- 1Department of General Ecology, Institute of Zoology, University of Cologne, Cologne, Germany
- 2Department of Biotechnology, Universidad de Antofagasta, Antofagasta, Chile
- 3Centre for Biotechnology and Bioengineering (CeBiB), Universidad de Antofagasta, Antofagasta, Chile
Introduction: Heterotrophic protists colonizing microbial mats have received little attention over the last few years, despite their importance in microbial food webs. A significant challenge originates from the fact that many protists remain uncultivable and their functions remain poorly understood.
Methods: Metabarcoding studies of protists in microbial mats across high-altitude lagoons of different salinities (4.3–34 practical salinity units) were carried out to provide insights into their vertical stratification at the millimeter scale. DNA and cDNA were analyzed for selected stations.
Results: Sequence variants classified as the amoeboid rhizarian Rhogostoma and the ciliate Euplotes were found to be common members of the heterotrophic protist communities. They were accompanied by diatoms and kinetoplastids. Correlation analyses point to the salinity of the water column as a main driver influencing the structure of the protist communities at the five studied microbial mats. The active part of the protist communities was detected to be higher at lower salinities (<20 practical salinity units).
Discussion: We found a restricted overlap of the protist community between the different microbial mats indicating the uniqueness of these different aquatic habitats. On the other hand, the dominating genotypes present in metabarcoding were similar and could be isolated and sequenced in comparative studies (Rhogostoma, Euplotes, Neobodo). Our results provide a snapshot of the unculturable protist diversity thriving the benthic zone of five athalossohaline lagoons across the Andean plateau.
Introduction
Microbial communities thriving in hostile environments, mainly composed of so-called extremophiles, represent hotspots of biodiversity and harbor invaluable genetic resources, which are the key to answering fundamental questions about the limits of life (Warren-Rhodes et al., 2006; Schulze-Makuch et al., 2018). Given the vast diversity of such communities, it is difficult to assess how they will respond to disturbances and to predict the possible functional consequences for the ecosystem. A significant challenge originates from the fact that many microbes remain uncultivable and their functions remain poorly understood (Solden et al., 2016). However, modern sequencing techniques allow for the exploration of the genetic pool of microbial communities, the study of their structure and functions, and the examination of biogeographic patterns in response to contemporary environmental factors and historical contingencies (Venter et al., 2017; Schiwitza et al., 2018, 2019, 2021; Arndt et al., 2020; Rybarski et al., 2023).
High-altitude lagoons in the Andean Altiplano (15°-22° S, up to 6.170 m a.s.l.) belong to endorheic basins and include diverse microbial ecosystems such as lagoons, brines, wetlands, and salars composed of plankton and benthos enduring hostile conditions (Albarracín et al., 2015; Aran et al., 2021). Part of these microbial guilds is formed by microbial mats, which colonize precipitating salts and rocks within such ecosystems. They interact with physical-chemical gradients, such as the varying mineral concentrations characteristic of Andean water bodies, distributed throughout the ancient Atacama Desert (Demergasso et al., 2003; Farías et al., 2014; Farias et al., 2017; Rasuk et al., 2014, 2016; Fernandez et al., 2016; Saghaï et al., 2017; Vignale et al., 2022). Microbial mats have been identified as important sites for greenhouse gas exchange and complex variable ecosystems in the athalossohaline Andean salt flats, linking these complex laminar assemblages to the cycling of carbon in the remote Altiplano plateau (Dorador et al., 2018; Molina et al., 2021). The microbiology of these stratified microhabitats has been linked mainly to primary producers (cyanobacteria) and is considered to be among Earth's earliest ecosystems (Margulis et al., 1980; Dupraz et al., 2009). Various Andean microbial mats have been studied, focusing on their prokaryotic communities, including archaea and a wide range of viruses, exhibiting changes along the vertical gradient (e.g., Dorador et al., 2008, 2013; Hernández et al., 2016; Eissler et al., 2019). Considering protists, unicellular eukaryotic biosignatures have mainly been described from lower altitudes (Salar de Llamará, 750 m a.s.l.), in saline microbial mats and hypersaline lagoons harboring biosignatures classified as diatoms, bicosoecids, and ciliates as detected by metabarcoding (Saghaï et al., 2017; Rybarski et al., 2023). Efforts to isolate and sequence the cultivable and uncultivable protist diversity from saline to hypersaline lagoons unveiled a novel endemic diversity including species from Opisthokonta (choanoflagellates), Rhizaria (Cercomonadida), Discoba (percolomonads), Stramenopiles (bicosoecids and placidids), and Alveolata (ciliates) (Schiwitza et al., 2018, 2019, 2021; Arndt et al., 2020; Carduck et al., 2021; Rybarski et al., 2021, 2023; Schoenle et al., 2022; Acosta et al., 2023; Hohlfeld et al., 2023).
The increasing focus on protists all over the planet has unveiled a heterogeneous and variable diversity that can be found across different biotopes (terrestrial to aquatic), unveiling biogeographical patterns even within ecosystems (Dillon et al., 2009; Mahé et al., 2017; Schoenle et al., 2021; Singer et al., 2021; Rybarski et al., 2023). Protists have been found to follow the vertical stratification characteristics in microbial mats, assembling differentially relative to vertical physic-chemical gradients (Saghaï et al., 2017). Under the current climate change scenario, water availability is being threatened, highlighting the importance of studying the structure of protist communities living in delicate ecosystems such as the Andean microbial mats. In this study, we assess the structure of these communities at the vertical millimeter scale in the microbial mats of five Andean Lagoons using metabarcoding and explore their differences and their interaction with the overlying water column.
Materials and methods
Sample collection and molecular biology procedure
Microbial mats were sampled from the sediments below the water line in five lagoons at different altitudes across the Chilean Altiplano (Table 1). The structure of the studied microbial mats differs visibly throughout the studied sites (Figure 1B). Microbial mat layers characterized by an upper green layer were found in Helada Lagoon, Amarilla Lagoon, and Salada Lagoon, while in Quepiaco Salar, microbial mats are covered by a layer of salt and the upper green layer is thinner. In contrast, the microbial mat sampled from Aguas Calientes Salar exhibits a structure of orange and red layers. Biological samples of the microbial mats were dissected in situ using sterile metalware and coveralls to maintain cross-contamination to a minimum. The microbial mats were dissected to obtain three biological samples from each layer, starting from the upper layer (first millimeter), the second layer (3 mm depth), and a third layer (5 mm depth; Figure 1). The environmental parameters of the water column, such as salinity (Practical Salinity Units, PSU), resistivity (Ohm/cm), conductivity (mS/cm), and total dissolved solids (parts per trillion), were measured using an Orion Star A322 Conductivity Portable Multiparameter (Thermo Scientific) (Table 1). The sediment and water temperatures were measured using a HI935002 Dual Channel K-Type Thermocouple Thermometer (Hanna Instruments). Each replicate was homogenized in RNA later to preserve the DNA and RNA from each sample (Wang et al., 2018) in sterile 2-ml Eppendorf tubes and stored at −20°C until further analyses. The total DNA and RNA were extracted from samples using Trizol (Invitrogen), according to the manufacturer's protocol with minor modifications: Samples were homogenized in 1 ml of Trizol reagent for 10 min as a cell lysis step using a vortex adaptor. The final DNA was resuspended in 100 μl of nuclease-free water and used immediately for the amplification of the target gene. RNA was resuspended in 50 μl of nuclease-free water for downstream analyses. Total RNA was used as a template for cDNA synthesis using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). Total concentrations of DNA, RNA, and cDNA were measured using the ND-1000 nanodrop (Peqlab Biotechnologie, Germany).
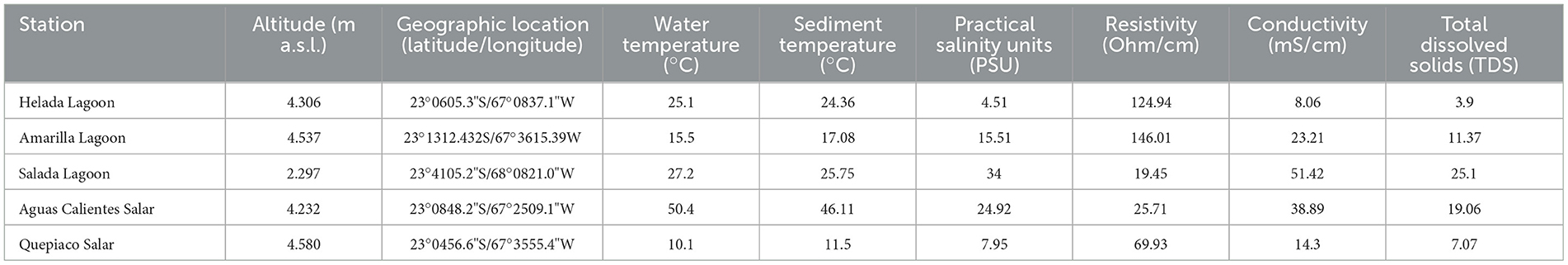
Table 1. Information of the stations indicating geographical and physic-chemical properties at each station.
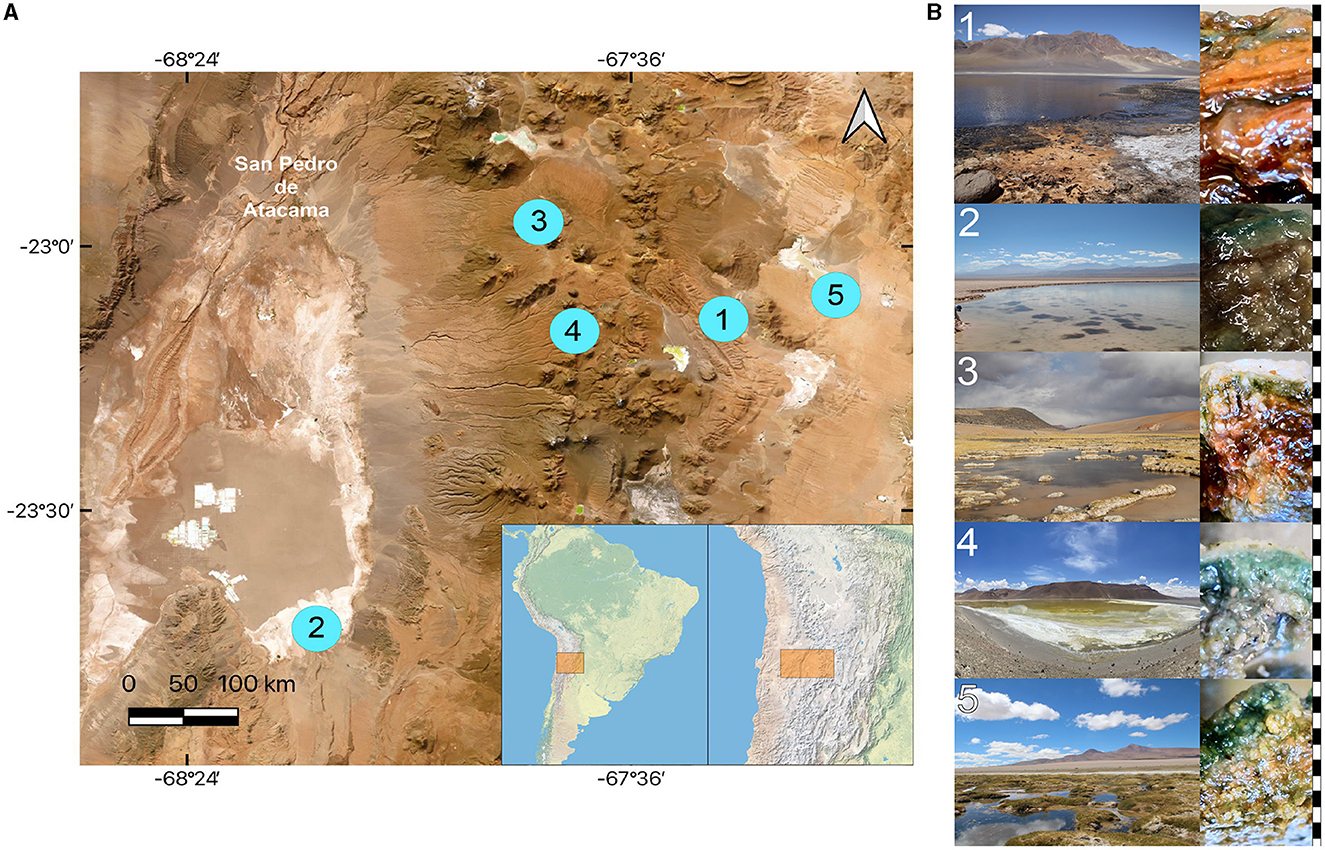
Figure 1. Sampling sites of microbial mat studies in the Atacama Desert. (A) Map showing the location of the stations. (B) Study sites and vertical structure of the corresponding microbial mats. 1, Aguas Calientes Salar; 2, Salada Lagoon. 3, Quepiaco Salar; 4, Amarilla Lagoon; 5, Helada Lagoon. Right side: scale in black and white indicating one millimeter intervals. Map produced and adapted from QGIS 3.28.5 - Firenze.
Mock community preparation and sequencing
Aiming to assess the accuracy of the used classification method for sequencing libraries, we included an artificial sample (a mock community). This sample was composed of equimolar concentrations of the PCR-amplified V9 region from eight species from the Heteroflagellate Collection Cologne (HFCC) (Schoenle et al., 2021). The control samples included the V9 region (rDNA) amplicon aliquots of strains from the six protist supergroups Alveolata, Ancyromonadida, Rhizaria, Discoba, Stramenopiles, and Opisthokonta: Protocruzia sp. (MT355146), Aristerostoma sp. (MT081566), Fabomonas tropica (MT355148), Massisteria sp. (MT355122), Rhynchomonadidae sp. (MT355133), Cafeteria burkhardae (MN315604), Bicosoecida sp. (MT355117), and Ministeria vibrans (MT355150) (Supplementary Table 1). For this, the DNA was extracted from the strains by transferring clonal cultures (30 ml) into 50-ml centrifuge tubes (Sarstedt, Nümbrecht, Germany) and centrifuging at 4,000 × g at 4°C for 20 min. After discarding the supernatant, the DNA was extracted from the cellular pellet using the Quick gDNA Mini-Prep Kit (Zymo Research Corporation, United States) following the manufacturer's instructions. An aliquot of 3 μl of DNA from each strain was used as a template in a PCR reaction using 5 μM of the universal primer set 18S: Forward (5-AACCTGGTTGATCCTGCCAGT-3) and Reverse (5-TGATCCTTCGCAGGTTCACCTAC-3) (Medlin et al., 1988). The PCR conditions were the following: a denaturation step at 96°C for 120 s, then 34 cycles of 96°C for 30 s, 55°C for 45 s, 72°C for 150 s, and a final elongation step for 420 s at 72°C. The PCR products were purified using the FastGene Gel/PCR Extraction Kit (Fast Gene, Japan), and the sanger was sequenced using the 18S rDNA primer sets at GATC Biotech, Germany. The assembled sequences of the 18S rDNA gene were analyzed using the Basic Local Alignment Search Tool (RRID:SCR_004870).
Amplification of the hypervariable region V9 SSU rDNA and next-generation sequencing
The aliquots of the DNA and cDNA from environmental samples as well as from the DNA isolated from the strains for the mock community were used as a template for the amplification of the hypervariable region V9 of the small sub-unit 18S rDNA. The amplification was carried out using the multiplex identifier-tagged (MIDs) primers 1389F and 1510R, following the PCR protocol of Amaral-Zettler et al. (2009). The PCR conditions were the following: 98°C for 30 s; 25 cycles of 10 s at 98°C, 30 s at 57°C, 30 s at 72°C, and a final step at 72°C for 600 s. The PCR products were purified using the FastGene Gel/PCR Extraction Kit (Fast Gene, Japan), and equimolar concentrations of all purified PCR products were pair-end sequenced (2 × 150 bp) in multiplexed libraries using an Illumina NovaSeq 6000 sequencer (Illumina, United States) at the Cologne Center for Genomics (CCG).
Bioinformatic analyses
Multiplexed libraries of the reads obtained through the metabarcoding of DNA and cDNA were processed in QIIME2 (RRID:SCR_021258, Bolyen et al., 2019). The MIDs and primer sequences were removed from the libraries using the “cutadapt trim-paired” function, and sequences with an error rate higher than 0 were filtered. The raw forward and reverse reads were trimmed to exclude the primer sequences, dereplicated, and denoised into amplicon sequence variants (ASVs), using the “denoise-paired” function from the DADA2 plugin (RRID:SCR_023519, Callahan et al., 2016). This method includes a chimera-filtering step on the ASVs using the “consensus” chimera detection method on all samples, and sequences found chimeric in a sufficient fraction of samples were removed (method: isBimeraDenovo). To discern between protist taxa, the ASVs were classified using a cut-off value of 0.8 and a 97% identity to the database through the function “feature-classifier classify-consensus-blast” (Camacho et al., 2009). For this, we used the Protist Ribosomal Reference (PR2) database version 4.14.0, amended with the V9 region of the 18 SSU rRNA gene sequences from the Heterotrophic Flagellate Collection Cologne (HFCC) (Schoenle et al., 2021). The ASVs classified as metazoa, archaeplastida, fungi, or unassigned were excluded from further analyses.
Statistics and reproducibility
The alpha and beta diversity metrics were obtained through the function “qiime diversity core-metrics-phylogenetic”, which relates to the lowest number of sequences. The difference in the alpha-diversity metrics (observed number of ASVs, the Shannon index) across the study sites was tested in a Kruskal-Wallis test for paired groups (study site-study site and DNA-cDNA) using the function “qiime diversity alpha-group-significance”. Additionally, we tested the dissimilarity in beta diversity metrics (Unifrac metrics, Jaccard, and Bray-Curtis distances) across study sites in a permutational analysis of variance (PERMANOVA, 999 permutations) through the function “qiime diversity beta-group-significance”. The output files, such as the frequency table, the sequences table, the phylogenetic tree, the assigned taxonomy, and the metadata used in QIIME2, were imported into R (version 4.3.0) and processed using the “microeco” package version 1.1.0 (Liu et al., 2021). The bar plots showing the relative abundance of the major protist lineages across study sites, as well as clustering patterns based on Euclidean distances, were plotted using the “plot_bar”, setting the clustering option “TRUE”. The beta diversity patterns observed through Jaccard distance were analyzed in a principal coordinate analysis (PCoA) for the DNA and DNA-cDNA-based protists community (97% of identity to the database), using the functions “cal_ordination” and “plot_ordination”. Venn diagrams were generated to depict the number of shared and unique ASVs across microbial mat layers. Furthermore, a canonical redundancy analysis (RDA) with feature selection was used to evaluate the association between the environmental parameters measured in the water bodies (independent variables) and the protist taxa (dependent variables), detected through DNA and cDNA, respectively. Due to the presence of null values, the data was normalized, scaling the abundance of the ASVs to unit variance and adding “scale = TRUE” in the “cal_ordination” function.
Results
Overall, all the studied water bodies are mesohaline, ranking from 4.51 PSU (Helada Lagoon) up to 34 PSU (Salada Lagoon, Atacama Salar). Other parameters measured in the water column are shown in Table 1.
Next-generation sequencing analysis
It was planned to have DNA and cDNA data sets from all sampling sites. However, cDNA could not be amplified from all stations. We successfully amplified the target phylogenetic marker mainly from the upper layers of a total of 23 microbial mat samples, distributed as follows: four samples from Helada Lagoon, three from the first layer and one from the second layer; a total of seven samples from Quepiaco Salar, three from the first layer, three from the second layer, and one from the third layer; a total of five samples from Amarilla Lagoon, three samples from the first layer and two samples from the second layer; from Aguas Calientes Salar, a total three samples from the first layer; and from Salada Lagoon, three samples from the first layer. We amplified the target gene from three out of five study sites in a total of 15 samples for cDNA. From Helada Lagoon, there were a total of four samples: one from the first layer, two from the second layer, and one from the third layer. From Quepiaco Salar, there were a total of seven samples: three from the first layer, three from the second layer, and one from the third layer. From Amarilla Lagoon, there were a total of four samples: two from the first layer and two from the second layer.
A total of over 54.6 million raw forward and reverse reads were obtained from the DNA metabarcoding of the five studied microbial mats, of which over 53.1 million (97.2%) were barcode-clipped. After quality control and denoising, a total of over 41.8 million sequences associated with 32,089 ASVs were obtained. For cDNA, a total of 24.7 million raw forward and reverse reads were obtained from three out of five of the studied microbial mats (Helada Lagoon, Amarilla Lagoon, and Quepiaco Salar). From these, ~17.5 million (70.82%) were barcode-clipped, and after quality control and denoising, we obtained a total of ~13.9 million sequences associated with 19,134 ASVs. As we included an artificial control sample of known composition (the so-called mock community), we could assess the accuracy of the taxonomic classification method used in this work. The sequences for each of the eight taxa of the mock community that could be assigned to the respective taxon contributed 3.1–24.4% to the total abundance of reads, using a diversity metric commonly used in environmental sequencing assessments (97% of identity to the database). Additionally, we detected low-frequency ASVs classified as taxa that were not included in the mock community. Such taxa were found at frequencies ranging from 0.032 to 0.047% in the different sequencing libraries. These percentages were used as a threshold for each library preparation, aiming to exclude potential spurious sequences of low abundance.
After applying the threshold from the mock community, our environmental samples include a total of over 37.7 million sequences (90.3% of the total sequences) associated with 4,868 ASVs (15.17% of the total ASVs), remaining from the DNA metabarcoding (Table 2), and a total of over 13.2 million sequences (94.8% of the total sequences) associated with 4,835 ASVs (25.2% of the total ASVs) remained from the cDNA metabarcoding. Using a 97% identity to the database, we identified 308 ASVs associated with 42 lineages (6.3% of the remaining ASVs after filtering) from the DNA metabarcoding, and these were associated with over 14.2 million sequences (17.6% of the remaining sequences after filtering). From the cDNA metabarcoding, we identified a total of 386 ASVs associated with 41 lineages (7.9% of the remaining ASVs after filtering), which were associated with 747,027 sequences (5.6% of the remaining sequences after filtering).
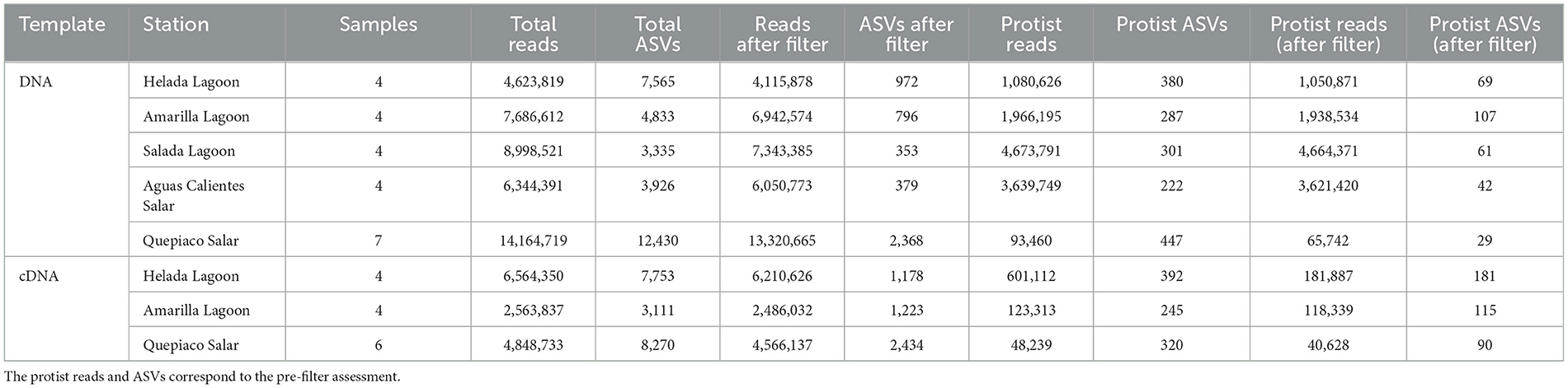
Table 2. The amounts of sequences and ASVs of DNA and cDNA origin obtained at critical steps of the bioinformatic analysis used in this study.
Protist community structure in Andean microbial mats
The protist communities determined by the DNA metabarcoding were mainly dominated by Rhizaria (15.1–95.6% of the reads), Stramenopiles (0.5–69.4%), Alveolata (5–22.3%), Amoebozoa (0.9–11.3%), and Haptista, which occurred only at Quepiaco Salar (61.5% of the reads) (Figure 2). The dominant taxa detected through cDNA metabarcoding (three study sites) were Stramenopiles, ranging between 19.8% of reads for Quepiaco Lagoon, 34.4% for Helada Lagoon, and up to 59.5% for Amarilla Lagoon. Other important taxa detected by cDNA metabarcoding were Rhizaria (22–51.6% of the reads), Alveolata (6.3–11.9%), and Discoba (0.4–5.5%). Further information on the relative abundance of protist taxa that could be detected across the stations studied by DNA and cDNA is shown in Figure 3.
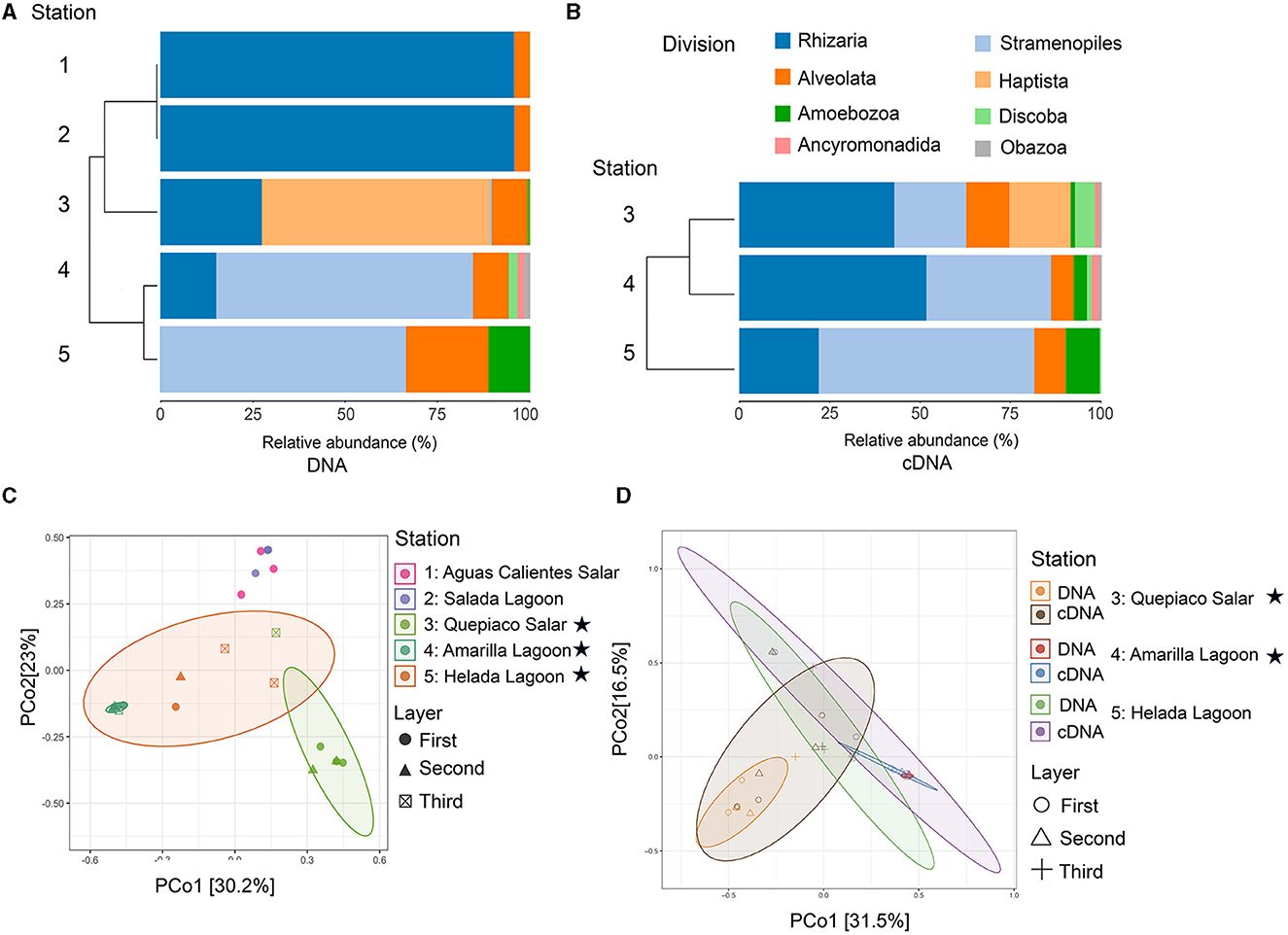
Figure 2. Diversity of protist taxa in microbial mats based on NGS data. (A) Cluster analysis and relative abundance of the major protist lineages detected through DNA (identity to the database > 97%) across the five stations. (B) Cluster analysis and relative abundance of the major protist lineages detected through cDNA (identity to the database > 97%) across the study sites 3, 4, and 5. Stations, 1, Aguas Calientes Salar; 2, Salada Lagoon; 3, Quepiaco Salar; 4, Amarilla Lagoon; 5, Helada Lagoon. (C) Principal coordinates analysis of the Jaccard distances based on DNA. (D) Principal coordinates analysis of the Jaccard distance between protists lineages detected through DNA and cDNA for three stations. Stars indicate stations with protist communities being significantly different from each other and from the non-labeled stations (for details see text).
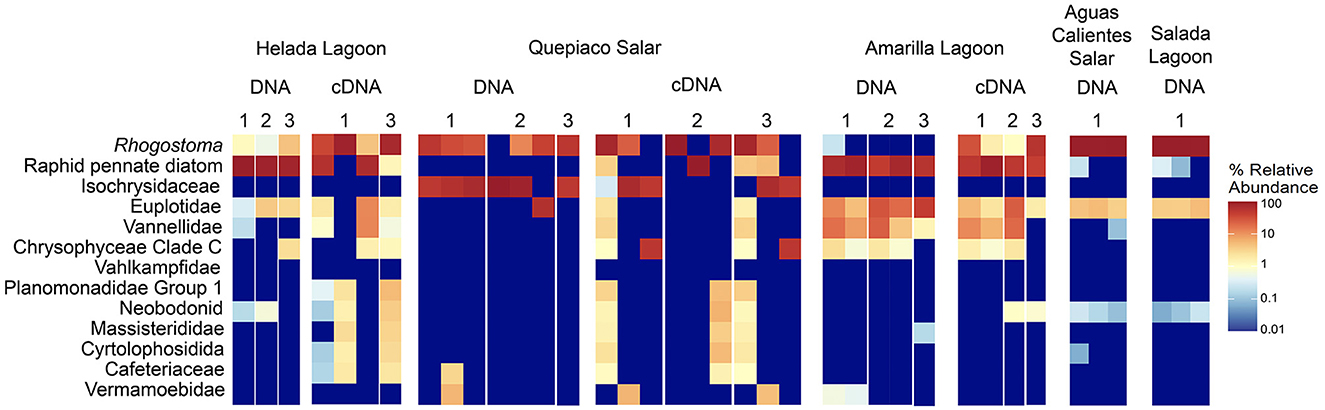
Figure 3. Heatmap of the relative abundance of the top 13 taxa detected in the studied layers from microbial mats based on DNA or cDNA.
The analysis of ASVs of the DNA data sets shared between the different stations revealed a relatively low number either when comparing the different sites or when comparing different layers (Figure 4). The relative number of shared sequences is relatively similar when comparing unfiltered filter data sets or when a rigid (mock) filter is applied.
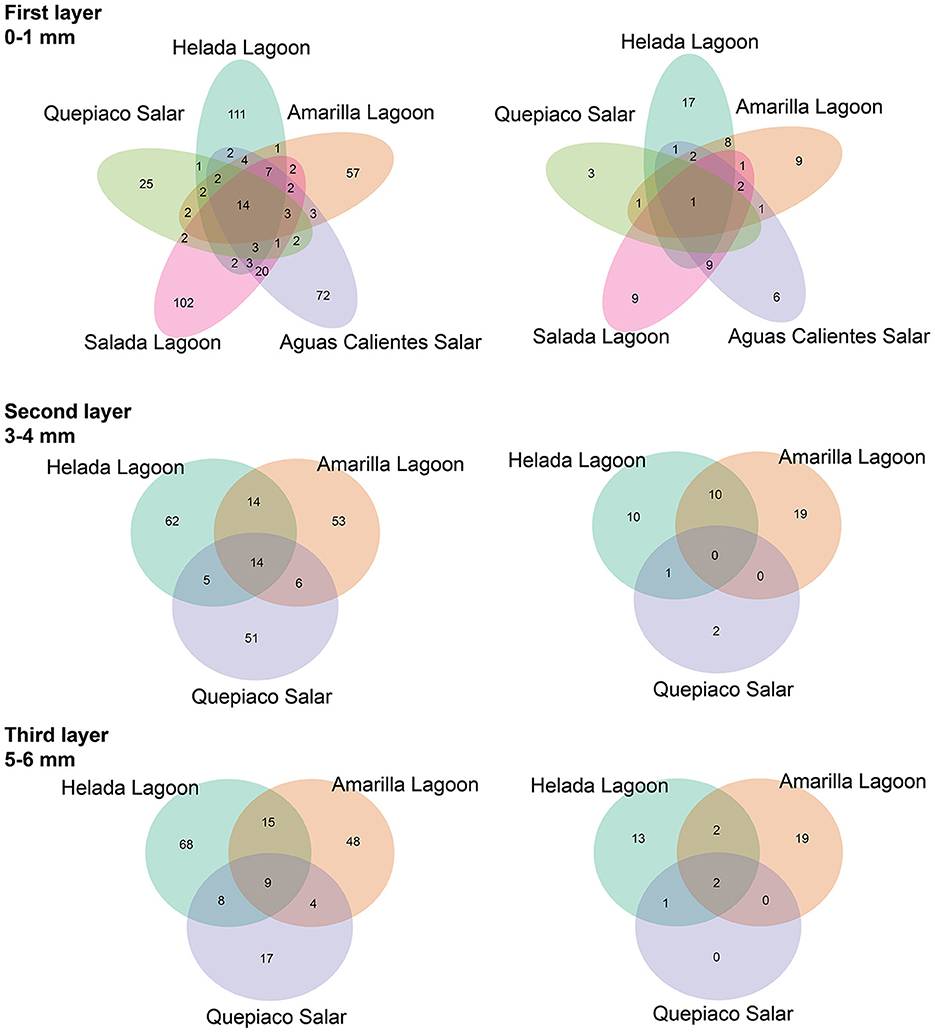
Figure 4. The shared amplicon sequence variants (ASVs) at each layer between stations are detected by DNA metabarcoding. (Left) Data without rigid filtering. (Right) Data filtered using the mock community-deduced threshold.
Alpha diversity across the five Andean microbial mats
The overall mean number of ASVs was 10.8 with a standard deviation (SD) of 7.1, with the highest mean ASVs obtained from Amarilla Lagoon (mean = 20.2, SD = 1.1) and the least from Quepiaco Salar (mean = 2.5, SD = 1.1) (Figure 5A). The overall mean Shannon diversity obtained was 1.2 with a standard deviation of 1.4, with the highest value for Amarilla Lagoon (mean = 3.3, SD = 0.2) and the lowest for Aguas Calientes Salar (mean = 0.5, SD = 0.06). The overall phylogenetic diversity mean value was 15.7 with a standard deviation of 5.3, with the highest mean value found for Amarilla Lagoon (mean = 18.6, SD = 6.05) and the lowest for Quepiaco Salar (mean = 10.7, SD = 0.6). We tested the dissimilarity between the groups of protists detected across the study sites, considering the observed ASV numbers and Shannon diversity calculated from the DNA as well as from cDNA data sets. The amount of observed ASVs was significantly different between Amarilla Lagoon and all other study sites, and that of Quepiaco Salar was also significantly different from all the other study sites (Figure 5A). When we compared the study sites regarding the Shannon index, significant differences were found between Quepiaco Salar and Helada Lagoon on the one side and the other three study sites on the other side (Figure 5A). Regarding the results of the cDNA metabarcoding data for the three study sites investigated, there were no significant differences regarding the number of observed ASVs, while the data from Quepiaco Salar differed from the other two study sites regarding the Shannon index. The cDNA values for one station clustered very similar to the respective DNA values when compared using the principal coordinate analysis (Figure 2D).
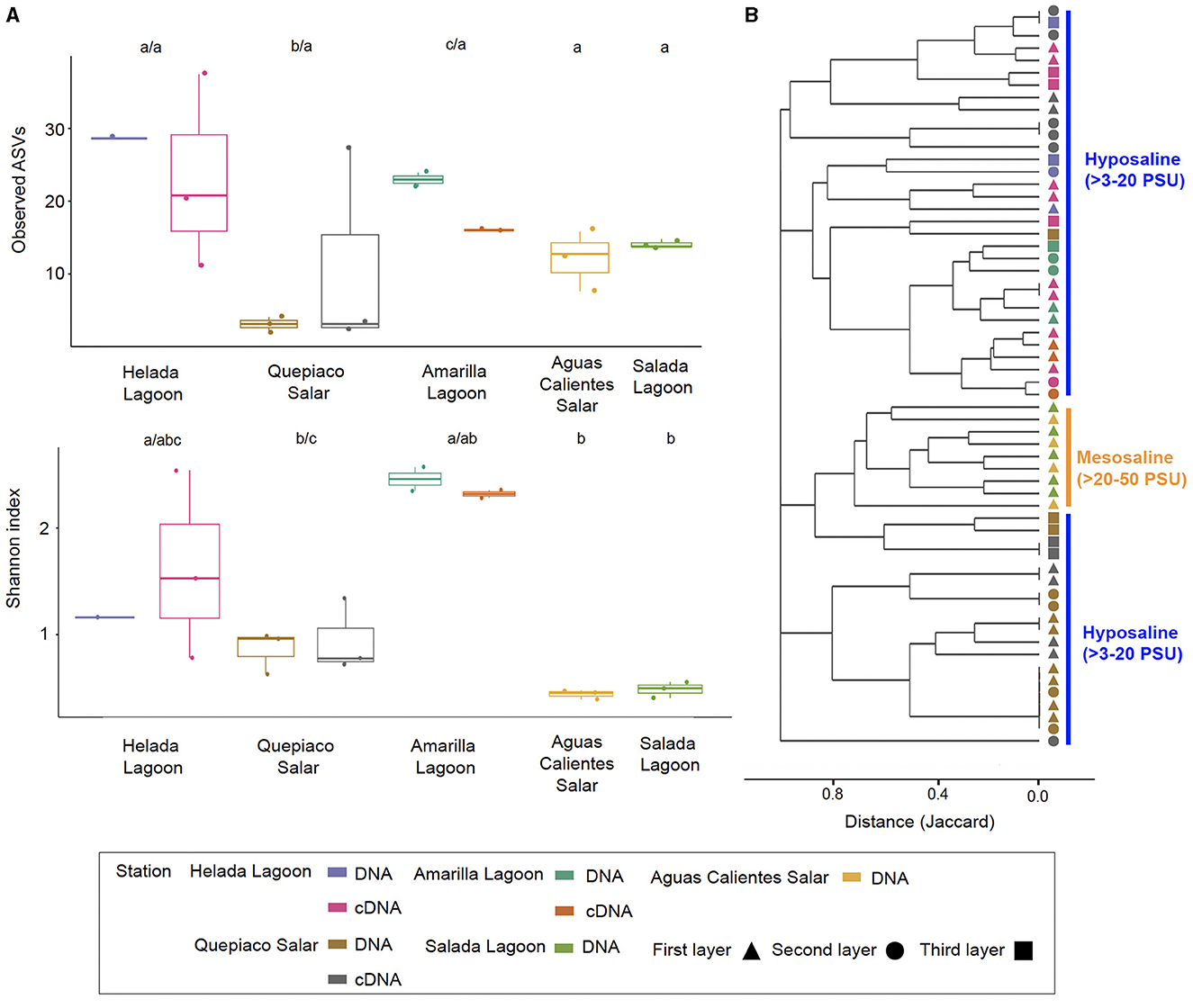
Figure 5. Alpha and beta diversity metrics were measured across the five stations. (A) Box and whiskers plots of the observed ASVs and the Shannon index measured in the first layer of microbial mats across stations. Different letters above the boxes mark significant differences, before the slash for DNA, and behind for cDNA data (Kruskal-Wallis, p < 0.05). (B) Cluster analysis based on the Jaccard distances between protist communities from each layer of microbial mats among stations (p < 0.05 for values >0.8). Salinity ranges are indicated on the right side.
The structure of protist communities detected through DNA and cDNA
The dissimilarity measured between the studied layers across stations is presented in a cladogram based on the Jaccard distance, which indicates four main groups (Figure 5). Two groups were composed of samples from Quepiaco Salar (DNA and cDNA), Aguas Calientes Salar, and Salada Lagoon (DNA), and two groups were composed of Helada Lagoon, Amarilla Lagoon, and Quepiaco Salar. This last group includes samples obtained from both DNA and cDNA from Helada Lagoon and Amarilla Lagoon and samples obtained from cDNA at Quepiaco Salar (Figure 5). The principal coordinate analysis (PCoA; Figure 2C) explains 53.2% (PCo1 = 30.2%, PCo2 = 23%) of the dissimilarity between the five study sites (DNA). On the other hand, the protist communities analyzed by the DNA and cDNA for three study sites (Figure 2D) showed a pattern of dissimilarity explained by 48% (PCo1 = 31.5%, PCo2 = 16.5%). However, the differences between the communities detected through DNA and cDNA within samples from one station were not significant in all three investigated cases.
Association of the abiotic factors and the protist communities
The results of the redundancy analysis (RDA), including the protist lineages of the DNA metabarcoding data sets in the first layer across the five stations, revealed significant correlations (Figure 6). On the other hand, the analysis of cDNA data detected no significant association between the protist taxa and environmental factors. The interpretation rate of the RDA for DNA metabarcoding was equal to 79.6% (RDA1 = 48.1%, RDA2 = 31.5%). The resistivity (R2 = 0.8818, Pr = 0.001), the conductivity (R2 = 0.7259, Pr = 0.003), and the salinity (R2 = 0.7231, Pr = 0.003) of the water bodies were selected as significant factors (p < 0.01) associated with the protist composition of the studied microbial mats. Furthermore, the protist taxa associated with these environmental parameters were chlorophytes, Lobosa (Amoebozoa), Cercozoa (Rhizaria), Ciliophora, Apicomplexa and Dinoflagellata (Alveolata), Ochrophyta, Haptophyta, Discicristata, and one unclassified division of Stramenopiles.
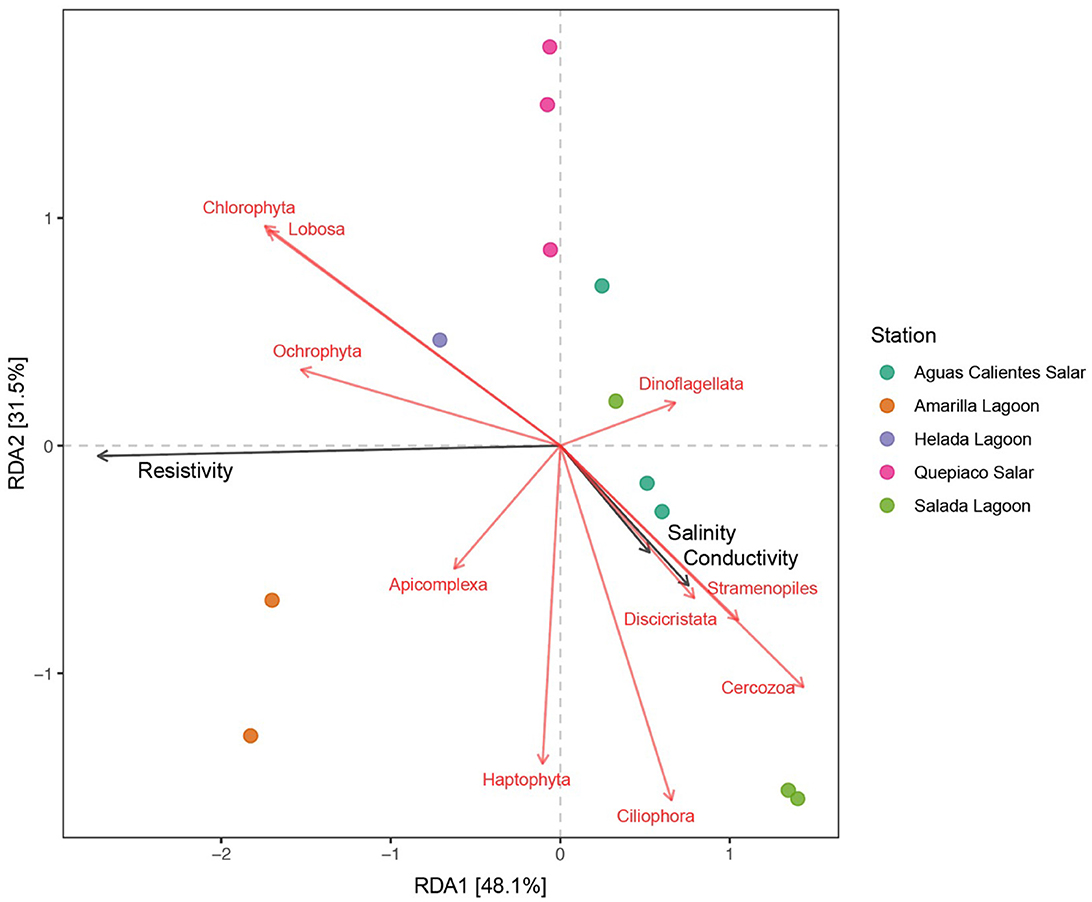
Figure 6. Canonical redundancy analysis (RDA) of the protist genotypes detected in the upper layer of the studied microbial mats (identity to the database > 97%). Black arrows represent the environmental variables that have a significant effect on the studied genotypes and the red arrows represent the genotypes associated with the environmental variables.
Discussion
In this study, we delve into the ecology of protists in the microbial mats of five Andean Lagoons with different salinities, characterizing the structure of their communities through the amplification of the V9 region (18S SSU). The primary goal was to assess, at the millimeter scale (Figure 1), the potentially resting stages as well as the active protist communities detected through the metabarcoding of DNA and cDNA in five stations. Partly, this could not be resolved, as the amplification of the target gene was successful mainly from the upper layers and from the DNA and cDNA templates only in three stations (Figure 3). Future studies aiming to study the stratification and activity of protist groups in Andean microbial mats should consider the use of both hypervariable regions (V4 and V9), as recommended for comprehensive diversity studies using environmental sequence variants (Choi and Park, 2020). Nevertheless, with our study, we could determine the ASVs shared across all stations at each studied layer (Figure 4). The restricted overlap between the different microbial mats indicates the specificity of the different aquatic habitats. The reasons for this specificity could not be fully explained in the course of the present study; however, salinity might be an important driver (see below). Overall, phototrophs and amoebae are frequent in the uppermost layers, and ciliates are common throughout all depths (DNA metabarcoding). The active protist groups (cDNA) were mainly diatoms shared between stations at the first (0–1 mm) and second (3–4 mm) layers, while Euplotes was the only genotype detected as active and shared by the deepest layer (5–6 mm). The functional diversity of microbes can vary across the layers of microbial mats due to changes in water and light availability, particularly for oxygen producers such as the cyanobacteria and diatoms studied in the saline lake of the Atacama Desert, the Salar de Llamará (Demergasso et al., 2003). Such variations could be responsible for the observed differences in the vertical structure of protist communities. As initially hypothesized, the studied ecosystems appeared to be highly underexplored regarding protists lineages. ASVs classified as eukaryotes accounted for < 1% of the total sequences (at 97% identity in databases). Moreover, using a lower identity threshold (80% of identity to the databases), only up to 7.7% of the total sequences could be assigned to protist lineages. This supports the hypothesis of a high novelty in such remote ecosystems. This is in agreement with Burki et al. (2021), who suggest that such remote and underexplored environments could constitute an untapped reservoir of potential new biodiversity. Our RDA analysis revealed that factors such as salinity, conductivity, and resistivity were correlated with the detected benthic communities, which included cercozoans, ciliates, lobose amoebae, diatoms, and kinetoplastids (Figure 6). Among these, salinity is a factor often related to desiccation, shaping the community structures of microbial communities under water fluctuations (Rothrock and Garcia-Pichel, 2005; Lozupone and Knight, 2007). Other abiotic factors, such as conductivity, solar radiation, and temperature, have been associated with the community composition of microbes in hypersaline lagoons, mainly linking salinity to microbial abundance (Bryanskaya et al., 2016). Based on our results, we underscore the potential of high-throughput sequencing approaches to explore the structure of protists communities thriving in delicate and complex habitats and the limitations of the study of protists living at elevated salinity.
One essential component of biodiversity surveys using metabarcoding has been the use of an artificial sample (a mock community), aiming to identify spurious sequences (Schloss et al., 2011). This has already been implemented in the study of aquatic ecosystems throughout the Atacama and the search for patterns at the community level without blurring the diversity patterns (Rybarski et al., 2023). In our study, we could assess the taxonomy of the artificial samples in three sequencing libraries for the studied microbial mats and could define a frequency threshold deduced from the frequency of reads with low abundances. We used the frequency of these reads to filter ASVs of small abundance in our environmental samples (frequency < 0.032–0.047%), affecting mainly the ASV numbers (Table 2). Approximately 9.6–18.9% of the total ASVs and between 82 and 95.2% of sequences passed this filter and were included in the diversity assessment in the downstream analyses. The correct identification of the eight strains in the mock communities additionally supported the classification method for the study of our environmental samples. With these results, we confirm the use of mock community-deduced thresholds as the backbone of studies on protist communities in different environments through metabarcoding (e.g., Fiore-Donno et al., 2019; Dünn and Arndt, 2023; Sachs et al., 2023).
Alpha diversity and taxonomy of protists across five microbial mats
Studies on the protist diversity using metabarcoding in the Atacama Desert and the Andean region are scarce. Our results are comparable to one study on the protists diversity (hypervariable region V4) in microbial mats from the hypersaline lagoons in Llamará Salar (Saghaï et al., 2017). We can also compare our work to one study of heterotrophic flagellate communities (hypervariable region V9), detected in the waters of diverse salars across the Atacama (Rybarski et al., 2023). Those authors found a diversity of protists, reaching a minimal amount of six and up to 214 OTUs per study site. In our study, we registered lower numbers ranging between 2 and 20 ASVs per replicate. The amount of shared and unique biosignatures per station was similar (our study: 1–11 ASVs; Rybarski et al., 2023: 1–44 OTUs). Our samples reached a mean number of ASVs that could be confirmed by cDNA studies at the referred stations, reaching similar values at each station (e.g., Figure 5). Furthermore, our analysis of the shared and unique communities, including raw sequences, showed similar numbers of observed shared and unique genotypes as obtained by Rybarski et al. (2023). The registered diversity in Llamará Salar microbial mats (Saghaï et al., 2017) is low at stations of higher salinity (Simpson diversity index 0.93). In accordance with this, our results showed that the protist communities detected through DNA and cDNA metabarcoding include a low diversity at stations with elevated salinity (Figure 5).
At the millimeter scale, we detected abundant raphid diatoms, mainly in the upper layers of the microbial mats in the lagoons of Helada and Amarilla. In the active community assessment (cDNA), they appear abundant in the upper layers (Supplementary Figure 2). Phototrophs are the main builders of microbial mats, contributing to their formation through the excretion of exopolymeric substances (EPSs) and by facilitating carbonate accumulation in organo-mineralization processes. In our dataset, we identified Amphora, Navicula, and Nitszchia as primary producers, which agrees with the findings from Saghaï et al. (2017). Their study documented a high occurrence of diatoms in the upper layers of microbial mats in another nearby Atacama saline lake, studied at the centimeter scale, as well as in other Andean microbial mats (Farías et al., 2014; Albarracín et al., 2015; Vignale et al., 2022). Among stramenopiles, we also detected ASVs closely related to genotypes of marine diatoms (e.g., Skeletonema, Thalassiosira, Phaeodactylum), freshwater diatoms (e.g., Cymbella, Pseudogomphonema), chrysomonads (e.g., Paraphysomonas, Ochromonas) and diverse unclassified raphid pennate diatoms. Saghaï et al. (2017) also found stramenopiles accompanied by choanoflagellates, bicosoecids, and ciliates, with small numbers of OTUs dominating the abundance of the protist communities. In deep layers, they detected amoebae, with Conosa reaching almost 10% of the relative abundance of protists. Interestingly, in our study in the deepest layer of Quepiaco Salar, amoebae were also detected as being active and abundant (Supplementary Figure 2, Quepiaco Salar cDNA). We assessed the shared communities of protists throughout the first, second, and third layers across the stations (Figure 4). In the first layer, a common amoeba in the DNA and cDNA sets is the thecate amoeba Rhogostoma. Other taxa detected through cDNA in the first layer (Helada Lagoon, Amarilla Lagoon, and Quepiaco Salar) include Amphora, Euplotes, Vannella, and raphid pennate diatoms. No shared protists were found in the second layer; however, Euplotes, Vannella, and Navicula genotypes occurred in the unfiltered data set among the shared taxa (Helada and Amarilla Lagoons, Quepiaco Salar). At the deepest layer, a genotype of the ciliate genus Euplotes appeared to be shared in both DNA and cDNA data sets (Supplementary Figure 2), being abundant and represented by highly similar ASVs (one nucleotide difference, Supplementary Table 3). One ASV was 100% identical to a strain isolated from the Amarilla Lagoon (HFCC988, Euplotes sp.). This speciose genotype, or at least a very similar one, was also reported from Llamará Salar by Saghaï et al. (2017).
A typical protistan genotype we found at the different stations belonged to the genus Rhogostoma, a common protist genotype that we identified through DNA and cDNA analysis and occurred mainly in the uppermost layers of the microbial mats. Species of this genus are common inhabitants of aquatic and terrestrial ecosystems (Öztoprak et al., 2020). Although we cannot indicate the presence of this genotype as a species, we can trace our result to one cercozoan genus registered earlier as an arboretum inhabitant (Howe et al., 2011) and recently to a species member of the phyllosphere of endemic cacti in the Atacama (Acosta et al., 2023). This thecofilose amoeba, Rhogostoma, likely predates phototrophs as the diatoms (e.g., Amphora) are detected in parallel. Other remarkable protist lineages are those taxa detected in Quepiaco Salar, represented by Cafeteria, Isochrysis, and Hartmannella, of which the first two include marine strains (Bendif et al., 2013; Schoenle et al., 2020). The occurrence of the genus Cafeteria has already been reported from the Atacama Desert (Schoenle et al., 2022).
Factors associated with the benthic protists
The clustering patterns of the detected protist communities are significant based on the distance metrics shown in our PCoA analyses (Figure 2). The active DNA pool (cDNA) differed between the stations but corresponded to the total DNA pool of the respective station. However, we found, at least in part, indications for layer specificity in the activity of protists (Figure 5). Phototrophs were mainly detected in the upper layers, which supports the existence of a vertical gradient reflected in the studied distribution patterns.
Furthermore, we found that salinity plays an important role in the community structure of protists registered by metabarcoding for the different microbial mats. Differences in the salinity of the water bodies originated from different catchment areas (Ritter et al., 2018), explaining in part the uniqueness of environmental conditions in each salar/lagoon and the differences in community structure (Figure 6). The salinity is specifically tolerated by protist species from various phylogenetic lineages isolated from hypersaline lagoons and salars of the Atacama (Schiwitza et al., 2018, 2019, 2021; Carduck et al., 2021; Rybarski et al., 2021; Schoenle et al., 2022).
Another important factor that might have governed the differences in the protist community structure observed in our studies could be the differences in the community structure of prokaryotes, which are known to have a significant effect on protist communities (e.g., Glücksman et al., 2010). It has been shown that the community structure of prokaryotes can differ between the different aquatic habitats across the Atacama and the Andes, as shown by Demergasso et al. (2003), Dorador et al. (2008), and in later studies by Rasuk et al. (2014) and Farias (2016). As salinity levels may vary due to changes in water availability, the composition of active and resting taxa within these ecosystems can change significantly, altering the biodiversity of these natural ecosystems. Future studies, including the metabarcoding of 16S and 18S rRNA phylogenetic markers, could help to elucidate overall changes in the community structures of protists detected co-occurring with the prokaryotes typically found in microbial mats at different salinities characterizing the Andean range.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI—PRJNA1062042.
Author contributions
EA: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. FN: Data curation, Conceptualization, Methodology, Supervision, Writing – review & editing. CD: Writing – review & editing, Resources. HA: Writing – review & editing, Conceptualization, Data curation, Methodology, Project administration, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was carried out in the frame of Collaborative Research Centre 1211 “Evolution at the dry limit”. This study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) to HA (project numbers 268236062 (SFB 1211; B03, B02) and to FN (SPP 1991, project NI 1097/4). EA was supported by the German Academic Exchange Service (DAAD) Program Research Scholarships “Promotionen in Deutschland, 2019/20” (57440921).
Acknowledgments
We would like to thank Jorge Muñoz Salinas, CEO of Gen Andino Company, for logistical support during sampling campaigns at remote latitudes. We are also grateful to “Corporación Nacional Forestal (CONAF)”, a governmental entity authorizing the sample collection and study of microbiology in protected areas in Chile.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1356977/full#supplementary-material
References
Acosta, E., Fincke, V., Nitsche, F., and Arndt, H. (2023). Novel cercozoan and heterolobosean protists from the rhizosphere and phyllosphere of two endemic cacti from the Atacama Desert. Eur. J. Protistol. 91:126034. doi: 10.1016/j.ejop.2023.126034
Albarracín, V. H., Kurth, D., Ordoñez, O. F., Belfiore, C., Luccini, E., Salum, G. M., et al. (2015). High-up: a remote reservoir of microbial extremophiles in central Andean wetlands. Front. Microbiol. 6:e01404. doi: 10.3389/fmicb.2015.01404
Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W., and Huse, S. M. (2009). A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 4:e6372. doi: 10.1371/annotation/50c43133-0df5-4b8b-8975-8cc37d4f2f26
Aran, P., Aguilar, P., Bowen, T., Farson, M., Farson, M., Tapia, J., et al. (2021). Insights into an undescribed high-elevation lake (6,170 m a.s.l.) on Volcán Llullaillaco: a physical and microbiological view. Aquat. Conserv. Mar. Freshw. Ecosyst. 31, 2293–2299. doi: 10.1002/aqc.3612
Arndt, H., Ritter, B., Lennartz, A. E., Schiwitza, S., Dunai, T. J., and Nitsche, F. (2020). Mirroring the effect of geological evolution: protist divergence in the Atacama Desert. Glob. Planet. Change 190:103193. doi: 10.1016/j.gloplacha.2020.103193
Bendif, E. M., Probert, I., Schroeder, D. C., and de Vargas, C. (2013). On the description of Tisochrysis lutea gen. nov. sp. nov. and Isochrysis nuda sp. nov. in the Isochrysidales. and the transfer of Dicrateria to the Prymnesiales (Haptophyta). J. Appl. Phycol. 25, 1763–1776. doi: 10.1007/s10811-013-0037-0
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bryanskaya, A. V., Malup, T. K., Lazareva, E. V., Taran, O. P., Rozanov, A. S., Efimov, V. M., et al. (2016). The role of environmental factors for the composition of microbial communities of saline lakes in the Novosibirsk region (Russia). BMC Microbiol. 16:S4. doi: 10.1186/s12866-015-0618-y
Burki, F., Sandin, M. M., and Jamy, M. (2021). Diversity and ecology of protists revealed by metabarcoding. Curr. Biol. 31, R1267–R1280. doi: 10.1016/j.cub.2021.07.066
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformat. 10:421. doi: 10.1186/1471-2105-10-421
Carduck, S., Nitsche, F., Rybarski, A., Hohlfeld, M., and Arndt, H. (2021). Diversity and phylogeny of percolomonads based on newly discovered species from hypersaline and marine waters. Eur. J. Protistol. 80:125808. doi: 10.1016/j.ejop.2021.125808
Choi, J., and Park, J. S. (2020). Comparative analyses of the V4 and V9 regions of 18S rDNA for the extant eukaryotic community using the Illumina platform. Sci. Rep. 10:6519. doi: 10.1038/s41598-020-63561-z
Demergasso, C., Chong, G., Galleguillos, P., Escudero, L., Martínez-Alonso, L., and Esteve, L. (2003). Tapetes microbianos del Salar de Llamará, norte de Chile. Rev. Chilena Hist. Nat. 76, 485–499. doi: 10.4067/S0716-078X2003000300012
Dillon, J. G., Miller, S., Bebout, B., Hullar, M., Pinel, N., and Stahl, D. A. (2009). Spatial and temporal variability in a stratified hypersaline microbial mat community: community variability in hypersaline microbial mat. FEMS Microbiol. Ecol. 68, 46–58. doi: 10.1111/j.1574-6941.2009.00647.x
Dorador, C., Fink, P., Hengst, M., Icaza, G., Villalobos, A. S., Vejar, D., et al. (2018). Microbial community composition and trophic role along a marked salinity gradient in Laguna Puilar, Salar de Atacama, Chile. Antonie Van Leeuwenhoek 111, 1361–1374. doi: 10.1007/s10482-018-1091-z
Dorador, C., Vila, I., Imhoff, J. F., and Witzel, K.-P. (2008). Cyanobacterial diversity in Salar de Huasco, a high altitude saline wetland in northern Chile: an example of geographical dispersion? FEMS Microbiol. Ecol. 64, 419–432. doi: 10.1111/j.1574-6941.2008.00483.x
Dorador, C., Vila, I., Witzel, K. P., and Imhoff, J. F. (2013). Bacterial and archaeal diversity in high altitude wetlands of the Chilean Altiplano. Fund. Appl. Limnol. 182, 135–159. doi: 10.1127/1863-9135/2013/0393
Dünn, M., and Arndt, H. (2023). Distribution patterns of benthic protist communities depending on depth revealed by environmental sequencing—from the sublittoral to the deep sea. Microorganisms 11:1664. doi: 10.3390/microorganisms11071664
Dupraz, C., Reid, R. P., Braissant, O., Decho, A. W., Norman, R. S., and Visscher, P. T. (2009). Processes of carbonate precipitation in modern microbial mats. Earth Sci. Rev. 96, 141–162. doi: 10.1016/j.earscirev.2008.10.005
Eissler, Y., Gálvez, M.-J., Dorador, C., Hengst, M., and Molina, V. (2019). Active microbiome structure and its association with environmental factors and viruses at different aquatic sites of a high-altitude wetland. Microbiol. Open 8:e00667. doi: 10.1002/mbo3.667
Farias, M. E. (2016). Microbial Ecosystems in Central Andes. Extreme Environments Biofilms, Microbial Mats, Microbialites and Endoevaporites: Biofilms, Microbial Mats, Microbialites and Endoevaporites. Cham: Springer. doi: 10.1007/978-3-030-36192-1
Farías, M. E., Contreras, M., Rasuk, M. C., Kurth, D., Flores, M. R., Poiré, D. G., et al. (2014). Characterization of bacterial diversity associated with microbial mats, gypsum evaporites and carbonate microbialites in thalassic wetlands: Tebenquiche and La Brava, Salar de Atacama, Chile. Extremophiles 18, 311–329. doi: 10.1007/s00792-013-0617-6
Farias, M. E., Rasuk, M. C., Gallagher, K. L., Contreras, M., Kurth, D., Fernandez, A. B., et al. (2017). Prokaryotic diversity and biogeochemical characteristics of benthic microbial ecosystems at La Brava, a hypersaline lake at Salar de Atacama, Chile. PLoS ONE 12:e0186867. doi: 10.1371/journal.pone.0186867
Fernandez, A. B., Rasuk, M. C., Visscher, P. T., Contreras, M., Novoa, F., Poire, D. G., et al. (2016). Microbial diversity in sediment ecosystems (evaporites domes, microbial mats, and crusts) of hypersaline Laguna Tebenquiche, Salar de Atacama, Chile. Front. Microbiol. 7:1284. doi: 10.3389/fmicb.2016.01284
Fiore-Donno, A. M., Richter-Heitmann, T., Degrune, F., Dumack, K., Regan, K. M., Marhan, S., et al. (2019). Functional traits and spatio-temporal structure of a major group of soil protists (Rhizaria: Cercozoa) in a temperate Grassland. Front. Microbiol. 10:1332. doi: 10.3389/fmicb.2019.01332
Glücksman, E., Bell, T., Griffiths, R. I., and Bass, D. (2010). Closely related protist strains have different grazing impacts on natural bacterial communities. Environ. Microbiol. 12, 3105–3113. doi: 10.1111/j.1462-2920.2010.02283.x
Hernández, K. L., Yannicelli, B., Olsen, L. M., Dorador, C., Menschel, E. J., Molina, V., et al. (2016). Microbial activity response to solar radiation across contrasting environmental conditions in Salar de Huasco, Northern Chilean Altiplano. Front. Microbiol. 7:1857. doi: 10.3389/fmicb.2016.01857
Hohlfeld, M., Meyer, C., Schoenle, A., Nitsche, F., and Arndt, H. (2023). Biogeography, autecology, and phylogeny of percolomonads based on newly described species. J. Eukaryot. Microbiol. 70:e12930. doi: 10.1111/jeu.12930
Howe, A. T., Bass, D., Scoble, J. M., Lewis, R., Vickerman, K., Arndt, H., et al. (2011). Novel cultured protists identify deep-branching environmental DNA clades of Cercozoa: new genera Tremula, Micrometopion, Minimassisteria, Nudifila, Peregrinia. Protist 162, 332–372. doi: 10.1016/j.protis.2010.10.002
Liu, C., Cui, Y., Li, X., and Yao, M. (2021). microeco : an R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 97:fiaa255. doi: 10.1093/femsec/fiaa255
Lozupone, C. A., and Knight, R. (2007). Global patterns in bacterial diversity. Proc. Nat. Acad. Sci. U. S. A. 104, 11436–11440. doi: 10.1073/pnas.0611525104
Mahé, F., de Vargas, C., Bass, D., Czech, L., Stamatakis, A., Lara, E., et al. (2017). Parasites dominate hyperdiverse soil protist communities in Neotropical rainforests. Nat. Ecol. Evol. 1:0091. doi: 10.1038/s41559-017-0091
Margulis, L., Barghoorn, E. S., Ashendorf, D., Banerjee, S., Chase, D., Francis, S., et al. (1980). The microbial community in the layered sediments at Laguna Figueroa, Baja California, Mexico: Does it have Precambrian analogues? Precambrian Res. 11, 93–123. doi: 10.1016/0301-9268(80)90042-X
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16!Wke rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Molina, V., Eissler, Y., Fernandez, C., Cornejo-D'Ottone, M., Dorador, C., Bebout, B. M., et al. (2021). Greenhouse gases and biogeochemical diel fluctuations in a high-altitude wetland. Sci. Total Environ. 768:144370. doi: 10.1016/j.scitotenv.2020.144370
Öztoprak, H., Walden, S., Heger, T., Bonkowski, M., and Dumack, K. (2020). What drives the diversity of the most abundant terrestrial cercozoan family (Rhogostomidae, Cercozoa, Rhizaria)? Microorganisms 8:1123. doi: 10.3390/microorganisms8081123
Rasuk, M. C., Fernández, A. B., Kurth, D., Contreras, M., Novoa, F., Poiré, D, et al. (2016). Bacterial diversity in microbial mats and sediments from the Atacama Desert. Microb. Ecol. 71, 44–56. doi: 10.1007/s00248-015-0649-9
Rasuk, M. C., Kurth, D., Flores, M. R., Contreras, M., Novoa, F., Poire, D., et al. (2014). Microbial characterization of microbial ecosystems associated to evaporites domes of Gypsum in Salar de Llamara in Atacama Desert. Microb. Ecol. 68, 483–494. doi: 10.1007/s00248-014-0431-4
Ritter, B., Binnie, S. A., Stuart, F. M., Wennrich, V., and Dunai, T. J. (2018). Evidence for multiple Plio-Pleistocene lake episodes in the hyperarid Atacama Desert. Quat. Geochronol. 44, 1–12. doi: 10.1016/j.quageo.2017.11.002
Rothrock, M. J., and Garcia-Pichel, F. (2005). Microbial diversity of benthic mats along a tidal desiccation gradient. Environ. Microbiol. 7, 593–601. doi: 10.1111/j.1462-2920.2005.00728.x
Rybarski, A. E. L. N., Nitsche, F., Schoenle, A., Voigt, C., Staubwasser, M., and Arndt, H. (2023). High diversity and isolated distribution of aquatic heterotrophic protists in salars of the Atacama Desert at different salinities. Eur. J. Protistol. 89:125987. doi: 10.1016/j.ejop.2023.125987
Rybarski, A. E., Nitsche, F., Park, J. S., Filz, P., Schmidt, P., Kondo, R., et al. (2021). Revision of the phylogeny of Placididea (Stramenopiles): molecular and morphological diversity of novel placidid protists from extreme aquatic environments. Eur. J. Protistol. 81:125809. doi: 10.1016/j.ejop.2021.125809
Sachs, M., Dünn, M., and Arndt, H. (2023). Benthic heterotrophic protist communities of the southern Baltic analyzed with the help of curated metabarcoding studies. Biology 12:1010. doi: 10.3390/biology12071010
Saghaï, A., Gutiérrez-Preciado, A., Deschamps, P., Moreira, D., Bertolino, P., Ragon, M., et al. (2017). Unveiling microbial interactions in stratified mat communities from a warm saline shallow pond: unveiling interactions in microbial mats. Environ. Microbiol. 19, 2405–2421. doi: 10.1111/1462-2920.13754
Schiwitza, S., Arndt, H., and Nitsche, F. (2018). Four new choanoflagellate species from extreme saline environments: indication for isolation-driven speciation exemplified by highly adapted Craspedida from salt flats in the Atacama Desert (Northern Chile). Eur. J. Protistol. 66, 86–96. doi: 10.1016/j.ejop.2018.08.001
Schiwitza, S., Arndt, H., and Nitsche, F. (2019). First description of an euryoecious acanthoecid choanoflagellate species, Enibas tolerabilis gen. et sp. nov. from a salar in the Chilean Andes based on morphological and transcriptomic data. Eur. J. Protistol. 67, 106–113. doi: 10.1016/j.ejop.2018.11.004
Schiwitza, S., Gutsche, L., Freches, E., Arndt, H., and Nitsche, F. (2021). Extended divergence estimates and species descriptions of new craspedid choanoflagellates from the Atacama Desert, Northern Chile. Eur. J. Protistol. 79:125798. doi: 10.1016/j.ejop.2021.125798
Schloss, P. D., Gevers, D., and Westcott, S. L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6:e27310. doi: 10.1371/journal.pone.0027310
Schoenle, A., Hohlfeld, M., Hermanns, K., Mahé, F., de Vargas, C., Nitsche, F., et al. (2021). High and specific diversity of protists in the deep-sea basins dominated by diplonemids, kinetoplastids, ciliates and foraminiferans. Commun. Biol. 4:501. doi: 10.1038/s42003-021-02012-5
Schoenle, A., Hohlfeld, M., Rosse, M., Filz, P., Wylezich, C., Nitsche, F., et al. (2020). Global comparison of bicosoecid Cafeteria-like flagellates from the deep ocean and surface waters, with reorganization of the family Cafeteriaceae. Eur. J. Protistol. 73:125665. doi: 10.1016/j.ejop.2019.125665
Schoenle, A., Hohlfeld, M., Rybarski, A., Sachs, M., Freches, E., Wiechmann, K., et al. (2022). Cafeteria in extreme environments: investigations on C. burkhardae and three new species from the Atacama Desert and the deep ocean. Eur. J. Protistol. 85:125905. doi: 10.1016/j.ejop.2022.125905
Schulze-Makuch, D., Wagner, D., Kounaves, S. P., Mangelsdorf, K., Devine, K. G., de Vera, J.-P., et al. (2018). Transitory microbial habitat in the hyperarid Atacama Desert. Proc. Nat. Acad. Sci. U. S. A. 115, 2670–2675. doi: 10.1073/pnas.1714341115
Singer, D., Seppey, C. V. W., Lentendu, G., Dunthorn, M., Bass, D., Belbahri, L., et al. (2021). Protist taxonomic and functional diversity in soil, freshwater and marine ecosystems. Environ. Int. 146:106262. doi: 10.1016/j.envint.2020.106262
Solden, L., Lloyd, K., and Wrighton, K. (2016). The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 31, 217–226. doi: 10.1016/j.mib.2016.04.020
Venter, P. C., Nitsche, F., Domonell, A., Heger, P., and Arndt, H. (2017). The protistan microbiome of grassland soil: diversity in the mesoscale. Protist 168, 546–564. doi: 10.1016/j.protis.2017.03.005
Vignale, F. A., Lencina, A. I., Stepanenko, T. M., Soria, M. N., Saona, L. A., Kurth, D., et al. (2022). Lithifying and non-lithifying microbial ecosystems in the wetlands and salt flats of the central Andes. Microb. Ecol. 83, 1–17. doi: 10.1007/s00248-021-01725-8
Wang, M., Ji, X., Wang, B., Li, Q., and Zhou, J. (2018). Simultaneous evaluation of the preservative effect of RNA later on different tissues by biomolecular and histological analysis. Biopreserv. Biobank. 16, 426–433. doi: 10.1089/bio.2018.0055
Keywords: microbial mats, metabarcoding, protists, co-occurrence, Rhogostoma, Euplotes, salinity
Citation: Acosta E, Nitsche F, Dorador C and Arndt H (2024) Protist communities of microbial mats from the extreme environments of five saline Andean lagoons at high altitudes in the Atacama Desert. Front. Microbiol. 15:1356977. doi: 10.3389/fmicb.2024.1356977
Received: 16 December 2023; Accepted: 08 February 2024;
Published: 20 March 2024.
Edited by:
Ian Hawes, University of Waikato, New ZealandReviewed by:
Sandi Orlic, Rudjer Boskovic Institute, CroatiaVirginia Helena Albarracín, CONICET Center for Electron Microscopy (CIME), Argentina
Copyright © 2024 Acosta, Nitsche, Dorador and Arndt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hartmut Arndt, hartmut.arndt@uni-koeln.de
 Eduardo Acosta
Eduardo Acosta Frank Nitsche
Frank Nitsche Cristina Dorador
Cristina Dorador Hartmut Arndt
Hartmut Arndt