- 1State Key Laboratory of Oral & Maxillofacial Reconstruction and Regeneration, Key Laboratory of Oral Biomedicine Ministry of Education, Hubei Key Laboratory of Stomatology, School & Hospital of Stomatology, Wuhan University, Wuhan, Hubei, China
- 2Xiamen Key Laboratory of Stomatological Disease Diagnosis and Treatment, Stomatological Hospital of Xiamen Medical College, Xiamen, China
- 3Guangdong Provincial Key Laboratory of Stomatology, Guanghua School of Stomatology, Hospital of Stomatology, Sun Yat-sen University, Guangdong, China
- 4Center for Cariology, Endodontics and Periodontics, School & Hospital of Stomatology, Wuhan University, Wuhan, Hubei, China
N6-methyladenosine (m6A), an abundant internal RNA modification in eukaryotes, serves as a dynamic post-transcriptional regulator of gene expression by influencing RNA splicing, stability, translation, and decay. This reversible epitranscriptomic mechanism, which is mediated by methyltransferase (writers), demethylase (erasers), and m6A-binding proteins (readers), is pivotal in diverse biological contexts. Among m6A erasers, alkylation repair homolog protein 5 (ALKBH5), an Fe(II)/α-ketoglutarate-dependent dioxygenase, is the second to be discovered and one of the most significant demethylases. Mounting evidence underscores ALKBH5’s role in modulating developmental programming, where it coordinates processes such as lineage specification, organogenesis, and tissue homeostasis. This review systematically deciphers the multifaceted contributions of ALKBH5-mediated m6A demethylation to developmental biology. We synthesize recent advances elucidating how ALKBH5-driven m6A erasure dynamically regulates transcriptomic rewiring during embryogenesis, reproductive development, cardiac development, central nervous system development, immune system development, pancreatic organogenesis, osteogenic/odontogenic differentiation, adipogenesis, and angiogenesis. These revelations not only deepen our understanding of epitranscriptomic regulation in ontogeny but also illuminate therapeutic avenues for developmental anomalies and regenerative medicine.
1 Introduction
Development is an incredibly intricate and elaborate process through which a single fertilized cell undergoes a series of remarkable transformations and eventually evolves into a highly complex multicellular organism (Shestopalov and Chen, 2008; Loseva and Gladyshev, 2024). Development is fundamental for life, guaranteeing survival, reproduction, and the continuity of species. This meticulously orchestrated journey encompasses cellular differentiation, tissue patterning, and organogenesis, driven by precise spatiotemporal regulation of gene expression (Warmflash et al., 2014; Mittnenzweig et al., 2021; Ohta and Yamada, 2023). An indispensable factor in development is epigenetics, which regulates gene expression without modifying the underlying DNA sequence (Bird, 2007; Taby and Issa, 2010). Key epigenetic processes, including DNA methylation, posttranslational modifications, and RNA-based mechanisms such as N6-methyladenosine (m6A) methylation, enable cells to interpret genetic information in a context-dependent manner (Matouk and Marsden, 2008). Disruptions in epigenetic regulation are linked to developmental disorders, aging, and cancer, underscoring their dual role as guardians of normal development and mediators of disease (Taby and Issa, 2010; Ashapkin et al., 2023; Wang et al., 2022). Consequently, understanding the epigenetic networks in development may contribute to unveil new frontiers in developmental biology and regenerative medicine.
The epitranscriptome, encompassing post-transcriptional chemical modifications of RNA, constitutes a fundamental regulatory layer in gene expression (Li C. et al., 2025). Among these modifications, m6A is the most abundant and dynamic internal modification, notably present in different RNA types including messenger RNAs (mRNAs), circular RNAs (circRNAs), micro RNAs (miRNAs), and long non-coding RNAs (lncRNAs) (Desrosiers et al., 1974; Xiao et al., 2023; Jiang et al., 2021). The deposition, removal, and recognition of m6A, which are respectively orchestrated by writer, eraser, and reader proteins, govern RNA metabolism at multiple levels, including splicing, stability, translation, and subcellular localization (Jiang et al., 2021; Sendinc and Shi, 2023; Zaccara et al., 2019). This reversible modification system is capable of responding to developmental cues and environmental stimuli, positioning m6A as a key regulator of cellular differentiation, tissue patterning and organismal development (Geula et al., 2015; Zheng et al., 2020). Among the enzymes responsible for m6A erasure, alkylation repair homolog protein 5 (ALKBH5), an Fe(II)/α-ketoglutarate-dependent dioxygenase, has garnered significant attention for its unique ability to selectively demethylate m6A in RNA species, including mRNA, circRNA, and lncRNA (Aik et al., 2014; Shao et al., 2023; Cai et al., 2024). Mounting evidence now implicates ALKBH5 as a key epigenetic regulator of development, where its demethylase activity influences embryogenesis, organogenesis, and tissue regeneration (Liang et al., 2024; Dong et al., 2023; Ma et al., 2022; Han et al., 2021a). However, there is no systematic article that comprehensively summarizes the role and regulatory mechanism of ALKBH5 in development.
This review synthesizes the current knowledge regarding the regulatory contributions of ALKBH5 to developmental biology, emphasizing its mechanistic interplay with m6A-modified transcripts. We first embark on a detailed description of the m6A modification, delving into its various characteristics and implications. Building upon this framework, we delineate the structural basis of ALKBH5’s enzymatic activity and substrate recognition, providing a molecular framework for its developmental functions. Subsequently, we dissect its subtle yet significant stage-specific influences in certain development processes such as embryogenesis, neurodevelopment, reproductive biology, and organogenesis, focusing on its regulation of key mRNA or signaling pathways (e.g., Wnt/β-catenin, PI3K/AKT) through selective m6A erasure. Finally, we briefly introduced the therapeutic strategies specifically targeting ALKBH5 and discussed its potential in treating developmental disorders.
2 m6A modifications: a dynamic regulatory layer in RNA biology
2.1 m6A modifications
RNA maturation requires a wide variety of enzymes for its chemical modification. To date, over 170 types of chemical modifications have been identified on RNA (Wang M. K. et al., 2023). Among them, m6A, which was first discovered in 1974 and is the most abundant internal chemical modification in eukaryotic mRNA, accounting for over 80% of all RNA methylation modifications, has emerged as a pivotal post-transcriptional regulator of gene expression (Desrosiers et al., 1974; Xiao et al., 2023; Zheng et al., 2020).
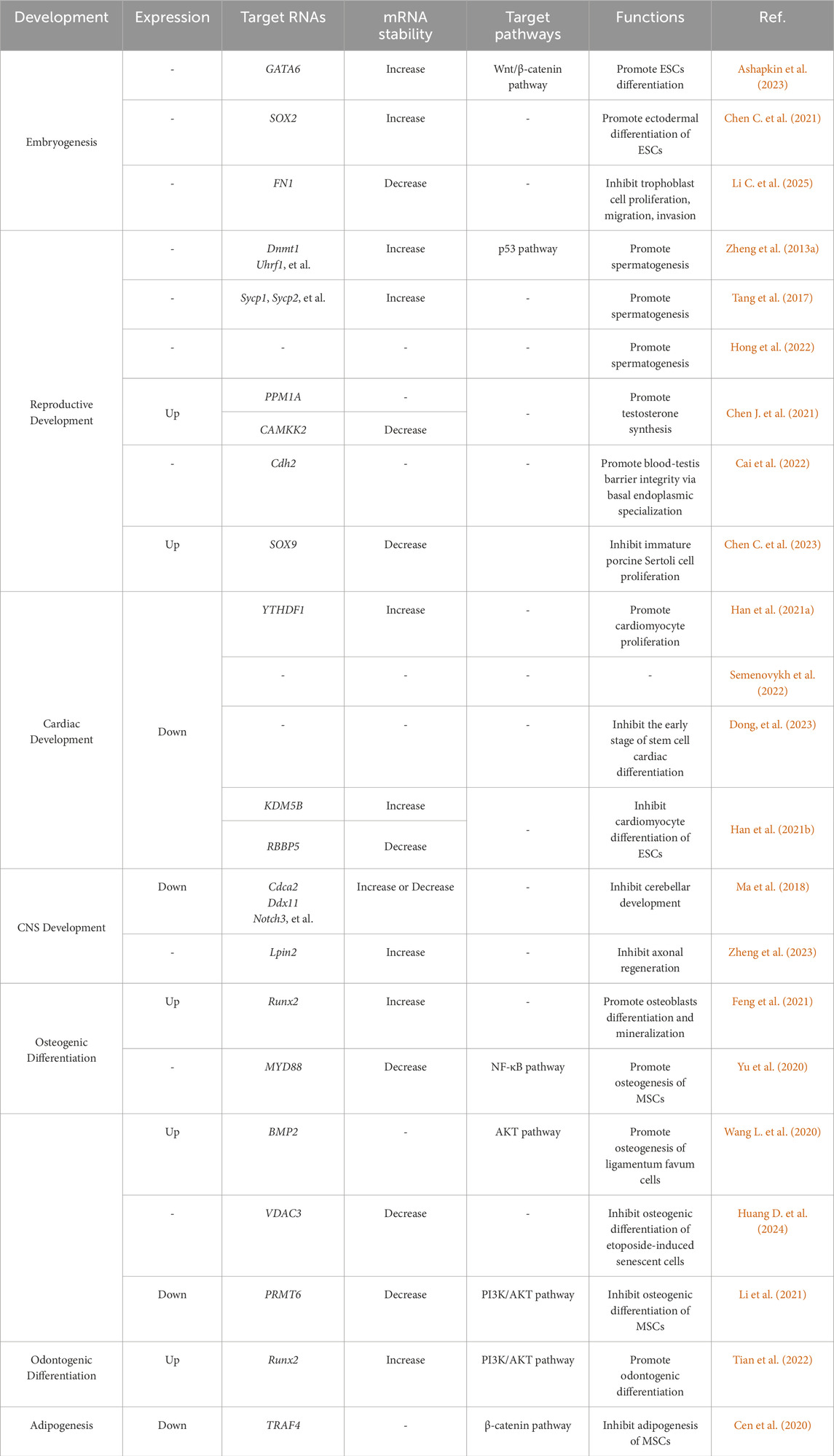
This reversible modification is dynamically deposited by methyltransferases (writers) and demethylases (erasers), and is recognized by specific binding proteins (readers) (Jiang et al., 2021; Sendinc and Shi, 2023). The methyltransferase complex, primarily comprising METTL3, METTL14, and WTAP, can install methyl groups on adenosine residues within consensus sequences (e.g., RRACH) to catalyze m6A modification (Xu and Ge, 2022). Conversely, demethylases, such as FTO and ALKBH5, mediate its removal, ensuring dynamic regulation (Gao et al., 2024). The “readers” including YTHDC and YTHDF families, decode m6A signals by binding to modified RNAs, thereby directing their fate (Yen and Chen, 2021). The m6A process governs diverse aspects of RNA metabolism, exerting a significant influence on a wide range of cellular processes, ranging from development to disease (Figure 1).
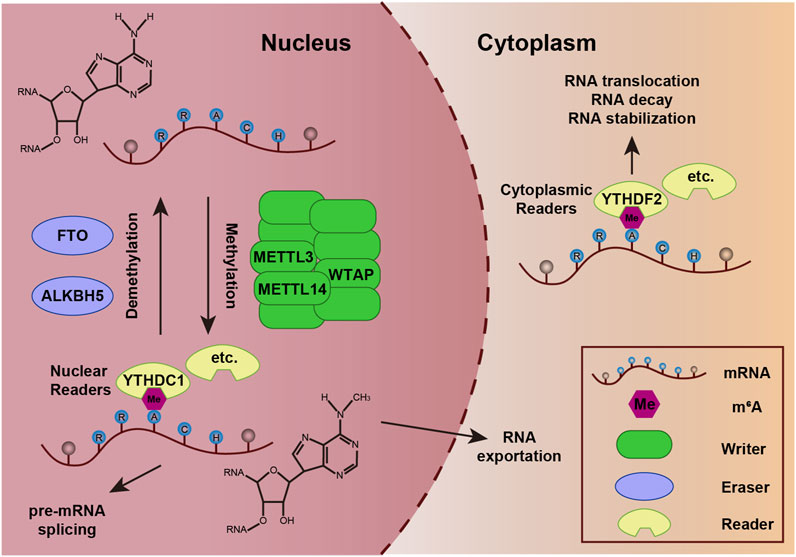
Figure 1. Dynamic regulation of m6A modification on mRNA. Methyltransferase complexes (e.g., METTL3-METTL14-WTAP) catalyze the addition of methyl groups (CH3) to adenine residues at conserved motifs (e.g., RRACH) on mRNA. Demethylases (FTO and ALKBH5) remove methyl groups, reversing m6A modification and enabling dynamic regulation of mRNA fate. m6A-binding proteins (YTHDC1, YTHDF2, etc.) recognize and bind to m6A sites, mediating downstream effects such as mRNA splicing, nuclear export, stability, degradation, or translation.
2.2 Biological functions of m6A
m6A encompasses roles in development, immune modulation, and disease pathogenesis. As a versatile modulator of RNA fate, m6A is significant in tuning transcript half-lives, guiding spliceosome assembly, licensing nuclear export, and reprogramming translation.
m6A dynamically controls mRNA stability through context-dependent interactions with reader proteins. For example, m6A can recruit YTHDF2 to participate in and promote the process of mRNA decay actively (Li et al., 2018; Zaccara and Jaffrey, 2020). YTHDF1 deletion prolongs the half-lives of mRNAs, thereby causing a degradation delay of mRNAs (Zaccara and Jaffrey, 2020; Li et al., 2022). m6A precisely fine-tunes alternative splicing events, playing a crucial role in regulating the diversity and complexity of gene expression. For instance, the reader YTHDC1 can recruit and modulate splicing factors to facilitate their access to the binding regions of targeted mRNAs (Xiao et al., 2016). m6A modification also impacts the RNA nuclear export. YTHDF3 has been proposed to act as an mRNA-transferring protein (Zaccara and Jaffrey, 2024). Additionally, YTHDC1 facilitates m6A-marked mRNA export from the nucleus, ensuring timely cytoplasmic translation (Roundtree et al., 2017). Furthermore, m6A plays a crucial role in regulating translation efficiency. 5′UTRs m6A can recruit eukaryotic initiation factor 3 to initiate cap-independent translation under stress conditions (Meyer et al., 2015). YTHDF1 can promote the translation of m6A-mRNAs (Zaccara and Jaffrey, 2024; Zou et al., 2023). In addition to the aforesaid functions, m6A extensively regulates non-coding RNAs (e.g., lncRNAs, miRNAs) to broaden its functional range (Huang et al., 2020).
3 ALKBH5: a key RNA demethylase in epitranscriptomic regulation
ALKBH5 is the second discovered demethylase and was first reported as a mammalian demethylase in 2013 (Zheng et al., 2013a; Zheng et al., 2013b). It belongs to the ALKB family of Fe(II)/α-ketoglutarate-dependent dioxygenases. Specifically, it plays a crucial role in catalyzing the oxidative demethylation of m6A on RNA substrates without generating any intermediate products (Shen et al., 2014).
The three-dimensional architecture of ALKBH5 exhibits a sophisticated organization featuring multiple α-helices, β-strands, and random coil that demonstrate precise spatial coordination (Figure 2A) (Aik et al., 2014). The active center of ALKBH5 is a highly conserved catalytic pocket known as the double-stranded β-helix (DSBH), which can coordinate Fe(II) and α-ketoglutarate (2-oxoglutarate, 2OG) to catalyze the oxidative demethylation of m6A in RNA (Qu et al., 2022; You et al., 2022). This remarkable structure has two distinct β-sheets: the major β-sheet, which is composed of strands β6, 8, 11, and 13, and the minor β-sheet, which consists of strands β7, 9, 10, and 12 (Figure 2B). The space between the two β-sheets serves as a passage, facilitating the substrate’s access to the active site, thereby enabling the efficient execution of the catalytic process. A loop extending from the DSBH is located between the β9 and β10 strands, conferring single-stranded RNA selectivity (Figure 2B) (Aik et al., 2014). Additionally, ALKBH5 contains a nucleotide recognition lid (NRL), which is a flexible loop region adjacent to its catalytic core and possesses β2, 3, 4, and 5 (Figure 2B). NRL is capable of interacting with m6A on RNA, ensuring precise positioning of the target nucleotide within the catalytic pocket, which plays a pivotal role in substrate binding (Qu et al., 2022; Feng et al., 2014). Collectively, the DSBH provides the enzymatic framework, whereas the NRL and βIV-V loop adjust substrate recognition and binding. These domains enable ALKBH5 to selectively demethylate m6A on RNA, impacting processes like mRNA splicing, stability, and translation.
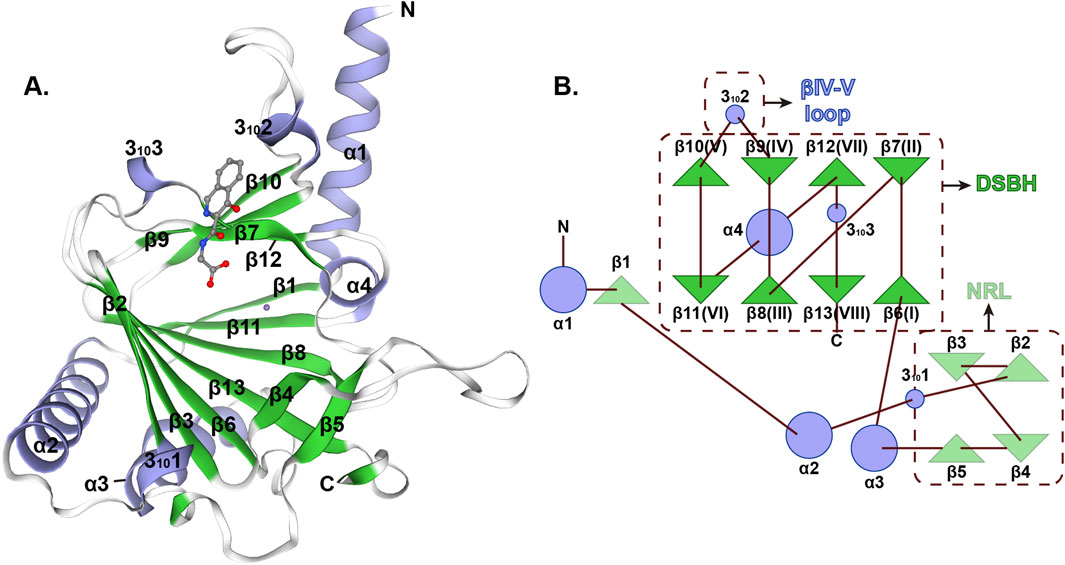
Figure 2. Structural characterization of human ALKBH5 protein (A) Overall three-dimensional structure of ALKBH5 (PDB ID: 4NJ4). (B) Topology of the structure of ALKBH566–292.
4 ALKBH5: orchestrating multifaceted developmental programs
Recent studies underscore that precise epitranscriptomic reprogramming, driven by context-dependent m6A demethylation, is essential for diverse developmental programs. Central to this reprogramming is the active removal of m6A marks by erasers, including ALKBH5, which enables rapid transcriptome rewiring in response to developmental cues. Building on this paradigm, the subsequent sections elaborate on how ALKBH5 functions in embryogenesis, reproductive development, cardiac development, central nervous system development, immune system development, pancreatic organogenesis, osteogenic differentiation, odontogenic differentiation, adipogenesis, and angiogenesis, uncovering the mechanisms for its role as a developmental modulator.
4.1 ALKBH5 in embryogenesis
Embryogenesis refers to the complicated developmental process by which a zygote undergoes cell proliferation, differentiation, and morphogenesis to form a structured embryo with functional organ systems (Ming et al., 2022; Du et al., 2022). A pivotal milestone in embryogenesis is gastrulation, during which the three germ layers (ectoderm, mesoderm, endoderm) are established (Lu et al., 2001). Notably, the commitment of the definitive endoderm represents a critical event in early cell fate specification (Robb et al., 2004).
ALKBH5 has been proved to possess the ability to influence the human endoderm fate (Liang et al., 2024). Knockout of ALKBH5 disrupts definitive differentiation and primitive streak specification in human embryonic stem cells (ESCs). Mechanistically, ALKBH5 deficiency destabilizes GATA6 mRNA in a YTHDF2-dependent manner. On the other hand, ALKBH5 can remove m6A modifications from GATA6 mRNA, enhancing its stability and translation efficiency. Then, GATA6 directly upregulates the expression of DKK1 and DKK4, which are key regulators of the Wnt/β-catenin signaling pathway, promoting the expression of endoderm-specific genes such as SOX17 and FOXA2 and guaranteeing the proper differentiation of human ESCs into definitive endoderm cells. At present, the doxycycline-inducible dCas13a system, when fused to the catalytic domain of ALKBH5, enables precise and reversible m6A demethylation at targeted mRNA sites (Chen X. et al., 2021). This engineered tool enhances mRNA stability while minimizing off-target effects, demonstrating high spatiotemporal specificity. Notably, site-specific m6A erasure at a single site of SOX2 mRNA suffices to regulate the differentiation of human ESCs. Another study highlights that the circRNA Hsa_circ_0069443 can bind to ALKBH5 in trophoblast cells (Li B. X. et al., 2025). It governs the stability and expression of FN1 through m6A methylation-dependent regulation, forming a functional epitranscriptomic axis essential for embryonic implantation and adhesion.
4.2 ALKBH5 in reproductive development
Reproductive development is a highly intricate and dynamic process initiated from the formation of the fertilized egg, encompassing the gradual formation, differentiation, and functional maturation of reproductive organs (Lochab and Extavour, 2017; Chan and Hirashima, 2022). This process plays a pivotal role in individual growth and development, as it not only dictates sexual characteristics and reproductive capacity but also profoundly influences hormonal regulation and systemic metabolic homeostasis (Du et al., 2022).
ALKBH5 plays a key role in male fertility by orchestrating spermatogenesis, while its dysregulation is linked to reproductive failure (Tang et al., 2017; Chen et al., 2022). This enzyme specifically regulates the m6A modification of mRNAs involved in spermatogenic processes, such as meiosis and spermatid differentiation. ALKBH5 is highly expressed in male mice testes, and its deficiency leads to abnormal spermatogenesis, reduced sperm count, impaired sperm motility, diminished testicular size, and male infertility (Zheng et al., 2013a; Tang et al., 2017; Hong et al., 2022). The underlying mechanism involves ALKBH5-mediated m6A demethylation of critical transcripts, particularly those spermatogenesis-related mRNAs involved in the p53 functional interaction network. This post-transcriptional regulation ensures both mRNA proper stability and efficient translation during spermatogenesis. Beyond its germ cell-autonomous functions, ALKBH5 exhibits essential roles in somatic niche maintenance. In the testicular interstitium, leydig cells (LCs) serve as the primary source of testosterone, which is a crucial hormone for male sexual development (Chen et al., 2009). ALKBH5 is upregulated during LC differentiation, where it regulates testosterone synthesis by promoting PPM1A translation and decreasing CAMKK2 stability (Chen Y. et al., 2021). In Sertoli cells, it maintains blood-testis barrier integrity through m6A-dependent regulation of Cdh2 mRNA translation, which is critical for basal ectoplasmic specialization dynamics (Cai et al., 2022). Additionally, ALKBH5 regulated the RNA methylation level and gene expression of SOX9 mRNA as well as negatively regulated the proliferation of immature porcine Sertoli cells (Chen C. et al., 2023).
In oocytes, ALKBH5 regulates the m6A modification of maternal mRNAs, which is critical for oocyte maturation and meiotic progression. Dysregulation of ALKBH5 can lead to defects in oocyte maturation and reduced fertility (Sun et al., 2022). ALKBH5 ensures timely maternal RNA degradation during oocyte maturation by dynamically erasing m6A marks, thereby preventing stabilization of transcripts via the m6A reader IGF2BP2; loss of ALKBH5 disrupts RNA clearance through persistent m6A-IGF2BP2 interactions, leading to defective meiosis and female infertility (Bai et al., 2023).
4.3 ALKBH5 in cardiac development and regeneration
The heart is the first functional organ to develop during organogenesis, which is precisely situated in the mediastinum, occupying a position behind the sternum and between the two lungs (Marano et al., 2011; Paige et al., 2015). The heart is irreplaceable in sustaining life. It acts as a muscular pump that continuously drives oxygenated blood to all tissues via systemic circulation and deoxygenated blood to the lungs for gas exchange through pulmonary circulation (Litviňuková et al., 2020). This dual-pump mechanism ensures oxygen, nutrients, and hormones are delivered to cells while eliminating metabolic waste.
Some studies confirmed a gradual decrease in the expression of ALKBH5 in cardiac tissue after birth and emphasized the significant role of ALKBH5 in the regulation of cardiomyocytes (Han et al., 2021a; Semenovykh et al., 2022). Alkbh5 knockout impaired cardiac regeneration and function in mice neonatal apex resection models, whereas its overexpression enhanced cardiomyocyte proliferation and restored cardiac function post-myocardial infarction (Han et al., 2021a). Mechanistically, the m6A modification mediated by ALKBH5 was of crucial importance as it enhanced the expression of YTHDF1 by regulating the stability of the corresponding mRNA. This modulation ultimately facilitated the translation of YAP, which is recognized as a core regulator governing cardiomyocyte proliferation and the process of heart regeneration. Other research discovered that ALKBH5 is responsible for the cardiomyocyte fate determination of human ESCs from mesoderm cells and mouse pluripotent stem cells (Dong et al., 2023; Han et al., 2021b). Mechanistically, the loss function of ALKBH5 regulated the mRNA stability of KDM5B and RBBP5, which in turn promoted the expression of GATA4 by enhancing histone H3 Lys4 trimethylation at its promoter region, thereby facilitating cardiac differentiation.
4.4 ALKBH5 in central nervous system development
The development of the central nervous system (CNS) refers to the process by which the brain and spinal cord form and mature from the early stages of embryonic development through childhood and adolescence (Yang et al., 2025). It involves a series of complex events such as neural tube formation, neuronal migration, axon guidance, synapse formation, and myelination. The proper development of the CNS is of utmost importance for an individual’s physical and mental wellbeing. It is the foundation for all cognitive functions including learning, memory, perception, and decision-making (Rice and Barone, 2000).
The development of the CNS requires precise spatiotemporal regulation of gene expression, with RNA methylation dynamics emerging as an important regulatory layer. The ALKBH5 protein exhibits widespread expression across brain regions, with predominant localization in neurons (Du et al., 2020). Its expression displays a dynamic developmental pattern: it is highly abundant during embryonic stages of brain development but declines progressively in late stages. Disrupted m6A methylation patterns can lead to developmental delays and functional abnormalities in the cerebellum. For instance, knockout of Alkbh5 under hypoxic conditions results in disordered m6A levels in a subset of cell fate determination genes (such as Cenpe, Cdca2, Ddx11, and Notch3), accelerated RNA nuclear export, causes abnormal cell proliferation and differentiation in the cerebellum, and significant cerebellar development delays (Ma et al., 2018). Notably, the cerebellar integrity preserved by ALKBH5 extends to aging populations (Fei et al., 2023). Additionally, ALKBH5 may be a potential target for promoting axon regeneration in both CNS and peripheral nervous systems. The study by Wang et al. demonstrated that Alkbh5 knockdown increased retinal ganglion cell survival rates and the number of regenerated axons (Zheng et al., 2023). The mechanism underlying this effect involves the regulation of lipid metabolism through the demethylation of Lpin2 mRNA. Alkbh5 knockdown reduces Lpin2 mRNA stability by increasing m6A modification on its 3′UTR, thereby enhancing axon regeneration. Concisely, ALKBH5 plays a key role in CNS development and function, regulating key processes such as cerebellar development, neuronal survival, and axonal regeneration.
4.5 ALKBH5 in immune system development
Proper immune system development is crucial for establishing immune competence, as it involves the maturation of immune cells and the establishment of immune tolerance, which together determine the body’s capacity to respond effectively to infections and prevent autoimmune diseases (Simon et al., 2015). T cells are essential for adaptive immunity, and their maturation in the thymus involves complex regulatory mechanisms (Thapa and Farber, 2019). Based on the expression of αβ and γδ receptors, T cells are mainly divided into αβ and γδ T cells.
ALKBH5 serves as an important regulator in T cell development, particularly influencing the differentiation and expansion of γδ T cells (Zhao et al., 2023). Specifically, Alkbh5 deficiency leads to a significant expansion of γδ T cells through enhanced proliferation and developmental programming, ultimately improving host defense against gastrointestinal Salmonella typhimurium infection, rather than affecting αβ T cells homeostasis (Zhao et al., 2023; Ding et al., 2022). The molecular mechanism involves m6A RNA modification dynamics: Alkbh5 deficiency elevates m6A levels, triggering specific mRNA degradation of key Notch signaling components including Jagged1 and Notch2. This mechanism elucidates the checkpoint function of m6A modification in T cell lineage commitment and unveils potential therapeutic targets for modulating γδ T cell-driven immune responses.
4.6 ALKBH5 in pancreatic organogenesis
The pancreas, an organ derived from the endoderm, is situated posterior to the stomach, with its head ensconced in the duodenal loop and its tail extending towards the spleen (Edlund, 2002). It has dual functions (the exocrine function and the endocrine function), which play a crucial role in glucose homeostasis and nutrient digestion (Alonge et al., 2023; Larsen and Grapin-Botton, 2017). In recent years, numerous transcription factors, such as MNX1, PDX1, NKX6.1, and SOX9, which play crucial roles in the organogenesis of the pancreas, have been identified (Cano et al., 2014).
Previous research has verified that ALKBH5 regulates pancreatic organogenesis by regulating RNA m6A demethylation (Ma et al., 2022). The research team discovered that ALKBH5 maintains the balance of m6A modifications on transcripts essential for pancreatic progenitor differentiation. Specifically, ALKBH5-mediated removal of m6A marks stabilizes key mRNAs encoding transcription factors like MNX1, SOX9, PDX1, and NKX6.1 to evade the YTHDF2-mediated mRNA decay pathway, thereby regulating human pancreatic differentiation. Additionally, the cofactor of ALKBH5, namely, α-ketoglutarate, could also exert functions in this organ differentiation.
4.7 ALKBH5 in osteogenic and odontogenic differentiation
Osteogenic differentiation refers to the fundamental biological process by which mesenchymal stem cells or osteoprogenitor cells (such as bone progenitor cells) undergo progressive maturation into functional osteoblasts under precise regulatory control (Valenti et al., 2016). As a cornerstone of skeletal development and homeostasis, this complex process involves a multifaceted cascade of biological events, including specific gene activation, coordinated signaling pathway regulation, extracellular matrix biosynthesis, and subsequent mineralization processes, ultimately culminating in the formation of functional bone tissue (Deng et al., 2008).
ALKBH5 exhibits context-dependent roles in osteogenic differentiation by dynamically regulating RNA of key osteogenic factors. For instance, ALKBH5 was upregulated during osteoblast differentiation and promotes osteogenesis by enhancing the stability of Runx2 mRNA, a master transcription factor for osteoblast differentiation (Feng et al., 2021). Additionally, ALKBH5 dynamically reverses the METTL3-driven m6A modification of MYD88 mRNA, thereby suppressing NF-κB signaling to facilitate osteogenic differentiation of mesenchymal stem cells (MSCs) (Yu et al., 2020). In the pathological context of ligamentum flavum ossification, an ectopic ossification disorder characterized by aberrant bone formation within spinal ligaments, ALKBH5 shows elevated expression and functionally drives the mineralization process in ligament flavum cells (Wang H.-F. et al., 2020). Mechanically, ALKBH5 facilitates osteogenesis by demethylating BMP2 and activating the AKT signaling pathway. These findings highlight ALKBH5’s role as a positive regulator of osteogenesis through m6A-dependent modulation of transcription factors and signaling pathways. However, ALKBH5 can also exert inhibitory effects on osteogenic differentiation. In senescent bone marrow mesenchymal stromal cells, ALKBH5 suppresses osteogenic differentiation by reducing m6A modification on VDAC3 mRNA and accelerating its degradation, which is a mitochondrial ROS sensor critical for counteracting cellular senescence (Huang Y. et al., 2024). Another study identified a distinct inhibitory axis where ALKBH5 destabilizes PRMT6 mRNA and enhances its decay, suppressing PI3K/AKT pathway and osteogenic differentiation (Li et al., 2021). These contrasting roles reflect ALKBH5’s functional duality, influenced by cellular senescence status, target mRNA specificity, and downstream signaling cross-talk.
Odontogenic differentiation constitutes a specialized cellular reprogramming event wherein dental pulp stem cells transition into polarized odontoblasts, the principal secretory cells governing dentin matrix synthesis (Wu et al., 2024). This differentiation cascade serves dual physiological imperatives: (1) establishing the primary dentin architecture during tooth development, and (2) mobilizing reparative dentinogenesis in response to carious or mechanical stimulis (Huang D. et al., 2024; Tian et al., 2022).
ALKBH5 may play a role analogous to its function in osteogenic differentiation during odontogenic differentiation. Experimental evidence indicates that ALKBH5 is upregulated during odontoblast differentiation (Tian et al., 2022). Conditional deficiency of Alkbh5 reduces odontoblast numbers and promotes pre-dentin formation, though it is important to note that the observed phenotype is not striking. Mechanistically, ALKBH5 promotes dentin matrix formation through a molecular strategy: epigenetic stabilization of Runx2 mRNA through m6A demethylation, and enhancement of the PI3K/AKT signaling pathway. However, research on the relationship between ALKBH5 and odontoblastic differentiation is currently limited, and more studies are needed in the future to fully understand its role in this process.
4.8 ALKBH5 in adipogenesis
Adipogenesis, a highly plastic and dynamic process, drives the phenotype of functionally mature adipocytes (the defining cell type of adipose tissue) (Fève, 2005). Adipose tissue serves as a critical site for lipid storage, systemic energy homeostasis, and insulin sensitivity regulation (Sarjeant and Stephens, 2012). m6A methylation has been demonstrated to regulate various aspects of mRNA metabolism during adipogenesis (Wang L. et al., 2020; Song et al., 2020).
During adipogenic differentiation, ALKBH5 expression progressively declines, leading to TRAF4 downregulation through its m6A RNA demethylation activity (Cen et al., 2020). Mechanistically, TRAF4 forms a functional complex with PKM2 to activate β-catenin signaling, thereby establishing an anti-adipogenic regulatory axis. Consequently, the depletion of ALKBH5 can enhance adipogenesis of MSCs.
4.9 ALKBH5 in angiogenesis
Angiogenesis, the growth of blood vessels from existing vasculature, is integral to development (organ formation) and adaptation (tissue repair) (Chen et al., 2017). ALKBH5 is regarded as a significant regulator of angiogenesis. Nevertheless, current research primarily focuses on its roles in pathological or hypoxic conditions, while its involvement in developmental angiogenesis remains underexplored. For instance, ALKBH5 acts as a negative regulator of post-ischemic angiogenesis through post-transcriptional modulation and destabilization of WNT5A mRNA in an m6A-dependent manner (Zhao et al., 2021). Conversely, it sustains angiogenesis in endothelial cells under acute ischemic stress by reducing m6A methylation of SPHK1 mRNA (Kumari et al., 2021). Furthermore, specific deletion of Alkbh5 in the murine hematopoietic system attenuates stress-induced hematopoietic fitness through regulation of Ogdh mRNA stability (Gao et al., 2023).
5 Therapeutic targeting of ALKBH5: from molecular inhibitors to clinical applications
As a pivotal m6A RNA demethylase, ALKBH5 has emerged as a therapeutic target due to its dysregulation in diverse pathological conditions. Current targeting strategies encompass inhibitors, proteolysis targeting chimera, programmable m6A-editing systems, compounds targeting the regulatory machinery of ALKBH5, as well as gene therapy approaches (Qu et al., 2022). Among these, pharmacological inhibition represents the most straightforward therapeutic paradigm.
A variety of ALKBH5 inhibitors have been developed, including natural, clinical pharmacological, and small-molecule inhibitors (Fang et al., 2025). For example, citrate, a natural inhibitor of ALKBH5, disrupts the demethylase activity of ALKBH5 by directly binding to it and replacing Fe(II) and 2OG (Xu et al., 2014). IOX1, the clinical pharmacological inhibitors of ALKBH5, competitively inhibits 2OG binding and suppresses ALKBH5, which demonstrates protective effects against acute kidney injury and sevoflurane-induced neuronal damage in the hippocampus (Li et al., 2016; Chen J. et al., 2023; Meng et al., 2024). In addition, several new small-molecule inhibitors of ALKBH5 have been developed. The binding site of imidazobenzoxazin-5-thione MV1035 in ALKBH5 partially overlaps with that of 2OG, inhibiting the demethylation activity of ALKBH5, which suppresses migration and invasion in glioma cell lines (Malacrida et al., 2020). Novel inhibitors Ena15 and Ena2 show differential inhibition modes (non-competitive or competitive 2OG binding) with efficacy against the growth activity of glioblastoma multiforme (Gao et al., 2024; Takahashi et al., 2022). In addition to the above ALKBH5 inhibitors, there are many unlisted, such as cmp-3, cmp-6, DO-2728 and so on (Selberg et al., 2021; Wang Y.-Z. et al., 2023). These compounds effectively modulate m6A level in target mRNAs, establishing ALKBH5-targeted therapy as a promising strategy for various human diseases.
Despite significant advances in ALKBH5 inhibitor development for certain disorders (such as oncology), their therapeutic potential in developmental pathologies remains an under-investigated frontier. During development, m6A modification plays a dynamic regulatory role in critical biological processes such as embryogenesis, neurogenesis, and organogenesis. ALKBH5 potentially influences these events by altering the expression of development-related genes. However, existing studies predominantly focus on post-developmental disease contexts, leaving the mechanistic and therapeutic implications of ALKBH5 inhibitors in developmental anomalies (e.g., neural tube defects, congenital malformations) unaddressed. Future investigations should integrate developmental models to elucidate how ALKBH5-mediated m6A remodeling governs developmental programs and evaluate the feasibility of pharmacological inhibition to intervene in abnormal developmental processes. This paradigm shift from disease treatment to developmental pathway modulation could broaden the clinical applicability of ALKBH5 inhibitors and offer novel strategies for targeting developmental disorders.
6 Conclusion and discussion
In summary, ALKBH5 has emerged as an important epigenetic regulator that intricately influences a multitude of developmental processes through the erasure of m6A RNA methylation (Figure 3; Table 1). This enzyme coordinates a hierarchical regulatory network across three fundamental dimensions: (1) foundational biological processes including embryogenesis and reproductive system maturation, where it maintains developmental plasticity; (2) organ-specific development spanning cardiac morphogenesis, pancreatic organogenesis, CNS development, and angiogenesis, demonstrating remarkable tissue adaptability; and (3) specialized cellular differentiation programs encompassing osteogenic differentiation, dentin matrix organization, adipogenesis, and thymic T cell differentiation. The precise regulation of RNA stability, splicing, and translation by ALKBH5-mediated m6A demethylation is crucial for orchestrating the complex molecular and cellular events underlying these developmental programs. The growing body of evidence underscores the significance of ALKBH5 in maintaining the integrity and functionality of various tissues and organs during development.
Figure 3. ALKBH5: a central regulator orchestrating multifaceted developmental programs. This schematic illustrates the pivotal role of ALKBH5, an m6A demethylase, in coordinating diverse developmental processes.
The therapeutic potential of targeting ALKBH5 in developmental disorders and regenerative medicine is promising. For instance, modulating ALKBH5 activity could offer strategies to enhance embryonic development, improve fertility, promote cardiac regeneration, and alleviate neuroinflammatory conditions. Moreover, understanding the specific m6A-modified transcripts and signaling pathways regulated by ALKBH5 in different developmental contexts may lead to the development of targeted therapies for various diseases associated with aberrant RNA methylation patterns.
However, several challenges remain: (1) ALKBH5 serves as a key regulator of mammalian development, as demonstrated by phenotypes in Alkbh5 knockout mouse models across germline development (Zheng et al., 2013a; Tang et al., 2017), cardiac repair (Han et al., 2021a), immune system development (Ding et al., 2022), cerebellar development (Ma et al., 2018), osteogenesis (Li et al., 2021), and odontogenesis (Tian et al., 2022). Although ALKBH5 expression peaks in testes, its functional impact in other non-testicular tissues demonstrates that physiologically relevant m6A demethylation occurs even at lower expression levels. However, proposed roles in broader developmental contexts, such as pancreatic organogenesis and adipogenesis, primarily derive from in vitro or correlative evidence, lacking direct validation in Alkbh5 knockout mice. This absence of in vivo confirmation suggests ALKBH5 might not be essential for these specific processes. Potential contributing factors may involve limited expression of ALKBH5 in relevant tissues, compensatory activity by related demethylases (e.g., FTO) or functional redundancy within epigenetic regulatory pathways. (2) The most commonly used method to map m6A and to detect changes in m6A is antibody-dependent techniques (e.g., MeRIP-seq/m6A-seq). In some cited studies, MeRIP-seq was performed to screen for molecules regulated by ALKBH5 (Cai et al., 2022; Tian et al., 2022). Although MeRIP-seq can indicate approximate sites of m6A, it can’t be used to quantitatively measure the precise fraction of transcript copies modified by m6A (McIntyre et al., 2020). And low-abundance m6A sites in critical genes may evade detection despite functional importance. Consequently, the need for further research and alternative assays (e.g., m6A-seq2 (Dierks et al., 2021), GLORI (Liu et al., 2023) and nanopore sequencing (Grigorev et al., 2024)) to resolve the ALKBH5-dependent changes at specific m6A sites. (3) The dynamic and context-dependent nature of m6A methylation requires a deeper understanding of the spatiotemporal regulation of ALKBH5 and its interplay with other epitranscriptomic factors. (4) Therapeutic targeting requires careful evaluation of off-target effects and interplay with epigenetic modifications. Future research should focus on elucidating the molecular mechanisms underlying ALKBH5’s functions in development, exploring its role in disease pathogenesis, and developing precise tools to modulate its activity for therapeutic benefits.
Author contributions
XZ: Conceptualization, Writing – review and editing, Writing – original draft. LZ: Writing – original draft. CT: Writing – review and editing. HT: Conceptualization, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the National Natural Science Foundation of China (82001000), and the Hubei Natural Science Foundation (2020CFB457).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
2OG, 2-oxoglutarate; ALKBH5, alkylation repair homolog protein 5; circRNAs, circular RNAs; CNS, central nervous system; DSBH, double-stranded β-helix; ESCs, embryonic stem cells; LCs, leydig cells; lncRNAs, long non-coding RNAs; m6A, N6-methyladenosine; mRNAs, messenger RNAs; miRNAs, micro RNAs; MSCs, mesenchymal stem cells; NRL, nucleotide recognition lid.
References
Aik, W., Scotti, J. S., Choi, H., Gong, L., Demetriades, M., Schofield, C. J., et al. (2014). Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 42 (7), 4741–4754. doi:10.1093/nar/gku085
Alonge, K. M., Porte, D., and Schwartz, M. W. (2023). Distinct roles for brain and pancreas in basal and postprandial glucose homeostasis. Diabetes 72 (5), 547–556. doi:10.2337/db22-0969
Ashapkin, V., Suvorov, A., Pilsner, J. R., Krawetz, S. A., and Sergeyev, O. (2023). Age-associated epigenetic changes in mammalian sperm: implications for offspring health and development. Hum. Reprod. Update 29 (1), 24–44. doi:10.1093/humupd/dmac033
Bai, L., Xiang, Y., Tang, M., Liu, S., Chen, Q., Chen, Q., et al. (2023). ALKBH5 controls the meiosis-coupled mRNA clearance in oocytes by removing the N 6-methyladenosine methylation. Nat. Commun. 14 (1), 6532. doi:10.1038/s41467-023-42302-6
Cai, B., Ma, M., Yuan, R., Zhou, Z., Zhang, J., Kong, S., et al. (2024). MYH1G-AS is a chromatin-associated lncRNA that regulates skeletal muscle development in chicken. Cell Mol. Biol. Lett. 29 (1), 9. doi:10.1186/s11658-023-00525-x
Cai, Z., Zhang, Y., Yang, L., Ma, C., Fei, Y., Ding, J., et al. (2022). ALKBH5 in mouse testicular sertoli cells regulates Cdh2 mRNA translation to maintain blood-testis barrier integrity. Cell Mol. Biol. Lett. 27 (1), 101. doi:10.1186/s11658-022-00404-x
Cano, D. A., Soria, B., Martín, F., and Rojas, A. (2014). Transcriptional control of mammalian pancreas organogenesis. Cell Mol. Life Sci. 71 (13), 2383–2402. doi:10.1007/s00018-013-1510-2
Cen, S., Li, J., Cai, Z., Pan, Y., Sun, Z., Li, Z., et al. (2020). TRAF4 acts as a fate checkpoint to regulate the adipogenic differentiation of MSCs by activating PKM2. EBioMedicine 54, 102722. doi:10.1016/j.ebiom.2020.102722
Chan, C. J., and Hirashima, T. (2022). Tissue hydraulics in reproduction. Semin. Cell Dev. Biol. 131, 124–133. doi:10.1016/j.semcdb.2022.05.008
Chen, C., Tang, X., Yan, S., Yang, A., Xiang, J., Deng, Y., et al. (2023). Comprehensive analysis of the transcriptome-wide m(6)A methylome in shaziling pig testicular development. Int. J. Mol. Sci. 24 (19), 14475. doi:10.3390/ijms241914475
Chen, H., Ge, R. S., and Zirkin, B. R. (2009). Leydig cells: from stem cells to aging. Mol. Cell Endocrinol. 306 (1-2), 9–16. doi:10.1016/j.mce.2009.01.023
Chen, H., Zhang, J., Yan, Y., Zhu, C., Wang, L., Fu, S., et al. (2022). N6-methyladenosine RNA demethylase ALKBH5 is testis-specifically downregulated in hybrid male sterile dzo and is a target gene of bta-miR-200a. Theriogenology 187, 51–57. doi:10.1016/j.theriogenology.2022.04.022
Chen, J., Fu, Y., Day, D. S., Sun, Y., Wang, S., Liang, X., et al. (2017). VEGF amplifies transcription through ETS1 acetylation to enable angiogenesis. Nat. Commun. 8 (1), 383. doi:10.1038/s41467-017-00405-x
Chen, J., Xu, C., Yang, K., Gao, R., Cao, Y., Liang, L., et al. (2023). Inhibition of ALKBH5 attenuates I/R-induced renal injury in male mice by promoting Ccl28 m6A modification and increasing treg recruitment. Nat. Commun. 14 (1), 1161. doi:10.1038/s41467-023-36747-y
Chen, X., Zhao, Q., Zhao, Y. L., Chai, G. S., Cheng, W., Zhao, Z., et al. (2021). Targeted RNA N(6) -Methyladenosine demethylation controls cell fate transition in human pluripotent stem cells. Adv. Sci. (Weinh) 8 (11), e2003902. doi:10.1002/advs.202003902
Chen, Y., Wang, J., Xu, D., Xiang, Z., Ding, J., Yang, X., et al. (2021). m(6)A mRNA methylation regulates testosterone synthesis through modulating autophagy in Leydig cells. Autophagy 17 (2), 457–475. doi:10.1080/15548627.2020.1720431
Deng, Z. L., Sharff, K. A., Tang, N., Song, W. X., Luo, J., Luo, X., et al. (2008). Regulation of osteogenic differentiation during skeletal development. Front. Biosci. 13, 2001–2021. doi:10.2741/2819
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Dierks, D., Garcia-Campos, M. A., Uzonyi, A., Safra, M., Edelheit, S., Rossi, A., et al. (2021). Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat. Methods 18 (9), 1060–1067. doi:10.1038/s41592-021-01242-z
Ding, C., Xu, H., Yu, Z., Roulis, M., Qu, R., Zhou, J., et al. (2022). RNA m(6)A demethylase ALKBH5 regulates the development of γδ T cells. Proc. Natl. Acad. Sci. U. S. A. 119 (33), e2203318119. doi:10.1073/pnas.2203318119
Dong, S., Sun, Y., Liu, C., Li, Y., Yu, S., Zhang, Q., et al. (2023). Stage-specific requirement for m(6)A RNA methylation during cardiac differentiation of pluripotent stem cells. Differentiation 133, 77–87. doi:10.1016/j.diff.2023.07.001
Du, T., Li, G., Yang, J., and Ma, K. (2020). RNA demethylase Alkbh5 is widely expressed in neurons and decreased during brain development. Brain Res. Bull. 163, 150–159. doi:10.1016/j.brainresbull.2020.07.018
Du, Z., Zhang, K., and Xie, W. (2022). Epigenetic reprogramming in early animal development. Cold Spring Harb. Perspect. Biol. 14 (6), a039677. doi:10.1101/cshperspect.a039677
Edlund, H. (2002). Pancreatic organogenesis--developmental mechanisms and implications for therapy. Nat. Rev. Genet. 3 (7), 524–532. doi:10.1038/nrg841
Fang, M., Ye, L., Zhu, Y., Huang, L., and Xu, S. (2025). M6A demethylase ALKBH5 in human diseases: from structure to mechanisms. Biomolecules 15 (2), 157. doi:10.3390/biom15020157
Fei, Y., Ma, C. H., Li, Q., Song, W., Tong, W. M., and Niu, Y. M. (2023). Effects of RNA M6A demethylase ALKBH5 gene deficiency on morphology and function of cerebellum in aged mice. Zhonghua Bing Li Xue Za Zhi 52 (6), 606–611. doi:10.3760/cma.j.cn112151-20221117-00966
Feng, C., Liu, Y., Wang, G., Deng, Z., Zhang, Q., Wu, W., et al. (2014). Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J. Biol. Chem. 289 (17), 11571–11583. doi:10.1074/jbc.M113.546168
Feng, L., Fan, Y., Zhou, J., Li, S., and Zhang, X. (2021). The RNA demethylase ALKBH5 promotes osteoblast differentiation by modulating Runx2 mRNA stability. FEBS Lett. 595 (15), 2007–2014. doi:10.1002/1873-3468.14145
Fève, B. (2005). Adipogenesis: cellular and molecular aspects. Best. Pract. Res. Clin. Endocrinol. Metab. 19 (4), 483–499. doi:10.1016/j.beem.2005.07.007
Gao, Y., Zimmer, J. T., Vasic, R., Liu, C., Gbyli, R., Zheng, S.-J., et al. (2023). ALKBH5 modulates hematopoietic stem and progenitor cell energy metabolism through m6A modification-mediated RNA stability control. Cell Rep. 42 (10), 113163. doi:10.1016/j.celrep.2023.113163
Gao, Z., Zha, X., Li, M., Xia, X., and Wang, S. (2024). Insights into the m(6)A demethylases FTO and ALKBH5: structural, biological function, and inhibitor development. Cell Biosci. 14 (1), 108. doi:10.1186/s13578-024-01286-6
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347 (6225), 1002–1006. doi:10.1126/science.1261417
Grigorev, K., Nelson, T. M., Overbey, E. G., Houerbi, N., Kim, J., Najjar, D., et al. (2024). Direct RNA sequencing of astronaut blood reveals spaceflight-associated m6A increases and hematopoietic transcriptional responses. Nat. Commun. 15 (1), 4950. doi:10.1038/s41467-024-48929-3
Han, Z., Wang, X., Xu, Z., Cao, Y., Gong, R., Yu, Y., et al. (2021a). ALKBH5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1. Theranostics 11 (6), 3000–3016. doi:10.7150/thno.47354
Han, Z., Xu, Z., Yu, Y., Cao, Y., Bao, Z., Gao, X., et al. (2021b). ALKBH5-mediated m(6)A mRNA methylation governs human embryonic stem cell cardiac commitment. Mol. Ther. Nucleic Acids 26, 22–33. doi:10.1016/j.omtn.2021.05.019
Hong, S., Shen, X., Luo, C., and Sun, F. (2022). Comparative analysis of the testes from wild-type and Alkbh5-knockout mice using single-cell RNA sequencing. G3 (Bethesda) 12 (8), jkac130. doi:10.1093/g3journal/jkac130
Huang, D., Li, Y., Han, J., Zuo, H., Liu, H., and Chen, Z. (2024). Xbp1 promotes odontoblastic differentiation through modulating mitochondrial homeostasis. Faseb J. 38 (7), e23600. doi:10.1096/fj.202400186R
Huang, H., Weng, H., and Chen, J. (2020). m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell 37 (3), 270–288. doi:10.1016/j.ccell.2020.02.004
Huang, Y., Wang, S., Hu, D., Zhang, L., and Shi, S. (2024). ALKBH5 regulates etoposide-induced cellular senescence and osteogenic differentiation in osteoporosis through mediating the m6A modification of VDAC3. Sci. Rep. 14 (1), 23461. doi:10.1038/s41598-024-75033-9
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target Ther. 6 (1), 74. doi:10.1038/s41392-020-00450-x
Kumari, R., Dutta, R., Ranjan, P., Suleiman, Z. G., Goswami, S. K., Li, J., et al. (2021). ALKBH5 regulates SPHK1-Dependent endothelial cell angiogenesis following ischemic stress. Front. Cardiovasc Med. 8, 817304. doi:10.3389/fcvm.2021.817304
Larsen, H. L., and Grapin-Botton, A. (2017). The molecular and morphogenetic basis of pancreas organogenesis. Semin. Cell Dev. Biol. 66, 51–68. doi:10.1016/j.semcdb.2017.01.005
Li, B. X., Wu, M. Y., Wang, Z. H., Zhou, D. M., Li, J. Q., Lu, B. F., et al. (2025). Mechanism of hsa_circ_0069443 promoting early pregnancy loss through ALKBH5/FN1 axis in trophoblast cells. iScience 28 (1), 111608. doi:10.1016/j.isci.2024.111608
Li, C., Chen, K., Li, X., and Xiong, X. (2025). Epitranscriptome-epigenome interactions in development and disease mechanisms. Trends Genet. 14 (25), S0168–S9525. doi:10.1016/j.tig.2025.04.009
Li, F., Kennedy, S., Hajian, T., Gibson, E., Seitova, A., Xu, C., et al. (2016). A radioactivity-based assay for screening human m6A-RNA methyltransferase, METTL3-METTL14 complex, and demethylase ALKBH5. SLAS Discov. 21 (3), 290–297. doi:10.1177/1087057115623264
Li, J., Chen, K., Dong, X., Xu, Y., Sun, Q., Wang, H., et al. (2022). YTHDF1 promotes mRNA degradation via YTHDF1-AGO2 interaction and phase separation. Cell Prolif. 55 (1), e13157. doi:10.1111/cpr.13157
Li, M., Zhao, X., Wang, W., Shi, H., Pan, Q., Lu, Z., et al. (2018). Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 19 (1), 69. doi:10.1186/s13059-018-1436-y
Li, Z., Wang, P., Li, J., Xie, Z., Cen, S., Li, M., et al. (2021). The N6-methyladenosine demethylase ALKBH5 negatively regulates the osteogenic differentiation of mesenchymal stem cells through PRMT6. Cell Death Dis. 12 (6), 578. doi:10.1038/s41419-021-03869-4
Liang, Z., Huang, T., Li, W., Ma, Z., Wang, K., Zhai, Z., et al. (2024). ALKBH5 governs human endoderm fate by regulating the DKK1/4-mediated Wnt/β-catenin activation. Nucleic Acids Res. 52 (18), 10879–10896. doi:10.1093/nar/gkae707
Litviňuková, M., Talavera-López, C., Maatz, H., Reichart, D., Worth, C. L., Lindberg, E. L., et al. (2020). Cells of the adult human heart. Nature 588 (7838), 466–472. doi:10.1038/s41586-020-2797-4
Liu, C., Sun, H., Yi, Y., Shen, W., Li, K., Xiao, Y., et al. (2023). Absolute quantification of single-base m(6)A methylation in the mammalian transcriptome using GLORI. Nat. Biotechnol. 41 (3), 355–366. doi:10.1038/s41587-022-01487-9
Lochab, A. K., and Extavour, C. G. (2017). Bone morphogenetic protein (BMP) signaling in animal reproductive system development and function. Dev. Biol. 427 (2), 258–269. doi:10.1016/j.ydbio.2017.03.002
Loseva, P. A., and Gladyshev, V. N. (2024). The beginning of becoming a human. Aging (Albany NY) 16 (9), 8378–8395. doi:10.18632/aging.205824
Lu, C. C., Brennan, J., and Robertson, E. J. (2001). From fertilization to gastrulation: axis formation in the mouse embryo. Curr. Opin. Genet. Dev. 11 (4), 384–392. doi:10.1016/s0959-437x(00)00208-2
Ma, C., Chang, M., Lv, H., Zhang, Z.-W., Zhang, W., He, X., et al. (2018). RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19 (1), 68. doi:10.1186/s13059-018-1435-z
Ma, X., Cao, J., Zhou, Z., Lu, Y., Li, Q., Jin, Y., et al. (2022). N(6)-methyladenosine modification-mediated mRNA metabolism is essential for human pancreatic lineage specification and islet organogenesis. Nat. Commun. 13 (1), 4148. doi:10.1038/s41467-022-31698-2
Malacrida, A., Rivara, M., Di Domizio, A., Cislaghi, G., Miloso, M., Zuliani, V., et al. (2020). 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg. and Med. Chem. 28 (4), 115300. doi:10.1016/j.bmc.2019.115300
Marano, R., Liguori, C., Savino, G., Merlino, B., Natale, L., and Bonomo, L. (2011). Cardiac silhouette findings and mediastinal lines and stripes: radiograph and CT scan correlation. Chest 139 (5), 1186–1196. doi:10.1378/chest.10-0660
Matouk, C. C., and Marsden, P. A. (2008). Epigenetic regulation of vascular endothelial gene expression. Circ. Res. 102 (8), 873–887. doi:10.1161/circresaha.107.171025
McIntyre, A. B. R., Gokhale, N. S., Cerchietti, L., Jaffrey, S. R., Horner, S. M., and Mason, C. E. (2020). Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci. Rep. 10 (1), 6590. doi:10.1038/s41598-020-63355-3
Meng, X., Wang, Y., Zhao, W., Chen, Y., Li, W., Peng, K., et al. (2024). Identification of differential m6A RNA methylomes and ALKBH5 as a potential prevention target in the developmental neurotoxicity induced by multiple sevoflurane exposures. Faseb J. 38 (14), e23793. doi:10.1096/fj.202400664R
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5' UTR m(6)A promotes cap-independent translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Ming, Z., Vining, B., Bagheri-Fam, S., and Harley, V. (2022). SOX9 in organogenesis: shared and unique transcriptional functions. Cell Mol. Life Sci. 79 (10), 522. doi:10.1007/s00018-022-04543-4
Mittnenzweig, M., Mayshar, Y., Cheng, S., Ben-Yair, R., Hadas, R., Rais, Y., et al. (2021). A single-embryo, single-cell time-resolved model for mouse gastrulation. Cell 184 (11), 2825–2842.e22. doi:10.1016/j.cell.2021.04.004
Ohta, S., and Yamada, Y. (2023). Exploring the potential of in vivo reprogramming for studying embryonic development, tissue regeneration, and organismal aging. Curr. Opin. Genet. Dev. 81, 102067. doi:10.1016/j.gde.2023.102067
Paige, S. L., Plonowska, K., Xu, A., and Wu, S. M. (2015). Molecular regulation of cardiomyocyte differentiation. Circ. Res. 116 (2), 341–353. doi:10.1161/circresaha.116.302752
Qu, J., Yan, H., Hou, Y., Cao, W., Liu, Y., Zhang, E., et al. (2022). RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential. J. Hematol. Oncol. 15 (1), 8. doi:10.1186/s13045-022-01224-4
Rice, D., and Barone, S. (2000). Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ. Health Perspect. 108 (Suppl. 3), 511–533. doi:10.1289/ehp.00108s3511
Robb, L., and Tam, P. P. (2004). Gastrula organiser and embryonic patterning in the mouse. Semin. Cell Dev. Biol. 15 (5), 543–554. doi:10.1016/j.semcdb.2004.04.005
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6, e31311. doi:10.7554/eLife.31311
Sarjeant, K., and Stephens, J. M. (2012). Cold Spring Harb. Perspect. Biol. 4 (9), a008417. doi:10.1101/cshperspect.a008417
Selberg, S., Seli, N., Kankuri, E., and Karelson, M. (2021). Rational design of novel anticancer small-molecule RNA m6A demethylase ALKBH5 inhibitors. ACS Omega 6 (20), 13310–13320. doi:10.1021/acsomega.1c01289
Semenovykh, D., Benak, D., Holzerova, K., Cerna, B., Telensky, P., Vavrikova, T., et al. (2022). Myocardial m6A regulators in postnatal development: effect of sex. Physiol. Res. 71 (6), 877–882. doi:10.33549/physiolres.934970
Sendinc, E., and Shi, Y. (2023). RNA m6A methylation across the transcriptome. Mol. Cell 83 (3), 428–441. doi:10.1016/j.molcel.2023.01.006
Shao, Y., Liu, Z., Song, X., Sun, R., Zhou, Y., Zhang, D., et al. (2023). ALKBH5/YTHDF2-mediated m6A modification of circAFF2 enhances radiosensitivity of colorectal cancer by inhibiting cullin neddylation. Clin. Transl. Med. 13 (7), e1318. doi:10.1002/ctm2.1318
Shen, L., Song, C. X., He, C., and Zhang, Y. (2014). Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 83, 585–614. doi:10.1146/annurev-biochem-060713-035513
Shestopalov, I. A., and Chen, J. K. (2008). Chemical technologies for probing embryonic development. Chem. Soc. Rev. 37 (7), 1294–1307. doi:10.1039/b703023c
Simon, A. K., Hollander, G. A., and McMichael, A. (2015). Evolution of the immune system in humans from infancy to old age. Proc. Biol. Sci. 282 (1821), 20143085. doi:10.1098/rspb.2014.3085
Song, T., Yang, Y., Jiang, S., and Peng, J. (2020). Novel insights into adipogenesis from the perspective of transcriptional and RNA N6-Methyladenosine-Mediated post-transcriptional regulation. Adv. Sci. (Weinh) 7 (21), 2001563. doi:10.1002/advs.202001563
Sun, X., Lu, J., Li, H., and Huang, B. (2022). The role of m6A on female reproduction and fertility: from gonad development to ovarian aging. Front. Cell Dev. Biol. 10, 884295. doi:10.3389/fcell.2022.884295
Taby, R., and Issa, J. P. (2010). Cancer epigenetics. CA Cancer J. Clin. 60 (6), 376–392. doi:10.3322/caac.20085
Takahashi, H., Hase, H., Yoshida, T., Tashiro, J., Hirade, Y., Kitae, K., et al. (2022). Discovery of two novel ALKBH5 selective inhibitors that exhibit uncompetitive or competitive type and suppress the growth activity of glioblastoma multiforme. Chem. Biol. Drug Des. 100 (1), 1–12. doi:10.1111/cbdd.14051
Tang, C., Klukovich, R., Peng, H., Wang, Z., Yu, T., Zhang, Y., et al. (2017). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U. S. A. 115 (2), E325–E333. doi:10.1073/pnas.1717794115
Thapa, P., and Farber, D. L. (2019). The role of the thymus in the immune response. Thorac. Surg. Clin. 29 (2), 123–131. doi:10.1016/j.thorsurg.2018.12.001
Tian, C., Chai, J., Liu, W., Zhang, X., Li, Y., Zuo, H., et al. (2022). Role of the demethylase AlkB homolog H5 in the promotion of dentinogenesis. Front. Physiol. 13, 923185. doi:10.3389/fphys.2022.923185
Valenti, M., Dalle, C. L., and Mottes, M. (2016). Osteogenic differentiation in healthy and pathological conditions. Int. J. Mol. Sci. 18 (1), 41. doi:10.3390/ijms18010041
Wang, H.-F., Kuang, M.-j., Han, S.-j., Wang, A.-b., Qiu, J., Wang, F., et al. (2020). BMP2 modified by the m6A demethylation enzyme ALKBH5 in the ossification of the ligamentum flavum through the AKT signaling pathway. Calcif. Tissue Int. 106 (5), 486–493. doi:10.1007/s00223-019-00654-6
Wang, K., Liu, H., Hu, Q., Wang, L., Liu, J., Zheng, Z., et al. (2022). Epigenetic regulation of aging: implications for interventions of aging and diseases. Signal Transduct. Target Ther. 7 (1), 374. doi:10.1038/s41392-022-01211-8
Wang, L., Song, C., Wang, N., Li, S., Liu, Q., Sun, Z., et al. (2020). NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 16 (12), 1394–1402. doi:10.1038/s41589-020-0601-2
Wang, M. K., Gao, C. C., and Yang, Y. G. (2023). Emerging roles of RNA methylation in development. Acc. Chem. Res. 56 (23), 3417–3427. doi:10.1021/acs.accounts.3c00448
Wang, Y.-Z., Li, H.-Y., Zhang, Y., Jiang, R.-X., Xu, J., Gu, J., et al. (2023). Discovery of Pyrazolo[1,5-a]pyrimidine derivative as a novel and selective ALKBH5 inhibitor for the treatment of AML. J. Med. Chem. 66 (23), 15944–15959. doi:10.1021/acs.jmedchem.3c01374
Warmflash, A., Sorre, B., Etoc, F., Siggia, E. D., and Brivanlou, A. H. (2014). A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11 (8), 847–854. doi:10.1038/nmeth.3016
Wu, S., Xu, X., Gao, S., Huo, S., Wan, M., Zhou, X., et al. (2024). MicroRNA-93-5p regulates odontogenic differentiation and dentin formation via KDM6B. J. Transl. Med. 22 (1), 54. doi:10.1186/s12967-024-04862-z
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xiao, Y. L., Liu, S., Ge, R., Wu, Y., He, C., Chen, M., et al. (2023). Transcriptome-wide profiling and quantification of N(6)-methyladenosine by enzyme-assisted adenosine deamination. Nat. Biotechnol. 41 (7), 993–1003. doi:10.1038/s41587-022-01587-6
Xu, C., Liu, K., Tempel, W., Demetriades, M., Aik, W., Schofield, C. J., et al. (2014). Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J. Biol. Chem. 289 (25), 17299–17311. doi:10.1074/jbc.M114.550350
Xu, P., and Ge, R. (2022). Roles and drug development of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur. J. Med. Chem. 230, 114118. doi:10.1016/j.ejmech.2022.114118
Yang, R., Ji, F., and Jiao, J. (2025). Early central nervous system development and neuron regeneration. Curr. Opin. Genet. Dev. 90, 102286. doi:10.1016/j.gde.2024.102286
Yen, Y. P., and Chen, J. A. (2021). The m(6)A epitranscriptome on neural development and degeneration. J. Biomed. Sci. 28 (1), 40. doi:10.1186/s12929-021-00734-6
You, Y., Fu, Y., Huang, M., Shen, D., Zhao, B., Liu, H., et al. (2022). Recent advances of m6A demethylases inhibitors and their biological functions in human diseases. Int. J. Mol. Sci. 23 (10), 5815. doi:10.3390/ijms23105815
Yu, J., Shen, L., Liu, Y., Ming, H., Zhu, X., Chu, M., et al. (2020). The m6A methyltransferase METTL3 cooperates with demethylase ALKBH5 to regulate osteogenic differentiation through NF-κB signaling. Mol. Cell Biochem. 463 (1-2), 203–210. doi:10.1007/s11010-019-03641-5
Zaccara, S., and Jaffrey, S. R. (2020). A unified model for the function of YTHDF proteins in regulating m6A-Modified mRNA. Cell 181 (7), 1582–1595. doi:10.1016/j.cell.2020.05.012
Zaccara, S., and Jaffrey, S. R. (2024). Understanding the redundant functions of the m(6)A-binding YTHDF proteins. Rna 30 (5), 468–481. doi:10.1261/rna.079988.124
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20 (10), 608–624. doi:10.1038/s41580-019-0168-5
Zhao, J., Ding, C., and Li, H. B. (2023). N6-Methyladenosine defines a new checkpoint in γδ T cell development. BioEssays 45 (5), e2300002. doi:10.1002/bies.202300002
Zhao, Y., Hu, J., Sun, X., Yang, K., Yang, L., Kong, L., et al. (2021). Loss of m6A demethylase ALKBH5 promotes post-ischemic angiogenesis via post-transcriptional stabilization of WNT5A. Clin. Transl. Med. 11 (5), e402. doi:10.1002/ctm2.402
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013a). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zheng, G., Dahl, J. A., Niu, Y., Fu, Y., Klungland, A., Yang, Y. G., et al. (2013b). Sprouts of RNA epigenetics: the discovery of mammalian RNA demethylases. RNA Biol. 10 (6), 915–918. doi:10.4161/rna.24711
Zheng, H. X., Zhang, X. S., and Sui, N. (2020). Advances in the profiling of N(6)-methyladenosine (m(6)A) modifications. Biotechnol. Adv. 45, 107656. doi:10.1016/j.biotechadv.2020.107656
Zheng, T., Wang, D., Zhou, S., Liu, M., Liu, Y., Gu, X., et al. (2023). Promoting axon regeneration by inhibiting RNA N6-methyladenosine demethylase ALKBH5. eLife 12, e85309. doi:10.7554/eLife.85309
Keywords: ALKBH5, m6A demethylation, development, organogenesis, therapeutics
Citation: Zhang X, Zhou L, Tian C and Tao H (2025) ALKBH5 in development: decoding the multifaceted roles of m6A demethylation in biological processes. Front. Mol. Biosci. 12:1599487. doi: 10.3389/fmolb.2025.1599487
Received: 29 March 2025; Accepted: 26 June 2025;
Published: 04 August 2025.
Edited by:
Marie-Pierre Golinelli, UPR2301 Institut de Chimie des Substances Naturelles (ICSN CNRS), FranceCopyright © 2025 Zhang, Zhou, Tian and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huangheng Tao, dGFvaGh3aHVAMTYzLmNvbQ==
†ORCID: Huangheng Tao, orcid.org/0000-0002-5530-0911; Xinye Zhang, orcid.org/0009-0006-1920-3002; Linfang Zhou, orcid.org/0009-0001-3637-6996; Cheng Tian, orcid.org/0009-0001-4498-5652
‡These authors have contributed equally to this work
 Xinye Zhang
Xinye Zhang Linfang Zhou
Linfang Zhou Cheng Tian3†
Cheng Tian3† Huangheng Tao
Huangheng Tao