- Cancer Centre and Institute of Translational Medicine, Faculty of Health Sciences, University of Macau, Macau, China
BRCA1 and BRCA2 (BRCA) play essential roles in maintaining genome stability. Rapidly evolving human BRCA generates oncogenic variants causing high cancer risk. BRCA variation is ethnic-specific in reflecting adaptation and/or effects of genetic drift. Taiwanese population of 23.8 million is an admixture of multiple ethnic origins; Taiwan’s subtropical and tropical climate and geographically islandic location provide a unique natural environment. Therefore, Taiwanese population provides a unique model to study human BRCA variation. Through collecting, standardizing, annotating, and classifying publicly available BRCA variants derived from Taiwanese general population and the cancer cohort, we identified 335 BRCA variants, of which 164 were from 1,517 non-cancer individuals, 126 from 2,665 cancer individuals, and 45 from both types of individuals. We compared the variant data with those from other ethnic populations such as mainland Chinese, Macau Chinese, Japanese, Korean, Indian, and non-Asians. We observed that the sharing rates with other Asian ethnic populations were correlated with its genetic relationship. Over 60% of the 335 Taiwanese BRCA variants were VUS, unclassified variants, or novel variants, reflecting the ethnic-specific features of Taiwanese BRCA variation. While it remains challenging to classify these variants, our structural and in silico analyses predicted their enrichment of BRCA deleterious variants. We further determined the 3.8% prevalence of BRCA pathogenic variants in the Taiwanese breast cancer cohort, and determined 0.53% prevalence of the BRCA pathogenic variants in Taiwanese general population, with the estimated 126,140 BRCA pathogenic variant carriers. We identified BRCA2 c.5164_5165delAG at BRCA2 BRC6 motif as a potential founder mutation in Taiwanese population. Our study on BRCA variation in Taiwanese and other East Asian populations demonstrates that ethnic specificity is a common phenomenon for BRCA variation in East Asian population; the data generated from the study provide a reference for clinical applications in BRCA-related cancer in Taiwanese population.
Introduction
BRCA1 and BRCA2 (hereafter refer as BRCA) play essential roles in maintaining genome stability by repairing double-strand DNA damage through homologous recombination (Roy et al., 2011). BRCA is under positive selection in the humans, leading to high variability (Lou et al., 2014). While the majority of variants can be beneficial or neutral, those occurred at specific positions can damage the function of BRCA, causing genome instability and increased risk of breast cancer, ovarian cancer, and other types of cancer (Kuchenbaecker et al., 2017). As BRCA variation is mostly of the germline nature, the later life stage of cancer occurrence provides a unique opportunity to prevent BRCA variation–caused cancer by early identification of the pathogenic variant carriers before cancer development (Burke et al., 1997). Furthermore, PARP inhibitors provide effective treatment of BRCA variant–caused cancer through synthetic lethal therapy (Jerez et al., 2020).
BRCA variation is well determined as highly ethnic specific in certain ethnic populations, such as the BRCA1 185delAG, 5382insC, and BRCA2 6174delT in Ashkenazi Jews population (Levy-Lahad et al., 1997). Restricted by the lack of BRCA variation data from non-Caucasian populations (Bhaskaran et al., 2019; Friebel et al., 2019), however, it remains unclear whether ethnic specificity is mainly in certain specific ethnic population or is a universal phenomenon across worldwide ethnic populations. Recently, we analyzed BRCA variation in Asian populations such as Indian, Chinese, Korean, and Japanese, and revealed that ethnic-specific BRCA variation is also widely present in these Asian populations (Bhaskaran et al., 2020). With a population size nearly 24 million, Taiwanese population consists of admixed ethnic origins across prehistory and current days. Although Taiwanese population included largely the ancestors from southern Han Chinese of Fujian and Guangdong regions of mainland China, it also included other ethnicities including the native Austronesians who also distributed to Pacific islands and Asian neighbors. Furthermore, the islandic location with subtropical and tropical climates in Taiwan Island provides a unique natural environment for Taiwanese population (Chen et al., 2016; Figure 1). Therefore, the Taiwanese population provides a unique model to study BRCA evolution and its impact on human health.
FIGURE 1. Geographic map and population density of Taiwan. The numbers show residents per square kilometer by village (from: https://en.wikipedia.org/wiki/Demographics_of_Taiwan).
In the current study, we performed a systematic analysis for BRCA variation in the Taiwanese general population and the cancer cohort. Of the BRCA variants identified, we observed that forty percent BRCA variants were Taiwanese specific; using the identified BRCA pathogenic variants as the reference, we determined the prevalence of BRCA pathogenic variation in Taiwanese general population and the cancer cohort. Data from our study provide further evidence to demonstrate that ethnic specificity of BRCA variation is a common phenomenon in East Asian populations.
Results
Data Collection
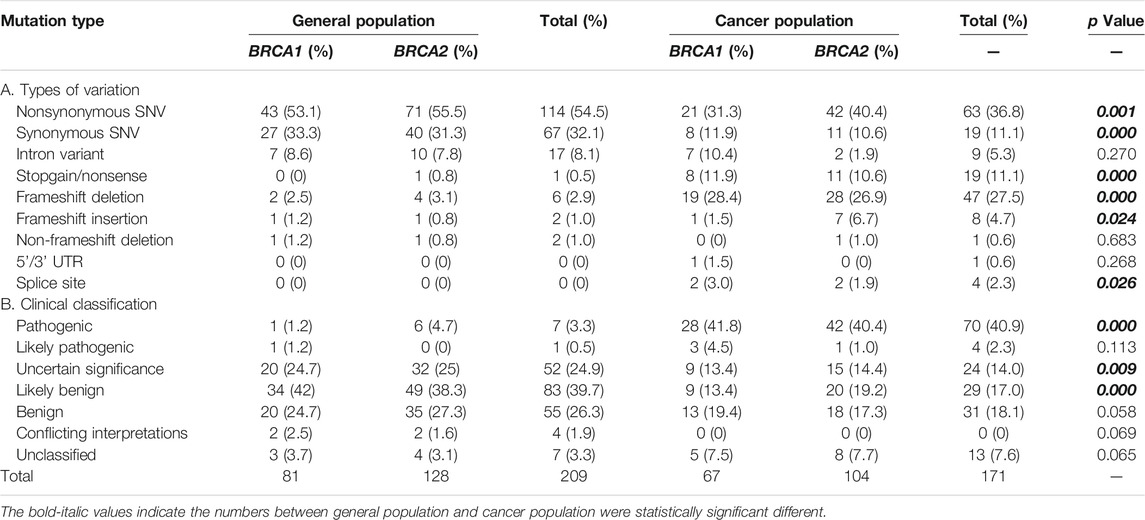
We collected a total of 335 BRCA variants derived from Taiwanese population, including 164 from general population, 126 from the Taiwanese cancer patient cohort, and 45 (19 in BRCA1 and 26 in BRCA2) from both groups. For the variants from cancer patients, nearly all were from breast cancer and ovarian cancer (Supplementary Table S1). We performed standardization, annotation, and clinical classification for all BRCA variants (Table 1, Supplementary Tables S2, S3).
Similarity and Differences Between General Population and the Cancer Cohort
Data from both general population and cancer patients gave a unique opportunity to compare the similarity and differences of BRCA variation between the two groups with the same ethnic background. Although the total number of BRCA variants at the individual level was similar, significant differences existed between the two groups. The types of BRCA variation between the two groups were significantly different, including nonsynonymous SNV, synonymous SNV, stopgain, frameshift deletion, frameshift insertion, and splice site; and the frequency of nonsynonymous SNV and synonymous SNV in general population was higher than that in the cancer cohort (54.5% vs. 36.8% and 32.1% vs. 11.1%, p < 0.001 and 0.000, accordingly), whereas the frequency of stopgain, frameshift deletion/insertion, and splice variants was higher in the cancer cohort than that in the general population (Table 1). Significant differences in the clinical classification were also present in between. For example, 40.9% of BRCA variants in the cancer cohort were pathogenic variants, which was much higher than the value of 3.3% in general population (p < 0.000); VUS (variants of uncertain significance) and likely benign were significantly higher in general population than those in the cancer cohort (24.9% vs. 14% and 39.7% vs. 17%, p < 0.009, 0.000 accordingly) (Table 1).
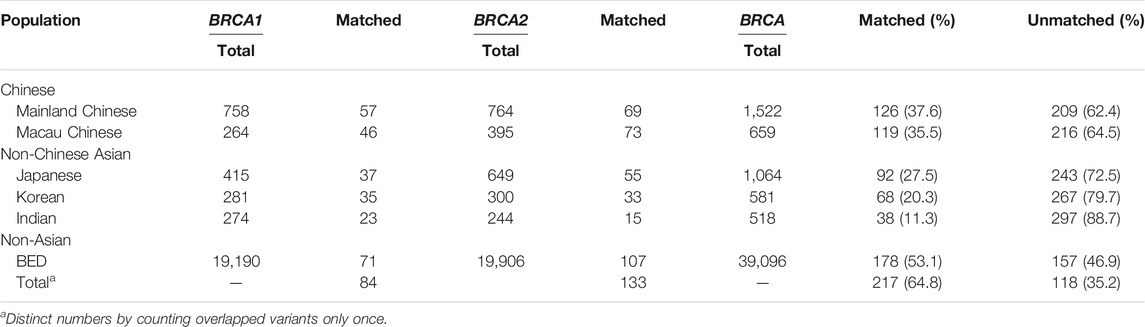
Similarity and Differences From Other Ethnic Populations
We compared BRCA variants between Taiwanese population and other populations including mainland Chinese (Bhaskaran et al., 2019); Macau Chinese representing southern Chinese (Qin et al., 2020); Asian populations including Korean, Japanese, and Indian (Bhaskaran et al., 2020); and non-Asian populations, of which the majority were Caucasians (Dutil et al., 2015; Rebbeck et al., 2018). The results show that 35.5 and 37.6% of the Taiwanese variants were shared with Macau Chinese and mainland Chinese, respectively, 27.5% with Japanese, 20.3% with Korean, 11.3% with Indian, and 53.1% of entire non-Asian populations. The different sharing rates reflected the evolutionary relationship of Taiwanese population with non-Taiwanese populations (Table 2). We also compared with the BRCA variants from Fujian Chinese, which has the closest genetic tie with the Taiwanese population. Of the 18 BRCA variants available for comparison, 8 (44.4%) were matched by Taiwanese variants.
VUS, Unclassified Variants, and Novel Variants
Of the BRCA variants identified, 20.7% were VUS (52 in BRCA1 and 24 in BRCA2, Table 1), 6.0% were unclassified variants (seven in BRCA1 and 13 in BRCA2, Table 1), and 35.2% (118 BRCA variants) were absent in the BRCA data from worldwide ethnic populations (Table 2). The combination of VUS, unclassified, and novel variants accounted for 61.9% of all 335 BRCA variants identified in the Taiwanese population. Although the definitive classification for these variants remains to be solved, they may enrich with the Taiwanese-specific pathogenic BRCA variants. For example, 64.4% of the 118 BRCA variants were nonsynonymous SNV, frameshift insertion/deletion/substitution, stopgain, and non-frameshift deletion (Table 2, Supplementary Tables S2, S3).
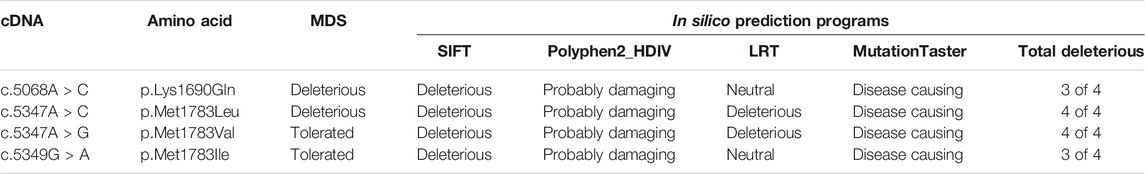
To further test this possibility, we used the molecular dynamic simulation (MDS) to measure the impact of the four BRCA1 unclassified variants (c.5068A > C p.Lys1690Gln, c.5347A > C p.Met1783Leu; c.5347A > G p.Met1783Val; c.5349G > A p.Met1783Ile) located at BRCA1 BRCT repeat on BRCT structural stability, and use the information as the indication for their potential deleterious effects. Of the four unclassified variants, c.5068A > C p.Lys1690Gln and c.5347A > C p.Met1783Leu were predicted to be deleterious (Figure 2). Taking c.5347A > C p.Met1783Leu as an example, p.Met1783 is located within the α’1 helix at C terminal near the edge of the inter repeat interface of the native BRCT structure. While p.Met1783Leu by c.5347A > C was sterically stable without physical contact or clashes with adjoining residues, it unfolded the structure of BRCT and destabilized the hydrophobic interface, causing reposition between the two BRCA1 BRCT repeats as reflected by the larger structure deviation and flexibility, reduced NH bond, and decreased structure compactness as measured by six different MDS programs (RMSD, RMSF, Rg, SASA, NH bond, and Covariance). The results showed that of the three missense variant-caused substitutions at the same position (p.Met1783Leu; p.Met1783Val; p.Met1783Ile), p.Met1783Leu was deleterious by disturbing BRCT structure stability.
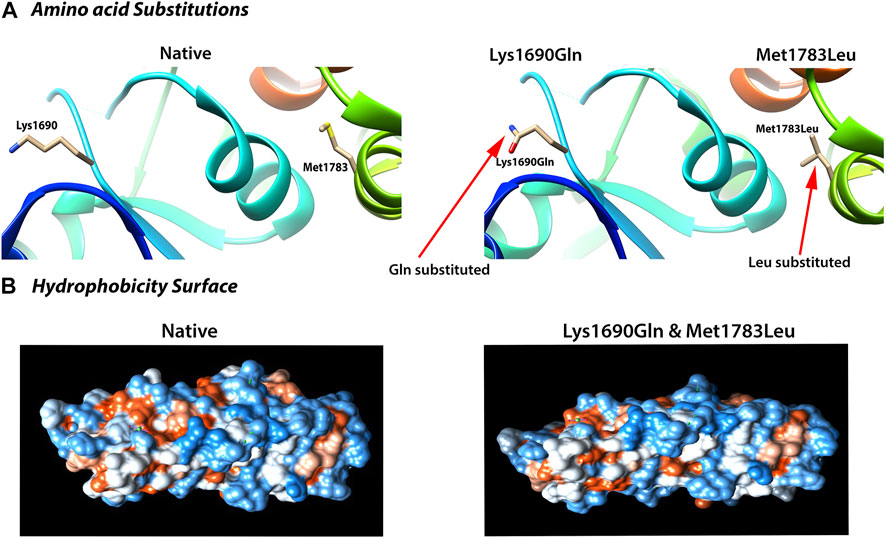
FIGURE 2. Deleterious impact of unclassified variants (c.5068 A > C; p.Lys1690Gln; c.5347 A > C; p.Met1783Leu) on BRCA1 BRCT structural stability. (A) Amino acid substitution showing the variant-caused amino acid change from Lys and Met (left) in the native structure to Gln and Leu (right) at the position of 1,690 and 1783, respectively. (B) Deleterious effects reflected by the change in hydrophobicity surface in the mutant BRCT. Both Lys1690Gln and Met1783Leu caused nearly identical change as shown here. The results were from 40 ns simulation (see text for detailed explanation).
We also used four different types of in silico prediction programs including SIFT, Polyphen2, LRT, and MutationTaster to predict the deleteriousness of the four unclassified variants. The results showed that the two deleterious variants (p.Lys1690Gln, p.Met1783Leu) predicted by MDS were also predicted as deleterious by at least three different programs. For example, p.Met1783Leu was predicted by all four programs as deleterious (Table 3).
The results from MDS and in silico prediction provide strong evidence for the enrichment of ethnic-specific deleterious variants in the unclassified variants.
Pathogenic Variants and Prevalence
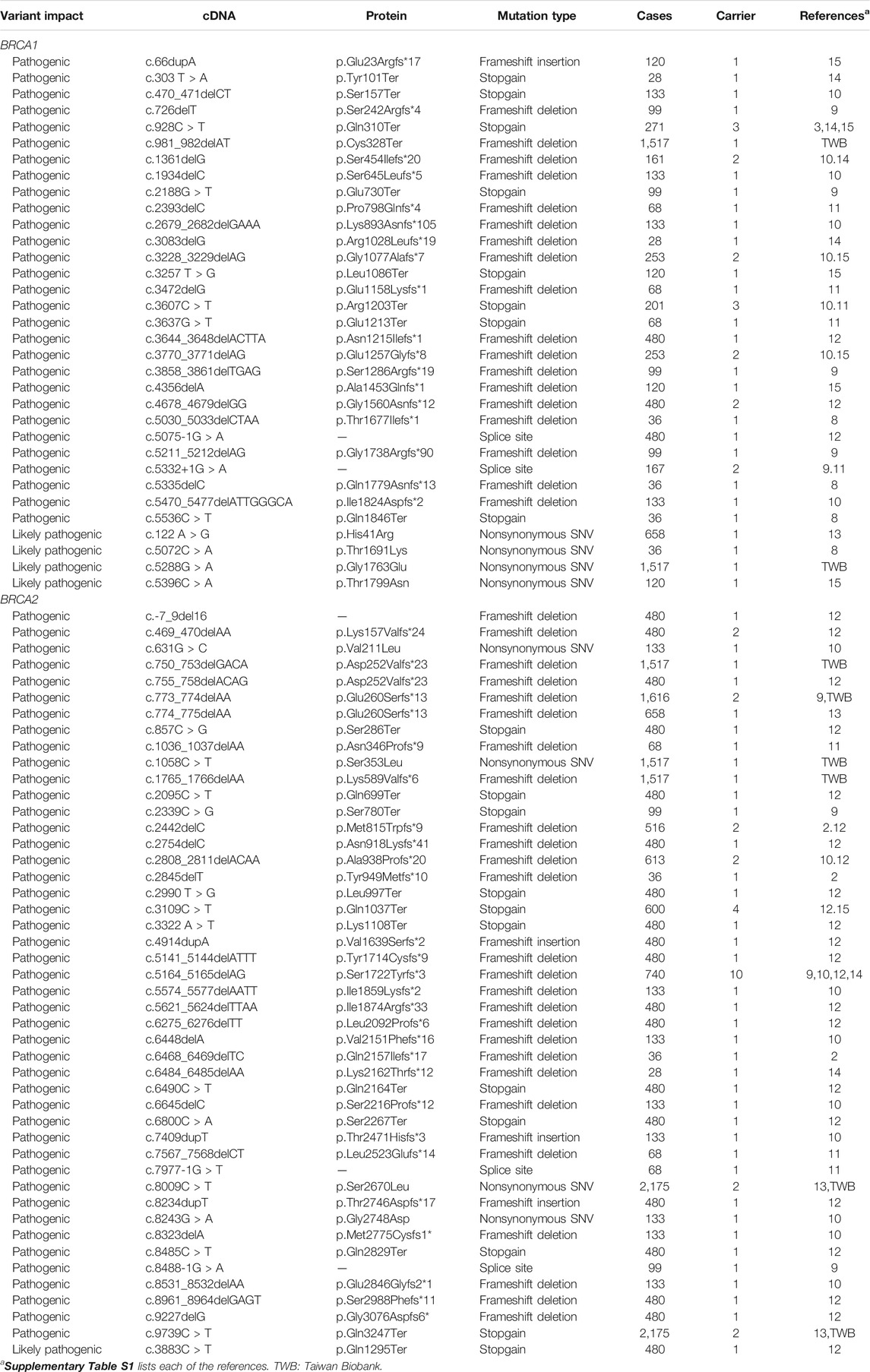
In the general population, we identified eight BRCA pathogenic and likely pathogenic variants, two in BRCA1 with two carriers and six in BRCA2 with six carriers. The eight pathogenic variant carriers in the 1,517 general individuals represent the prevalence of 0.53% BRCA pathogenic variants (0.13% in BRCA1 and 0.40% in BRCA2) in Taiwanese population. The higher prevalence of BRCA2 than BRCA1 is consistent with the pattern in other Asian ethnic populations (Bhaskaran et al., 2020). With 0.53% prevalence, there are estimated 126,140 BRCA pathogenic variant carriers (30,940 in BRCA1 and 95,200 in BRCA2) estimated in the Taiwanese population of 23.8 million or one BRCA pathogenic variant carrier in every 189 Taiwanese individuals. In the cancer cohort of 2,665 cases, we identified 74 BRCA pathogenic variants, 31 in BRCA1 with 40 carriers (2.1% in 1,880 cases) and 43 in BRCA2 with 61 carriers (2.5% in 2,417 cases), resulting in the prevalence of 3.8% in the Taiwanese cancer cohort of breast/ovarian cancer. Five pathogenic variants (two in BRCA1 and three in BRCA2) were present only in general population with five carriers, and three BRCA2 pathogenic variants were present in both general population and the cancer cohort with six carriers. BRCA2 c.5164_5165delAG was a known pathogenic variant in Chinese cancer patients (Kwong et al., 2012). This pathogenic variant was present in the cancer cohort with 10 carriers but not in the general population. This variant is a potential founder mutation in the Taiwanese population and need to be validated by the haplotype test (Table 4).
Discussion
It is well known that the number of benign variants is higher in the general population than in the cancer cohort, and the number of pathogenic mutations is higher in the cancer cohort than in the general population. Our current study aimed to obtain the detailed variant information including position, frequency, classification, and ethnic specificity in the Taiwanese healthy population and the cancer cohort in order to understand the genetic basis of BRCA variation in the population and to develop a precise reference to guide clinical applications.
Taiwanese population has its unique genetic features in reflecting its evolutionary and admixture history (Chen et al., 2016). With a population size of 23.8 million, BRCA variation information provides a unique source to understand its genetic variation in adaptation to the unique environment and the pathogenic variation causing cancer risk in the population. Data from our study provide an overview for BRCA variation and pathogenicity in this population, and further confirms the highly ethnic-specific nature of BRCA variation in eastern Asian population (Bhaskaran et al., 2020).
The availability of BRCA variant data from both general population and the cancer cohort allows comparison of the similarity and differences of BRCA variation between the two groups with the same ethnic background under the same geological environment. The higher rate of BRCA variation in its general population over other ethnic populations may reflect the rapidly evolving BRCA in Taiwanese population for better adaptation in Taiwan’s natural environment (Chen et al., 2016). This could be a factor contributing to higher prevalence of pathogenic variation in Taiwanese general population by increased probability of generating more pathogenic variants. The prevalence of 0.53% of pathogenic variation in the general Taiwanese population is the highest in Asian ethnic populations, comparing to 0.26% in Japanese (Momozawa et al., 2018), 0.29% in southern Chinese (Qin et al., 2020), 0.38% in mainland Chinese (Dong et al., 2020), and 0.39% in Malaysia (Wen et al., 2018), and has reached the same level of 0.53% as in Caucasian populations (Kurian et al., 2019). One BRCA pathogenic variant carrier in every 189 Taiwanese individuals represents a serious threat for public health in Taiwanese population, justifying the inclusion of BRCA-related cancer diagnosis, treatment, and prevention in the healthcare system in Taiwan. Considering its impact on population health, further confirmation of the result with a larger sample size will be necessary to validate the observations. The prevalence of 3.8% in the cancer cohort was lower than that in other ethnic cancer patient groups, such as 5.4% in Caucasians (Sun et al., 2017) and 5.3% in mainland Chinese (Bhaskaran et al., 2019).
All the pathogenic variants identified in Taiwanese population are present in public BRCA databases. Similar situation exists for the pathogenic variants identified in other Asian populations (Bhaskaran et al., 2019; Bhaskaran et al., 2020; Dong et al., 2020; Qin et al., 2020; Zhang et al., 2020). In the meantime, 44.1% of the BRCA variants identified in Taiwanese population remain as novel, VUS, and unclassified variants. Our BRCA study across multiple ethnic Asian populations also showed that 30–50% of variants present in each population were novel, VUS, and unclassified variants. The distribution patterns of pathogenic and unclassified variants seem to suggest that pathogenic variants are universally shared between human populations, whereas non-pathogenic variants are largely ethnic specific. However, such assumption does not have a biological sense. Considering that BRCA variation is highly ethnic specific and a large portion of the BRCA variants identified in ethnic population remain unclassified, it will be logical to consider that ethnic-specific pathogenic variants should also exist, and these are likely enriched within the unclassified variants. The pathogenic variants highly shared between the human populations represent the common pathogenic variants inherited from their common ancestors. They are identifiable by referring to the current well-annotated BRCA pathogenic data predominately derived from Caucasian populations (Rebbeck et al., 2018; Bhaskaran et al., 2019). As these reference databases lack the pathogenic variant data from the non-Caucasian populations, the ethnic-specific pathogenic variants in the non-Caucasian populations are not identifiable by referring to these databases. The ethnic-specific pathogenic variants can be highly enriched within the ethnic-specific novel, VUS, and unclassified variants, as evidenced from our MDS and in silico analyses. However, it remains a challenge in cancer genetic study to develop extensive ethnic-specific pathogenic variant references.
In summary, the data generated from the study provide a comprehensive view for BRCA variation in the Taiwanese population and a reference for clinical applications in BRCA-related cancer in the Taiwanese population.
Materials and Methods
Data Sources
Studies were selected by the following inclusion criteria: 1) cancer patients should be pathologically confirmed, 2) germline variants in BRCA1/2 should be genotyped, and 3) studies on nonhuman or cell line were excluded. Thoroughly searching PubMed and Google Scholar using the keywords such as “BRCA1” “BRCA2” “Taiwan” “Taiwanese” and “cancer predisposition” we identified 15 publications reporting the BRCA data from Taiwanese cancer patients between 1997 and 2020 (Liu et al., 1997; Li et al., 1999; Wang et al., 2000; Chen et al., 2003; Lin et al., 2003; Chen et al., 2005; Chang et al., 2006; Kuo et al., 2012; Chao et al., 2016; Lin et al., 2016; Sung et al., 2017; Wang et al., 2018; Lin et al., 2019; Chao et al., 2020; Lin et al., 2020). Searching Taiwan Biobank (https://taiwanview.twbiobank.org.tw/index; accessed December 15, 2020) using the keywords “BRCA1” and “BRCA2” we obtained the BRCA variation data derived from Taiwanese general population.
Data Analysis
The following details were extracted from the filtered publications, including first author, year of publication, BRCA variants, mutation type of variants, study population, and the number of cases in the study. We standardized the collected BRCA variation data following the Human Genome Variation Society (HGVS) guidelines (den Dunnen et al., 2016). The following reference sequences were used for the mapping analysis: BRCA1: cDNA NM_007294.3, protein NP_009225.1, and genome hg19 NC_000017.10; BRCA2; cDNA NM_000059.3, protein NP_000050.2, and genome hg19 NC_000013.10. We annotated the variants using the ANNOVAR program (Wang et al., 2010). The population frequency was referred to East Asian variants (EAC) from the 1,000 Genome Project (Fairley et al., 2020), the Exome Aggregation Consortium (ExAC) (Lek et al., 2016), and the Genome Aggregation Database (gnomAD) (Karczewski et al., 2020). The variants were compared with the following two BRCA databases: the BRCA Exchange Database (BED, http://brcaexchange.org, accessed December 15, 2020) and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/, accessed December 15, 2020). The variants present in BRCA databases were classified as known variants by referring to the existing classification of pathogenic, likely pathogenic, uncertain significance, likely benign, and benign. The classes for those variants not present in existing BRCA databases were predicted using the InterVar program with default parameters (Li and Wang, 2017). The Fujianese BRCA variants were extracted from the whole genome sequences of Fujian individuals (Yang et al., 2020).
Molecular Dynamics Simulations and in silico Prediction
We utilized molecular dynamics simulations (MDS) to measure the impact of the four BRCA1 unclassified variants in the Taiwanese population (c.5068A > C p.Lys1690Gln, c.5347A > C p.Met1783Leu; c.5347A > G p.Met1783Val; and c.5349G > A p.Met1783Ile) on the stability of the BRCA1 BRCT structure. The MDS system was developed for BRCA1 BRCT variant classification as described in details (Sinha and Wang, 2020). In brief, the process included two major steps: 1) modeling mutant structure. Using the wild-type BRCT structure as the template, each mutant structure was constructed using the Modeller program (version 9.22, UCSF, CA, United States), and further evaluated using the PROCHECK (Sippl, 1993) and PROSA (Wiederstein et al., 2007) programs following the instructions; 2) analyzing the impact of variants on BRCT structural stability by using MDS (Karplus, 2002). Using the wild-type structure as the reference, MDS analyzes the trajectory of the mutant structure over a time period through multiple parameters including RMSD (root mean square deviation) to measure the average deviation in the backbone of Cα trace (Dong et al., 2018), RMSF (root mean square fluctuations) to measure the residue flexibility of the structure (Benson et al., 2012), Rg (radius of gyration) to measure the distance of the atoms of the structure from its center of gravity and axis for the compactness of each structure (Daidone et al., 2003), SASA (solvent accessible surface area) to measure the surface accessibility (Sheu et al., 2003), NH-bond (number of hydrogen bonds) to measure the overall change in the compactness of the mutant structures, and covariance analysis to compare the overall protein motions (Amadei et al., 1993).
Four in silico prediction methods of SIFT (Kumar et al., 2009), Polyphen2_HDIV (Adzhubei et al., 2010), LRT (Chun et al., 2009), and MutationTaster (Schwarz et al., 2010) were used to predict the deleteriousness for the four unclassified variants in BRCT repeats following the default setting in each method.
Statistical Analysis
A chi-square test was used to compare the differences of BRCA variant data between different populations using SPSS (version 26.0, IBM, NY, United States). A p value lower than 0.05 was considered as statistically significant.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JC: data collection, annotation, analysis, and manuscript writing; SS: structural analysis; ZQ: data analysis; SW: funding, experimental design, data analysis and interpretation, and manuscript writing and revision. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Macau Science and Technology Development Fund (No. 085/2017/A2, 0,077/2019/AMJ); the University of Macau (No. SRG2017-00097-FHS, MYRG2019-00018-FHS); and Faculty of Health Sciences, the University of Macau (No. FHSIG/SW/0007/2020P, startup fund) (SW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Taiwan Biobank for providing the BRCA data from Taiwanese general population. We thank the Information and Communication Technology Office (ICTO), the University of Macau for providing the High-Performance Computing Cluster (HPCC) resource and facilities for the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.685174/full#supplementary-material
References
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P., et al. (2010). A Method and Server for Predicting Damaging Missense Mutations. Nat. Methods 7 (4), 248–249. doi:10.1038/nmeth0410-248
Amadei, A., Linssen, A. B. M., and Berendsen, H. J. C. (1993). Essential Dynamics of Proteins. Proteins 17 (4), 412–425. doi:10.1002/prot.340170408
Benson, N. C., and Daggett, V. (2012). A Comparison of Multiscale Methods for the Analysis of Molecular Dynamics Simulations. J. Phys. Chem. B 116 (29), 8722–8731. doi:10.1021/jp302103t
Bhaskaran, S. P., Chandratre, K., Gupta, H., Zhang, L., Wang, X., Cui, J., et al. (2019). Germline Variation in BRCA1/2 Is Highly Ethnic‐specific: Evidence from over 30,000 Chinese Hereditary Breast and Ovarian Cancer Patients. Int. J. Cancer 145 (4), 962–973. doi:10.1002/ijc.32176
Bhaskaran, S. P., Huang, T., Rajendran, B. K., Guo, M., Luo, J., Qin, Z., et al. (2020). Ethnic-specific BRCA1/2 Variation within Asia Population: Evidence from over 78 000 Cancer and 40 000 Non-cancer Cases of Indian, Chinese, Korean and Japanese Populations. J. Med. Genet. [Epub ahead of print]. doi:10.1136/jmedgenet-2020-107299
Burke, W., Daly, M., Garber, J., Botkin, J., Kahn, M. J., Lynch, P., et al. (1997). Recommendations for Follow-Up Care of Individuals with an Inherited Predisposition to Cancer. JAMA 277 (12), 997–1003. doi:10.1001/jama.1997.03540360065034
Chang, T.-W., Wang, S.-M., Guo, Y.-L. L., Tsai, P.-C., Huang, C.-J., and Huang, W. (2006). Glutathione S-Transferase Polymorphisms Associated with Risk of Breast Cancer in Southern Taiwan. The Breast 15 (6), 754–761. doi:10.1016/j.breast.2006.03.008
Chao, A., Chang, T.-C., Lapke, N., Jung, S.-M., Chi, P., Chen, C.-H., et al. (2016). Prevalence and Clinical Significance of BRCA1/2 Germline and Somatic Mutations in Taiwanese Patients with Ovarian Cancer. Oncotarget 7 (51), 85529–85541. doi:10.18632/oncotarget.13456
Chao, A., Lin, Y.-H., Yang, L.-Y., Wu, R.-C., Chang, W.-Y., Chang, P.-Y., et al. (2020). BRCA1/2mutation Status in Patients with Metachronous Breast and Ovarian Malignancies: Clues towards the Implementation of Genetic Counseling. J. Gynecol. Oncol. 31 (3), e24. doi:10.3802/jgo.2020.31.e24
Chen, C.-H., Yang, J.-H., Chiang, C. W. K., Hsiung, C.-N., Wu, P.-E., Chang, L.-C., et al. (2016). Population Structure of Han Chinese in the Modern Taiwanese Population Based on 10,000 Participants in the Taiwan Biobank Project. Hum. Mol. Genet. 25, ddw346–5331. doi:10.1093/hmg/ddw346
Chen, F.-M., Hou, M.-F., Chang, M.-Y., Wang, J.-Y., Hsieh, J.-S., Ou-Yang, F., et al. (2005). High Frequency of Somatic Missense Mutation of BRCA2 in Female Breast Cancer from Taiwan. Cancer Lett. 220 (2), 177–184. doi:10.1016/j.canlet.2004.10.024
Chen, S.-T., Chen, R.-A., Kuo, S.-J., and Chien, Y.-C. (2003). Mutational Screening of Breast Cancer Susceptibility Gene 1 from Early Onset, Bi-lateral, and Familial Breast Cancer Patients in Taiwan. Breast Cancer Res. Treat. 77 (2), 133–143. doi:10.1023/a:1021386026051
Chun, S., and Fay, J. C. (2009). Identification of Deleterious Mutations within Three Human Genomes. Genome Res. 19 (9), 1553–1561. doi:10.1101/gr.092619.109
Daidone, I., Amadei, A., Roccatano, D., and Nola, A. D. (2003). Molecular Dynamics Simulation of Protein Folding by Essential Dynamics Sampling: Folding Landscape of Horse Heart Cytochrome C. Biophysical J. 85 (5), 2865–2871. doi:10.1016/S0006-3495(03)74709-2
den Dunnen, J. T., Dalgleish, R., Maglott, D. R., Hart, R. K., Greenblatt, M. S., McGowan-Jordan, J., et al. (2016). HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 37 (6), 564–569. doi:10.1002/humu.22981
Dong, H., Chandratre, K., Qin, Y., Zhang, J., Tian, X., Rong, C., et al. (2020). Prevalence of BRCA1/BRCA2 Pathogenic Variation in Chinese Han Population. J. Med. Genet. [Epub ahead of print]. doi:10.1136/jmedgenet-2020-106970
Dong, Y.-w., Liao, M.-l., Meng, X.-l., and Somero, G. N. (2018). Structural Flexibility and Protein Adaptation to Temperature: Molecular Dynamics Analysis of Malate Dehydrogenases of marine Molluscs. Proc. Natl. Acad. Sci. USA 115 (6), 1274–1279. doi:10.1073/pnas.1718910115
Dutil, J., Golubeva, V. A., Pacheco-Torres, A. L., Diaz-Zabala, H. J., Matta, J. L., and Monteiro, A. N. (2015). The Spectrum of BRCA1 and BRCA2 Alleles in Latin America and the Caribbean: a Clinical Perspective. Breast Cancer Res. Treat. 154 (3), 441–453. doi:10.1007/s10549-015-3629-3
Fairley, S., Lowy-Gallego, E., Perry, E., and Flicek, P. (2020). The International Genome Sample Resource (IGSR) Collection of Open Human Genomic Variation Resources. Nucleic Acids Res. 48 (D1), D941–D947. doi:10.1093/nar/gkz836
Friebel, T. M., Andrulis, I. L., Balmaña, J., Blanco, A. M., Couch, F. J., Daly, M. B., et al. (2019). BRCA1 and BRCA2 Pathogenic Sequence Variants in Women of African Origin or Ancestry. Hum. Mutat. 40 (10), 1781–1796. doi:10.1002/humu.23804
Jerez, Y., Márquez-Rodas, I., Aparicio, I., Alva, M., Martín, M., and López-Tarruella, S. (2020). Poly (ADP-Ribose) Polymerase Inhibition in Patients with Breast Cancer and BRCA 1 and 2 Mutations. Drugs 80 (2), 131–146. doi:10.1007/s40265-019-01235-5
Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alföldi, J., Wang, Q., et al. (2020). The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 581, 434–443. doi:10.1038/s41586-020-2308-7
Karplus, M. (2002). Molecular Dynamics Simulations of Biomolecules. Acc. Chem. Res. 35, 321–323. doi:10.1021/ar020082r
Kuchenbaecker, K. B., Hopper, J. L., Barnes, D. R., Phillips, K. A., Mooij, T. M., Roos-Blom, M. J., et al. (2017). Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 317 (23), 2402–2416. doi:10.1001/jama.2017.7112
Kumar, P., Henikoff, S., and Ng, P. C. (2009). Predicting the Effects of Coding Non-synonymous Variants on Protein Function Using the SIFT Algorithm. Nat. Protoc. 4 (7), 1073–1081. doi:10.1038/nprot.2009.86
Kuo, W.-H., Lin, P.-H., Huang, A.-C., Chien, Y.-H., Liu, T.-P., Lu, Y.-S., et al. (2012). Multimodel Assessment of BRCA1 Mutations in Taiwanese (Ethnic Chinese) Women with Early-Onset, Bilateral or Familial Breast Cancer. J. Hum. Genet. 57(2), 130–138. doi: doi:10.1038/jhg.2011.142
Kurian, A. W., Ward, K. C., Howlader, N., Deapen, D., Hamilton, A. S., Mariotto, A., et al. (2019). Genetic Testing and Results in a Population-Based Cohort of Breast Cancer Patients and Ovarian Cancer Patients. Jco 37, 1305–1315. doi:10.1200/JCO.18.01854
Kwong, A., Ng, E. K. O., Wong, C. L. P., Law, F. B. F., Au, T., Wong, H. N., et al. (2012). Identification of BRCA1/2 Founder Mutations in Southern Chinese Breast Cancer Patients Using Gene Sequencing and High Resolution DNA Melting Analysis. PLoS One 7 (9), e43994. doi:10.1371/journal.pone.0043994
Lek, M., Karczewski, K. J., Minikel, E. V., Samocha, K. E., Banks, E., Fennell, T., et al. (2016). Analysis of Protein-Coding Genetic Variation in 60,706 Humans. Nature 536, 285–291. doi:10.1038/nature19057
Levy-Lahad, E., Catane, R., Eisenberg, S., Kaufman, B., Hornreich, G., Lishinsky, E., et al. (1997). Founder BRCA1 and BRCA2 Mutations in Ashkenazi Jews in Israel: Frequency and Differential Penetrance in Ovarian Cancer and in Breast-Ovarian Cancer Families. Am. J. Hum. Genet. 60, 1059–1067.
Li, Q., and Wang, K. (2017). InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 100 (2), 267–280. doi:10.1016/j.ajhg.2017.01.004
Li, S. S.-L., Tseng, H.-M., Yang, T.-P., Liu, C.-H., Teng, S.-J., Huang, H.-W., et al. (1999). Molecular Characterization of Germline Mutations in the BRCA1 and BRCA2 Genes from Breast Cancer Families in Taiwan. Hum. Genet. 104, 201–204. doi:10.1007/s004390050936
Lin, P.-C., Yeh, Y.-M., Wu, P.-Y., Hsu, K.-F., Chang, J.-Y., and Shen, M.-R. (2019). Germline Susceptibility Variants Impact Clinical Outcome and Therapeutic Strategies for Stage III Colorectal Cancer. Sci. Rep. 9, 3931. doi:10.1038/s41598-019-40571-0
Lin, P.-H., Kuo, W.-H., Huang, A.-C., Lu, Y.-S., Lin, C.-H., Kuo, S.-H., et al. (2016). Multiple Gene Sequencing for Risk Assessment in Patients with Early-Onset or Familial Breast Cancer. Oncotarget 7, 8310–8320. doi:10.18632/oncotarget.7027
Lin, P. H., Chen, M., Tsai, L. W., Lo, C., Yen, T. C., Huang, T. Y., et al. (2020). Using Next‐generation Sequencing to Redefine BRCAness in Triple‐negative Breast Cancer. Cancer Sci. 111 (4), 1375–1384. doi:10.1111/cas.14313
Lin, Y. P., Chen, Y. L., Chang, H. T., and Li, S. S. L. (2003). Nature of Genetic Variants in the BRCA1 and BRCA2 Genes from Breast Cancer Families in Taiwan. Life Sci. J. 6, 93–97. doi:10.7537/marslsj060309.15
Liu, F.-S., Ho, E. S.-C., Shih, R. T.-P., and Shih, A. (1997). Mutational Analysis of the BRCA1 Tumor Suppressor Gene in Endometrial Carcinoma. Gynecol. Oncol. 66 (3), 449–453. doi:10.1006/gyno.1997.4800
Lou, D. I., McBee, R. M., Le, U. Q., Stone, A. C., Wilkerson, G. K., Demogines, A. M., et al. (2014). Rapid Evolution of BRCA1 and BRCA2 in Humans and Other Primates. BMC Evol. Biol. 14 (1), 155. doi:10.1186/1471-2148-14-155
Momozawa, Y., Iwasaki, Y., Parsons, M. T., Kamatani, Y., Takahashi, A., Tamura, C., et al. (2018). Germline Pathogenic Variants of 11 Breast Cancer Genes in 7,051 Japanese Patients and 11,241 Controls. Nat. Commun. 9 (1), 4083. doi:10.1038/s41467-018-06581-8
Qin, Z., Kuok, C. N., Dong, H., Jiang, L., Zhang, L., Guo, M., et al. (2020). Can Population BRCA Screening Be Applied in Non-ashkenazi Jewish Populations? Experience in Macau Population. J. Med. Genet. [Epub ahead of print]. doi:10.1136/jmedgenet-2020-107181
Rebbeck, T. R., Friebel, T. M., Friedman, E., Hamann, U., Huo, D., Kwong, A., et al. (2018). Mutational Spectrum in a Worldwide Study of 29,700 Families with BRCA1 or BRCA2 Mutations. Hum. Mutat. 39, 593–620. doi:10.1002/humu.23406
Roy, R., Chun, J., and Powell, S. N. (2011). BRCA1 and BRCA2: Different Roles in a Common Pathway of Genome protection. Nat. Rev. Cancer 12 (1), 68–78. doi:10.1038/nrc3181
Schwarz, J. M., Rödelsperger, C., Schuelke, M., and Seelow, D. (2010). MutationTaster Evaluates Disease-Causing Potential of Sequence Alterations. Nat. Methods 7 (8), 575–576. doi:10.1038/nmeth0810-575
Sheu, S.-Y., Yang, D.-Y., Selzle, H. L., and Schlag, E. W. (2003). Energetics of Hydrogen Bonds in Peptides. Proc. Natl. Acad. Sci. 100, 12683–12687. doi:10.1073/pnas.2133366100
Sinha, S., and Wang, S. M. (2020). Classification of VUS and Unclassified Variants in BRCA1 BRCT Repeats by Molecular Dynamics Simulation. Comput. Struct. Biotechnol. J. 18, 723–736. doi:10.1016/j.csbj.2020.03.013
Sippl, M. J. (1993). Recognition of Errors in Three-Dimensional Structures of Proteins. Proteins 17, 355–362. doi:10.1002/prot.340170404
Sun, J., Meng, H., Yao, L., Lv, M., Bai, J., Zhang, J., et al. (2017). Germline Mutations in Cancer Susceptibility Genes in a Large Series of Unselected Breast Cancer Patients. Clin. Cancer Res. 23 (20), 6113–6119. doi:10.1158/1078-0432.CCR-16-3227
Sung, P.-L., Wen, K.-C., Chen, Y.-J., Chao, T.-C., Tsai, Y.-F., Tseng, L.-M., et al. (2017). The Frequency of Cancer Predisposition Gene Mutations in Hereditary Breast and Ovarian Cancer Patients in Taiwan: From BRCA1/2 to Multi-Gene Panels. PLoS One 12, e0185615. doi:10.1371/journal.pone.0185615
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 38 (16), e164. doi:10.1093/nar/gkq603
Wang, P.-H., Shyong, W. Y., Li, Y. F., Lee, H. H., Tsai, W. Y., Chao, H. T., et al. (2000). BRCA1 Mutations in Taiwanese with Epithelial Ovarian Carcinoma and Sporadic Primary Serous Peritoneal Carcinoma. Jpn. J. Clin. Oncol. 30 (8), 343–348. doi:10.1093/jjco/hyd092
Wang, Y. A., Jian, J.-W., Hung, C.-F., Peng, H.-P., Yang, C.-F., Cheng, H.-C. S., et al. (2018). Germline Breast Cancer Susceptibility Gene Mutations and Breast Cancer Outcomes. BMC Cancer 18, 315. doi:10.1186/s12885-018-4229-5
Wen, W. X., Allen, J., Lai, K. N., Mariapun, S., Hasan, S. N., Ng, P. S., et al. (2018). Inherited Mutations in BRCA1 and BRCA2 in an Unselected Multiethnic Cohort of Asian Patients with Breast Cancer and Healthy Controls from Malaysia. J. Med. Genet. 55 (2), 97–103. doi:10.1136/jmedgenet-2017-104947
Wiederstein, M., and Sippl, M. J. (2007). ProSA-web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 35 (Web Server), W407–W410. doi:10.1093/nar/gkm290
Yang, M. A., Fan, X., Sun, B., Chen, C., Lang, J., Ko, Y.-C., et al. (2020). Ancient DNA Indicates Human Population Shifts and Admixture in Northern and Southern China. Science 369 (6501), 282–288. doi:10.1126/science.aba0909
Keywords: BRCA1, BRCA2, prevalence, population, ethnicity, pathogenic
Citation: Chian J, Sinha S, Qin Z and Wang SM (2021) BRCA1 and BRCA2 Variation in Taiwanese General Population and the Cancer Cohort. Front. Mol. Biosci. 8:685174. doi: 10.3389/fmolb.2021.685174
Received: 24 March 2021; Accepted: 28 May 2021;
Published: 21 June 2021.
Edited by:
Xin Wang, City University of Hong, Hong KongReviewed by:
Kajal Biswas, National Cancer Institute, United StatesAngelo Minucci, Agostino Gemelli University Hospital Foundation, Italy
Copyright © 2021 Chian, Sinha, Qin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: San Ming Wang, c2FubWluZ3dhbmdAdW0uZWR1Lm1v
 Jiasheng Chian
Jiasheng Chian San Ming Wang
San Ming Wang