- 1College of Agronomy, Guangxi University, Nanning, Guangxi, China
- 2Key Laboratory of Agro-Environment and Agro-Product Safety, Guangxi University, Nanning, China
- 3Guangxi Academy of Agricultural Sciences, Cash Crops Research Institute, Nanning, Guangxi, China
- 4Center for Ecological Civilization Research, Chinese Research Academy of Environmental Sciences, Beijing, China
Introduction: Microbial carbon (C) and nutrient limitation exert key influences on soil organic carbon (SOC) and nutrient cycling through enzyme production for C and nutrient acquisition. However, the intercropping effects on microbial C and nutrient limitation and its driving factors between rhizosphere and bulk soil are unclear.
Methods: Therefore, we conducted a field experiment that covered sugarcane–peanut intercropping with sole sugarcane and peanut as controls and to explore microbial C and nutrient limitation based on the vector analysis of enzyme stoichiometry; in addition, microbial diversity was investigated in the rhizosphere and bulk soil. High throughput sequencing was used to analyze soil bacterial and fungal diversity through the 16S rRNA gene and internal transcribed spacer (ITS) gene at a phylum level.
Results: Our results showed that sugarcane–peanut intercropping alleviated microbial C limitation in all soils, whereas enhanced microbial phosphorus (P) limitation solely in bulk soil. Microbial P limitation was also stronger in the rhizosphere than in bulk soil. These results revealed that sugarcane-peanut intercropping and rhizosphere promoted soil P decomposition and facilitated soil nutrient cycles. The Pearson correlation results showed that microbial C limitation was primarily correlated with fungal diversity and fungal rare taxa (Rozellomycota, Chyltridiomycota, and Calcarisporiellomycota) in rhizosphere soil and was correlated with bacterial diversity and most rare taxa in bulk soil. Microbial P limitation was solely related to rare taxa (Patescibacteria and Glomeromycota) in rhizosphere soil and related to microbial diversity and most rare taxa in bulk soil. The variation partitioning analysis further indicated that microbial C and P limitation was explained by rare taxa (7%–35%) and the interactions of rare and abundant taxa (65%–93%).
Conclusion: This study indicated the different intercropping effects on microbial C and nutrient limitation in the rhizosphere and bulk soil and emphasized the importance of microbial diversity, particularly rare taxa.
1 Introduction
Microorganisms are the main biota groups in soil and play a crucial role in agroecosystems by delivering essential functions such as storage of soil organic carbon (SOC) and nutrient cycling (Wieder, 2014). They produce diverse enzymes for the breakdown of soil organic matter (SOM) and the mineralization of soil nutrients, such as nitrogen (N) and phosphorus (P), to maintain plant and microbial growth (Sinsabaugh et al., 2008, 2009). The enzyme production is ascribed to soil nutrient availability and microbial biomass demands, with high enzyme activity in resource-poor environments (Gomez et al., 2020; Zhou L. H. et al., 2020). The C-, N-, and P-acquiring enzyme activity and their stoichiometry thus reflect the magnitudes of microbial C and nutrient limitation (N vs. P) (Qiu et al., 2021; Zheng et al., 2022). For example, higher C:N:P-requiring enzymes show stronger microbial C limitation relative to nutrients, which regulates SOM decomposition and soil nutrient cycling (Zheng et al., 2022). Thus, the response of microbial C and nutrient limitation to different agricultural management practices is of great importance in understanding agroecosystem nutrient cycling and supporting the development of sustainable agricultural management practices.
Intercropping is a sustainable agricultural practice to promote belowground productivity, yield, and ecosystem services, such as soil C, N, and P cycles (Cong et al., 2015; Li et al., 2020). This is due to the functional compensability of multiple crops to increase the land equivalent ratio with high water, air, and sunlight, as well as nutrient use efficiency by increasing the soil enzyme activities in the same land compared to sole crops (Li et al., 2020; Curtright and Tiemann, 2021). Compared with sole crop, on the one hand, the increase in soil nutrient and root-derived C (root exudates) induced by intercropping might alleviate microbial C and nutrient limitation by increasing fertilizer use efficiency (Steinauer et al., 2015; Tang X. Y. et al., 2021). On the other hand, intercropping-induced frequent interactions in root–soil–microbes systems and fast-growing microbes appear to enhance nutrient competition between microbes–microbes and microbes–crops, which is likely to regulate microbial C and nutrient limitation (Hilbig and Allen, 2015; Kuster et al., 2016; Feng et al., 2021). It is therefore essential to explore the intercropping effects on microbial C and nutrient limitation, which could further affect soil nutrient uptake and plant biomass productivity and yields (Tang X. Y. et al., 2021). Although many studies observed the intercropping effects on enzyme activity, most of them considered the bulk soil and few studies focused on the rhizosphere, particularly, the differences in intercropping effects on microbial C and nutrient limitation between rhizosphere and bulk soil (Curtright and Tiemann, 2021; Shi et al., 2022; dos Santos Bastos et al., 2023).
The rhizosphere, as the interface of soil–microbes–plant, directly controls nutrient and C decomposition and reutilization, thereby regulating plant productivity. Given the root disturbance and selection, the rhizosphere, as a microbial hotspot, has higher microbial activity and diversity than those in bulk soil (Elmajdoub et al., 2014; Pang et al., 2021; Chen et al., 2023). Additionally, the changes in enzyme activities between rhizosphere and bulk soil are dependent on the differences in nutrient and labile C in these two soils (Ren et al., 2021; Chen et al., 2023). For example, the larger intercropping effect on increasing enzyme activity is visible in nutrient-poor soils, such as bulk soil (Curtright and Tiemann, 2021). This implies that microbial C and nutrient limitation in the rhizosphere and bulk soil would have divergent responses to intercropping through changes in nutrient and labile C. Therefore, assessing the intercropping effects on microbial C and nutrient limitation in both rhizosphere and bulk soil would improve the understanding of soil nutrient and C cycling in root-soil systems in response to intercropping, which offers benefits for the development of sustainable agricultural management practices.
Microbial C and nutrient limitation are highly controlled by the microbial community (Creamer et al., 2015; Yao et al., 2018), such as rare and abundant taxa, which are different between rhizosphere and bulk soil (Wang et al., 2022; Zhang G. Z. et al., 2022). Rare taxa, groups with a narrower niche breadth, are more sensitive to environmental disturbances and might be present at specific locations or be exclusive to arable farming practices (Banerjee et al., 2024). Many studies show that compared with abundant taxa, rare taxa (e.g., Actinobacteria and Cyanobacteria) play a more important role in C and nutrient cycles, especially in rhizosphere owing to the root-induced selection and disturbance effects (Wei et al., 2019; Zhang et al., 2021; Wang et al., 2022). Furthermore, soil property, such as nutrient availability, is another vital factor controlling microbial resource limitation (Chen et al., 2019; Cui et al., 2021). Microbes are more C- or nutrient-limited in resource-poor environments with higher soil C:N ratio (Cui et al., 2021). This is because microbes, especially for rare taxa, prefer fertile soils with low soil C:N ratio substrates, which are easily decomposed and utilized (Liu et al., 2023). However, the intercropping effects on rare and abundant taxa, as well as the relative contributions of the microbial community (rare and abundant taxa) and soil properties to microbial C and nutrient limitation in the rhizosphere and bulk soil, are limited. This limitation would shed insights into the contributions of biotic and abiotic factors to microbial nutrient limitation in root–soil systems under intercropping practices.
Sugarcane–peanut intercropping is widely introduced to increase crop yield and soil fertility (Tang et al., 2021a, 2022). In this study, we aimed to compare the intercropping effects on microbial C and nutrient limitation and the relative contributions of rare and abundant taxa in the rhizosphere and bulk soil. We hypothesize that (1) sugarcane/peanut intercropping might decrease microbial C and nutrient limitation due to more C and nutrient input via legume biological N2-fixing than monoculture; (2) the intercropping effects on microbial C and nutrient limitation would be lower in rhizosphere soil than in bulk soil with poor resource; and (3) microbial C and nutrient limitation might be mostly regulated by rare taxa in rhizosphere soil and abundant taxa in bulk soil. This is because compared to abundant taxa, rare taxa depend on nutrient availability to a larger extent (Xu et al., 2021a; Lin et al., 2022), and thus rare taxa might primarily regulate microbial C and nutrient limitation in rhizosphere with richer root-derived substrates input.
2 Materials and methods
2.1 Site description
This field experiment was conducted from 2021 to 2023 at the Lijian Scientific Base of the Guangxi Academy of Agricultural Sciences (E108°3′40″, N23°14′58″), Nanning City, Guangxi Zhuang Autonomous Region (GZAR), China. This study site is located in the southwest of China with a subtropical climate zone. The mean annual temperature and precipitation were 22°C yr−1 and 1322 mm yr−1, respectively, obtained from the World Weather Information Service (https://worldw-eather.wmo.int/en/home.html) based on the longitude and latitude of the studied location. The soil type was classified as red soil with a loam texture. Before the field experiment, soil water content (SWC) was 14.39%, pH (H2O) was 6.75, and SOC, total N (TN), and P (TP) were 18.52, 1.29, and 1.26 g kg−1 in 0–20 cm soil layer, respectively. The provided sugarcane and peanut varieties were, respectively, “Guitang44” and “Guihua376” from the Cash Crops Research Institute of the Guangxi Academy of Agricultural Sciences (Tang et al., 2022).
2.2 Experiment design
This study included four treatments of cropping systems, including (1) monoculture peanut (MP); (2) monoculture sugarcane (MS); (3) sugarcane soil in sugarcane–peanut intercropping systems (IS); and (4) peanut soil in sugarcane–peanut intercropping systems (IP). For MS, sugarcane was planted with a row spacing of 1.2 m and a plant width of 50 cm. For MP, the row spacing of peanut was 30 cm with a plant spacing of 12 cm. For IS and IP, the line spacing between sugarcane and peanut was 65 cm (sugarcane: peanut = 2:4) in intercropping systems. The line spacing for sugarcanes was 1.2 m and that for the intercropped peanuts was 30 cm (Figure 1). The experiment was conducted in plots (10 m × 11.7 m) in a randomized design with six replicates in each treatment. All peanut treatments received 450 kg ha−1 special compound fertilizer (N-P2O5-K2O = 15-15-15) and 750 kg ha−1 fused calcium–magnesium phosphate fertilizer (P2O5 = 18%). All sugarcane treatments were fertilized with 2,250 kg ha−1 special compound fertilizer (N-P2O5-K2O = 15-15-15). The input rates of N–P2O5-K2O fertilizers were widely recommended to promote the rapid growth of sugarcane and peanut cultivation, which was described in detail in the previous studies (Pang et al., 2021; Tang et al., 2021b, 2022).
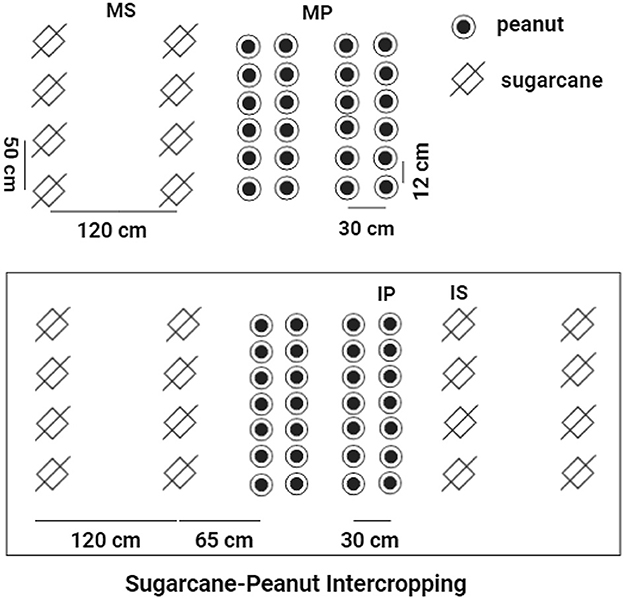
Figure 1. The schematic diagram of sugarcane–peanut intercropping. MS, monoculture sugarcane soil; IS, sugarcane soil peanut soil in sugarcane–peanut system; MP, monoculture peanut soil; IP, peanut soil in sugarcane–peanut system.
2.3 Soil sampling
Soils were sampled after the peanut harvest on 11 July 2023. We collected two soils: rhizosphere and bulk soil (0–20 cm soil layer) from peanut and sugarcane from the above four cropping systems. The bulk soil was collected far away from 20 cm of crops. Rhizosphere soil that lightly adhered to plant roots was obtained with a brush (Jiang et al., 2017). In total, 48 soil samples were finally collected (four treatments × two soils × six replicates). All soil samples were delivered to the laboratory immediately and were sieved into 2 mm with the removal of visible stones and root residues. In total, 20 g of soil were stored at −20°C for microbial analysis, and the rest of the soil was stored at 4°C for soil physicochemical analysis.
2.4 Physicochemical properties of the soil
The water content of the soil was determined by oven-drying. Soil pH was extracted by a 1:5 fresh soil:water ratio (mass:volume) and was measured using a pH meter. SOC was determined by the potassium dichromate oxidation method. Soil TN was measured by the Kjeldahl method (Bremner and Mulvaney, 1982). Soil total P (TP) was extracted by H2SO4-HClO4 and then was measured by the molybdenum blue method at 700 nm emission (Page, 1982) using an ultraviolet spectrophotometer (UV2600, Shimadzu, Japan).
2.5 Soil microbial biomass and community composition
Soil microbial biomass C (MBC), N (MBN), and P (MBP) were analyzed with fresh soil samples using the chloroform fumigation–extraction method (Vance et al., 1987).
Soil total DNA was extracted from 1 g of frozen soil by the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA). A NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA) and 2% agarose gel electrophoresis were used to assess the quantity and quality of DNA. The primer pairs 515F/907R (5′-GTGCCAGCMGCCGCGG-3′/5′-CCGTCAATTCMTTTRAGTTT-3′) (Xu et al., 2021b) and ITS 1F/ITS 2R (5′-CTTGGTCATTTAGAGGAAGTAA-3′/5′-GCTGCGTTCTTCATCGATGC-3′) (Zhou et al., 2023) were used to amplify the bacterial 16S rRNA gene and fungal ITS gene, respectively. Polymerase chain reaction (PCR) was performed in a thermal cycler (ABI GeneAmp 9700) with a volume of 20 μl containing 4 μl of FastPfu Buffer (5 × ), 0.2 μl of BSA, 0.8 μl of forward primer (5 μM), 0.8 μl of reverse primer (5 μM), 2 μl of dNTPs (2.5 mM), 10 ng of DNA template, 0.4 μl of FastPfu Polymerase, and 11.8 μl of ddH2O. The reaction conditions were as follows: 3 min of initial denaturation at 95°C, 35 cycles of 30 s at 95°C, 30 s at 55°C, and 45 s at 72°C, and a final elongation for 10 min at 72°C.
Purified amplicons were pooled in equimolar amounts and paired-end sequenced on an Illumina PE250 platform (Illumina, San Diego, CA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). DADA2 denoising was used to remove low-quality reads (Callahan et al., 2016), and then the eligible merged sequences were clustered into amplicon sequence variants (ASVs). Taxonomic assignment of ASVs was performed using the basic local alignment tool (BLAST) consensus taxonomy classifier implemented in Qiime2 and the SILVA 16S rRNA database (v138). To minimize the effects of sequencing depth on alpha and beta diversity measure, the number of sequences from each sample was rarefied to 96,908 sequences and 6,738 sequences per sample randomly selected from bacterial 16S rRNA and fungal ITS datasets, respectively, which still yielded an average Good's coverage of 99.0%. Shannon indices, as microbial alpha diversity, were calculated according to the 97% amplicon sequence variant (ASV) similarity of the sequences. The raw sequencing reads were deposited into the NCBI Sequence Read Archive (SRA) database under accession number: PRJNA1051854.
2.6 Soil enzyme activity
The frequently determined C, N, and P acquisition enzymes, including β-1,4-glucosidase (BG, C-acquiring enzyme), β-1,4-Nacetylglucosaminidase (NAG, N-acquiring enzyme), leucine aminopeptidase (LAP, N-acquiring enzyme), and acid phosphatase (AP, P-acquiring), were considered in this study (Sinsabaugh et al., 2008, 2009). The potential activities of BG, NAG, LAP, and AP were determined following the protocol of our previous study (Sun et al., 2023). First, a 1.50 g of fresh soil sample was suspended in 150 ml of sodium acetate buffer (30 mmol L−1, pH = 5.3) and completely homogenized in a blender for 2 min. The homogenized soil slurries (200 μl) were next added to a black 96-well microplate with eight replicates for each sample. Then, 50 μl of 200 μmol L−1 substrates labeled by 4-methylumbelliferone (MUB) for BG, NAG, and AP, or 7-amino-4-methylcoumarin (AMC) for LAP were added to each well and mixed with soil slurries slightly. The standard curve was conducted for each soil slurry sample with a concentration gradient of MUB or AMC (0, 0.1, 0.2, 0.5, 1, 2, 2.5, and 5 mmol L−1). All microplates were incubated at 25°C in the dark for 3 h. Finally, the intensity of fluorescence was determined at 365 nm excitation and 460 nm emission by using a microplate reader (Infinite M2000, Tecan, Männedorf, Switzerland). Enzyme activities were calculated and expressed as nmol g−1 dry soil h−1.
2.7 Statistical analysis
The soil microbial resource limitation was calculated by the enzymatic vector analysis (Moorhead et al., 2016) according to Equations 1 and 2 as follows:
x indicates the proportional activity of C:P acquiring enzymes [ln(BG): ln(BG + AP)], and y indicates the proportional activity of C:N acquiring enzymes [ln(BG): ln(BG + NAG + LAP)]. The vector length represents microbial C limitation, and the vector angle indicates microbial P vs. N limitation (Moorhead et al., 2013, 2016). This approach has been widely used to explore microbial resource limitation in response to climate changes and agricultural management (Jing et al., 2020; Zhang S. H. et al., 2022; Sun et al., 2023). Microorganisms are more C-limited with higher vector length, whereas they are relatively P-limited when vector angle >45°, and conversely, N-limited.
Microbial alpha diversity was represented by Shannon indices, which were common indicators of microbial species richness and diversity and were widely used in previous studies (Větrovský et al., 2019; Xu et al., 2021b). Microbial beta diversity was conducted by the non-metric multidimensional scaling (NMDS) analysis based on the Bray–Curtis dissimilarity matrix using the “vegan” package and the first component (NMDS1) was used for the latter analysis (Bahram et al., 2018; Domeignoz-Horta et al., 2020). An analysis of similarities (ANOSIM) was used to evaluate the significant differences in soils (rhizosphere and bulk soil) and cropping systems (monoculture and intercropping) presented in NMDS (Ju et al., 2023; Zhou et al., 2023). The abundant and rare taxa were grouped by the relative abundance approach with the threshold of 1% at the phylum level, which was widely discussed in previous studies (Xu et al., 2021a; Zhang G. Z. et al., 2022; Liu et al., 2023; Wang et al., 2024). The comparisons of the intercropping effects on soil properties, microbial alpha diversity, microbial C, and nutrient limitation between rhizosphere and bulk soil were conducted by one-way ANOVA. The multiple-way ANOVA test was further approached to explore the interactions between crop types (sugarcane vs. peanut), soils (rhizosphere vs. bulk soil), and cropping systems (intercropping vs. monoculture) on soil and microbial properties. Pearson's correlation was used to explore the relationships of biotic (soil properties) and abiotic factors (microbial biomass, microbial community composition, and rare and abundant taxa) with microbial C and nutrient limitation. Furthermore, the variation partitioning analysis (VPA) was conducted to assess the relative contributions of rare and abundant taxa on microbial C and nutrient limitation by using multiple linear regression models due to the weak impact of soil properties (Jing et al., 2020; Sun et al., 2023). All analyses were conducted with R (version 4.2.0) by using the “ggpmisc” package for linear regression models and “modEvA” packages for VPA analyses (Jing et al., 2020; Sun et al., 2023).
3 Results
3.1 Intercropping effects on soil properties in the rhizosphere and bulk soil
Compared with monoculture systems, sugarcane–peanut intercropping decreased SWC content and soil pH in all soils, especially in the rhizosphere soil with a reduction of 2 units of pH (Supplementary Table S1). Soil pH was also reduced by 0.8 units in the rhizosphere than that in the bulk soil (Supplementary Table S1). Intercropping increased SOC by 16.18%−22.73% relative to monoculture treatment in sugarcane rhizosphere and bulk soil. Furthermore, intercropping increased TN and TP, especially in peanut soils, with an increase of 112.69%−115.27% of TN and 10.37%−33.88% of TP in the rhizosphere and bulk soil. Intercropping did not change soil C:N, C:P, and N:P ratios in all soils, except for MP and IP treatments in bulk soil.
3.2 Intercropping effects on microbial community composition and biomass in the rhizosphere and bulk soil
Compared with monoculture systems, intercropping increased MBC and MBC:MBN in the rhizosphere and bulk soil but did not alter MBN, MBP, MBC:MBP, and MBN:MBP (Supplementary Table S2). Intercropping increased abundant and total bacterial Shannon diversity in both rhizosphere and bulk soil (Figure 2). The Shannon diversity of abundant and total fungal taxa was higher in sugarcane than in peanut systems (Figure 2). The NMDS analysis showed that intercropping and rhizosphere affected rare and abundant taxa beta diversity at the phylum level, and the intercropping effects were stronger than rhizosphere effects (Figure 3). Across all soils, fungi were dominated by Ascomycota (>75%), Basidiomycota, Mortierellomycota, and unclassified phyla (Supplementary Figure S1A). Bacteria were dominated by Actinobacteriota, Proteobacteria, Chloroflexi, and Acidobacteriota, accounting for ~75% in total (Supplementary Figure S1B). Intercropping significantly altered the relative abundance of abundant taxa, including Planctomycetota and Gemmatimonadota in bulk soil and Ascomycota and Actinobacteriota in rhizosphere soil (Supplementary Figure S2). As for rare taxa, intercropping significantly affected the relative abundance of Rozellomycota, Bdellovibrionota, and Armatimonadota in bulk soil, and the relative abundance of Methylomirabilota and Armatimonadota in rhizosphere soil (Supplementary Figure S3). In addition, only the beta diversity of bacterial composition differed between rhizosphere and bulk soil (P < 0.05, Supplementary Table S3).
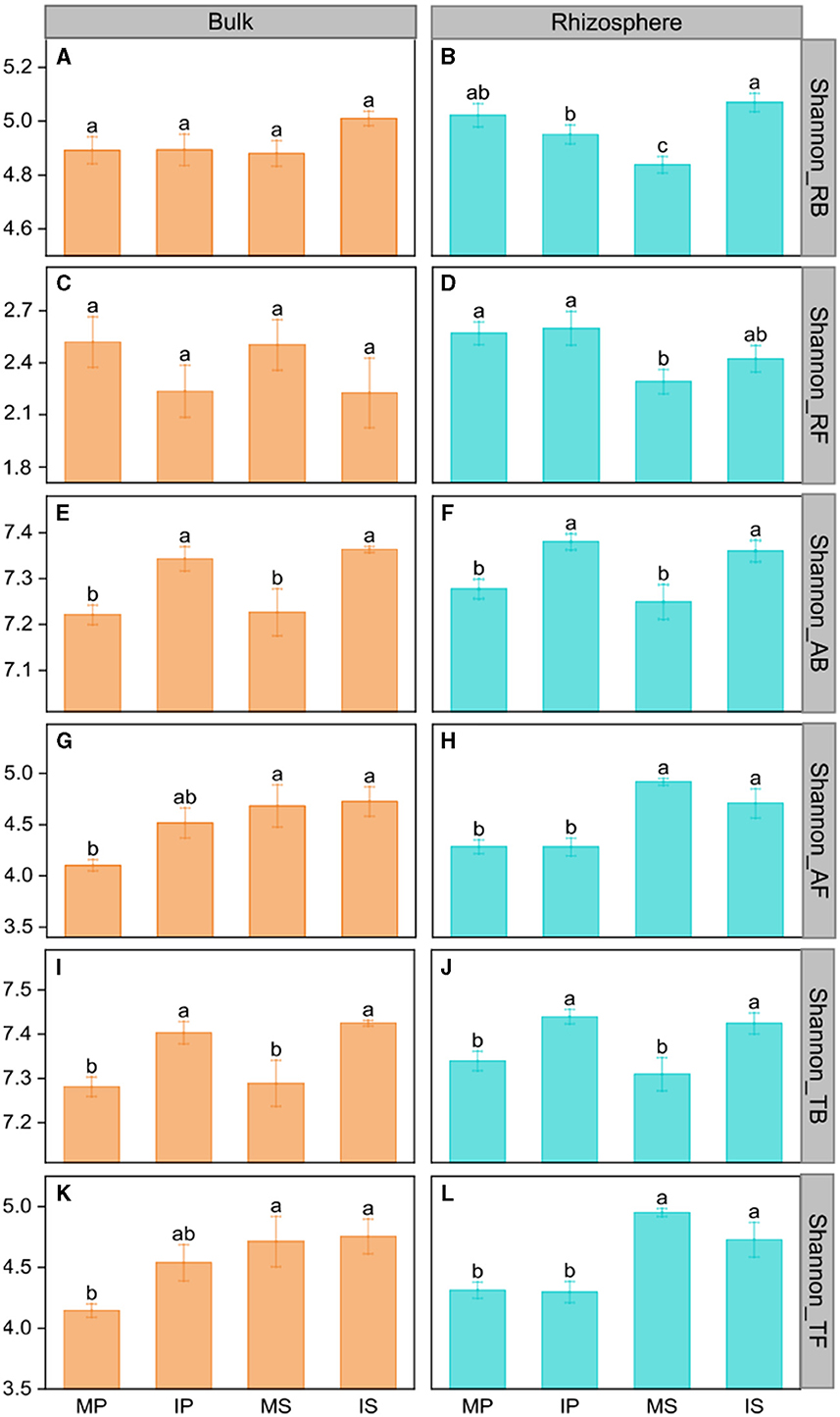
Figure 2. The intercropping effects on Shannon diversity of the microbial community in the rhizosphere (B, D, F, H, J, L) and bulk soil (A, C, E, G, I, K) at the phylum level. MS, monoculture sugarcane; IS, intercropping sugarcane; MP, monoculture peanut; IP, intercropping peanut. Shannon_RB, Shannon_RF, Shannon_AB, Shannon_AF, Shannon_TB, and Shannon_TF indicate the Shannon diversity of rare bacteria and fungi, abundant bacteria and fungi, and total bacteria and fungi, respectively. The lowercase letters indicate significant differences among different cropping patterns in both rhizosphere and bulk soil by LSD test at P < 0.05, respectively.
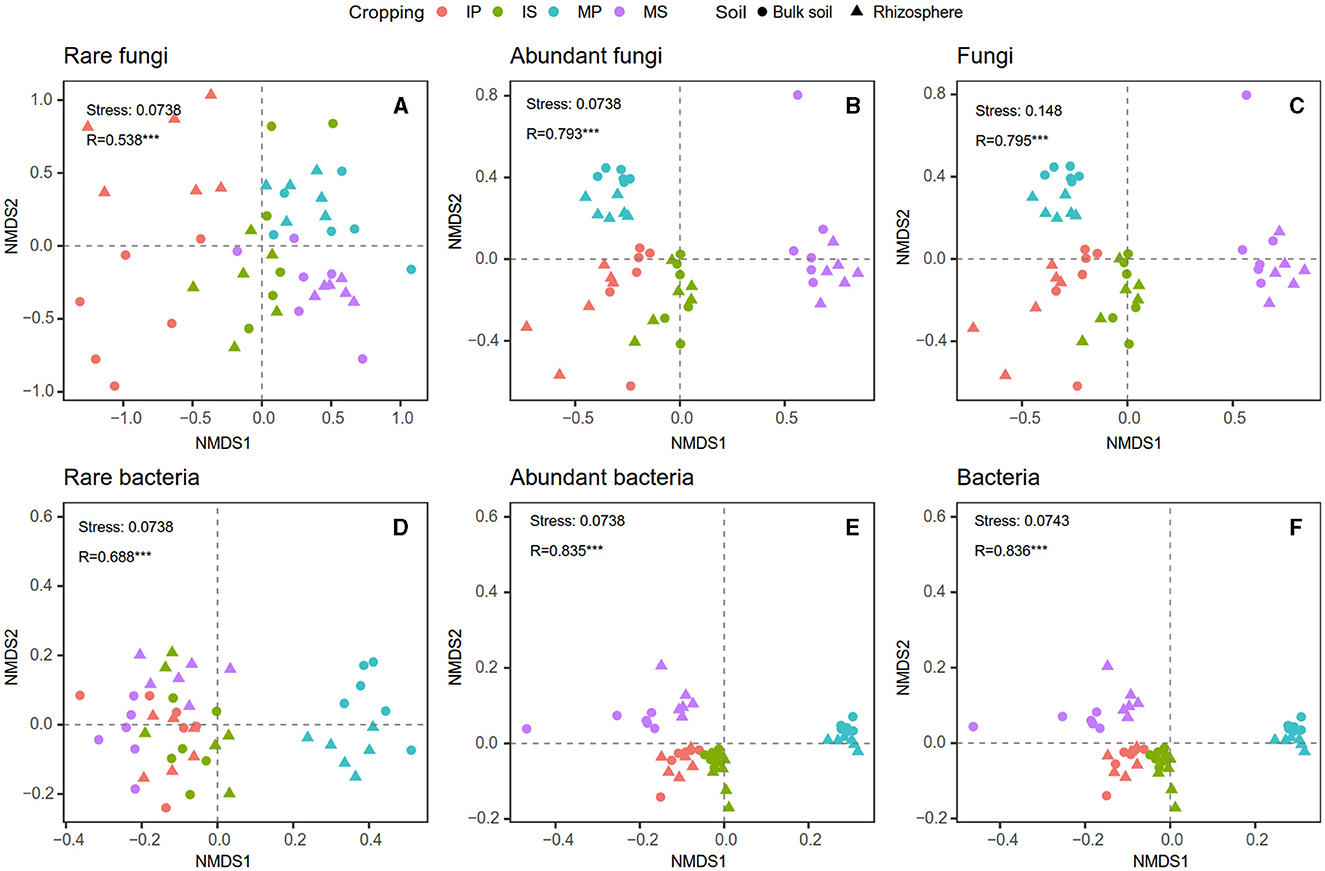
Figure 3. Intercropping effects on fungal (A–C) and bacterial (D–F) community beta diversity at phylum level based on non-metric multidimensional scaling (NMDS) analysis in the rhizosphere and bulk soil. MS, monoculture sugarcane; IS, intercropping sugarcane; MP, monoculture peanut; IP, intercropping peanut.
3.3 Intercropping effects on soil enzyme activity and stoichiometry in the rhizosphere and bulk soils
Intercropping affected AP activity in bulk soil, but its effect differed in crop types. Intercropping decreased LAP activity in both rhizosphere and bulk soil, whereas decreased BG activity solely in rhizosphere soil (Supplementary Table S3). The ln (BG): ln (NAG+LAP) was more than 1:1, indicating microbes were limited by C relative to N. Based on the vector analysis of enzyme stoichiometry, intercropping decreased vector length in both rhizosphere and bulk soil (Figure 4A), and the vector angle was more than 45°, indicating microbes were limited by P relative to N (Figure 4B). Intercropping primarily impacted enzyme stoichiometry, vector length, and angle in bulk soil but not in rhizosphere soil (Supplementary Tables S3, S4, Figure 4). The enzyme activity, stoichiometry, and vector angle were also different between the rhizosphere and bulk soil, with higher BG, AP, ln (NAG+LAP): ln AP, and vector angle, as well as lower LAP and ln BG:ln AP in rhizosphere than those in bulk soil (Supplementary Tables S3, S4). Furthermore, crop type exerted an influence on vector length and angle, with higher vector length in sugarcane rhizosphere soil and higher vector angle in peanut bulk soil, respectively (Supplementary Table S4).
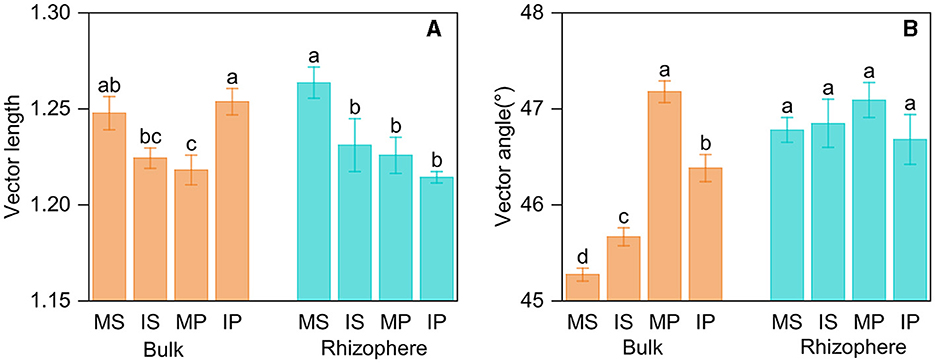
Figure 4. Vector length (A) and vector angle (B) based on soil enzyme stoichiometry in intercropping and monoculture systems. MS, monoculture sugarcane; IS, intercropping sugarcane; MP, monoculture peanut; IP, intercropping peanut. The lowercase and uppercase letters indicate significant intercropping effects in the rhizosphere and bulk soil by LSD test at P < 0.05, respectively.
3.4 Relationship among soil properties, microbial community, and enzymes in the rhizosphere and bulk soils
The Pearson correlation showed that microbial communities of rare and abundant taxa and vector angle were significantly related to soil C:N and N:P ratio in bulk soil alone (Supplementary Figure S6). In rhizosphere soil, vector length and angle were primarily related to rare taxa, fungal Shannon, and beta diversity (Figures 5, 6). Vector angle was solely negatively related to Patescibacteria and Glomeromycota (Figure 6). In bulk soil, vector length was primarily and negatively related to bacterial diversity (Figure 5A) and was significantly related to abundant taxa (Mortierellomycota and Gemmatimonadota) and rare taxa (Figure 6A). Vector angle was primarily related to fungal and bacterial beta diversity, abundant taxa (Nitrospirota and Mortierellomycota), and most of the rare taxa at the phylum level in bulk soil (Figures 5, 6). Furthermore, the VPA analysis showed that the individual effects of rare taxa explained 7–35% of the variations, and the interactions of rare and abundant taxa explained 65–93% of variations in vector length and angle in both rhizosphere and bulk soil (Figure 7). The vector length had a negative correlation with vector angle in bulk soil but not in rhizosphere soil, especially in the sugarcane system ( = 0.52, P = 0.005; Figure 8).
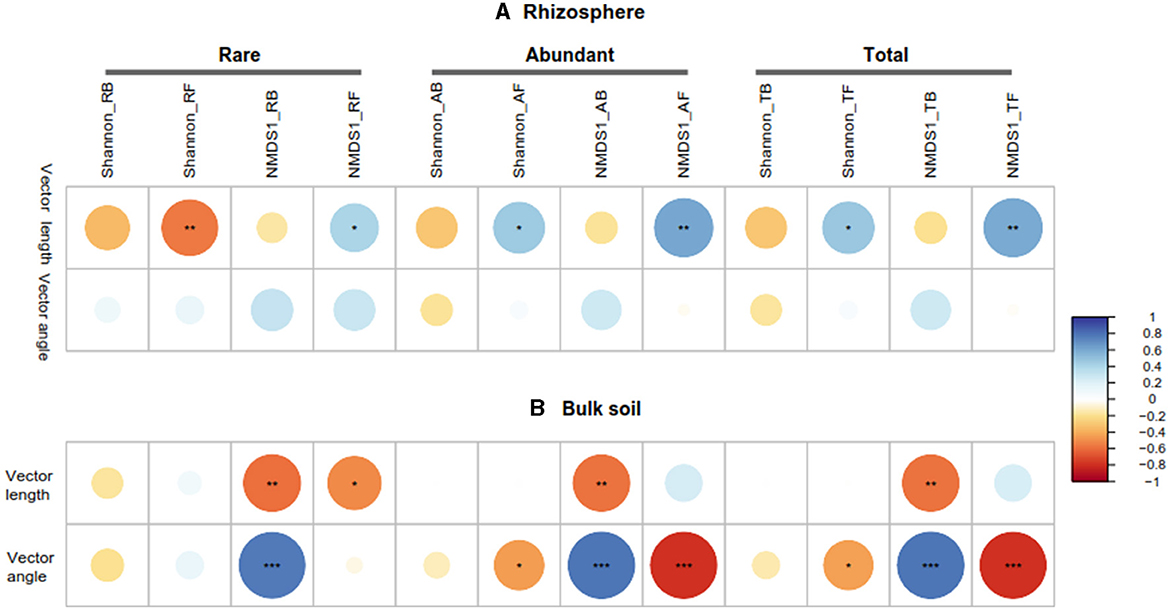
Figure 5. Pearson correlations between microbial community (rare, abundant, and total), vector length, and vector angle in rhizosphere (A) and bulk soil (B). RB, RF, AB, and AF: the rare bacteria and fungi, abundant bacteria and fungi, respectively; NMDS1, the first component of the non-metric multidimensional scaling (NMDS) analysis of microbial community by bray-distance. *P < 0.05; **P < 0.01; ***P < 0.001.
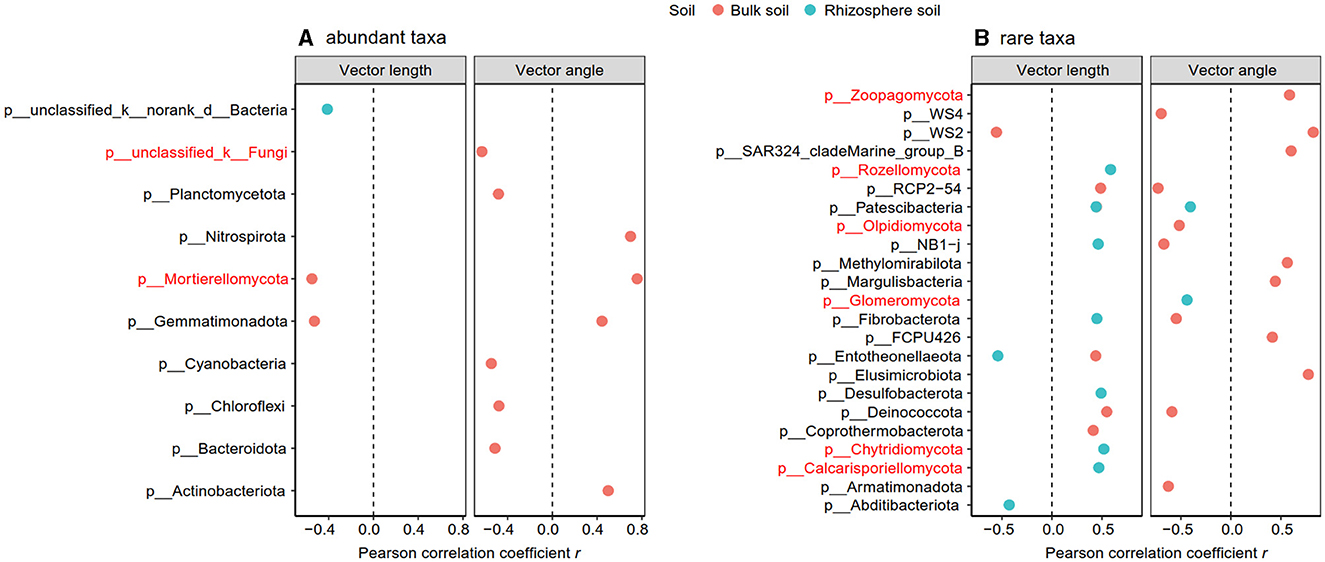
Figure 6. Significant Pearson correlation between abundant (A) and rare taxa (B) with vector length and angle in the rhizosphere and bulk soil. The taxa in red indicated the fungal group.
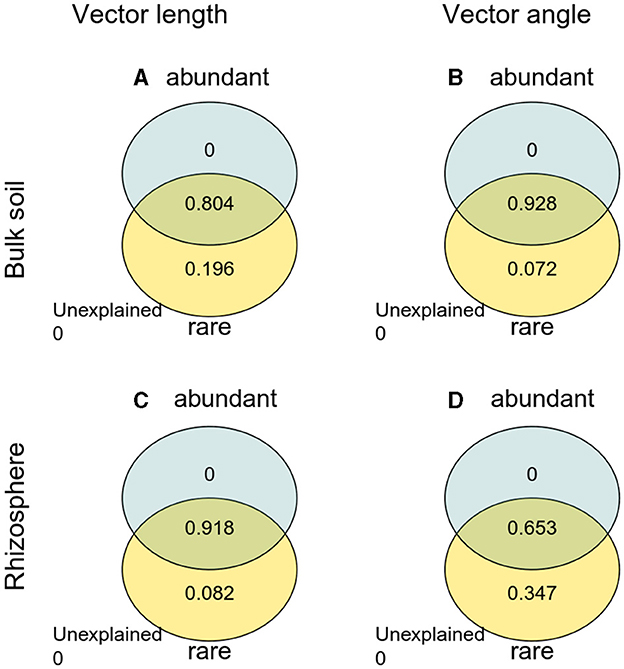
Figure 7. Variation partitioning analysis (VPA) of abundant and rare taxa with vector length and angle. (A) Vector length in bulk soil; (B) vector angle in bulk soil; (C) vector length in rhizosphere soil; (D) vector angle in rhizosphere soil.
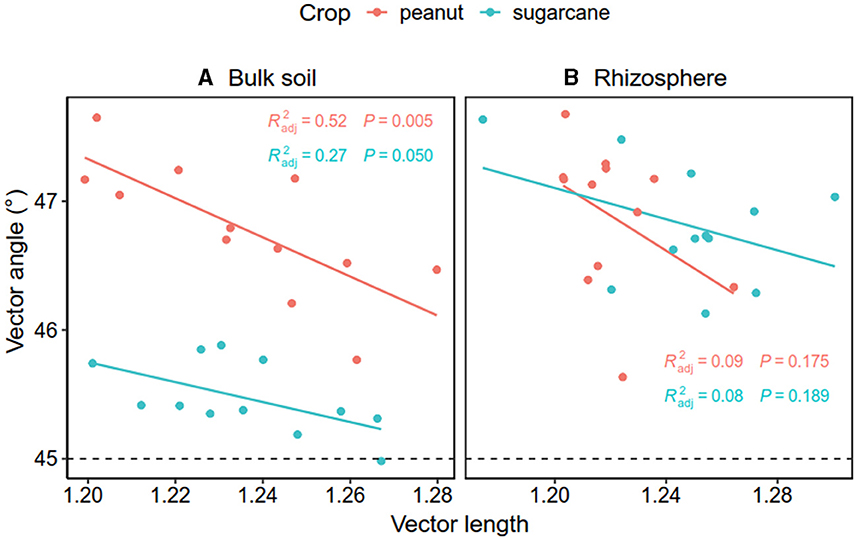
Figure 8. Relationship between vector length and angle in bulk soil (A) and the rhizosphere soil (B). The red and green dots and lines indicate peanut and sugarcane soils, respectively.
4 Discussion
4.1 Intercropping effects on microbial C and nutrient limitation between rhizosphere and bulk soil
Rhizo-deposition significantly affects microbial metabolisms due to high nutrient availability and microbial activity (Gong et al., 2019; Cui et al., 2020). In this study, we measured microbial C and nutrient metabolism limitation in the rhizosphere and bulk soil based on enzyme stoichiometry and vector analysis. In this study, microbes were limited by C and P in both bulk and rhizosphere soil because of ln(BG)/ln(NAG+LAP) >1 and vector angle >45°. In contrast, microbial N limitation was alleviated especially intercropping with peanut through its biological N2 fixation function, which supports the view that intercropping peanut helps to promote soil N availability. We also observed that microbial P limitation was stronger in rhizosphere soil than in bulk soil, suggesting a faster P cycle in rhizosphere soil, in agreement with a previous study (Cui et al., 2021). This was ascribed to high demands of microbes on soil available P in rhizosphere soil where microbes are r-strategist copiotrophs and fast-growing, which require more P investments for biomass synthesis (ribosomal RNA), and thus have higher enzyme production for P-acquisition (Ren et al., 2021). The rhizosphere effects are stronger and regulate the intercropping effects on microbial P limitation. Intercropping had significant effects on microbial P limitation in bulk soils but not rhizosphere soils. Bulk soil is generally limited by nutrient and microbial activity, and the soil nutrient status is regulated by rhizosphere nutrient release and migration. Intercropping might shift bulk soil from oligotrophic into eutrophic soils and regulate bulk soil microbial community through stronger rhizo-deposition effects compared to sole crops. This result can be supported by higher bulk soil nutrient status and microbial diversity when intercropped than sole crops.
Microbial C limitation was alleviated by intercropping peanuts in the rhizosphere and bulk soil, suggesting the dependence of microbial C metabolisms on intercropping. Compared to sole crops, intercropping peanuts can increase soil N availability through biological N2 fixation and promote crop and microbial growth, consequently, increasing soil labile-C inputs and decreasing microbial C limitation. In contrast, microbial C limitation was almost unchanged between rhizosphere and bulk soil. Previous studies have revealed that the rhizosphere has various effects on microbial C limitation with positive, negative, and no impacts relative to bulk soil (Cui et al., 2021; Bi et al., 2022; Kang et al., 2022). These different rhizosphere effects on microbial C limitation might be attributed to the interactions of soil and microbial properties.
Crop type exerted an important influence on microbial C and P limitation (Supplementary Table S4). Microbial C limitation was alleviated in the peanut system than in the sugarcane system. In contrast, microbial P limitation was enhanced in the peanut system compared to the sugarcane system. This is because the peanut system has high N availability due to its biological N2-fixing function, thereby increasing microbial P demands. Meanwhile, the coupled soil C and N might explain the alleviated microbial C limitation in peanut systems where soil N is rich (Moorhead et al., 2016). We also found that the intercropping effects on microbial C and P limitation were crop-related in bulk soil. When intercropped, the N2 fixed by the peanut is gradually depleted by the surrounding soil, leading to a decrease in soil N availability within the peanut system. In contrast, sugarcane–peanut intercropping (IS treatment) increased soil N availability and promoted microbial growth and metabolism, particularly in bulk soil. Taken together, the differences in microbial C and P limitation across crops might be explained by the interspecific competition on soil N in the intercropping system (Feng et al., 2021).
4.2 Microbial community mediates the intercropping effects on microbial C and nutrient limitation between rhizosphere and bulk soil
The microbial community is the key to regulating ecosystem functions and services, such as soil nutrient and carbon cycles via altering microbial metabolisms (Delgado-Baquerizo et al., 2016; Maron et al., 2018). Supported by our hypothesis, both rare and abundant taxa diversity are vital to affecting microbial P and C limitation in bulk soil, which agreed with a previous study (Yang et al., 2021). Microbial P limitation was negatively related to fungal beta diversity in bulk soil, indicating that higher fungal community composition diversity helps to alleviate microbial P limitation. This may be attributed to the fact that complex microbial community structures can have diverse functionalities, such as decomposing and utilizing diverse soil substrates (Zhou Z. H. et al., 2020), which increase the decomposition of P compounds and satisfy microbial P requirements. In rhizosphere soil, however, microbial diversity did not affect microbial P limitation. One reason may be ascribed to the functional redundancy hypothesis in rhizosphere soil (Wertz et al., 2006). The rhizosphere, as a hotspot in soil, has high microbial diversity and multi-functionality in soil nutrient and C cycle processes (Ling et al., 2022). This hotspot leads to diverse microbial groups that share similar and even the same functions (functional redundancy), thereby weakening the contributions of microbial diversity to microbial metabolisms. Another possible reason was that microbial P limitation in rhizosphere soil was primarily regulated by rare taxa, including Patescibacteria and Glomeromycota at the phylum level. Phylum Glomeromycota (fungal community) generally increases soil P availability and P uptake, as well as improves the stress tolerance of their host plants (Liu et al., 2012; Procter et al., 2014).
The rare taxa, but not abundant ones, regulate microbial C and P limitation in both rhizosphere and bulk soil, indicating the importance of rare taxa in soil C and nutrient cycles (Han and Wang, 2023; Xu et al., 2024). This is because rare taxa have faster responses to stress and disturbance than abundant taxa, which can be a sensitive indicator of environmental changes (Xu et al., 2021a; Lin et al., 2022). In this study, compared to abundant taxa, rare taxa had a stronger correlation with microbial C and P limitation in all soils (Figure 5). Furthermore, microbial C limitation was primarily affected by rare bacterial taxa in bulk soil and by rare fungal taxa (Rozellomycota, Chytridiomycota, and Calcarisporiellomycota) in the rhizosphere (Figure 5). Fungi prefer plant-derived C compounds (e.g., root exudates), whereas bacteria prefer small molecular substrates (e.g., sugars) (de Boer et al., 2005; Paterson et al., 2008). Microbial substrate preferences lead to that fungi are prior to utilizing soil substrates in rhizosphere soil where root-derived C is rich (Sylvia et al., 2005). In bulk soil, however, bacterial beta diversity was strongly dependent on soil C:N and N:P ratios because of the nutrient imbalance (Supplementary Figure S6B). The imbalance of soil nutrients leads to changes in the bacterial community to utilize a wider variety of substrates through enzyme production (Liu et al., 2020). Thus, the substrate preferences explained the different responses of bacterial and fungal rare taxa to C acquisition in bulk and rhizosphere soil.
4.3 Relationships between microbial C and nutrient limitation in the rhizosphere and bulk soil
The breakdown of SOM often releases both C and P; therefore, soil C and P are generally coupled, and in turn, microbes produce C-related enzymes to meet C acquisition demands and alleviate P limitation as well. However, this coupling relationship varies with soil nutrient availability (Cui et al., 2020; Sun et al., 2023). For instance, a recent study observed that P/C–acquisition enzymes have a negative correlation in bulk soil but a positive correlation in rhizosphere soil where labile C is rich (Liu et al., 2021), which is partly consistent with our study. In this study, we observed that microbial P limitation (vector angle) had a negative correlation with microbial C limitation (vector length) solely in bulk soils. This implies that microbial P/C acquisition is coupled in bulk soil but not in rhizosphere soil. Root acidifying rhizosphere soil might be one possible reason for the no relationship between microbial C and P limitation. A meta-analysis indicates that compared with bulk soil, legumes induce a decrease of 0.44 unit in rhizosphere soil pH during biological N2 fixation (Liu et al., 2022), which could enhance soil P availability via the dissolution of Fe–P and Al–P complexes in acidic soils (Haynes, 1983; Liu et al., 1989) and reduces microbial P limitation. This is supported by the positive relationship between soil pH and microbial P limitation. Therefore, intercropping legumes alleviate microbial P limitation through enzyme production and soil acidification in rhizosphere soil (Liu et al., 2021), which explains the no correlation between microbial C and P limitation in rhizosphere soil.
5 Conclusion
In this study, microbes were limited by C and P in all soils. Microbial C limitation was alleviated by sugarcane–peanut intercropping, microbial P limitation was stronger in rhizosphere soil than in bulk soil, and was enhanced by sugarcane–peanut intercropping in bulk soil but not in rhizosphere soil due to root acidification. These results indicate that strong microbial P limitation promotes soil P decomposition and thus enhances P turnover in rhizosphere and intercropping systems. The intercropping effects on microbial C limitation were primarily regulated by microbial beta diversity and rare taxa in the rhizosphere and bulk soil. Microbial P limitation was mostly modified by the interactions of rare and abundant taxa in bulk soil and by rare taxa in rhizosphere soil. This study emphasizes the positive effects of intercropping on soil P cycles, and the importance of microbial community, in particular, rare taxa, in microbial nutrient limitation. In conclusion, intercropping peanuts facilitates soil nutrient cycles through accelerating microbial metabolisms (e.g., rare taxa), especially in P-limited soils.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/, PRJNA1051854.
Author contributions
YF: Formal analysis, Investigation, Resources, Visualization, Writing—review & editing. XT: Methodology, Supervision, Writing—review & editing. TS: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Visualization, Writing—original draft, Writing—review & editing. LL: Investigation, Writing—original draft, Writing—review & editing. LW: Writing—original draft, Writing—review & editing. TZ: Writing—original draft, Writing—review & editing. YG: Writing—original draft, Writing—review & editing. YL: Writing—original draft, Writing—review & editing. HW: Writing—original draft. JX: Writing—original draft. RT: Supervision, Writing—original draft, Writing—review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research work was supported by the start-up funding for introduced talents in Guangxi University (A3310051077 and EE101734) and National Natural Science Foundation Project (32260544), the earmarked fund for CARS-13.
Acknowledgments
We are grateful to Muqing Zhang at Guangxi University for providing support and other team members who participated in the field survey and for their help in laboratory analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1403338/full#supplementary-material
References
Bahram, M., Hildebrand, F., Forslund, S.K., Anderson, J.L., Soudzilovskaia, N.A., Bodegom, P.M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Banerjee, S., Zhao, C., Garland, G., Edlinger, A., García-Palacios, P., Romdhane, S., et al. (2024). Biotic homogenization, lower soil fungal diversity and fewer rare taxa in arable soils across Europe. Nat. Commun. 15:327. doi: 10.1038/s41467-023-44073-6
Bi, B. Y., Wang, Y., Wang, K., Zhang, H., Fei, H. Y., Pan, R. P., et al. (2022). Changes in microbial metabolic C- and N- limitations in the rhizosphere and bulk soils along afforestation chronosequence in desertified ecosystems. J. Environ. Manage. 303:114215. doi: 10.1016/j.jenvman.2021.114215
Bremner, J. M., and Mulvaney, C. S. (1982). Methods of soil analysis, chemical and microbial properties. Agron. Soc. Am. Monogr. 9, 595–624. doi: 10.2134/agronmonogr9.2.2ed.c31
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., Holmes, S. P., et al. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, H., Zheng, M. H., Mao, Q. G., Xiao, K. C., Wang, K. L., Li, D. J., et al. (2019). Cropland conversion changes the status of microbial resource limitation in degraded karst soil. Geoderma 352, 197–203. doi: 10.1016/j.geoderma.2019.06.018
Chen, Y., Xia, A. Q., Zhang, Z. J., Wang, F., Chen, J. H., Hao, Y. B., et al. (2023). Extracellular enzyme activities response to nitrogen addition in the rhizosphere and bulk soil: a global meta-analysis. Agric. Ecosyst. Environ. 356:108630. doi: 10.1016/j.agee.2023.108630
Cong, W. F., Hoffland, E., Li, L., Six, J., Sun, J. H., Bao, X. G., et al. (2015). Intercropping enhances soil carbon and nitrogen. Global Change Biol. 21, 1715–1726. doi: 10.1111/gcb.12738
Creamer, C. A., Krull, A. B., Sanderman, E. S., Newton-Walters, J., Farrell, R., et al. (2015). Microbial community structure mediates response of soil C decomposition to litter addition and warming. Soil Biol. Biochem. 80, 175–188. doi: 10.1016/j.soilbio.2014.10.008
Cui, Y. X., Bing, H. J., Fang, L. C., Jiang, M., Shen, G. T., Yu, J. L., et al. (2021). Extracellular enzyme stoichiometry reveals the carbon and phosphorus limitations of microbial metabolisms in the rhizosphere and bulk soils in alpine ecosystems. Plant Soil 458, 7–20. doi: 10.1007/s11104-019-04159-x
Cui, Y. X., Wang, X., Zhang, X. C., Ju, W. L., Duan, C. J., Guo, X. B., et al. (2020). Soil moisture mediates microbial carbon and phosphorus metabolism during vegetation succession in a semiarid region. Soil Biol. Biochem. 147:107814. doi: 10.1016/j.soilbio.2020.107814
Curtright, A. J., and Tiemann, L. K. (2021). Intercropping increases soil extracellular enzyme activity: a meta-analysis. Agric. Ecosyst. Environ. 319:107489. doi: 10.1016/j.agee.2021.107489
de Boer, D., Folman, W., Summerbell, L. B., and Boddy, R. C. L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. Fems Microbiol. Rev. 29, 795–811. doi: 10.1016/j.femsre.2004.11.005
Delgado-Baquerizo, M., Giaramida, L., Reich, P. B., Khachane, A. N., Hamonts, K., Edwards, C., et al. (2016). Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J. Ecol. 104, 936–946. doi: 10.1111/1365-2745.12585
Domeignoz-Horta, L. A., Pold, G., Liu, X. J. A., Frey, S. D., and Melillo, J. M. (2020). Microbial diversity drives carbon use efficiency in a model soil. Nat. Commun. 11:3684. doi: 10.1038/s41467-020-17502-z
dos Santos Bastos, T. R., Barreto-Garcia, P. A. B., de Carvalho Mendes, I., Monroe, P. H. M., and de Carvalho, F. F. (2023). Response of soil microbial biomass and enzyme activity in coffee-based agroforestry systems in a high-altitude tropical climate region of Brazil. Catena 230:107270. doi: 10.1016/j.catena.2023.107270
Elmajdoub, B., Barnett, S., and Marschner, P. (2014). Response of microbial activity and biomass in rhizosphere and bulk soils to increasing salinity. Plant Soil 381, 297–306. doi: 10.1007/s11104-014-2127-4
Feng, L., Yang, W. T., Zhou, Q., Tang, H. Y., Ma, Q. Y., Huang, G. Q., et al. (2021). Effects of interspecific competition on crop yield and nitrogen utilization in maize-soybean intercropping system. Plant Soil Environ. 67, 460–467. doi: 10.17221/665/2020-PSE
Gomez, E. J., Delgado, J. A., and Gonzalez, J. M. (2020). Persistence of microbial extracellular enzymes in soils under different temperatures and water availabilities. Ecol. Evol. 10, 10167–10176. doi: 10.1002/ece3.6677
Gong, X., Liu, C., Li, J., Luo, Y., Yang, Q., Zhang, W., et al. (2019). Responses of rhizosphere soil properties, enzyme activities and microbial diversity to intercropping patterns on the Loess Plateau of China. Soil Tillage Res. 195:104355. doi: 10.1016/j.still.2019.104355
Han, S., and Wang, A. (2023). Belowground bacterial communities and carbon components contribute to soil respiration in a subtropical forest. Plant Soil 1, 1–3. doi: 10.1007/s11104-023-06257-3
Haynes, R. J. (1983). Soil acidification induced by leguminous crops. Grass Forage Sci. 38, 1–11. doi: 10.1111/j.1365-2494.1983.tb01614.x
Hilbig, B. E., and Allen, E. B. (2015). Plant-soil feedbacks and competitive interactions between invasive Bromus diandrus and native forb species. Plant Soil 392, 191–203. doi: 10.1007/s11104-015-2451-3
Jiang, Y. J., Liu, M. Q., Zhang, J. B., Chen, Y., Chen, X. Y., Chen, L. J., et al. (2017). Nematode grazing promotes bacterial community dynamics in soil at the aggregate level. ISME J. 11, 2705–2717. doi: 10.1038/ismej.2017.120
Jing, X., Chen, X., Fang, J. Y., Ji, C. J., Shen, H. H., Zheng, C. Y., et al. (2020). Soil microbial carbon and nutrient constraints are driven more by climate and soil physicochemical properties than by nutrient addition in forest ecosystems. Soil Biol. Biochem. 141:107657. doi: 10.1016/j.soilbio.2019.107657
Ju, W. L., Fang, L. C., Shen, G. T., Delgado-Baquerizo, M., Chen, J., Zhou, G. Y., et al. (2023). New perspectives on microbiome and nutrient sequestration in soil aggregates during long-term grazing exclusion. Global Change Biol. 30:e17027. doi: 10.1111/gcb.17027
Kang, H. B., Xue, Y., Yan, C. L., Lu, S., Yang, H., Zhu, J. Q., et al. (2022). Contrasting patterns of microbial nutrient limitations between rhizosphere and bulk soil during stump sprout restoration in a clear-cut oak forest. Forest Ecol. Manage. 515:120241. doi: 10.1016/j.foreco.2022.120241
Kuster, T. M., Wilkinson, A., Hill, P. W., Jones, D. L., and Bardgett, R. D. (2016). Warming alters competition for organic and inorganic nitrogen between co-existing grassland plant species. Plant Soil 406, 117–129. doi: 10.1007/s11104-016-2856-7
Li, C. J., Hoffland, E., Kuyper, T. W., Yu, Y., Zhang, C. C., Li, H. G., et al. (2020). Syndromes of production in intercropping impact yield gains. Nat. Plants 6, 653–660. doi: 10.1038/s41477-020-0680-9
Lin, L. T., Jing, X., Lucas-Borja, M. E., Shen, C. C., Wang, Y. G., Feng, W. T., et al. (2022). Rare taxa drive the response of soil fungal guilds to soil salinization in the taklamakan desert. Front. Microbiol. 13:862245. doi: 10.3389/fmicb.2022.862245
Ling, N., Wang, T. T., and Kuzyakov, Y. (2022). Rhizosphere bacteriome structure and functions. Nat. Commun. 13:836. doi: 10.1038/s41467-022-28448-9
Liu, C. J., Gong, X. W., Dang, K., Li, J., Yang, P., Gao, X. L., et al. (2020). Linkages between nutrient ratio and the microbial community in rhizosphere soil following fertilizer management. Environ. Res. 184:109261. doi: 10.1016/j.envres.2020.109261
Liu, S. B., He, F. K., Kuzyakov, Y., Xiao, H. X., Hoang, D. T. T., Pu, S. Y., et al. (2022). Nutrients in the rhizosphere: a meta-analysis of content, availability, and influencing factors. Sci. Total Environ. 826:153908. doi: 10.1016/j.scitotenv.2022.153908
Liu, W. C., Lund, L. J., and Page, A. L. (1989). Acidity produced by leguminous plants through symbiotic dinitrogen fixation. J. Environ. Q. 18, 529–534. doi: 10.2134/jeq1989.00472425001800040025x
Liu, Y. H., Shahbaz, M., Fang, Y. Y., Li, B. Z., Wei, X. M., Zhu, Z. K., et al. (2021). Stoichiometric theory shapes enzyme kinetics in paddy bulk soil but not in rhizosphere soil. Land Degr. Dev. 33, 246–256. doi: 10.1002/ldr.4141
Liu, Y. J., Shi, G. X., Mao, L., Cheng, G., Jiang, S. J., Ma, X. J., et al. (2012). Direct and indirect influences of 8 yr of nitrogen and phosphorus fertilization on Glomeromycota in an alpine meadow ecosystem. New Phytol. 194, 523–535. doi: 10.1111/j.1469-8137.2012.04050.x
Liu, Y. Y., Dong, L. Z., Zhang, H. J., Deng, Y. Y., Hu, B., Wang, W., et al. (2023). Distinct roles of bacteria and fungi in mediating soil extracellular enzymes under long-term nitrogen deposition in temperate plantations. Forest Ecol. Manage. 529:120658. doi: 10.1016/j.foreco.2022.120658
Maron, P. A., Sarr, A., Kaisermann, A., Lévêque, J., Mathieu, O., Guigue, J., et al. (2018). High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 84, e02738–e02717. doi: 10.1128/AEM.02738-17
Moorhead, D. L., Rinkes, Z. L., Sinsabaugh, R. L., and Weintraub, M. N. (2013). Dynamic relationships between microbial biomass, respiration, inorganic nutrients and enzyme activities: informing enzyme-based decomposition models. Front. Microbiol. 4:00223. doi: 10.3389/fmicb.2013.00223
Moorhead, D. L., Sinsabaugh, R. L., Hill, B. H., and Weintraub, M. N. (2016). Vector analysis of ecoenzyme activities reveal constraints on coupled C, N and P dynamics. Soil Biol. Biochem. 93, 1–7. doi: 10.1016/j.soilbio.2015.10.019
Page, A. L. (Ed.). (1982). Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties. Madison, WI: American Society of Agronomy, Soil Science Society of America 1159.
Pang, Z. Q., Fallah, N., Weng, P. Y., Zhou, Y. M., Tang, X. M., Tayyab, M., et al. (2021). Sugarcane-peanut intercropping system enhances bacteria abundance, diversity, and sugarcane parameters in rhizospheric and bulk soils. Front. Microbiol. 12:815129. doi: 10.3389/fmicb.2021.815129
Paterson, E., Osler, G., Dawson, L. A., Gebbing, T., Sim, A., Ord, B., et al. (2008). Labile and recalcitrant plant fractions are utilised by distinct microbial communities in soil: independent of the presence of roots and mycorrhizal fungi. Soil Biol. Biochem. 40, 1103–1113. doi: 10.1016/j.soilbio.2007.12.003
Procter, A. C., Ellis, J. C., Fay, P. A., Polley, H. W., Jackson, R. B., Cullen, D., et al. (2014). Fungal community responses to past and future atmospheric CO2 differ by soil type. Appl. Environ. Microbiol. 80, 7364–7377. doi: 10.1128/AEM.02083-14
Qiu, X. C., Peng, D. L., Tian, H. X., Wang, H. B., Liu, X., Cao, L., et al. (2021). Soil ecoenzymatic stoichiometry and microbial resource limitation driven by thinning practices and season types in Larix principis-rupprechtii plantations in North China. Forest Ecol. Manage. 482:118880. doi: 10.1016/j.foreco.2020.118880
Ren, C. J., Zhou, Z. H., Guo, Y. X., Yang, G. H., Zhao, F. Z., Wei, G. H., et al. (2021). Contrasting patterns of microbial community and enzyme activity between rhizosphere and bulk soil along an elevation gradient. Catena 196:104921. doi: 10.1016/j.catena.2020.104921
Shi, X. L., Zhou, Y. F., Guo, P., Ren, J. Y., Zhang, H., Dong, Q. Q., et al. (2022). Peanut/sorghum intercropping drives specific variation in peanut rhizosphere soil properties and microbiomes under salt stress. Land Degr. Dev. 34, 736–750. doi: 10.1002/ldr.4490
Sinsabaugh, R. L., Hill, B. H., and Follstad Shah, J. J. (2009). Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 462, 795–798. doi: 10.1038/nature08632
Sinsabaugh, R. L., Lauber, C. L., Weintraub, M. N., Ahmed, B., Allison, S. D., Crenshaw, C., et al. (2008). Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 11, 1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x
Steinauer, K., Tilman, D., Wragg, P. D., Cesarz, S., Cowles, J. M., Pritsch, K., et al. (2015). Plant diversity effects on soil microbial functions and enzymes are stronger than warming in a grassland experiment. Ecology 96, 99–112. doi: 10.1890/14-0088.1
Sun, T. T., Wang, Y. G., Guo, Y. F., Jing, X., and Feng, W. T. (2023). Contrasting elevational patterns of microbial carbon and nutrient limitation in soil from alpine meadow to desert. Catena 223:106901. doi: 10.1016/j.catena.2022.106901
Sylvia, D. M., Fuhrmann, J. J., Hartel, P. G., and Zuberer, D. A. (2005). Principles and Applications of Soil Microbiology. Upper Saddle River, NJ: Pearson Prentice Hall.
Tang, X. M., He, Y. L., Zhang, Z., Wu, H. N., He, L. Q., Jiang, J., et al. (2022). Beneficial shift of rhizosphere soil nutrients and metabolites under a sugarcane/peanut intercropping system. Front. Plant Sci. 13:1018727. doi: 10.3389/fpls.2022.1018727
Tang, X. M., Jiang, J., Huang, Z. P., Wu, H. N., Wang, J., He, L. Q., et al. (2021a). Sugarcane/peanut intercropping system improves the soil quality and increases the abundance of beneficial microbes. J. Basic Microbiol. 61, 165–176. doi: 10.1002/jobm.202000750
Tang, X. M., Zhang, Y. X., Jiang, J., Meng, X. Z., Huang, Z. P., Wu, H. N., et al. (2021b). Sugarcane/peanut intercropping system improves physicochemical properties by changing N and P cycling and organic matter turnover in root zone soil. Peerj 9:e10880. doi: 10.7717/peerj.10880
Tang, X. Y., Zhang, C. C., Yu, Y., Shen, J. B., van der Werf, W., Zhang, F. S., et al. (2021). Intercropping legumes and cereals increases phosphorus use efficiency; a meta-analysis. Plant Soil 460, 89–104. doi: 10.1007/s11104-020-04768-x
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass-C. Soil Biol. Biochem. 19, 703–707. doi: 10.1016/0038-0717(87)90052-6
Větrovský, T., Kohout, P., Kopecký, M., Machac, A., Man, M., Bahnmann, B. D., et al. (2019). A meta-analysis of global fungal distribution reveals climate-driven patterns. Nat. Commun. 10:5142. doi: 10.1038/s41467-019-13164-8
Wang, P. C., Ding, L. L., Zou, C., Zhang, Y. J., and Wang, M. Y. (2022). Rhizosphere element circling, multifunctionality, aboveground productivity and trade-offs are better predicted by rhizosphere rare taxa. Front. Plant Sci. 13:985574. doi: 10.3389/fpls.2022.985574
Wang, X. X., Zhou, L. Y., Fu, Y. L., Jiang, Z., Jia, S. X., Song, B. Q., et al. (2024). Drought-induced changes in rare microbial community promoted contribution of microbial necromass C to SOC in a subtropical forest. Soil Biol. Biochem. 189:109252. doi: 10.1016/j.soilbio.2023.109252
Wei, X. M., Hu, Y. J., Razavi, B. S., Zhou, J., Shen, J. L., Nannipieri, P., et al. (2019). Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol. Biochem. 131, 62–70. doi: 10.1016/j.soilbio.2018.12.025
Wertz, S., Degrange, V., Prosser, J. I., Poly, F., Commeaux, C., Freitag, T., et al. (2006). Maintenance of soil functioning following erosion of microbial diversity. Environ. Microbiol. 8, 2162–2169. doi: 10.1111/j.1462-2920.2006.01098.x
Wieder, W. (2014). Microbes, roots and global carbon. Nat. Clim. Change 4, 1052–1053. doi: 10.1038/nclimate2454
Xu, M., Huang, Q. Y., Xiong, Z. Q., Liao, H., Lv, Z. G., Chen, W. L., et al. (2021a). Distinct responses of rare and abundant microbial taxa to in situ chemical stabilization of cadmium-contaminated soil. Msystems 6, e01040–e01021. doi: 10.1128/mSystems.01040-21
Xu, M., Li, X. L., Kuyper, T. W., Xu, M., Li, X. L., Zhang, J. L., et al. (2021b). High microbial diversity stabilizes the responses of soil organic carbon decomposition to warming in the subsoil on the Tibetan Plateau. Global Change Biol. 27, 2061–2075. doi: 10.1111/gcb.15553
Xu, Z. W., Wang, Y. T., Li, H. K., Dong, Y. M., Wang, Z. C., Liu, Z. P., et al. (2024). Soil moisture and bacterial carbon limitation regulate the soil organic carbon in mountain peatlands. Catena 234:107610. doi: 10.1016/j.catena.2023.107610
Yang, Y., Li, T., Wang, Y. Q., Dou, Y. X., Cheng, H., Liu, L. X., et al. (2021). Linkage between soil ectoenzyme stoichiometry ratios and microbial diversity following the conversion of cropland into grassland. Agric. Ecosyst. Environ. 314, 107418. doi: 10.1016/j.agee.2021.107418
Yao, Q. M., Li, Z., Song, Y., Wright, S. J., Guo, X., Tringe, S. G., et al. (2018). Community proteogenomics reveals the systemic impact of phosphorus availability on microbial functions in tropical soil. Nature Ecol. Evol. 2, 499–509. doi: 10.1038/s41559-017-0463-5
Zhang, G. Z., Wei, F. G., Chen, Z. J., Wang, Y., Zheng, Y. Q., Wu, L., et al. (2022). Rare biosphere in cultivated Panax rhizosphere shows deterministic assembly and cross-plant similarity. Ecol. Indic. 142:109215. doi: 10.1016/j.ecolind.2022.109215
Zhang, G. Z., Wei, G. F., Wei, F. G., Chen, Z. J., He, M. J., Jiao, S., et al. (2021). Homogeneous selection shapes rare biosphere in rhizosphere of medicinal plant. Ecol. Indic. 129:107981. doi: 10.1016/j.ecolind.2021.107981
Zhang, S. H., Pan, Y., Zhou, Z. H., Deng, J., Zhao, F. Z., Guo, Y. X., et al. (2022). Resource limitation and modeled microbial metabolism along an elevation gradient. Catena 209:105807. doi: 10.1016/j.catena.2021.105807
Zheng, H. F., Vesterdal, L., Schmidt, I. K., and Rousk, J. (2022). Ecoenzymatic stoichiometry can reflect microbial resource limitation, substrate quality, or both in forest soils. Soil Biol. Biochem. 167:108613. doi: 10.1016/j.soilbio.2022.108613
Zhou, L. H., Liu, S. S., Shen, H. H., Zhao, M. Y., Xu, L. C., Xing, A. J., et al. (2020). Soil extracellular enzyme activity and stoichiometry in China's forests. Funct. Ecol. 34, 1461–1471. doi: 10.1111/1365-2435.13555
Zhou, Y. Y., Yang, Z., Liu, J. G., Li, X. D., Wang, X. X., Dai, C. C., et al. (2023). Crop rotation and native microbiome inoculation restore soil capacity to suppress a root disease. Nat. Commun. 14:8126. doi: 10.1038/s41467-023-43926-4
Keywords: microbial nutrient limitation, intercropping, rhizosphere, microbial community, rare taxa
Citation: Fu Y, Tang X, Sun T, Lin L, Wu L, Zhang T, Gong Y, Li Y, Wu H, Xiong J and Tang R (2024) Rare taxa mediate microbial carbon and nutrient limitation in the rhizosphere and bulk soil under sugarcane–peanut intercropping systems. Front. Microbiol. 15:1403338. doi: 10.3389/fmicb.2024.1403338
Received: 19 March 2024; Accepted: 08 May 2024;
Published: 30 May 2024.
Edited by:
Yongxing Cui, Free University of Berlin, GermanyReviewed by:
Jiwen Cui, Chinese Academy of Agricultural Sciences (CAAS), ChinaJunxiang Ding, Zhengzhou University, China
Copyright © 2024 Fu, Tang, Sun, Lin, Wu, Zhang, Gong, Li, Wu, Xiong and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingting Sun, ttsun9@gxu.edu.cn; Ronghua Tang, tronghua@163.com
†These authors have contributed equally to this work
 Yue Fu
Yue Fu Xiumei Tang3†
Xiumei Tang3† Tingting Sun
Tingting Sun