- 1Research Laboratories, Bambino Gesù Children’s Hospital, IRCCS, Rome, Italy
- 2Department of Public Health and Infectious Diseases, Sapienza University of Rome, Rome, Italy
- 3Laboratory Affiliated to Institute Pasteur Italia-Cenci Bolognetti Foundation, Department of Public Health and Infectious Diseases, Sapienza University of Rome, Rome, Italy
- 4Department of Infectious Diseases, National Institute of Health, Rome, Italy
- 5Department of Human Sciences and Promotion of the Quality of Life, San Raffaele Roma Open University, Rome, Italy
- 6IRCCS San Raffaele Roma, Rome, Italy
Bacterial small RNAs (sRNAs) research has accelerated over the past decade, boosted by advances in RNA-seq technologies and methodologies for capturing both protein–RNA and RNA–RNA interactions. The emerging picture is that these regulatory sRNAs play important roles in controlling complex physiological processes and are required to survive the antimicrobial challenge. In recent years, the RNA content of OMVs/EVs has also gained increasing attention, particularly in the context of infection. Secreted RNAs from several bacterial pathogens have been characterized but the exact mechanisms promoting pathogenicity remain elusive. In this review, we briefly discuss how secreted sRNAs interact with targets in infected cells, thus representing a novel perspective of host cell manipulation during bacterial infection. During the last decade, Acinetobacter baumannii became clinically relevant emerging pathogens responsible for nosocomial and community-acquired infections. Therefore, we also summarize recent findings of regulation by sRNAs in A. baumannii and discuss how this emerging bacterium utilizes many of these sRNAs to adapt to its niche and become successful human pathogen.
Introduction
Both coding and non-coding RNA molecules (ncRNAs) have attracted great interest for their ubiquitary roles in almost all cellular and evolutionary processes of living organisms (Wu et al., 2021; Yates et al., 2021). By definition, both coding and ncRNAs are transcribed from DNA but ncRNAs are not translated into functional proteins (Han and Chen, 2015; Diamantopoulos et al., 2018). Remarkably, ncRNAs are expressed at specific stages of cell’s development, and involved in gene modulation in response to external stimuli (Szymański et al., 2003; Li et al., 2021). Long (>200 nt) and short (<200 nt) ncRNAs were found in both eukaryotes and prokaryotes (de la Fuente et al., 2012; Deogharia & Gurha, 2022).
Discovered as early as the 1950s, ncRNAs were initially deemed as by-products of huge transcripts with negligible biological roles (i.e., “junk” RNAs). Thereafter, the development of advanced technologies revolutionized the exploration of ncRNAs, resulting in the discovery of distinctive ncRNA species, both cellular and circulating, with different biological roles in many human diseases (Cech & Steitz, 2014; Felli et al., 2017; Hung et al., 2018; Slack & Chinnaiyan, 2019; Tait et al., 2020; Toden et al., 2021). Among all, the best-studied family of ncRNAs is miRNAs that able to interact with other ncRNAs as well as various types of mRNA transcripts in a regulatory crosstalk known as competing endogenous RNAs (ceRNAs) leading to an additional post-transcriptional levels (Agirre & Eyras, 2011; Ala, 2020; Li et al., 2020).
Recently, Seal and colleagues have provided a guide to the nomenclature of human ncRNAs genes by reviewing each major class based on the HUGO Gene Nomenclature Committee (HGNC; http://www.genenames.org), providing an updated database and useful resources available online (Da Sacco et al., 2011; Seal et al., 2020). They function as post-transcriptional regulators of their mRNA targets resulting in the translational repression and/or degradation (Djebali et al., 2012; Catalanotto et al., 2016; Hung et al., 2018; Sun et al., 2018). Moreover, different kinds of ncRNAs can interact with each other and/or with DNA, as well as other RNA classes and proteins (Baldassarre & Masotti, 2012; RamónCajal et al., 2019; Sun et al., 2020). Various ncRNAs have been discovered to be encapsulated and transported through exosomes, microvesicles, and apoptotic bodies, all referred to as extracellular vesicles (EVs), ranging from ∼30 to 150 nm in diameter (Fischer & Deindl, 2021; Li et al., 2021). Exosomal ncRNAs, mainly miRNAs, lncRNAs, and circRNAs, have been reported to be expressed by different cells in several physiological and pathological conditions (Li et al., 2021). Once secreted, they are transported by body fluids, such as serum, plasma, urine, saliva, etc, to recipient cells where they could influence different functionalities (Li et al., 2021; Qiu et al., 2021; Wang et al., 2022) or can be employed as useful diagnostic biomarkers of paediatric diseases (Masotti et al., 2017a; Masotti et al., 2017b; Paolini et al., 2022a; Paolini et al., 2022b; Felli et al., 2022).
Insights into bacterial sRNAs
Further investigations have led to explore bacterial RNA-dependent mechanisms associated with ncRNAs. In bacteria, a class of small ncRNAs (i.e., sRNAs), are gene expression mediators at both transcriptional and post-transcriptional levels. So far, hundreds of sRNA molecules ranging from approximately 50–400 nt were found ubiquitously in different bacterial species, among which sRNAs from Escherichia coli (E. coli) are the most studied (Storz et al., 2011; Kang et al., 2013; Pérez-Reytor et al., 2017; Mediati et al., 2021). Using next-generation sequencing (NGS), Lee and Hong identified several short sRNAs (ca. 26 nt) in Streptococcus mutans, and proposed the term “microRNA-size,” small RNA (msRNA) (Lee & Hong, 2012). Furthermore, very small RNAs (vsRNAs <16 nt) as well as tRNA-derived fragments (tRFs), belonging to the category of vsRNAs, were recently discovered in bacteria (Diallo et al., 2022). Interestingly, tRFs seem to represent the majority of sncRNA types in bacteria as well as in eukaryotes (Plante et al., 2012; Lambert et al., 2021) suggesting a conserved evolutionary strategy to modulate gene expression from bacteria to eukaryotes.
Nowadays, thanks to the development of RNA sequencing, it is known that production and regulation of bacterial sRNAs is coordinated through other components, including others sRNAs, mRNAs, and remarkably sRNA-binding proteins (sRBPs) (Adams & Storz, 2020; Goldberger et al., 2021). Several sRBPs (e.g., Hfq, ProQ, and CsrA) are involved in facilitating sRNA-mRNA base-pairing in Enterobacteriaceae, mainly investigated in E. coli and Salmonella enterica serovar Typhimurium (Storz et al., 2011; Kim & Kwon, 2013; Klein & Raina, 2017; Goldberger et al., 2021). In E. coli, the Hfq protein, an RNA chaperone, enhances base pairing between a specific sRNA and its mRNA target (Waters & Storz, 2009; De Lay et al., 2013; Vazquez-Anderson & Contreras, 2013). Hfq interacts with at least 40% of the known sRNAs and has the ability to interact not only with other ncRNAs, but also with longer RNAs (i.e., rRNAs and tRNAs) and most importantly with many different mRNAs (Pichon & Felden, 2007; Bandyra et al., 2012; Waters et al., 2017; Quendera et al., 2020). The functional importance of sRBPs in “sRNA-mediated” regulation of gene expression in bacteria was summarized previously (Pichon & Felden, 2007).
Host-pathogen interactions mediated by sRNAs
sRNAs secreted by bacteria may play important roles not only in microbe–microbe but also in host–microbe interactions (Ahmadi Badi et al., 2020). In both Gram-negative and Gram-positive bacteria, two major characteristics of sRNAs have been identified, the cis- and trans-encoded sRNAs. These sRNAs, once coupled to their RNA target(s) lead to inhibition or activation of target gene expression (e.g., genes responsible for controlling bacterial adaptation to environmental changes and virulence) through a variety of post-transcriptional gene regulation mechanisms (Li et al., 2012; Felden & Gilot, 2018; Chakravarty & Massé, 2019; Felden & Augagneur, 2021; Millar & Raghavan, 2021). Usually, cis-encoded sRNAs are transcribed from the same genetic locus but in the opposite direction to their single RNA target against which they pair, whereas trans-encoded sRNAs are transcribed in a distant locus (respect to theis targets) and exert their regulatory functions by partially pairing to their RNA targets (Adams & Storz, 2020; Carrier et al., 2020; Dell’Annunziata et al., 2020; Jørgensen et al., 2020). Several bacterial sRNAs are considered as master regulators in that they are able to modulate the expression of transcriptional factors responsible for the activation of virulence genes or they can target the quorum-sensing regulatory components by regulating multiple mRNA targets (Quorum Regulatory RNAs, QRR) (Broach et al., 2012; Pérez-Reytor et al., 2017; Felden & Cattoir, 2018; Janssen et al., 2020; Lalaouna et al., 2021). Hence, sRNAs are emerging as key controllers of central regulatory circuits determining the bacterial lifestyle. sRNAs encoded on pathogenicity islands (PAIs) or virulence plasmids are prime candidates to directly control the expression of virulence genes (Papenfort & Vogel, 2010; Papenfort & Vogel, 2014). Several types of RNA-based regulatory mechanisms as well as their interactome in pathogenic bacteria have been reported so far (Sridhar & Gunasekaran, 2013; Waters et al., 2017; Chakravarty & Massé, 2019). The most intriguing types of bacterial sRNAs are those targeting human host mRNAs. Some sRNAs adopt secondary structures that allow them to interact with human host mRNAs during bacterial infection. Although the function of this class of RNAs has not been fully elucidated yet, their secondary structure could bind not only to specific human mRNAs but also could regulate the expression of individual bacterial genes (Choi et al., 2017). Bacterial sRNAs that mimic eukaryotic miRNAs could target the host immune response during infection. A tRF from Pseudomonas aeruginosa downregulates the inflammatory response in both in vitro and in vivo pulmonary infection models. Indeed, it was shown that this sRNA could downregulate the expression of specific MAPKs involved in the activation of NF-kB, thereby leading to a decreased expression of IL-8 (Koeppen et al., 2016). Recently, Sahr and colleagues reported that the sRNAs RsmY and tRNA-Phe from Legionella pneumophila (L. pneumophila) could base pair with eukaryotic mRNAs either in the coding region or the untranslated region (UTR) (Sahr et al., 2022). This binding leads to the inhibition of proteins involved in the formation of the RIG-I-like receptor as well as those involved in the Toll-like receptor signalling pathway, such as RIG-I, cRel, and IRAK1 (Sahr et al., 2022).
Furthermore, it has been reported that several bacterial pathogens produce sRNAs that can be secreted within EVs produced by Gram-positive bacteria or outer membrane vesicles (OMVs) produced by Gram-negative bacteria and, by these means, transferred into eukaryotic cells and/or to other bacteria (Ahmadi Badi et al., 2017; Choi et al., 2017; Ahmadi Badi et al., 2020; Stanton, 2021). For instance, L. pneumophila releases EVs containing sRNAs both in vitro and in vitro during infection; these sRNAs modulate the host innate immune response in argonaute-2 (Ago2)-dependent manner (Sahr et al., 2022). Ago2 as a component of the RNA-induced silencing complex (RISC), involved in several cellular processes and functions in RNA-mediated gene silencing (RNAi) (Müller et al., 2020). Taken together, in the host–pathogen interaction scenario, pathogens could actively alter the host machinery for their own benefits by changing the pattern of host mRNA expression (Sarshar et al., 2022a). However, additional researches are necessary to elucidate how bacterial sRNAs delivered by OMVs/EVs mediate inter-kingdom communication upon entering human cells.
The role of A. baumannii sRNAs in different networks of interactions
Hospital-acquired infections caused by multidrug resistant (MDR) bacteria remain an unresolved problem in healthcare settings worldwide. Recently, A. baumannii emerged as one of the major opportunistic nosocomial and community-acquired pathogens; its propensity to acquire multidrug, extensive drug and even pandrug resistance phenotypes reduced effective therapies and increased mortality rates (Dehbanipour and Ghalavand, 2022). Apart from pneumonia, several others infections such as wound infections, meningitis, endocarditis, osteomyelitis, endophthalmitis and urinary tract infections (UTIs) have been reported in adults as well as children (Hu & Robinson, 2010; Di Venanzio et al., 2019; Sarshar et al., 2021). The A. baumannii ability to adhere and form biofilms on both biotic and abiotic surfaces is the cause of several biofilm-mediated infections such as catheter-associated UTIs (CAUTIs) in clinical settings (Di Venanzio et al., 2019). Within the last decade, several studies tried to unravel the complex mechanisms that led to the emergence of A. baumannii as a formidable human pathogen, from its pathogenicity to resistance strategies (Ambrosi et al., 2017; Scribano et al., 2019; Ambrosi et al., 2020; Pompilio et al., 2021; Marazzato et al., 2022). However, these researches have been hampered by the wide heterogeneity of A. baumannii isolates owing to its high genome plasticity (i.e., capability to acquire extracellular DNA, high frequency of homologous recombination and elevated presence of mobile elements), that widens its ability to adapt and persist in hospital settings where the presence of these kind of pathogens is constantly monitored (Gaiarsa et al., 2019; Leal et al., 2020; Whiteway et al., 2022). Gene expression modulation by transcription factors (TFs), two-component systems (TCSs), σ factors and sRNA-mediated mechanisms are among the preferred bacterial strategies to fight environmental stresses (i.e., oxidation, acidic environment, osmotic or temperature gradients, nutrient starvation, and antibiotic exposure) (Felden & Cattoir, 2018; Allen et al., 2020). In particular, it was recently shown that bacterial trans-encoded sRNAs could modulate antibiotic resistance mechanisms through several strategies, such as by acting on efflux pumps, membrane transporters, LPS and/or cell wall biosynthetic modifications, DNA mutagenesis and biofilm synthesis, thereby fine tuning the responsiveness to a broad spectrum of antibiotics. Detailed reviews on this topic have appeared in the last few years (Dersch et al., 2017; Mediati et al., 2021). However, the overall properties of trans-encoded sRNAs in A. baumannii remain elusive compared to other Gram-negative species. Of note, carbapenem-resistant A. baumannii is a major concern owing to the enhanced expression of carbapenamases, several classes of efflux pumps and outer membrane proteins (OMPs), which have been reported to be correlated with the capability of this pathogen to form biofilm (Pompilio et al., 2021; Sarshar et al., 2021). Conversely, the sRNAs regulating efflux pumps (EPs) have been extensively studied in E. coli, Clostridium acetobutylicum, Mycobacterium tuberculosis, Shigella sonnei and S. enterica ser. Typhimurium (Yu & Schneiders, 2012; Chan et al., 2017; Felden & Cattoir, 2018; Gan & Tan, 2019). For example, in Enterobacteriaceae, the homeostasis of major OMPs, such as OmpA, OmpC, OmpD, and OmpF, is controlled by the regulatory action of several sRNAs (Guillier et al., 2006; Papenfort et al., 2006; Klein & Raina, 2017; Matera et al., 2022). As these sRNAs have a pivotal role as virulence factors and in drug resistance of several OMPs in A. baumannii, mainly OmpA, BamA, LptD, Omp33–36, OmpW, CarO, and OprD, it would be interesting to investigate whether specific sRNAs could either suppress or enhance the expression of these OMPs in A. baumannii (Figure 1, top left).
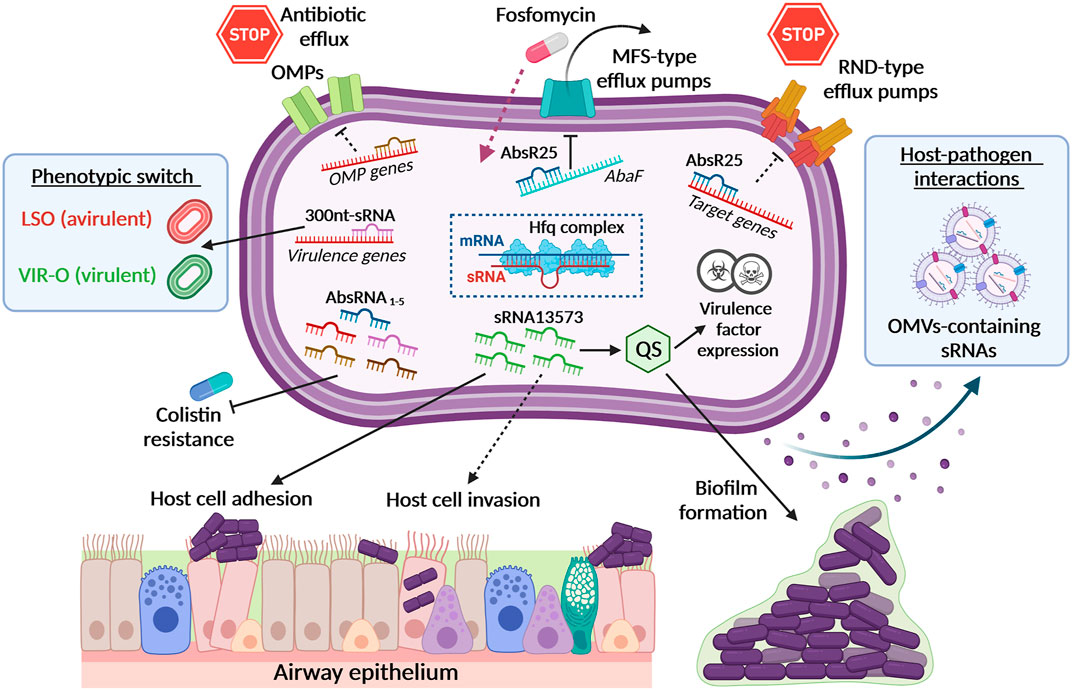
FIGURE 1. Small RNAs (sRNAs) in A. baumannii physiology, antibiotic-resistance and virulence. The figure summarizes A. baumannii sRNAs associated with their targets and relative functions. Black dotted lines represent putative or underexplored roles. By the downregulation of the abaF gene, sRNA AbsR25 affects the function of the major facilitator superfamily (MFS) transporter, thereby leading to an increased susceptibility to fosfomycin. Other sRNAs can regulate the expression of other efflux pumps and outer membrane proteins (OMPs). The sRNA 13573 was found to be overexpressed during A. baumannii biofilm formation as well as during airway epithelial cell adhesion, probably belonging to the Quorum Sensing (QS) Regulatory RNAs. Additional sRNAs are believed to control the expression of A. baumannii virulence factors. The identification of differentially regulated sRNAs linked to colistin resistance in COLR A. baumannii strains provides the basis for their use as diagnostic markers. A 300 nt-long sRNA has been found to be overexpressed in a subpopulation of opaque virulent colonies showing low rate of translucent switch, named low-switching opaque variants (LSO). Additionally, A. baumannii can release sRNAs-containing outer membrane vesicles (OMVs) that may have a role in host-pathogen interactions. A. baumannii Hfq complex (i.e., Hfq, mRNA and sRNA) depicted in the centered dotted square represents the main regulatory machinery able to modulate the expression of several fundamental proteins, such as OMPs or to impair sRNA stability and influence bacterial pathogenicity. All the items depicted in this picture have been fully described in the main text.
Pathania and colleagues were the first group who investigated the presence of regulatory sRNAs and their functionality in A. baumannii. They identified distinct sets of novel differentially expressed sRNAs in A. baumannii strain MTCC 1425 compared to ATCC 17978, suggesting that in A. baumannii the expression of sRNAs is strain-specific (Sharma et al., 2014). In this screening, the authors found a sRNA, designated AbsR25. By target gene prediction, AbsR25 was initially recognized as the regulator of an efflux pump in A. baumannii, therefore suggesting that it might be either directly or indirectly involved in the expression of a transporter and potentially also in drug resistance mechanisms (Sharma et al., 2014) (Figure 1, top right). Later on, Sharma and colleagues demonstrated that the sRNA AbsR25 negatively regulates the expression of the abaF gene, which belongs to the major facilitator superfamily (MFS) transporter. AbaF actively effluxes fosfomycin, rendering the cells resistant. Accordingly, its expression is upregulated upon exposure to this antibiotic (Figure 1, top center). Inhibition of abaF by AbsR25 lowered fosfomycin minimum inhibitory concentration (MIC) by eightfold but also decreased biofilm formation and virulence in A. baumannii (Sharma et al., 2016). Additionally, Alvarez-Fraga and colleagues analyzed the regulatory effects of A. baumannii sRNAs on biofilm formation both in vitro and in vivo (Alvarez-Fraga et al., 2017). Among the differentially expressed sRNAs, the expression level of the sRNA 13573 was significantly higher in biofilm as well as during adhesion to A549 human alveolar epithelial cells, compared to planktonic A. baumannii ATCC 17978 cells (Figure 1, middle center and bottom center) (Alvarez-Fraga et al., 2017). Although the mechanism(s) underlying this regulation remains to be fully determined, the involvement of sRNAs in the adhesion to epithelial cells highlights the unrealized key role of sRNAs in A. baumannii-host interaction to establish a successful infection.
So far, the comprehensive transcriptomic analysis of Acinetobacter spp. led to the discovery of 110 potential sRNAs, the majority of which are conserved in A. baumannii and, to a lesser extent, in Acinetobacter nosocomialis and Acinetobacter pittii (Kröger et al., 2018). Kröger’s group improved the resolution of sRNA de novo discovery by differential RNA-seq pooled from sixteen different growth conditions in A. baumannii ATCC 17978. Their results indicated that the majority of sRNAs were located within intergenic regions of the bacterial genome or antisense with respect to coding regions or within the 3′ regions of coding genes. Interestingly, 22 sRNAs were found within this latter location and all possessed their own promoters located upstream of the coding gene (Kröger et al., 2018). However, the authors did not explore the possible targets of these 22 sRNAs.
Recently, Cafiso et al. identified several msRNAs that are differentially expressed in colistin resistant (COL R) A. baumannii strains through both experimental and computational analyses (Figure 1, bottom left). Although identified as precursors of their mature microRNA-size small RNA form (pre-microAbsRNA), computational prediction of their targets revealed their involvement in the regulation of different biological functions (i.e., biofilm production, virulence and aminoglycoside-resistance), highlighting their role in host-microbe interactions (Cafiso et al., 2020). The innovative aspect of this work is the identification of downregulated sRNAs in COLR vs. colistin susceptible A. baumannii strains that could be used to rapidly identify resistant isolates. Most importantly, despite several sRNAs have been identified in the MDR A. baumannii strain AB5075, their physiological function(s) remain uncertain so far. Weiss and colleagues identified 78 novel short and conserved strain-specific sRNAs that are present in large copy numbers within the AB5075 genome, using a RNA-seq-based approach (Weiss et al., 2016). Two more sRNAs, Aar and AbsR28 were previously reported in Acinetobacter baylyi and A. baumannii ATCC 17978, respectively (Schilling et al., 2010; Sharma et al., 2014). Among the others and based on internal homology, they identified six groups of sRNAs with one sRNA particularly abundant and homologous to regulatory C4 antisense RNAs found in bacteriophages P1 and P7. Four additional highly conserved RNA species were identified in strain AB5075, including the signal recognition particle (SRP) RNA, 6S RNA, tmRNA and RNase P RNA, annotated as ABUWs030, ABUWs053, ABUWs059, and ABUWs062. Interestingly, three of them, ABUWs030, ABUWs053, and ABUWs059, were among the most highly expressed ncRNAs in A. baumannii, suggesting their potential physiological relevance in this pathogen (Weiss et al., 2016).
Recently, the expression of a ∼300-nt sRNA has been found to be involved in controlling the phenotypic switch from the virulent opaque (VIR-O) to the avirulent translucent (AV-T) phenotype in strain AB5075 (Anderson et al., 2020). This sRNA was encoded at the 5′ end of the aadB gene within resistance island two and showed a variable expression. In fact, gene expression analysis revealed that bacterial opacity switching rate varied from VIR-O to the low-switching opaque variants (LSO) as a function of the copy number of this locus (Figure 1, middle left). The LSO represents a subpopulation of cells that exhibit dramatically reduced levels of switching to AV-T relative to that for the VIR-O. This identified sRNA affected virulence, as the LSO cells exhibited decreased virulence in vivo. In addition, more than 100 genes were identified as differentially expressed between VIR-O and LSO, suggesting that the ∼300-nt sRNA may act as a global regulator (Anderson et al., 2020).
As already outlined, Hfq can regulate the interactions between mRNA and sRNA, both positively and negatively. Among RNA chaperones, Hfq has been the only one functionally studied in Acinetobacter spp. (Figure 1, middle center). Indeed, recent analysis of Hfq in A. baylyi and A. baumannii ATCC 17978 revealed that deletion of hfq in A. baumannii impaired growth with respect to the wild-type strain, increased susceptibility to environmental stressors, including desiccation, decreased carbon metabolism and reduced host cell adhesion and virulence (Kuo et al., 2017). The same pleotropic effects observed in A. baylyi and A. baumannii ATCC 17978 were also shown in Salmonella and E. coli hfq mutants, including reduced growth rates, elevated sensitivity to environmental stresses, defeated OMV production, fimbriae, biofilm formation and adhesion, invasion and survival in eukaryotic cells (Sittka et al., 2008; Hayashi-Nishino et al., 2012; Kuo et al., 2017). While it appears that Hfq is a vital virulence factor in A. baumannii, it remains unclear how this chaperone is involved in mediating sRNA-mRNA interactions, a function that is worth to be explored.
Conclusion and future perspectives
Host-pathogen interactions are based on the production of specific types of communication molecules. Among them, non-coding small RNAs, sRNAs, received an increased attention as they are directly involved in the regulation of gene expression, resulting in a rapid phenotypic change and fast adaptation to the environment. The study of eukaryotic miRNAs started about 30 years ago and a huge knowledge on their targets and functions are nowadays available. Owing to this, several miRNAs are currently used as disease biomarkers, they are targets for new therapeutics (anti-miR compounds) or under therapeutic evaluation in several clinical trials. Vice versa, the massive discovery of bacterial sRNAs, the similarity with human miRNAs, and detailed functional studies are very recent concepts and the role of bacteria sRNAs in host-pathogen interaction is still an unexplored field. Only few studies reported that secreted bacterial RNAs can be stably transported either in OMVs or as complexes with proteins/biomolecules (Ghosal, 2018; Quendera, et al., 2020) or found in free-form circulating in the human plasma of healthy donors (Wang and Fu, 2019). sRNA-dependent gene regulation influences bacterial lifestyle, modulating the switch from a “harmless” colonizer to a pathogen. However, the study of the expression profiles of bacterial sRNAs has not been investigated in details yet, neither in healthy individuals nor in infected patients. We think that further studies about bacterial sRNAs could help to better understand the role of these molecules as a diverse toolkit for bacterial adaptation to the host environment (Sarshar et al., 2017; Ambrosi et al., 2019; Sarshar et al., 2022b) or as potentially novel therapeutics (i.e., administration of natural or synthetic oligonucleotides) (Markelova et al., 2021; Ozoline and Shavkunov, 2021). Being an opportunistic pathogen, A. baumannii represents an excellent model to characterize bacterial sRNAs and evaluate their role in A. baumannii antibiotic-resistance and pathogenesis. This is particularly true for those sRNAs packed into OMVs that can be directly absorbed by the host cells (Figure 1, middle right). However, the studies on the functional role of bacterial secreted RNAs in infected host cells has only recently started to appear and achieved results are insufficient for depicting the whole crosstalk during bacterial infection. Indeed, the majority of the studies were conducted using in vitro or animal models that do not consider the human tissue architecture and function. Hence, application of human-derived advanced cell cultures, such as organoids, together with high resolution RNA-seq techniques will enable us to understand the impact of sRNAs on infectious diseases. These models will allow the molecular characterization of the interactions between bacterial RNAs and host factors and will clarify their roles and importance in bacterial pathogenicity, thereby including them under the general concept of “RNA effector.” Last but not least, owing to the presence of specific sRNAs in MDR bacteria and/or their differential expression upon antibiotic stress, this field of research opens the possibility to develop alternative and innovative therapeutic strategies based on sRNAs.
Author contributions
MS conceived and planned the study and organized the manuscript; MS, DS, and CA performed a literature search and prepared the first draft of the manuscript. DS and AM illustrated the picture. MS, DS, CA, AP, and AM provided expert input, discussed the conception of the review, and contributed to the final manuscript preparation. MS and AM supervised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Italian Ministry of Health (Ricerca Corrente IRCCS San Raffaele Roma to CA and 5 × 1000 to AM). Salary of MS was funded by the Italian Ministry of Health (starting grant SG-2018-12365432). The research was also supported by the Bandi Ateneo Sapienza, grant number RP120172B7FF9E6F to CA. Salary of DS was funded by POR Lazio FSE 2014–2020 and Sapienza Ateneo funding.
Acknowledgments
Image has been created with Biorender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, P. P., and Storz, G. (2020). Prevalence of small base-pairing RNAs derived from diverse genomic loci. Biochimica Biophysica Acta (BBA)-Gene Regul. Mech. 1863, 194524. doi:10.1016/j.bbagrm.2020.194524
Agirre, E., and Eyras, E. (2011). Databases and resources for human small non-coding RNAs. Hum. Genomics 5, 1–8. doi:10.1186/1479-7364-5-3-192
Ahmadi Badi, S., Bruno, S. P., Moshiri, A., Tarashi, S., Siadat, S. D., and Masotti, A. (2020). Small RNAs in outer membrane vesicles and their function in host-microbe interactions. Front. Microbiol. 11, 1209. doi:10.3389/fmicb.2020.01209
Ahmadi Badi, S., Moshiri, A., Fateh, A., Rahimi Jamnani, F., Sarshar, M., Vaziri, F., et al. (2017). Microbiota-derived extracellular vesicles as new systemic regulators. Front. Microbiol. 8, 1610. doi:10.3389/fmicb.2017.01610
Ala, U. (2020). Competing endogenous RNAs, non-coding RNAs and diseases: An intertwined story. Cells 9, 1574. doi:10.3390/cells9071574
Allen, J. L., Tomlinson, B. R., Casella, L. G., and Shaw, L. N. (2020). Regulatory networks important for survival of Acinetobacter baumannii within the host. Curr. Opin. Microbiol. 55, 74–80. doi:10.1016/j.mib.2020.03.001
Alvarez-Fraga, L., Rumbo-Feal, S., Perez, A., Gomez, M. J., Gayoso, C., Vallejo, J. A., et al. (2017). Global assessment of small RNAs reveals a non-coding transcript involved in biofilm formation and attachment in Acinetobacter baumannii ATCC 17978. PloS one 12, e0182084. doi:10.1371/journal.pone.0182084
Ambrosi, C., Sarshar, M., Aprea, M. R., Pompilio, A., Di Bonaventura, G., Strati, F., et al. (2019). Colonic adenoma-associated Escherichia coli express specific phenotypes. Microbes Infect. 21 (7), 305–312. doi:10.1016/j.micinf.2019.02.001
Ambrosi, C., Scribano, D., Aleandri, M., Zagaglia, C., Di Francesco, L., Putignani, L., et al. (2017). Acinetobacter baumannii virulence traits: A comparative study of a novel sequence type with other Italian endemic international clones. Front. Microbiol. 8, 1977. doi:10.3389/fmicb.2017.01977
Ambrosi, C., Scribano, D., Sarshar, M., Zagaglia, C., Singer, B. B., and Palamara, A. T. (2020). Acinetobacter baumannii targets human carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) for invasion of pneumocytes. mSystems 5, e00604–e00620. doi:10.1128/mSystems.00604-20
Anderson, S. E., Chin, C. Y., Weiss, D. S., and Rather, P. N. (2020). Copy number of an integron-encoded antibiotic resistance locus regulates a virulence and opacity switch in Acinetobacter baumannii AB5075. mBio 11, e02338–e02320. doi:10.1128/mBio.02338-20
Baldassarre, A., and Masotti, A. (2012). Long non-coding RNAs and p53 regulation. Int. J. Mol. Sci. 13, 16708–16717. doi:10.3390/ijms131216708
Bandyra, K. J., Said, N., Pfeiffer, V., Górna, M. W., Vogel, J., and Luisi, B. F. (2012). The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell. 47, 943–953. doi:10.1016/j.molcel.2012.07.015
Broach, W. H., Egan, N., Wing, H. J., Payne, S. M., and Murphy, E. R. (2012). VirF-independent regulation of Shigella virB transcription is mediated by the small RNA RyhB. PloS one 7, e38592. doi:10.1371/journal.pone.0038592
Cafiso, V., Stracquadanio, S., Lo Verde, F., Dovere, V., Zega, A., Pigola, G., et al. (2020). COLR acinetobacter baumannii sRNA signatures: Computational comparative identification and biological targets. Front. Microbiol. 10, 3075. doi:10.3389/fmicb.2019.03075
Carrier, M-C., Ng Kwan Lim, E., Jeannotte, G., and Massé, E. (2020). Trans-acting effectors versus RNA cis-elements: A tightly knit regulatory mesh. Front. Microbiol. 11, 609237. doi:10.3389/fmicb.2020.609237
Catalanotto, C., Cogoni, C., and Zardo, G. (2016). MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 17, 1712. doi:10.3390/ijms17101712
Cech, T. R., and Steitz, J. A. (2014). The noncoding RNA revolution—Trashing old rules to forge new ones. Cell. 157, 77–94. doi:10.1016/j.cell.2014.03.008
Chakravarty, S., and Massé, E. (2019). RNA-dependent regulation of virulence in pathogenic bacteria. Front. Cell. Infect. Microbiol. 9, 337. doi:10.3389/fcimb.2019.00337
Chan, H., Ho, J., Liu, X., Zhang, L., Wong, S. H., Chan, M. T., et al. (2017). Potential and use of bacterial small RNAs to combat drug resistance: A systematic review. Infect. Drug Resist. 10, 521–532. doi:10.2147/IDR.S148444
Choi, J-W., Kim, S-C., Hong, S-H., and Lee, H-J. (2017). Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J. Dent. Res. 96, 458–466. doi:10.1177/0022034516685071
Da Sacco, L., Baldassarre, A., and Masotti, A. (2011). Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int. J. Mol. Sci. 13, 97–114. doi:10.3390/ijms13010097
de la Fuente, M., Valera, S., and Martínez-Guitarte, J. L. (2012). ncRNAs and thermoregulation: a view in prokaryotes and eukaryotes. FEBS Lett. 586, 4061–4069. doi:10.1016/j.febslet.2012.10.018
De Lay, N., Schu, D. J., and Gottesman, S. (2013). Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 288, 7996–8003. doi:10.1074/jbc.R112.441386
Dehbanipour, R., and Ghalavand, Z. (2022). Acinetobacter baumannii: Pathogenesis, virulence factors, novel therapeutic options and mechanisms of resistance to antimicrobial agents with emphasis on tigecycline. J. Clin. Pharm. Ther. 47, 1875–1884. doi:10.1111/jcpt.13787
Dell’Annunziata, F., Ilisso, C. P., Dell’Aversana, C., Greco, G., Coppola, A., Martora, F., et al. (2020). Outer membrane vesicles derived from klebsiella pneumoniae influence the mirna expression profile in human bronchial epithelial beas-2b cells. Microorganisms 8, 1985. doi:10.3390/microorganisms8121985
Deogharia, M., and Gurha, P. (2022). The “guiding” principles of noncoding RNA function. Wiley Interdiscip. Rev. RNA 13, e1704. doi:10.1002/wrna.1704
Dersch, P., Khan, M. A., Mühlen, S., and Görke, B. (2017). Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 8, 803. doi:10.3389/fmicb.2017.00803
Di Venanzio, G., Flores-Mireles, A. L., Calix, J. J., Haurat, M. F., Scott, N. E., Palmer, L. D., et al. (2019). Urinary tract colonization is enhanced by a plasmid that regulates uropathogenic Acinetobacter baumannii chromosomal genes. Nat. Commun. 10, 2763–2813. doi:10.1038/s41467-019-10706-y
Diallo, I., Ho, J., Lalaouna, D., Massé, E., and Provost, P. (2022). RNA sequencing unveils very small RNAs with potential regulatory functions in bacteria. Front. Mol. Biosci. 9, 914991. doi:10.3389/fmolb.2022.914991
Diamantopoulos, M. A., Tsiakanikas, P., and Scorilas, A. (2018). Non-coding RNAs: The riddle of the transcriptome and their perspectives in cancer. Ann. Transl. Med. 6, 241. doi:10.21037/atm.2018.06.10
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi:10.1038/nature11233
Felden, B., and Augagneur, Y. (2021). Diversity and versatility in small RNA-mediated regulation in bacterial pathogens. Front. Microbiol. 2273, 719977. doi:10.3389/fmicb.2021.719977
Felden, B., and Cattoir, V. (2018). Bacterial adaptation to antibiotics through regulatory RNAs. Antimicrob. Agents Chemother. 62, e02503–e02517. doi:10.1128/AAC.02503-17
Felden, B., and Gilot, D. (2018). Modulation of bacterial sRNAs activity by epigenetic modifications: Inputs from the eukaryotic miRNAs. Genes. 10, 22. doi:10.3390/genes10010022
Felli, C., Baldassarre, A., and Masotti, A. (2017). Intestinal and circulating microRNAs in coeliac disease. Int. J. Mol. Sci. 18, 1907. doi:10.3390/ijms18091907
Felli, C., Baldassarre, A., Uva, P., Alisi, A., Cangelosi, D., Ancinelli, M., et al. (2022). Circulating microRNAs as novel non-invasive biomarkers of paediatric celiac disease and adherence to gluten-free diet. EBioMedicine 76, 103851. doi:10.1016/j.ebiom.2022.103851
Fischer, S., and Deindl, E. (2021). Characterization of RNA in extracellular vesicles. Appl. Sci. 11, 7520. doi:10.3390/app11167520
Gaiarsa, S., Bitar, I., Comandatore, F., Corbella, M., Piazza, A., Scaltriti, E., et al. (2019). Can insertion sequences proliferation influence genomic plasticity? Comparative analysis of acinetobacter baumannii sequence type 78, a persistent clone in Italian hospitals. Front. Microbiol. 10, 2080. doi:10.3389/fmicb.2019.02080
Gan, I., and Tan, H. S. (2019). A small RNA decreases the sensitivity of Shigella sonnei to norfloxacin. BMC Res. Notes 12, 97–103. doi:10.1186/s13104-019-4124-4
Ghosal, A. (2018). Secreted bacterial RNA: An unexplored avenue. FEMS Microbiol. Lett. 365 (7). doi:10.1093/femsle/fny036
Goldberger, O., Livny, J., Bhattacharyya, R., and Amster-Choder, O. (2021). Wisdom of the crowds: A suggested polygenic plan for small-RNA-mediated regulation in bacteria. Iscience 24, 103096. doi:10.1016/j.isci.2021.103096
Guillier, M., Gottesman, S., and Storz, G. (2006). Modulating the outer membrane with small RNAs. Genes. Dev. 20, 2338–2348. doi:10.1101/gad.1457506
Han, Li C., and Chen, Y. (2015). Small and long non-coding RNAs: Novel targets in perspective cancer therapy. Curr. Genomics 16, 319–326. doi:10.2174/1389202916666150707155851
Hayashi-Nishino, M., Fukushima, A., and Nishino, K. (2012). Impact of Hfq on the intrinsic drug resistance of Salmonella enterica serovar typhimurium. Front. Microbiol. 3, 205. doi:10.3389/fmicb.2012.00205
Hu, J., and Robinson, J. L. (2010). Systematic review of invasive Acinetobacter infections in children. Can. J. Infect. Dis. Med. Microbiol. 21, 83–88. doi:10.1155/2010/690715
Hung, J., Miscianinov, V., Sluimer, J. C., Newby, D. E., and Baker, A. H. (2018). Targeting non-coding RNA in vascular biology and disease. Front. Physiol. 9, 1655. doi:10.3389/fphys.2018.01655
Janssen, K. H., Corley, J. M., Djapgne, L., Cribbs, J., Voelker, D., Slusher, Z., et al. (2020). Hfq and sRNA 179 inhibit expression of the Pseudomonas aeruginosa cAMP-Vfr and type III secretion regulons. mBio 11, e00363–e00320. doi:10.1128/mBio.00363-20
Jørgensen, M. G., Pettersen, J. S., and Kallipolitis, B. H. (2020). sRNA-mediated control in bacteria: An increasing diversity of regulatory mechanisms. Biochim. Biophys. Acta. Gene Regul. Mech. 1863, 194504. doi:10.1016/j.bbagrm.2020.194504
Kang, S-M., Choi, J-W., Lee, Y., Hong, S-H., and Lee, H-J. (2013). Identification of microRNA-size, small RNAs in Escherichia coli. Curr. Microbiol. 67, 609–613. doi:10.1007/s00284-013-0411-9
Kim, J. N., and Kwon, Y. M. (2013). Genetic and phenotypic characterization of the RyhB regulon in Salmonella Typhimurium. Microbiol. Res. 168, 41–49. doi:10.1016/j.micres.2012.06.007
Klein, G., and Raina, S. (2017). Small regulatory bacterial RNAs regulating the envelope stress response. Biochem. Soc. Trans. 45, 417–425. doi:10.1042/BST20160367
Koeppen, K., Hampton, T. H., Jarek, M., Scharfe, M., Gerber, S. A., Mielcarz, D. W., et al. (2016). A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 12, e1005672. doi:10.1371/journal.ppat.1005672
Kröger, C., MacKenzie, K. D., Alshabib, E. Y., Kirzinger, M. W. B., Suchan, D. M., Chao, T-C., et al. (2018). The primary transcriptome, small RNAs and regulation of antimicrobial resistance in Acinetobacter baumannii ATCC 17978. Nucleic Acids Res. 46, 9684–9698. doi:10.1093/nar/gky603
Kuo, H. Y., Chao, H. H., Liao, P. C., Hsu, L., Chang, K. C., Tung, C. H., et al. (2017). Functional characterization of acinetobacter baumannii lacking the RNA chaperone hfq. Front. Microbiol. 8, 2068. doi:10.3389/fmicb.2017.02068
Lalaouna, D., Fochesato, S., Harir, M., Ortet, P., Schmitt-Kopplin, P., Heulin, T., et al. (2021). Amplifying and fine-tuning rsm sRNAs expression and stability to optimize the survival of Pseudomonas brassicacerum in nutrient-poor environments. Microorganisms 9, 250. doi:10.3390/microorganisms9020250
Lambert, M., Benmoussa, A., Diallo, I., Ouellet-Boutin, K., Dorval, V., Majeau, N., et al. (2021). Identification of abundant and functional dodecaRNAs (doRNAs) derived from ribosomal RNA. Int. J. Mol. Sci. 22, 9757. doi:10.3390/ijms22189757
Leal, N. C., Campos, T. L., Rezende, A. M., Docena, C., Mendes-Marques, C. L., de Sá Cavalcanti, F. L., et al. (2020). Comparative genomics of Acinetobacter baumannii clinical strains from Brazil reveals polyclonal dissemination and selective exchange of mobile genetic elements associated with resistance genes. Front. Microbiol. 1176, 1176. doi:10.3389/fmicb.2020.01176
Lee, H-J., and Hong, S-H. (2012). Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol. Lett. 326, 131–136. doi:10.1111/j.1574-6968.2011.02441.x
Li, C., Ni, Y-Q., Xu, H., Xiang, Q-Y., Zhao, Y., Zhan, J-K., et al. (2021). Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target. Ther. 6, 383–431. doi:10.1038/s41392-021-00779-x
Li, W., Ying, X., Lu, Q., and Chen, L. (2012). Predicting sRNAs and their targets in bacteria. Genomics Proteomics Bioinforma. 10, 276–284. doi:10.1016/j.gpb.2012.09.004
Li, Y., Shan, G., Teng, Z-Q., and Wingo, T. S. (2020) Editorial: Non-Coding RNAs and Human Diseases. Front. Genet. 11, 523. doi:10.3389/fgene.2020.00523
Marazzato, M., Scribano, D., Sarshar, M., Brunetti, F., Fillo, S., Fortunato, A., et al. (2022). Genetic diversity of antimicrobial resistance and key virulence features in two extensively drug-resistant acinetobacter baumannii isolates. Int. J. Environ. Res. Public Health 19, 2870. doi:10.3390/ijerph19052870
Markelova, N., Glazunova, O., Alikina, O., Panyukov, V., Shavkunov, K., and Ozoline, O. (2021). Suppression of Escherichia coli growth dynamics via RNAs secreted by competing bacteria. Front. Mol. Biosci. 8, 609979. doi:10.3389/fmolb.2021.609979
Masotti, A., Baldassarre, A., Fabrizi, M., Olivero, G., Loreti, M., Giammaria, P., et al. (2017b). Oral glucose tolerance test unravels circulating miRNAs associated with insulin resistance in obese preschoolers. Pediatr. Obes. 12, 229–238. doi:10.1111/ijpo.12133
Masotti, A., Baldassarre, A., Guzzo, M. P., Iannuccelli, C., Barbato, C., and Di Franco, M. (2017a). Circulating microRNA profiles as liquid biopsies for the characterization and diagnosis of fibromyalgia syndrome. Mol. Neurobiol. 54, 7129–7136. doi:10.1007/s12035-016-0235-2
Matera, G., Altuvia, Y., Gerovac, M., El Mouali, Y., Margalit, H., and Vogel, J. (2022). Global RNA interactome of Salmonella discovers a 5′ UTR sponge for the MicF small RNA that connects membrane permeability to transport capacity. Mol. Cell. 82, 629–644.e4. doi:10.1016/j.molcel.2021.12.030
Mediati, D. G., Wu, S., Wu, W., and Tree, J. J. (2021). Networks of resistance: Small RNA control of antibiotic resistance. Trends Genet. 37, 35–45. doi:10.1016/j.tig.2020.08.016
Millar, J. A., and Raghavan, R. (2021). Modulation of bacterial fitness and virulence through antisense RNAs. Front. Cell. Infect. Microbiol. 10, 596277. doi:10.3389/fcimb.2020.596277
Müller, M., Fazi, F., and Ciaudo, C. (2020). Argonaute proteins: From structure to function in development and pathological cell fate determination. Front. Cell. Dev. Biol. 7, 360. doi:10.3389/fcell.2019.00360
Ozoline, O., and Shavkunov, K. (2021). Editorial: Small RNAs as a diverse toolkit for bacteria. Front. Mol. Biosci. 8, 791021. doi:10.3389/fmolb.2021.791021
Paolini, A., Baldassarre, A., Bruno, S. P., Felli, C., Muzi, C., Badi, S. A., et al. (2022b). Improving the diagnostic potential of extracellular miRNAs coupled to multiomics data by exploiting the power of artificial intelligence. Front. Microbiol. 13, 888414. doi:10.3389/fmicb.2022.888414
Paolini, A., Sarshar, M., Felli, C., Bruno, S. P., Rostami-Nejad, M., Ferretti, F., et al. (2022a). Biomarkers to monitor adherence to gluten-free diet by celiac disease patients: Gluten immunogenic peptides and urinary miRNAs. Foods 11, 1380. doi:10.3390/foods11101380
Papenfort, K., Pfeiffer, V., Mika, F., Lucchini, S., Hinton, J. C., and Vogel, J. (2006). SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688. doi:10.1111/j.1365-2958.2006.05524.x
Papenfort, K., and Vogel, J. (2010). Regulatory RNA in bacterial pathogens. Cell. Host Microbe 8, 116–127. doi:10.1016/j.chom.2010.06.008
Papenfort, K., and Vogel, J. (2014). Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front. Cell. Infect. Microbiol. 4, 91. doi:10.3389/fcimb.2014.00091
Pérez-Reytor, D., Plaza, N., Espejo, R. T., Navarrete, P., Bastías, R., and Garcia, K. (2017). Role of non-coding regulatory RNA in the virulence of human pathogenic Vibrios. Front. Microbiol. 7, 2160. doi:10.3389/fmicb.2016.02160
Pichon, C., and Felden, B. (2007). Proteins that interact with bacterial small RNA regulators. FEMS Microbiol. Rev. 31, 614–625. doi:10.1111/j.1574-6976.2007.00079.x
Plante, I., Plé, H., Landry, P., Gunaratne, P. H., and Provost, P. (2012). Modulation of microRNA activity by semi-microRNAs. Front. Genet. 3, 99. doi:10.3389/fgene.2012.00099
Pompilio, A., Scribano, D., Sarshar, M., Di Bonaventura, G., Palamara, A. T., and Ambrosi, C. (2021). Gram-negative bacteria holding together in a biofilm: The acinetobacter baumannii way. Microorganisms 9, 1353. doi:10.3390/microorganisms9071353
Qiu, Y., Li, P., Zhang, Z., and Wu, M. (2021). Corrigendum: Insights into exosomal non-coding RNAs sorting mechanism and clinical application. Front. Oncol. 11, 781714. doi:10.3389/fonc.2021.781714
Quendera, A. P., Seixas, A. F., Dos Santos, R. F., Santos, I., Silva, J. P., Arraiano, C. M., et al. (2020). RNA-binding proteins driving the regulatory activity of small non-coding RNAs in bacteria. Front. Mol. Biosci. 7, 78. doi:10.3389/fmolb.2020.00078
Ramón, y., Cajal, S., Segura, M. F., and Hümmer, S. (2019). Interplay between ncRNAs and cellular communication: A proposal for understanding cell-specific signaling pathways. Front. Genet. 10, 281. doi:10.3389/fgene.2019.00281
Sahr, T., Escoll, P., Rusniok, C., Bui, S., Pehau-Arnaudet, G., Lavieu, G., et al. (2022). Translocated Legionella pneumophila small RNAs mimic eukaryotic microRNAs targeting the host immune response. Nat. Commun. 13, 762–818. doi:10.1038/s41467-022-28454-x
Sarshar, M., Behzadi, P., Scribano, D., Palamara, A. T., and Ambrosi, C. (2021). Acinetobacter baumannii: An ancient commensal with weapons of a pathogen. Pathogens 10 (4), 387. doi:10.3390/pathogens10040387
Sarshar, M., Scribano, D., Behzadi, P., Masotti, A., and Ambrosi, C. (2022a). Outer membrane vesicles are the powerful cell-to-cell communication vehicles that allow bacteria to monitor extracellular milieu. ExRNA 4, 25. doi:10.21037/exrna-22-18
Sarshar, M., Scribano, D., Limongi, D., Zagaglia, C., Palamara, A. T., and Ambrosi, C. (2022b). Adaptive strategies of uropathogenic Escherichia coli CFT073: From growth in lab media to virulence during host cell adhesion. Int. Microbiol. 25 (3), 481–494. doi:10.1007/s10123-022-00235-y
Sarshar, M., Scribano, D., Marazzato, M., Ambrosi, C., Aprea, M. R., Aleandri, M., et al. (2017). Genetic diversity, phylogroup distribution and virulence gene profile of pks positive Escherichia coli colonizing human intestinal polyps. Microb. Pathog. 112, 274–278. doi:10.1016/j.micpath.2017.10.009
Schilling, D., Findeiß, S., Richter, A. S., Taylor, J. A., and Gerischer, U. (2010). The small RNA aar in acinetobacter baylyi: A putative regulator of amino acid metabolism. Arch. Microbiol. 192, 691–702. doi:10.1007/s00203-010-0592-6
Scribano, D., Marzano, V., Levi Mortera, S., Sarshar, M., Vernocchi, P., Zagaglia, C., et al. (2019). Insights into the periplasmic proteins of acinetobacter baumannii AB5075 and the impact of imipenem exposure: A proteomic approach. Int. J. Mol. Sci. 20, 3451. doi:10.3390/ijms20143451
Seal, R. L., Chen, L. L., Griffiths-Jones, S., Lowe, T. M., Mathews, M. B., O'Reilly, D., et al. (2020). A guide to naming human non-coding RNA genes. EMBO J. 39, e103777. doi:10.15252/embj.2019103777
Sharma, A., Sharma, R., Bhattacharyya, T., Bhando, T., and Pathania, R. (2016). Fosfomycin resistance in acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter—AbaF. J. Antimicrob. Chemother. 72, 68–74. doi:10.1093/jac/dkw382
Sharma, R., Arya, S., Patil, S. D., Sharma, A., Jain, P. K., Navani, N. K., et al. (2014). Identification of novel regulatory small RNAs in Acinetobacter baumannii. PloS one 9, e93833. doi:10.1371/journal.pone.0093833
Sittka, A., Lucchini, S., Papenfort, K., Sharma, C. M., Rolle, K., Binnewies, T. T., et al. (2008). Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4, e1000163. doi:10.1371/journal.pgen.1000163
Slack, F. J., and Chinnaiyan, A. M. (2019). The role of non-coding RNAs in oncology. Cell. 179, 1033–1055. doi:10.1016/j.cell.2019.10.017
Sridhar, J., and Gunasekaran, P. (2013). Computational small RNA prediction in bacteria. Bioinforma. Biol. insights 7, 83–95. doi:10.4137/BBI.S11213
Stanton, B. A. (2021). Extracellular vesicles and host–pathogen interactions: A review of inter-kingdom signaling by small noncoding RNA. Genes. 12, 1010. doi:10.3390/genes12071010
Storz, G., Vogel, J., and Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell. 43, 880–891. doi:10.1016/j.molcel.2011.08.022
Sun, B., Liu, C., Li, H., Zhang, L., Luo, G., Liang, S., et al. (2020). Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol. Lett. 19, 595–605. doi:10.3892/ol.2019.11182
Sun, Q., Hao, Q., and Prasanth, K. V. (2018). Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 34, 142–157. doi:10.1016/j.tig.2017.11.005
Szymański, M., Erdmann, V. A., and Barciszewski, J. (2003). Noncoding regulatory RNAs database. Nucleic Acids Res. 31, 429–431. doi:10.1093/nar/gkg124
Tait, S., Baldassarre, A., Masotti, A., Calura, E., Martini, P., Varì, R., et al. (2020). Integrated transcriptome analysis of human visceral adipocytes unravels dysregulated microRNA-long non-coding RNA-mRNA networks in obesity and colorectal cancer. Front. Oncol. 10, 1089. doi:10.3389/fonc.2020.01089
Toden, S., Zumwalt, T. J., and Goel, A. (2021). Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys. Acta. Rev. Cancer 1875, 188491. doi:10.1016/j.bbcan.2020.188491
Vazquez-Anderson, J., and Contreras, L. M. (2013). Regulatory RNAs: Charming gene management styles for synthetic biology applications. RNA Biol. 10, 1778–1797. doi:10.4161/rna.27102
Wang, J., Yue, B-L., Huang, Y-Z., Lan, X-Y., Liu, W-J., and Chen, H. (2022). Exosomal RNAs: Novel potential biomarkers for diseases—a review. Int. J. Mol. Sci. 23, 2461. doi:10.3390/ijms23052461
Wang, Y. F., and Fu, Y (2019). Secretory and circulating bacterial small RNAs: A mini-review of the literature. ExRNA 1, 14. doi:10.1186/s41544-019-0015-z
Waters, L. S., and Storz, G. (2009). Regulatory RNAs in bacteria. Cell. 136, 615–628. doi:10.1016/j.cell.2009.01.043
Waters, S. A., McAteer, S. P., Kudla, G., Pang, I., Deshpande, N. P., Amos, T. G., et al. (2017). Small RNA interactome of pathogenic E. coli revealed through crosslinking of RN ase E. EMBO J. 36, 374–387. doi:10.15252/embj.201694639
Weiss, A., Broach, W. H., Lee, M. C., and Shaw, L. N. (2016). Towards the complete small RNome of Acinetobacter baumannii. Microb. Genom. 2, e000045. doi:10.1099/mgen.0.000045
Whiteway, C., Breine, A., Philippe, C., and Van der Henst, C. (2022). Acinetobacter baumannii. Trends Microbiol. 30, 199–200. doi:10.1016/j.tim.2021.11.008
Wu, J. Y., Ma, X. C., Ma, L., Fang, Y., Zhang, Y. H., Liu, L. J., et al. (2021). Complete chloroplast genome sequence and phylogenetic analysis of winter oil rapeseed (Brassica rapa L.). Mitochondrial DNA. B Resour. 6, 723–731. doi:10.1080/23802359.2020.1860697
Yates, M. C., Derry, A. M., and Cristescu, M. E. (2021). Environmental RNA: A revolution in ecological resolution? Trends Ecol. Evol. 36, 601–609. doi:10.1016/j.tree.2021.03.001
Keywords: Acinetobacter baumannii, small RNAs (sRNAs), host-pathogen interactions, non-coding RNAs, antibiotic-resistance, outer membrane vesicles (OMVs)
Citation: Sarshar M, Scribano D, Palamara AT, Ambrosi C and Masotti A (2022) The Acinetobacter baumannii model can explain the role of small non-coding RNAs as potential mediators of host-pathogen interactions. Front. Mol. Biosci. 9:1088783. doi: 10.3389/fmolb.2022.1088783
Received: 03 November 2022; Accepted: 07 December 2022;
Published: 21 December 2022.
Edited by:
Olga N. Ozoline, Institute of Cell Biophysics (RAS), RussiaReviewed by:
Jonathan Perreault, Université du Québec, CanadaCopyright © 2022 Sarshar, Scribano, Palamara, Ambrosi and Masotti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meysam Sarshar, bWV5c2FtLnNhcnNoYXJAdW5pcm9tYTEuaXQ=; Andrea Masotti, YW5kcmVhLm1hc290dGlAb3BiZy5uZXQ=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Meysam Sarshar
Meysam Sarshar Daniela Scribano
Daniela Scribano Anna Teresa Palamara3,4
Anna Teresa Palamara3,4 Cecilia Ambrosi
Cecilia Ambrosi Andrea Masotti
Andrea Masotti