- Department of Microbiology, University of Georgia, Athens, GA, USA
Transcription sigma factors direct the selective binding of RNA polymerase holoenzyme (Eσ) to specific promoters. Two families of sigma factors determine promoter specificity, the σ70 (RpoD) family and the σ54 (RpoN) family. In transcription controlled by σ54, the Eσ54-promoter closed complex requires ATP hydrolysis by an associated bacterial enhancer-binding protein (bEBP) for the transition to open complex and transcription initiation. Given the wide host range of Salmonella enterica serovar Typhimurium, it is an excellent model system for investigating the roles of RpoN and its bEBPs in modulating the lifestyle of bacteria. The genome of S. Typhimurium encodes 13 known or predicted bEBPs, each responding to a unique intracellular or extracellular signal. While the regulons of most alternative sigma factors respond to a specific environmental or developmental signal, the RpoN regulon is very diverse, controlling genes for response to nitrogen limitation, nitric oxide stress, availability of alternative carbon sources, phage shock/envelope stress, toxic levels of zinc, nucleic acid damage, and other stressors. This review explores how bEBPs respond to environmental changes encountered by S. Typhimurium during transmission/infection and influence adaptation through control of transcription of different components of the S. Typhimurium RpoN regulon.
Introduction
Salmonella enterica subsp. enterica serovar Typhimurium is the most common serotype of Salmonella enterica subspecies, which causes tens of millions of cases of salmonellosis and more than 100,000 deaths worldwide each year (Majowicz et al., 2010). S. Typhimurium has been extensively studied to reveal the virulence factors and strategies that lead to morbidity and mortality, defining novel mechanisms of bacterial transmission and pathogenesis (reviewed in Fàbrega and Vila, 2013). The response of S. Typhimurium to the stresses it encounters in its infectious pathway—from the external environment to the host's intestines—is controlled largely by overlapping transcriptional regulatory systems (reviewed in Runkel et al., 2013).
Transcription in bacteria is carried out by the RNA polymerase core enzyme (RNAP; α2ββ′ω). However, the core enzyme alone cannot recognize specific promoter sequences; the variable sigma (σ) subunit confers DNA-binding specificity to ensure that transcription starts at the appropriate promoter sequence (reviewed in Feklístov et al., 2014). RNAP and σ together make up the holoenzyme (Eσ). There are two families of sigma factors: the σ70 (RpoD) family and the σ54 (RpoN) family. The σ70 family includes the housekeeping sigma factor (σ70/D) and all of the alternative sigma factors, except σ54. These σ70-type sigma factors, which in Salmonella include σ70/D, σ24/E, σ32/H, σ38/S, and σ28, exhibit similar structure and recognize promoter sequences with −35 (TTGACA) and −10 (TATAAT) promoter elements that are conserved to varying extents. When Eσ70 binds to promoter sequences, it initially forms a closed complex, where no DNA melting has occurred. Free energy from specific interactions of Eσ70 with promoter DNA activate conformational changes in both Eσ70 and DNA to form a stable open complex in which duplex DNA is opened at the +1 transcription start site and the template strand moves into the active site of RNAP (reviewed in Saecker et al., 2011).
σ54 is structurally distinct from the σ70-type sigma factors (Yang et al., 2015), thus Eσ54 recognizes very different promoter elements located at −24 (GC) and −12 (GG) upstream of the transcription start site (Morett and Buck, 1989). When Eσ54 binds to a promoter, it forms a stable closed complex due to direct interaction of Eσ54 with two bases within a DNA distortion immediately downstream of the −12 element (Morris et al., 1994). Open complex formation by Eσ54 requires an activator protein (bacterial enhancer-binding protein; bEBP; Yang et al., 2015). bEBPs are typically found as dimers in the cell but, upon receiving the appropriate cellular signal, they oligomerize into complexes that are competent to bind ATP and interact with enhancer sequences usually located 80–150 bp upstream of the promoter (Figure 1A). A DNA-looping event, often facilitated by integration host factor, brings the bEBP oligomer in contact with Eσ54 at the promoter (Wedel et al., 1990); bEBP then hydrolyzes ATP, which causes conformational changes in bEBP that trigger remodeling of Eσ54 and stimulate open complex formation (Chen et al., 2010). Bacteria often have multiple bEBPs that are responsive to different environmental signals and activate transcription of different sets of genes (Francke et al., 2011).
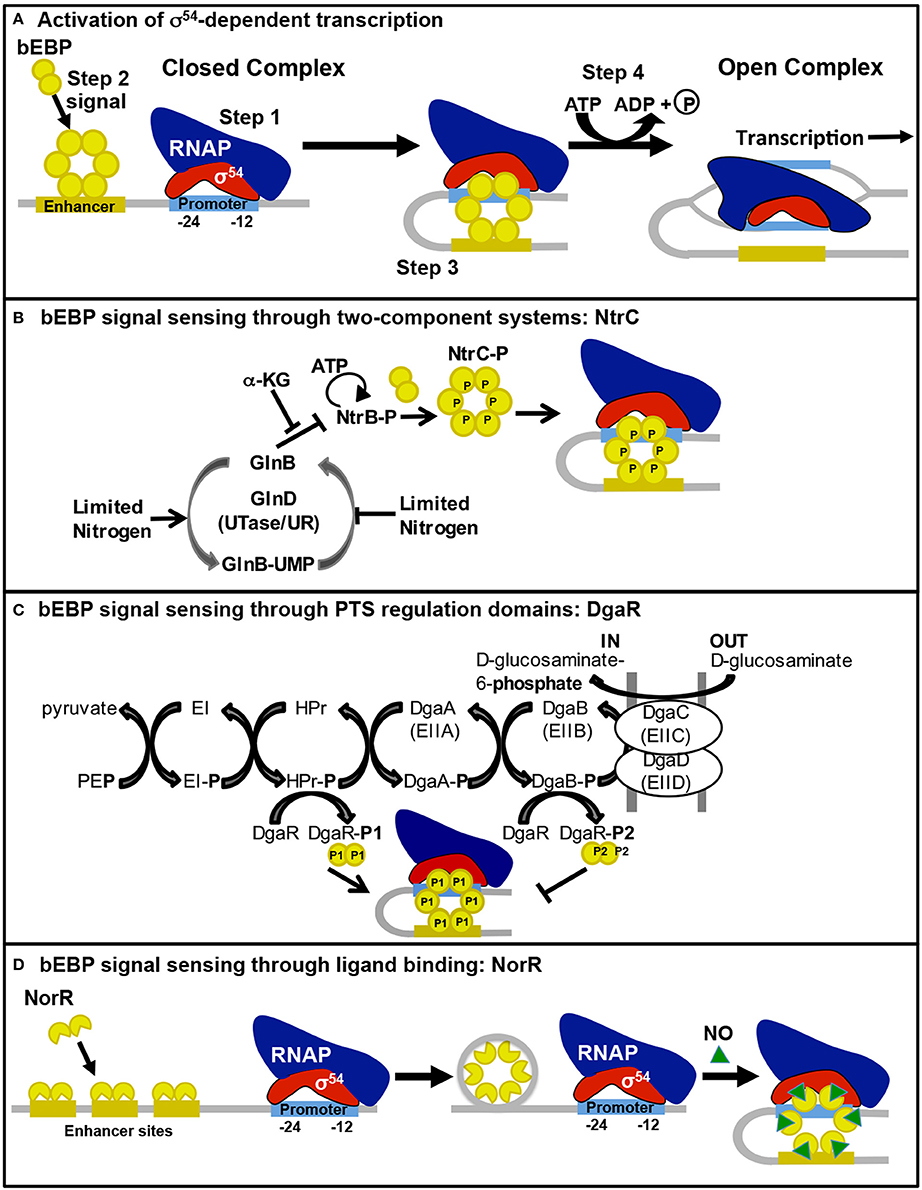
Figure 1. Bacterial enhancer-binding protein sensing of environmental signals and activation of σ54-dependent transcription. The process for bEBP activation of σ54-dependent transcription is illustrated in (A). Step 1, Eσ54 binds to the promoter in a stable closed complex. Step 2, the bEBP receives a signal from the internal or external environment, becomes active, and binds to an enhancer sequence. Step 3, DNA looping brings the bEBP in contact with Eσ54. Step 4, the bEBP hydrolyzes ATP to promote open complex formation. The mechanism for bEBP sensing of environmental signals through (B) two-component systems, (C) PTS regulatory domains, and (D) ligand binding are illustrated here and described in the text.
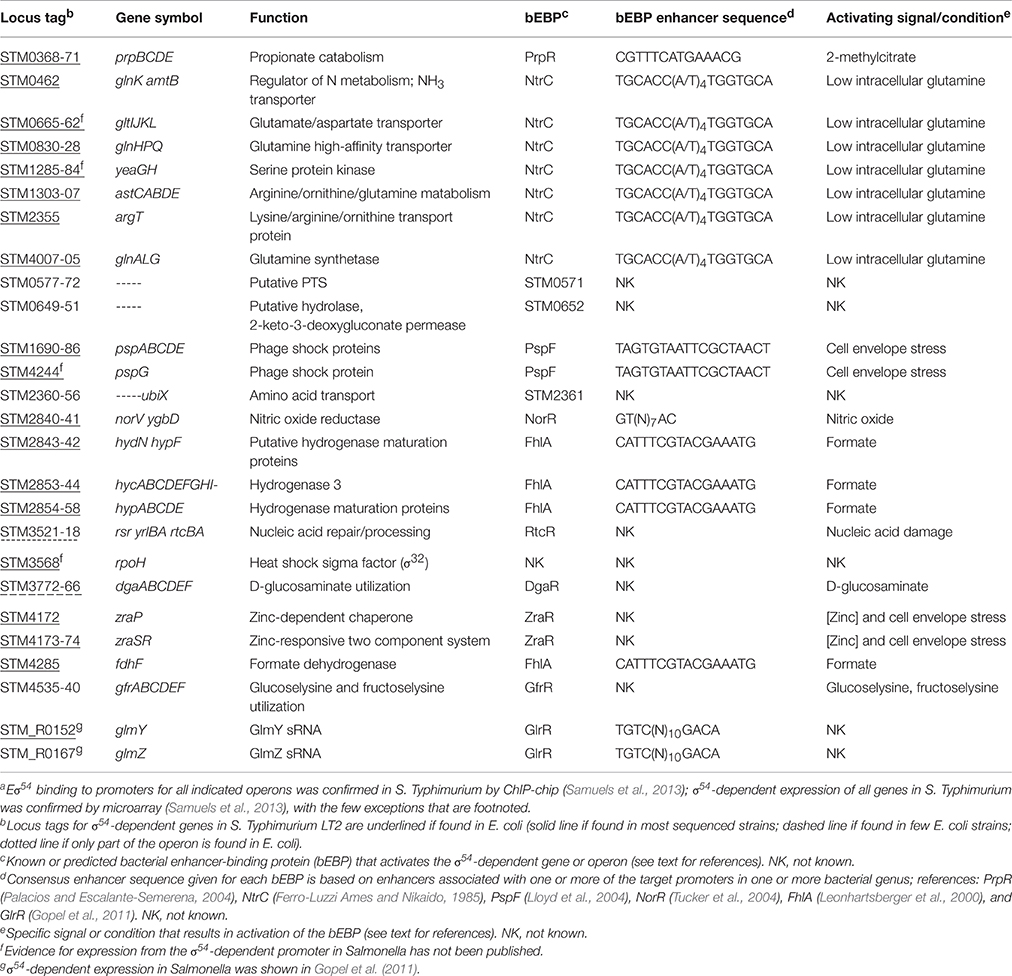
The global RpoN regulon of S. Typhimurium, including σ54-dependent transcripts and Eσ54 chromosomal DNA-binding sites, was characterized in the presence of a promiscuous, constitutively-active bEBP using microarray and ChIP-chip analyses (Samuels et al., 2013). Promoters of this extensive and diverse RpoN regulon in S. Typhimurium respond to 1 of 13 known or predicted bEBPs (Table 1; Studholme, 2002). The target promoters and activating environmental stimuli for most of these bEBPs have been demonstrated experimentally or inferred from studies with orthologs in E. coli (Table 1). RpoN regulons of S. Typhimurium (Samuels et al., 2013) and E. coli (Zhao et al., 2010; Bonocora et al., 2015) share many genes/operons (see Table 1); significant differences include the absence in Salmonella of nac, the LysR-type regulator of multiple operons involved in nitrogen assimilation (Zimmer et al., 2000), and the absence in E. coli of the gfr operon and rsr-yrlBA of the Salmonella RNA repair operon (see below). Cellular processes regulated by σ54-dependent bEBPs in S. Typhimurium include nitrogen metabolism in response to limiting nitrogen conditions [NtrC (GlnG, NRI), Keener and Kustu, 1988; Zimmer et al., 2000], transport and catabolism of D-glucosaminate (DgaR, Miller et al., 2013) and glucoselysine/fructoselysine (GfrR, Miller et al., 2015), regulation of cytoplasmic pH homeostasis during fermentative growth by the formate-hydrogen lyase system (FhlA, Hopper and Böck, 1995; Lamichhane-Khadka et al., 2015), response to assaults to the cell envelope (PspF, Karlinsey et al., 2010; Flores-Kim and Darwin, 2015 and zinc-dependent ZraR, Appia-Ayme et al., 2012), reduction of nitric oxide under anaerobic conditions (NorR, Hutchings et al., 2002; Mills et al., 2005), propionate catabolism (PrpR, Palacios and Escalante-Semerena, 2000), regulation of amino-sugar synthesis by sRNAs (GlrR; Gopel et al., 2011), and RNA repair/processing (RtcR, Samuels, 2014; Engl et al., 2016). A comprehensive study of the genes that are required for infection of animal hosts by S. Typhimurium identified RpoN as important in colonization of chicks, pigs, cattle and mice; transposon mutants in bEBP genes ntrC (glnG) and prpR were attenuated in at least two animal hosts and RpoN-regulated genes argT, glnA, glnL, and gfrACDEF (SL1344_4466, 4468–4471) were attenuated in at least two animal hosts (Chaudhuri et al., 2013).
Bacterial Enhancer-Binding Proteins of S. Typhimurium Sense and Respond to Signals for Adaptation in a Changing Environment
bEBPs typically consist of three domains: an N-terminal regulatory domain, a central AAA+ ATPase/transcriptional activation domain, and a C-terminal DNA-binding domain. The N-terminal regulatory domain responds to cellular signals and negatively or positively controls AAA+ domain oligomerization, ATPase activity, and/or interaction with σ54. The central AAA+ ATPase domain is responsible for bEBP oligomerization; association of two AAA+ domains within the bEBP oligomer forms the ATP hydrolysis site. This domain also includes the highly conserved GAFTGA motif that mediates the interaction with σ54. The C-terminal DNA-binding domain contains a helix-turn-helix DNA-binding motif, which determines bEBP specificity for an enhancer. For some bEBPs binding to the enhancer facilitates or stabilizes oligomerization. Consensus enhancer sequences for bEBPs found in S. Typhimurium are given in Table 1. Further details on bEBP structure and function are reviewed in (Bush and Dixon, 2012).
The regulatory domains of S. Typhimurium bEBPs can function as response-regulator domains of two-component systems (TCS), phosphotransferase regulation domains (PRDs) or ligand-binding domains. One bEBP, PspF, lacks a regulatory domain, but a separate protein, PspA, controls PspF activity. The PspF-PspA system, which is required for S. Typimurium virulence in a mouse model (Karlinsey et al., 2010), is not further discussed in this review, but two recent studies provide insight into this anti-activator mechanism for regulating bEBP activity (Flores-Kim and Darwin, 2015; Osadnik et al., 2015). Representative examples for the different mechanisms by which the regulatory domains of bEBPs from S. Typhimurium respond to extracellular or intracellular signals are considered here.
Signal Sensing through Two-Component Systems
S. Typhimurium has three bEBPs (NtrC, ZraR, and GlrR) that are response regulators of TCSs, in which a sensor kinase protein recognizes the cellular signal, autophosphorylates, and transfers the phosphate to a conserved aspartate residue of the response regulator. Phosphorylation of the regulatory domain stimulates the bEBP to interact with enhancer sequence(s) and the Eσ54 closed complex, activating open complex formation (Figure 1B).
NtrC (GlnG)
The NtrB-NtrC TCS is activated in response to limited nitrogen conditions. NtrB is the sensor kinase of the TCS. Nitrogen limitation is perceived by the cell as low intracellular levels of glutamine (Ikeda et al., 1996), which stimulates the uridylyltransferase GlnD to uridylylate the PII protein GlnB (Jiang et al., 1998). Unmodified GlnB inhibits NtrB kinase activity but GlnB-UMP cannot interact with NtrB, thus allowing autophosphorylation of NtrB and transfer of the phosphate to NtrC (Reitzer, 2003). GlnB also responds to α-ketoglutarate. During nitrogen limitation, the level of α-ketoglutarate is high and inhibits GlnB interaction with NtrB, thereby increasing NtrC phosphorylation (Schumacher et al., 2013). Phosphorylation of NtrC dimers results in oligomerization and enhancer binding (Weiss et al., 1991). NtrC-dependent transcription of target genes (Table 1) allows the cell to assimilate low levels of ammonia and utilize alternative nitrogen sources in nutrient-limited environments; NtrC-regulated glnA (glutamine synthetase) and glnHQ (glutamine transport) together contribute to S. Typhimurium virulence in a mouse model and increased survival in macrophages (Klose and Mekalanos, 1997).
ZraR (HydG)
In S. Typhimurium, ZraR is a response regulator, activated by its sensor kinase ZraS in a zinc-dependent response to envelope stress (Leonhartsberger et al., 2001; Appia-Ayme et al., 2012). ZraR controls expression from divergent σ54-dependent promoters for zraSR and zraP. ZraP encodes a zinc-binding periplasmic protein that acts as a zinc-dependent chaperone in both S. Typhimurium and E. coli; ZraP responds to misfolding of periplasmic and outer membrane proteins due to envelope stress, such as disruption of the outer membrane by antimicrobial cationic peptides that may be encountered in the environment and/or the host (Appia-Ayme et al., 2012; Petit-Härtlein et al., 2015).
Signal Sensing through Phosphotransferase Regulation Domains
The bEBPs DgaR, GfrR, and STM0571 of S. Typhimurium are members of the family of LevR-like regulators, which previously have only been described in Gram-positive bacteria controlling transcription of the genes for permease components of phosphotransferase systems (PTSs) and enzymes required for utilization of the imported sugar/amino sugar (reviewed in Deutscher et al., 2014). PTSs import and phosphorylate sugars through the Enzyme II complex (EII) membrane-bound components that are linked to a cascade of phosphoryl transfer, beginning with phosphoenolpyruvate as the donor and continuing through Enzyme I (EI), HPr, and finally the EII complex (Figure 1C). These PTS enzymes control the activity of the LevR-like bEBPs through phosphorylation of the regulatory domain. In contrast to most bEBPs, the regulatory domains of LevR-like bEBPs are found at the C-terminus. These regulatory domains contain two PTS regulation domains (PRDs) with competing activities. HPr-mediated phosphorylation of a conserved histidine residue adjacent to PRD1 leads to activation while EII-mediated phosphorylation of a conserved histidine residue within PRD2 is inhibitory (Martin-Verstraete et al., 1998).
DgaR
The LevR-like bEBP DgaR is phosphorylated by PTS HPr~P (DgaR-P1), resulting in expression of dgaABCDEF, which encodes the permease and catabolic enzymes for D-glucosaminate (Miller et al., 2013). When D-glucosaminate is present, EII preferentially phosphorylates the sugar, instead of DgaR, to complete the PTS cascade; but in the absence of D-glucosaminate, DgaR is phosphorylated by EII (DgaR-P2), which inhibits DgaR activation (Figure 1C; Miller et al., 2013).
S. Typhimurium can utilize D-glucosaminate as both a carbon and nitrogen source (Miller et al., 2013), so it is likely that this PTS system gives S. Typhmurium a competitive advantage over competing microbes under nutrient-limited conditions; the source of D-glucosaminate in the environment/host is likely to be other bacteria containing D-glucosaminate in lipid A or glucose oxidase that effectively oxidizes D-glucosamine (Miller et al., 2013).
GfrR
GfrR activates σ54-dependent transcription of the gfrABCDEF operon (Miller et al., 2015). GfrR differs from DgaR and other LevR-like bEBPs in its regulatory domain by substitution with tyrosine of the conserved histidine that is normally phosphorylated by HPr~P. By analogy to another LevR-like bEBP, MtlR (Joyet et al., 2015), GfrR is likely controlled solely by the repressive EII-mediated phosphorylation of PRD2; this results in GfrR being insensitive to the catabolite repression observed for DgaR (Miller et al., 2013), in which EI and HPr phosphorylation activity is directed to the uptake of another primary carbon source (glucose) instead of phosphorylation of the bEBP. Thus, S. Typhimurium is able to utilize glucose and fructoselysine (or glucoselysine) simultaneously (Miller et al., 2015).
Enzymes encoded by the gfrABCDEF operon enable glucoselysine and fructoselysine uptake and catabolism. Glucoselysine and frustoselysine, as well as other Maillard reaction products, are found at varying levels in the gut of human and animal hosts depending on the diet and microbiota (reviewed in Tuohy et al., 2006). The PTS permease and dual deglycases encoded by gfrABCDEF give S. Typhimurium flexibility in carbon and nitrogen sources, improving persistence in animal hosts (Chaudhuri et al., 2013).
Signal Sensing through Ligand Binding
In S. Typhimurium there are four bEBPs that are known, or predicted, to be regulated by the binding of an effector molecule to the regulatory domain: NorR, FhlA, PrpR, and RtcR. Although the regulatory domain structure is different for each of these bEBPs, in each case ligand binding alters the bEBP structure such that repression of AAA+ domain oligomerization, ATPase activity, and/or interaction with σ54 by the regulatory domain is relieved (Figure 1D).
NorR
NorR stimulates expression of nitric oxide (NO) reductase genes, norVW, in response to NO under anaerobic conditions (Gardner et al., 2003) The N-terminal region of NorR contains a GAF (cyclic GMP-specific and stimulated phosphodiesterases, Anabaena adenylate cyclases, and E. coli FhlA) domain with a non-heme iron center that recognizes NO (D'Autréaux et al., 2005). Binding of NO to the GAF domain relieves repression of the ATPase activity of the AAA+ domain, allowing activation of transcription from the σ54-dependent promoter for norVW (D'Autréaux et al., 2005). NorR recognizes three enhancer sequences upstream of the norVW operon, all of which are required for transcriptional activation (Tucker et al., 2010). As illustrated in Figure 1D, unlike many bEBPs, NorR is able to multimerize in the absence of the activating ligand, forming hexamers through assembly of dimers that are bound to the enhancer sequences. The hexamer-enhancer complex is unable to hydrolyze ATP until activated by NO binding (Bush et al., 2015). It has been suggested that this “pre-activated” complex may exist to enable rapid response to the presence of NO (Bush et al., 2015). NO and other reactive nitrogen species are generated by macrophages during the immune response to infection and have bactericidal and bacteriostatic effects on Salmonella (Vazquez-Torres et al., 2000). Transient increased sensitivity of a norV mutant to NO suggests that the NorR-regulated NO reductase is part of a multiple enzyme response to NO stress during the infection process (Mills et al., 2005).
RtcR
RtcR controls σ54-dependent transcription of putative RNA repair operons of S. Typhimurium (rsr-yrlBA-rtcBA; Chen et al., 2013; Samuels, 2014) and E. coli (rtcBA; Genschik et al., 1998; Engl et al., 2016). rtcB and rtcA encode homologs of the metazoan and archaeal RNA ligase and RNA 3′-phosphate cyclase, respectively (Das and Shuman, 2013). rsr and yrlBA of Salmonella encode homologs of metazoan Ro60 and Y-RNAs that form ribonucleoprotein complexes involved in noncoding-RNA quality control (Chen et al., 2013; Wolin et al., 2013). The regulatory domain of RtcR exhibits significant sequence similarity with the CRISPR-associated Rossmann fold (CARF) domains (Makarova et al., 2014). CARF domains are predicted to bind nucleotides, but the RtcR regulatory domain lacks a positively-charged residue involved in nucleotide binding (Makarova et al., 2014). The lack of this residue suggests that RtcR utilizes a different ligand, possibly a nucleoside or modified nucleotide (Makarova et al., 2014).
Metazoan RtcB functions in repair of xbp-1 mRNA, which is required for the unfolded protein response (Jurkin et al., 2014), as well as tRNA splicing (Popow et al., 2011). RtcA repairs 3′-phosphate or 2′-phosphate ends of cleaved RNA to 2′,3′-cyclic phosphates, which can serve as substrates for RtcB-mediated ligation (Remus and Shuman, 2013). RtcB and RtcA from E. coli exhibit the same biochemical activities as the metazoan homologs in vitro (Genschik et al., 1998; Tanaka et al., 2011). In addition, E. coli RtcB and RtcA utilize DNA substrates; RtcB adds a guanylyl “cap” to a 3′-phosphate end of nicked DNA (Das et al., 2013, 2014), and RtcA adenylylates DNA 5′-phosphate ends (Chakravarty and Shuman, 2011). In S. Typhimurium, the Rsr-YrlA complex associates with PNPase (polynucleotide phosphorylase; Chen et al., 2013); this is consistent with the activity of Rsr in Deinococcus radiodurans, where Rsr forms a ribonucleoprotein complex with YrlA and PNPase and is involved in starvation-induced rRNA degradation (Wurtmann and Wolin, 2010). Additionally, Rsr works with RNase PH and RNase II to fully process 23S rRNAs during growth at elevated temperature (37°C; Chen et al., 2007).
RtcR is activated in S. Typhimurium upon exposure to the antibiotic mitomycin C (MMC), stimulating transcription of the rsr-yrlBA-rtcBA operon (Samuels, 2014). MMC is an alkylating agent that causes intra- and inter-strand crosslinking in nucleic acids, and results in the formation of DNA-MMC (Bizanek et al., 1992) and RNA-MMC adducts (Snodgrass et al., 2010). MMC induces the SOS response (Kenyon and Walker, 1980), and RtcR activation by MMC is RecA-dependent, suggesting involvement of the SOS response in the activation of RtcR (Samuels, 2014). In E. coli RtcR is activated by conditions that disrupt translation, including VapC-mediated cleavage of tRNAfmet and treatment with tetracycline (Engl et al., 2016). The signal that is recognized by RtcR in either bacterium is unknown, but candidate signal molecules include: alkylated bases or DNA-MMC adducts removed by nucleotide excision repair in the SOS response (reviewed in Kisker et al., 2013); MMC-modified nucleotides from rRNA or increased free nucleotide/nucleoside intracellular pools upon MMC-induced rRNA degradation (Suzuki and Kilgore, 1967a,b); 2′,3′-cyclic NMPs released from RNAs cleaved by toxins of toxin-antitoxin systems, which leave 2′,3′-cyclic phosphate at the 3′-end of cleaved RNA (reviewed in Sofos et al., 2015); or modified nucleotides of tRNAs (reviewed in Motorin and Helm, 2010) released by cleavage/degradation. The substrates for RtcA, RtcB, and Rsr-YrlA/B are unidentified in both S. Typhimurim and E. coli, although ribosome analysis in an E. coli rtcB mutant suggests a role in 16s rRNA stability (Engl et al., 2016).
Conclusion
The σ54 regulon of S. Typhimurium is involved in a range of potential stress responses, including nitrogen/carbon limitation, cell envelope stress, nitric oxide stress, and nucleic acid damage/turnover. As summarized in this mini-review, the response to these stresses and the resulting modulation of the S. Typhimurium lifestyle are often mediated through bEBPs, which receive signals from the environment through a variety of mechanisms and activate the appropriate components of the σ54 regulon. Further characterization of RtcR activation by nucleic acid damage/modification and of the three currently uncharacterized bEBPs (STM0571, STM0652, and STM2361) will give a clearer picture of how bEBPs can alter the lifestyle of S. Typhimurium and other pathogens to improve their chances of survival during the infection process.
Author Contributions
CH, AK, and DS each made substantial intellectual contributions to the work, participated in the writing of the mini-review, and approved it for publication.
Funding
NSF (MCB-1051175) and NIH (R21 AI117102-01A1) grants (to ACK) funded the study of the Salmonella RpoN regulon and putative RNA repair operon, respectively.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tim Hoover and Maureen Powers for their edits and suggestions for this review.
References
Appia-Ayme, C., Hall, A., Patrick, E., Rajadurai, S., Clarke, T. A., and Rowley, G. (2012). ZraP is a periplasmic molecular chaperone and a repressor of the zinc-responsive two-component regulator ZraSR. Biochem. J. 442, 85–93. doi: 10.1042/BJ20111639
Bizanek, R., McGuinness, B. F., Nakanishi, K., and Tomasz, M. (1992). Isolation and structure of an intrastrand cross-link adduct of mitomycin C and DNA. Biochemistry 31, 3084–3091. doi: 10.1021/bi00127a008
Bonocora, R. P., Smith, C., Lapierre, P., and Wade, J. T. (2015). Genome-Scale mapping of Escherichia coli σ54 reveals widespread, conserved intragenic binding. PLoS Genet. 11:e1005552. doi: 10.1371/journal.pgen.1005552
Bush, M., and Dixon, R. (2012). The role of bacterial enhancer binding proteins as specialized activators of sigma54-dependent transcription. Microbiol. Mol. Biol. Rev. 76, 497–529. doi: 10.1128/MMBR.00006-12
Bush, M., Ghosh, T., Sawicka, M., Moal, I. H., Bates, P. A., Dixon, R., et al. (2015). The structural basis for enhancer-dependent assembly and activation of the AAA transcriptional activator NorR. Mol. Microbiol. 95, 17–30. doi: 10.1111/mmi.12844
Chakravarty, A. K., and Shuman, S. (2011). RNA 3′-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5'-monophosphate ends. J. Biol. Chem. 286, 4117–4122. doi: 10.1074/jbc.M110.196766
Chaudhuri, R. R., Morgan, E., Peters, S. E., Pleasance, S. J., Hudson, D. L., Davies, H. M., et al. (2013). Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 9:e1003456. doi: 10.1371/journal.pgen.1003456
Chen, B., Sysoeva, T. A., Chowdhury, S., Guo, L., De Carlo, S., Hanson, J. A., et al. (2010). Engagement of arginine finger to ATP triggers large conformational changes in NtrC1 AAA+ ATPase for remodeling bacterial RNA polymerase. Structure 18, 1420–1430. doi: 10.1016/j.str.2010.08.018
Chen, X., Taylor, D. W., Fowler, C. C., Galan, J. E., Wang, H. W., and Wolin, S. L. (2013). An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell 153, 166–177. doi: 10.1016/j.cell.2013.02.037
Chen, X., Wurtmann, E. J., Van Batavia, J., Zybailov, B., Washburn, M. P., and Wolin, S. L. (2007). An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev. 21, 1328–1339. doi: 10.1101/gad.1548207
Das, U., Chakravarty, A. K., Remus, B. S., and Shuman, S. (2013). Rewriting the rules for end joining via enzymatic splicing of DNA 3′-PO4 and 5′-OH ends. Proc. Natl. Acad. Sci. U.S.A. 110, 20437–20442. doi: 10.1073/pnas.1314289110
Das, U., Chauleau, M., Ordonez, H., and Shuman, S. (2014). Impact of DNA3′pp5′G capping on repair reactions at DNA 3′ ends. Proc. Natl. Acad. Sci. U.S.A. 111, 11317–11322. doi: 10.1073/pnas.1409203111
Das, U., and Shuman, S. (2013). 2′-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA 19, 1355–1362. doi: 10.1261/rna.039917.113
D'Autréaux, B., Tucker, N. P., Dixon, R., and Spiro, S. (2005). A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 437, 769–772. doi: 10.1038/nature03953
Deutscher, J., Aké, F. M., Derkaoui, M., Zébré, A. C., Cao, T. N., Bouraoui, H., et al. (2014). The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256. doi: 10.1128/MMBR.00001-14
Engl, C., Schaefer, J., Kotta-Loizou, I., and Buck, M. (2016). Cellular and molecular phenotypes depending upon the RNA repair system RtcAB of Escherichia coli. Nucleic Acids Res. doi: 10.1093/nar/gkw628. [Epub ahead of print].
Fàbrega, A., and Vila, J. (2013). Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341. doi: 10.1128/CMR.00066-12
Feklístov, A., Sharon, B. D., Darst, S. A., and Gross, C. A. (2014). Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376. doi: 10.1146/annurev-micro-092412-155737
Ferro-Luzzi Ames, G., and Nikaido, K. (1985). Nitrogen regulation in Salmonella typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J. 4, 539–547.
Flores-Kim, J., and Darwin, A. J. (2015). Activity of a bacterial cell envelope stress response is controlled by the interaction of a protein binding domain with different partners. J. Biol. Chem. 290, 11417–11430. doi: 10.1074/jbc.M114.614107
Francke, C., Groot Kormelink, T., Hagemeijer, Y., Overmars, L., Sluijter, V., Moezelaar, R., et al. (2011). Comparative analyses imply that the enigmatic Sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. doi: 10.1186/1471-2164-12-385
Gardner, A. M., Gessner, C. R., and Gardner, P. R. (2003). Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and sigma54 in the nitric oxide stress response. J. Biol. Chem. 278, 10081–10086. doi: 10.1074/jbc.M212462200
Genschik, P., Drabikowski, K., and Filipowicz, W. (1998). Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its sigma54-regulated operon. J. Biol. Chem. 273, 25516–25526. doi: 10.1074/jbc.273.39.25516
Göpel, Y., Lüttmann, D., Heroven, A. K., Reichenbach, B., Dersch, P., and Görke, B. (2011). Common and divergent features in transcriptional control of the homologous small RNAs GlmY and GlmZ in Enterobacteriaceae. Nucleic Acids Res. 39, 1294–1309. doi: 10.1093/nar/gkq986
Hopper, S., and Böck, A. (1995). Effector-mediated stimulation of ATPase activity by the sigma 54-dependent transcriptional activator FHLA from Escherichia coli. J. Bacteriol. 177, 2798–2803.
Hutchings, M. I., Mandhana, N., and Spiro, S. (2002). The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 184, 4640–4643. doi: 10.1128/JB.184.16.4640-4643.2002
Ikeda, T. P., Shauger, A. E., and Kustu, S. (1996). Salmonella Typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259, 589–607. doi: 10.1006/jmbi.1996.0342
Jiang, P., Peliska, J. A., and Ninfa, A. J. (1998). Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37, 12782–12794. doi: 10.1021/bi980667m
Joyet, P., Derkaoui, M., Bouraoui, H., and Deutscher, J. (2015). PTS-Mediated regulation of the transcription activator mtlr from different species: surprising differences despite strong sequence conservation. J. Mol. Microbiol. Biotechnol. 25, 94–105. doi: 10.1159/000369619
Jurkin, J., Henkel, T., Nielsen, A. F., Minnich, M., Popow, J., Kaufmann, T., et al. (2014). The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 33, 2922–2936. doi: 10.15252/embj.201490332
Karlinsey, J. E., Maguire, M. E., Becker, L. A., Crouch, M. L., and Fang, F. C. (2010). The phage shock protein PspA facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium. Mol. Microbiol. 78, 669–685. doi: 10.1111/j.1365-2958.2010.07357.x
Keener, J., and Kustu, S. (1988). Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NtrB and NtrC of enteric bacteria: roles of the conserved amino-terminal domain of NtrC. Proc. Natl. Acad. Sci. U.S.A. 85, 4976–4980. doi: 10.1073/pnas.85.14.4976
Kenyon, C. J., and Walker, G. C. (1980). DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 77, 2819–2823. doi: 10.1073/pnas.77.5.2819
Kisker, C., Kuper, J., and Van Houten, B. (2013). Prokaryotic nucleotide excision repair. Cold Spring Harb. Perspect. Biol. 5:a012591. doi: 10.1101/cshperspect.a012591
Klose, K. E., and Mekalanos, J. J. (1997). Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella Typhimurium virulence. Infect. Immun. 65, 587–596.
Lamichhane-Khadka, R., Benoit, S. L., Miller-Parks, E. F., and Maier, R. J. (2015). Host hydrogen rather than that produced by the pathogen is important for Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 83, 311–316. doi: 10.1128/IAI.02611-14
Leonhartsberger, S., Ehrenreich, A., and Böck, A. (2000). Analysis of the domain structure and the DNA binding site of the transcriptional activator FhlA. Eur. J. Biochem. 267, 3672–3684. doi: 10.1046/j.1432-1327.2000.01399.x
Leonhartsberger, S., Huber, A., Lottspeich, F., and Böck, A. (2001). The hydH/G Genes from Escherichia coli code for a zinc and lead responsive two-component regulatory system. J. Mol. Biol. 307, 93–105. doi: 10.1006/jmbi.2000.4451
Lloyd, L. J., Jones, S. E., Jovanovic, G., Gyaneshwar, P., Rolfe, M. D., Thompson, A., et al. (2004). Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein G (PspG). J. Biol. Chem. 279, 55707–55714. doi: 10.1074/jbc.M408994200
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O'Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Makarova, K. S., Anantharaman, V., Grishin, N. V., Koonin, E. V., and Aravind, L. (2014). CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 5:102. doi: 10.3389/fgene.2014.00102
Martin-Verstraete, I., Charrier, V., Stülke, J., Galinier, A., Erni, B., Rapoport, G., et al. (1998). Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol. Microbiol. 28, 293–303. doi: 10.1046/j.1365-2958.1998.00781.x
Miller, K. A., Phillips, R. S., Kilgore, P. B., Smith, G. L., and Hoover, T. R. (2015). A mannose family phosphotransferase system permease and associated enzymes are required for utilization of Fructoselysine and Glucoselysine in Salmonella enterica serovar Typhimurium. J. Bacteriol. 197, 2831–2839. doi: 10.1128/JB.00339-15
Miller, K. A., Phillips, R. S., Mrázek, J., and Hoover, T. R. (2013). Salmonella utilizes D-glucosaminate via a mannose family phosphotransferase system permease and associated enzymes. J. Bacteriol. 195, 4057–4066. doi: 10.1128/JB.00290-13
Mills, P. C., Richardson, D. J., Hinton, J. C., and Spiro, S. (2005). Detoxification of nitric oxide by the flavorubredoxin of Salmonella enterica serovar Typhimurium. Biochem. Soc. Trans. 33(Pt 1), 198–199. doi: 10.1042/BST0330198
Morett, E., and Buck, M. (1989). In vivo studies on the interaction of RNA polymerase-sigma 54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J. Mol. Biol. 210, 65–77. doi: 10.1016/0022-2836(89)90291-X
Morris, L., Cannon, W., Claverie-Martin, F., Austin, S., and Buck, M. (1994). DNA distortion and nucleation of local DNA unwinding within sigma-54 (sigma N) holoenzyme closed promoter complexes. J. Biol. Chem. 269, 11563–11571.
Motorin, Y., and Helm, M. (2010). tRNA stabilization by modified nucleotides. Biochemistry 49, 4934–4944. doi: 10.1021/bi100408z
Osadnik, H., Schöpfel, M., Heidrich, E., Mehner, D., Lilie, H., Parthier, C., et al. (2015). PspF-binding domain PspA1-144 and the PspA.F complex: new insights into the coiled-coil-dependent regulation of AAA+ proteins. Mol. Microbiol. 98, 743–759. doi: 10.1111/mmi.13154
Palacios, S., and Escalante-Semerena, J. C. (2000). prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182, 905–910. doi: 10.1128/JB.182.4.905-910.2000
Palacios, S., and Escalante-Semerena, J. C. (2004). 2-Methylcitrate-dependent activation of thepropionate catabolic operon (prpBCDE) of Salmonella enterica by the PrpR protein. Microbiology 150(Pt 11), 3877–3887. doi: 10.1099/mic.0.27299-0
Petit-Härtlein, I., Rome, K., de Rosny, E., Molton, F., Duboc, C., Gueguen, E., et al. (2015). Biophysical and physiological characterization of ZraP from Escherichia coli, the periplasmic accessory protein of the atypical ZraSR two-component system. Biochem. J. 472, 205–216. doi: 10.1042/BJ20150827
Popow, J., Englert, M., Weitzer, S., Schleiffer, A., Mierzwa, B., Mechtler, K., et al. (2011). HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764. doi: 10.1126/science.1197847
Reitzer, L. (2003). Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57, 155–176. doi: 10.1146/annurev.micro.57.030502.090820
Remus, B. S., and Shuman, S. (2013). A kinetic framework for tRNA ligase and enforcement of a 2'-phosphate requirement for ligation highlights the design logic of an RNA repair machine. RNA 19, 659–669. doi: 10.1261/rna.038406.113
Runkel, S., Wells, H. C., and Rowley, G. (2013). Living with stress: a lesson from the enteric pathogen Salmonella enterica. Adv. Appl. Microbiol. 83, 87–144. doi: 10.1016/B978-0-12-407678-5.00003-9
Saecker, R. M., Record, M. T. Jr., and Dehaseth, P. L. (2011). Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 412, 754–771. doi: 10.1016/j.jmb.2011.01.018
Samuels, D. J. (2014). The RpoN Regulon of Salmonella and its Component RNA Repair System. Dissertation, University of Georgia, Athens, GA.
Samuels, D. J., Frye, J. G., Porwollik, S., McClelland, M., Mrázek, J., Hoover, T. R., et al. (2013). Use of a promiscuous, constitutively-active bacterial enhancer-binding protein to define the sigma(5)(4) (RpoN) regulon of Salmonella Typhimurium LT2. BMC Genomics 14:602. doi: 10.1186/1471-2164-14-602
Schumacher, J., Behrends, V., Pan, Z., Brown, D. R., Heydenreich, F., Lewis, M. R., et al. (2013). Nitrogen and carbon status are integrated at the transcriptional level by the nitrogen regulator NtrC in vivo. MBio 4, e00881–e00813. doi: 10.1128/mBio.00881-13
Snodgrass, R. G., Collier, A. C., Coon, A. E., and Pritsos, C. A. (2010). Mitomycin C inhibits ribosomal RNA: a novel cytotoxic mechanism for bioreductive drugs. J. Biol. Chem. 285, 19068–19075. doi: 10.1074/jbc.M109.040477
Sofos, N., Xu, K., Dedic, E., and Brodersen, D. E. (2015). Cut to the chase–Regulating translation through RNA cleavage. Biochimie 114, 10–17. doi: 10.1016/j.biochi.2015.01.009
Studholme, D. J. (2002). Enhancer-dependent transcription in Salmonella enterica Typhimurium: new members of the sigmaN regulon inferred from protein sequence homology and predicted promoter sites. J. Mol. Microbiol. Biotechnol. 4, 367–374.
Suzuki, H., and Kilgore, W. W. (1967a). Decomposition of ribosomal particles in Escherichia coli treated with mitomycin C. J. Bacteriol. 94, 666–676.
Suzuki, H., and Kilgore, W. W. (1967b). Effects of mitomycin C on macromolecular synthesis in Escherichia coli. J. Bacteriol. 93, 675–682.
Tanaka, N., Chakravarty, A. K., Maughan, B., and Shuman, S. (2011). Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 286, 43134–43143. doi: 10.1074/jbc.M111.302133
Tucker, N. P., D'Autréaux, B., Studholme, D. J., Spiro, S., and Dixon, R. (2004). DNA binding activity of the Escherichia coli nitric oxide sensor NorR suggests a conserved target sequence in diverse proteobacteria. J. Bacteriol. 186, 6656–6660. doi: 10.1128/JB.186.19.6656-6660.2004
Tucker, N. P., Ghosh, T., Bush, M., Zhang, X., and Dixon, R. (2010). Essential roles of three enhancer sites in sigma54-dependent transcription by the nitric oxide sensing regulatory protein NorR. Nucleic Acids Res. 38, 1182–1194. doi: 10.1093/nar/gkp1065
Tuohy, K. M., Hinton, D. J., Davies, S. J., Crabbe, M. J., Gibson, G. R., and Ames, J. M. (2006). Metabolism of Maillard reaction products by the human gut microbiota–implications for health. Mol. Nutr. Food Res. 50, 847–857. doi: 10.1002/mnfr.200500126
Vazquez-Torres, A., Jones-Carson, J., Mastroeni, P., Ischiropoulos, H., and Fang, F. C. (2000). Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192, 227–236. doi: 10.1084/jem.192.2.227
Wedel, A., Weiss, D. S., Popham, D., Dröge, P., and Kustu, S. (1990). A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science 248, 486–490. doi: 10.1126/science.1970441
Weiss, D. S., Batut, J., Klose, K. E., Keener, J., and Kustu, S. (1991). The phosphorylated form of the enhancer-binding protein NtrC has an ATPase activity that is essential for activation of transcription. Cell 67, 155–167. doi: 10.1016/0092-8674(91)90579-N
Wolin, S. L., Belair, C., Boccitto, M., Chen, X., Sim, S., Taylor, D. W., et al. (2013). Non-coding Y RNAs as tethers and gates: insights from bacteria. RNA Biol. 10, 1602–1608. doi: 10.4161/rna.26166
Wurtmann, E. J., and Wolin, S. L. (2010). A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc. Natl. Acad. Sci. U.S.A. 107, 4022–4027. doi: 10.1073/pnas.1000307107
Yang, Y., Darbari, V. C., Zhang, N., Lu, D., Glyde, R., Wang, Y. P., et al. (2015). Structures of the RNA polymerase-sigma54 reveal new and conserved regulatory strategies. Science 349, 882–885. doi: 10.1126/science.aab1478
Zhao, K., Liu, M., and Burgess, R. R. (2010). Promoter and regulon analysis of nitrogen assimilation factor, sigma54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 38, 1273–1283. doi: 10.1093/nar/gkp1123
Zimmer, D. P., Soupene, E., Lee, H. L., Wendisch, V. F., Khodursky, A. B., Peter, B. J., et al. (2000). Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. U.S.A. 97, 14674–14679. doi: 10.1073/pnas.97.26.14674
Keywords: Salmonella RpoN regulon, sigma 54, bacterial enhancer-binding protein, bEBP, transcription activation, stress adaptation
Citation: Hartman CE, Samuels DJ and Karls AC (2016) Modulating Salmonella Typhimurium's Response to a Changing Environment through Bacterial Enhancer-Binding Proteins and the RpoN Regulon. Front. Mol. Biosci. 3:41. doi: 10.3389/fmolb.2016.00041
Received: 27 June 2016; Accepted: 28 July 2016;
Published: 17 August 2016.
Edited by:
Tatiana Venkova, University of Texas Medical Branch at Galveston, USAReviewed by:
David John Studholme, University of Exeter, UKLarry Reitzer, University of Texas at Dallas, USA
Copyright © 2016 Hartman, Samuels and Karls. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna C. Karls, YWthcmxzQHVnYS5lZHU=
†Present Address: Christine E. Hartman, Office for Teaching and Learning, Wayne State University, Detroit, MI, USA
David J. Samuels, Department of Biology, Georgetown University, Washington, DC, USA
 Christine E. Hartman
Christine E. Hartman David J. Samuels
David J. Samuels Anna C. Karls
Anna C. Karls