- 1State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, China
Arbuscular mycorrhizal (AM) fungi are soil-borne fungi belonging to the ancient phylum Glomeromycota and are important symbionts of the arbuscular mycorrhiza, enhancing plant nutrient acquisition and resistance to various abiotic stresses. In contrast to their significant physiological implications, the molecular basis involved is poorly understood, largely due to their obligate biotrophism and complicated genetics. Here, we identify and characterize three genes termed Fm201, Ri14-3-3 and RiBMH2 that encode 14-3-3-like proteins in the AM fungi Funneliformis mosseae and Rhizophagus irregularis, respectively. The transcriptional levels of Fm201, Ri14-3-3 and RiBMH2 are strongly induced in the pre-symbiotic and symbiotic phases, including germinating spores, intraradical hyphae- and arbuscules-enriched roots. To functionally characterize the Fm201, Ri14-3-3 and RiBMH2 genes, we took advantage of a yeast heterologous system owing to the lack of AM fungal transformation systems. Our data suggest that all three genes can restore the lethal Saccharomyces cerevisiae bmh1 bmh2 double mutant on galactose-containing media. Importantly, yeast one-hybrid analysis suggests that the transcription factor RiMsn2 is able to recognize the STRE (CCCCT/AGGGG) element present in the promoter region of Fm201 gene. More importantly, Host-Induced Gene Silencing of both Ri14-3-3 and RiBMH2 in Rhizophagus irregularis impairs the arbuscule formation in AM symbiosis and inhibits the expression of symbiotic PT4 and MST2 genes from plant and fungal partners, respectively. We further subjected the AM fungus-Medicago truncatula association system to drought or salinity stress. Accordingly, the expression profiles in both mycorrhizal roots and extraradical hyphae reveal that these three 14-3-3-like genes are involved in response to drought or salinity stress. Collectively, our results provide new insights into molecular functions of the AM fungal 14-3-3 proteins in abiotic stress responses and arbuscule formation during AM symbiosis.
Introduction
Arbuscular mycorrhizal (AM) fungi, belonging to the ancient phylum Glomeromycota, are soil-borne microbes and capable of establishing the most widespread mutualistic association, namely AM symbiosis, with more than 80% terrestrial flowering plant species (Simon et al., 1993; Remy et al., 1994). Due to the obligate biotrophic nature, AM fungi need to consume plant photosynthates (Bago et al., 2000) and lipids to complete their life cycle (Bravo et al., 2017; Jiang et al., 2017), and reciprocally AM fungi significantly contribute to plant growth not only by enhancing mineral nutrient uptake and water acquisition from surrounding soil, but also protecting plants against fungal pathogens (Smith and Read, 2008; Jung et al., 2012; Chitarra et al., 2016) and a variety of abiotic stresses (Augé, 2001; Schützendübel and Polle, 2002; Lenoir et al., 2016). Therefore, AM fungi are key endosymbionts of the plant symbiosis and have significant impacts on plant productivity and ecosystem function (Van der Heijden et al., 1998), and are of great interest for the sustainable agricultural development (Gianinazzi et al., 2010).
The formation of a functional AM symbiosis requires successive stages between AM fungal and host symbionts at both physiological and molecular levels (Genre et al., 2005; Bonfante and Genre, 2010). Specifically, the development of arbuscular mycorrhiza consists of three major distinct stages through the progression of AM fungal hyphae during root colonization (Genre et al., 2005; Harrison, 2012; Gutjahr and Parniske, 2013). Arbuscules are generally thought to be the primary sites for nutrients exchange between the two symbionts (Parniske, 2008; Bonfante and Genre, 2010). In this symbiotic interface, the host membrane surrounding an arbuscule, known as the periarbuscular membrane (PAM), harbors AM-specific Pi transporters that acquire Pi released from the arbuscule (Harrison et al., 2002; Javot et al., 2007a). Outside the roots, the extraradical mycelia of AM fungi can extend the soil substratum beyond the depletion zone of the rhizosphere to uptake nutrients (particularly Pi and N) and water from the surrounding soils (Govindarajulu et al., 2005; Javot et al., 2007b; Li et al., 2013).
Despite their great importance, the underlying signaling events during initiation and formation of AM symbiosis are not well understood (Paszkowski, 2006; Bonfante and Requena, 2011; Gutjahr and Parniske, 2013; Oldroyd, 2013; Schmitz and Harrison, 2014; Bonfante and Genre, 2015). In contrast to a plethora of discoveries on morphological and chemical features in AM fungi, the molecular basis involved is still largely unknown, partially due to the limited available genomic resources. Many genome-wide gene expression analysis have been employed recently in order to understand the underlying molecular mechanisms of the AM formation. These studies mainly focused on the host plants (recently reviewed in Salvioli and Bonfante, 2013), whereas only a few investigations addressed the fungi partners (Requena et al., 2002; Breuninger and Requena, 2004; Cappellazzo et al., 2007; Kikuchi et al., 2014). Major progress has been recently achieved using transcriptomics and genomics data of Rhizophagus irregularis (Tisserant et al., 2012; Tisserant et al., 2013; Lin et al., 2014) and Gigaspora genus (Salvioli et al., 2016; Tang et al., 2016).
Using the suppression subtractive hybridization library (SSH) strategy, Breuninger and Requena (2004) firstly found some ESTs of fungal genes which were induced in the appressorium stage may display potential roles in this stage of Funneliformis mosseae. In this case, an EST tag termed 201, which encodes a 14-3-3 like protein in fungi, shows a significant up-regulation in the appressorium stage of AM symbiosis (Breuninger and Requena, 2004). Recently, Tisserant et al. (2012) released the first genome-wide overview of the transcriptional profiles of the various fungal tissues of R. irregularis. Particularly, a large number of fungal non-redundantly expressed transcripts was investigated in spores, intraradical mycelia (IRM), extraradical mycelia (ERM), and arbuscules. Interestingly, the transcripts encoding R. irregularis 14-3-3 proteins were inducible in both IRM and ERM.
14-3-3 proteins are highly conserved and dimeric proteins with a subunit mass of approximate 30 KDa (van Heusden and Steensma, 2006). These proteins are named based on the fraction number after EDTA-cellulose chromatography and the position after subsequent starch gel-electrophoresis (Moore, 1967). The first description of the function of 14-3-3 protein is substantially comparable to the ‘activator’ protein, that is important in the regulation of serotonin and noradrenaline biosynthesis in the brain (Ichimura et al., 1987). Moreover, 14-3-3 proteins form homo- or hetero-dimers by two subunits harboring the independent ligand-binding channels. Until now, it is extensively studied that these proteins generally serve as adapters, chaperones, activators, or repressors in the regulation of signal transduction pathways by reorganization of specific phosphoserine/phosphothreonine-inclusive binding motifs phosphorylated by protein kinase A (Smith et al., 1998; van Heusden, 2009; Smith et al., 2011; Parua and Young, 2014). Additionally, 14-3-3 proteins also play important roles in the pseudohyphal growth of Saccharomyces cerevisiae and the pathogenic fungal infection, such as Ustilago maydis (Gancedo, 2001; Rispail et al., 2009; Ballou et al., 2013; Liu et al., 2015). These known 14-3-3 proteins have also been implicated in several signaling cascades responding to biotic and abiotic stresses in plants (Roberts et al., 2002; Lozano-Duran and Robatzek, 2015; Li et al., 2016), suggesting that these proteins may display distinct roles during eukaryotes life cycle (Liu et al., 2015). So far, at least two distinct 14-3-3 subunits have been characterized in fungi (Darling et al., 2005; Hermeking and Benzinger, 2006). Porcel et al. (2006) identified a gene Gi14-3-3 (currently Ri14-3-3) from the AM fungus R. irregularis, encoding a 14-3-3 protein subunit that is enhanced under drought stress during AM symbiosis, being the first 14-3-3 protein from AM fungus reported so far. Additionally, recent work has provided new evidence for the potential involvement of Ri14-3-3 gene in the interaction between maize and R. irregularis under drought stress (Li et al., 2016). However, the molecular mechanisms of Ri14-3-3 gene in enhancing plant resistance to drought stress are still unclear.
To further advance our understanding of the roles of 14-3-3 proteins in fungal symbionts during AM symbiosis, we here report three novel fungal genes, so called Fm201, Ri14-3-3 and RiBMH2, which encode 14-3-3-like proteins from F. mosseae (BEG12) and R. irregularis (DAOM197198), respectively. 14-3-3 genes are strongly induced in the early stage of AM symbiosis. Moreover, the expression of 14-3-3 genes are regulated in response to drought and osmotic stresses. To further characterize these AM fungal 14-3-3 genes, we validated the capability of these genes to complement the metabolic deficient Δbmhs mutant (bmh1 and bmh2 double mutant) in a yeast heterologous expression system. We also provided insights into the regulatory mechanism between 14-3-3 protein and Msn2 transcription factor from AM fungi and further proved the existence of two distinct 14-3-3 subunits in AM fungi. More importantly, in the absence of stable transformation protocols for AM fungi (Helber and Requena, 2008; Helber et al., 2011), host-induced gene silencing (HIGS) of the two 14-3-3 genes in R. irregularis, whereby these genes are silenced in the AM fungal symbiont by expressing an RNA interference construct in the host, provides a potential tool to address the function of 14-3-3 proteins in obligate biotrophic AM fungi. Collectively, our results provide new insights into molecular functions of the AM fungal 14-3-3 proteins in stress responses and arbuscule formation during AM symbiosis.
Results
Identification of Fm201 Gene From Funneliformis mosseae
In the previous study, transcript abundance of 201-tag was significantly enhanced at the early appressorium stage of AM symbiosis (Breuninger and Requena, 2004). The amino acid sequence of 201-tag exhibits a high similarity (∼97%) with the Ri14-3-3 protein from R. irregularis (Breuninger and Requena, 2004; Tisserant et al., 2012). With the aim to confirm if this fungal 14-3-3 protein is involved in AM symbiosis, a DNA clone of 1.5 kb in length was obtained by inverse PCR from the cloning procedures on F. mosseae genomic DNA based on the 201-tag. The isolated fragment with 5′ end and upstream region was highly similar to the sequence of Ri14-3-3 gene from R. irregularis (Porcel et al., 2006). Since the 5′ and 3′ end sequences of this gene are not available, 5′ and 3′ RACE experiments on RNA pools of F. mosseae germinating spores were subsequently performed to obtain the full-length CDS sequence. A 1,401 bp full-length cDNA sequence of Fm201, covering the 5′UTR (188 bp) and 3′UTR (411 bp), was thus identified (Accession number: KM258580). The corresponding genomic sequence of Fm201 gene is 1,685 bp in length, containing seven exons and seven introns (Supplementary Figure S1). Interestingly, Fm201 gene contains a 100 nt intron in the 5′ UTR and two transcriptional variants of 3′UTRs (61 and 411 nt in length, respectively). These unusual features of AM fungal 14-3-3 gene firstly reported in the present study may suggest important roles in the regulation of Fm201 expression during AM symbiosis.
Fm201 Protein Is Conserved Among Eukaryotes
To further investigate the phylogenetic and structural features of the Fm201 protein from AM fungi, we exploited the phylogenetic placement and 3D structure of Fm201 protein using bioinformatics strategy. The in silico analysis revealed that the open reading frame (ORF) of Fm201 gene consists of 804 bp corresponding to 267 amino acids with a predicted molecular weight of approximate 30 kDa. A phylogeny of basal fungi and 14-3-3 proteins from Homo sapiens clearly supports Fm201 as a sister clade to Ri14-3-3 (Figure 1A), indicative of the conserved evolutionary origin of the 14-3-3 genes in AM fungi, whereas the RiBMH2 protein from R. irregularis belongs to the closer relative of the yeast BMH2 (Figure 1A). Compared to the 14-3-3 proteins from H. sapiens, Fm201 protein still shares a very high homology. This also demonstrates that 14-3-3 proteins are highly conserved in eukaryotes. As a conserved protein, Fm201 protein shares 97% similarities with Ri14-3-3 protein from AM fungi. The amino acid sequence of Fm201 was compared with BMHs from S. cerevisiae, 14-3-3s from R. irregularis and H. sapiens 14-3-3 epsilon and a high homology with over 72% identity at the amino acid level was observed (Supplementary Figure S2).
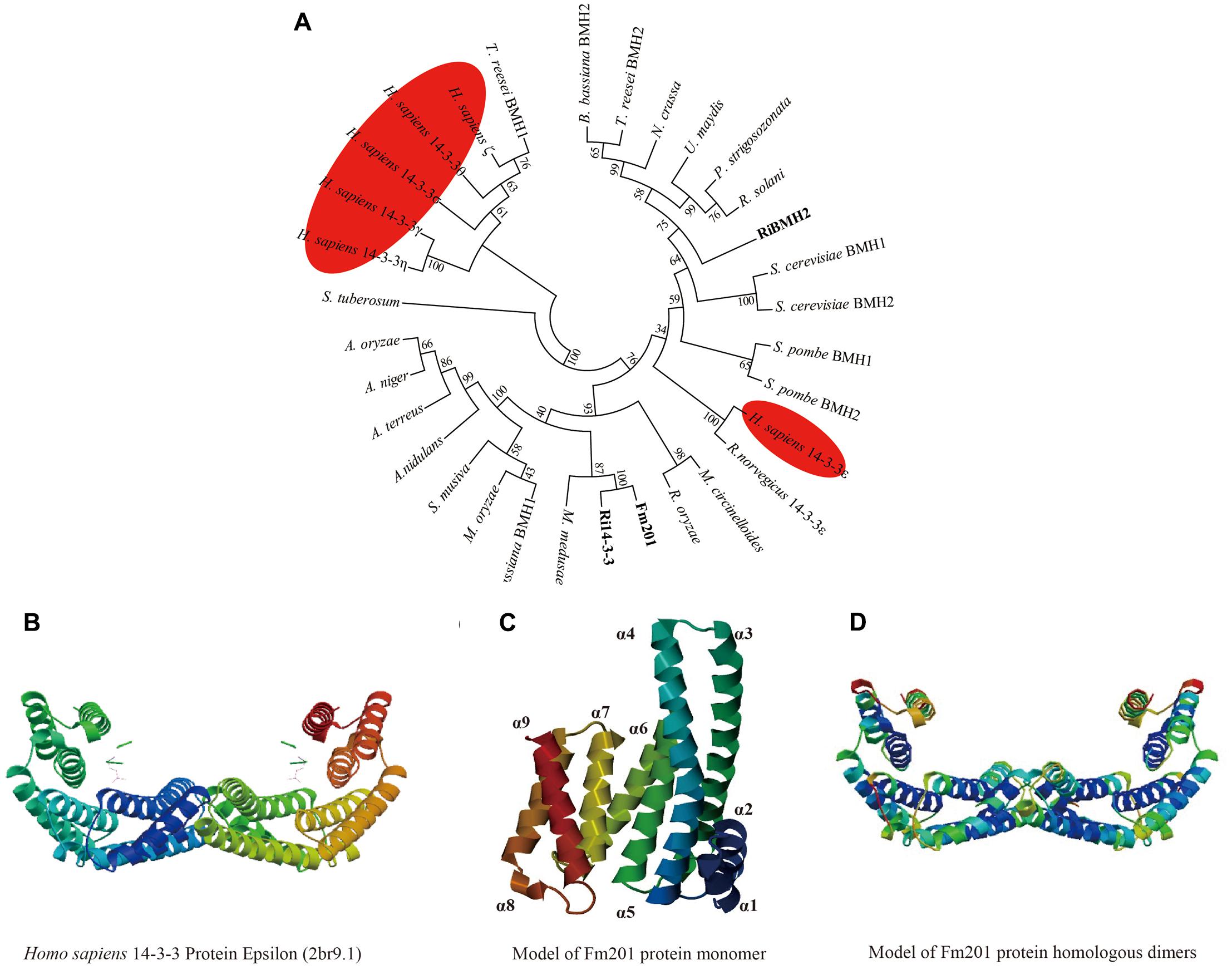
FIGURE 1. In silico analysis of Fm201 protein and its homologs from AM fungi. (A) The unrooted phylogenetic tree was constructed based on multiple sequence alignment of 14-3-3 proteins in fungal species and Homo sapiens. The phylogenetic relationships were analyzed by Neighbor-joining method with MEGA v6.0 software. Bootstrap values were calculated using 1,000 replicates. 14-3-3 proteins from mycorrhizal fungi are highlighted in bold and their homologs from H. sapiens are colored red. (B) Predicted structure of Homo sapiens 14-3-3 epsilon (2br9A) dimers. (C,D) Homology modeling of Fm201 monomer and homodimer using Homo sapiens 14-3-3 epsilon (2br9A) as reference.
We further carried out the Homology modeling using Homo sapiens 14-3-3 epsilon (80.52% identity in amino acid sequences) as a model. The predicted three-dimensional conformation of Fm201 indicates that Fm201 is a typical 14-3-3 protein with 9 alpha helices and 8 loops, with the highly homologous α3, α5, α7, and α9 putatively forming its amphipathic ligand-binding grooves (Figure 1C). Fm201 homologous dimers could form a typical C-shape cup, which provides a basic structure of 14-3-3 dimers for implementing its function (Figure 1D).
14-3-3 Proteins Restore Metabolic Activity of S. cerevisiae Δbmhs Mutant
To gain further insights into the function of 14-3-3, a S. cerevisiae heterologous expression system was exploited. Since Fm201 shares 82.3% identity at the amino acid sequence with both BMH1 and BMH2 in S. cerevisiae (see Supplementary Figure S2), the ORF of Fm201 was cloned into pMR-12 under the control of the Gal7 promoter and replaced S. cerevisiae BMH1. To test if Fm201 can restore the metabolic activity of S. cerevisiae Δbmhs mutant, as referred in Materials and Methods (also see Supplementary Table S1). S. cerevisiae Δbmhs mutant with pMR-12-Fm201 cannot grow on YPD with 2% glucose as the sole carbon source. However, cell growth was recovered when using 2% galactose as the sole carbon source (Figure 2). Similar results were observed when replacing Fm201 with Ri14-3-3 or RiBMH2 (Figure 2). These data confirmed that Fm201 has similar function as BMH1 in S. cerevisiae.
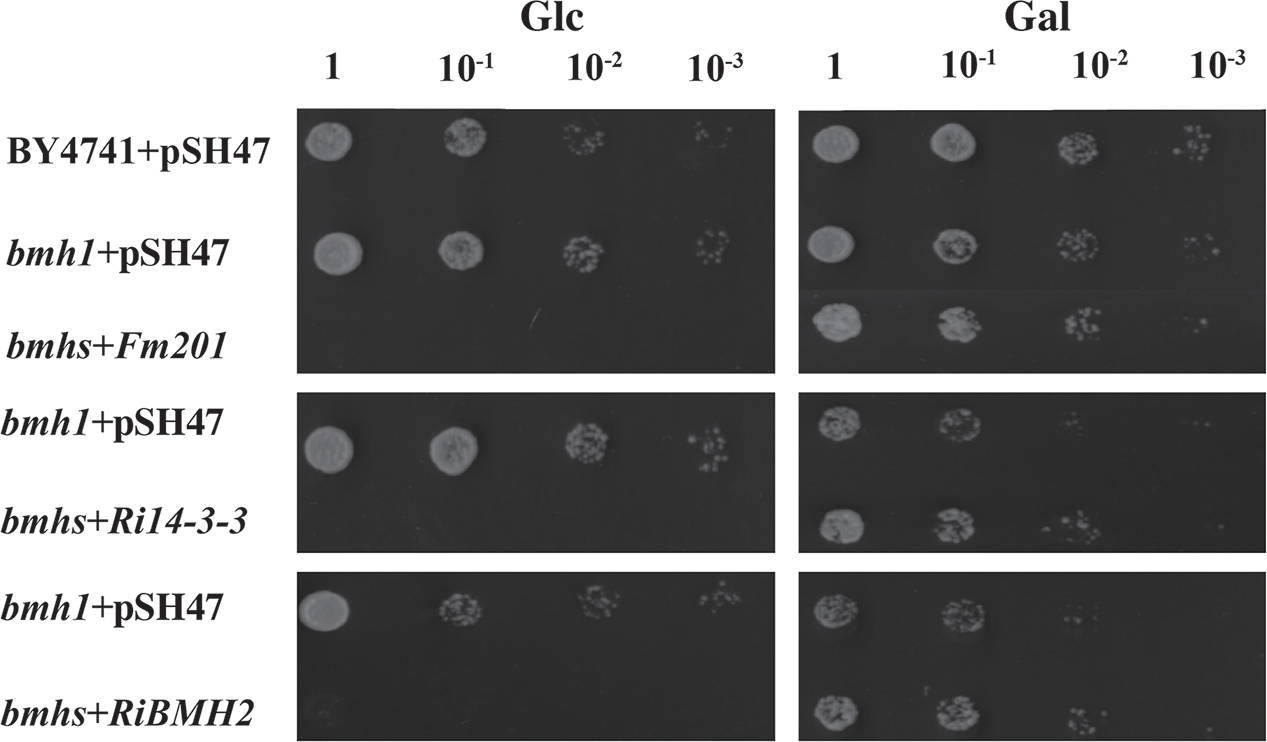
FIGURE 2. 14-3-3 proteins from mycorrhizal fungi restore the growth of S. cerevisiae Δbmhs mutant on galactose-contained media. Ten-fold serial dilutions of yeast cells (the wild-type BY4741, Δbmh1 mutant or Δbmhs mutant) carrying different constructs (the empty vector pSH47, the full-length cDNA of Fm201, Ri14-3-3, or RiBMH2) were grown on SD/-Ura plates with 2% glucose or galactose as carbon source. Expression of Fm201, Ri14-3-3, or RiBMH2 gene is controlled by galactose-inducible Gal7 promoter.
Functional Dissection of CREs on Promoter of Fm201 (pFm201) in S. cerevisiae
14-3-3 proteins have been reported to participate in pseudohyphal growth and resistance in yeast (Roberts et al., 1997; Hurtado and Rachubinski, 2002). However, the roles of these 14-3-3 proteins in mycorrhizal fungi are largely unknown. It has been observed that expression specificity of plant 14-3-3 genes in response to various stresses is largely promoter dependent (Aksamit et al., 2005). To analyze the putative CREs (Cis-Regulatory Elements) located in the promoter region of Fm201, a 1.5 kb length promoter sequence upstream of Fm201 coding region (pFm201) was analyzed via Yeastract database1. The CREs present in pFm201 were compared with pBMH1, pBMH2 from S. cerevisiae and pRiBMH2 in R. irregularis (van Heusden, 2009) (Table 1). Many common CREs in corresponding sites shared by pFm201 and pBMHs have been shown to be recognized by many transcriptional factors (Bruckmann et al., 2004; van Heusden, 2009). In the present study, two CREs possiblely recognized by Msn2 and STE12 were chosen for further investigations. Msn2 is an STRE element (AGGGG/CCCCT) binding transcription factor, which is supposed to be related to fungal infection and resistance to abiotic stress in other filamentous fungi (Schmitt and Mcentee, 1996; Seidl et al., 2004; Elfving et al., 2014; Zhang et al., 2014). STE12, so-called GintSTE, is the transcriptional factor that has been reported in mycorrhizal fungi and is believed to be an indispensable component in the early process of mycorrhizal fungi infection (Tollot et al., 2009; Tang et al., 2016). The common and shared CREs present upstream of Fm201, RiBMH2 and S. cerevisiae BMHs imply that 14-3-3 proteins in AM fungi may be involved in the regulation of resistance to abiotic stress and hyphal growth in AM fungi as BMHs in S. cerevisiae.
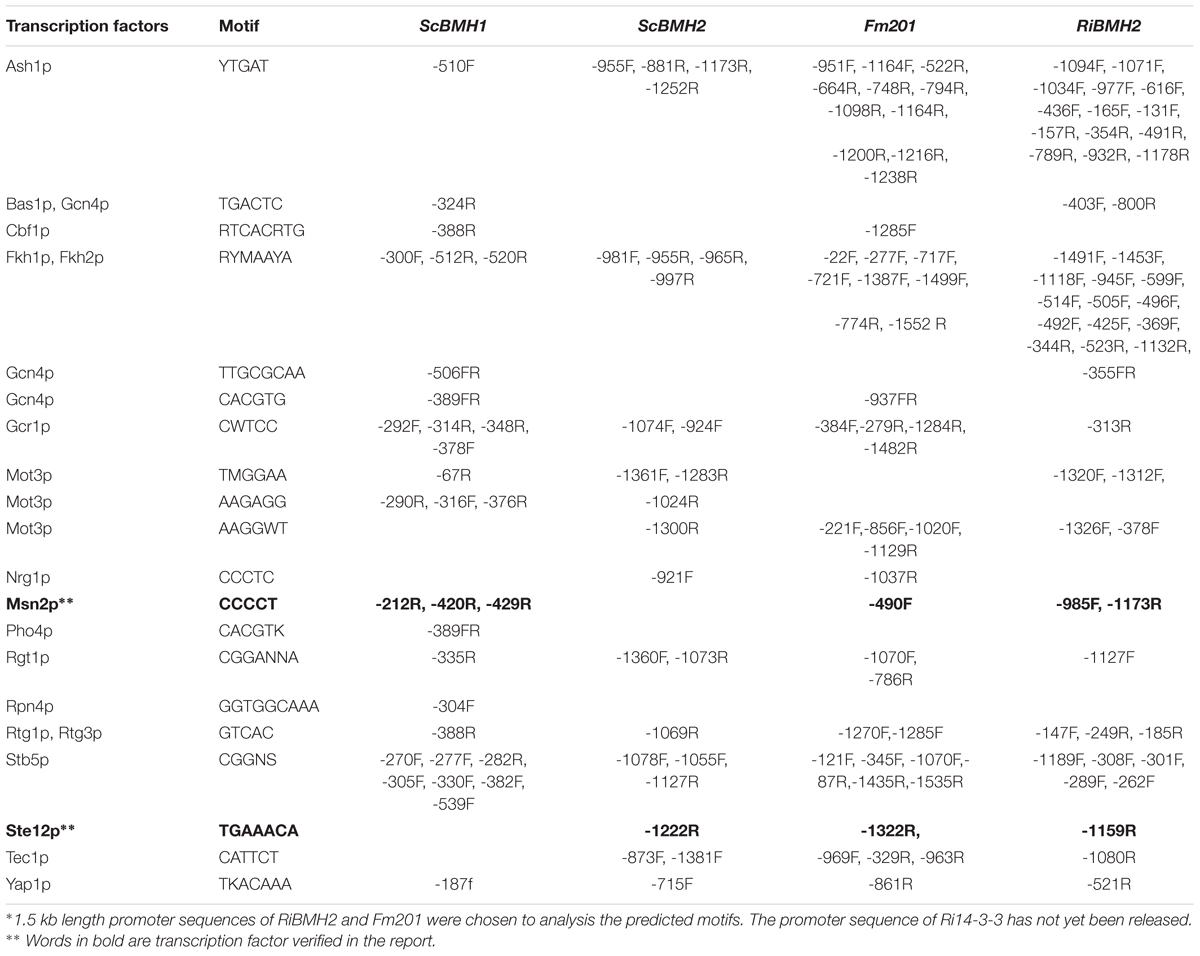
TABLE 1. Predicted motifs on the Fm201 gene promoter compared with that of RiBMH2, ScBMH1 and ScBMH2∗
Due to the lack of stable genetic transformation approaches in the AM fungi, it is technically challenging to knockout Fm201 gene to confirm the biological function in the early stages during symbiosis (Sanders, 1999; Maldonado-Mendoza et al., 2001). To probe the possible function of Fm201 protein, we employed the site-specific mutagenesis and yeast one-hybrid system to initially explore the essential region of Fm201 promoter. Compared to the site-specific mutagenesis of STRE (CCCCT/AGGGG) located in pFm201(pFm201-ΔSTRE), pFm201 is more sensitive to abiotic stresses, osmotic pressure and drought stress (Figure 3A). This result indicates that the STRE element of pFm201 could be recognized by Msn2 in S. cerevisiae. Although there is no any report, to our knowledge, about the functional properties of RiMsn2 factor mentioned as RiMsn4 in R. irregularis, it may play a major role in eukaryotic abiotic stress response and hyphae differentiation (Tisserant et al., 2013; Zhang et al., 2014).
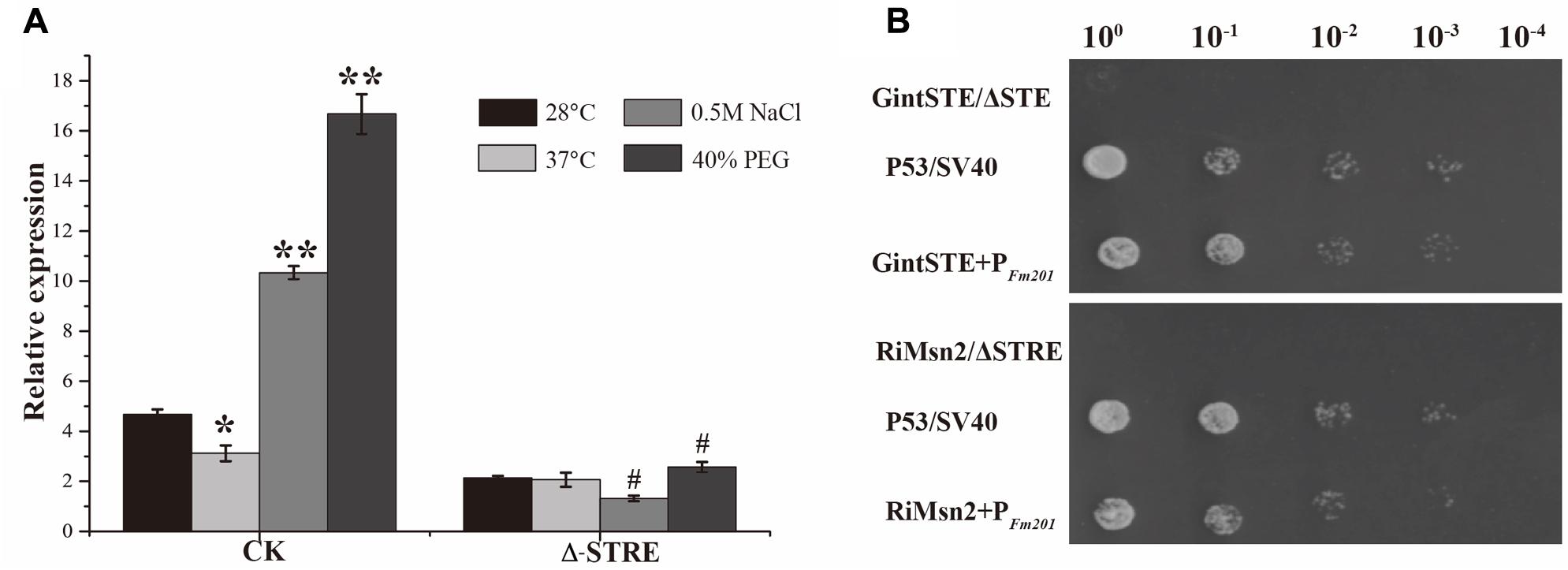
FIGURE 3. Promoter activity analysis of Fm201 gene in Yeast. (A) Real-time RT-PCR quantification of Fm201 gene expression in yeast under different abiotic stress treatments. CK indicates the full-length promoter sequence of Fm201 gene. Lines with a significant ratio to the express rate of each group in 28°C. Error bars indicate the means of three biological replicates with SD values. Data shown are averages ± SD; n = 3. (#, ∗p < 0.05, ∗∗p < 0.01). (B) Yeast one-hybrid analysis of the interaction between GintSTE or RiMsn2 with Fm201 promoter (pFm201). Yeast carrying both pGBKT7-P53 and pGADT7-SV40 was used as the positive control. pHis2- ΔSTRE with pGADT7-Rec2-RiMsn2 and pHis2- ΔSTE with pGADT7-Rec2-GintSTE were used as negative controls. Δ-STRE indicates the deletion of STRE elements located in pFm201 promoter, Δ-STE indicates the deletion of GintSTE binding sites located in pFm201 promoter. 10-fold serial dilutions of yeast cells were spotted on plates containing 2% Glc as carbon source.
A yeast one-hybrid system was also performed to confirm the interaction between GintSTE and RiMsn2 with CREs on pFm201 (Figure 3B). As shown in Figure 3B, yeast cells harboring pFm201 with STRE element and RiMsn2 or GintSTE protein grew well, whereas growth of the cells carrying RiMsn2 or GintSTE and Fm201 promoter without STRE element as negative controls were severly inhibited under the same conditions. These data suggest that GintSTE and RiMsn2 proteins interact with the STRE element of Fm201 gene, the similar results were also acquired from the promoter of RiBMH2 (data not shown). It is therefore reasonable to speculate that GintSTE and RiMsn2 proteins positively regulate the expression of 14-3-3 genes in presymbiotic stage through the binding between GintSTE/RiMsn2 and pFm201.
14-3-3 Genes Are Highly Induced in Germinating Spores and Early Stages of Symbiosis
Due to the obligate biotrophic and asexual multinucleate nature of the AM fungi (Sanders, 1999; Maldonado-Mendoza et al., 2001), it is difficult to generate mutants and overexpression strains to analyze the biological functions of Fm201 protein during AM fungal infection. To obtain further insights into the expression profile of Fm201 gene during the colonization process, we performed a time-course analysis of Medicago truncatula roots inoculated with F. mosseae in the pot system, then sampled at 12, 18, 25, and 50 days post-inoculation (dpi) and also collected the quiescent spores and germinated spores. Morphological analyses of mycorrhizal roots showed almost majority of the appressoria and intraradical hyphae at 12–18 dpi. More arbuscules were obviously detected starting from 25 dpi, while the abundance of arbuscules decreased at 50 dpi (Figure 4A), the mycorrhizal colonization of the root samples was also calculated as described by Trouvelot et al. (2015) (data not shown). The transcript abundance of Fm201 gene in F. mosseae at different stages were also analyzed by qRT-PCR. As shown in Figure 4B, the transcript abundance of Fm201 was obviously higher in germinating spores than in quiescent spores, the transcript abundance of Fm201 is ∼40% lower in 50 dpi than in 25 dpi when arbuscules decreased. This expression pattern was similar with fungal colonization in early stages of symbiosis, especially during the hyphopodium formation and root penetration (see Figures 4A,D). The similar results of RiBMH2 and Ri14-3-3 were also obtained from quiescent spores, germinated spores and sampled at 8, 12, 18, 40 days post-inoculation (dpi) by R. irregularis (Figure 4C). The expression levels of both RiBMH2 and Ri14-3-3 obviously increased accompanying the infection process and arbuscules initiation, as demonstrated by the parallel increased transcriptional levels of MtStbM1 (Figure 4E), the host plant subtilase-encoding gene which is considered as a molecular marker of arbuscular mycorrhiza development (Wegel et al., 2007; Takeda et al., 2009).
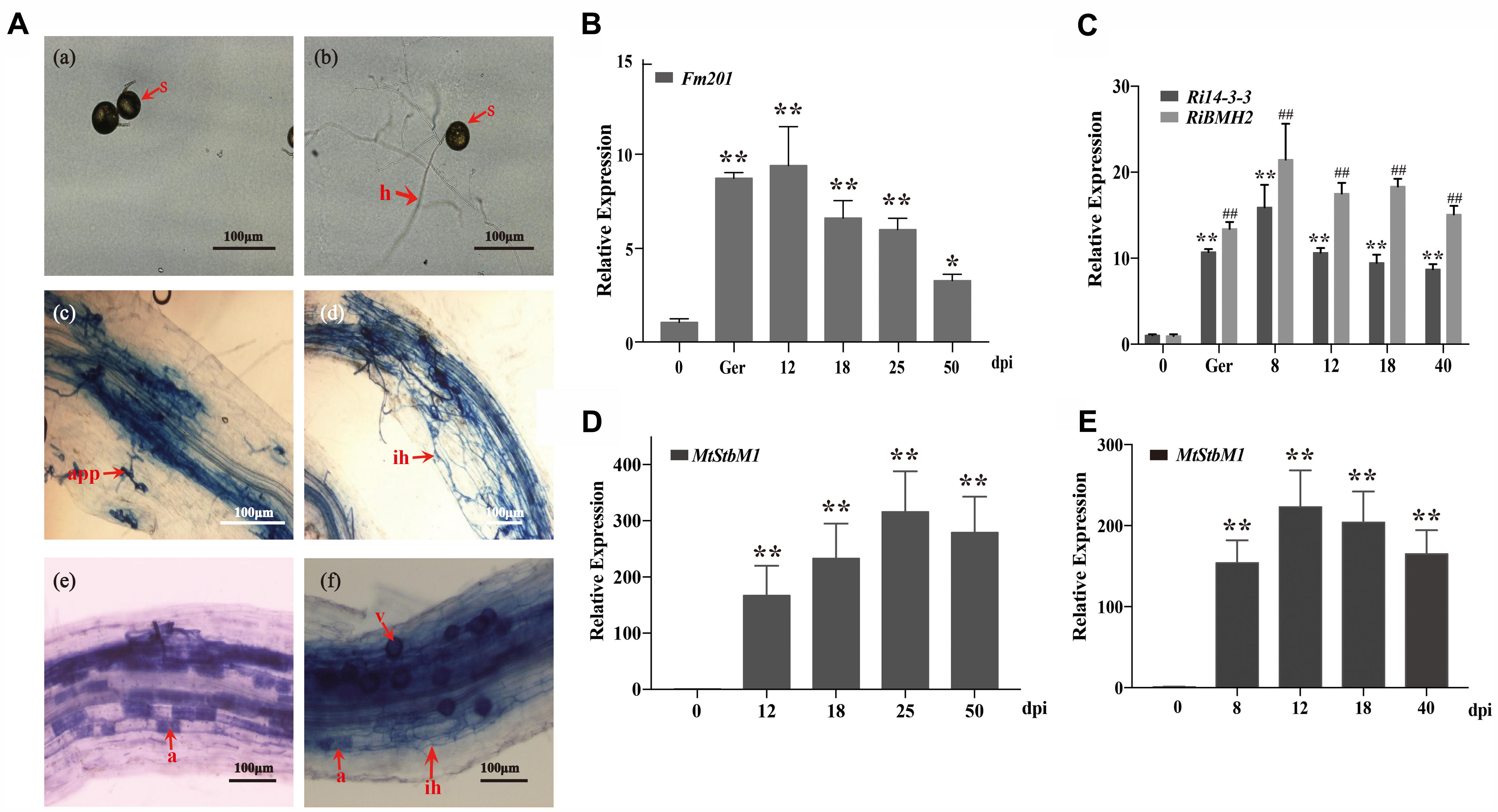
FIGURE 4. Transcript profiles of 14-3-3 genes in quiescent spores, germinating spores and during different symbiotic stages. (A) Optical micrographs of the morphological structures of F. mosseae BEG12 during different stages of mycorrhizal symbiosis. Spore (s), hyphae (h), appressorium (app), arbuscule (a), and Vesicle (v) are shown. Scale bars represent 100 μm. (B) Expression fold change of Fm201 from the AM fungus F. mosseae. Transcript abundance of Fm201 was evaluated by real time RT-PCR in different fungal tissues: quiescent spores (0), germinating spores (ger), uninfected M. truncatula roots (Uninfected) and mycorrhizal roots (12–50 dpi). Fm201 gene is expressed as a ratio relative to FmActin gene from F. mosseae. Lines with a significant ratio to the express rate of Fm201 in quiescent spores. (C) Expression fold change of Ri14-3-3 and RiBMH2 from R. irregularis. Transcript abundance of Ri14-3-3 and RiBMH2 was evaluated by qRT-PCR in different fungal tissues: quiescent spores (0), germinating spores (ger), uninfected M. truncatula roots (Uninfected) and mycorrhizal roots (8–40 dpi). Ri14-3-3 and RiBMH2 gene is expressed as a ratio relative to RiActin gene from R. irregularis. Lines with a significant ratio to the express rate of Ri14-3-3 or RiBMH2 in quiescent spores. (D) The transcript abundance of MtSbtM1 in M. truncatula relative to MtTEF in mycorrhizal roots infected by F. mosseae (12–50 dpi). (E) The transcript abundance of MtSbtM1 in M. truncatula relative to MtTEF in mycorrhizal roots infected by R. irregularis (8–40 dpi). Error bars indicate the means of three biological replicates with SD values. Data shown are averages ± SD; n = 3. (#, ∗p < 0.05, ##, ∗∗p < 0.01).
Knock-Down of 14-3-3 Impairs the Arbuscule Formation in AM Symbiosis
Because of the obligate property of mycorrhizal fungi, the effect of ΔFm201 mutant on the establishment and maintenance of mycorrhizal symbiosis cannot be confirmed in vivo. RNAi technique has been successfully utilized to inhibit Ri14-3-3 encoding a homologous protein of Fm201 as described above in R. irregularis induced by hairy root lines of M. truncatula during symbiosis (Nowara et al., 2010; Helber et al., 2011). A 262 bp cDNA sequence from R. irregularis Ri14-3-3 gene was cloned into pK7GWIWG2 (II) RR according to the approach mentioned in Materials and Methods. The Host-Induced Gene Silencing (HIGS) results of Ri14-3-3 showed no significant influence on the intraradical structures of R. irregularis within the roots, when compared with the control roots (Figure 5A).
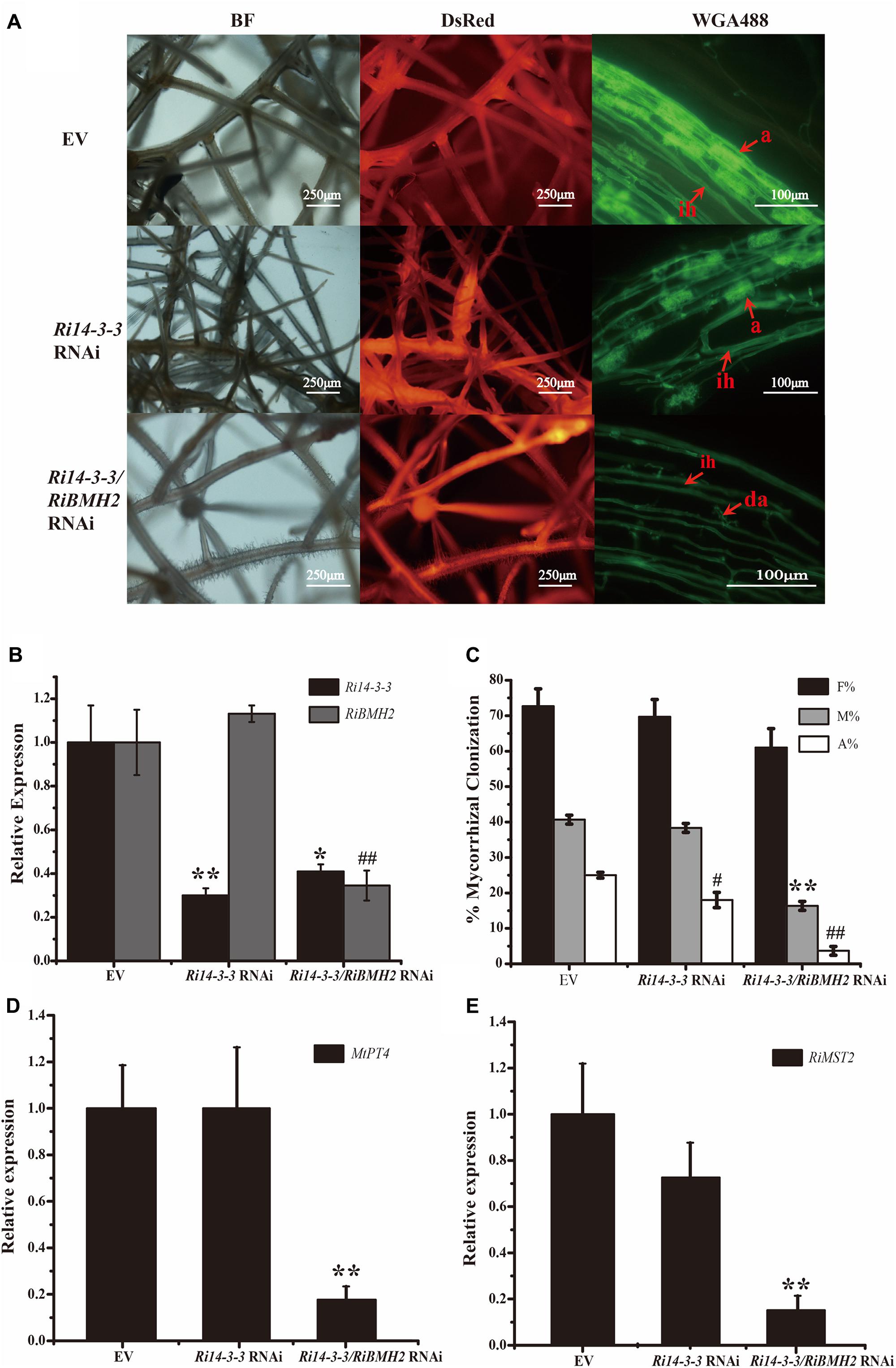
FIGURE 5. Mycorrhizal symbiotic phenotypes of Host-Induced Gene Silencing of Ri14-3-3 and RiBMH2. (A) Hairy root transformation of M. truncatula with empty vector (EV), Ri14-3-3 RNAi vector, or Ri14-3-3/BMH2 RNAi vector. Transgenic hairy roots were infected by AM fungi and the mycorrhizal phenotypes were observed with fluorescence microscope. a, mature arbuscules; ad, arbuscule degradation; ih, internal hyphae. (B) Transcript abundance change of Ri14-3-3 and RiBMH2 in transgenic hairy roots as measured by qRT-PCR using RiActin gene as the reference gene. (C) Mycorrhization level was analyzed by WGA 488 staining of hairy roots at 30 dpi with R. irregularis. F%, frequency of colonization; M%, intensity of mycorrhiza; A%, arbuscule abundance. (D) Expression levels of MtPT4 in control (EV) and RNAi lines were determined by real-time RT-PCR. The M. truncatula MtTEF gene was used as the reference gene. (E) Transcript accumulation of RiMST2 in control (EV) and RNAi mycorrhizal roots measured by real-time RT-PCR. The R. irregularis RiActin gene was used as endogenous control. Three technical replicates were analyzed. Asterisks indicate statistically significant differences from respective control lines. Error bars indicate the means of three biological replicates with SD values. Data shown are averages ± SD; n = 3. (#, ∗p < 0.05, ##, ∗∗p < 0.01).
Since the draft of R. irregularis genome was recently released (Tisserant et al., 2013; Lin et al., 2014), we cloned and identified the coding sequence of another 14-3-3 protein subunit termed RiBMH2 (EXX69786.1). The existence of this novel 14-3-3 protein subunit may explain the nice arbuscule observed in HIGS of Ri14-3-3. Thus, the HIGS experiment targeting both Ri14-3-3 and RiBMH2 was designed to address this issue. Mycorrhizal phenotype analysis uncovered that the arbuscules are defective. The almost collapsed arbuscules were present in the hairy root of Ri14-3-3/RiBMH2 RNAi plants, when these two genes were both strongly repressed. Furthermore, the mycorrhizal colonization of each group was also calculated. The data suggests that the abundance of arbuscules in RNAi roots was also significantly lower than that in the control roots (Figures 5A–C). Moreover, the expression levels of symbiotic MtPT4 and RiMST2, which are considered as molecular markers of the functioning of arbuscules (Harrison et al., 2002; Helber et al., 2011), are significantly reduced in the Ri14-3-3/RiBMH2 RNAi roots relative to the control roots (Figures 5D,E), indicating that knock-down of both Ri14-3-3 and RiBMH2 has a significant effect on the symbiotic phenotype of AM symbiosis. These results also suggest that RiBMH2 may be required for arbuscule formation in AM symbiosis. It also provides a direct evidence that AM fungal 14-3-3 proteins play important roles during AM symbiosis.
14-3-3 Genes Are Up-Regulated in Response to Salinity and Drought Stresses During AM Symbiosis
To further investigate the potential roles of 14-3-3 proteins in response to salinity and drought stresses, the transcript profiles of 14-3-3s in mycorrhizal roots and external hyphae were analyzed by qRT-PCR after 150 mM NaCl treatment for various time (Figures 6A,C). The transcript abundance of 14-3-3 shows slight but significant increase after 1.5 h and relatively stable within 24 h in mycorrhizal roots. In addition, the transcription profile of Fm201 in extraradical hyphae treated with NaCl shows more rapid induction than in intraradical mycelia. To determine whether 14-3-3 genes are responsive to drought stress, the transcript abundance of 14-3-3 genes under 1/2 water holding capacity of drought treatment was also compared (Figures 6B,D). Unlike salinity stress treatment, the transcript abundance of 14-3-3 genes show a ∼4 fold and ∼7 fold up regulation in mycorrhizal roots and extraradical hyphae, respectively. These findings suggest that Fm201 may be responsible for the crosstalk between plant and R. intraradices under salinity and/or drought stresses.
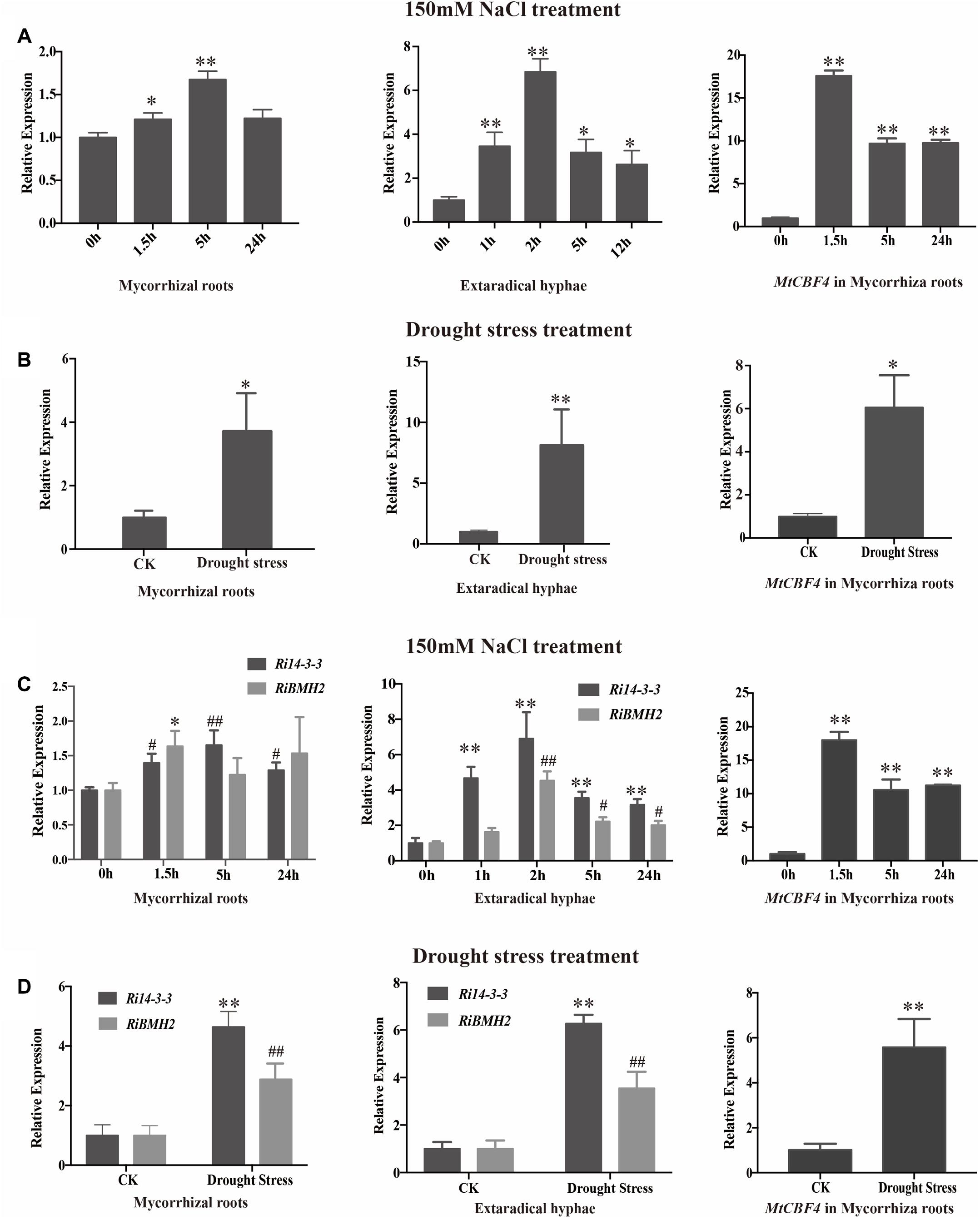
FIGURE 6. The transcript profile of 14-3-3 in AM symbiosis under drought and osmotic stresses. (A) Expression fold change of Fm201 and MtCBF4 in mycorrhizal roots, external hyphae or after exposure to osmotic stress treated by 150 mM NaCl for different hours. (B) Expression fold change of Fm201 and MtCBF4 in mycorrhizal roots, external hyphae under drought stress. (C) Expression fold change of Ri14-3-3, RiBMH2 and MtCBF4 in mycorrhizal roots, external hyphae after exposure to osmotic stress treated by 150 mM NaCl for different hours. Lines with a significant ratio to the express rate of Ri14-3-3, RiBMH2 or MtCBF4 in 0 h. (D) Expression fold change of Ri14-3-3, RiBMH2, MtCBF4 in mycorrhizal roots, external hyphae. The FmActin, RiActin or MtTEF was used as the reference gene. Three biological replicates were analyzed. Asterisks indicate statistically significant differences from respective control lines. Lines with a significant ratio to the express rate of Ri14-3-3, RiBMH2 or MtCBF4 in CK. Error bars indicate the means of three biological replicates with SD values. Data shown are averages ± SD; n = 3. (#, ∗p < 0.05, ##, ∗∗p < 0.01).
Discussion
In terrestrial ecosystems, AM symbiosis is considered to be the most widespread ecologically and agriculturally mutualistic beneficial association among plant symbioses. Despite their great importance in both ecology and agriculture, advance in understanding the molecular basis of AM symbiosis from the fungal aspect is slow until the release of the transcriptomic data of several AM fungal species (Tisserant et al., 2012; Salvioli et al., 2016; Tang et al., 2016) and genomic data of R. irregularis (Tisserant et al., 2013; Lin et al., 2014), biological functions of only a few genes have been characterized during interaction with plants. In such a context, we focus on the characterization of the 14-3-3 genes from AM fungi based on its higher expression during the colonization process.
AM Fungal 14-3-3 Proteins Are a Conserved Feature of Glomeromycota
According to bioinformatics analyses, 14-3-3s from AM fungi are typical 14-3-3 proteins with higher similarity to the known 14-3-3 sequences of yeast and human. Additionally, Fm201 protein is conserved across eukaryotes based on the phylogenetic relationships among AM fungi and other basal eukaryotic species as well as the conserved 3D homology structures between F. mosseae and human (Yang et al., 2006). Therefore, it is of interest to find that two similar sequences were found in the recently released genome and transcriptome of another AM fungus, R. irregularis (Tisserant et al., 2012, 2013). Since the sequences of the two additional genes, the so called Ri14-3-3 and RiBMH2, are complete with the full-length of CDSs, and the percentage of identity is relatively high (∼97%), the three AM fungal proteins share the same nine α-helix domain topologies. Among them, Ri14-3-3 gene from R. irregularis has been firstly reported by Porcel et al. (2006). Moreover, RNA-seq data presented a significant induction in planta phase compared to spores (Tisserant et al., 2012). Only the investigation within genomic and transcriptomic data in AM fungi will clarify whether Fm201-related sequences are a general feature among fungi. Consistent with the previous in silico analyses, these three Fm201, Ri14-3-3 and RiBMH2 are able to complement the yeast BMH1 and BMH2 double mutants. This finding is in agreement with those data reported in the earlier studies (van Heusden et al., 1995, 1996), indicating that these genes identified above encode the functional 14-3-3-like proteins in AM fungi. Further studies need to be carried out to confirm whether these 14-3-3-like proteins identified are a conserved feature of Glomeromycota and whether they may have an essential role in the intraradical phase during interaction with the host plants.
The transcription of 14-3-3 genes show a clear increase in the germinating spores as well as the intraradical phase in both R. irregularis and F. mosseae. The data stemming from the time-course experiment presented that the relatively higher transcription levels were achieved in the phases of root penetration and arbuscules formation, while the expression levels of Fm201 and Ri14-3-3 are obviously reduced compared with RiBMH2 in the degenerating mycorrhizal roots. In addition, we also correlated the Fm201 mRNA abundance with the morphological structures of F. mosseae inside the roots (at 12–50 dpi). The results of Fm201 transcription patterns also suggest that it may play an important role in the germination and hyphopodium formation of F. mosseae, which was also proposed by Breuninger and Requena (2004) through SSH of AM symbiosis at the early stage. In addition, transcript levels of Fm201 remain higher during the symbiotic stage (see Figure 4B), suggesting that this 14-3-3 protein may also play important roles during AM symbiosis, especially the formation of arbuscule besides the root penetration stage. It is thus speculated that the expression of 14-3-3s are, to some extent, related to root penetration and arbuscules formation. This hypothesis is supported by the evidence that 14-3-3 transcripts were present in both the laser micro-dissected arbuscule-containing cells and the IRM including intercellular hyphae (Tisserant et al., 2012). Overall these data implicate a relationship between AM fungal 14-3-3 related genes and intraradical hyphal growth and arbuscule differentiation.
Two AM Fungal 14-3-3 Protein Subunits Have the Impacts on the Success of Arbuscular Mycorrhizal Colonization and Arbuscule Formation
The potential involvement of AM fungal 14-3-3 genes Ri14-3-3 and RiBMH2 in the in planta phase of the colonization process was also supported by the HIGS of Ri14-3-3 and/or RiBMH2 during the M. truncatula–R. irregularis mycorrhizal symbiosis. Lacking the stable genetic transformation protocols for AM fungi, HIGS was confined to AM fungi (Helber et al., 2011; Xie et al., 2016).
The data of the knock-down of both Ri14-3-3 and RiBMH2 genes by HIGS resulting in the impaired arbuscule formation of R. irregularis suggest the significance of these AM fungal 14-3-3 proteins for AM symbiosis. Connecting with the transcripts of RiBMH2 during M. truncatula–R. irregularis mycorrhizal symbiosis, RiBMH2 may be required for the development of AM symbioses and the arbuscule differentiation within roots. However, the Ri14-3-3 RNAi roots colonized by R. irregularis exhibited a considerable arbuscule abundance as compared with control mycorrhizal roots. These findings suggest that the AM functionality or arbuscule formation is redundantly regulated by the two 14-3-3-like genes in R. irregularis. Nevertheless, we here propose that RiBMH2 is essential for arbuscule formation, whereas Ri14-3-3 could be involved in the colonization process but not AM functionality. This hypothesis is supported by the evidence that the transcripts of MtPT4 and RiMST2, two symbiotic genes responsible for arbuscule functionality, were strongly reduced in Ri14-3-3/RiBMH2 RNAi roots, while they were not repressed in Ri14-3-3 RNAi roots. Although Ri14-3-3 homologous gene RiBMH2 is identified in the R. irregularis draft genome (Tisserant et al., 2013; Lin et al., 2014) and RiBMH2 was not down-regulated in Ri14-3-3 RNAi roots (see Figure 5B), the normal AM fungal structures observed in this HIGS system indicate a novel but unknown role for Ri14-3-3 in the establishment of AM symbiosis. Based on the above findings and the previous study (Liu et al., 2015), we hypothesize that RiBMH2-mediated signal could be an important signal in the control of arbuscules formation and R. irregularis hyphal growth within roots. This unknown signal relayed by RiBMH2 serves as the essential signal to ensure the metabolic activity of R. irregularis in the hyphal growth and/or arbuscule differentiation during symbiosis. In the absence of this RiBMH2-mediated signal, the arbuscules are impaired, and growth of the fungus is prevented. The R. irregularis itself needs to activate 14-3-3 protein RiBMH2 in response to the environmental clues to meet demands during fungal growth and division. In addition, our functional analysis in yeast cells suggested that Ri14-3-3 and RiBMH2 encode functional signal proteins involved in growth induction (see Figures 2B,C), indicating that these two proteins may play potential roles in signal transduction during the colonization process and arbuscule formation, respectively. Thus, we can speculate the involvement of Ri14-3-3 in fine-tuning fungal growth in the intraradical phase responding to the external stimuli, moreover, RiBMH2 may be indispensable for arbuscules differentiation. This complex mechanism by which arbuscular mycorrhizas are formed in roots requires the elaborate control of the two AM fungal 14-3-3 proteins in the intraradical phase during cross-talk with host plant.
Remarkably, these results from the HIGS experiments revealed that one 14-3-3 protein subunit can adjust its own expression quantity to offset the adverse influence caused by the lack of another 14-3-3 protein subunit. This conclusion is consistent with the previous results derived from yeast system (van Heusden et al., 1995). Based on this point, it is reasonable to hypothesize that the AM fungal 14-3-3 proteins are indispensable for the symbiosis functioning.
Involvement of AM Fungal 14-3-3 Proteins in Msn2/STRE Element-Mediated Signaling Pathway
The knockdown of both Ri14-3-3 and RiBMH2 by HIGS exhibits somewhat distinct phenotypes, i.e., fewer arbuscule abundance and impaired arbuscules (see Figures 5A,C), repression of the endosymbiosis functioning with regard to transcription of the symbiotic MtPT4 and MST2 genes (see Figures 5D,E). We hypothesize that there exists a positive feedback mechanism in the potential signaling pathway in R. irregularis. It is also proposed that the CREs upstream of a gene always show close relationship with its function, especially for the regulatory proteins (Carey et al., 2012; Petrov et al., 2012), although most CREs are composed of short sequences which may be very abundant in eukaryotic genomes (van Heusden, 2009). As expected, we observed some conserved motifs including STRE elements in the promoters of two AM fungal 14-3-3 genes Fm201 and RiBMH2 (see Table 1), as predicted by YEASTRACT database, in comparison with the promoters of yeast BMH1 and BMH2. Interestingly, the deletion of STRE (CCCCT/AGGGG) element in the promoter of Fm201 showed significantly reduced levels of the reporter gene mRNAs when expressed in yeast cells (see Figure 3A). These hypotheses mentioned above are also supported by the fact that the orthologous Fm201 gene promoter with STRE element (pFm201) directly interacts with transcription factor Msn2 in yeast cells (see Figure 3B). Furthermore, this recognition between RiMsn2 and pFm201 may contribute to the induction of Fm201 in extraradical hyphae in response to the salinity (150 mM NaCl treatment) stress (see Figure 6A). Therefore, based on the site-specific mutagenesis and the yeast one-hybrid analyses, the transcription of AM fungal 14-3-3-like genes during AM symbiosis is Msn2/STRE-element dependent. The zinc finger DNA-binding proteins Msn2 and Msn4 serve as the key factors that controlling fungal growth and stress responses in different fungal species (Martinez-Pastor et al., 1996; Schmitt and Mcentee, 1996; Liu et al., 2013; Zhang et al., 2014). In addition, the Msn2-controlled and STRE-driven gene Fm201 and RiBMH2 from F. mosseae and R. irregularis, respectively, are positively regulated in response to drought stress during AM symbiosis (see Figures 6B,D), reinforcing that AM fungal 14-3-3 genes participate in the Msn2/STRE element-mediated signaling pathway in AM fungal symbiont during AM symbiosis.
Overall these data presented in this study provided new insights into the signaling function of the 14-3-3 proteins in AM fungal cells during crosstalk with host plants. Based on the aforementioned data, we also propose the hypothesis that abiotic stresses such as salinity and drought affect a Msn2/STRE-mediated signaling pathway governing the expression of AM fungal 14-3-3 proteins that promoted fungal colonization and arbuscule formation within roots (see Figure 7). In the first version of the scheme for abiotic stresses induced signaling, it has been proposed that 14-3-3 proteins preferentially expressed in the intraradical phase are involved in AM fungal colonization process and arbuscule functionality by the regulation of Msn2/STRE-mediated signaling pathway that may control the fungal growth and arbuscule lifespan during AM symbiosis.
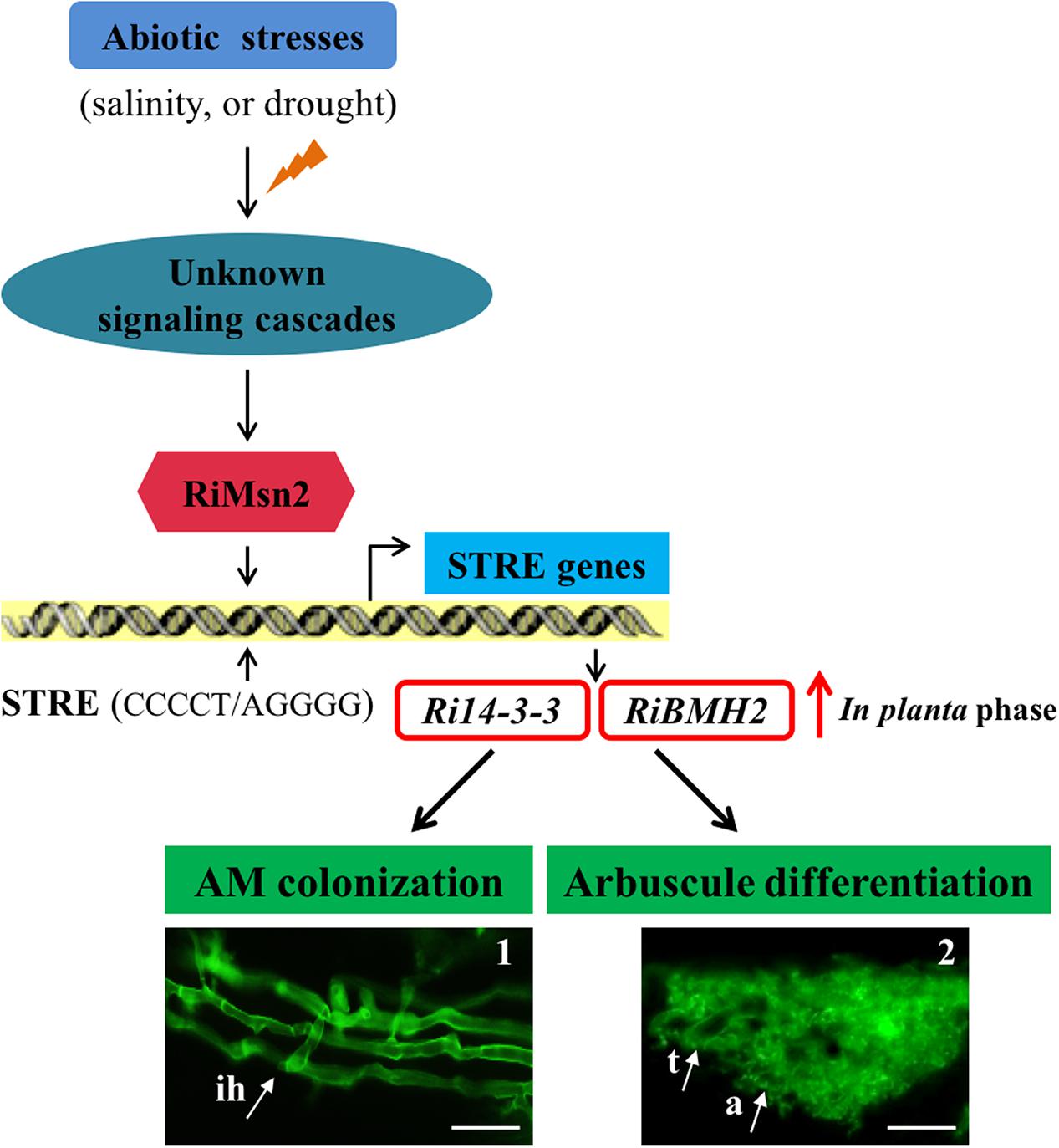
FIGURE 7. Proposed Working Model of two 14-3-3 proteins in R. irregularis. Schematic representation of the abiotic stresses-induced signaling cascades in AM fungi and the involvement of Ri14-3-3 and RiBMH2 genes in the RiMsn2/STRE-mediated signaling pathway during AM symbiosis. In AM fungi, the unknown signaling cascades are triggered by the external stress stimuli such as salinity or drought; then the core component RiMsn2 factor is activated by the upstream of potential cascades. The functional RiMsn2 is translocated to the nuclear to recognize the STRE (Stress response element) (CCCCT/AGGGG) on the AM fungal genome. Meanwhile, in the in planta phase, transcription of the STRE genes, including Ri14-3-3 and RiBMH2, are clearly induced after this interaction between RiMsn2 and STRE. Thus, the expression of Ri14-3-3 and RiBMH2 proteins are involved in or essential for AM colonization and arbuscule differentiation within roots. The black arrows indicate the positive interactions, while the red arrow suggests the induction of Ri14-3-3 and RiBMH2 genes in the in planta phase. The images 1 and 2 represent the intraradical hyphae and arbuscule from WGA488 staining. ih, intraradical hyphae; t, trunk; a, arbuscule. Scale bars represent 25 μm.
Further studies, such as characterizing the precise roles of the novel RiMsn2 gene identified in this work, validating the protein-protein interactions in the Msn2-mediated signaling pathway and the biochemical functions of core components of this pathway, and determining the direct evidence of Msn2-dependent mechanisms in R. irregularis, are needed to define the underlying stress response mechanisms in AM symbionts. Furthermore, the RNA-seq data and gene expression analyses show that both R. irregularis and Gigaspora margarita contain multiple distinct MAPK (Mitogen-activated protein kinase)-related proteins (Tisserant et al., 2012; Salvioli et al., 2016; Xie et al., 2016), indicative of the presence of MAPK signaling cascade in AM fungi to respond to external stresses stimuli and adapt to environmental fluctuation. Thus, a major goal in this field will be to uncover a master MAPK protein regulating the AM fungal growth and differentiation during symbiosis under various abiotic stresses.
In summary, we showed that Fm201, Ri14-3-3 and RiBMH2, three genes from two different AM fungi, are preferentially expressed in the intraradical phase and may have impacts on the success of AM colonization and arbuscule formation. Our data also presented that Msn2 protein governs the Fm201 gene transcription, indicating that AM fungal 14-3-3 gene identified is involved in Msn2 factor/STRE element-mediated signaling pathway. Importantly, host-induced gene silencing of both Ri14-3-3 and RiBMH2 impairs the arbuscule differentiation within roots, indicating that the two AM fungal 14-3-3 protein subunits are required for arbuscule formation. Additionally, these AM fungal 14-3-3 genes are up-regulated in response to salinity and drought stresses during AM symbiosis. Based on these new findings, we propose that the AM fungal 14-3-3 genes are essential for the interaction between AM fungi and host plants, and are potentially involved in enhancing plant salinity and drought tolerance by Msn2/STRE element-controlled signaling.
Materials and Methods
Biological Materials and Growth Conditions
A-grade spores of R. irregularis DAOM197198 were purchased from Agronutrition (Carbonne, France). Spores of F. mosseae BEG12 were kindly provided by the International Bank of Glomeromycota (IBG, Dijon, France) and collected from Medicago truncatula pot cultures by wet sieving to isolate genomic DNA and total RNA. Spores surface sterilized by 2% chloramine T, and then immersed in a solution containing 0.02% streptomycin and 0.02% gentamycin for 10 min (Besserer et al., 2008). Germinated spores of F. mosseae were selected from acetone solution containing 10-8mol/L GR24 (10-9mol/L GR24 for R. irregularis in two days) in 25°C dark incubator. Quiescent spores, germination spores and mycorrhizal roots (at 10, 18 25, 50 dpi for F. mosseae; 8, 12, 18, 40 dpi for R. irregularis) were harvested. After washing in sterile water, all materials described above were immediately frozen in liquid nitrogen and stored at -80°C before nucleic acid extraction.
The water-holding capacity of the soil was computated before planting. The soil was weighed before plant, watered uniformly until flowed from the bottom. Keep the pot suspended in midair for 1 day before weighing. The increase weight of soil after waterd is water-holding capacity. M. truncatula mycorrhizal roots inoculated with AM fungi were treated with NaCl (0.5 M) to 150 mM in final concentration (Calculate according to 70% of water holding capacity in soil) (Giovannetti et al., 2001; Li et al., 2011). Drought treatment was measured by 1/2 water-holding capacity treatment which was proved drought stress treatment to M. truncatula in previous experiments. Mycorrhizal roots and extraradical hyphae from sandwich system were harvested at 1.5, 5, 24 h after treatments (Abdel Latef and Chaoxing, 2011; Estrada et al., 2012) to monitor the transcript profiles of 14-3-3 genes under different abiotic stresses by qRT-PCR analysis.
Fm201 promoter-YGFP chimeric gene in S. cerevisiae BY4741 was treated with NaCl (500 mM), CuSO4 (50 mM), CdCl2 (0.2 mM), PEG4000 (25%), and 37°C abiotic stresses in YPD medium and then harvested after 1h treatments (Vido et al., 2001).
DNA and RNA Extraction, RT-PCR and Real Time RT-PCR
The total DNA was isolated from AM fungal sporescarps as described by Zézé et al. (1994). Total RNA of different AM fungal tissues was extracted with TRIzol reagent (Invitrogen) according to the protocol. Surface-sterilized spores were placed into 1.5 ml RNase free microtube and then frozen in liquid nitrogen, 0.3ml TRIzol solution was immediately added to the microtube. Electric mill (TIANGEN OSE-Y20, Beijing, China) and Phase Lock Gel (TIANGEN, Beijing, China) were used to make sure the quality of RNA. Total RNA yields and concentrations were measured by the Thermo NanoDrop 2000 spectrophotometer (Thermo). To remove residual genomic DNA, each total RNA sample was treated with RNase-free DNaseI (Thermo) according to the manufacturer’s instructions. The first cDNA strand was synthesized as described in RevertAid First Strand cDNA Synthesis Kit (Thermo).
Transcript profiles of AM fungal genes Fm201, Ri14-3-3, and RiBMH2 as well as host plant genes MtSbtM1 and MtCBF4 in different symbiotic stages and under abiotic stresses were studied by qRT-PCR using ViiA 7 system (Life Technologies, United States), three biological replications were performed. The expression levels were normalized to transcripts of the β-actin gene of F. mosseae or R. irregularis and to transcripts of the MtTEF gene of M. truncatula (Hohnjec et al., 2005). Before real time RT-PCR, gene-specific primers for all target genes were validated on genomic DNA and cDNA. Total RNA was isolated from AM roots comprised plant and fungal materials. The specificity of the primer pairs were also confirmed via PCR method on M. truncatula total DNA. No amplification signals were present on plant DNA. The primers sequences for all genes studied in this work are provided in Supplementary Table S2. qRT-PCR was performed using SYBR Green Real-time PCR Master Mix (TOYOBO, Japan) according to the manufacturer’s instructions. Each 10 μl reaction contained 1 μl of the synthesized cDNA (cDNA pool was diluted to 200 μl), 5 μl SYBR Green Real-time PCR Master Mix, 0.5 μl each primer(10 μM), 3 μl ddH2O. PCR program consisted of a 30 s incubation at 95°C to active the hot-start recombinant Taq DNA polymerase, followed by 40 cycles of 10 s at 95°C, 15 s at 57°C, and 20 s at 72°C. The relative levels of transcripts were calculated by using the 2-ΔΔct method (Livak and Schmittgen, 2001).
Cloning of Fm201 Gene From F. mosseae
The Fm201 EST sequence was obtained from NCBI (Accession number: CF803281), (Breuninger and Requena, 2004). Reverse PCR was utilized to get the 5′ flanking sequence of Fm201 gene. The gene-specific primers 201F and 201R were designed to amplify the partial DNA fragment of Fm201 according to the available sequence of Fm201 EST. Genomic DNA of F. mosseae was digested by FastDigest restriction enzyme XhoI (Thermo). DNA fragments were self-ligated by T4 DNA ligase, and the reaction was carried out in a final volume of 20 μl containing 0.5 μl digested DNA fragments, 2 μl 10× buffer, 0.5 μl T4 DNA ligase (Thermo), 17 μl ddH2O, incubated for 12 h at 10°C. Nest-PCR was performed in this experiment, 0.5 μl ligated production was used as PCR template, the specific primers used in the first PCR reaction were 201RF1 and 201RR1, products from first PCR reaction were diluted to 1/1000 as the template for the second PCR reaction, and specific primer 201RF2 and 201RR2 were used.
RACE as a classic method to rapidly obtain the 5′ and 3′ ends of the Fm201 gene (Scotto-Lavino et al., 2006a,b). 3′RACE was carried out on the total RNA from F. mosseae sporecarps by using primer QT. Two pairs of primers RACE201F/QO and RACE201F2/QI were used for the subsequent nest PCR reactions, respectively. Due to the high A/T containing feature of AM fungi genomic DNA, the dGTP and QC replaced the dATP and QT used in classic 5′RACE, responsively. The first cDNA strand was obtained from F. mosseae sporocarps by using specific primer RACE201R1. Primers QC, Qo and RACE201R2 were used as the first PCR cycle primers, while primers QI and RACE201R3 were used as the second PCR cycle. Transfast pfu DNA polymerase (Transgen, Beijing, China) was used in the PCR reactions mentioned above, PCR products were cloned into pEASY-Blunt vector (Transgen, Beijing, China) and sequenced.
Plasmids Construction
Plasmid pMR12 was generated from pMRI-11(Xie et al., 2014), the promoter of Gal7 was amplified from the genome DNA of S. cerevisiae BY4741 by PCR using the specific primers PGal7F/PGal7R. PGal7 and pMR-11 were digested with both SacI and SpeI, respectively, then the digests were cloned into target vector pMR-11. To address the regulation of CREs located in the promoter of Fm201, the expression profiles of YGFP reporter were conducted in S. cerevisiae BY4741. Monoclonal vector pUG35 carrying a YGFP reporter is used in yeast heterologous systems (Cormack et al., 1997). Two restriction sites were SacI and XbaI, which were added to the start codon upstream sequence of Fm201 (pFm201) by using primers 201PFn (n = 1,2,3,4) and 201PR1, then cloned into pUG35 to replace PMET-25 to produce a series of 5′ truncated promoters-reporter vectors. pFm201 targeted deletion of cis-element was conducted by using SOE-PCR method (Ho et al., 1989). The specific primers used are provided in Supplementary Table S2). The site-directed mutation promoter sequences were also cloned into pUG35 as the promoter truncated verification vectors. qRT-PCR was used for monitoring transcriptional efficiency of Fm201, the Ura gene of pUG35 was chosen as the internal standard, primers UraF, UraR, YGFPF, YGFPR were used in this experiment (Peter et al., 2006).
Yeast One-Hybrid Screening
Yeast one hybrid experiment was carried out using the Matchmaker One-hybrid System (Clontech), the ORF of GintSTE was cloned from cDNA of Rhizophagus irregularis by using primers RiSTE12F and RiSTE12R, and cloned into pGADT7-rec2. Two same 272 bp length promoter fragments contained STE12 targeted cis-element STRE were cloned into pHIS2 in tandem, the same fragment only lacking the cis-element STRE was also inserted into the pHIS2 as a negative control. Yeast one-hybrid experiment of Msn2 was carried out in the same way and the primers RiMsn2F and RiMsn2R were used as mentioned in Supplementary Table S2.
Plasmids for yeast one-hybrid were co-transformed into yeast Y187 strain. Y187 cells carrying the target plasmids were cultivated in SD medium lacking leucine and tryptophan, and were also gradiently inoculated at 1.0 OD600 on SD medium lacking leucine, histidine and tryptophan and supplemented with 30 mM 3-AT, which is a competitive inhibitor of the His3 protein.
HIGS of R. irregularis 14-3-3 Genes in Hairy Root Lines of M. truncatula
The RNAi-target sequences of Ri14-3-3 and RiBMH2 were amplified by the specific primers Ri14-3-3F/Ri14-3-3R and RiBMH2ATG/RiBMH2F. The PCR products were cloned to the linearized pDONR221 used CloneExpressII (Vazyme, Nanjing, China), then the LR reaction was done to recombine the target sequences into the pK7GWIWG2(II)RR according to the instructions in Gateway protocol.
Agrobacterium rhizogenes Msu440-mediated root transformation was performed following the method as described in Medicago Truncatula Handbook (International Committee, 2006). In vitro hairy roots were cultured on EM plates containing Benzyl penicillin (200 mg/L) for three times. The root tip (2∼3 cm in length) was used for each subculture. The root was re-cultured in the M medium without antibiotics for half a month. The hairy root lines without bacteria were re-cultured in new M medium for mycorrhization. Mycorrhizal hairy root of P. crispum without DsRed tag was cut into small pieces (∼3 mm) and placed around the hairy root of M. truncatula harboring DsRed marker as described in Supplementary Figure S3. The mycorrhizal roots with red fluorescence were harvested in one month until the external hyphae of R. irregularis beyond the hairy root surface of M. truncatula.
Quantification of Arbuscular Mycorrhizal Colonization
Mycorrhizal roots collected from pot cultures were stained with 0.1% Typan blue, while the mycorrhizal hairy roots expressing red fluorescence grown on plates were stained with WGA488, and the estimation of AM colonization was performed as described by Trouvelot et al. (2015) using MYCOCALC program2.
In Silico Analysis of Fm201 Protein
The deduced amino acid sequence of Fm201 was analyzed with the computer program DNAstar. Multiple sequence alignments were performed by DNAMAN8. The unrooted phylogenetic tree constructed by neighbor-joining algorithm was carried out using MEGA6. The computation of physical and chemical parameters was conducted by using ProtParam tool3. Homology modeling of the three-dimensional structure of Fm201 protein was done with the program Swiss Model4 using Homo sapiens 14-3-3ε protein (2br9A) as the template (Figure 1B; Yang et al., 2006). The cis-elements of the promoters Fm201 and RiBMH2 were analyzed on YEASTRACT5 using S. cerevisiae S288c as the reference.
Yeast mutant strains used in this article are constructed with the methods mentioned by van Heusden (van Heusden et al., 1995, 1996). The detail information for each strain is available at Supplementary Table S1. In the construction of bhms-Fm201(MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; BMH1::KanMX (Gal7[Fm201]); BMH2::ura3), fragments of pMRI-12 which contain a KanMX and a Gal7 promoter and pSH47 which contain a Ura3 marker were used to replace BY4741, BMH1 and BMH2, respectively, by primers PMRI-12F1, PMRI-12R1 and PSH47F, PSH47R. The bmhs-Ri14-3-3 and bmhs-RiBMH2 were also built in the same way.
Statistical Analyses
Statistical analyses were performed through one-way ANOVA. Following ANOVA, Tukey’s test was performed to make comparisons between treatments, using a probability level of p < 0.05(∗, #), 0.05 ≤p < 0.01 (∗∗, ##). All statistical analyses were performed using SPSS statistical package (version 23.0, SPSS Inc., United States).
Accession Numbers
The sequence data can be found in the GenBank data libraries under accession numbers. Nucleic acid sequence: MtCBF4 (HQ110079.1), MtStbM1 (XM_003611148.1), MtPT4 (AY116211.1), Fmactin (KM360085.1), Fm201 (KM258580.1), Riactin (EXX64987.1), RiBMH2 (JEMT01016782.1), Ri14-3-3 (AM049264.1), RiMST2 (HM143864.1), Ri14-3-3 (CAJ16742.1). Amino acid sequence: Fm201(KM258580), RiBMH2 (EXX69786.1), R. oryzae 14-3-3 (EIE87660.1), M. medusa 14-3-3 (ABS86241.1), M. circinelloides 14-3-3 (EPB82885.1), A. oryzae 14-3-3 (XP_001819291.2), A. niger 14-3-3 (XP_001399080.1), S. borealis 14-3-3 (ESZ95350.1), M. oryzae 14-3-3 (XP_003710925.1), A. nidulans (CBF81292.1), A. terreus 14-3-3 (XP_001212078.1), P. strigosozonata 14-3-3 (XP_007382290.1), S. musiva 14-3-3 (EMF09853.1), B. bassiana 14-3-3 (XP_008601347.1), R. solani 14-3-3 (CCO32840.1), S. cerevisiae BMH1 (CAA46959.1), S. tuberosum 14-3-3 (XP_004250139.1), O. sativa 14-3-3 (NP_001047234.1), S. cerevisiae BMH2 (CAA59275.1), R. norvegicus 14-3-3ε (NP_113791.1), H. sapiens 14-3-3ε (NP_006752.1).
Author Contributions
BZ and XXie conceived this research. ZS and JS prepared the biological material for gene expression analysis. ZS and XXin performed the data analysis. ZS and XXie wrote the manuscript. BZ and XXie revised the manuscript.
Funding
This study was financially supported by grant from the Natural Science Foundation of China (Grant No. 31270159).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Professor Ton Bisseling, Ph.D. U. Gueldener and Ph.D. Wenping Xie for kindly providing the pK7GWIWG2 (II) RR, pUG35 and pMR-11 plasmids, respectively. We also thank Professor Deqiang Duanmu for the constructive discussions and language corrections during the manuscript preparation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00091/full#supplementary-material
Footnotes
- ^http://www.yeastract.com/
- ^http://www2.dijon.inra.fr/mychintec/Mycocalc-prg/download.html
- ^http://www.expasy.ch/tools/protparam.html
- ^http://swissmodel.expasy.org/
- ^http://www.yeastract.com/
References
Abdel Latef, A. A. H., and Chaoxing, H. (2011). Effect of arbuscular mycorrhizal fungi on growth, mineral nutrition, antioxidant enzymes activity and fruit yield of tomato grown under salinity stress. Sci. Hortic. 127, 228–233. doi: 10.1016/j.scienta.2010.09.020
Aksamit, A., Korobczak, A., Skala, J., Lukaszewicz, M., and Szopa, J. (2005). The 14-3-3 gene expression specificity in response to stress is promoter-dependent. Plant Cell Physiol. 46, 1635–1645. doi: 10.1093/pcp/pci179
Augé, R. M. (2001). Water relations, drought and vesicular-arbuscular mycorrhizal symbiosis. Mycorrhiza 11, 3–42. doi: 10.1007/s005720100097
Bago, B., Pfeffer, P. E., and Shachar-Hill, Y. (2000). Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol. 124, 949–958. doi: 10.1104/pp.124.3.949
Ballou, E. R., Kozubowski, L., Nichols, C. B., and Alspaugh, J. A. (2013). Ras1 acts through duplicated Cdc42 and Rac proteins to regulate morphogenesis and pathogenesis in the human fungal pathogen Cryptococcus neoformans. PLoS Genet. 9:e1003687. doi: 10.1371/journal.pgen.1003687
Besserer, A., Bécard, G., Jauneau, A., Roux, C., and Séjalon-Delmas, N. (2008). GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148, 402–413. doi: 10.1104/pp.108.121400
Bonfante, P., and Genre, A. (2010). Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1:48. doi: 10.1038/ncomms1046
Bonfante, P., and Genre, A. (2015). Arbuscular mycorrhizal dialogues: do you speak ‘plantish’or ‘fungish’? Trends Plant Sci. 20, 150–154. doi: 10.1016/j.tplants.2014.12.002
Bonfante, P., and Requena, N. (2011). Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 14, 451–457. doi: 10.1016/j.pbi.2011.03.014
Bravo, A., Brands, M., Wewer, V., Dörmann, P., and Harrison, M. J. (2017). Arbuscular mycorrhiza-specific enzymes fatm and ram2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 214, 1631–1645. doi: 10.1111/nph.14533
Breuninger, M., and Requena, N. (2004). Recognition events in AM symbiosis: analysis of fungal gene expression at the early appressorium stage. Fungal Genet. Biol. 41, 794–804. doi: 10.1016/j.fgb.2004.04.002
Bruckmann, A., Steensma, H. Y., Mj, T. D. M., and Van Heusden, G. P. (2004). Regulation of transcription by Saccharomyces cerevisiae 14-3-3 proteins. Biochem. J. 382, 867–875. doi: 10.1042/BJ20031885
Cappellazzo, G., Lanfranco, L., and Bonfante, P. (2007). A limiting source of organic nitrogen induces specific transcriptional responses in the extraradical structures of the endomycorrhizal fungus Glomus intraradices. Curr. Genet. 51:59. doi: 10.1007/s00294-006-0101-2
Carey, M. F., Peterson, C. L., and Smale, S. T. (2012). Identifying cis-acting DNA elements within a control region. Cold Spring Harb. Protoc. 2012, 279–296. doi: 10.1101/pdb.top068171
Chitarra, W., Pagliarani, C., Maserti, B., Lumini, E., Siciliano, I., Cascone, P., et al. (2016). Insights on the impact of arbuscular mycorrhizal symbiosis on tomato tolerance to water stress. Plant Physiol. 171, 00307.2016. doi: 10.1104/pp.16.00307
Cormack, B. P., Bertram, G., Egerton, M., Gow, N. A., Falkow, S., and Brown, A. J. (1997). Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143, 303–311.
Darling, D. L., Yingling, J., and Wynshaw-Boris, A. (2005). Role of 1433 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 68, 281–315. doi: 10.1016/S0070-2153(05)68010-68016
Elfving, N., Chereji, R. V., Bharatula, V., Bjorklund, S., Morozov, A. V., and Broach, J. R. (2014). A dynamic interplay of nucleosome and Msn2 binding regulates kinetics of gene activation and repression following stress. Nucleic Acids Res. 42, 5468–5482. doi: 10.1093/nar/gku176
Estrada, B., Barea, J. M., Aroca, R., and Ruiz-Lozano, J. M. (2012). A native Glomus intraradices strain from a Mediterranean saline area exhibits salt tolerance and enhanced symbiotic efficiency with maize plants under salt stress conditions. Plant Soil 366, 333–349. doi: 10.1007/s11104-012-1409-y
Gancedo, J. M. (2001). Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25, 107–123.
Genre, A., Chabaud, M., Timmers, T., Bonfante, P., and Barker, D. G. (2005). Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17, 3489–3499. doi: 10.1105/tpc.105.035410
Gianinazzi, S., Gollotte, A., Binet, M. N., van Tuinen, D., Redecker, D., and Wipf, D. (2010). Agroecology: the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 20, 519–530. doi: 10.1007/s00572-010-0333-3
Giovannetti, M., Fortuna, P., Citernesi, A. S., Morini, S., and Nuti, M. P. (2001). The occurrence of anastomosis formation and nuclear exchange in intact arbuscular mycorrhizal networks. New Phytol. 151, 717–724. doi: 10.1046/j.0028-646x.2001.00216.x
Govindarajulu, M., Pfeffer, P. E., Jin, H., Abubaker, J., Douds, D. D., Bücking, H., et al. (2005). Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435, 819–823. doi: 10.1038/nature03610
Gutjahr, C., and Parniske, M. (2013). Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 29, 593–617. doi: 10.1146/annurev-cellbio-101512-122413
Harrison, M. J. (2012). Cellular programs for arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 15, 691–698. doi: 10.1016/j.pbi.2012.08.010
Harrison, M. J., Dewbre, G. R., and Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14, 2413–2429. doi: 10.1105/tpc.004861
Helber, N., and Requena, N. (2008). Expression of the fluorescence markers dsred and gfp fused to a nuclear localization signal in the arbuscular mycorrhizal fungus glomus intraradices. New Phytol. 177, 537–548. doi: 10.1111/j.1469-8137.2007.02257.x
Helber, N., Wippel, K., Sauer, N., Schaarschmidt, S., Hause, B., and Requena, N. (2011). A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 23, 3812–3823. doi: 10.1105/tpc.111.089813
Hermeking, H., and Benzinger, A. (2006). 14-3-3 proteins in cell cycle regulation. Semin. Cancer Biol. 16, 183–192. doi: 10.1016/j.semcancer.2006.03.002
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. doi: 10.1007/978-1-4939-6472-7_27
Hohnjec, N., Vieweg, M. F., Pühler, A., Becker, A., and Küster, H. (2005). Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 137, 1283–1301. doi: 10.1104/pp.104.056572
Hurtado, C. A., and Rachubinski, R. A. (2002). YlBMH1 encodes a 14-3-3 protein that promotes filamentous growth in the dimorphic yeast Yarrowia lipolytica. Microbiology 148, 3725–3735.
Ichimura, T., Isobe, T., Okuyama, T., Yamauchi, T., and Fujisawa, H. (1987). Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+,calmodulin-dependent protein kinase II. FEBS Lett. 219, 79–82. doi: 10.1155/2017/3682752
International Committee (2006). Medicago truncatula Handbook. Ardmore, OK: Samuel Roberts Noble Foundation.
Javot, H., Penmetsa, R. V., Terzaghi, N., Cook, D. R., and Harrison, M. J. (2007a). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104, 1720–1725. doi: 10.1073/pnas.0608136104
Javot, H., Pumplin, N., and Harrison, M. J. (2007b). Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ. 30, 310–322. doi: 10.1111/j.1365-3040.2006.01617.x
Jiang, Y., Wang, W., Xie, Q., Liu, N., Liu, L., Wang, D., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Jung, S. C., Martinez-Medina, A., Lopez-Raez, J. A., and Pozo, M. J. (2012). Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 38, 651–664. doi: 10.1007/s10886-012-0134-6
Kikuchi, Y., Hijikata, N., Yokoyama, K., Ohtomo, R., Handa, Y., Kawaguchi, M., et al. (2014). Polyphosphate accumulation is driven by transcriptome alterations that lead to near-synchronous and near-equivalent uptake of inorganic cations in an arbuscular mycorrhizal fungus. New Phytol. 204, 638–649. doi: 10.1111/nph.12937
Lenoir, I., Fontaine, J., and Lounèshadj, S. A. (2016). Arbuscular mycorrhizal fungal responses to abiotic stresses: a review. Phytochemistry 123, 4–15. doi: 10.1016/j.phytochem.2016.01.002
Li, D., Zhang, Y., Hu, X., Shen, X., Ma, L., Su, Z., et al. (2011). Transcriptional profiling of Medicago truncatula under salt stress identified a novel cbf transcription factor mtcbf4 that plays an important role in abiotic stress responses. BMC Plant Biol. 11:109. doi: 10.1186/1471-2229-11-109
Li, T., Hu, Y. J., Hao, Z. P., Li, H., Wang, Y. S., and Chen, B. D. (2013). First cloning and characterization of two functional aquaporin genes from an arbuscular mycorrhizal fungus glomus intraradices. New Phytol. 197, 617–630. doi: 10.1111/nph.12011
Li, T., Sun, Y., Ruan, Y., Xu, L., Hu, Y., Hao, Z., et al. (2016). Potential role of d-myo-inositol-3-phosphate synthase and 14-3-3 genes in the crosstalk between zea mays and rhizophagus intraradices under drought stress. Mycorrhiza 26, 1–15. doi: 10.1007/s00572-016-0723-2
Lin, K., Limpens, E., Zhang, Z., Ivanov, S., Saunders, D. G., Mu, D., et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 10:e1004078. doi: 10.1371/journal.pgen.1004078
Liu, Q., Li, J. G., Ying, S. H., Wang, J. J., Sun, W. L., Tian, C. G., et al. (2015). Unveiling equal importance of two 14-3-3 proteins for morphogenesis, conidiation, stress tolerance and virulence of an insect pathogen. Environ. Microbiol. 17, 1444–1462. doi: 10.1111/1462-2920.12634
Liu, Q., Ying, S. H., Li, J. G., Tian, C. G., and Feng, M. G. (2013). Insight into the transcriptional regulation of msn2 required for conidiation, multi-stress responses and virulence of two entomopathogenic fungi. Fungal Genet. Biol. 54, 42–51. doi: 10.1016/j.fgb.2013.02.008
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lozano-Duran, R., and Robatzek, S. (2015). 14-3-3 proteins in plant-pathogen interactions. Mol. Plant Microbe Interact. 28, 511–518. doi: 10.1094/MPMI-10-14-0322-CR
Maldonado-Mendoza, I. E., Dewbre, G. R., and Harrison, M. J. (2001). A phosphate transporter gene from the extra-radical mycelium of an arbuscular mycorrhizal fungus Glomus intraradices is regulated in response to phosphate in the environment. Mol. Plant Microbe Interact. 14, 1140–1148. doi: 10.1094/MPMI.2001.14.10.1140
Martinez-Pastor, M. T., Marchler, G., Schuller, C., Marchler-Bauer, A., Ruis, H., and Estruch, F. (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235.
Moore, B. W. (1967). “Specific proteins of the nervous system,” in Physiological and Biochemical Aspects of Nervous Integration, ed. F. D. Carlson (Englewood Cliffs, NJ: Prentice-Hall), 343–359.
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Parua, P. K., and Young, E. T. (2014). Binding and transcriptional regulation by 14-3-3 (Bmh) proteins requires residues outside of the canonical motif. Eukaryot. Cell 13, 21–30. doi: 10.1128/EC.00240-13
Paszkowski, U. (2006). A journey through signaling in arbuscular mycorrhizal symbiosis 2006. New Phytol. 172, 35–46. doi: 10.1111/j.1469-8137.2006.01840.x
Peter, G. J., Düring, L., and Ahmed, A. (2006). Carbon catabolite repression regulates amino acid permeases in Saccharomyces cerevisiae via the TOR signaling pathway. J. Biol. Chem. 281, 5546–5552. doi: 10.1074/jbc.M513842200
Petrov, V., Vermeirssen, V., De Clercq, I., Van Breusegem, F., Minkov, I., Vandepoele, K., et al. (2012). Identification of cis-regulatory elements specific for different types of reactive oxygen species in Arabidopsis thaliana. Gene 499, 52–60. doi: 10.1016/j.gene.2012.02.035
Porcel, R., Aroca, R., Cano, C., Bago, A., and Ruiz-Lozano, J. M. (2006). Identification of a gene from the arbuscular mycorrhizal fungus glomus intraradices encoding for a 14-3-3 protein that is up-regulated by drought stress during the AM symbiosis. Microb. Ecol. 52, 575–582. doi: 10.1007/s00248-006-9015-2
Remy, W., Taylor, T. N., Hass, H., and Kerp, H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U.S.A. 91, 11841–11843. doi: 10.1073/pnas.91.25.11841
Requena, N., Mann, P., Hampp, R., and Franken, P. (2002). Early developmentally regulated genes in the arbuscular mycorrhizal fungus glomus mosseae : identification of gmgin1, a novel gene with homology to the c-terminus of metazoan hedgehog proteins. Plant Soil 244, 129–139. doi: 10.1023/A:1020249932310
Rispail, N., Soanes, D. M., Ant, C., Czajkowski, R., Grunler, A., Huguet, R., et al. (2009). Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46, 287–298. doi: 10.1016/j.fgb.2009.01.002
Roberts, M. R., Salinas, J., and Collinge, D. B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50, 1031–1039. doi: 10.1023/A:1021261614491
Roberts, R. L., Mösch, H.-U., and Fink, G. R. (1997). 14-3-3 proteins are essential for Ras/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89, 1055–1065. doi: 10.1016/S0092-8674(00)80293-7
Salvioli, A., and Bonfante, P. (2013). Systems biology and “omics” tools: a cooperation for next-generation mycorrhizal studies. Plant Sci. 203–204, 107–114. doi: 10.1016/j.plantsci.2013.01.001
Salvioli, A., Ghignone, S., Novero, M., Navazio, L., Bagnaresi, P., and Bonfante, P. (2016). Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J. 10, 130–144. doi: 10.1038/ismej.2015.91
Sanders, I. R. (1999). Evolutionary genetics: no sex please, we’re fungi. Nature 399, 737–739. doi: 10.1038/21544
Schmitt, A. P., and Mcentee, K. (1996). Msn2p, a zinc finger dna-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 93, 5777–5782. doi: 10.1073/pnas.93.12.5777
Schmitz, A. M., and Harrison, M. J. (2014). Signaling events during initiation of arbuscular mycorrhizal symbiosis. J. Integr. Plant Biol. 56, 250–261. doi: 10.1111/jipb.12155
Schützendübel, A., and Polle, A. (2002). Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 53, 1351–1365. doi: 10.1093/jxb/53.372.1351
Scotto-Lavino, E., Du, G., and Frohman, M. A. (2006a). 3′ end cdna amplification using classic RACE. Nat. Protoc. 1, 2742–2745. doi: 10.1101/pdb.prot4130
Scotto-Lavino, E., Du, G., and Frohman, M. A. (2006b). 5′ end cDNA amplification using classic RACE. Nat. Protoc. 1, 2555–2562. doi: 10.1101/pdb.prot4131
Seidl, V., Seiboth, B., Karaffa, L., and Kubicek, C. (2004). The fungal STRE-element-binding protein Seb1 is involved but not essential for glycerol dehydrogenase (gld1) gene expression and glycerol accumulation in Trichoderma atroviride during osmotic stress. Fungal Genet. Biol. 41, 1132–1140. doi: 10.1016/j.fgb.2004.09.002
Simon, L., Bousquet, J., Levesque, R. C., and Lalonde, M. (1993). Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 363, 67–69. doi: 10.1038/363067a0
Smith, A., Ward, M. P., and Garrett, S. (1998). Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17, 3556–3564. doi: 10.1093/emboj/17.13.3556
Smith, A. J., Daut, J., and Schwappach, B. (2011). Membrane proteins as 14-3-3 clients in functional regulation and intracellular transport. Physiology 26, 181–191. doi: 10.1152/physiol.00042.2010
Takeda, N., Sato, S., Asamizu, E., Tabata, S., and Parniske, M. (2009). Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58, 766–777. doi: 10.1111/j.1365-313X.2009.03824.x
Tang, N., San Clemente, H., Roy, S., Becard, G., Zhao, B., and Roux, C. (2016). A survey of the gene repertoire of Gigaspora rosea unravels conserved features among glomeromycota for obligate biotrophy. Front. Microbiol. 7:233. doi: 10.3389/fmicb.2016.00233
Tisserant, E., Kohler, A., Dozolme-Seddas, P., Balestrini, R., Benabdellah, K., Colard, A., et al. (2012). The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 193, 755–769. doi: 10.1111/j.1469-8137.2011.03948.x
Tisserant, E., Malbreil, M., Kuo, A., Kohler, A., Symeonidi, A., Balestrini, R., et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. U.S.A. 110, 20117–20122. doi: 10.1073/pnas.1313452110
Tollot, M., Wong Sak Hoi, J., Van Tuinen, D., Arnould, C., Chatagnier, O., Dumas, B., et al. (2009). An STE12 gene identified in the mycorrhizal fungus Glomus intraradices restores infectivity of a hemibiotrophic plant pathogen. New Phytol. 181, 693–707. doi: 10.1111/j.1469-8137.2008.02696.x
Trouvelot, S., Bonneau, L., Redecker, D., van Tuinen, D., Adrian, M., and Wipf, D. (2015). Arbuscular mycorrhiza symbiosis in viticulture: a review. Agron. Sustain. Dev. 35, 1449–1467. doi: 10.1007/s13593-015-0329-7
Van der Heijden, M. G., Klironomos, J. N., Ursic, M., Moutoglis, P., Streitwolf-Engel, R., Boller, T., et al. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396, 69–72. doi: 10.1038/23932
van Heusden, G. P. (2009). 14-3-3 Proteins: insights from genome-wide studies in yeast. Genomics 94, 287–293. doi: 10.1016/j.ygeno.2009.07.004
van Heusden, G. P., Griffiths, D. J., Ford, J. C., Chin, A. W. T. F., Schrader, P. A., Carr, A. M., et al. (1995). The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 229, 45–53.
van Heusden, G. P., and Steensma, H. Y. (2006). Yeast 14-3-3 proteins. Yeast 23, 159–171. doi: 10.1002/yea.1338
van Heusden, G. P. H., van der Zanden, A. L., Ferl, R. J., and Steensma, H. Y. (1996). Four Arabidopsis thaliana 14-3-3 protein isoforms can complement the lethal yeast bmh1 bmh2 double disruption. FEBS Lett. 391, 252–256. doi: 10.1016/0014-5793(96)00746-6
Vido, K., Spector, D., Lagniel, G., Lopez, S., Toledano, M. B., and Labarre, J. (2001). A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 276, 8469–8474. doi: 10.1074/jbc.M008708200
Wegel, E., Schauser, L., Sandal, N., Stougaard, J., and Parniske, M. (2007). Mycorrhiza mutants of Lotus japonicus define genetically independent steps during symbiotic infection. Mol. Plant Microbe Interact. 11, 933–936. doi: 10.1094/MPMI.1998.11.9.933
Xie, W., Liu, M., Lv, X., Lu, W., Gu, J., and Yu, H. (2014). Construction of a controllable β-carotene biosynthetic pathway by decentralized assembly strategy in Saccharomyces cerevisiae. Biotechnol. Bioeng. 111, 125–133. doi: 10.1002/bit.25002
Xie, X., Lin, H., Peng, X., Xu, C., Sun, Z., Jiang, K., et al. (2016). Arbuscular mycorrhizal symbiosis requires a phosphate transceptor in the Gigaspora margarita fungal symbiont. Mol. Plant 9, 1583–1608. doi: 10.1016/j.molp.2016.08.011
Yang, X., Wen, H. L., Sobott, F., Papagrigoriou, E., Robinson, C. V., et al. (2006). Structural basis for protein–protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. U.S.A. 103, 17237–17242.
Zézé, A., Dulieu, H., and Gianinazzi-Pearson, V. (1994). DNA cloning and screening of a partial genomic library from an arbuscular mycorrhizal fungus Scutellospora castanea. Mycorrhiza 4, 251–254. doi: 10.1007/BF00206773
Keywords: arbuscular mycorrhiza, abiotic stresses, Funneliformis mosseae, Rhizophagus irregularis, Fm201, host-induced gene silencing, 14-3-3 proteins
Citation: Sun Z, Song J, Xin X, Xie X and Zhao B (2018) Arbuscular Mycorrhizal Fungal 14-3-3 Proteins Are Involved in Arbuscule Formation and Responses to Abiotic Stresses During AM Symbiosis. Front. Microbiol. 9:91. doi: 10.3389/fmicb.2018.00091
Received: 18 September 2017; Accepted: 16 January 2018;
Published: 05 March 2018.
Edited by:
Erika Kothe, Friedrich-Schiller-Universität Jena, GermanyReviewed by:
Raffaella Balestrini, Consiglio Nazionale delle Ricerche (CNR), ItalyMaria Rapala-Kozik, Jagiellonian University, Poland
Copyright © 2018 Sun, Song, Xin, Xie and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Zhao, binzhao@mail.hzau.edu.cn Xianan Xie, 30004537@scau.edu.cn
 Zhongfeng Sun
Zhongfeng Sun Jiabin Song1
Jiabin Song1 Xi’an Xin
Xi’an Xin