- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, Guangxi Research Center for Microbial and Enzyme Engineering Technology, College of Life Science and Technology, Guangxi University, Nanning, China
- 2Guangxi Key Laboratory of Sugarcane Biology, College of Agriculture, Guangxi University, Nanning, China
Introduction: SUMOylation is an important post-translational modification that regulates the expression, localization, and activity of substrate proteins, thereby participating in various important cellular processes such as the cell cycle, cell metabolism, gene transcription, and antiviral activity. However, the function of SUMOylation in phytopathogenic fungi has not yet been adequately explored.
Methods: A comprehensive analysis composed of proteomics, affinity pull-down, molecular and cellular approaches was performed to explore the roles of SUMOylation in Cryphonectria parasitica, the fungal pathogen responsible for chestnut blight.
Results and discussion: CpSmt3, the gene encoding the SUMO protein CpSmt3 in C. parasitica was identified and characterized. Deletion of the CpSmt3 gene resulted in defects in mycelial growth and hyphal morphology, suppression of sporulation, attenuation of virulence, weakening of stress tolerance, and elevated accumulation of hypovirus dsRNA. The ΔCpSmt3 deletion mutant exhibited an increase in mitochondrial ROS, swollen mitochondria, excess autophagy, and thickened cell walls. About 500 putative SUMO substrate proteins were identified by affinity pull-down, among which many were implicated in the cell cycle, ribosome, translation, and virulence. Proteomics and SUMO substrate analyses further revealed that deletion of CpSmt3 reduced the accumulation of CpRho1, an important protein that is involved in TOR signal transduction. Silencing of CpRho1 resulted in a phenotype similar to that of ΔCpSmt3, while overexpression of CpRho1 could partly rescue some of the prominent defects in ΔCpSmt3. Together, these findings demonstrate that SUMOylation by CpSmt3 is vitally important and provide new insights into the SUMOylation-related regulatory mechanisms in C. parasitica.
1 Introduction
Small ubiquitin-related modifier (SUMO) is an important protein that modifies substrate proteins and thereby plays a crucial role in various biological processes such as the cell cycle, secondary metabolism, transcription, DNA damage repair, and other cellular processes (Eifler and Vertegaal, 2015; Yau et al., 2021). The process of SUMOylation is similar to that of ubiquitination of a protein. First, the precursor of the SUMO molecule is cleaved at the C-terminus by SUMO protease, exposing the di-glycine motif. With the participation of ATP, the mature SUMO molecule is then connected to the cysteine residue of the E1 enzyme (composed of Aos1 and Uba2), forming a high-energy thioester bond and activating the SUMO molecule. The SUMO molecule is then transferred to the cysteine residue of the E2 enzyme (Ubc9) to form a second thioester bond between SUMO and the E2 enzyme. The E2 enzyme then recognizes the SUMO consensus motif of the target protein and the E3 SUMO ligase facilitates the transfer of the activated SUMO to a lysine residue of the target protein (Chang and Yeh, 2020; Vertegaal, 2022). Although most E3 ligases (Siz1, Siz2, Mms21, and Zip3) are characterized by an SP-RING domain (Cheng et al., 2006; Reindle et al., 2006; Pasupala et al., 2012; Kim et al., 2016), some E3 ligases do not have this canonical domain for catalytic activity (Johnson and Gupta, 2001; Kahyo et al., 2001; Pichler et al., 2002; Kagey et al., 2003).
Recent studies indicate that protein SUMOylation is important for fungi. In Saccharomyces cerevisiae, SUMOylation is involved in DNA replication, hypoxia, and protein folding. Disruption of the SUMO-encoding gene Smt3 results in a lethal phenotype (Takahashi et al., 1999; Newman et al., 2017). In model filamentous fungus Aspergillus nidulans, SUMO protein is non-essential for vegetative growth, but required for cellular differentiation (Gupta et al., 2020). In the phytopathogenic fungus Aspergillus flavus, deletion of the SUMO gene adversely affected colony forming ability and pathogenicity (Nie et al., 2016).
Cryphonectria parasitica is the fungal pathogen responsible for the chestnut blight (Alfen, 1982). It can be infected by Cryphonectria hypovirus 1 (CHV1)-EP713. The virus–fungus interaction regulates fungal virulence and virulence-associated traits so the virus could serve as a potent biocontrol agent (Nuss, 2005). The virus exploits the trans-Golgi network and ribosomes of C. parasitica for replication. Viral infection alters its host’s physiological processes by regulating the MAPK pathway, the citric acid synthesis pathway, and cellular glutamate biogenesis to bring about a cellular environment that is conducive to viral survival and replication (Jacob-Wilk et al., 2006; Yao et al., 2013; So et al., 2017; So and Kim, 2017; Chiba et al., 2018; Chun et al., 2020). However, the impact of SUMOylation on the virus–fungus interaction has not been investigated.
In this study, we characterized the SUMO protein ortholog CpSmt3 in C. parasitica. CpSmt3 was found to be involved in mycelial growth, hyphal morphology, sporulation, organelle function, stress response, virulence, and antiviral defense. Among the proteins that interact with CpSmt3 in vivo, CpRho1 seemed to be an essential target of CpSmt3, as strains with silenced CpRho1 had a phenotype very similar to that of the ΔCpSmt3 deletion mutant. Collectively, these results demonstrate the key role of SUMOylation in C. parasitica growth and provide insights into the mechanisms underlying the diverse functions of SUMOylation in C. parasitica.
2 Materials and methods
2.1 Strains and culture conditions
The C. parasitica wildtype strain EP155 (ATCC 38755), its isogenic hypovirus (CHV1-EP713)-infected strain EP155/CHV1-EP713 (designated EP155/+virus; ATCC 52571) (Chen et al., 1994), highly efficient homologous recombinant strain KU80 (ΔCpku80 of EP155) (Lan et al., 2008), and derived mutant strains were incubated on potato glucose agar (PDA) medium at 26°C with a 12/12-h light/dark cycle for phenotypic analyses. PDA medium was also used for DNA, RNA, and protein extraction, as previously described (Li et al., 2022).
2.2 Generation of gene deletion, complementation and overexpression strains
The ΔCpSmt3 deletion mutant was constructed using a homologous recombination method based on the KU80 strain. Using EP155 genome DNA as the template, the upstream (1,051 bp) and downstream (1,150 bp) regions flanking the CpSmt3 gene were amplified using the primers smt3-left-F/smt3-left-R and smt3-right-F/smt3-right-R, respectively. The hygromycin B resistance gene hph was amplified using the primers hph-F/hph-R, and then the three amplification products were ligated by fusion polymerase chain reaction (PCR) to obtain a 4.1-kb cassette. The purified products were transformed into KU80 protoplasts mediated by polyethylene glycol (PEG), which were regenerated and screened on medium containing 30 μg/mL hygromycin B (Thermo Fisher Scientific, USA). The transformed strains were consecutively cultured on PDA containing hygromycin B for three generations, and were subsequently identified. According to the standard protocol described by Sambrook and Russell (2001), Southern blot and PCR methods were used to identify the ΔCpSmt3 deletion mutant. The construction of ΔCpUbc2 and ΔCpUbc9 deletion mutants were performed using the same protocol with corresponding primer sets.
The ΔCpSmt3-Com complementation strain was generated by amplifying the entire CpSmt3 gene (open reading frame and promoter sequence) by PCR. Subsequently, the amplified fragment was cloned into the transformation vector pCPXG418, with a geneticin resistance (G418) cassette. The resulting construct pCPXG418-CpSmt3 was transformed into ΔCpSmt3. ΔCpSmt3-Com was then confirmed by PCR using primer smt3-qPCR-F/smt3-qPCR-R.
To construct the CpSmt3-OE overexpression strain, the CpSmt3 open reading frame was amplified and then inserted into the vector pCPXG418, with the gpd promoter for the transgene. The resulting construct pCPXG418-CpSmt3 was then introduced into wildtype (EP155) protoplasts. Positive clones were confirmed by PCR and quantitative reverse transcription PCR (qRT-PCR). ΔCpSmt3 overexpressing CpRho1 (designated ΔCpSmt3/CpRho1-OE) was constructed using the same protocol with a corresponding primer set. Primers used in this study are compiled in Supplementary Table 1.
2.3 Construction of RNA interference (RNAi) strains
CpRho1 fragment (including 182 bp intron and 412 bp exon) was amplified by PCR and cloned into the short hairpin RNA (shRNA) expression vector WRNAIPG (constructed by our group), between the Pgpd promoter and TrapC terminator, with hygromycin B resistance as the selection marker. The RNAi construct was then transformed into wildtype (EP155) protoplasts, and confirmed by qRT-PCR. Primers used in this study are compiled in Supplementary Table 1.
2.4 Characterization of fungal phenotype
Phenotypic traits (growth rate, pigmentation, sporulation, and stress tolerance) were assessed using previously established methods (Kim et al., 1995). Briefly, for assessing sporulation, fungal strains were cultured on PDA medium at 26°C for 14 days. Conidia were collected and quantified using a hemacytometer. For assessing stress tolerance, strains were cultured on PDA media supplemented with stress chemicals. Three independent replicates were performed for each experiment. For mycelium dry weight assays, the same size blocks (0.5 mm2× 0.5 mm2) cut from 5-day-old PDA cultures were inoculated into EP liquid medium (Puhalla, 1971). After incubation at 26°C for 7 days, the mycelia in EP liquid medium were filtered and washed. Then, the mycelium was dried at 60°C for 48 h. The dry weight of each mycelium was determined by an electronic balance (Sartorius, Germany).
2.5 Microscopy
Light microscopy was performed using an Olympus BX51 fluorescent microscope (Olympus, Japan). Transmission electron microscopy (TEM) was performed using a JEM-1400-FLASH Transmission Electron Microscope (JEOL, Japan). For in situ mitochondria analysis, fungal hyphae were cultured in PDA medium for 10 days, scraped, and fixed with 2.5% glutaraldehyde in 100 mM phosphate buffer (pH 7.2) at 4°C overnight. After rinsing with phosphate buffer (50 mM, pH 6.8), the samples were dehydrated, embedded with Epon 812, then sliced into ultrathin sections, stained with uranium acetate (2%), and poststained with lead citrate (Shi et al., 2019). Three biological replicates were performed for each sample.
2.6 Assessment of mitochondrial membrane potential (MMP)
Mitochondrial membrane potential was assessed using a JC-1 fluorescent probe (Beyotime, China), and the ratio of JC-1 red/green fluorescence intensity was utilized as a representation of MMP. Briefly, the strains were cultured on PDA medium with a piece of cellophane for 10 days. The harvested mycelia were placed in JC-1 staining buffer containing 10 μM JC-1, incubated at 37°C in the dark for 20 min, and then washed with JC-1 washing buffer twice. The red fluorescence of JC-1 dimers represents normal MMP, while the green fluorescence of JC-1 monomers signifies a depolarized MMP. A transition from red to green fluorescence indicates a reduction in MMP.
2.7 Virulence assays
The virulence of the strains was analyzed using dormant stems of Chinese chestnut (Castanea mollissima), as described previously (Shi et al., 2014). Canker sizes were measured and photographed 28 days post inoculation. The virulence assay was repeated three times for each fungal strain.
2.8 Detection of reactive oxygen species (ROS)
Total fungal ROS production was detected by 2′, 7′-dichlorodihydrofluorescein diacetate (DCFH-DA) staining. The fungal mycelia were incubated with 10 μM DCFH-DA (Beyotime, China) in the dark at 37°C for 30 min, and then washed with PBS (10 mM, pH 7.5) three times. At a maximum excitation wavelength of 480 nm and a maximum emission wavelength of 525 nm, fluorescence microscopy was used to detect fluorescence signals. For mitochondrial ROS (mtROS) level detection, MitoSOX red mitochondrial superoxide dismutase indicator (MEC, USA) was used as described previously (Luo et al., 2021). Briefly, mycelia collected from PDA medium were washed three times with PBS, incubated with 10 μM MitoSOX red for 30 min at room temperature, washed three times with PBS again, and observed using fluorescence microscopy.
2.9 Purification of SUMO conjugates
To obtain SUMO conjugates, a plasmid expressing 3 × FLAG–CpSmt3 fusion protein was constructed and transformed into the wildtype strain, and transformants were identified based on G418 resistance. A transformant was confirmed by western blot analysis using an anti-FLAG antibody (1: 2000, ABclonal) as the primary antibody. The verified strain was incubated in liquid EP medium for 3 days then harvested. Total protein was extracted from the cells using NP-40 Lysis Buffer (Beyotime, China) with 1 mM phenylmethylsulfonyl fluoride (PMSF), incubated with anti-FLAG antibody (ABclonal) overnight at 4°C and then mixed with protein A/G agarose (Sangon, China) for 2 h, which was then washed using 1× immunoprecipitation (IP) buffer and 1 × 0.1 IP buffer three and six times, respectively. Next, the lysates were incubated with loading buffer at 95°C for 5 min and then centrifuged at 25°C and 1,200 rpm for 1 min. The proteins were collected, digested using trypsin, and analyzed by LC-MS/MS (Q Exactive HF, Thermo Scientific). The resulting MS/MS data were used to search against the nonredundant C. parasitica protein database of the Joint Genome Institute (JGI).1 Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using eggnog-mapper2 and Tbtools software (Chen et al., 2020).
2.10 Proteomics analysis
Fungal strains were cultured on PDA medium with a piece of cellophane for 10 days, and then the mycelium was collected for proteome sequencing. Protein isolation was conducted using a fungal protein extraction kit (ProExcell™, China). Next, 20 μg protein was digested overnight using 1 μg sequencing-grade trypsin (Promega Corp., USA). Thereafter, the peptides were prepared for nano-LC-MS/MS by C18 Zip-Tip purification according to the manufacturer’s protocol (Millipore Inc., USA). Three biological replicate samples were then suspended in water with 0.1% formic acid (v/v) and subjected to nano-LC-MS/MS (Q Exactive HF, Thermo Scientific, USA). Briefly, 1 μg peptide sample was injected into a reverse-phase BEH C18 column (100 μm × 100 mm; particle size: 1.7 μm; pore size: 300 Å) (Waters Corp., Massachusetts, USA) for LC using a Waters nanoACQUITY LC system. Peptides eluting from the column were analyzed by data-dependent MS/MS on a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific Inc., Massachusetts, USA). The data were searched against the C. parasitica genome database and a decoy database using the Sequest HT search engine in Proteome Discoverer 1.4 software (Thermo Fisher Scientific Inc., Massachusetts, USA).
Differentially expressed proteins (DEPs) between the wildtype strain and ΔCpSmt3 were determined using the t-test function in R language, with fold change > 1.2 and p < 0.05 indicating statistical significance. The number of DEPs was then calculated. GO enrichment analysis of the DEPs was conducted using DAVID software,3 with p < 0.05 indicating statistical significance. DEPs were also annotated using KEGG Mapper.4 The KEGG enrichment analysis of the DEPs was conducted using DAVID software.5 Fisher’s exact test was employed as the statistical test, with p < 0.05 indicating statistical significance.
3 Results
3.1 Identification of CpSmt3 gene in C. parasitica
The SUMO ortholog of C. parasitica was identified by searching the C. parasitica genome database6 using the S. cerevisiae Smt3 protein sequence as a query (GenBank: QHB07964). A protein comprising 106 amino acids, designated CpSmt3 (JGI ID: 356715), was found to share 68.3% similarity with the S. cerevisiae Smt3. The CpSmt3 gene contains three exons and two introns (Supplementary Figure 1A). Domain analysis revealed that it has a conserved Ubl_Smt3_like domain (coordinates 25–97). Phylogenetic analysis showed that CpSmt3 is most closely related to the SUMO proteins of Valsa mali and Neurospora crassa (Supplementary Figure 1B).
3.2 CpSmt3 is required for fungal development, stress response, and virulence
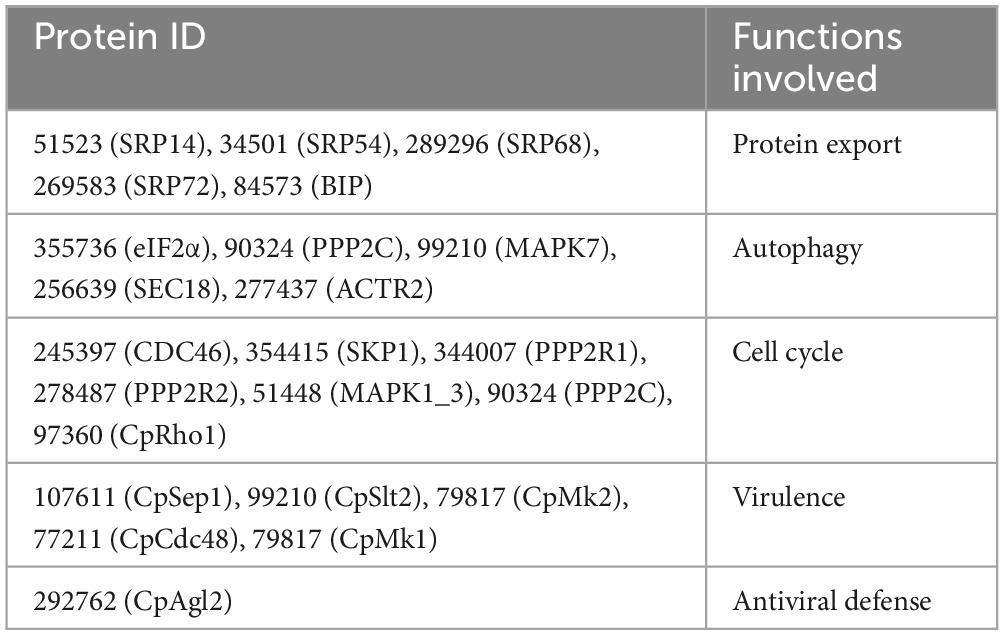
To investigate the function of the CpSmt3 gene in C. parasitica, we constructed a CpSmt3 deletion mutant (ΔCpSmt3) and a CpSmt3 overexpression strain (CpSmt3-OE) (Supplementary Figure 2). Southern blot and RT-PCR analyses of ΔCpSmt3 showed that CpSmt3 was successfully deleted from the genome of C. parasitica (Supplementary Figures 2B–D). In addition, western blot analysis using anti-SUMO1 antibody showed that the wildtype and complementation strain (ΔCpSmt3-Com) had a ∼16 kDa band, while ΔCpSmt3 did not (Supplementary Figure 2E). This indicated that the CpSmt3 was involved in SUMOylation of C. parasitica. Compared to the wildtype strain EP155 and the parental strain KU80, ΔCpSmt3 exhibited reduced aerial hyphae, intense pigmentation, slow colony growth, and no spore formation (Figures 1A–C). Microscopy showed that the hyphae of ΔCpSmt3 were swollen and accompanied by multinucleated cells, and the hyphal diameter was also significantly larger than that of the wildtype strain (Figure 1D and Supplementary Figure 2F). However, no significant changes were observed in CpSmt3-OE (Supplementary Figures 2G, H).
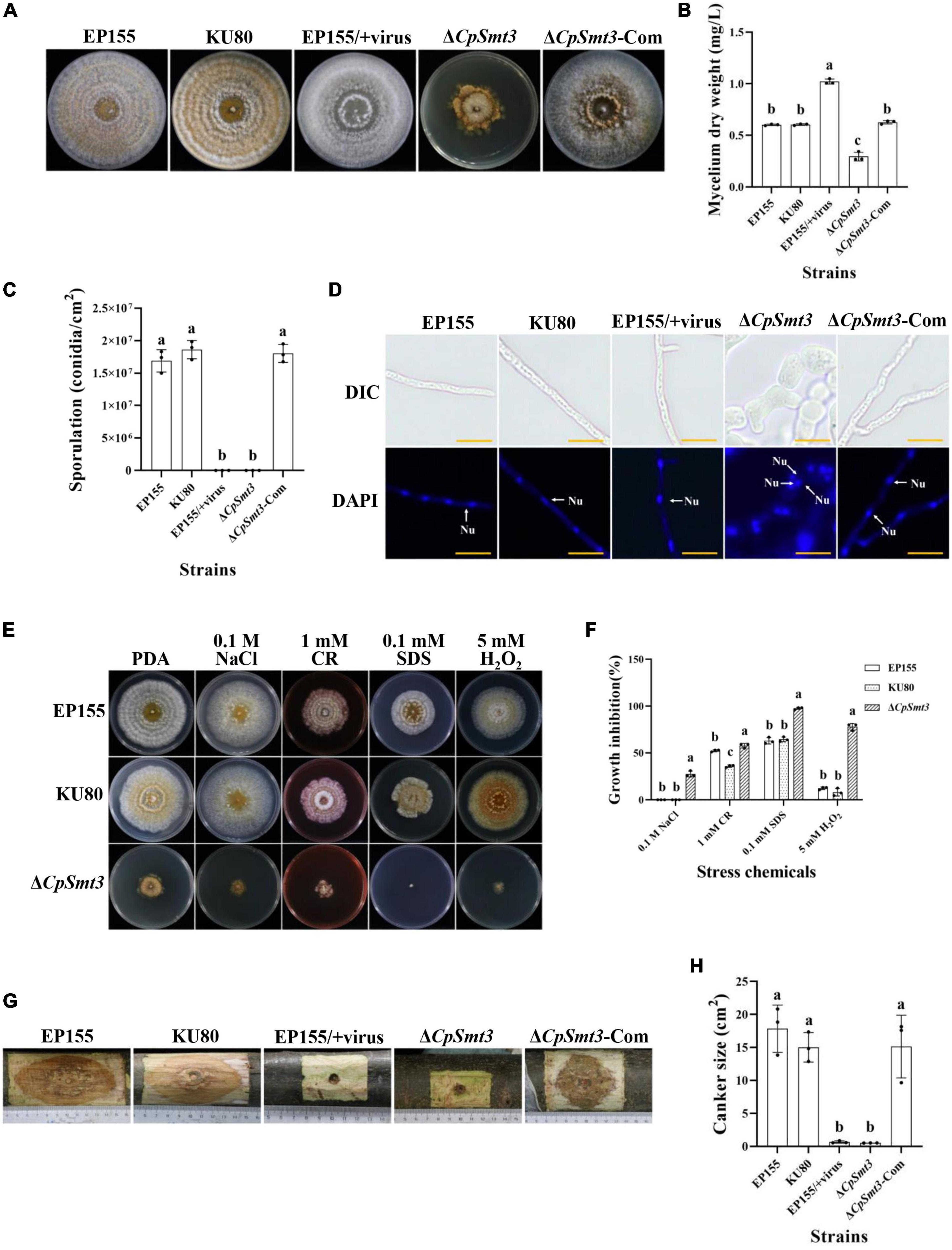
Figure 1. Analysis of colony phenotypes, sporulation, stress tolerance and virulence of CpSmt3 deletion mutants. (A) Colonial morphologies of EP155, KU80, EP155/CHV1-EP713, ΔCpSmt3, and ΔCpSmt3-Com strains cultured on PDA for 14 days. (B) Mycelium dry weight statistics of the tested strains in (A). (C) Sporulation levels of the tested strains. (D) DAPI staining indicates aberrant nuclear segregation in ΔCpSmt3 mutant. White arrow represents nucleus. Scale bar = 10 μm. (E) Colony morphologies of the tested strains on cultured PDA medium supplemented with stress agents at 26°C for 7 days. (F) Colony area statistics of the tested strains in (E). (G,H) Cankers induced by the tested strains on dormant stems of Chinese chestnut and statistics of the canker sizes. The inoculated stems were kept at 26°C and cankers were measured and photographed 28 days post inoculation. Error bars represent the standard deviation based on three independent experiments. Different letters on the bars indicate significant differences (p < 0.05).
To determine the role of SUMO in the stress response, wildtype and ΔCpSmt3 were inoculated on PDA media supplemented with cell stress agents Congo red, sodium dodecyl sulfate (SDS), NaCl, and H2O2. ΔCpSmt3 was more sensitive to the stress than the wildtype strain, especially to H2O2 and SDS, with inhibition rates of 78.02 and 97.69%, respectively (Figures 1E, F).
To explore the role of CpSmt3 in C. parasitica virulence, chestnut stems were inoculated with the wildtype strain, KU80, EP155/+virus, ΔCpSmt3, and complementation strain to assess their virulence. The canker formed by ΔCpSmt3 was significantly smaller than that of wildtype strain or KU80, but comparable to the canker formed by the hypovirus-infected wildtype strain. The reduced virulence of ΔCpSmt3 was fully restored by re-induction of the wildtype copy of CpSmt3 (Figures 1G, H).
3.3 Deletion of CpSmt3 results in organelle defects and excessed autophagy
As SUMOylation has been reported to be involved in mitochondrial and cell wall morphology (Yamada et al., 2021; Azizullah et al., 2023), we wondered whether deletion of CpSmt3 would have similar effects in C. parasitica. TEM revealed that ΔCpSmt3 exhibited swollen mitochondria and a thickened cell wall compared to the wildtype strain (Figures 2A, B).
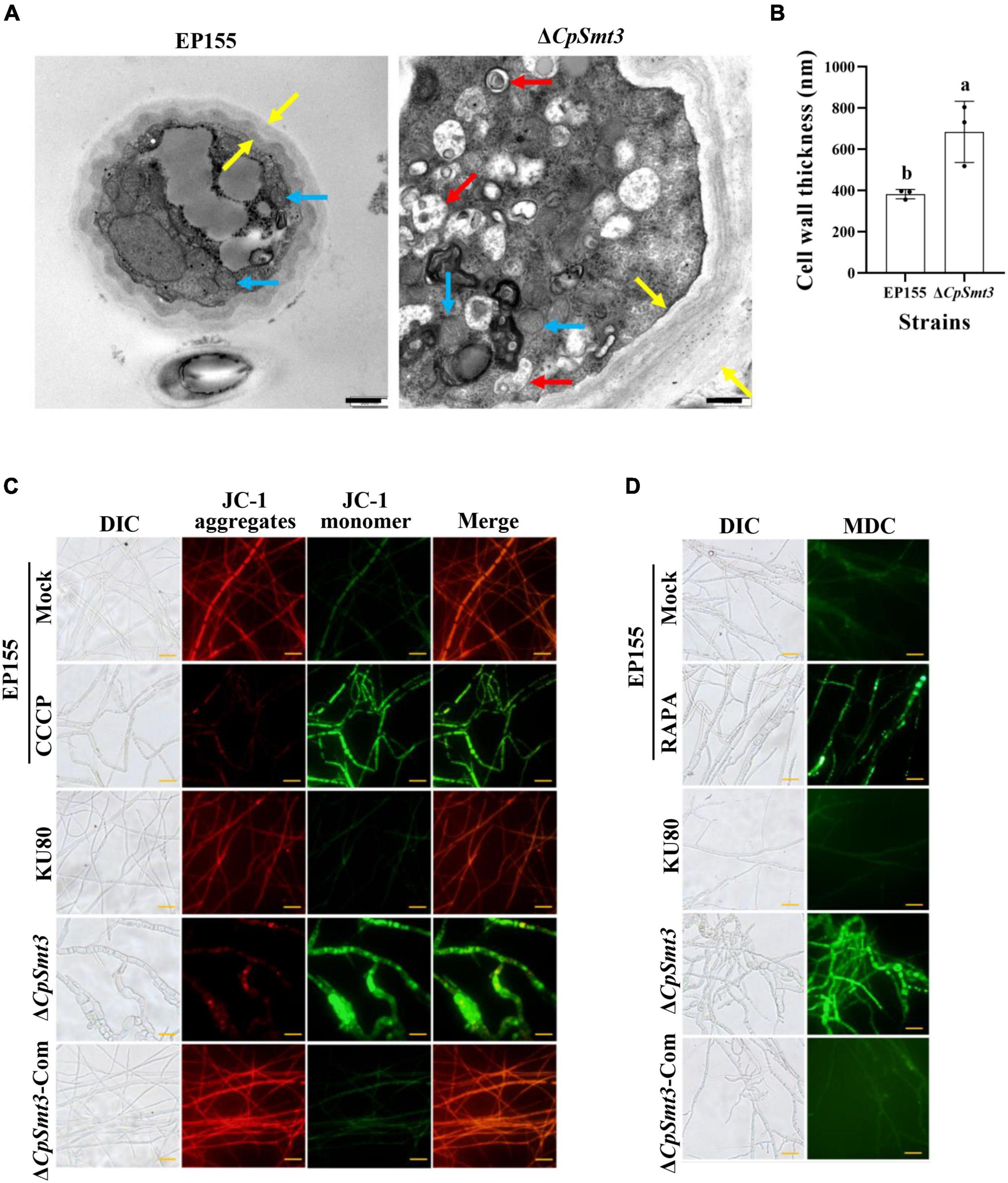
Figure 2. Deletion of CpSmt3 caused organelle defects and fungal autophagy. (A) Morphology of mitochondria, autophagic vacuoles, and cell walls under transmission electron microscope. Blue arrow represents mitochondrion, red arrow represents autophagic vacuole, and yellow arrow represents cell wall. Scale bar = 500 nm. (B) Cell wall thickness was measured by ImageJ software. Error bars represent the standard deviation based on three independent experiments. Different letters on the bars indicate significant differences (p < 0.05). (C) Assay of mitochondrial membrane potential (MMP). Mycelium was subjected to JC-1 staining and viewed using an Olympus fluorescent microscope. Wildtype mycelium treated with 10 μM CCCP for 24 h was used as the positive control. In healthy mitochondria, JC-1 forms a polymer and emits red fluorescence. When MMP decreases, JC-1 is stored as a monomer and emits green fluorescence. (D) Assay of autophagy. Mycelium was incubated with MDC at 37°C for 10 min. Wildtype strain treated with 10 nM rapamycin (RAPA) for 48 h was used as the positive control. Autophagic vesicles emit green fluorescence by excitation of ultraviolet light. Scale bar = 20 μm.
To further explore the mitochondrial dysfunction caused by CpSmt3 deletion, JC-1 staining was used to measure the MMP. There was a significant decrease in MMP upon CpSmt3 deletion (Figure 2C). In addition, TEM analysis showed that autophagic bodies were evident in the mycelia of ΔCpSmt3 (Figure 2A). Monodansylcadaverine (MDC) staining further proved that autophagic bodies accumulated in ΔCpSmt3 cells, but not in the wildtype strain (Figure 2D). Together, these results suggested that deletion of CpSmt3 caused organelle defects and autophagy.
3.4 Deletion of CpSmt3 causes mitochondrial ROS burst
Previous studies have shown that SUMOylation play essential roles in the regulation of ROS homeostasis by controlling ROS production and clearance, and reduced SUMOylation may lead to higher ROS production (Stankovic-Valentin and Melchior, 2018). As deletion of CpSmt3 caused decreased MMP, we measured the intracellular ROS in ΔCpSmt3 using the DCFH-DA fluorescence assay. Compared to the wildtype strain, ΔCpSmt3 had a significantly higher ROS level (Figures 3A, B). To further identify the source of ROS in ΔCpSmt3, we assessed the mtROS by using MitoSOX Red staining. A large amount of mtROS was observed in ΔCpSmt3 (Figures 3C, D), suggesting that the ROS burst was derived from mitochondria. To confirm this speculation, we supplemented the PDA medium with the ROS inhibitor N-Acetyl-L-cysteine (NAC) or the mtROS-specific inhibitor Mito-TEMPO. After the treatment, the ΔCpSmt3 colonies became larger and the ROS level was significantly reduced. In contrast, treatment with the ERO1-α inhibitor EN460 failed to increase the ΔCpSmt3 colony size or reduce the ROS level (Figures 3E, F).
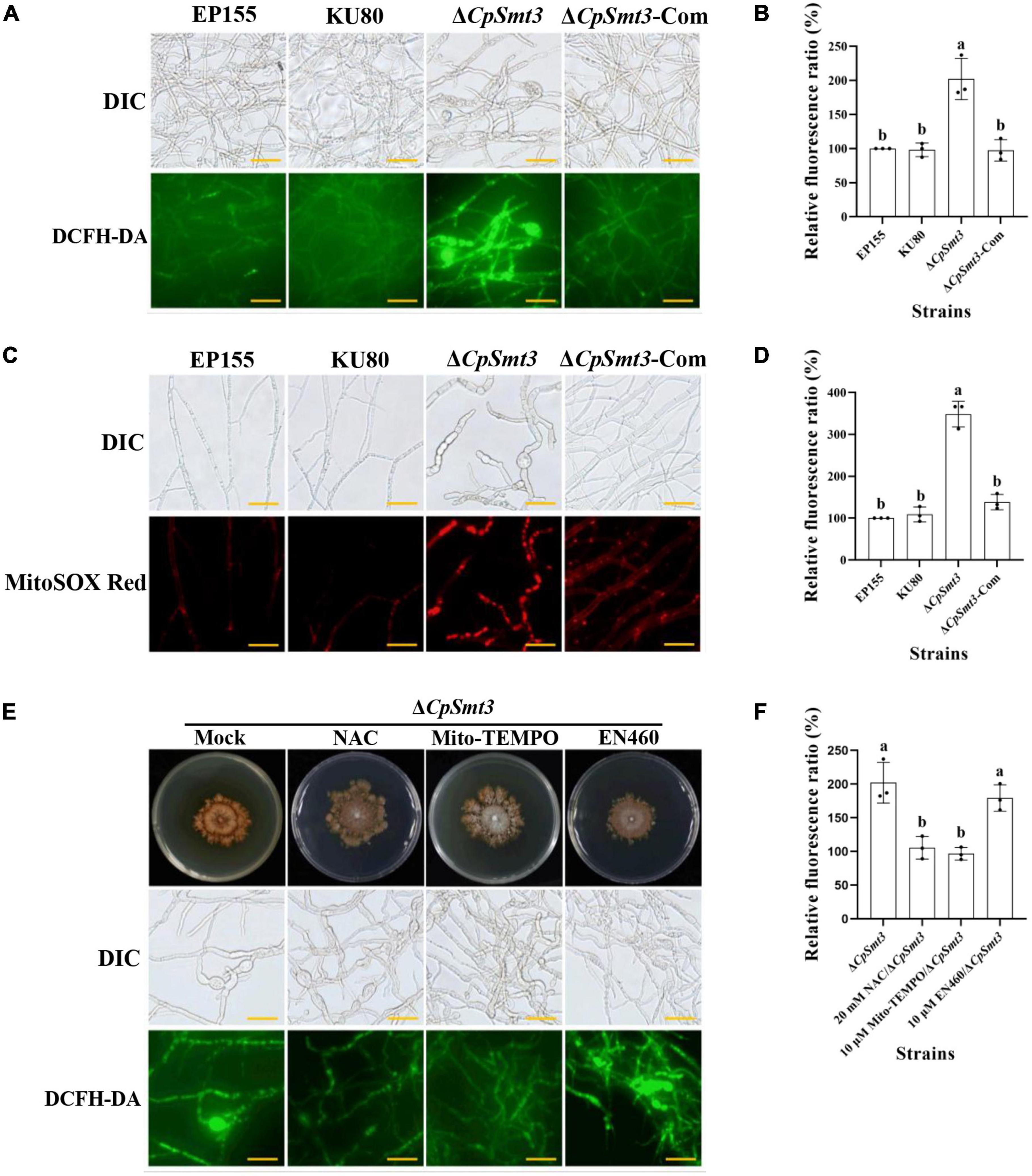
Figure 3. (A) ROS production of the tested strains was detected by DCFH-DA staining. Scale bar = 20 μm. (B) Relative ROS fluorescence ratio of the tested strains was analyzed by ImageJ software. (C) mtROS production of the tested strains was detected by MitoSOX red staining. Red fluorescence indicates mtROS production. Scale bar = 20 μm. (D) Relative mtROS fluorescence ratio of the tested strains was analyzed by ImageJ software. (E) Changes in ROS production in ΔCpSmt3 treated with 20 mM NAC, 10 μM Mito-TEMPO, or 10 μM EN460. Scale bar = 20 μm. (F) Relative ROS fluorescence of tested strains was analysis by Image J software. Error bars represent the standard deviation based on three independent experiments. Different letters on the bars indicate significant differences (p < 0.05).
3.5 CpSmt3 inhibits CHV1 replication by CpDcl2 upregulation
In animals, SUMOylation has been reported to activate the immune system to counteract viral infections (Imbert and Langford, 2021). To examine the role of SUMOylation in the interaction between C. parasitica and the hypovirus CHV1-EP713, the hypovirus was introduced into ΔCpSmt3 (ΔCpSmt3/CHV1-EP713, designated ΔCpSmt3/+virus) via hyphal anastomosis with the isogenic hypovirus-infected wildtype strain (EP155/+virus). The resultant colony size was significantly increased compared to ΔCpSmt3, suggesting that hypovirus infection partially reversed the growth impairment resulting from CpSmt3 deletion. In contrast, the hypovirus-infected CpSmt3-OE did not exhibit a change in colony size compared to the hypovirus-infected wildtype strain (Figures 4A and Supplementary Figure 3D). However, when ΔCpSmt3 and ΔCpSmt3/+virus inoculated on chestnut branches, the canker sizes of them were the same (Supplementary Figure 3A). Interestingly, hypovirus-infected ΔCpSmt3 accumulated about five times more viral dsRNA than the hypovirus-infected wildtype strain, while hypovirus-infected ΔCpSmt3-Com regained the ability to inhibit viral dsRNA accumulation. Hypovirus-infected CpSmt3-OE did not exhibit an altered viral dsRNA accumulation compared to the hypovirus-infected wildtype strain (Figures 4B and Supplementary Figure 3E).
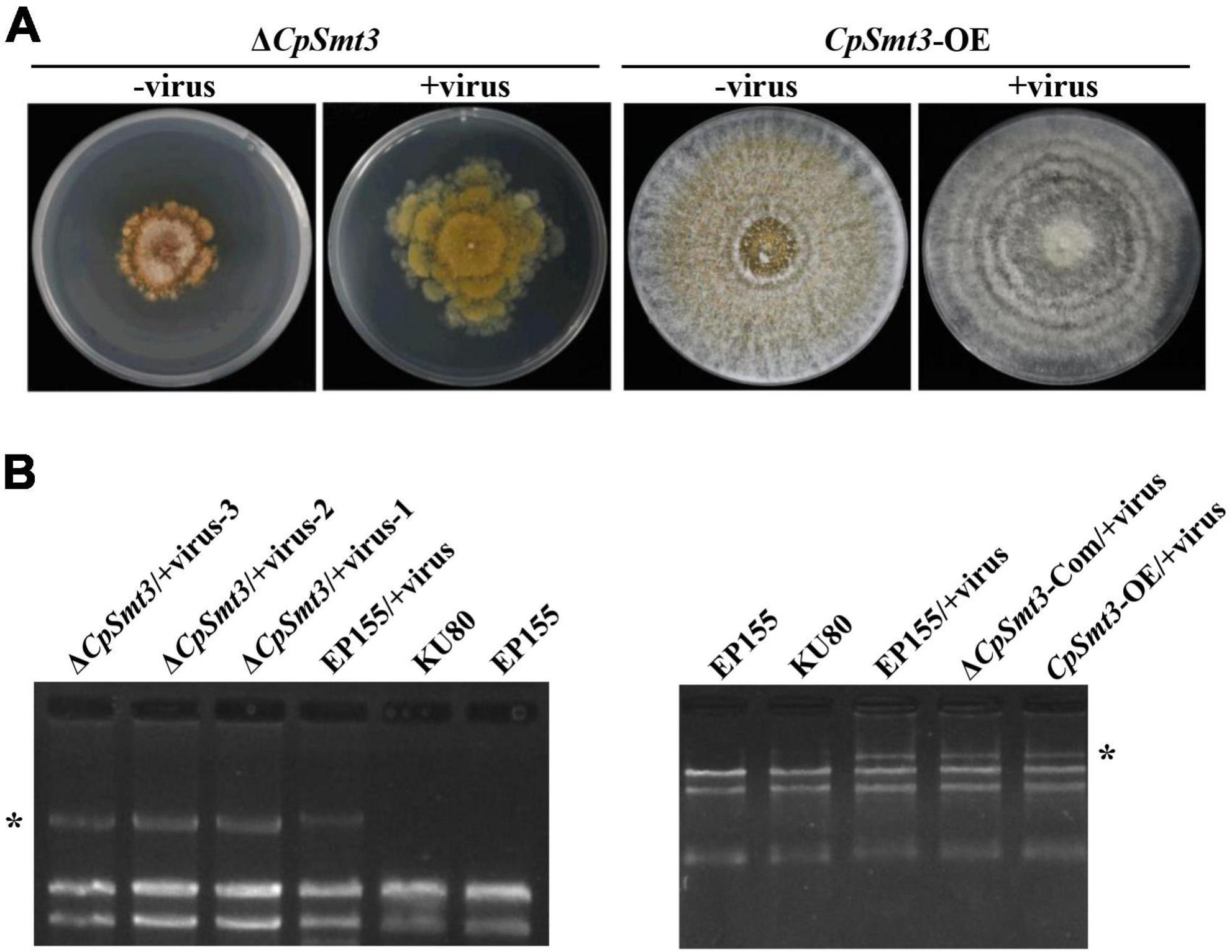
Figure 4. SUMOylation is involved in regulating CHV1 RNA accumulation. (A) Colony morphologies of ΔCpSmt3 and CpSmt3-OE, and their CHV1-infected strains. (B) Agarose gel electrophoresis of total RNA without gDNA. * indicates position of full-length CHV1 RNA.
To verify whether SUMOylation influences hypovirus replication, we constructed SUMO E1 subunit Uba2 and E2 enzyme Ubc9 mutant strains of C. parasitica, designated ΔCpUba2 (JGI ID: 255521) and ΔCpUbc9 (JGI ID: 324396), respectively (Supplementary Figures 3B, C). They both exhibited a phenotype very similar to that of ΔCpSmt3, and hypovirus infection of both strains partially rescued the colony size and growth (Figure 5A and Supplementary Figure 3D) but viral dsRNA accumulation was elevated five- and ten-fold, respectively (Figures 5B and Supplementary Figure 3E).

Figure 5. Deletion of CpUba2 or CpUbc9 increased CHV1 RNA accumulation. (A) Colony morphologies of ΔCpSmt3, ΔCpUba2, and ΔCpUbc9 and their CHV1-infected strains. (B) Agarose gel electrophoresis of viral dsRNA accumulation in the tested strains. * represents CHV1 RNA. (C) qRTPCR analysis of the relative expression of CpDcl2 gene. The expression of CpDcl2 in EP155 was set at 1.0, and the levels of indicated strains were expressed as fold change relative to those in EP155. Error bars represent the standard deviation from three independent experiments. Different letters on the bars indicate significant differences (p < 0.05).
To investigate the molecular mechanism underlying the increased viral dsRNA, we measured the transcript level of CpDcl2, a key gene in the antiviral RNA silencing pathway. There was no significant difference in CpDcl2 expression between the wildtype strain and ΔCpSmt3. However, CpDcl2 was highly upregulated (13-fold) in the hypovirus-infected wildtype strain, but only upregulated ∼4-fold in the hypovirus-infected ΔCpSmt3 strain (Figure 5C). These results suggest that SUMOylation is required for strong inhibition of viral dsRNA accumulation, likely by upregulating CpDcl2 to suppress viral replication.
3.6 Deletion of CpSmt3 alters protein expression pattern in C. parasitica
It is generally believed that SUMOylation increases protein stability through competitive binding to the ubiquitination modification sites in the substrate proteins (Bae et al., 2004; Prudent et al., 2015; Nakagawa et al., 2016). To explore the molecular mechanism of SUMOylation in phenotype, we performed a label-free proteomic analysis of wildtype vs. ΔCpSmt3 mycelia. A total of 3,970 and 3,254 proteins were identified in these strains, respectively, representing 35.82% (4,158 proteins) of the 11,609 annotated proteins from the whole genome (Supplementary Table 2). Compared to the wildtype strain, 1,869 differentially expressed proteins (DEPs), 1,702 downregulated and 167 upregulated, were observed in ΔCpSmt3 (Supplementary Table 2).
To define the SUMOylation target spectrum of CpSmt3 in C. parasitica, we enriched the SUMOylated proteins in a wildtype (EP155) transformant expressing 3 × FLAG–CpSmt3 fusion protein, using affinity pull-down assays involving anti-FLAG beads. Western blot analysis with anti-FLAG antibody showed that the fusion protein was successfully expressed (Supplementary Figure 4A). The anti-FLAG bead-captured proteins were then trypsin-digested and subjected to LC-MS/MS analysis. A total of 500 SUMOylated proteins were identified in three independent biological replicates. A total of 1,229 SUMOylation sites and 644 SUMO interaction motifs (SIM) were identified within these proteins using the SUMO site prediction software GPS-SUMO 2.0 online tool7 (Supplementary Figures 4B, C and Supplementary Table 3). Interestingly, the majority (408 out of 500) of these potential SUMOylated proteins were the DEPs identified in ΔCpSmt3 (Figure 6A). Intriguingly, most of these SUMOylated DEPs (397 out of 408) were downregulated in ΔCpSmt3, including the five pathogenesis-related proteins listed in Supplementary Table 3.
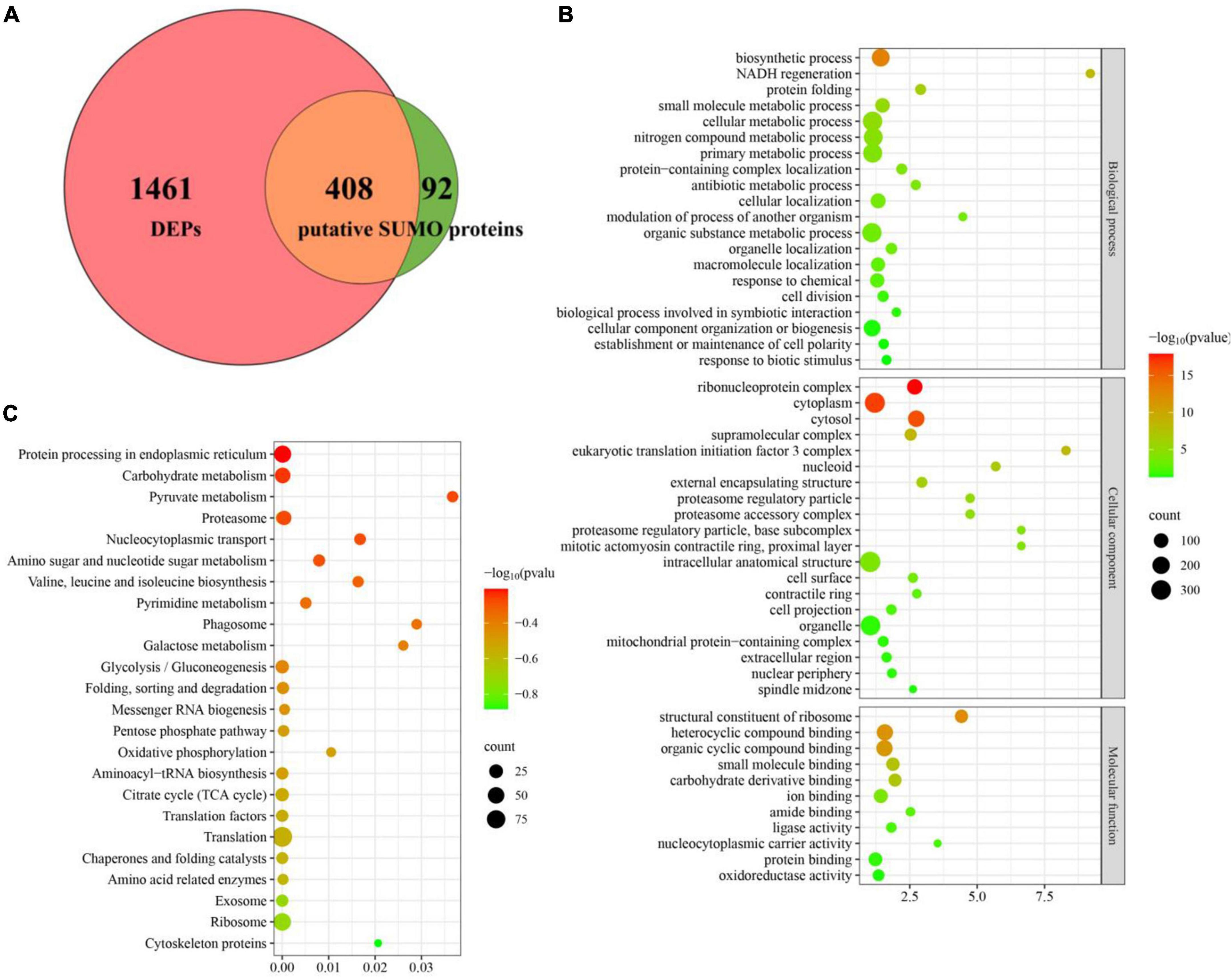
Figure 6. Deletion of CpSmt3 results in a reduction of total SUMOylated proteins. (A) Venn diagram of proteomics and co-immunoprecipitation data. The overlapping orange area indicates that there were 408 SUMOylated proteins whose expression levels changed significantly in the proteomics analysis of ΔCpSmt3 vs. wildtype strain. (B) GO enrichment analysis of 408 overlapping proteins shown in (A). (C) KEGG enrichment analysis of 408 overlapping proteins shown in (A). All the pathways shown in maps were significantly enriched (p < 0.01).
Gene Ontology enrichment analysis indicated that the downregulated SUMOylated proteins in ΔCpSmt3 were significantly enriched in structural constituent of ribosome, ribonucleoprotein complex, cytoplasm, cytosol, protein folding, and response to stress (Figure 6B and Supplementary Table 4). KEGG enrichment analysis showed that these proteins were significantly enriched in translation, ribosome, exosome, citrate cycle, translation factors, and oxidative phosphorylation pathway (Figure 6C and Supplementary Table 5), implying that SUMOylation is essential for maintaining ribosomal and mitochondrial function and survival in response to environmental stress.
3.7 CpRho1 is the key control point for CpSmt3-regulated traits
To further explore the regulated mechanism of CpSmt3, we attempted to find out the proteins related to phenotypic defects of ΔCpSmt3 mutant from SUMO substrates with known functions (Table 1). We found Rho1 was related to cell cycle, which has also been reported in Aspergillus fumigatus that regulates cell wall integrity and stress response to H2O2 stress (Zhang et al., 2018). In addition, CpRho1 (JGI ID: 97360) was downregulated SUMOylated DEPs in ΔCpSmt3 (Supplementary Tables 2, 3). Therefore, we selected CpRho1 as the target for further study. To examine the contribution of this protein in C. parasitica, we attempted to generate a CpRho1 deletion mutant. However, we failed to obtain a CpRho1-knockout strain after screening hundreds of candidate strains. Thus, we opted to generate a CpRho1-silenced strain. Three independent strains, CpRho1-RNAi-1, CpRho1-RNAi-2, and CpRho1-RNAi-3, with 64, 73, and 51% CpRho1 mRNA levels, respectively (Supplementary Figures 5A, B), were selected for further analysis. They exhibited 84 to 93% reduction in colony size, had no spore formation, and their hyphae were swollen, similar to that of ΔCpSmt3. However, CpRho1 overexpression in ΔCpSmt3 (ΔCpSmt3/CpRho1-OE) partially restored the reduced colony size of ΔCpSmt3 (Figures 7A, B and Supplementary Figure 5C), suggesting a link between CpSmt3 and CpRho1 in modulating fungal traits. TEM showed autophagic vacuoles and significantly thickened hyphal cell walls in the CpRho1-RNAi mutants, but the mitochondrial morphology remained normal (Figures 7C, D). Cellular analysis showed that the CpRho1-RNAi mutants had increased accumulation of autophagic vacuoles, elevated ROS production, and multinucleation. Importantly, these aberrations were greatly ameliorated by CpRho1 overexpression in ΔCpSmt3 (ΔCpSmt3/CpRho1-OE) (Figures 7E–H), implying that CpSmt3 may exert its effects by maintaining a proper CpRho1 level to ensure the normal cellular functions of the fungus.
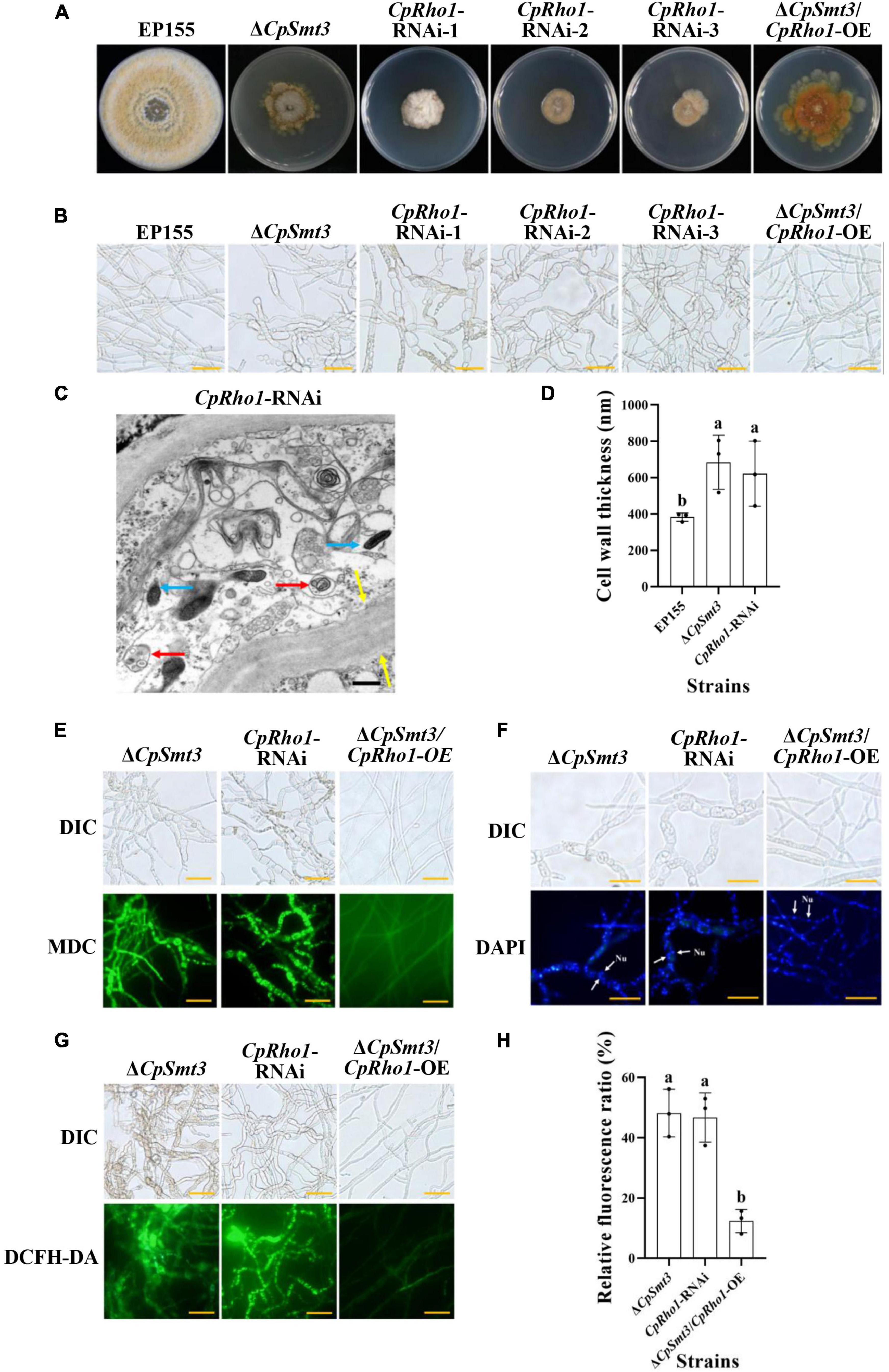
Figure 7. CpRho1 is essential for growth, organelle function, and the cell cycle. (A) Colony morphologies of wildtype strain, ΔCpSmt3, CpRho1 RNA interference strains (CpRho1-RNAi), and CpRho1-overexpressing ΔCpSmt3 (ΔCpSmt3+CpRho1-OE). (B) Mycelial morphologies of the tested strains. (C) Organelle structure of CpRho1-RNAi was observed by TEM. Blue arrow represents mitochondrion, red arrow represents autophagic vacuole, and yellow arrow represents cell wall. Scale bar = 500 nm. (D) Cell wall thickness statistics of wildtype, ΔCpSmt3, and CpRho1-RNAi strains. Cell wall thickness was measured by ImageJ software. (E) MDC staining showed autophagy in ΔCpSmt3 and CpRho1-RNAi, but not ΔCpSmt3+CpRho1-OE. Scale bar = 20 μm. (F) DAPI staining showed aberrant nuclear segregation in ΔCpSmt3 and CpRho1-RNAi, but not ΔCpSmt3+CpRho1-OE. Scale bar = 20 μm. (G) DCFH-DA staining showed the ROS burst in ΔCpSmt3 and CpRho1-RNAi, but not ΔCpSmt3+CpRho1-OE. (H) Relative ROS fluorescence ratio of the tested strains in (G) measured by ImageJ software. Error bars represent the standard deviation from three independent experiments. Different letters on the bars indicate significant differences (p < 0.05).
4 Discussion
In fungi, SUMO is encoded by Smt3 and deletion of this gene leads to alterations in many important phenotypic traits, such as abnormal distribution of chitin in the cell wall, septum formation defect, cell cycle disturbance, impairment of appressorium development, and loss of pathogenicity (Leach et al., 2011; Nie et al., 2016; Wotton et al., 2017; Liu et al., 2018). In this study, we characterized the SUMO-encoding gene, CpSmt3, in the chestnut blight fungus C. parasitica, and investigated its functions using a gene knockout strategy. CpSmt3 shares 85.1% similarity with Smt3 of the model filamentous fungus N. crassa and 68.3% similarity with Smt3 of the yeast S. cerevisiae, and it has the characteristic domains of a SUMO protein (Supplementary Figure 1B). Deletion of CpSmt3 in C. parasitica resulted in colony, hyphae, and growth defects, abnormalities of important cellular organelles, and attenuation of virulence and ROS burst (Figures 1–3). In the rice blast fungus Magnaporthe oryzae, SUMOylation has been shown to be involved in translation, ribosome biogenesis, the cell cycle, and nuclear division (Liu et al., 2018). Similarly, our comparative proteomics analysis showed that essential cellular processes were significantly altered in ΔCpSmt3 compared to the wildtype strain (Figure 6 and Supplementary Tables 2–5).
SUMOylation involves a large number of substrate proteins and promotes protein stability by competing with ubiquitin for binding sites, thereby protecting the proteins from degradation (Chang and Yeh, 2020). In line with this, we found that 97% (397 out of 408) of SUMOylated DEPs were downregulated in ΔCpSmt3 (Figure 6A and Supplementary Table 3). These downregulated proteins were implicated in a wide range of biological processes, including translation, ribosome biogenesis, the cell cycle, oxidative phosphorylation and TCA cycle, protein processing in endoplasmic reticulum, and exosome (Supplementary Tables 4, 5). Abnormalities of these cellular processes jeopardize cell growth and functions, from energy metabolism to pathogenicity (Fernández-Álvarez et al., 2013; Koh and Sarin, 2018; Richardson, 2019; Zhao et al., 2021; Yuan et al., 2023). Of the SUMOylated proteins, CpSep1, CpSlt2, CpMK2, CpMK1, and Cdc48 have been shown to be virulence factors in C. parasitica (Park et al., 2004; Choi et al., 2005; Ko et al., 2016; So et al., 2017; Jo et al., 2019). Therefore, we speculate that more SUMOylated proteins may also contribute to the phenotypic traits and virulence of C. parasitica.
Rho1, a member of the Rho GTPase family, is a component of the TOR signaling pathway, capable of regulating the CREB phosphorylation and involved in actin polarization and cell wall biosynthesis (Qadota et al., 1996; Drgonová et al., 1999). In mammals, Rho1 regulates mitochondria distribution and function, and the defect of Rho1 can lead to abnormality of oxidative phosphorylation, reduced MMP, and increased ROS production (Minin et al., 2006; Zhang and Jiang, 2017). In the fungus Schizosaccharomyces pombe, Rho1 is essential for viability, cell cycle, and cell wall integrity (Drgonová et al., 1999; Zhang et al., 2018; Vicente-Soler et al., 2021). In this study, we showed that CpRho1 was a SUMOylation target and it was significantly downregulated in ΔCpSmt3, indicating that CpSmt3 may regulate the stability of CpRho1. As the hyphal morphology, and cell wall thickness of the CpRho1-RNAi mutants were similar to those of ΔCpSmt3, and CpRho1 overexpression rescued some prominent defects of ΔCpSmt3 (Figure 7), we propose that the impact of CpSmt3 on the C. parasitica phenotype may largely depend on its regulation of CpRho1, likely by stabilizing the CpRho1 protein. Interestingly, the phenotype of ΔCpSmt3/+virus mutant was similar to that of the ΔCpSmt3/CpRho1-OE mutant, and neither of them had ROS burst or autophagy, indicating that CHV1 infection might rescue the phenotype of the ΔCpSmt3 mutant by upregulated the expression of CpRho1. Thus, CpRho1 is a key control point for regulation of important traits in C. parasitica, and this may offer a new opportunity to combat the pathogen by targeting this protein or its pathway.
SUMOylation plays an important role in antiviral immunity. In both mammals and plants, SUMOylation enhances the host immunity against virus infection by regulating immune signaling pathways (Saleh et al., 2015; El Motiam et al., 2020). In C. parasitica, CpDcl2 and CpAgl2 have been shown to play crucial roles in inhibiting the replication of hypovirus CHV1-EP713 (Segers et al., 2007). In this study, we found that CpAgl2 was a SUMOylation target and it was significantly downregulated in ΔCpSmt3. It was sharply upregulated upon hypovirus infection in the wildtype strain but highly downregulated in hypovirus-infected ΔCpSmt3 (Figure 5C). This discrepancy may compromise the antiviral activity in ΔCpSmt3 and result in more viral dsRNA accumulation (Figure 4B). These findings suggest that SUMOylation regulates antiviral activity at the transcription level in C. parasitica. Relatedly, previous studies have shown that Spt–Ada–Gcn5 acetyltransferase (SAGA), a universal transcriptional coactivator, upregulates CpDcl2 upon hypovirus infection, and knockout of SAGA subunits Gcn5, Sgf73, and Ada2 abolished any induction of CpDcl2 transcription upon viral infection (Andika et al., 2017, 2019). As Gcn5 is a SUMOylated protein (Sterner et al., 2006; Espinola-Lopez and Tan, 2021), we hypothesize that SUMOylation of the SAGA complex might be used to modulate resistance to viral infection in C. parasitica.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors. The data presented in the study are deposited in iPROX (https://www.iprox.cn/), accession number: PXD051810 and (https://www.ebi.ac.uk/pride), accession number: PXD051781.
Author contributions
SL: Data curation, Formal analysis, Methodology, Software, Writing – original draft. FC: Data curation, Methodology, Software, Writing – review & editing. XW: Data curation, Methodology, Software, Writing – review & editing. LY: Data curation, Methodology, Software, Writing – review & editing. JQ: Data curation, Methodology, Software, Writing – review & editing. RL: Conceptualization, Data curation, Formal analysis, Project administration, Supervision, Writing – review & editing. BC: Conceptualization, Formal analysis, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant Nos. 31960030 and 31370173) and Guangxi Natural Science Foundation (2021GXNSFAA196036).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1391855/full#supplementary-material
Footnotes
- ^ http://genome.jgi-psf.org/Crypa1/Crypa1.home.html
- ^ http://eggnog-mapper.embl.de/
- ^ https://david-d.ncifcrf.gov/
- ^ https://www.kegg.jp/kegg/mapper/
- ^ https://david.ncifcrf.gov/
- ^ http://genome.jgi-psf.org/Crypa2/Crypa2.home.html
- ^ http://sumosp.biocuckoo.org/online.php
References
Alfen, N. K. V. (1982). Biology and potential for disease control of hypovirulence of Endothia parasitica. Annu. Rev. Phytopathol. 20, 349–362. doi: 10.1146/annurev.py.20.090182.002025
Andika, I. B., Jamal, A., Kondo, H., and Suzuki, N. (2017). SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc. Natl. Acad. Sci. U.S.A. 114, e3499–e3506. doi: 10.1073/pnas.1701196114
Andika, I. B., Kondo, H., and Suzuki, N. (2019). Dicer functions transcriptionally and post transcriptionally in a multilayer antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 116, 2274–2281. doi: 10.1073/pnas.1812407116
Azizullah, Noman, M., Gao, Y., Wang, H., Xiong, X., Wang, J., et al. (2023). The SUMOylation pathway components are required for vegetative growth, asexual development, cytotoxic responses, and programmed cell death events in Fusarium oxysporum f. sp. niveum. J. Fungi (Basel) 9:94. doi: 10.3390/jof9010094
Bae, S. H., Jeong, J. W., Park, J. A., Kim, S. H., Bae, M. K., Choi, S. J., et al. (2004). Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem. Biophys. Res. Commun. 324, 394–400. doi: 10.1016/j.bbrc.2004.09.068
Chang, H. M., and Yeh, E. T. H. (2020). SUMO: From bench to bedside. Physiol. Rev. 100, 1599–1619. doi: 10.1152/physrev.00025.2019
Chen, B., Choi, G. H., and Nuss, D. L. (1994). Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264, 1762–1764. doi: 10.1126/science.8209256
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, C. H., Lo, Y. H., Liang, S. S., Ti, S. C., Lin, F. M., Yeh, C. H., et al. (2006). SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20, 2067–2081. doi: 10.1101/gad.1430406
Chiba, S., Jamal, A., and Suzuki, N. (2018). First evidence for internal ribosomal entry sites in diverse fungal virus genomes. mBio 9, e2350–e2317. doi: 10.1128/mBio.02350-17
Choi, E. S., Chung, H. J., Kim, M. J., Park, S. M., Cha, B. J., Yang, M. S., et al. (2005). Characterization of the ERK homologue CpMK2 from the chestnut blight fungus Cryphonectria parasitica. Microbiology 151, 1349–1358. doi: 10.1099/mic.0.27796-0
Chun, J., Ko, Y. H., and Kim, D. H. (2020). Transcriptome analysis of Cryphonectria parasitica infected with Cryphonectria hypovirus 1 (CHV1) reveals distinct genes related to fungal metabolites, virulence, antiviral RNA-silencing, and their regulation. Front. Microbiol. 11:1711. doi: 10.3389/fmicb.2020.01711
Drgonová, J., Drgon, T., Roh, D. H., and Cabib, E. (1999). The GTP-binding protein Rho1p is required for cell cycle progression and polarization of the yeast cell. J. Cell Biol. 146, 373–387. doi: 10.1083/jcb.146.2.373
Eifler, K., and Vertegaal, A. C. O. (2015). SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem. Sci. 40, 779–793.
El Motiam, A., Vidal, S., Seoane, R., Bouzaher, Y. H., González-Santamaría, J., and Rivas, C. (2020). SUMO and cytoplasmic RNA viruses: From enemies to best friends. Adv. Exp. Med. Biol. 1233, 263–277. doi: 10.1007/978-3-030-38266-7_11
Espinola-Lopez, J. M., and Tan, S. (2021). The Ada2/Ada3/Gcn5/Sgf29 histone acetyltransferase module. Biochim. Biophys. Acta Gene Regul. Mech. 1864:194629. doi: 10.1016/j.bbagrm.2020.194629
Fernández-Álvarez, A., Elías-Villalobos, A., Jiménez-Martín, A., Marín-Menguiano, M., and Ibeas, J. I. (2013). Endoplasmic reticulum glucosidases and protein quality control factors cooperate to establish biotrophy in Ustilago maydis. Plant Cell 25, 4676–4690. doi: 10.1105/tpc.113.115691
Gupta, D., Garapati, H. S., Kakumanu, A. V. S., Shukla, R., and Mishra, K. (2020). SUMOylation in fungi: A potential target for intervention. Comput. Struct. Biotechnol. J. 18, 3484–3493. doi: 10.1016/j.csbj.2020.10.037
Imbert, F., and Langford, D. (2021). Viruses, SUMO, and immunity: The interplay between viruses and the host SUMOylation system. J. Neuro Virol. 27, 531–541. doi: 10.1007/s13365-021-00995-9
Jacob-Wilk, D., Turina, M., and Van Alfen, N. K. (2006). Mycovirus Cryphonectria hypovirus 1 elements cofractionate with trans-Golgi network membranes of the fungal host Cryphonectria parasitica. J. Virol. 80, 6588–6596. doi: 10.1128/jvi.02519-05
Jo, M., So, K. K., Ko, Y. H., Chun, J., Kim, J. M., and Kim, D. H. (2019). Characterization of a hypovirus-regulated Septin Cdc11 ortholog, CpSep1, from the chestnut blight fungus Cryphonectria parasitica. Mol. Plant Microbe Interact. 32, 286–295. doi: 10.1094/mpmi-07-18-0194-r
Johnson, E. S., and Gupta, A. A. (2001). An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744. doi: 10.1016/s0092-8674(01)00491-3
Kagey, M. H., Melhuish, T. A., and Wotton, D. (2003). The polycomb protein Pc2 is a SUMO E3. Cell 113, 127–137. doi: 10.1016/S0092-8674(03)00159-4
Kahyo, T., Nishida, T., and Yasuda, H. (2001). Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718. doi: 10.1016/s1097-2765(01)00349-5
Kim, D., Rigling, D., Zhang, L., and Alfen, N. (1995). A new extracellular laccase of Cryphonectha parasitica is revealed by deletion of Lac1. MPMI 8, 259–266. doi: 10.1094/MPMI-8-0259
Kim, D.-H., Harris, B., Wang, F., Seidel, C., McCroskey, S., and Gerton, J. L. (2016). Mms21 SUMO ligase activity promotes nucleolar function in Saccharomyces cerevisiae. Genetics 204, 645–658. doi: 10.1534/genetics.115.181750
Ko, Y. H., So, K. K., Kim, J. M., and Kim, D. H. (2016). Heterokaryon analysis of a Cdc48-like gene, CpCdc48, from the chestnut blight fungus Cryphonectria parasitica demonstrates it is essential for cell division and growth. Fungal Genet. Biol. 88:10. doi: 10.1016/j.fgb.2016.01.010
Koh, C. S., and Sarin, L. P. (2018). Transfer RNA modification and infection - Implications for pathogenicity and host responses. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 419–432. doi: 10.1016/j.bbagrm.2018.01.015
Lan, X., Yao, Z., Zhou, Y., Shang, J., Lin, H., Nuss, D. L., et al. (2008). Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr. Genet. 53, 59–66. doi: 10.1007/s00294-007-0162-x
Leach, M. D., Stead, D. A., Argo, E., and Brown, A. J. (2011). Identification of sumoylation targets, combined with inactivation of SMT3, reveals the impact of sumoylation upon growth, morphology, and stress resistance in the pathogen Candida albicans. Mol. Biol. Cell 22, 687–702. doi: 10.1091/mbc.E10-07-0632
Li, R., Zhao, L., Li, S., Chen, F., Qiu, J., Bai, L., et al. (2022). The autophagy-related gene CpAtg4 is required for fungal phenotypic traits, stress tolerance, and virulence in Cryphonectria parasitica. Phytopathology 112, 299–307. doi: 10.1094/phyto-01-21-0015-r
Liu, C., Li, Z., Xing, J., Yang, J., Wang, Z., Zhang, H., et al. (2018). Global analysis of sumoylation function reveals novel insights into development and appressorium-mediated infection of the rice blast fungus. New Phytol. 219, 1031–1047. doi: 10.1111/nph.15141
Luo, Q., Song, Y., Kang, J., Wu, Y., Wu, F., Li, Y., et al. (2021). mtROS-mediated Akt/AMPK/mTOR pathway was involved in Copper-induced autophagy and it attenuates Copper-induced apoptosis in RAW264.7 mouse monocytes. Redox Biol. 41, 101912. doi: 10.1016/j.redox.2021.101912
Minin, A. A., Kulik, A. V., Gyoeva, F. K., Li, Y., Goshima, G., and Gelfand, V. I. (2006). Regulation of mitochondria distribution by RhoA and formins. J. Cell Sci. 119, 659–670. doi: 10.1242/jcs.02762
Nakagawa, K., Kohara, T., Uehata, Y., Miyakawa, Y., Sato-Ueshima, M., Okubo, N., et al. (2016). PIAS3 enhances the transcriptional activity of HIF-1α by increasing its protein stability. Biochem. Biophys. Res. Commun. 469, 470–476. doi: 10.1016/j.bbrc.2015.12.047
Newman, H. A., Meluh, P. B., Lu, J., Vidal, J., Carson, C., Lagesse, E., et al. (2017). A high throughput mutagenic analysis of yeast sumo structure and function. PLoS Genet. 13:e1006612. doi: 10.1371/journal.pgen.1006612
Nie, X., Yu, S., Qiu, M., Wang, X., Wang, Y., Bai, Y., et al. (2016). Aspergillus flavus SUMO contributes to fungal virulence and toxin attributes. J. Agric. Food Chem. 64, 6772–6782. doi: 10.1021/acs.jafc.6b02199
Nuss, D. L. (2005). Hypovirulence: Mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3, 632–642. doi: 10.1038/nrmicro1206
Park, S. M., Choi, E. S., Kim, M. J., Cha, B. J., Yang, M. S., and Kim, D. H. (2004). Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol. Microbiol. 51, 1267–1277. doi: 10.1111/j.1365-2958.2004.03919.x
Pasupala, N., Easwaran, S., Hannan, A., Shore, D., and Mishra, K. (2012). The SUMO E3 ligase Siz2 exerts a locus-dependent effect on gene silencing in Saccharomyces cerevisiae. Eukaryotic Cell 11, 452–462. doi: 10.1128/ec.05243-11
Pichler, A., Gast, A., Seeler, J. S., Dejean, A., and Melchior, F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120. doi: 10.1016/s0092-8674(01)00633-x
Prudent, J., Zunino, R., Sugiura, A., Mattie, S., Shore, G. C., and McBride, H. M. (2015). MAPL SUMOylation of Drp1 stabilizes an ER/mitochondrial platform required for cell death. Mol. Cell 59, 941–955. doi: 10.1016/j.molcel.2015.08.001
Puhalla, J. E. (1971). Genetics and nutritional requirements of Endothia parasitica. Phytopathology 61, 169–173.
Qadota, H., Python, C. P., Inoue, S. B., Arisawa, M., Anraku, Y., Zheng, Y., et al. (1996). Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 272, 279–281. doi: 10.1126/science.272.5259.279
Reindle, A., Belichenko, I., Bylebyl, G. R., Chen, X. L., Gandhi, N., and Johnson, E. S. (2006). Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 119, 4749–4757. doi: 10.1242/jcs.03243
Richardson, A. R. (2019). Virulence and metabolism. Microbiol. Spectr. 7:1689. doi: 10.1128/microbiolspec.GPP3-0011-2018
Saleh, A., Withers, J., Mohan, R., Marqués, J., Gu, Y., Yan, S., et al. (2015). Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18, 169–182. doi: 10.1016/j.chom.2015.07.005
Sambrook, J., and Russell, D. W. (2001). Molecular cloning: A laboratory manual. New York, NY: Cold Spring Harbor Press.
Segers, G. C., Zhang, X., Deng, F., Sun, Q., and Nuss, D. L. (2007). Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U.S.A. 104, 12902–12906. doi: 10.1073/pnas.0702500104
Shi, L., Li, R., Liao, S., Bai, L., Lu, Q., and Chen, B. (2014). Prb1, a subtilisin-like protease, is required for virulence and phenotypical traits in the chestnut blight fungus. FEMS Microbiol. Lett. 359, 26–33. doi: 10.1111/1574-6968.12547
Shi, L., Wang, J., Quan, R., Yang, F., Shang, J., and Chen, B. (2019). CpATG8, a homolog of yeast autophagy protein ATG8, is required for pathogenesis and hypovirus accumulation in the chest blight fungus. Front. Cell Infect. Microbiol. 9:222. doi: 10.3389/fcimb.2019.00222
So, K. K., and Kim, D. H. (2017). Role of MAPK signaling pathways in regulating the hydrophobin cryparin in the chestnut blight fungus Cryphonectria parasitica. Mycobiology 45, 362–369. doi: 10.5941/myco.2017.45.4.362
So, K. K., Ko, Y. H., Chun, J., Kim, J. M., and Kim, D. H. (2017). Mutation of the Slt2 ortholog from Cryphonectria parasitica results in abnormal cell wall integrity and sectorization with impaired pathogenicity. Sci. Rep. 7:9038. doi: 10.1038/s41598-017-09383-y
Stankovic-Valentin, N., and Melchior, F. (2018). Control of SUMO and Ubiquitin by ROS: Signaling and disease implications. Mol. Aspects Med. 63, 3–17. doi: 10.1016/j.mam.2018.07.002
Sterner, D. E., Nathan, D., Reindle, A., Johnson, E. S., and Berger, S. L. (2006). Sumoylation of the yeast Gcn5 protein. Biochemistry 45, 1035–1042. doi: 10.1021/bi051624q
Takahashi, Y., Iwase, M., Konishi, M., Tanaka, M., Toh-e, A., and Kikuchi, Y. (1999). Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem. Biophys. Res. Commun. 259, 582–587. doi: 10.1006/bbrc.1999.0821
Vertegaal, A. C. O. (2022). Signalling mechanisms and cellular functions of SUMO. Nat. Rev. Mol. Cell Biol. 23, 715–731. doi: 10.1038/s41580-022-00500-y
Vicente-Soler, J., Soto, T., Franco, A., Cansado, J., and Madrid, M. (2021). The multiple functions of Rho GTPases in fission yeasts. Cells 10:1422. doi: 10.3390/cells10061422
Wotton, D., Pemberton, L. F., and Merrill-Schools, J. (2017). SUMO and chromatin remodeling. Adv. Exp. Med. Biol. 963, 35–50. doi: 10.1007/978-3-319-50044-7_3
Yamada, S., Sato, A., Ishihara, N., Akiyama, H., and Sakakibara, S. I. (2021). Drp1 SUMO/deSUMOylation by Senp5 isoforms influences ER tubulation and mitochondrial dynamics to regulate brain development. iScience 24, 103484. doi: 10.1016/j.isci.2021.103484
Yao, Z., Zou, C., Zhou, H., Wang, J., Lu, L., Li, Y., et al. (2013). Δ(1)-pyrroline-5-carboxylate/glutamate biogenesis is required for fungal virulence and sporulation. PLoS One 8:e73483. doi: 10.1371/journal.pone.0073483
Yau, T. Y., Sander, W., Eidson, C., and Courey, A. J. (2021). SUMO Interacting Motifs: Structure and function. Cells 10:2825. doi: 10.3390/cells10112825
Yuan, Y., Mao, X., Abubakar, Y. S., Zheng, W., Wang, Z., Zhou, J., et al. (2023). Genome-wide characterization of the RNA exosome complex in relation to growth, development, and pathogenicity of Fusarium graminearum. Microbiol. Spectr. 11, e0505822. doi: 10.1128/spectrum.05058-22
Zhang, K., and Jiang, D. (2017). RhoA inhibits the hypoxia-induced apoptosis and mitochondrial dysfunction in chondrocytes via positively regulating the CREB phosphorylation. Biosci. Rep. 37:BSR20160622. doi: 10.1042/BSR20160622
Zhang, X., Jia, X., Tian, S., Zhang, C., Lu, Z., Chen, Y., et al. (2018). Role of the small GTPase Rho1 in cell wall integrity, stress response, and pathogenesis of Aspergillus fumigatus. Fungal Genet. Biol. 120, 30–41. doi: 10.1016/j.fgb.2018.09.003
Keywords: Cryphonectria parasitica, CpSmt3, SUMOylation, virulence, hypovirus
Citation: Li S, Chen F, Wei X, Yuan L, Qin J, Li R and Chen B (2024) CpSmt3, an ortholog of small ubiquitin-like modifier, is essential for growth, organelle function, virulence, and antiviral defense in Cryphonectria parasitica. Front. Microbiol. 15:1391855. doi: 10.3389/fmicb.2024.1391855
Received: 26 February 2024; Accepted: 15 April 2024;
Published: 09 May 2024.
Edited by:
Fei Yan, Ningbo University, ChinaReviewed by:
Jiaoyu Wang, Zhejiang Academy of Agricultural Sciences, ChinaKrishnaveni Mishra, University of Hyderabad, India
Copyright © 2024 Li, Chen, Wei, Yuan, Qin, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ru Li, liruonly@163.com; Baoshan Chen, chenyaoj@gxu.edu.cn
 Shuangcai Li
Shuangcai Li Fengyue Chen
Fengyue Chen Xiangyu Wei1
Xiangyu Wei1 Luying Yuan
Luying Yuan Ru Li
Ru Li Baoshan Chen
Baoshan Chen