- 1Institute of Animal Science and Veterinary Medicine, Shandong Academy of Agricultural Sciences, Shandong, China
- 2College of Life Sciences, Shandong Normal University, Shandong, China
The objective of this study was to evaluate the prevalence and characteristics of avian-origin mcr-1-harbouring Escherichia coli in Shandong Province, China. During 2017—2018, a total of 668 non-duplicate E. coli isolates were separately collected from 8eight large intensive poultry farms in Shandong Province. Antimicrobial susceptibility testing for 10 antimicrobial agents commonly used in farms was performed on all E. coli isolates by the agar dilution method; the mobile colistin resistance gene (mcr-1) gene was screened by PCR, and mcr-1 positive isolates were PCR-screened for antimicrobial resistance genes and typed by multi-locus sequence typing (MLST). Among the 668 E. coli, 102 (15.3%) harbored the mcr-1 gene; high antimicrobial resistance rates were observed for ampicillin (100/102, 98.0%), followed by amoxicillin (99/102, 97.1%) and florfenicol (97/102, 95.1%), and a low level of resistance was found for amoxicillin/clavulanic acid (24/102, 23.5%). Five ESBL genes were detected, and all isolates carried blaTEM (102/102, 100%), followed by blaCTX–M (90/102, 88.2%). Four PMQR genes were detected; aac(6)-Ib-cr (40/102, 39.2%) was the most commonly isolated PMQR gene, followed by qnrA (10/102, 9.8%). Thirty-eight different kinds of STs were identified, and the dominant ST was ST93 (19/102, 18.6%), followed by ST48 (9/102, 8.8%). In summary, E. coli from poultry in Shandong could be a reservoir for the mcr-1 gene, which could pose serious risks to human public health.
Introduction
Antimicrobial resistance is an ongoing severe public and animal health problem (Poirel et al., 2011). This issue occurs not only in pathogenic bacteria but also in non-pathogenic bacteria. The emergence of multidrug-resistant bacteria (MDR) has increased in the last decade and has caused public concerns around the world (Huang et al., 2018). Many studies have been reported that in Brazil, about 71.0% of the E. coli isolates from commercial chicken carcasses were MDR (Magiorakos et al., 2012); in Italy, 66.9% of the E. coli isolates from chicken meat were MDR (Ghodousi et al., 2015); in Egypt, 100% of the E. coli isolates from avian source were MDR (Enany et al., 2019), which demonstrated the high antimicrobial resistance. The addition of extensive subtherapeutic doses of antibiotics in animal feed is a very important reason for the rapid dissemination of antibiotic-resistant bacteria (Johnson et al., 2017).
Polymyxins are used to treat food-producing animals and have been considered the last resort antibiotics for the rapidly increasing MDR gram-negative pathogens, especially carbapenem-resistant Enterobacteriaceae (Pardon et al., 2012; Nation et al., 2015; Kaye et al., 2016). However, polymyxin use in animals and humans has promoted the selection and spread of mcr-1 drug-resistant strains (Sun et al., 2018). Since mcr-1 was discovered in food, animal and human isolates from Southern China, it has already spread to over 40 countries, implying that it played a prevalent role in the transferability of polymyxin resistance (Teo et al., 2017; Liu et al., 2016). In addition, mcr-1 positive plasmids could coexist with other resistance genes, notably ESBL genes; this has probably led to the emergence of pan-drug resistant strains and treatment failure (Haenni et al., 2016).
It has been reported that mcr-1 was restricted mainly to the Enterobacteriaceae, including E. coli (Olaitan et al., 2016). E. coli is a common inhabitant of the vertebrate intestinal tract and warm-blooded animals (Van den bogaard and Stobberingh, 2000). Moreover, avian-origin E. coli might hold zoonotic potential to cause human infections (Mellata et al., 2018).
China is one of the largest producers of chickens, especially in Shandong Province. In 2018, 1.65 billion commercial broilers were produced in Shandong Province, accounting for 41.2% of the country. An epidemiological survey of E. coli isolates from chickens from the early 1970s to 2014 suggested that the carriage rate of mcr-1 in China increased from 5.2% in 2009 to 30.0% in 2014 (Shen et al., 2016). However, in recent years, few studies have been performed on the prevalence and characteristics of avian-origin mcr-1-harboring E. coli in Shandong Province. Here, we aimed to determine the occurrence of the mcr-1-harboring E. coli isolated from poultry and their resistance profile between 2017 and 2018 in Shandong Province.
Materials and Methods
Ethics Statement
All procedures were approved by the Animal Care and Use Committee of Shandong Academy of Agricultural Sciences (SAAS-2019-021).
Samples and Bacteria Isolation
From August 2017 to May 2018, 722 rectal swabs were collected from 8 large intensive poultry farms (farms 1-8) located in Weifang, Dezhou, Yantai, Qingdao, and Liaocheng regions in Shandong Province. The samples were independently collected from individual animals. The stock of poultry per farm was more than 200,000 heads. All the samples were transported in an ice box to our laboratory within 6 h for further analysis.
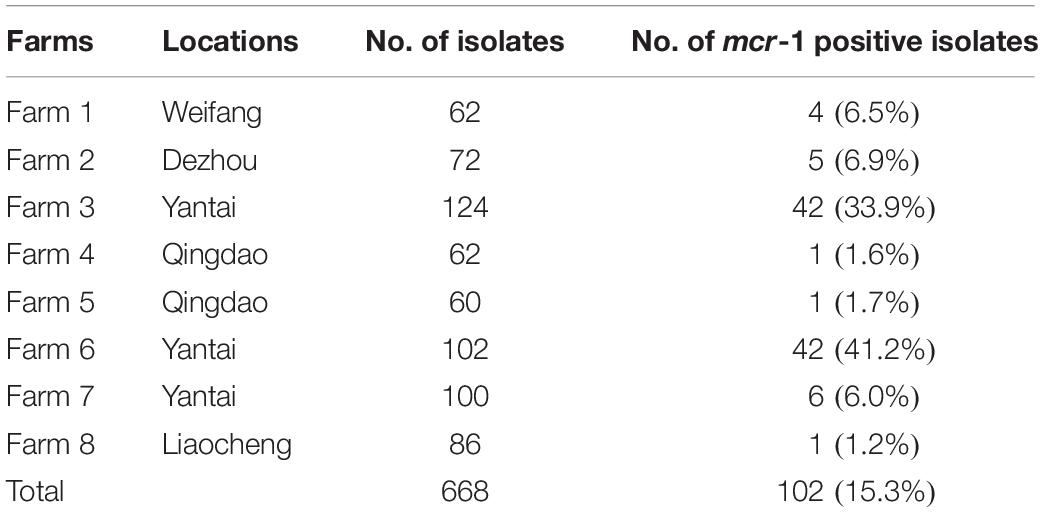
Each swab sample was inoculated in peptone water and cultured at 37°C for 24 h. A loop-full from incubated broth culture was streaked onto MacConkey’s agar and EMB plates. After incubation at 37°C for 24 h, suspected colonies were chosen by microscopical examination and further biochemical identification using the API 20E system (Sysmex bioMerieux, Tokyo, Japan). A total of 668 non-duplicate E. coli isolates were collected, and all of them were stored in Tryptic Soy broth plus glycerol (20%) at −20°C until further analysis (Table 1).
mcr-1 Gene Detection
All E. coli isolates were grown in Tryptic Soy broth with shaking overnight at 37°C. According to the manufacturer’s protocol, genomic DNA was extracted with a TIANamp Bacteria DNA Kit (Tiangen, Beijing, China). All isolates were screened for the mcr-1 gene by polymerase chain reaction (PCR) using primers and conditions as previously described (Liu et al., 2016). The PCR amplicons were purified and subsequently sequenced (Sangon, Beijing, China), and the obtained DNA sequences were compared with those in GenBank using Basic Local Alignment Search Tool (BLAST).
Antimicrobial Susceptibility Test
Antimicrobial susceptibility testing was performed on all E. coli isolates for 10 antimicrobial agents commonly used in farms including amoxicillin/clavulanic acid (AC), amoxicillin (AMO), ampicillin (AMP), ceftiofur (CEF), enrofloxacin (ENR), neomycin (NEO), doxycycline (DOX), florfenicol (FLO), gentamicin (GEN), and polymyxin (PB) by the agar dilution method recommended by the Clinical and Laboratory Standards Institute (CLSI, 2017); the susceptibility to colistin was interpreted according to the EUCAST guidelines1. The E. coli isolate ATCC 25922 was used as a quality control in this study.
Detection of Antimicrobial Resistance Genes
mcr-1 positive isolates were characterized for extended-spectrum β-lactamase genes (ESBL) (blaTEM, blaCTX–M, blaOXA, blaSHV and blaPSE) and plasmid-mediated quinolone resistance genes (PMQR) [aac(6)-Ib-cr, qnrA, qnrB, qnrC, qnrD, qepA, oqxA, and oqxB] by PCR using previously reported primers and conditions (Ahmed et al., 2013; Li et al., 2013). PCR products were visualized on 1.5% agarose gels, and the images were captured using image Capture Systems (Tanon-2500, Shanghai, China).
MLST
The mcr-1 positive isolates were characterized using the MLST scheme. MLST analysis was carried out by sequencing fragments of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) as described online2. The PCR amplicons were purified and subsequently sequenced (Sangon, Beijing, China), and the alleles and sequence types (STs) were assigned according to the MLST database3. A minimum spanning tree was created using BioNumerics software 6.5 (Applied Maths, Kortrijk, Belgium) to analyze the distribution of STs.
Statistical Analysis
Statistical comparisons of resistance to the 10 antimicrobial agents between mcr-1 positive isolates and mcr-1 negative isolates were analyzed using Fisher’s exact test or the chi-square test, which were performed using GraphPad INSTAT (GraphPad Software, Inc., United States). A value of P < 0.01 was considered significant.
Results
Prevalence of mcr-1 in Poultry Farms
Over the course of the study, 668 E. coli isolates were collected, and 102 (15.3%) of these were found to harbor the mcr-1 gene. The 102 mcr-1 positive isolates originated from poultry of farm 1 (n = 4), farm 2 (n = 5), farm 3 (n = 42), farm 4 (n = 1), farm 5 (n = 1), farm 6 (n = 42), farm 7 (n = 6), and farm 8 (n = 1). The prevalence of mcr-1 was 15.3% (102/668) (Table 1).
Antimicrobial Susceptibility Test
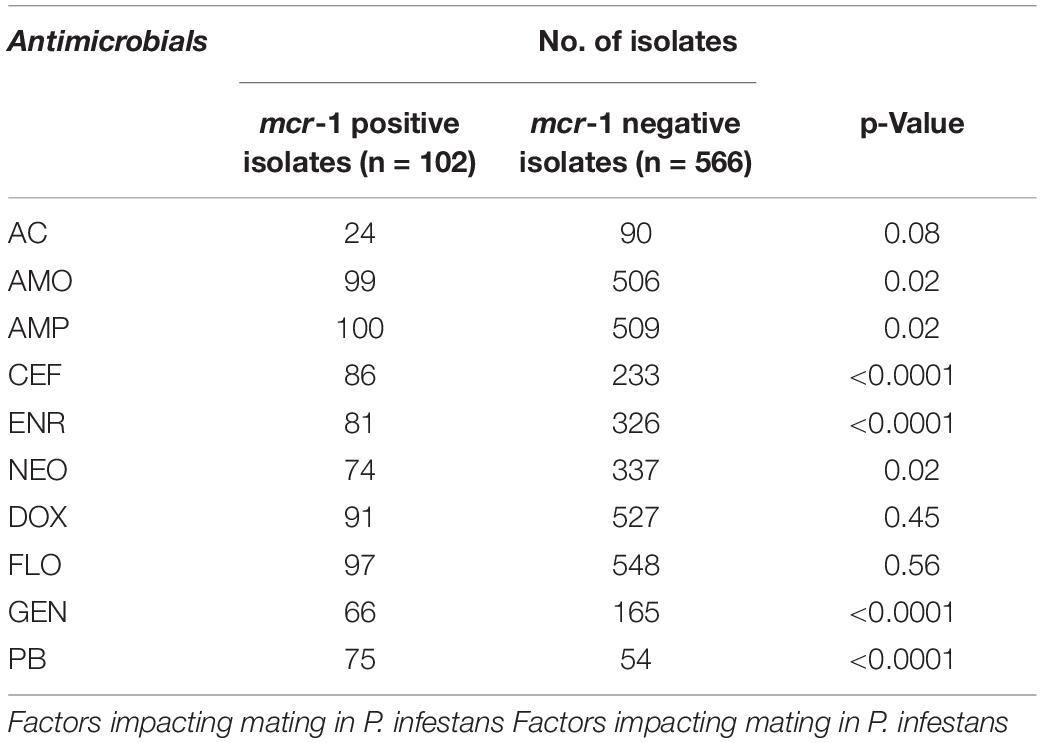
Of the 102 mcr-1 positive isolates, resistance to AMP was common (100/102, 98.0%), followed by resistance to AMO (99/102, 97.1%) and FLO (97/102, 95.1%), and a low level of resistance was found for AC (24/102, 23.5%). In addition, the 102 mcr-1 positive isolates showed higher resistance to CEF, ENR, GEN and PB than the mcr-1 negative E. coli isolates (Table 2).
Detection of Antimicrobial Resistance Genes
Among the 102 mcr-1 positive isolates, five ESBL genes were detected, all the isolates carried blaTEM (102/102, 100%), followed by blaCTX–M (90/102, 88.2%), blaPSE (30/102, 29.4%), blaOXA (16/102, 15.7%) and blaSHV (4/102, 3.9%). Four PMQR genes were detected; aac(6)-Ib-cr (40/102, 39.2%) was the most commonly isolated PMQR gene, followed by qnrA (10/102, 9.8%), qnrB (4/102, 3.9%), and qnrC (1/102, 0.9%), and no other PMQR genes were detected (Table 3).
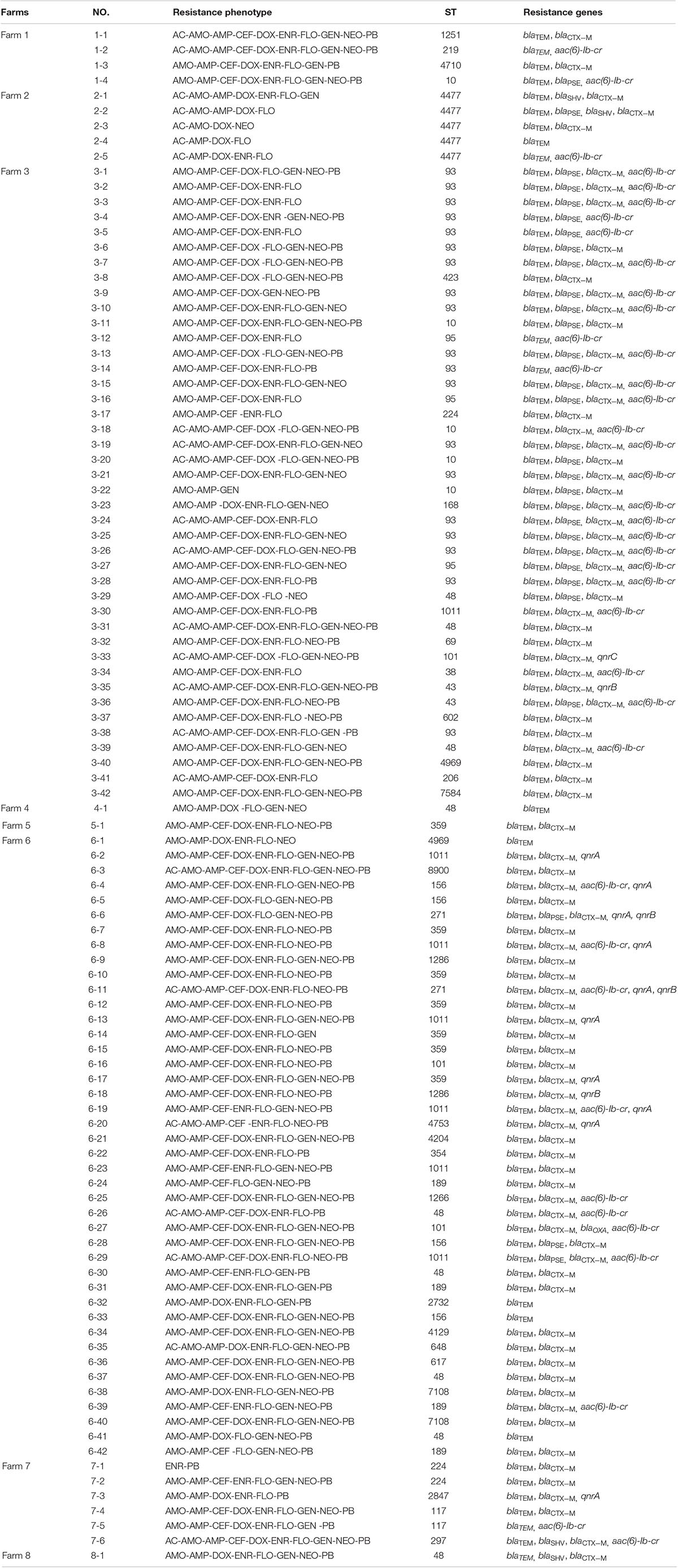
Table 3. Resistance phenotype, STs and resistance genes in the mcr-1 positive E. coli isolated from poultry farms.
MLST
Among the 102 mcr-1 positive isolates, 38 different kinds of STs were identified. The dominant ST was ST93 (19/102, 18.6%), followed by ST48 (9/102, 8.8%), ST1011 (7/102, 6.9%) and ST359 (7/102, 6.9%). Of note, 73.7% (14/19) of ST93 isolates carried blaTEM, blaPSE, blaCTX–M, aac(6)-Ib-cr. BioNumerics software version 6.5 was used to generate a minimum-spanning tree based on all the sources of STs. As a result, ST2732 and ST4129 belonged to one clone complex, ST2847 and ST4969 belonged to one clone complex, and ST7108, ST48, ST1286, ST10, ST43, ST4204, and ST8900 belonged to one complex (Figure 1).
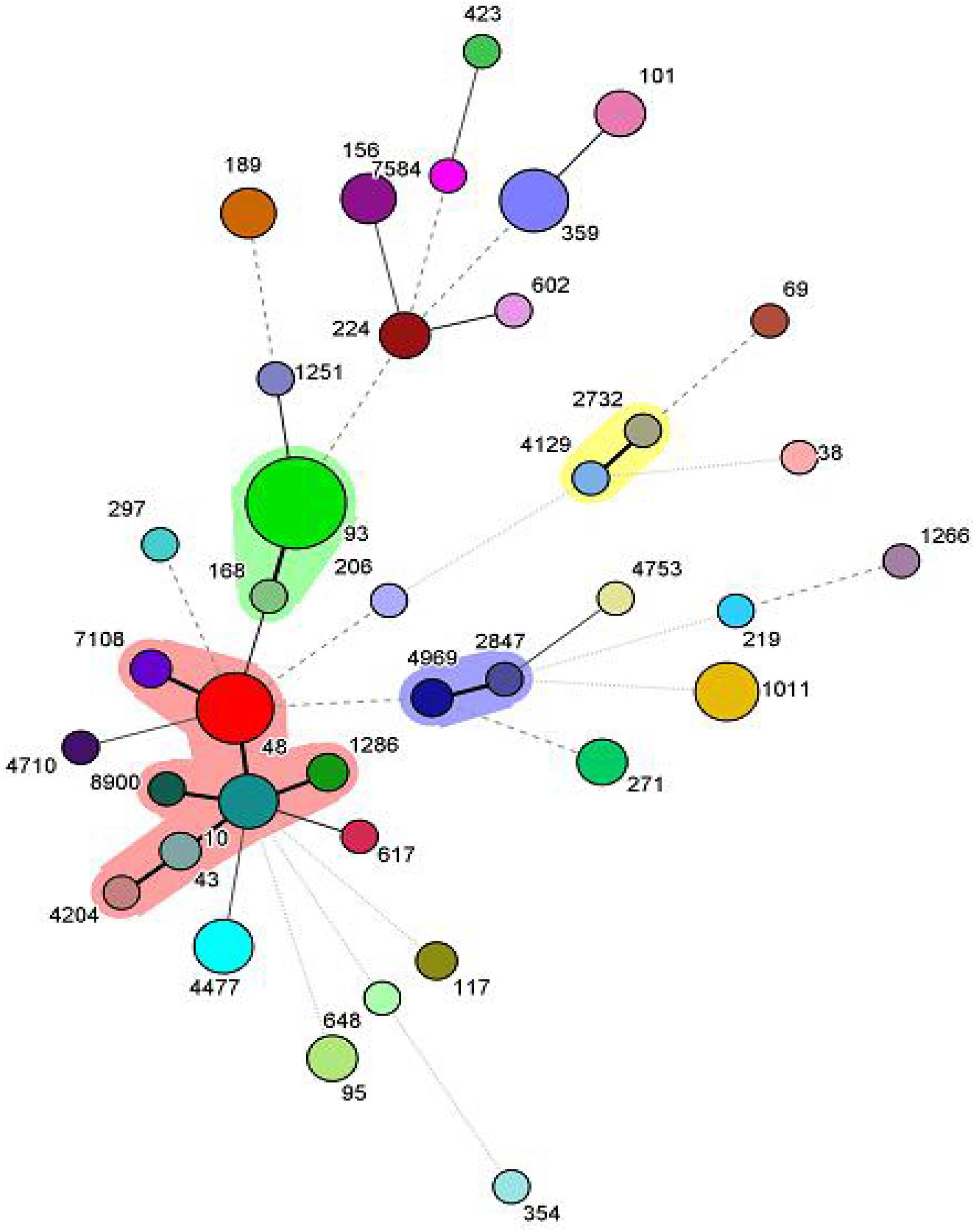
Figure 1. Minimum-spanning tree analysis of the 102 mcr-1 positive isolates. Each circle represents one ST, and the area of the circle corresponds to the number of isolates, the shadow region indicates that ST2732 and ST4129 belonged to one clone complex, ST2847 and ST4969 belonged to one clone complex, ST7108, ST48, ST1286, ST10, ST43, ST4204, and ST8900 belonged to one complex.
Discussion
The colibacillosis of poultry caused by E. coli is the most common infectious disease worldwide (Mitchell et al., 2015; Liu et al., 2018). The growing selective pressure of antimicrobial treatments may contribute to the rapid spread of antimicrobial resistance genes (Guardabassi et al., 2004). In this study, we detected the mcr-1 gene in 102 (15.3%) E. coli isolates. The prevalence of mcr-1 (15.3%) in this study was higher than that in Japan; 9,303 E. coli were isolated from healthy animals between 2000 and 2014, and 39 (0.4%) of them were carried the mcr-1 gene (Kawanishi et al., 2017). In Europe, 10,206 E. coli were isolated from cattle, pigs and chickens between 2008 and 2014, 68 (0.7%) of which carried the mcr-1 gene (Farid et al., 2018). Such a high prevalence may be due to the selection of the farms tested and the methodology used. In addition, the different prevalence of mcr-1 in each farm might be associated with the different use of drugs and management in this study. Of note, in the same region, the prevalence of mcr-1 from farm 3 and farm 6 was higher than farm 7, this probably due to the different dosage of the polymyxin used in the farms.
In the current study, we noticed a high rate of resistance to AMP, AMO, and FLO in the mcr-1 positive isolates similar to a report in Eastern China from 2015 to 2017 (Zhuge et al., 2019), suggesting that these drugs may have been widely used in poultry farming. Most of the mcr-1 positive isolates showed susceptibility to AC, which was associated with the infrequent use of this drug, and the result was consistent with other reports (Dominguez et al., 2018). In addition, the mcr-1 positive isolates showed higher resistance to PB than the mcr-1 negative E. coli isolates in this study, which suggested that there was a close positive correlation between the resistance phenotypes and genotypes of the mcr-1 positive isolates. This result is consistent with other reports (Huang et al., 2017; Shen et al., 2018; Zhuge et al., 2019).
Extended-spectrum β-lactamase genes and mcr-1 phosphoethanolamine transferase enzymes have been identified as the main plasmid-mediated mechanisms of resistance to third-generation cephalosporins and colistin, respectively, and are currently considered a major concern both in human and veterinary medicine. Many studies of E. coli strains, most of which were isolated from animals, have demonstrated the presence of the mcr-1 gene together with ESBL genes (Rhouma and Letellier, 2017). In this study, all the mcr-1 positive isolates carried blaTEM, which was consistent with the result that 98.0% of the mcr-1 positive isolates were resistant to AMP. CTX-M-producing enterobacteria are widespread among human populations, but an increasing number of reports have described their presence in food animals and their environment (Li et al., 2007; Ewers et al., 2012; Lazarus et al., 2015). In the present study, 88.2% of the mcr-1 positive isolates carried blaCTX–M, which was similar to a previous study in which more than half of the chicken-origin mcr-1 positive strains presented the coexistence of the mcr-1 and blaCTX–M genes (Yang et al., 2017), but higher than the 43.1% mcr-1 positive isolates carrying blaCTX–M genes. The increasing prevalence and dissemination of co-carriage of ESBL genes in mcr-1 positive E. coli isolates found in our study suggested that it may add challenge to the global problem of difficult to treat and untreatable infections.
Quinolones are one of the main choices for the treatment of colibacillosis that occurs in humans and animals (Moawad et al., 2017). Thus, quinolone resistance is a public health issue that can lead to treatment failures and the use of alternative agents with greater side effects. In this study, aac(6)-Ib-cr was the most commonly isolated PMQR gene among the mcr-1 positive isolates, which was consistent with a previous report (Zhao et al., 2018). However, it was different from a report in Argentina that did not detect aac(6)-Ib-cr from mcr-1 positive isolates from poultry, and the most prevalent PMQR gene was qnrB (Dominguez et al., 2018). In addition, most isolates carried aac(6)-Ib-cr, which confer resistance to enrofloxacin and gentamicin. In this study, we also found the qnrA, qnrB, and qnrC genes. According to the results of this study, we suggest that E. coli from poultry could be a reservoir not only for the mcr-1 gene but also for PMQR and ESBL genes.
In the current study, the MLST results revealed 38 distinct STs, which showed abundant diversity among mcr-1 positive isolates from different geographic locations. Notably, ST93 was the most common. It was different from that of previous studies that identified 40 distinct STs in 102 mcr-1 positive isolates, and ST48 was the most common (Yang et al., 2017). In addition, 73.7% of ST93 isolates carried blaTEM, blaPSE, blaCTX–M, aac(6)-Ib-cr, which were all from the same farm, indicating that the majority of the mcr-1 isolates from the same farm were probably clonal.
Conclusion
Our results showed that the mcr-1 gene was present at a high prevalence in Shandong Province, and poultry can be an important reservoir of mcr-1 carrying E. coli strains. The wide range of phenotypic resistance to antibiotics in veterinary medicine was identified, and the high rate of antibiotic resistance gene-positive isolates detected suggest that defining avian-source mcr-1 positive isolates is critical for public health purposes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
YL and XZ conceived and designed the study, wrote and revised the manuscript. YZ collected the samples. XZ, ZL, XY, and MH performed the experiments and analyzed the data.
Funding
This work was supported by the National key Research and Development Program of China (2018YFD0500500) and Shandong Agricultural Major Applied Technology Innovation Project (SD2019XM009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ www.EUCAST.org
- ^ https://enterobase.readthedocs.io/en/latest/mlst/mlst-legacy-info-ecoli.html
- ^ http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search
References
Ahmed, A. M., Shimamoto, T., and Shimamoto, T. (2013). Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. Int. J. Med. Microbiol. 303, 475–483. doi: 10.1016/j.ijmm.2013.06.009
CLSI, (2017). “Performance standards for antimicrobial susceptibility testing,” in Clinical and Laboratory Standards Institute Document M100-S27, 27th Edn, (Wayne, PA: CLSI).
Dominguez, J. E., Redondo, L. M., Espinosa, R. F., Cejas, D., Gutkind, G. O., Chacana, P. A., et al. (2018). Simultaneous carriage of mcr-1 and other antimicrobial resistance determinants in Escherichia coli from poultry. Front. Microbiol. 9:1679. doi: 10.3389/fmicb.2018.01679
Enany, M. E., Algammal, A. M., Nasef, S. A., Abo-Eillil, S. A. M., Jumah, M. B., Taha, A. E., et al. (2019). The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli. AMB Exp. 9:192. doi: 10.1186/s13568-019-0920-4
Ewers, C., Bethe, A., Semmler, T., Guenther, S., and Wieler, L. H. (2012). Extended-spectrum β-lactamase producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18, 646–655. doi: 10.1111/j.1469-0691.2012.03850.x
Farid, E. G., Anno, D. J., Xavier, B., Didier, H., and Marlene, S. (2018). mcr-1-like detection in commensal Escherichia coli and Salmonella spp. From food-producing animals at slaughter in Europe. Vet. Microbiol. 213, 42–46. doi: 10.1016/j.vetmic.2017.11.014
Ghodousi, A., Bonura, C., Noto, A. M. D., and Mammina, C. (2015). Extended-spectrum ß-lactamase, AmpC-Producing, and fluoroquinolone-resistant Escherichia coli in retail broiler chicken meat, Italy. Foodborne Pathog. Dis. 12, 619–625. doi: 10.1089/fpd.2015.1936
Guardabassi, L., Schwarz, S., and Lloyd, D. H. (2004). Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemoth. 54, 321–332. doi: 10.1093/jac/dkh332
Haenni, M., Poirel, L., Kiefer, N., Chatre, P., Saras, E., Metayer, V., et al. (2016). Co-occurrence of extended spectrum beta lactamase and MCR-1 encoding genes on plasmids. Lancet Infect. Dis. 16, 281–282. doi: 10.1016/s1473-3099(16)00007-4
Huang, Y. H., Chou, S. H., Liang, S. W., Ni, C. E., Lin, Y. T., Huang, Y. W., et al. (2018). Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J. Antimicrob. Chemother. 73, 2039–2046. doi: 10.1093/jac/dky164
Huang, Y. H., Yu, L. F., Chen, X. J., Zhi, C. P., Yao, X., Liu, Y. Y., et al. (2017). High prevalence of colistin resistance and mcr-1 gene in Escherichia coli isolated from food animals in China. Front. Microbiol. 8:562. doi: 10.3389/fmicb.2017.00562
Johnson, J. R., Porter, S. B., Johnston, B., Thuras, P., Clock, S., Crupain, M., et al. (2017). Extraintestinal pathogenic and antimicrobial-resistant Escherichia coli, including sequence type 131 (ST131), from retail chicken breasts in the United States in 2013. Appl. Environ. Microbiol. 81, 4498–4596. doi: 10.1128/AEM.02956-16
Kawanishi, M., Abo, H., Ozawa, M., Uchiyama, M., Shirakawa, T., Suzuki, S., et al. (2017). Prevalence of colistin resistance gene mcr-1 and Absence of mcr-2 in Escherichia coli isolated from healthy food-producing animals in Japan. Antimicrob. Agents Chem 61:e02057-16. doi: 10.1128/AAC.02057-16
Kaye, K. S., Pogue, J. M., Tran, T. B., Nation, R. L., and Li, J. (2016). Agents of last resort: polymyxin resistance. Infect Dis. Clin. N. Am. 30, 391–414. doi: 10.1016/j.idc.2016.02.005
Lazarus, B., Paterson, D. L., Mollinger, J. L., and Rogers, B. A. (2015). Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food producing animals? A systematic review. Clin. Infect. Dis. 60, 439–452. doi: 10.1093/cid/ciu785
Li, R. C., Lai, J., Wang, Y., Liu, S. L., Li, Y., Liu, K. Y., et al. (2013). Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province. China. Int. J. Food Microbiol. 163, 14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020
Li, X.-Z., Mehrotra, M., Ghimire, S., and Adewoye, L. (2007). β-lactam resistance and β-lactamases in bacteria of animal origin. Vet. Microbiol. 121, 197–214. doi: 10.1016/j.vetmic.2007.01.015
Liu, C., Diao, Y., Wang, D., Chen, H., and Tang, Y. (2018). Duck viral infection escalated the incidence of avian pathogenic Escherichia coli in China. Transbound Emerg. Dis. 66, 929–938. doi: 10.1111/tbed.13107
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mellata, M., Johnson, J. R., and Curtiss, R. (2018). Escherichia coli isolates from commercial chicken meat and eggs cause sepsis, meningitis and urinary tract infection in rodent models of human infections. Zoonoses Public Health 65, 103–113. doi: 10.1111/zph.12376
Mitchell, N. M., Johnson, J. R., Johnston, B., Curtiss, R., and Mellata, M. (2015). Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl. Environ. Microbiol. 81, 1177–1187. doi: 10.1128/AEM.03524-14
Moawad, A. A., Hotzel, H., Awad, O., Tomaso, H., Heinrich, N., Hafez, H. M., et al. (2017). Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 9:57. doi: 10.1186/s13099-017-0206-9
Nation, R. L., Li, J., Cars, O., Couet, W., Dudley, M. N., Kaye, K. S., et al. (2015). Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect. Dis. 15, 225–234. doi: 10.1016/S1473-3099(14)70850-3
Olaitan, A. O., Chabou, S., Okdah, L., Morand, S., and Rolain, J. M. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16:147. doi: 10.1016/s1473-3099(15)00540-x
Pardon, B., Catry, B., Dewulf, J., Persoons, D., Hostens, M., De Bleecker, K., et al. (2012). Prospective study on quantitative and qualitative antimicrobial and anti-inflammatory drug use in white veal calves. J. Antimicrob. Chemother. 67, 1027–1038. doi: 10.1093/jac/dkr570
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Rhouma, M., and Letellier, A. (2017). Extended-spectrum β-lactamases, carbapenemases and the mcr-1 gene: is there a historical link? Int. J. Antimicrob. Agents. 49, 269–271. doi: 10.1016/j.ijantimicag.2016.11.026
Shen, Z. Q., Wang, Y., Shen, Y. B., Shen, J. Z., and Wu, C. M. (2016). Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 16:293. doi: 10.1016/s1473-3099(16)00061-x
Shen, Y., Wu, Z., Wang, Y., Zhang, R., Zhou, H. W., Wang, S. M., et al. (2018). Heterogeneous and flexible transmission of mcr-1 in hospital-associated Escherichia coli. MBio 9:e00943-18
Sun, J., Zhang, H., Liu, Y. H., and Feng, Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
Teo, J. W. P., Kalisvar, M., Venkatachalam, I., Ng, O. T., Lin, T. P., and Octavia, S. (2017). mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J. Clin. Microbiol. 56:e01562-17.
Van den bogaard, A. E., and Stobberingh, E. E. (2000). Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicro Chem. 14, 327–335. doi: 10.1016/s0924-8579(00)00145-x
Yang, Y. Q., Li, Y. X., Song, T., Yang, Y. X., Jiang, W., Zhang, A. Y., et al. (2017). Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob. Agents Chem. 61:e01204-16. doi: 10.1128/AAC.01204-16
Zhao, X. N., Yang, J., Jv, Z. J., Chang, W. S., and Sun, S. H. (2018). Molecular characterization of antimicrobial resistance in escherichia coli from rabbit farms in Tai’an, China. BioMed Res. Int. 2018:8607647.
Keywords: Escherichia coli, mcr-1, poultry, resistance, MLST
Citation: Zhao X, Liu Z, Zhang Y, Yuan X, Hu M and Liu Y (2020) Prevalence and Molecular Characteristics of Avian-Origin mcr-1-Harboring Escherichia coli in Shandong Province, China. Front. Microbiol. 11:255. doi: 10.3389/fmicb.2020.00255
Received: 25 October 2019; Accepted: 03 February 2020;
Published: 20 February 2020.
Edited by:
Hemda Garelick, Middlesex University, United KingdomReviewed by:
Carlos Henrique Camargo, Adolfo Lutz Institute, BrazilJianmin Zhang, South China Agricultural University, China
Copyright © 2020 Zhao, Liu, Zhang, Yuan, Hu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuqing Liu, liuiuqing@163.com
†These authors have contributed equally to this work
 Xiaonan Zhao
Xiaonan Zhao Zhengjie Liu1†
Zhengjie Liu1†