- 1Biotechnology Centre, National Veterinary Research Institute, Jos, Nigeria
- 2Fleming Laboratory Microbiology Department, National Veterinary Research Institute, Jos, Nigeria
- 3Menzies School of Public Health, Charles Darwin University, Darwin, NT, Australia
- 4Regional Laboratory for Animal Influenza & Transboundary Animal Diseases, National Veterinary Research Institute, Jos, Nigeria
Introduction: Antimicrobial resistance is increasingly becoming a global health concern. This study aimed to investigate and report MDR Escherichia coli (E. coli) prevalence, resistance, and virulence genes from poultry in Jos, Plateau State, Nigeria.
Methods: The samples were analyzed using microbiological standard methods and polymerase chain reactions (PCRs).
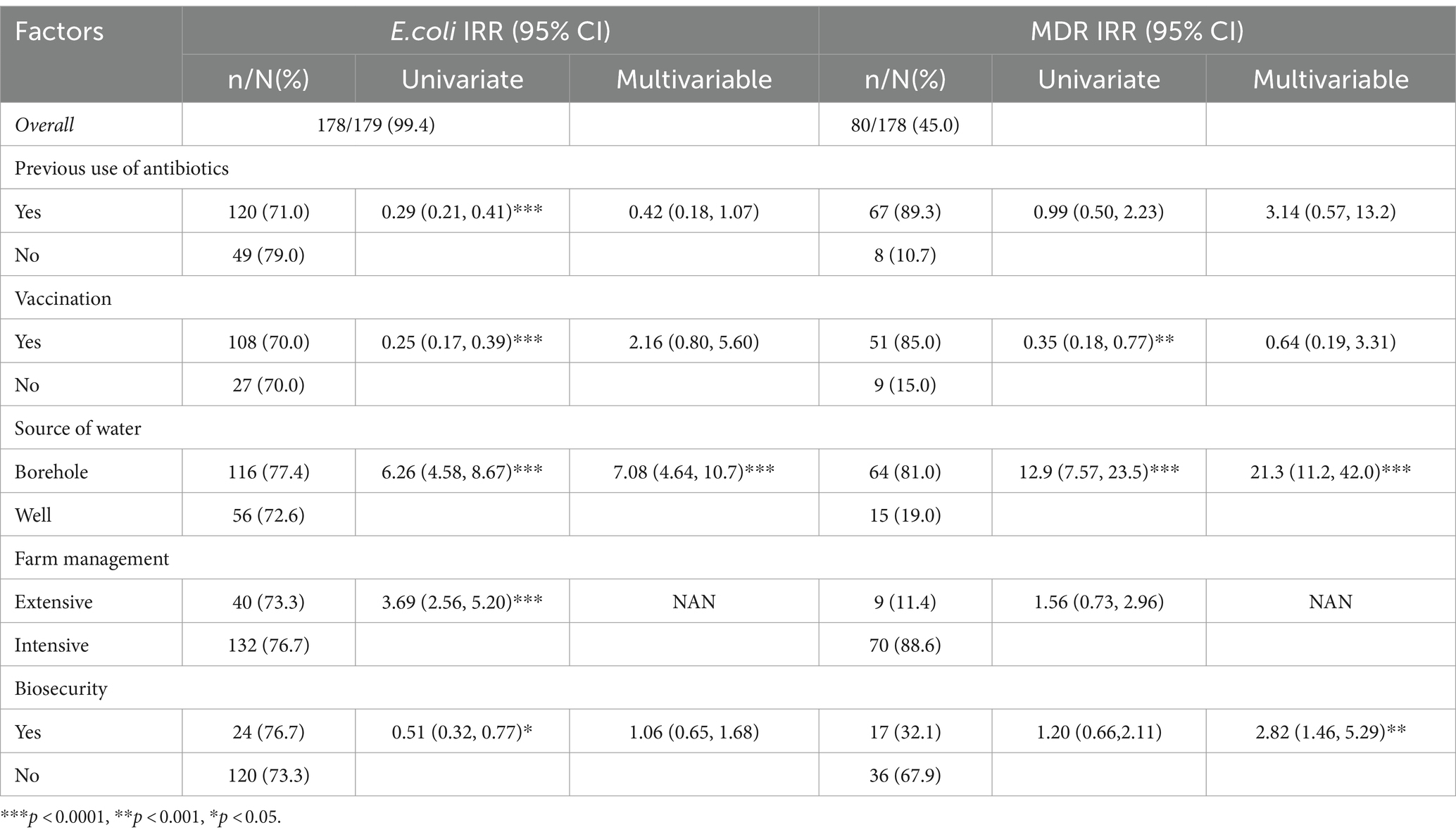
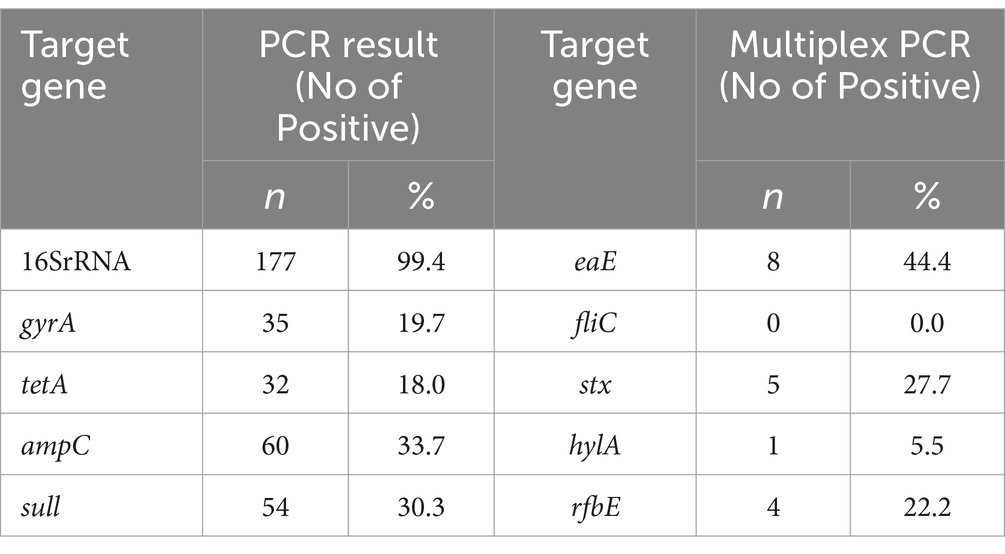
Results: A total of 179 cloacal swabs were collected from bothlocal and exotic poultry breeds, of which 99.4% (178/179) tested positive for E. coli. Among these culturally identified samples, 99.4% (177/178) were furtherconfirmed Escherichia coli with a molecular weight of 401 bp. Multidrugresistance of 45% (80/178) was observed from the confirmed isolates. PCR assays were conducted to detect genes associated with resistance to antibiotics, specifically, tetracycline (tetA gene), sulfonamide (sul1 gene), ampicillin (ampC gene), and quinolone (gyrA gene). Antimicrobial susceptibility test (AST) results revealed substantial antibiotic resistance, with 81.9% (145/177) of the isolates being resistant to tetracycline, 80.2% (142/177) to quinolone, 69.5% (123/177) to sulfonamide, and 66.1% (117/177) to ampicillin. Further analysis on 18 isolates that showed resistance to up to four different antibiotics was carried out using multiplex PCR to detect eae, hlyA, rfbE, fliC, and fstx virulence genes. The study found that 44.4% (15/18) of the isolates were positive for the eae gene, 27.7% (5/18) for stx, 22.2% (4/18) for rfbe gene, and 5.5% (1) for hlya gene, and none tested positive for fliC gene.
Conclusion: These results showed high antibiotic resistance, virulent genes, and significant levels of MDR in E. coli from poultry. This study highlights the urgent need for antimicrobial stewardship practices within the poultry industry due to their profound implications for food safety and public health. This issue is particularly critical in Nigeria, where poultry farming constitutes a significant portion of smallholder farming practices.
Highlights
What is already known about this topic
• Antimicrobial resistance is a growing concern for One Health, especially in the livestock industry.
• Poultry farms close to humans pose a risk of transmission episodes of zoonotic MDRs
• Up to 70% of antibiotics are excreted by animals as unmetabolized substances, which means an increase in selective pressure leading to environmental antimicrobial resistance.
What this study adds
• The presence of resistant genes in free-range poultry in the communities suggests environmental contamination by antimicrobial genes.
• There is a high prevalence of drug-resistant Escherichia coli in poultry from Jos.
Introduction
Multidrug resistance (MDR), a phenomenon often used to describe bacteria that are resistant to three or more antibiotic classes, has become increasingly common (Kallau et al., 2018). Recently, antimicrobial resistance (AMR) has gained global recognition due to the rise in multidrug-resistant organisms such as Escherichia coli (E. coli), which has led to increased economic burden and mortality, especially in Nigeria. Aworh et al. (2019) posited that MDR among many organisms has become a huge challenge to infectious disease management, and it is increasingly being reported in bacteria, often mediated by genetic mobile elements, such as integrons, transposons, and plasmids (Nnaji et al., 2021).
E. coli strains that are multidrug-resistant from animal and human isolates are ubiquitous in different parts of the world (O’Neill, 2019), with the non-pathogenic strains having the likelihood of being an important reservoir of resistance genes. Extensive usage of antibiotics in livestock production for disease prevention (prophylaxis and metaphylaxis), treatment, and growth promotion is one of the principal factors in the spread of antibiotic resistance (Brower et al., 2017; Hassan et al., 2021). Antimicrobials are routinely added to animal feeds, and bacterial populations are repeatedly exposed to sub-therapeutic doses required for the development and spread of MDR (Brower et al., 2017). In addition, in many developing countries in Asia and Africa, there is little or no restriction on antibiotic usage, and antimicrobial agents are less restricted in local drug stores without prescription. Such practice has led to the misuse of antibiotics, leading to resistance among isolates from animals, food sources, and the environment (Agada et al., 2014).
E. coli is one of the commensal bacteria found in the gastrointestinal tracts of humans, poultry, and mammals (Akond, et al., 2009; Olatoye et al., 2012) but may become opportunistic, leading to disease states (Akond, et al., 2009). Although most E. coli strains are commensal, a few highly pathogenic ones carry stx2 and eae genes, causing watery and bloody diarrhea associated with life-threatening diseases, such as thrombocytopenic purpura, hemolytic uremic syndrome, hemorrhagic colitis (Al Azad et al., 2019), and extra-intestinal diseases. This bacterium can be acquired through contaminated water, soil, and food (Okorie-Kanu et al., 2016), with most human infections occurring from contaminated food products of poultry origin (Hamisi et al., 2014). Globally, E. coli effects on poultry production can be severe (Hamisi et al., 2014), and it is a public health threat in sub-Saharan African countries, including Nigeria. Some drug-resistant E. coli strains, which may not directly cause disease, remain significant in public health as a reservoir of drug-resistant genes that can be transferred to humans (Aworh et al., 2019).
Poultry is a good source of high-quality animal protein for the growing population of Nigeria and a vital part of the Nigerian economy, which provides income and livelihood for smallholder farmers (Awoyomi et al., 2022). It accounts for approximately 25% of local meat from livestock produced in Nigeria (Anosike et al., 2018). Due to the increased consumption of poultry products in Nigeria, there is dependence on poultry production by many families for income and livelihood (Agada et al., 2014; Aworh et al., 2019), as well as antimicrobial agents to boost production by minimizing diseases. These antimicrobial agents are used for prophylaxis, treatment, and growth promotion (Brower et al., 2017; Hassan et al., 2021). The addition of these antimicrobial agents poses a significant risk to the emergence of some resistant bacteria either through genetic or non-genetic means, thereby making poultry a major reservoir of antimicrobial-resistant E. coli (Okorie-Kanu et al., 2016).
Although poultry may serve as a significant reservoir of resistant bacteria in food animals, there is a paucity of information on the multidrug resistance pattern of E. coli in poultry in the study area. This study was designed to detect E. coli from local and exotic poultry breeds in Jos, Plateau State, and to examine their antimicrobial susceptibility pattern and the presence of multidrug-resistant and virulence genes from the isolated bacteria.
Materials and methods
Data collection
Between November 2019 and February 2020, cloacal swabs were collected from birds in 26 randomly selected farms in Jos South Local Government Area, Plateau State, Nigeria. Birds were grouped into two categories: caged (intensively managed) and free-ranged (extensively managed) local birds. Additionally, farm-level data such as biosecurity, vaccination status, source of water, previous use of antibiotics, and farm management (intensive or extensive) were obtained using interviewer-administered questionnaires.
Sample size determination
The sample size for the study was calculated based on the prevalence of E. coli, aiming for a 95% confidence level and a 5% margin of error. Utilizing the formula, N=Z^2×P(1−P) /L^2, where N represents the required sample size, Z is the Z-statistic corresponding to a 95% confidence level (1.96 for two-tailed tests), P is the prevalence of E. coli, and L is the allowable error margin at 5%. For this study, P was taken as 13.4% based on previous findings by Shecho et al. (2017). Substituting these values into the formula, we computed N as (1.96^2*0.134*0.866)/(0.05^2) which resulted in approximately 178. To accommodate the study design and ensure robustness, the sample size was rounded up to 180 cloacal swabs.
Isolation of E. coli
Samples were inoculated into bacteriological peptone water and incubated for 24 h at 37 ± 0.5°C before streaked onto MacConkey agar. E. coli was isolated as Gram-negative short rods from lactose-fermenting colonies that were positive for indole produced acid and gas in the triple sugar iron agar test but negative for citrate and urea. To further confirm isolates, they were cultured on eosin methylene blue agar (EMB) (FLUMEDIA, United Kingdom), which is a selective medium for E. coli. Only colonies that produced the green metallic sheen on EMB were used for the antimicrobial susceptibility test. The isolated E. coli were subjected to molecular confirmation using E. coli genus-specific primer targeting the 16S rRNA (Sabat et al., 2000).
Antimicrobial susceptibility testing
The agar disk diffusion method was used to determine resistant, intermediate, or susceptible isolates to the antibiotics. A total of seven antibiotics spanning across five classes of antibiotics were used (cefoxitin 30 μg, oxacillin 10 μg, erythromycin 10 μg, sulfamethoxazole/trimethoprim 1.25/23.7 μg, nalidixic 30 μg acid, ampicillin 10 μg, and ciprofloxacin 5 μg). A turbidity of 0.5 McFarland standard of the isolate was streaked onto a Mueller–Hinton agar (HiMEDIA, Bombay, India) plate before the respective antibiotic disks were placed aseptically on the surface. Zone clearing was measured in millimeters and recorded as resistance, intermediate, or susceptible following CLSI standards. Isolates that showed resistance to at least three different classes of antibiotics will be classified as MDR E.coli.
PCR confirmation of isolates and detection of virulence and antimicrobial genes
We conducted a microbiological standard culture of cloacal swab samples collected from both exotic breeds and local birds. Subsequently, samples that showed positive indications for E. coli were further analyzed through molecular techniques using a 16S rRNA primer. To identify antibiotic resistance genes, we employed a selection process for PCR analysis based on the outcomes of antibiotic susceptibility testing methods. This approach aimed to ascertain the presence of resistance genes to various antibiotics, such as quinolone, tetracycline, ampicillin, and sulfonamide, each identified by their distinct molecular weights. Amplicons were separated on a 1.5% agarose gel at 80 V for 40 min.
To dedect the virulence genes in the isolates, we applied the method previously described by Fusaro et al. (2009) and utilized primer as per the guidelines of Wang et al. (2002). The reaction conditions were first optimized to obtain optimum reaction and temperature conditions. After optimization, eae, hlyA, rfbE, fliC, and stx2 genes for virulence genes were performed using multiplex PCR.
Ethical clearance
All procedures involving animals in the study were conducted following guidelines obtained from the ethics committee of the National Veterinary Research Institute NVRI, Vom. Approval reference number AEC/03/76/19.
Data analysis
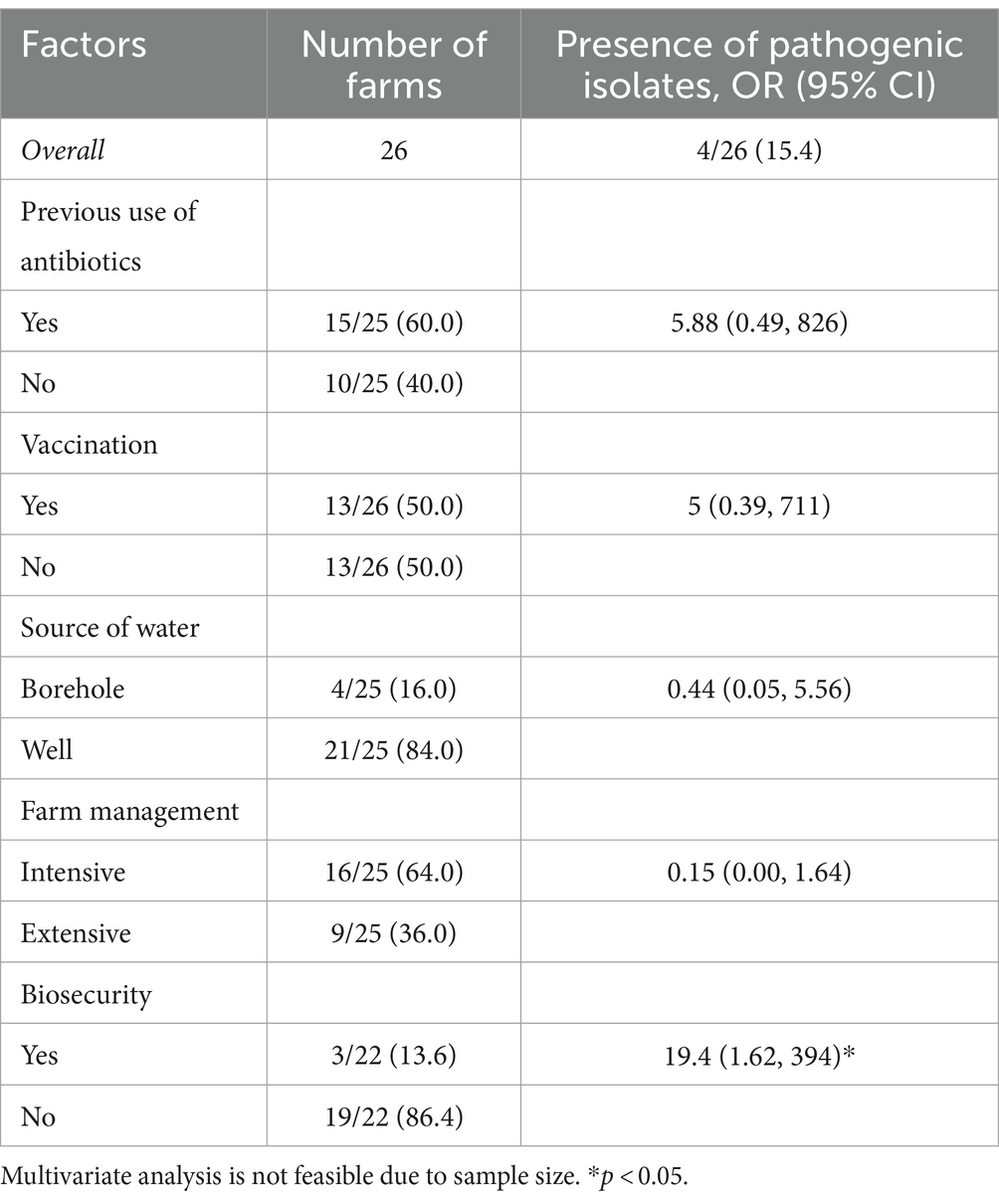
Descriptive statistics such as frequencies and percentages were used to describe the data. Group frequencies were compared using the chi-squared or Fisher’s exact test where applicable. Poisson regression was utilized to assess the association between farm-level characteristics and the presence of E. coli and MDR, while Firth’s logistic regression model that corrects for bias in rare events was used to investigate the effect of farm-level factors on the presence of pathogenic isolates (Beaujean and Grant, 2016; Puhr et al., 2021).
Results
Isolation and prevalence of E. coli
Of the 179 cloacal swab samples from exotic breeds and local birds from 26 farms subjected to microbiological standard culture, 178 (99.44%) were positive for E. coli (Table 1). Molecular analysis confirmed 177 of the samples to be positive for E. coli with a molecular weight of 401 bp. In total, 36 isolates that showed a strong bright band (Figure 1A) were further screened.
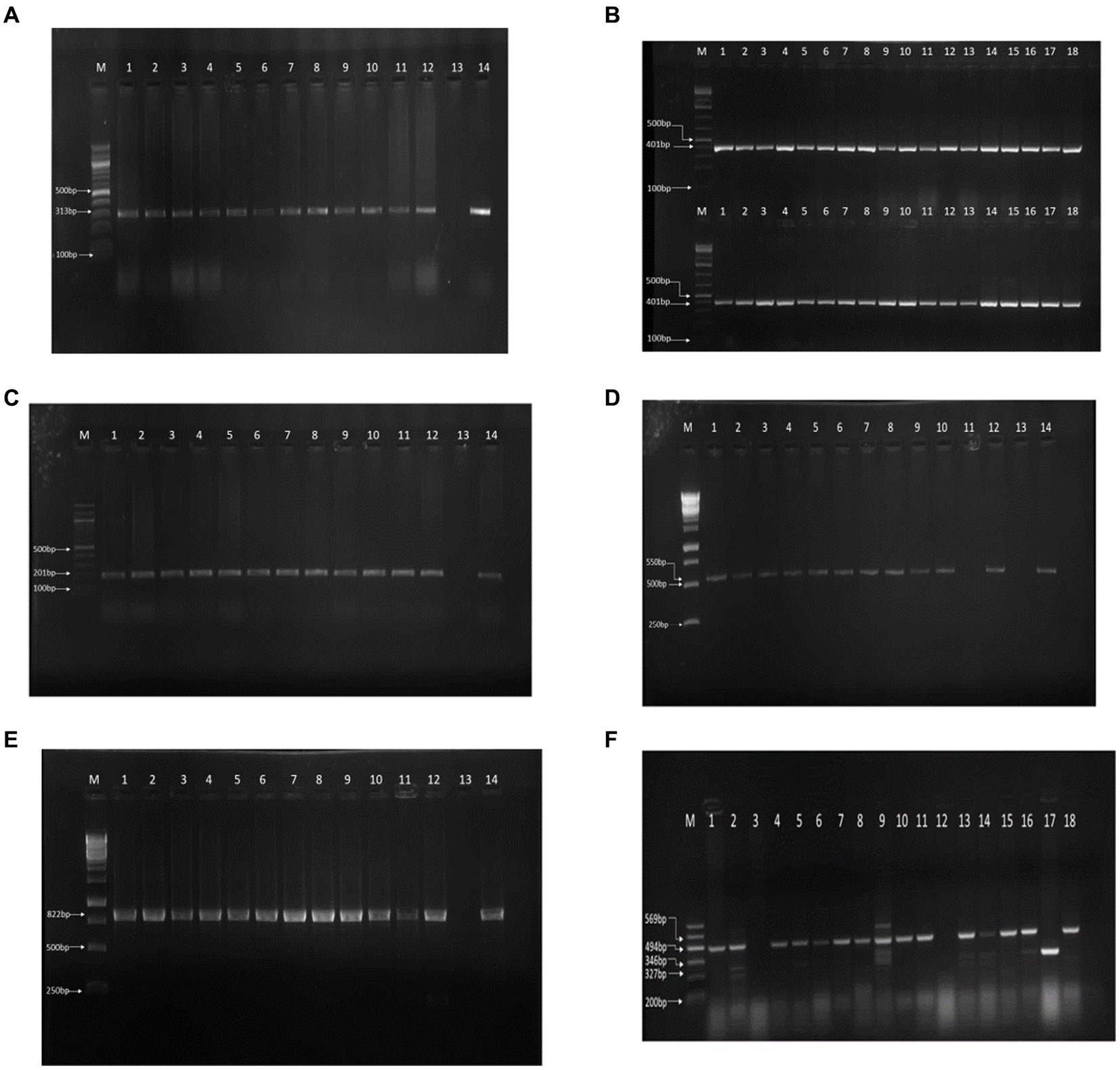
Figure 1. PCR results on gel electrophoresis. (A) 16S rRNA PCR result. (B) gyrA-resistant gene. (C) tetA-resistant gene. (D) ampC resistant gene PCR resistant gene PCR result. (E) sul1 resistant gene PCR result. (F) Multiplex PCR of the virulence genes.
Antimicrobial susceptibility
We reported resistance to more than 3 classes of antibiotics in 80 out of 178 cases (45.0%) and found at least 1 MDR isolate in all poultry farms sampled. Further exploration into antibiotic resistance revealed significant findings. PCR analysis, informed by antibiotic susceptibility testing, indicated that 80.23% (142 out of 177) of the samples were resistant to quinolone (313 bp), 81.92% (145 out of 177) to tetracycline (201 bp), 66.10% (117 out of 177) to ampicillin (550 bp), and 69.50% (123 out of 177) to sulfonamide (822 bp), as detailed in Figures 1B–E. Consequently, 18 isolates, which were positive for all four antibiotic resistance genes, were selected for more in-depth analysis. The study also employed optimized multiplex PCR for the detection of virulence genes, revealing the presence of the eae gene (494 bp) in 8 out of 18 analyzed isolates, the hlyA gene in 1 isolate, the rfbE gene in 4 isolates, and stx2 gene in 5 isolates, with the fliC gene being absent in all tested isolates.
Amplicons were analyzed using agarose gel electrophoresis to separate DNA fragments based on size. A 1.5% agarose gel was utilized, and the electrophoresis was run at 80 V for 40 min. In the analysis of the 16S rRNA gene, lanes 1 through 18 contained isolates with a 401-bp fragment, and lane (M) served as the molecular size marker, indicated by a 100-bp ladder, to facilitate the estimation of DNA fragment sizes (Figure 1A). Similarly, for the detection of the gyrA resistance gene, lanes 1 to 12 harbored isolates that contained the gyrA gene, characterized by a 313-bp fragment. Lane 13 was designated as the negative control (containing no DNA), and lane 14 was the positive control, with a known positive isolate exhibiting the same 313-bp gyrA gene fragment (Figure 1B). The analysis for the tetA resistance gene followed the same procedure, where lanes 1 to 12 contained isolates with the tetA gene (201 bp), and lanes 13 and 14 served as the negative and positive controls, respectively, mirroring the setup for gyrA (Figure 1C).
Furthermore, multiplex PCR targeting various virulence genes revealed that among the isolates analyzed, 8 harbored the eae gene (494-bp), 1 contained the hlyA gene (569-bp), 4 possessed the rfbe gene (327-bp), and 5 had the stx2 gene (346-bp), as separated on a 1.5% agarose gel run at 80 V for 40 min. The molecular marker lane (M) again featured a 100-bp ladder to assist in the sizing of these amplicons (Figure 1F). This comprehensive analysis provided a detailed overview of the presence and distribution of resistance and virulence genes among the isolates, leveraging both single and multiplex PCR approaches for the identification of specific gene fragments indicative of bacterial resistance and virulence.
Association between farm-level risk factors and the presence of E. coli and MDR
Table 1 represents farm-level data and its association with the prevalence of E. coli and MDR, while Table 2 presents the association between various farm-level characteristics and the presence of pathogenic isolates across 26 farms. In the univariate analysis, we found a significant reduction in E. coli among farms that previously used antibiotics, with an IRR of 0.29 (95% CI: 0.21, 0.41). However, this association was not statistically significant in the multivariable model after adjusting for other farm factors (IRR: 0.42, 95% CI: 0.18, 1.07).
Similarly, vaccination was initially associated with a lower prevalence of E. coli (IRR: 0.25, 95% CI: 0.17, 0.39) in univariate analysis, but the significance diminished in the multivariable context (IRR: 2.16, 95% CI: 0.80, 5.60). Borehole water usage was strongly associated with higher E. coli presence (IRR: 6.26, 95% CI: 4.58, 8.67) in the univariate model, an association that remained robust and even more pronounced in the multivariable analysis (IRR: 7.08, 95% CI: 4.64, 10.7), underscoring borehole water as a critical factor for bacterial contamination.
The analysis of MDR prevalence also yielded similar significant results. The source of water again stood out, with borehole water associated with a substantially higher prevalence of MDR organisms (univariate IRR: 12.9, 95% CI: 7.57, 23.5), which further increased in the multivariable model (IRR: 21.3, 95% CI: 11.2, 42.0). On the other hand, biosecurity measures showed a protective effect against MDR in the multivariable model (IRR: 2.82, 95% CI: 1.46, 5.29), suggesting effective biosecurity practices that can significantly reduce the prevalence of MDR bacteria despite the univariate model showing a less pronounced association (IRR: 1.20, 95% CI: 0.66, 2.11).
Table 2 examines the association between various farm-level characteristics and the presence of pathogenic isolates across 26 farms. The overall prevalence of pathogenic isolates is reported as 15.4%. Although most of the farm-level factors (except biosecurity measures) were not significantly associated with the presence of pathogenic isolates, they showed interesting associations. For example, farms with a history of antibiotic use exhibited higher odds, farms using borehole water had lower odds, and intensive farm management practices were associated with notably lower odds. In contrast, farms implementing biosecurity practices had significantly higher odds of pathogen presence (OR = 19.4; CI: 1.62 to 394). This counterintuitive finding could reflect complexities not captured by the data, such as the possibility that farms with higher risk profiles or prior pathogen issues are more likely to implement biosecurity measures.
Virulence detection and antimicrobial genes
Table 3 presents the summary of the PCR gel and multiplex PCR of the virulent genes. For single PCR, the highest detection rate was observed for the 16SrRNA gene, with 177 (99.4%) positive cases. Other genes detected with varying frequencies include gyrA (19.7%), tetA (18.0%), ampC (33.7%), and sull (30.3%), reflecting a diverse presence of virulence factors. In contrast, multiplex PCR, which allows for the simultaneous detection of multiple genes, showed a considerably lower detection rate across different genes, with eae detected in 8 cases (4.5%), stx in 5 cases (2.8%), and rfbe in 4 cases (2.2%). Notably, fliC was not detected in any samples (0.0%), and hyla was found in only 1 case (0.6%).
Discussion
In this study, a comprehensive analysis was conducted on 179 cloacal swab samples from both exotic breeds and local birds to investigate the presence of E. coli and associated antibiotic resistance in farms in Jos Plateau, Nigeria. Microbiological standard culture techniques initially identified 178 (99.4%) samples as positive for E. coli, which was further confirmed through molecular analysis using a 16S rRNA primer for 177 samples, each exhibiting a molecular weight of 401 bp. Among these, 36 isolates were particularly notable for displaying very strong bright bands. The high occurrence of the positive E. coli isolates by PCR from this study supports the findings of Al Azad et al. (2019). As a ubiquitous microbe often isolated from the gastrointestinal tract of warm-blooded animals (Desvaux et al., 2020), its highly promiscuous nature facilitates its acquisition of antimicrobial resistomes (Nhung et al., 2016), thereby making it a tool for the spread of antimicrobial-resistant genes in the food chain and the environment. Against this backdrop, a high prevalence (99.44%) of E. coli may have deleterious consequences for both human and animal health (Kwoji et al., 2019). Consequently, infections caused by resistant pathogenic E. coli depict high morbidity and negative economic downturn for livelihood and poultry businesses. Environmental contamination by antibiotic residue and AMR genes are a major driver of AMR in livestock because up to 70% of antimicrobials administered to livestock have been reported to be released as unmetabolized agents (Bhushan et al., 2017).
At least one MDR isolate was reported in all poultry farms sampled, which is consistent with the study by Adebowale et al. (2022), which showed a lesser MDR value of 56.3%. The higher prevalence of MDR in this study may be connected to poor biosecurity on farms and the excessive use of antibiotics by farmers. The resistance to more than three classes of antibiotics is indicative of extensive antimicrobial usage. A study by Enany et al. (2019) showed the connection between the frequent use of antibiotics in farm production and treatment and the emergence of multidrug resistance in different bacterial pathogens. Similarly, other studies have emphasized the rising trend in AMR and MDR in low- and middle-income countries such as Nigeria (Osman et al., 2018; Kabantiyok et al., 2023).
What are the implications of a high MDR ubiquitous microbe on the environment, humans, and food value chain? First, there have been reports of AMR and antimicrobial-resistant genes (ARGs) from the environment around animal farms, including poultry and other biological spaces (Mazhar et al., 2021; Mkpuma et al., 2021) supported by the isolation of similar antibiotic-resistant genes simultaneously from human and poultry samples (Johnson et al., 2007). Apart from prophylactic administration, poultry farming benefits from the use of heavy metals in animal feed as growth promoters and has served as a tool for selective pressure in the emergence and maintenance of ARGs in the environment (Muhammad et al., 2020). Furthermore, Manyi-Loh et al. (2018) suggested that the inclusion of antimicrobial agents in feed production as growth promoters by farmers to meet the high demand for animal protein in developing countries may also cause bacteria to develop resistance to the antibiotics used. The excessive use of antibiotics and heavy metals expose humans to a cocktail of antimicrobial-resistant zoonosis, which is a fall-out of selective pressure from unmetabolized residues of antibiotics and heavy metals in the environment. Second, this leads to acquired resistance to newly developed and last-resort antibiotics, which presents a challenge for humans and animals, especially in the management of health outcomes. The emergence of resistance limits the choice of drugs and makes treatment more expensive. In a recent report by Laxminarayan (2022), the estimated number of deaths in 2019 due to AMR was 1.27 million, a figure greater than deaths due to malaria (627,000) and HIV (680,000) put together and an astounding 4.95 million deaths due to AMR-related complications. Antimicrobial resistance is seen to be a concern for the economy as it threatens food security, animal welfare, longer treatment cycles, and public health worldwide (Selaledi et al., 2020).
We also screened for antimicrobial resistance genes in some selected isolates to understand the genetic basis for their antimicrobial resistance. The genes identified include tetracycline (tetA), fluoroquinolone (gyrA), sulfonamide (sul1), and ampicillin (blaTEM ). The resistance to tetracycline was 68.6%, fluoroquinolone was 52.8%, sulfonamide 53.2%, and ampicillin 49.4%. The most common resistant antibiotic gene from this study was observed to be the tetracycline gene with 68.6%. The presence of these resistant genes has been recorded in previous studies (Momtaz et al., 2012; Dehkordi et al., 2014; Al Azad et al., 2019). Antimicrobial-resistant genes play a vital role in the spread of environmental resistance (Massé et al., 2014) especially in the food chain because food doubles as a vector for the transfer of residual antibiotics and for the transfer of antibiotic genes carried by zoonotic pathogens thus constituting public health and One Health concern. The misuse of antibiotics for prophylactic treatment and as growth promoters, poor biosecurity, and close clustering of different poultry in the same area are drivers for the emergence and spread of antimicrobial resistance. According to Manyi-Loh et al. (2018), commonly used antibiotics such as tetracyclines, aminoglycosides, β-lactams, lincosamides, macrolides, pleuromutilins, and sulfonamides by farmers worldwide can have negative effects on health. This raises concerns about disease control and risk management. We showed the intricate relationship between farm management practice and biosecurity.
Another important finding in this study is the presence of some virulence genes found in the isolates. One or more isolates were positive for each virulence gene tested. Of the 18 selected E. coli-positive isolates, 8 (44.44%) isolates contained the eaeA gene, and this is high when compared to similar data from two other studies on the eaeA gene in poultry conducted by Wani et al. (2004) and Enany et al., (2019), which recorded 2.49 and 20.3%, respectively. This is noteworthy because of the invaluable role the gene plays in the attachment and effacing of enteropathogenic E. coli (EPEC) in the intestinal mucosa (Rahimi et al., 2022); 1 (5.55%) out of the 18 isolates also contained the hylA gene, which is in line with the study by Wani et al. (2004), reporting a hylA prevalence of 1.49% of the E. coli isolates in chicken. These genes responsible for virulence and resistance in Escherichia coli have been identified in humans, especially poultry workers (Adelowo et al., 2014; Joshua et al., 2018), thus contributing to the spread of antimicrobial resistance. In their study, AMR is more frequent in pathogenic than in other commensal (non-pathogenic) E. coli strains. This suggests that the association between resistance and virulence genes is one that self-preserves the pathogen.
It has been projected that by 2050, almost 10 million people would die from bacterial infections that are resistant to antibiotics (O’Neill, 2019). Due to the threat of antibiotic resistance on health outcomes globally, the World Health Organization (WHO) considers antimicrobial resistance to be one of the major threats to public health as more bacteria are becoming resistant to more antibiotics (WHO, 2021). It is worrisome that most of these resistant bacteria are zoonotic, and infections triggered by these pathogens can be difficult to treat because previously potent drugs become less efficacious against the same pathogen. The resistance reported from this study may have emanated from lay farm owners, but it generally reflects poorly on a system of underperforming antimicrobial stewardship.
Although we set out to detect MDR and resistant genes in intensive and extensively managed poultry, our study was limited in terms of statistical power to make a compelling statement on MDR in local birds; the detection of resistant genes in local birds sampled indicates environmental contamination by resistant genes since all local birds sampled are not administered antibiotics compared to extensively managed birds. This study illuminates the complex interplay of farm practices, environmental conditions, and biosecurity measures in influencing the prevalence and spread of pathogenic and resistant bacteria. Notably, the significance of certain associations changes from univariate to multivariable models, highlighting the importance of considering a multifactorial approach to manage and mitigate bacterial risks on farms effectively.
Conclusion
Antibiotics are of importance in agriculture, veterinary, and clinical settings. Notwithstanding the beneficial roles of antibiotics, their abuse may lead to serious public health complications. Results from this study indicate that there are significant levels of MDR and antibiotic resistance of E. coli in poultry farms, suggesting that there is a lot of antibiotic abuse in the poultry farming process. The presence of virulence genes associated with these isolates makes it more worrisome for public health, and this underscores the need for farmers to be educated on the appropriate use of antibiotics and stewardship.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal studies were approved by Animal ethics committee of the National Veterinary Research Institute NVRI, Vom. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
EA: Writing – review & editing, Writing – original draft, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. DK: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. NM: Writing – original draft, Methodology, Investigation, Conceptualization. RA: Writing – original draft, Methodology, Investigation, Conceptualization. CO-E: Writing – review & editing, Methodology, Investigation, Conceptualization. EB: Writing – review & editing, Methodology, Investigation, Conceptualization. JB: Writing – review & editing, Methodology, Investigation, Conceptualization. KS: Writing – review & editing, Methodology, Investigation. RR: Writing – review & editing, Methodology, Investigation, Conceptualization. GJ: Writing – review & editing, Methodology, Investigation, Conceptualization. BA: Writing – review & editing, Visualization, Validation, Supervision. GA: Writing – review & editing, Visualization, Validation, Supervision. OA: Methodology, Investigation, Formal analysis, Writing – review & editing, Visualization, Supervision. CM: Writing – review & editing, Visualization, Validation, Supervision.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was partly funded by the Research Officers Association, National Veterinary Research Vom, Plateau State, Nigeria (Grant numbers NVRI/ARO001).
Acknowledgments
The authors would like to acknowledge the support of the research/academic committee of the Association of Research Officers (ARO), National Veterinary Research Institute (NVRI) Vom, for supporting this project with a seed grant. The authors acknowledge the assistance of Francis Maisaje, Mark Samson, Patrick Gabriel Nyango, Bitrus Yakubu, and Anthony Chukwuedo for facilitating this project through field support and mentorship. This article is presently uploaded as a preprint.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1298582/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | A Condition Typical of Escherichia coli Septicaemia as seen in one of the Farms.
SUPPLEMENTARY FIGURE S2 | Gram Positive Short Rods Typical of Escherichia coli seen under Oil Immersion.
SUPPLEMENTARY FIGURE S3 | Pathogenic isolates on Sorbitol Macconkey Agar Appear as yellow colonies and Escherichia coli on Eosin Methylene Blue Agar. The PCR reactions were performed in a final volume of 25 μl containing 7 μl nuclease free water, 12.5 μl 2x green master mix, 20 pmole of 0.25 μl of primer and 5 μl of DNA template. Tetracycline and quinolones had 0.25 μl of 20 pmole of each primer, while sulphonamide and ampicillin had 0.5 μl of each primer and 6.5 μl of nuclease free water respectively. PCR amplification cycle of Tetracycline resistant genes consisted of an initial denaturation temperature at 94 OC for 5 min, followed by denaturation temperature at 94 OC 1 min, annealing temperature at 55 OC for 1 min and final elongation temperature at 72 OC for 90 sec. PCR amplification cycle of quinolones resistant genes consisted of an initial denaturation temperature at 95 OC for 5 min, followed by denaturation temperature at 94 OC 1 min, annealing temperature at 56 OC for 1 min and final elongation temperature at 72 OC for 1 min. PCR amplification cycle of sulphonamide resistant genes consisted of an initial denaturation temperature at 94 OC for 5 min, followed by denaturation temperature at 94 OC for 1 min, annealing temperature at 55 OC for 1 min and final elongation temperature at 72 OC for 5 min. PCR amplification cycle of ampicillin resistant genes consisted of an initial denaturation temperature at 94 OC for 5 min, followed by denaturation temperature at 94 OC for 30 sec, annealing temperature at 50 OC for 30 sec, and final elongation temperature at 72 OC for 90 sec. Electrophoresis were carried out on the amplicons using 1.5% agarose powder in 100 ml of 1X TBE at 100v for 35 min. the gel were visualized using Gel Documentation System (Synegene®). A molecular weight marker with 100bp and 1kb plus was used as a size standard. An in-house positive control, extracted from E. coli as used by Biotechnology Centre of the National Veterinary Research Institute, Vom, Plateau State, Nigeria, was used as positive control, while reaction with no template DNA added was used as negative control.
References
Adebowale, O., Makanjuola, M., Bankole, N., Olasoju, M., Alamu, A., Kperegbeyi, E., et al. (2022). Multi-drug resistant Escherichia coli, biosecurity and anti-microbial use in live bird markets, Abeokuta, Nigeria. Antibiotics 11:253. doi: 10.3390/antibiotics11020253
Adelowo, O. O., Fagade, O. E., and Agersø, Y. (2014). Antibiotic resistance and resistance genes in Escherichia coli from poultry farms, southwest Nigeria. J. Infect. Dev. Ctries. 8, 1103–1112. doi: 10.3855/JIDC.4222
Agada, G. O., Abdullahi, I. O., Moses, O., Chollom, M., Kumbish, P. R., and Okwori, A. E. J. (2014). Prevalence and antibiotic resistance profile of Salmonella isolates from commercial poultry and poultry farm-handlers in Jos, plateau state, Nigeria. Br. Microbiol. Res. J. 4, 462–479. doi: 10.9734/BMRJ/2014/5872
Akond, M. A., Hassan, S. M. R., Alam, S., and Momena, S. (2009). Antibiotic Resistance of Escherichia Coli Isolated From Poultry and Poultry Environment of Bangladesh. Am. J. Environ. Sci. 11. doi: 10.3844/ajessp.2009.47.52
Al Azad, M. A. R., Rahman, M. M., Amin, R., Begum, M. I. A., Fries, R., Husna, A., et al. (2019). Susceptibility and multidrug resistance patterns of Escherichia coli isolated from cloacal swabs of live broiler chickens in Bangladesh. Pathogens 8:118. doi: 10.3390/PATHOGENS8030118
Anosike, F. U., Rekwot, G. Z., Owoshagba, O. B., Ahmed, S., and Atiku, J. A. (2018). Challenges of poultry production in Nigeria: a review. Niger. J. Anim. Prod. 45, 252–258. doi: 10.51791/NJAP.V45I1.335
Aworh, M. K., Kwaga, J., Okolocha, E., Mba, N., and Thakur, S. (2019). Prevalence and risk factors for multi-drug resistant Escherichia coli among poultry workers in the Federal Capital Territory, Abuja, Nigeria. PLoS One 14:e0225379. doi: 10.1371/JOURNAL.PONE.0225379
Awoyomi, O. J., Olasoju, M. I., Kehinde, O. O., Adebowale, O. O., Awoyomi, F. S. O., Agbajelola, V. I., et al. (2022). Impact of COVID-19 on livestock production and marketing in Nigeria. Niger. J. Anim. Prod. 49, 215–232. doi: 10.51791/NJAP.V49I3.3552
Beaujean, A. A., and Grant, M. B. (2016). Tutorial on using regression models with count outcomes using R. Pract. Assess. Res. Eval. 21:2. doi: 10.7275/PJ8C-H254
Bhushan, C., Khurana, A., Sinha, R., and Nagaraju, M. (2017). Antibiotic resistance in poultry environment: Spread of resistance from poultry farm to agricultural field. Centre for Science and Environment, 36.
Brower, C. H., Mandal, S., Hayer, S., Sran, M., Zehra, A., Patel, S. J., et al. (2017). The prevalence of extended-spectrum beta-lactamase-producing multidrug-resistant Escherichia coli in poultry chickens and variation according to farming practices in Punjab, India. Environ. Health Perspect. 125:077015. doi: 10.1289/EHP292
Dehkordi, F. S., Yazdani, F., Mozafari, J., and Valizadeh, Y. (2014). Virulence factors, serogroups and antimicrobial resistance properties of Escherichia coli strains in fermented dairy products. BMC. Res. Notes 7, 1–8. doi: 10.1186/1756-0500-7-217/TABLES/6
Desvaux, M., Dalmasso, G., Beyrouthy, R., Barnich, N., Delmas, J., and Bonnet, R. (2020). Pathogenicity factors of Genomic Islands in intestinal and Extraintestinal Escherichia coli. Front. Microbiol. 11:558927. doi: 10.3389/FMICB.2020.02065/BIBTEX
Enany, M. E., Algammal, A. M., Nasef, S. A., Abo-Eillil, S. A. M., Bin-Jumah, M., Taha, A. E., et al. (2019). The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Express 9, 1–9. doi: 10.1186/S13568-019-0920-4/FIGURES/2
Fusaro, A., Joannis, T., Monne, I., Salviato, A., Yakubu, B., Meseko, C., et al. (2009). Introduction into Nigeria of a distinct genotype of avian influenza virus (H5N1). Emerg. Infect. Dis. 15, 445–447. doi: 10.3201/EID1503.081161
Hamisi, Z., Tuntufye, H., and Shahada, F. (2014). Antimicrobial resistance phenotypes of Escherichia coli isolated from tropical free range chickens. Int. J. Sci. Res. Available at: www.ijsr.net
Hassan, M. M., El Zowalaty, M. E., Lundkvist, Å., Järhult, J. D., Khan Nayem, M. R., Tanzin, A. Z., et al. (2021). Residual antimicrobial agents in food originating from animals. Trends Food Sci. Technol. 111, 141–150. doi: 10.1016/j.tifs.2021.01.075
Joshua, A., Moses, A., and Ezekiel Olugbenga, A. (2018). A Survey of Antimicrobial Agents Usage in Poultry Farms and Antibiotic Resistance in Escherichia Coli and Staphylococci Isolates from the Poultry in Ile-Ife, Nigeria. J. Infect. Dis. Epidemiol. 4. doi: 10.23937/2474-3658/1510047
Johnson, J. R., Sannes, M. R., Croy, C., Johnston, B., Clabots, C., Kuskowski, M. A., et al. (2007). Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002-2004. Emerg. Infect. Dis. 13, 838–846. doi: 10.3201/EID1306.061576
Kabantiyok, D., Gyang, M. D., Agada, G. O., Ogundeji, A., Nyam, D., Uhiara, U. G., et al. (2023). Analysis of retrospective Laboratory data on the burden of bacterial pathogens isolated at the National Veterinary Research Institute Nigeria, 2018–2021. Vet. Sci. 10:505. doi: 10.3390/vetsci10080505
Kallau, N. H. G., Wibawan, I. W. T., Lukman, D. W., and Sudarwanto, M. B. (2018). Detection of multi-drug resistant (MDR) Escherichia coli and tet gene prevalence at a pig farm in Kupang, Indonesia. J. Adv. Vet. Anim. Res. 5, 388–396. doi: 10.5455/JAVAR.2018.E289
Kwoji, I., Musa, J. A., Daniel, N., and Mohzo, D. L. (2019). Extended-spectrum β-lactamase-producing Escherichia coli in chickens from small-scale (backyard) poultry farms in Maiduguri, Nigeria. Article in International Journal of One Health. 5, 26–30. doi: 10.14202/IJOH.2019.26-30
Laxminarayan, R. (2022). The overlooked pandemic of antimicrobial resistance. The Lancet, 399, 606–607. doi: 10.1016/S0140-6736(22)00087-3
Manyi-Loh, C., Mamphweli, S., Meyer, E., and Okoh, A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:795. doi: 10.3390/MOLECULES23040795
Massé, D. I., Saady, N. M. C., and Gilbert, Y. (2014). Potential of biological processes to eliminate antibiotics in livestock manure: an overview. Animals 4, 146–163. doi: 10.3390/ANI4020146
Mazhar, S. H., Li, X., Rashid, A., Su, J. M., Xu, J., Brejnrod, A. D., et al. (2021). Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 755:142702. doi: 10.1016/J.SCITOTENV.2020.142702
Mkpuma, N., Ebere, R. A., Kabantiyok, D., Isioma, D. C., Sati, N., and Meseko, C. A. (2021). Interfering microbes in virological processes and their antimicrobial resistance pattern. J. Pharm. Allied Sci. 18, 3530–3536. Available at: https://www.ajol.info/index.php/jophas/article/view/218547
Momtaz, H., Rahimi, E., and Moshkelani, S. (2012). Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Vet. Med. 57, 193–197. doi: 10.17221/5916-VETMED
Muhammad, J., Khan, S., Su, J. Q., Hesham, A. E. L., Ditta, A., Nawab, J., et al. (2020). Antibiotics in poultry manure and their associated health issues: a systematic review. J. Soils Sediments 20, 486–497. doi: 10.1007/s11368-019-02360-0
Nhung, N. T., Cuong, N. V., Thwaites, G., and Carrique-Mas, J. (2016). Antimicrobial usage and antimicrobial resistance in animal production in Southeast Asia: a review. Antibiotics 5:37. doi: 10.3390/ANTIBIOTICS5040037
Nnaji, J. O., Moses, I. B., Ejikeugwu, P. C., Nwakaeze, E. A., Ude-Ude, I., and Iroha, I. R. (2021). Antibiogram and molecular characterization of AmpC and ESBL-producing gram-negative Bacteria from poultry and abattoir samples. Pak. J. Biol. Sci. 24, 193–198. doi: 10.3923/PJBS.2021.193.198
O’Neill, J. (2019). Review on antimicrobial resistance: tackling drug-resistant infections globally: final report and recommendations. In Centre for Agriculture and Bioscience International. Available at: https://www.cabdirect.org/globalhealth/abstract/20163354200
Okorie-Kanu, O. J., Ezenduka, E. V., Okorie-Kanu, C. O., Ugwu, L. C., and Nnamani, U. J. (2016). Occurrence and antimicrobial resistance of pathogenic Escherichia coli and Salmonella spp. in retail raw table eggs sold for human consumption in Enugu state, Nigeria. Vet. World 9, 1312–1319. doi: 10.14202/VETWORLD.2016.1312-1319
Olatoye, I. O., Amosun, E. A., and Ogundipe, G. (2012). Contamination of beef and chicken in municipal abattoirs of Southwest Nigeria. Nat. Sci. Available at: https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=Olatoye+IO%2C+Amosun+EA%2C+Ogundipe+GAT.+Multidrug+Resistant+Escherichia+coli+O157+Contamination+%09of+%09Beef+and+Chicken+in+Municipal+Abattoirs+of+Southwest+Nigeria.+Nature+and+Science%2C+%09%28201
Osman, K. M., Kappell, A. D., Elhadidy, M., Elmougy, F., El-Ghany, W. A. A., Orabi, A., et al. (2018). Poultry hatcheries as potential reservoirs for antimicrobial-resistant Escherichia coli: a risk to public health and food safety. Sci. Rep. 8, 5859–5814. doi: 10.1038/s41598-018-23962-7
Puhr, R., Heinze, G., Nold, M., Lusa, L., and Geroldinger, A. (2021). Firth’s logistic regression with rare events: Accurate effect estimates AND predictions? Stat. Med. 36, 2302–2317. doi: 10.1002/sim.7273
Rahimi, P., Islam, M. S., Duarte, P. M., Tazerji, S. S., Sobur, M. A., El Zowalaty, M. E., et al. (2022). Impact of the COVID-19 pandemic on food production and animal health. Trends Food Sci. Technol. 121, 105–113. doi: 10.1016/J.TIFS.2021.12.003
Sabat, G., Rose, P., Hickey, W. J., and Harkin, J. M. (2000). Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl. Environ. Microbiol. 66, 844–849. doi: 10.1128/AEM.66.2.844-849.2000
Selaledi, L. A., Hassan, Z. M., Manyelo, T. G., and Mabelebele, M. (2020). The Current Status of the Alternative Use to Antibiotics in Poultry Production: An African Perspective. Antibiotics 9, 594. doi: 10.3390/ANTIBIOTICS9090594
Shecho, M., Thomas, N., Kemal, J., and Muktar, Y. (2017). Cloacael carriage and multidrug resistance Escherichia coli O157:H7 from poultry farms, eastern Ethiopia. J. Vet. Med. 2017, 1–9. doi: 10.1155/2017/8264583
Wang, G., Clark, C. G., and Rodgerst, F. G. (2002). Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40, 3613–3619. doi: 10.1128/JCM.40.10.3613-3619.2002
Wani, S. A., Samanta, I., Bhat, M. A., and Nishikawa, Y. (2004). Investigation of Shiga toxin-producing Escherichia coli in avian species in India. Lett. Appl. Microbiol. 39, 389–394. doi: 10.1111/J.1472-765X.2004.01586.X
WHO. (2021). Antimicrobial resistance. Available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance
Keywords: One Health, biosecurity, multidrug resistance, virulence genes, livestock, Escherichia coli
Citation: Agusi ER, Kabantiyok D, Mkpuma N, Atai RB, Okongwu-Ejike C, Bakare EL, Budaye J, Sule KG, Rindaps RJ, James GK, Audu BJ, Agada GO, Adegboye O and Meseko CA (2024) Prevalence of multidrug-resistant Escherichia coli isolates and virulence gene expression in poultry farms in Jos, Nigeria. Front. Microbiol. 15:1298582. doi: 10.3389/fmicb.2024.1298582
Edited by:
Ioana Cristina Marinas, University of Bucharest, RomaniaReviewed by:
Mabel Kamweli Aworh, North Carolina State University, United StatesSteward Mudenda, University of Zambia, Zambia
Copyright © 2024 Agusi, Kabantiyok, Mkpuma, Atai, Okongwu-Ejike, Bakare, Budaye, Sule, Rindaps, James, Audu, Agada, Adegboye and Meseko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ebere Roseann Agusi, roseebere8@gmail.com; Oyelola Adegboye, oyelola.adegboye@menzies.edu.au
 Ebere Roseann Agusi
Ebere Roseann Agusi Dennis Kabantiyok
Dennis Kabantiyok Nicodemus Mkpuma
Nicodemus Mkpuma Rebecca Bitiyong Atai2
Rebecca Bitiyong Atai2 Godwin Ojonugwa Agada
Godwin Ojonugwa Agada Oyelola Adegboye
Oyelola Adegboye Clement Adebajo Meseko
Clement Adebajo Meseko