3SS genes was at relatively higher level in the Δpnp strain (Rosenzweig et al., 2007). Similar to Yersinia, the expression T3SS genes were relatively higher in Δpnp mutant compared to the wild-type strain in S. enterica (Rosenzweig et al., 2007; Rosenzweig and Chopra, 2013). It seems that the roles of PNPase in the regulation of T3SS are different in Yersinia, Salmonella and P. aeruginosa, which might be due to different regulatory pathways or mechanisms.
Our transcriptomic analysis on the P. aeruginosa ΔKH-S1 mutant reveals multiple RNAs are under the influence of PNPase. Overexpression of exsA driven by a lac promoter restored the T3SS expression in the ΔKH-S1 mutant. On the chromosome, exsA was considered to be in the operon of exsCEBA (Hovey and Frank, 1995). However, a transcriptomic analysis revealed a gap between the RNAs of exsA and exsCEB, suggesting an independent promoter of exsA (Wurtzel et al., 2012). And a recent study demonstrated that an RNA helicases, DeaD is required for the translation of exsA (Intile et al., 2015). Based on these results, one possibility is that the exsCEBA transcript is subject to cleavage between exsB and exsA by endonuclease and PNPase, which is required for efficient translation of exsA. Other possible roles of PNPase on exsA expression might be that PNPase inhibits the translation of a negative regulator of exsA through its KH-S1 domains or PNPase degrades a sRNA that represses exsA expression.
In our study, we demonstrated that the levels of RsmY and RsmZ were increased in the ΔKH-S1 mutant, which was due to increased RNA stability. The degradation rates of RsmY and RsmZ in the ΔKH-S1 mutant were slower after rifampicin treatment. However, the RsmY/Z levels gradually dropped in the ΔKH-S1 mutant. These results suggest that PNPase might play an important role in the degradation process, while other RNAses are also involved in the degradation. The RNA EMSA demonstrated a direct binding between PNPase and RsmY/Z. Since structured RNAs are poor substrates of PNPase, we suspect that PNPase might bind to target RNAs, such as RsmY/Z through its KH-S1 domains. And RNA helicases are recruited to facilitate the degradation. It could be possible that under different environmental conditions, PNPase binds to different RNA helicases.
Overall, our results suggest that PNPase is involved in the regulation of multiple phenotypes, and an important mechanism is through the regulation of RsmY/Z stability. It would be interesting to screen the known sRNAs for PNPase targets. And the KH and S1 fragment of PNPase could be a useful tool to identify novel sRNAs as well as verify potential sRNAs. In addition, the mRNA targets of PNPase could also be identified by the KH-S1-RNA co-purification experiment.
Author Contributions
Conceived and designed the experiments: WW, SJ, RC, ZC. Performed the experiments: RC, YW, FZ, YJ, CL, XP, BX. Analyzed the data: RC, ZC, SJ, WW. Wrote the paper: RC, SJ, WW.
Funding
This work was supported by National Science Foundation of China (31370168 to WW, and 31170128, 31370167 to SJ); National Basic Research Program of China (973 Program, 2012CB518700 and 2015DFG32500 to SJ) and Science and Technology Committee of Tianjin (13JCYBJC36700 to WW, 15JCZDJC33000 to SJ).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00247
Table S1. Strains and plasmids used in this study.
Table S2. Primers used in real time PCR Strains and plasmids used in this study.
Table S3. Transcriptome analysis: differentially regulated genes.
Figure S1. (A) Constructs of NTD-His. (B) Expression of NTD-His in the indicated strains. Wild type PAK and the ΔKH-S1 mutant containing a NTD-His in their chromosomes were grown for 12 h in LB. Samples from equivalent numbers of bacterial cells were loaded onto a SDS-PAGE gel and probed with an anti-His antibody.
Figure S2. Secretion and expression of ExoS in indicated strains. Bacteria were grown to an OD600 of 1.0 in LB with or without 5 mM EGTA. Lysates from equivalent bacterial cells were loaded onto SDS-PAGE gels and probed with the ExoS antibody.
Figure S3. Relative RNA levels of retS, ladS gacS, and gacA in wild type PAK and the ΔKH-S1 mutant. Bacteria were grown to an OD600 of 1.0. Total RNAs were purified and the relative mRNA levels were determined with real time PCR. *p < 0.05 compared to wild type PAK by Student's t-test.
Figure S4. Levels of Hfq mRNA in wild type PAK and the ΔKH-S1 mutant. Bacteria were grown to an OD600 of 1.0. Total RNA from each strain was purified and analyzed with real time PCR.
Figure S5. Growth of indicated strains. The wild type PA14 containing pMMB67EH or the Δpnp mutant containing pMMB67EH-pnp was grown in the presence of 100 μM IPTG for 16 h. Each bacterial culture was diluted into fresh LB to an OD600 of 0.03. The bacteria were then grown in the absence of presence of 100 μM IPTG with agitation. The bacteria concentration was determined by serial dilution and plating every hour.
References
AJ, C. (2007). The RNA degradosome of Escherichia coli an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61, 71–87. doi: 10.1146/annurev.micro.61.080706.093440
Anderson, K. L., and Dunman, P. M. (2009). Messenger RNA turnover processes in Escherichia coli, Bacillus subtilis, and emerging studies in Staphylococcus aureus. Int. J. Microbiol. 2009:525491. doi: 10.1155/2009/52549
Andrade, J. M., Pobre, V., Matos, A. M., and Arraiano, C. M. (2012). The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18, 844–855. doi: 10.1261/rna.029413.111
Aubert, D. F., Flannagan, R. S., and Valvano, M. A. (2008). A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 76, 1979–1991. doi: 10.1128/IAI.01338-07
Bermúdez-Cruz, R. M., Ramírez, F., Kameyama-Kawabe, L., and Montañez, C. (2005). Conserved domains in polynucleotide phosphorylase among eubacteria. Biochimie 87, 737–745. doi: 10.1016/j.biochi.2005.03.005
Bordi, C., Lamy, M. C., Ventre, I., Termine, E., Hachani, A., Fillet, S., et al. (2010). Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 76, 1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x
Brencic, A., and Lory, S. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72, 612–632. doi: 10.1111/j.1365-2958.2009.06670.x
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Briani, F., Del Favero, M., Capizzuto, R., Consonni, C., Zangrossi, S., Greco, C., et al. (2007). Genetic analysis of polynucleotide phosphorylase structure and functions. Biochimie 89, 145–157. doi: 10.1016/j.biochi.2006.09.020
Cairrão, F., Cruz, A., Mori, H., and Arraiano, C. M. (2003). Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 50, 1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x
Carzaniga, T., Dehò, G., and Briani, F. (2015). RNase III-independent autogenous regulation of Escherichia coli polynucleotide phosphorylase via translational repression. J. Bacteriol. 197, 1931–1938. doi: 10.1128/JB.00105-15
Choi, K.-H., and Schweizer, H. P. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161. doi: 10.1038/nprot.2006.24
Clements, M. O., Eriksson, S., Thompson, A., Lucchini, S., Hinton, J. C., Normark, S., et al. (2002). Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A. 99, 8784–8789. doi: 10.1073/pnas.132047099
De Lay, N., and Gottesman, S. (2011). Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17, 1172–1189. doi: 10.1261/rna.2531211
Deretic, V., Schurr, M. J., and Yu, H. (1995). Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 3, 351–356. doi: 10.1016/S0966-842X(00)88974-X
Driscoll, J. A., Brody, S. L., and Kollef, M. H. (2007). The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368. doi: 10.2165/00003495-200767030-00003
Fernández-Ramírez, F., Bermúdez-Cruz, R. M., and Montañez, C. (2010). Nucleic acid and protein factors involved in Escherichia coli polynucleotide phosphorylase function on RNA. Biochimie 92, 445–454. doi: 10.1016/j.biochi.2010.01.004
Goverde, R. L., Huis in't Veld, J. H., Kusters, J. G., and Mooi, F. R. (1998). The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5 C). Mol. Microbiol. 28, 555–569. doi: 10.1046/j.1365-2958.1998.00816.x
Guarneros, G., and Portier, C. (1991). Different specificities of ribonuclease II and polynucleotide phosphorylase in 3′ mRNA decay. Biochimie 73, 543–549. doi: 10.1016/0300-9084(91)90021-R
Haddad, N., Burns, C. M., Bolla, J. M., Prévost, H., Fédérighi, M., Drider, D., et al. (2009). Long-term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity. Appl. Environ. Microbiol. 75, 7310–7318. doi: 10.1128/AEM.01366-09
Hauser, A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665. doi: 10.1038/nrmicro2199
Hovey, A. K., and Frank, D. W. (1995). Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 177, 4427–4436.
Howell, H. A., Logan, L. K., and Hauser, A. R. (2013). Type III secretion of ExoU is critical during early Pseudomonas aeruginosa pneumonia. MBio 4, e00032–e00013. doi: 10.1128/mbio.00032-13
Hueck, C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433.
Intile, P. J., Balzer, G. J., Wolfgang, M. C., and Yahr, T. L. (2015). The RNA helicase dead stimulates exsa translation to promote expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 197, 2664–2674. doi: 10.1128/JB.00231-15
Irie, Y., Starkey, M., Edwards, A. N., Wozniak, D. J., Romeo, T., and Parsek, M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172. doi: 10.1111/j.1365-2958.2010.07320.x
Jarrige, A.-C., Bréchemier-Baey, D., Mathy, N., Duché, O., and Portier, C. (2002). Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J. Mol. Biol. 321, 397–409. doi: 10.1016/S0022-2836(02)00645-9
Jarrige, A. C., Mathy, N., and Portier, C. (2001). PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 20, 6845–6855. doi: 10.1093/emboj/20.23.6845
Jones, P. G., VanBogelen, R. A., and Neidhardt, F. C. (1987). Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 169, 2092–2095.
Khajanchi, B. K., Sha, J., Kozlova, E. V., Erova, T. E., Suarez, G., Sierra, J. C., et al. (2009). N-acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology 155, 3518–3531. doi: 10.1099/mic.0.031575-0
Khemici, V., Toesca, I., Poljak, L., Vanzo, N. F., and Carpousis, A. J. (2004). The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 54, 1422–1430. doi: 10.1111/j.1365-2958.2004.04361.x
Li, K., Xu, C., Jin, Y., Sun, Z., Liu, C., Shi, J., et al. (2013). SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. MBio 4, e00419–e00413. doi: 10.1128/mbio.00419-13
MacIntyre, D. L., Miyata, S. T., Kitaoka, M., and Pukatzki, S. (2010). The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U.S.A. 107, 19520–19524. doi: 10.1073/pnas.1012931107
Miczak, A., Kaberdin, V. R. C., Wei, L., and Lin-Chao, S. (1996). Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U.S.A. 93, 3865–3869. doi: 10.1073/pnas.93.9.3865
Mulcahy, H., O'Callaghan, J., O'Grady, E. P., Maciá, M. D., Borrell, N., GÂĺ®mez, C., et al. (2008). Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect. Immun. 76, 632–638. doi: 10.1128/IAI.01132-07
Neuhaus, K., Rapposch, S., Francis, K. P., and Scherer, S. (2000). Restart of exponential growth of cold-shocked Yersinia enterocolitica occurs after down-regulation ofcspA1/A2 mRNA. J. Bacteriol. 182, 3285–3288. doi: 10.1128/JB.182.11.3285-3288.2000
Nurmohamed, S., Vaidialingam, B., Callaghan, A. J., and Luisi, B. F. (2009). Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J. Mol. Biol. 389, 17–33. doi: 10.1016/j.jmb.2009.03.051
Polissi, A., De Laurentis, W., Zangrossi, S., Briani, F., Longhi, V., Pesole, G., et al. (2003). Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154, 573–580. doi: 10.1016/S0923-2508(03)00167-0
Robert-Le Meur, M., and Portier, C. (1994). Polynucleotide phosphorylase of Escherichia coli induces the degradation of its RNase III processed messenger by preventing its translation. Nucleic Acids Res. 22, 397–403. doi: 10.1093/nar/22.3.397
Rosenzweig, J. A., and Chopra, A. K. (2013). The exoribonuclease Polynucleotide Phosphorylase influences the virulence and stress responses of yersiniae and many other pathogens. Front. Cell. Infect. Microbiol. 3:81. doi: 10.3389/fcimb.2013.00081
Rosenzweig, J. A., Chromy, B., Echeverry, A., Yang, J., Adkins, B., Plano, G. V., et al. (2007). Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol. Lett. 270, 255–264. doi: 10.1111/j.1574-6968.2007.00689.x
Rosenzweig, J. A., and Schesser, K. (2007). Polynucleotide phosphorylase and the T3SS. Adv. Exp. Med. Biol. 603, 217–224. doi: 10.1007/978-0-387-72124-8_19
Rosenzweig, J. A., Weltman, G., Plano, G. V., and Schesser, K. (2005). Modulation of Yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280, 156–163. doi: 10.1074/jbc.M405662200
Roy-Burman, A., Savel, R. H., Racine, S., Swanson, B. L., Revadigar, N. S., Fujimoto, J., et al. (2001). Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183, 1767–1774. doi: 10.1086/320737
Russell, A. B., Hood, R. D., Bui, N. K., LeRoux, M., Vollmer, W., and Mougous, J. D. (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. doi: 10.1038/nature10244
Sadikot, R. T., Blackwell, T. S., Christman, J. W., and Prince, A. S. (2005). Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171, 1209–1223. doi: 10.1164/rccm.200408-1044SO
Schubert, M., Edge, R. E., Lario, P., Cook, M. A., Strynadka, N. C., Mackie, G. A., et al. (2004). Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J. Mol. Biol. 341, 37–54. doi: 10.1016/j.jmb.2004.05.061
Sheahan, K.-L., and Isberg, R. R. (2015). Identification of mammalian proteins that collaborate with type III secretion system function: involvement of a chemokine receptor in supporting translocon activity. MBio 6, e02023–e02014. doi: 10.1128/mbio.02023-14
Sonnleitner, E., Schuster, M. T., Sorger-Domenigg, Greenberg, E. P., and Bläsi, U. (2006). Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59, 1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x
Sorger-Domenigg, T., Sonnleitner, E., Kaberdin, V. R., and Bläsi, U. (2007). Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 352, 769–773. doi: 10.1016/j.bbrc.2006.11.084
Stover, C., Pham, X., Erwin, A., Mizoguchi, S., Warrener, P., Hickey, M., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Sun, Z., Shi, J., Liu, C., Jin, Y., Li, K., Chen, R., et al. (2014). PrtR homeostasis contributes to Pseudomonas aeruginosa pathogenesis and resistance against ciprofloxacin. Infect. Immun. 82, 1638–1647. doi: 10.1128/IAI.01388-13
Symmons, M. F., Jones, G. H., and Luisi, B. F. (2000). A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 8, 1215–1226. doi: 10.1016/S0969-2126(00)00521-9
Vanzo, N. F., Li, Y. S., Py, B., Blum, E., Higgins, C. F., Raynal, L. C., et al. (1998). Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12, 2770–2781. doi: 10.1101/gad.12.17.2770
Wang, W., and Bechhofer, D. H. (1996). Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 178, 2375–2382.
Wong, A. G., McBurney, K. L., Thompson, K. J., Stickney, L. M., and Mackie, G. A. (2013). S1 and KH domains of polynucleotide phosphorylase determine the efficiency of RNA binding and autoregulation. J. Bacteriol. 195, 2021–2031. doi: 10.1128/JB.00062-13
Keywords: PNPase, small RNA, gene regulation, pathogenesis, P. aeruginosa
Citation: Chen R, Weng Y, Zhu F, Jin Y, Liu C, Pan X, Xia B, Cheng Z, Jin S and Wu W (2016) Polynucleotide Phosphorylase Regulates Multiple Virulence Factors and the Stabilities of Small RNAs RsmY/Z in Pseudomonas aeruginosa. Front. Microbiol. 7:247. doi: 10.3389/fmicb.2016.00247
Received: 28 October 2015; Accepted: 15 February 2016;
Published: 02 March 2016.
Edited by:
Awdhesh Kalia, University of Texas MD Anderson Cancer Center, USAReviewed by:
Sascha Brunke, Hans-Knöll-Institut, GermanyParas Jain, Albert Einstein College of Medicine, USA
Copyright © 2016 Chen, Weng, Zhu, Jin, Liu, Pan, Xia, Cheng, Jin and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihui Cheng, zhihuicheng@nankai.edu.cn;
Shouguang Jin, sjin@ufl.edu;
Weihui Wu, wuweihui@nankai.edu.cn
 Ronghao Chen
Ronghao Chen Yuding Weng
Yuding Weng Feng Zhu
Feng Zhu Yongxin Jin
Yongxin Jin Chang Liu
Chang Liu Xiaolei Pan
Xiaolei Pan Bin Xia
Bin Xia Zhihui Cheng
Zhihui Cheng Shouguang Jin1,2*
Shouguang Jin1,2* Weihui Wu
Weihui Wu