- 1State Key Laboratory of Grassland Agro-Ecosystems, Key Laboratory of Grassland Livestock Industry Innovation, Ministry of Agriculture, College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou, Gansu, China
- 2Agri-Food and Biosciences Institute, Hillsborough, United Kingdom
Compared to traditional herbage, functional native herbage is playing more important role in ruminant agriculture through improving digestion, metabolism and health of livestock; however, their effects on rumen microbial communities and hindgut fermentation are still not well understood. The objective of present study was to evaluate the effects of dietary addition of Allium mongolicum on bacterial communities in rumen and feces of claves. Sixteen 7-month-old male calves were randomly divided into four groups (n = 4). All calves were fed a basal ration containing roughage (alfalfa and oats) and mixed concentrate in a ratio of 60:40 on dry matter basis. In each group, the basal ration was supplemented with Allium mongolicum 0 (SL0), 200 (SL200), 400 (SL400), and 800 (SL800) mg/kg BW. The experiment lasted for 58 days. Rumen fluid and feces in rectum were collected, Rumen fluid and hindgut fecal were collected for analyzing bacterial community. In the rumen, Compared with SL0, there was a greater relative abundance of phylum Proteobacteria (p < 0.05) and genera Rikenellaceae_RC9_gut_group (p < 0.01) in SL800 treatment. In hindgut, compared with SL0, supplementation of A. mongolicum (SL200, SL400, or SL800) decreased in the relative abundances of Ruminococcaceae_UCG-014 (p < 0.01), Ruminiclostridium_5 (p < 0.01), Eubacterium_coprostanoligenes_group (p < 0.05), and Alistipes (p < 0.05) in feces; Whereas, the relative abundances of Christensenellaceae_R-7_group (p < 0.05), and Prevotella_1 (p < 0.01) in SL800 were higher in feces, to maintain hindgut stability. This study provided evidence that A. mongolicum affects the gastrointestinal of calves, by influencing microbiota in their rumen and feces.
1. Introduction
Some plants (e.g., Gentiana straminea, Ligularia virgaurea, and Cistanche deserticola) are often not favored by herbivores because of their unique taste or poor palatability (Liu X. et al., 2020; Xie et al., 2022; Cui et al., 2023). These undesirable plants are abundant all year round and thus can be used as non-conventional feed resources or “functional native herbage (FNH).” The whole plant or some organs of FNH are rich in secondary metabolites throughout the whole year or during the growing season. Compared with traditional herbage, FNH are rarely ingest by livestock. It is usually intake during non-growing season, or unavoidably intake by grazing livestock, or only actively intake in exceptional circumstances such as injury or illness (Wang et al., 2022). FNH can regulate digestion and metabolism, productivity and health of livestock. It plays an irreplaceable role in livestock production (Zhang et al., 2022). FNH comprises 16.8–26.8 and 13.5% of plant species and biomass in global terrestrial ecosystems, respectively (Richards et al., 2015; Jamshidi-Kia et al., 2018; Hou et al., 2021). The FNH contributes up to 15% of the livestock feed from grazing (Zhang and Zhang, 2017). Feeding amount and function of FNH are similar to fresh grass and additives, respectively (Harris et al., 1968). Multiple secondary metabolites (e.g., phenylethanoid glycoside, volatile components, iridoids, flavonoids, alkaloids, simple secoiridoid glycoside, flavonoids, and iridoid glycosides) in Cistanche deserticola and Gentiana straminea can directly alter enteric microbe involved in livestock digestion and metabolism (Liu X. et al., 2020; Xie et al., 2022).
Rumen bacterial community play a vital role in maintaining the normal digestion and absorption in the host (Liu et al., 2016). The bacterial community structure in feces plays an important role in fermentation of carbohydrates and proteins; and the composition of bacterial population is influenced by the chemical composition of diet (Petri et al., 2019). Although FNH was used as safe and effective feed additive in ruminant agriculture (Ahmad et al., 2020), and the effects of supplementing extracts from FNHs on the rumen microbiome have been studied; the effect of whole plant FNHs have not been thoroughly evaluated. Therefore, it is important from the ruminant nutritionists to gain a better understanding of the relationship between the rumen and fecal microbes and the potential contribution of FNH to gut nutrient metabolism and function.
The rumen and hindgut microbial communities play an important role in the degradation of plant fiber and protein absorption (Cancino-Padilla et al., 2021), and their subsequent conversion into volatile fatty acids (VFA; Wang et al., 2016; Toral et al., 2018). Microbial fermentation of carbohydrates accounts for 5–10% of the total-tract digestibility of carbohydrates in the entire alimentary canal of cattle (Gressley et al., 2011). In previous studies (Panthee et al., 2017; Redoy et al., 2020), the effects of allicin, garlic powder, garlic oil, onion, and Allium sativum extracts have been examined on rumen fermentation and microbes in vitro. However, the effects of these additives have not been studied on gastrointestinal microbes in other parts of the digestive tract. Some secondary metabolites (condensed tannins, saponins, essential oils, and flavonoids) of FNHs, (Cichorium L., Allium L., Gentiana L., and Morus L.), have been proved to improved the intestinal absorption of nutrients (Patra et al., 2017; Ibrahim et al., 2021). Gentiana straminea and Ligularia virgaurea contain many secondary metabolites, including monoterpenoids, diterpenoids, triterpenes, sesquiterpenes, simple secoiridoid glycoside, flavonoids, and iridoid glycosides, which may have antagonistic or synergistic on livestock performance and gastrointestinal flora (Xie et al., 2022; Cui et al., 2023), the whole herbage additions could enhance livestock performance and rumen microbes simultaneously. The “combined effect” consists of three aspects: firstly, a FNH normally contains a variety of active components which have a kind of synergistic or antagonistic effect on livestock performance as a wholeness; secondly, there were differences in the types and contents of various FNH and active components in diets, which had positive, negative or no effect on livestock; finally, the productive performance of livestock responds to the active component of FNH, which is reflected in the regulation of immune performance, digestion and metabolism, and growth performance, etc. The “dilution effect” consists of three aspects: firstly, due to the active component or the special odor of FNH, FNH increases or decreases the proportion of FNH in dry matter intake (DMI) by influencing DMI of livestock base diets; secondly, the bright color or active components of FNH regulate neuroendocrine feeding centers, improve the selective feeding of livestock, promote feed intake of forage with high water content (based on the some dry matter), stimulate gastric secretion, and reduce the concentration of FNH or its active components in internal organs and tissues; finally, The “combined effects” of multiple FNHS, or between FNHS and diet, cause the active component to break down and reduce the concentration in the body. As a result, the whole herbage of FNH also has the potential to reduce methane emissions through a “combined effect” of multiple secondary metabolites (Nobler et al., 2019; Xie et al., 2020), or indirectly by promoting intake of other diets by ingesting whole forage. This “dilution effect” of multiple secondary metabolites also has a positive effect on livestock performance and the hindgut microbiome (Neave et al., 2018). Consequently, a comprehensive analysis of whole forage on ruminants’ microbiota is crucial for a realistic comparison of the differences in secondary metabolism. Therefore, it was hypothesized that whole herbage had a “combined effect” or “dilution effect” on the gastrointestinal microbes of ruminants (Figure 1).
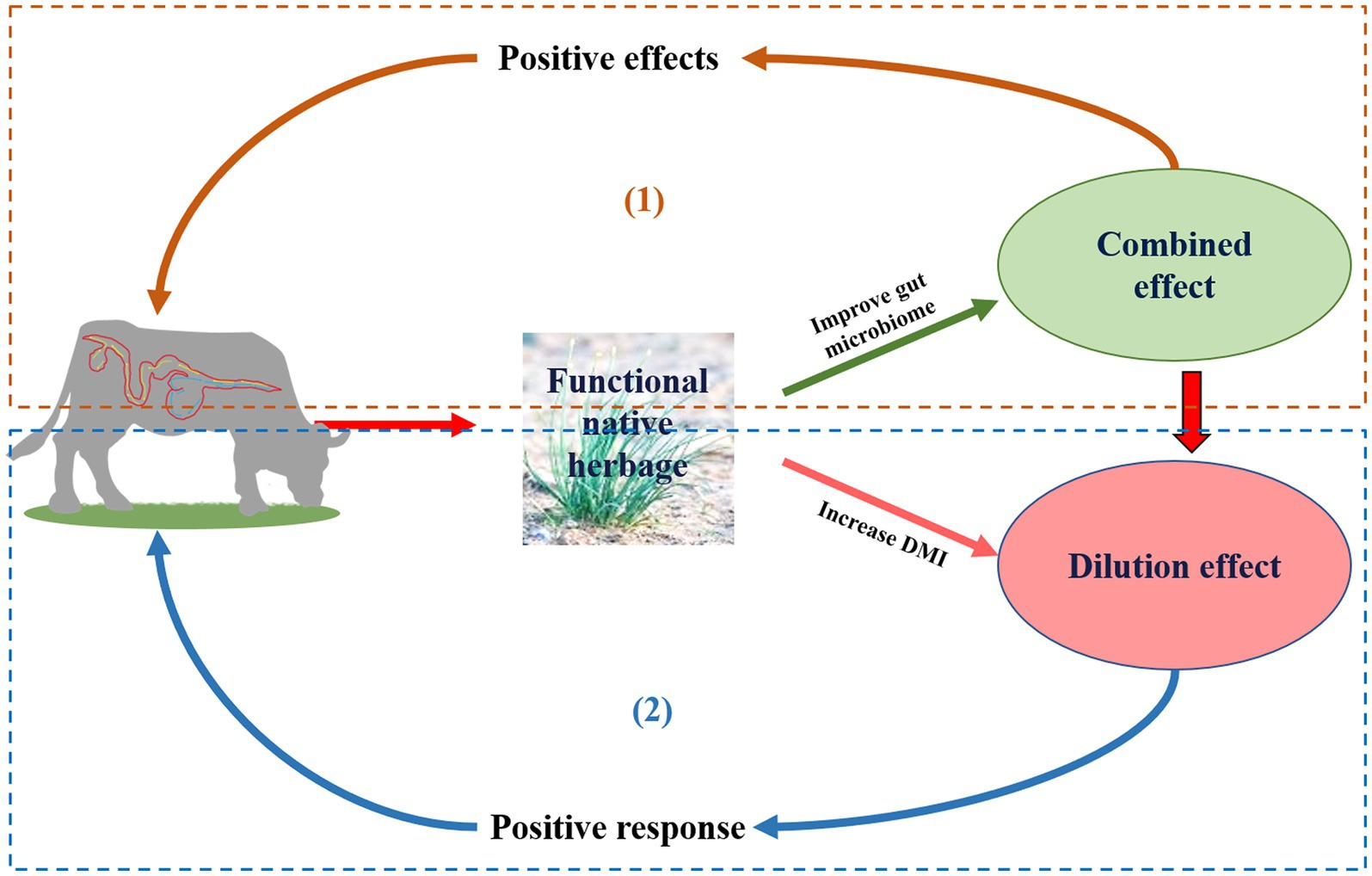
Figure 1. Conceptual sketch of effect of the whole functional native herbage (FNH) feed on gastrointestinal microorganisms of ruminants. (1) Combined effect: Multiple secondary metabolites in FNH have a positive effect on animals by combining to simultaneously produce beneficial effects on gastrointestinal microbes; (2) Dilution effect: Indirectly by promoting intake of other diets by ingesting FNH.
Allium, a worldwide genus belonging to FNH, contains garlic oil, flavonoids, polysaccharides, alkaloids, etc. (Wingard et al., 2011), which are fed to ruminant livestock (Wanapat et al., 2015). Allium mongolicum grows widely in the Euro-Asia steppe zone (Huang et al., 2011), which is one of the most expansive nomadic regions of the world (Jeong et al., 2020). Despite A. mongolicum has potential to improve growth performance, nutrient digestibility, immunity, and rumen fermentation parameters of Simmental calf (Patra et al., 2017; Xie et al., 2020), the effects of whole plant feed supplements on the gastrointestinal microbiome of ruminants are not well understood. The present study was designed to evaluate the effect of A. mongolicum on the relationship of the microbes between the rumen and feces, to identify the relationship between intestinal function and rumen-hindgut microbes changes, and to provide evidence for the application of A.mongolicum on the diet of Simmental calf.
2. Materials and methods
2.1. Study site
The experiment was conducted at the Linze Pastoral Agriculture Station of Lanzhou University, in Linze County, Gansu Province, China (39.24°N, 100.06°E) from July to September 2017. The area is classified as a temperate arid, with an average annual temperature and precipitation of 7.7°C and 117.2 mm, respectively. The dominant agricultural systems here are intensively and extensively specialized crop and livestock production systems (Hou et al., 2021).
2.2. Feeding and management of animals
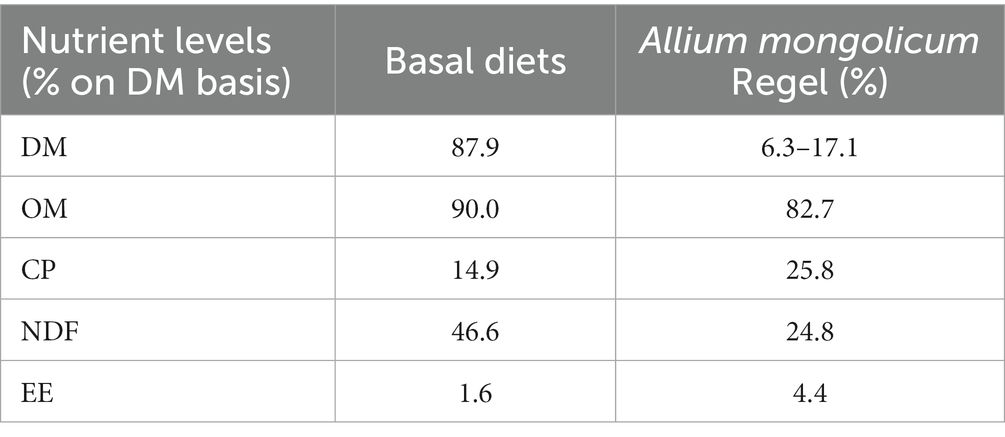
Sixteen 7-month-olds male Simmental calves (175 ± 10 kg), were randomly divided into four groups with four calves in each group. All calves were fed a basal ration containing roughage (alfalfa and oats) and mixed concentrate in a ratio of 60:40 on dry matter basis. The basal diet fulfilled the maintenance and growth requirements of calves to support 1 kg daily gain for fattening cattle (175 kg BW) according to the Chinese recommended beef cattle breeding standards (NY/T815-2004). In each group, the basal ration was supplemented with A. mongolicum 0 (SL0), 200 (SL200), 400 (SL400), or 800 mg/kg BW (SL800). The experiment lasted for 58 days. The nutrients of fresh A. mongolicum and basal diets (oat and alfalfa hay, soybean meal, wheat barn, and corn kernel) were determined (Table 1). Roughage was fed separately twice daily in equal amounts at 7 a.m. and 7 p.m. Fresh A. mongolicum was mixed with concentrates and offered at 2 pm. daily. All the calves were kept in single-pens (1.6 m × 3.6 m) and had free access to drinking water around the clock. A detailed description of the animal digestion and metabolism trial, laboratory analyses and calculation of DMI, body weight (BW), nutrient digestibility, fecal nutrient content, methane (CH4), volatile fatty acids (acetate, propionate, butyrate, and valerate) can be found in a complementary paper on Simmental claves (Xie et al., 2020).
2.3. Fecal collection
Samples from rumen fluid and hindgut feces were taken on the last day of the experiment for analyze bacterial community. Fecal samples were collected and divided into three parts: one portion was dried at 65°C for 96 h to determine the neutral detergent fiber (NDF), acid detergent fiber (ADF), organic matter (OM) and ether extract (EE) content; other portion of fresh feces (100 g) was combined with 10 mL of 10% (v/v) sulfuric acid for N fixation and frozen at −20°C; and the final portion of the feces was placed in a vacuum tube and immediately stored in liquid nitrogen for bacterial determination.
Fecal collection, for subsequent analysis of its microbiota, was done following the method described by Petri et al. (2019). Specifically, fecal samples were collected from rectum of the calves by hand, using a new palpation sleeve for each calf at the 58 day of the experiment, between 600 to 630 h. The fecal sample collection and rumen collection were on the same day to reduce the stress of animals. The use of rectally collected fecal samples, for assessing hindgut fermentation profiles and microbial activity has been considered a reasonable approach (Gressley et al., 2011; Castillo-Lopez et al., 2020).
2.4. Rumen fluid sampling
Rumen fluid samples of 16 calves were taken via the orogastric tube technique on day 58. Two hours after morning feeding, a ruminal fluid collection tube (Anscitech Co., Ltd. Winnipeg, Manitoba, Canada) was inserted into the rumen and the rumen fluid was aspirated using a 150 mL syringe. After cleaning the orogastric tube with clean water, 100 mL of rumen fluid was discarded, preventing saliva contamination (Fan et al., 2021). The rumen fluid was collected in cryovials after filtration through double layers of gauze and immediately flash frozen in liquid nitrogen for subsequent bacterial analysis.
2.5. DNA isolation, 16S rRNA sequencing, and bioinformatics analyses
Genomic DNA was extracted from rumen fluid and feces by hexadecyl trimethyl ammonium Bromide (Minas et al., 2011). Purity and concentration of DNA was assessed by atomic spectrophotometry and 1% agarose gel electrophoresis. An appropriate amount of sample was aliquoted into a centrifuge tubes, and diluted to 1 ng/μL with sterile water. The V3-V4 hypervariable region of the bacterial 16S rRNA gene was amplified using universal primers forward primer (515F) 5′ -GTGCCAGCMGCCGCGG-3′ and reverse primer (806R) 5′-GGACTACHVGGGTWTCTAAT-3′. The PCR amplification reaction cycles for subsequent high-throughput sequencing were as follows: initial denaturation was done at 95°C for 2 min, followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 60 s at 72°C, with a final extension at 72°C for 5 min. The PCR products were purified using 2% agarose gel electrophoresis. The TruSeq® DNA PCR-Free Sample Preparation Kit was used for library construction. The constructed library was quantified using a Qubit and q-PCR. After qualification of the library, HiSeq2500 PE250 was used for on-machine sequencing. The 16-rumen fluid and 16 fecal samples were sequenced. Paired-end reads were assigned to samples based on their unique barcodes. The reads of each sample were spliced using FLASH (V1.2.7; Magoč and Steven, 2011) after truncating the barcode and primer sequences, and the splicing sequence obtained was considered the original Tag data (Raw Tags).1 The spliced Raw Tags underwent strict filtering for obtaining high-quality Tag data (Clean Tags; Bokulich et al., 2013). Low-quality raw tags were filtered using QIIME (V1.7.0; Caporaso et al., 2010).2 The tag sequence (UCHIME Algorithm)3 were compared with the database (Gold database)4 for detecting the chimera sequence, which were then removed for subsequent analyses (Effective Tags; Edgar et al., 2011; Haas et al., 2021). Sequence analyses was clustered by UPARSE into operational taxonomic units (OTUs) based on 97% similarity (Edgar, 2013). Representative sequences of OTUs were subjected to species annotation analysis using the SSUrRNA database of SILVA (Wang et al., 2007). Alpha diversity analysis for Observed-species, Chao1, Shannon, Simpson, and Goods-coverage indices was performed using the QIIME software (Version 1.7.0). The principal coordinates analysis (PCoA) was analyzed done using R software’s WGCNA, stats and ggplot2 packages using UniFrac distance-based principal coordinate analysis.
2.6. Statistical analysis
The data were statistically analyzed Normal distributions were checked using the Shapiro–Wilk test, using one-way analysis of variance using ANOVA procedures of SAS (SAS v 9.2, SAS Institute Inc., Cary, NC, United States). Treatment means were compared using LSD test and differences were considered statistically significant at p < 0.05.
Spearman correlation coefficients between chemical composition of herbage, nutrient digestibility, CH4, bacterial genus, and ruminal fermentation parameters was performed using R software (version 4.0.2).5 Microbial networks were generated to calculate the correlations between predominant taxa using Gephi software (version 0.9.2).6 The putative drivers of keystone taxa in the rumen and feces microbial communities of Simmental calves from the gastrointestinal region were estimated using combined score of the high mean degree, high closeness centrality, and low betweenness centrality. Spearman correlation coefficients between the relative abundances of the rumen bacteria (genus) and nutrient digestibility were calculated using the heatmap package in R software (version 4.0.2, see text footnote 5). The R.4.1.2 (Gemplus software package) was used to perform Aggregate Boosted Tree Analysis (ABT) to quantitatively analyze the correlations of nutrient digestibility, rumen fermentation parameters and fecal nutrient nutrients, and dominant bacteria in rumen and feces. Redundancy analysis (RDA) in CANOCO (Windows 4.5) was used to analyze the correlations of nutrient digestibility, rumen fermentation parameters, fecal nutrient nutrients, and dominant bacteria in rumen and feces.
3. Results
3.1. Microbial community richness, diversity, and distribution in rumen and feces
Chao 1 index in SL200 was significantly higher than that in SL0 (p = 0.036). The Goods_coverage value of the rumen and feces were 99.3 ± 0.2 and 99.5 ± 0.1%, respectively (Table 2). In feces, the bacterial species on the SL200 treatment was the lowest of all the treatment groups (p = 0.031). The Shannon and Simpson diversity indices of rumen and feces were not significantly different among the four treatment groups. In the rumen, each of the four treatments had a cluster of microorganisms (Figure 2A). However, the principal coordinate analysis showed clear separations of fecal bacteriome in SL0 and SL200 treatments. The observed spatial heterogeneity of the fecal bacterial community was similar in SL400 and SL800 treatment groups (Figure 2B).
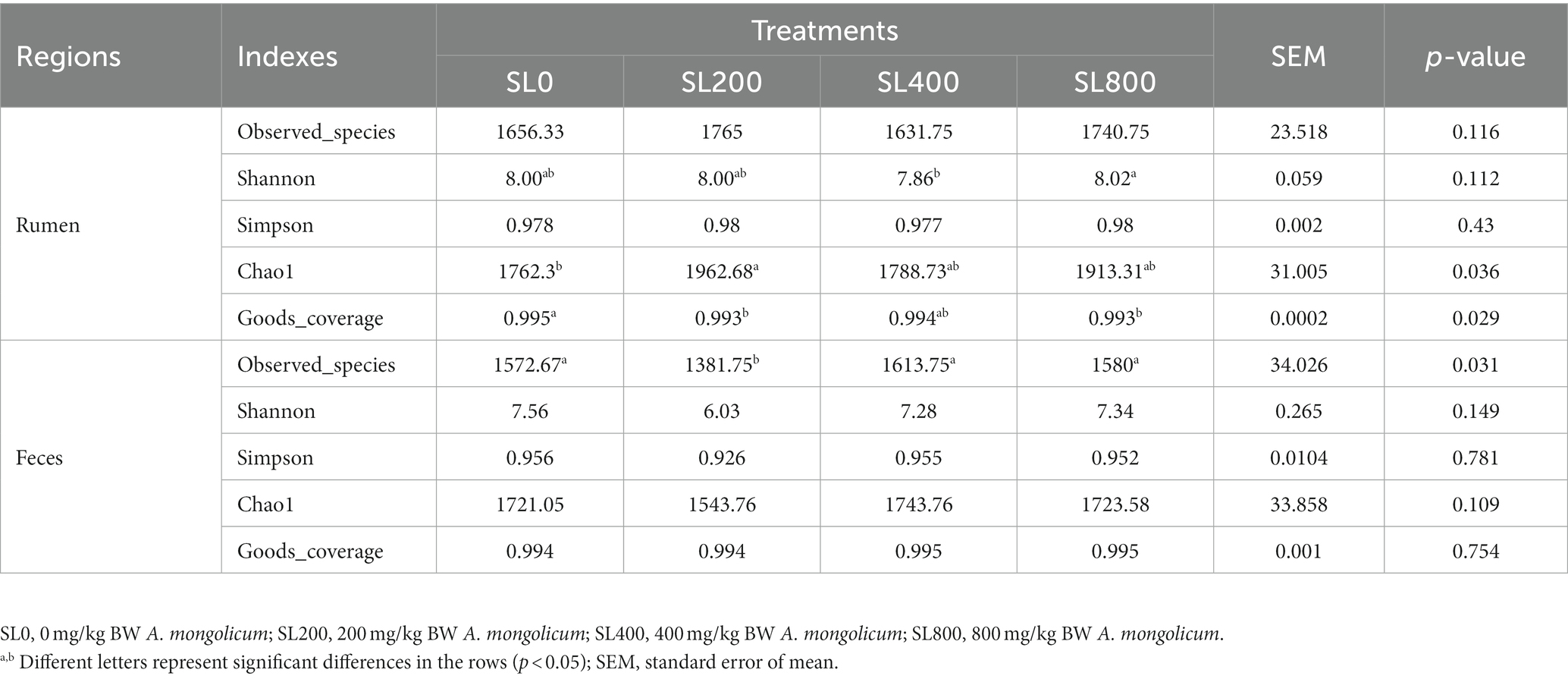
Table 2. Alpha-diversity of the microbiome of rumen and fecal sample of calves fed different amounts of A. mongolicum.
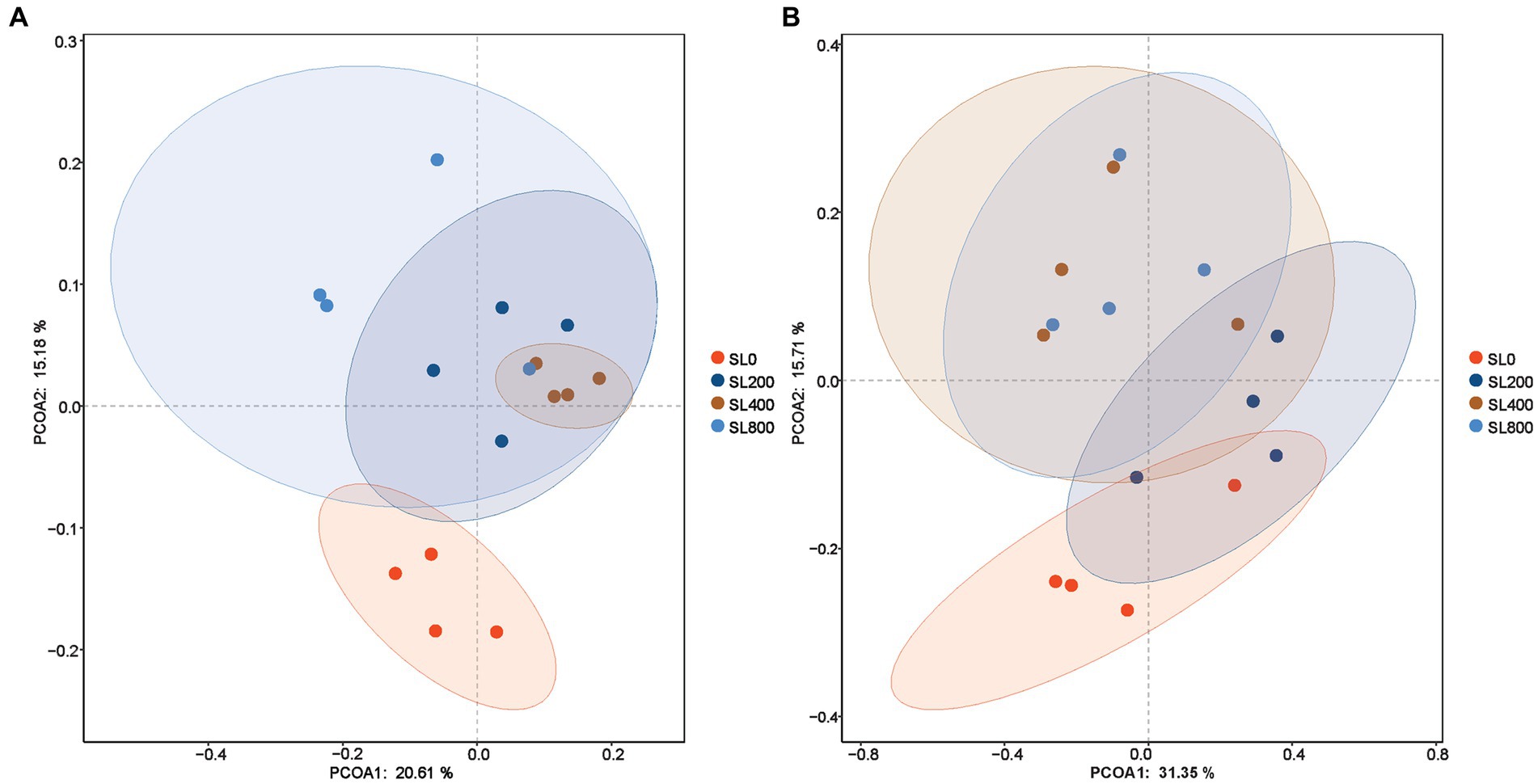
Figure 2. Alpha-diversity of rumen and fecal microbiome in Simmental calves fed different levels of A. mongolicum. (A) Rumen; (B) feces. SL0, 0 mg/kg BW A. mongolicum; SL200, 200 mg/kg BW A. mongolicum; SL400, 400 mg/kg BW A. mongolicum; SL800, 800 mg/kg BW A. mongolicum.
3.2. Bacterial community composition in the rumen and feces of Simmental calves
There were 26 and 24 bacterial phyla observed in the rumen and fecal microbiota, respectively (Figure 3). At the phylum level, the relative abundance of the two predominant phyla Firmicutes and Bacteroidetes in rumen accounted for 52.9 and 40.3%, respectively (Figure 3A). Compared to SL0 treatment, the relative abundance of Proteobacteria were higher and of Bacteroidetes was lower (p < 0.05) in the SL800 treatment (Supplementary Table S1) in rumen.
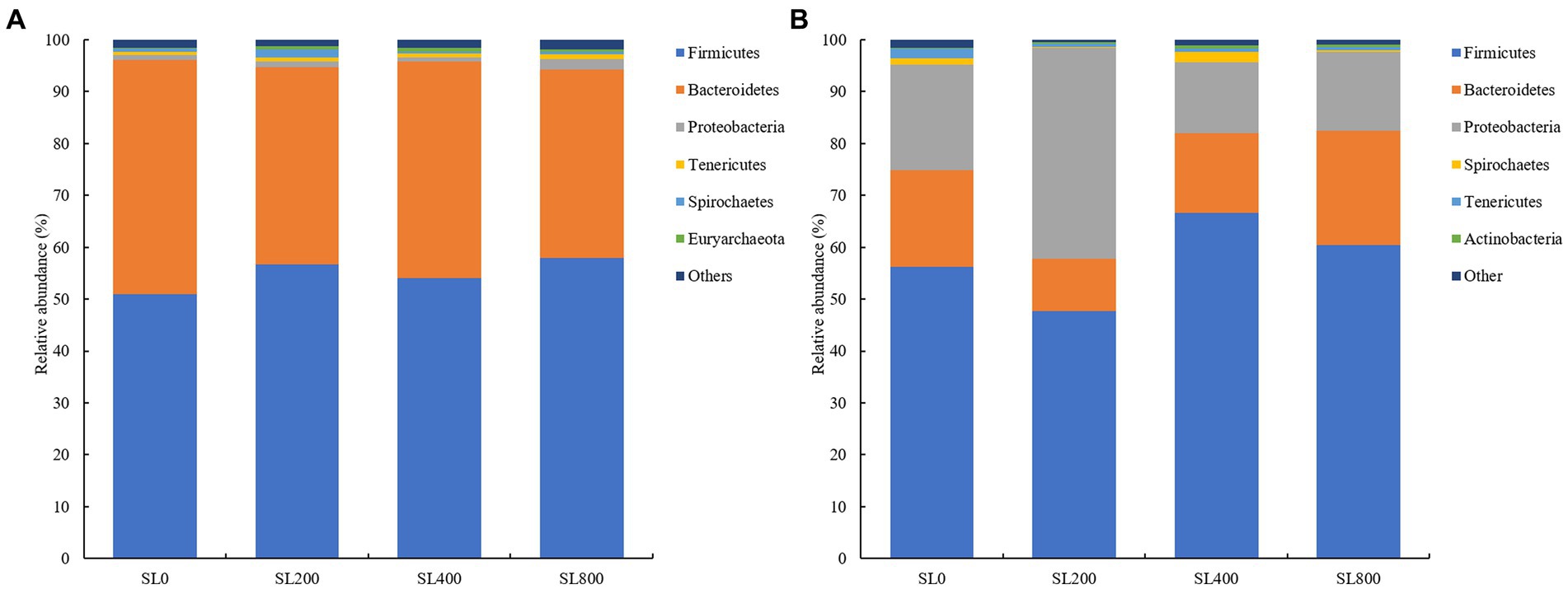
Figure 3. Composition of bacterial communities in Simmental calves at phylum levels in rumen (A) and feces [(B); only taxa with an average relative abundance >0.5% are shown]. SL0, 0 mg/kg BW A. mongolicum; SL200, 200 mg/kg BW A. mongolicum; SL400, 400 mg/kg BW A. mongolicum; SL800, 800 mg/kg BW A. mongolicum.
In feces, Firmicutes (57.7% ± 4.4), Proteobacteria (22.5% ± 5.8), and Bacteroidetes (16. 6% ± 2.1) were the predominant bacteria, accounting for more than 96.7% of the total bacteria (Figure 3B). The relative abundances of Firmicutes (p < 0.01) were higher in the SL400 and SL800 than those in the SL0 treatment. The relative abundances of Spirochaetes (p < 0.01) were lower in the SL200 and SL800 than those in the SL0 and SL400 treatment groups (Supplementary Table S2) in feces. Relative abundance of Tenericutes (p < 0.05) was lower in the groups receiving Allium mongolicum than SL0 in feces. The relative abundances of Actinobacteria (p < 0.05) were higher in the SL400 than those in the SL0 treatment group. The ratio of Firmicutes to Bacteroidetes (p > 0.05) was not significantly different among the four treatment groups.
At the genus level, the relative abundance of Prevotella_1 was the most dominant in ruminal fluid, followed by the Christensenellaceae_R-7_group, Rikenellaceae_RC9_gut_group, and Ruminococcaceae_NK4A214_group (Figure 4A). Compared with SL0, the relative abundances of Prevotella_1 was lower (p < 0.05), representing 16.8 and 14.7% of the populations in the SL200 and SL800 groups, respectively (Supplementary Table S1). The relative abundance of Rikenellaceae_RC9_gut_group of the SL800 treatment was higher (p < 0.01) than in the SL0 and SL200 group, while SL400 was not significantly different than SL0, SL200, and SL800.
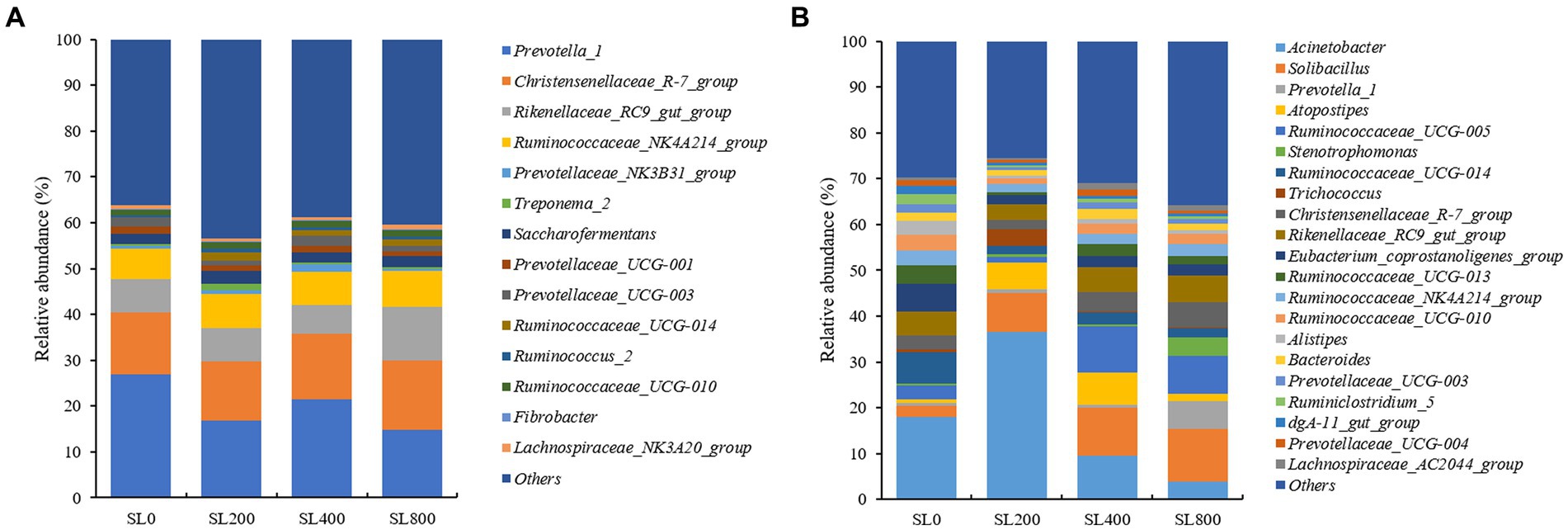
Figure 4. Composition of bacterial communities in Simmental calves at genus levels in rumen (A) and feces [(B); only taxa with an average relative abundance >0.5% are shown]. SL0, 0 mg/kg BW A. mongolicum; SL200, 200 mg/kg BW A. mongolicum; SL400, 400 mg/kg BW A. mongolicum; SL800, 800 mg/kg BW A. mongolicum.
In feces, Acinetobacter, Solibacillus, and Ruminococcaceae_UCG-005 were the dominant genera (Figure 4B). The relative abundances of Prevotella_1 (p < 0.01), Stenotrophomonas (p < 0.01), and Christensenellaceae_R-7_group (p < 0.05) were significantly higher in the SL800 group than the SL0 group (Supplementary Table S2). Ruminococcaceae_UCG-005 (p < 0.05) had the greatest relative abundance in the SL400 group. The relative abundances of Ruminococcaceae_UCG-014 (p < 0.01), Ruminiclostridium_5 (p < 0.01), Alistipes (p < 0.05), Eubacterium_coprostanoligenes_group (p < 0.05), and dgA-11_gut_group (p < 0.01) were lower in feces of claves supplemented with Allium Mongolicum (at any rate) than observed in SL0. The highest relative abundance of Trichococcus (p < 0.01) was greatest in animals receiving SL200, while Acinetobacter (p < 0.05) was in the lowest abundance in feces of claves receiving SL400 and SL800 treatments.
The relative abundances of Ruminiclostridium_5, Alistipes, and dgA-11_gut_group had highest variation in the regions (p < 0.001) in feces of calves given different supplementation of A. mongolicum (p < 0.05) and supplementary level and regions interaction (p < 0.05; Supplementary Table S3). The relative abundances of Acinetobacter (p < 0.01), Christensenellaceae_R-7_group (p < 0.001), Solibacillus (p < 0.05), Ruminococcaceae_UCG-010 (p < 0.01), Prevotellaceae_NK3B31_group (p < 0.01), Saccharofermentans (p < 0.001), Prevotellaceae_UCG-001 (p < 0.001), Fibrobacter (p < 0.001), Lachnospiraceae_NK3A20_group (p < 0.001), Ruminococcaceae_UCG-013 (p < 0.001), Ruminococcaceae_NK4A214_group (p < 0.001), Bacteroides (p < 0.001), and Prevotellaceae_UCG-004 (p < 0.001) were affected by the gastrointestinal region.
3.3. The linkage between dry matter intake, nutrient digestibility, total VFA, methane, fecal nutrient, and rumen and feces microbial community
Redundancy analysis was also conducted on the relative abundance of rumen bacteria and the DMI, nutrient digestibility, total VFA and CH4 (Figure 5A), RDA 1 and RDA 2 accounted for 58.7 and 15.1% of the variation, respectively. In the rumen bacteria community structure, the fecal nutrient levels of RAD 1 and RDA 2 represented 31.8 and 24.2%, respectively (Figure 5B). Emission of CH4 was the most important factor related to rumen and fecal bacterial structure (p < 0.05; Figures 5A,B; Supplementary Table S4). Butyrate and valerate were negatively correlated with bacterial community structure (p < 0.05) in the rumen, while the total VFA and acetate were positively correlated with bacterial community structure in feces (p < 0.05). In the rumen bacteria community structure, the DMI, nutrient digestibility, total VFA, and CH4 of RAD 1 and RDA 2 represented 20.34 and 10.79% (Figure 5C), the fecal nutrient levels of RAD 1 and RDA 2 represented 18.74 and 15.11% (Figure 5D). Fecal ADF content was negatively correlated with the bacterial community structure in rumen. Fecal EE and OM contents were negatively correlated with bacterial community structure in feces (Supplementary Table S4).
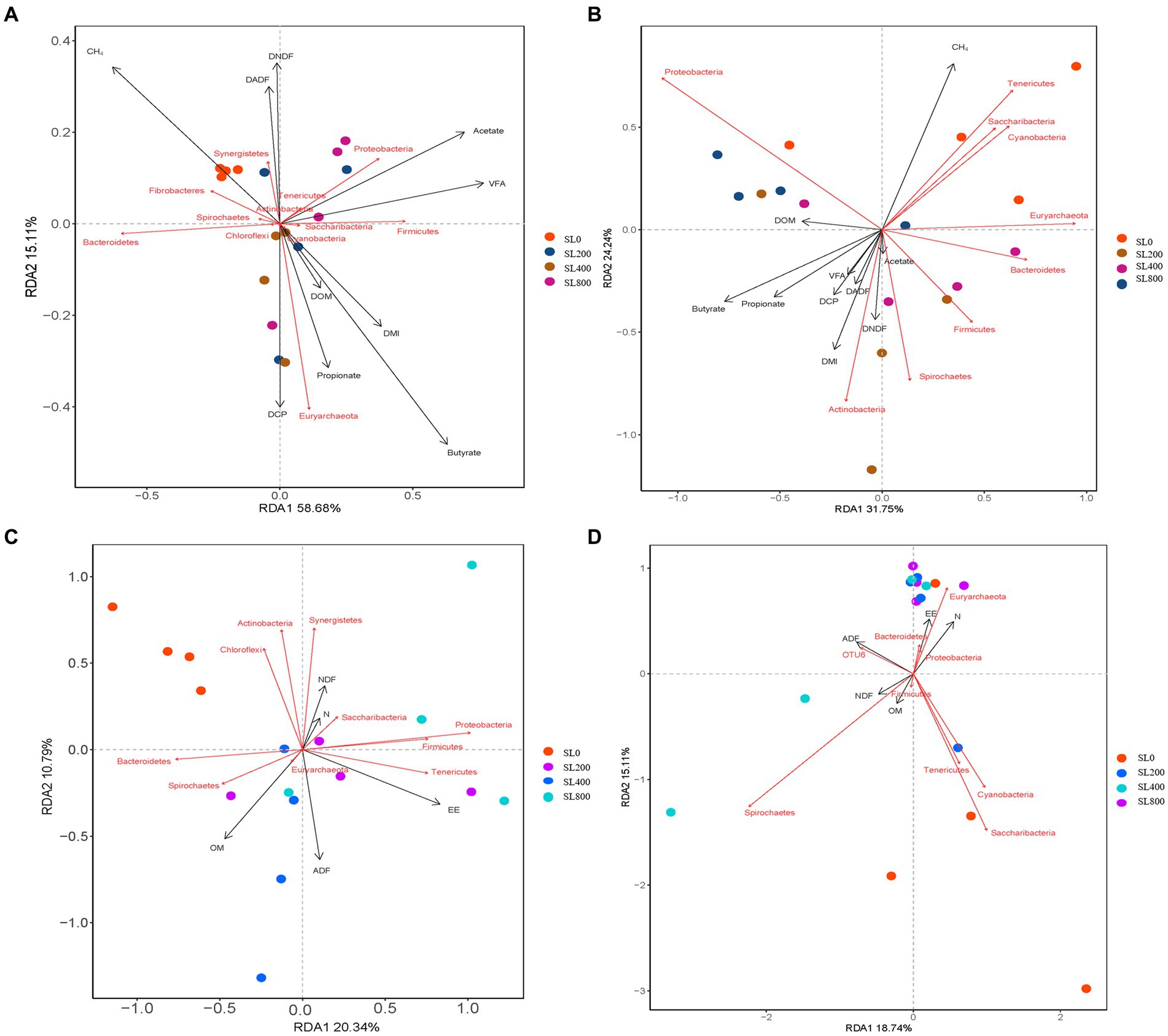
Figure 5. Redundancy analysis (RDA) for nutrient digestibility, rumen fermentation parameters, and fecal nutrients (black arrows) and rumen and fecal main bacterial (red arrows) phyla at varying supplement level of A. mongolicum. The length of the arrow indicates the correlation between nutrient digestibility, rumen fermentation parameters, fecal nutrients, and rumen or fecal main bacterial with the distribution of samples at different supplemental levels of A. mongolicum; the longer the arrow length, the greater the correlation. The direction of the arrow indicates trends in nutrient digestibility, rumen fermentation parameters, fecal nutrients, and major bacteria in the rumen or fecal. (A) Relations between rumen bacterial taxa (phylum) and nutrient digestibility and rumen fermentation parameters. (B) Relations between rumen bacterial taxa (phylum) and fecal nutrients. (C) Relations between fecal bacterial taxa (phylum) and nutrient digestibility and rumen fermentation parameters. (D) Relations between fecal bacterial taxa (phylum) and fecal nutrients. DMI, dry matter intake; OM, organic matter; ADF, acid detergent fiber; EE, ether extract; N, nitrogen; NDF, neutral detergent fiber; DOM, OM digestibility; DADF, ADF digestibility; DEE, EE digestibility; DCP, Crude protein digestibility; DNDF, NDF digestibility; VFA, total volatile fatty acids.
3.4. Correlations between DMI, nutrient digestibility, total VFA, and bacterial community diversity in rumen and feces
In the rumen, DMI was positively correlated with the genera Ruminococcaceae_UCG-005 (r = 0.620) (Figure 6A). Dry matter digestibility was positively correlated with genera Roseburia (r = 0.699) and negatively correlated with Lachnospiraceae_XPB1014_group (r = −0.663), Ruminococcus_1 (r = −0.669), and Lachnospiraceae_NK3A20_group (r = −0.652). Daily CH4 emission was positively correlated with genera Fusobacterium (r = 0.563) and negatively correlated with Lachnospiraceae_ND3007_group (r = 0.536). The CH4/DMI ratio was positively correlated with the genera Butyrivibrio_2 (r = 0.529) and negatively correlated with Methanobrevibacter (r = −0.638) and Fibrobacter (r = −0.540). The CH4/BW0.75 was positively correlated with genera Roseburia (r = 0.527) and Lachnospiraceae_ND3007_group (r = 0.536) and negatively correlated with Ruminococcus_1 (r = −0.517) and Lachnospiraceae_NK3A20_group (r = −0.607). Nitrogen digestibility was positively correlated with genera Saccharofermentans (r = 0.551), Ruminococcus_2 (r = 0.540), Trichococcus (r = 0.539), Fusobacterium (r = 0.611) and Lachnospiraceae_AC2044_group (r = 0.598) and negatively correlated with Butyrivibrio_2 (r = −0.556) and Ruminiclostridium_5 (r = −0.563). The NDF digestibility was positively correlated with the genera Saccharofermentans (r = 0.538), Ruminococcus_2 (r = 0.565), Trichococcus (r = 0.532), Fusobacterium (r = 0.644) and Lachnospiraceae_AC2044_group (r = 0.627). Whereas OM digestibility was positively correlated with genera Ruminococcus_1 (r = 0.570). The total VFA concentration was positively correlated with genera Solibacillus (r = 0.577), Ruminococcus_1 (r = 0.566), and Ruminococcaceae_UCG-010 (r = 0.540) and negatively correlated with Trichococcus (r = −0.566). Acetate was positively correlated with genera Prevotella_1 (r = 0.667) and negatively correlated with Eubacterium_coprostanoligenes_group (r = −0.626). Butyrate was negatively correlated with genus Roseburia (r = −0.547). Valerate was positively correlated with genera Solibacillus (r = 0.584) and Ruminococcaceae_UCG-010 (r = 0.515) and negatively correlated with Prevotella_1 (r = −0.523) and Trichococcus (r = −0.580). The acetate to propionate ratio was positively correlated with Roseburia (r = 0.582) and negatively correlated with Christensenellaceae_R-7_group (r = −0.553), Prevotellaceae_UCG-003 (r = −0.609), Lachnospiraceae_XPB1014_group (r = 0.603), and Lachnospiraceae_NK3A20_group (r = 0.568) genera.
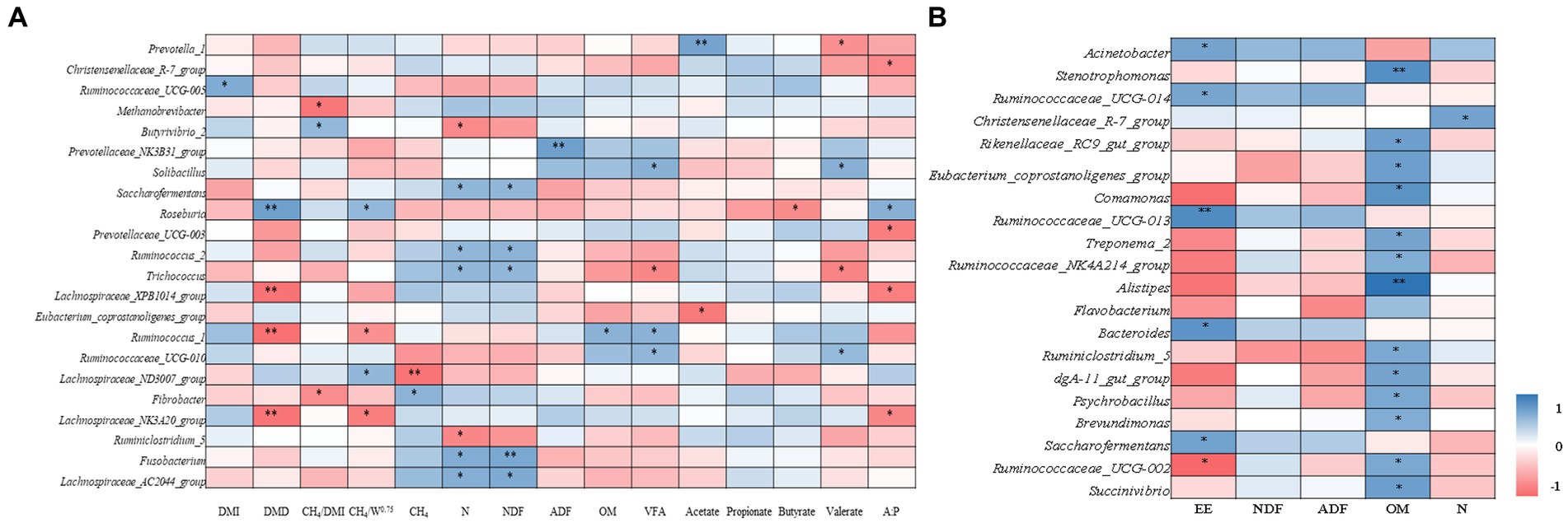
Figure 6. Relationship among DMI, nutrient digestibility, total VFA, CH4, and main bacterial communities at genus level in Rumen (A). DMI, dry matter intake; DMD, dry matter digestibility; N, nitrogen; NDF, neutral detergent fiber; ADF, acid detergent fiber; OM, organic matter; VFA, total volatile fatty acids; A:P, acetate to propionate ratio. Relationship among nutrient and main bacterial communities at genus level in feces (B). EE, fecal ether extract; NDF, fecal neutral detergent fiber; ADF, fecal acid detergent fiber; OM, fecal organic matter; N, fecal nitrogen. ** and * indicate significance levels at 0.01 and 0.05, respectively.
In feces, the EE content was positively correlated with genera Acinetobacter (r = 0.400), Ruminococcaceae_UCG-014 (r = 0.393), Ruminococcaceae_UCG-013 (r = 0.494), Bacteroides (r = 0.464), and Saccharofermentans (r = 0.410), and negatively correlated with Ruminococcaceae_UCG-002 (r = −0.367; Figure 6B). The OM content was positively correlated with the genera Stenotrophomonas (r = 0.483), Rikenellaceae_RC9_gut_group (r = 0.417), Eubacterium_coprostanoligenes_group (r = 0.417), Comamonas (r = 0.467), Treponema_2 (r = 0.400), Ruminococcaceae_NK4A214_group (r = 0.367), Alistipes (r = 0.583), Ruminiclostridium_5 (r = 0.383), dgA-11_gut_group (r = 0.400), Psychrobacillus (r = 0.383), and Brevundimonas (r = 0.367). The N content was positively correlated with the genus Christensenellaceae_R-7_group (r = 0.407).
3.5. Network analysis of bacterial communities in rumen and feces
Microbial interactions between rumen and fecal bacterial communities were analyzed using microbial networks (Figure 7). Positive correlation within ruminal bacteria accounted for 68.2 and of the feces accounted for 79% of the total correlation. The putative drivers of keystone taxa in rumen and fecal microbial communities of Simmental calves from the gastrointestinal region were estimated through use of the combined score of the high mean degree, high closeness centrality, and low betweenness centrality (Supplementary Table S5). Prevotella_1 and Prevotellaceae_UCG-003 in rumen and Ruminococcaceae_NK4A214_group, Christensenellaceae_R-7_group and Ruminococcaceae_UCG-010 in feces could be considered as keystone taxa in Simmental calves.
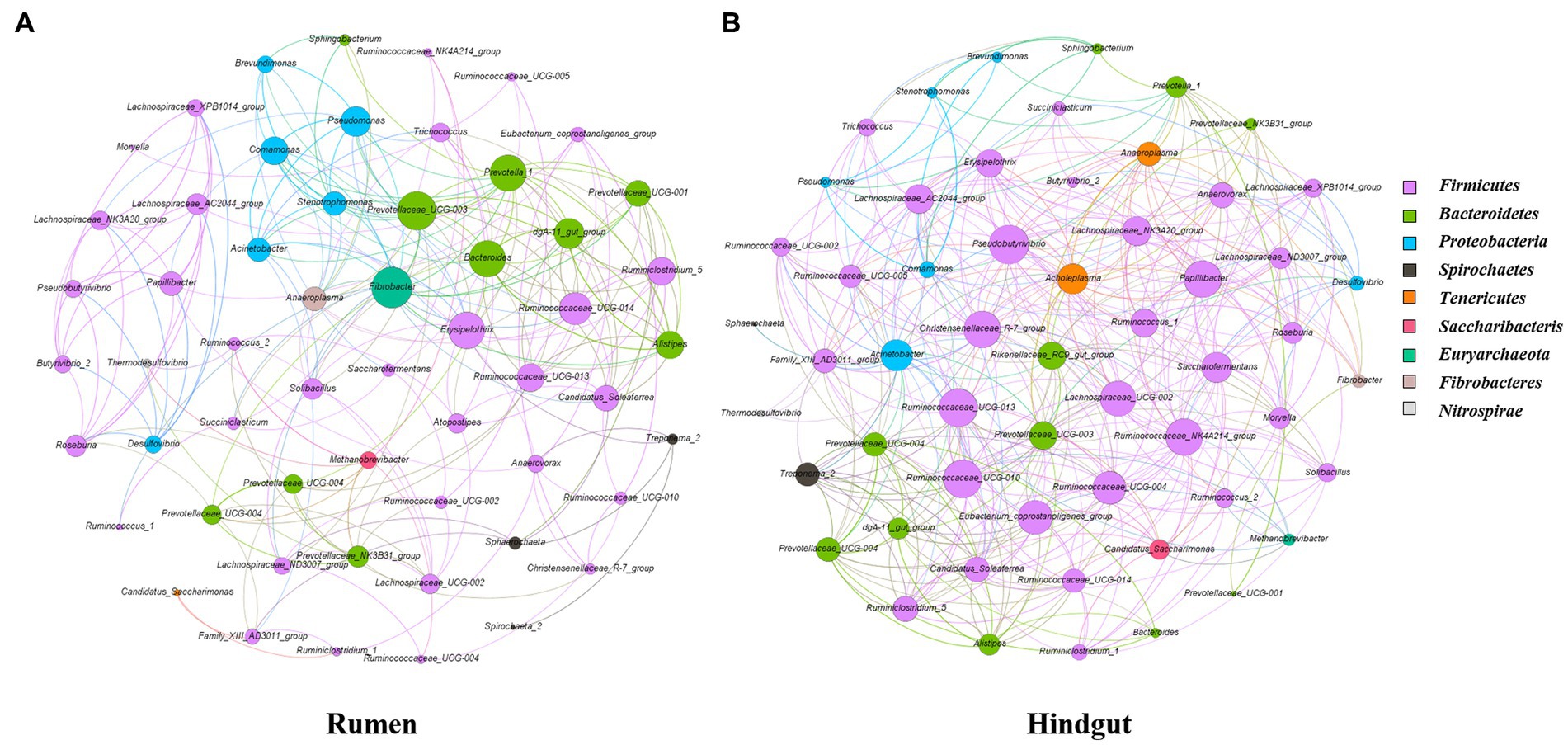
Figure 7. Interaction network of the rumen (A) and feces (B) microbiota. 16S rRNA gene-based correlation network of the rumen microbiota, displaying statistically significant interactions with absolute value of correlation coefficients >0.6. Node be colored by phylum. The node size was scaled based on the overall abundance of each taxa in the microbiota.
4. Discussion
Extracts and other by-products of A. mongolicum have the potential to improve average daily gain, feed efficiency, and body antioxidant properties in ruminant species (Panthee et al., 2017; Redoy et al., 2020; Xie et al., 2020). To identify the effect of A. mongolicum on the gastrointestinal tract of ruminants, following analyses are necessary for a deep understanding: (1) relationship of microbe functions between rumen and hindgut; (2) the integrated effects to gastrointestinal, and (3) exploring the “gray box” of the intestine through analysis of the microbe in rumen and feces (Figure 8).
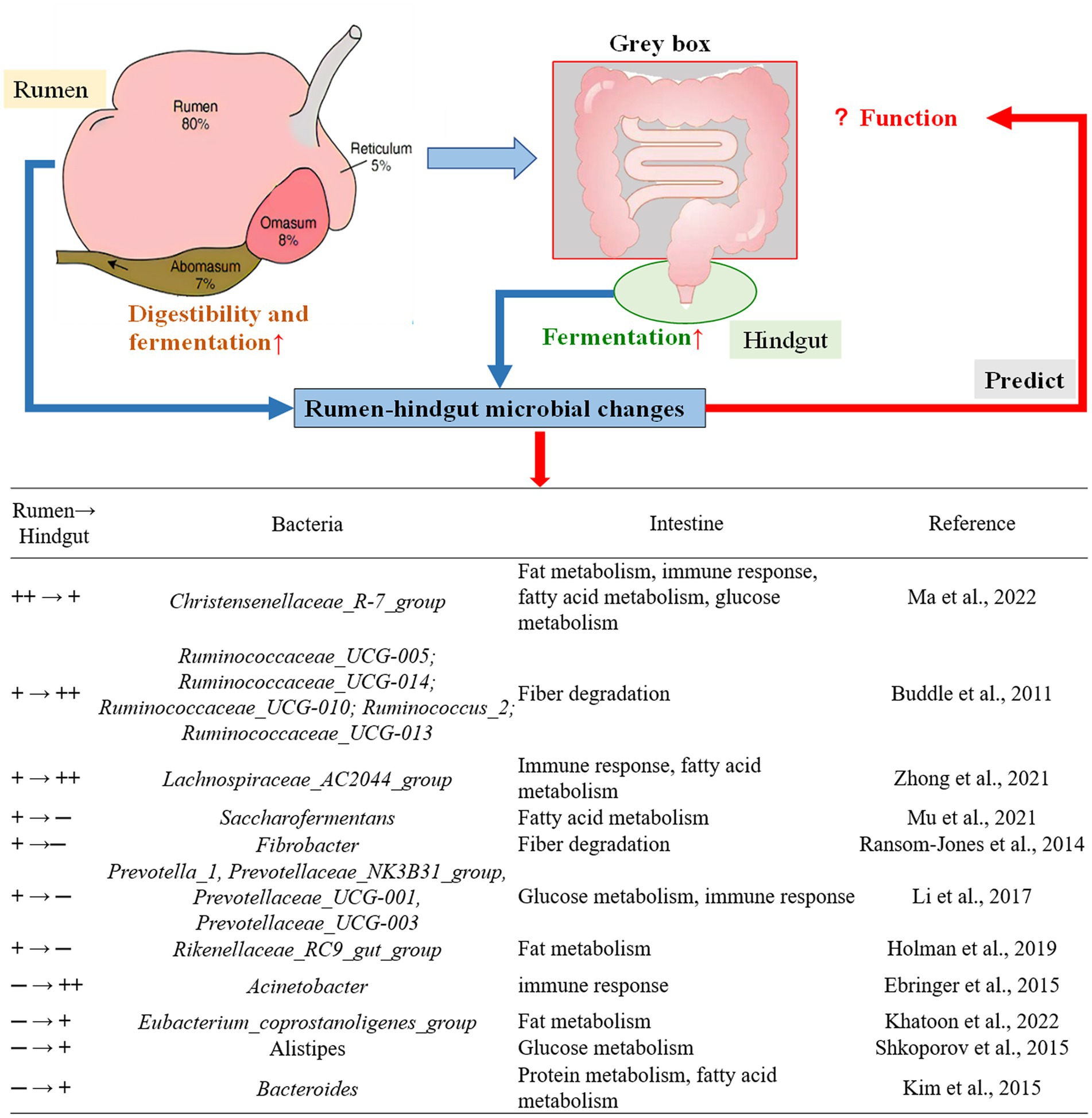
Figure 8. Rumen-hindgut microbial differences predict the corresponding function of small intestine. ++→+, The relative abundance of microorganisms in rumen was greater than in feces (p < 0.001); +→++, the relative abundance of microorganisms in feces was greater than in rumen (p < 0.05); +→─, microbes in the rumen only; ─→+, microbes in feces only. According to published studies of microbiota change and intestinal function, intestinal function was predicted by rumen-hindgut bacterial differences at the genus level. Gray box, intestinal function was predicted by analyzing rumen-hindgut intestinal changes.
The rumen bacteriome supplemented with A. mongolicum showed significantly clear separations (Figure 2). Furthermore, significant differences in the bacterial communities between the supplemented and non-supplemented A. mongolicum treatments were related to nutrient digestibility (Coasta et al., 2012). Firmicutes and Bacteroidetes constituted the predominant bacteria in rumen and feces, whether A. mongolicum supplement was supplemented or not (Figure 3). Firmicutes, which degrade cellulose to total VFAs, constituted the main bacterial type. This bacterium has also been implicated in host immune responses, inhibiting the invasion of opportunistic pathogens, and preventing of inflammation (Gruninger et al., 2014; Zhang et al., 2015) of the intestinal tract. Different regions of the gastrointestinal tract have different effects on the body, facilitating feed energy conversion and harvesting (Chen et al., 2015), and indirectly promoting the physiological function of digestion in calves. Dietary supplementation with A. mongolicum promoted feedback mechanism of physiological metabolism through the regulation of Firmicutes, thereby increasing feed intake and ultimately resulting in a decreased proportion of FNH in the DMI (Xie et al., 2020). Supplementation of A. mongolicum significantly increased average daily gain of calves (Xie et al., 2020), probably because the ratio of Bacteroidetes to Firmicutes in the rumen tended to be increased by A. mongolicum supplementation (p = 0.063, Supplementary Table S1), and the ratio of Bacteroidetes to Firmicutes was positively associated with growth rate and feed efficiency (Myer et al., 2015). Therefore, a boosting effect on energy metabolism in cattle was observed. However, the effects of different levels of A. mongolicum supplementation on Bacteroidetes were inconsistent. Lower levels (SL200 and SL400) of A. mongolicum supplementation did not affect Bacteroidetes significantly; however, SL800 significantly decreased the expansion of Bacteroidetes. This suggested that a critical dose of A. mongolicum must be achieved to affect the relative abundance of Bacteroidetes in rumen. It is possible that elevated levels of A. mongolicum containing higher concentrations of unsaturated fatty acids may be harmful to the Bacteroidetes (Pitta et al., 2018). The low abundance of the Bacteroidetes may have been compensated through the higher abundance of Firmicutes (Matthews et al., 2019). There were more Bacteroides in feces than in rumen, possibly playing a role in degrading fiber from the rumen in cattle feces (Kim et al., 2015). Thus, it is possible that A. mongolicum modulated the relative abundance of Firmicutes and Bacteroidetes to maintain a stable rumen environment, which maintained intestinal stability via balancing the Acinetobacter populations in feces. These may be “combined effects” of multiple secondary metabolites of A. mongolicum, which can simultaneously regulate Firmicutes and Bacteroidetes in rumen to maintain a healthy gastrointestinal tract.
Ruminococcaceae and Fibrobacter were well-described genera in the rumen (Figure 4A). The Ruminococcaceae was abundant in the rumen and four genera of Ruminococcaceae were relatively abundant in fecal microflora (Figure 4B). More efficient ruminants had greater bacterial richness and diversity in their guts, which resulted in more fiber-degrading bacteria in their rumen (Welch et al., 2020). Ruminococcaceae is the best-known fiber-degrading bacteria (Biddle et al., 2013), and the relative abundance of Ruminococcaceae_UCG-005 in SL400 was increased significantly. This observation suggested that supplementation of A. mongolicum may improve the feed conversion efficiency of Simmental calves. Ruminococcaceae also acted synergistically with other rumen microorganisms (Bloemendaal et al., 2021). For example, the relative abundance of Fibrobacter was greater in rumen than in feces, which may be the main rumen fibrinolytic bacteria that can decompose cellulose and pentosan into usable organic acids (acetate, propionate, butyrate, etc.; Ransom-Jones et al., 2012). An increase in these organic acids (acetate, propionate, butyrate, etc) were associated with negative effects on fibrinolytic bacteria and biohydrogenation. A reduction in biohydrogenation leads to greater flow of organic acids to the gut. Thus, absorbing and storing these fatty acids in adipose tissue as body fat (Ransom-Jones et al., 2014) and indirectly promoting intestinal lipid metabolism. CH4 emission was the most important factor related to rumen and fecal bacterial structure (Figure 5). Fibrobacter in the rumen was positively correlated with CH4 emissions (Figure 6A), and the main metabolite of fiber degradation was hydrogen, which ultimately increased the hydrogen ions required for the process of CH4 production (Xie et al., 2022). The genus Ruminococcus in rumen was positively correlated with NDF and OM digestibility (Figure 6A), thus promoting sufficient degradation of fiber in the rumen (Firkins, 2021).
Although the genera Prevotella and Alistipes were commonly found in cattle feces, their relative abundance were more significantly affected by the level A. mongolicum supplementation and the region of the gut sampled. Prevotella_1 was a potential biomarker for individualized nutritional management of obesity. Decrease in the relative abundance of Prevotella_1 enhanced the immune performance of ruminant by reducing local and systemic diseases (Xie et al., 2020). Similar results due to A. mongolicum supplementation have already been reported (Kholy et al., 2015). To further support this hypothesis, it has been reported that A. mongolicum supplementation inhibited the body’s metabolism by reducing the abundance of Prevotella_1 in rumen, thereby increasing the daily gain of ruminants (Tett et al., 2021). Flavonoids in A. mongolicum can promote the decomposition of plant polysaccharides by increasing the relative abundance of Prevotella, while most complex plant polysaccharides from diet are not digested by livestock and enter the colon as a potential nutrient source for the microbiota (Briggs et al., 2020; Ghimire et al., 2022), therefore, A. mongolicum supplementation increased the relative abundance of Prevotella in feces in SL800 group. Alistipes are often glycolytic (Shkoporov et al., 2015), which may explain their association with hindgut rather than with rumen.
Acinetobacter is a conditioned pathogen, so the relative abundance of Acinetobacter in feces may have been reduced due to the antibacterial activity of flavonoids in A. mongolicum, which may have required a critical mass dosage of A. mongolicum (SL400 and SL800) to exert such effects. A. mongolicum supplementation maintains the stability of the intestinal flora (Klevenhusen et al., 2011; Ebringer et al., 2015). Conversely, if the number of pathogenic bacteria can be reduced, and therefore less excreted in feces, the infectious environmental pollution can be reduced (Wilson et al., 2020). Antioxidant index (superoxide dismutase and glutathione peroxidase) in serum were improved in Simmental calves given SL400 (Xie et al., 2020), which may be due to the increased relative abundance of Actinobacteria (Supplementary Table S2), thereby helping to maintain intestinal barrier functions, increasing tight junctions’ expression, regulating mucin biosynthesis and catabolism, providing energy for epithelial cells proliferation and stimulating the immune system (D’Argenio and Salvatore, 2015; Zhu et al., 2016). Rikenellaceae_RC9_gut_group abundance was significantly higher in SL800, which may play an important role in rumen lipid metabolism (Holman and Gzyl, 2019), thereby promoting the role of A. mongolicum in intestinal metabolism. Additionally, A. mongolicum supplementation could contribute to glucose metabolism to produce acetate and butyrate via Rikenellaceae_RC9_gut_group (Seshadri et al., 2018). A large amount of cellulose can be converted into propionic acid by the action of Rikenellaceae_R9_gut_group in the digestive tract of ruminants. The relative abundance of Rikenellaceae_R9_gut_group in SL800 was increased in feces, thereby increasing the propionic concentration to promote gluconeogenesis in Simmental calves (Aschenbach et al., 2010; Graf, 2014). Thus, in this study, the regulation mechanism of intestine microbiota in the metabolism of glucose, protein, amino acid, as well as lipid metabolism were explored by analyzing the microbial community structure of the rumen and feces and their corresponding functions.
The important role of the gastrointestinal microbiota in intestinal function and animal production has been clarified. This study identified a trapezoid taxon with network topological properties in the gastrointestinal tract of Simmental cattle (Figure 7). The balance and stability of microbial community in the gastrointestinal tract contribute to host health, with key genera differing between the rumen and feces. Network analyses can reveal interactions between species (Liu H. et al., 2020). Both rumen and fecal microbes had higher of positive correlations than negative correlations. The positive interactions might strengthen and negative interaction might weaken competitive (Fan et al., 2021). Microbes in the rumen were more positively correlated than in feces. It is reasonable to consider that the rumen is a unique digestive chamber of the gut for digestion, absorption and associated immune responses. In on-farm greenhouse gas emissions models, microorganisms can make full use of limited feed resources by strengthening cooperation (Vibart et al., 2021). The main function of the back end of the digestive tract was digestion, decomposition and absorption, which contains a large number of enzymes and microorganisms.
5. Conclusion
Dietary supplementation with A. mongolicum altered the rumen and fecal microflora of Simmental calves, which positively affected microbial community structure in rumen and feces. The microbiome diversity of rumen and feces significantly affected under the SL400 and SL800 treatments, but not under SL200. The addition of A. mongolicum increased relative abundance Rikenellaceae_RC9_gut_group and Ruminococcaceae_UCG-010 in rumen, and decreased relative abundance of Bacteroidetes and Prevotella_1 in rumen, which enhanced diet digestibility and fermentation. Both the increase of Firmicutes, Prevotella_1, Ruminococcaceae_UCG-005 Christensenellaceae_R-7_group abundance, and the decrease of Acinetobacter, Ruminiclostridium_5, and Ruminococcaceae_UCG-014 abundance could promote fecal fermentation with a positive effect on Simmental calf health. This study demonstrated that rumen-hindgut microbial were affected by supplementation of a certain amount of A. mongolicum, further studies are needed to determine the mechanisms of observed effects.
Data availability statement
The data presented in this study are deposited in the NCBI repository, accession number PRJNA941479.
Ethics statement
All animal experimental procedures were authorized by the Laboratory Animal Management and Welfare Committee of Lanzhou University (file No: 2010-1 and 2010-2).
Author contributions
FH: conceptualization, supervision, and experimental design. KX: data analysis and original draft writing. KX, YG, and CZ: animal feeding experiment. JN and CZ: laboratory sample analyses. FH, SC, and TY: review and editing. All authors contributed to the article and approved the submitted version.
Funding
FH acknowledges the scholarship by the National Natural Science Foundation of China (32161143028); the Program of National Science and Technology Assistance (KY202002011); the Lanzhou City’s Scientifc Research Funding Subsidy to Lanzhou University; and the Program for Innovative Research Team of Ministry of Education (IRT17R50).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1174740/full#supplementary-material
Footnotes
1. ^ http://ccb.jhu.edu/software/FLASH/
2. ^ http://qiime.org/scripts/split_libraries_fastq.html
3. ^ http://www.drive5.com/usearch/manual/uchime_algo.html
4. ^ http://drive5.com/uchime/uchime_download.html
References
Ahmad, A. A., Yang, C., Zhang, J., Kalwar, Q., Liang, Z., Li, C., et al. (2020). Effects of dietary energy levels on rumen fermentation, microbial diversity, and feed efficiency of yaks (Bos grunniens). Front. Microbiol. 11:625. doi: 10.3389/fmicb.2020.00625
Aschenbach, J. R., Kristensen, N. B., Donkin, S. S., Hammon, H. M., and Penner, G. B. (2010). Gluconeogenesis in dairy cows: the secret of making sweet milk from sour dough. IUBM Life 62, 869–877. doi: 10.1002/iub.400
Biddle, A., Stewart, L., Blanchard, J., and Leschine, S. (2013). Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5, 627–640. doi: 10.3390/d5030627
Bloemendaal, M., Szopinska-Tokov, J., Belzer, C., Boverhoff, D., Papalini, S., Michels, F., et al. (2021). Probiotics-induced changes in gut microbial composition and its effects on cognitive performance after stress: exploratory analyses. Transl. Psychiatry 11, 1–11. doi: 10.1101/2020.11.23.20236836
Bokulich, N. A., Subramanian, S., Faith, J. J., Gever, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Briggs, J. A., Grondin, J. M., and Brumer, H. (2020). Communal living: glycan utilization by the human gut microbiota. Environ. Microbiol. 23, 15–35. doi: 10.1111/1462-2920.15317
Cancino-Padilla, N., Catalán, N., Siu-Ting, K., Creevey, C. J., Huws, S. A., Romero, J., et al. (2021). Long-term effects of dietary supplementation with olive oil and hydrogenated vegetable oil on the rumen microbiome of dairy cows. Microorganisms 9, 1–12. doi: 10.3390/microorganisms9061121
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Castillo-Lopez, E., Haselmann, A., Petri, R. M., Knaus, W., and Zebeli, Q. (2020). Evaluation of fecal fermentation profile and bacterial community in organically fed dairy cows consuming forage-rich diets with different particle sizes. J. Dairy Sci. 103, 8020–8033. doi: 10.3168/jds.2019-18036
Chen, Y. B., Lan, D. L., Tang, C., Yang, X. N., and Li, J. (2015). Effect of DNA extraction methods on the apparent structure of yak rumen microbial communities as revealed by 16S rDNA sequencing. Pol. J. Microbiol. 64, 29–36. doi: 10.33073/pjm-2015-004
Coasta, M., Arroyo, L. G., Allen-Vercoe, E., Stämpfli, H. R., Kim, P. T., Sturgeon, A., et al. (2012). Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS One 7:e41484. doi: 10.1371/journal.pone.0041484
Cui, X., Wang, Z., Fan, Q., Chang, S., Yan, T., and Hou, F. (2023). Ligularia virgaurea improved nutrient digestion, ruminal fermentation, and bacterial composition in Tibetan sheep grazing on the Qinghai–Tibetan plateau in winter. Anim. Feed Sci. Technol. 299:115628. doi: 10.1016/j.anifeedsci.2023.115628
D’Argenio, V., and Salvatore, F. (2015). The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta 451, 97–102. doi: 10.1016/j.cca.2015.01.003
Ebringer, A., Rashid, T., and Wilson, C. (2015). Chapter 29—Multiple sclerosis and creutzfeldt-jakob disease are autoimmune diseases probably caused by exposure to the nasal microbe Acinetobacter. In Infection and autoimmunity, 2nd Ed.; Y. Shoenfeld, N. Agmon-Levin, and N.R Rose., Eds.; Academic Press: Amsterdam, Netherlands, 505–517
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fan, Q., Cui, X., Wang, Z., Chang, S., Wanapat, M., Yan, T., et al. (2021). Rumen microbiota of Tibetan sheep (Ovis aries) adaptation to extremely cold season on the Qinghai-Tibetan plateau. Fron. VeT. Sci. 8:673822. doi: 10.3389/fvets.2021.673822
Firkins, J. L. (2021). Invited review: advances in rumen efficiency. Appl. Anim. Sci. 37, 388–403. doi: 10.15232/aas.2021-02163
Ghimire, S., Cady, N. M., Lehman, P., Peterson, S. R., Shahi, S. K., Rashid, F., et al. (2022). Dietary isoflavones alter gut microbiota and lipopolysaccharide biosynthesis to reduce inflammation. Gut Microbes 14:2127446. doi: 10.1080/19490976.2022.2127446
Graf, J. (2014). “The family Rikenellaceae” in The Prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 857–859.
Gressley, T. F., Hall, M. B., and Armentano, L. E. (2011). Ruminant nutrition symposium: productivity, digestion, and health responses to hindgut acidosis in ruminants. J. Anim. Sci. 89, 1120–1130. doi: 10.2527/jas.2010-3460
Gruninger, R. J., Sensen, C. W., McAllister, T. A., and Forster, R. J. (2014). Diversity of rumen bacteria in Canadian cervids. PLoS One 9:e89682. doi: 10.1371/journal.pone.0089682
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2021). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Harris, L., Asplund, J., and Crampton, E. (1968). International feed nomenclature and methods for summarizing and using feed data to calculate diets. Logan, UT: Agricultural Experiment Station, Utah State University. (1968), pp: 117–120
Holman, D. B., and Gzyl, K. E. (2019). A meta-analysis of the bovine gastrointestinal tract microbiota. FEMS Microbiol. Ecol. 95, 1–9. doi: 10.1093/femsec/fiz072
Hou, F. J., Jia, Q. M., Lou, S. N., Yang, C. T., and Fan, Q. S. (2021). Grassland agriculture in China—a review. Front. Agr. Sci. Eng. 8, 35–44. doi: 10.15302/J-FASE-2020378
Huang, Q. C., Chu, Y. L., Zhao, Y. H., Zhao, H. T., Liu, J. M., and Zhou, R. Y. (2011). Status and prospect of plant fiber industry in Guangxi province. Plant Fiber Sci. 33, 202–205. doi: 10.3969/453 j. issn.1671-3532.2011.04.010
Ibrahim, N. A., Alimon, A. R., Yaakub, H., Samsudin, A. A., Candyrine, S. C. L., Mohamed, W. N. W., et al. (2021). Effects of vegetable oil supplementation on rumen fermentation and microbial population in ruminant: a review. Trop. Anim. Health Prod. 53:422. doi: 10.1007/s11250-021-02863-4
Jamshidi-Kia, F., Lorigooini, Z., and Amini-Khoei, H. (2018). Medicinal plants: history and future perspective. J. Herbmed. Pharmacol. 7, 1–7. doi: 10.15171/jhp.2018.01
Jeong, C., Wang, K., Wilkin, S., Taylor, W., and Warinner, C. (2020). A dynamic 6,000-year genetic history of Eurasia's eastern steppe. Cells 183, 890–904.e29. doi: 10.1016/j.cell.2020.10.015
Kholy, E., Karim, G., Robert, J., Dyke, V., and Thomas, E. (2015). Oral infections and cardiovascular disease. Trends Endocrinol. Metab. 26, 315–321. doi: 10.1016/j.tem.2015.03.001
Kim, E. T., Guan, L. L., Lee, S. J., Lee, S. M., and Lee, S. S. (2015). Effects of flavonoid-rich plant extracts on in vitro ruminal methanogenesis, microbial populations and fermentation characteristics. Asian-Australas J Anim. Sci. 28, 530–537. doi: 10.5713/ajas.14.0692
Klevenhusen, F., Zeitz, J. O., Duval, S., Kreuzer, M., and Soliva, C. R. (2011). Garlic oil and its principal component diallyl disulfide fail to mitigate methane, but improve digestibility in sheep. Anim. Feed. Sci. Tech. 166-167, 356–363. doi: 10.1016/j.anifeedsci.2011.04.071
Liu, H., Hu, L., Han, X., Zhao, N., Xu, T., Ma, L., et al. (2020). Tibetan sheep adapt to plant phenology in alpine meadows by changing rumen microbial community structure and function. Front. Microbiol. 11:587558. doi: 10.3389/fmicb.2020.587558
Liu, X., Liu, F., Yan, T., Chang, S. H., Wanapat, M., and Hou, F. J. (2020). Cistanche deserticola addition improves growth, digestibility, and metabolism of sheep fed on fresh forage from alfalfa/tall fescue pasture. Animals 10:668. doi: 10.3390/ani10040668
Liu, K., Xu, Q., Wang, L., Wang, J., Guo, W., and Zhou, M. (2016). The impact of diet on the composition and relative abundance of rumen microbes in goat. Asian-Australas J. Anim. Sci. 30, 531–537. doi: 10.5713/ajas.16.0353
Magoč, T. J., and Steven, L. S. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Matthews, C., Crispie, F., Lewis, E., Reid, M., O’Toole, P. W., and Cotter, P. D. (2019). The rumen microbiome: a crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 10, 115–132. doi: 10.1080/19490976.2018.1505176
Minas, K., McEwan, N. R., Newbold, C. J., and Scott, K. J. (2011). Optimization of a high-throughput ctab-based protocol for the extraction of qpcr-grade dna from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol. Lett. 325, 162–169. doi: 10.1111/j.1574-6968.2011.02424.x
Myer, P. R., Smith, T. P. L., Wells, J. E., Kuehn, L. A., and Freetly, H. C. (2015). Rumen microbiome from steers differing in feed efficiency. PLoS One 10:e0129174. doi: 10.1371/journal.pone.0129174
Neave, H. W., Weary, D. M., and von Keyserlingk, M. A. G. (2018). Individual variability in feeding behaviour of domesticated ruminants. Animal 12, s419–s430. doi: 10.1017/S1751731118001325
Nobler, J. D., Camp, M. J., Crowell, M. M., Shipley, L. A., Dadabay, C., Rachlow, J. L., et al. (2019). Preferences of specialist and generalist mammalian herbivores for mixtures versus individual plant secondary metabolites. J. Chem. Ecol. 45, 74–85. doi: 10.1007/s10886-018-1030-5
Panthee, A., Matsun, A., Al-Mamun, M., and Sano, H. (2017). Effect of feeding garlic leaves on rumen fermentation, methane emission, plasma glucose kinetics, and nitrogen utilization in sheep. J. Anim. Sci. Technol. 59:14. doi: 10.1186/s40781-017-0139-3
Patra, A., Park, T., Kim, M., and Yu, Z. (2017). Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Technol. 8, 13–18. doi: 10.1186/s40104-017-0145-9
Petri, R. M., Münnich, M., Zebeli, Q., and Klevenhusen, F. (2019). Graded replacement of corn grain with molassed sugar beet pulp modulates the fecal microbial community and hindgut fermentation profile in lactating dairy cows. J. Dairy Sci. 102, 5019–5030. doi: 10.3168/jds.2018-15704
Pitta, D. W., Indugu, N., Vecchiarelli, B., Rico, D. E., and Harvatine, K. J. (2018). Alterations in ruminal bacterial populations at induction and recovery from diet-induced milk fat depression in dairy cows. J. Dairy Sci. 101, 295–309. doi: 10.3168/jds.2016-12514
Ransom-Jones, E., Jones, D. L., Edwards, A., and Mcdonald, J. E. (2014). Distribution and diversity of members of the bacterial phylum fibrobacteres in environments where cellulose degradation occurs. Syst. Appl. Microbiol. 37, 502–509. doi: 10.1016/j.syapm.2014.06.001
Ransom-Jones, E., Jones, D. L., Mccarthy, A. J., and Mcdonald, J. E. (2012). The fibromatoses: an important phylum of cellulose-degrading bacteria. Microb. Ecol. 63, 267–281. doi: 10.1007/s00248-011-9998-1
Redoy, M. R. A., Shuvo, A. A. S., Cheng, L., and Al-Mamun, M. (2020). Effect of herbal supplementation on growth, immunity, rumen histology, serum antioxidants and meat quality of sheep. Animal 14, 2433–2441. doi: 10.1017/S1751731120001196
Richards, L. A., Dyer, L. A., Forister, M. L., Smilanich, A. M., Dodson, C. D., Leonard, M. D., et al. (2015). Phytochemical diversity drives plant–insect community diversity. Proc. Natl. Acad. Sci. U. S. A. 112, 10973–10978. doi: 10.1073/pnas.1504977112
Seshadri, R., Leahy, S. C., Attwood, G. T., Attwood, G. T., The, K. H., Lambie, S. C., et al. (2018). Cultivation and sequencing of rumen microbiome members from the Hungate 1000 collection. Nat. Biotechnol. 36, 359–367. doi: 10.1038/nbt.4110
Shkoporov, A. N., Chaplin, A. V., Khokhlova, E. V., Shcherbakova, V. A., Motuzova, O. V., Bozhenko, V. K., et al. (2015). Alistipes inops sp. nov. and Coprobacter secundus sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 65, 4580–4588. doi: 10.1099/ijsem.0.000617
Tett, A., Pasolli, E., Masetti, G., Ercolini, D., and Segata, N. (2021). Prevotella diversity, niches and interactions with the human host. Nat. Rev. Microbiol. 19, 585–599. doi: 10.1038/s41579-021-00559-y
Toral, P. G., Monahan, F. J., Hervas, G., Frutos, P., and Moloney, A. P. (2018). Review: modulating ruminal lipid metabolism to improve the fatty acid composition of meat and milk challenges and opportunities. Animal 12, S272–S281. doi: 10.1017/S1751731118001994
Vibart, R., Klein, C. D., Jonker, A., Weerden, T., Bannink, A., Bayat, R., et al. (2021). Challenges and opportunities to capture dietary effects in on-farm greenhouse gas emissions models of ruminant systems. Sci. Total Environ. 769:144989. doi: 10.1016/j.scitotenv.2021.144989
Wanapat, M., Cherdthong, A., Phesatcha, K., and Kang, S. (2015). Dietary sources and their effects on animal production and environmental sustainability. Anim. Nutr. 1, 96–103. doi: 10.1016/j.aninu.2015.07.004
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, Y., Sun, Y., Liu, Y., Wang, Z., Chang, S., Qian, Y., et al. (2022). Ecological thresholds of toxic plants for sheep production and ecosystem multifunctionality and their trade-off in an alpine meadow. J. Environ. Manag. 323:116167. doi: 10.1016/j.jenvman.2022.116167
Wang, M., Wang, R., Xie, T. Y., Janssen, P. H., Zhao, S. X., Beauchemin, K. A., et al. (2016). Shifts in rumen fermentation and microbiota are associated with dissolved ruminal hydrogen concentrations in lactating dairy cows fed different types of carbohydrates. J. Nutr. 146, 1714–1721. doi: 10.3945/jn.116.232462
Welch, C. B., Lourenco, J. M., Davis, D. B., Krause, T. R., Carmichael, M. N., Rothrock, M. J., et al. (2020). The impact of feed efficiency selection on the ruminal, cecal, and fecal microbiomes of angus steers from a commercial feedlot. J. Anim. Sci. 98:skaa230. doi: 10.1093/jas/skaa230Xi
Wilson, B. M., Perez, F., Pan, Q., Jiang, Y., Evans, S. R., and Bonomo, R. A. (2020). Acinetobacter infections. Clin. Infect. Dis. 71, 1357–1358. doi: 10.1093/cid/ciz1099
Wingard, G. J., Harris, R. B., Pletscher, D. H., Bedunah, D. J., Mandakh, B., Amgalanbaatar, S., et al. (2011). Argali food habits and dietary overlap with domestic livestock in ikh nart nature reserve, Mongolia. J. Arid. Environ. 75, 138–145. doi: 10.1016/j.jaridenv.2010.09.014
Xie, K. L., Wang, Z. F., Guo, Y. R., Zhang, C., Zhu, W. H., and Hou, F. J. (2022). Gentiana straminea supplementation improves feed intake, nitrogen and energy utilization, and methane emission among Simmental calves in Northwest China. Anim. Biosci. 35, 838–836. doi: 10.5713/ab.21.0263
Xie, K., Wang, Z. F., Wang, Y. J., Wang, C. M., Hua, C. S., Zhang, C., et al. (2020). Effects of Allium mongolicum Regel supplementation on the digestibility, methane production, and antioxidant capacity of Simmental calves in Northwest China. Anim. Sci. J. 91:e13392. doi: 10.1111/asj.13392
Zhang, J., Guo, Z., Xue, Z., Sun, Z., Zhang, M., Wang, L., et al. (2015). A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 9, 1979–1990. doi: 10.1038/ismej.2015.11
Zhang, X. Y., Liu, X. L., Chang, S. H., Zhang, C., Du, W. C., and Hou, F. J. (2022). Effect of Cistanche deserticola on rumen microbiota and rumen function in grazing sheep. Front. Microbiol. 13:840725. doi: 10.3389/fmicb.2022.840725
Zhang, J., and Zhang, J. (2017). Grazing strategies and suggestions for sustainable utilization of alpine meadow resources in the Qinghai-Tibet Plateau. China Herbivores. Sci. 37, 63–67.
Zhu, D. Q., Sun, Y., Hou, G. C., Yang, L. M., Liu, F., Li, A., et al. (2016). Complete genome sequence of Bifidobacterium animalis subsp lactis klds 2.0603, a probiotic strain with digestive tract resistance and adhesion to the intestinal epithelial cells. J. Biotechnol. 220, 49–50. doi: 10.1016/j.jbiotec.2016.01.013
Keywords: functional native herbage, community, gastrointestinal, ruminant, microorganism
Citation: Xie K, Chang S, Ning J, Guo Y, Zhang C, Yan T and Hou F (2023) Dietary supplementation of Allium mongolicum modulates rumen-hindgut microbial community structure in Simmental calves. Front. Microbiol. 14:1174740. doi: 10.3389/fmicb.2023.1174740
Edited by:
Yanfen Cheng, Nanjing Agricultural University, ChinaReviewed by:
Hao Lizhuang, Qinghai University, ChinaMiao Lin, Yangzhou University, China
Qianming Jiang, University of Illinois at Urbana-Champaign, United States
Copyright © 2023 Xie, Chang, Ning, Guo, Zhang, Yan and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fujiang Hou, Y3lob3VmakBsenUuZWR1LmNu
†ORCID: Shenghua Chang, https://orcid.org/0000-0003-1561-7030
Jiao Ning, https://orcid.org/0000-0002-1529-6321
Yarong Guo, https://orcid.org/0000-0003-3303-6889
Cheng Zhang, https://orcid.org/0000-0002-5949-7192
Tianhai Yan, https://orcid.org/0000-0002-1994-5202
 Kaili Xie
Kaili Xie Shenghua Chang1†
Shenghua Chang1† Fujiang Hou
Fujiang Hou