- 1Department of Neurology, Chengdu Second People’s Hospital, Chengdu, China
- 2Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, China
Variants in the apolipoprotein E (APOE) gene play an important role in the development of Alzheimer’s disease (AD). Specifically, the APOE ε4 allele is an established genetic risk factor for AD, while the APOE ε2 allele is a protective factor against AD. However, the mechanism underlying this impact of APOE genotype on the pathogenesis of AD remain unclear. This study sought to investigate the influence of APOE genotype on cognition and neuroimaging features in cognitively normal (CN) elderly individuals and patients with mild cognitive impairment (MCI). A total of 177 participants were selected from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database, including 101 MCI patients and 76 CN individuals. A 2 × 3 (consisting of two groups and three APOE genotypes) analysis of covariance was carried out to measure the influences of diagnosis and APOE genotype on cognition and brain features, assessed based on global functional connectivity density (gFCD) and hippocampal volume. In addition, a mediation analysis was carried out to investigate the indirect influence of neuroimaging features on the relationship between APOE genotype and cognitive performance in the MCI group. This analysis revealed that APOE genotype had an influence on brain function in the bilateral precentral gyrus, right thalamus, and posterior cingulate cortex (PCC). In addition, an interactive influence between diagnosis and APOE genotype was found on general cognition, immediate memory, executive function, hippocampal volume, and gFCD in the right dorsolateral prefrontal cortex and medial prefrontal cortex (MPFC). Finally, this mediation analysis revealed that hippocampal volume and gFCD in the thalamus may mediate the relationship between APOE genotype and cognitive performance in the MCI group. Taken together, our findings provide novel insights into the neural mechanisms underlying the genetically guided pathogenic mechanisms of AD.
Introduction
Apolipoprotein E (APOE) is a major genetic risk factor contributing to the development of late-onset Alzheimer’s disease (AD). Individuals with the ε4 allele have increased risks, while those with the ε2 allele show decreased risk of developing AD (Seripa et al., 2009; Fei and Jianhua, 2013; Liu et al., 2013). However, the neuropathological mechanism underlying the opposite outcomes on cognitive function is still unclear. In the last few decades, many studies investigated the impact of APOE genotype variations on brain structure and function in individuals with AD or mild cognitive impairment (MCI) and healthy elders using neuroimaging and genetic analysis (Spampinato et al., 2011; Troyer et al., 2012; Wang et al., 2015). Most of the studies focused on the neural basis of the APOE ε4 allele and investigated the group differences in brain structure and function between ε4 and non-ε4 carriers. Because of the relative rarity of the ε2 allele, studies examining the influences of the APOE ε2 allele on the neuropathology of AD are still lacking, and the mechanisms underlying the impact of the ε2 allele on AD development remain unclear (Suri et al., 2013).
With recent advances in neuroimaging, genetic imaging approaches have been widely used to explore the genetic impact on the brain, of which structural MRI and functional MRI (fMRI) are the most widely used technologies. A large body of work has focused on the relationship between APOE genotype and AD pathology. Many prior studies have examined the structural and functional differences in the brain, particularly in the hippocampus, resulting from the presence of ε4 (Schuff et al., 2009; Wang et al., 2015; Sinclair et al., 2017). More recently, researchers have paid more attention to the impact of the APOE ε2 allele on the brain. Using both task-based and resting-state fMRI (rs-fMRI), Trachtenberg et al. (2012a; 2012b). found that hippocampus activation and functional connectivity were similarly changed in ε4 and ε2 carriers when compared with those in ε3 carriers in cognitively normal (CN) elders. Shu et al. (2014) also demonstrated that APOE ε2 and ε4 carriers both showed decreased functional connectivity compared with carriers of homozygous ε3 in CN elders. However, Chen et al. (2016) found opposite outcomes of APOE ε2 and ε4 on the intrinsic entorhinal cortex functional connectivity in the middle temporal gyrus. Altered functional connectivity was correlated with episodic memory in the preclinical stage of AD (amnestic MCI) (Chen et al., 2016). In addition, the influences of APOE genotypes on hippocampal volumes have been frequently examined. Hostage et al. (2013) found dose-dependent influences of ε4 alleles on hippocampal volumes in AD and MCI patients, as well as those of ε2 alleles in CN elders. Therefore, the hippocampus is the most widely studied and affected part of the brain in early stages of AD (Schuff et al., 2009). However, whether and how altered hippocampus volume and brain function is implicated in the opposite role of APOE genotypes in cognitive performance in the development of AD remains unclear.
In the present study, we used global functional connectivity density (gFCD) mapping, a new data-driven, voxel-wise method, to measure the function of the whole brain based on rs-fMRI data. We investigated the impact of the APOE genotype (ε2+, ε3/ε3, and ε4+) and its interaction with disease status on cognitive function, hippocampal volume, and gFCD in CN elderly and MCI patients. Importantly, mediation analysis was used to explore potential intermediate phenotypes in the relationship between the APOE genotype and cognitive performance in the CN and MCI population. We hypothesized that APOE ε2 will have a particular impact on cognitive performance, hippocampal volume, and brain function in CN elders and MCI participants compared to the APOE ε4 allele. In addition, we hypothesized that the gene–brain–cognition model may potentially be able to elucidate the mechanisms underlying APOE-associated cognitive impairment in the MCI group.
Materials and Methods
The Alzheimer’s Disease Neuroimaging Initiative
Data used in this study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database1. The database was launched in 2003 as part of a public–private partnership led by principal investigator Michael W. Weiner, MD, United States. The primary goal of ADNI is to test whether serial MRI, PET, other biological markers, and findings of clinical and neuropsychological examinations can be combined to assess the progression of MCI and early AD. Updated information can be obtained at www.adni-info.org. Ethical approval was obtained by the investigators of the ADNI2. The institutional review boards of the respective institutions at all participating sites approved the study. All ADNI participants provided written informed consent before the start of the study.
Participants
The inclusion and exclusion criteria for all ADNI participants are available at http://www.adni-info.org. Participants were selected in the present study according to the following inclusion criteria: Caucasian ethnicity, availability of rs-fMRI and 3D T1-weighted MRI images, and underwent genotyping of the APOE gene. A battery of neuropsychological tests were used in this study, including the Mini-Mental State Examination (MMSE), AD assessment scale – 13-item cognitive subscale (ADAS13) for general cognition. The Rey Auditory Verbal Learning Test (RAVLT) is a powerful tool for assessing memory and learning (Schmidt, 1996). In this study, we used the RAVLT-immediate (sum of scores of trials 1–5) to assess participants’ immediate memory, the RAVLT learning (score of trial 5 minus trial 1) to assess learning, and the RAVLT-forgetting (score of trial 5 minus the score of delayed recall) and Logical Memory test (delayed recall score) to assess delay memory. The scores of the Trails B test were used to assess participants’ executive function. The naming of the neuropsychological test used in our study will be available from the merged table provided by the ADNI, and the derived summary scores obtained from the RAVLT measurements can be found in the ADNI data dictionary3. The diagnosis of MCI was based on the guidelines described in the ADNI protocol, including the following: (1) complaints with regard to memory-related problem or observation of such problems by a partner; (2) a specified education-adjusted cutoff score on the Logical Memory test; an MMSE score between 24 and 30; a Clinical Dementia Rating score of 0.5; a Memory Box score of at least 0.5; and (3) relatively well-preserved daily life activities. Detailed inclusion and exclusion criteria are available at http://adni.loni.usc.edu/wp-content/uploads/2010/09/ADNI_GeneralProceduresManual.pdf. Accordingly, 185 participants were included in the first step; however, 8 participants were excluded because of poor signals in the rs-fMRI images (N = 3) or excessive head motion (translations >3 mm or rotation > 2°) (N = 5). Finally, the remaining 177 participants, including 76 CN elders and 101 patients with MCI, were included in the analysis.
Genotyping
All selected participants underwent genotyping to assess their APOE allele status. APOE genotyping was performed using DNA extracted from peripheral blood cells which were collected in plastic tubes with ethylenediaminetetraacetic acid (10 ml). The University of Pennsylvania AD Biomarker Fluid Bank Laboratory will receive and store the biomarker samples, which will be processed at the University of Pennsylvania. More detailed information is available in the ADNI-1 Procedures manual4. In addition, five participants with the ε2/ε4 genotype were excluded from the analysis because of the opposing outcomes of the ε2 and ε4 alleles (Shaw et al., 2007). APOE ε2 is a rare genotype; therefore, our study had only one CN participant who was homozygous for APOE ε2. Therefore, the ε2 homozygotes and heterozygotes were pooled into a single ε2 + carrier category. Furthermore, the study had only six patients with MCI who were homozygous for APOE ε4, and five CN participants who were homozygous for APOE ε4. Therefore, we also pooled the ε4 homozygotes and heterozygotes into a single ε4 + carrier category. In total, 31 APOE ε2 + (ε2ε2/ε2ε3), 85 ε3ε3, and 60 ε4 + (ε3ε4/ε4ε4) participants were included in the final analysis.
MRI Data Acquisition
All rs-fMRIs were performed using a single-shot T2 star-weighted echo-planar imaging pulse sequence with a Philips 3T MRI scanner with an eight-channel head coil. The rs-fMRI parameters were as follows: repetition time, 3,000 ms; echo time, 30 ms; flip angle, 80°; acquisition matrix, 64 × 64; field of view, 240 × 240 mm; thickness, 3.3 mm; gap, 0 mm; and number of slices, 48. A total of 140 image volumes were acquired in the rs-fMRI scans. In addition, 3D T1-weighted MRI images were obtained using a magnetization-prepared gradient echo sequence with a spatial resolution of 1 mm × 1 mm × 1.2 mm. The detailed MRI protocols can be found at http://adni.loni.usc.edu/methods/documents/mri-protocols/.
Structural Image Analysis
Cortical reconstruction and volumetric segmentation were performed using the FreeSurfer version 5.1 image analysis suite5. The technical details for using the FreeSurfer with the ADNI data have been described by many studies previously (Holland et al., 2009; Chiang et al., 2010; Jack et al., 2010). Briefly, the image processing protocol included motion correction and registration, non-uniform intensity normalization, Talairach transform computation, intensity normalization 1, skull strip, segmentation of the subcortical white matter and the deep gray matter volumetric structures, tessellation of the gray matter/white matter boundary, automated topology correction, and surface deformation with intensity gradients to optimally place the gray/white and gray/cerebrospinal fluid borders at the location where the greatest shift in intensity defines the transition to other tissue class. The image data were processed based on the 2010 Desikan–Killiany atlas (Fischl et al., 2004). We selected the hippocampal and entorhinal cortical volumes for structural analysis, as these two regions are usually affected in the early stages of AD and MCI. The fusiform was selected as a reference region in the structural analysis. The bilateral hippocampal, entorhinal, and fusiform volumes were summed in the analyses to increase the statistical power of the analysis. Because an interactive outcome (diagnosis × APOE) was found in the hippocampal volume (see section “Results”), an additional analysis was carried out using the volumes of the left and right hippocampus separately (see the Supplementary Material for detailed information).
Rs-fMRI Data Preprocessing
Functional data were preprocessed using the SPM12 toolkit6 and MATLAB version 7.10 (The MathWorks, Inc., Natick, MA, United States). The preprocessing steps were conducted using the BRAinNetome Toolkit software7. Structural images were segmented (VBM toolbox in SPM) and coregistered with the resting functional images. The fMRI images were preprocessed as follows: The first 10 volumes in the scanning session were discarded to attain equilibration. The remaining 130 volumes were corrected for slice timing, realigned, and subsequently normalized spatially using diffeomorphic high-dimensional registration as implemented in the DARTEL toolbox using default settings, and the volumes were resampled to 3 mm × 3 mm × 3 mm cubic voxels. The blood oxygenation level dependent signal was low-pass filtered (0.01–0.1 Hz) and detrended. We also calculated the frame-wise displacement (FD), which evaluates the mismatch of volume-to-volume superimposed head position (Power et al., 2013). The mean FD was also used as a covariate in the image analyses. The FD did not differ significantly among the different groups (p > 0.05). Subsequently, the six motion parameters and the white matter and cerebrospinal fluid signals were excluded from the data analysis by linear regression.
gFCD Calculation
We calculated the gFCD of each voxel using an in-house script according to the method described by Tomasi and Volkow (2010). The gFCD at a given voxel (x0) was computed as the global number of functional connections using Pearson’s linear correlation between x0 and all the other voxels; two voxels with a correlation coefficient of >0.6 were considered functionally connected. In addition, the gFCD calculation was restricted to the gray matter regions with a signal/noise ratio of >50% to minimize unwanted effects from susceptibility-related signal-loss artifacts (Tomasi and Volkow, 2010). To increase the normality of the distribution, grand mean scaling of the gFCD values of each voxel was divided by the mean value of the qualified voxels of the whole brain. Finally, the normalized gFCD maps were spatially smoothed with 6 mm × 6 mm × 6 mm Gaussian kernels.
Statistical Analyses
The 2 × 3 (two groups, three APOE genotypes) analysis of covariance (ANCOVA) and the Kruskal–Wallis tests were used to compare the demographic data, neuropsychological performances and structural brain maps among the different groups using the Statistical Package for the Social Sciences software version 24.0 (SPSS, Inc., Chicago, IL, United States). The effects of age, sex, and years of education were regressed out in the analyses of cognitive function, and the effect of intracranial volume was regressed out in the structural comparison of the brain. The statistical threshold was set at p < 0.05.
The voxel-wise comparisons of the global FCD maps were conducted using a 2 × 3 (diagnosis × APOE genotype) ANCOVA from the SPM12 toolkit with age, sex, years of education, and mean FD as the nuisance covariates. The voxel-level significant threshold was set at p < 0.005, corrected for multiple comparisons at the cluster level with the latest version of the 3dClustSim program in AFNI_16.3.00 software [gray matter mask correction (67,541 voxels), voxel-level p < 0.005, cluster level α < 0.001, κ > 50, cluster size >1,647 mm3]8. Subsequently, the average gFCD strength in each cluster was extracted from each subject using the masks generated from the ANCOVA analysis for quantitative illustration and additional mediation analyses.
Furthermore, the mediation analysis was used to examine whether the generated gFCD maps and brain volumes could mediate the influences of the APOE genotypes on cognitive performances in the MCI group. First, multivariate linear regression analyses were performed to explore the influence of the APOE genotype on cognitive performances in the MCI and CN groups, separately, after adjusting for the effects of age, sex, and years of education. Subsequently, the APOE genotype was set as an independent variable (X), and the APOE genotype associated with cognition performances (the MMSE and ADAS scores) was set as the dependent variables (Y). Because a significant association between X and Y is not a prerequisite to testing a mediation hypothesis (Hayes and Rockwood, 2017), other indices of cognitive performance, including the RAVLT, Trail B, and Logical Memory scores, were also set as dependent variables (Y). Second, the extracted gFCD values in the brain regions exhibiting the primary outcome of the APOE genotypes and the hippocampus volume were set as the mediators (M). Third, we used the simple mediation model from the PROCESS macro in SPSS (Model 4, version 2.16.3) (Hayes, 2013; Gong et al., 2017b) and adjusted the effects of sex, age, and years of education as covariates. This model was based on 10,000 bootstrap samples for assessing the bias-corrected bootstrap confidential interval (CI). The indirect influence was determined as significant at 95% CI, with a null hypothesis without indirect influence. Linear regression analyses showed no influence of the APOE genotypes on any cognitive performances in the CN group; therefore, the mediation analysis was only conducted in the MCI group.
Results
Demographic Information and Neuropsychological Characteristics
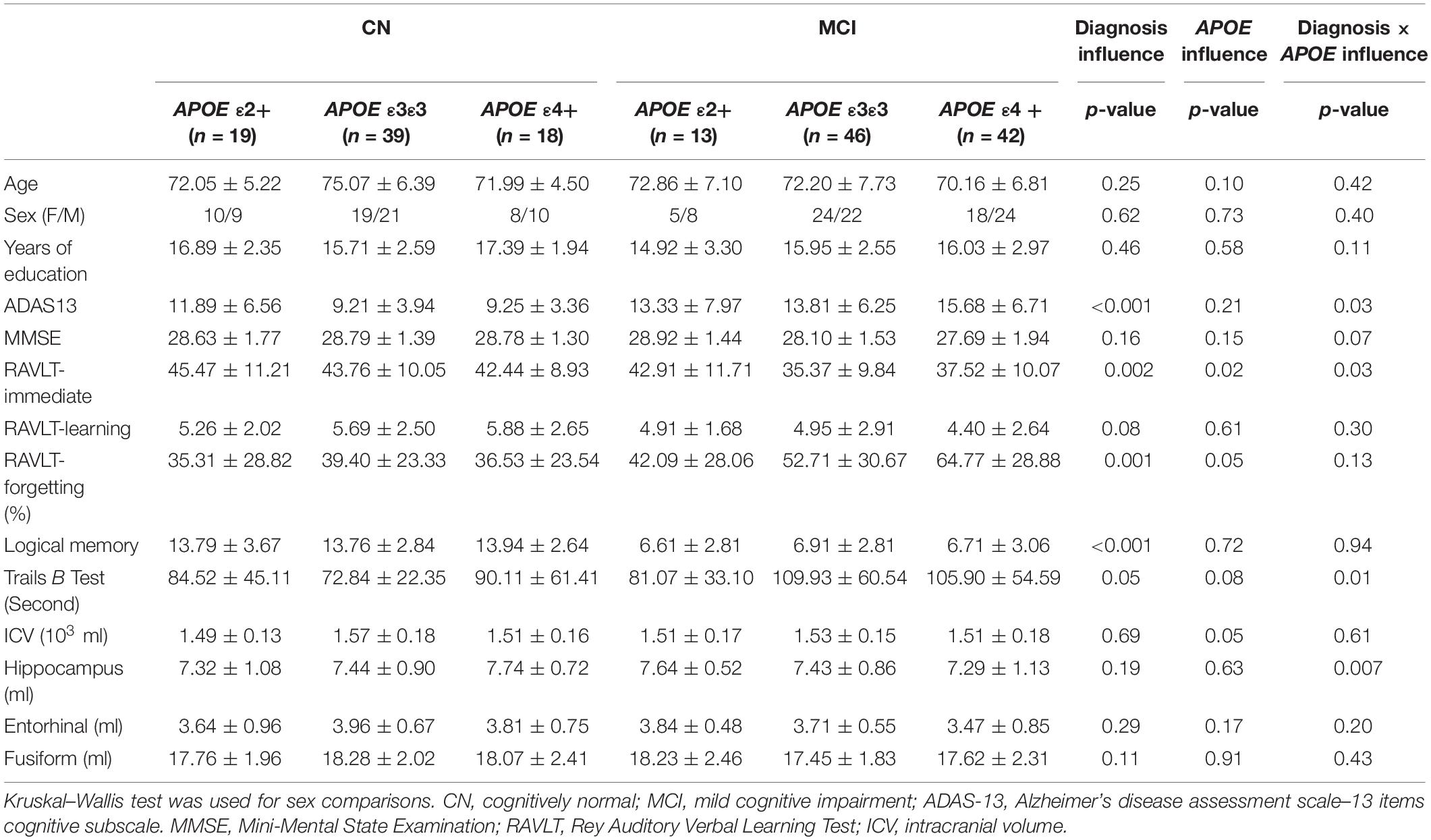
Demographic information and neuropsychological evaluations for each group are shown in Table 1. In total, 31 APOE ε2 +(ε2ε2/ε2ε3), 85 ε3ε3, and 60 ε4+ (ε3ε4/ε4ε4) participants were included in the final analysis. There were no significant differences in age, sex, and years of education between the CN and MCI groups with different APOE genotypes (all p > 0.05). No significant main and interactive influences of diagnosis and APOE genotypes on intracranial volume and mean FD were found between the groups. Compared with the CN participants, patients with MCI had poor cognitive performances in ADAS13 (Figure 1A, F = 13.59, p < 0.001), RAVLT-immediate (Figure 1B, F = 9.54, p = 0.002), RAVLT-forgetting (Figure 1C, F = 7.71, p = 0.006), and Logical Memory (Figure 1D, F = 200.39, p < 0.001). The APOE genotype influence was found in RAVLT-immediate (F = 3.69, p = 0.02). Post hoc analysis revealed that the RAVLT-immediate score in the APOE ε2+ group (44.48) was higher than that in the APOE ε3/ε3 (39.22) and ε4+ (39.00) groups (p = 0.02), while the RAVLT-immediate score was similar in the ε3/ε3 and ε4+ carriers. In addition, the interactive influences of diagnosis and the APOE genotypes on cognitive function, as assessed by ADAS13 (F = 3.35, p = 0.03), RAVLT-immediate (F = 3.35, p = 0.03), and Trails B test (F = 4.57, p = 0.01), and the hippocampal volume (F = 5.78, p = 0.007) were observed. More specifically, as shown in Figures 1A,B,E, the interactive influence was mainly observed in the APOE ε2+ genotype and APOE ε4+ genotype between the CN and MCI groups. Participants with the APOE ε2+ genotype showed more stabilized cognitive function, including general cognition (ADAS13 score), immediate memory (RAVLT-immediate score), and executive function (Trails B test) in the progression from CN to MCI. In addition, the hippocampal volume in MCI with the APOE ε2+ genotype was higher than CN with APOE ε2+ genotype, while the hippocampal volume in MCI with the APOE ε4+ genotype was lower than that in CN participants with the APOE ε4+ genotype (Figure 1F). Notably, no significant main or interactive outcome of disease and APOE genotypes was found in the volumes of the entorhinal cortex and fusiform gyrus (all p > 0.05).
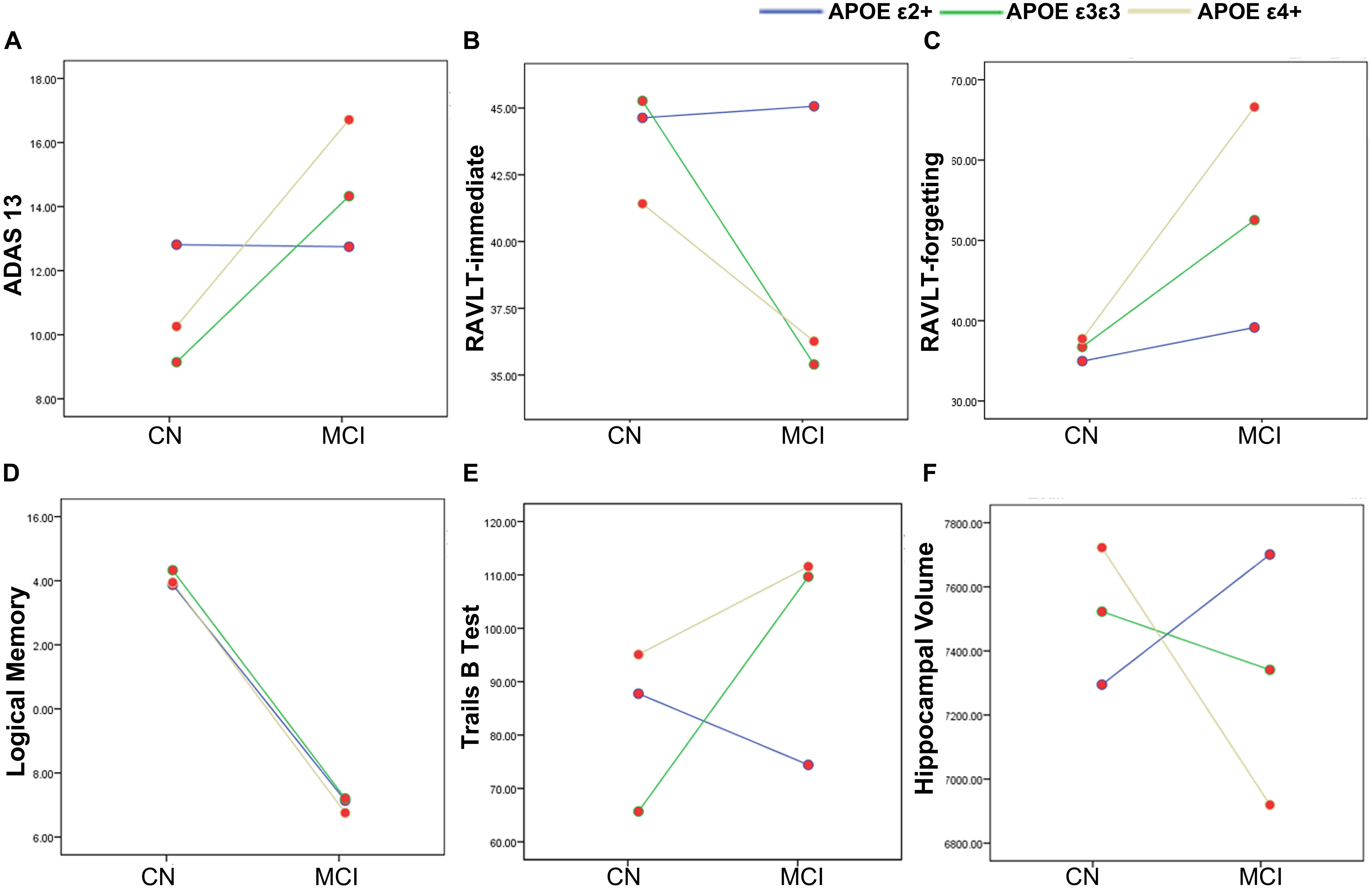
Figure 1. The main and interactive influences of diagnosis and apolipoprotein E (APOE) genotype on cognitive function and hippocampal volume. (A,B,E,F) Interactive influence of diagnosis and APOE genotype on the hippocampal volume and scores of ADAS13, RAVLT-immediate, and Trails B test. Compared to the APOE ε3ε3 and ε4 + genotypes, the APOE ε2+ genotype show more stable cognitive function and hippocampus volume during the CN to MCI period. (C,D) Main influences of diagnosis on the scores of RAVLT-forgetting and Logical Memory tests. The MCI group show lower logical memory and more RAVLT-forgetting rate as compared to the CN group. CN, cognitively normal; MCI, mild cognitive impairment; ADAS13, Alzheimer’s disease (AD) assessment scale–13-item cognitive subscale; RAVLT, Rey Auditory Verbal Learning Test.
Main Influence of the Diagnosis and the APOE Genotype on gFCD
As illustrated in Figure 2 and Table 2, the main influence of diagnosis on the gFCD was found in the bilateral dorsolateral prefrontal cortex (DLPFC). Particularly, the gFCD of the bilateral DLPFC was lower in the MCI group than in the CN group. The main influence of the APOE genotype on the gFCD was found in the bilateral precentral gyrus, right thalamus, and posterior cingulate cortex (PCC) (Figures 3A–C,E–G), which is located in the posterior default mode network (DMN, PCC) and the sensorimotor network (SMN, thalamus, and precentral gyrus) (Biswal et al., 1995; Raichle, 2015). Post hoc analyses revealed that the ε2+ genotype was the primary factor affecting the brain function. Particularly, compared with ε3 and ε4 carriers, ε2 carriers showed a higher gFCD in the PCC and the right thalamus and a lower gFCD in the bilateral precentral gyrus (Figures 3D,H). No significant difference was found between the ε3 and ε4 carriers.
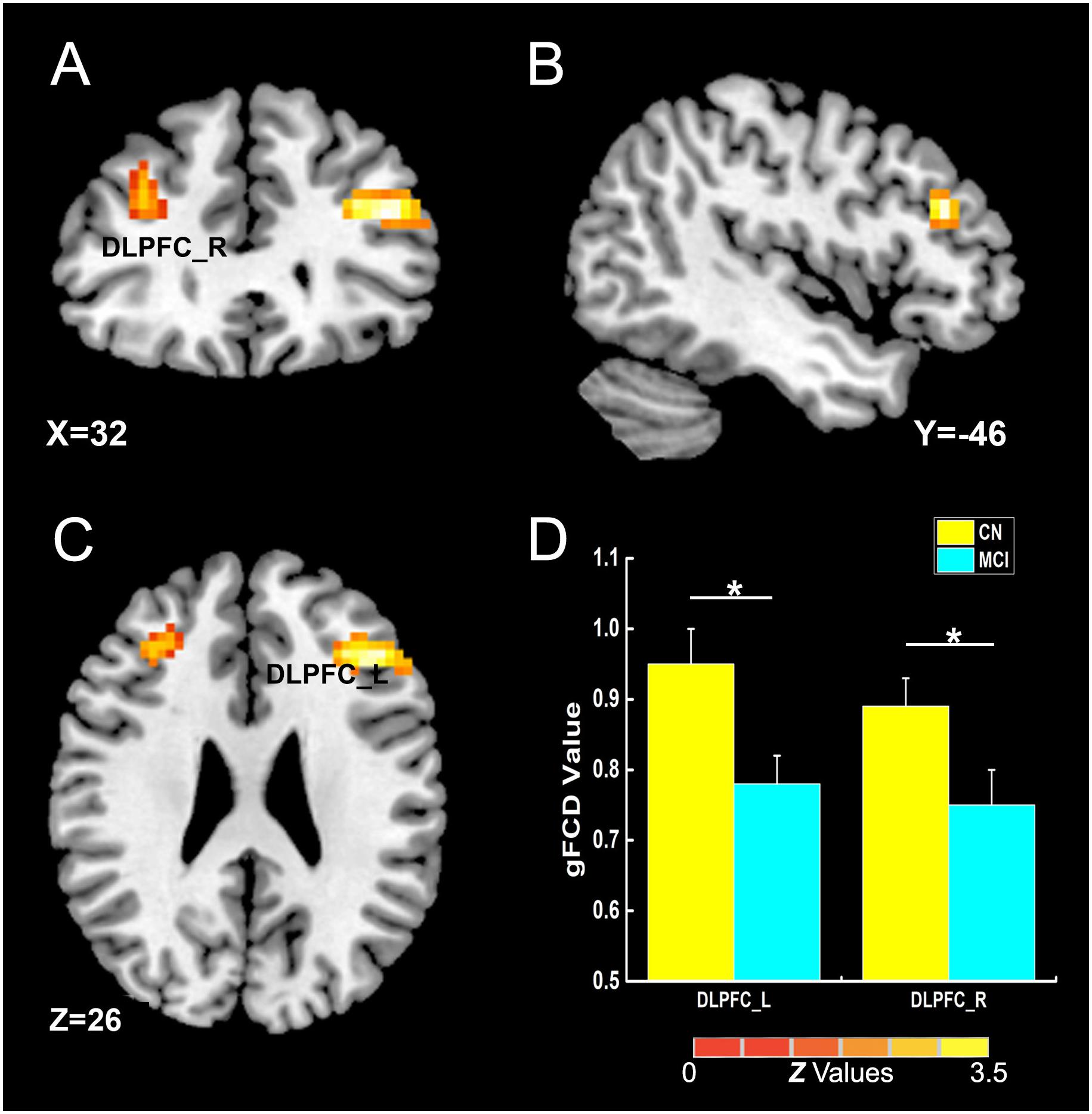
Figure 2. The main influence of diagnosis on gFCD (p < 0.005; α < 0.001; 3dClustSim corrected). (A–C) The brain regions show the differences in gFCD values between the two groups; (D) the histogram quantitatively illustrates that the values of gFCD in the bilateral DLPFC in the MCI group was lower than that in the CN group. The asterisks mean significant difference between two groups. gFCD, global functional connectivity density; CN, cognitively normal; MCI, mild cognitive impairment; DLPFC, dorsolateral prefrontal cortex.
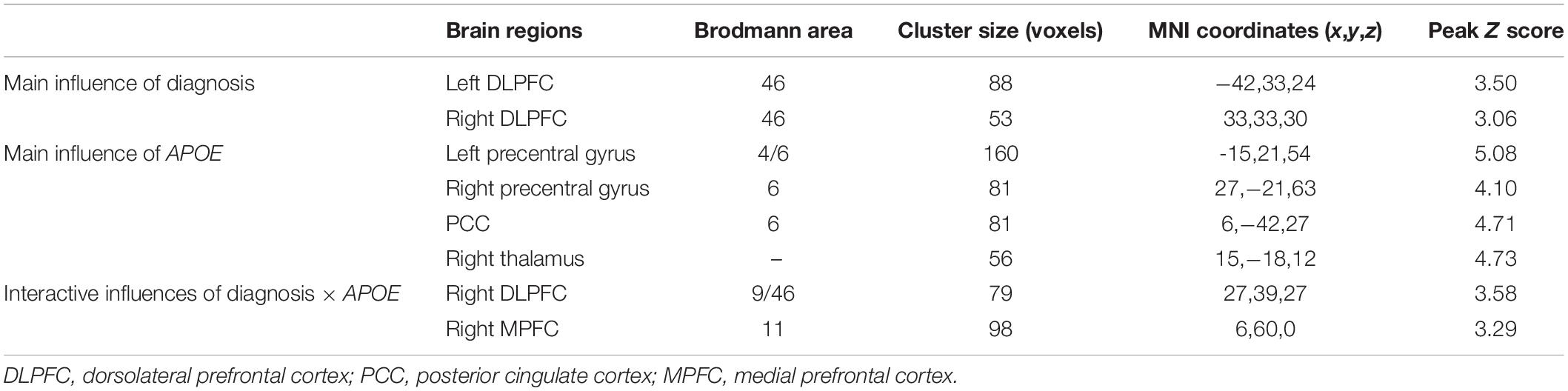
Table 2. Brain areas with significant diagnosis, APOE, and diagnosis × APOE influences on global functional connectivity density.
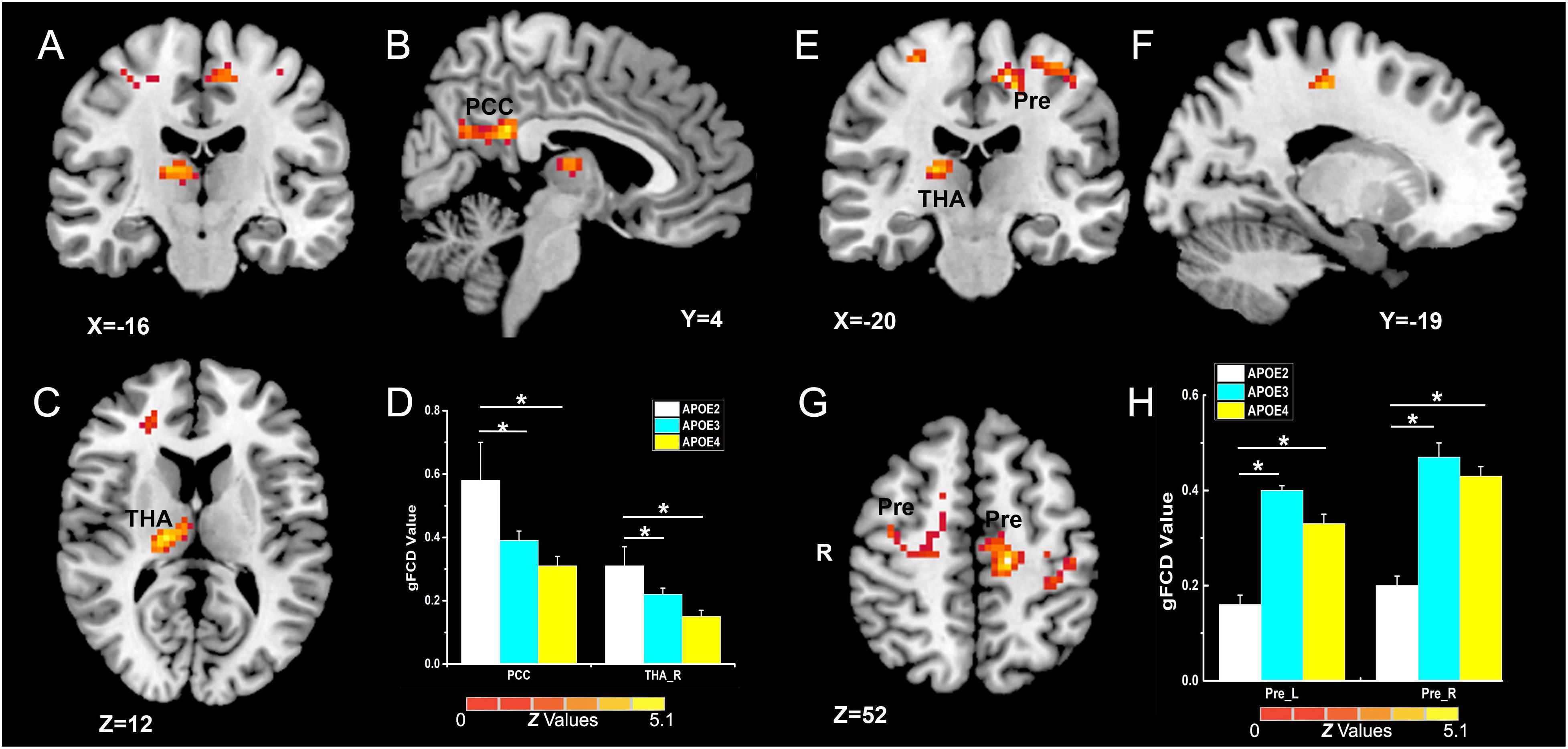
Figure 3. Primary outcome of the APOE genotypes on gFCD (p < 0.005; α < 0.001; 3dClustSim corrected). (A–C,E–G) Influence of APOE genotype on the brain functions of different brain regions. (D,H) The histograms quantitatively illustrate the gFCD differences among the three APOE genotypes. The asterisks mean significant difference between two APOE genotype groups. Compared to APOE ε3ε3 and ε4+ genotypes, the APOE ε2 + genotype show higher gFCD values in the PCC and right thalamus (D), and lower gFCD values in the bilateral precentral gyrus (H). No significant difference was found in brain functions between the two APOE ε3ε3 and ε4+ genotype groups. gFCD, global functional connectivity density; PCC, posterior cingulate cortex; THA, thalamus; Pre, precentral gyrus.
Interactive Influence Between the Diagnosis and the APOE Genotype on gFCD
The interactive influence between the diagnosis and the APOE genotype on gFCD was observed in the right DLPFC and medial prefrontal cortex (MPFC) (Figures 4A–C,E–G), which is located in the executive control network (ECN) (DLPFC) (Vincent et al., 2008) and anterior DMN (MPFC) (Raichle, 2015). As shown in Figures 4D,H, the interaction was observed in the alteration line between the APOE ε2 and ε4 carriers. Particularly, when compared with the CN group, the gFCDs in the right DLPFC and MPFC were decreased in MCI with APOE ε2 genotypes, but increased in MCI with ε4 carriers.
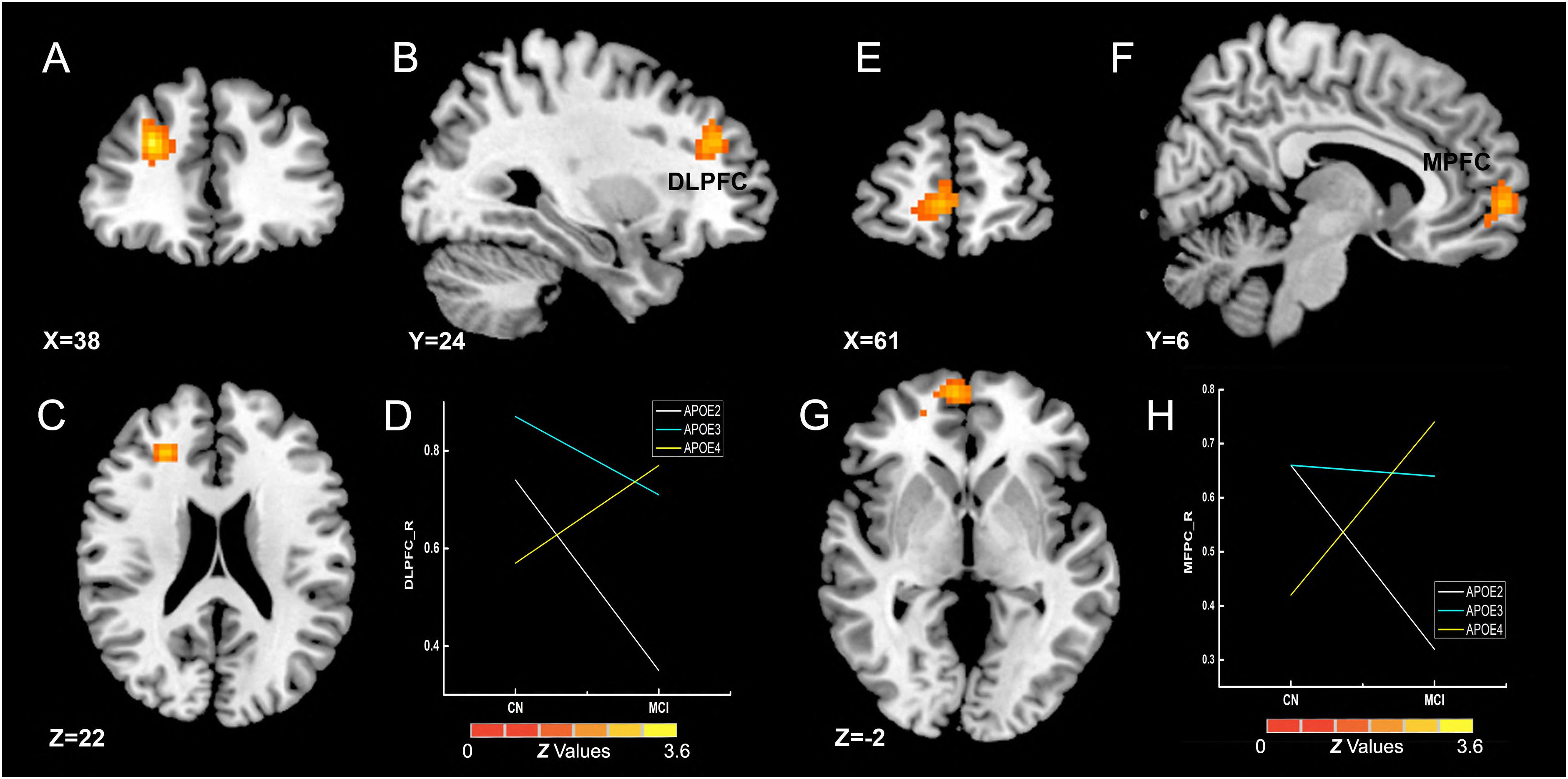
Figure 4. Interactive influence of diagnosis and APOE genotype on gFCD (p < 0.005; α < 0.001; 3dClustSim corrected). (A–C,E–G) Interactive influence of diagnosis and APOE genotype on the brain functions in the right DLPFC and MPFC. (D,H) The diagrams illustrate the interactive influence of diagnosis and APOE genotype in the DLPFC and MPFC. Compared to the CN group, the gFCD values in the DLPFC and MPFC was higher in patients with MCI who had the APOE ε2+ genotype and was lower in those who had the APOE ε4+ genotype; no significant alteration was noted in the MCI group with the APOE ε3ε3 genotype. gFCD, global functional connectivity density; CN, cognitively normal; MCI, mild cognitive impairment; DLPFC, dorsolateral prefrontal cortex; MPFC, medial prefrontal cortex.
Mediation Analyses
Multivariate linear regression analyses revealed significant relationships between the APOE genotypes and MMSE scores (F = 4.69, p = 0.03) or ADAS 13 scores (F = 4.83, p = 0.01) in the MCI group. No other significant association between the APOE genotype and cognition function was found in either the MCI and CN group (all p > 0.05). We assessed the potential mediating role of the gFCD in APOE genotype-associated regions and hippocampal volumes in the relationship between the APOE genotype and cognitive performance in the MCI group. As shown in Figure 5, three significant mediators were found in the study. First, the hippocampal volume negatively mediated the APOE genotype influence on the MMSE score [Figure 5A, indirect influence, β = -0.147, 95% CI = (−0.425, −0.002)]. Second, the gFCD in the right thalamus also negatively mediated the APOE genotype influence on ADAS scores in MCI patients [Figure 5B, indirect influence, β = −1.123, 95% CI = (−3.053, −0.201)]. Third, the hippocampal volume could positively mediate the APOE genotype influence on the RAVLT-forgetting score in the MCI group [Figure 5C, indirect influence, β = 6.285, 95% CI = (2.161, 12.352)], although the direct influence of the APOE genotype on the RAVLT-forgetting score is not significant. No other significant mediator was found in the mediation analyses.
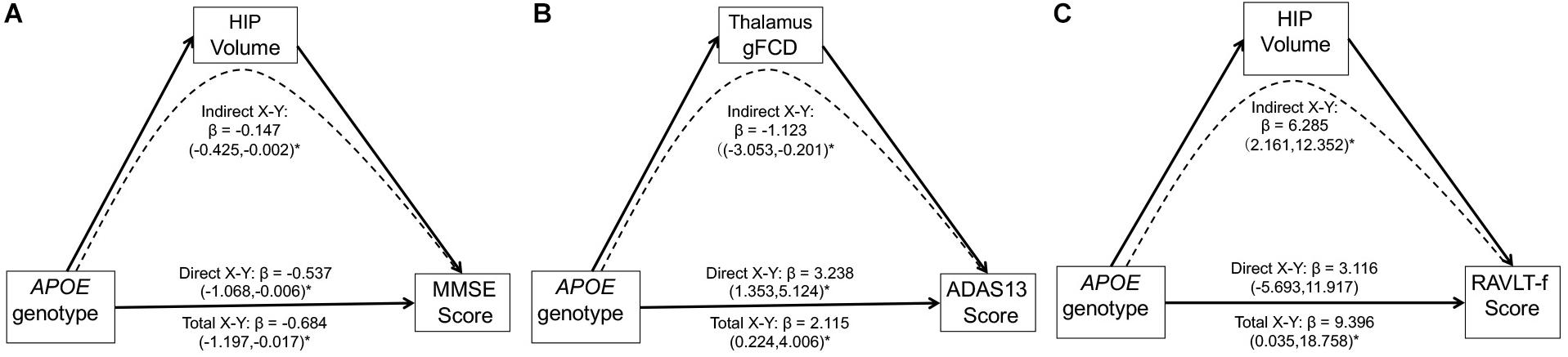
Figure 5. Mediation analyses show that the hippocampal volume and gFCD of the thalamus mediate the relationship between the APOE genotypes and cognitive performance in the MCI group. The asterisks mean the direct/indirect effect is significant. (A) The hippocampal volume negatively mediates the influence of APOE genotypes on the MMSE score in the MCI group. (B) gFCD values in the right thalamus negatively mediates the influence of APOE genotype on the ADAS score in the MCI group. (C) The hippocampal volume positively mediates the influence of APOE genotype on the RAVLT-forgetting score in the MCI group. gFCD, global functional connectivity density; MCI, mild cognitive impairment; HIP, hippocampus; MMSE, Mini-Mental State Examination; ADAS, Alzheimer’s disease assessment scale; RAVLT, Rey Auditory Verbal Learning Test.
Discussion
The present study aimed to explore the impact of APOE genotypes, especially the opposite ε2 and ε4 alleles, on cognitive function and brain in the CN and MCI groups. Four main findings were observed in our study. First, the APOE ε2 allele had a protective outcome on immediate memory, executive function, and hippocampal volume in the development from normal aging to MCI. Second, compared to APOE ε4 carriers, APOE ε2 carriers showed increased gFCD in the thalamus network, posterior DMN, and decreased gFCD in motor network. Third, the diagnosis state and APOE genotype synergistically contribute to brain functional alterations, especially in executive control network (ECN) and anterior DMN. Fourth, using the gene–brain–cognition model, we found that the hippocampal volume and thalamus function mediated the APOE genotype-associated cognitive performance in MCI patients. Taken together, these findings further extend our understanding of the influence of different APOE genotypes on cognition and brain and suggest that the gene–brain–cognition model may be used to determine the complex neural mechanisms underlying the pathogenesis and progression of AD.
The association between APOE ε4 and cognitive decline has been demonstrated in AD, MCI, and normal aging in many studies (Whitehair et al., 2010; Wisdom et al., 2011; Prada et al., 2014; Suarez et al., 2014), and the protective outcome of APOE ε2 on cognitive decline has been observed in AD patients (Corder et al., 1994; Suri et al., 2013). However, the positive outcome of the ε2 allele on cognitive function has not been replicated in healthy individuals. The present largest single-sample study has not found any association between the ε2 allele and special cognitive ability across all age ranges (Marioni et al., 2016). Sinclair et al. (2017) found that the ε2 allele showed positive outcomes on episode memory and executive function in early to mid-adult life (ages 23–67). In the present study, the APOE genotype was associated with general cognitive performance (MMSE and ADAS13 scores) in the MCI group, but was not associated with any cognitive domains in CN elders or MCI patients; this can be attributed to the small sample size in this study. Interestingly, the interactive influence of diagnosis and APOE genotype was found on the measure of immediate memory (RAVLT-immediate) and executive function (Trails B test) (Figures 1B,E). Both immediate memory and executive function are thought to be dependent on the brain function in the regions including the hippocampus and prefrontal cortex (Penney, 1989; Zelazo, 2015). These findings indicate the influence of APOE genotype on special cognition domains and brain regions during AD progression.
Changes in hippocampal volume serve as a sensitive and early biomarker of neurodegeneration in AD patients, and APOE ε4 is associated with atrophic hippocampal volume in AD (Liu et al., 2015) and MCI patients, but not in CN elders (Farlow et al., 2004; Morra et al., 2009). A previous multicohort study found a linear reduction in hippocampal volumes in AD and MCI patients with different APOE genotypes, i.e., ε4 carriers < ε3 carriers < ε2 carriers (Khan et al., 2017). Our study did not find differences in the hippocampus between the CN and MCI groups and the influence of APOE on hippocampal volume in the pooled group. However, the interactive influence of diagnosis and gene was found in the hippocampal volume (Figure 1F), and the result indicated that the ε2 and ε4 alleles had different influences at CN and MCI stages. The opposite outcomes of ε2 and ε4 on hippocampal volumes also point to the early pathological underpinnings of the APOE genotypes on cognitive function.
Consistent with previously reported abnormal brain functions in MCI patients, the MCI patients showed dysfunctional gFCD in the bilateral DLPFC (Wu et al., 2014), the core hub in the ECN (Vincent et al., 2008). These results support the idea that the abnormal functional connectivity in ECN may be an early biomarker of AD progression. The influence of APOE genotype on the intrinsic functional brain was found in the SMN (the bilateral precentral gyrus and thalamus) and posterior default mode network (PCC) (Biswal et al., 1995; Raichle, 2015; Figure 3). A previous study reported that the APOE ε2 carriers have robust white matter integrity (fractional anisotropy) in the right thalamus when compared to ε3/ε3 carriers in CN elderly individuals (Chiang et al., 2012) and that the APOE ε4 allele shows additive gray matter volume reduction in the precentral gyrus (Cacciaglia et al., 2018). Our findings showed the influence of APOE on brain structure, and the reverse influence of APOE ε2 and ε4 on SMN might indicate a counterbalance of brain function in SMN in the preclinical stage of AD. Regarding the impact of APOE on DMN, some studies showed increased DMN in young ε4 carriers and decreased DMN in older ε4 carriers (Filippini et al., 2009; Sheline et al., 2010), but other studies found similar DMN alterations in ε4 and ε2 carriers among CN elderly individuals and MCI patients (Trachtenberg et al., 2012a; Shu et al., 2014; Gong et al., 2017a). In our study, the gFCD in the posterior DMN was higher in ε2 carriers than in ε4 carriers. It is possible that the APOE genotype has different influences at different ages and disease development stages. Therefore, exploring the interactive influence of diagnosis and APOE genotypes is important for elucidating the influence of genes on the brain at different stages of AD development. We found that the interactive influence of diagnosis and APOE genotype on brain gFCD was located in the ECN and anterior DMN (MPFC). There were opposite alterations of ECN and DMN from CN to MCI in ε4 and ε2 carriers, and, unexpectedly, brain function was decreased in ε2 carriers and increased in ε4 carriers in MCI patients compared to CN individuals (Figure 4). As the DLPFC plays an essential role for executive function and cognition control processing, while the MPFC is related to the self-referential mental activity (Andrews-Hanna et al., 2014), we speculate that ε2 and ε4 alleles play opposite roles in executive control and self-referential mental processing during the AD development.
Importantly, the mediation analyses showed that the link between APOE genotype and cognitive performances was mediated through the hippocampal volume and gFCD in the thalamus in the MCI group. In other words, the protective outcome in APOE ε2 carriers and detrimental influence in ε4 carriers on cognitive performance might be attributed to their different influences on hippocampal volume and thalamus function in MCI. In particular, even the direct association between APOE and immediate memory is not significant, and the APOE genotype could significantly influence memory performance through its influence on hippocampal volume in the MCI group. The hippocampus is the core region for memory processes, and this region is altered early in the development of cognition decline. Recent evidence indicated that APOE might regulate neurogenesis in the hippocampus (Hong et al., 2016; Tensaouti et al., 2018). The thalamus, which integrates multimodal information across different cortical networks, has been considered the hub of the functional network and plays a critical role in cognitive flexibility (Halassa and Kastner, 2017; Hwang et al., 2017). The mediator role of the hippocampus and thalamus in APOE-related cognition performances in MCI might indicate the neuroplasticity and cognitive flexibility protection mechanism underlying the APOE ε2 allele-mediated outcome on cognition. In this regard, our findings suggest that hippocampal volume and thalamus function could be predictors of cognition performance and potential treatment targets in MCI with different APOE genotypes. Thus, we propose that the gene–brain–cognition model could be a useful tool to reveal the neural mechanisms underlying the gene influence on cognition.
The present study has several limitations. First, our study had a small sample size and was a cross-sectional study because the longitudinal data of fMRI are limited in ADNI dataset. We did not include the AD group in the study because of the small number of patients in the AD subgroup with the APOE ε2 allele. Second, Iacono and Feltis recently reviewed the newly analyzed data and proposed that the APOE ε2 and ε4 alleles have antagonistic pleotropic influences on cognitive performance and brain structure and function across the human lifespan (Iacono and Feltis, 2019). The present study explored only the influence of the APOE genotype in elderly individuals; further studies should investigate the influence of APOE during different periods of life. Third, the cognition domains in the present study included only episode memory and executive function but did not include declarative memory, attention, and visuospatial function. Therefore, further studies should involve neuropsychological tests with special referential cognition domains. Fourth, homozygous and heterozygous ε2 and ε4 allele participants were pooled together in the present study since there were so few participants in the homozygote groups. We have provided this information in the Supplementary Material. However, we believe that the current study provides a useful starting point for further exploring APOE-associated behavior and for imaging investigations in patients with AD.
Conclusion
In conclusion, the present findings indicate that the APOE ε2 allele has specific protective influences on immediate memory, executive function, and hippocampal volume during AD progression. In addition, the hippocampus and thalamus mediate the influence of APOE on cognitive performance in MCI. The gene–brain–cognition model may provide novel insights on the complex neural underpinnings of the genetically guided pathogenic mechanisms and progression of AD.
Data Availability Statement
The datasets generated for this study can be found in the http://www.adni-info.org.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical approval was obtained by the ADNI investigators. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LG and CZ designed the study, analyzed, and drafted the manuscript. RX, LL, DL, and BZ revised the manuscript and interpreted the data. All authors read and approved the final manuscript.
Funding
This work was partially supported by the Chengdu Medical Research Project (2019115), the Sichuan Medical Research Project (S18030), “Xinglin Scholar” Talent Program of Chengdu University of Traditional Chinese Medicine (XSGG2019016), and National Traditional Chinese Medicine Innovation Training Program for Talents ([2019] No. 91).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The study was obtained by the ADNI investigators (http://www.adni-info.org/pdfs/adni_protocol_9_19_08.pdf). The group acknowledgments list of ADNI could be found in this document (http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf). We would like to thank Editage (http://www.editage.com) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2019.00374/full#supplementary-material
Footnotes
- ^ adni.loni.usc.edu; accessed on April 10, 2019
- ^ http://www.adni-info.org/pdfs/adni_protocol_9_19_08.pdf
- ^ http://adni.loni.usc.edu/data-dictionary-search/?q=avlt
- ^ http://adni.loni.usc.edu/wp-content/themes/freshnews-dev-v2/documents/clinical/ADNI-1_Protocol.pdf
- ^ http://surfer.nmr.mgh.harvard.edu/
- ^ http://www.fil.ion.ucl.ac.uk/spm
- ^ http://www.brainnetome.org
- ^ https://afni.nimh.nih.gov/pub/dist/doc/program_help/3dClustSim.html
References
Andrews-Hanna, J. R., Smallwood, J., and Spreng, R. N. (2014). The default network and self-generated thought: component processes, dynamic control, and clinical relevance. Ann. N. Y. Acad. Sci. 1316, 29–52. doi: 10.1111/nyas.12360
Biswal, B., Zerrin Yetkin, F., Haughton, V. M., and Hyde, J. S. (1995). Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541. doi: 10.1002/mrm.1910340409
Cacciaglia, R., Molinuevo, J. L., Falcón, C., Brugulat-Serrat, A., Sánchez-Benavides, G., Gramunt, N., et al. (2018). Effects of APOE-ε4 allele load on brain morphology in a cohort of middle-aged healthy individuals with enriched genetic risk for Alzheimer’s disease. Alzheimers Dement. 14, 902–912. doi: 10.1016/j.jalz.2018.01.016
Chen, J., Shu, H., Wang, Z., Liu, D., Shi, Y., Xu, L., et al. (2016). Protective effect of APOE epsilon 2 on intrinsic functional connectivity of the entorhinal cortex is associated with better episodic memory in elderly individuals with risk factors for Alzheimer’s disease. Oncotarget 7, 58789–58801. doi: 10.18632/oncotarget.11289
Chiang, G., Insel, P., Tosun, D., Schuff, N., Truran-Sacrey, D., Raptentsetsang, S., et al. (2010). Hippocampal atrophy rates and CSF biomarkers in elderly APOE2 normal subjects. Neurology 75, 1976–1981. doi: 10.1212/WNL.0b013e3181ffe4d1
Chiang, G., Zhan, W., Schuff, N., and Weiner, M. (2012). White matter alterations in cognitively normal apoe ε2 carriers: insight into Alzheimer resistance? Am. J. Neuroradiol. 33, 1392–1397. doi: 10.3174/ajnr.A2984
Corder, E. H., Saunders, A. M., Risch, N. J., Strittmatter, W. J., Schmechel, D. E., Gaskell, P. C., et al. (1994). Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 7, 180–184. doi: 10.1038/ng0694-180
Farlow, M. R., He, Y., Tekin, S., Xu, J., Lane, R., and Charles, H. C. (2004). Impact of APOE in mild cognitive impairment. Neurology 63, 1898–1901. doi: 10.1212/01.wnl.0000144279.21502.b7
Fei, M., and Jianhua, W. (2013). Apolipoprotein epsilon4-allele as a significant risk factor for conversion from mild cognitive impairment to Alzheimer’s disease: a meta-analysis of prospective studies. J. Mol. Neurosci. 50, 257–263. doi: 10.1007/s12031-012-9934-y
Filippini, N., Macintosh, B. J., Hough, M. G., Goodwin, G. M., Frisoni, G. B., Smith, S. M., et al. (2009). Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. Proc. Natl. Acad. Sci. U.S.A. 106, 7209–7214. doi: 10.1073/pnas.0811879106
Fischl, B., Van Der Kouwe, A., Destrieux, C., Halgren, E., Ségonne, F., Salat, D. H., et al. (2004). Automatically parcellating the human cerebral cortex. Cereb. Cortex 14, 11–22. doi: 10.1093/cercor/bhg087
Gong, L., Shu, H., He, C., Ye, Q., Bai, F., Xie, C., et al. (2017a). Convergent and divergent effects of apolipoprotein E ε4 and ε2 alleles on amygdala functional networks in non-demented older adults. Neurobiol. Aging 54, 31–39. doi: 10.1016/j.neurobiolaging.2017.02.013
Gong, L., Yin, Y., He, C., Ye, Q., Bai, F., Yuan, Y., et al. (2017b). Disrupted reward circuits is associated with cognitive deficits and depression severity in major depressive disorder. J. Psychiatric Res. 84, 9–17. doi: 10.1016/j.jpsychires.2016.09.016
Halassa, M. M., and Kastner, S. (2017). Thalamic functions in distributed cognitive control. Nat. Neurosci. 20, 1669–1679. doi: 10.1038/s41593-017-0020-1
Hayes, A. F. (2013). Introduction to Mediation, Moderation, and Conditional Process Analysis: A Regression-Based Approach. New York, NY: The Guilford Press.
Hayes, A. F., and Rockwood, N. J. (2017). Regression-based statistical mediation and moderation analysis in clinical research: observations, recommendations, and implementation. Behav. Res. Ther. 98, 39–57. doi: 10.1016/j.brat.2016.11.001
Holland, D., Brewer, J. B., Hagler, D. J., Fennema-Notestine, C., and Dale, A. M., and Alzheimer’s Disease Neuroimaging Initiative, (2009). Subregional neuroanatomical change as a biomarker for Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 106, 20954–20959. doi: 10.1073/pnas.0906053106
Hong, S., Washington, P. M., Kim, A., Yang, C. P., Yu, T. S., and Kernie, S. G. (2016). Apolipoprotein E regulates injury-induced activation of hippocampal neural stem and progenitor cells. J. Neurotrauma 33, 362–374. doi: 10.1089/neu.2014.3860
Hostage, C. A., Roy Choudhury, K., Doraiswamy, P. M., and Petrella, J. R. (2013). Dissecting the gene dose-effects of the APOE ε4 and ε2 alleles on hippocampal volumes in aging and Alzheimer’s disease. PLoS One 8:e54483. doi: 10.1371/journal.pone.0054483
Hwang, K., Bertolero, M. A., Liu, W. B., and D’esposito, M. (2017). The human thalamus is an integrative hub for functional brain networks. J. Neurosci. 37, 5594–5607. doi: 10.1523/JNEUROSCI.0067-17.2017
Iacono, D., and Feltis, G. C. (2019). Impact of Apolipoprotein E gene polymorphism during normal and pathological conditions of the brain across the lifespan. Aging 11, 787–816. doi: 10.18632/aging.101757
Jack, C. R. Jr., Bernstein, M. A., Borowski, B. J., Gunter, J. L., Fox, N. C., Thompson, P. M., et al. (2010). Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative. Alzheimer’s Dement. 6, 212–220. doi: 10.1016/j.jalz.2010.03.004
Khan, W., Giampietro, V., Banaschewski, T., Barker, G. J., Bokde, A. L., Büchel, C., et al. (2017). A multi-cohort study of apoE ε4 and Amyloid-β effects on the hippocampus in Alzheimer’s disease. J. Alzheimers Dis. 56, 1159–1174. doi: 10.3233/JAD-161097
Liu, C. C., Kanekiyo, T., Xu, H., and Bu, G. (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118. doi: 10.1038/nrneurol.2012.263
Liu, Y., Yu, J. T., Wang, H. F., Han, P. R., Tan, C. C., Wang, C., et al. (2015). APOE genotype and neuroimaging markers of Alzheimer’s disease: systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 86, 127–134. doi: 10.1136/jnnp-2014-307719
Marioni, R. E., Campbell, A., Scotland, G., Hayward, C., Porteous, D. J., and Deary, I. J. (2016). Differential effects of the APOE e4 allele on different domains of cognitive ability across the life-course. Eur. J. Hum. Genet. 24:919. doi: 10.1038/ejhg.2015.210
Morra, J. H., Tu, Z., Apostolova, L. G., Green, A. E., Avedissian, C., Madsen, S. K., et al. (2009). Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Neuroimage 45, S3–S15. doi: 10.1016/j.neuroimage.2008.10.043
Penney, C. G. (1989). Modality effects and the structure of short-term verbal memory. Mem. Cogn. 17, 398–422. doi: 10.3758/bf03202613
Power, J. D., Barnes, K. A., Snyder, A. Z., Schlaggar, B. L., and Petersen, S. E. (2013). Steps toward optimizing motion artifact removal in functional connectivity MRI; a reply to Carp. Neuroimage 76, 439–441. doi: 10.1016/j.neuroimage.2012.03.017
Prada, D., Colicino, E., Power, M. C., Cox, D. G., Weisskopf, M. G., Hou, L., et al. (2014). Influence of multiple APOE genetic variants on cognitive function in a cohort of older men - results from the Normative Aging Study. BMC Psychiatry 14:223. doi: 10.1186/s12888-014-0223-x
Raichle, M. E. (2015). The brain’s default mode network. Ann. Rev. Neurosci. 38, 433–447. doi: 10.1146/annurev-neuro-071013-014030
Schmidt, M. (1996). Rey Auditory Verbal Learning Test: A Handbook. Los Angeles, CA: Western Psychological Services.
Schuff, N., Woerner, N., Boreta, L., Kornfield, T., Shaw, L., Trojanowski, J., et al. (2009). MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain 132, 1067–1077. doi: 10.1093/brain/awp007
Seripa, D., Panza, F., Franceschi, M., D’onofrio, G., Solfrizzi, V., Dallapiccola, B., et al. (2009). Non-apolipoprotein E and apolipoprotein E genetics of sporadic Alzheimer’s disease. Ageing Res. Rev. 8, 214–236. doi: 10.1016/j.arr.2008.12.003
Shaw, P., Lerch, J. P., Pruessner, J. C., Taylor, K. N., Rose, A. B., Greenstein, D., et al. (2007). Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 6, 494–500. doi: 10.1016/s1474-4422(07)70106-0
Sheline, Y. I., Morris, J. C., Snyder, A. Z., Price, J. L., Yan, Z., D’angelo, G., et al. (2010). APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Aβ42. J. Neurosci. 30, 17035–17040. doi: 10.1523/jneurosci.3987-10.2010
Shu, H., Shi, Y., Chen, G., Wang, Z., Liu, D., Yue, C., et al. (2014). Opposite neural trajectories of apolipoprotein E 4 and 2 alleles with aging associated with different risks of Alzheimer’s disease. Cereb. Cortex. 26, 1421–1429. doi: 10.1093/cercor/bhu237
Sinclair, L. I., Pleydell-Pearce, C. W., and Day, I. N. M. (2017). Possible positive effect of the APOE ε2 allele on cognition in early to mid-adult life. Neurobiol. Learn. Mem. 146, 37–46. doi: 10.1016/j.nlm.2017.10.008
Spampinato, M. V., Rumboldt, Z., Hosker, R. J., and Mintzer, J. E., and Alzheimer’s Disease Neuroimaging Initiative, (2011). Apolipoprotein E and gray matter volume loss in patients with mild cognitive impairment and Alzheimer disease. Radiology 258, 843–852. doi: 10.1148/radiol.10100307
Suarez, V. M., Fernandez, Y., Lopez Edel, C., Clarke, D. H., Bobes, M. A., Riveron, A. M., et al. (2014). Apolipoprotein E alleles in Cuban patients with mild cognitive impairment. Am. J. Alzheimers Dis. Other Dement. 29, 236–241. doi: 10.1177/1533317513517037
Suri, S., Heise, V., Trachtenberg, A. J., and Mackay, C. E. (2013). The forgotten APOE allele: a review of the evidence and suggested mechanisms for the protective effect of APOE varepsilon2. Neurosci. Biobehav. Rev. 37, 2878–2886. doi: 10.1016/j.neubiorev.2013.10.010
Tensaouti, Y., Stephanz, E. P., Yu, T. S., and Kernie, S. G. (2018). ApoE regulates the development of adult newborn hippocampal neurons. eNeuro 5:ENEURO.155–ENEURO.118.
Tomasi, D., and Volkow, N. D. (2010). Functional connectivity density mapping. Proc. Natl. Acad. Sci. U.S.A. 107, 9885–9890. doi: 10.1073/pnas.1001414107
Trachtenberg, A. J., Filippini, N., Cheeseman, J., Duff, E. P., Neville, M. J., Ebmeier, K. P., et al. (2012a). The effects of APOE on brain activity do not simply reflect the risk of Alzheimer’s disease. Neurobiol. Aging 61:e613. doi: 10.1016/j.neurobiolaging.2010.11.011
Trachtenberg, A. J., Filippini, N., Ebmeier, K. P., Smith, S. M., Karpe, F., and Mackay, C. E. (2012b). The effects of APOE on the functional architecture of the resting brain. Neuroimage 59, 565–572. doi: 10.1016/j.neuroimage.2011.07.059
Troyer, A. K., Murphy, K. J., Anderson, N. D., Craik, F. I., Moscovitch, M., Maione, A., et al. (2012). Associative recognition in mild cognitive impairment: relationship to hippocampal volume and apolipoprotein E. Neuropsychologia 50, 3721–3728. doi: 10.1016/j.neuropsychologia.2012.10.018
Vincent, J. L., Kahn, I., Snyder, A. Z., Raichle, M. E., and Buckner, R. L. (2008). Evidence for a frontoparietal control system revealed by intrinsic functional connectivity. J. Neurophysiol. 100, 3328–3342. doi: 10.1152/jn.90355.2008
Wang, J., Wang, X., He, Y., Yu, X., Wang, H., and He, Y. (2015). Apolipoprotein E epsilon4 modulates functional brain connectome in Alzheimer’s disease. Hum. Brain Mapp. 36, 1828–1846. doi: 10.1002/hbm.22740
Whitehair, D. C., Sherzai, A., Emond, J., Raman, R., Aisen, P. S., Petersen, R. C., et al. (2010). Influence of apolipoprotein E varepsilon4 on rates of cognitive and functional decline in mild cognitive impairment. Alzheimers Dement. 6, 412–419. doi: 10.1016/j.jalz.2009.12.003
Wisdom, N. M., Callahan, J. L., and Hawkins, K. A. (2011). The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol. Aging 32, 63–74. doi: 10.1016/j.neurobiolaging.2009.02.003
Wu, L., Soder, R. B., Schoemaker, D., Carbonnell, F., Sziklas, V., Rowley, J., et al. (2014). Resting state executive control network adaptations in amnestic mild cognitive impairment. J. Alzheimer’s Dis. 40, 993–1004. doi: 10.3233/JAD-131574
Keywords: apolipoprotein E, Alzheimer’s disease, functional connectivity density, imaging genetics, hippocampus
Citation: Gong L, Xu R, Liu D, Lan L, Zhang B and Zhang C (2020) The Specific Impact of Apolipoprotein E Epsilon 2 on Cognition and Brain Function in Cognitively Normal Elders and Mild Cognitive Impairment Patients. Front. Aging Neurosci. 11:374. doi: 10.3389/fnagi.2019.00374
Received: 07 June 2019; Accepted: 19 December 2019;
Published: 28 January 2020.
Edited by:
Johannes Schröder, Heidelberg University, GermanyReviewed by:
Xi-jian Dai, The Chinese University of Hong Kong, ChinaFernando Goni, New York University, United States
Katherine Reiter, Cleveland Clinic, United States
Copyright © 2020 Gong, Xu, Liu, Lan, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuantao Zhang, emhhbmdjaHVhbnRhb0BjZHV0Y20uZWR1LmNu
†Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete list of members of the ADNI is available at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
 Liang Gong
Liang Gong Ronghua Xu1
Ronghua Xu1 for the Alzheimer’s Disease Neuroimaging Initiative†
for the Alzheimer’s Disease Neuroimaging Initiative†