- Cancer Immunology and Immunotherapy Research Unit, Cancer Research Malaysia, Subang Jaya, Malaysia
With the regulatory approval of Provenge and Talimogene laherparepvec (T-VEC) for the treatment of metastatic prostate cancer and advanced melanoma respectively, and other promising clinical trials outcomes, cancer vaccine is gaining prominence as a cancer therapeutic agent. Cancer vaccine works to induce T cell priming, expansion, and infiltration resulting in antigen-specific cytotoxicity. Such an approach that can drive cytotoxicity within the tumor could complement the success of checkpoint inhibitors as tumors shown to have high immune cell infiltration are those that would respond well to these antibodies. With the advancements in cancer vaccine, methods to monitor and understand how cancer vaccines modify the immune milieu is under rapid development. This includes using ELISpot and intracellular staining to detect cytokine secretion by activated T cells; tetramer and CyTOF to quantitate the level of antigen specific T cells; proliferation and cell killing assay to detect the expansion of T cell and specific killing activity. More recently, T cell profiling has provided unprecedented detail on immune cell subsets and providing clues to the mechanism involved in immune activation. Here, we reviewed cancer vaccines currently in clinical trials and highlight available techniques in monitoring the clinical response in patients.
Introduction
Vaccination against infectious diseases such as smallpox, polio, mumps and measles have markedly reduced mortality rates from these diseases and have been adopted as a standard practice across the world. The idea of cancer vaccination was first discovered in late 1800, when Dr. William Coley demonstrated better patient outcomes after the administration of Coley toxin. However, due to the absence of good manufacturing protocol, this approach yielded varied outcomes as different institutions used different formulations. Notably, the mechanism of action of Coley toxin was not fully understood, as the knowledge underlying immune activation was limited at that time. Radiation therapy was introduced at the same time and this demonstrated clear and consistent results in all patients, hence the development of Coley toxin was neglected. Only until recently, to further improve the success of immunotherapy in the form of checkpoint inhibitors, the interest in using vaccination to treat cancer is reinvigorated. This is mainly due to the ability of vaccines to stimulate the proliferation of antigen specific T cells, the main effector cells in fighting and killing cancer cells. Generally, two approaches have been developed, the first, is to use attenuated cancer cells or unique antigens (in the form of neoantigen, mRNA, DNA, peptide, or dendritic cell) expressed by cancer cells to stimulate immune responses to recognize cancer cells; the second approach is to generate immune response in situ using oncolytic viruses (either virus alone or virus-expressing cancer antigens) that selectively replicate in tumor tissue, which will then enhance antigen presentation and different immune responses (Conry et al., 2018).
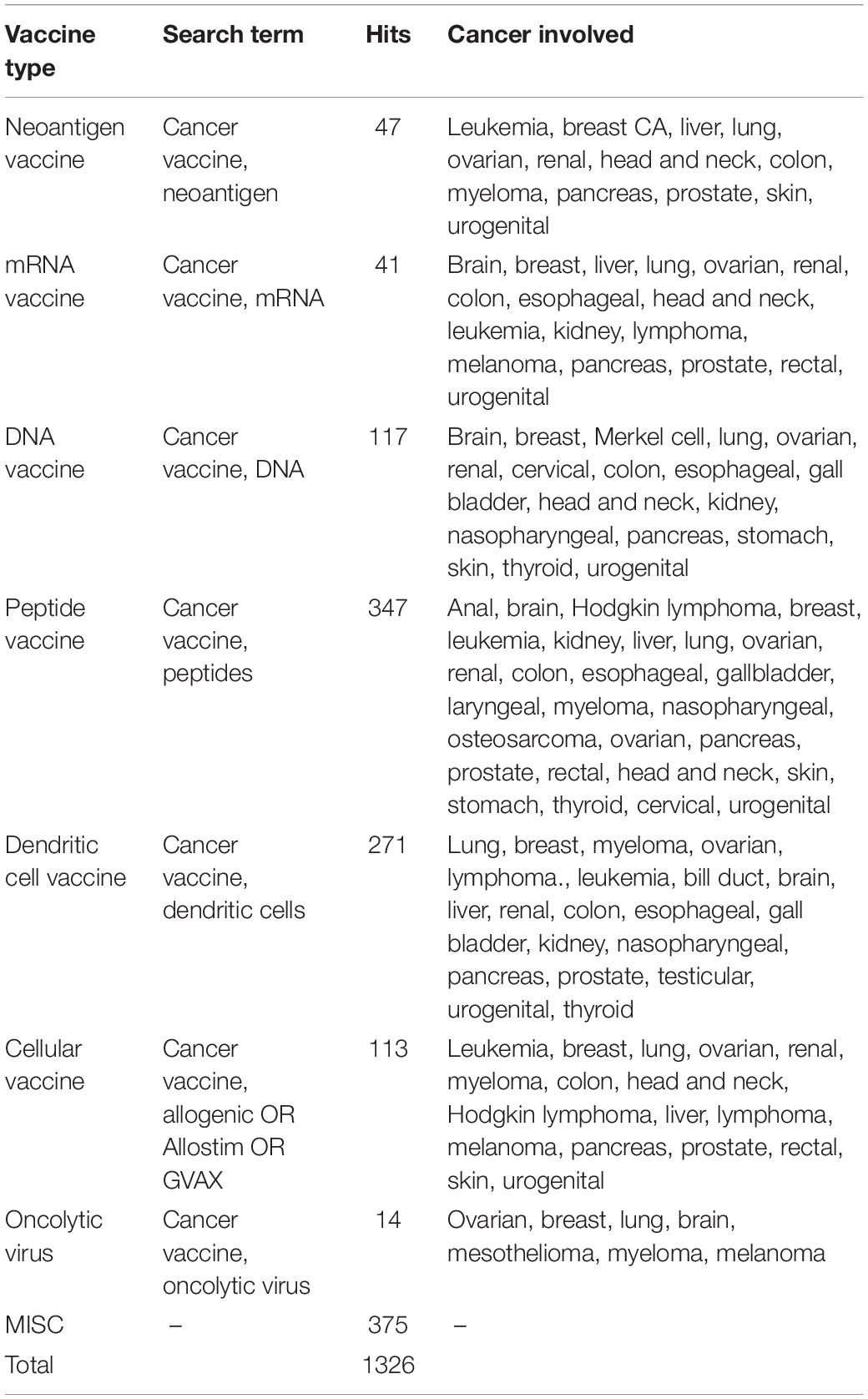
Cancer vaccine is predicted to be one of the fastest growing areas in immunotherapy development worldwide. To investigate the up-to-date development in the cancer vaccine field, we have conducted a thorough search using the ClinicalTrials.gov database. Using “cancer” as disease and “cancer vaccine” as other terms, yielding 1326 interventional studies after excluding studies that are suspended, terminated, withdrawn, with unknown status or involved children. From these 1326 studies, we further sub-classified cancer vaccine into seven types according to the format of the cancer vaccine at the time of administration, (1) neoantigen, (2) mRNA, (3) DNA, (4) peptide, (5) dendritic cell, (6) cellular vaccines, and (7) oncolytic virus (Table 1). Notably, cancer vaccines in the form of peptide and dendritic cell have the highest number of on-going or completed trials. We also noted some overlapped due to the similarity of the format of the vaccine, for example, neoantigen vaccine can also be classified as peptide vaccine as the mutated epitope was actually a string of amino acids. We focus on the newly published clinical trials on cancer vaccines and their respective immune monitoring approaches in subsequent sections.
The Landscape of Cancer Vaccines
DNA/RNA/Peptide-Based Vaccine
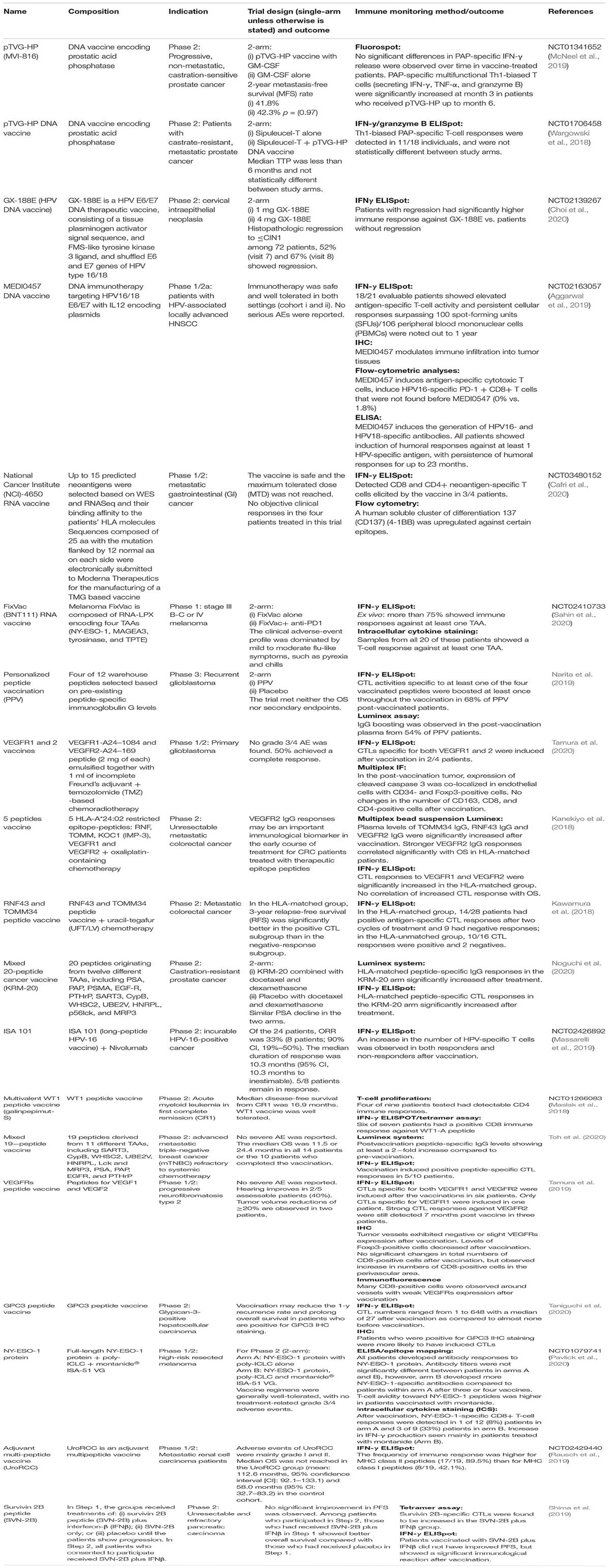
DNA/RNA/peptide-based vaccines are different vaccine formats of how antigens are presented or delivered into the body to elicit antigen-specific immune responses. All of which have their advantages and disadvantages; DNA is stable and can be easily manipulated (codon-optimization, adding on sequences that could code for immune adjuvants, etc.) and produced. Whilst RNA is similar to DNA, it is less stable and requires a cold-chain for transportation. However, both DNA and RNA have limited delivery efficacy (Jou et al., 2021). Peptide vaccines are peptides of 9–25 amino acid in length designed based on the prediction of specific regions of an antigen that can bind to MHC-I and II molecules. As compared to DNA and RNA vaccine, peptide vaccine is safer as it does not contain any extraneous regions like promoter or antibiotic resistance regions in the expression vector but the usage is often limited to patients with specific MHC subtypes. Table 2 presents an overview of the DNA/RNA/peptide vaccines that have resulted in a publication from 2018 to 2020.
The use of DNA vaccine encoding HPV16/18 in cancer prevention has remarkably caused more than 50% of patients with cervical intraepithelial neoplasia to experience lesion regression (Choi et al., 2020). Subsequently, this vaccine is also being evaluated as a therapeutic vaccine in HPV positive head and neck cancer and was shown to elicit strong T cells and B cells responses in majority of patients (Aggarwal et al., 2019). DNA Vaccine Encoding Prostatic Acid Phosphatase (pTVG-HP) has been tested in both castration-sensitive and castration-resistant prostate cancer. However, it did not demonstrate immunological and clinical benefits (Scurr et al., 2017; Wargowski et al., 2018). Recently, promising clinical response was observed in melanoma patients who failed anti-PD1 treatment by using the of RNA-lipoplex vaccine (targeting four tumor associated antigens (TAA) namely NY-ESO-1, MAGEA3, tyrosinase and TPTE; FixVac) (Sahin et al., 2020). This encouraging result has provided hope for patients who failed checkpoint immunotherapy. In addition to TAA, neoepitopes resulting from cancer-specific mutation are also a preferred target for cancer vaccine as the mutation is not present during the selection in the thymus and thus exempt from central tolerance (Wickström et al., 2019). The use of mRNA vaccine encoding up to 15 neoepitopes was evaluated in patients with metastatic gastrointestinal cancer where positive mutation-specific T cell responses were detected in patients after vaccination (Cafri et al., 2020). However, the use of neoantigen-specific vaccine that requires routine next-generation sequencing and the need for timely vaccine design and manufacture pipeline has hindered this from wide-scale implementation.
The use of MAGE-A3 peptide vaccine as cancer therapeutic agent has resulted in a big disappointment as no significant clinical benefit was observed in two Phase III clinical trials (MAGRIT and DERMA) for patients with non-small cell lung cancer and melanoma (Vansteenkiste et al., 2016; Dreno et al., 2018). To circumvent the lack of response to peptide vaccine, new studies are designed to target more than one antigen to avoid the risk for tumor escape due to antigen loss and to combine these vaccines with other approved therapies, for example, chemotherapy and checkpoint blockade inhibitors (Kanekiyo et al., 2018; Kawamura et al., 2018; Noguchi et al., 2020; Tamura et al., 2020). The use of RNF43/TOMM3 peptide vaccine in combination with chemotherapy in metastatic colorectal cancer has demonstrated significantly better 3-year relapse-free survival in patients who have higher T cell responses compared to the negative response subgroup (Kawamura et al., 2018). The use of HPV 16 synthetic long peptide in combination with nivolumab resulted in an overall response rate of 33% and median overall survival of 17.5 months in patients with oropharyngeal carcinoma, which is better than PD-1 inhibition alone in a similar patient cohort (Massarelli et al., 2019).
Cellular-Based Vaccine
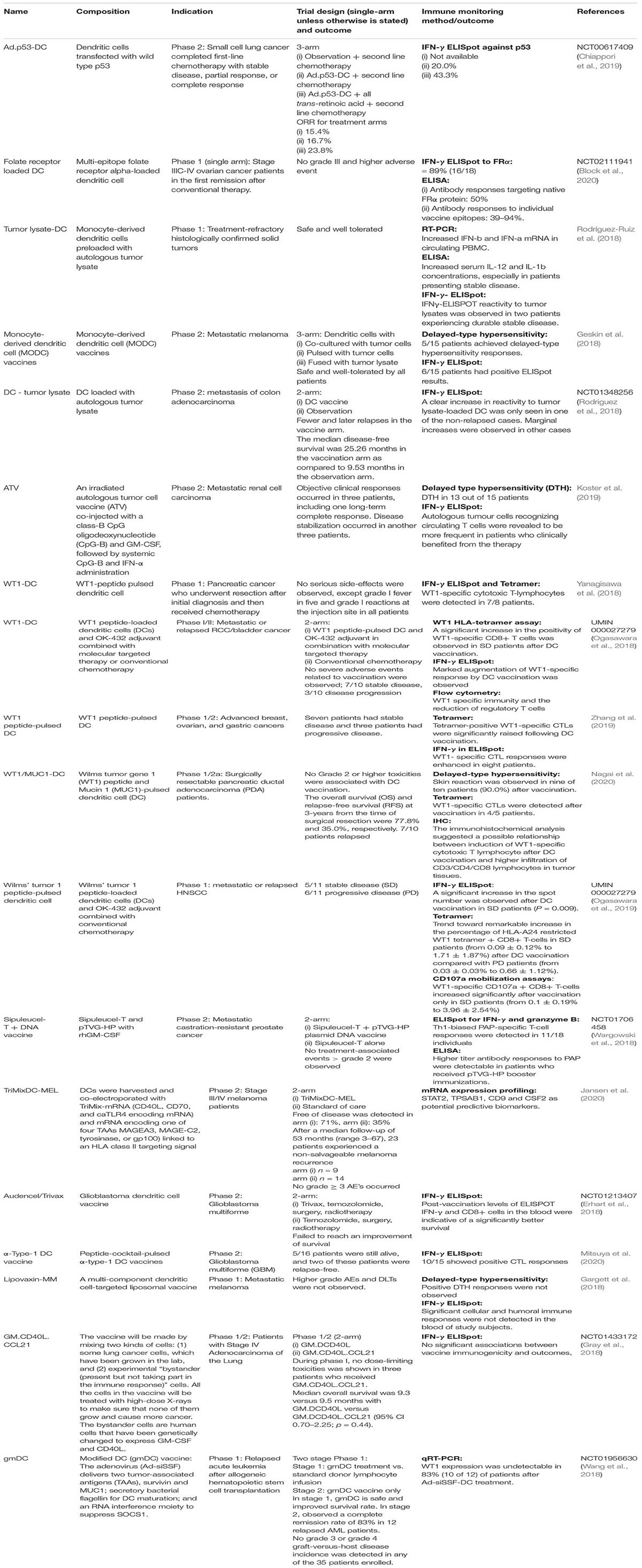
Cellular vaccine refers to cell-based vaccines that use autologous tumor cells or antigen-presenting dendritic cells to stimulate the immune system. Sipuleucel-T, an autologous dendritic cell targeting prostatic acid phosphatase (PAP) has improved survival for men with castration-resistant prostate cancer, is the first cancer vaccine approved by FDA in 2010 (Kantoff et al., 2010). To further improve the clinical response of Sipuleucel-T, DNA vaccine (pTVG-HP) targeting PAP was administered to patients who received Sipuleucel-T. Despite demonstrating higher antibody titer in patients who received pTVG-HP booster immunization as compared to Sipuleucel-T alone, the median time to progression was similar for both arms (Wargowski et al., 2018). Wilm’s tumor (WT1) protein is overexpressed in hematologic tumor and various solid tumors and it is recognized as one of the most promising targets for cancer immunotherapy (Cheever et al., 2009). Dendritic cell targeting WT1 has been evaluated in a number of early phase clinical trials where 7/10 bladder cancer patients, 7/10 breast/ovarian/gastric cancer patients, and 5/11 head and neck cancer patients achieved stable disease. Notably, patients who demonstrated clinical benefit developed higher WT1 specific cytotoxic T cell responses (Ogasawara et al., 2019; Zhang et al., 2019; Nagai et al., 2020). Recently, the use of genetically modified dendritic cells has been reported. Adenovirus was employed to deliver tumor-associated antigens (survivin, MUC1), secretory bacterial flagellin and an RNA interference moiety to suppress SOCS1 in dendritic cells (gmDC). This novel engineered gmDC was shown to be safe and resulted in 10/12 relapsed acute myeloid leukemia patients to experience complete remission (Wang et al., 2018).
The use of tumor cell lysate as vaccine is also a fast-growing field. The use of GVAX, a GMCSF secreting allogeneic pancreatic tumor cells has resulted in clinical response in metastatic pancreatic adenocarcinoma and is being tested in combination with CRS-207, a recombinant live-attenuated double-deleted Listeria monocytogenes (Lutz et al., 2011; Le et al., 2015; Nair et al., 2020). The use of dendritic cell presenting tumor lysate is also being evaluated in melanoma and colon cancer and data showed that this approach is well tolerated (Geskin et al., 2018; Rodriguez et al., 2018). Another study in renal cell carcinoma was using dendritic cell presenting tumor lysate with addition of systemic CpG and IFN-α. Encouragingly 6/15 patients achieved clinical response (1 complete response, 2 partial response, and 3 stable disease) (Koster et al., 2019). Table 3 presents an overview of the cellular vaccines that have resulted in a publication from 2018 to 2020.
Viral-Based Vaccine
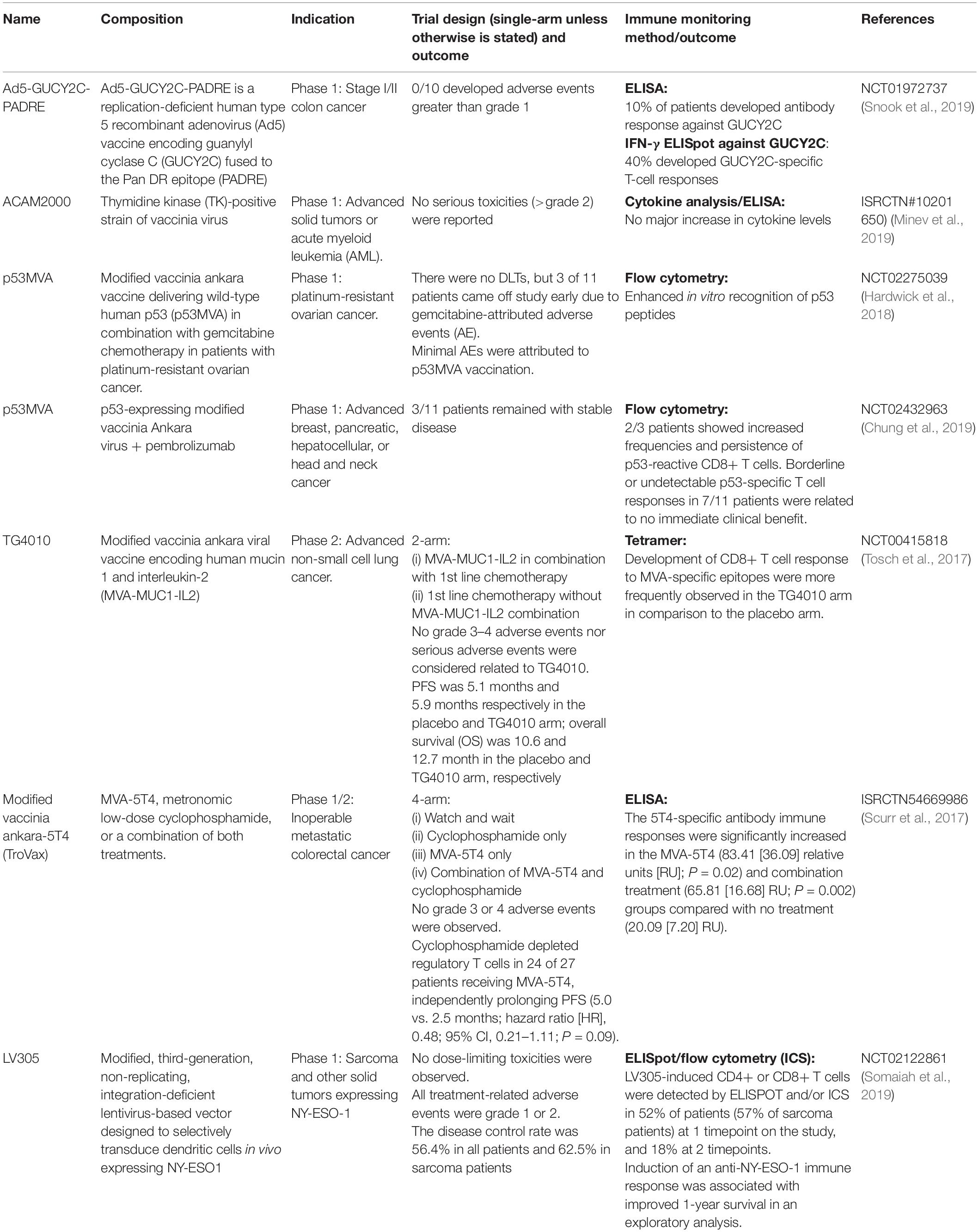
As cancer vaccines targeting tumor associated antigen or protein often induce central tolerance where clinical benefit may be limited (Jou et al., 2021), packaging the antigen of interest into oncolytic viruses is known to improve its immunogenicity and efficacy. Genetically modified oncolytic viruses can replicate selectively in tumor cells and promote tumor cell lysis, at the same time promote both the innate and adaptive immune responses (Harrington et al., 2019). In 2015, FDA approved the first oncolytic virus therapy, Talimogene laherparepvec (T-VEC) for the treatment of advanced melanoma that cannot be removed by surgery. This approval was based on a Phase III OPTiM study that demonstrated significant improvement in durable response rate and overall survival when compared to granulocyte macrophage colony-stimulating factor (Andtbacka et al., 2015, 2019). In addition to viral vaccine monotherapy, a combination of viral vaccine with other approved treatments have been studied extensively in early phase clinical trials in the past few years. For example, the use of the modified vaccinia ankara (MVA) virus expressing p53 is now used in combination with chemotherapy to treat ovarian cancer and with pembrolizumab to treat advanced breast/pancreatic/liver/head and neck cancer (Hardwick et al., 2018; Chung et al., 2019). Further, MVA is engineered to expressed oncofetal antigen 5T4 (MVA-5T4, TroVax) or simultaneously expressed tumor antigen MUC1 and immune modulator IL2 (TG4010) for the treatment of metastatic colorectal cancer and advanced non-small cell lung cancer respectively (Scurr et al., 2017; Tosch et al., 2017). The use of replication-deficient human type 5 recombinant adenovirus (Ad5) vaccine to express target of interest has gained a lot of interest lately as it is the vector of choice for many COVID-19 vaccines. Ad5 is used to expressed guanylyl cyclase C (GUCY2C) fused to a CD4+ T cell epitope (the pan-DR epitope, PARDE) that is selectively expressed by intestinal epithelial cells to treat colon cancer (Snook et al., 2019). Table 4 presents an overview of the viral-based vaccine that have resulted in a publication from 2016 to 2020.
The ultimate aim of a cancer vaccine is to generate sufficient T-cell responses. Upon antigen exposure, naïve T cells will be differentiated into effector T cells. The immune cells can be harvested while they are traveling to the tumor bed through blood vessels or when they have reached the tumor bed as indicated in Figure 1. The quantity or functionality of these antigen-specific T cells are the most important immunogenicity and efficacy readouts. Patients who ultimately demonstrating clinical benefit are those who have elevated specific immune responses. Referring to the immune monitoring methods as detailed in Tables 2–4, we will specifically discuss some of the common techniques to quantify antigen-specific T cells, and discuss their respective advantages and disadvantages in the following sections.
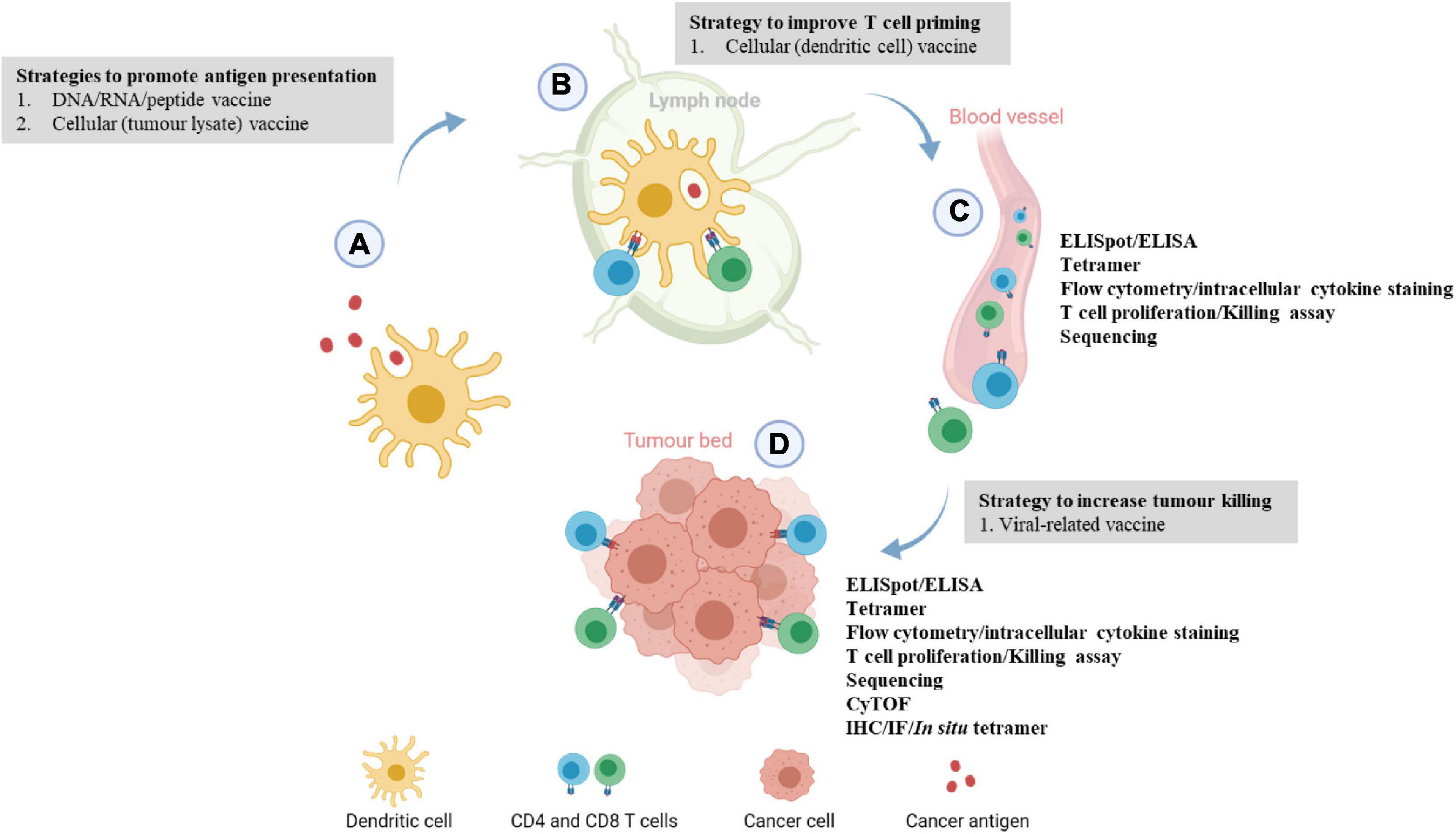
Figure 1. Immune monitoring of cancer vaccination at different phases of the cancer immunity cycle. (A) Upon vaccination, peptides or cancer antigens are digested and internalized by antigen-presenting cells, usually dendritic cells (DCs). (B) Antigen-presenting DCs travel to the lymph nodes to prime and activate naïve T cells through T cell receptor signaling and co-stimulation. (C) Upon activation, antigen-specific CD4 or CD8 T cells circulate through the blood to locate the tumor, this juncture allows the development of ex vivo immunoassays where antigen-specific T cells can be detected. Blood/peripheral blood mononuclear cell from cancer patients are usually harvested and antigen-specific immune responses are evaluated by ELISpot, T cell proliferation, killing assay, intracellular cytokine staining, and flow cytometry phenotyping. (D) At the tumor bed, activated immune cells will bind to tumor cells expressing the target of interest and induces cell lysis. Localization and levels of infiltrating antigen-specific T cells can be examined through multiplexed IHC/IF and ELISpot assay. More recently, tumor-infiltrating lymphocytes from patients’ tumors are extracted to evaluate the phenotypic changes or functions of the immune cells via RNA sequencing or scRNA sequencing. This image was created with BioRender.com.
Methods for Immune Monitoring
Enzyme-Linked Immune Absorbent Spot (ELISpot)
When CD8 T cell encounters a pathogen or an antigen it recognizes, it becomes activated. This activated T cells will then secrete cytokines like TNF-α, IFN-γ, perforin or granzyme that have anti-microbial or anti-tumor effects. The enzyme-linked immune absorbent spot (ELISpot) is an assay to detect the frequency of cytokine secretion in different immune types especially T cells. In brief, T cells or co-culture of immune cells stimulated with a stimulant (antigen/peptide) will be seeded on a PVDF or nitrocellulose membrane in a 96-well plate precoated with an antibody specific to the secreted cytokine. Secreted cytokine will be captured and further detected using biotinylated antibody. This technique is regarded as a gold standard for quantitating antigen-specific cellular responses after vaccination in clinical trials (Ranieri et al., 2014), where every single spot represents a single T cell that is activated. The most widely used ELISpot in vaccine studies is the IFN-γ assay by which the secretion is augmented following immune activation, hence representing a good biomarker of successful vaccination. This assay is robust (low intra-assay, inter-assay and inter-operator variability) (Barabas et al., 2017), economical and can be conducted in a high-throughput manner. Precoated plates for different species including human, mouse and non-human primates are commercially available. Importantly, the seeded immune cells can be recovered for in vitro stimulation assay or other relevant assays to corroborate the findings from ELISpot reading (Saade et al., 2012). Disadvantages of ELISpot assay include the lack of information about cell phenotype and it is normally a single parameter readout. To circumvent the limitation of ELISpot assay, dual and triple color fluorospot assays allow an analysis of multiple cytokines, and commercial kits are now available (Calarota and Baldanti, 2013). Currently, several new techniques, for example, Luminex, LegendPlex and Meso-Scale Discovery (MSD) systems are being used to quantify multiple cytokines simultaneously in the cell culture supernatant, tissue homogenate, plasma, or serum. However, due to the high running cost, these are not as widely used as compared to ELISpot.
Intracellular Cytokine Staining (ICS)
Another method to detect cytokine secretion by immune cells is intracellular cytokine staining (ICS) followed by flow cytometry analysis. This technique involves culturing immune cells in the presence of Golgi block to inhibit intracellular protein transportation. These proteins will then be retained intracellularly and therefore are available for antibody staining after fixation and permeabilization (Jung et al., 1993; Prussin and Metcalfe, 1995; Barabas et al., 2017). ICS allows both single and multiparameter cytokine analysis. In addition to detect specific cytokines, this can be coupled with markers to define specific cellular subtypes such as effector cells or memory T cells, which is not possible with the ELISpot assay (Lovelace and Maecker, 2011). This multiplexing is possible with the availability of flow cytometer that simultaneously analyzes 18 or more parameters. However, a detailed selection of antibody panel coupled with appropriate fluorochrome and careful compensation is required to ensure accurate readouts. Sometimes, signal spillover issues could compromise detection especially when there is a need for high-resolution sensitivity for example when detecting rare cell types. ICS has good sensitivity but will not be able to detect very low frequency responses as compared to the ELISpot (Karlsson et al., 2003; Maecker et al., 2005). Furthermore, ICS is more labor extensive, involves a higher cost per sample and required more expertise in detailed data analysis compared to ELISpot (Freer and Rindi, 2013). Notably, assay variability is a challenge as standardization across instruments is difficult and variability could also be introduced by different data processing methods and subjective interpretation of the researchers.
Tetramer/Multimer Assay
In addition to quantitating the cytokine secretion by activated T cells, quantitating the expression level or the number of antigen-specific T cells using tetramer/multimer assay is also a widely used method to measure immune activation. Tetramers are synthetic structures of HLA molecules, four or more identical versions of which are linked together to form a multimeric complex that is then loaded with antigen-specific peptide (Altman et al., 1996). Antigen-loaded tetramer will be used to bind the T-cell receptors (TCR) of antigen-specific T cells, allowing the detection and quantification of this rare class of T cells through fluorescent molecules conjugated to the tetramer structure.
The major advantage of tetramer staining is the reproducibility of the assay on either fresh or cryopreserved samples when compared with ICS and ELISPOT (Maecker et al., 2008); Further, the assay time is also relatively shorter compared to ICS and ELISPOT. The downside of tetramer assay is that (i) these molecules are peptide-MHC/HLA specific, only T cells recognizing a specific MHC-peptide pair can be detected by a single tetramer. This potentially underestimates the total immune response to a pathogen or vaccine that could lead to an over interpretation of results for that particular epitope (Saade et al., 2012; Abdelaal et al., 2019); (ii) limited availability of tetramer for specific MHC molecules has limit the detection of antigen specific T cells especially in patient harboring rare MHC alleles (Abdelaal et al., 2019); (iii) tetramer is a custom-made molecule and is not available off the shelf, expertise in developing this is needed; (iv) the affinity threshold required for staining peptide-MHC complex with tetramer is higher than what is required for actual T cell activation in vivo, which biases tetramer to detect high affinity T cells but missing low affinity responses that could also have contributed to the immune responses (Martinez et al., 2016; Rius et al., 2018); (v) tetramer staining can inform on the phenotypes of antigen specific T cells but is unable to indicate the specific tissue location of these cells as this protocol is applied on harvested immune cells and not directly on tissue specimens (Abdelaal et al., 2019).
The ability to expand antigen-specific T cells in vivo is one of the direct measurements of successful cancer vaccination. However, to accurately quantitate the differences of this rare population before and after vaccination is challenging. Different innovations have been reported to increase the sensitivity of tetramer staining. Recently, there is a report demonstrating the use of magnetic nanoparticles that presented neoantigen-loaded MHC tetramers by barcoded DNA linkers which have shown to enhance the T cells-capture. The presence of magnetic nanoparticles allows the isolation of these antigen-specific T cell populations and DNA barcoding allows the multiplexing detection of different antigen-specific T cells (Peng et al., 2019).
In situ Tetramer Staining
One of the limitations of the tetramer staining described in the previous section is the lack of information on the localization of the immune cells. The spatial information is gaining more importance as the presence of immune cells that can travel to the tumor bed is of therapeutic and prognostic significance (Tumeh et al., 2014). Recently, the development of in situ tetramer staining has provided information on the location of antigen-specific T cells and insights on the mechanisms of the anti-tumor response. This is achieved by staining tissues (FFPE or frozen section) with tetramer, followed by anti-CD8 antibody and subsequently these are detected through secondary antibodies conjugated with specific fluorophores. One of the advantages of in situ tetramer staining is that the signal intensity can be amplified using a relevant secondary antibody, hence lesser tetramer is needed when compared to the direct staining method, making it more cost-effective compared to direct tetramer staining (Skinner and Haase, 2002). However, in situ tetramer staining involves a multi-step procedure that is more time-consuming. Ideally, in situ tetramer staining should be performed using unfixed, fresh tissue sections to maintain the structure and mobility of T cell receptors to interact with tetramer molecule (Haanen et al., 2000; Skinner et al., 2000). Recently, this technique was used to determine antigen-specific CD8 T cells in melanoma patients post-administration of a dendritic cell vaccine and revealed the location of where these antigen-specific T cells resided (De Vries et al., 2007).
Mass Cytometry (CyTOF)
One of the drawbacks of tetramer staining is the number of phenotypic markers that can be included in the assay. Although some flow cytometers can analyze more than 18 markers simultaneously, assay compensation has proven to be difficult. Time of flight (TOF) mass cytometry allows the evaluation of more than 40 markers per cell. Instead of labeling with fluorescently conjugated antibodies, cells are labeled with different metal isotope-tagged antibodies. When running samples on the dedicated mass cytometer, each cell is vaporized and metal ions are resolved to produce a mass spectrum. Each unique metal ion relates to the specific markers like an emission spectrum does in flow cytometry. The advantage of CyTOF stems from the minimal overlap in metal signals hence afford for detecting more than 40 parameters in a single cell resolution (Spitzer and Nolan, 2016). The disadvantages of this technique include lower flow rate as compared to flow cytometer; cells are destroyed by ionization and cannot be recovered for further analysis and the cost of running is much more compared to flow cytometry (Palit et al., 2019). Notably, the data generated by CyTOF is more complex and multidimensional and special software and expertise are needed for data analysis.
Proliferation Assay
In addition to cytokine secretion, immune cell proliferation upon stimulation by antigen or peptide is an indication of successful cellular immunity. For example, the ability of PBMC to proliferate upon stimulation with antigen or peptide in culture has been used as a measurement of immunogenicity. In a previous section, we described the detection of immune cell expansion by quantitating antigen-specific T cells using tetramer staining. In this section, we will describe the use of dye incorporation method to track the proliferation of immune cells. Traditionally, cell proliferation is measured using the incorporation of radioisotope 3H-thymidine and this was regarded as the gold standard for measuring proliferative responses (Goodell et al., 2007). However, the use of radioisotopes requires additional safety measures and safety containment that is not available in many laboratories and 3H-thymidine incorporation assays do not provide information on the identity or function of the proliferating cells. Recently, carboxyfluorescein succinimidyl ester (CFSE) assay has been widely used to evaluate antigen-specific T-cell proliferation (Lyons, 2000). CSFE assay has eliminated the need for radioactivity and has greater flexibility, where additional antibodies can be added to identify the immune cell type that is expanding and the corresponding cytokines produced by these cells through flow cytometry analysis. PBMC can be easily stained with CFSE and T-cell proliferation following antigen stimulation is measured through the halving of fluorescence in daughter cells for 7–8 cell division. Importantly, CFSE dye incorporation can be used to track the proliferation and migration of lymphocytes in vivo (Parish et al., 2009). However, CFSE assays require careful optimization as a high concentration of CFSE is toxic to cells (Last’ovicka et al., 2009). Further, this assay has a high degree of variability which makes reproducibility challenging (Leroux-Roels et al., 1994). This is mainly due to differences in initial cell count, media and culture conditions (Ten Brinke et al., 2018).
Killing Assay
In the previous sections, we have described assays to measure immune activation post-exposure to antigen/peptide, through measuring the expansion of immune cells and secretion of cytotoxic cytokines. During the antigen stimulation process and in the presence of co-stimulatory signals and CD4 helper T cells, precursor T cells will also differentiate into specialized T cells, the cytotoxic CD8 T cells (Saade et al., 2012). These cytotoxic T cells can kill target cells through two mechanisms, first is to release cytokines including perforin and granzymes, these enzymes will enter target cells and trigger a caspase cascade that eventually causes cells to undergo apoptosis; the second mechanism is through the interaction of target cell death-receptors such as Fas leading to apoptosis (Krähenbühl et al., 1988; Lowin et al., 1994). This induction of apoptosis post-exposure to antigen/peptide has made the quantification of target cell death a meaningful measurement of a successful cancer vaccine therapy in vitro. Traditionally, chromium (51Cr) release assay was used to quantify cell death, where chromium is released after the cytotoxic T cells induce lysis of 51Cr-labeled target cells loaded/expressed with an antigen of interest. Despite being a gold standard for measuring cell death, the use of radioisotopes is slowly being phased out due to safety reasons. Further, high interexperiment variability in labeling efficacy and high background due to the non-specific release of 51Cr from target cells are also some drawbacks of this technique (Zaritskaya et al., 2010).
Several non-radioactive tests have been developed to measure cell killing activity as alternatives to the chromium release assay. One of the widely used methods is the colorimetric measurement of lactate dehydrogenase (LDH) activity in the culture medium. LDH is present in the cytoplasm of all eukaryotic cells and is released into the culture media after cell death (Korzeniewski and Callewaert, 1983). LDH assay is easy to perform and has low inter-operator variability as it generates output through absorbance reading and does not involve sophisticated data interpretation. However, phenotypic identification of antigen-specific CD8 T cells and other parameters is not possible.
RNA Sequencing
Moving into the high-dimensional analysis, RNA sequencing is a technique used to profile the cellular transcriptome quantitatively. Owing to the significant cost reduction over the years and the high throughput approach of this platform, RNA sequencing is now a viable approach in evaluating T cell responses in immunotherapy studies. Bulk RNA sequencing is widely used to characterize the cellular composition of immune infiltration (Chen et al., 2016; Sade-Feldman et al., 2019). The gene expression of immune cells within a tumor can inform us of the presence or absence of key immune cell types such as T cells, B cells, dendritic cells, natural killer cells and regulatory T cells. Additionally, bulk RNA sequencing analysis enables us to identify gene signatures that are associated with response to immunotherapies (Rooney et al., 2015; Ayers et al., 2017; Hugo et al., 2017; Cristescu et al., 2018). This knowledge will help to provide mechanistic insights into therapeutic cancer vaccines and inform on patient selection criteria for the use of cancer vaccines in the clinic. For example, RNA sequencing on PBMC samples of pediatric glioma patients revealed a response biomarker of a peptide-based vaccine where low expressions of IDO1 and PDL-1 before treatment were associated with prolonged progression free survival and patients with elevated T cell activation markers followed by cytotoxic T cell signatures are those demonstrated clinical response (Müller et al., 2018). Importantly, the vast information resulting from sequencing will provide information on possible mechanisms of treatment resistance that are not/least possibly made by all methods described previously. Challenges of RNA sequencing include the need for fresh tissue or body fluid to achieve high-quality RNA (Gallego Romero et al., 2014), and the possible generation of biases and artifacts introduced from the various sample preparation strategies (Ozsolak and Milos, 2011).
Single Cell RNA Sequencing (scRNA-Seq)
More recently, single-cell RNA sequencing (scRNA-seq) technology has been developed to enable transcriptomic profiling analysis that is beyond the scope of bulk RNA sequencing. A major limitation of bulk RNA sequencing is that the sequence library represents the entire cell population within the tumor which obscures the heterogeneity of the transcriptomic data. Unlike bulk RNA, scRNA-seq can isolate and capture transcripts from single cells leading to the generation of sequence libraries that can be mapped to individual cells. scRNA-seq platform is currently used as a tool to examine the phenotype of key immune pathways in cancer patients. For instance, single-cell transcriptomic profiling revealed the activation and exhaustion state of infiltrated CD8 T cells in liver cancer patients (Zheng et al., 2017). Notably, the use of scRNA-seq has successfully identified a subpopulation of cells known as immunotherapy persister cells (IPCs) that resisted CD8+ T cell-mediated killing, contributing to the immune escape from anti-PD1 treatment (Sehgal et al., 2021). scRNA-seq can be applied for a more in-depth analysis of the immune cell’s functionality as opposed to only focusing on immune cell composition achieved through the conventional bulk RNA sequencing. Additionally, the use of scRNA sequencing platform will also provide the opportunity to characterize rare or unknown immune cell types or signatures that may be associated with therapeutic responses. Currently, published data on single cell transcriptomic responses in the context of therapeutic cancer vaccination are still lacking and there remains much to learn from other available pilot studies on data analysis and interpretation.
TCR Sequencing
The T cell receptors (TCRs) on the surface of T cells are responsible for recognizing antigens presented by the major histocompatibility complex in human. The availability of RNA sequencing technologies has made it possible to profile T cell receptors (TCR) at the transcriptomic level. The TCR repertoire is one of the predictive biomarkers that is associated with effective immunotherapy responses. For example, the TCR repertoire of immunotherapy-treated pancreatic ductal adenocarcinoma was shown to be associated with a patient’s survival (Hopkins et al., 2018). In response to antigen stimulation, T cells will undergo proliferation and clonal expansion. Therefore, identifying vaccine-responsive T cell clonotypes is informative for the monitoring of therapeutic cancer vaccines. In a trial evaluating the combination of dsRNA analog and tumor-lysate loaded dendritic cells vaccine, TCR sequencing performed on the patient’s PBMC indicated some clones to be enriched 42 days post-vaccination, suggesting the clonality of T cells was induced by the immunotherapy (Rodríguez-Ruiz et al., 2018). A study also demonstrated patients who underwent tremelimumab treatment have broadened their TCR diversity, suggesting the use of CTLA4 checkpoint blockade has expanded the pool of circulating T cells (Robert et al., 2014). Additionally, TCR sequencing is also used to evaluate the infiltration of functional peptide-specific cytotoxic T cells through the detection of an identical sequence of peptide-specific TCRs from T cells derived from both tumor tissue and PBMC (Mackensen et al., 1997; Daiko et al., 2020). Importantly, TCR sequencing has also revealed that patients who responded to checkpoint blockade are those with less clonotype loss during treatment (Cha et al., 2014). TCR sequencing is the only available technique that can provide insights into the T cell clonotypes and diversity post immunotherapy treatment. The disadvantages of this technique are mainly due to the inconsistency of input materials (DNA, mRNA or PCR enriched DNA) and variations resulting from different data analysis pipelines (Liu and Wu, 2018).
Summary
In summary, when a naïve T cell encounters its cognate antigen, it will get activated and differentiate into specialized T cells, followed by clonal expansion. These specialized T cells will then migrate from the lymph node to tumor bed, where they will hunt and kill target cells expressing the same antigen. The induction of this series of processes represents an active stimulation by a cancer vaccine. The ability to detect these processes individually has provided insight into successful vaccinations. In this paper, we have discussed 10 different techniques that are currently being used to evaluate immune responses triggered by cancer vaccination in both preclinical and clinical studies. As clinical immune monitoring is a continuous process, despite the advances in new technologies, ELISpot and flow cytometry are still the preferred methods due to their affordability and robustness. Other methods such as in situ tetramer staining and sequencing (RNA, sc-RNA, and TCR sequencing) are currently used in the discovery setting rather than as a clinical monitoring tool.
Important factors that need to be taken into account for the successful detection and measurement of T-cell immunity in response to immunization are the timings of when each assay is performed and when samplings are made. Worth noting that immune response is a dynamic response, different vaccine candidates will confer different kinetic profiles. It is advisable to conduct a timepoint measurement to identify a time point that confers the highest immune activity before a study is implemented. In some instances, even at the time of maximum response, the frequency of antigen-specific T cells in the peripheral blood may be below the limits of detection (Saade et al., 2012). Despite the advancement in detecting immune responses at single-cell level, defining T cell responses that are reflective of conferring protection remains the biggest challenge for the field.
Author Contributions
KPL contributed to the concept and manuscript writing. NSZ contributed to the manuscript writing. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by sponsors of Cancer Research Malaysia. Cancer Research Malaysia is a non-profit research organization, committed to an understanding of cancer prevention, diagnosis and treatment through a fundamental research program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thanked Prof. Dr. Sok Ching Cheong for the critical review of the manuscript.
References
Abdelaal, H. M., Cartwright, E. K., and Skinner, P. J. (2019). Detection of antigen-specific T cells using in situ MHC tetramer staining. Int. J. Mol. Sci. 20:5165. doi: 10.3390/ijms20205165
Aggarwal, C., Cohen, R. B., Morrow, M. P., Kraynyak, K. A., Sylvester, A. J., Knoblock, D. M., et al. (2019). Immunotherapy targeting HPV16/18 generates potent immune responses in HPV-associated head and neck cancer. Clin. Cancer Res. 25, 110–124. doi: 10.1158/1078-0432.ccr-18-1763
Altman, J. D., Moss, P. A., Goulder, P. J., Barouch, D. H., McHeyzer-Williams, M. G., Bell, J. I., et al. (1996). Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96.
Andtbacka, R. H. I., Collichio, F., Harrington, K. J., Middleton, M. R., Downey, G., ?hrling, K., et al. (2019). Final analyses of OPTiM: a randomized phase III trial of talimogene laherparepvec versus granulocyte-macrophage colony-stimulating factor in unresectable stage III-IV melanoma. J. Immunother. Cancer 7:145.
Andtbacka, R. H., Kaufman, H. L., Collichio, F., Amatruda, T., Senzer, N., Chesney, J., et al. (2015). Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J. Clin. Oncol. 33, 2780–2788.
Ayers, M., Lunceford, J., Nebozhyn, M., Murphy, E., Loboda, A., Kaufman, D. R., et al. (2017). IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940. doi: 10.1172/jci91190
Barabas, S., Spindler, T., Kiener, R., Tonar, C., Lugner, T., Batzilla, J., et al. (2017). An optimized IFN-gamma ELISpot assay for the sensitive and standardized monitoring of CMV protein-reactive effector cells of cell-mediated immunity. BMC Immunol. 18:14. doi: 10.1186/s12865-017-0195-y
Block, M. S., Dietz, A. B., Gustafson, M. P., Kalli, K. R., Erskine, C. L., Youssef, B., et al. (2020). Th17-inducing autologous dendritic cell vaccination promotes antigen-specific cellular and humoral immunity in ovarian cancer patients. Nat. Commun. 11:5173.
Cafri, G., Gartner, J. J., Zaks, T., Hopson, K., Levin, N., Paria, B. C., et al. (2020). mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J. Clin. Invest. 130, 5976–5988. doi: 10.1172/jci134915
Calarota, S. A., and Baldanti, F. (2013). Enumeration and characterization of human memory T cells by enzyme-linked immunospot assays. Clin. Dev. Immunol. 2013:637649.
Cha, E., Klinger, M., Hou, Y., Cummings, C., Ribas, A., Faham, M., et al. (2014). Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci. Transl. Med. 6:238ra70. doi: 10.1126/scitranslmed.3008211
Cheever, M. A., Allison, J. P., Ferris, A. S., Finn, O. J., Hastings, B. M., Hecht, T. T., et al. (2009). The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 15, 5323–5337. doi: 10.1158/1078-0432.ccr-09-0737
Chen, P. L., Roh, W., Reuben, A., Cooper, Z. A., Spencer, C. N., Prieto, P. A., et al. (2016). Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837.
Chiappori, A. A., Williams, C. C., Gray, J. E., Tanvetyanon, T., Haura, E. B., Creelan, B. C., et al. (2019). Randomized-controlled phase II trial of salvage chemotherapy after immunization with a TP53-transfected dendritic cell-based vaccine (Ad.p53-DC) in patients with recurrent small cell lung cancer. Cancer Immunol. Immunother. 68, 517–527. doi: 10.1007/s00262-018-2287-9
Choi, Y. J., Hur, S. Y., Kim, T. J., Hong, S. R., Lee, J. K., Cho, C. H., et al. (2020). A phase II, prospective, randomized, multicenter, open-label study of GX-188E, an HPV DNA vaccine, in patients with cervical intraepithelial neoplasia 3. Clin. Cancer Res. 26, 1616–1623. doi: 10.1158/1078-0432.ccr-19-1513
Chung, V., Kos, F. J., Hardwick, N., Yuan, Y., Chao, J., Li, D., et al. (2019). Evaluation of safety and efficacy of p53MVA vaccine combined with pembrolizumab in patients with advanced solid cancers. Clin. Transl. Oncol. 21, 363–372. doi: 10.1007/s12094-018-1932-2
Conry, R. M., Westbrook, B., McKee, S., and Norwood, T. G. (2018). Talimogene laherparepvec: First in class oncolytic virotherapy. Hum. Vaccin. Immunother. 14, 839–846. doi: 10.1080/21645515.2017.1412896
Cristescu, R., Mogg, R., Ayers, M., Albright, A., Murphy, E., Yearley, J., et al. (2018). Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362:eaar3593. doi: 10.1126/science.aar3593
Daiko, H., Marafioti, T., Fujiwara, T., Shirakawa, Y., Nakatsura, T., Kato, K., et al. (2020). Exploratory open-label clinical study to determine the S-588410 cancer peptide vaccine-induced tumor-infiltrating lymphocytes and changes in the tumor microenvironment in esophageal cancer patients. Cancer Immunol. Immunother. 69, 2247–2257. doi: 10.1007/s00262-020-02619-3
De Vries, I. J., Bernsen, M. R., van Geloof, W. L., Scharenborg, N. M., Lesterhuis, W. J., Rombout, P. D., et al. (2007). In situ detection of antigen-specific T cells in cryo-sections using MHC class I tetramers after dendritic cell vaccination of melanoma patients. Cancer Immunol. Immunother. 56, 1667–1676. doi: 10.1007/s00262-007-0304-5
Dreno, B., Thompson, J. F., Smithers, B. M., Santinami, M., Jouary, T., Gutzmer, R., et al. (2018). MAGE-A3 immunotherapeutic as adjuvant therapy for patients with resected, MAGE-A3-positive, stage III melanoma (DERMA): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 19, 916–929. doi: 10.1016/s1470-2045(18)30254-7
Erhart, F., Buchroithner, J., Reitermaier, R., Fischhuber, K., Klingenbrunner, S., Sloma, I., et al. (2018). Immunological analysis of phase II glioblastoma dendritic cell vaccine (Audencel) trial: immune system characteristics influence outcome and Audencel up-regulates Th1-related immunovariables. Acta Neuropathol. Commun. 6:135.
Freer, G., and Rindi, L. (2013). Intracellular cytokine detection by fluorescence-activated flow cytometry: basic principles and recent advances. Methods 61, 30–38. doi: 10.1016/j.ymeth.2013.03.035
Gallego Romero, I., Pai, A. A., Tung, J., and Gilad, Y. (2014). RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol. 12:42. doi: 10.1186/1741-7007-12-42
Gargett, T., Abbas, M. N., Rolan, P., Price, J. D., Gosling, K. M., Ferrante, A., et al. (2018). Phase I trial of Lipovaxin-MM, a novel dendritic cell-targeted liposomal vaccine for malignant melanoma. Cancer Immunol. Immunother. 67, 1461–1472. doi: 10.1007/s00262-018-2207-z
Geskin, L. J., Damiano, J. J., Patrone, C. C., Butterfield, L. H., Kirkwood, J. M., and Falo, L. D. (2018). Three antigen-loading methods in dendritic cell vaccines for metastatic melanoma. Melanoma Res. 28, 211–221. doi: 10.1097/cmr.0000000000000441
Goodell, V., dela Rosa, C., Slota, M., MacLeod, B., and Disis, M. L. (2007). Sensitivity and specificity of tritiated thymidine incorporation and ELISPOT assays in identifying antigen specific T cell immune responses. BMC Immunol. 8:21. doi: 10.1186/1471-2172-8-21
Gray, J. E., Chiappori, A., Williams, C. C., Tanvetyanon, T., Haura, E. B., Creelan, B. C., et al. (2018). A phase I/randomized phase II study of GM.CD40L vaccine in combination with CCL21 in patients with advanced lung adenocarcinoma. Cancer Immunol. Immunother. 67, 1853–1862. doi: 10.1007/s00262-018-2236-7
Haanen, J. B., van Oijen, M. G., Tirion, F., Oomen, L. C., Kruisbeek, A. M., Vyth-Dreese, F. A., et al. (2000). In situ detection of virus- and tumor-specific T-cell immunity. Nat. Med. 6, 1056–1060. doi: 10.1038/79573
Hardwick, N. R., Frankel, P., Ruel, C., Kilpatrick, J., Tsai, W., Kos, F., et al. (2018). p53-Reactive T cells are associated with clinical benefit in patients with platinum-resistant epithelial ovarian cancer after treatment with a p53 vaccine and gemcitabine chemotherapy. Clin. Cancer Res. 24, 1315–1325. doi: 10.1158/1078-0432.ccr-17-2709
Harrington, K., Freeman, D. J., Kelly, B., Harper, J., and Soria, J. C. (2019). Optimizing oncolytic virotherapy in cancer treatment. Nat. Rev. Drug Discov. 18, 689–706.
Hopkins, A. C., Yarchoan, M., Durham, J. N., Yusko, E. C., Rytlewski, J. A., Robins, H. S., et al. (2018). T cell receptor repertoire features associated with survival in immunotherapy-treated pancreatic ductal adenocarcinoma. JCI Insight 3:e122092.
Hugo, W., Zaretsky, J. M., Sun, L., Song, C., Moreno, B. H., Hu-Lieskovan, S., et al. (2017). Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell 168:542. doi: 10.1016/j.cell.2017.01.010
Jansen, Y., Kruse, V., Corthals, J., Schats, K., van Dam, P. J., Seremet, T., et al. (2020). A randomized controlled phase II clinical trial on mRNA electroporated autologous monocyte-derived dendritic cells (TriMixDC-MEL) as adjuvant treatment for stage III/IV melanoma patients who are disease-free following the resection of macrometastases. Cancer Immunol. Immunother. 69, 2589–2598. doi: 10.1007/s00262-020-02618-4
Jou, J., Harrington, K. J., Zocca, M. B., Ehrnrooth, E., and Cohen, E. E. W. (2021). The changing landscape of therapeutic cancer vaccines-novel platforms and neoantigen identification. Clin. Cancer Res. 27, 689–703. doi: 10.1158/1078-0432.ccr-20-0245
Jung, T., Schauer, U., Heusser, C., Neumann, C., and Rieger, C. (1993). Detection of intracellular cytokines by flow cytometry. J. Immunol. Methods 159, 197–207. doi: 10.1016/0022-1759(93)90158-4
Kanekiyo, S., Hazama, S., Takenouchi, H., Nakajima, M., Shindo, Y., Matsui, H., et al. (2018). IgG response to MHC class I epitope peptides is a quantitative predictive biomarker in the early course of treatment of colorectal cancer using therapeutic peptides. Oncol. Rep. 39, 2385–2392.
Kantoff, P. W., Higano, C. S., Shore, N. D., Berger, E. R., Small, E. J., Penson, D. F., et al. (2010). Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 363, 411–422.
Karlsson, A. C., Martin, J. N., Younger, S. R., Bredt, B. M., Epling, L., Ronquillo, R., et al. (2003). Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J. Immunol. Methods 283, 141–153. doi: 10.1016/j.jim.2003.09.001
Kawamura, J., Sugiura, F., Sukegawa, Y., Yoshioka, Y., Hida, J. I., Hazama, S., et al. (2018). Cytotoxic T lymphocyte response to peptide vaccination predicts survival in stage III colorectal cancer. Cancer Sci. 109, 1545–1551. doi: 10.1111/cas.13547
Korzeniewski, C., and Callewaert, D. M. (1983). An enzyme-release assay for natural cytotoxicity. J. Immunol. Methods 64, 313–320. doi: 10.1016/0022-1759(83)90438-6
Koster, B. D., Santegoets, S., Harting, J., Baars, A., van Ham, S. M., Scheper, R. J., et al. (2019). Autologous tumor cell vaccination combined with systemic CpG-B and IFN-α promotes immune activation and induces clinical responses in patients with metastatic renal cell carcinoma: a phase II trial. Cancer Immunol. Immunother. 68, 1025–1035. doi: 10.1007/s00262-019-02320-0
Krähenbühl, O., Rey, C., Jenne, D., Lanzavecchia, A., Groscurth, P., Carrel, S., et al. (1988). Characterization of granzymes A and B isolated from granules of cloned human cytotoxic T lymphocytes. J. Immunol. 141, 3471–3477.
Last’ovicka, J., Budinský, V., Spísek, R., and Bartůnková, J. (2009). Assessment of lymphocyte proliferation: CFSE kills dividing cells and modulates expression of activation markers. Cell. Immunol. 256, 79–85. doi: 10.1016/j.cellimm.2009.01.007
Le, D. T., Wang-Gillam, A., Picozzi, V., Greten, T. F., Crocenzi, T., Springett, G., et al. (2015). Safety and survival with GVAX pancreas prime and Listeria Monocytogenes-expressing mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J. Clin. Oncol. 33, 1325–1333. doi: 10.1200/jco.2014.57.4244
Leroux-Roels, G., Van Hecke, E., Michielsen, W., Voet, P., Hauser, P., and Pêtre, J. (1994). Correlation between in vivo humoral and in vitro cellular immune responses following immunization with hepatitis B surface antigen (HBsAg) vaccines. Vaccine 12, 812–818. doi: 10.1016/0264-410x(94)90290-9
Liu, X., and Wu, J. (2018). History, applications, and challenges of immune repertoire research. Cell Biol. Toxicol. 34, 441–457. doi: 10.1007/s10565-018-9426-0
Lovelace, P., and Maecker, H. T. (2011). Multiparameter intracellular cytokine staining. Methods Mol. Biol. 699, 165–178. doi: 10.1007/978-1-61737-950-5_8
Lowin, B., Hahne, M., Mattmann, C., and Tschopp, J. (1994). Cytolytic T-cell cytotoxicity is mediated through perforin and Fas lytic pathways. Nature 370, 650–652. doi: 10.1038/370650a0
Lutz, E., Yeo, C. J., Lillemoe, K. D., Biedrzycki, B., Kobrin, B., Herman, J., et al. (2011). A lethally irradiated allogeneic granulocyte-macrophage colony stimulating factor-secreting tumor vaccine for pancreatic adenocarcinoma. A Phase II trial of safety, efficacy, and immune activation. Ann. Surg. 253, 328–335. doi: 10.1097/sla.0b013e3181fd271c
Lyons, A. B. (2000). Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J. Immunol. Methods 243, 147–154. doi: 10.1016/s0022-1759(00)00231-3
Mackensen, A., Veelken, H., Lahn, M., Wittnebel, S., Becker, D., Köhler, G., et al. (1997). Induction of tumor-specific cytotoxic T lymphocytes by immunization with autologous tumor cells and interleukin-2 gene transfected fibroblasts. J. Mol. Med. (Berl) 75, 290–296. doi: 10.1007/s001090050114
Maecker, H. T., Hassler, J., Payne, J. K., Summers, A., Comatas, K., Ghanayem, M., et al. (2008). Precision and linearity targets for validation of an IFNgamma ELISPOT, cytokine flow cytometry, and tetramer assay using CMV peptides. BMC Immunol. 9:9. doi: 10.1186/1471-2172-9-9
Maecker, H. T., Rinfret, A., D’Souza, P., Darden, J., Roig, E., Landry, C., et al. (2005). Standardization of cytokine flow cytometry assays. BMC Immunol. 6:13. doi: 10.1186/1471-2172-6-13
Martinez, R. J., Andargachew, R., Martinez, H. A., and Evavold, B. D. (2016). Low-affinity CD4+ T cells are major responders in the primary immune response. Nat. Commun. 7:13848.
Maslak, P. G., Dao, T., Bernal, Y., Chanel, S. M., Zhang, R., Frattini, M., et al. (2018). Phase 2 trial of a multivalent WT1 peptide vaccine (galinpepimut-S) in acute myeloid leukemia. Blood Adv. 2, 224–234. doi: 10.1182/bloodadvances.2017014175
Massarelli, E., William, W., Johnson, F., Kies, M., Ferrarotto, R., Guo, M., et al. (2019). Combining immune checkpoint blockade and tumor-specific vaccine for patients with incurable human papillomavirus 16-related cancer: a phase 2 clinical trial. JAMA Oncol. 5, 67–73. doi: 10.1001/jamaoncol.2018.4051
McNeel, D. G., Eickhoff, J. C., Johnson, L. E., Roth, A. R., Perk, T. G., Fong, L., et al. (2019). Phase II trial of a DNA vaccine encoding prostatic acid phosphatase (pTVG-HP [MVI-816]) in patients with progressive, nonmetastatic, castration-sensitive prostate cancer. J. Clin. Oncol. 37, 3507–3517. doi: 10.1200/jco.19.01701
Minev, B. R., Lander, E., Feller, J. F., Berman, M., Greenwood, B. M., Minev, I., et al. (2019). First-in-human study of TK-positive oncolytic vaccinia virus delivered by adipose stromal vascular fraction cells. J. Transl. Med. 17:271.
Mitsuya, K., Akiyama, Y., Iizuka, A., Miyata, H., Deguchi, S., Hayashi, N., et al. (2020). Alpha-type-1 polarized dendritic cell-based vaccination in newly diagnosed high-grade glioma: a phase II clinical trial. Anticancer Res. 40, 6473–6484. doi: 10.21873/anticanres.14669
Müller, S., Agnihotri, S., Shoger, K. E., Myers, M. I., Smith, N., Chaparala, S., et al. (2018). Peptide vaccine immunotherapy biomarkers and response patterns in pediatric gliomas. JCI Insight 3:e98791.
Nagai, K., Adachi, T., Harada, H., Eguchi, S., Sugiyama, H., and Miyazaki, Y. (2020). Dendritic cell-based immunotherapy pulsed with wilms tumor 1 peptide and mucin 1 as an adjuvant therapy for pancreatic ductal adenocarcinoma after curative resection: a phase I/IIa clinical trial. Anticancer Res. 40, 5765–5776. doi: 10.21873/anticanres.14593
Nair, N., Chen, S. Y., Lemmens, E., Chang, S., Le, D. T., Jaffee, E. M., et al. (2020). Single-Cell immune competency signatures associate with survival in phase II GVAX and CRS-207 randomized studies in patients with metastatic pancreatic cancer. Cancer Immunol. Res. 8, 609–617. doi: 10.1158/2326-6066.cir-19-0650
Narita, Y., Arakawa, Y., Yamasaki, F., Nishikawa, R., Aoki, T., Kanamori, M., et al. (2019). A randomized, double-blind, phase III trial of personalized peptide vaccination for recurrent glioblastoma. Neuro Oncol. 21, 348–359. doi: 10.1093/neuonc/noy200
Noguchi, M., Arai, G., Egawa, S., Ohyama, C., Naito, S., Matsumoto, K., et al. (2020). Mixed 20-peptide cancer vaccine in combination with docetaxel and dexamethasone for castration-resistant prostate cancer: a randomized phase II trial. Cancer Immunol. Immunother. 69, 847–857. doi: 10.1007/s00262-020-02498-8
Ogasawara, M., Miyashita, M., and Ota, S. (2018). Vaccination of urological cancer patients with WT1 peptide-pulsed dendritic cells in combination with molecular targeted therapy or conventional chemotherapy induces immunological and clinical responses. Ther. Apher. Dial. 22, 266–277. doi: 10.1111/1744-9987.12694
Ogasawara, M., Miyashita, M., Yamagishi, Y., and Ota, S. (2019). Phase I/II pilot study of Wilms’ tumor 1 peptide-pulsed dendritic cell vaccination combined with conventional chemotherapy in patients with head and neck cancer. Ther. Apher. Dial. 23, 279–288. doi: 10.1111/1744-9987.12831
Ozsolak, F., and Milos, P. M. (2011). RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98. doi: 10.1038/nrg2934
Palit, S., Heuser, C., de Almeida, G. P., Theis, F. J., and Zielinski, C. E. (2019). Meeting the challenges of high-dimensional single-cell data analysis in immunology. Front. Immunol. 10:1515. doi: 10.3389/fimmu.2019.01515
Parish, C. R., Glidden, M. H., Quah, B. J., and Warren, H. S. (2009). Use of the intracellular fluorescent dye CFSE to monitor lymphocyte migration and proliferation. Curr. Protoc. Immunol. Chapter 4, Unit49.
Pavlick, A., Blazquez, A. B., Meseck, M., Lattanzi, M., Ott, P. A., Marron, T. U., et al. (2020). Combined vaccination with NY-ESO-1 protein, Poly-ICLC, and montanide improves humoral and cellular immune responses in patients with high-risk melanoma. Cancer Immunol. Res. 8, 70–80. doi: 10.1158/2326-6066.cir-19-0545
Peng, S., Zaretsky, J. M., Ng, A. H. C., Chour, W., Bethune, M. T., Choi, J., et al. (2019). Sensitive detection and analysis of neoantigen-specific T cell populations from tumors and blood. Cell Rep. 28, 2728–2738.e7.
Prussin, C., and Metcalfe, D. D. (1995). Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J. Immunol. Methods 188, 117–128. doi: 10.1016/0022-1759(95)00209-x
Ranieri, E., Popescu, I., and Gigante, M. (2014). CTL ELISPOT assay. Methods Mol. Biol. 1186, 75–86. doi: 10.1007/978-1-4939-1158-5_6
Rausch, S., Gouttefangeas, C., Hennenlotter, J., Laske, K., Walter, K., Feyerabend, S., et al. (2019). Results of a phase 1/2 study in metastatic renal cell carcinoma patients treated with a patient-specific adjuvant multi-peptide vaccine after resection of metastases. Eur. Urol. Focus 5, 604–607. doi: 10.1016/j.euf.2017.09.009
Rius, C., Attaf, M., Tungatt, K., Bianchi, V., Legut, M., Bovay, A., et al. (2018). Peptide-MHC class I tetramers can fail to detect relevant functional T cell clonotypes and underestimate antigen-reactive T cell populations. J. Immunol. 200, 2263–2279. doi: 10.4049/jimmunol.1700242
Robert, L., Tsoi, J., Wang, X., Emerson, R., Homet, B., Chodon, T., et al. (2014). CTLA4 blockade broadens the peripheral T-cell receptor repertoire. Clin. Cancer Res. 20, 2424–2432. doi: 10.1158/1078-0432.ccr-13-2648
Rodriguez, J., Castañón, E., Perez-Gracia, J. L., Rodriguez, I., Viudez, A., Alfaro, C., et al. (2018). A randomized phase II clinical trial of dendritic cell vaccination following complete resection of colon cancer liver metastasis. J. Immunother. Cancer 6:96.
Rodríguez-Ruiz, M. E., Perez-Gracia, J. L., Rodríguez, I., Alfaro, C., Oñate, C., Pérez, G., et al. (2018). Combined immunotherapy encompassing intratumoral poly-ICLC, dendritic-cell vaccination and radiotherapy in advanced cancer patients. Ann. Oncol. 29, 1312–1319. doi: 10.1093/annonc/mdy089
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G., and Hacohen, N. (2015). Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61. doi: 10.1016/j.cell.2014.12.033
Saade, F., Gorski, S. A., and Petrovsky, N. (2012). Pushing the frontiers of T-cell vaccines: accurate measurement of human T-cell responses. Expert Rev. Vaccines 11, 1459–1470. doi: 10.1586/erv.12.125
Sade-Feldman, M., Yizhak, K., Bjorgaard, S. L., Ray, J. P., de Boer, C. G., Jenkins, R. W., et al. (2019). Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 176:404.
Sahin, U., Oehm, P., Derhovanessian, E., Jabulowsky, R. A., Vormehr, M., Gold, M., et al. (2020). An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 585, 107–112. doi: 10.1038/s41586-020-2537-9
Scurr, M., Pembroke, T., Bloom, A., Roberts, D., Thomson, A., Smart, K., et al. (2017). Effect of modified vaccinia ankara-5T4 and low-dose cyclophosphamide on antitumor immunity in metastatic colorectal cancer: a randomized clinical trial. JAMA Oncol. 3:e172579. doi: 10.1001/jamaoncol.2017.2579
Sehgal, K., Portell, A., Ivanova, E. V., Lizotte, P. H., Mahadevan, N. R., Greene, J. R., et al. (2021). Dynamic single-cell RNA sequencing identifies immunotherapy persister cells following PD-1 blockade. J. Clin. Invest. 131:e135038.
Shima, H., Tsurita, G., Wada, S., Hirohashi, Y., Yasui, H., Hayashi, H., et al. (2019). Randomized phase II trial of survivin 2B peptide vaccination for patients with HLA-A24-positive pancreatic adenocarcinoma. Cancer Sci. 110, 2378–2385. doi: 10.1111/cas.14106
Skinner, P. J., and Haase, A. T. (2002). In situ tetramer staining. J. Immunol. Methods 268, 29–34. doi: 10.1016/s0022-1759(02)00197-7
Skinner, P. J., Daniels, M. A., Schmidt, C. S., Jameson, S. C., and Haase, A. T. (2000). Cutting edge: In situ tetramer staining of antigen-specific T cells in tissues. J. Immunol. 165, 613–617. doi: 10.4049/jimmunol.165.2.613
Snook, A. E., Baybutt, T. R., Xiang, B., Abraham, T. S., Flickinger, J. C. Jr., Hyslop, T., et al. (2019). Split tolerance permits safe Ad5-GUCY2C-PADRE vaccine-induced T-cell responses in colon cancer patients. J. Immunother. Cancer. 7:104.
Somaiah, N., Block, M. S., Kim, J. W., Shapiro, G. I., Do, K. T., Hwu, P., et al. (2019). First-in-Class, first-in-human study evaluating LV305, a dendritic-cell tropic lentiviral vector, in sarcoma and other solid tumors expressing NY-ESO-1. Clin. Cancer Res. 25, 5808–5817. doi: 10.1158/1078-0432.ccr-19-1025
Spitzer, M. H., and Nolan, G. P. (2016). Mass cytometry: single cells, many features. Cell 165, 780–791. doi: 10.1016/j.cell.2016.04.019
Tamura, R., Fujioka, M., Morimoto, Y., Ohara, K., Kosugi, K., Oishi, Y., et al. (2019). A VEGF receptor vaccine demonstrates preliminary efficacy in neurofibromatosis type 2. Nat. Commun. 10:5758.
Tamura, R., Morimoto, Y., Kosugi, K., Sato, M., Oishi, Y., Ueda, R., et al. (2020). Clinical and histopathological analyses of VEGF receptors peptide vaccine in patients with primary glioblastoma – a case series. BMC Cancer 20:196. doi: 10.1186/s12885-020-6589-x
Taniguchi, M., Mizuno, S., Yoshikawa, T., Fujinami, N., Sugimoto, M., Kobayashi, S., et al. (2020). Peptide vaccine as an adjuvant therapy for glypican-3-positive hepatocellular carcinoma induces peptide-specific CTLs and improves long prognosis. Cancer Sci. 111, 2747–2759. doi: 10.1111/cas.14497
Ten Brinke, A., Marek-Trzonkowska, N., Mansilla, M. J., Turksma, A. W., Piekarska, K., Iwaszkiewicz-Grześ, D., et al. (2018). Corrigendum: monitoring T-cell responses in translational studies: optimization of dye-based proliferation assay for evaluation of antigen-specific responses. Front. Immunol. 9:343. doi: 10.3389/fimmu.2018.00343
Toh, U., Sakurai, S., Saku, S., Takao, Y., Okabe, M., Iwakuma, N., et al. (2020). Early phase II study of mixed 19-peptide vaccine monotherapy for refractory triple-negative breast cancer. Cancer Sci. 111, 2760–2769. doi: 10.1111/cas.14510
Tosch, C., Bastien, B., Barraud, L., Grellier, B., Nourtier, V., Gantzer, M., et al. (2017). Viral based vaccine TG4010 induces broadening of specific immune response and improves outcome in advanced NSCLC. J. Immunother. Cancer 5:70.
Tumeh, P. C., Harview, C. L., Yearley, J. H., Shintaku, I. P., Taylor, E. J., Robert, L., et al. (2014). PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571.
Vansteenkiste, J. F., Cho, B. C., Vanakesa, T., Pas, T. De, Zielinski, M., Kim, M. S., et al. (2016). Efficacy of the MAGE-A3 cancer immunotherapeutic as adjuvant therapy in patients with resected MAGE-A3-positive non-small-cell lung cancer (MAGRIT): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 17, 822–835. doi: 10.1016/s1470-2045(16)00099-1
Wang, D., Huang, X. F., Hong, B., Song, X. T., Hu, L., Jiang, M., et al. (2018). Efficacy of intracellular immune checkpoint-silenced DC vaccine. JCI Insight 3:e98368.
Wargowski, E., Johnson, L. E., Eickhoff, J. C., Delmastro, L., Staab, M. J., Liu, G., et al. (2018). Prime-boost vaccination targeting prostatic acid phosphatase (PAP) in patients with metastatic castration-resistant prostate cancer (mCRPC) using Sipuleucel-T and a DNA vaccine. J. Immunother. Cancer 6:21.
Wickström, S. L., Lövgren, T., Volkmar, M., Reinhold, B., Duke-Cohan, J. S., Hartmann, L., et al. (2019). Cancer neoepitopes for immunotherapy: discordance between tumor-infiltrating T cell reactivity and tumor MHC peptidome display. Front. Immunol. 10:2766. doi: 10.3389/fimmu.2019.02766
Yanagisawa, R., Koizumi, T., Koya, T., Sano, K., Koido, S., Nagai, K., et al. (2018). WT1-pulsed dendritic cell vaccine combined with chemotherapy for resected pancreatic cancer in a phase I study. Anticancer Res. 38, 2217–2225.
Zaritskaya, L., Shurin, M. R., Sayers, T. J., and Malyguine, A. M. (2010). New flow cytometric assays for monitoring cell-mediated cytotoxicity. Expert Rev. Vaccines 9, 601–616. doi: 10.1586/erv.10.49
Zhang, W., Lu, X., Cui, P., Piao, C., Xiao, M., Liu, X., et al. (2019). Phase I/II clinical trial of a Wilms’ tumor 1-targeted dendritic cell vaccination-based immunotherapy in patients with advanced cancer. Cancer Immunol. Immunother. 68, 121–130. doi: 10.1007/s00262-018-2257-2
Keywords: cancer vaccine, immunotherapy, T cell response, tetramer, immune monitoring, immune activation
Citation: Lim KP and Zainal NS (2021) Monitoring T Cells Responses Mounted by Therapeutic Cancer Vaccines. Front. Mol. Biosci. 8:623475. doi: 10.3389/fmolb.2021.623475
Received: 30 October 2020; Accepted: 24 March 2021;
Published: 15 April 2021.
Edited by:
Joe Yeong, Institute of Molecular and Cell Biology (A∗STAR), SingaporeReviewed by:
Chwee Ming Lim, Singapore General Hospital, SingaporeShawn Michael Jensen, Providence Cancer Center, Providence Portland Medical Center, United States
Copyright © 2021 Lim and Zainal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kue Peng Lim, a3VlcGVuZy5saW1AY2FuY2VycmVzZWFyY2gubXk=
 Kue Peng Lim
Kue Peng Lim Nur Syafinaz Zainal
Nur Syafinaz Zainal