- 1ENT Institute and Otorhinolaryngology Department of Affiliated Eye and ENT Hospital, State Key Laboratory of Medical Neurobiology, Fudan University, Shanghai, China
- 2Key Laboratory for Developmental Genes and Human Disease, Ministry of Education, Institute of Life Sciences, Southeast University, Nanjing, China
- 3Department of Otolaryngology Head and Neck Surgery, Jiangsu Provincial Key Medical Discipline Laboratory, Affiliated Drum Tower Hospital, Nanjing University Medical School, Nanjing University, Nanjing, China
- 4Jiangsu Province Hi-Tech Key Laboratory for Bio-Medical Research, Southeast University, Nanjing, China
- 5Co-innovation Center of Neuroregeneration, Nantong University, Nantong, China
- 6Key Laboratory of Hearing Medicine, National Health and Family Planning Commission (NHFPC), Shanghai, China
- 7Shanghai Engineering Research Center of Cochlear Implant, Shanghai, China
- 8The Institutes of Brain Science and the Collaborative Innovation Center for Brain Science, Fudan University, Shanghai, China
- 9Institutes of Biomedical Sciences, Fudan University, Shanghai, China
Hair cell (HC) loss is irreversible because only very limited HC regeneration has been observed in the adult mammalian cochlea. Wnt/β-catenin signaling regulates prosensory cell proliferation and differentiation during cochlear development, and Wnt activation promotes the proliferation of Lgr5+ cochlear HC progenitors in newborn mice. Similar to Lgr5, Lgr6 is also a Wnt downstream target gene. Lgr6 is reported to be present in adult stem cells in the skin, nail, tongue, lung, and mammary gland, and this protein is very important for adult stem cell maintenance in rapidly proliferating organs. Our previous studies showed that Lgr6+ cells are a subpopulation of Lgr5+ progenitor cells and that both Lgr6+ and Lgr5+ progenitors can generate Myosin7a+ HCs in vitro. Thus we hypothesized that Lgr6+ cells are an enriched population of cochlear progenitor cells. However, the detailed distinctions between the Lgr5+ and Lgr6+ progenitors are unclear. Here, we systematically compared the proliferation, HC differentiation, and detailed transcriptome expression profiles of these two progenitor populations. We found that the same number of isolated Lgr6+ progenitors generated significantly more Myosin7a+ HCs compared to Lgr5+ progenitors; however, Lgr5+ progenitors formed more epithelial colonies and more spheres than Lgr6+ progenitors in vitro. Using RNA-Seq, we compared the transcriptome differences between Lgr5+ and Lgr6+ progenitors and identified a list of significantly differential expressed genes that might regulate the proliferation and differentiation of these HC progenitors, including 4 cell cycle genes, 9 cell signaling pathway genes, and 54 transcription factors. In conclusion, we demonstrate that Lgr6+ progenitors are an enriched population of inner ear progenitors that generate more HCs compared to Lgr5+ progenitors in the newborn mouse cochlea, and the our research provides a series of genes that might regulate the proliferation of progenitors and HC generation.
Introduction
Hearing loss is a permanent sensory disorder affecting all populations throughout the world, and sensory hair cell (HC) loss is the main reason for permanent hearing loss (Bohne et al., 1976; Kelley, 2006). In non-mammalian vertebrates, HCs are able to regenerate to recover lost hearing function through direct differentiation and mitotic regeneration (Corwin and Cotanche, 1988; Ryals and Rubel, 1988; Stone and Cotanche, 2007). In mammals, the vestibular system has the capacity for only very limited spontaneous HC regeneration (Forge et al., 1993; Warchol et al., 1993; Kawamoto et al., 2009; Burns et al., 2012; Golub et al., 2012), and the neonatal mouse cochlea still has some progenitors that have a limited ability to regenerate HCs if they are damaged during early neonatal development (Jan et al., 2013; Cox et al., 2014). However, upon maturation, HC loss in mammals tends to be irreversible because HC progenitors gradually lose their regenerative capacity as the animal ages and because of the complex organization of progenitors and other cells within the fully developed organ of Corti (Oesterle et al., 2008; Warchol, 2011).
During cochlear development, both HCs and supporting cells (SCs) originate from prosensory progenitor cells that express the cyclin dependent kinase inhibitor p27 kip1 (Cdkn1b) (Chen and Segil, 1999). Prosensory regions can be identified at embryonic day (E)11. The progenitors of HCs and SCs are still proliferating at E12 (Ruben, 1967), but between E13 and E14 most of the cells in the prosensory region begin to exit the cell cycle (Ruben, 1967). The progenitors located in the apical cochlear duct exit the cell cycle earlier than those in the basal cochlear duct (Ruben, 1967). Starting at E13.5, the progenitors in the mid-basal cochlear duct begin to differentiate into HCs and to spread bidirectionally (Chen et al., 2002); the formation of HCs is finished when the entire length of the cochlear epithelium is patterned into three outer rows of HCs and one inner row between E17 and E18 (Chen and Segil, 1999; Chen et al., 2002). HC maturation is finished when the key proteins, such as prestin, are expressed in the outer HCs (Belyantseva et al., 2000). It was reported that isolated SCs in the neonatal cochlea have the ability to proliferate and differentiate into HCs in vitro (White et al., 2006; Sinkkonen et al., 2011), although the postnatal sensory HCs and SCs are postmitotic in vivo. SCs in the mouse cochlea and utricle have been regarded as a reliable source of HC regeneration after damage (Li et al., 2003, 2015; White et al., 2006; Lin et al., 2011; Sinkkonen et al., 2011; Golub et al., 2012; Cox et al., 2014; Wang et al., 2015). Wnt/β-catenin signaling plays important roles by regulating the differentiation and proliferation of prosensory cells in cochlear development (Chai et al., 2012; Shi et al., 2012), and when Wnt/β-catenin signaling is knocked out or inhibited sensory progenitor cells cannot proliferate and differentiate into HCs (Jacques et al., 2012; Shi et al., 2014). Lgr5, one of the Wnt downstream target genes, is expressed in a subset of SCs in the cochlea (Chai et al., 2011), and Lgr5+ cells have been identified as an enriched HC progenitor population compared to the rest of the SC population in many studies (Chai et al., 2012; Shi et al., 2012).
Similar to Lgr5, Lgr6 is also a Wnt downstream target gene, and it is present in adult stem cells in the skin, nail, tongue, lung, and mammary gland (Snippert et al., 2010; Oeztuerk-Winder et al., 2012; Ren et al., 2014; Lehoczky and Tabin, 2015; Blaas et al., 2016). Lgr6 is very important for adult stem cell maintenance in rapidly proliferating organs. In the adult mouse skin, Lgr6+ cells can proliferate and differentiate into all skin cell lineages, and they function in wound repair (Snippert et al., 2010). Lgr6+ cells give rise to the nails during homeostatic growth, and they play a role during digit tip regeneration (Lehoczky and Tabin, 2015). In the tongue, Lgr6+ stem cells can generate mature taste cells within the taste papillae in vitro (Ren et al., 2014). In the lung, E-Cad/Lgr6+ cells can self-renew and differentiate into bronchial and alveolar tissue (Oeztuerk-Winder et al., 2012). In the mammary gland, adult Lgr6+ stem cells can sustain alveologenesis throughout multiple pregnancies (Blaas et al., 2016).
In our previous study, we found that Lgr6 was only expressed in the inner pillar cells (IPs) from the embryonic to the neonatal period in the mouse cochlea and that these cells were a distinct subpopulation of Lgr5+ progenitors (Zhang et al., 2015). When we isolated the Lgr6+ cells by flow cytometry, they could generate Myosin7a+ HCs in vitro, and thus we hypothesized that Lgr6+ cells are an enriched cochlear progenitor population that generate HCs more efficiently than Lgr5+ progenitors. However, the detailed proliferation and differentiation ability of Lgr6+ progenitors has not been studied yet, and the differences between the Lgr5+ and Lgr6+ progenitors has been unclear.
In this study, we undertook a comprehensive comparison between the Lgr5+ and Lgr6+ progenitors. We found that Lgr6+ progenitors had a significantly greater capacity to generate Myosin7a+ HCs than Lgr5+ progenitors, while Lgr5+ progenitors had a greater capacity to proliferate than Lgr6+ progenitors. RNA-seq was performed to explore the differences of transcriptome expression profiles between these two HC progenitors, and we identified a set of significantly differentially expressed genes that might play roles in the proliferation and differentiation of HC progenitors. In summary, our data demonstrate that Lgr6+ progenitors are an enriched population of HC progenitor cells, and we provide a set of candidate genes that might serve as new potential therapeutic targets for HC generation.
Materials and Methods
Experimental Animals
Transgenic mice were purchased from Jackson Laboratory, including Lgr5-EGFP-IRES-creERT2 (Stock #008875) and Lgr6-EGFP-IRES-CreERT2 mice (Stock #016934). DirectPCR Lysis Reagent (Mouse Tail) (102-T, Viagen) was used to isolate genomic DNA from mouse tail tips. Genotyping of transgenic mice was performed as in our previous study (Zhang et al., 2015). All protocols were approved by the Animal Care and Use Committee of Fudan University and were consistent with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. We made all efforts to minimize the number of animals used and alleviate their suffering.
Isolation of Lgr5+ Progenitors and Lgr6+ Progenitors via Flow Cytometry
The isolation of HC progenitors was performed as in previous studies (Zhang et al., 2015; Waqas et al., 2016; Cheng et al., 2017). Briefly, postnatal day (P)0–P3 mouse sensory epithelium samples were digested by 0.125% trypsin/EDTA (Invitrogen) in 0.01M PBS for 9 min at 37°C. The reaction was terminated by soybean trypsin inhibitor (10 mg/ml; Worthington Biochem). Following mechanical trituration with about 80–100 strokes with blunt tips (Eppendorf, #22491245), isolated cells were percolated through a 40 μm cell strainer (BD Biosciences). Non-viable cells were labeled by propidium iodide (1 μg/ml, Sigma). Dissociated cells were then sorted using the GFP channel on a MoFlo® SX FACS cytometer (Beckman Coulter), and the EGFP+ cells were collected. Isolated cells were lysed in RLT buffer (Qiagen) for quantitative real-time PCR (q-PCR) assay or stored at -80°C until RNA-Seq analysis. For each RNA-Seq library, nearly 10,000 sorted cells were used. Re-sort analysis, immunofluorescence, and q-PCR were used to confirm the identity of the sorted cells.
Sphere-Forming Assay and Differentiation Assay
The sphere-forming assay and differentiation assay were performed as in previous studies (Waqas et al., 2016; Cheng et al., 2017). For the sphere-forming assay, sorted Lgr5+ and Lgr6+ cells were cultured separately at a density of 2 cells/μl in low attachment dishes (Costar, 3599) in DMEM/F12 medium supplemented with N2 (1%, Invitrogen, 17502), B27 (2%, Invitrogen, 17504), EGF (20 ng/ml, Sigma, E9644), IGF (50 ng/ml, Sigma, I8779), heparan sulfate (20 ng/ml, Sigma, H4777), β-FGF (10 ng/ml, Sigma, F0291), and ampicillin (0.1%, Sigma, A9518). After 5 days in culture, the numbers of spheres were quantified. Then the spheres were plated on laminin/polylysine (1:1)-coated 4-well dishes and cultured for 10 days in DMEM/F12 media with N2, B27, and 0.1% ampicillin for the differentiation assay. For the direct differentiation assay, flow cytometry-isolated Lgr5+ and Lgr6+ cells (at a density of 20 cells/μl) were plated on laminin/polylysine coated dishes and cultured for 10 days in DMEM/F12 media with N2, B27, and 0.1% ampicillin. EdU (10 ng/ml, Sigma, F0291) was added in the medium to measure the proliferation of the Lgr5+ and Lgr6+ cells. After 10 days, the cells and spheres were fixed and immunostained for different markers.
Cryosection, Immunostaining, and Image Acquisition
Cryosections were made according to a previous study (Zhang et al., 2015). In brief, P3 cochleae from Lgr6-EGFP-Ires-CreERT2 and Lgr5-EGFP-Ires-CreERT2 mice were fixed in 4% paraformaldehyde (PFA), and a Leica CM3050S microtome was used to make the cryosections. The spheres and isolated cells were fixed overnight at 4°C in 4% PFA. Non-specific binding was blocked with 10% donkey serum in 0.01 M PBS for 1 h at 37°C. Samples were incubated overnight at 4°C with primary antibodies, including anti-Myo7a (Proteus Bioscience, #25-6790, 1:1,000 dilution), anti-Sox2 (Santa Cruz, #sc-17320, 1:500 dilution), anti-EGFP (Abcam, 1:1,000 dilution), and anti-cytokeratin (Sigma-Aldrich, C2562, 1:200 dilution). After rinsing with 0.01 M PBS three times, the samples were incubated for 1 h at 37°C with corresponding secondary antibodies, including antibodies conjugated with FITC, Cy3, or Cy5 (1:500 dilution, Jackson ImmunoResearch). 4,6-diamidino-2-phenylindole (DAPI, 1:800 dilution, Sigma-Aldrich) was used to label cell nuclei. Cell proliferation was labeled with the Click-it EdU imaging kit (Invitrogen). Images were captured with a Leica SP8 confocal microscope. ImageJ (NIH) and Photoshop CS4 (Adobe System) were used to analyze the images.
Quantitative Real-Time PCR
The total RNA from the flow cytometry-isolated GFP+ cells was extracted with an RNeasy micro kit (Cat. #74004, Qiagen). A PrimerScript II 1st Strand cDNA Synthesis Kit (Cat. #6210A, TaKaRa) was used to synthesize the cDNA. q-PCR was performed using SYBR Premix Ex Taq II (Tli RNaseH Plus) (Cat. #RR820A, TaKaRa) on an AB 7500 Real-Time PCR System. The ΔΔCT method was used to analyze the gene expression level. Primers for the q-PCR assay are listed in Supplementary Table S1.
RNA-Seq and Data Analysis
Approximately 10,000 FACS-isolated Lgr5+ or Lgr6+ cells from P3 cochleae were dissolved in RNALater. The extracted RNA from Lgr5+ or Lgr6+ cells was split into three fractions for separate replicates. The SMART-Seq v4 Ultra Low Input RNA Kit and Illumina mRNA-Seq Sample Prep Kit were used for library preparation, and library quality was analyzed using an Agilent Bioanalyzer. An Illumina HiSeq2500 Platform was used to generate 150-bp paired-end sequences. Fastq format RNA-Seq reads were trimmed using Trimmomatic. We mapped the clean reads to the mouse reference genome (mm9) using TopHat (Trapnell and Schatz, 2009). Transcripts and genes were annotated according to the RefGene database (NCBI). Differential gene expression analysis was performed using Cufflinks. Genes with p-values < 0.05 were noted as significantly differentially expressed.
Statistical Analyses
GraphPad Prism software was used for statistical analyses. Two-tailed, unpaired Student’s t-tests were used to determine statistical significance. Data are shown as the means ± SE, and p < 0.05 was considered statistically significant.
Results
Lgr6 Was Expressed in a Subpopulation of Lgr5+ Progenitors in P3 Cochleae
First, we assessed the expression pattern of Lgr5 and Lgr6 in the P3 mouse sensory epithelium using Lgr5-EGFP-Ires-CreERT2 and Lgr6-EGFP-Ires-CreERT2 mice. Consistent with our previous studies (Chai et al., 2011; Zhang et al., 2015), immunohistochemical results showed that Lgr5 was expressed in the IPs, the inner phalangeal cells (IPCs), the third row of Deiters’ cells (DC3), and the lateral greater epithelial ridge (GER) in the whole mounts and cryosections of the sensory epithelium (Figures 1A,B), and the Lgr5 expression pattern was similar from the apex to the base in the cochlear duct (Supplementary Figure S2). Lgr6 was only expressed in a subset of the IPs, which are a subpopulation of Lgr5+ progenitors (Figures 1A,B), and Lgr6 was only expressed in the basal and middle turns of the organ of Corti (Supplementary Figure S2).
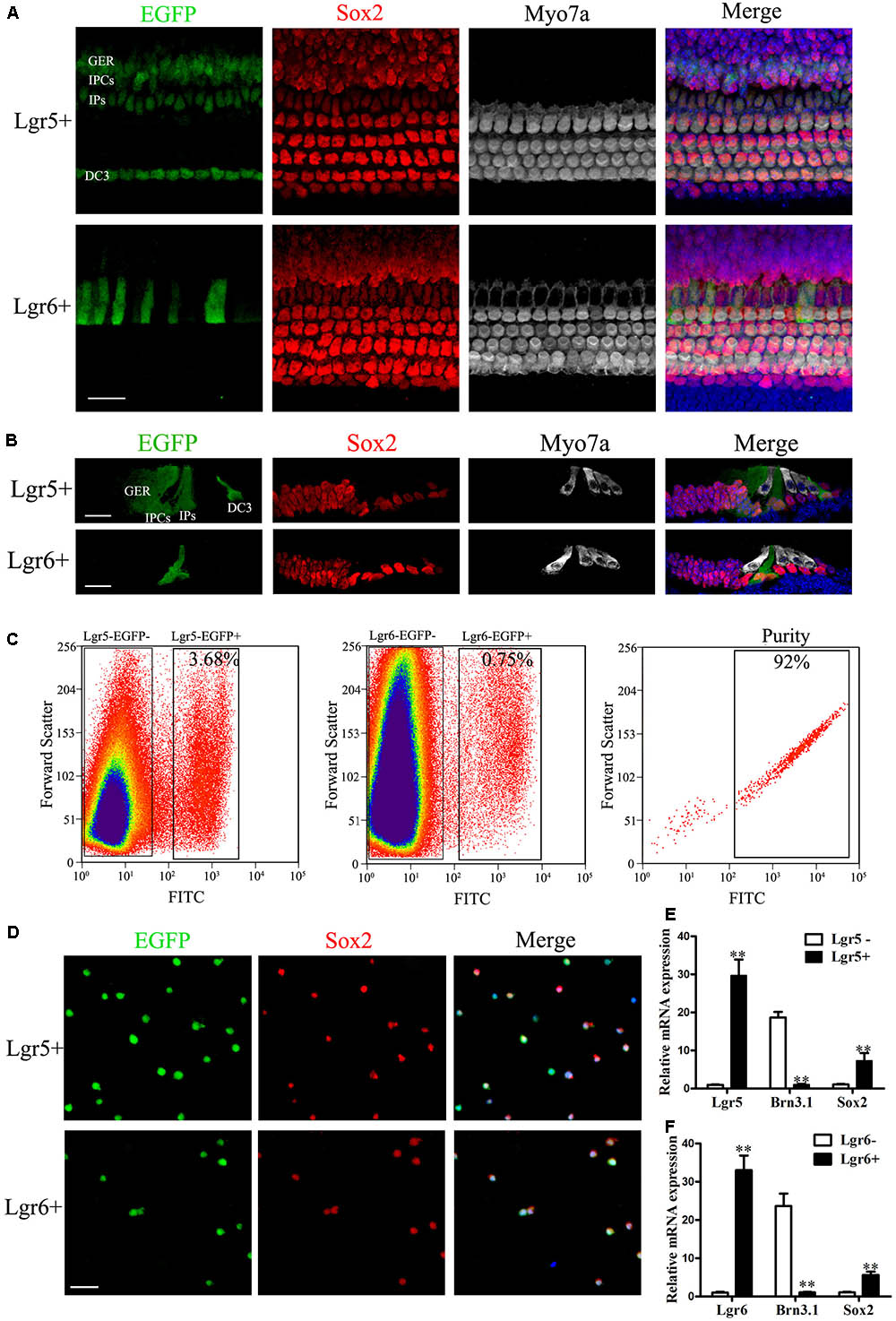
FIGURE 1. Re-sort analysis, immunostaining, and q-PCR of flow-sorted Lgr5+ and Lgr6+ cells from the postnatal cochlea. (A) At P3, Lgr5 was expressed in the third row of Deiters’ cells (DC3), the inner pillar cells (IPs), the inner phalangeal cells (IPCs), and the lateral GER, while Lgr6 was only expressed in the IPs. (B) Cryosection showed that Lgr5 was expressed in DC3s, IPs, IPCs and the GER, and Lgr6 was only expressed in a subset of IPs in the P3 organ of Corti. (C) GFP+ cells and GFP– cells were isolated using flow cytometry. Re-sort analysis of GFP+ cells demonstrated > 90% purity. (D) Immunostaining of Lgr5+ cells and Lgr6+ cells from the cochlea showed a high percentage of Sox2+ (95.4% and 95.2%, respectively) and GFP+ (95.8% and 96.6%, respectively) cells, and no Myo7a+ cells, among the sorted cells. (E,F) q-PCR showed that isolated Lgr5+ cells and Lgr6+ cells had significantly higher Lgr5 and Lgr6 expression, slightly higher Sox2 expression, and significantly lower Brn3.1 expression compared to the Lgr5- cells and Lgr6– cells, respectively. Scale bars are 20 μm. ∗∗p < 0.01.
Previous studies showed that both Lgr5+ and Lgr6+ progenitors can generate HCs in vitro (Chai et al., 2012; Zhang et al., 2015). In order to determine the HC generation ability and proliferation ability of these two progenitor populations, we isolated the Lgr5+ and Lgr6+ progenitors via flow cytometry from P0–P3 Lgr5-EGFP-Ires-CreERT2 and Lgr6-EGFP-Ires-CreERT2 transgenic mice. We found that around 3.68% of the isolated cells were Lgr5+ and around 0.75% of the isolated cells were Lgr6+ (Figure 1C). Immunofluorescent staining showed that 95.8% of the Lgr5+ progenitors (Figure 1D) and 95.6% of the Lgr6+ progenitors (Figure 1D) sorted by flow cytometry were GFP+. Isolated Lgr5+ and Lgr6+ progenitors showed no staining for the HC marker Myo7a (data not shown), but nearly all cells showed staining for the SC marker Sox2 (95.4 and 95.2%, respectively) (Figure 1D). q-PCR was performed immediately after FACS to determine the purity of FACS-isolated Lgr5+ and Lgr6+ cells, and the results showed that the Lgr5+ and Lgr6+ progenitors had significantly higher Lgr5 and Lgr6 expression, respectively, and both populations had higher Sox2 expression and lower HC marker Brn3.1 expression compared to Lgr5- and Lgr6- cells (p < 0.05, Figures 1E,F). These results confirmed the high purity of the sorted Lgr5+ and Lgr6+ progenitors.
Lgr6+ Progenitors Generate Significantly More HCs Than Lgr5+ Progenitors in Vitro
The differentiation assay used in this study was a standard method to investigate the differentiation and proliferation ability of HC progenitors as previously reported (Chai et al., 2012; Shi et al., 2012; Waqas et al., 2016; Cheng et al., 2017). To explore the HC generation capability of Lgr5+ and Lgr6+ progenitors, 2,000 sorted GFP+ cells were cultured in polylysine-laminin coated 4-well dishes at a density of 20 cells/μl in serum-free medium for 10 days. EdU was added to label the mitotically generated cells from day 3 to day 8, and the cells were then immunostained with antibodies against the HC marker Myo7a (Figure 2A). The results showed that both Lgr5+ and Lgr6+ progenitors could generate Myo7a+ cells and Myo7a+ colonies (n = 4, Figures 2B–D), and we found that the same number of Lgr6+ progenitors generated significantly more HCs than Lgr5+ progenitors (361.8 ± 28.35 HCs and 137.5 ± 12.7 HCs, respectively) (p < 0.01, n = 4, Figures 2D,E). We also used the epithelium marker cytokeratin to label the colonies and found that compared with the Lgr5+ progenitors, the Lgr6+ progenitors generated more HCs outside of the colonies, which represent the directly differentiated HCs (104 ± 13.72 HCs and 325.5 ± 29.85 HCs, respectively, per 2,000 cells) (p < 0.01, n = 4, Figure 2E). However, when we counted the cytokeratin-positive colonies, we found that the Lgr5+ progenitors generated significantly more colonies than the Lgr6+ progenitors (20.5 ± 0.65 colonies and 9.75 ± 1.11 colonies, respectively, per 2,000 cells) (p < 0.01, n = 4, Figure 2F). Moreover, the number of Myo7a+ colonies generated from the Lgr5+ progenitors was also significantly greater than the number of Myo7a+ colonies generated from the Lgr6+ progenitors (13.25 ± 1.65 Myo7a+ colonies and 5.75 ± 0.85 Myo7a+ colonies, respectively, per 2,000 cells) (p < 0.01, n = 4, Figure 2F). However, each Myo7a+ colony from the Lgr6+ progenitors generated significantly more HCs than the Myo7a+ colonies from Lgr5+ progenitors (6.30 ± 0.68 HCs and 2.5 ± 0.58 HCs, respectively, per colony) (p < 0.01, n = 4, Figure 2H). When we counted the Myo7a and EdU double positive (Myo7a+/EdU+) cells, we observed Myo7a+/EdU+ cells inside of the colonies in both Lgr5+ and Lgr6+ progenitors. Meanwhile, Lgr5+ progenitors generated significantly more Myo7a+/EdU+ HCs than the Lgr6+ progenitors (11.0 ± 0.91 HCs and 1.5 ± 0.29 HCs, respectively, per 2,000 cells) (n = 4, Figure 2G). Thus, together the total number of Myo7a+ HCs inside of the colony, which represents the mitotically generated HCs, was not statistically different between the Lgr5+ and Lgr6+ progenitors (33.0 ± 3.67 HCs and 36.25 ± 2.18 HCs, respectively, per 2,000 cells) (n = 4, Figure 2E). When we calculated the percentages, we found that the majority of the HCs generated from Lgr6+ progenitors were outside of the colony (89.78%), while the HCs generated from Lgr5+ progenitors were 75.91% outside of the colony and 24.09% inside of the colony. Together, these results demonstrated that Lgr6+ progenitors generated significantly more HCs than Lgr5+ progenitors and that Lgr6+ progenitors generated HCs mainly through direct differentiation, while Lgr5+ progenitors generated HCs through both direct differentiation and mitotic generation. This also indicates that the Lgr5+ populations other than inner pillar cells are responsible for the mitotic HC generation.
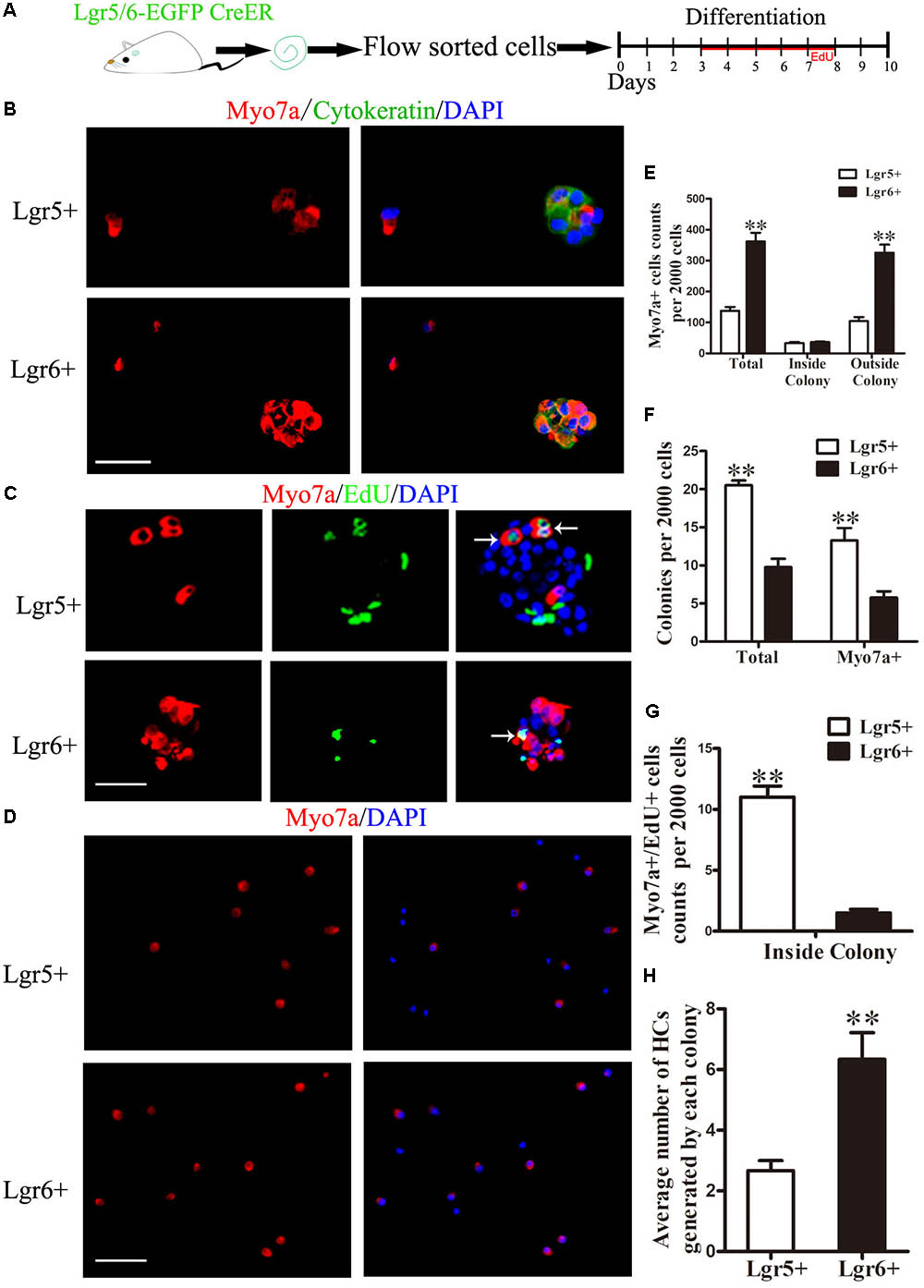
FIGURE 2. Lgr6+ progenitors generate more HCs compared to Lgr5+ progenitors in vitro. (A) Schematic depicting the experimental process. We dissected the cochleae and then cultured the sorted GFP+ cells at 20 cells/μl for 10 days. (B) Both Lgr5+ and Lgr6+ progenitors could generate Myo7a+ colonies. (C) Both Lgr5+ and Lgr6+ progenitors could generate Myo7a+/EdU+ cells inside of the colonies. (D) Both Lgr5+ and Lgr6+ progenitors could generate Myo7a+ cells outside the colonies. (E) The total number of Myo7a+ cells in each well per 2,000 cells and the numbers of Myo7a+ cells inside and outside the colonies. The Lgr5+ progenitors formed around 140 Myo7a+ cells per 2,000 cells and about 33 Myo7a+ cells inside the colonies, while the Lgr6+ progenitors formed around 360 Myo7a+ cells per 2,000 cells and about 36 Myo7a+ cells inside the colonies. (F) The numbers of colonies in each well per 2,000 cells. The Lgr5+ progenitors formed around 20 colonies and 10 Myo7a+ colonies, while the Lgr6+ progenitors formed around 13 colonies and 6 Myo7a+ colonies. (G) Lgr5+ progenitors generated significantly more Myo7a+/EdU+ cells than Lgr5+ progenitors inside of the colonies (11.0 ± 0.91 and 1.5 ± 0.29 HCs, respectively). (H) Each Myo7a+ colony from the Lgr6+ progenitors generated significantly more HCs than the Myo7a+ colonies from Lgr5+ progenitors. ∗∗p < 0.01, n = 4. Scale bars are 20 μm.
Lgr5+ Progenitors Have Greater Sphere-Forming Ability, While Lgr6+ Progenitors Have Greater HC Generation Ability in Vitro
It has been reported that postnatal Lgr5+ progenitors can proliferate and form spheres when cultured in serum-free medium (Chai et al., 2012; Shi et al., 2012). In order to specifically compare the proliferative ability between Lgr5+ and Lgr6+ progenitors, we performed a sphere-forming assay. A total of 2,000 isolated Lgr5+ and Lgr6+ progenitors (2 cells/μl) were cultured in 24-well ultra low-attachment plates for 5 days in the medium that is suitable for sphere forming (Figure 3A). The number and the diameter of spheres generated from Lgr5+ or Lgr6+ progenitors were measured to determine the proliferation capacity of HC progenitors (Figure 3B). Both the numbers and the diameters of the spheres generated from Lgr5+ progenitors were significantly greater than spheres generated from the Lgr6+ progenitors (p < 0.05, n = 4, Figure 3C). To further evaluate the HC generation capacity of the spheres derived from Lgr5+ and Lgr6+ progenitors, we collected the spheres and allowed them to differentiate in serum-free medium for another 10 days (Figure 3A). Then we quantified the number of Myo7a+ HCs in all differentiated spheres generating from the 2,000 isolated cells and in each differentiated sphere (Figures 3F,G). Among the spheres generated from Lgr5+ progenitors, around 60.47% contained Myo7a+ HCs, while 45.61% of the spheres generated from Lgr6+ progenitors contained Myo7a+ HCs (Figures 3D,E). Moreover, in the spheres containing Myo7a+ HCs, our results showed that each Lgr6+ sphere generated significantly more Myo7a+ HCs compared to the Lgr5+ spheres (9.77 ± 0.45 HCs/sphere and 4.38 ± 0.48 HCs/sphere, respectively) (p < 0.01, n = 4, Figure 3F). In total, the 2,000 Lgr6+ progenitor cells generated significantly more Myo7a+ HCs than the same number of Lgr5+ progenitors (263.7 ± 5.78 HCs and 227.3 ± 11.36 HCs, respectively) (p < 0.05, n = 4, Figure 3G). These results indicated that Lgr5+ progenitors have greater proliferation and sphere-forming capacity, while Lgr6+ progenitors have greater HC generation ability in vitro.
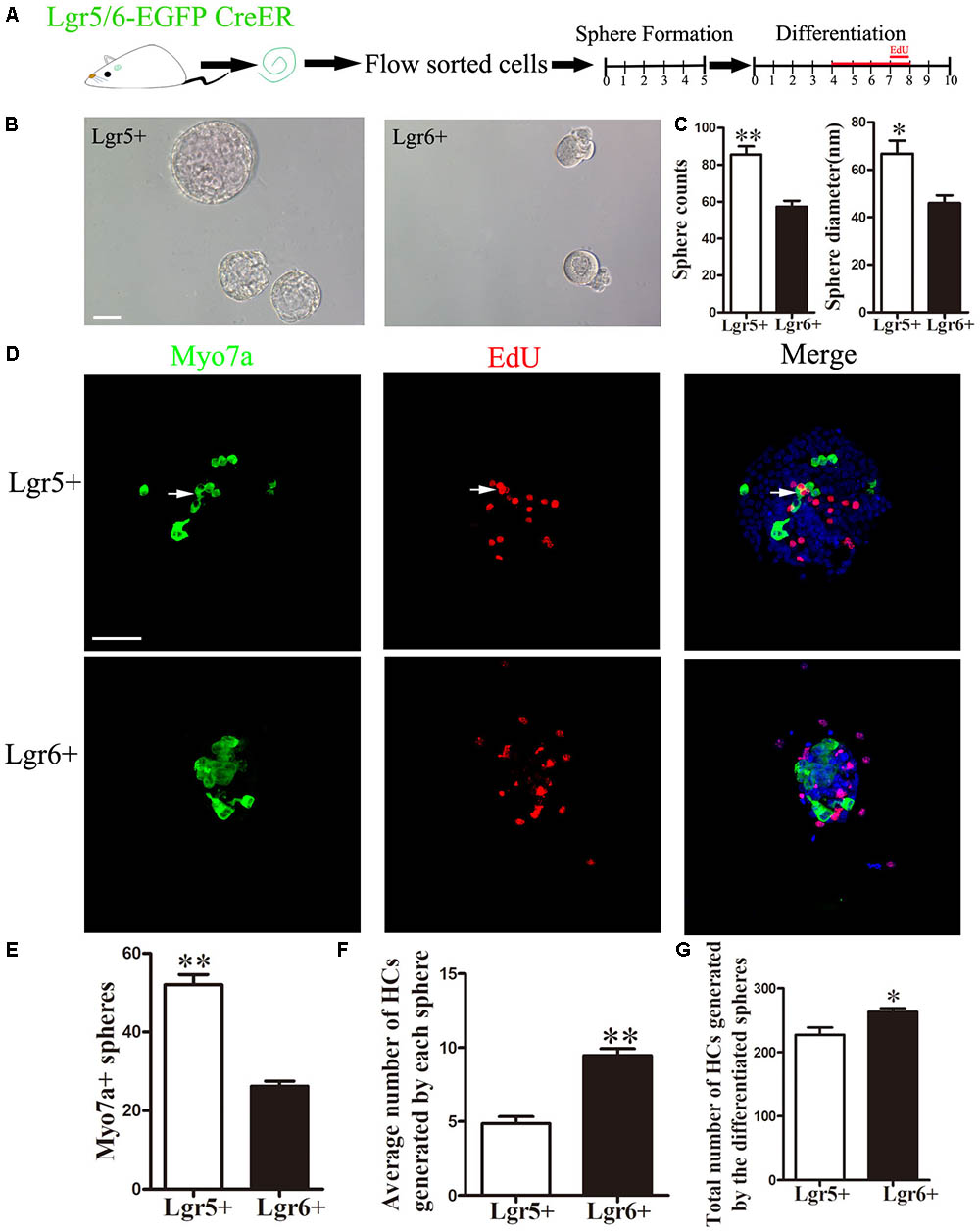
FIGURE 3. Lgr5+ progenitors have greater sphere-forming ability than Lgr6+ progenitors, and Lgr6+ progenitors have greater differentiation ability than Lgr5+ progenitors (A) Schematic depicting the experimental procedure. Cochleae were dissected, and flow-sorted Lgr5+ and Lgr6+ progenitors were cultured for 5 days in ultra low-attachment dishes, after which the spheres were transferred to laminin/polylysine (1:1)-coated 4-well dishes and cultured for another 10 days with EdU added to the medium from day 4 to day 8. (B) Both Lgr5+ and Lgr6+ progenitors could generate spheres. (C) Lgr5+ progenitors generated significantly more spheres than Lgr6+ progenitors, and spheres from Lgr5+ progenitors had a significantly larger diameter than those from the Lgr6+ progenitors. (D) Differentiated spheres from Lgr5+ progenitors generated Myo7a+ HCs and Myo7a+/EdU+ double-positive cells (arrows). Differentiated spheres from Lgr6+ progenitors also generated Myo7a+ HCs. (E) Lgr5+ progenitors generated significantly more Myo7a+ spheres than those from Lgr6+ progenitors. (F) Each differentiated sphere from Lgr6+ progenitors generated significantly more Myo7a+ HCs than those from Lgr5+ progenitors. (G) The 2,000 differentiated spheres from Lgr6+ progenitors generated significantly more Myo7a+ HCs than those from Lgr5+ progenitors. ∗p < 0.05; ∗∗p < 0.01, n = 4. Scale bars are 50 μm.
Analysis of RNA-Seq Results
In order to shed light on the mechanism involved in the proliferation and differentiation of Lgr5+ and Lgr6+ progenitors, we collected 10,000 FACS-isolated Lgr5+ and Lgr6+ cells from P3 cochleae to perform gene expression analysis using RNA-Seq to investigate differentially expressed genes between Lgr5+ and Lgr6+ progenitors. Between 11.9 million and 28.0 million paired-end reads were obtained for each sample, in which 51.0–75.7% of the reads were mapped correctly to the reference genome (mouse mm9). Every gene expression was measured by FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) as in a previous report (Chen et al., 2017). The top 200 most abundant genes in the Lgr5+ and Lgr6+ progenitors are shown in Supplementary Figure S1.
Differentially Expressed Genes in Lgr5+ and Lgr6+ Progenitors
Top differentially expressed genes in these two HC progenitors were selected for further analysis. The expression levels above background and at least 1.5-fold different between these two groups were defined as differentially expressed genes (Figure 4A, p < 0.05). We found that there were 1,157 and 862 differentially highly expressed genes in Lgr5+ progenitors and Lgr6+ progenitors, respectively. The top 100 differentially expressed genes between these two progenitors are shown in Figures 4B,C. The functions of some genes have been studied in the sensory epithelium, including Hes5, Hey2, Gata2, Fgfr2, and Stox1, which were highly expressed in the Lgr5+ progenitors, and Adk and Otoa, which were highly expressed in the Lgr6+ progenitors. However, the expression of most differentially expressed genes and their functions in the ear have not been reported.
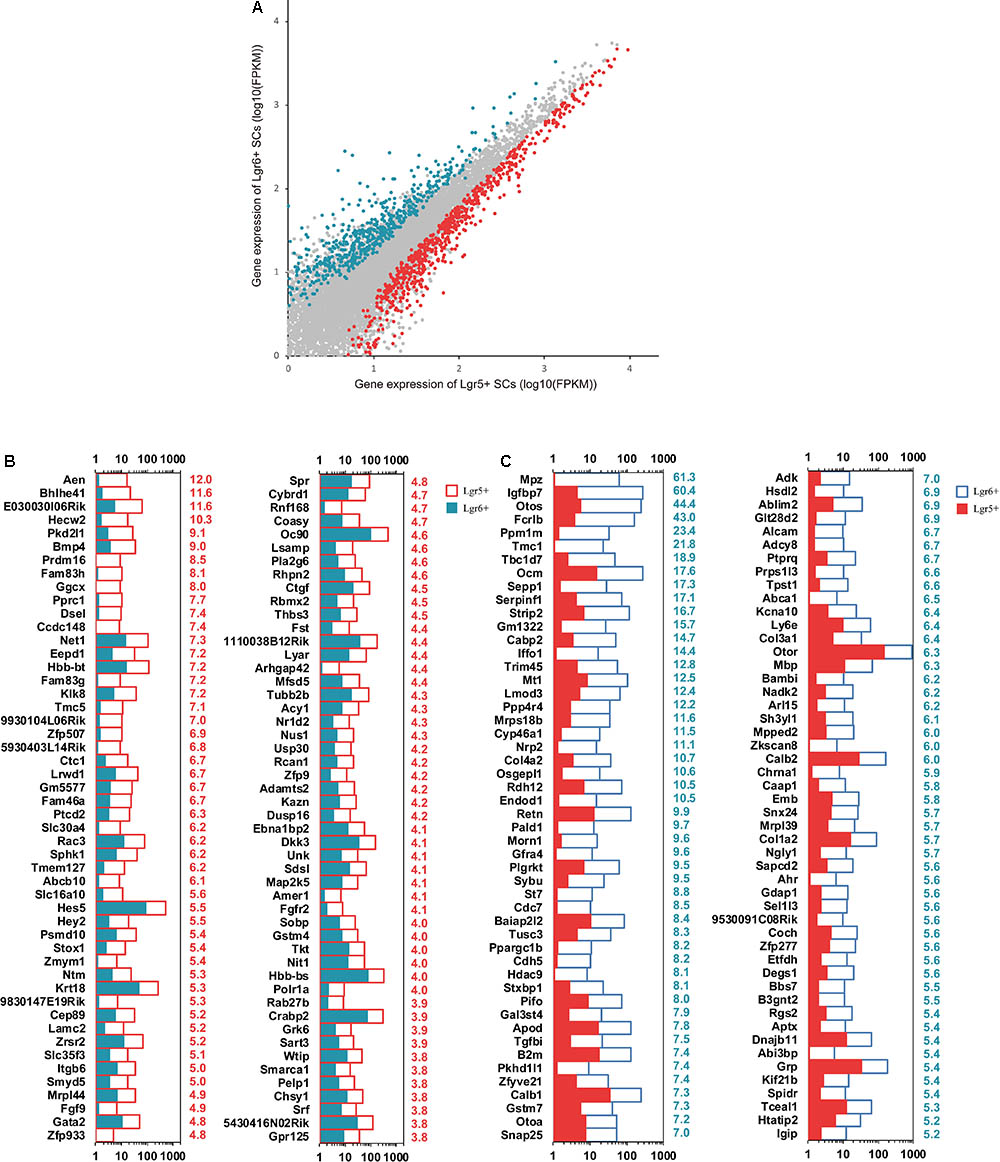
FIGURE 4. Differentially expressed genes in Lgr5+ progenitors and Lgr6+ progenitors. (A) X-Y scatter plot showing differentially expressed genes in Lgr5+ progenitors and Lgr6+ progenitors. The red dots represent highly expressed genes in Lgr5+ progenitors, and the blue dots represent highly expressed genes in Lgr6+ progenitors. (B) The 100 most differentially expressed genes in Lgr5+ progenitors. The numerical values in red on the right side of each panel represent the fold difference in expression in Lgr5+ progenitors versus Lgr6+ progenitors. (C) The 100 most differentially expressed genes in Lgr6+ progenitors. The numerical values in blue on the right side of each panel represent the fold difference in expression in Lgr6+ progenitors versus Lgr5+ progenitors.
Transcription Factor Analysis
Transcription factors (TFs) play vital roles during sensory epithelium development and the differentiation of progenitor cells into HCs. In order to determine which TFs might play roles in regulating the HC generation ability of these two HC progenitors, we examined 1,301 TF genes and found 54 significantly differentially expressed TF genes (p < 0.05). Esr2 was significantly more highly expressed in Lgr6+ progenitors compared to the Lgr5+ progenitors (Figure 5A). Hey2, Hes1, Hes5, Sox4, Id1, and Nr2f1 were significantly highly expressed in Lgr5+ progenitors. Hey2 (Doetzlhofer et al., 2009), Hes1, and Hes5 (Zine et al., 2001; Kelley, 2006) are transcriptional repressors that negatively regulate HC differentiation, and Sox4 (Gnedeva and Hudspeth, 2015), Id1 (Ozeki et al., 2007), and Nr2f1 (Tang et al., 2006) play roles in the developing sensory tissues of the mammalian inner ear by regulating cell proliferation and HC formation. To verify the RNA-Seq data, qPCR was performed and the results were consistent with above RNA-Seq analysis (Figure 5B). The functions of many differentially expressed TFs in the proliferation and differentiation of these two HC progenitors are unclear and need to be researched in the future.
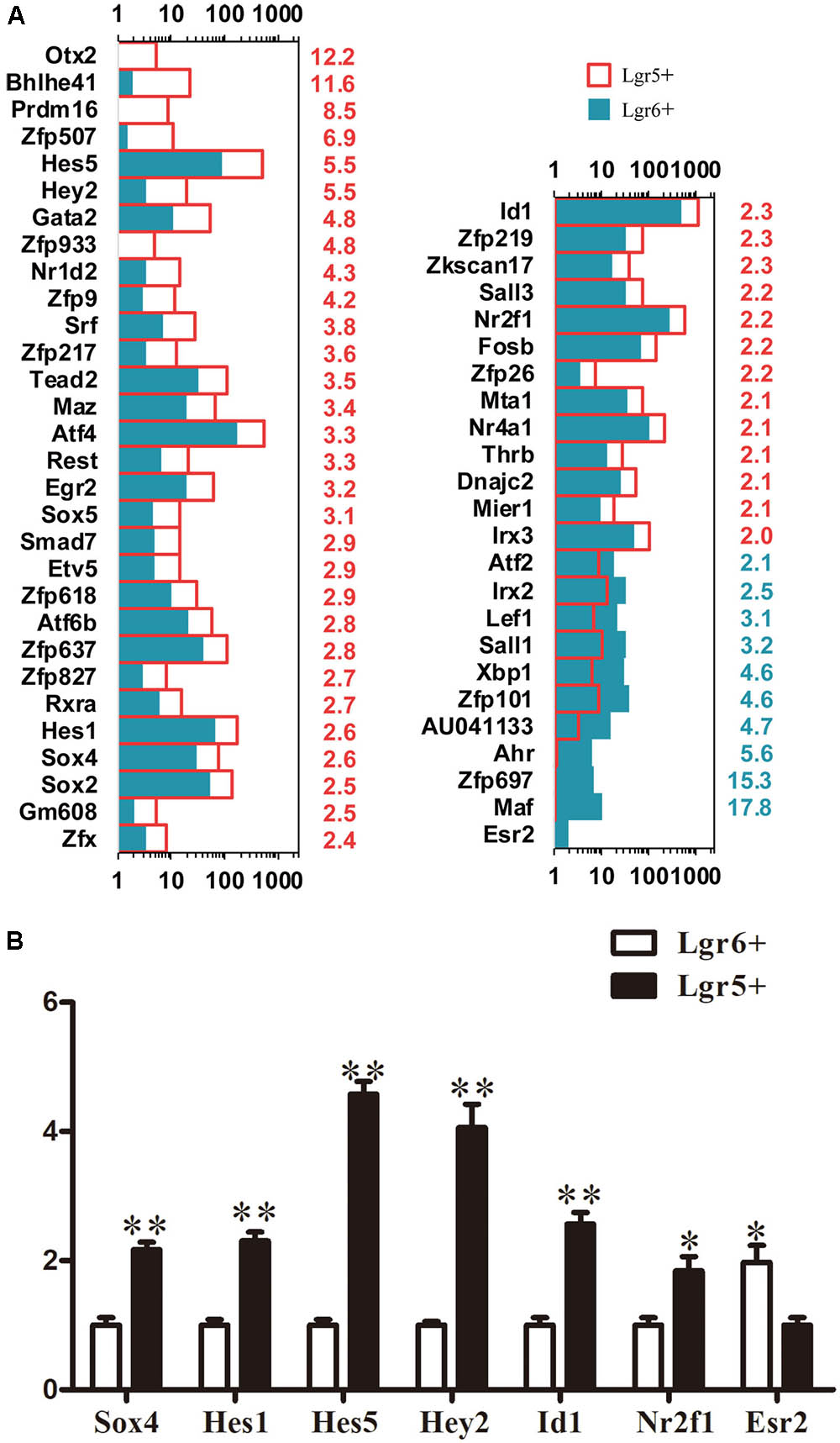
FIGURE 5. Genes regulating transcription factors. (A) Expression levels of differentially expressed TFs. The number in red on the right of each panel represents the fold difference in expression in Lgr5+ progenitors versus Lgr6+ progenitors, and the number in blue on the right of each panel represents the fold difference in expression in Lgr6+ progenitors versus Lgr5+ progenitors. (B) q-PCR analysis of the TF genes. ∗p < 0.05, ∗∗p < 0.01, n = 3.
Cell Cycle Analysis
Cell cycle analysis was performed to explore the mechanism behind the difference of proliferation ability between these two HC progenitors. A total of 41 cell cycle genes were examined, and 4 significantly differentially expressed cell cycle genes were found (p < 0.05). Dst and Cdc7 were highly expressed in Lgr6+ HC progenitors (Figure 6A), while Mcm3 and Skp2 were highly expressed in Lgr5+ HC progenitors (Figure 6A). The qPCR data were consistent with the above RNA-Seq results (Figure 6B).
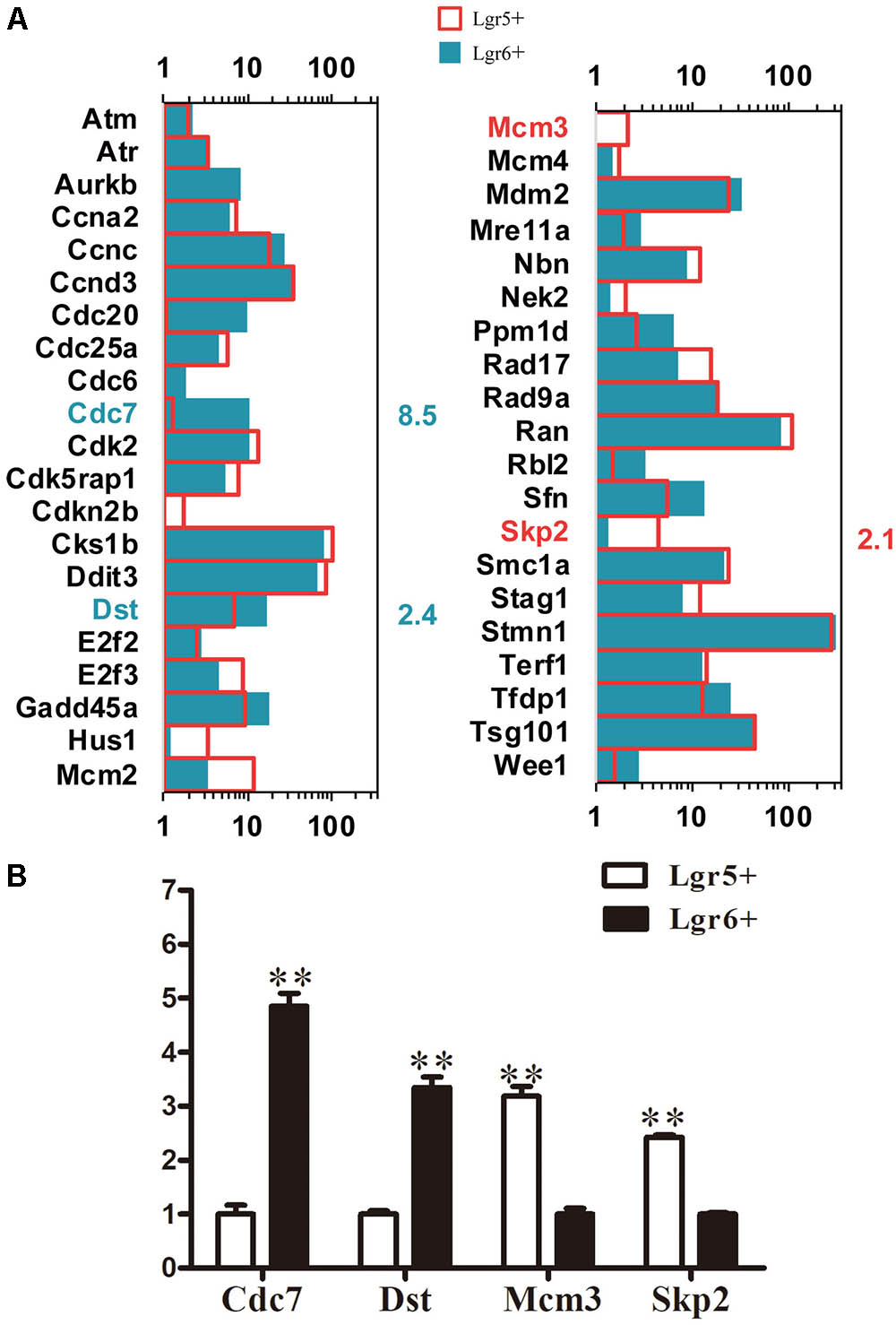
FIGURE 6. Cell cycle genes in Lgr5+ progenitors and Lgr6+ progenitors. (A) The expression of genes involved in the cell cycle in Lgr5+ progenitors and Lgr6+ progenitors. The significantly differentially expressed gene names in Lgr5+ progenitors and Lgr6+ progenitors are labeled in red and blue, respectively. The number in red on the right of each panel represents the fold difference in expression in Lgr5+ progenitors versus Lgr6+ progenitors, and the number in blue on the right of each panel represents the fold difference in expression in Lgr6+ progenitors versus Lgr5+ progenitors. (B) q-PCR analysis of the cell cycle genes. ∗∗p < 0.01, n = 3.
Signaling Pathway Analysis
Lgr5 and Lgr6 are both downstream target genes of the Wnt/β-catenin signaling pathway. We measured the expression levels of genes involved in the Notch and Wnt pathways and found 54 significantly differentially expressed genes between these two HC progenitors (p < 0.05, Figure 7A). The genes significantly increased in Lgr5+ progenitors compared to Lgr6+ progenitors included Dkk3, Fzd8, Sfrp1, Hes1, Hes5, Hey2, and Id1. Hes1, Hes5, Hey2, and Id1 are target genes of Notch signaling, and this suggests that a lower level of Notch signaling in Lgr6+ progenitors might be responsible for their greater capacity for HC generation. The expression of the Notch signaling pathway genes Maml2 and Mfng was significantly greater in Lgr6+ progenitors compared to Lgr5+ progenitors, but only Mfng has been reported in the development of the cochlea (Basch et al., 2016). Again, the q-PCR data were consistent with the above RNA-Seq analysis (Figure 7B).
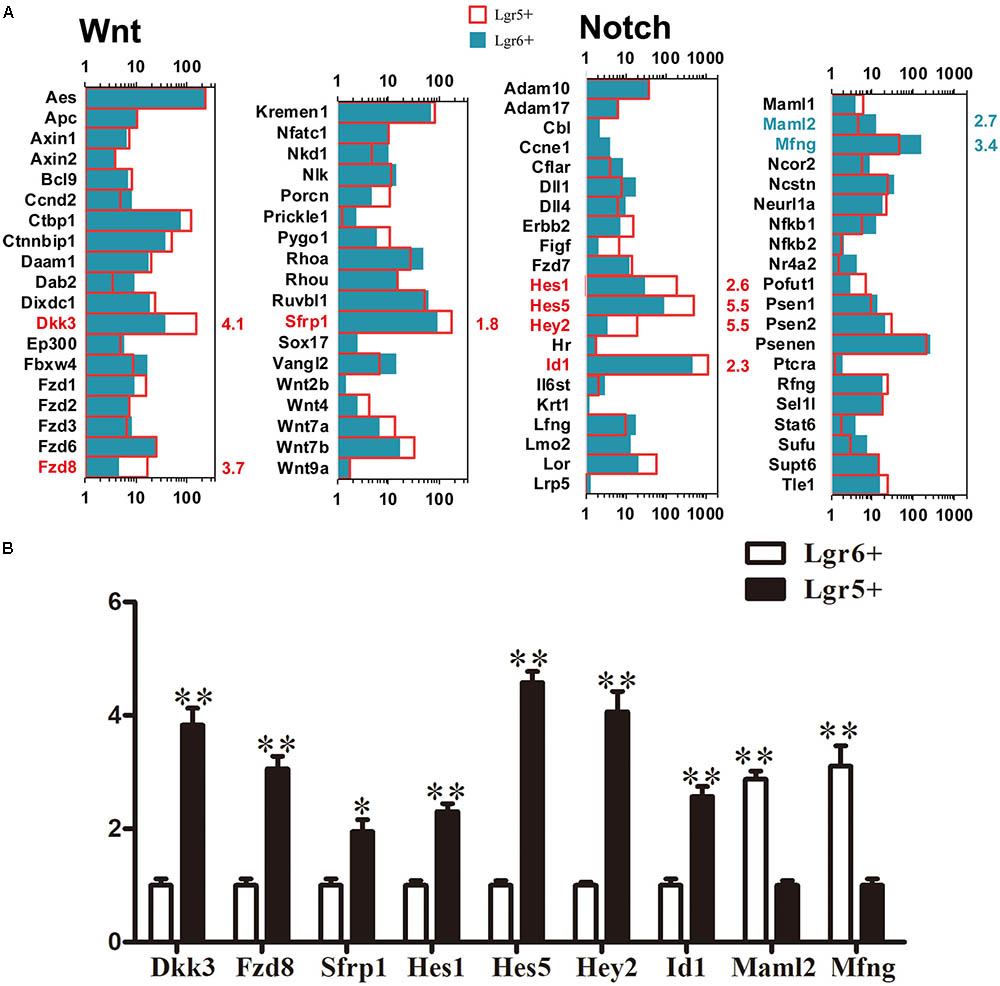
FIGURE 7. Signaling pathway genes in Lgr5+ progenitors and Lgr6+ progenitors. (A) The differentially expressed genes in Lgr5+ progenitors and Lgr6+ progenitors that are involved in the Wnt and Notch signaling pathways. The significantly differentially expressed genes in Lgr5+ progenitors and Lgr6+ progenitors are labeled in red and blue, respectively. The number in red on the right of each panel represents the fold difference in expression in Lgr5+ progenitors versus Lgr6+ progenitors, and the number in blue on the right of each panel represents the fold difference in expression in Lgr6+ progenitors versus Lgr5+ progenitors. (B) q-PCR analysis of the Notch and Wnt signaling pathway genes. ∗∗p < 0.01, n = 3.
Discussion
Lgr5 and Lgr6, which are encoded by Wnt target genes, are markers of adult stem/progenitor cells in many tissues. Lgr5 has been reported to mark stem cells in the small intestine, colon (Barker et al., 2007), stomach (Barker et al., 2010), hair follicle (Jaks et al., 2008), and cochlea (Chai et al., 2012); Meanwhile, in the skin (Snippert et al., 2010), nails (Lehoczky and Tabin, 2015), tongue (Ren et al., 2014), lung (Oeztuerk-Winder et al., 2012), and mammary gland (Blaas et al., 2016), Lgr6 is known to be a stem cell marker.
Our previous studies have reported the expression of Lgr5 (Chai et al., 2011) and Lgr6 (Zhang et al., 2015) in the developing inner ear. The expression of Lgr5 gradually decreases during the development and maturation of the cochlea, and the expression of Lgr5 in the IPs, the IPCs, and the lateral GER gradually disappears as the neonatal cochlea matures; in the adult cochlea, Lgr5 is expressed only in D3s. In the embryotic and neonatal mouse cochlea, Lgr6 is expressed in the IPs. With the maturation of the cochlea, Lgr6 expression is seen in both the IPs and the inner border cells (IBCs) (Zhang et al., 2015). No Lgr6 expression was observed in the mature cochlea (Zhang et al., 2015). Although the expression patterns of Lgr6 and Lgr5 share many differences, as well as some similarities, in the development of the cochlea, in general Lgr6 is expressed in a subpopulation of Lgr5+ progenitors in the embryonic and neonatal cochlea.
Multiple studies have demonstrated that Lgr5 marks the cochlear progenitors (Chai et al., 2012; Shi et al., 2013), and after HC damage Lgr5+ progenitors can generate new cochlear HCs via mitotic regeneration and/or direct differentiation in the neonatal stage (Chai et al., 2012; Shi et al., 2012, 2013; Jan et al., 2013; Cox et al., 2014). Our previous studies showed that Lgr6+ progenitors isolated from neonatal cochleae by flow cytometry are capable of generating HCs in vitro, which indicates that Lgr6+ cells are also cochlear HC progenitors (Zhang et al., 2015). However, a detailed comparison between Lgr6+ progenitors and Lgr5+ progenitors has not yet been undertaken. In this study, we found that isolated Lgr6+ progenitors showed a much greater capacity to generate HCs than Lgr5+ progenitors, while Lgr5+ progenitors showed greater capacity for proliferation and sphere-forming compared with Lgr6+ progenitors. In order to explore the detailed mechanisms leading to the differences of Lgr5+ and Lgr6+ progenitors, we analyzed the complete gene expression profiles of these two progenitor populations.
Differentially Expressed Genes in Lgr5+ Progenitors and Lgr6+ Progenitors
Most of the top 100 differentially expressed genes between these two HC progenitors were not reported in the inner ear before. Five reported genes, including Hes5, Hey2, Gata2, Fgfr2, and Stox1, were highly expressed in Lgr5+ HC progenitors. Hes5 and Hes1 encode two inhibitory basic helix-loop-helix (bHLH) proteins that down-regulate the expression level of prosensory genes, including Atoh1, in differentiating cochlear SCs (Kelley, 2006). Atoh1 is essential for the formation of HCs in the development of the cochlear epithelium (Bermingham et al., 1999), thus the high expression of Hes5 in Lgr5+ progenitors might suppress the differentiation of progenitors into HCs. Similarly, Hey2 has been reported to block Atoh1-induced differentiation of SCs into HCs (Doetzlhofer et al., 2009). Fgfr2 plays a key role in mesenchymal-epithelial signaling during early organogenesis, and Fgfr2-null mice have developmental defects in the inner ear (De Moerlooze et al., 2000). Gata2 is critical for vestibular organ morphogenesis (Haugas et al., 2010). Stox1 is selectively expressed in the cochlear sensory epithelium, and overexpression of Stox1 enhances cell proliferation and sphere formation (Nie et al., 2015), suggesting that Stox1 might be one of the genes leading to the greater proliferation ability of Lgr5+ progenitors. Genes highly expressed in Lgr6+ progenitors compared to Lgr5+ progenitors include Adk and Otoa. Inhibition of Adk in the cochlea postpones the onset of age-related hearing loss (Vlajkovic et al., 2011). Otoa mutations are connected with autosomal recessive deafness type 22 (Zwaenepoel et al., 2002). Other than these, the rest of the top 100 differentially expressed genes have not been reported and their roles in the sensory epithelium need further research in the future.
Transcription Factor Analysis
We examined 1,301 TF genes and found 54 significantly differentially expressed TF genes. Some reported TF genes, including Hes1, Hes5, Hey2, Sox4, Id1, and Nr2f1, are highly expressed in the Lgr5+ progenitors compared to Lgr6+ progenitors. Sox4, which is a member of the SoxC family of TF genes, has effects on the proliferation and differentiation process of the prosensory epithelium, and enhanced expression of SoxC genes restores proliferation in the adult utricular sensory epithelia (Gnedeva and Hudspeth, 2015). To confirm the qPCR verification of Sox4 expression, we also performed immunostaining experiments using the Sox4 antibody. We found that Sox4 was expressed in all of the SCs, including the first row of Deiters’ cells (DC1), the second row of Deiters’ cells (DC2), DC3, IPs, outer pillar cells(OPs), the lesser epithelial ridge (LER) and the GER in P3 cochlea, and there was no obvious expression difference between DCs and IPs in our immunostaining result (Supplementary Figure S3). However, immunostaining is not an accurate method to quantify the exact expression level of the protein. Id proteins are bHLH factors that inhibit cell differentiation and promote cell proliferation by negatively regulating many bHLH TFs, including Math1 (Benezra et al., 1990; Norton, 2000). Id1 promotes the proliferation of OC1 cells and is involved in cochlear development (Ozeki et al., 2007). Nr2f1 is expressed during cochlear development prior to and during the differentiation process of HCs and SCs (Tang et al., 2006). Nr2f1 deficiency results in supernumerary HCs and SCs in the cochlear duct (Tang et al., 2006). In general, these reported TF genes that have higher expression in Lgr5+ progenitors are involved in cell proliferation and prosensory epithelial differentiation, and upregulation of these TF genes inhibits the differentiation of SCs into HCs. This might explain why the Lgr6+ progenitors have greater HC differentiation ability compared to Lgr5+ progenitors. Among the previously characterized TFs, Esr2 is highly expressed in Lgr6+ progenitors compared to Lgr5+ progenitors. Esr2 plays a role in preventing age-related hearing loss (Simonoska et al., 2009). However, the majority of the significantly differentially expressed TF genes need to be studied in detail in the inner ear.
Cell Cycle Analysis
Two cell cycle-related genes, Mcm3 and Skp2, are highly expressed in Lgr5+ progenitors. Skp2 plays a critical role in cell proliferation and differentiation of the developing auditory system. Skp2 positively regulates the G1-S transition through regulating p27 and thus triggers the proliferation of quiescent cochlear precursor cells (Dong et al., 2003; Minoda et al., 2007); however, during differentiation Skp2 expression is down-regulated in the auditory epithelia. This suggests that up-regulation of Skp2 might result in the significantly higher proliferation seen in Lgr5+ progenitors compared to Lgr6+ progenitors. In this study, we also found that some positive regulatory genes of the cell cycle (Ccna2, Ccnd3, Ran, Stmn1, and Smc1a) and negative regulatory genes of the cell cycle (Cadkrap1 and Mdm2) are expressed in cochlear Lgr5+ progenitors and Lgr6+ progenitors. These newly discovered cell cycle regulatory genes need detailed characterization in the future.
Signaling Pathway Analysis
Most of the genes for the Wnt and Notch signaling factors are expressed at similar levels in Lgr5+ progenitors and Lgr6+ progenitors. Dkk3, Fzd8, and Sfrp1, which are Wnt signaling pathway-related genes, have higher expression levels in Lgr5+ progenitors. Dkk3 encodes a secreted protein that can interact with the Wnt signaling pathway and thus plays important roles in embryonic development processes (Ang et al., 2004; Diep et al., 2004). Sfrp1 can directly interact with frizzled (Bafico et al., 1999) and activate Wnt/β-catenin signaling (Uren et al., 2000). Fzd8, a Wnt receptor that can activate both the canonical Wnt/β-catenin pathway and non-canonical Wnt signaling pathway, is reported to regulate the polarity and motility of neuroblasts (Kikuchi et al., 2007). However, the functions of Dkk3, Fzd8, and Sfrp1 in the cochlea require further studies in greater detail. Hes5, Hes1, Hey2, and Id1, which are Notch signaling-related genes, have higher expression levels in Lgr5+ progenitors. The functions of these genes have been discussed above. The increased expression of Hes1, Hes5, and Hey2 in Lgr5+ progenitors might suppress the differentiation of Lgr5+ progenitors into HCs (Doetzlhofer et al., 2009). Id1 was shown to positively regulate the proliferation of cochlear sensory epithelial cells (Ozeki et al., 2007), and the upregulation of Id1 in Lgr5+ progenitors might promote the proliferation of Lgr5+ progenitors. The Notch signaling pathway-related genes that have higher expression levels in Lgr6+ progenitors include Mam2 and Mfng. Mfng, which is one of the Notch modifiers, is transiently expressed specifically in the future inner HC region and is required to regulate the differentiation of inner HCs and IPCs (Basch et al., 2016), but the functions of Mam2 in the inner ear need to be studied further. Activating Wnt signaling can enhanced SC proliferation (Chai et al., 2012; Shi et al., 2014), while inhibiting the Notch pathway can promote HC differentiation and activating the Notch pathway can lead to the overproduction of SCs (Kiernan, 2013). Our results suggest that it might be the combined functions of Wnt and Notch signaling that lead to the increased proliferation capacity of Lgr5+ progenitors and the increased differentiation capacity of Lgr6+ progenitors (Murata et al., 2012; Munnamalai and Fekete, 2013).
Conclusion
Our results show that compared to the Lgr5+ progenitors, Lgr6+ progenitors have significantly greater capacity to generate HCs and thus are an enriched HC progenitor population, while Lgr5+ progenitors have greater capacity to proliferate than Lgr6+ progenitors. Furthermore, our RNA-Seq data provide a handful of genes that might play roles in the proliferation and differentiation of cochlear progenitors, and these data will be very useful in designing strategies for promoting hair cell regeneration and hearing loss therapy.
Data Accession
The raw data are available on NCBI GEO database, accession number GSE114034.
Author Contributions
YZ, HL, and RC designed the study. YZ, LG, and XL performed the laboratory experiments. YZ, HL, RC, LG, XL, YC, SZ, MT, SS, and LZ contributed to critical discussion and data analysis. YZ, HL, LG, YC, and RC wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (Nos. 2017YFA0103900, 2016YFC0905200, and 2015CB965000), the National Natural Science Foundation of China (Nos. 81700906, 81771010, 81670928, 81470687, 81622013, 81470692, 81500790, 81570921, 31500852, 31501194, and 81670938), the Natural Science Foundation of Jiangsu Province (BK20150022, BK20150598, and BK20160125), Boehringer Ingelheim Pharma GmbH, the Fundamental Research Funds for the Central Universities (2242017K41040 and 2242017K3DN25), and the Huo Yingdong Education Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2018.00147/full#supplementary-material
FIGURE S1 | Expression levels of the top 200 genes in Lgr5+ progenitors and Lgr6+ progenitors. (A) Expression levels of the top 200 genes in Lgr5+ progenitors in descending order. Numbers in red on the right side of each panel represent the ranking of the same genes in Lgr6+ progenitors. (B) Expression levels of the top 200 genes in Lgr6+ progenitors in descending order. Numbers in blue on the right side of each panel represent the ranking of the same genes in Lgr5+ progenitors.
FIGURE S2 | Lgr5 and Lgr6 expression in the sensory epithelium of P3 cochleae. At P3, there was only one row of Lgr6+ cells in the middle and basal turns of the cochlea, but Lgr6+ did not expressed in the apical turn of cochlea, while Lgr5 expression was similar from the apex to the base in the sensory epithelium.
FIGURE S3 | Sox4 expression in the sensory epithelium at P3 cochlea. At P3, Sox4 expressed in all of SCs, including first row of Deiters’ cells (DC1), the second row of Deiters’ cells (DC2), DC3, IP, outer pillar cells(OP), the lesser epithelial ridge (LER) and the GER.
TABLE S1 | The Primers for the q-PCR assay.
References
Ang, S. J., Stump, R. J., Lovicu, F. J., and McAvoy, J. W. (2004). Spatial and temporal expression of Wnt and Dickkopf genes during murine lens development. Gene Expr. Patterns 4, 289–295. doi: 10.1016/j.modgep.2003.11.002
Bafico, A., Gazit, A., Pramila, T., Finch, P. W., Yaniv, A., and Aaronson, S. A. (1999). Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J. Biol. Chem. 274, 16180–16187. doi: 10.1074/jbc.274.23.16180
Barker, N., Huch, M., Kujala, P., van de Wetering, M., Snippert, H. J., van Es, J. H., et al. (2010). Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36. doi: 10.1016/j.stem.2009.11.013
Barker, N., van Es, J. H., Kuipers, J., Kujala, P., van den Born, M., Cozijnsen, M., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. doi: 10.1038/nature06196
Basch, M. L., Brown, R. M., Jen, H. I., Semerci, F., Depreux, F., Edlund, R. K., et al. (2016). Fine-tuning of Notch signaling sets the boundary of the organ of Corti and establishes sensory cell fates. Elife 5:e19921. doi: 10.7554/eLife.19921
Belyantseva, I. A., Adler, H. J., Curi, R., Frolenkov, G. I., and Kachar, B. (2000). Expression and localization of prestin and the sugar transporter GLUT-5 during development of electromotility in cochlear outer hair cells. J. Neurosci. 20:RC116. doi: 10.1523/JNEUROSCI.20-24-j0002.2000
Benezra, R., Davis, R. L., Lassar, A., Tapscott, S., Thayer, M., Lockshon, D., et al. (1990). Id: a negative regulator of helix-loop-helix DNA binding proteins. Control of terminal myogenic differentiation. Ann. N. Y. Acad. Sci. 599, 1–11. doi: 10.1111/j.1749-6632.1990.tb42359.x
Bermingham, N. A., Hassan, B. A., Price, S. D., Vollrath, M. A., Ben-Arie, N., Eatock, R. A., et al. (1999). Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837–1841. doi: 10.1126/science.284.5421.1837
Blaas, L., Pucci, F., Messal, H. A., Andersson, A. B., Josue Ruiz, E., Gerling, M., et al. (2016). Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat. Cell Biol. 18, 1346–1356. doi: 10.1038/ncb3434
Bohne, B. A., Ward, P. H., and Fernandez, C. (1976). Irreversible inner ear damage from rock music. Trans. Sect. Otolaryngol. Am. Acad. Ophthalmol. Otolaryngol. 82, ORL50–ORL59.
Burns, J. C., Cox, B. C., Thiede, B. R., Zuo, J., and Corwin, J. T. (2012). In vivo proliferative regeneration of balance hair cells in newborn mice. J. Neurosci. 32, 6570–6577. doi: 10.1523/JNEUROSCI.6274-11.2012
Chai, R., Kuo, B., Wang, T., Liaw, E. J., Xia, A., Jan, T. A., et al. (2012). Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc. Natl. Acad. Sci. U.S.A. 109, 8167–8172. doi: 10.1073/pnas.1202774109
Chai, R., Xia, A., Wang, T., Jan, T. A., Hayashi, T., Bermingham-McDonogh, O., et al. (2011). Dynamic expression of Lgr5, a Wnt target gene, in the developing and mature mouse cochlea. J. Assoc. Res. Otolaryngol. 12, 455–469. doi: 10.1007/s10162-011-0267-2
Chen, P., Johnson, J. E., Zoghbi, H. Y., and Segil, N. (2002). The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129, 2495–2505.
Chen, P., and Segil, N. (1999). p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development 126, 1581–1590.
Chen, Y., Lu, X., Guo, L., Ni, W., Zhang, Y., Zhao, L., et al. (2017). hedgehog signaling promotes the proliferation and subsequent hair cell formation of progenitor cells in the neonatal mouse cochlea. Front. Mol. Neurosci. 10:426. doi: 10.3389/fnmol.2017.00426
Cheng, C., Guo, L., Lu, L., Xu, X., Zhang, S., Gao, J., et al. (2017). Characterization of the transcriptomes of Lgr5+ hair cell progenitors and Lgr5- supporting cells in the mouse cochlea. Front. Mol. Neurosci. 10:122. doi: 10.3389/fnmol.2017.00122
Corwin, J. T., and Cotanche, D. A. (1988). Regeneration of sensory hair cells after acoustic trauma. Science 240, 1772–1774. doi: 10.1126/science.3381100
Cox, B. C., Chai, R., Lenoir, A., Liu, Z., Zhang, L., Nguyen, D., et al. (2014). Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141, 816–829. doi: 10.1242/dev.103036
De Moerlooze, L., Spencer-Dene, B., Revest, J. M., Hajihosseini, M., Rosewell, I., and Dickson, C. (2000). An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483–492.
Diep, D. B., Hoen, N., Backman, M., Machon, O., and Krauss, S. (2004). Characterisation of the Wnt antagonists and their response to conditionally activated Wnt signalling in the developing mouse forebrain. Brain Res. Dev. Brain Res. 153, 261–270. doi: 10.1016/j.devbrainres.2004.09.008
Doetzlhofer, A., Basch, M. L., Ohyama, T., Gessler, M., Groves, A. K., and Segil, N. (2009). Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 16, 58–69. doi: 10.1016/j.devcel.2008.11.008
Dong, Y., Nakagawa, T., Endo, T., Kim, T. S., Iguchi, F., Yamamoto, N., et al. (2003). Role of the F-box protein Skp2 in cell proliferation in the developing auditory system in mice. Neuroreport 14, 759–761. doi: 10.1097/00001756-200304150-00020
Forge, A., Li, L., Corwin, J. T., and Nevill, G. (1993). Ultrastructural evidence for hair cell regeneration in the mammalian inner ear. Science 259, 1616–1619. doi: 10.1126/science.8456284
Gnedeva, K., and Hudspeth, A. J. (2015). SoxC transcription factors are essential for the development of the inner ear. Proc. Natl. Acad. Sci. U.S.A. 112, 14066–14071. doi: 10.1073/pnas.1517371112
Golub, J. S., Tong, L., Ngyuen, T. B., Hume, C. R., Palmiter, R. D., Rubel, E. W., et al. (2012). Hair cell replacement in adult mouse utricles after targeted ablation of hair cells with diphtheria toxin. J. Neurosci. 32, 15093–15105. doi: 10.1523/JNEUROSCI.1709-12.2012
Haugas, M., Lillevali, K., Hakanen, J., and Salminen, M. (2010). Gata2 is required for the development of inner ear semicircular ducts and the surrounding perilymphatic space. Dev. Dyn. 239, 2452–2469. doi: 10.1002/dvdy.22373
Jacques, B. E., Puligilla, C., Weichert, R. M., Ferrer-Vaquer, A., Hadjantonakis, A. K., Kelley, M. W., et al. (2012). A dual function for canonical Wnt/beta-catenin signaling in the developing mammalian cochlea. Development 139, 4395–4404. doi: 10.1242/dev.080358
Jaks, V., Barker, N., Kasper, M., van Es, J. H., Snippert, H. J., Clevers, H., et al. (2008). Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291–1299. doi: 10.1038/ng.239
Jan, T. A., Chai, R., Sayyid, Z. N., van Amerongen, R., Xia, A., Wang, T., et al. (2013). Tympanic border cells are Wnt-responsive and can act as progenitors for postnatal mouse cochlear cells. Development 140, 1196–1206. doi: 10.1242/dev.087528
Kawamoto, K., Izumikawa, M., Beyer, L. A., Atkin, G. M., and Raphael, Y. (2009). Spontaneous hair cell regeneration in the mouse utricle following gentamicin ototoxicity. Hear. Res. 247, 17–26. doi: 10.1016/j.heares.2008.08.010
Kelley, M. W. (2006). Regulation of cell fate in the sensory epithelia of the inner ear. Nat. Rev. Neurosci. 7, 837–849. doi: 10.1038/nrn1987
Kiernan, A. E. (2013). Notch signaling during cell fate determination in the inner ear. Semin. Cell Dev. Biol. 24, 470–479. doi: 10.1016/j.semcdb.2013.04.002
Kikuchi, A., Yamamoto, H., and Kishida, S. (2007). Multiplicity of the interactions of Wnt proteins and their receptors. Cell. Signal. 19, 659–671. doi: 10.1016/j.cellsig.2006.11.001
Lehoczky, J. A., and Tabin, C. J. (2015). Lgr6 marks nail stem cells and is required for digit tip regeneration. Proc. Natl. Acad. Sci. U.S.A. 112, 13249–13254. doi: 10.1073/pnas.1518874112
Li, H., Liu, H., and Heller, S. (2003). Pluripotent stem cells from the adult mouse inner ear. Nat. Med. 9, 1293–1299. doi: 10.1038/nm925
Li, W., Wu, J., Yang, J., Sun, S., Chai, R., Chen, Z. Y., et al. (2015). Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 166–171. doi: 10.1073/pnas.1415901112
Lin, V., Golub, J. S., Nguyen, T. B., Hume, C. R., Oesterle, E. C., and Stone, J. S. (2011). Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J. Neurosci. 31, 15329–15339. doi: 10.1523/JNEUROSCI.2057-11.2011
Minoda, R., Izumikawa, M., Kawamoto, K., Zhang, H., and Raphael, Y. (2007). Manipulating cell cycle regulation in the mature cochlea. Hear. Res. 232, 44–51. doi: 10.1016/j.heares.2007.06.005
Munnamalai, V., and Fekete, D. M. (2013). Wnt signaling during cochlear development. Semin. Cell Dev. Biol. 24, 480–489. doi: 10.1016/j.semcdb.2013.03.008
Murata, J., Ikeda, K., and Okano, H. (2012). Notch signaling and the developing inner ear. Adv. Exp. Med. Biol. 727, 161–173. doi: 10.1007/978-1-4614-0899-4_12
Nie, X., Zhang, K., Wang, L., Ou, G., Zhu, H., and Gao, W. Q. (2015). Transcription factor STOX1 regulates proliferation of inner ear epithelial cells via the AKT pathway. Cell Prolif. 48, 209–220. doi: 10.1111/cpr.12174
Norton, J. D. (2000). ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 113( Pt 22), 3897–3905.
Oesterle, E. C., Campbell, S., Taylor, R. R., Forge, A., and Hume, C. R. (2008). Sox2 and JAGGED1 expression in normal and drug-damaged adult mouse inner ear. J. Assoc. Res. Otolaryngol. 9, 65–89. doi: 10.1007/s10162-007-0106-7
Oeztuerk-Winder, F., Guinot, A., Ochalek, A., and Ventura, J. J. (2012). Regulation of human lung alveolar multipotent cells by a novel p38alpha MAPK/miR-17-92 axis. EMBO J. 31, 3431–3441. doi: 10.1038/emboj.2012.192
Ozeki, M., Hamajima, Y., Feng, L., Ondrey, F. G., Schlentz, E., and Lin, J. (2007). Id1 induces the proliferation of cochlear sensory epithelial cells via the nuclear factor-kappaB/cyclin D1 pathway in vitro. J. Neurosci. Res. 85, 515–524. doi: 10.1002/jnr.21133
Ren, W., Lewandowski, B. C., Watson, J., Aihara, E., Iwatsuki, K., Bachmanov, A. A., et al. (2014). Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. U.S.A. 111, 16401–16406. doi: 10.1073/pnas.1409064111
Ruben, R. J. (1967). Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. 220, 221–244.
Ryals, B. M., and Rubel, E. W. (1988). Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science 240, 1774–1776.
Shi, F., Hu, L., and Edge, A. S. (2013). Generation of hair cells in neonatal mice by beta-catenin overexpression in Lgr5-positive cochlear progenitors. Proc. Natl. Acad. Sci. U.S.A. 110, 13851–13856. doi: 10.1073/pnas.1219952110
Shi, F., Hu, L., Jacques, B. E., Mulvaney, J. F., Dabdoub, A., and Edge, A. S. (2014). beta-Catenin is required for hair-cell differentiation in the cochlea. J. Neurosci. 34, 6470–6479. doi: 10.1523/JNEUROSCI.4305-13.2014
Shi, F., Kempfle, J. S., and Edge, A. S. (2012). Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J. Neurosci. 32, 9639–9648. doi: 10.1523/JNEUROSCI.1064-12.2012
Simonoska, R., Stenberg, A. E., Duan, M., Yakimchuk, K., Fridberger, A., Sahlin, L., et al. (2009). Inner ear pathology and loss of hearing in estrogen receptor-beta deficient mice. J. Endocrinol. 201, 397–406. doi: 10.1677/JOE-09-0060
Sinkkonen, S. T., Chai, R., Jan, T. A., Hartman, B. H., Laske, R. D., Gahlen, F., et al. (2011). Intrinsic regenerative potential of murine cochlear supporting cells. Sci. Rep. 1:26. doi: 10.1038/srep00026
Snippert, H. J., Haegebarth, A., Kasper, M., Jaks, V., van Es, J. H., Barker, N., et al. (2010). Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389. doi: 10.1126/science.1184733
Stone, J. S., and Cotanche, D. A. (2007). Hair cell regeneration in the avian auditory epithelium. Int. J. Dev. Biol. 51, 633–647.
Tang, L. S., Alger, H. M., and Pereira, F. A. (2006). COUP-TFI controls Notch regulation of hair cell and support cell differentiation. Development 133, 3683–3693. doi: 10.1242/dev.02536
Trapnell, C., and Schatz, M. C. (2009). Optimizing data intensive GPGPU computations for DNA sequence alignment. Parallel Comput. 35, 429–440. doi: 10.1016/j.parco.2009.05.002
Uren, A., Reichsman, F., Anest, V., Taylor, W. G., Muraiso, K., Bottaro, D. P., et al. (2000). Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J. Biol. Chem. 275, 4374–4382. doi: 10.1074/jbc.275.6.4374
Vlajkovic, S. M., Guo, C. X., Telang, R., Wong, A. C., Paramananthasivam, V., Boison, D., et al. (2011). Adenosine kinase inhibition in the cochlea delays the onset of age-related hearing loss. Exp. Gerontol. 46, 905–914. doi: 10.1016/j.exger.2011.08.001
Wang, T., Chai, R., Kim, G., Pham, X., and Cheng, A. (2015). Damage-recruited Lgr5+ cells regenerate hair cells via proliferation and direct transdifferentiation in neonatal mouse utricle. Nat. Commun. 6:6613. doi: 10.1038/ncomms7613
Waqas, M., Guo, L., Zhang, S., Chen, Y., Zhang, X., Wang, L., et al. (2016). Characterization of Lgr5+ progenitor cell transcriptomes in the apical and basal turns of the mouse cochlea. Oncotarget 7, 41123–41141. doi: 10.18632/oncotarget.8636
Warchol, M. E. (2011). Sensory regeneration in the vertebrate inner ear: differences at the levels of cells and species. Hear. Res. 273, 72–79. doi: 10.1016/j.heares.2010.05.004
Warchol, M. E., Lambert, P. R., Goldstein, B. J., Forge, A., and Corwin, J. T. (1993). Regenerative proliferation in inner ear sensory epithelia from adult guinea pigs and humans. Science 259, 1619–1622. doi: 10.1126/science.8456285
White, P. M., Doetzlhofer, A., Lee, Y. S., Groves, A. K., and Segil, N. (2006). Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441, 984–987. doi: 10.1038/nature04849
Zhang, Y., Chen, Y., Ni, W., Guo, L., Lu, X., Liu, L., et al. (2015). Dynamic expression of Lgr6 in the developing and mature mouse cochlea. Front. Cell. Neurosci. 9:165. doi: 10.3389/fncel.2015.00165
Zine, A., Aubert, A., Qiu, J., Therianos, S., Guillemot, F., Kageyama, R., et al. (2001). Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J. Neurosci. 21, 4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001
Zwaenepoel, I., Mustapha, M., Leibovici, M., Verpy, E., Goodyear, R., Liu, X. Z., et al. (2002). Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc. Natl. Acad. Sci. U.S.A. 99, 6240–6245. doi: 10.1073/pnas.082515999
Keywords: Wnt, Lgr5, Lgr6, proliferation, differentiation, hair cell, inner ear, RNA-Seq
Citation: Zhang Y, Guo L, Lu X, Cheng C, Sun S, Li W, Zhao L, Lai C, Zhang S, Yu C, Tang M, Chen Y, Chai R and Li H (2018) Characterization of Lgr6+ Cells as an Enriched Population of Hair Cell Progenitors Compared to Lgr5+ Cells for Hair Cell Generation in the Neonatal Mouse Cochlea. Front. Mol. Neurosci. 11:147. doi: 10.3389/fnmol.2018.00147
Received: 05 December 2017; Accepted: 12 April 2018;
Published: 14 May 2018.
Edited by:
Fabio Mammano, Istituto di Biologia Cellulare e Neurobiologia (CNR), ItalyReviewed by:
Matthew William Kelley, National Institutes of Health (NIH), United StatesStefano Farioli Vecchioli, Consiglio Nazionale delle Ricerche (CNR), Italy
Copyright © 2018 Zhang, Guo, Lu, Cheng, Sun, Li, Zhao, Lai, Zhang, Yu, Tang, Chen, Chai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renjie Chai, cmVuamllY0BzZXUuZWR1LmNu Huawei Li, aHdsaUBzaG11LmVkdS5jbg==
†These authors have contributed equally to this work.
 Yanping Zhang
Yanping Zhang Luo Guo
Luo Guo Xiaoling Lu
Xiaoling Lu Cheng Cheng2
Cheng Cheng2 Shan Sun
Shan Sun Chenjie Yu
Chenjie Yu Mingliang Tang
Mingliang Tang Yan Chen
Yan Chen Renjie Chai
Renjie Chai Huawei Li
Huawei Li