- 1Department of Biomedicine, Neurosciences and Advanced Diagnostics (Bi.N.D), University of Palermo, Section of Biology and Genetics, Palermo, Italy
- 2Institute for Sustainable Plant Protection, National Research Council of Italy (IPSP-CNR), Torino, Italy
- 3National Research Council, Institute of Biosciences and Bioresources (IBBR), Palermo, Italy
- 4Navhetec s.r.l., Spinoff of the University of Palermo, Palermo, Italy
- 5ATeN (Advanced Technologies Network) Center, University of Palermo, Palermo, Italy
- 6Institute for Biomedical Research and Innovation (IRIB), National Research Council (CNR), Palermo, Italy
Plant-derived nanovesicles (PDNVs) are emerging as a novel class of biological messengers, capable of crossing biological barriers and transferring bioactive molecules to human cells. We have previously isolated and characterized nanovesicles from lemon juice (LNVs) that interact with human cells, modulating the mechanisms of oxidative stress and inflammation. However, the mechanisms through which LNVs exert their effect are still poorly understood. Extensive researches have been conducted on the microRNA content of PDNVs, however the profile and role of long noncoding RNAs (lncRNAs) within the vesicles remain unexplored still. In the present study, the lncRNA cargo of LNVs and its role was investigated; highly conserved lncRNAs among Citrus species was highlighted, with a notable enrichment of LM_XLOC_013494 within the vesicles. The lncRNA was successfully transferred to human hepatic (THLE-2) and intestinal (CACO-2) cells treated with LNVs, as confirmed by RT-qPCR and RNA in situ hybridization. Bioinformatic prediction analyses coupled with experimental validation revealed that the isolated lncRNA acts as a molecular sponge, specifically targeting human miR-181b-3p and miR-4420. Importantly, LNV-treated cells showed a statistically significant downregulation of these miRNAs (p ≤ 0.05), suggesting a cross-kingdom regulatory role for the plant lncRNA in modulating human gene expression. Overall, to our knowledge, this study provides novel insights into the trans-kingdom transfer of plant-derived lncRNAs, expanding upon previous findings and offering new experimental evidence.
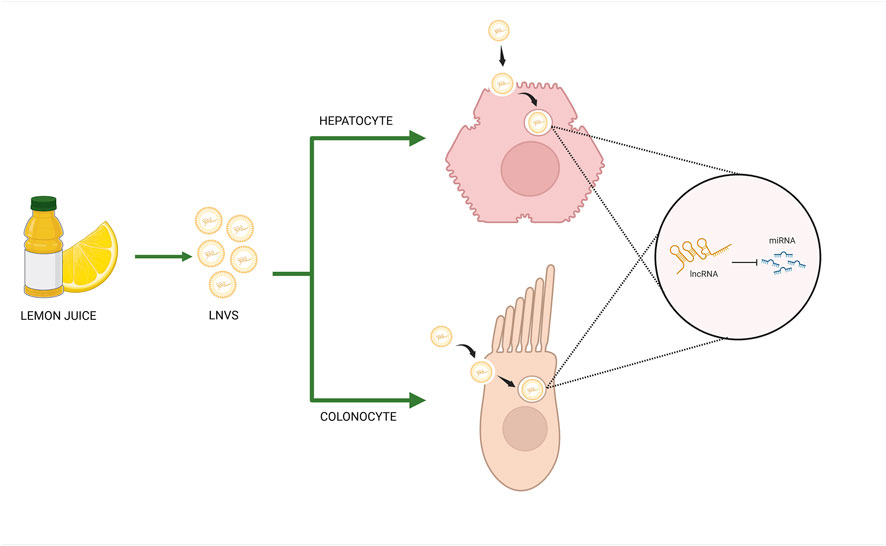
GRAPHICAL ABSTRACT | Lemon-derived nanovesicles (LNVs) deliver long noncoding RNAs (lncRNAs) into human hepatic and intestinal cells, enabling cross-kingdom regulation of gene expression via miRNA sponging mechanism (Created in BioRender. RAIMONDO, S. (2025) https://BioRender.com/yhwk2aq).
1 Introduction
Extracellular vesicles (EVs) represent a pivotal system of intercellular and trans-kingdom communication, orchestrating the transfer of biological signals between cells (van Niel et al., 2018; Raposo and Stahl, 2019). While the molecular mechanisms underlying selective packaging and targeted delivery of EV cargo are not fully understood, there is increasing evidence that EV cargo, consisting of a wide array of biomolecules, plays a central role in determining EV’s functional properties (Dixson et al., 2023; Mir and Goettsch, 2020). In the frame of increasing interest in EVs, plant-derived nanovesicles have recently emerged as a class of biological vesicles, exhibiting promising potential in human health applications. PDNVs, isolated from different plant species, are characterized by high biocompatibility, structural stability, and the ability to cross biological barriers (Lian et al., 2022). Emerging evidences indicate that PDNVs show several bioactive properties, including anti-inflammatory (Ye et al., 2024; Urzì et al., 2023), immunomodulatory (Nguye et al., 2023), and antioxidant activities (Perut et al., 2021). These functional characteristics suggest a potential role of PDNVs in modulating cellular physiology and signaling pathways in mammalian systems. However, despite the encouraging findings, the molecular mechanisms underlying the beneficial effects of PDNVs remain often unclear. The biomolecules encapsulated and protected by the vesicles are thought to play a crucial role in mediating these effects (Pocsfalvi et al., 2018). Among them, non-coding RNAs (ncRNAs) have emerged as critical regulators of cellular processes. They are involved in the regulation mechanisms of gene expression at multiple levels, including chromatin remodeling as well as transcriptional and post-transcriptional modifications (Panni et al., 2020; Mattick and Makunin, 2006). Among ncRNAs, long non-coding RNAs (lncRNAs) have attracted significant interest due to their complex regulatory role in important biological processes (Statello et al., 2021). These molecules, with a length of more than 200 nucleotides, can be transcribed from exons, introns, intergenic regions or 5′/3′untranslated regions and fold into intricate secondary structures that facilitate their interactions with DNA, RNA and proteins (Gut et al., 2012; Batista and Chang, 2013). LncRNAs regulate gene expression through multiple mechanisms. Indeed, they can act as molecular sponges or decoys for miRNAs, preventing degradation of target mRNA. Additionally, specific lncRNAs can modulate transcription factors, enabling their binding to promoters and thereby regulating the transcriptional regulation of target genes (Yan and Bu, 2021).
Based on these premises, we hypothesized that the beneficial effects of PDNVs may partially be attributed to the presence of specific lncRNAs. Given our extensive experience in the lemon-derived nanovesicles (LNVs) (Urzì et al., 2023; Raimondo et al., 2015; Raimondo et al., 2022), in the present work we focused our efforts to: (i) characterize the lncRNA cargo of LNVs, (ii) investigate whether these lncRNAs can be transferred to human cells, and (iii) assess their potential regulatory function in human cells. This work addresses a critical gap in the current understanding of PDNV-mediated interspecies communication and creates new opportunities for the therapeutic exploitation of plant-derived nanovesicles.
2 Materials and methods
2.1 LNV isolation
Lemon-derived nanovesicles (LNVs) were isolated from the juice of Citrus limon L. according to procedures previously described (Raimondo et al., 2015). The fruits, sourced from a private farmer, were thoroughly washed with water and manually squeezed. The juice obtained was subjected to two sequential centrifugations at 3,000 × g for 15 min, followed by two centrifugations at 10,000 × g for 30 min. The supernatant was filtered with 0.8 μm pore size membranes, then centrifuged at 16,500 × g for 1 h and again subjected to a filtration step with 0.45 μm pore size membranes, then centrifuged at 16,500 × g for 3 h. Finally, the resulting supernatant was subjected to a final ultracentrifugation at 100,000 × g for 1 h 45 min using a fixed-angle rotor (Type 70 Ti). The obtained pellet was washed and suspended in phosphate-buffered saline (PBS) (Raimondo et al., 2015). The isolated vesicles have been previously characterized by our research group at the proteomic, metabolomic, morphological, and size distribution levels, ensuring the reproducibility and quality of the preparations used in this study (Urzì et al., 2023; Raimondo et al., 2015; Raimondo et al., 2022).
2.2 Cell culture
The THLE-2 cell line (ATCC CRL-2706™, LGC Standards, Manassas, VA, USA) was used as an in vitro model of healthy human hepatocytes. Cells were cultured on a coating of 0.03 mg/mL bovine collagen type I (Advanced Biomatrix, San Diego, CA, USA) and 0.01 mg/mL bovine serum albumin (Sigma-Aldrich, St. Louis, MO, USA). The culture medium consisted of RPMI 1640 (Euroclone, UK) supplemented with 10% fetal bovine serum (FBS, Euroclone, UK), 1% penicillin (100 U/mL) and streptomycin (100 μg/mL), as well as 0.3 mL recombinant human epidermal growth factor (EGF, 10 μg/mL) and 0.4 mL phosphoethanolamine (PEA, 100 μg/mL). CACO-2 cell line (ATCC HTB-37™, LGC Standards, Manassas, VA, USA) was used as an in vitro model of human enterocytes. Cells were maintained in Eagle’s Minimum Essential Medium (EMEM) (ATCC, Manassas, VA, USA) supplemented with 20% fetal bovine serum (FBS, Euroclone, UK), 1% penicillin (100 U/mL), and streptomycin (100 μg/mL). Cell cultures were incubated in a humidified atmosphere of 5% CO2 at 37 °C.
2.3 RNA purification and cDNA synthesis
Total RNA was purified from lemon juice, LNVs, and cell lines.
2.3.1 RNA purification from lemon juice and LNVs and RNase protection assays
RNA was isolated from the previously lyophilized lemon juice. To extract only RNAs located inside EVs, an RNase protection assay was carried out following the procedure described in Karimi et al. (Zand Karimi et al., 2022), using RNase A (Qiagen, Hilden, Germany; diluted in 15-mM NaCl, 10-mM Tris–HCl pH 7.5) with a final concentration of 5 μg/mL and an incubation step at room temperature (RT) for 30 min. Immediately after RNase A treatment, 0.5 mL of cold (4 °C) PureLink™ Plant RNA Reagent (Thermo Fisher Scientific, Waltham, MA, USA) was added to 100 µL of the mixture. The solution was mixed by vortex until the sample was thoroughly resuspended, followed by 5 min of incubation at room temperature (RT) and then centrifuged at 12,000 x g for 2 min at RT. The supernatant was transferred to a clean RNase-free tube, adding 0.1 mL of 5 M NaCl to the clarified extract. The solution was then shaken by tapping, followed by the addition of 0.3 mL of chloroform, mixed thoroughly by inverting the tube for 30 s, and then centrifuged at 12,000 x g for 10 min at 4 °C. The aqueous phase was recovered and mixed with an equal volume of cold isopropyl alcohol to precipitate the RNA and let stand at room temperature for 10 min. RNA pellet was washed using cold 75% ethanol and then, after centrifugation at 12,000 x g for 1 min at RT, the supernatant was carefully decanted. RNA was finally resuspended in 30 µL of RNase-free Water. To inhibit RNase A activity, used in the preliminary step, a mixture of 10 mg/mL RNase Inhibitor (Thermo Fisher Scientific, Waltham, MA, USA) and 40 units/mL of RNase Out (Invitrogen, Waltham, MA, USA) was added to the extracted RNAs, which were stored at −80 °C until use. The RNA quality and quantity were assessed using either a ThermoFisher NanoDrop One spectrophotometer and an Agilent 2100 Bioanalyzer Instrument. Total RNA was subsequently reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA).
2.3.2 RNA isolation from human cell lines
THLE-2 and CACO-2 cells were seeded in 12-well plates at a density of 100,000 cells per well. 24 hours after seeding, cells were treated with two different LNVs concentrations, 25 and 50 μg/mL respectively, and incubated at different time points (6, 24, 48 h). At the end of each treatment, RNA was isolated using the commercially available Nucleospin miRNA Kit (Macherey-Nagel, Düren, Germany), which enables the isolation of both total and microRNA, according to the manufacturer’s instructions. The concentration and purity of extracted RNA were determined by a Nanodrop spectrophotometer (NanoDrop Technologies, USA). Total RNA was subsequently reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). The miRNAs of interest were reverse transcribed using the TaqMan™ microRNA reverse transcription kit (Applied Biosystems™, USA) employing specific primers for U6, and mir-4420.
2.4 Polymerase chain reaction (PCR)
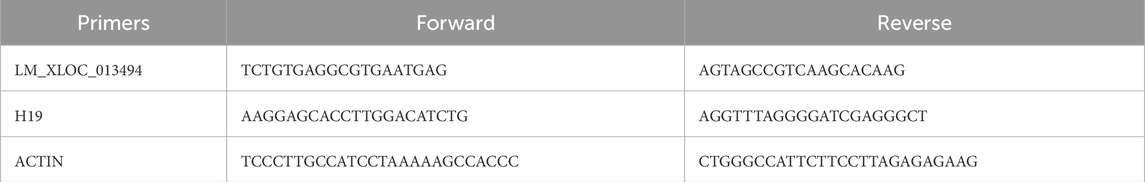
Total cDNA was used to amplify the specific plant lncRNA LM_XLOC_013494 and the human lncRNA H19 by polymerase chain reaction (PCR). Primer sequences are reported in Table 1. The PCR amplification reaction was conducted, using yourSial Taq mix kit (Sial Group, Italy) and following the instructions provided by the kit. The reaction mixture for LM_XLOC_013494 was initially heated at 95 °C for 1 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 120s. The reaction mixture for H19 was initially heated at 95 °C for 1 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 58 °C for 15 s, and extension at 72 °C for 60s. After amplification, the PCR products were subjected to electrophoresis on a 2.5% (LM_XLOC_013494) and 1.5% (H19) agarose gels and visualized by Chemidoc acquisition instrument (Bio-Rad, USA).
2.5 Real time PCR
The presence of plant lncRNAs in LNVs and human THLE-2 cells treated with LNVs was confirmed by RT-PCR (Step One Real-time PCR system, Applied Biosystem) in a 20-μL reaction containing 300 nM of each primer, 2 μL of template cDNA, and 18 μL of 2X SYBR Green I Master Mix. Actin was used as an endogenous control. Changes in lncRNA levels between control and treated samples were determined by the 2−ΔCt method.
The list of primers used is reported in Table 1.
To assess the levels of miRNAs in human cell lines treated with LNVs, each RT-PCR reaction was prepared with TaqMan™ Fast Universal PCR Master Mix (Applied Biosystems™, USA). The primers used were hsa-miR-4420, has-miR-181b-30, has-miR-199a-3p and U6 snRNA (all from Applied Biosystems™, USA). The levels of miRNAs were normalized to U6 snRNA, and data were analyzed using the 2−ΔΔCt method.
2.6 In situ hybridization analysis
The presence of investigated plant lncRNAs in human THLE-2 and CACO-2 cells treated with LNVs was evaluated using the BaseScope Detection Reagent Kit v2 - RED. Briefly, the treated THLE-2 and CACO-2 cells were seeded in a 8-well chamber and, after 24 h, treated with 70 μg/mL LNVs for 24 h. This concentration was selected specifically for the RNA scope assay to enhance image resolution and signal clarity during qualitative visualization of RNA molecules. At the end of treatment, cells were fixed with Neutral Buffered Formalin 10% and then dehydrated and rehydrated with ethanol scale at different concentrations. Then, a dilute protease III solution was applied for 10 min. After two washes, a probe specifically designed to bind the plant lncRNA sequence of interest was used in the cell hybridizetion for 2 h at 40 °C. Subsequently, eight amplification steps and signal detection reactions were performed according to the kit instructions, as described in the BaseScope Detection Reagent Kit v2 - RED User Manual. Finally, for better bright-field visualization, the cells were stained with Gill’s 50% hematoxylin solution and the slides mounted with Ecomount (Advanced Cell Diagnostics). To ensure the accuracy of the assay, internal positive and negative control provided by the BaseScope™ kit was included in all experiments. All images were acquired at 40× magnification.
2.7 Bioinformatic analysis
To identify potential homologous sequences in different species, the sequence homology of the lncRNA LM_XLOC_013494 was investigated using RNAcentral (http://rnacentral.org/), a comprehensive database of noncoding RNA sequences (The RNAcentral Consortium et al., 2017); homology was evaluated according to sequence similarity, E-value, percent identity, query and target coverage, and presence of gaps. The subcellular localization of the lncRNA LM_XLOC_013494 was predicted using lncLocator 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator2/) (Lin et al., 2021).
The bioinformatic tools miRDB (https://mirdb.org) and linc2function (https://bioinformaticslab.erc.monash.edu/linc2function) (Chen and Wang, 2020; Ramakrishnaiah et al., 2023) were used to explore the RNA interactome and predict potential miRNAs interacting with the lncRNA LM_XLOC_013494. The results of both analyses were cross-referenced, and miRNAs with the highest prediction scores in both analyses were selected for further analyses.
2.8 Statistical analysis
Data are reported as mean ± standard deviation (SD) of biological replicates. Statistical analysis was conducted using GraphPad Prism software (GraphPad software, Inc., La Jolla, CA). The normality of data distribution was assessed by the Shapiro-Wilk test. For normally distributed data, the statistical significance of the differences was analyzed using a two-tailed Student’s T-test; otherwise, a non-parametric method (Mann-Whitney test) was applied to compare the groups. For the statistical evaluation of miRNA levels in cells treated with LNVs, a one-sample t-test was performed. A p-value ≤ 0.05 was considered statistically significant. The statistical details for each experiment are provided in the corresponding figure legends.
3 Results
3.1 LncRNAs conserved in Citrus species are present in LNVs
Through an extensive available literature (Ke et al., 2019), we identified three lncRNAs that are highly conserved in Citrus species: LM_XLOC_031833, LM_XLOC_013494, and LM_XLOC_018060. The presence and levels of these selected lncRNAs were evaluated in Citrus limon juice and LNVs isolated from it. Two out of the three lncRNAs studied - LM_XLOC_013494 and LM_XLOC_018060 - were detected in both LNVs and juice. Interestingly, a more significant abundant level (p ≤ 0.05) in LNVs compared to the juice was detected, suggesting an enrichment of these lncRNAs within the vesicles (Figure 1A). To confirm that these specific lncRNAs are localized within LNVs rather than being on their surface, we performed RNase A treatment on the isolated LNVs. This approach selectively degrades the external RNA while leaving the encapsulated RNA protected. Both lncRNAs remained detectable by RT-PCR after RNase A treatment, indicating that they are encapsulated within the vesicles and not simply bound to the vesicle surface (Figure 1B).
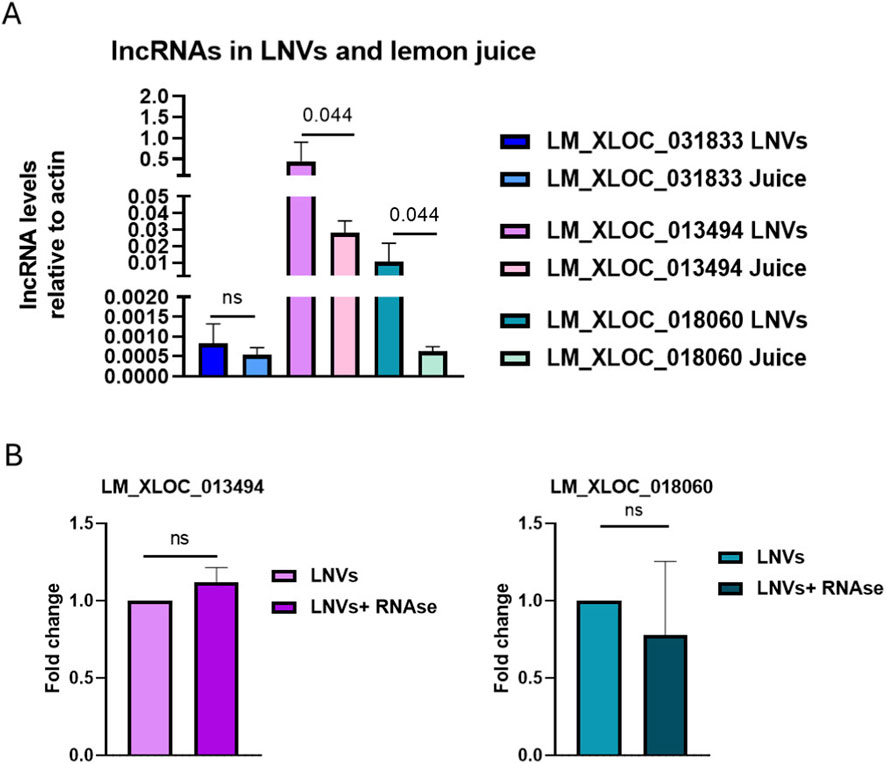
Figure 1. LncRNAs conserved in Citrus species are present in LNVs (A) RT-PCR analysis was performed to evaluate the presence and relative abundance of three lncRNAs in Citrus limon juice and LNVs (LM_XLOC_031833, LM_XLOC_013494, and LM_XLOC_018060). LncRNAs levels were normalized to actin. (LNV samples n = 3–8; juice samples n = 2); (B) RT-PCR analysis of the levels of the two most abundant lncRNAs (LM_XLOC_013494 and LM_XLOC_018060) was performed on LNVs, either untreated or treated with RNase A. LncRNA levels were normalized to actin (LNV samples n = 3; LNV + RNAse samples n = 3). Data is reported as mean ± SD. Statistical significance was assessed by a nonparametric test (Mann-Whitney test).
Since LM_XLOC_013494 was the most abundant among the three studied lncRNAs in our LNVs, we focused our work on this specific lncRNA. Homology search performed by RNAcentral highlighted a high degree of sequence similarity with the Citrus sinensis lncRNA CSIN_LNC000747. The analysis also highlighted homology, even if limited identity and coverage, with the Homo sapiens lncRNA HSALNT0205494 (Table 2).
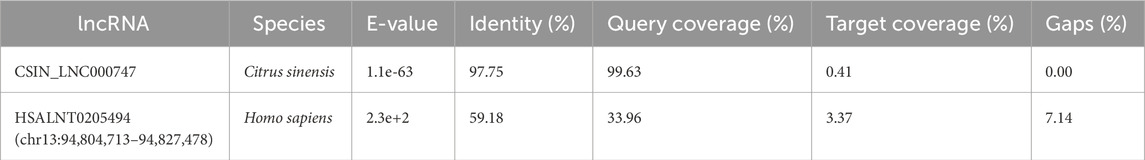
Table 2. Alignment parameters and similarity metrics identified during the analysis with RNAcentral.
Given the observed sequence similarity between LM_XLOC_013494 and the human lncRNA HSALNT0205494, a bioinformatic in silico analysis was conducted to verify that the primers designed for LM_XLOC_013494 did not recognize human sequences present in our samples. The BLAST analysis confirmed that the primer binding sites are specifically located within the plant lncRNA sequence without any overlapping in the homology region with human lncRNA, thus ensuring the specificity of our assays (data not shown). In addition, the full sequence of LM_XLOC_013494 was used to predict its localization in the different cellular compartments, including nucleus, cytoplasm, exosome, ribosome, and cytosol. The analysis performed with lncLocator predicted that LM_XLOC_013494 is predominantly localized in the nucleus with a score of 0.77 (Supplementary Figure 1).
3.2 THLE-2 cells treated with LNVs showed an increase in the presence of lncRNA
After the LM_XLOC_013494 identification and characterization, we evaluated whether LNVs could effectively transport and deliver this specific lncRNA to mammalian cells, thereby mediating trans-kingdom communication. Therefore, human hepatocyte THLE-2 cells were treated with two concentrations of isolated LNVs (25 and 50 μg/mL, respectively) at different time points (6, 24, and 48 h). PCR amplification of LM_XLOC_013494 in THLE-2 treated cells, followed by agarose gel electrophoresis revealed the presence of the lncRNA in LNV-treated cells (Figure 2A). To validate the specificity of the plant-derived lncRNA detection, we also assessed the expression of the human lncRNA H19 as an internal control. As shown in Supplementary Figure 2A, H19 was consistently expressed across all samples, regardless of LNV treatment.
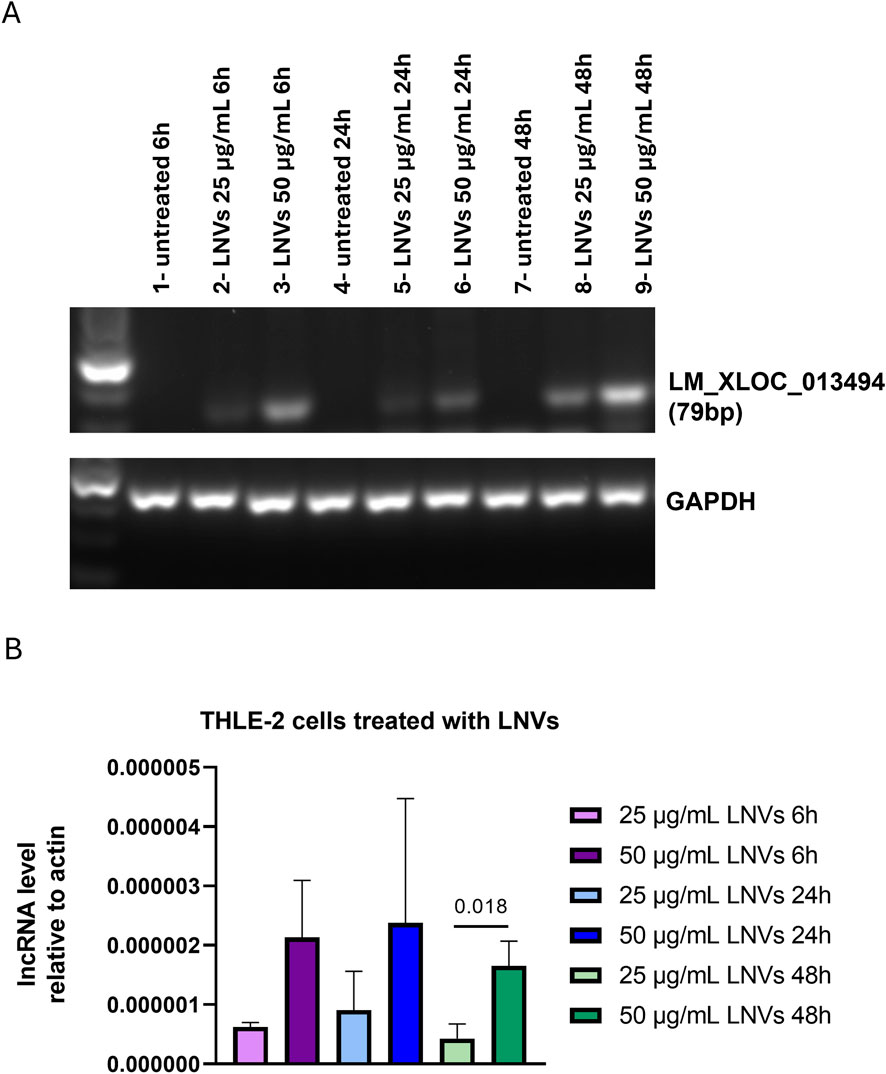
Figure 2. Cells treated with LNVs showed an increase in lncRNA levels. (A) PCR amplification of plant-derived lncRNA LM_XLOC_013494 was performed on cDNA from THLE-2 cells treated with LNVs (25 and 50 μg/mL for 6, 24 and 48 h). Amplified products were separated on a 2.5% agarose gel and visualized with a ChemiDoc imaging system. GAPDH was used as a loading control. (B) Levels of plant-derived lncRNA LM_XLOC_013494 were measured by RT-PCR in THLE-2 cells treated with LNVs (25 and 50 μg/mL for 6, 24, and 48 h). LncRNAs levels were normalized to actin (n = 3–6). Data is reported as mean ± SD. Statistical significance was assessed by unpaired Student’s t test (the normal data distribution was analyzed by Shapiro-Wilk test).
To further confirm these findings, we performed RT-qPCR; as shown in Figure 2B, we observed an increase in LM_XLOC_013494 levels with a dose-dependent effect observed at 6, 24, and 48 h.
Additionally, the in situ hybridization was performed to qualitatively detect the presence of plant-derived LM_XLOC_013494 in the human LNV-treated cells, both THLE-2 and CACO-2 cells using an additional LNVs concentration of 70 μg/mL, for 24 h. By using target-specific probes, the assay allowed to detect the lncRNA as distinct red dots in the cytoplasm of treated cells. To assess whether LM_XLOC_013494 was encapsulated within the vesicles or simply associated with their surface, the in situ hybridization was also carried out using LNVs pre-treated with RNase A. A marked increase in red dots underlined the presence of LM_XLOC_013494 in both THLE-2 and CACO-2 cells treated with LNVs, compared to untreated ones (Figure 3; Supplementary Figure 2B). Notably, no differences were observed between LNV-treated cells and LNV-treated samples with preliminary RNase digestion step; this finding supports the conclusion that the lncRNA is encapsulated within the plant-derived nanovesicles, thereby protected from enzymatic degradation.
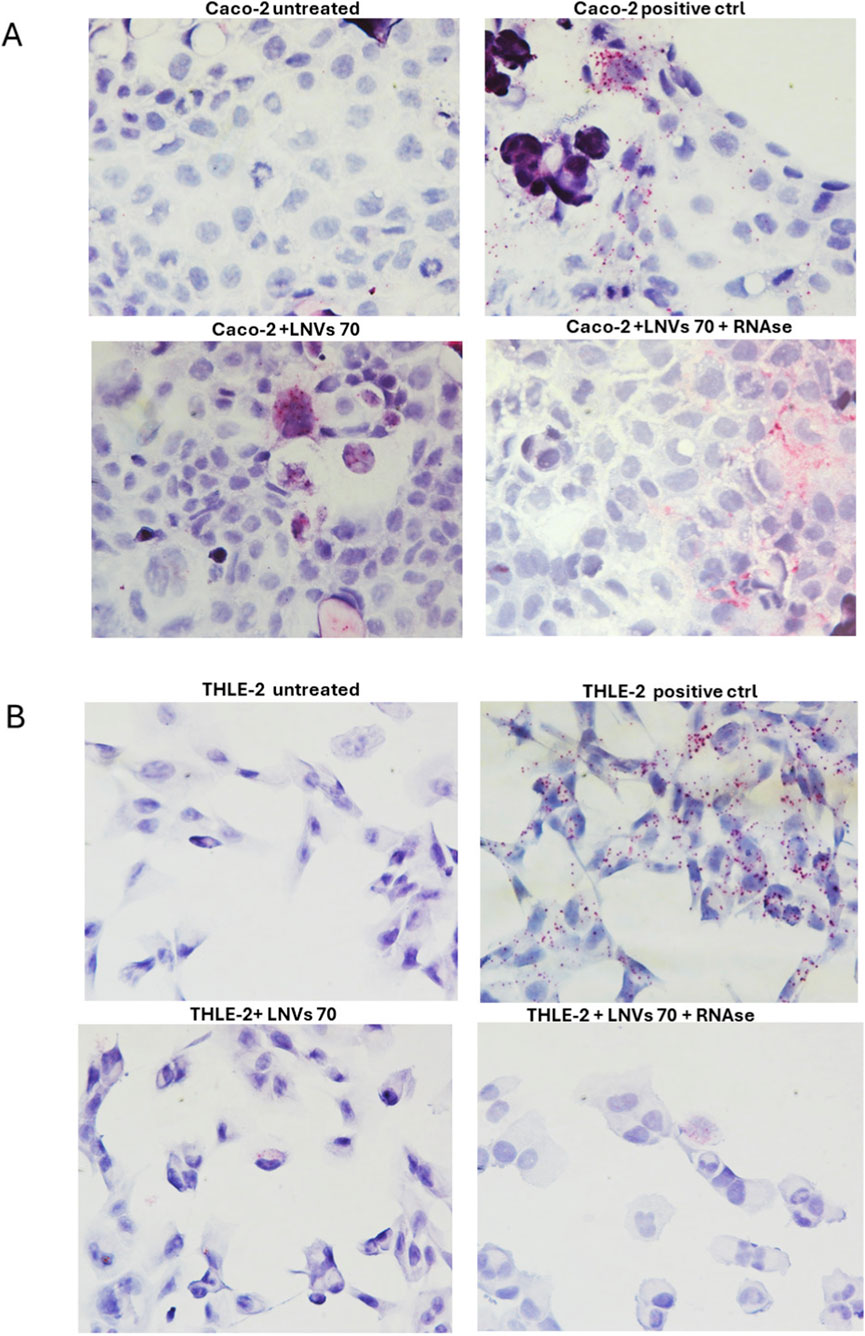
Figure 3. Detection of plant-derived lncRNA LM_XLOC_013494 in human cells by BaseScope™ in situ hybridization. (A) Caco-2 cells and (B) THLE-2 cells were treated for 24 h with 70 μg/mL of LNVs, pretreated or not with RNase A. The red/purple dotted signals indicate the intracellular presence of the plant-derived lncRNA LM_XLOC_013494. Nuclei were counterstained with hematoxylin. Internal positive controls supplied with the BaseScope™ kit were included to ensure the specificity and accuracy of the assay. All images were acquired at 40× magnification under bright-field microscopy.
3.3 THLE-2 and CACO-2 cells treated with LNVs showed a reduction in the level of miRNA sponged by lncRNA
In cells, lncRNAs can act as sponges of miRNAs, reducing their regulatory effect on mRNAs. To assess whether the plant-derived lncRNA LM_XLOC_013494 has functional activity after being delivered into human cells via LNVs, we first conducted a in silico analysis to identify miRNAs that could potentially interact with LM_XLOC_013494 based on sequence complementarity, and binding site prediction algorithms. Two bioinformatics tools, miRDB and linc2function, were employed to perform this analysis. The miRDB analysis revealed 11 possible miRNAs that could interact with LM_XLOC_013494 based on sequence complementarity and prediction of binding sites. To further explore the RNA interactome, linc2function was used, and more than 250 potential interactions were identified. The complementary approach allowed to reduce the potential targets. Indeed, both tools (Supplementary Figure 3) converged on a specific miRNA, miR-181b-2-3p, which shares the same seed sequence with miR-181b-3p and miR-4420. To validate the bioinformatic prediction and verify if the plant-derived lncRNA LM_XLOC_013494 can interact with the selected miRNAs and sponge them, the levels of miR-4420 were evaluated in both THLE-2 and CACO-2 cells, after LNVs treatment. Starting from the results described above, for this experimental step only the cells treated with 50 μg/mL LNVs, showing higher LM_XLOC_013494 transcription level, for 6, 24, and 48 h, were used. Interestingly, except at 48h, both THLE-2 and CACO-2 LNVs-treated cells showed a reduction in miR-4420 levels at each experimental time, comparing untreated ones (Figure 4). To further support the proposed sponging mechanism, we also validated the expression of miR-181b-3p, the second predicted target, which showed a reduction upon LNV treatment, consistent with the predicted interaction with the lncRNA (Supplementary Figure 4A). This findings suggested that the lncRNA LM_XLOC_013494 may function as a molecular sponge for the tested miRNAs. As a further control, the levels of a not targeted miRNA by LM_XLOC_013494 was also assessed. As expected, no modulation was observed upon LNV treatment (Supplementary Figure 4B).
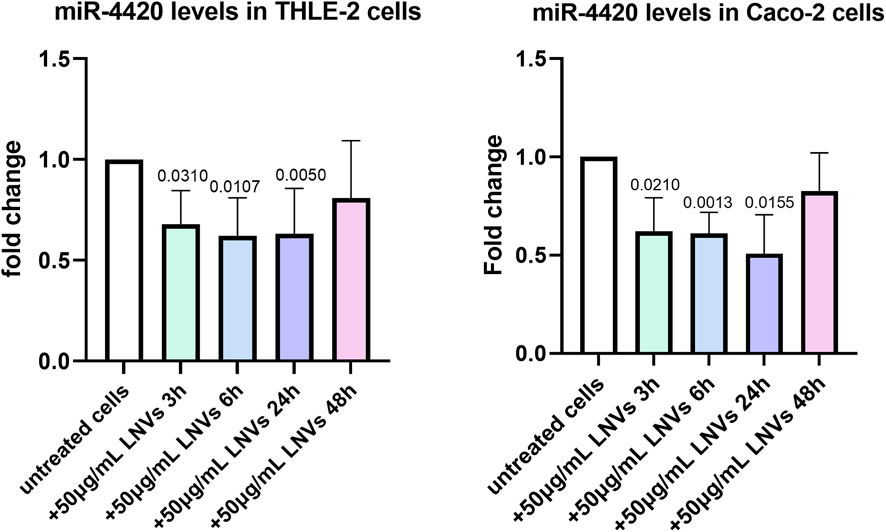
Figure 4. THLE-2 and CACO-2 cells treated with LNVs showed a reduction in the level of miRNA sponged by lncRNA. THLE-2 (left panel) and Caco-2 (right panel) cells were treated with 50 μg/mL of LNVs for 3, 6, 24 and 48 h. Levels of miR-4420 were measured by RT-PCR and normalized by U6 snRNA. Values are presented as fold change relative to untreated cells. THLE-2: n = 4 -6; Caco-2: n = 3 -5. Data is reported as a mean SD. Statistical significance was assessed by one-sample t-test.
4 Discussion
The growing interest in PDNVs has driven the scientific community to investigate the molecular mechanisms underlying their remarkable effects on mammalian cells. Although several studies demonstrated the properties of PDNVs derived from different plant sources, the specific pathways responsible for these activities remain largely undefined (Di Gioia et al., 2020).
PDNVs act as carriers of a wide range of bioactive molecules, naturally occurring in plants, encapsulated within a protective lipid bilayer. This lipid structure not only enhances the stability and bioavailability of these compounds but also facilitates their targeted delivery to recipient cells (Lian et al., 2022). In our previous studies, we demonstrated the biological properties of LNVs, highlighting their strong anti-inflammatory, antioxidant, and anti-tumor activities (Urzì et al., 2023; Raimondo et al., 2015; Gasparro et al., 2024). Based on the previous findings, the present work aimed to further investigate the molecular cargo of LNVs, with a specific focus on their non-coding RNA component. Most of the existing literature analyzing the non-coding RNA component of PDNVs focuses primarily on plant physiology or plant-microbe, and plant-parasite interactions (He et al., 2021; Zhao et al., 2024). In the context of human health, research has largely concentrated on the presence and potential roles of plant-derived miRNAs. For instance, Xiao et al. identified several miRNAs in vesicles isolated from 11 different plant species, most of which were associated with inflammatory and tumor-related pathways (Xiao et al., 2018). An intriguing additional study demonstrated that ginger-derived extracellular vesicles are internalized by the gut microbiota and contain miRNAs capable of altering their composition, thereby influencing host physiology (Teng et al., 2018). Furthermore, Baldrich and collaborators (Baldrich et al., 2019) compared the RNA profiles of EVs isolated from the apoplastic fluid of Arabidopsis with those of the leaves and the EV-depleted wash fluid. This study revealed that the EVs are enriched in small RNAs ranging from 10 to 17 nucleotides in length, referred to as 'tiny RNAs’ (Baldrich et al., 2019). Limited attention has been given to other classes of non-coding RNAs, such as lncRNAs (Xiao et al., 2018; Zhu et al., 2023). This gap highlights the novelty of our study, which shifts the focus toward exploring lncRNAs within LNVs and their potential functional impact on mammalian cells, offering new insights into the molecular mechanisms underlying PDNV-mediated interspecies communication.
Our study successfully identified ncRNAs highly conserved in Citrus species, with LM_XLOC_013494 enriched within Citrus limon-derived nanovesicles (LNVs). The enrichment of these lncRNAs in LNVs compared to citrus juice suggested that the vesicles may play a role in their stabilization and targeted delivery. This evidence is in agreement to other studies showing that specific types of non-coding RNAs are preferentially packaged into extracellular vesicles (Baldrich et al., 2019; Borniego and Innes, 2023). Moreover, we demonstrated that LNVs can deliver lncRNAs into human cells in a dose-dependent manner. The successful transfer was confirmed by evaluating lncRNA levels in LNV-treated cells using both RT-qPCR and in situ hybridization. These findings are consistent with previous studies demonstrating the role of PDNVs in facilitating trans-kingdom communication through the horizontal transfer of functional RNAs (Teng et al., 2018; Teng et al., 2021; Potestà et al., 2020).
In addition, to demonstrating RNA delivery, our study provides strong evidence that LM_XLOC_013494 maintains functional activity within human cells. Among the predicted targets of LM_XLOC_013494, in silico analysis identified two homologous micro-RNAs, miR-181b-3p and miR-4420. These miRNAs are known to be involved in regulations of important biological processes. Indeed, it has been implicated in oncogenic processes (Liu et al., 2014), including tumor initiation and progression in colorectal cancer (Jiang et al., 2023), showing an upregulation in hepatocellular carcinoma, where it serves as a marker of poor prognosis (Baysal et al., 2024; Ji et al., 2009; Ji et al., 2011; Chen et al., 2024). In addition, miR-181b-3p promotes inflammation and oxidative stress in liver disease, and its inhibition has been associated with protective effects in liver injury models (Wang et al., 2021; Azar et al., 2020; Cai et al., 2019). In parallel, miR-4420 can be considered particularly interesting, as no functional information is currently available in the literature, making it a new and promising candidate for uncovering novel regulatory pathways. Due to its strong sequence similarity with miR-181b-3p, which is the better-characterized homolog, miR-4420 was selected as a representative target to validate this predicted interaction. Our results revealed a reduction levels of miR-4420 following the LNV treatment in THLE-2 and CACO-2 cells, consistent with a “sponging” mechanism. However, while the reported findings suggest an interaction between the plant lncRNA LM_XLOC_013494 and specific human miRNAs, direct evidence of physical binding was not obtained in this study.
5 Conclusion, limitation and future directions
Our results provide new insights into the role of plant-derived lncRNAs in interspecies and trans-kingdom communication. LM_XLOC_013494, naturally occurring in LNVs, is not only efficiently delivered into human cells but also shows functional regulatory effects by modulating human miRNA levels. While these findings support the potential of LNVs as carriers of bioactive RNA molecules, some limitation of the current study should be mentioned. First, the precise intracellular fate of the delivered lncRNA remains to be elucidated. Second, although miRNA downregulation was observed, the downstream effects on target gene expression and cellular phenotypes, were not explored. Additionally, in vivo studies will be essential to validate the potential of LNV-delivered lncRNAs.
Overall, this study has enhanced our understanding of the therapeutic potential of LNVs, laying the foundation for the development of innovative plant nanovesicle-based compounds for the management of human diseases.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
VT: Methodology, Writing – original draft, Investigation. RG: Methodology, Writing – review and editing, Investigation. GD: Writing – review and editing, Methodology, Investigation. LM: Methodology, Writing – review and editing, Investigation. SR: Methodology, Investigation, Writing – review and editing. FM: Writing – review and editing, Methodology, Investigation. AC: Writing – review and editing. RA: Funding acquisition, Writing – review and editing. SR: Writing – original draft, Data curation, Investigation, Funding acquisition, Resources, Conceptualization, Writing – review and editing, Supervision.
Funding
The authors declare that financial support was received for the research and/or publication of this article. This work was funded by a grant from the University of Palermo to Stefania Raimondo (Unione europea – NextGenerationEU – fondi MUR D.M. 737/2021 – project title: “Le nanovescicole vegetali nella comunicazione tra regni: il ruolo degli RNA non codificanti”, PRJ-1969 and PRJ-0998). The work was also partially funded by a grant to Riccardo Alessandro (“Pubblicazione articolo scientifico – PRIN 2022, Prof. Alessandro- 2022MF9H4F_001, CUP B53D23017880006. - Cod. progetto U-GOV PRJ-1402 - Titolo progetto Exploring the effects of a nutraceutical supplement containing plant-derived extracellular vesicles in primary Sjrogen syndrome: an in vivo and in vitro study - Investimento finanziato dall’Unione Europea - NextGenerationEU”).
Acknowledgments
AcknowledgementsRG is the recipient of a Postdoctoral fellowship funded by Fondazione Umberto Veronesi, GD is a PhD student in “Biomedicina, Neuroscienze e Diagnostica Avanzata” at the University of Palermo. The authors acknowledge the COST Action CA20110 – exRNAPATH – for fostering a collaborative network in the field of exRNA.
Conflict of interest
Authors SR, AC and RA are cofounders of Navhetec, an Academic Spin-Off of the University of Palermo that did not have any role in the conceptualization of this article and the analysis of the results. The same authors are inventors of the Italian patents 102019000005090 and 102015902344749.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2025.1697575/full#supplementary-material
References
Azar, F., Courtet, K., Dekky, B., Bonnier, D., Dameron, O., Colige, A., et al. (2020). Integration of miRNA-regulatory networks in hepatic stellate cells identifies TIMP3 as a key factor in chronic liver disease. Liver Int. official J. Int. Assoc. Study Liver 40, 2021–2033. doi:10.1111/liv.14476
Baldrich, P., Rutter, B. D., Karimi, H. Z., Podicheti, R., Meyers, B. C., and Innes, R. W. (2019). Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-Nucleotide ‘tiny’ RNAs. Plant Cell 31, 315–324. doi:10.1105/tpc.18.00872
Batista, P. J., and Chang, H. Y. (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307. doi:10.1016/j.cell.2013.02.012
Baysal, İ., Örsten, S., Cengiz, G., Ünal, E., Doğrul, A. B., Çiftçi, T., et al. (2024). Assessing the potential apoptotic effects of different hydatid cyst fluids on human healthy hepatocytes and hepatocellular carcinoma cells. Acta Parasitol. 69, 700–709. doi:10.1007/s11686-024-00797-z
Borniego, M. L., and Innes, R. W. (2023). Extracellular RNA: mechanisms of secretion and potential functions. J. Exp. Bot. 74, 2389–2404. doi:10.1093/jxb/erac512
Cai, Y., Zhang, C., Zhan, L., Cheng, L., Lu, D., Wang, X., et al. (2019). Anticancer effects of Gleditsia sinensis extract in rats transplanted with hepatocellular carcinoma cells. Oncol. Res. 27, 889–899. doi:10.3727/096504018X15482423944678
Chen, Y., and Wang, X. (2020). miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 48, D127-D131–D131. doi:10.1093/nar/gkz757
Chen, J., Liu, K., Vadas, M. A., Gamble, J. R., and McCaughan, G. W. (2024). The role of the MiR-181 family in hepatocellular carcinoma. Cells 13, 1289. doi:10.3390/cells13151289
Di Gioia, S., Hossain, M. N., and Conese, M. (2020). Biological properties and therapeutic effects of plant-derived nanovesicles. Open Med. Wars. Pol. 15, 1096–1122. doi:10.1515/med-2020-0160
Dixson, A. C., Dawson, T. R., Di Vizio, D., and Weaver, A. M. (2023). Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat. Rev. Mol. Cell Biol. 24, 454–476. doi:10.1038/s41580-023-00576-0
Gasparro, R., Gambino, G., Duca, G., Majo, D. D., Di Liberto, V., Tinnirello, V., et al. (2024). Protective effects of lemon nanovesicles: evidence of the Nrf2/HO-1 pathway contribution from in vitro hepatocytes and in vivo high-fat diet-fed rats. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 180, 117532. doi:10.1016/j.biopha.2024.117532
Guttman, M., and Rinn, J. L. (2012). Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346. doi:10.1038/nature10887
He, B., Cai, Q., Qiao, L., Huang, C. Y., Wang, S., Miao, W., et al. (2021). RNA-Binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. plants 7, 342–352. doi:10.1038/s41477-021-00863-8
Ji, J., Yamashita, T., Budhu, A., Forgues, M., Jia, H. L., Li, C., et al. (2009). “Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells,”, 50. Baltimore, Md, 472–480. doi:10.1002/hep.22989
Ji, J., Yamashita, T., and Wang, X. W. (2011). Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell and Biosci. 1, 4. doi:10.1186/2045-3701-1-4
Jiang, Y., Qiu, Q., Jing, X., Song, Z., Zhang, Y., Wang, C., et al. (2023). Cancer-associated fibroblast-derived exosome miR-181b-3p promotes the occurrence and development of colorectal cancer by regulating SNX2 expression. Biochem. Biophysical Res. Commun. 641, 177–185. doi:10.1016/j.bbrc.2022.12.026
Ke, L., Zhou, Z., Xu, X. W., Wang, X., Liu, Y., Xu, Y., et al. (2019). Evolutionary dynamics of lincRNA transcription in nine citrus species. Plant J. Cell Mol. Biol. 98, 912–927. doi:10.1111/tpj.14279
Lian, M. Q., Chng, W. H., Liang, J., Yeo, H. Q., Lee, C. K., Belaid, M., et al. (2022). Plant-derived extracellular vesicles: recent advancements and current challenges on their use for biomedical applications. J. Extracell. Vesicles 11, e12283. doi:10.1002/jev2.12283
Lin, Y., Pan, X., and Shen, H.-B. (2021). lncLocator 2.0: a cell-line-specific subcellular localization predictor for long non-coding RNAs with interpretable deep learning. Bioinforma. Oxf. Engl. 37, 2308–2316. doi:10.1093/bioinformatics/btab127
Liu, J., Shi, W., Wu, C., Ju, J., and Jiang, J. (2014). miR-181b as a key regulator of the oncogenic process and its clinical implications in cancer (Review). Biomed. Rep. 2, 7–11. doi:10.3892/br.2013.199
Mattick, J. S., and Makunin, I. V. (2006). Non-coding RNA. Hum. Mol. Genet.15 Spec. No 1, R17–R29. doi:10.1093/hmg/ddl046
Mir, B., and Goettsch, C. (2020). Extracellular vesicles as delivery vehicles of specific cellular cargo. Cells 9, 1601. doi:10.3390/cells9071601
Nguyen, T. N.-G., Pham, C. V., Chowdhury, R., Patel, S., Jaysawal, S. K., Hou, Y., et al. (2023). Development of blueberry-derived extracellular nanovesicles for immunomodulatory therapy. Pharmaceutics 15, 2115. doi:10.3390/pharmaceutics15082115
Panni, S., Lovering, R. C., Porras, P., and Orchard, S. (2020). Non-coding RNA regulatory networks. Biochimica biophysica acta. Gene Regul. Mech. 1863, 194417. doi:10.1016/j.bbagrm.2019.194417
Perut, F., Roncuzzi, L., Avnet, S., Massa, A., Zini, N., Sabbadini, S., et al. (2021). Strawberry-Derived exosome-like nanoparticles prevent oxidative stress in human mesenchymal stromal cells. Biomolecules 11, 87. doi:10.3390/biom11010087
Pocsfalvi, G., Turiák, L., Ambrosone, A., Del Gaudio, P., Puska, G., Fiume, I., et al. (2018). Protein biocargo of citrus fruit-derived vesicles reveals heterogeneous transport and extracellular vesicle populations. J. Plant Physiology 229, 111–121. doi:10.1016/j.jplph.2018.07.006
Potestà, M., Roglia, V., Fanelli, M., Pietrobono, E., Gismondi, A., Vumbaca, S., et al. (2020). Effect of microvesicles from Moringa oleifera containing miRNA on proliferation and apoptosis in tumor cell lines. Cell Death Discov. 6, 43. doi:10.1038/s41420-020-0271-6
Raimondo, S., Naselli, F., Fontana, S., Monteleone, F., Lo Dico, A., Saieva, L., et al. (2015). Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 6, 19514–19527. doi:10.18632/oncotarget.4004
Raimondo, S., Urzì, O., Meraviglia, S., Di Simone, M., Corsale, A. M., Rabienezhad Ganji, N., et al. (2022). Anti-inflammatory properties of lemon-derived extracellular vesicles are achieved through the inhibition of ERK/NF-κB signalling pathways. J. Cell. Mol. Med. 26, 4195–4209. doi:10.1111/jcmm.17404
Ramakrishnaiah, Y., Morris, A. P., Dhaliwal, J., Philip, M., Kuhlmann, L., and Tyagi, S. (2023). Linc2function: a comprehensive pipeline and webserver for long non-coding RNA (lncRNA) identification and functional predictions using deep learning approaches. Epigenomes 7, 22. doi:10.3390/epigenomes7030022
Raposo, G., and Stahl, P. D. (2019). Extracellular vesicles: a new communication paradigm? Nat. Rev. Mol. Cell Biol. 20, 509–510. doi:10.1038/s41580-019-0158-7
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Teng, Y., Ren, Y., Sayed, M., Hu, X., Lei, C., Kumar, A., et al. (2018). Plant-derived exosomal MicroRNAs shape the Gut Microbiota. Cell host and microbe 24, 637–652. doi:10.1016/j.chom.2018.10.001
Teng, Y., Xu, F., Zhang, X., Mu, J., Sayed, M., Hu, X., et al. (2021). Plant-derived exosomal microRNAs inhibit lung inflammation induced by exosomes SARS-CoV-2 Nsp12. Mol. Ther. J. Am. Soc. Gene Ther. 29, 2424–2440. doi:10.1016/j.ymthe.2021.05.005
The RNAcentral Consortium Petrov, A. I., Kay, S. J. E., Kalvari, I., Howe, K. L., Gray, K. A., et al. (2017). RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 45, D128–D134. doi:10.1093/nar/gkw1008
Urzì, O., Cafora, M., Ganji, N. R., Tinnirello, V., Gasparro, R., Raccosta, S., et al. (2023). Lemon-derived nanovesicles achieve antioxidant and anti-inflammatory effects activating the AhR/Nrf2 signaling pathway. iScience 26, 107041. doi:10.1016/j.isci.2023.107041
van Niel, G., D’Angelo, G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228. doi:10.1038/nrm.2017.125
Wang, W., Zhong, G. Z., Long, K. B., Liu, Y., Liu, Y. Q., and Xu, A. L. (2021). Silencing miR-181b-5p upregulates PIAS1 to repress oxidative stress and inflammatory response in rats with alcoholic fatty liver disease through inhibiting PRMT1. Int. Immunopharmacol. 101, 108151. doi:10.1016/j.intimp.2021.108151
Xiao, J., Feng, S., Wang, X., Long, K., Luo, Y., Wang, Y., et al. (2018). Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ 6, e5186. doi:10.7717/peerj.5186
Yan, H., and Bu, P. (2021). Non-coding RNA in cancer. Essays Biochem. 65, 625–639. doi:10.1042/EBC20200032
Ye, L., Gao, Y., Mok, S. W. F., Liao, W., Wang, Y., Chen, C., et al. (2024). Modulation of alveolar macrophage and mitochondrial fitness by medicinal plant-derived nanovesicles to mitigate acute lung injury and viral pneumonia. J. nanobiotechnology 22, 190. doi:10.1186/s12951-024-02473-w
Zand Karimi, H., Baldrich, P., Rutter, B. D., Borniego, L., Zajt, K. K., Meyers, B. C., et al. (2022). Arabidopsis apoplastic fluid contains sRNA- and circular RNA-protein complexes that are located outside extracellular vesicles. Plant Cell 34, 1863–1881. doi:10.1093/plcell/koac043
Zhao, Y., Zhou, Y., Xu, J., Fan, S., Zhu, N., Meng, Q., et al. (2024). Cross-Kingdom RNA transport based on extracellular vesicles provides innovative tools for plant protection. Plants Basel, Switz. 13, 2712. doi:10.3390/plants13192712
Keywords: plant-derived nanovesicles (PDNVs), lemon nanovesicles (LNVs), long noncoding RNAs (lncRNAs), cross-kingdom regulation, miRNA sponging
Citation: Tinnirello V, Gasparro R, Duca G, Miozzi L, Rotunno S, Mercati F, Conigliaro A, Alessandro R and Raimondo S (2025) Lemon-derived nanovesicles facilitate trans-kingdom transfer of lncRNAs to human cells. Front. Mol. Biosci. 12:1697575. doi: 10.3389/fmolb.2025.1697575
Received: 02 September 2025; Accepted: 06 November 2025;
Published: 17 November 2025.
Edited by:
Souichi Oe, Kansai Medical University, JapanReviewed by:
Uday Chintapula, University of Pennsylvania, United StatesChuan Shen, Ankang University, China
Copyright © 2025 Tinnirello, Gasparro, Duca, Miozzi, Rotunno, Mercati, Conigliaro, Alessandro and Raimondo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Riccardo Alessandro, cmljY2FyZG8uYWxlc3NhbmRyb0B1bmlwYS5pdA==; Stefania Raimondo, c3RlZmFuaWEucmFpbW9uZG9AdW5pcGEuaXQ=
 Vincenza Tinnirello
Vincenza Tinnirello Roberta Gasparro1
Roberta Gasparro1 Laura Miozzi
Laura Miozzi Silvia Rotunno
Silvia Rotunno Alice Conigliaro
Alice Conigliaro Riccardo Alessandro
Riccardo Alessandro Stefania Raimondo
Stefania Raimondo