- 1School of Biological Sciences, University of Aberdeen, Aberdeen, United Kingdom
- 2Centre for Sustainable Aquatic Research, Swansea University, Swansea, United Kingdom
- 3College of Life and Environmental Sciences, University of Exeter, Exeter, United Kingdom
- 4Centre for Environment, Fisheries and Aquaculture Science (Cefas), Weymouth, United Kingdom
- 5Department of Biosciences, Faculty of Science and Engineering, Swansea University, Swansea, United Kingdom
With an ever-growing human population, the need for sustainable production of nutritional food sources has never been greater. Aquaculture is a key industry engaged in active development to increase production in line with this need while remaining sustainable in terms of environmental impact and promoting good welfare and health in farmed species. Microbiomes fundamentally underpin animal health, being a key part of their digestive, metabolic and defense systems, in the latter case protecting against opportunistic pathogens in the environment. The potential to manipulate the microbiome to the advantage of enhancing health, welfare and production is an intriguing prospect that has gained considerable traction in recent years. In this review we first set out what is known about the role of the microbiome in aquaculture production systems across the phylogenetic spectrum of cultured animals, from invertebrates to finfish. With a view to reducing environmental footprint and tightening biological and physical control, investment in “closed” aquaculture systems is on the rise, but little is known about how the microbial systems of these closed systems affect the health of cultured organisms. Through comparisons of the microbiomes and their dynamics across phylogenetically distinct animals and different aquaculture systems, we focus on microbial communities in terms of their functionality in order to identify what features within these microbiomes need to be harnessed for optimizing healthy intensified production in support of a sustainable future for aquaculture.
1. Introduction
As the global population continues to expand rapidly, so too does the need for sustainably produced food. The world’s water bodies cover over 70% of the planet and are home to a bounty of protein-rich seafood (edible fish, invertebrates and algae from marine, brackish and freshwater environments). Global consumption of seafood (from capture and culture production in inland and marine waters) is growing at an annual rate of 3.1% (1961–2017), which is faster than for any livestock and animal production sector (FAO, 2020). Concurrent with stagnation of wild fisheries, aquaculture has progressively increased its contribution to fish and shellfish production with a mean annual growth of 5.3% between 2001 and 2018 (FAO, 2020), accounting for 52% of all seafood produced for human consumption in 2018 (FAO, 2020). The continued expansion of aquaculture has however been hampered by outbreaks of infectious disease, concerns over environmental footprint, and impacts of climate change.
Health management in aquaculture has historically relied upon antibiotics and other chemotherapeutics (de Bruijn et al., 2018), but continued development of targeted vaccines (Ma et al., 2019) and selective breeding to generate disease resistant lines (Houston et al., 2020) are now coming to the forefront as environmentally sustainable alternatives. A more holistic approach to health management in aquaculture is to consider the cultured animal and its surrounding environment together. Emerging evidence indicates that microbial communities both within and surrounding an animal in aquaculture can contribute directly to productivity in terms of growth, disease resistance and animal welfare (Perry et al., 2020; Rajeev et al., 2021; Dai et al., 2022; Murphy et al., 2022). Microbial communities play dynamic functional roles with both commonalities and contrasts across cultured animal groups from invertebrates such as mollusks and crustaceans to vertebrates—predominantly finfish—and should be viewed as central components in the development of a sustainable aquaculture industry.
The associated microbial communities both within and surrounding farmed aquatic animals are commonly referred to as the microbiome (Figure 1). Bacterial communities are the focal point of the vast majority of microbiome studies and a paucity of information exists relating to the communities of other microbes including viruses, fungi, archaea and protozoa. The fungal community has been characterized in telelosts including zebrafish, the gut of which contained fungal taxa belonging to more than 15 classes across the phyla Ascomycota, Basidiomycota, and Zygomycota, and distinct profiles in wild versus laboratory reared fish (Siriyappagouder et al., 2018). In the wood-eating Amazonian catfish (Panaque nigrolineatus), the mycobiome varied in different gut regions, and as a function of diet (Marden et al., 2017). Using a metatranscriptomic approach in the intestine of gilthead sea bream (Sparus aurata), fungal transcripts were found to be as abundant as bacterial, and while bacteria were important in vitamin and amino acid metabolism as well as rhythmic and symbiotic pathways, fungi were determined to play a role in host immunity, digestion and endocrine processes (Naya-Català et al., 2022). In green-lipped mussels (Perna canaliculus), the fungal community was determined in gills, hemolymph, digestive gland and stomach, and the majority of amplicon sequence variants (ASVs) belonged to the phylum Ascomycota in line with other studies (Li et al., 2022). However, more than 50% of ASVs for fungal taxa were only classified at the phylum level, highlighting the lack of fungal studies and thus lack of reference sequences, a hurdle which will need to be overcome to fully characterize complete microbial communities.
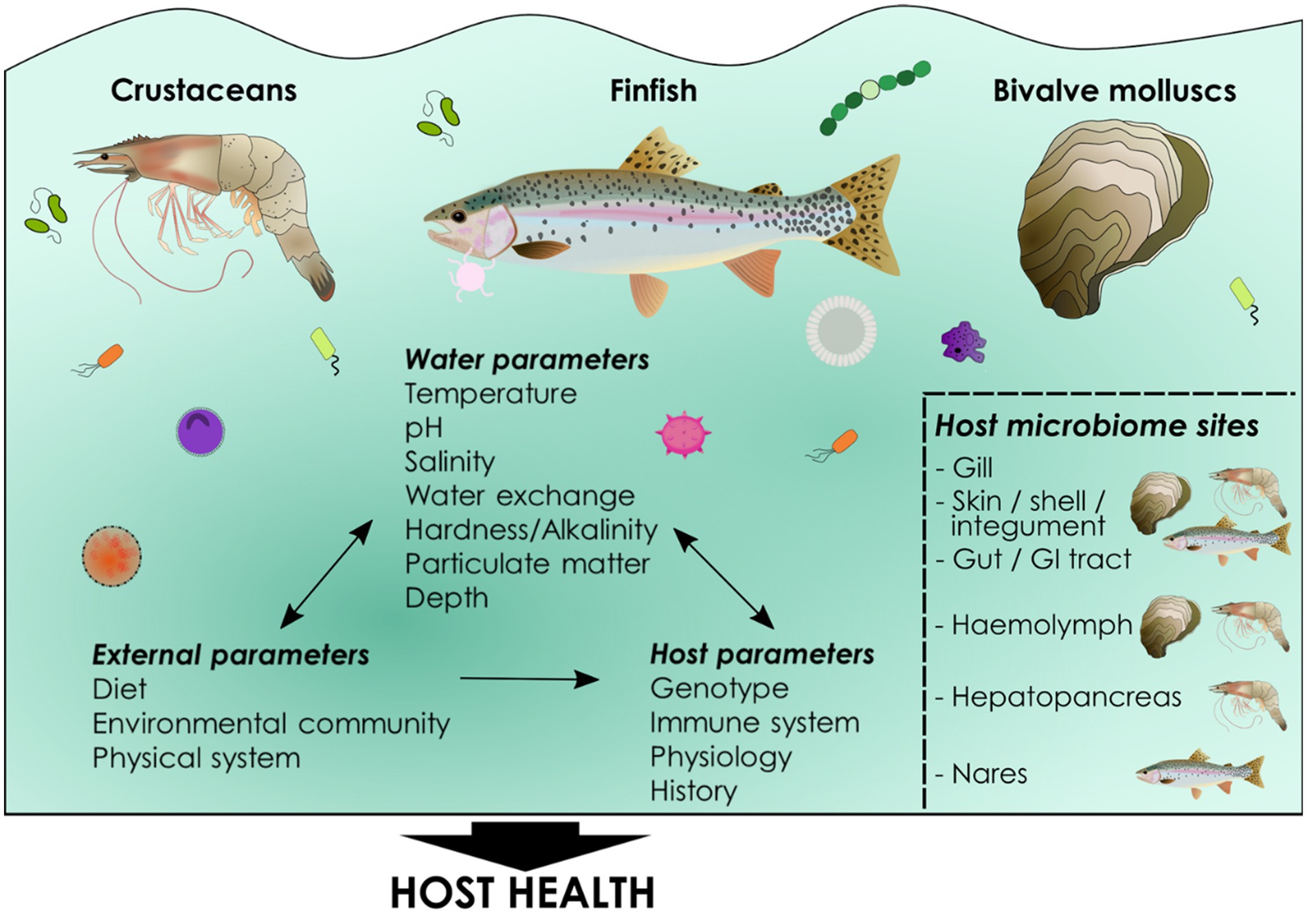
Figure 1. What is a microbiome? A microbiome can be defined simply as “a community of commensal, symbiotic, and pathogenic microorganisms within a body space or other environment” (Lederberg and McCray, 2001), although many varying definitions exist (Berg et al., 2020). A microbiome comprises a community of microbes including bacteria, viruses, fungi, microeukaryotic and metazoan parasites and Archaea (collectively known as the microbiota) as well as the downstream products and functionality of the microbiota. Cultured aquatic species harbor communities of microbes in the gut or gastrointestinal tract, as well as in the skin, nasal and oral cavities, gills, hemolymph, shells, integument and hepatopancreas, some of which are common across all cultured animal phyla (e.g., gills). The established microbiome composition is complex and involves not only host factors such as genotype and the host immune system, but also external environmental influences such as diet, physical parameters of culture systems, microbes in the aquatic environment and an array of water physiochemical parameters, including temperature, salinity, and dissolved oxygen. Together these parameters determine microbiome composition in host niches and these communities of commensals influence host health. Key determinants of host microbiome composition in cultured aquatic animals. Host microbiomes are determined by both environmental and host-associated factors. Environmental parameters can also reciprocally impact each other, as well as the host, particularly the microbial community within the water. Environmental parameters can be measured and, in some cases, controlled in aquaculture as can host factors such as genetics. Microbiomes occur in all cultured taxa including finfish, bivalve mollusks and crustaceans. Common microbial niches are found in a number of mucosal tissues including gut, gill and skin while others are specific to particular phyla, for example invertebrate hemolymph. Host microbiome composition itself has important implications for host health, but the functional and mechanistic pathways underlying these associations are poorly understood in aquaculture.
Across cultured phyla there are common tissues which host microbial communities including gut and gill, but microbiomes also occur in fish on their skin, and in their nasal and oral cavities, and in crustaceans and mollusks in their hemolymph (blood), digestive glands, shells (mollusks) and integument (crustaceans). Differences in microbiome composition in different tissues is driven by niche specialization and host selective pressures (Kelly and Salinas, 2017). A common feature across phylogenetically distinct animals in aquaculture is the primary role of internal mucosal microbiomes in digestion and nutrient absorption while external mucosal communities assume a primarily defensive role from the perspective of outcomes for the host. Yet microbiomes can also contain opportunistic pathogens, and to some extent are determined by the surrounding environment and not regulated by the host. In this perspective we explore how microbiomes contribute to productivity and health in aquaculture, with a particular focus on the commonalities and differences between phylogenetically diverse cultured animals and culture systems. We also present future prospects on how we may harness the power of the microbiome to promote sustainable aquaculture as production seeks to be intensified.
2. Establishing the host microbiome—where do commensal microbes come from in aquaculture?
Distinct from their terrestrial counterparts, aquatic organisms are in constant contact with (potentially) microbially rich water and this constant exposure may offer adaptive advantages, in addition to some inherent threats. In aquaculture systems, the aquatic environment is highly dynamic and can change both frequently and rapidly in response to many factors, including weather, temperature, and husbandry practices. The majority of aquaculture species, both invertebrate and vertebrate, reproduce by external fertilization, producing an independent egg exposed to the environment. External mucosal surfaces are first colonized by environmental microbes shortly after hatching (Giatsis et al., 2014; Stephens et al., 2016). As initial colonizers originate from the environment, hatchery conditions play a strong role in determining the diversity and richness of the initial colonizing community (Minich et al., 2021). In contrast, the internal gut mucosa is not colonized until the time of mouth opening and first feeding, exposing this internal niche to a potentially different pool of colonizers from the environment as well as diet-associated microbes. Critically, a sterile or partially sanitized environment restricts the pool of microbes from which to establish resident communities.
2.1. Selectivity in colonization—how to tell friend from foe?
Interactions between mucosal microbiota and the host immune system is particularly intense during initial colonization by commensals. Commonalities exist between invertebrates and vertebrates during the first phase of colonization in that acceptance or rejection of microbial taxa is controlled by a triad of the environmental conditions, the microbes themselves, and the host innate immune system. Germline-encoded cell receptors, where they exist, can play an early role in community selection and the innate immune system must have the ability for tolerance and allow settlement of commensals while selectively excluding opportunists or undesirable taxa (Kelly and Salinas, 2017). These cell receptors are likely to differ between mucosal surfaces, which raises interesting mechanistic questions. However, it is not solely the case that the immune system “selects” symbionts, but also that microbes from the environment may successfully colonize as they are best adapted to survive in the nutrient rich biochemical environment of the mucosal surface, independent of host influences.
In contrast with crustaceans and mollusks, the immune systems of which are devoid of lymphocytes and immunoglobulins (Igs), the acquired immune system or immunological memory capability of fish develops at first-feeding (Castro et al., 2015), while initial colonization of the gut occurs between mouth opening and first-feeding. As such, in addition to being exposed to a temporally distinct group of potential colonizers, colonization of the gut may be regulated by different immune pathways in invertebrates and vertebrates. In fish, as in other vertebrates, Igs are a key feature in acquired mucosal immunity, coating resident microbes to aid in selectivity (Woof and Mestecky, 2005; Kelly and Salinas, 2017). Dominant Ig isotypes have been identified at different mucosal sites, suggesting a role in determining site specific microbiota (Salinas et al., 2021). Although mollusks and crustaceans lack immunological memory in terms of Ig-mediated mechanisms, they harbor complex repertoires of lectins and other binding molecules which enable recognition of diverse microbes, resulting in control of undesirable species for successful innate immune protection (Vasta et al., 2007, 2012; Gerdol et al., 2018; Coates et al., 2022). Thus, successful colonizers are microbes in the environment which are suited to a mucosal niche and evade exclusion by the host immune system. This has been well studied in bobtail squid, Euprymna scolopes which acquire a specific strain of Vibrio fischeri from the surrounding environment into a specialized light organ, excluding all other bacterial taxa (Kostic et al., 2013). An additional facet of the immune system to consider when trying to understand microbial colonization are antimicrobial peptides (AMPs). AMPs refer to small peptides also known as bacteriocins including hepcidins, beta-defensins, cathelicidins and fish-specific piscidins (Masso-Silva and Diamond, 2014; Katzenback, 2015). AMPs are generally toxic to closely-related strains, although wider target ranges also occur (Rodney et al., 2014) and thus have the potential to impact colonization and succession by making an inhospitable environment for certain taxa. Less well understood in aquaculture is the interaction between environmental microbes and host genetics in microbiome colonization.
2.2. How do host genetics impact microbial colonization?
Host genetics may play a role in the establishment of microbial associations through microbial recognition, immune selection, and determination of the biochemical niche (Spor et al., 2011). Fish of different species reared in the same or closely related environments can differ in their microbiome composition (Nikouli et al., 2021). Additionally, a genetic component may persist within species. In genetically divergent three-spined stickleback (Gasterosteus aculeatus) populations, divergence in the intestinal microbiota is greater than could be accounted for by environment and ecology, supporting a role for host genetics in the selection of bacterial species (Smith et al., 2015; Steury et al., 2019). Even within populations, an interaction between major histocompatibility complex class IIb (MHC class IIb) polymorphisms and genotype could impact microbiome diversity (Bolnick et al., 2014). However, other studies in Atlantic salmon (Salmo salar) identified a relatively minor role of host genetics (Dvergedal et al., 2020), particularly when compared to environmental effects (Uren Webster et al., 2018). Yet using hybrid populations of Chinook salmon, compositional differences in microbiota were attributed to quantitative genetic architecture (Ziab et al., 2023). It may be the case that, in common with mammalian systems, genotype-dependent selection may influence specific microbes, as opposed to the whole microbial community (Tabrett and Horton, 2020). Some animals in aquaculture, particularly salmonids, generally originate from brood-stock under genetic selection for production traits, notably related to growth, which may influence the contribution of environmental relative to genetic factors. In the case of shrimp farming, traits selected for are almost entirely focused on disease resistance. Importantly, genetic processes select microbes after the host has come into contact with bacterial communities in the environment, an important consideration in trait selection for different environments.
Little is known about the interactions between host genetics, microbiome stability, and health outcomes in relation to animals in aquaculture. Pacific oyster [Crassostrea (Magallana) gigas] families susceptible to Ostreid herpesvirus-1 have been shown to have differences in bacterial community structure and evenness compared to resistant families (King et al., 2019; Clerissi et al., 2020) and there are similar observations for the gill and gut of susceptible and resistant rainbow trout (Oncorhynchus mykiss) in response to Flavobacterium psychrophilum infection (Brown et al., 2019). In addition to genetic influences, epigenetic processes provide a link between host and microbiota. Metabolites resulting from cellular metabolic processes can impact the activity of enzymes involved in histone and DNA methylation and demethylation (Narne et al., 2017; Reid et al., 2017). The environmental microbiome is important in host immune system maturation, including the development of pathogen recognition systems. In the Pacific oyster, early life exposure to microbes improved survival during a challenge with Pacific oyster mortality syndrome (POMS) not just in the exposed generation, but in the following generation also. A combination of microbiota, transcriptomics, genetics and epigenetic analyses determined a distinct change in epigenetic methylation marks during microbial exposure leading to an altered immune gene expression and long-lasting, intergenerational immune protection against POMS (Fallet et al., 2022).
Overall, host genetics appear to play a central role in microbiome establishment, but little is known about the genomics x environment interaction for establishment and maintenance of microbial communities. The interactive nature of genetics and environment raises the potential for targeted environmental conditioning in aquaculture. To consider such approaches, it is important to first understand the role of physical aquaculture systems in dictating parameters which influence both host and environmental microbes.
3. Aquaculture system dynamics and host microbiome determination
Water has many variable properties including temperature, salinity, pH, ionic composition and chemistry related to dissolved gasses. Additionally, physical aquaculture systems dictate water exchange rate, retention time, and a level of biological sterility, particularly in hatcheries, a key stage for community establishment. Water harbors an array of microbes which provide ecosystem services within wild, and aquaculture systems and system parameters are important determinants of microbial dynamics. In low diversity microbial environments with input of organic material, fast-growing opportunistic r-strategists characteristic of instability can outcompete slow-growing K-strategists which are markers of stability. However, if water retention time in the system is short, opportunists are less able to replicate and stability of the system is more likely maintained by virtue of this management practice (Attramadal et al., 2012; Vadstein et al., 2018). A key end-goal of system optimization in aquaculture is to define environmental parameters which are conducive to desirable microbiomes in terms of sustainability and productivity, and to then have the ability to monitor and control these parameters to ensure optimum performance. However, due to the wide range of aquaculture system types (categorized as flow-through, open, or recirculating) and their associated temporal dynamics, a “one-size fits all” approach to determining optimum parameters is unlikely to succeed.
3.1. Aquaculture systems and evolving microbial stability
The nature of the aquaculture system can have major impacts on microbiomes of finfish (Minich et al., 2020, 2021; Uren Webster et al., 2020) and invertebrates, as illustrated for oyster (Arfken et al., 2021) and shrimp (Tepaamorndech et al., 2020). Such impacts are determined by physical and environmental parameters within systems and can influence production outcomes including growth and survival (Table 1). Culture systems come in an array of shapes and sizes and can be broadly described on a scale from open to closed (Figure 2). Larval stages are generally cultivated in highly controlled flow-through or recirculating aquaculture system (RAS) hatcheries with many species requiring planktonic organisms as feed before being transferred to on-growing facilities that vary between species. On-growing occurs in tanks, ponds and cages, or on other structures such longlines and rafts for bivalves (Tidwell, 2012). Most commonly these on-growing stages are exposed to the natural environment and generally employ the use of processed diets for finfish and crustaceans, or natural feed for filter feeding animals. Most open systems such as sea cages are exposed to natural fluctuations in the environment and may experience temperature extremes, exposure to harmful algal blooms, jellyfish (Clinton et al., 2021), and pollutants (Trabal Fernández et al., 2014). Such challenges have direct impacts on microbial communities and exposure to pathogens. The desire to have greater control over environmental parameters with a view to optimizing water quality to promote growth and minimize pathogen exposure, has led to an expansion of closed or semi-closed systems such as RAS. However, although it is more feasible in closed environments to maintain a high degree of stability, water chemistry can change over time in closed systems, depending on how they are maintained, and this can affect the biological load (Fossmark et al., 2020) and in turn the environmental microbiome. How the physiochemistry of the different aquaculture systems may affect microbial stability in aquaculture is now a significant area of research interest. Although the physical systems in which invertebrates and vertebrates are reared in and on-grown differ, commonalities can be defined in terms of the nature of the systems on the scale from open to closed.
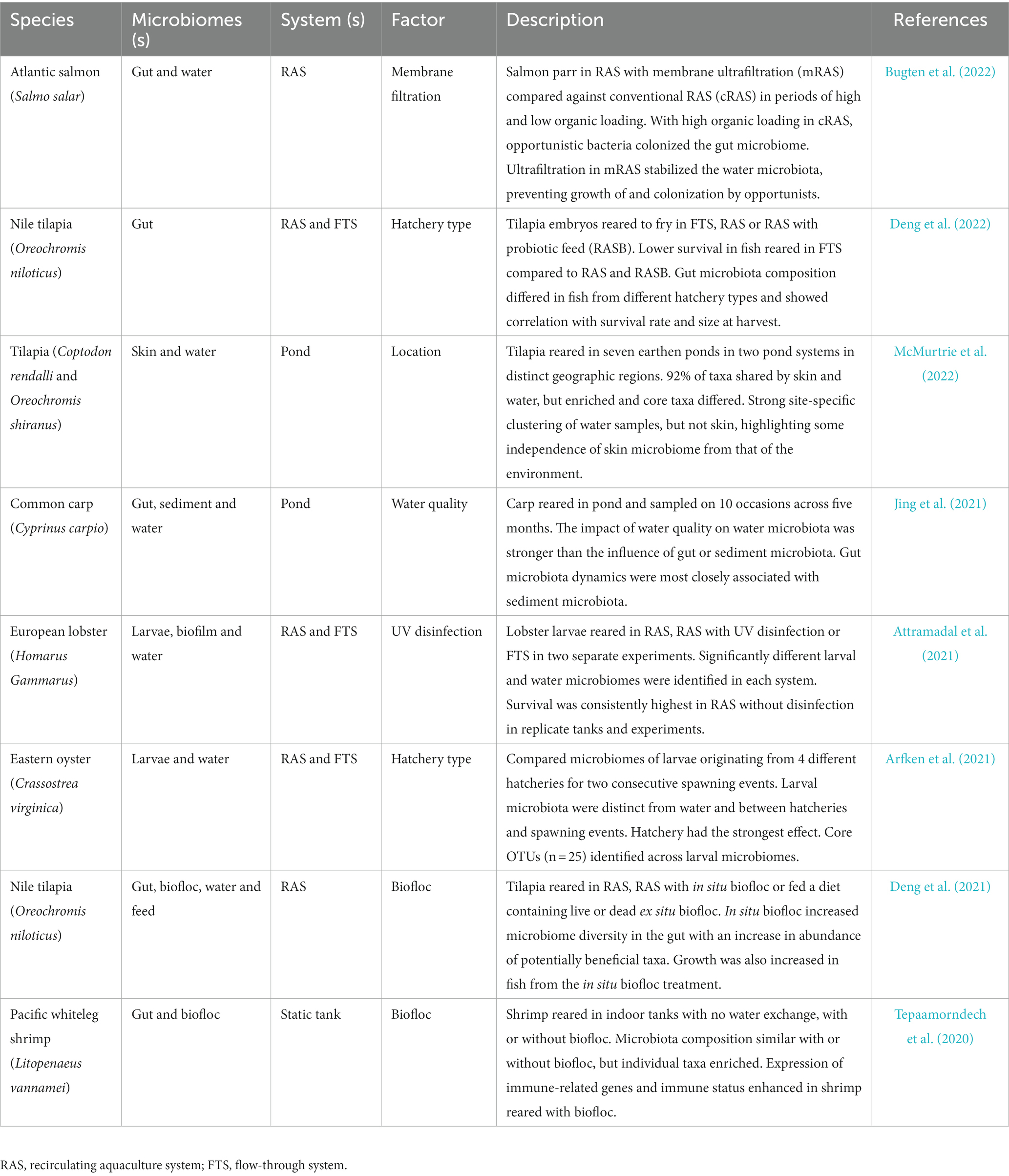
Table 1. Key published literature describing system parameter effects on animal microbiomes and production outcomes.
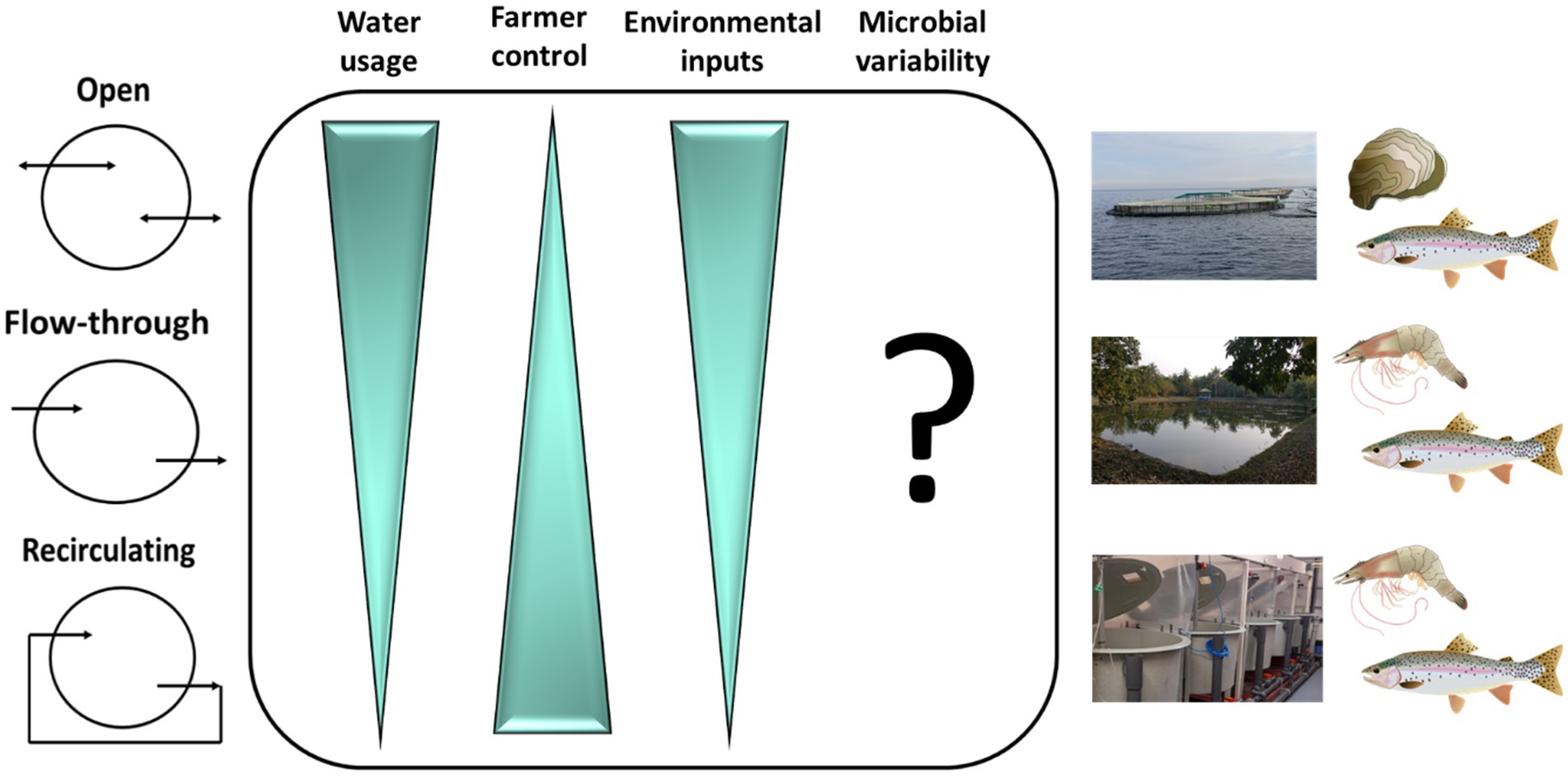
Figure 2. Microbiome dynamics in different aquaculture systems. Water usage, farmer control, environmental inputs and microbial variability in aquaculture systems. An important consideration in microbial maturation within aquaculture systems is water usage, particularly in terms of water flow and exchange. Systems vary from open (e.g., Atlantic salmon pens in lochs or fjords) to closed (e.g., land-based recirculating aquaculture systems; RAS) with systems between the two extremes referred to as flow-through (e.g., tilapia or shrimp ponds). Farmer control over environmental parameters including temperature and photoperiod are greater moving from open to recirculating systems and environmental factors such as natural seasonality and exposure to pollutants decrease. These dynamics may suggest a more stable microbial environment in closed systems, but in reality microbial variability is unlikely to follow a gradient from open to closed systems. Open systems are exposed to a rich pool of natural microbes which vary with season and geographic location. Contrastingly, strict husbandry practices to ensure pathogen exclusion in RAS, may result in periods of near sterility in the system, effectively re-setting the microbial community and introducing variability. In both cases, the result is potential instability in the system, but it is not known if stability is essential to a functioning microbiome.
3.2. Open versus closed—what do aquaculture system structures mean for the microbiome?
In open aquaculture systems with constant water exchange—such as those used in shellfish cultivation and finfish cage culture—water quality, chemistry and microbial composition vary with geographical location, time, and season, with temperature having a strong impact on the microbiota (Sunagawa et al., 2015). For example, in response to increased water temperature, a surge in opportunistic pathogenic taxa in the Pacific oyster, C. gigas with dominance of Vibrionaceae occurs (Green et al., 2019). Similarly in marine Atlantic salmon, gut microbiomes are dominated by Leuconostoc and Weissella in cold water temperatures, but by Vibrio, Allivibrio, and Photobacterium in warmer water (Zarkasi et al., 2014). Circadian rhythmicity is naturally determined by latitude and influences microbiome composition in rainbow trout skin (Ellison et al., 2021), but photoperiod is also often manipulated in aquaculture to promote growth and enable year-round production (Mero et al., 2019; Wang et al., 2020; Pino Martinez et al., 2021), and is likely to impact on immune function with consequences in mucosal immune activity (West et al., 2021).
In RAS, microbial processes are vital in maintaining water quality (Attramadal et al., 2012; Blancheton et al., 2013). Biofilters in the RAS loop host essential communities of bacteria which regulate conversion of waste nutrients to prevent build-up of toxic metabolites such as ammonia and hydrogen sulfide resulting from uneaten feed and fecal matter. Build-up of organic matter or introduction of pathogens result in mass mortalities, and therefore various forms and combinations of filtration and disinfection are often carried out in a RAS loop (Summerfelt et al., 2009). Filtration in the RAS loop assists in stabilizing carrying capacity (Fossmark et al., 2020), while disinfection is an effective means of removing pathogens, but may effectively “re-set” the community. Such dynamics raise questions about how, where, and when RAS water should be subject to disinfection. Simply put, how clean is too clean? At the other end of the sterility scale, the use of biofloc technology, a microbial feed source added to culture water to maintain microbial communities in culture water, has associated benefits for immune reactivity in shrimp (Tepaamorndech et al., 2020), and faster growth in tilapia (Nguyen et al., 2021). Importantly, despite the relative stability of RAS in comparison to open systems, microbiomes in Atlantic salmon gut in RAS are still temporally dynamic, possibly related to biological load in the system (Lorgen-Ritchie et al., 2021).
Intermediate between open and closed states, pond systems are the dominant forms of global aquaculture, particularly prominent in low-middle income countries of Asia, responsible for farming carp, tilapia and shrimp (FAO, 2020). Similar to open systems, ponds are exposed to fluctuating external environmental parameters, but the physiochemical and biotic condition of ponds can shift rapidly due to their shallow depth and limited water exchange, especially when compared to the large water body buffer of open cage culture. During the rainy season, rapid salinity fluxes occur due to rainwater diluting ion concentrations. In intensive culture, algal blooms can form due to excessive food and fertilizer (e.g., manure) inputs providing the key phytoplankton nutrients phosphorous and nitrogen (Huang et al., 2016). In summer months overgrowth of photoautotrophic bacteria can lead to highly turbid pond water, limiting oxygen production (Sriyasak et al., 2015; Dien et al., 2019). In the oriental river prawn (Macrobrachium nipponense) hypoxia induced changes in the intestinal microbiome composition with a deviation toward higher levels of pathogenic bacteria (Aeromonas) (Sun et al., 2020). Mitigation strategies for anoxic conditions include pond aeration (Boyd, 1998), and multi-trophic farming with mollusks that filter feed on phytoplankton resulting in a net increase of dissolved oxygen (Cunha et al., 2019). As aquaculture continues to expand, it would be beneficial to employ an ecosystem-based approach when designing and optimizing aquaculture systems, attempting to integrate interactions between physical environment, host, and microbiome to promote establishment of desirable microbiomes for productivity and host health.
4. Which microbiomes promote animal health in aquaculture?
Productivity and welfare in all areas of the aquaculture industry are under constant threat from opportunistic pathogens, infectious disease and syndromes which result in poor health. A growing body of evidence challenges the idea of a single pathogen causing a single disease; the concept of a “pathobiome” implies that a triad of interactions between environmental microbes, host-associated commensals and the host itself steer overall host health (Bass et al., 2019). For example, in skin mucus of brook charr (Salvelinus fontinalis), host stress was associated with a decreased abundance of beneficial commensals including Methylobacterium and Propionibacterium, known to produce pathogen growth inhibitors poly-β-hydroxybutyrate and bacteriocin, coincident with a rise in abundance of opportunistic pathogenic genera including Aeromonas, Psychrobacter and Acientobacter (Boutin et al., 2013). Additionally, cryptic infection is possible whereby pathogens with dormant, or non-pathogenic life stages may infect a host asymptomatically, but be activated as a pathogen in response to a change in the environment, for example, seasonal changes in temperature (Overstreet and Lotz, 2016; Bass et al., 2019). Mucosal surfaces and their associated microbiota are the first line of defense against pathogens and are equipped with a range of protective molecules such as antimicrobial peptides and immune-related cell components (Reverter et al., 2018) which may induce competitive advantage directly or indirectly. It is becoming increasingly clear that disease impacts host microbiomes, but also that host micorbiomes can mediate disease severity or progression (Table 2). For example, in Atlantic salmon infected with amoebic gill disease (AGD), a significant reduction in bacterial diversity was apparent when compared with healthy individuals and a particular taxon (Tenacibaculum dicentrarchi) was particularly prevalent in lesions of infected gill tissue (Slinger et al., 2020). In red abalone suffering from Withering syndrome (WS), Mycoplasma was replaced as the dominant taxon in the digestive gland by Candidatus Xenohaliotis californiensis, but C. X. californiensis was also a member of the microbiome of healthy individuals, suggesting that in this case, the ratio between the two taxa may be key in determining pathogenicity of the disease (Villasante et al., 2020). The array of life history strategies, species and systems across which aquatic animals are farmed represent a challenge in identifying desirable microbiome signatures at the taxonomic level, yet at a functional level, commonalities can be identified. In this line of thinking, it is the ability of the microbiome to maintain functionality in the face of environmental change that promotes host health, rather than consistency of the taxonomic composition of microbial communities.
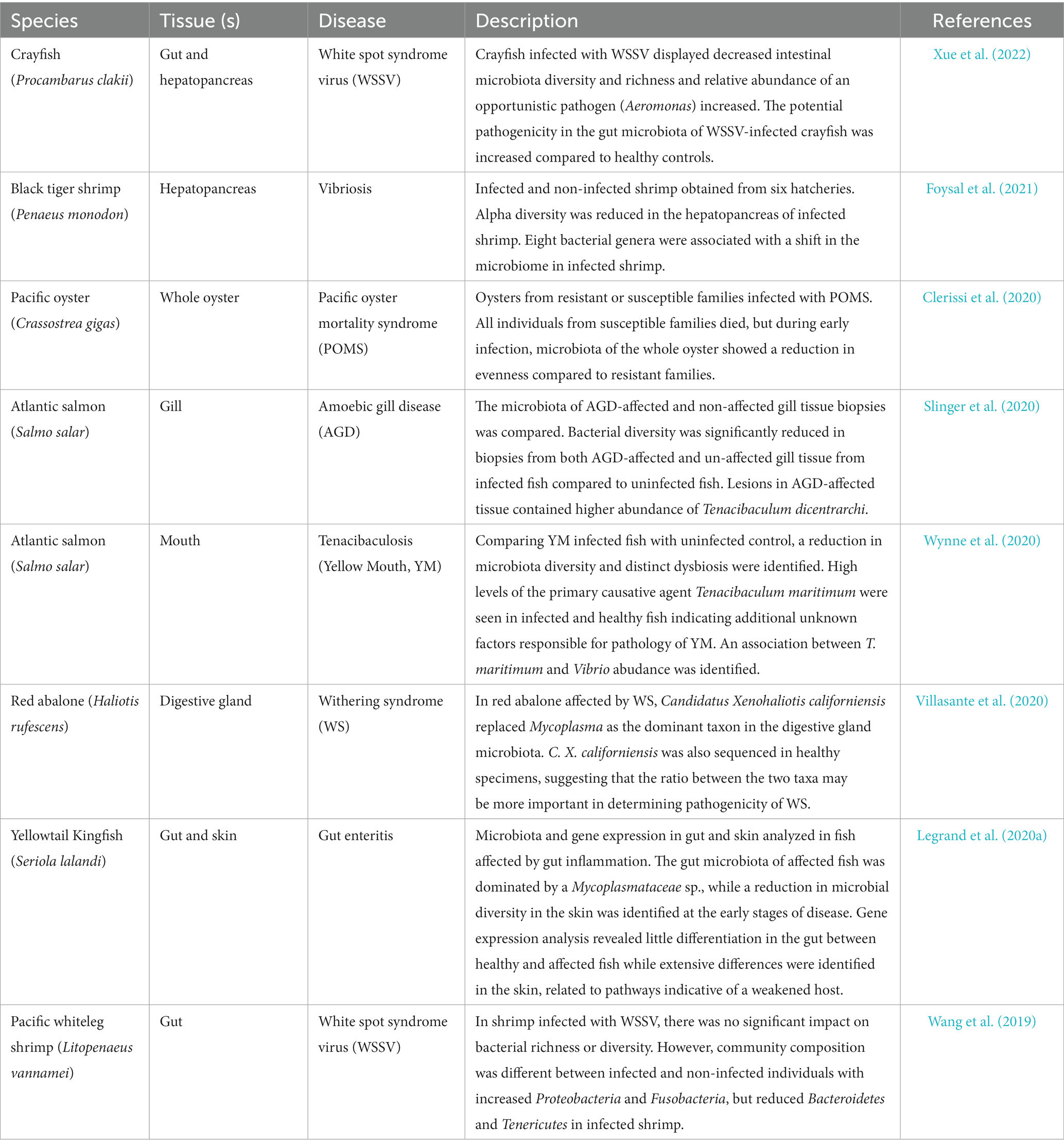
Table 2. Key published literature exploring the interaction between disease and host microbiome composition.
4.1. Can a “healthy” microbiome be defined for aquaculture practices?
Attempts have been made in the past to define a “healthy” microbiome in terms of aquaculture but realistically a single consistent “healthy” microbiome is unlikely to exist, even within a single species in a single culture system (Infante-Villamil et al., 2021; Rajeev et al., 2021). Thus, perhaps a desirable microbiome for aquaculture should instead be defined as one that has the capacity to adjust to its environment, and in doing so maintain a beneficial symbiotic relationship with the host. Maintenance of community homeostasis historically has been considered important, but this should be expanded to considering community homeostasis in terms of functionality rather than taxonomic identity. Microbial communities can be remarkably dynamic in response to changes in their environment and a highly plastic, dynamic microbiome may also contribute to host phenotypic plasticity, and thus host ability to tolerate environmental stressors. The “Anna Karenina principle” (Zaneveld et al., 2017) infers that stressors create more stochastic microbiomes as a consequence of a dampening of the host’s ability to regulate its microbial communities, resulting in increased inter-individual variation in microbiomes of hosts experiencing stress (Boutin et al., 2013). However, the contrasting pattern has also been observed, with remarkably similar microbiome responses to environmental disturbance which have been attributed to reduced competition allowing increased dominance of certain taxa (Webster et al., 2019). Such differences may well reflect differences in external environment. In open systems with more variation in environmental microbes, we might expect greater stochasticity, but in closed systems with less environmental microbial variation, we might expect to see the domination of certain taxa.
Although maintaining low levels of stress in aquaculture is a central tenet of stock management, even routine husbandry practices can induce stress, which may increase disease susceptibility (Eissa and Wang, 2016). A microbiome with a high adaptive plasticity is less likely to be disturbed by environmental stressors and therefore there are less likely to be knock-on adverse effects of microbiome disturbance on host health. A plastic microbiome may also confer specific fitness benefits to the host (e.g., an ability to metabolize or sequester dietary toxins, adapt to novel nutrient or feed sources, or to cope with changing water temperatures) (Alberdi et al., 2016; Macke et al., 2017). It is important to consider that the nature and outcomes of stress responses will depend on the nature of the existing microbial community, with different mucosal sites displaying differing responses to stress in terms of both cortisol levels and microbiome dynamics (Webster et al., 2019). Despite variable responses, identification of markers of stress or dysbiosis across datasets could enable these to be leveraged in real-time microbiome monitoring in aquaculture through non-invasive sampling of water, feces, or external mucus to detect microbial signatures of potential ill-health. Going a step further, the future ideal would be to actively intervene and manipulate the microbiome during these periods of dysbiosis to help re-balance disruptions and maintain optimal host health.
4.2. Dysbiosis and rebiosis—an opportunity to condition microbiomes for sustainable aquaculture?
As explained above, the first microbial colonizers are key in determining succession via competitive exclusion or “colonization resistance,” competing for nutrients and physical space (Vonaesch et al., 2018). Post-colonization, development of the fish microbiome is characterized by periods of establishment and proliferation which are especially dynamic and strongly influenced by environment, and periods of stabilization which are strongly influenced by the host. Husbandry practices can introduce instability to the environment and these transitional periods are often accompanied by high levels of mortality or increased disease susceptibility (Paul-Pont et al., 2013; Johansson et al., 2016). As an example, on-growing of Atlantic salmon requires transition from a freshwater to a saltwater environment, exposing commensal microbes to a distinct shift in salinity and ultimately creating a new niche for salt-tolerant taxa to the detriment of freshwater commensals. Such distinct shifts in environmental parameters are accompanied by rapid remodeling of host microbiomes (Dehler et al., 2017; Lorgen-Ritchie et al., 2021) characterized by an initial loss of community structure, a process known as dysbiosis, followed by re-establishment of a new community—rebiosis. Periods of rapid change in the microbiome are especially sensitive to external influences of incoming microbes that may later be more inhibited by colonization resistance, and this may have a lasting effect on microbiome composition and function via priority effects (Debray et al., 2021). This could have important implications for aquaculture as these periods may be especially sensitive to disruption but may also represent an opportunity for increased sensitivity to conditioning.
Dysbiosis in the naturally occurring microbiome of the host often coincides with infection or poor health, from invertebrates to humans. Dysbiotic states may manifest as (i) loss of beneficial organisms, (ii) expansion of pathobionts and/or (iii) loss of microbial diversity (Petersen and Round, 2014). It is important to consider community interactions when trying to understand these processes. Ecological theory suggests diverse communities are more resistant to dysbiosis and invasion by opportunistic pathogens (Levine and D'Antonio, 1999) as they have more diverse functionality and more effective competitive exclusion. Salmon experimentally infected with the copepod “louse” ectoparasite, Lepeophtherius salmonis, have been shown to display a reduced bacterial richness in their skin microbiome (Llewellyn et al., 2017). A similar pattern has been observed in the gut of shrimp suffering white feces syndrome (Hou et al., 2018). However, in shrimp suffering from white spot syndrome virus (WSSV), no significant difference in diversity was observed although community composition was distinct in control vs. diseased animals (Wang et al., 2019). Moving forward, defining dysbiosis in terms of loss of functional integrity and knock-on effects on host health may be more useful when it comes to potential development of therapeutics.
It remains unclear whether dysbiosis occurs in response to infection or if opportunists exploit an existing or incipient dysbiotic state, perhaps induced by change in environmental conditions or host physiology or immune capabilities, in order to infect. A combination of both is probably most likely—infection may well exacerbate an already dysbiotic state. To illustrate this scenario, production of the Pacific oyster (C. gigas) is hampered by disease, especially Pacific oyster mortality syndrome (POMS) that has caused major economic losses (Segarra et al., 2010). POMS is a polymicrobial disease where oysters become infected by osterid herpes virus 1 μvar (OSHV-1 μvar) and this leads to immune suppression facilitating dysbiotic shifts in the oyster’s microbiome. This altered microbiome, the pathobiome, is characterized by dominance in taxa including Vibrio, Aliivibrio, Arcobacter, Marinobacterium, Marinomonas, Photobacterium, Psychrobium, and Pschromonas (de Lorgeril et al., 2018; Clerissi et al., 2020; Petton et al., 2021; Richard et al., 2021). Affected oysters show bacteremia where opportunistic vibrios including V. aestuarianus cause significant mortality. Oyster families displaying greater microbial diversity within individuals tend to be less likely to suffer from POMS (Clerissi et al., 2020) and selective breeding programs may improve disease resistance and hence reduce the impact of this condition (Dégremont et al., 2020). Similarly in Atlantic salmon, amoebic gill disease (AGD) is a increasing gill health issue, the etiological agent of which is the free-living amoeba Neoparamoeba perurans (Crosbie et al., 2012). AGD infection is initiated by adherence of N. perurans to the gill mucosa where commensal bacteria persist (Embar-Gopinath et al., 2005). Microbial profiling revealed lower bacterial diversity and a moderate positive correlation between N. perurans and Tenacibaculum dicentarchi in infected tissues (Slinger et al., 2020). Altering bacterial load on the gills prior to N. perurans infection using antimicrobial treatments resulted in differing progression of AGD with significantly higher severity observed in chloramine-trihydrate (CI-T) treated fish vs. control fish. High levels of Tenacibaculum were observed in the CI-T group again suggesting a protagonistic role for this taxon in AGD infection highlighting the potential for dysbiosis to allow expansion of potentially pathogenic communities which may affect subsequent disease progression (Slinger et al., 2021). Future research is needed to identify useful commonalities in these intertwined relationships between dysbiosis, disease progression, and host health.
5. Moving from microbiome characterization to beneficial manipulation in aquaculture
Identifying commensal microbes and their dynamics is the first step in understanding the value of microbiomes for aquaculture productivity and sustainability. If “the microbiome” (accepting there are many), is to be used as a tool to improve health and performance of the animals, we need to know not just which microbes are present, but how these microbial communities function (Gibbons, 2017). In recent years there has been a distinct shift in microbiome research to “function over phylogeny.” Functionality has historically been inferred from high-throughput amplicon sequencing, but predictions are limited in their use in aquaculture studies due to poor taxonomic resolution and low numbers of characterized bacterial genomes across aquaculture environments (Infante-Villamil et al., 2021). Additionally, such predictions relate to a specific set of conditions, and functional analysis tools assume that all genes are transcriptionally active in all microbial taxa at that moment in time, which more often than not is simply not the case. More holistic ‘omics strategies such as metagenomics (Tepaamorndech et al., 2020), metatranscriptomics (Nam et al., 2018), metaproteomics (Jose et al., 2020), and metabolomics (Roques et al., 2020), as well as integration of these approaches (multi-omics) (Uengwetwanit et al., 2020) are increasingly being adopted. A step further is to consider the “hologenome,” that is the collection of genes in the microbiomes along with those of the host (Rosenberg et al., 2007; Rosenberg and Zilber-Rosenberg, 2018). The strength of these approaches is the ability to examine microbiomes in terms of community ecology, considering interactions among microbiota, with the host and with the environment, but integration requires well designed and executed sampling strategies, sufficient levels of sampling (generally producing very large datasets), and complex bioinformatics analyses. Omics approaches are key to uncovering contrasts and commonalities between microbiome function across aquaculture but must be applied in combination with carefully designed mechanistic studies.
5.1. Modeling a desirable microbiome function for aquaculture
One approach to understanding functionality is to use germ-free (GF) or gnotobiotic models. Gnotobiotic refers to animals hatched and reared in a sterile environment which provide a “blank canvas” on which to test the impact of introducing a single microbe or a community of microbes. Although gnotobiotic research has been applied to aquaculture species since the 1940s (Zhang et al., 2020), full use of this approach as a tool to understand microbiome functionality is only now being realized. Nile tilapia (Oreochromis niloticus) models have been used to test the efficacy of therapeutics and to study colonization by free-living microbes (Situmorang et al., 2014; Giatsis et al., 2016) while in gnotobiotic rainbow trout larvae commensal taxa which offered protection against pathogenic infection have been identified (Pérez-Pascual et al., 2021). Furthermore, cod (Gadus morhua) models have recently been used to study the role of microbiota in innate immune development and gut morphology (Vestrum et al., 2022). Gnotobiotic oyster (Crassostrea gigas) models have also been utilized to detect effects of pathogens (Arzul et al., 2001) while in crustacea, germ-free Artemia brine-shrimp (Baruah et al., 2017) and Daphnia (Macke et al., 2020) models are widely used to study host-microbiome-environment interactions.
Pioneering work in zebrafish (reviewed in Murdoch and Rawls, 2019) established a germ-free model, but also a conventionalized model by harvesting bacteria from tank water of conventionally reared zebrafish and adding this community to gnotobiotic zebrafish medium to colonize germ-free animals (Rawls et al., 2004). Using these models and transcriptomic analysis, 212 genes regulated by the microbiota were identified in the intestine, and 59 responses were conserved in the mouse intestine indicative of an evolutionary conserved role of the vertebrate gut microbiome. Similarly, exposure of germ free zebrafish to yeast in the form of Pseudozyma sp. elucidated differential expression of 59 genes compared to naive controls, many involved in metabolism and immune response, indicating that commensal fungi also have the potential to influence the early development of fish larvae (Siriyappagouder et al., 2020). This work was extended using reciprocal gut microbiome transplants from zebrafish and mice to germ-free recipients and identified the importance of host habitat selection (Rawls et al., 2006). Transplanted communities resembled the community of origin in terms of taxonomic lineages present, but relative abundances were modified to resemble the normal microbial community composition of the recipient host. These transplantation studies are important in understanding microbiome function and selection pressures during colonization, but also have the potential to be adapted for therapeutic means similar to fecal microbiota transplantation (FMT) (Petersen and Round, 2014; Vargas-Albores et al., 2021). Such approaches may well be used in aquaculture as understanding of functionality of microbiomes improves.
Limitations exist in terms of long-term germ-free husbandry and untangling complexities of microbe-microbe interactions. Genetic manipulations including knock-out, knock-in and clonal approaches are additional avenues by which to uncover mechanistic microbiome pathways and potential genetic influences on microbiome composition. IgT, a teleost mucosal immunoglobulin believed to be an important adaptive immune determinant of microbial homeostasis, was depleted in a rainbow trout model. This depletion resulted in dysbiosis characterized by an increased presence of pathobionts, tissue damage and inflammation as well as increased susceptibility to mucosal parasites, highlighting the importance of this immunoglobulin in microbiome homeostasis in finfish (Xu et al., 2020). Clonal lines of aquaculture species are an additional tool that can be utilized to uncover genetic contributions to microbiome composition under varying environmental conditions. Clonal lines have advantages over inbred lines by avoiding artificially low levels of heterozygosity and inbreeding depression. The first clonal line of Atlantic salmon has recently been verified (Hansen et al., 2020) adding to previous lines in aquaculture species, and future work could potentially see the extension of aquaculture breeding programs to select for resistant microbiome types if these can be determined. These tools take time to develop, but with the increasing availability of fully sequenced genomes of aquaculture species, genetic manipulations to enhance functional understanding are becoming more accessible. Despite the evolution of ‘omic tools, the practice of culturing microbes is also making a resurgence as a tool for understanding functionality by enabling mechanistic experiments to determine physiological processes of microbes, or how they interact with their host. Culturing also allows for more in depth genomic analyses of taxa and discovery and description of novel taxa which can then feed into sequencing databases (Hitch et al., 2021).
5.2. Applying microbial manipulation effectively in aquaculture systems
Potential therapeutics for the aquaculture industry must be biologically- and cost-effective to manufacture and apply. Evidence-based procedures should be adopted with a view to identifying pivotal taxa, whether that is a single species which plays a role in a specific disease or resistance to infection provided by controlled communities of bacteria and other microbes.
Gut microbiomes of many organisms play an important role in aiding digestion and supporting gut (and organismal) health. Perhaps the most well-known or widely-applied example of microbial therapeutics in aquaculture (and human) health are the inclusion of pre- pro- or synbiotics in the diet. Probiotics refer to live microbial feed supplements which promote host health (Hill et al., 2014) whereas pre-biotics are non-digestible feed ingredients which selectively stimulate the growth of one or a limited number of particular microbes (Yadav et al., 2022). Synbiotics refers to products containing both pre- and pro-biotics working in synergy (Schrezenmeir and de Vrese, 2001). Recent reviews on prebiotic and probiotic use have shown favorable outcomes including increased growth or survival, improved immune responses, increased digestive efficiency, and improved water quality (López Nadal et al., 2020; El-Saadony et al., 2021; Knipe et al., 2021), but caution that more knowledge is needed in fish to promote efficacy (Brugman et al., 2018).
Probiotics can exert their function via the principles of competitive exclusion, be that exploitation or interference competition (Knipe et al., 2021). In exploitation, competition is indirect and characterized by consumption of resources while in interference, direct harm is caused, for example via the production of antimicrobial compounds. For example, the lactic acid bacteria (LAB) family of probiotics produce antimicrobial substances including lactic acid, acetic acid, hydrogen peroxide and bacteriocins interfering with the growth of other bacteria (Ringø, 2020; Ringø et al., 2020). Feeding shrimp (Litopenaeus stylirostris) with a diet containing the probiotic Pediococcus acidilactici resulted in the purging of potentially pathogenic vibrios from the gut (Castex et al., 2008). Similarly, in freshwater marron crayfish (Cherax cainii), feeding with Clostridium butyricum resulted in a reduction of Vibrio and Aeromonas counts in the hindgut (Foysal et al., 2019). In Atlantic salmon, feeding with Peiococcus acidilactici resulted in distinct reduction of Fusobacteriia, Clostridiales, Actinomycetales, Pasteurellales, and Streptococcacea, and an increase in Bradyrhizobiaceae in gut mucosa, but also identified that this probiotic effect is dependent on the water habitat (fresh vs. seawater) (Jaramillo-Torres et al., 2019). Contrastingly, in Nile tilapia, probiotic feeding with a mix of three Bacillus species did not modify gut microbiota to any large extent (Adeoye et al., 2016). Despite differences in efficacy, intervention with probiotics, prebiotics or synbiotics in an aquaculture setting more often than not illicit changes in the host microbiome (Table 3).The complex and dynamic nature of changing environments in aquaculture systems has confounded wide usage of probiotics to date. Variability in efficacy may be a consequence of therapeutics often being derived from non-aquatic hosts or systems, resulting in poor tolerance of biophysical conditions within the host and subsequently poor survival and establishment. In reality, to ensure efficacy, any potential pro- or pre-biotic therapeutic needs to be evaluated under different environmental circumstances to target optimum windows of application for maximum impact, particularly in aquaculture where the environment itself can often be manipulated. Although pre- and pro-biotics are generally applied via feed, the opportunity also exists in aquaculture systems to apply such therapeutics directly to the water with a view to directly influencing external microbial communities (Rowley, 2022). Utilizing specific microbial probiotics targeted at specific pathogens carries a degree of risk whether it be the invasion of secondary pathogens or evolution of target pathogen to evade the mechanism of exclusion (Knipe et al., 2021). AMPs are an intriguing route toward developing therapeutic compounds in aquaculture as they have the power to modulate the immune system while maintaining a low probability of the development of bacterial resistance (Chaturvedi et al., 2020). The gut microbiome represents an environment which may harbor AMPs with therapeutic and microbiome sequencing datasets can be mined to uncover candidate AMPs (Dong et al., 2017).
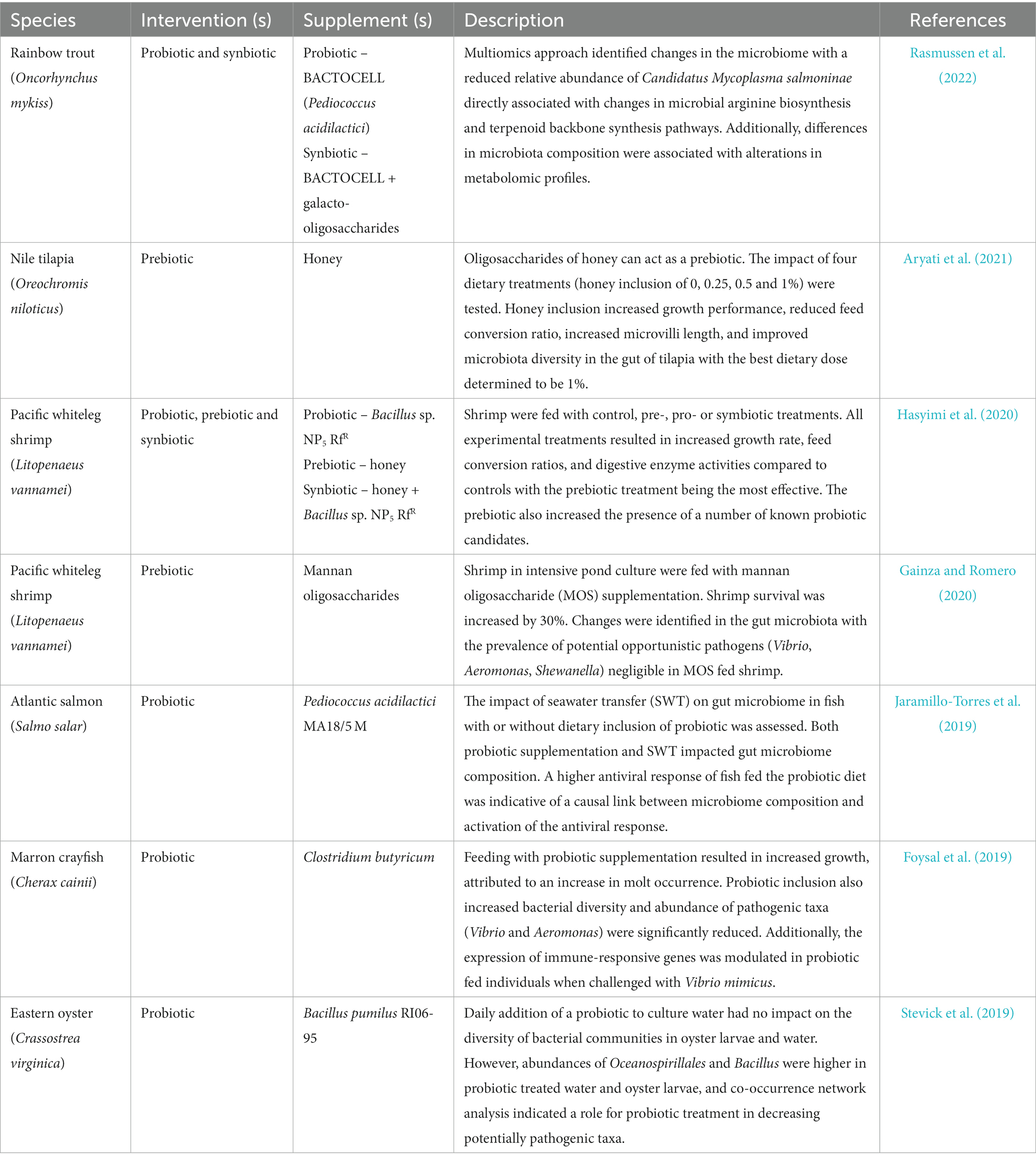
Table 3. Key published literature analyzing the impact of probiotic, prebiotic, and synbiotic interventions on microbiome composition.
6. Future prospects
The importance of the microbiome in the future development of sustainable aquaculture systems is clearly evident considering the vast outpouring of research in this area over the past 5–10 years, including a raft of recent reviews (de Bruijn et al., 2018; Perry et al., 2020; Legrand et al., 2020b; Infante-Villamil et al., 2021; Parata et al., 2021; Rajeev et al., 2021; Vargas-Albores et al., 2021; Diwan et al., 2022; Paillard et al., 2022). Despite this great interest, few focused studies have been conducted relating to the impact of microbiomes on host health and subsequent productivity, and the vast majority of studies have focused only on bacteria, mainly due to technical difficulties or cost associated with characterizing eukaryotes and viruses. Understanding more about these other communities and their interactions is imperative and also holds many opportunities for microbiome engineering, for example using phage to control opportunistic pathogens (Donati et al., 2021). There is now a critical need for better understanding of microbiome function in the context of aquaculture for enabling application to enhancing improved production systems. Poor animal welfare and loss of revenue due to disease remains one of the most restrictive issues in aquaculture development. One future focus should be pinpointing optimum periods for microbiome conditioning or manipulation, both during immune system development at larval stages and within periods of dysbiosis caused by disease or husbandry practices. Directing studies also toward functionality over identity will also be key in unlocking the full therapeutic potential of the microbiome in the aquaculture sector and employing next generation sequencing technologies in tandem with culture-based approaches will be essential in making this transition.
Author contributions
SM, CT, AR, and ML-R conceived the original concept. TW, JM, and DB expanded on scientific areas. JM and ML-R created the figures. ML-R wrote the initial draft and managed. All authors edited and contributed equally to the final manuscript.
Funding
This work was supported by BBSRC grants BB/P017223/1 and BB/P017215/1 for the Aquaculture Research Hub (ARCH-UK).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adeoye, A. A., Yomla, R., Jaramillo-Torres, A., Rodiles, A., Merrifield, D. L., and Davies, S. J. (2016). Combined effects of exogenous enzymes and probiotic on Nile tilapia (Oreochromis niloticus) growth, intestinal morphology and microbiome. Aquaculture 463, 61–70. doi: 10.1016/j.aquaculture.2016.05.028
Alberdi, A., Aizpurua, O., Bohmann, K., Zepeda-Mendoza, M. L., and Gilbert, M. T. P. (2016). Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689–699. doi: 10.1016/j.tree.2016.06.008
Arfken, A., Song, B., Allen, S. K., and Carnegie, R. B. (2021). Comparing larval microbiomes of the eastern oyster (Crassostrea virginica) raised in different hatcheries. Aquaculture 531:735955. doi: 10.1016/j.aquaculture.2020.735955
Aryati, Y., Widanarni, W., Wahjuningrum, D., Rusmana, I., and Lusiastuti, A. M. (2021). The effect of dietary honey prebiotic on microbiota diversity in the digestive tract of Nile tilapia (Oreochromis niloticus) and its growth performance. Aquac. Res. 52, 1215–1226. doi: 10.1111/are.14980
Arzul, I., Renault, T., and Lipart, C. (2001). Experimental herpes-like viral infections in marine bivalves: demonstration of interspecies transmission. Dis. Aquat. Org. 46, 1–6. doi: 10.3354/dao046001
Attramadal, K. J. K., Øien, J. V., Kristensen, E., Evjemo, J. O., Kjørsvik, E., Vadstein, O., et al. (2021). UV treatment in RAS influences the rearing water microbiota and reduces the survival of european lobster larvae (Homarus gammarus). Aquac. Eng. 94:102176. doi: 10.1016/j.aquaeng.2021.102176
Attramadal, K. J. K., Salvesen, I., Xue, R., Øie, G., Størseth, T. R., Vadstein, O., et al. (2012). Recirculation as a possible microbial control strategy in the production of marine larvae. Aquac. Eng. 46, 27–39. doi: 10.1016/j.aquaeng.2011.10.003
Baruah, K., Norouzitallab, P., Phong, H. P. P. D., Smagghe, G., and Bossier, P. (2017). Enhanced resistance against Vibrio harveyi infection by carvacrol and its association with the induction of heat shock protein 72 in gnotobiotic Artemia franciscana. Cell Stress Chaperones 22, 377–387. doi: 10.1007/s12192-017-0775-z
Bass, D., Stentiford, G. D., Wang, H., Koskella, B., and Tyler, C. R. (2019). The pathobiome in animal and plant diseases. Trends Ecol. Evol. 34, 996–1008. doi: 10.1016/j.tree.2019.07.012
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0
Blancheton, J. P., Attramadal, K. J. K., Michaud, L., d’Orbcastel, E. R., and Vadstein, O. (2013). Insight into bacterial population in aquaculture systems and its implication. Aquac. Eng. 53, 30–39. doi: 10.1016/j.aquaeng.2012.11.009
Bolnick, D. I., Snowberg, L. K., Caporaso, J. G., Lauber, C., Knight, R., and Stutz, W. E. (2014). Major histocompatibility complex class IIb polymorphism influences gut microbiota composition and diversity. Mol. Ecol. 23, 4831–4845. doi: 10.1111/mec.12846
Boutin, S., Bernatchez, L., Audet, C., and Derôme, N. (2013). Network analysis highlights complex interactions between pathogen, host and commensal microbiota. PLoS One 8:e84772. doi: 10.1371/journal.pone.0084772
Boyd, C. E. (1998). Pond water aeration systems. Aquac. Eng. 18, 9–40. doi: 10.1016/S0144-8609(98)00019-3
Brown, R. M., Wiens, G. D., and Salinas, I. (2019). Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 86, 497–506. doi: 10.1016/j.fsi.2018.11.079
Brugman, S., Ikeda-Ohtsubo, W., Braber, S., Folkerts, G., Pieterse, C. M., and Bakker, P. A. (2018). A comparative review on microbiota manipulation: lessons from fish, plants, livestock, and human research. Front. Nutr. 5:80. doi: 10.3389/fnut.2018.00080
Bugten, A. V., Attramadal, K. J. K., Fossmark, R. O., Rosten, T. W., Vadstein, O., and Bakke, I. (2022). Changes in rearing water microbiomes in RAS induced by membrane filtration alters the hindgut microbiomes of Atlantic salmon (Salmo salar) parr. Aquaculture 548:737661. doi: 10.1016/j.aquaculture.2021.737661
Castex, M., Chim, L., Pham, D., Lemaire, P., Wabete, N., Nicolas, J., et al. (2008). Probiotic P. acidilactici application in shrimp Litopenaeus stylirostris culture subject to vibriosis in New Caledonia. Aquaculture 275, 182–193. doi: 10.1016/j.aquaculture.2008.01.011
Castro, R., Jouneau, L., Tacchi, L., Macqueen, D. J., Alzaid, A., Secombes, C. J., et al. (2015). Disparate developmental patterns of immune responses to bacterial and viral infections in fish. Sci. Rep. 5:15458. doi: 10.1038/srep15458
Chaturvedi, P., Bhat, R. A. H., and Pande, A. (2020). Antimicrobial peptides of fish: innocuous alternatives to antibiotics. Rev. Aquacult. 12, 85–106. doi: 10.1111/raq.12306
Clerissi, C., de Lorgeril, J., Petton, B., Lucasson, A., Escoubas, J., Gueguen, Y., et al. (2020). Microbiota composition and evenness predict survival rate of oysters confronted to Pacific oyster mortality syndrome. Front. Microbiol. 11:311. doi: 10.3389/fmicb.2020.00311
Clinton, M., Ferrier, D. E. K., Martin, S. A. M., and Brierley, A. S. (2021). Impacts of jellyfish on marine cage aquaculture: an overview of existing knowledge and the challenges to finfish health. ICES J. Mar. Sci. 78, 1557–1573. doi: 10.1093/icesjms/fsaa254
Coates, C., Rowley, A., Smith, L., and Whitten, M. (2022). “Host defences of invertebrates to pathogens and parasites” in Invertebrate pathology. eds. A. F. Rowley, C. J. Coates, and M. W. Whitten (Oxford: Oxford University Press), 3–40.
Crosbie, P. B. B., Bridle, A. R., Cadoret, K., and Nowak, B. F. (2012). In vitro cultured Neoparamoeba perurans causes amoebic gill disease in Atlantic salmon and fulfils Koch’s postulates. Int. J. Parasitol. 42, 511–515. doi: 10.1016/j.ijpara.2012.04.002
Cunha, M. E., Quental-Ferreira, H., Parejo, A., Gamito, S., Ribeiro, L., Moreira, M., et al. (2019). Understanding the individual role of fish, oyster, phytoplankton and macroalgae in the ecology of integrated production in earthen ponds. Aquaculture 512:734297. doi: 10.1016/j.aquaculture.2019.734297
Dai, W., Dong, Y., Ye, J., Xue, Q., and Lin, Z. (2022). Gut microbiome composition likely affects the growth of razor clam Sinonovacula constricta. Aquaculture 550:737847. doi: 10.1016/j.aquaculture.2021.737847
de Bruijn, I., Liu, Y., Wiegertjes, G. F., and Raaijmakers, J. M. (2018). Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol. Ecol. 94:fix161. doi: 10.1093/femsec/fix161
de Lorgeril, J., Lucasson, A., Petton, B., Toulza, E., Montagnani, C., Clerissi, C., et al. (2018). Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat. Commun. 9:4215. doi: 10.1038/s41467-018-06659-3
Debray, R., Herbert, R. A., Jaffe, A. L., Crits-Christoph, A., Power, M. E., and Koskella, B. (2021). Priority effects in microbiome assembly. Nat. Rev. Microbiol. 20, 109–121. doi: 10.1038/s41579-021-00604-w
Dégremont, L., Azéma, P., Maurouard, E., and Travers, M. (2020). Enhancing resistance to Vibrio aestuarianus in Crassostrea gigas by selection. Aquaculture 526:735429. doi: 10.1016/j.aquaculture.2020.735429
Dehler, C. E., Secombes, C. J., and Martin, S. A. M. (2017). Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.). Sci. Rep. 7:13877. doi: 10.1038/s41598-017-13249-8
Deng, Y., Borewicz, K., van Loo, J., Olabarrieta, M. Z., Kokou, F., Sipkema, D., et al. (2021). In-situ biofloc affects the core prokaryotes community composition in gut and enhances growth of Nile tilapia (Oreochromis niloticus). Microb. Ecol. 84, 879–892. doi: 10.1007/s00248-021-01880-y
Deng, Y., Verdegem, M. C. J., Eding, E., and Kokou, F. (2022). Effect of rearing systems and dietary probiotic supplementation on the growth and gut microbiota of Nile tilapia (Oreochromis niloticus) larvae. Aquaculture 546:737297. doi: 10.1016/j.aquaculture.2021.737297
Dien, L. D., Hiep, L. H., Faggotter, S. J., Chen, C., Sammut, J., and Burford, M. A. (2019). Factors driving low oxygen conditions in integrated rice-shrimp ponds. Aquaculture 512:734315. doi: 10.1016/j.aquaculture.2019.734315
Diwan, A. D., Harke, S. N., Gopalkrishna,, and Panche, A. N. (2022). Aquaculture industry prospective from gut microbiome of fish and shellfish: an overview. J. Anim. Physiol. Anim. Nutr. 106, 441–469. doi: 10.1111/jpn.13619
Donati, V. L., Dalsgaard, I., Sundell, K., Castillo, D., Er-Rafik, M., Clark, J., et al. (2021). Phage-mediated control of Flavobacterium psychrophilum in aquaculture: in vivo experiments to compare delivery methods. Front. Microbiol. 12:628309. doi: 10.3389/fmicb.2021.628309
Dong, B., Yi, Y., Liang, L., and Shi, Q. (2017). High throughput identification of antimicrobial peptides from fish gastrointestinal microbiota. Toxins 9:266. doi: 10.3390/toxins9090266
Dvergedal, H., Sandve, S. R., Angell, I. L., Klemetsdal, G., and Rudi, K. (2020). Association of gut microbiota with metabolism in juvenile Atlantic salmon. Microbiome 8:160. doi: 10.1186/s40168-020-00938-2
Eissa, N., and Wang, H. (2016). Transcriptional stress responses to environmental and husbandry stressors in aquaculture species. Rev. Aquac. 8, 61–88. doi: 10.1111/raq.12081
Ellison, A. R., Wilcockson, D., and Cable, J. (2021). Circadian dynamics of the teleost skin immune-microbiome interface. Microbiome 9:222. doi: 10.1186/s40168-021-01160-4
El-Saadony, M., Alagawany, M., Patra, A. K., Kar, I., Tiwari, R., Dawood, M. A. O., et al. (2021). The functionality of probiotics in aquaculture: an overview. Fish Shellfish Immunol. 117, 36–52. doi: 10.1016/j.fsi.2021.07.007
Embar-Gopinath, S., Butler, R., and Nowak, B. (2005). Influence of salmonid gill bacteria on development and severity of amoebic gill disease. Dis. Aquat. Org. 67, 55–60. doi: 10.3354/dao067055
Fallet, M., Montagnani, C., Petton, B., Dantan, L., de Lorgeril, J., Comarmond, S., et al. (2022). Early life microbial exposures shape the Crassostrea gigas immune system for lifelong and intergenerational disease protection. Microbiome 10:85. doi: 10.1186/s40168-022-01280-5
FAO . (2020). The state of world fisheries and aquaculture: sustainability in action. Food and Agriculture Organization of the United Nations, Rome.
Fossmark, R. O., Vadstein, O., Rosten, T. W., Bakke, I., Košeto, D., Bugten, A. V., et al. (2020). Effects of reduced organic matter loading through membrane filtration on the microbial community dynamics in recirculating aquaculture systems (RAS) with Atlantic salmon parr (Salmo salar). Aquaculture 524:735268. doi: 10.1016/j.aquaculture.2020.735268
Foysal, M. J., Momtaz, F., Kawser, A. Q. M. R., Ali, M. H., Raihan, T., Siddik, M. A. B., et al. (2021). Amplicon sequencing reveals significantly increased vibrio abundance and associated gene functions in vibriosis-infected black tiger shrimp (Penaeus monodon). J. Fish Dis. 44, 591–599. doi: 10.1111/jfd.13304
Foysal, M. J., Nguyen, T. T. T., Chaklader, M. R., Siddik, M. A. B., Tay, C., Fotedar, R., et al. (2019). Marked variations in gut microbiota and some innate immune responses of fresh water crayfish, marron (Cherax cainii, Austin 2002) fed dietary supplementation of Clostridium butyricum. PeerJ 7:e7553. doi: 10.7717/peerj.7553
Gainza, O., and Romero, J. (2020). Effect of mannan oligosaccharides on the microbiota and productivity parameters of Litopenaeus vannamei shrimp under intensive cultivation in Ecuador. Sci. Rep. 10:2719. doi: 10.1038/s41598-020-59587-y
Gerdol, M., Gomez-Chiarri, M., Castillo, M. G., Figueras, A., Fiorito, G., Moreira, R., et al. (2018). “Immunity in molluscs: recognition and effector mechanisms, with a focus on Bivalvia” in Advances in comparative immunology. ed. E. L. Cooper (Cham: Springer International Publishing), 225–341.
Giatsis, C., Sipkema, D., Ramiro-Garcia, J., Bacanu, G. M., Abernathy, J., Verreth, J., et al. (2016). Probiotic legacy effects on gut microbial assembly in tilapia larvae. Sci. Rep. 6:33965. doi: 10.1038/srep33965
Giatsis, C., Sipkema, D., Smidt, H., Verreth, J., and Verdegem, M. (2014). The colonization dynamics of the gut microbiota in tilapia larvae. PLoS One 9:e103641. doi: 10.1371/journal.pone.0103641
Gibbons, S. M. (2017). Microbial community ecology: function over phylogeny. Nat. Ecol. Evol. 1:32. doi: 10.1038/s41559-016-0032
Green, T. J., Siboni, N., King, W. L., Labbate, M., Seymour, J. R., and Raftos, D. (2019). Simulated marine heat wave alters abundance and structure of Vibrio populations associated with the Pacific oyster resulting in a mass mortality event. Microb. Ecol. 77, 736–747. doi: 10.1007/s00248-018-1242-9
Hansen, T. J., Penman, D., Glover, K. A., Fraser, T. W. K., Vågseth, T., Thorsen, A., et al. (2020). Production and verification of the first Atlantic salmon (Salmo salar L.) clonal lines. BMC Genet. 21:71. doi: 10.1186/s12863-020-00878-8
Hasyimi, W., Widanarni, W., and Yuhana, M. (2020). Growth performance and intestinal microbiota diversity in pacific white shrimp Litopenaeus vannamei fed with a probiotic bacterium, honey prebiotic, and synbiotic. Curr. Microbiol. 77, 2982–2990. doi: 10.1007/s00284-020-02117-w
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hitch, T. C. A., Afrizal, A., Riedel, T., Kioukis, A., Haller, D., Lagkouvardos, I., et al. (2021). Recent advances in culture-based gut microbiome research. Int. J. Med. Microbiol. 311:151485. doi: 10.1016/j.ijmm.2021.151485
Hou, D., Huang, Z., Zeng, S., Liu, J., Wei, D., Deng, X., et al. (2018). Intestinal bacterial signatures of white feces syndrome in shrimp. Appl. Microbiol. Biotechnol. 102, 3701–3709. doi: 10.1007/s00253-018-8855-2
Houston, R. D., Bean, T. P., Macqueen, D. J., Gundappa, M. K., Jin, Y. H., Jenkins, T. L., et al. (2020). Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 21, 389–409. doi: 10.1038/s41576-020-0227-y
Huang, S., Wu, M., Zang, C., Du, S., Domagalski, J., Gajewska, M., et al. (2016). Dynamics of algae growth and nutrients in experimental enclosures culturing bighead carp and common carp: phosphorus dynamics. Int. J. Sediment Res. 31, 173–180. doi: 10.1016/j.ijsrc.2016.01.003
Infante-Villamil, S., Huerlimann, R., and Jerry, D. R. (2021). Microbiome diversity and dysbiosis in aquaculture. Rev. Aquac. 13, 1077–1096. doi: 10.1111/raq.12513
Jaramillo-Torres, A., Rawling, M. D., Rodiles, A., Mikalsen, H. E., Johansen, L., Tinsley, J., et al. (2019). Influence of dietary supplementation of probiotic Pediococcus acidilactici MA18/5M during the transition from freshwater to seawater on intestinal health and microbiota of Atlantic salmon (Salmo salar L.). Front. Microbiol. 10:2243. doi: 10.3389/fmicb.2019.02243
Jing, X., Su, S., Zhang, C., Zhu, J., Hou, Y., Li, Z., et al. (2021). Dynamic changes in microbial community structure in farming pond water and their effect on the intestinal microbial community profile in juvenile common carp (Cyprinus carpio L.). Genomics 113, 2547–2560. doi: 10.1016/j.ygeno.2021.05.024
Johansson, L., Timmerhaus, G., Afanasyev, S., Jørgensen, S. M., and Krasnov, A. (2016). Smoltification and seawater transfer of Atlantic salmon (Salmo salar L.) is associated with systemic repression of the immune transcriptome. Fish Shellfish Immunol. 58, 33–41. doi: 10.1016/j.fsi.2016.09.026
Jose, D., Preena, P. G., Kumar, V. J. R., Philip, R., and Singh, I. S. B. (2020). Metaproteomic insights into ammonia oxidising bacterial consortium developed for bioaugmenting nitrification in aquaculture systems. Biologia 75, 1751–1757. doi: 10.2478/s11756-020-00481-3
Katzenback, B. A. (2015). Antimicrobial peptides as mediators of innate immunity in teleosts. Biology 4, 607–639. doi: 10.3390/biology/4040607
Kelly, C., and Salinas, I. (2017). Under pressure: interactions between commensal microbiota and the teleost immune system. Front. Immunol. 8:559. doi: 10.3389/fimmu.2017.00559
King, W. L., Siboni, N., Williams, N. L. R., Kahlke, T., Nguyen, K. V., Jenkins, C., et al. (2019). Variability in the composition of Pacific oyster microbiomes across oyster families exhibiting different levels of susceptibility to OsHV-1 μvar disease. Front. Microbiol. 10:473. doi: 10.3389/fmicb.2019.00473
Knipe, H., Temperton, B., Lange, A., Bass, D., and Tyler, C. R. (2021). Probiotics and competitive exclusion of pathogens in shrimp aquaculture. Rev. Aquac. 13, 324–352. doi: 10.1111/raq.12477
Kostic, A. D., Howitt, M. R., and Garrett, W. S. (2013). Exploring host–microbiota interactions in animal models and humans. Genes Dev. 27, 701–718. doi: 10.1101/gad.212522.112
Lederberg, J., and McCray, A. T. (2001). ‘Ome sweet ‘omics - a genealogical treasury of words. Scientist 15:8.
Legrand, T. P. R. A., Wynne, J. W., Weyrich, L. S., and Oxley, A. P. A. (2020a). Investigating both mucosal immunity and microbiota in response to gut enteritis in yellowtail kingfish. Microorganisms 8:1267. doi: 10.3390/microorganisms8091267
Legrand, T. P. R. A., Wynne, J. W., Weyrich, L. S., and Oxley, A. P. A. (2020b). A microbial sea of possibilities: current knowledge and prospects for an improved understanding of the fish microbiome. Rev. Aquac. 12, 1101–1134. doi: 10.1111/raq.12375
Levine, J. M., and D'Antonio, C. M. (1999). Elton revisited: a review of evidence linking diversity and invasibility. Oikos 87, 15–26. doi: 10.2307/3546992
Li, S., Young, T., Archer, S., Lee, K., Sharma, S., and Alfaro, A. C. (2022). Mapping the Green-lipped mussel (Perna canaliculus) microbiome: a multi-tissue analysis of bacterial and fungal diversity. Curr. Microbiol. 79:76. doi: 10.1007/s00284-021-02758-5
Llewellyn, M. S., Leadbeater, S., Garcia, C., Sylvain, F. -E., Custodio, M., Ang, K. P., et al. (2017). Parasitism perturbs the mucosal microbiome of Atlantic salmon. Sci. Rep. 7:43465. doi: 10.1038/srep43465
López Nadal, A., Ikeda-Ohtsubo, W., Sipkema, D., Peggs, D., McGurk, C., Forlenza, M., et al. (2020). Feed, microbiota, and gut immunity: using the zebrafish model to understand fish health. Front. Immunol. 11:114. doi: 10.3389/fimmu.2020.00114
Lorgen-Ritchie, M., Clarkson, M., Chalmers, L., Taylor, J. F., Migaud, H., and Martin, S. A. M. (2021). A temporally dynamic gut microbiome in Atlantic salmon during freshwater recirculating aquaculture system (RAS) production and post-seawater transfer. Front. Mar. Sci. 8:869. doi: 10.3389/fmars.2021.711797
Ma, J., Bruce, T. J., Jones, E. M., and Cain, K. D. (2019). A review of fish vaccine development strategies: conventional methods and modern biotechnological approaches. Microorganisms 7:569. doi: 10.3390/microorganisms7110569
Macke, E., Callens, M., De Meester, L., and Decaestecker, E. (2017). Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat. Commun. 8:1608. doi: 10.1038/s41467-017-01714-x
Macke, E., Callens, M., Massol, F., Vanoverberghe, I., De Meester, L., and Decaestecker, E. (2020). Diet and genotype of an aquatic invertebrate affect the composition of free-living microbial communities. Front. Microbiol. 11:380. doi: 10.3389/fmicb.2020.00380
Marden, C. L., McDonald, R., Schreier, H. J., and Watts, J. E. M. (2017). Investigation into the fungal diversity within different regions of the gastrointestinal tract of Panaque nigrolineatus, a wood-eating fish. AIMS Microbiol. 3, 749–761. doi: 10.3934/microbiol.2017.4.749
Masso-Silva, J. A., and Diamond, G. (2014). Antimicrobial peptides from fish. Pharmaceuticals 7, 265–310. doi: 10.3390/ph7030265
McMurtrie, J., Alathari, S., Chaput, D. L., Bass, D., Ghambi, C., Nagoli, J., et al. (2022). Relationships between pond water and tilapia skin microbiomes in aquaculture ponds in Malawi. Aquaculture 558:738367. doi: 10.1016/j.aquaculture.2022.738367
Mero, F. F. C., Pedroso, F. L., Apines-Amar, M., Cadangin, J. F., Rendaje, D. C., Verde, C. S., et al. (2019). Influence of water management, photoperiod and aeration on growth, survival, and early spat settlement of the hatchery-reared green mussel, Perna viridis. Int. Aquat. Res. 11, 159–172. doi: 10.1007/s40071-019-0226-9
Minich, J. J., Nowak, B., Elizur, A., Knight, R., Fielder, S., and Allen, E. E. (2021). Impacts of the marine hatchery built environment, water and feed on mucosal microbiome colonization across ontogeny in yellowtail kingfish, Seriola lalandi. Front. Mar. Sci. 8:516. doi: 10.3389/fmars.2021.676731
Minich, J. J., Poore, G. D., Jantawongsri, K., Johnston, C., Bowie, K., Bowman, J., et al. (2020). Microbial ecology of Atlantic salmon (Salmo salar) hatcheries: impacts of the built environment on fish mucosal microbiota. Appl. Environ. Microbiol. 86, e00411–e00420. doi: 10.1101/828749
Murdoch, C. C., and Rawls, J. F. (2019). Commensal microbiota regulate vertebrate innate immunity-insights from the zebrafish. Front. Immunol. 10:2100. doi: 10.3389/fimmu.2019.02100
Murphy, T. R., Xiao, R., Brooks, M. L., Rader, B. A., and Hamilton-Brehm, S. (2022). Aquaculture production of hatchling Hawaiian bobtail squid (Euprymna scolopes) is negatively impacted by decreasing environmental microbiome diversity. J. Appli. Microbiol. 132, 1724–1737. doi: 10.1111/jam.15350
Nam, B., Jang, J., Caetano-Anolles, K., Kim, Y., Park, J. Y., Sohn, H., et al. (2018). Microbial community and functions associated with digestion of algal polysaccharides in the visceral tract of Haliotis discus hannai: insights from metagenome and metatranscriptome analysis. PLoS One 13:e0205594. doi: 10.1371/journal.pone.0205594
Narne, P., Pandey, V., and Phanithi, P. B. (2017). Interplay between mitochondrial metabolism and oxidative stress in ischemic stroke: an epigenetic connection. Mol. Cell. Neurosci. 82, 176–194. doi: 10.1016/j.mcn.2017.05.008
Naya-Català, F., Piazzon, M. C., Calduch-Giner, J. A., Sitjà-Bobadilla, A., and Pérez-Sánchez, J. (2022). Diet and host genetics drive the bacterial and fungal intestinal metatranscriptome of Gilthead Sea bream. Front. Microbiol. 13:883738. doi: 10.3389/fmicb.2022.883738
Nguyen, H. Y. N., Trinh, T. L., Baruah, K., Lundh, T., and Kiessling, A. (2021). Growth and feed utilisation of Nile tilapia (Oreochromis niloticus) fed different protein levels in a clear-water or biofloc-RAS system. Aquaculture 536:736404. doi: 10.1016/j.aquaculture.2021.736404
Nikouli, E., Meziti, A., Smeti, E., Antonopoulou, E., Mente, E., and Kormas, K. A. (2021). Gut microbiota of five sympatrically farmed marine fish species in the Aegean Sea. Micriob. Ecol. 81, 460–470. doi: 10.1007/s00248-020-01580-z
Overstreet, R. M., and Lotz, J. M. (2016). “Host-symbiont relationships: understanding the change from guest to pest” in The Rasputin effect; when commensals and symbionts become parasitic. Advances in environmental microbiology. ed. C. Hurst , vol. 3 (Cham: Springer), 27–64.
Paillard, C., Gueguen, Y., Wegner, K. M., Bass, D., Pallavicini, A., Vezzulli, L., et al. (2022). Recent advances in bivalve-microbiota interactions for disease prevention in aquaculture. Curr. Opin. Biotechnol. 73, 225–232. doi: 10.1016/j.copbio.2021.07.026
Parata, L., Sammut, J., and Egan, S. (2021). Opportunities for microbiome research to enhance farmed freshwater fish quality and production. Rev. Aquac. 13, 2027–2037. doi: 10.1111/raq.12556
Paul-Pont, I., Dhand, N. K., and Whittington, R. J. (2013). Influence of husbandry practices on OsHV-1 associated mortality of Pacific oysters Crassostrea gigas. Aquaculture 412-413, 202–214. doi: 10.1016/j.aquaculture.2013.07.038
Pérez-Pascual, D., Vendrell-Fernández, S., Audrain, B., Bernal-Bayard, J., Patiño-Navarrete, R., Petit, V., et al. (2021). Gnotobiotic rainbow trout (Oncorhynchus mykiss) model reveals endogenous bacteria that protect against Flavobacterium columnare infection. PLoS Pathog. 17:e1009302. doi: 10.1371/journal.ppat.1009302
Perry, W. B., Lindsay, E., Payne, C. J., Brodie, C., and Kazlauskaite, R. (2020). The role of the gut microbiome in sustainable teleost aquaculture. Proc. Royal Soc. B 287:20200184. doi: 10.1098/rspb.2020.0184
Petersen, C., and Round, J. L. (2014). Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 16, 1024–1033. doi: 10.1111/cmi.12308
Petton, B., Destoumieux-Garzón, D., Pernet, F., Toulza, E., de Lorgeril, J., Degremont, L., et al. (2021). The Pacific oyster mortality syndrome, a polymicrobial and multifactorial disease: state of knowledge and future directions. Front. Immunol. 12:630343. doi: 10.3389/fimmu.2021.630343
Pino Martinez, E., Balseiro, P., Pedrosa, C., Haugen, T. S., Fleming, M. S., and Handeland, S. O. (2021). The effect of photoperiod manipulation on Atlantic salmon growth, smoltification and sexual maturation: a case study of a commercial RAS. Aquac. Res. 52, 2593–2608. doi: 10.1111/are.15107
Rajeev, R., Adithya, K. K., Kiran, G. S., and Selvin, J. (2021). Healthy microbiome: a key to successful and sustainable shrimp aquaculture. Rev. Aquac. 13, 238–258. doi: 10.1111/raq.12471
Rasmussen, J. A., Villumsen, K. R., Ernst, M., Hansen, M., Forberg, T., Gopalakrishnan, S., et al. (2022). A multi-omics approach unravels metagenomic and metabolic alterations of a probiotic and synbiotic additive in rainbow trout (Oncorhynchus mykiss). Microbiome 10:21. doi: 10.1186/s40168-021-01221-8
Rawls, J. F., Mahowald, M. A., Ley, R. E., and Gordon, J. I. (2006). Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cells 127, 423–433. doi: 10.1016/j.cell.2006.08.043
Rawls, J. F., Samuel, B. S., and Gordon, J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. U. S. A. 101:4596. doi: 10.1073/pnas.0400706101
Reid, M. A., Dai, Z., and Locasale, J. W. (2017). The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 19, 1298–1306. doi: 10.1038/ncb3629
Reverter, M., Tapissier-Bontemps, N., Lecchini, D., Banaigs, B., and Sasal, P. (2018). Biological and ecological roles of external fish mucus: a review. Aust. Fish. 3:41. doi: 10.3390/fishes3040041
Richard, M., Rolland, J. L., Gueguen, Y., de Lorgeril, J., Pouzadoux, J., Mostajir, B., et al. (2021). In situ characterisation of pathogen dynamics during a Pacific oyster mortality syndrome episode. Mar. Environ. Res. 165:105251. doi: 10.1016/j.marenvres.2020.105251
Ringø, E. (2020). Probiotics in shellfish aquaculture. Aquac. Fisheries 5, 1–27. doi: 10.1016/j.aaf.2019.12.001
Ringø, E., Doan, H. V., Lee, S., and Song, S. K. (2020). Lactic acid bacteria in shellfish: possibilities and challenges. Rev. Fisheries Sci. Aquac. 28, 139–169. doi: 10.1080/23308249.2019.1683151
Rodney, H. P., Takeshi, Z., and Kenji, S. (2014). Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Factories 13:S3. doi: 10.1186/1475-2859-13-S1-S3
Roques, S., Deborde, C., Richard, N., Skiba-Cassy, S., Moing, A., and Fauconneau, B. (2020). Metabolomics and fish nutrition: a review in the context of sustainable feed development. Rev. Aquac. 12, 261–282. doi: 10.1111/raq.12316
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Rosenberg, E., and Zilber-Rosenberg, I. (2018). The hologenome concept of evolution after 10 years. Microbiome 6:78. doi: 10.1186/s40168-018-0457-9
Rowley, A. F. (2022). “Bacterial diseases of crustaceans” in Invertebrate pathology. eds. A. F. Rowley, C. J. Coates, and M. W. Whitten (Oxford: Oxford University Press), 400–435.
Salinas, I., Fernández-Montero, Á., Ding, Y., and Sunyer, J. O. (2021). Mucosal immunoglobulins of teleost fish: a decade of advances. Dev. Comp. Immunol. 121:104079. doi: 10.1016/j.dci.2021.104079
Schrezenmeir, J., and de Vrese, M. (2001). Probiotics, prebiotics, and synbiotics—approaching a definition. Am. J. Clin. Nutr. 73, 361s–364s. doi: 10.1093/ajcn/73.2.361s
Segarra, A., Pépin, J. F., Arzul, I., Morga, B., Faury, N., and Renault, T. (2010). Detection and description of a particular ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 153, 92–99. doi: 10.1016/j.virusres.2010.07.011
Siriyappagouder, P., Galindo-Villegas, J., Dhanasiri, A. K. S., Zhang, Q., Mulero, V., Kiron, V., et al. (2020). Pseudozyma priming influences expression of genes involved in metabolic pathways and immunity in zebrafish larvae. Front. Immunol. 11:987. doi: 10.3389/fimmu.200.00978
Siriyappagouder, P., Kiron, V., Lokesh, J., Rajeish, M., Kopp, M., and Fernandes, J. (2018). The intestinal mycobiota in wild zebrafish comprises mainly Dothideomycetes while Saccharomycetes predominate in their laboratory-reared counterparts. Front. Microbiol. 9:387. doi: 10.3389/fmicb.2018.00387
Situmorang, M. L., Dierckens, K., Mlingi, F. T., Van Delsen, B., and Bossier, P. (2014). Development of a bacterial challenge test for gnotobiotic Nile tilapia Oreochromis niloticus larvae. Dis. Aquat. Org. 109, 23–33. doi: 10.3354/dao02721
Slinger, J., Adams, M. B., Stratford, C. N., Rigby, M., and Wynne, J. W. (2021). The effect of antimicrobial treatment upon the gill bacteriome of Atlantic salmon (Salmo salar L.) and progression of amoebic gill disease (AGD) in vivo. Microorganisms 9:987. doi: 10.3390/microorganisms9050987
Slinger, J., Adams, M. B., and Wynne, J. W. (2020). Bacteriomic profiling of branchial lesions induced by Neoparamoeba perurans challenge reveals commensal dysbiosis and an association with Tenacibaculum dicentrarchi in agd-affected Atlantic salmon (Salmo salar L.). Microorganisms 8, 1–17. doi: 10.3390/microorganisms8081189
Smith, C. C. R., Snowberg, L. K., Gregory Caporaso, J., Knight, R., and Bolnick, D. I. (2015). Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 9, 2515–2526. doi: 10.1038/ismej.2015.64
Spor, A., Koren, O., and Ley, R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290. doi: 10.1038/nrmicro2540
Sriyasak, P., Chitmanat, C., Whangchai, N., Promya, J., and Lebel, L. (2015). Effect of water de-stratification on dissolved oxygen and ammonia in tilapia ponds in northern Thailand. Int. Aquat. Res. 7, 287–299. doi: 10.1007/s40071-015-0113-y
Stephens, W. Z., Burns, A. R., Stagaman, K., Wong, S., Rawls, J. F., Guillemin, K., et al. (2016). The composition of the zebrafish intestinal microbial community varies across development. ISME J. 10, 644–654. doi: 10.1038/ismej.2015.140
Steury, R. A., Currey, M. C., Cresko, W. A., and Bohannan, B. J. M. (2019). Population genetic divergence and environment influence the gut microbiome in Oregon threespine stickleback. Genes 10:484. doi: 10.3390/genes10070484
Stevick, R. J., Sohn, S., Modak, T. H., Nelson, D. R., Rowley, D. C., Tammi, K., et al. (2019). Bacterial community dynamics in an oyster hatchery in response to probiotic treatment. Front. Microbiol. 10:1060. doi: 10.3389/fmicb.2019.01060
Summerfelt, S. T., Sharrer, M. J., Tsukuda, S. M., and Gearheart, M. (2009). Process requirements for achieving full-flow disinfection of recirculating water using ozonation and UV irradiation. Aquac. Eng. 40, 17–27. doi: 10.1016/j.aquaeng.2008.10.002
Sun, S., Yang, M., Fu, H., Ge, X., and Zou, J. (2020). Altered intestinal microbiota induced by chronic hypoxia drives the effects on lipid metabolism and the immune response of oriental river prawn Macrobrachium nipponense. Aquaculture 526:735431. doi: 10.1016/j.aquaculture.2020.735431
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Ocean plankton. Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359
Tabrett, A., and Horton, M. W. (2020). The influence of host genetics on the microbiome. F1000Res 9:84. doi: 10.12688/f1000research.20835.1
Tepaamorndech, S., Nookaew, I., Higdon, S. M., Santiyanont, P., Phromson, M., Chantarasakha, K., et al. (2020). Metagenomics in bioflocs and their effects on gut microbiome and immune responses in Pacific white shrimp. Fish Shellfish Immunol. 106, 733–741. doi: 10.1016/j.fsi.2020.08.042
Trabal Fernández, N., Mazón-Suástegui, J. M., Vázquez-Juárez, R., Ascencio-Valle, F., and Romero, J. (2014). Changes in the composition and diversity of the bacterial microbiota associated with oysters (Crassostrea corteziensis, Crassostrea gigas and Crassostrea sikamea) during commercial production. FEMS Microbiol. Ecol. 88, 69–83. doi: 10.1111/1574-6941.12270
Uengwetwanit, T., Uawisetwathana, U., Arayamethakorn, S., Khudet, J., Chaiyapechara, S., Karoonuthaisiri, N., et al. (2020). Multi-omics analysis to examine microbiota, host gene expression and metabolites in the intestine of black tiger shrimp (Penaeus monodon) with different growth performance. PeerJ 8:e9646. doi: 10.7717/peerj.9646
Uren Webster, T. M., Consuegra, S., Hitchings, M., and Garcia de Leaniz, C. (2018). Interpopulation variation in the Atlantic salmon microbiome reflects environmental and genetic diversity. Appl. Environ. Microbiol. 84, e00691–e00618. doi: 10.1128/AEM.00691-18
Uren Webster, T. M., Rodriguez-Barreto, D., Castaldo, G., Gough, P., Consuegra, S., and Garcia de Leaniz, C. (2020). Environmental plasticity and colonisation history in the Atlantic salmon microbiome: a translocation experiment. Mol. Ecol. 29, 886–898. doi: 10.1111/mec.15369
Vadstein, O., Attramadal, K. J. K., Bakke, I., and Olsen, Y. (2018). K-selection as microbial community management strategy: a method for improved viability of larvae in aquaculture. Front. Microbiol. 9:2730. doi: 10.3389/fmicb.2018.02730
Vargas-Albores, F., Martínez-Córdova, L. R., Hernández-Mendoza, A., Cicala, F., Lago-Lestón, A., and Martínez-Porchas, M. (2021). Therapeutic modulation of fish gut microbiota, a feasible strategy for aquaculture? Aquaculture 544:737050. doi: 10.1016/j.aquaculture.2021.737050
Vasta, G. R., Ahmed, H., Bianchet, M. A., Fernández-Robledo, J. A., and Amzel, L. M. (2012). Diversity in recognition of glycans by F-type lectins and galectins: molecular, structural, and biophysical aspects. Ann. N. Y. Acad. Sci. 1253, E14–E26. doi: 10.1111/j.1749-6632.2012.06698.x
Vasta, G. R., Ahmed, H., Tasumi, S., Odom, E. W., and Saito, K. (2007). Biological roles of lectins in innate immunity: molecular and structural basis for diversity in self/non-self recognition. Adv. Exp. Med. Biol. 598, 389–406. doi: 10.1007/978-0-387-71767-8_27
Vestrum, R. I., Forberg, T., Luef, B., Bakke, I., Winge, P., Olsen, Y., et al. (2022). Commensal and opportunistic bacteria present in the microbiota in Atlantic cod (Gadus morhua) larvae differentially alter the hosts’ innate immune responses. Microorganisms 10:24. doi: 10.3390/microorganisms10010024
Villasante, A., Catalán, N., Rojas, R., Lohrmann, K. B., and Romero, J. (2020). Microbiota of the digestive gland of red abalone (Haliotis rufescens) is affected by withering syndrome. Microorganisms 8:1411. doi: 10.3390/microorganisms8091411
Vonaesch, P., Anderson, M., and Sansonetti, P. J. (2018). Pathogens, microbiome and the host: emergence of the ecological Koch's postulates. FEMS Microbiol. Rev. 42, 273–292. doi: 10.1093/femsre/fuy003
Wang, J., Huang, Y., Xu, K., Zhang, X., Sun, H., Fan, L., et al. (2019). White spot syndrome virus (WSSV) infection impacts intestinal microbiota composition and function in Litopenaeus vannamei. Fish Shellfish Immunol. 84, 130–137. doi: 10.1016/j.fsi.2018.09.076
Wang, K., Li, K., Liu, L., Tanase, C., Mols, R., and van der Meer, M. (2020). Effects of light intensity and photoperiod on the growth and stress response of juvenile Nile tilapia (Oreochromis niloticus) in a recirculating aquaculture system. Aquac. Fish. 8, 85–90. doi: 10.1016/j.aaf.2020.03.001
Webster, T. M. U., Rodriguez-Barreto, D., Consuegra, S., and Garcia de Leaniz, C. (2019). Cortisol-induced signatures of stress in the fish microbiome. bioRxiv :826503. doi: 10.1101/826503
West, A. C., Mizoro, Y., Wood, S. H., Ince, L. M., Iversen, M., Jørgensen, E. H., et al. (2021). Immunologic profiling of the Atlantic salmon gill by single nuclei transcriptomics. Front. Immunol. 12:669889. doi: 10.3389/fimmu.2021.669889
Woof, J. M., and Mestecky, J. (2005). Mucosal immunoglobulins. Immunol. Rev. 206, 64–82. doi: 10.1111/j.0105-2896.2005.00290.x
Wynne, J. W., Thakur, K. K., Slinger, J., Samsing, F., Milligan, B., Powell, J. F. F., et al. (2020). Microbiome profiling reveals a microbial dysbiosis during a natural outbreak of Tenacibaculosis (yellow mouth) in Atlantic salmon. Front. Microbiol. 11:586387. doi: 10.3389/fmicb.2020.586387
Xu, Z., Takizawa, F., Casadei, E., Shibasaki, Y., Ding, Y., Sauters, T. J. C., et al. (2020). Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol. 5:eaay3254. doi: 10.1126/sciimmunol.aay3254
Xue, M., Jiang, N., Fan, Y., Yang, T., Li, M., Liu, W., et al. (2022). White spot syndrome virus (WSSV) infection alters gut histopathology and microbiota composition in crayfish (Procambarus clarkii). Aquac. Rep. 22:101006. doi: 10.1016/j.aqrep.2022.101006
Yadav, M. K., Kumari, I., Singh, B., Sharma, K. K., and Tiwari, S. K. (2022). Probiotics, prebiotics and synbiotics: safe options for next-generation therapeutics. Appl. Microbiol. Biotechnol. 106, 505–521. doi: 10.1007/s00253-021-11646-8
Zaneveld, J. R., McMinds, R., and Vega Thurber, R. (2017). Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Zarkasi, K. Z., Abell, G. C. J., Taylor, R. S., Neuman, C., Hatje, E., Tamplin, M. L., et al. (2014). Pyrosequencing-based characterization of gastrointestinal bacteria of Atlantic salmon (Salmo salar L.) within a commercial mariculture system. J. Appl. Microbiol. 117, 18–27. doi: 10.1111/jam.12514
Zhang, M., Shan, C., Tan, F., Limbu, S. M., Chen, L., and Du, Z. (2020). Gnotobiotic models: powerful tools for deeply understanding intestinal microbiota-host interactions in aquaculture. Aquaculture 517:734800. doi: 10.1016/j.aquaculture.2019.734800
Keywords: functionality, health, immune system, microbiota, sustainability
Citation: Lorgen-Ritchie M, Uren Webster T, McMurtrie J, Bass D, Tyler CR, Rowley A and Martin SAM (2023) Microbiomes in the context of developing sustainable intensified aquaculture. Front. Microbiol. 14:1200997. doi: 10.3389/fmicb.2023.1200997
Edited by:
Xiaodan Huang, Lanzhou University, ChinaReviewed by:
Prabhugouda Siriyappagouder, Nord University, NorwayMehdi Raissy, Azad University, Iran
Copyright © 2023 Lorgen-Ritchie, Uren Webster, McMurtrie, Bass, Tyler, Rowley and Martin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samuel A. M. Martin, sam.martin@abdn.ac.uk
†ORCID: Samuel A. M. Martin https://orcid.org/0000-0003-0289-6317
 Marlene Lorgen-Ritchie
Marlene Lorgen-Ritchie Tamsyn Uren Webster
Tamsyn Uren Webster Jamie McMurtrie
Jamie McMurtrie David Bass3,4
David Bass3,4 Charles R. Tyler
Charles R. Tyler Andrew Rowley
Andrew Rowley Samuel A. M. Martin
Samuel A. M. Martin