- 1Department of Biology, Baylor University, Waco, TX, USA
- 2School of Biological Sciences, University of Nebraska-Lincoln, Lincoln, NE, USA
The nonsense-mediated mRNA decay (NMD) pathway is a specialized pathway that triggers the rapid degradation of select mRNAs. Initially, identified as a pathway that degrades mRNAs with premature termination codons, NMD is now recognized as a pathway that also regulates some natural mRNAs. Since natural mRNAs do not typically contain premature termination codons, these mRNAs contain features that target them to NMD. In Saccharomyces cerevisiae mRNAs with atypically long 3′-UTRs are usually degraded by NMD, however in some conditions a constitutively expressed SSY5 mRNA with multiple NMD targeting signals including an atypically long 3′-UTR is an exception. We investigated the features of the SSY5 mRNAs that confer immunity to NMD. We found that the SSY5 mRNA 3′-UTRs are sufficient to target NMD insensitive mRNA to the pathway. Replacing the SSY5 3′-UTRs with the cyc1-512 3′-UTRs, known to target mRNAs to NMD or with the CYC1 3′-UTR, known not to target mRNAs to NMD, resulted in production of SSY5 mRNAs that were regulated by NMD. These observations suggest that the SSY5 mRNAs require sequences both within the 5′-UTR and/or ORF as well as the 3′-UTR to escape decay by NMD.
Introduction
The nonsense-mediated mRNA decay (NMD) pathway is conserved in all eukaryotic organisms that have been examined so far. It elicits the rapid degradation of mRNAs with premature termination codons and, importantly, some natural mRNAs as well (Reviewed in Culbertson and Leeds, 2003; Amrani et al., 2006). Three core trans-acting factors are required for NMD in all eukaryotes. These are the up-frameshift proteins Upf1p, Upf2p, and Upf3p. The essential role of these proteins in this pathway was initially discovered in the yeast Saccharomyces cerevisiae and later found in multicellular eukaryotes. Elimination of any one of these three proteins selectively stabilizes mRNAs that are targeted for NMD-mediated degradation.
The regulation of natural mRNAs by NMD has been observed in multiple organisms ranging from yeast to humans. In global analysis studies of the effect of NMD on transcript levels in S. cerevisiae (Lelivelt and Culbertson, 1999; He et al., 2003; Guan et al., 2006; Johansson et al., 2007), D. melanogaster (Rehwinkel et al., 2005) and humans (Mendell et al., 2004; Whittmann et al., 2006; Yepiskoposyan et al., 2011) ~10% of the transcriptome is affected when NMD is non-functional. The majority of the affected mRNAs accumulate in nmd mutants (Lelivelt and Culbertson, 1999; He et al., 2003; Guan et al., 2006; Johansson et al., 2007).
There are several signals that are known to activate the decay of natural mRNAs by NMD. They include: (1) Translated upstream open reading frames (uORFs) (Gaba et al., 2005; Guan et al., 2006). (2) Translation initiation at an out-of-frame AUG within the main open reading frame, also referred to as leaky scanning (Welch and Jacobson, 1999; Guan et al., 2006), (3) Inefficiently spliced pre-mRNAs (He et al., 1993; Guan et al., 2006). (4) Some non-productive alternatively spliced transcripts (Kawashima et al., 2014). (5) ribosomal frameshifts (Belew et al., 2010). (6) an Upf1p-dependent destabilizing element (UDE) has been shown to cause degradation of the PPR1 mRNA by NMD (Kebaara et al., 2003b), and (7) atypically long 3′-UTRs (Muhlrad and Parker, 1999; Singh et al., 2008; Kebaara and Atkin, 2009; Yepiskoposyan et al., 2011). Collectively, these studies show that there are a variety of known signals that induce degradation of mRNAs by NMD. Most of these NMD-targeting features are conserved in other organisms.
Currently three natural S. cerevisiae mRNAs containing NMD targeting signals, are known to be immune to the pathway (Ruiz-Echevarria and Peltz, 2000; Kebaara and Atkin, 2009). These NMD-resistant mRNAs include SSY5 mRNA, a natural mRNA with an atypically long 3′-UTR (Kebaara and Atkin, 2009), as well as GCN4 and YAP1 mRNAs which have uORFs (Ruiz-Echevarria and Peltz, 2000). That some endogenous mRNAs with NMD targeting features can escape NMD suggests that these mRNAs have evolved mechanisms to evade degradation by the pathway.
We originally identified SSY5 mRNA as a potential NMD substrate in a screen for S. cerevisiae mRNA with atypically long 3′-UTRs. SSY5 mRNAs have 3′-UTRs of ~475 nucleotides. This is atypically long for S. cerevisiae 3′-UTRs. In S. cerevisiae 3′-UTRs typically range in size from 50 to 200 nt, with a median length of 121 nt. The atypically long SSY5 3′-UTRs would be expected to target the mRNAs to the NMD pathway. However, in the conditions examined initially, we found that SSY5 mRNAs are not degraded by the pathway (Kebaara and Atkin, 2009).
Here, we investigated the immunity of the SSY5 mRNAs to the NMD pathway. We show that the SSY5 mRNA is immune to NMD in some conditions. Upon exposure to heat stress this SSY5 mRNA becomes sensitive to the NMD pathway in some genetic backgrounds, while a second, shorter SSY5 mRNA is insensitive to the pathway. However, the long SSY5 mRNA 3′-UTR is sufficient to target an NMD-insensitive mRNA for NMD induced degradation in all conditions tested. Further, we demonstrate that replacing the SSY5 3′-UTR with the cyc1-512 3′-UTR, which has been shown to target NMD-insensitive mRNAs to the pathway, results in production of fusion mRNAs that were regulated in an NMD-dependent manner. Additionally, replacement of the SSY5 3′-UTR with the CYC1 3′-UTR, which does not target mRNAs to the NMD pathway, also resulted in mRNAs that were regulated by NMD. These observations suggest that the SSY5 mRNA requires sequences within both the 5′-UTR and/or ORF as well as the 3′-UTR to escape decay by the NMD pathway.
Materials and Methods
Yeast Strains
The S. cerevisiae strains used in this study were, W303a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100), AAY320 (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 upf1-Δ2), AAY334 (MATa ura3-1 his3-11,15 trp1-1 leu2-3,112 rpb1-1), AAY335 (MATa ura3-1 his3-11,15 trp1-1 leu2-3,112 rpb1-1 upf1-Δ2) and B4060 (MATa lys2 his1 trp1 cyc1-512) (Wente et al., 1992; Kebaara et al., 2003a). Yeast strains were grown and maintained using standard methods (Ausubel et al., 1998). Plasmids were maintained in E. coli DH5α and transformed into yeast strains using Lithium Acetate mediated transformation (Gietz and Woods, 1998).
Heat Shock Treatment
S. cerevisiae strains W303a (UPF1), AAY320 (upf1Δ), AAY334 (UPF1 rpb1-1) and AAY335 (upf1Δ rpb1-1) were grown at 28°C in rich medium (YPD) to mid-log phase according to our mRNA steady-state accumulation protocol (Kebaara et al., 2012). The yeast strains were then transferred to a 39°C water bath for a 1 min heat shock before harvesting. Subsequently, RNA was extracted from the yeast strains as described below in the RNA methods.
DNA Methods
The DNA constructs used in this study were generated using cloning free PCR (Erdeniz et al., 1997). The primers used to generate the PCR fragments are listed in Table 1. Yeast genomic DNA from W303 or B4060 was used as the template to generate the PCR fragments. For each construct two separate PCR fragments were generated. One fragment encompassed the 5′-UTR and ORF and the second fragment contained the 3′-UTR. The 5′-UTR and ORF fragments were then fused to the 3′-UTR fragment using cloning free PCR. The 5′-UTR and ORF fragments generated by PCR were for CYC1 and SSY5 genes using the primers listed in Table 1. The CYC1 5′-UTR and ORF fragment was fused to the SSY5 3′-UTR to generate the CYC1-SSY5 3′-UTR fragment. The SSY5 5′-UTR and ORF was fused to either the CYC1 3′-UTR or the cyc1-512 3′-UTR to generate the SSY5-CYC1 3′-UTR fragment and the SSY5-cyc1-512 3′-UTR fragment respectively. The fusion DNA fragments were then inserted into the TOPO-TA cloning vector according to the manufacturer's instructions (Life Technologies, Grand Island, NY). All DNA fusion constructs generated by PCR were sequenced to confirm that they were error free and that precise fusions were generated. After sequencing the fusion constructs were subcloned out of the TOPO-TA cloning vector and inserted into the high copy yeast vector pRS425 (Sirkosky and Heiter, 1989).
RNA Methods
Yeast total RNA used for mRNA steady-state accumulation measurements was extracted by the hot phenol method from yeast strains, W303 and AAY320 harvested at mid-log phase (Kebaara et al., 2003a). Total RNA used to measure mRNA decay rates, was harvested as described in Kebaara et al. (2012). Briefly, yeast strains AAY334 (UPF1, rpb1-1) and AAY335 (upf1Δ, rpb1-1) were grown at 28°C to mid-log phase. The yeast cells were then transferred to 39°C, the non-permissive temperature. Yeast cells were successively harvested at different time points after transcription was inhibited by the temperature shift and total RNA extracted.
Yeast total RNA was resolved on a 1% agarose-formaldehyde gel followed by transfer of the RNA to Genescreen Plus membranes. The northern blots were probed with oligolabeled DNA probes. DNA templates for probe synthesis were generated by PCR. The probes were labeled with 32P using an Oligolabeling kit (Life Technologies, Grand Island, NY). Northern blots were phosphorImaged™ with a Typhoon phosphorImager (Amersham Pharmacia Biotech, Inc.). mRNA decay rates were determined by graphing log10 of the percent mRNA remaining vs. time using SigmaPlot 2000, Version 6.10 (SPSS Science, Chicago, IL). The mRNA steady-state levels and decay rates were normalized using SCR1 RNA, an RNA polymerase III transcript.
3′-RACE Analysis
3′-RACE analysis was done as described in Kebaara et al. (2012). Briefly, 5 μg of yeast total RNA from W303 or AAY320 used for quantitative northern analysis was utilized to make cDNA using SuperScript™ II RT (Life Technologies, Grand Island, NY). The cDNA was subsequently used as template DNA for the primary PCR reactions using the Abridged Universal Amplification Primer (AUAP) provided with the 3′-RACE kit and a SSY5 or CYC1 gene-specific primer. Following the primary PCR reactions, a nested PCR reaction was done using the initial PCR product (primary reaction) as the template and a nested SSY5 or CYC1 primer. Both the primary and nested PCR products were run on 1.5% agarose gels for visualization of the 3′-RACE PCR products.
Results
The SSY5 mRNA Produced in Wild-Type and NMD Mutant Yeast Strains has a Long 3′-UTR, but is not Degraded by the NMD Pathway While it is Regulated by the NMD Pathway in Identical Yeast Strains with the rpb1-1 Allele
Our previous study demonstrated that most natural mRNAs with atypically long 3′-UTRs are degraded by the NMD pathway (Kebaara and Atkin, 2009). In sharp contrast, we found that SSY5 mRNAs have atypically long 3′-UTRs but did not accumulate in nmd mutants. SSY5 3′-UTRs are predicted to range in size from 384 to 464 nt by the mRNA 3′-processing site predictor (http://harlequin.jax.org/drupal/?q=polyA; Graber et al., 2002). This 3′-UTR length was experimentally confirmed by 3′-RACE and determined to be ~475 nt, using steady-state accumulation total RNA (Kebaara and Atkin, 2009 and Supplementary Figure S1, lane 5). This 3′-UTR is atypically long for S. cerevisiae and should target the SSY5 mRNAs to the NMD pathway.
SSY5 mRNA steady-state levels in isogenic wild-type (UPF1) and nmd mutant (upf1Δ) yeast strains was measured by quantitative northern blotting (Figure 1A). One SSY5 transcript of ~2.8 kb was detected on steady-state northern blots. SSY5 mRNA levels were 0.88 ± 0.24 (n = 3) in nmd mutants (upf1Δ) relative to wild-type (UPF1) yeast strains. Subsequently, SSY5 mRNA half-lives were measured in wild-type and nmd mutant strains. Unlike the steady-state northern blot, two SSY5 transcripts were detected on the half-life northerns (Figure 1B). The faster migrating band was slightly shorter than the 2.8 kb SSY5 transcript. This SSY5 mRNA had the same half-life in the wild-type (UPF1) and nmd mutant (upf1Δ) strains (Figure 1B). The second SSY5 transcript detected on the half-life northern was the ~2.8 kb transcript detected on the steady-state northerns and it was now sensitive to the NMD pathway. Notably, we only observed the two SSY5 transcripts in the conditions and yeast strains used to measure mRNA half-lives. The half-life of the longer SSY5 transcript was 4.3 ± 1.53 (n = 3) min in the wild-type strains and 12.50 ± 0.7 (n = 2) min in nmd mutant strains (Figure 1B).
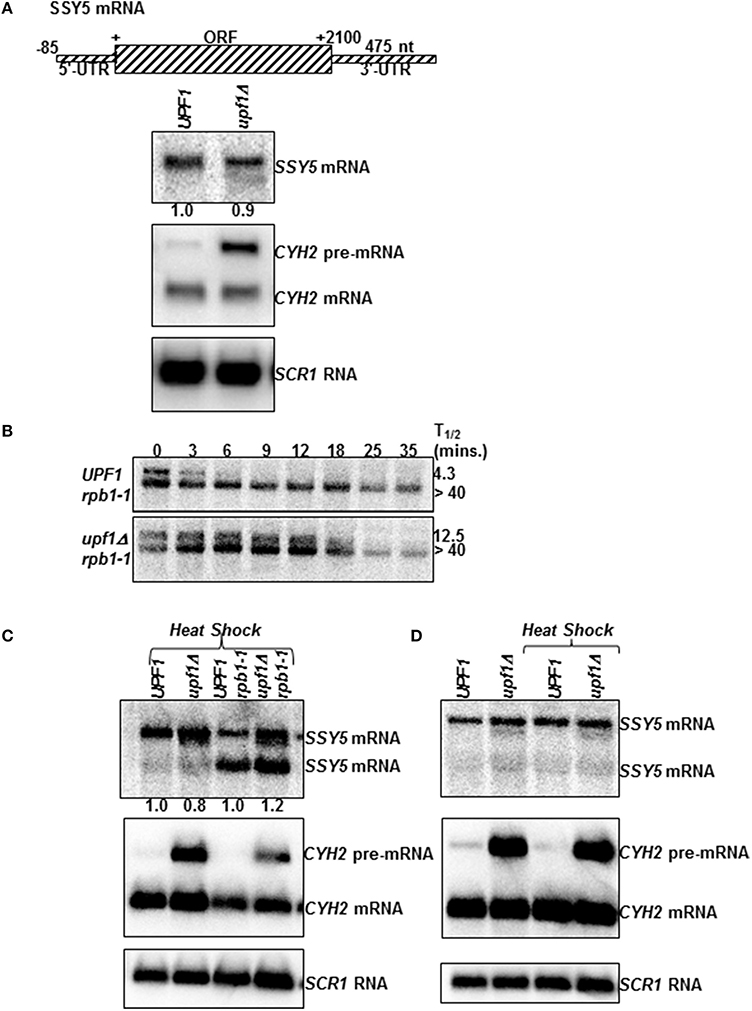
Figure 1. The SSY5 mRNA with an atypically long 3′-UTR can evade degradation by the NMD pathway. mRNA schematic (A) and northern blot analysis of the SSY5 mRNA steady-state levels (A), and half-lives (B) in wild-type (UPF1) and yeast strains with a non-functional NMD pathway (upf1Δ). (A) The SSY5 mRNA steady-state levels were measured from total RNA isolated from W303 (UPF1, Wente et al., 1992) and AAY320 (upf1Δ, Kebaara et al., 2003a). CYH2 and SCR1 were used as controls. CYH2 pre-mRNA is degraded by NMD and is used to confirm the NMD phenotype of the yeast strains. SCR1 is a loading control. It is transcribed by RNA polymerase III and is not degraded by the NMD pathway (Kebaara and Atkin, 2009). (B) The mRNA half-lives were measured using total RNA isolated from AAY334 (UPF1 rpb1-1) and AAY335 (upf1Δ rpb1-1) (Kebaara et al., 2003a). mRNA half-life measurements are shown on the right of the representative northern blots. Time after inhibition of transcription is shown above the northern blots. The mRNA half-lives are an average of three independent experiments. (C) The SSY5 mRNA steady-state levels following heat shock were measured from total RNA isolated from W303 (UPF1), AAY320 (upf1Δ), AAY334 (UPF1 rpb1-1) and AAY335 (upf1Δ rpb1-1) after exposure to 39°C for 1 min. The steady-state levels of the longer SSY5 mRNA (top band) are shown below the steady-state northern blot. (D) The SSY5 mRNA steady-state levels from W303 (UPF1) and AAY320 (upf1Δ) without exposing the yeast strains to heat shock (left two lanes) or after exposure to heat shock (the right two lanes).
SSY5 mRNA decay rate measurements were done by inhibiting transcription in yeast strains with a temperature sensitive allele of RNA polymerase II (Nonet et al., 1987). The additional SSY5 transcript could have been produced as a result of the heat shock used to inhibit transcription or due to the yeast strain having a mutation in RNA polymerase II (Nonet et al., 1987). To distinguish between these possibilities, isogenic wild-type and nmd mutants with or without the temperature sensitive allele of RNA polymerase II were exposed to a 1 min heat shock at 39°C. 1 min was used because the second SSY5 band was detected at the zero time point of the half-life measurements (Figure 1B). There was a faint faster migrating band that accumulated to approximately the same levels in wild-type and nmd mutant strains (Figure 1C left two lanes). This mRNA was not observed in Figure 1A. In addition the slower migrating SSY5 mRNA observed in Figure 1A was present. This SSY5 mRNA was insensitive to the NMD pathway in three independent experiments in this strain background (Figure 1C). As a control, SSY5 mRNA accumulation levels were compared in wild-type and nmd mutant strain background with or without heat shock (Figure 1D). The ~2.8 kb SSY5 mRNA was the major transcript observed in these conditions and it was insensitive to NMD. These results show that the shorter, novel SSY5 mRNA is present primarily in the rpb1-1 genetic background and is immune to degradation by the NMD.
As discussed above, exposure of the wild-type (UPF1 rpb1-1) and nmd mutant (upf1Δ rpb1-1) with the temperature sensitive allele of RNA polymerase II to heat shock resulted in the production of two transcripts (Figures 1B,C). As observed with the half-life analysis, the longer ~2.8 kb SSY5 transcript consistently accumulated to slightly higher levels in the nmd mutant relative to wild-type strains in the rpb1-1 background (1.2 ± 0.14 fold, Figure 1C), and the shorter transcript accumulated 0.8 ± 0.4 in the same strain background (Figure 1C). In addition, analysis of the relative accumulation levels of the ~2.8 kb SSY5 transcript at time zero on the half-life northerns support these results. The ~2.8 kb SSY5 transcript accumulated 1.4 ± 0.35 fold in nmd mutants (upf1Δ rpb1-1) relative to wild-type (UPF1 rpb1-1) yeast strains, while the shorter transcript accumulated 0.87 ± 0.34 fold in nmd mutants (Figure 1B, 0 time point). These results show that the production of an NMD sensitive SSY5 transcript is associated with the temperature sensitive RNA polymerase II in the rpb1-1 mutant strains. This is not surprising since RNA polymerase II plays an important role in RNA processing, including 3′-end cleavage and polyadenylation.
These SSY5 transcripts are differentially sensitive to NMD. The shorter transcript, which has elevated levels in the rpb1-1 yeast strain after heat shock, is degraded at the same rate in wild-type and nmd mutant strains (Figures 1B,C). In contrast, the longer SSY5 transcript which is the major transcript in wild-type strains is degraded more rapidly in wild-type than nmd mutant strains that have the temperature sensitive allele of RNA polymerase II (Figures 1B,C). Thus, this transcript becomes sensitive to NMD in this strain background and under these conditions. The reason why this SSY5 transcript becomes sensitive to NMD in these conditions is not clear but it could be due to the increased expression of the shorter transcript or due to some indirect effect that alleviates the accumulation of this transcript in nmd mutants. Thus, these results demonstrate that, the SSY5 mRNA 3′-UTR is insufficient to target all SSY5 mRNAs for NMD in all conditions tested, while the longer transcript is degraded by the NMD pathway in the rpb1-1 strain background. The shorter transcript could escape NMD in this strain background because its 3′-UTR may not be sufficiently long to act as an NMD targeting signal even though it is atypically long, or because it is protected from degradation by the NMD pathway.
The SSY5 mRNA 3′-UTR is Sufficient to Target an NMD-Insensitive mRNA for Decay by the Pathway in All Conditions and Yeast Strains Tested
To determine the extent to which the atypically long SSY5 mRNA 3′-UTR is sufficient to target an NMD insensitive mRNA to the pathway, the 3′-UTR of the CYC1 mRNA was replaced with the SSY5 3′-UTR (Figure 2A). The CYC1 mRNA, which encodes iso 1 cytochrome C, is normally not degraded by the NMD pathway (Biswadip et al., 2000). The CYC1 mRNA was selected because it has previously been used to show that abnormally 3′-extended mRNAs resulting from a mutation in the poly(A) site are degraded by the NMD pathway (Muhlrad and Parker, 1999). 3′-RACE and northern blot analysis were utilized to measure 3′-UTR length and determine the steady-state levels of the CYC1-SSY5 3′-UTR mRNA. The 3′-UTR of the CYC1 mRNA was specifically replaced with the SSY5 3′-UTR (Figure 2C and Supplementary Figure S1, compare lanes 3 and 8).
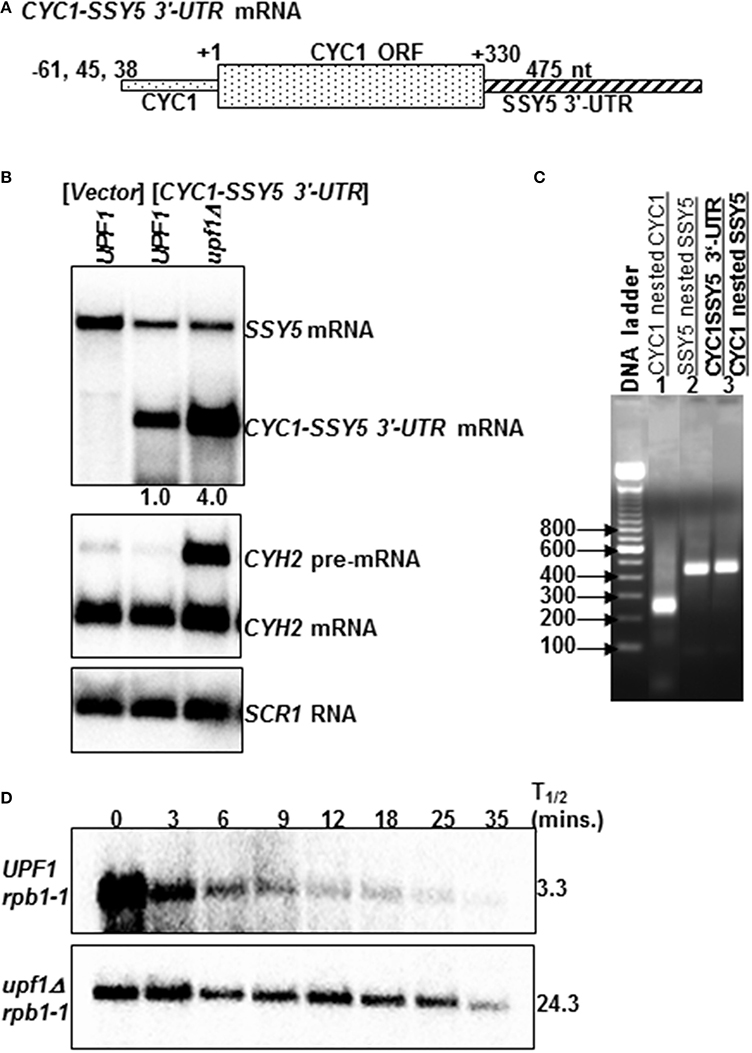
Figure 2. The atypically long SSY5 3′-UTR is sufficient to target an NMD-insensitive mRNA to the NMD pathway. The schematic diagram represents the CYC1-SSY5 3'-UTR mRNA (A). This mRNA was used to assess the requirement for the SSY5 mRNA atypically long 3′-UTR for decay by the NMD pathway (A). CYC1-SSY5 3′-UTR mRNA contains the 5′-UTR and ORF of CYC1, and the 3′-UTR of SSY5 (A). Representative steady-state northerns of total RNA isolated from wild-type (UPF1) lacking the CYC1-SSY5 3'-UTR mRNA and wild-type (UPF1) and nmd mutants (upf1Δ) containing the CYC1-SSY5 3′-UTR construct (B). The northern blot was probed with DNA from the SSY5 3′-UTR. The relative CYC1-SSY5 3′-UTR steady-state levels are shown below the northern blot (B). An agarose gel of 3′ RACE nested PCR products from wild-type yeast strains lacking the CYC1-SSY5 3′-UTR mRNA (lanes 1–2) and strains expressing the CYC1-SSY5 3′-UTR mRNA (lane 3). Primers used for the 3′ RACE PCR reactions are listed above the lane numbers (C). Half-life measurements of the CYC1-SSY5 3′-UTR mRNA are shown in (D) and were done as described in Figure 1. mRNA half-life measurements are shown to the right of the northern blots. The steady-state levels and half-life measurements are an average of three independent experiments.
The steady-state levels of the CYC1-SSY5 3′-UTR mRNA were 4.06 ± 1.42 (n = 3) fold higher in nmd mutant yeast strains than in wild-type strains (Figure 2B). Additionally, the half-life of the CYC1-SSY5 3′-UTR mRNA is longer in the nmd mutant strain than the wild-type strain 24.33 ± 4.04 (n = 3) min vs. 3.3 ± 1.53 (n = 3) min; (Figure 2D). We interpret these results to mean that the SSY5 mRNA 3′-UTR is sufficient to target the NMD-insensitive CYC1 mRNA to the NMD pathway and this indicates that the short SSY5 mRNA is protected from degradation by the NMD pathway.
The SSY5 mRNAs with Abnormally Extended cyc1 3′-UTRs are Degraded by the NMD Pathway
We next tested whether the SSY5 5′-UTR and ORF are sufficient to protect the mRNA from degradation by the NMD pathway by replacing the SSY5 3′-UTR with the cyc1-512 3′-UTR. cyc1-512 has a 38 nt deletion that results in defective 3′ end processing leading to the production of multiple transcripts with aberrant extended 3′-UTRs (Zaret and Sherman, 1982). These aberrantly extended cyc1 transcripts are degraded by the NMD pathway (Muhlrad and Parker, 1999). 3′-RACE was used to show that the 3′-UTR of the SSY5 mRNA was specifically replaced with the cyc1-512 3′-UTR (Figure 3C and Supplementary Figure S2, compare lanes 5 and 11).
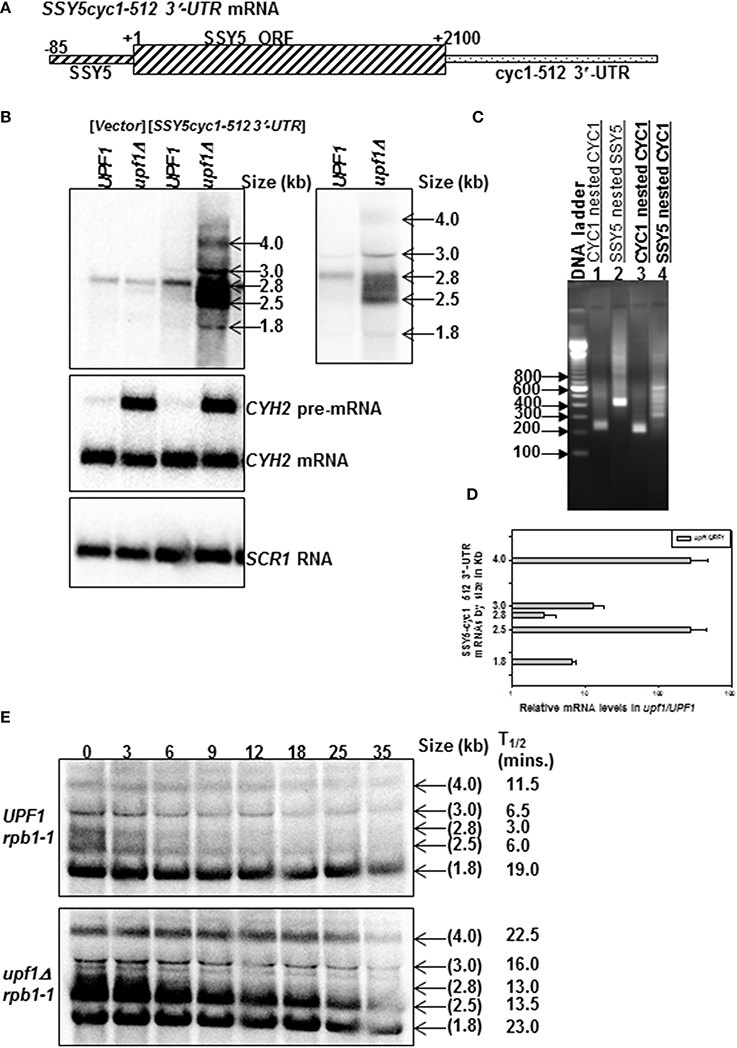
Figure 3. SSY5 mRNA requires the 3′-UTR to escape from NMD-mediated degradation. Replacement of the SSY5 3′-UTR with the cyc1-512 3′-UTR results in the production of multiple transcripts that a degraded by the NMD pathway. Schematic representation of the SSY5cyc1-512 3′-UTR fusion mRNAs (A). Representative steady-state northerns of total RNA isolated from wild-type (UPF1) and nmd mutants (upf1Δ) are shown below the mRNA schematic (B). A shorter exposure of the steady-state northern blot is shown to the right of the steady-state accumulation northern for clearer visualization of the bands. The two right lanes of the northern blot contain RNA from yeast strains expressing the SSY5cyc1-512 3′-UTR construct (B). The sizes of the predominant bands are shown to the right of the northern blot. The relative SSY5cyc1-512 3′-UTR steady-state levels are represented graphically in log scale (D). An agarose gel of 3′ RACE nested PCR products from an nmd mutant yeast strain lacking the SSY5cyc1-512 3′-UTR mRNAs (lanes 1–2) and expressing the SSY5cyc1-512 3′-UTR mRNAs (lane 3–4). Primers used for the 3′ RACE PCR reactions are listed above the lane numbers (C). Half-life measurements of the SSY5cyc1-512 3′-UTR mRNAs are shown in (E) and are an average of at least two independent experiments. The northern blots were probed with DNA from the SSY5 5'-UTR-ORF, CYH2 and SCR1.
SSY5 is an essential gene. Consequently the SSY5cyc1-512 3′-UTR transcripts were expressed from a high copy plasmid in wild-type and nmd mutant strains with SSY5. Multiple SSY5cyc1-512 3′-UTR transcripts were observed on northerns (Figures 3B,E). The SSY5 transcript is ~2.8 kb (Figure 3B, vector control; Kebaara and Atkin, 2009). The endogenous SSY5 mRNA accumulated 2.8-fold and ~133-fold lower than the most abundant SSY5cyc1-512 3′-UTR transcript in nmd mutant strains (Figures 3B,D). The SSY5cyc1-512 3′-UTR transcripts are at different positions on the northern blots because they have 3′-UTRs that differ in length correlating with the heterogeneous bands observed by 3′-RACE (Figures 3B,C, lane 4). The major transcripts ranged in size from ~1.8 to 4 kb (Figure 3B). The 1.8 kb transcript could be a non-stop transcript or a decay intermediate because the SSY5 ORF is 2100 nt. Notably, while the levels of these transcripts varied within a strain, they consistently accumulated to higher levels in nmd mutant strains relative to wild-type strains. The relative fold changes of the SSY5cyc1-512 3′-UTR transcripts are represented graphically in Figure 3D.
All but the ~1.8 kb major SSY5cyc1-512 3′-UTR fusion mRNAs had shorter half lives in the wild-type cells than the nmd mutant (Figure 3E). The shortest transcript detected of ~1.8 kb decayed at comparable rates in wild-type and nmd mutants. This transcript is unlikely to contain the entire SSY5 ORF of 2100 nt. The band that corresponds in size to the endogenous SSY5 transcript also showed sensitivity to the NMD pathway (Figure 3E). The half-life of the SSY5 mRNA in the nmd mutant strain was 3.0 ± 1.41 (n = 2) min vs. 13.0 ± 2.83 (n = 2) min in wild-type stains (Figure 3E). These half-life measurements are comparable to those observed for the longer ~2.8 kb SSY5 transcript (Figure 1B). Thus, replacement of the SSY5 3′-UTR with a 3′-UTR known to target mRNAs to the NMD pathway results in the production of multiple mRNAs that were sensitive to NMD. These results demonstrate that the SSY5 5′-UTR and ORF are insufficient to stabilize the mRNA in the presence of the cyc1-512 3′-UTR and suggest that additional sequences within the SSY5 3′-UTR are required to escape degradation by the NMD pathway.
The SSY5 mRNA Requires the 5′-UTR, ORF and the 3′-UTR to Escape the NMD Pathway
The observation that replacement of the SSY5 3′-UTR with a 3′-UTR known to target mRNA to the NMD pathway results in the production of multiple transcripts the majority of which are sensitive to NMD, led us to generate an additional fusion mRNA as a control. This control mRNA was utilized to determine whether removal of the SSY5 3′-UTR from the mRNA would result in stabilization of the mRNA. The SSY5-CYC1 3′-UTR mRNA was synthesized using cloning free PCR to precisely replace the SSY5 3′-UTR with the CYC1 3′-UTR (Figure 4A) and verified by 3′-RACE (Figure 4C, compare lanes 1 and 3). The SSY5-CYC1 3′-UTR transcripts were expressed from a high copy plasmid in wild-type and nmd mutant strains with SSY5. Three SSY5-CYC1 3′-UTR-specific bands, a major one of ~2.6 kb and two minor ones of ~1.8 kb and ~3.3 kb were detected on northern blots (Figure 4B). These bands were not expressed in the vector control and are much more abundant than the endogenous SSY5 transcript (Figure 4B compare lanes 1 and 2 to 3 and 4).
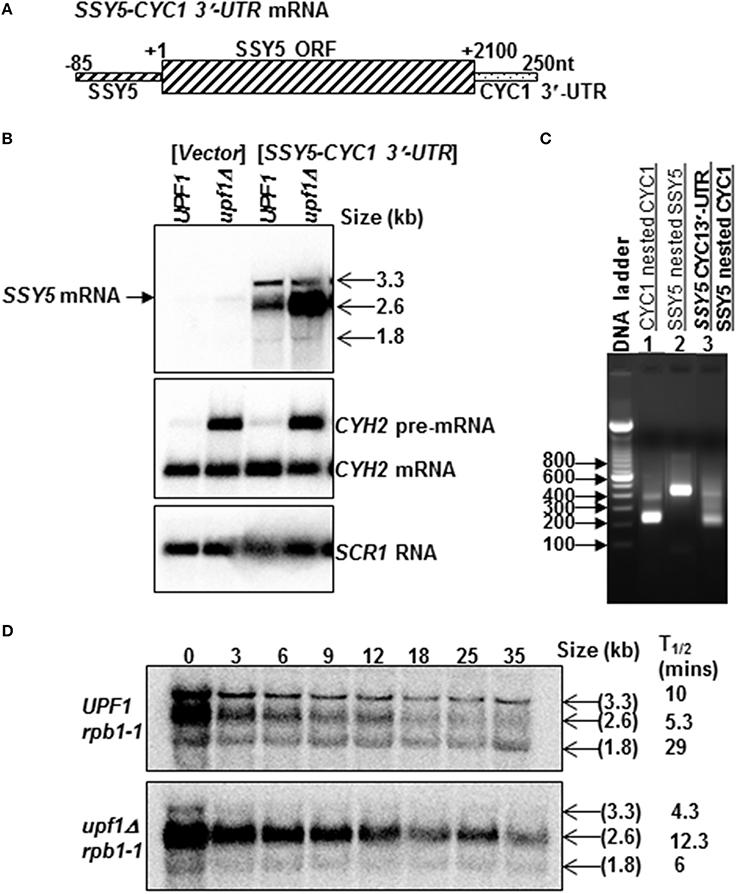
Figure 4. Some mRNAs expressed from yeast strains transformed with the SSY5-CYC1 3′-UTR mRNA are sensitive to the NMD pathway. Replacement of the SSY5 3′-UTR with the CYC1 3′-UTR resulted in the production of three transcripts that are differentially regulated by the NMD pathway. Schematic diagram of the SSY5-CYC1 3′-UTR fusion mRNA (A). Representative steady-state northern blot of total RNA isolated from wild-type (UPF1) and nmd mutants (upf1Δ) are shown below the mRNA schematic. The first 2 lanes of the northern blot are from yeast strains lacking the SSY5-CYC1 3′-UTR mRNA (B). The sizes of the SSY5-CYC1 3′-UTR transcripts are shown to the right of the northern blot (B). An agarose gel of 3′ RACE nested PCR products from a wild-type (UPF1) yeast strain lacking the SSY5-CYC1 3′-UTR mRNA (lanes 1–2) and expressing the SSY5-CYC1 3′-UTR mRNA (lane 3). Primers used for the 3′ RACE PCR reactions are listed above the lane numbers (C). Half-life measurements of the SSY5-CYC1 3′-UTR mRNAs are shown in (D) and are an average of three independent experiments. The northern blots were probed with DNA from the SSY5 5'-UTR-ORF, CYH2, and SCR1.
We expected the SSY5-CYC1 3′-UTR mRNA to be equally stable in wild-type and nmd mutant cells, but this was not the case. The predominant ~2.6 kb fusion mRNA accumulated 7.59 ± 3.7 (n = 3) fold more in the nmd mutants than wild-type (UPF1) strains. The levels of the less abundant 3.3 kb and 1.8 kb transcripts were not significantly different in nmd mutant or wild-type strains [0.57 ± 0.09 (n = 3) and 1.5 ± 0.1 (n = 2), respectively]. The ~1.8 kb transcript was shorter than the expected size of the SSY5-CYC1 3′-UTR fusion mRNA, although it was unique to yeast strains expressing the SSY5-CYC1 3′-UTR fusion construct (Figure 4B).
The half-lives of all three SSY5-CYC1 3′-UTR mRNAs were determined (Figure 4D). The predominant ~2.6 kb transcript had half-lives of 5.3 ± 1.53 (n = 3) min in wild-type strains and 12.3 ± 0.57 (n = 3) min in nmd mutants and was thus degraded faster in wild-type cells than nmd mutants. The ~3.3 kb and shortest ~1.8 kb transcripts were not stabilized in nmd mutants. The half-lives of the 3.3 kb transcript were 10 ± 2.65 (n = 3) in wild-type strains and 4.3 ± 1.53 (n = 3) min in nmd mutants and the ~1.8 kb transcript had half-lives of 29 ± 5.29 (n = 3) min in wild-type strains and 6 ± 2 (n = 3) min in nmd mutants. Thus, both of these mRNAs had shorter half-lives in the nmd mutant than wild-type cells. This is a novel phenotype. These results show that the yeast strains expressing the SSY5-CYC1 3′-UTR fusion construct produce multiple transcripts. These transcripts are differentially affected by the NMD pathway; the 2.6 kb transcript was degraded by the NMD pathway, while the ~3.3 and 1.8 kb targets were stabilized by inactivation of the NMD pathway. As far as we know this is the first example of transcripts that are destabilized by inactivation of the NMD pathway.
Degradation of the 2.6 kb SSY5-CYC1 3′-UTR transcript by the NMD pathway suggests that the SSY5 mRNA may contain alternative NMD targeting features besides the atypically long 3′-UTR, since in the absence of the long 3′-UTR from the SSY5 transcripts generates some transcripts that are still sensitive to the NMD pathway. Examination of the SSY5 mRNA showed that the start codon is in a suboptimal context and is followed by an out-of-frame AUG at +71, which is in an optimal context (Welch and Jacobson, 1999). This feature could potentially target the SSY5 mRNA to NMD through leaky scanning. Additionally, the SSY5 mRNA contains ribosomal frameshifting signals and has a translated uORF (Ingolia et al., 2009; Belew et al., 2010). These three features have been shown to target natural mRNAs to the NMD pathway in S. cerevisiae (Welch and Jacobson, 1999; Guan et al., 2006; Belew et al., 2010). Since these features are still present in the SSY5-CYC1 3′-UTR mRNA, one or more of these features could be targeting the SSY5-CYC1 3′-UTR mRNA for decay by the NMD pathway.
Discussion
Here, we show that there are two SSY5 transcripts. One that is expressed constitutively and a second that was only observed under specific conditions and yeast genetic backround. The constitutively expressed SSY5 mRNAs has an atypically long 3′-UTR of ~475 nts and can escape degradation by NMD in some conditions (Figure 1A). The SSY5 mRNA produced in rpb1-1 genetic background is shorter than the constitutively expressed SSY5 mRNA and is immune to the NMD pathway. SSY5 mRNAs contains multiple NMD targeting signals including a translated uORF, leaky scanning, programmed ribosome frameshift sites and a long 3′-UTR (Welch and Jacobson, 1999; Guan et al., 2006; Kebaara and Atkin, 2009; Belew et al., 2010). We showed that the ~475 nt 3′-UTR of the constitutive SSY5 mRNA is sufficient to target an mRNA that is normally NMD-insensitive for decay by the pathway (Figure 2). And finally, we show that protection of the SSY5 mRNA from decay by the NMD pathway requires sequences in the 5′-UTR-ORF and the 3′-UTR because SSY5 cyc1 512 3′-UTR and SSY5-CYC1 3′-UTR fusion RNAs were both degraded by the NMD pathway (Figures 3, 4). Collectively, we show that the SSY5 mRNA contains multiple NMD targeting features; however this mRNA is immune to NMD-mediated degradation in some conditions and this immunity is due to the presence of elements in the 5′-UTR and/or ORF and the 3′-UTR.
Ssy5p is a component of the SPS (Ssy1-Ptr3-Ssy5) extracellular amino acid sensing system. It is a protease that cleaves the cytosolic precursor forms of the transcription factors Stp1p and Stp2p to activate transcription of amino acid permease genes. The SSY5 mRNA is protected from NMD in yeast cells that are grown at normal growth temperature (30°C) and in rich medium (Kebaara and Atkin, 2009; Figure 1). We detected an additional transcript on Northern blots probed with a SSY5 probe in rpb1-1 yeast strains subjected to heat stress (Figures 1B,C). Surprisingly in these conditions the constitutively expressed SSY5 transcript was degraded by the NMD pathway. In addition, manipulation of the SSY5 mRNA coding and 3′ regulatory sequences leads to the production of multiple transcripts (Figures 3, 4). Some of these transcripts were regulated in an NMD-dependent manner (Figures 1, 4). Sensitivity of the SSY5 mRNA to the NMD pathway is influenced by both the mRNA secondary structure and trans-acting factors that bind to the mRNA. Future studies on the SSY5 messenger ribonucleoprotein (mRNP) composition will elucidate the role trans acting factor(s) function in the evasion of this mRNA from NMD. We hypothesize that the SSY5 mRNP composition in different conditions is important in determining the fate of the SSY5 mRNA, specifically whether the SSY5 mRNAs will be sensitive to the NMD pathway.
A Previous study identified direct NMD targets in S. cerevisiae based on their association with Upf1p, a core component of the NMD pathway. This study found that the SSY5 mRNAs associates with Upf1p (Johansson et al., 2007). Whether Upf1p binds to the SSY5 mRNA before or after the decision to degrade the mRNAs by NMD is unclear. In mammalian cells it has been proposed that Upf1p binds to mRNAs before the decision on whether an mRNA will be degraded by the pathway or escape degradation by the pathway (Zund et al., 2013). If this scenario applies to the SSY5 mRNA, then the decision on whether the SSY5 mRNAs are degraded by NMD could occur after Upf1p binding.
Replacement of the SSY5 3′-UTR with the cyc1-512 3′-UTR resulted in the production of multiple transcripts the majority of which were regulated in an NMD dependent manner (Figure 3). If the SSY5 mRNA contains a universal NMD stabilizer element in the 5′-UTR and/or ORF, we would expect that the transcripts generated from the SSY5cyc1-512 3′-UTR fusion mRNA would be insensitive to the NMD pathway, if proper 3′-end processing is not required to stabilize the transcripts. Regulation of these transcripts by the NMD pathway suggests that the SSY5 mRNA stabilizer element requires sequences in both the 5′-UTR-ORF region and the 3′-UTR since neither regions was sufficient to stabilize the fusion RNAs, or that the functionality of the stabilizer element is context specific. Specifically, the stabilizer element stabilizes the SSY5 mRNA in the presence of the SSY5 3′-UTR but not in the presence of the cyc1-512 3′-UTR.
Replacement of the SSY5 3′-UTR with the CYC1 3′-UTR resulted in the production of three transcripts. Two of the SSY5-CYC1 3′-UTR transcripts were unexpectedly degraded more rapidly in nmd mutants relative to wild-type yeast strains. NMD targets are normally degraded faster in wild-type yeast strains and stabilized in nmd mutants. The only transcript that was sensitive to the NMD pathway was the ~2.6 kb transcript, which was expressed at higher levels than the other two mRNAs and migrated slightly faster than the endogenous SSY5 mRNA. These data show that fusion mRNAs produced from the same fusion construct can be differentially regulated by the NMD pathway, where some transcripts are sensitive to the pathway and others are immune to the pathway (Figure 4B). The explanation for the differential sensitivity of the SSY5-CYC1 3′-UTR transcripts to the NMD pathway is unclear but there are some possible explanations. First, the loss of part of the stabilizer element present in the SSY5 3′-UTR and also the presence of three additional NMD targeting features within the NMD sensitive transcript may result in differential regulation. Alternatively, the sensitivity of ~2.6 kb SSY5-CYC1 3′-UTR to the NMD pathway could be due to the titration of trans-acting factors required for decay of these mRNAs in wild-type cells. Interestingly, a recent study by Geisberg et al., showed that mRNAs isoforms whose 3′-ends differ by as few as 3 nucleotides have relatively different half-lives (Geisberg et al., 2014).
The role the SSY5 ORF plays in the immunity of the mRNA to the NMD pathway was not specifically examined here. However, the SSY5 ORF may contribute to the escape of the mRNA from the pathway due to a couple of observations. First, in our previous studies examining endogenous mRNAs with atypically long 3′-UTRs that are regulated by the NMD pathway, the SSY5 mRNA was the only mRNA out of 11 with an atypically long 3′-UTR that was immune to the NMD pathway (Kebaara and Atkin, 2009). Notably, the SSY5 mRNA also had the longest ORF, suggesting that the ORF length may play a role in the escape of the mRNA from the NMD. This is consistent with a recent study in S. cerevisiae that reported that mRNAs subject to NMD due to the presence of a long 3′-UTR had short ORFs and that increasing the ORF length of these mRNAs had a stabilizing effect (Decourty et al., 2014). The SSY5 ORF length however does not fully account for all our observations because the SSY5cyc512 3′-UTR and SSY5-CYC1 3′-UTRs fusions contained the full length SSY5 ORF, and yet the predominant transcripts encoded by these fusions were degraded by the NMD pathway. This indicates that there are additional requirement for protection.
In S. cerevisiae two additional mRNAs with NMD targeting features have been reported to escape degradation by the pathway, GCN4 and YAP1 mRNAs (Ruiz-Echevarria and Peltz, 2000). These mRNA have uORFs. mRNAs containing uORFs in the 5′-UTR or overlapping the ORF are normally degraded by the NMD pathway (Ruiz-Echevarria and Peltz, 2000; Gaba et al., 2005; Guan et al., 2006; Yun et al., 2012). The immunity of GCN4 and YAP1 mRNA to NMD is reported to be dependent on the presence of Pub1p (Ruiz-Echevarria and Peltz, 2000). We hypothesize that specific trans-acting factors may regulate the degradation of SSY5 mRNA by NMD. In mammalian cells, a number of mRNAs with atypically long 3′-UTRs have been reported to escape NMD-mediated degradation (Singh et al., 2008). Similarly, these mRNAs may have also evolved mechanisms to avoid degradation by NMD. Collectively, the results presented here demonstrate that naturally occurring transcripts with multiple NMD targeting features can escape NMD-mediated degradation in some conditions. These mRNAs escape degradation because they have evolved mechanisms to prevent degradation by NMD. These NMD evasion mechanisms maybe particular for specific transcripts and may also be context specific.
Author Contributions
Jesseeca Obenoskey and Dakota Lane contributed to the acquisition and analysis of data. Audrey Atkin contributed to the conception of the research and revising the manuscript. Bessie Kebaara contributed to conception, acquisition and analysis of data, drafting and revising the manuscript and final approval of the version to be published.
Funding
This work was supported by grants from the National Science Foundation (MCB-0444333 to Audrey L. Atkin, RIG-0642154 to Bessie W. Kebaara) and start-up funds from Baylor University. Any opinions, findings, conclusions, or recommendations expressed in this report are ours, and do not necessarily reflect the views of the National Science Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the molecular Biology Core (MBC) facility at Baylor University for equipment and supplies. We thank Megan C. Peccarelli for critically evaluating the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmolb.2014.00025/abstract
References
Amrani, N., Dong, S., He, F., Ganesan, R., Ghosh, S., Kervestin, S., et al. (2006). Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 34(Pt 1), 39–42. doi: 10.1042/BST20060039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. (eds.) et al. (1998). Current Protocols in Molecular Biology. New York, NY: John Wiley and Sons, Inc.
Belew, A. T., Advani, V. M., and Dinman, J. D. (2010). Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 39, 2799–2808. doi: 10.1093/nar/gkq1220
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Biswadip, D., Guo, Z., Russo, P., Chartrand, P., and Sherman, F. (2000). The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Bio. 20, 2827–2838. doi: 10.1128/MCB.20.8.2827-2838.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Culbertson, M. R., and Leeds, P. F. (2003). Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13, 207–214. doi: 10.1016/S0959-437X(03)00014-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Decourty, L., Doyen, A., Malabat, C., Frachon, E., Rispal, D., Seraphin, B., et al. (2014). Long open reading frame transcripts escape nonsense-mediated mRNA decay in yeast. Cell Rep. 6, 1–6. doi: 10.1016/j.celrep.2014.01.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Erdeniz, N., Mortensen, U. H., and Rothstein, R. (1997). Cloning-free PCR based allele replacement methods. Genome Res. 7, 1174–1183.
Gaba, A., Jacobson, A., and Sachs, M. S. (2005). Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20, 449–460. doi: 10.1016/j.molcel.2005.09.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geisberg, J. V., Moqtaderi, Z., Fan, X., Ozsolak, F., and Struhl, K. (2014). Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156, 812–824. doi: 10.1016/j.cell.2013.12.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gietz, R. D., and Woods, R. A. (1998). Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method. Methods Microbiol. 26, 53–66. doi: 10.1016/S0580-9517(08)70325-8
Graber, J. H., McAllister, G. D., and Smith, T. F. (2002). Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30, 1851–1858. doi: 10.1093/nar/30.8.1851
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guan, Q., Zheng, W., Tang, S., Liu, X., Zinkel, R. A., Tsui, K., et al. (2006). Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 24:e2003. doi: 10.1371/journal.pgen.0020203
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, F., Li, X., Spatrick, P., Castillo, R., Dong, S., and Jacobson, A. (2003). Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12, 1439–1452. doi: 10.1016/S1097-2765(03)00446-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, F., Peltz, S. W., Donahue, J. L., Rosbash, M., and Jacobson, A. (1993). Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. U.S.A. 90, 7034–7038. doi: 10.1073/pnas.90.15.7034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ingolia, T. N., Ghaemmaghami, S., Newman, J. R. S., and Weissman, J. S. (2009). Genome-Wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. doi: 10.1126/science.1168978
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Johansson, M. J. O., He, F., Spatrick, P., Li, C., and Jacobson, A. (2007). Association of yeast Upf1p with direct substrates of the NMD pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 20872–20877. doi: 10.1073/pnas.0709257105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kawashima, T., Douglas, S., Gabunilas, J., Pellegrini, M., and Chanfreau, G. F. (2014). Widespread use of non-productive alternative splice sites in Saccharomyces cerevisiae. PLoS Genet. 10:e1004249. doi: 10.1371/journal.pgen.1004249
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kebaara, B., and Atkin, A. L. (2009). Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 2771–2778. doi: 10.1093/nar/gkp146
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kebaara, B., Nazarenus, T., Taylor, R., and Atkin, A. L. (2003a). Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains. Curr. Genet. 43, 171–177. doi: 10.1007/s00294-003-0386-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kebaara, B., Nazarenus, T., Taylor, R., Forch, A., and Atkin, A. L. (2003b). The Upf1-dependent decay of wild-type PPR1 mRNA depends on its 5′-UTR and the first 92 ORF nucleotides. Nucleic Acids Res. 31, 3157–3165. doi: 10.1093/nar/gkg430
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kebaara, B. W., Baker, K. E., Patfield, K. D., and Atkin, A. L. (2012). Analysis of nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Curr. Protoc. Cell Biol. Chapter 27:Unit 27.23. doi: 10.1002/0471143030.cb2703s54
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lelivelt, M. J., and Culbertson, M. R. (1999). Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 19, 6710–6719.
Mendell, J. T., Sharifi, N. A., Meyers, J. L., Martinez-Murillo, F., and Dietz, H. C. (2004). Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36, 1073–1078. doi: 10.1038/ng1429
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muhlrad, D., and Parker, R. (1999). Aberrant mRNAs with extended 3′-UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5, 1299–1307. doi: 10.1017/S1355838299990829
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nonet, M., Scafe, C., Sexton, J., and Young, R. (1987). Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7, 1602–1611.
Rehwinkel, J., Letunic Raes, J., Bork, P., and Izaurralde, E. (2005). Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11, 1530–1544. doi: 10.1261/rna.2160905
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ruiz-Echevarria, M. J., and Peltz, S. W. (2000). The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101, 741–751. doi: 10.1016/S0092-8674(00)80886-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, G., Rebbapragada, I., and Lykke-Andersen, J. (2008). A Competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 6:e111. doi: 10.1371/journal.pbio.0060111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sirkosky, R. S., and Heiter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27.
Welch, E. M., and Jacobson, A. (1999). An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18, 6134–6145. doi: 10.1093/emboj/18.21.6134
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wente, S. R., Rout, M. P., and Blobel, G. (1992). A new family of yeast nuclear pore complex proteins. J. Cell Biol. 119, 705–723. doi: 10.1083/jcb.119.4.705
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Whittmann, J., Hol, E. M., and Jack, H. M. (2006). hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 26, 1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yepiskoposyan, H., Aeschimann, F., Nilsson, D., Okoniewski, M., and Mühlemann, O. (2011). Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17, 2108–2118. doi: 10.1261/rna.030247.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yun, Y., Adesanya, T. M. A., and Mitra, R. D. (2012). A systematic study of gene expression variation at single-nucleotide resolution reveals widespread regulatory roles for uAUGs. Genome Res. 22, 1089–1097. doi: 10.1101/gr.117366.110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zaret, K. S., and Sherman, F. (1982). DNA sequence required for efficient transcription termination in yeast. Cell 28, 563–573. doi: 10.1016/0092-8674(82)90211-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zund, D., Gruber, A. R., Zavolan, M., and Muhlemann, O. (2013). Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in the 3′-UTRs. Nat. Struct. Mol. Biol. 20, 936–943. doi: 10.1038/nsmb.2635
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: 3′-untranslated regions, nonsense-mediated mRNA decay, SSY5, mRNA decay, mRNA stability
Citation: Obenoskey J, Lane DR, Atkin AL and Kebaara BW (2014) Immunity of the Saccharomyces cerevisiae SSY5 mRNA to nonsense-mediated mRNA decay. Front. Mol. Biosci. 1:25. doi: 10.3389/fmolb.2014.00025
Received: 29 August 2014; Accepted: 13 November 2014;
Published online: 08 December 2014.
Edited by:
P. Hemachandra Reddy, Texas Tech University, USAReviewed by:
Arun Bhardwaj, University of South Alabama Mitchell Cancer Institute, USAKoteswara Rao Valasani, The University of Kansas, USA
Copyright © 2014 Obenoskey, Lane, Atkin and Kebaara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bessie W. Kebaara, Department of Biology, Baylor University, One Bear Place # 97388, Waco, TX 76798-7388, USA e-mail:YmVzc2llX2tlYmFhcmFAYmF5bG9yLmVkdQ==
 Jesseeca Obenoskey
Jesseeca Obenoskey Dakota R. Lane1
Dakota R. Lane1 Bessie W. Kebaara
Bessie W. Kebaara