- Dipartimento di Biotecnologie e Bioscienze, Università degli Studi di Milano-Bicocca, Milan, Italy
DNA double-strand breaks (DSBs) are highly cytotoxic lesions that must be repaired to ensure genomic stability and avoid cell death. The cellular response to DSBs is initiated by the evolutionarily conserved Mre11-Rad50-Xrs2/NBS1 (MRX/MRN) complex that has structural and catalytic functions. Furthermore, it is responsible for DSB signaling through the activation of the checkpoint kinase Tel1/ATM. Here, we review functions and regulation of the MRX/MRN complex in DSB processing in a chromatin context, as well as its interplay with Tel1/ATM.
Introduction
Chromosomal DNA double-strand breaks (DSBs) are potentially lethal DNA lesions that can form accidentally during DNA replication and transcription, or upon exposure to genotoxic agents, such as ionizing radiation or chemicals. Failure to repair them can result in loss of genetic information or cell death, whereas inaccurate repair can lead to chromosome rearrangements (Jackson and Bartek, 2009; Liu et al., 2012). Even though DSBs pose a significant threat to genome stability, DSBs are programmed recombination intermediates during gametogenesis or antigen-receptor diversity in lymphocyte development (Lam and Keeney, 2014; Arya and Bassing, 2017). In all cases, DSBs need to be repaired to preserve genomic integrity.
Eukaryotic cells possess two main mechanisms for repairing DSBs: non-homologous end-joining (NHEJ) and homologous recombination (HR). Repair by NHEJ requires the Ku70–80 heterodimer (hereafter referred to as Ku) that recruits the DNA ligase IV complex (Lig4/Dnl4 in Saccharomyces cerevisiae), which directly religates the two broken ends (Chang et al., 2017). By contrast, HR is a more complex process that uses DNA information stored in a homologous double-stranded DNA (dsDNA) as template to reconstitute any missing genetic information at the break site (Mehta and Haber, 2014; Kowalczykowski, 2015).
The key process in determining which pathway is used to repair DSBs is the initial processing of the DSB ends. While NHEJ requires little or no DNA end processing, HR is initiated by nucleolytic degradation of the 5′ terminated strands at both DNA ends by a concerted action of nucleases in a process termed DNA end resection (Bonetti et al., 2018). The preferential degradation of the 5′-terminated strands results in formation of 3′-ended single-stranded DNA (ssDNA) ends that are first coated by the Replication Protein A (RPA) complex. RPA is subsequently replaced by Rad51 to form a nucleoprotein filament that is used to search for a homologous dsDNA sequence (Kowalczykowski, 2015). Repair can then proceed via synthesis-dependent strand annealing or the canonical recombination pathway that involves formation of a double Holliday junction (Mehta and Haber, 2014).
Extended resection of the DSB ends not only commits DSB repair to HR, but it makes the DNA ends non-ligatable by NHEJ. In vegetatively growing cells, HR uses the sister chromatid as repair template and this restricts recombination to the S and G2 phases of the cell cycle when the sister chromatid is available. This cell-cycle control of recombination is based on activation of key resection proteins by cyclin-dependent kinase (CDK)-catalyzed phosphorylation events (Aylon et al., 2004; Ira et al., 2004; Huertas et al., 2008; Chen et al., 2011).
The evolutionarily conserved Mre11-Rad50-Xrs2/NBS1 complex (MRX in S. cerevisiae, MRN in humans) recognizes, signals and initiates repair of DSBs. MRX is rapidly recruited to DSBs, where it has structural and enzymatic activities to initiate DSB resection and to maintain the DSB ends tethered to each other for their repair (Syed and Tainer, 2018). MRX also recruits and activates the checkpoint protein Tel1 (ATM in mammals) to coordinate DSB repair with cell cycle progression (Villa et al., 2016). Germline hypomorphic mutations of human MRN complex components are associated with Ataxia Telangiectasia-like disorder (ATLD), Nijmegen Breakage Syndrome (NBS) and NBS-like disorder, which are characterized by cellular radiosensitivity, immune deficiency and cancer predisposition (O'Driscoll, 2012). Here we review structure, functions and regulation of the MRX complex in sensing, signaling and processing DSBs within a chromatin context, focusing mainly on the work done in the budding yeast S. cerevisiae.
Structural and Biochemical Properties of MRX
In both yeast and mammals, the MRX complex exists as a hetero-hexameric assembly, in which the Mre11 subunit interacts independently with both Rad50 and Xrs2 (NBS1 in mammals), and dimerizes with itself. Mre11 has five phosphodiesterase motifs in the N-terminal region and exhibits 3′-5′ dsDNA exonuclease and ssDNA endonuclease activities in vitro (Bressan et al., 1998; Paull and Gellert, 1998; Trujillo et al., 1998; Usui et al., 1998). The Sae2 protein (CtIP in mammals) stimulates Mre11 endonuclease activity to cleave the 5′-terminated DNA strands at both DSB ends (Cannavo and Cejka, 2014;Reginato et al., 2017; Wang et al., 2017).
Rad50 is characterized by ATPase motifs at the N− and C− terminal regions of the protein, with the sequence in between forming two long coiled-coil domains that are separated by a zinc binding CXXC motif referred to as zinc hook (Syed and Tainer, 2018; Figure 1). The two ATPase motifs associate together to generate an ATP nucleotide binding domain and the coiled-coil domains fold back on themselves to form antiparallel intramolecular coiled coils (Hopfner et al., 2001; Moncalian et al., 2004; Williams et al., 2008; Figure 1). The zinc hook at the apex of the coiled-coil domains can form intralinked or interlinked complexes via tetrahedral coordination of a zinc2+ atom and the interlinked assembly can account for the MRX ability to maintain the DSB ends in close proximity (de Jager et al., 2001; Hopfner et al., 2002; Kaye et al., 2004; Lobachev et al., 2004; Wiltzius et al., 2005; Hohl et al., 2011; Nakai et al., 2011; He et al., 2012). Recently, crystal structure and X-ray scattering analyses of human RAD50 Zn-hook with a portion of the coiled-coil domain indicate the existence of a novel eukaryotic-specific interface that stabilizes Rad50 coiled coils in an intramolecular dimer assembly (Park et al., 2017), suggesting that the intralinked arrangement is the predominant form of the complex.
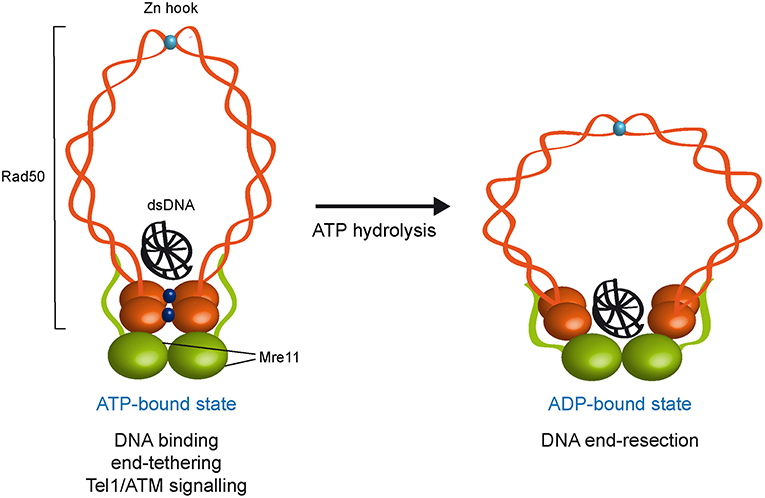
Figure 1. ATP- and ADP-bound state of the MRX complex. The Mre11 dimer (green) is bound to Rad50 dimer (orange) with a double-stranded DNA molecule located on the top surface of Rad50. The ATP-bound state of Rad50 supports DNA binding, end-tethering, and Tel1/ATM signaling, whereas it renders the dsDNA inaccessible to the Mre11 nuclease active sites and therefore negatively regulates Mre11 nuclease activity. ATP hydrolysis by Rad50 opens the complex to allow the Mre11 active sites to access DNA. Whether the ADP-bound state maintains an interlinked assembly is unknown. ATP molecules are indicated as blue dots. Zn2+ atoms are indicated as light blue dots. Xrs2 is not represented.
Several studies have shown that ATP binding and hydrolysis activities of Rad50 are crucial to regulate DNA binding, tethering and nuclease functions of the MRX complex. Structural studies of Mre11 in complex with Rad50 core domains from bacteria and archaea indicate that, upon ATP binding, Rad50 closes into a rigid conformation, in which the N- and C-terminal domains interact with each other and form a central groove that can accommodate dsDNA. This closed ATP-bound state of Rad50 renders dsDNA inaccessible to the Mre11 nuclease active site (Lammens et al., 2011; Lim et al., 2011; Williams et al., 2011; Möckel et al., 2012; Liu et al., 2016; Seifert et al., 2016). Point mutations that stabilize the ATP-bound conformation of Rad50 increase DNA binding, NHEJ and end-tethering (Deshpande et al., 2014), suggesting that MRX exerts these functions when it is present in the ATP-bound state. By contrast, in the ATP-free or hydrolyzed state, the Rad50 ATPase subunits are flexible and relatively open, suggesting that ATP hydrolysis drives the rotation of the two nucleotide binding domains of Rad50 and the disengagement of the Rad50 dimer that makes DNA accessible to the Mre11 nuclease active sites (Lammens et al., 2011; Lim et al., 2011; Williams et al., 2011; Möckel et al., 2012; Deshpande et al., 2014). Consistent with this hypothesis, biochemical analyses demonstrate that ATP hydrolysis by Rad50 is a prerequisite for Mre11/Rad50-mediated nuclease activity on dsDNA molecules (Paull and Gellert, 1999; Hopfner et al., 2000; Trujillo and Sung, 2001; Herdendorf et al., 2011). Altogether, these findings lead to a model whereby these ATP-driven transitions regulate the balance between MRX functions in NHEJ and end-tethering, which require ATP binding, and those in resection and HR, which require ATP hydrolysis (Figure 1).
Rad50 has a slow ATP hydrolysis rate (Herdendorf et al., 2011; Majka et al., 2012; Deshpande et al., 2017; Saathoff et al., 2018), suggesting that other proteins can promote its ATP hydrolysis activity within a cell. In S. cerevisiae, MRX is known to interact with Rif2, which is recruited to telomeric DNA ends and negatively regulates telomerase-mediated telomere elongation (Wotton and Shore, 1997; Levy and Blackburn, 2004; Hirano et al., 2009; Martina et al., 2012). Interestingly, Rif2, which is recruited to DSBs in a manner partially dependent on MRX, enhances ATP hydrolysis by Rad50 (Cassani et al., 2016). This observation, together with the finding that the lack of Rif2 increases the efficiency of both end-tethering and NHEJ (Cassani et al., 2016), suggests that Rif2 can regulate MRX ATP-driven transitions.
While Mre11 and Rad50 are conserved in bacteria and archaea, only eukaryotes possess Xrs2, which is the only MRX component that harbors a nuclear localization signal and is necessary for translocation of the Mre11-Rad50 subcomplex into the nucleus (Desai-Mehta et al., 2001; Tsukamoto et al., 2005). Localization of Mre11 into the nucleus in the absence of Xrs2 restores Mre11-Rad50 functions in DSB resection, hairpin resolution and meiotic recombination, but not in NHEJ and Tel1 activation (Oh et al., 2016), indicating an essential role for Xrs2 in these two latter processes. This finding is consistent with the observation that stimulation of the Mre11 endonucleolytic clipping activity by Sae2 requires Rad50 but not Xrs2 (Cannavo and Cejka, 2014).
By contrast, human NBS1 is required to promote MRE11 endonuclease activity on blocked DNA ends and hairpin substrates (Paull and Gellert, 1999; Deshpande et al., 2016). Using a reconstituted system, it has been recently shown that human NBS1 stimulates the MRE11-RAD50 nuclease by directly interacting with the MRE11 subunit and this stimulation requires CtIP phosphorylation (Anand et al., 2019). By contrast, in the absence of NBS1, MRE11-RAD50 subcomplex exhibits a weak nuclease activity that requires CtIP but not its phosphorylation (Anand et al., 2019). These findings lead to a model in which CtIP promotes MRE11 nuclease activity in a phosphorylation-dependent mode in the presence of NBS1 and in a phosphorylation-independent mode in the absence of NBS1, suggesting a role for NBS1 in restricting the MRE11-RAD50 nuclease to S and G2 phases of the cell cycle when CtIP is phosphorylated by CDKs.
Role of MRX in DSB Resection
The obligate step that initiates all recombination pathways is the degradation of the 5′-terminated DNA strands at both DSB ends to generate 3′-ended ssDNA overhangs that catalyze homologous pairing and strand exchange (Bonetti et al., 2018). In both yeast and mammals, DNA end resection occurs in two main steps (Garcia et al., 2011; Shibata et al., 2014; Figure 2). In the first step, Sae2 activates the endonuclease activity of Mre11 within the context of the MRX complex to cleave the 5′-terminated DNA strands at both DSB DNA ends (Cannavo and Cejka, 2014). This step is followed by 3′-5′ nucleolytic degradation by Mre11 that proceeds back toward the DNA ends (Reginato et al., 2017; Wang et al., 2017). The MRX-Sae2 ensemble can degrade the 5′-terminated strands up to ~300 nucleotides away from the end and this processing is thus referred to as short-range resection. The resulting nick/gap provides an internal entry site for either Exo1 or the combined activities of the Sgs1 helicase and the Dna2 nuclease (Mimitou and Symington, 2008; Zhu et al., 2008; Cejka et al., 2010; Nicolette et al., 2010; Niu et al., 2010; Cannavo et al., 2013; Reginato et al., 2017; Wang et al., 2017). Exo1 and Dna2 are capable of resecting thousands of nucleotides in length in the 5′-3′ direction and this nucleolytic degradation is thus referred to as long-range resection.
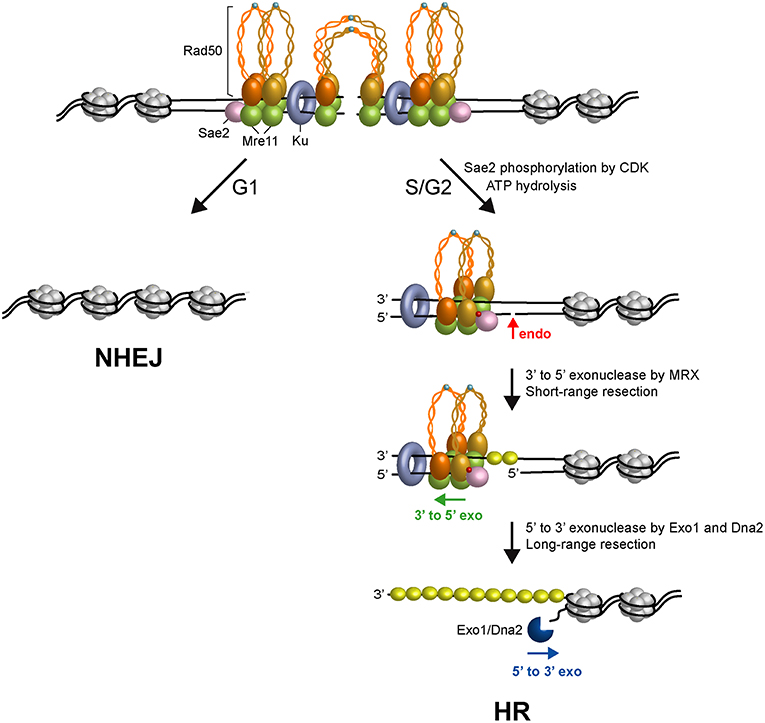
Figure 2. Model for initiation of DSB repair. Two ATP-bound MRX complexes are loaded onto both sides of the DSB, together with Ku and Sae2 proteins. The Rad50 subunits interact through the Zn-hook to form intralinked complexes. Owing to their proximity, the Zn-hook and coiled-coil domain may switch to form interlinked complexes that maintain the DSB ends tethered to each other. In the interlinked assembly, the Mre11 and Rad50 molecules are pictured separated from each other to visualize the DNA interruption. In G1, the DSB is repaired mainly by NHEJ because Sae2 is not phosphorylated, Rad50 is an ATP-bound state that blocks the Mre11 nuclease and Ku inhibits Exo1. In the S and G2 phases of the cell cycle, upon Sae2 phosphorylation by CDK and ATP hydrolysis by Rad50, Rad50 dimerization interface opens and dsDNA becomes accessible to the Mre11 nuclease active sites. Phosphorylated Sae2 then stimulates the Mre11 endonuclease to incise the 5′-terminated strands (red arrows) at Ku-bound DNA ends or adjacent to nucleosomes. MRX proceeds back toward the DSB end using the Mre11 3′-5′ exonuclease activity. Exo1 or Sgs1-Dna2 nuclease then can degrade DNA in the 5′-3′ direction. ssDNA generated by resection is coated by RPA to initiate HR. Phosphorylation is indicated as red dots. Zn2+ atoms are indicated as light blue dots. The Rad50 subunits belonging to a dimeric assembly are indicated with the same color (orange or gold). Xrs2 is not represented.
Short-Range Resection
Sae2 is known to be phosphorylated by multiple kinases, including CDKs and Mec1/Tel1 in a cell cycle- and DNA damage-dependent manner, respectively (Baroni et al., 2004; Cartagena-Lirola et al., 2006; Huertas et al., 2008; Manfrini et al., 2010). Using a reconstituted system, it has been shown that the ability of Sae2 to promote Mre11 endonuclease activity requires CDK-mediated Sae2 phosphorylation, and this control represents one of the key mechanisms that allow DSB resection to take place only during the S and G2 phases of the cell cycle when sister chromatids are available as repair templates (Huertas et al., 2008; Huertas and Jackson, 2009; Cannavo and Cejka, 2014; Anand et al., 2016). The phosphorylation state of Sae2 was shown to affect its oligomeric state that is critical for its activity (Kim et al., 2008; Fu et al., 2014; Andres et al., 2015; Davies et al., 2015). In particular, during the G1 phase of the cell cycle, Sae2 exists as unphosphorylated inactive soluble multimeric complexes (Cannavo et al., 2018). During S and G2 cell cycle phases or after DNA damage, phosphorylation at multiple Sae2 sites promotes formation of active Sae2 tetramers, which promote the Mre11 nuclease within the MRX complex (Cannavo et al., 2018). Furthermore, phosphorylation of the Sae2 C-terminus is necessary for a direct physical interaction between Sae2 and Rad50 (Cannavo et al., 2018). Since stimulation of Mre11 nuclease activity by Sae2 is dependent on ATP hydrolysis by Rad50 (Cannavo and Cejka, 2014; Wang et al., 2017), phosphorylated Sae2 might control the Mre11 nuclease by coupling ATP hydrolysis by Rad50 with Mre11 processing activity.
Genetic experiments have shown that MRX-Sae2-catalyzed cleavage is dispensable for resection of endonuclease-induced “clean” DSBs (Llorente and Symington, 2004), as Exo1 and Sgs1-Dna2 can directly access and resect the 5′-terminated strands of these DNA ends, although less efficiently. By contrast, MRX-Sae2-mediated cleavage is essential for removing hairpin-capped DSBs or protein blocks that render DNA ends refractory to Exo1- and Sgs1-Dna2-mediated resection (Lobachev et al., 2002; Neale et al., 2005). These end-binding factors can include trapped topoisomerases (Hoa et al., 2016) or Spo11, a meiosis-specific type II topoisomerase-like that generates programmed DSBs in meiosis by forming a covalent linkage between a conserved tyrosine residue and the 5′ end of the cleaved strand (Bergerat et al., 1997; Keeney et al., 1997). Spo11 is then removed endonucleolytically by Mre11, which introduces internal incisions at short distance from Spo11-bound DNA ends and releases short Spo11-attached oligonucleotides (Neale et al., 2005; Garcia et al., 2011).
Interestingly, using a reconstituted system, it has been shown that phosphorylated Sae2, or CtIP in humans, promotes the Mre11 nuclease within the MRX/MRN complex to cleave endonucleolytically the 5′-terminated DNA strand ~15–20 nucleotides away from a streptavidin block located at the end of a linear duplex DNA molecule (Cannavo and Cejka, 2014; Anand et al., 2016; Deshpande et al., 2016). Phosphorylated Sae2 was shown also to stimulate the MRX endonuclease activity on linear dsDNA substrates harboring either a streptavidin block or a catalytic inactive EcoRI restriction enzyme located at sites internal to the DSB end (Reginato et al., 2017; Wang et al., 2017). These findings suggest that any stable protein obstacle bound either internally or at the end of a DNA molecule can activate the 5′ DNA strand cleavage activity of MRX-Sae2.
The above observations raised the question of whether physiological protein blocks would also stimulate MRX-Sae2-catalyzed endonucleolytic cleavage. The Ku complex is rapidly recruited to DNA ends and protects them from degradation, particularly in the G1 phase of the cell cycle (Lisby et al., 2004; Clerici et al., 2008; Zierhut and Diffley, 2008). The lack of Ku partially restores DNA damage resistance in sae2Δ and mre11 nuclease-deficient alleles (Clerici et al., 2008; Bonetti et al., 2010; Mimitou and Symington, 2010; Shim et al., 2010; Foster et al., 2011; Langerak et al., 2011), indicating that Ku bound to the DSB ends acts as a block to resection. Remarkably, Ku is as effective as a streptavidin block in stimulating the endonucleolytic cleavage by MRX in a manner that depends on phosphorylated Sae2 and ATP hydrolysis by Rad50 (Reginato et al., 2017; Wang et al., 2017). Furthermore, Ku shields DNA ends from the Mre11-catalyzed 3′-5′ degradation (Reginato et al., 2017; Wang et al., 2017). As MRX and Ku also promote NHEJ, these results support a model in which the presence of both MRX and Ku at the DSB ends in the G1 phase of the cell cycle first channels DSB repair into NHEJ (Figure 2). In S and G2 phases of the cell cycle, when Sae2 is phosphorylated by CDK and ATP hydrolysis by Rad50 is allowed, the presence of Ku at the DSB ends renders the 5′ DNA strand susceptible to endonucleolytic cleavage by MRX-Sae2 that directs the repair toward HR (Figure 2).
In any case, as Ku preferentially binds dsDNA ends over ssDNA (Griffith et al., 1992), the 3′-5′ MRX-Sae2 processing activity should cause the removal of Ku from DNA ends (Mimitou and Symington, 2010; Langerak et al., 2011; Chanut et al., 2016), raising the possibility that other proteins could stimulate 5′ strand scission by MRX-Sae2 to overcome any obstacles present not only at DNA ends but also at sites internal to the DSB. Interestingly, similar to Ku, binding of the RPA complex to either partially resected DNA ends or terminal hairpin structures also stimulates MRX-Sae2 cleavage of the 5′ strand (Wang et al., 2017), suggesting that RPA can allow MRX-Sae2 to generate an entry site in case the long-range resection machinery is disassembled from partially resected DNA ends. Furthermore, a recent reconstitution of the S. cerevisiae short-range resection machinery has shown that the Mre11-Rad50 subcomplex and phosphorylated Sae2 can cleave a 5′-terminated DNA strand by stepwise incision without the requirement for a separate protein block (Cannavo et al., 2019). Altogether, these data lead to a model (Figure 3), in which Ku bound to DNA ends acts as a protein block to stimulate MRX-Sae2 cleavage. 3′-5′ Mre11 exonuclease proceeds back toward the DSB end and removes Ku from the DSB. Then, MRX-mediated degradation can proceed by stepwise endonucleolytic incisions, in which one MRX-Sae2 ensemble can act by its own as protein block to stimulate DNA cleavage by another MRX-Sae2 ensemble that is bound at adjacent sites internal to the DSB. The endonucleolytic cuts are followed by 3′-5′ exonucleolytic degradation by Mre11 exonuclease of the short DNA fragments between the incision sites.
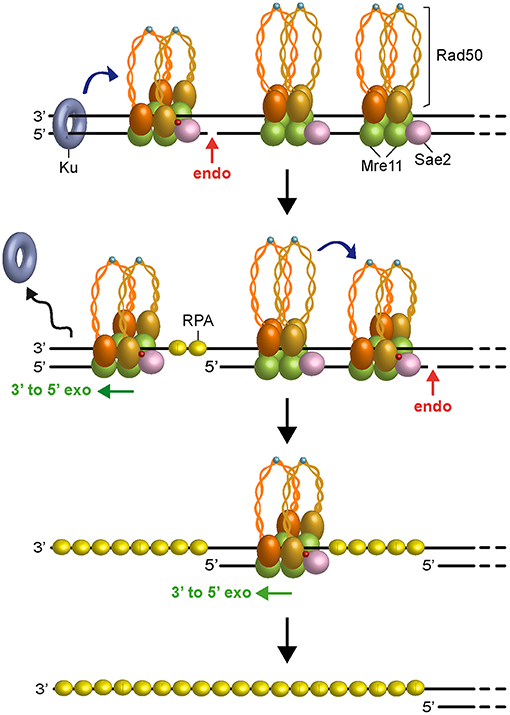
Figure 3. Model for short-range resection. Upon Sae2 phosphorylation and ATP hydrolysis by Rad50, MRX-Sae2 introduces an endonucleolytic cleavage (red arrow) on DNA ends that are bound by Ku, which acts as protein block to stimulate MRX-Sae2 cleavage (blue arrow). Mre11 3′-5′ exonuclease proceeds back toward the DSB end to generate ssDNA that removes Ku from DNA ends. Degradation proceeds by stepwise endonucleolytic incisions, in which one MRX complex can promote (blue arrow) cleavage by another MRX complex that is bound at an adjacent site. The endonucleolytic cleavage is followed by Mre11 3′-5′ exonucleolytic degradation of the DNA fragments between the incision sites. Zn2+ atoms are indicated as light blue dots. Phosphorylation is indicated as red dots.
Rad50 prevents degradation of the 3′-terminated DNA strand by limiting Mre11 exonuclease activity in an ATP-binding-dependent manner, thus explaining why the 3′-5′ exonuclease activity of Mre11 does not resect 3′-terminated strands at DSB sites (Cannavo et al., 2019). By contrast, phosphorylated Sae2 can partially overcome this inhibition by stimulating Mre11 exonuclease when ATP hydrolysis is allowed (Cannavo et al., 2019). However, because phosphorylated Sae2 also promotes the endonuclease of MRX, the exonuclease and endonuclease activities of MRX-Sae2 likely compete with each other.
Long-Range Resection
Long-range resection can be carried out by either of two partially overlapping pathways, dependent on the enzymatic activities of Dna2 and Exo1 nucleases (Mimitou and Symington, 2008; Zhu et al., 2008; Cejka et al., 2010; Nicolette et al., 2010; Niu et al., 2010; Cannavo et al., 2013; Reginato et al., 2017; Wang et al., 2017). Inactivation of a single pathway results in only a minor resection defect, whereas major resection defects are only observed when both pathways are inactivated simultaneously (Mimitou and Symington, 2008; Zhu et al., 2008). While Exo1 is a dsDNA-specific exonuclease capable to degrade 5′-terminated DNA strands within a duplex DNA molecule (Tran et al., 2002), Dna2 is loaded on ssDNA ends and degrades them endonucleolytically, resulting in products of ~5–10 nucleotides in length (Kao et al., 2004). Dna2 resection activity requires an helicase activity that is provided by Sgs1 in yeast and by either BLM or WRN in human cells (Zhu et al., 2008; Sturzenegger et al., 2014; Pinto et al., 2016). In both yeast and mammals, Dna2 was shown to stimulate degradation of long ssDNA molecules by acting as a ssDNA translocase with 5′-3′ polarity (Levikova et al., 2017; Miller et al., 2017). This finding suggests that Sgs1 unwinds DNA in a 3′-5′ direction to provide Dna2 with ssDNA, and Dna2 translocates in a 5′-3′ direction to degrade the unwound 5′-terminated ssDNA strand.
In addition to provide an entry site for Dna2 and Exo1, MRX has also a structural role in promoting their resection activity, thus explaining why the resection defect of mre11Δ cells is more severe than that of sae2Δ or mre11 nuclease defective mutants. Biochemical reconstitution experiments in both yeast and mammals have shown that MRX enhances the ability of Sgs1 to unwind dsDNA independently of Mre11 nuclease, possibly by increasing Sgs1 association to DNA ends (Cejka et al., 2010; Nicolette et al., 2010; Niu et al., 2010; Nimonkar et al., 2011; Cannavo et al., 2013). Furthermore, MRX/MRN enhances both the affinity to DNA ends and the processivity of Exo1 (Cejka et al., 2010; Nicolette et al., 2010; Niu et al., 2010; Nimonkar et al., 2011; Cannavo et al., 2013). Although Exo1 is a processive nuclease in vitro, single-molecule fluorescence imaging has shown that RPA strips Exo1 from DNA (Myler et al., 2016), implying that efficient resection requires multiple cycles of Exo1 rebinding at the same DNA end. Interestingly, MRX was shown to possess a weak ATP-dependent unwinding activity on dsDNA (Paull and Gellert, 1999; Cannon et al., 2013), which was proposed to be dependent on a rotation of the Rad50 nucleotide-binding domains (Liu et al., 2016). The recent identification of the hypermorphic mre11-R10T mutation, which increases Exo1 resection activity, has allowed us to demonstrate that this strand-separation function of MRX is important to stimulate Exo1 resection activity (Gobbini et al., 2018). In fact, molecular dynamic simulations have shown that the capping domains of wild type Mre11 dimer rapidly interact with the DNA ends and cause a partial unwinding of the dsDNA molecule, whereas the mutant Mre11-R10T dimer undergoes an abnormal rotation that leads one of the capping domain to wedge in between the two DNA strands and to persistently melt the dsDNA ends (Gobbini et al., 2018).
Role of MRX in Tel1/ATM Activation
In both yeast and mammals, MRX is necessary for activation of the protein kinase Tel1/ATM (Carson et al., 2003; Uziel et al., 2003; Lee and Paull, 2004), which is a member of a serine/threonine protein kinase family with an N-terminal HEAT repeat domain and C-terminal kinase domain (Ciccia and Elledge, 2010; Gobbini et al., 2013). Mutations in the ATM gene are associated with the human syndrome Ataxia Telangiectasia (AT), whose clinical phenotypes are similar to those of ATLD and include neurodegeneration, sensitivity to IR, immunodeficiency, premature aging, radiosensitivity and predisposition to cancer (Shiloh and Ziv, 2013; Rothblum-Oviatt et al., 2016).
The exact mechanism of Tel1/ATM activation by MRX/MRN is mechanistically poorly understood. Indeed, in both yeast and mammals, MRX is required to recruit Tel1/ATM to DSBs through direct interaction between the N-terminal HEAT domain of Tel1/ATM and the C-terminal domain of the Xrs2/NBS1 subunit (Nakada et al., 2003; Falck et al., 2005; Lee and Paull, 2005; You et al., 2005). In S. cerevisiae, MRX and Tel1 association to DSBs is counteracted by Rif2, whose lack increases the association of MRX to DSBs in a Tel1-dependent manner (Hirano et al., 2009; Cassani et al., 2016). Co-immunoprecipitation experiments have shown that the C terminus of Xrs2 interacts with Rif2. As Tel1 also binds this Xrs2 region, Rif2 can limit Tel1 association to DSBs by interfering with MRX-Tel1 interaction (Hirano et al., 2009). Once Tel1 is recruited to DSBs by MRX, it plays a structural role in stabilizing the association of MRX to the DSB ends in a manner independently of its kinase activity (Cassani et al., 2016). This Tel1-mediated regulation of MRX retention on DNA ends is important to allow proper MRX-DNA binding that is needed for end-tethering and DSB repair (Cassani et al., 2016).
In any case, in vitro activation of human ATM by MRN requires ATP binding but not ATP hydrolysis (Lee et al., 2013), raising the possibility that MRX activates Tel1/ATM when it is present in the ATP-bound state. This hypothesis is supported by the identification of the separation-of-function S. cerevisiae rad50-A78T mutant allele, which specifically abolishes Tel1 activation without impairing MRX functions in DSB repair (Cassani et al., 2019). Molecular dynamics simulations have revealed that the mutant Mre11-Rad50A78T subcomplex bound to ATP undergoes conformational rearrangements similar to those observed when wild type Mre11-Rad50 subcomplex is bound to ADP (Cassani et al., 2019), suggesting that failure of Mre11-Rad50A78T to activate Tel1 is due to the inability of the mutant complex to maintain the closed conformation.
In S. cerevisiae, the lack of Sae2 increases MRX and therefore Tel1 persistence at DSBs (Lisby et al., 2004; Clerici et al., 2006, 2014). mre11-nd cells also exhibit persistent MRX and Tel1 association at DSB ends (Lisby et al., 2004; Yu et al., 2018; Colombo et al., 2019). These findings suggest that MRX-Sae2 processing activity contributes to eliminate MRX bound to DNA ends and this MRX displacement limits Tel1 signaling activity. However, sae2Δ cells, but not mre11-nd cells, exhibit increased accumulation of the Rad9 protein at DSBs and enhanced activity of the Rad53 checkpoint kinase, both of which inhibit the resection activity of Dna2-Sgs1 and Exo1 (Usui et al., 2001; Bonetti et al., 2015; Ferrari et al., 2015; Yu et al., 2018; Colombo et al., 2019). Mutations that decrease either MRX/Rad9 association to DSBs or Rad53/Tel1 signaling restores DNA damage resistance in Sae2-deficient cells (Bonetti et al., 2015; Chen et al., 2015; Ferrari et al., 2015; Gobbini et al., 2015; Puddu et al., 2015; Yu et al., 2018). These findings indicate that Sae2 has an Mre11 nuclease-independent function in resection that counteracts the inhibition that Rad9 and Rad53 exert on Exo1 and Dna2-Sgs1. The identification of the sae2-ms allele, which upregulates MRX and Tel1 signaling activities at DSBs but does not cause increased Rad9 association at DSBs and persistent Rad53 activation, suggests that Sae2 functions in dampening MRX-Tel1 and Rad53 signaling activities can be uncoupled (Colombo et al., 2019). These findings lead to a model whereby Sae2 removes MRX and Tel1 from DNA ends by promoting Mre11 nuclease activity, whereas it limits Rad9 accumulation to DSBs independently of Mre11 nuclease activity. Both these Sae2 functions contribute to downregulate Rad53 activation, with the control of Rad9 association playing the major role in supporting DNA damage resistance and checkpoint activation (Colombo et al., 2019).
DSB Resection in A CHROMATIN Context
DNA is packaged through histone and non-histone proteins into a higher order structure called chromatin, which raises the question as to how DNA end resection occurs in the context of chromatin. Chromatin surrounding DSBs undergoes extensive modification and several highly conserved nucleosome remodelers are recruited to DNA DSBs. While some of them deposit covalent modifications on histone tails to facilitate DNA damage signaling and recruitment of repair factor, others alter chromatin structure either by replacing canonical histones with histone variants or by moving or evicting nucleosomes (Hauer and Gasser, 2017). These latter functions are carried out by proteins that use the energy of ATP hydrolysis to translocate on dsDNA and to disrupt histone-DNA contacts by nucleosome sliding, eviction or histone exchange (Osley et al., 2007).
Chromatin immunoprecipitation experiments support nucleosome disassembly near DSBs in both yeast and human cells (Li and Tyler, 2016; Tsabar et al., 2016), suggesting that nucleosome eviction occurs during resection. A key question is whether nucleosomes are evicted prior to the onset of resection or whether chromatin remodelers help the resection machinery to navigate through chromatin, with nucleosome loss occurring as a consequence of nucleolytic degradation. Genome-wide studies in meiotic cells suggest that MRX-Sae2 catalyzes the endonucleolytic cleavage preferentially on an internucleosomal DNA region at +1 and +2 nucleosomes proximal to meiotic DSB ends (Mimitou et al., 2017). Furthermore, MRX-Sae2 endonucleolytically cleaves the 5′ DNA strand bordering a nucleosome (Wang et al., 2017), thus explaining the ~100-nucleotide incremental cleavages detected at endonuclease-induced DSBs in sgs1Δ exo1Δ cells (Zhu et al., 2008). Thus, if nucleosomes are evicted near a DSB, their removal might occur after Mre11-dependent incision of the 5′-terminated strands. Consistent with a coexistence of both nucleosomes and MRX bound at DSB ends, single-molecule imaging studies have shown that MRX can diffuse along dsDNA even in the presence of nucleosomes (Myler et al., 2017).
Interestingly, by using an in vitro-reconstituted chromatin assay, it has been shown that the presence of nucleosomes impedes resection by both Exo1 and Sgs1-Dna2, with Exo1-dependent resection much more strongly affected (Adkins et al., 2013). This finding suggests that nucleosome destabilization or removal occurs before nucleolytic processing by Exo1, with a constraint on resection length being how many nucleosomes are removed (Mimitou et al., 2017). In any case, removal of H2A-H2B dimers from nucleosomes was shown to enhance Exo1 activity (Adkins et al., 2013). Furthermore, biochemical and genetic evidence reveals that nucleosomes harboring H2AZ, an H2A variant that has been linked to DSB repair, are more accessible to Exo1 (Adkins et al., 2013). These findings suggest that ATP-dependent chromatin-remodeling enzymes promote Exo1-mediated resection in vivo.
Several chromatin remodelers are recruited to chromatin regions adjacent to DSBs and are candidates for nucleosome destabilization during DSB resection (Hauer and Gasser, 2017). Both the RSC and the SWI/SNF complexes appear to promote MRX association to DSBs and subsequent DSB processing by catalyzing eviction or mobilization of nucleosomes adjacent to a DSB (Chai et al., 2005; Shim et al., 2007; Wiest et al., 2017). Also the INO80 complex is recruited to DSBs and participates in eviction of nucleosomes to facilitate Rad51 nucleoprotein filament formation (Morrison et al., 2004; van Attikum et al., 2004, 2007; Tsukuda et al., 2009). Furthermore, two other remodelers have been shown to facilitate long-range resection. Both the SWR-C complex, which replaces the H2A/H2B dimers with H2A.Z in an ATP-dependent manner (Mizuguchi et al., 2004), and the Fun30/SMARCAD1 nucleosome remodeler promote Exo1-mediated degradation (Morillo-Huesca et al., 2010; Chen et al., 2012; Costelloe et al., 2012; Eapen et al., 2012; Adkins et al., 2013). Interestingly, the resection defect of fun30Δ cells is suppressed by elimination of Rad9, suggesting that Fun30 stimulates Exo1 resection activity by alleviating a Rad9-dependent chromatin barrier (Chen et al., 2012; Eapen et al., 2012). Finally, mammalian CHD1, which belongs to the chromodomain helicase DNA-binding CHD family of chromatin remodelers, is recruited to chromatin in response to DSBs in an MRE11-dependent manner and promotes the loading of CtIP onto damaged DNA (Kari et al., 2016).
Conclusions
Work in the last years has advanced our understanding of the structure, biochemical activities, and regulation of the MRX complex. However, we still do not know at the mechanistic level how the functions of Sae2 and Rad50 ATPase integrate to regulate Mre11 nuclease activity, how the endonuclease activity of MRX is targeted locally, or how chromatin structure influence the MRX/Sae2-mediated DNA incision. Given the importance of this protein complex in ensuring genome stability and therefore in preventing carcinogenesis, answering these questions will be strongly relevant to human diseases.
Author Contributions
MPL conceptualized the work. EC, CR, and MPL wrote the manuscript. AM, MG, CVC, and DB revised and edited the manuscript.
Funding
This article was supported by Fondazione AIRC under IG 2017—ID. 19783 project—P.I. MPL, and Progetti di Ricerca di Interesse Nazionale (PRIN) 2015 to MPL (IG Grant 19783). EC was supported by a fellowship from the Italian Ministry of University and Research (MIUR) through grant Dipartimenti di Eccellenza−2017.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the MPL lab for helpful discussions.
References
Adkins, N. L., Niu, H., Sung, P., and Peterson, C. L. (2013). Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 20, 836–842. doi: 10.1038/nsmb.2585
Anand, R., Jasrotia, A., Bundschuh, D., Howard, S. M., Ranjha, L., Stucki, M., et al. (2019). NBS1 promotes the endonuclease activity of the MRE11-RAD50 complex by sensing CtIP phosphorylation. EMBO J. 38:e101005. doi: 10.15252/embj.2018101005
Anand, R., Ranjha, L., Cannavo, E., and Cejka, P. (2016). Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell 64, 940–950. doi: 10.1016/j.molcel.2016.10.017
Andres, S. N., Appel, C. D., Westmoreland, J. W., Williams, J. S., Nguyen, Y., Robertson, P. D., et al. (2015). Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat. Struct. Mol. Biol. 22, 158–166. doi: 10.1038/nsmb.2945
Arya, R., and Bassing, C. H. (2017). V(D)J recombination exploits DNA damage responses to promote immunity. Trends Genet. 33, 479–489. doi: 10.1016/j.tig.2017.04.006
Aylon, Y., Liefshitz, B., and Kupiec, M. (2004). The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23, 4868–4875. doi: 10.1038/sj.emboj.7600469
Baroni, E., Viscardi, V., Cartagena-Lirola, H., Lucchini, G., and Longhese, M. P. (2004). The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Cell. Biol. 24, 4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004
Bergerat, A., de Massy, B., Gadelle, D., Varoutas, P. C., Nicolas, A., and Forterre, P. (1997). An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417. doi: 10.1038/386414a0
Bonetti, D., Clerici, M., Manfrini, N., Lucchini, G., and Longhese, M. P. (2010). The MRX complex plays multiple functions in resection of Yku- and Rif2-protected DNA ends. PLoS ONE 5:e14142. doi: 10.1371/journal.pone.0014142
Bonetti, D., Colombo, C. V., Clerici, M., and Longhese, M. P. (2018). Processing of DNA ends in the maintenance of genome stability. Front. Genet. 9:390. doi: 10.3389/fgene.2018.00390
Bonetti, D., Villa, M., Gobbini, E., Cassani, C., Tedeschi, G., and Longhese, M. P. (2015). Escape of Sgs1 from Rad9 inhibition reduces the requirement for Sae2 and functional MRX in DNA end resection. EMBO Rep. 16, 351–361. doi: 10.15252/embr.201439764
Bressan, D. A., Olivares, H. A., Nelms, B. E., and Petrini, J. H. (1998). Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150, 591–600.
Cannavo, E., and Cejka, P. (2014). Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125. doi: 10.1038/nature13771
Cannavo, E., Cejka, P., and Kowalczykowski, S. C. (2013). Relationship of DNA degradation by Saccharomyces cerevisiae Exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc. Natl. Acad. Sci. U.S.A. 110, E1661–E1668. doi: 10.1073/pnas.1305166110
Cannavo, E., Johnson, D., Andres, S. N., Kissling, V. M., Reinert, J. K., Garcia, V., et al. (2018). Regulatory control of DNA end resection by Sae2 phosphorylation. Nat. Commun. 9:4016. doi: 10.1038/s41467-018-06417-5
Cannavo, E., Reginato, G., and Cejka, P. (2019). Stepwise 5' DNA end-specific resection of DNA breaks by the Mre11-Rad50-Xrs2 and Sae2 nuclease ensemble. Proc. Natl. Acad. Sci. U.S.A. 16, 5505–5513. doi: 10.1073/pnas.1820157116
Cannon, B., Kuhnlein, J., Yang, S. H., Cheng, A., Schindler, D., Stark, J. M., et al. (2013). Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET. Proc. Natl. Acad. Sci. U.S.A. 110, 18868–18873. doi: 10.1073/pnas.1309816110
Carson, C. T., Schwartz, R. A., Stracker, T. H., Lilley, C. E., Lee, D. V., and Weitzman, M. D. (2003). The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22, 6610–6620. doi: 10.1093/emboj/cdg630
Cartagena-Lirola, H., Guerini, I., Viscardi, V., Lucchini, G., and Longhese, M. P. (2006). Budding yeast Sae2 is an in vivo target of the Mec1 and Tel1 checkpoint kinases during meiosis. Cell Cycle 5, 1549–1559. doi: 10.4161/cc.5.14.2916
Cassani, C., Gobbini, E., Wang, W., Niu, H., Clerici, M., Sung, P., et al. (2016). Tel1 and Rif2 regulate MRX functions in end-tethering and repair of DNA double-strand breaks. PLoS Biol. 14:e1002387. doi: 10.1371/journal.pbio.1002387
Cassani, C., Vertemara, J., Bassani, M., Marsella, A., Tisi, R., Zampella, G., et al. (2019). The ATP-bound conformation of the Mre11-Rad50 complex is essential for Tel1/ATM activation. Nucleic Acids Res. 47, 3550–3567. doi: 10.1093/nar/gkz038
Cejka, P., Cannavo, E., Polaczek, P., Masuda-Sasa, T., Pokharel, S., Campbell, J. L., et al. (2010). DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116. doi: 10.1038/nature09355
Chai, B., Huang, J., Cairns, B. R., and Laurent, B. C. (2005). Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19, 1656–1661. doi: 10.1101/gad.1273105
Chang, H. H. Y., Pannunzio, N. R., Adachi, N., and Lieber, M. R. (2017). Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell. Biol. 18, 495–506. doi: 10.1038/nrm.2017.48
Chanut, P., Britton, S., Coates, J., Jackson, S. P., and Calsou, P. (2016). Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat. Commun. 7:12889. doi: 10.1038/ncomms12889
Chen, H., Donnianni, R. A., Handa, N., Deng, S. K., Oh, J., Timashev, L. A., et al. (2015). Sae2 promotes DNA damage resistance by removing the Mre11–Rad50–Xrs2 complex from DNA and attenuating Rad53 signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 1880–1887. doi: 10.1073/pnas.1503331112
Chen, X., Cui, D., Papusha, A., Zhang, X., Chu, C. D., Tang, J., et al. (2012). The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489, 576–580. doi: 10.1038/nature11355
Chen, X., Niu, H., Chung, W. H., Zhu, Z., Papusha, A., Shim, E. Y., et al. (2011). Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat. Struct. Mol. Biol. 18, 1015–1019. doi: 10.1038/nsmb.2105
Ciccia, A., and Elledge, S. J. (2010). The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204. doi: 10.1016/j.molcel.2010.09.019
Clerici, M., Mantiero, D., Guerini, I., Lucchini, G., and Longhese, M. P. (2008). The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 9, 810–818. doi: 10.1038/embor.2008.121
Clerici, M., Mantiero, D., Lucchini, G., and Longhese, M. P. (2006). The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 7, 212–218. doi: 10.1038/sj.embor.7400593
Clerici, M., Trovesi, C., Galbiati, A., Lucchini, G., and Longhese, M. P. (2014). Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J. 33, 198–216. doi: 10.1002/embj.201386041
Colombo, C. V., Menin, L., Ranieri, R., Bonetti, D., Clerici, M., and Longhese, M. P. (2019). Uncoupling Sae2 functions in downregulation of Tel1 and Rad53 signaling activities. Genetics 211, 515–530. doi: 10.1534/genetics.118.301830
Costelloe, T., Louge, R., Tomimatsu, N., Mukherjee, B., Martini, E., Khadaroo, B., et al. (2012). The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489, 581–584. doi: 10.1038/nature11353
Davies, O. R., Forment, J. V., Sun, M., Belotserkovskaya, R., Coates, J., Galanty, Y., et al. (2015). CtIP tetramer assembly is required for DNA-end resection and repair. Nat. Struct. Mol. Biol. 22, 150–157. doi: 10.1038/nsmb.2937
de Jager, M., van Noort, J., van Gent, D. C., Dekker, C., Kanaar, R., and Wyman, C. (2001). Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135. doi: 10.1016/S1097-2765(01)00381-1
Desai-Mehta, A., Cerosaletti, K. M., and Concannon, P. (2001). Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol. Cell. Biol. 21, 2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001
Deshpande, R. A., Lee, J. H., Arora, S., and Paull, T. T. (2016). Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol. Cell 64, 593–606. doi: 10.1016/j.molcel.2016.10.010
Deshpande, R. A., Lee, J. H., and Paull, T. T. (2017). Rad50 ATPase activity is regulated by DNA ends and requires coordination of both active sites. Nucleic Acids Res. 45, 5255–5268. doi: 10.1093/nar/gkx173
Deshpande, R. A., Williams, G. J., Limbo, O., Williams, R. S., Kuhnlein, J., Lee, J. H., et al. (2014). ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 33, 482–500. doi: 10.1002/embj.201386100
Eapen, V. V., Sugawara, N., Tsabar, M., Wu, W. H., and Haber, J. E. (2012). The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol. Cell. Biol. 32, 4727–4740. doi: 10.1128/MCB.00566-12
Falck, J., Coates, J., and Jackson, S. P. (2005). Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611. doi: 10.1038/nature03442
Ferrari, M., Dibitetto, D., De Gregorio, G., Eapen, V. V., Rawal, C. C., Lazzaro, F., et al. (2015). Functional interplay between the 53BP1-ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS Genetics 11:e1004928. doi: 10.1371/journal.pgen.1004928
Foster, S. S., Balestrini, A., and Petrini, J. H. J. (2011). Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol. Cell. Biol. 31, 4379–4389. doi: 10.1128/MCB.05854-11
Fu, Q., Chow, J., Bernstein, K. A., Makharashvili, N., Arora, S., Lee, C. F., et al. (2014). Phosphorylation-regulated transitions in an oligomeric state control the activity of the Sae2 DNA repair enzyme. Mol. Cell. Biol. 34, 778–793. doi: 10.1128/MCB.00963-13
Garcia, V., Phelps, S. E. L., Gray, S., and Neale, M. J. (2011). Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479, 241–244. doi: 10.1038/nature10515
Gobbini, E., Cassani, C., Vertemara, J., Wang, W., Mambretti, F., Casari, E., et al. (2018). The MRX complex regulates Exo1 resection activity by altering DNA end structure. EMBO J. 37:e98588. doi: 10.15252/embj.201798588
Gobbini, E., Cesena, D., Galbiati, A., Lockhart, A., and Longhese, M. P. (2013). Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair 12, 791–799. doi: 10.1016/j.dnarep.2013.07.009
Gobbini, E., Villa, M., Gnugnoli, M., Menin, L., Clerici, M., and Longhese, M. P. (2015). Sae2 function at DNA double-strand breaks is bypassed by dampening Tel1 or Rad53 activity. PLoS Genetics 11:e1005685. doi: 10.1371/journal.pgen.1005685
Griffith, A. J., Blier, P. R., Mimori, T., and Hardin, J. A. (1992). Ku polypeptides synthesized in vitro assemble into complexes which recognize ends of double-stranded DNA. J. Biol. Chem. 267, 331–338.
Hauer, M. H., and Gasser, S. M. (2017). Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 31, 2204–2221. doi: 10.1101/gad.307702.117
He, J., Shi, L. Z., Truong, L. N., Lu, C. S., Razavian, N., Li, Y., et al. (2012). Rad50 zinc hook is important for the Mre11 complex to bind chromosomal DNA double-stranded breaks and initiate various DNA damage responses. J. Biol. Chem. 287, 31747–31756. doi: 10.1074/jbc.M112.384750
Herdendorf, T. J., Albrecht, D. W., Benkovic, S. J., and Nelson, S. W. (2011). Biochemical characterization of bacteriophage T4 Mre11-Rad50 complex. J. Biol. Chem. 286, 2382–2392. doi: 10.1074/jbc.M110.178871
Hirano, Y., Fukunaga, K., and Sugimoto, K. (2009). Rif1 and Rif2 inhibit localization of Tel1 to DNA ends. Mol. Cell 33, 312–322. doi: 10.1016/j.molcel.2008.12.027
Hoa, N. N., Shimizu, T., Zhou, Z. W., Wang, Z. Q., Deshpande, R. A., Paull, T. T., et al. (2016). Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell 64, 580–592. doi: 10.1016/j.molcel.2016.10.011
Hohl, M., Kwon, Y., Galván, S. M., Xue, X., Tous, C., Aguilera, A., et al. (2011). The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 18, 1124–1131. doi: 10.1038/nsmb.2116
Hopfner, K. P., Craig, L., Moncalian, G., Zinkel, R. A., Usui, T., Owen, B. A., et al. (2002). The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418, 562–566. doi: 10.1038/nature00922
Hopfner, K. P., Karcher, A., Craig, L., Woo, T. T., Carney, J. P., and Tainer, J. A. (2001). Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105, 473–485. doi: 10.1016/S0092-8674(01)00335-X
Hopfner, K. P., Karcher, A., Shin, D. S., Craig, L., Arthur, L. M., Carney, J. P., et al. (2000). Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800. doi: 10.1016/S0092-8674(00)80890-9
Huertas, P., Cortés-Ledesma, F., Sartori, A. A., Aguilera, A., and Jackson, S. P. (2008). CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692. doi: 10.1038/nature07215
Huertas, P., and Jackson, S. P. (2009). Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565. doi: 10.1074/jbc.M808906200
Ira, G., Pellicioli, A., Balijja, A., Wang, X., Fiorani, S., Carotenuto, W., et al. (2004). DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431, 1011–1017. doi: 10.1038/nature02964
Jackson, S. P., and Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078. doi: 10.1038/nature08467
Kao, H. I., Campbell, J. L., and Bambara, R. A. (2004). Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 279, 50840–55089. doi: 10.1074/jbc.M409231200
Kari, V., Mansour, W. Y., Raul, S. K., Baumgart, S. J., Mund, A., Grade, M., et al. (2016). Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 17, 1609–1623. doi: 10.15252/embr.201642352
Kaye, J. A., Melo, J. A., Cheung, S. K., Vaze, M. B., Haber, J. E., and Toczyski, D. P. (2004). DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14, 2096–2106. doi: 10.1016/j.cub.2004.10.051
Keeney, S., Giroux, C. N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. doi: 10.1016/S0092-8674(00)81876-0
Kim, H. S., Vijayakumar, S., Reger, M., Harrison, J. C., Haber, J. E., Weil, C., et al. (2008). Functional interactions between Sae2 and the Mre11 complex. Genetics 178, 711–723. doi: 10.1534/genetics.107.081331
Kowalczykowski, S. C. (2015). An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7:a016410. doi: 10.1101/cshperspect.a016410
Lam, I., and Keeney, S. (2014). Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 7:a016634. doi: 10.1101/cshperspect.a016634
Lammens, K., Bemeleit, D. J., Möckel, C., Clausing, E., Schele, A., Hartung, S., et al. (2011). The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145, 54–66. doi: 10.1016/j.cell.2011.02.038
Langerak, P., Mejia-Ramirez, E., Limbo, O., and Russell, P. (2011). Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7:e1002271. doi: 10.1371/journal.pgen.1002271
Lee, J. H., Mand, M. R., Deshpande, R. A., Kinoshita, E., Yang, S. H., Wyman, C., et al. (2013). Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J. Biol. Chem. 288, 12840–12851. doi: 10.1074/jbc.M113.460378
Lee, J. H., and Paull, T. T. (2004). Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304, 93–96. doi: 10.1126/science.1091496
Lee, J. H., and Paull, T. T. (2005). ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308, 551–554. doi: 10.1126/science.1108297
Levikova, M., Pinto, C., and Cejka, P. (2017). The motor activity of DNA2 functions as an ssDNA translocase to promote DNA end resection. Genes Dev. 31, 493–502. doi: 10.1101/gad.295196.116
Levy, D. L., and Blackburn, E. H. (2004). Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol. Cell. Biol. 24, 10857–10867. doi: 10.1128/MCB.24.24.10857-10867.2004
Li, X., and Tyler, J. K. (2016). Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. Elife 5:e15129. doi: 10.7554/eLife.15129
Lim, H. S., Kim, J. S., Park, Y. B., Gwon, G. H., and Cho, Y. (2011). Crystal structure of the Mre11-Rad50-ATPγS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 25, 1091–1104. doi: 10.1101/gad.2037811
Lisby, M., Barlow, J. H., Burgess, R. C., and Rothstein, R. (2004). Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713. doi: 10.1016/j.cell.2004.08.015
Liu, P., Carvalho, C. M., Hastings, P. J., and Lupski, J. R. (2012). Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 22, 211–220. doi: 10.1016/j.gde.2012.02.012
Liu, Y., Sung, S., Kim, Y., Li, F., Gwon, G., Jo, A., et al. (2016). ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 35, 743–758. doi: 10.15252/embj.201592462
Llorente, B., and Symington, L. S. (2004). The Mre11 nuclease is not required for 5' to 3' resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 24, 9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004
Lobachev, K., Vitriol, E., Stemple, J., Resnick, M. A., and Bloom, K. (2004). Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 14, 2107–2112. doi: 10.1016/j.cub.2004.11.051
Lobachev, K. S., Gordenin, D. A., and Resnick, M. A. (2002). The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108, 183–193. doi: 10.1016/S0092-8674(02)00614-1
Majka, J., Alford, B., Ausio, J., Finn, R. M., and McMurray, C. T. (2012). ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. J. Biol. Chem. 287, 2328–2341. doi: 10.1074/jbc.M111.307041
Manfrini, N., Guerini, I., Citterio, A., Lucchini, G., and Longhese, M. P. (2010). Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases. J. Biol. Chem. 285, 11628–11637. doi: 10.1074/jbc.M110.104083
Martina, M., Clerici, M., Baldo, V., Bonetti, D., Lucchini, G., and Longhese, M. P. (2012). A balance between Tel1 and Rif2 activities regulates nucleolytic processing and elongation at telomeres. Mol. Cell. Biol. 32, 1604–1617. doi: 10.1128/MCB.06547-11
Mehta, A., and Haber, J. E. (2014). Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6:a016428. doi: 10.1101/cshperspect.a016428
Miller, A. S., Daley, J. M., Pham, N. T., Niu, H., Xue, X., Ira, G., et al. (2017). A novel role of the Dna2 translocase function in DNA break resection. Genes Dev. 31, 503–510. doi: 10.1101/gad.295659.116
Mimitou, E. P., and Symington, L. S. (2008). Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774. doi: 10.1038/nature07312
Mimitou, E. P., and Symington, L. S. (2010). Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29, 3358–3369. doi: 10.1038/emboj.2010.193
Mimitou, E. P., Yamada, S., and Keeney, S. (2017). A global view of meiotic double-strand break end resection. Science 355, 40–45. doi: 10.1126/science.aak9704
Mizuguchi, G., Shen, X., Landry, J., Wu, W. H., Sen, S., and Wu, C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. doi: 10.1126/science.1090701
Möckel, C., Lammens, K., Schele, A., and Hopfner, K. P. (2012). ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 40, 914–927. doi: 10.1093/nar/gkr749
Moncalian, G., Lengsfeld, B., Bhaskara, V., Hopfner, K. P., Karcher, A., Alden, E., et al. (2004). The Rad50 signature motif: essential to ATP binding and biological function. J. Mol. Biol. 335, 937–951. doi: 10.1016/j.jmb.2003.11.026
Morillo-Huesca, M., Clemente-Ruiz, M., Andújar, E., and Prado, F. (2010). The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS ONE 5:e12143. doi: 10.1371/journal.pone.0012143
Morrison, A. J., Highland, J., Krogan, N. J., Arbel-Eden, A., Greenblatt, J. F., Haber, J. E., et al. (2004). INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119, 767–775. doi: 10.1016/j.cell.2004.11.037
Myler, L. R., Gallardo, I. F., Soniat, M. M., Deshpande, R. A., Gonzalez, X. B., Kim, Y., et al. (2017). Single-molecule imaging reveals how Mre11-Rad50-Nbs1 initiates DNA break repair. Mol. Cell 67, 891–898.e4. doi: 10.1016/j.molcel.2017.08.002
Myler, L. R., Gallardo, I. F., Zhou, Y., Gong, F., Yang, S. H., Wold, M. S., et al. (2016). Single-molecule imaging reveals the mechanism of Exo1 regulation by single-stranded DNA binding proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E1170–E1179. doi: 10.1073/pnas.1516674113
Nakada, D., Matsumoto, K., and Sugimoto, K. (2003). ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17, 1957–1962. doi: 10.1101/gad.1099003
Nakai, W., Westmoreland, J., Yeh, E., Bloom, K., and Resnick, M. A. (2011). Chromosome integrity at a double-strand break requires exonuclease 1 and MRX. DNA Repair 10, 102–110. doi: 10.1016/j.dnarep.2010.10.004
Neale, M. J., Pan, J., and Keeney, S. (2005). Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057. doi: 10.1038/nature03872
Nicolette, M. L., Lee, K., Guo, Z., Rani, M., Chow, J. M., Lee, S. E., et al. (2010). Mre11-Rad50-Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 17, 1478–1485. doi: 10.1038/nsmb.1957
Nimonkar, A. V., Genschel, J., Kinoshita, E., Polaczek, P., Campbell, J. L., Wyman, C., et al. (2011). BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362. doi: 10.1101/gad.2003811
Niu, H., Chung, W. H., Zhu, Z., Kwon, Y., Zhao, W., Chi, P., et al. (2010). Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–111. doi: 10.1038/nature09318
O'Driscoll, M. (2012). Diseases associated with defective responses to DNA damage. Cold Spring Harb. Perspect. Biol. 4:a012773. doi: 10.1101/cshperspect.a012773
Oh, J., Al-Zain, A., Cannavo, E., Cejka, P., and Symington, L. S. (2016). Xrs2 dependent and independent functions of the Mre11-Rad50 complex. Mol. Cell 64, 405–415. doi: 10.1016/j.molcel.2016.09.011
Osley, M. A., Tsukuda, T., and Nickoloff, J. A. (2007). ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res. 618, 65–80. doi: 10.1016/j.mrfmmm.2006.07.011
Park, Y. B., Hohl, M., Padjasek, M., Jeong, E., Jin, K. S., Krezel, A., et al. (2017). Eukaryotic Rad50 functions as a rod-shaped dimer. Nat. Struct. Mol. Biol. 24, 248–257. doi: 10.1038/nsmb.3369
Paull, T. T., and Gellert, M. (1998). The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979. doi: 10.1016/S1097-2765(00)80097-0
Paull, T. T., and Gellert, M. (1999). Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13, 1276–1288. doi: 10.1101/gad.13.10.1276
Pinto, C., Kasaciunaite, K., Seidel, R., and Cejka, P. (2016). Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases. Elife 5:e18574. doi: 10.7554/eLife.18574
Puddu, F., Oelschlaegel, T., Guerini, I., Geisler, N. J., Niu, H., Herzog, M., et al. (2015). Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J. 34, 1509–1522. doi: 10.15252/embj.201590973
Reginato, G., Cannavo, E., and Cejka, P. (2017). Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 31, 2325–2330. doi: 10.1101/gad.308254.117
Rothblum-Oviatt, C., Wright, J., Lefton-Greif, M. A., McGrath-Morrow, S. A., Crawford, T. O., and Lederman, H. M. (2016). Ataxia telangiectasia: a review. Orphanet J. Rare Dis. 11:159. doi: 10.1186/s13023-016-0543-7
Saathoff, J. H., Käshammer, L., Lammens, K., Byrne, R. T., and Hopfner, K. P. (2018). The bacterial Mre11-Rad50 homolog SbcCD cleaves opposing strands of DNA by two chemically distinct nuclease reactions. Nucleic Acids Res. 46, 11303–11314. doi: 10.1093/nar/gky878
Seifert, F. U., Lammens, K., Stoehr, G., Kessler, B., and Hopfner, K. P. (2016). Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 35, 759–772. doi: 10.15252/embj.201592934
Shibata, A., Moiani, D., Arvai, A. S., Perry, J., Harding, S. M., Genois, M. M., et al. (2014). DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18. doi: 10.1016/j.molcel.2013.11.003
Shiloh, Y., and Ziv, Y. (2013). The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell. Biol. 14, 197–210. doi: 10.1038/nrm3546
Shim, E. Y., Chung, W. H., Nicolette, M. L., Zhang, Y., Davis, M., and Zhu, Z. (2010). Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380. doi: 10.1038/emboj.2010.219
Shim, E. Y., Hong, S. J., Oum, J. H., Yanez, Y., Zhang, Y., and Lee, S. E. (2007). RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 27, 1602–1613. doi: 10.1128/MCB.01956-06
Sturzenegger, A., Burdova, K., Kanagaraj, R., Levikova, M., Pinto, C., Cejka, P., et al. (2014). DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 289, 27314–27326. doi: 10.1074/jbc.M114.578823
Syed, A., and Tainer, J. A. (2018). The MRE11–RAD50–NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 87, 263–294. doi: 10.1146/annurev-biochem-062917-012415
Tran, P. T., Erdeniz, N., Dudley, S., and Liskay, R. M. (2002). Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair 1, 895–912. doi: 10.1016/S1568-7864(02)00114-3
Trujillo, K. M., and Sung, P. (2001). DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276, 35458–35464. doi: 10.1074/jbc.M105482200
Trujillo, K. M., Yuan, S. S., Lee, E. Y., and Sung, P. (1998). Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273, 21447–21450. doi: 10.1074/jbc.273.34.21447
Tsabar, M., Hicks, W. M., Tsaponina, O., and Haber, J. E. (2016). Re-establishment of nucleosome occupancy during double-strand break repair in budding yeast. DNA Repair 47, 21–29. doi: 10.1016/j.dnarep.2016.09.005
Tsukamoto, Y., Mitsuoka, C., Terasawa, M., Ogawa, H., and Ogawa, T. (2005). Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol. Biol. Cell. 16, 597–608. doi: 10.1091/mbc.e04-09-0782
Tsukuda, T., Lo, Y. C., Krishna, S., Sterk, R., Osley, M. A., and Nickoloff, J. A. (2009). INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair 8, 360–369. doi: 10.1016/j.dnarep.2008.11.014
Usui, T., Ogawa, H., and Petrini, J. H. J. (2001). A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7, 1255–1266. doi: 10.1016/S1097-2765(01)00270-2
Usui, T., Ohta, T., Oshiumi, H., Tomizawa, J., Ogawa, H., and Ogawa, T. (1998). Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95, 705–716. doi: 10.1016/S0092-8674(00)81640-2
Uziel, T., Lerenthal, Y., Moyal, L., Andegeko, Y., Mittelman, L., and Shiloh, Y. (2003). Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22, 5612–5621. doi: 10.1093/emboj/cdg541
van Attikum, H., Fritsch, O., and Gasser, S. M. (2007). Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113–4125. doi: 10.1038/sj.emboj.7601835
van Attikum, H., Fritsch, O., Hohn, B., and Gasser, S. M. (2004). Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788. doi: 10.1016/j.cell.2004.11.033
Villa, M., Cassani, C., Gobbini, E., Bonetti, D., and Longhese, M. P. (2016). Coupling end resection with the checkpoint response at DNA double-strand breaks. Cell. Mol. Life Sci. 73, 3655–3663. doi: 10.1007/s00018-016-2262-6
Wang, W., Daley, J. M., Kwon, Y., Krasner, D. S., and Sung, P. (2017). Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 31, 2331–2336. doi: 10.1101/gad.307900.117
Wiest, N. E., Houghtaling, S., Sanchez, J. C., Tomkinson, A. E., and Osley, M. A. (2017). The SWI/SNF ATP-dependent nucleosome remodeler promotes resection initiation at a DNA double-strand break in yeast. Nucleic Acids Res. 45, 5887–5900. doi: 10.1093/nar/gkx221
Williams, G. J., Williams, R. S., Williams, J. S., Moncalian, G., Arvai, A. S., Limbo, O., et al. (2011). ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 18, 423–431. doi: 10.1038/nsmb.2038
Williams, R. S., Moncalian, G., Williams, J. S., Yamada, Y., Limbo, O., Shin, D. S., et al. (2008). Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135, 97–109. doi: 10.1016/j.cell.2008.08.017
Wiltzius, J. J., Hohl, M., Fleming, J. C., and Petrini, J. H. (2005). The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12, 403–407. doi: 10.1038/nsmb928
Wotton, D., and Shore, D. (1997). A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748–760. doi: 10.1101/gad.11.6.748
You, Z., Chahwan, C., Bailis, J., Hunter, T., and Russell, P. (2005). ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25, 5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005
Yu, T. Y., Kimble, M. T., and Symington, L. S. (2018). Sae2 antagonizes Rad9 accumulation at DNA double-strand breaks to attenuate checkpoint signaling and facilitate end resection. Proc. Natl. Acad. Sci. U.S.A. 115, E11961–E11969. doi: 10.1073/pnas.1816539115
Zhu, Z., Chung, W. H., Shim, E. Y., Lee, S. E., and Ira, G. (2008). Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994. doi: 10.1016/j.cell.2008.08.037
Keywords: Mre11, Rad50, Xrs2/NBS1, Sae2/CtIP, Tel1/ATM, MRX complex, double-strand break, resection
Citation: Casari E, Rinaldi C, Marsella A, Gnugnoli M, Colombo CV, Bonetti D and Longhese MP (2019) Processing of DNA Double-Strand Breaks by the MRX Complex in a Chromatin Context. Front. Mol. Biosci. 6:43. doi: 10.3389/fmolb.2019.00043
Received: 22 March 2019; Accepted: 21 May 2019;
Published: 07 June 2019.
Edited by:
Pablo Huertas, Centro Andaluz de Biología Molecular y Medicina Regenerativa (CABIMER), SpainReviewed by:
Tanya Paull, University of Texas at Austin, United StatesRichard Chahwan, University of Zurich, Switzerland
Copyright © 2019 Casari, Rinaldi, Marsella, Gnugnoli, Colombo, Bonetti and Longhese. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Pia Longhese, bWFyaWFwaWEubG9uZ2hlc2VAdW5pbWliLml0
†These authors have contributed equally to this work
 Erika Casari†
Erika Casari† Diego Bonetti
Diego Bonetti Maria Pia Longhese
Maria Pia Longhese