- State Key Laboratory of Agro-Biotechnology, College of Biological Sciences, China Agricultural University, Beijing, China
Sld3/Treslin is an evolutionarily conserved protein essential for activation of DNA helicase Mcm2-7 and replication initiation in all eukaryotes. Nevertheless, it remains elusive how Sld3 is recruited to origins. Here, we have identified the direct physical association of Sld3 with Mcm2 and Mcm6 subunits in vitro, which is significantly enhanced by DDK in vivo. The Sld3-binding domain (SBD) is mapped to the N-termini of Mcm2 and Mcm6, both of them are essential for cell viability and enriched with the DDK phosphorylation sites. Glutamic acid substitution of four conserved positively charged residues of Sld3 (sld3-4E), near the Cdc45-binding region, interrupts its interaction with Mcm2/6 and causes cell death. By using a temperature-inducible degron (td), we show that deletion of Mcm6 SBD (mcm6ΔN122) abolishes not only Sld3 enrichment at early origins in G1 phase, but also subsequent recruitment of GINS and RPA during S phase. These findings elucidate the in vivo molecular details of the DDK-dependent Sld3-MCM association, which plays a crucial role in MCM helicase activation and origin unwinding.
Introduction
The initiation of eukaryotic chromosome replication is spatial-temporally controlled by the assembly and activation of DNA helicase mini-chromosome maintenance (MCM) composed by six paralogous subunits (Mcm2-7) in a multi-step fashion (Remus and Diffley, 2009; Masai et al., 2010; Vijayraghavan and Schwacha, 2012; Bell and Kaguni, 2013; Li and Araki, 2013; Tognetti et al., 2014).
From late M to G1 phase, two Mcm2-7 heterohexameric rings are loaded onto each origin as a double hexamer, which results in the assembly of the pre-replication complex (pre-RC) in yeast (Evrin et al., 2009; Remus et al., 2009; Duzdevich et al., 2015; Ticau et al., 2015). Mcm2-7 helicase remains inactive during this licensing step. When cells enter the S phase, the Mcm2-7 complex must undergo multiple sophisticated changes prior to executing the origin unwinding task. First, Sld3, Sld7 and Cdc45 are recruited to the pre-RC and assembled into the Cdc45-MCM-Sld3 (CMS) complex with the elevated levels of Dbf4-dependent kinase (DDK; Labib, 2010; Heller et al., 2011; Tanaka et al., 2011). Second, the S-phase cyclin-dependent kinases (S-CDKs) phosphorylate Sld3 and Sld2 to stimulate their interactions with Dbp11 (the scaffold of the pre-loading complex containing Sld2, GINS and DNA polymerase 𝜀), which leads to the assembly of the pre-initiative complex (pre-IC; Tanaka et al., 2007; Zegerman and Diffley, 2007; Muramatsu et al., 2010). Then, Sld3 is replaced by GINS (Go, Ichi, Nii and San, for Sld5, Psf1, Psf2, Psf3), eventually leading to the formation of the active holo-helicase Cdc45-MCM-GINS (CMG) complex (Kanemaki and Labib, 2006; Bruck and Kaplan, 2011; Tanaka et al., 2011; Ilves et al., 2012; Costa et al., 2014; Bruck and Kaplan, 2015b; Sheu et al., 2016).
Firstly discovered in budding yeast as a synthetic lethal mutant with dpb11-1, Sld3/Treslin is a conserved initiation factor from fungi to higher eukaryotes (Kamimura et al., 2001; Kumagai et al., 2010; Boos et al., 2011). The carboxy-terminus of Sld3 is phosphorylated by S-CDK and then binds to the amino-terminal BRCT repeats of Dpb11(Tanaka et al., 2007; Zegerman and Diffley, 2007), while its amino-terminus comprises a Cdc45-binding domain (CBD) as shown in the crystal structure of Sld3-Cdc45 complex from budding yeast (Itou et al., 2014). During the process of MCM helicase activation, the binding of Sld3-Cdc45 to pre-RC is a pivotal step (Kamimura et al., 2001; Kanemaki and Labib, 2006). The Mcm2-7 complex is thought to be the docking platform for Sld3-Cdc45 recruitment. Mcm2-7 phosphorylation by DDK is also presumed to be involved in Sld3-Cdc45 loading albeit with unknown mechanisms (Labib, 2010; Tanaka et al., 2011; Heller et al., 2011; Bruck and Kaplan, 2015a,b; Yeeles et al., 2015).
In this study, we report that Sld3 is recruited through binding directly to the N-termini of Mcm2 and Mcm6 subunits in vitro, which are enriched with the phosphorylation sites of DDK. Indeed, their interactions are pronouncedly enhanced by DDK in vivo. According to the crystal structure of Sld3, we are able to identify that four highly conserved positively charged residues of Sld3, near the Cdc45-binding region, are important for associating with Mcm2/6 and cell viability. By utilizing a temperature-inducible degron (td) strategy, we show that the interaction defective mutations in Mcm6 (mcm6Δ122) interrupt the origin association of Sld3, but not Mcm2-7 per se. The subsequent recruitment of GINS to Mcm2-7 becomes abolished, which in turn compromises the Mcm2-7 helicase-driven origin unwinding as evidenced by the RPA-ChIP analysis. These data provide the detailed mechanism by which Sld3 is recruited to the Mcm2-7 complex in a DDK-dependent manner, which defines an upstream step in helicase activation and replication initiation in eukaryotes.
Results and Discussion
Sld3 Interacts with Mcm2-7 in a DDK-Dependent Manner
To investigate the molecular mechanism of Sld3 recruitment and CMS assembly, we set out to characterize the protein–protein interactions mediated by Sld3. In the yeast two hybrid assay, Sld3 interacted with Mcm2 as previously reported (Figure 1A; Herrera et al., 2015). Meanwhile, we found that Sld3 also displayed robust interaction with Mcm6 subunit.
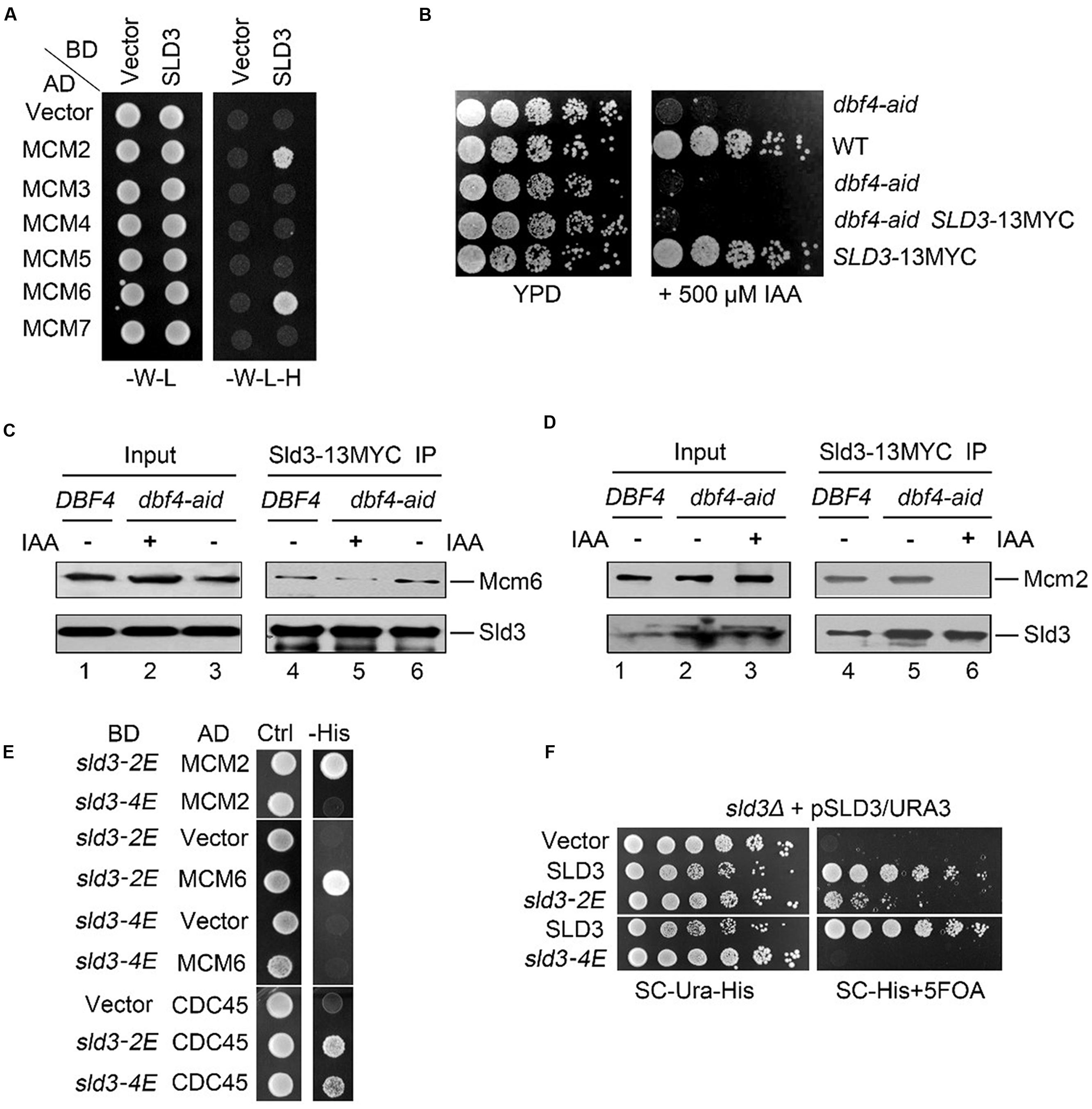
FIGURE 1. DDK-dependent Sld3–MCM interaction is essential for viability. (A) Sld3 displays robust interactions with Mcm2 and Mcm6 subunits in the yeast two hybrid assay. Yeast cells transformed with the indicated plasmids were grown at 30°C on either SC-W-L or SC-W-L-H plates as described in Experimental Procedures. (BD: Gal4 DNA binding domain fusion; AD: Gal4-activation domain fusion). (B) An auxin-inducible degron (aid) system to deplete the essential protein Dbf4 in yeast cells. The aid system was turned on by addition of 500 μM IAA. (C,D) The interaction between Sld3 and Mcm6 (C) or Mcm2 (D) is significantly impaired in the absence of Dbf4. Cells were cultured with or without IAA treatment for 2 h. Cell lysates were subjected to Sld3-13MYC immunoprecipitation. The precipitates were analyzed by western blot with the indicated antibodies. (E) Glu-substitution of a conserved basic patch at the N-terminus of Sld3 disrupts the Sld3–MCM interaction. (F) The MCM-interaction defective mutant sld3-4E fails to grow. Plasmid shuffling was carried out as described in Experimental Procedures. The strains expressing the indicated plasmid were plated on selective medium, or supplemented with 5-FOA, then grown for 2 days at 30°C.
Since DDK has been shown to be involved in Sld3 recruitment (Heller et al., 2011; Tanaka et al., 2011), we next tested whether DDK promotes Sld3–MCM interaction. To this end, we adopted an auxin-inducible degron (aid) method to deplete cellular Dbf4 proteins (Nishimura et al., 2009). When 500 μM indole-3-acetic acid (IAA) was supplemented in the medium, yeast cells carrying the carboxy-terminal aid tagged DBF4 (dbf4-aid) failed to grow, suggesting the efficient degradation of Dbf4 proteins which are essential for cell growth (Figure 1B). Meanwhile, SLD3 gene was tagged with 13MYC at the genomic locus, which did not alter the normal cell growth. Sld3-13MYC was immunoprecipitated with an anti-MYC antibody and detected by western blotting following SDS-PAGE. In the absence of IAA, Mcm6 was detected in the immunoprecipitate of Sld3-13MYC from dbf4-aid cells as well as from wild-type (WT) cells (Figure 1C, lanes 4 and 6). These results corroborate the physical association between Sld3 and Mcm6 in vivo. However, after IAA treatment for 2 h, the levels of Mcm6 co-precipitated with Sld3 were significantly decreased (Figure 1C, compare lane 5 to 6). In an independent experiment, co-precipitation of Mcm2 with Sld3 was also abrogated by the auxin-induced Dbf4 depletion (Figure 1D, compare lane 6 to 5). These data indicate that Sld3-MCM association is facilitated by DDK, through catalyzing the phosphorylation of Mcm2-7 subunits as demonstrated in a very recent study (Deegan et al., 2016).
Sld3-MCM Interaction is Essential for Cell Growth
Sld3 does not bear any apparent known phosphopeptide-binding motifs. Interestingly, it comprises two conserved positively charged regions close to each other as revealed by the crystal structure (Itou et al., 2014). One basic patch (Sld3-BP1, a.a. 301–330) mediates the interaction with an acidic region of Cdc45. Interestingly, the second basic patch (Sld3-BP2, K181, R186, K188, R192, K404, K405) is not involved in Cdc45 binding, but is also important for cell growth albeit with unknown function (Itou et al., 2014). We hypothesized that Sld3-BP2 might be involved in binding to phosphorylated MCM subunits. To test this, we first constructed the Glu-substitution mutants of the positively charged residues within Sld3-BP2. In the yeast two hybrid assay, the interaction with either Cdc45 or Mcm2/6 was not dramatically affected in the Sld3-2E(K188E, R192E) mutants (Figure 1E). Sld3-4E (K188E, R192E, K404E, K405E) also retained positive interaction with Cdc45 (Figure 1E, lower panel), which is consistent with the previous co-purification results (Itou et al., 2014). In contrast, Sld3-4E completely lost the interactions with both Mcm2 and Mcm6 (Figure 1E). These results indicate that the DDK-dependent Sld3–MCM interaction is mainly mediated by the positively charged Sld3-BP2 close to the Cdc45 binding interface, which might be an evolutionarily conserved event given that these basic residues are highly conserved between Sld3 and Treslin.
We next examined the physiological role of Sld3–MCM interaction in yeast. Since SLD3 is essential for cell growth, we constructed the sld3 mutants via plasmid shuffling. Briefly, WT SLD3 was introduced by a plasmid with URA3 selective marker. The genomic copy of SLD3 was then knocked out. The sld3 mutants were expressed from a HIS3 plasmid. The URA3 plasmid can be counter-selected on a 5-fluoro-orotic acid (5-FOA) plate. Therefore, cell growth on the 5-FOA plates reflects the physiological function of the remaining sld3 copy in the HIS3 plasmid. Strikingly, sld3-4E was not able to support cell growth, whereas sld3-2E showed moderate sick growth (Figure 1F), correlating with their ability to interact with MCM. These data suggest that Sld3–MCM interaction is essential for cell viability.
Sld3 Binds Directly with the N-Termini of Mcm2 and Mcm6
Then, we mapped the Sld3-binding domain (SBD) in Mcm2. Through construction of a series of Mcm2 truncations, we identified that a small region (a.a. 300–390) close to its N-terminus was required for interaction with Sld3 (Figure 2A). Moreover, the very N-terminal 299 amino acids were sufficient to bind Dbf4, which is consistent with a previous report that devoid of the N-terminal 63 amino acids in Mcm2 abolishes the interaction with Dbf4 (Ramer et al., 2013). These results indicate that Sld3 and Dbf4 interact with two adjacent regions within the Mcm2 amino terminus (Figure 2B). Similarly, when the N-terminal 122 amino acids were deleted, Mcm6 lost the interaction with Sld3. Meanwhile, the interactions of Mcm6 with its neighbor MCM subunits (Mcm2 or Mcm4) were not affected (Figures 2C,D), indicating that the SBD of Mcm6 is separable from the interface of the Mcm2-7 hexameric complex. When pull-down experiments were conducted with purified recombinant proteins, Sld3 was successfully detected together with both GST-Mcm2N (1–390) and GST-Mcm6N (1–439), indicating a direct physical association between Sld3 and Mcm2/6 N-termini (Figure 2E). Taken together, these data suggest that both N-termini of Mcm2 and Mcm6 mediate interaction with Sld3, which is enriched with the DDK phosphorylation sites (Randell et al., 2010; Sheu and Stillman, 2010). These results are in agreement with the notion that Sld3–MCM interaction can be facilitated by DDK as shown in Figure 1 and other studies (Heller et al., 2011; Tanaka et al., 2011; Deegan et al., 2016).
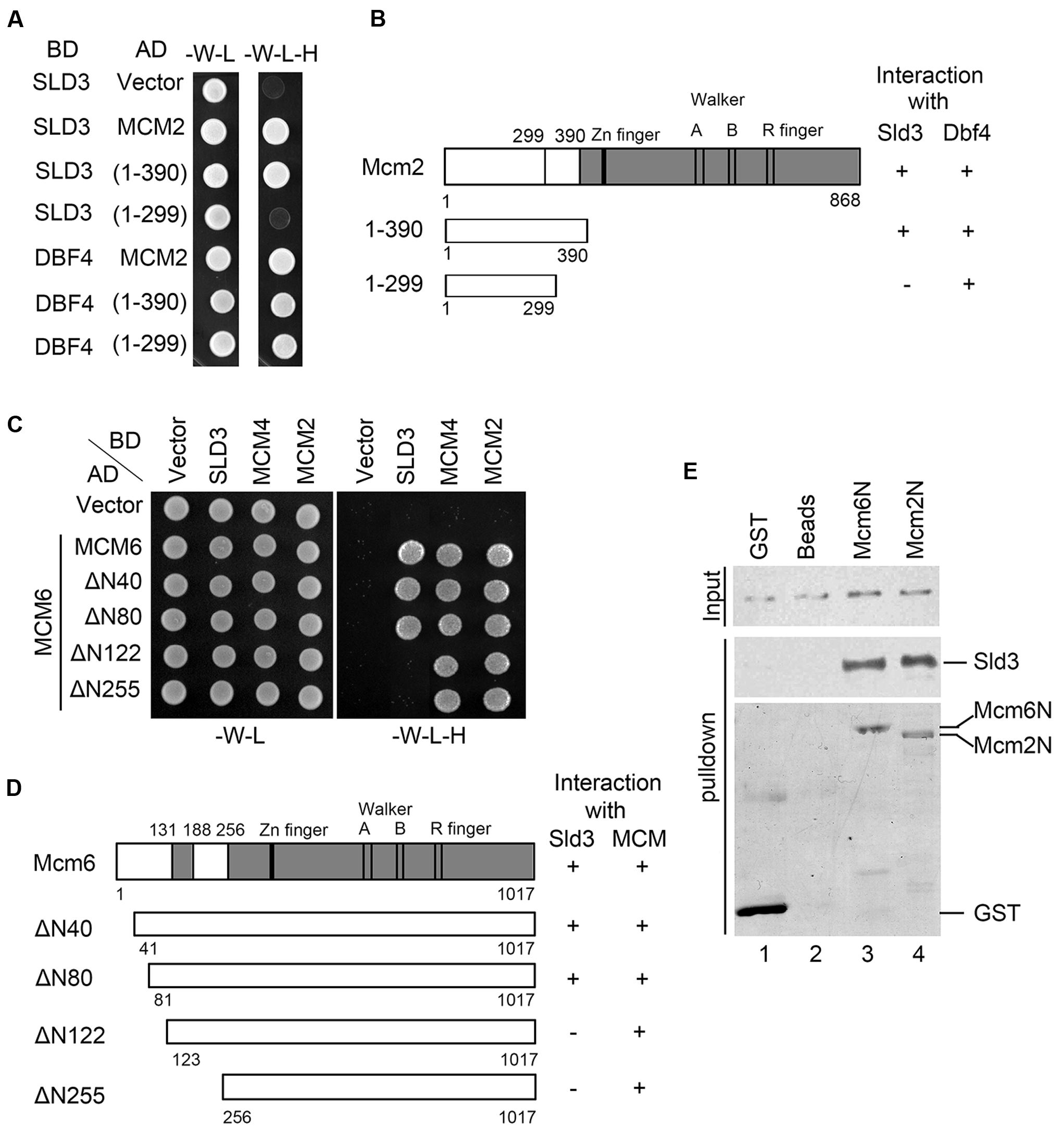
FIGURE 2. Sld3 binds directly to the N-terminal regions of Mcm2 and Mcm6. (A,B) Sld3 interacts with a short region near the Mcm2 N-terminus (300–390) in the yeast two hybrid assay as described in Figure 1A. (C) Mapping the domain of Mcm6 required for interaction with Sld3. Mcm6 truncations were constructed and tested in the yeast two hybrid assay as described above. (D) A summary of Mcm6 truncations and their ability to interact with Sld3 and other MCM subunits, respectively. (E) The N-termini of Mcm6 and Mcm2 are sufficient to bind directly to Sld3 in vitro. Purified recombinant GST-Mcm6 (1–439) or GST-Mcm2 (1–390) and 6His-Sld3 were incubated with glutathione Sepharose beads in the binding buffer containing 1 μg/μl BSA. The protein bands were detected by immunoblots with anti-His and anti-GST antibodies, respectively.
Mcm6 SBD is Indispensable for Replication Initiation
Next, we asked whether the Sld3 interaction defective mutations in Mcm2/6 affect normal cell growth. To this end, we adopted a temperature-inducible degron (td) to deplete the endogenous Mcm2 proteins (Kanemaki et al., 2003). The expression of ubiquitin ligase E3 Ubr1 for td-labeled protein degradation is under control of a galactose-inducible promoter. The separation-of-function mutations were introduced in a plasmid copy of MCM2. When cells were switched to the galactose plates incubated at 37°C, the endogenous td-Mcm2 proteins were degraded and resulted in cell death (Figure 3A). Strikingly, the lethality could be rescued by an extra copy of WT MCM2, but not an mcm2Δ(300–390) allele. Similarly, a Mcm6 mutant devoid of SBD, the N-terminal 122 amino acids (mcm6ΔN122), was not able to support yeast growth either (Figure 3B). These results are consistent with the phenotype of the interaction defective Sld3-4E mutants described in Figure 1F. Recently, Itou et al reported a hetero-tetrameric structure of Sld3-Sld7 (Itou et al., 2015), which provides one possible scenario that two Sld3 molecules bind to Mcm2 and Mcm6, respectively. Putting together, these data suggest that both Mcm2 and Mcm6 N-termini mediated interactions with Sld3 are essential requirements for cell viability.
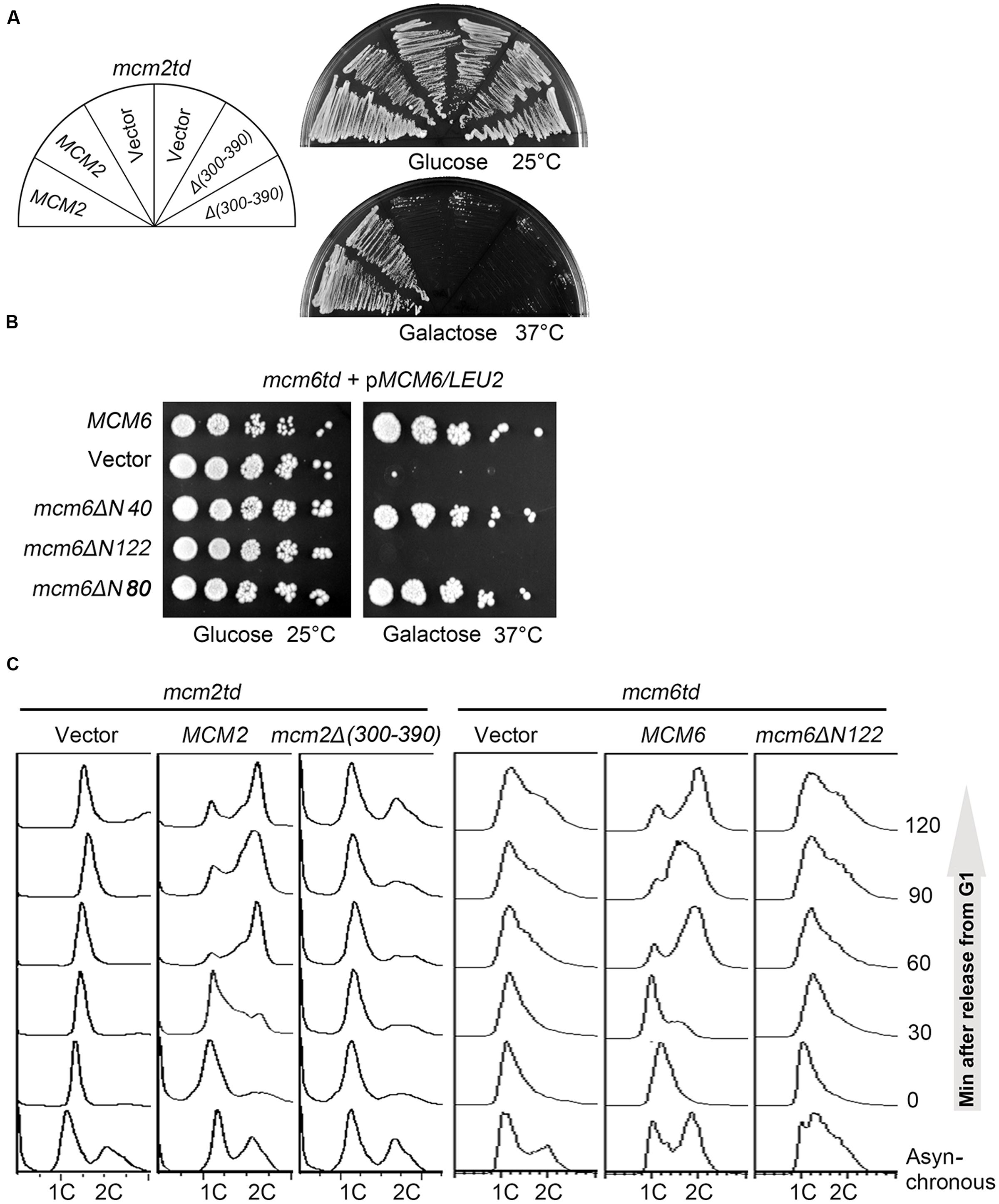
FIGURE 3. The SBDs of Mcm2 and Mcm6 are indispensable for replication initiation and cell viability. (A) Yeast cells are not able to grow in the absence of the Sld3 interaction motif of Mcm2. A plasmid expressing the indicated MCM2 alleles was introduced into the td-MCM2 background strain. The endogenous Mcm2 was depleted by a temperature-inducible degron at 37°C in the presence of galactose. (B) The N-terminal 122 amino acids deletion of Mcm6 causes cell death. 5-fold serial dilutions of td-MCM6 cells harboring the indicated plasmids were spotted on either glucose or galactose plates. The endogenous Mcm6 was depleted as described above. (C) Disruption of the Sld3 interaction regions of Mcm2 or Mcm6 compromises the S phase entry. Cells were released from α factor synchronization and analyzed for DNA content by flow cytometry.
Since both MCM and Sld3 play essential roles in DNA replication initiation. We then examined whether the lethality of the Sld3–MCM interaction defective mutant is due to the failure in DNA replication by flow cytometry. Notably, both mcm2Δ(300–390) and mcm6ΔN122 mutant cells were significantly compromised in S phase progression, thus implying that both Mcm2- and Mcm6- mediated interactions with Sld3 are crucial for replication initiation (Figure 3C).
Mcm6 SBD Participates in Sld3 Recruitment and Origin Unwinding
To investigate the exact role of Mcm6 N-terminus medicated interaction with Sld3, we carried out three sets of experiments. First, a chromatin immunoprecipitation (ChIP) assay was performed to evaluate the enrichment levels of Sld3 at early origins in G1-arrested cells. The endogenous Sld3 proteins carrying a 13MYC tag were immunoprecipitated by an anti-MYC antibody from the formaldehyde cross-linked cell lysates. The co-immunoprecipitated DNA was analyzed by quantitative PCR (ChIP-qPCR). A significant amount of ARS607 DNA was detected in WT cells, but not in mcm6ΔN122 mutant (Figure 4A). Meanwhile, the origin localization of Mcm6 itself was not changed after deletion of Mcm6 SBD (Figure 4B). These results indicate that the origin recruitment of Sld3 is inhibited when Mcm6 loses Sld3 binding capacity.
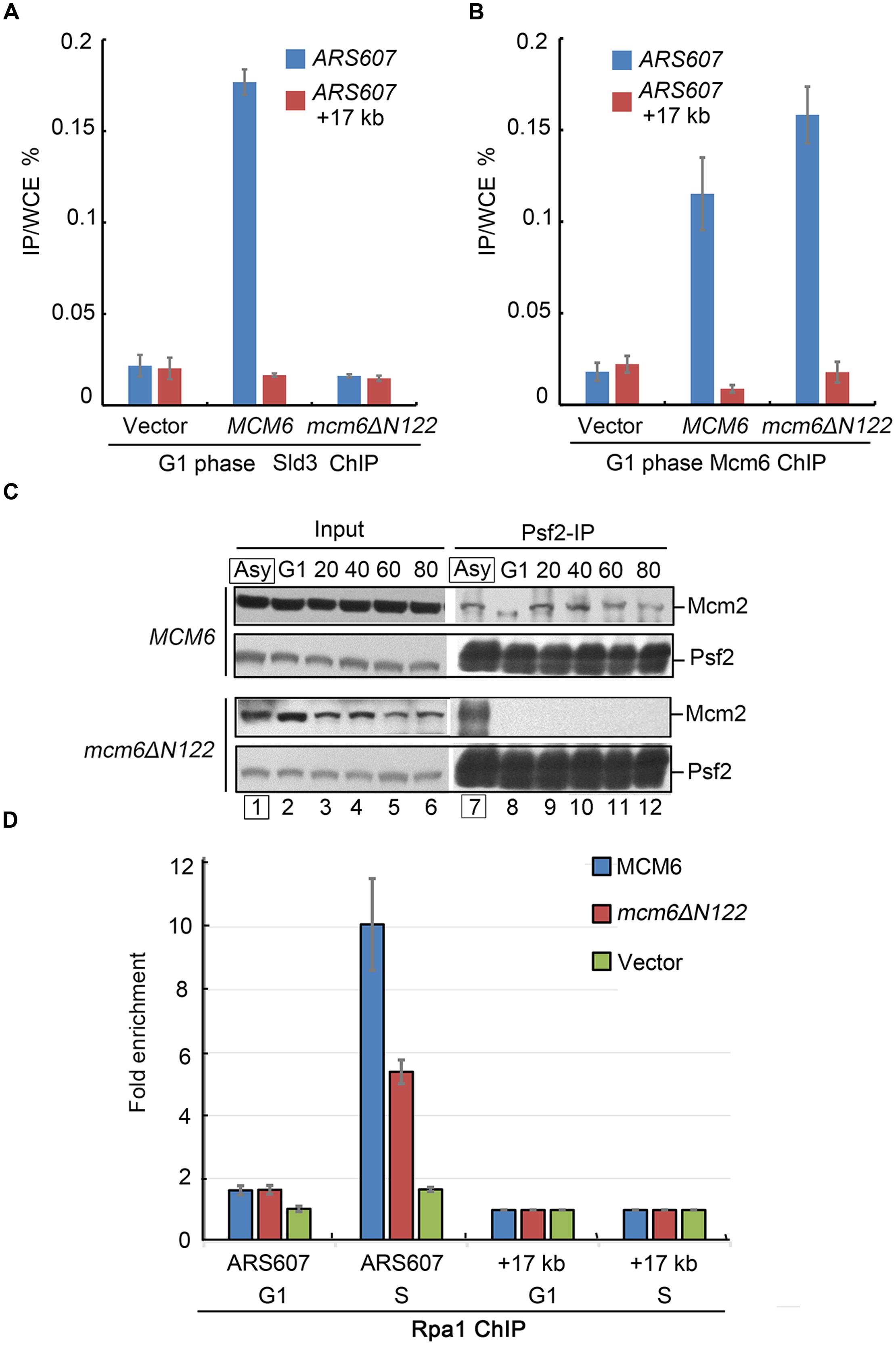
FIGURE 4. Sld3 recruitment and GINS–MCM interaction are compromised in mcm6ΔN122. (A) Sld3 recruitment at early origin ARS607 is abrogated in mcm6ΔN122. Sld3-13MYC ChIP-qPCR was conducted as described in Experimental Procedures. A non-origin region, 17 kb downstream of ARS607, is detected as a negative control. (B) Enrichment of Mcm6 at ARS607 is not affected in mcm6ΔN122. Anti-Mcm6 ChIP was conducted basically as described above. (C) Interaction between Psf2 (a GINS subunit) and Mcm2 is abolished in the absence of the Sld3 binding module of Mcm6. After cultured to OD600 0.3 at 23°C, yeast cells were arrested by α-factor in G1 at 23°C for 3 h and released into galactose media with α-factor at 37°C for 1 h, then released into fresh galactose media for the indicated times (S phase). The endogenous td-Mcm6 proteins were depleted when cells were shifted to 37°C during synchronization, but not in the asynchronized samples (Asy in the box). Cell lysates were subjected to Psf2 immunoprecipitation and analyzed by western blot. (D) The origin association of Rfa1 becomes compromised in the interaction defective mutant mcm6ΔN122. Cells were arrested in G1 and the endogenous td-Mcm6 proteins were depleted as above. After released into fresh galactose media for another 20 min (early S phase), cells expressing WT MCM6 or mcm6ΔN122 were withdrawn from the culture and subjected to Rfa1-13MYC ChIP-qPCR as described in (A).
The recruitment of Sld3 to pre-RC has been shown to be required for subsequent GINS recruitment (Kanemaki and Labib, 2006; Bruck and Kaplan, 2015a,b). Therefore, Psf2, a subunit of GINS complex, was examined for its association with MCM during the cell cycle. G1-arrested cells were harvested every 20 min after release into the S phase. Cell lysates were subjected to anti-Psf2 immunoprecipitation. In WT, the amounts of Mcm2 co-precipitated with Psf2 were gradually increased with the S phase progression (Figure 4C). Strikingly, there was barely detectable Mcm2 in the precipitate of mcm6ΔN122 (Figure 4C). On the contrary, if the endogenous Mcm6 was not depleted like in the asynchronized cells, Mcm2 was detected normally (lane 7). These results suggest a severe defect in the assembly of active CMG.
Third, we asked whether origin unwinding by MCM helicase depends on Mcm6 SBD. When duplex origin DNA is unwound by activated MCM helicase, the single-stranded DNA (ssDNA) is immediately protected by RPA. Therefore, RPA localization at origins can reflect the MCM-created ssDNA levels in vivo. The lysates were prepared from cells synchronized at G1 or early S phase (20 min after release from G1). Rfa1-3HA was precipitated from the lysates of G1 or S phase cells by an anti-HA antibody. Coprecipitated DNA was measured by qPCR. The endogenous Mcm6 was depleted by the td degron as described above. In the presence of WT MCM6, the relative enrichment of Rpa1 at early origin ARS607 substantially increased after release from G1 phase for 20 min (Figure 4D). Nevertheless, in mcm6ΔN122, the signals of Rpa1 at this origin were about 50% lower compared to the ones in WT, indicating a severe defect in the origin recruitment of RPA (Figure 4D). These results suggest that origin unwinding by MCM helicase activity is dependent on the Sld3–MCM interaction.
In this study, we provide the molecular details of the DDK-dependent recruitment of Sld3 to the MCM complex. These findings, together with the previous notion of Sld3-dependent Cdc45 and GINS loading (Kanemaki and Labib, 2006; Bruck and Kaplan, 2015b; Herrera et al., 2015; Deegan et al., 2016), shed new light on the assembly of the CMS complex, which defines a crucial transient step upstream of the recruitment of GINS and assembly of the active form of the replicative helicase, the CMG complex, for duplex DNA unwinding and replication initiation.
Experimental Procedures
Yeast Strains and Basic Methods
The yeast strains used in this study are derived from W303-1a (Table 1) and constructed basically as previously described (Lou et al., 2008). Plasmids constructed for this study are listed in Table 2 and validated by sequencing. Yeast two-hybrid, plasmid shuffling, cell synchronization and flow cytometry analysis were performed as described in (Quan et al., 2015).
Immunoprecipitation (IP)
Immunoprecipitation (IP) was carried out basically as described previously with some modifications (Lou et al., 2008). Briefly, yeast cultures were arrest at G1 phase with α-factor for 3 h. Cells (4 × 108) were crosslinked and collected and lysed at 4°C with glass beads (BeadBeater) in IP buffer [45 mM HEPES, pH 7.2, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.2% NP-40, 2 mM DTT, 1 mM PMSF, 1 × Protease Inhibitor Cocktail tablet (Roche), 1 × PhosSTOP tablet (Roche)]. Whole cell extract was mixed with 2 μl indicated antibodies and rotated for 3 h at 4°C, and then 20 μl of protein G beads were added and incubated for another hour. Beads were then washed three times with 1 ml IP buffer and boiled in 50 μl SDS-sample buffer. Western analysis was performed to detect specific proteins. Blots were probed with the indicated antibodies in phosphate-buffered saline containing 0.1% Tween and 2% dried milk. 9E10 (1:1000) was used to detect an MYC tag, M2 (1:1000) to detect Flag-tagged proteins, 12CA5 (1:1000) to detect an HA epitope. Polyclonal sera against Mcm2 (1:10000) and Mcm6 (1:10000) was used to detect corresponding proteins.
Protein Expression, Purification
All recombinant proteins were overexpressed in Escherichia coli BL21 (DE3) CodonPlus RIL (Stratagene). Cells were sonicated in lysis buffer containing 1% Triton X-100. 6His-Sld3 was purified by Ni2+ columns (GE Healthcare). The elution was concentrated to 3 mg/ml in the storage buffer 15 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM dithiothreitol, and 15% glycerol. GST-Mcm2, GST-Mcm6 and their derivatives were expressed and purified as previously described (Quan et al., 2015).
GST-Pull Down
For GST pull-downs, 2 μg 6His-Sld3 and 5 μg GST-Mcm6 (or GST-Mcm2) were incubated with glutathione-Sepharose beads in the presence of binding buffer (40 mM Tris-HCl, pH7.5, 100 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 1 mg/ml BSA, 1 mM PMSF, and protease inhibitors) for 1 h at 4°C. The glutathione agarose beads were washed extensively and bound proteins were separated on 8% SDS-PAGE gels. Blots were probed with monoclonal antibody against GST (1:1000) or 6His (1:1000).
Chromatin Immunoprecipitation (ChIP-qPCR)
ChIP experiments were performed with extracts of formaldehyde cross-linked cells, using 1.0 μg of purified anti-Sld3 or anti-Mcm6 antibodies (Tanaka et al., 2011). After The DNA fragments in precipitates were quantified by real-time PCR for each genomic locus. The ratio of immunoprecipitated DNA to total DNA in input was normalized and shown as fold enrichment (Natsume et al., 2013).
Author Contributions
HL and DF conceived and designed research. DF and QC performed experiments. HL analyzed the data and wrote the paper with inputs from all other authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Drs. Karim Labib and Toyoaki Natsume (University of Dundee) for sharing yeast strains. We thank members of the Lou lab for helpful discussion. This work was supported by National Natural Science Foundation of China 31271331 to HL; Research Fund for the Doctoral Program of Higher Education of China 20120008110017; Chinese Universities Scientific Fund 2015TC039 and 2014JD075.
References
Bell, S. P., and Kaguni, J. M. (2013). Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 5:a010124. doi: 10.1101/cshperspect.a010124
Boos, D., Sanchez-Pulido, L., Rappas, M., Pearl, L. H., Oliver, A. W., Ponting, C. P., et al. (2011). Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr. Biol. 21, 1152–1157. doi: 10.1016/j.cub.2011.05.057
Bruck, I., and Kaplan, D. L. (2011). GINS and Sld3 compete with one another for Mcm2-7 and Cdc45 binding. J. Biol. Chem. 286, 14157–14167. doi: 10.1074/jbc.M111.218305
Bruck, I., and Kaplan, D. L. (2015a). Conserved mechanism for coordinating replication fork helicase assembly with phosphorylation of the helicase. Proc. Natl. Acad. Sci. U.S.A. 112, 11223–11228. doi: 10.1073/pnas.1509608112
Bruck, I., and Kaplan, D. L. (2015b). The replication initiation protein Sld3/Treslin orchestrates the assembly of the replication fork helicase during S phase. J. Biol. Chem. 290, 27414–27424. doi: 10.1074/jbc.M115.688424
Costa, A., Renault, L., Swuec, P., Petojevic, T., Pesavento, J. J., Ilves, I., et al. (2014). DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife 3:e03273. doi: 10.7554/eLife.03273
Deegan, T. D., Yeeles, J. T., and Diffley, J. F. (2016). Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 35, 961–973. doi: 10.15252/embj.201593552
Duzdevich, D., Warner, M. D., Ticau, S., Ivica, N. A., Bell, S. P., and Greene, E. C. (2015). The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level. Mol. Cell 58, 483–494. doi: 10.1016/j.molcel.2015.03.017
Evrin, C., Clarke, P., Zech, J., Lurz, R., Sun, J., Uhle, S., et al. (2009). A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. U.S.A. 106, 20240–20245. doi: 10.1073/pnas.0911500106
Heller, R. C., Kang, S., Lam, W. M., Chen, S., Chan, C. S., and Bell, S. P. (2011). Eukaryotic origin-dependent dna replication in vitro reveals sequential action of ddk and s-cdk kinases. Cell 146, 80–91. doi: 10.1016/j.cell.2011.06.012
Herrera, M. C., Tognetti, S., Riera, A., Zech, J., Clarke, P., Fernandez-Cid, A., et al. (2015). A reconstituted system reveals how activating and inhibitory interactions control DDK dependent assembly of the eukaryotic replicative helicase. Nucleic Acids Res. 43, 10238–10250. doi: 10.1093/nar/gkv881
Ilves, I., Tamberg, N., and Botchan, M. R. (2012). Checkpoint kinase 2 (Chk2) inhibits the activity of the Cdc45/MCM2-7/GINS (CMG) replicative helicase complex. Proc. Natl. Acad. Sci. U.S.A. 109, 13163–13170. doi: 10.1073/pnas.1211525109
Itou, H., Muramatsu, S., Shirakihara, Y., and Araki, H. (2014). Crystal structure of the homology domain of the eukaryotic dna replication proteins sld3/treslin. Structure 22, 1341–1347. doi: 10.1016/j.str.2014.07.001
Itou, H., Shirakihara, Y., and Araki, H. (2015). The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3. Acta Crystallogr. D Biol. Crystallogr. 71, 1649–1656. doi: 10.1107/S1399004715010457
Kamimura, Y., Tak, Y. S., Sugino, A., and Araki, H. (2001). Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20, 2097–2107. doi: 10.1093/emboj/20.8.2097
Kanemaki, M., and Labib, K. (2006). Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 25, 1753–1763. doi: 10.1038/sj.emboj.7601063
Kanemaki, M., Sanchez-Diaz, A., Gambus, A., and Labib, K. (2003). Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423, 720–725. doi: 10.1038/nature01692
Kumagai, A., Shevchenko, A., Shevchenko, A., and Dunphy, W. G. (2010). Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140, 349–359. doi: 10.1016/j.cell.2009.12.049
Labib, K. (2010). How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24, 1208–1219. doi: 10.1101/gad.1933010
Li, Y., and Araki, H. (2013). Loading and activation of DNA replicative helicases: the key step of initiation of DNA replication. Genes Cells 18, 266–277. doi: 10.1111/gtc.12040
Lou, H., Komata, M., Katou, Y., Guan, Z., Reis, C. C., Budd, M., et al. (2008). Mrc1 and DNA Polymerase 𝜀 Function Together in Linking DNA Replication and the S Phase Checkpoint. Mol. Cell 32, 106–117. doi: 10.1016/j.molcel.2008.08.020
Masai, H., Matsumoto, S., You, Z., Yoshizawa-Sugata, N., and Oda, M. (2010). Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79, 89–130. doi: 10.1146/annurev.biochem.052308.103205
Muramatsu, S., Hirai, K., Tak, Y.-S., Kamimura, Y., and Araki, H. (2010). CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol 𝜀, and GINS in budding yeast. Genes Dev. 24, 602–612. doi: 10.1101/gad.1883410
Natsume, T., Muller, C. A., Katou, Y., Retkute, R., Gierlinski, M., Araki, H., et al. (2013). Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol. Cell 50, 661–674. doi: 10.1016/j.molcel.2013.05.011
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T., and Kanemaki, M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922. doi: 10.1038/nmeth.1401
Quan, Y., Xia, Y., Liu, L., Cui, J., Li, Z., Cao, Q., et al. (2015). Cell-cycle-regulated interaction between mcm10 and double hexameric mcm2-7 is required for helicase splitting and activation during S phase. Cell Rep. 13, 2576–2586. doi: 10.1016/j.celrep.2015.11.018
Ramer, M. D., Suman, E. S., Richter, H., Stanger, K., Spranger, M., Bieberstein, N., et al. (2013). Dbf4 and Cdc7 promote DNA replication through interactions with distinct Mcm2-7 subunits. J. Biol. Chem. 288, 14926–14935. doi: 10.1074/jbc.M112.392910
Randell, J. C., Fan, A., Chan, C., Francis, L. I., Heller, R. C., Galani, K., et al. (2010). Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol. Cell 40, 353–363. doi: 10.1016/j.molcel.2010.10.017
Remus, D., Beuron, F., Tolun, G., Griffith, J. D., Morris, E. P., and Diffley, J. F. (2009). Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–730. doi: 10.1016/j.cell.2009.10.015
Remus, D., and Diffley, J. F. (2009). Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21, 771–777. doi: 10.1016/j.ceb.2009.08.002
Sheu, Y.-J., Kinney, J. B., and Stillman, B. (2016). Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin activation and DNA replication fork progression. Genome Res. 26, 315–330. doi: 10.1101/gr.195248.115
Sheu, Y. J., and Stillman, B. (2010). The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463, 113–117. doi: 10.1038/nature08647
Tanaka, S., Nakato, R., Katou, Y., Shirahige, K., and Araki, H. (2011). Origin association of sld3, sld7, and cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 21, 2055–2063. doi: 10.1016/j.cub.2011.11.038
Tanaka, S., Umemori, T., Hirai, K., Muramatsu, S., Kamimura, Y., and Araki, H. (2007). CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445, 328–332. doi: 10.1038/nature05465
Ticau, S., Friedman, L. J., Ivica, N. A., Gelles, J., and Bell, S. P. (2015). Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 161, 513–525. doi: 10.1016/j.cell.2015.03.012
Tognetti, S., Riera, A., and Speck, C. (2014). Switch on the engine: how the eukaryotic replicative helicase MCM2–7 becomes activated. Chromosoma 124, 13–26. doi: 10.1007/s00412-014-0489-2
Vijayraghavan, S., and Schwacha, A. (2012). “The eukaryotic Mcm2-7 replicative helicase,” in The Eukaryotic Replisome: A Guide to Protein Structure and Function, ed. S. MacNeill (Dordrecht: Springer), 113–134.
Yeeles, J. T., Deegan, T. D., Janska, A., Early, A., and Diffley, J. F. (2015). Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519, 431–435. doi: 10.1038/nature14285
Keywords: budding yeast, DNA replication, Sld3/Treslin, DNA helicase, Mcm2-7
Citation: Fang D, Cao Q and Lou H (2016) Sld3-MCM Interaction Facilitated by Dbf4-Dependent Kinase Defines an Essential Step in Eukaryotic DNA Replication Initiation. Front. Microbiol. 7:885. doi: 10.3389/fmicb.2016.00885
Received: 13 April 2016; Accepted: 25 May 2016;
Published: 10 June 2016.
Edited by:
Vijai Kumar Gupta, NUI Galway, IrelandReviewed by:
Philippe Pasero, Centre National de la Recherche Scientifique, FranceBing Li, University of Texas Southwestern Medical Center, USA
Copyright © 2016 Fang, Cao and Lou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiqiang Lou, bG91QGNhdS5lZHUuY24=
 Dingqiang Fang
Dingqiang Fang Qinhong Cao
Qinhong Cao Huiqiang Lou
Huiqiang Lou
