- 1Center for Autoimmune Genomics and Etiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 2Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 3Department of Pediatrics, University of Cincinnati, Cincinnati, OH, United States
- 4Department of Internal Medicine, University of Cincinnati, Cincinnati, OH, United States
- 5Cincinnati Veterans Affairs Medical Center, Cincinnati, OH, United States
- 6Department of Chemistry, University of Cincinnati, Cincinnati, OH, United States
Introduction: Pneumocystis species are pathogenic fungi known to cause pneumonia in immunocompromised mammals. They are obligate to their host, replicate extracellularly in lung alveoli and thrive in the copper-enriched environment of mammalian lungs. In this study, we investigated the proteome of Pneumocystis murina, a model organism that infects mice, in the context of its copper sensing and tolerance.
Methods and results: The query for copper-associated annotations in FungiDB followed by a manual curation identified only 21 genes in P. murina, significantly fewer compared to other clinically relevant fungal pathogens or phylogenetically similar free-living fungi. We then employed instrumental analyses, including Size-Exclusion Chromatography Inductively Coupled Plasma Mass Spectrometry (SEC-ICP-MS), Immobilized Metal Affinity Chromatography (IMAC), and Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS), to isolate and identify copper-binding proteins from freshly extracted organisms, revealing 29 distinct cuproproteins. The RNA sequencing (RNA-seq) analysis of P. murina exposed to various CuSO4 concentrations at three temporal intervals (0.5, 2, and 5 h) indicated that significant gene expression changes occurred only under the highest CuSO4 concentration probed (100 μM) and the longest exposure duration (5 h). This stimulus led to the upregulation of 43 genes and downregulation of 27 genes compared to untreated controls. Quantitative PCR (qPCR) confirmed the expression of four out of eight selected upregulated genes, including three assumed transcription factors (PNEG_01236, PNEG_01675, and PNEG_01730) and a putative copper transporter (PNEG_02609). Notably, the three applied methodologies — homology-based annotation, SEC-ICP-MS/IMAC/LC–MS/MS, and RNA-seq — yielded largely distinct findings, with only four genes (PNEG_02587, PNEG_03319, PNEG_02584, and PNEG_02989) identified by both instrumental methods.
Discussion: The insights contribute to the broader knowledge of Pneumocystis copper homeostasis and provide novel facets of host-pathogen interactions for extracellular pathogens. We suggest that future studies of Pneumocystis pathogenicity and copper stress survival should consider the entire spectrum of identified genes.
Introduction
Copper is a biochemically versatile and indispensable trace element that plays a fundamental role in various biological processes (Festa and Thiele, 2011). This transition metal is a critical cofactor for a diverse array of enzymes, functioning as a catalytic center in redox reactions. Among its numerous roles, copper is essential for electron transfer processes in mitochondrial cytochrome c oxidase, which is vital for cellular respiration and adenosine triphosphate (ATP) production. Copper contributes to the organism’s antioxidant defense system by serving as a cofactor for Cu/Zn superoxide dismutase, an enzyme that neutralizes harmful superoxide radicals. The multifaceted involvement of copper in cellular activities is particularly pronounced in fungi, where it serves as an indispensable component for copper-dependent enzymes. Such enzymes are essential for virulence and antioxidant defense in pathogenic fungi (Gerwien et al., 2018). Copper homeostasis is intricately linked to the regulation of fungal growth, development, and pathogenicity (Smith et al., 2017).
Conversely, an excessive accumulation of copper beyond homeostatic requirements can result in toxicity in various scenarios. First, copper cytotoxicity stems from its redox-active nature and its capacity to oxidize lipids, nucleic acids, and proteins by generating reactive oxygen species (ROS) such as hydroxyl radicals through Fenton-like reaction (Valko et al., 2005). Second, copper has a higher affinity to proteins that normally bind zinc, iron, or other transition metals and can replace these trace metals at their protein binding sites (Waldron et al., 2009). Since numerous metalloproteins are involved in amino acid biosynthesis, DNA replication and repair, telomere maintenance, and other critical cellular functions, these proteins may be significant targets in copper toxicity due to the loss of catalytic function.
The dual nature of copper is harnessed by animals in their host-pathogen interactions to combat invasive threats. Lungs are the natural route of infection and the initial infectious niche in mammals for many infectious agents. Host innate immune cells, particularly macrophages in the lungs, actively accumulate and intricately compartmentalize copper as an effective defense against invaders. When encountering pathogens, macrophages engulf and confine these microorganisms within a specialized compartment known as the phagolysosome. This microenvironment is uniquely inhospitable, characterized by heightened levels of ROS, reactive nitrogen compounds, low pH, substantial copper concentrations, and proteases, all meticulously orchestrated to eradicate these intruders. High copper levels are also found in plasma, where copper is bound primarily to ceruloplasmin, but also available from albumin, extracellular Cu/Zn-superoxide dismutase (SOD), and plasma metallothioneins. In contrast, the host’s nutritional immunity strategies aim to deprive invading pathogens of copper (among other important trace metals). For instance, the invasion of the kidney by the fungal pathogen Candida albicans during the later stages of infection results in a reduction of total kidney copper. Similarly, infection by Cryptococcus neoformans in the brain is associated with clear signs of copper limitation stress in the fungus. Further details on the copper-related host defense tactics against invading pathogens can be found in the following reviews (Garcia-Santamarina and Thiele, 2015; Besold et al., 2016).
While most bacterial pathogens do not exhibit a need for cytoplasmic copper, fungal pathogens and humans share common Cu-binding proteins located in various cellular compartments, emphasizing a commonality in their Cu homeostasis mechanisms. For example, in response to elevated pulmonary Cu concentrations during infection, C. neoformans employs a time-dependent induction of the expression of two metallothionein (MT) genes, CMT1 and CMT2. These genes act as primary and redundant defense mechanisms, protecting C. neoformans from otherwise toxic Cu levels (Ding et al., 2011, 2013). In contrast to the MTs of C. neoformans, C. albicans relies on the Cu-exporting pump CRP1 to play a dominant role in Cu detoxification and Cu-related virulence in this fungal pathogen (Weissman et al., 2000). When faced with copper deprivation in the kidney, the yeast shifts to induce copper uptake through the fungal copper transporter CTR1 and transitions from expressing Cu/Zn Sod1 to Mn Sod3 (Li et al., 2015). Likewise, C. neoformans initiates the CUF1/CTR4 axis when access to copper is limited in the brain (Waterman et al., 2007). A similar strategy was found in another fungal pathogen, Aspergillus fumigatus, that responds to low copper through AfMac1 (a homolog to Mac1, a copper-responsive transcription factor in Saccharomyces cerevisiae) by upregulating the ctrC and ctrA2 genes that encode high-affinity copper transporters (Cai et al., 2017). The fungus utilizes the copper efflux pump (CrpA) to detoxify excess copper (Cai et al., 2018). More details on copper homeostasis in A. fumigatus can be found in the review (Song et al., 2019). Histoplasma capsulatum, on the other hand, encounters dichotomous copper conditions during macrophage interaction, necessitating adaptive responses to fluctuating copper availability. Initially, at the stage of innate immunity, the fungus is exposed to an excess of copper, prompting upregulation of the copper efflux pump CRP1 (Moraes et al., 2024) and activation of antioxidant enzymes, including catalase B (CATB), peroxidase, and Cu/Zn SOD1 (Moraes et al., 2023), to mitigate ROS stress. As the infection progresses, at the stage of adaptive immunity, H. capsulatum transitions to coping with copper scarcity, implementing a strategy for high-affinity copper acquisition via the Ctr3 transporter. This uptake mechanism is modulated by the copper-sensitive transcriptional regulator Mac1 (Ray and Rappleye, 2022). For bacterial responses to copper, the reader is referred to the following reviews (Arguello et al., 2013; Becker and Skaar, 2014; Andrei et al., 2020).
The multifaceted role of copper, serving both as a host defense mechanism and a pathogen vulnerability, underscores the intricate and dynamic nature of host-pathogen interactions in the battle against infectious agents. While Histoplasma fungi or other microorganisms are engulfed by macrophages and cope with macrophage-imposed hostile environments such as phagosomes, Pneumocystis spp. are extracellular organisms that grow within the lung alveoli, and as such, we studied their coping mechanisms outside macrophages. Our study explores the adaptive strategies employed by the fungal Pneumocystis species to flourish in the copper-enriched environment of mammalian lungs. These host-obligate parasitic extracellular organisms, marked by significant gene loss relative to free-living fungi (Hauser et al., 2010; Porollo et al., 2014; Ma et al., 2016), notably lack homologs to the majority of known fungal copper-control proteins. Intriguingly, recent findings have demonstrated a remarkable increase in the viability of P. jirovecii organisms ex vivo with addition of copper(II) sulfate to the culture medium (Riebold et al., 2023). Consequently, it becomes imperative to unravel the mechanisms governing copper homeostasis in these pathogens, with the aims of refining culture conditions and identifying potential new drug targets. This study represents the first endeavor to shed light on how these pathogenic fungi sense and tolerate elevated copper levels.
Materials and methods
Sampling of pathogen organisms and host tissue
P. murina organisms for these studies were obtained from 6 to 8-week old Bagg Albino (BALB/c) mice (Charles River Wilmington, MA). The mice were handled in strict accordance with good animal practice, as defined by the Institutional Animal Care and Use Committee at the Veterans Affairs Medical Center, Cincinnati, OH. To safeguard against environmental exposure of P. murina and other microbes, mice were housed under barrier conditions with autoclaved food, acidified water, and bedding in sterilized shoebox cages equipped with sterile microfilter lids.
Mice were immunosuppressed by adding dexamethasone (4 mg/L) to acidified drinking water and infected with P. murina organisms isolated from previously infected mice and stored in liquid nitrogen. A dose of 2 × 106 organisms / 50 μL was instilled by intranasal inoculation under anesthesia. The mice were continued on immunosuppression for 6 weeks and then euthanized by CO2 for isolation of organisms.
Lungs were removed, homogenized (gentleMACS™ Dissociator, Miltenyi Biotec, Auburn, CA), and filtered through a 40 μm pore mesh, and cells were recovered by centrifugation at 3400 rpm for 10 min. Host cells were removed by a 2nd centrifugation at 250 rpm for 5 min. Slides were then prepared to enumerate organisms for the downstream CuSO4 treatment, as previously described (Cushion et al., 2010).
Treatment with copper(II) sulfate
Two experiments were conducted to evaluate the impact of copper(II) sulfate on P. murina. Experiment 1, a short-term assessment, focused on differential gene expression in response to CuSO4 exposure. Experiment 2, a long-term study, aimed to determine the viability of P. murina under sustained CuSO4 exposure.
During the isolation process, P. murina were purified from lung cells by washing at least 3 times with phosphate buffered saline (PBS) as well as a short incubation period with 0.85% aqueous ammonium chloride, after which they were washed again with PBS. After rinsing with PBS, the cells were centrifuged to form a pellet, reconstituted in PBS for the next washing.
P. murina cultures were prepared at 2.5 × 106 cells/mL using Roswell Park Memorial Institute (RPMI) 1,640 medium (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) (Cytiva, Marlborough, MA), 1,000 U/mL penicillin, 1,000 μg/mL streptomycin (Gibco, Grand Island, NY), 1% minimum essential medium (MEM) vitamin solution, and 1% MEM non-essential amino acid solution (Gibco, Grand Island, NY). Cultures were distributed in quadruplicate into 48-well polystyrene plates (Corning Costar, Glendale, AZ). Each well was inoculated with 0.25 mL of either media (control) or CuSO4 solutions at final concentrations of 1, 10, and 100 μM for both experiments, and an additional concentration of 625 μM for Experiment 2. The plates were incubated at 36°C in a 5% CO2 atmosphere.
Sampling for ATP quantification was conducted at designated time points: 30 min, 2 h, and 5 h for Experiment 1; and 1, 3, 5, and 7 days for Experiment 2. For this, 50 μL samples were extracted from each well and transferred to a Nunclon Delta Surface white 96-well plate (Nunc A/S, Roskilde, Denmark). ATP levels were quantified using an ATP-lite luciferin-luciferase assay (Perkin Elmer, Waltham, MA), with luminescence readings obtained via a Synergy HTX spectrophotometer (BioTek, Winooski, Vermont). Background luminescence was measured in control wells with blank culture medium. To confirm sample purity, microscopic examinations for microbial contamination were performed and the cultures were microscopically monitored over the sampling periods to detect any bacterial or fungal outgrowth.
Instrumental analysis
Cell pellets were lysed on ice using a lysis buffer composed of 0.1% SDS, 1% digitonin, 50 mM Tris–HCl, and 50 mM NaCl (pH 7.4) for 5 min, followed by 30 s of vortexing at maximum speed. The resulting cell lysates were then filtered through a 0.45 μm filter. Protein concentrations were determined using the Bradford assay, and the lysis buffer in each sample was adjusted accordingly to standardize protein levels across all samples. These lysates underwent SEC-ICP-MS analysis to detect copper across different molecular size fractions. The separation utilized a TSKgel G3000SWxl column (7.8 × 300 mm, 10 μm particle size, Tosoh Corporation, Tokyo, Japan) connected to an Agilent 1200 HPLC system. The mobile phase consisted of 50 mM ammonium acetate and 0.5% methanol (pH 7.4), with a flow rate of 0.6 mL/min. Copper quantification in each fraction employed three 50 μL injections, calibrated against a Cu standard curve (5 to 100 ng/mL) derived from human recombinant ceruloplasmin. Total metal content was assessed in triplicate aliquots via microwave digestion, comparing SEC-derived copper concentrations to total copper levels to ascertain a 94 ± 2% recovery efficiency.
Post-copper profiling, remaining samples (approximately 20 mL, excluding 100 μL for whole-lysate proteomics and 500 μL for non-fractionated IMAC enrichment) were fractionated via multiple 100 μL HPLC injections, with column performance verified every 20 injections using standards (Bio-Rad gel filtration standard mix and ceruloplasmin for Cu signal). Offline fraction collection from SEC was timed at intervals for high (10–12 min), medium (12.5–17.5 and 17.5–20 min), and low (20–25 min) molecular weight fractions. Collected fractions were lyophilized prior to IMAC processing.
To prepare for Cu-based IMAC protein enrichment, native copper was removed from proteins using Chelex 100 resin (Bio-Rad, CA) to expose Cu-binding sites. Proteins, either from whole lysate or dried fractions, were resuspended in 2 mL of 50 mM Na2HPO4 and 300 mM NaCl (pH 7.4), treated with 0.5 g of Chelex resin, and agitated at 4°C for 1 h, centrifuged at 5,000 g for 2 min. This process was repeated two more times to maximize copper removal. The clarified supernatant was then subjected to IMAC enrichment using the Cu-loaded IMAC resin.
An IMAC copper resin (Geno Technology, Inc., MO) was used to enrich the SEC-fractionated samples for Cu-binding proteins. The manufacturer instructions were followed, including the rinsing of the resin with milli-Q water, the conditioning of the resin with the loading buffer that contains 10 mM imidazole, loading of the sample followed by 20 min of agitation as incubation, the removal of the supernatant, washing of the resin with binding buffer, and the final step consisted on the elution of the Cu-binding proteins with 3 consecutive volumes of 50 mM Na2HPO4, 0.3 M NaCl, 0.25 M imidazole, pH 8.0. The amount of resin was adjusted according to the Cu concentration in each fraction, with the nominal Cu concentration of 30 μmol per ml of resin as reported by the vendor. As a positive control, Cu-depleted ceruloplasmin was introduced to the sample blank, while Zn-depleted carbonic anhydrase served as the negative control in a separate sample blank. The IMAC procedure was applied to the controls in a manner identical to that of the fractionated sample. The enriched sample fractions obtained were lyophilized to dryness and re-suspended in 50 mM ammonium carbonate for future tryptic digestion. Protein identification was conducted at the University of Cincinnati proteomics core facility utilizing liquid chromatography–tandem mass spectrometry (LC–MS/MS), following the previously published protocol (Green et al., 2013). Samples submitted for proteomics analysis included the high, medium, and low molecular weight fractions derived from the uninfected host lungs, the host lungs infected with P. murina, and the purified pathogen.
RNA sequencing and analysis
P. murina organisms were resuspended in 500 μL of mirVana miRNA lysis buffer (Invitrogen, Waltham, MA) and promptly snap-frozen at −80°C for subsequent storage and transfer to a sequencing core. Directional polyA RNA sequencing (RNA-seq) was conducted at the Genomics, Epigenomics, and Sequencing Core at the University of Cincinnati, employing established protocols detailed in previous publications (Walsh et al., 2019; Rapp et al., 2020) with updates. Briefly, total RNA quality was assessed using the Bioanalyzer (Agilent, Santa Clara, CA). The NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs, Ipswich, MA) was employed for the isolation of polyA RNA during library preparation, utilizing 400 ng of high-quality total RNA. The enrichment of polyA RNA was performed using the SMARTer Apollo automated NGS library prep system (Takara Bio United States, Mountain View, CA). Subsequently, the NEBNext Ultra II Directional RNA Library Prep kit (New England BioLabs) was utilized for library preparation, employing a Polymerase Chain Reaction (PCR) cycle number of 10. Following library QC and quantification via Qubit quantification (ThermoFisher, Waltham, MA), individually indexed libraries were proportionally pooled and subjected to sequencing on the NextSeq 550 Sequencer (Illumina, San Diego, CA) with a sequencing setting of SR 1 × 85 bp, generating approximately 25 million reads. Upon completion of the sequencing run, FASTQ files for subsequent data analysis were automatically generated through Illumina BaseSpace Sequence Hub.
Raw sequencing files were first preprocessed by FASTP (Chen et al., 2018) to remove or prune low quality reads, and trim adapter sequences. Then, transcript abundance was quantified by SALMON (Patro et al., 2017) using the Pneumocystis murina B123 strain’s genome as the reference (NCBI RefSeq Assembly ID: GCF_000349005). Differential gene expression analysis was conducted through DESeq2 (Love et al., 2014). Differentially expressed genes were identified based on a fold change (FC) of ≥2 and an adjusted p-value (q-value) of <0.05.
RT-qPCR
Genes of interest were selected for validation by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The oligonucleotide primers to these genes were designed based on sequences from NCBI/GenBank. Primers were designed to span an intron junction to detect amplification of genomic DNA. cDNA was synthesized from the extracted RNA using SuperScript IV VILO master mix with ezDNase enzyme (Invitrogen, Grand Island, NY) per the manufacturer’s protocol. The remaining RNA and the freshly synthesized complementary DNA (cDNA) were stored at −80°C.
Real-time PCR was performed in an Applied Biosystems 7,500 fast real-time PCR system (Life Technologies, Grand Island, NY). The reactions were performed in triplicate in a final volume of 20 μL containing 1 μL PowerUp SYBR green master mix (Life Technologies, Grand Island, NY) and 500 nM each primer pair. The reaction mixtures were initially incubated at 50°C for 2 min and 95°C for 2 min, followed by 40 cycles of 15 s at 95°C, 59°C for 15 s, and 72°C for 1 min. Fluorescence data were captured during the 72°C step. Disassociation curves were also calculated for the reactions to determine the melting temperature (Tm) of the products. The reference gene used was thymidylate synthase (TS). The threshold cycle (ΔCT) value between the validation gene and the TS gene was calculated by subtracting the average CT value of TS from the average CT value of the validation gene. ΔΔCT values were calculated by subtracting the ΔCT value of the TS gene from the ΔCT value of the validation sample.
Results
Annotated copper related proteins in Pneumocystis species
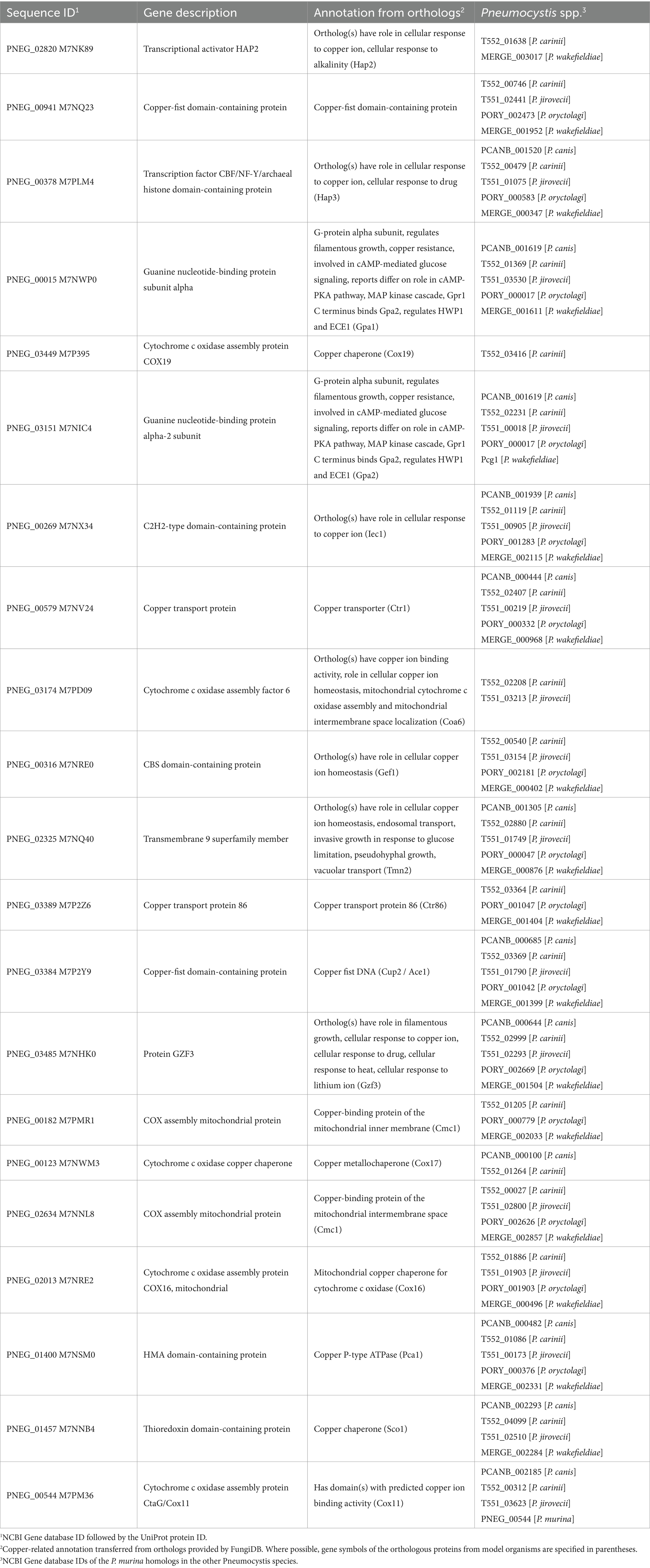
Copper associated protein annotations were retrieved from the FungiDB database (Amos et al., 2022) for sequenced Pneumocystis species and other clinically significant and model fungi (as of August 2023). Since FungiDB transfers annotations inclusively, even from non-top homologs, an additional manual curation of Pneumocystis proteins was conducted using a homology search by BLAST against the NCBI nr database (top 250 hits with E-value<0.001, restricted to the fungi taxon) in conjunction with the UniProt and Pfam annotations. Table 1 provides a summary of the number of proteins identified using the keyword “copper.” Notably, Pneumocystis species exhibit approximately three to five times fewer copper-associated proteins compared to other fungi. Given that this study employed P. murina as the experimental model organism, Table 2 presents a list of respective proteins, derived from FungiDB annotations, either through direct gene description (second column) or annotations of homologous genes (third column), followed by the manual curation.
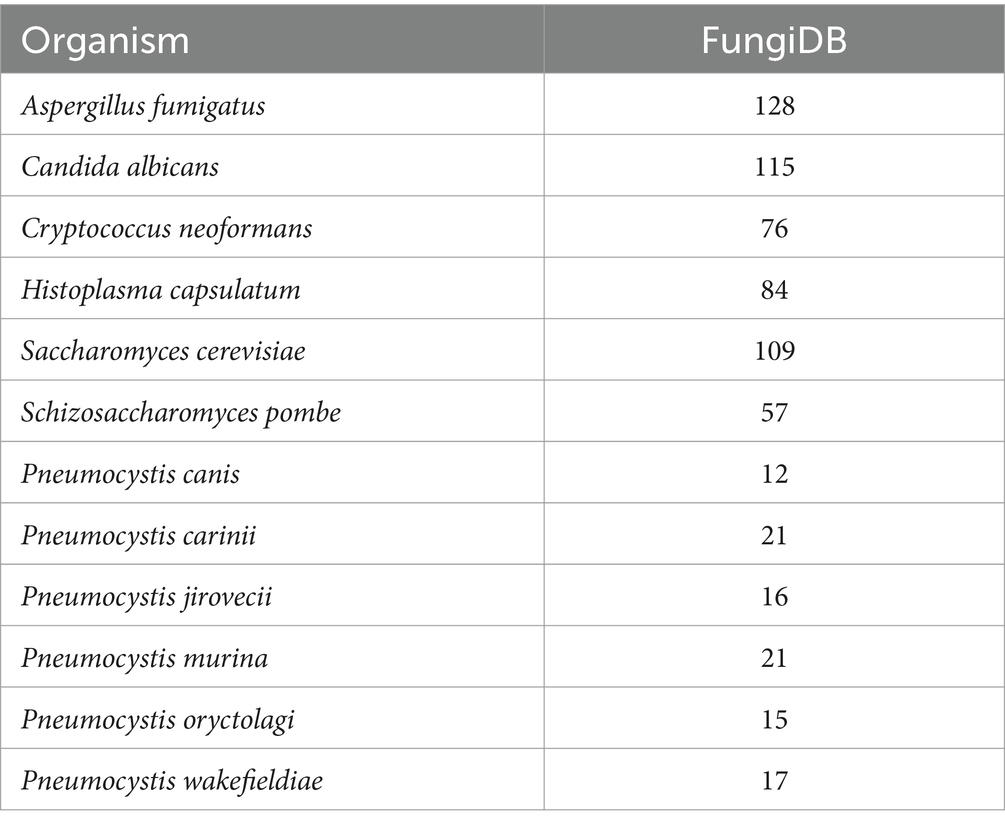
Table 1. Summary of copper-associated proteins in select clinically important and model fungi in FungiDB.
Instrumentally identified copper-associated proteins in Pneumocystis murina
P. murina cell lysates and lung homogenates from both infected and uninfected mice underwent SEC-ICP-MS analysis to detect Cu. Figure 1 presents the chromatograms for Cu profiling, where the area under the curve correlates directly with the Cu concentration in each fraction. Since the total protein concentration of each cell lysate was normalized, the chromatograms allow for a direct comparison of Cu content and distribution. Notably, the Cu distribution in host cell lysates remained consistent regardless of P. murina infection. However, the Cu distribution in the isolated pathogen markedly differed, underscoring the effectiveness of the pathogen isolation procedure.
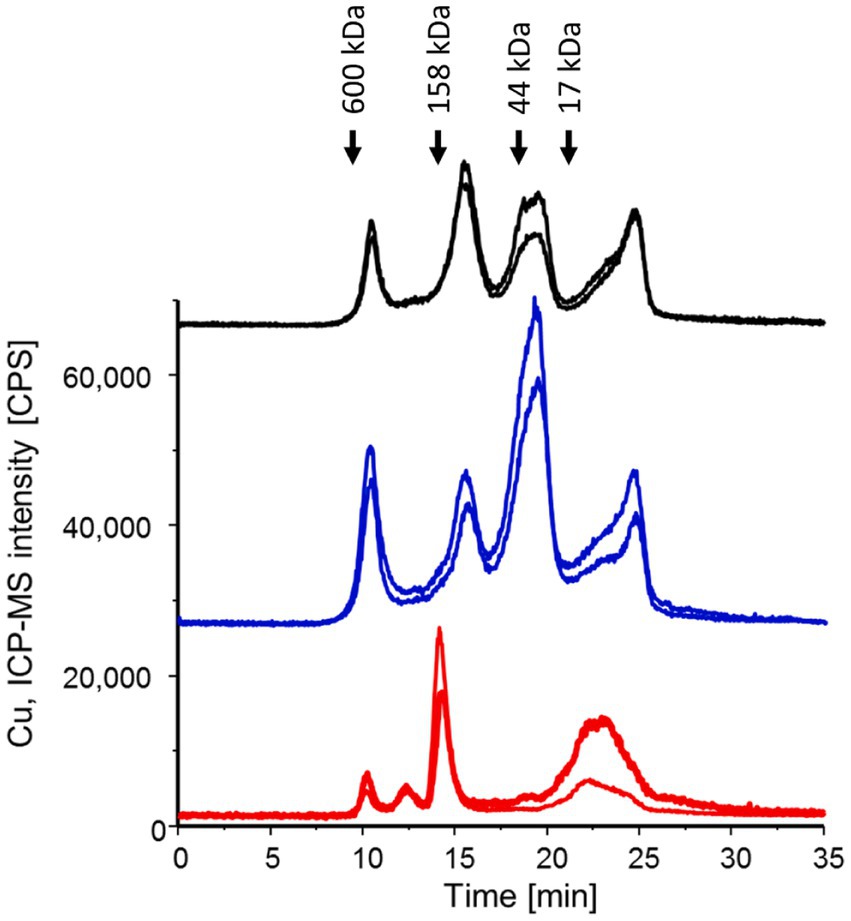
Figure 1. SEC-ICP-MS chromatograms were generated for P. murina samples (in red), lung homogenates from infected mice (in black), and lung homogenates from uninfected mice (in blue) to investigate the Cu metalloproteome. Each analysis was conducted in two technical replicates, and the chromatograms have been offset for clarity. The peaks observed at 23–24 min correspond to free copper. Molecular weight markers are indicated based on the Bio-Rad Gel filtration standard mixture containing thyroglobulin (bovine), g-globulin (bovine), ovalbumin (chicken) and myoglobin (horse).
A summary of the subsequent analysis of the fractions and the identification of the IMAC enriched Cu-binding proteins via LC–MS/MS is presented in Table 3. Of note, since the pathogen cannot be cultivated, any samples with extracted and purified organisms are still contaminated with the host cells. Table 4 provides the list of P. murina proteins identified in all molecular weight factions. The complete list of the identified proteins associated with copper, both for the pathogen and the host, can be found in Supplementary File S1.
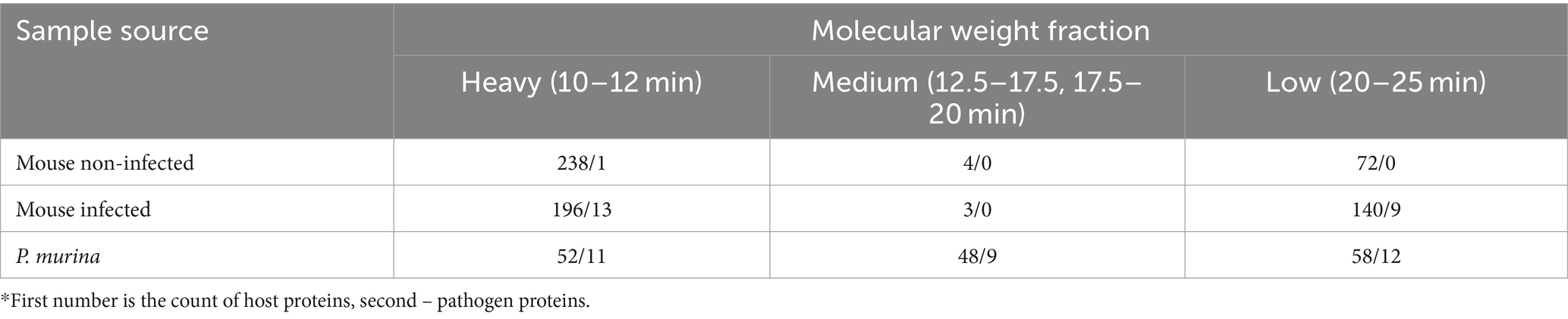
Table 3. Counts of Cu-associated proteins in host and pathogen identified by proteomics.*
The proteomic identification through the IMAC enrichment method likely includes a range of metalloproteins typically linked with zinc, manganese, or other divalent metal ions, alongside the intended copper-binding proteins. This diversity arises from the demetallation process using Chelex 100 resin, which also extracts these metals while primarily removing Cu to generate apo-forms of the existing Cu-binding proteins. The predictability of differing affinities for Cu across various apo-forms of metalloproteins is challenging due to the IMAC resin’s nature and the attempt to limit available Cu to minimize nonspecific binding. Consequently, achieving complete selectivity for Cu-binding proteins is difficult. To enhance method specificity, optimization of incubation times, rinse volumes, and elution buffer volumes was undertaken to minimize the retention of carbonic anhydrase, a common off-target zinc protein, employing ceruloplasmin as a positive control due to its multifaceted copper-binding sites. Further refinement was evidenced by re-analyzing demetallated SEC fractions with ICP-MS prior to IMAC enrichment, revealing an 89 ± 6% reduction in the original Cu content (data not shown). This adjustment significantly improved the selectivity for ceruloplasmin over carbonic anhydrase, potentially decreasing false negatives in Cu-proteome analysis.
Gene expression in Pneumocystis murina upon copper stimulus
To gain insights into the copper sensing mechanisms in P. murina, we exposed freshly extracted organisms to copper(II) sulfate for 0.5, 2, and 5 h, followed by RNA-seq analysis. In the initial batch, extracted from 6 mice, we applied concentrations of 1 ng/mL and 100 ng/mL of CuSO4 for comparison with untreated organisms. Notably, no statistically significant differential gene expression was observed (Supplementary Figure S1A).
In a subsequent batch, we exposed the organisms to elevated CuSO4 concentrations: 1, 10, and 100 μM (0.1596, 1.596, and 15.96 μg/mL, respectively), at the same time points as the first batch. In this case, organisms from three out of five animals exhibited differential gene expression patterns (Supplementary Figure S1B), leading to their selection for further RNA-seq analysis.
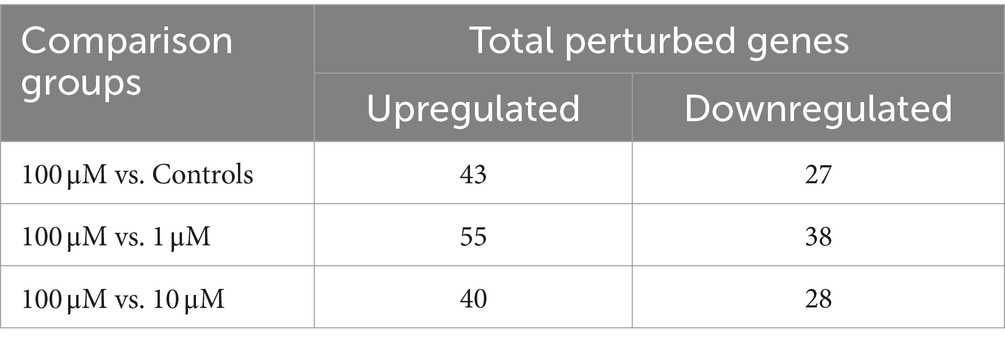
Figure 2 depicts the temporal dynamics of gene perturbations, with notable changes occurring at the 5-h time point. The most significant perturbation was evident during treatment with 100 μM CuSO4. Table 5 provides a summary of the genes that were statistically significantly perturbed. Comprehensive gene lists can be accessed in Supplementary File S2.
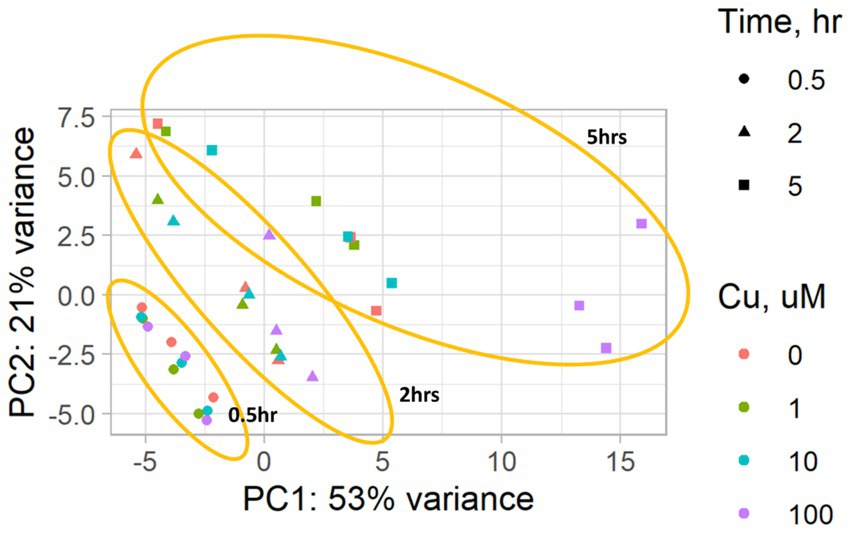
Figure 2. A PCA plot of the RNA-seq data from P. murina organisms exposed to CuSO4 at concentrations of 0 (controls), 1, 10, and 100 μM over time durations of 0.5, 2, and 5 h. Results are based on three biological replicates, i.e., each derived from organisms originating from three distinct animals. The PCA plot highlights the grouping of samples by the duration of exposure to CuSO4, marked in orange.
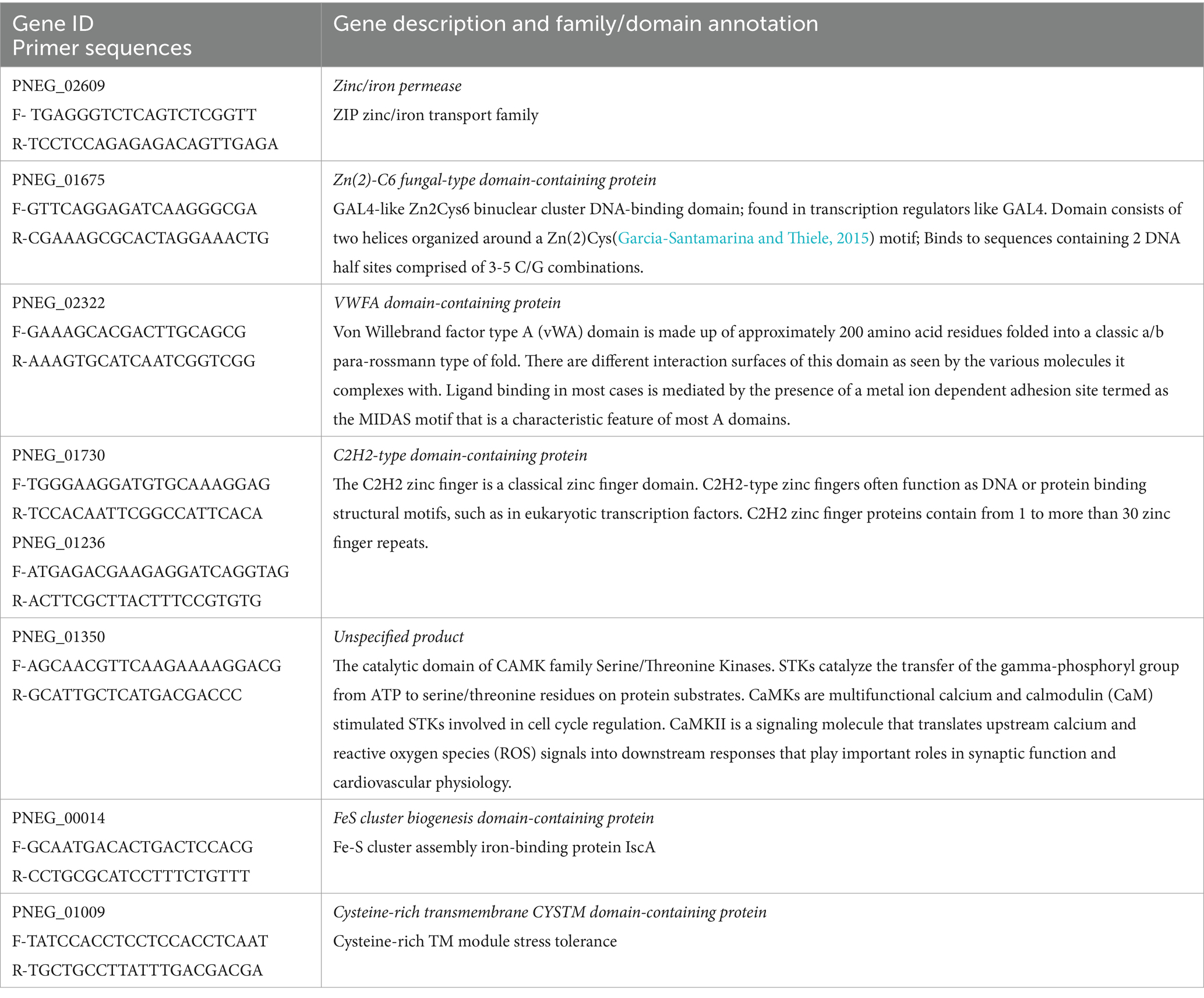
To validate some genes as potential candidates responding to copper excess in P. murina, we selected eight genes with descriptions or domain annotations related to copper or other metal ions (Table 6). Figure 3 illustrates the outcomes of RT-qPCR analysis for these genes using samples treated with 100 μM CuSO4, in comparison to control samples. Notably, Figure 3 reveals that four out of the eight selected genes exhibit statistically significant upregulation. Among these four genes, PNEG_02609 is indicative of a metal ion transporter, while PNEG_01236, PNEG_01675, and PNEG_01730 are likely transcription factors.
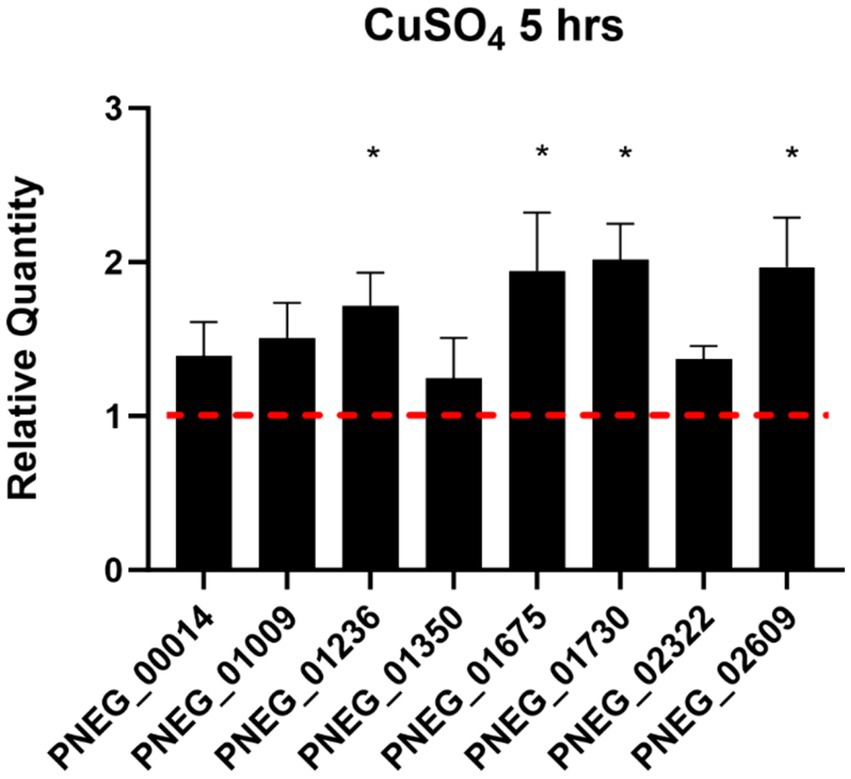
Figure 3. RT-qPCR of select genes found over-expressed in P. murina treated with 100 μM CuSO4 at 5 h. Reference gene used is thymidylate synthase. Measurements were made in triplicates. Relative quantity is shown as 2ΔΔCT. * – statistical significance at p-value <0.05.
Extended treatment of Pneumocystis murina with copper
To assess the tolerance of P. murina to free labile copper, viability assays were conducted. Cryopreserved samples of the organisms were exposed to varying concentrations of Cu(II) sulfate for seven days, including both the concentrations employed in differential gene expression studies above and the concentration (625 μM) reported as beneficial in axenic culture media (Riebold et al., 2023). The results, depicted in Figure 4, reveal that P. murina exhibits comparable viability to untreated controls at lower CuSO4 concentrations (1 and 10 μM). However, at higher concentrations, Cu(II) sulfate demonstrates a toxic effect, indicated by a rapid decline in the number of metabolically active organisms.
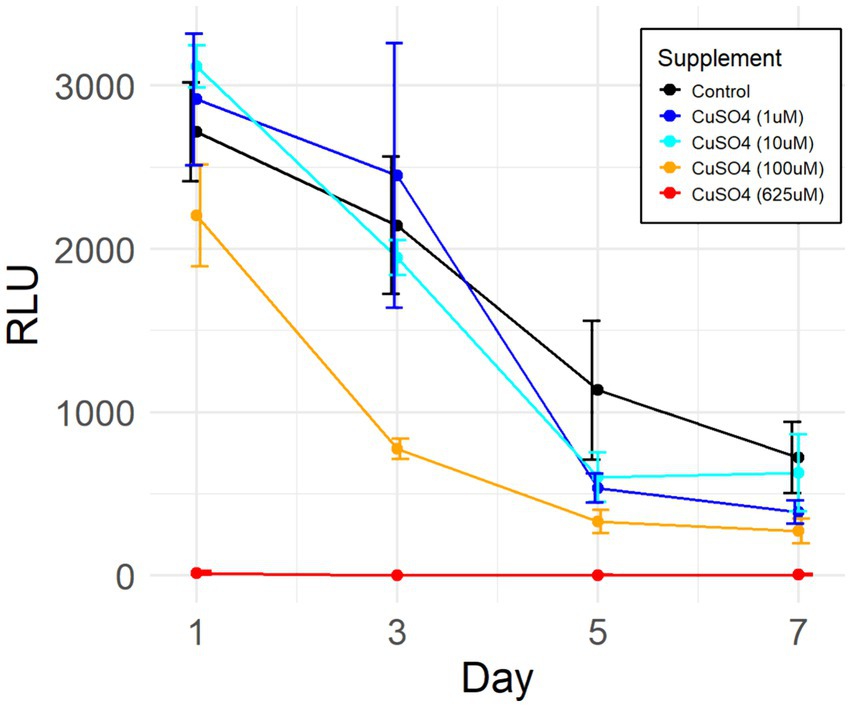
Figure 4. Efficacy of Cu(II) sulfate on P. murina viability. Viability assessed via ATP luciferase assay, indicated by relative light units (RLU) which correlate with the quantity of metabolically active organisms. Data derived from four biological replicates in each group. Adjustments made by deducting mean RLU of blank medium wells from all other values. Error bars represent standard deviations. Control comprises untreated organisms.
The two-way ANOVA test reveals that both individual variables (days post-treatment and copper sulfate concentration) as well as their interaction exhibit statistically significant differences in results. Further analysis through Tukey’s post-hoc test demonstrates that treatments utilizing 100 μM and 625 μM CuSO4 significantly differ from the controls and treatments with lower concentrations. It should be noted that there is no continuous culture/growth method for any Pneumocystis species outside the host lungs. The decline in ATP in the short-term culture we used is typical for such in vitro systems.
Discussion
This study explores the adaptive mechanisms of Pneumocystis species in a copper-rich environment. Due to their reduced genomic complexity, owing to an obligate parasitic lifestyle, these species exhibit a marked reduction (3–5 fold) in genes associated with copper binding (including sensing and tolerance) compared to other pathogenic and closely related free-living fungi (Table 1). Free labile copper exerts cytotoxic effects at elevated concentrations (Borkow and Gabbay, 2005; Valko et al., 2005). Higher organisms leverage this property for bactericidal and fungicidal purposes as part of innate immunity (Festa and Thiele, 2012; Djoko et al., 2015; Culbertson and Culotta, 2021), a phenomenon exemplified by the high concentrations of free copper in mammalian lungs (Garcia-Santamarina and Thiele, 2015; Besold et al., 2016). Our SEC-LC–MS/IMAC analysis of murine lung homogenates also showed that both infected and uninfected mice exhibited substantial levels of free labile copper in the lungs (Figure 1).
The identification of Cu-associated proteins and the genes differentially expressed under copper stress represents significant advancement in Pneumocystis biology. While prior studies have focused on Cu homeostasis in other clinically important fungal pathogens, such as Candida albicans (Schwartz et al., 2013; Khemiri et al., 2020), Cryptococcus neoformans (Ding et al., 2013; Festa et al., 2014), and Aspergillus species (Wiemann et al., 2017; Yang et al., 2018; Kang et al., 2020), our work offers unique insights into the much less-studied Pneumocystis species. Notably, we observed that significant gene expression changes in P. murina only occurred at elevated Cu(II) sulfate concentrations (100 μM, Table 5), suggesting a tolerance to lower copper ion levels. While it is established that mammalian lung environments can exhibit elevated copper levels, particularly under inflammatory conditions, we concede that our study did not directly measure these levels in infected lungs. Consequently, the copper concentrations used in our in vitro experiments (1, 10, and 100 μM) were chosen based on existing literature rather than empirical data from the specific conditions of Pneumocystis infection. This represents a limitation as these concentrations may not accurately mirror the in vivo environment.
Our study employed three orthogonal methods to identify Cu-related proteins in P. murina: sequence homology-based annotations, IMAC enrichment followed by proteomics, and RNA-seq analysis of gene expression changes after Cu exposure. Interestingly, these methods yielded largely non-overlapping results, with only four overlapping genes identified by the latter two approaches: PNEG_02587 (Hsp72-like protein), PNEG_03319 (Histidine kinase/HSP90-like ATPase domain-containing protein), PNEG_02584 (Hsp60-like protein), and PNEG_02989 (Major surface glycoprotein 2 C-terminal domain-containing protein). This discrepancy may arise from limitations in automated protein annotations based on sequence homology, which may not capture all protein molecular functions and binding sites, the low sensitivity and possible off-target selectivity of IMAC enrichment, or the constitutive overexpression of Cu-binding genes unaltered by additional CuSO4 treatment. Consequently, we recommend that future studies on Pneumocystis pathogenicity and survival under Cu stress consider all gene pools identified by these approaches.
The genes identified in our study provide the foundation for novel antifungal therapies targeting Pneumocystis-specific mechanisms of copper tolerance. This strategy is supported by literature indicating that disruption of copper uptake or detoxification processes compromises virulence in various fungal pathogens, such as C. albicans (Mackie et al., 2016), A. fumigatus (Cai et al., 2017), and C. neoformans (Waterman et al., 2007; Ding et al., 2013), underscoring the promise of targeting copper metabolism for antifungal interventions. Moreover, the conditional activation of copper ionophores, as demonstrated by the nontoxic boronic ester-masked 8-hydroxyquinoline derivative (QBP) against C. neoformans, illustrates a novel method for inducing copper-dependent cytotoxicity in fungal pathogens within host tissues (Festa et al., 2014). Collectively, these findings advocate for the exploration of copper homeostasis regulators as viable targets for antifungal drug development, aiming to discover and develop agents that specifically disrupt fungal copper regulation pathways while minimizing host impact, thereby advancing the treatment of fungal infections through a targeted approach to pathogen adaptation mechanisms.
Intriguingly, recent literature on axenic cultivation of Pneumocystis carinii recommends a medium composition (coined DMEM-O3) containing 100 μg/mL Cu(II) sulfate (Riebold et al., 2023). However, our CuSO4 supplementation viability assay indicates that such a concentration is exceedingly toxic, leading to rapid organismal death during ex vivo cultivation (Figures 4, 625 μM). This difference may stem from discrepancy in aliquot sizes used in the studies. We added a 0.25 mL aliquot of CuSO4 (625 μM) to the medium in the viability assay (see Methods), whereas the other study did not specify the actual amount of copper sulfate of the same concentration added to the culture medium. Another possibility in discrepancy is that P. jirovecii, the human-infecting species used in the other study, may have higher tolerance to the copper stress.
It should be noted that in contrast to other fungal pathogens affecting humans, Pneumocystis species are less studied and annotated due to challenges in cultivation and the application of traditional gene functional studies, such as knockouts or mutagenesis. Consequently, a significant proportion of their genes remain uncharacterized, with many proteins identified only as “uncharacterized,” “hypothetical,” or “predicted.” This limitation extends to Gene Ontology (GO) terms, as well as the cellular processes these genes participate in. Therefore, our ability to perform traditional functional enrichment analysis and identify enriched GO categories or biological pathways is hindered. Despite these obstacles, our study lays the groundwork for future research. As methodologies evolve and allow for the detailed study of these organisms, the genes we report could become focal points for understanding copper tolerance mechanisms and identifying new drug targets in Pneumocystis.
Conclusion
This study provides new insights into the copper sensing and tolerance mechanisms of Pneumocystis species, with a particular focus on P. murina. Despite considerable gene loss compared to free-living fungi, Pneumocystis species have developed unique strategies for managing copper homeostasis. The discovery of specific copper-binding proteins and the differential gene expression in response to excessive copper exposure are significant contributions to the growing understanding of the adaptation mechanisms of extracellular pathogens in copper-rich environments, such as mammalian lungs. This preliminary study investigated Pneumocystis murina genes that may play a role in copper tolerance. These findings pave the way for future research into copper-binding proteins and their mechanisms of action once in vitro cultivation of the organisms becomes feasible. The findings also hold potential for the development of new therapeutic strategies targeting the distinctive aspects of Pneumocystis copper homeostasis.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/bioproject/; PRJNA1076335.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee at the Veterans Affairs Medical Center, Cincinnati, OH. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AP: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. SS: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. AA: Data curation, Investigation, Methodology, Resources, Validation, Writing – original draft. SR: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft. JL: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft. MC: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Institutes of Health via the following grants: R21AI143467, R01HL146266, and by the Veterans Affairs grant I01BX004441. MC is a Senior Research Career Scientist supported by IK6BX005232 Department of Veterans Affairs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at:
https://www.frontiersin.org/articles/10.3389/fmicb.2024.1383737/full#supplementary-material
SUPPLEMENTARY FILE S1 | Proteins identified by proteomics after the Cu-IMAC enrichment.
SUPPLEMENTARY FILE S2 | Differentially expressed genes of P. murina exposed to CuSO4.
SUPPLEMENTARY FIGURE S1 | PCA plots of the RNA-seq data from P. murina organisms exposed to CuSO4 over time durations of 0.5, 2, and 5 h. (A) The initial batch of treatment with CuSO4 at concentrations of 0 (controls), 1 ng/mL and 100 ng/mL. The organisms were derived from 6 animals. No differentially expressed genes were found. (B) The second batch of treatment with CuSO4 at concentrations of 0 (controls), 1, 10, and 100 μM over time durations of 0.5, 2, and 5 h. The organisms were derived from 5 animals, three of which (1, 5, and 6) were used in this study.
References
Amos, B., Aurrecoechea, C., Barba, M., Barreto, A., Basenko, E. Y., Bazant, W., et al. (2022). VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 50, D898–D911. doi: 10.1093/nar/gkab929
Andrei, A., Ozturk, Y., Khalfaoui-Hassani, B., Rauch, J., Marckmann, D., Trasnea, P. I., et al. (2020). Cu homeostasis in Bacteria: the ins and outs. Membranes (Basel). 10, 1–46. doi: 10.3390/membranes10090242
Arguello, J. M., Raimunda, D., and Padilla-Benavides, T. (2013). Mechanisms of copper homeostasis in bacteria. Front. Cell. Infect. Microbiol. 3:73. doi: 10.3389/fcimb.2013.00073
Becker, K. W., and Skaar, E. P. (2014). Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 38, 1235–1249. doi: 10.1111/1574-6976.12087
Besold, A. N., Culbertson, E. M., and Culotta, V. C. (2016). The Yin and Yang of copper during infection. J. Biol. Inorg. Chem. 21, 137–144. doi: 10.1007/s00775-016-1335-1
Borkow, G., and Gabbay, J. (2005). Copper as a biocidal tool. Curr. Med. Chem. 12, 2163–2175. doi: 10.2174/0929867054637617
Cai, Z., Du, W., Zeng, Q., Long, N., Dai, C., and Lu, L. (2017). Cu-sensing transcription factor Mac 1 coordinates with the Ctr transporter family to regulate cu acquisition and virulence in Aspergillus fumigatus. Fungal Genet. Biol. 107, 31–43. doi: 10.1016/j.fgb.2017.08.003
Cai, Z., Du, W., Zhang, Z., Guan, L., Zeng, Q., Chai, Y., et al. (2018). The Aspergillus fumigatus transcription factor ace a is involved not only in cu but also in Zn detoxification through regulating transporters Crp a and Zrc a. Cell. Microbiol. 20:e12864. doi: 10.1111/cmi.12864
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Culbertson, E. M., and Culotta, V. C. (2021). Copper in infectious disease: using both sides of the penny. Semin. Cell Dev. Biol. 115, 19–26. doi: 10.1016/j.semcdb.2020.12.003
Cushion, M. T., Linke, M. J., Ashbaugh, A., Sesterhenn, T., Collins, M. S., Lynch, K., et al. (2010). Echinocandin treatment of pneumocystis pneumonia in rodent models depletes cysts leaving trophic burdens that cannot transmit the infection. PLoS One 5, 1–12. doi: 10.1371/journal.pone.0008524
Ding, C., Festa, R. A., Chen, Y. L., Espart, A., Palacios, O., Espin, J., et al. (2013). Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe 13, 265–276. doi: 10.1016/j.chom.2013.02.002
Ding, C., Yin, J., Tovar, E. M., Fitzpatrick, D. A., Higgins, D. G., and Thiele, D. J. (2011). The copper regulon of the human fungal pathogen Cryptococcus neoformans H99. Mol. Microbiol. 81, 1560–1576. doi: 10.1111/j.1365-2958.2011.07794.x
Djoko, K. Y., Ong, C. L., Walker, M. J., and McEwan, A. G. (2015). The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J. Biol. Chem. 290, 18954–18961. doi: 10.1074/jbc.R115.647099
Festa, R. A., Helsel, M. E., Franz, K. J., and Thiele, D. J. (2014). Exploiting innate immune cell activation of a copper-dependent antimicrobial agent during infection. Chem. Biol. 21, 977–987. doi: 10.1016/j.chembiol.2014.06.009
Festa, R. A., and Thiele, D. J. (2011). Copper: an essential metal in biology. Curr. Biol. 21, R877–R883. doi: 10.1016/j.cub.2011.09.040
Festa, R. A., and Thiele, D. J. (2012). Copper at the front line of the host-pathogen battle. PLoS Pathog. 8:e1002887. doi: 10.1371/journal.ppat.1002887
Garcia-Santamarina, S., and Thiele, D. J. (2015). Copper at the fungal pathogen-host Axis. J. Biol. Chem. 290, 18945–18953. doi: 10.1074/jbc.R115.649129
Gerwien, F., Skrahina, V., Kasper, L., Hube, B., and Brunke, S. (2018). Metals in fungal virulence. FEMS Microbiol. Rev. 42, 1–21. doi: 10.1093/femsre/fux050
Green, J. V., Orsborn, K. I., Zhang, M., Tan, Q. K. G., Greis, K. D., Porollo, A., et al. (2013). Heparin-binding motifs and biofilm formation by Candida albicans. J. Infect. Dis. 208, 1695–1704. doi: 10.1093/infdis/jit391
Hauser, P. M., Burdet, F. X., Cisse, O. H., Keller, L., Taffe, P., Sanglard, D., et al. (2010). Comparative genomics suggests that the fungal pathogen Pneumocystis is an obligate parasite scavenging amino acids from its host's lungs. PLoS One 5:e15152. doi: 10.1371/journal.pone.0015152
Kang, S., Seo, H., Moon, H. S., Kwon, J. H., Park, Y. S., and Yun, C. W. (2020). The role of zinc in copper homeostasis of Aspergillus fumigatus. Int. J. Mol. Sci. 21, 1–15. doi: 10.3390/ijms21207665
Khemiri, I., Tebbji, F., and Sellam, A. (2020). Transcriptome analysis uncovers a link between copper metabolism, and both fungal fitness and antifungal sensitivity in the opportunistic yeast Candida albicans. Front. Microbiol. 11:935. doi: 10.3389/fmicb.2020.00935
Li, C. X., Gleason, J. E., Zhang, S. X., Bruno, V. M., Cormack, B. P., and Culotta, V. C. (2015). Candida albicans adapts to host copper during infection by swapping metal cofactors for superoxide dismutase. Proc. Natl. Acad. Sci. USA 112, E5336–E5342. doi: 10.1073/pnas.1513447112
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ma, L., Chen, Z., Huang da, W., Kutty, G., Ishihara, M., Wang, H., et al. (2016). Genome analysis of three Pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat. Commun. 7:10740. doi: 10.1038/ncomms10740
Mackie, J., Szabo, E. K., Urgast, D. S., Ballou, E. R., Childers, D. S., Mac Callum, D. M., et al. (2016). Host-imposed copper poisoning impacts fungal micronutrient acquisition during systemic Candida albicans infections. PLoS One 11:e0158683. doi: 10.1371/journal.pone.0158683
Moraes, D., Assuncao, L. D. P., Silva, K., Soares, C. M. A., Silva-Bailao, M. G., and Bailao, A. M. (2023). High copper promotes cell wall remodeling and oxidative stress in Histoplasma capsulatum, as revealed by proteomics. Fungal Biol. 127, 1551–1565. doi: 10.1016/j.funbio.2023.05.004
Moraes, D., Tristao, G. B., Rappleye, C. A., Ray, S. C., Ribeiro-Dias, F., Gomes, R. S., et al. (2024). The influence of a copper efflux pump in Histoplasma capsulatum virulence. FEBS J. 291, 744–760. doi: 10.1111/febs.16999
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Porollo, A., Sesterhenn, T. M., Collins, M. S., Welge, J. A., and Cushion, M. T. (2014). Comparative genomics of Pneumocystis species suggests the absence of genes for myo-inositol synthesis and reliance on inositol transport and metabolism. MBio 5:e01834. doi: 10.1128/mBio.01834-14
Rapp, S. J., Dershem, V., Zhang, X., Schutte, S. C., and Chariker, M. E. (2020). Varying negative pressure wound therapy acute effects on human Split-thickness autografts. J. Burn Care Res. 41, 104–112. doi: 10.1093/jbcr/irz122
Ray, S. C., and Rappleye, C. A. (2022). Mac 1-dependent copper sensing promotes Histoplasma adaptation to the phagosome during adaptive immunity. MBio 13:e0377321. doi: 10.1128/mbio.03773-21
Riebold, D., Mahnkopf, M., Wicht, K., Zubiria-Barrera, C., Heise, J., Frank, M., et al. (2023). Axenic Long-term cultivation of Pneumocystis jirovecii. J. Fungi (Basel) 9, 1–35. doi: 10.3390/jof9090903
Schwartz, J. A., Olarte, K. T., Michalek, J. L., Jandu, G. S., Michel, S. L., and Bruno, V. M. (2013). Regulation of copper toxicity by Candida albicans GPA2. Eukaryot. Cell 12, 954–961. doi: 10.1128/EC.00344-12
Smith, A. D., Logeman, B. L., and Thiele, D. J. (2017). Copper acquisition and utilization in Fungi. Ann. Rev. Microbiol. 71, 597–623. doi: 10.1146/annurev-micro-030117-020444
Song, J., Li, R., and Jiang, J. (2019). Copper homeostasis in Aspergillus fumigatus: opportunities for therapeutic development. Front. Microbiol. 10:774. doi: 10.3389/fmicb.2019.00774
Valko, M., Morris, H., and Cronin, M. T. (2005). Metals, toxicity and oxidative stress. Curr. Med. Chem. 12, 1161–1208. doi: 10.2174/0929867053764635
Waldron, K. J., Rutherford, J. C., Ford, D., and Robinson, N. J. (2009). Metalloproteins and metal sensing. Nature 460, 823–830. doi: 10.1038/nature08300
Walsh, K. B., Zhang, X., Zhu, X., Wohleb, E., Woo, D., Lu, L., et al. (2019). Intracerebral hemorrhage induces inflammatory gene expression in peripheral blood: global transcriptional profiling in intracerebral hemorrhage patients. DNA Cell Biol. 38, 660–669. doi: 10.1089/dna.2018.4550
Waterman, S. R., Hacham, M., Hu, G., Zhu, X., Park, Y. D., Shin, S., et al. (2007). Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J. Clin. Invest. 117, 794–802. doi: 10.1172/JCI30006
Weissman, Z., Berdicevsky, I., Cavari, B. Z., and Kornitzer, D. (2000). The high copper tolerance of Candida albicans is mediated by a P-type ATPase. Proc. Natl. Acad. Sci. USA 97, 3520–3525. doi: 10.1073/pnas.97.7.3520
Wiemann, P., Perevitsky, A., Lim, F. Y., Shadkchan, Y., Knox, B. P., Landero Figueora, J. A., et al. (2017). Aspergillus fumigatus copper export machinery and reactive oxygen intermediate defense counter host copper-mediated oxidative antimicrobial offense. Cell Rep. 19, 1008–1021. doi: 10.1016/j.celrep.2017.04.019
Keywords: Pneumocystis , copper sensing, copper tolerance, metalloproteome, cuproproteins, host-pathogen interaction
Citation: Porollo A, Sayson SG, Ashbaugh A, Rebholz S, Landero Figueroa JA and Cushion MT (2024) Insights into copper sensing and tolerance in Pneumocystis species. Front. Microbiol. 15:1383737. doi: 10.3389/fmicb.2024.1383737
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Alexandre Melo BAILAO, Universidade Federal de Goiás, BrazilLimper Harold Andrew, Mayo Clinic, United States
Olga Matos, New University of Lisbon, Portugal
Hubertus Haas, Innsbruck Medical University, Austria
Copyright © 2024 Porollo, Sayson, Ashbaugh, Rebholz, Landero Figueroa and Cushion. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleksey Porollo, Alexey.Porollo@cchmc.org; Melanie T. Cushion, Melanie.Cushion@uc.edu
 Aleksey Porollo
Aleksey Porollo Steven G. Sayson
Steven G. Sayson Alan Ashbaugh
Alan Ashbaugh Sandra Rebholz
Sandra Rebholz Julio A. Landero Figueroa6
Julio A. Landero Figueroa6 Melanie T. Cushion
Melanie T. Cushion