- 1First Clinical College, Shandong University of Traditional Chinese Medicine, Jinan, China
- 2Department of Geriatrics, Xiyuan Hospital, China Academy of Chinese Medical Sciences, Beijing, China
Alzheimer’s disease (AD) is the most common form of dementia. Although the incidence of AD is high, the rates of diagnosis and treatment are relatively low. Moreover, effective means for the diagnosis and treatment of AD are still lacking. MicroRNAs (miRNAs, miRs) are non-coding RNAs that play regulatory roles by targeting mRNAs. The expression of miRNAs is conserved, temporal, and tissue-specific. Impairment of microRNA function is closely related to AD pathogenesis, including the beta-amyloid and tau hallmarks of AD, and there is evidence that the expression of some microRNAs differs significantly between healthy people and AD patients. These properties of miRNAs endow them with potential diagnostic and therapeutic value in the treatment of this debilitating disease. This review provides comprehensive information about the regulatory function of miRNAs in AD, as well as potential applications as diagnostic biomarkers.
Introduction
Alzheimer’s disease (AD) is the most frequently occurring dementia in the elderly. It is a multifactorial and heterogeneous neurodegenerative disease, clinically manifested as progressive cognitive dysfunction and behavioral impairment (Lane et al., 2018). The typical pathological features are essentially present with amyloid plaques and neurofibrillary tangles that are associated with beta-amyloid (Aβ) metabolism and the hyperphosphorylation of tau protein, respectively, as the core pathological mechanisms. Moreover, AD pathogenesis is closely related to impaired synaptic plasticity, immune-inflammatory responses, and numerous other processes associated with the central nervous system (CNS). MicroRNAs (miRNAs, miRs) are abundantly present in the CNS, and involve in the complicated pathogenesis of AD through a variety of mechanisms. The diagnosis of AD has great limitations currently. There is an urgent need for reliable biomarkers, especially in the early stages of the disease so that interventions can be promptly instituted to improve clinical outcome.
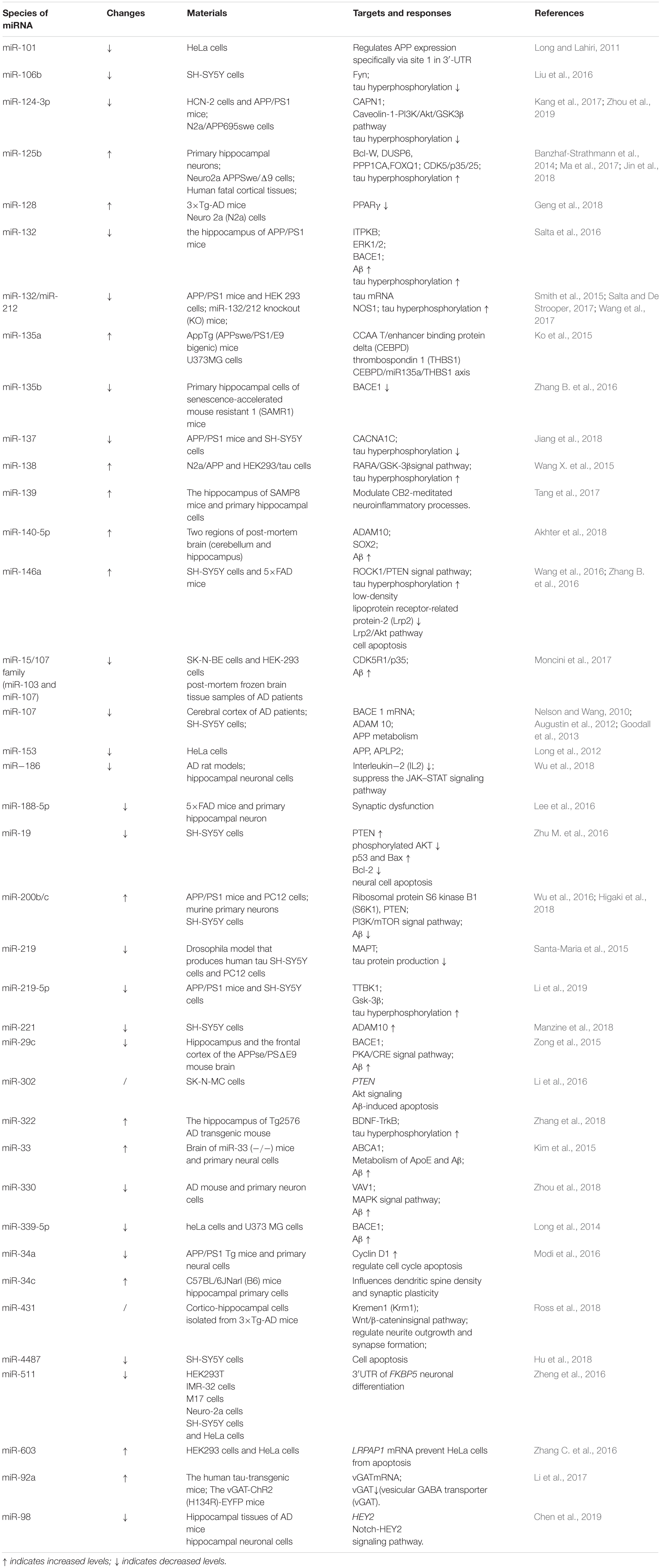
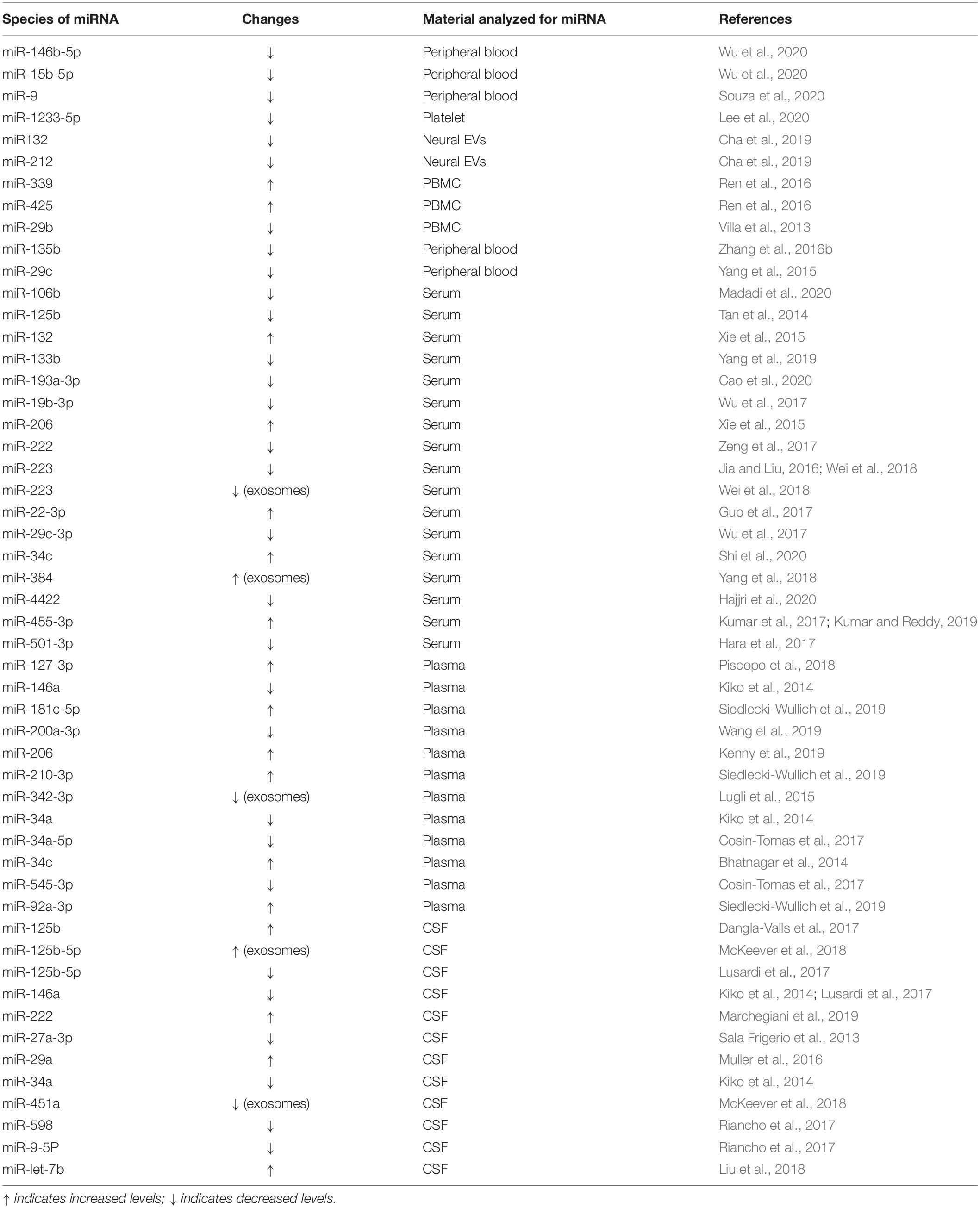
In this review, we summarize the evidence relating to how miRNAs modulate the onset and pathological progression of AD. Part of the summary is shown in Figure 1 and Table 1. We also review the potential of using miRNAs as diagnostic biomarkers for AD (Table 2), thereby providing a perspective of the clinical applications of miRNAs for AD management (Figure 2).
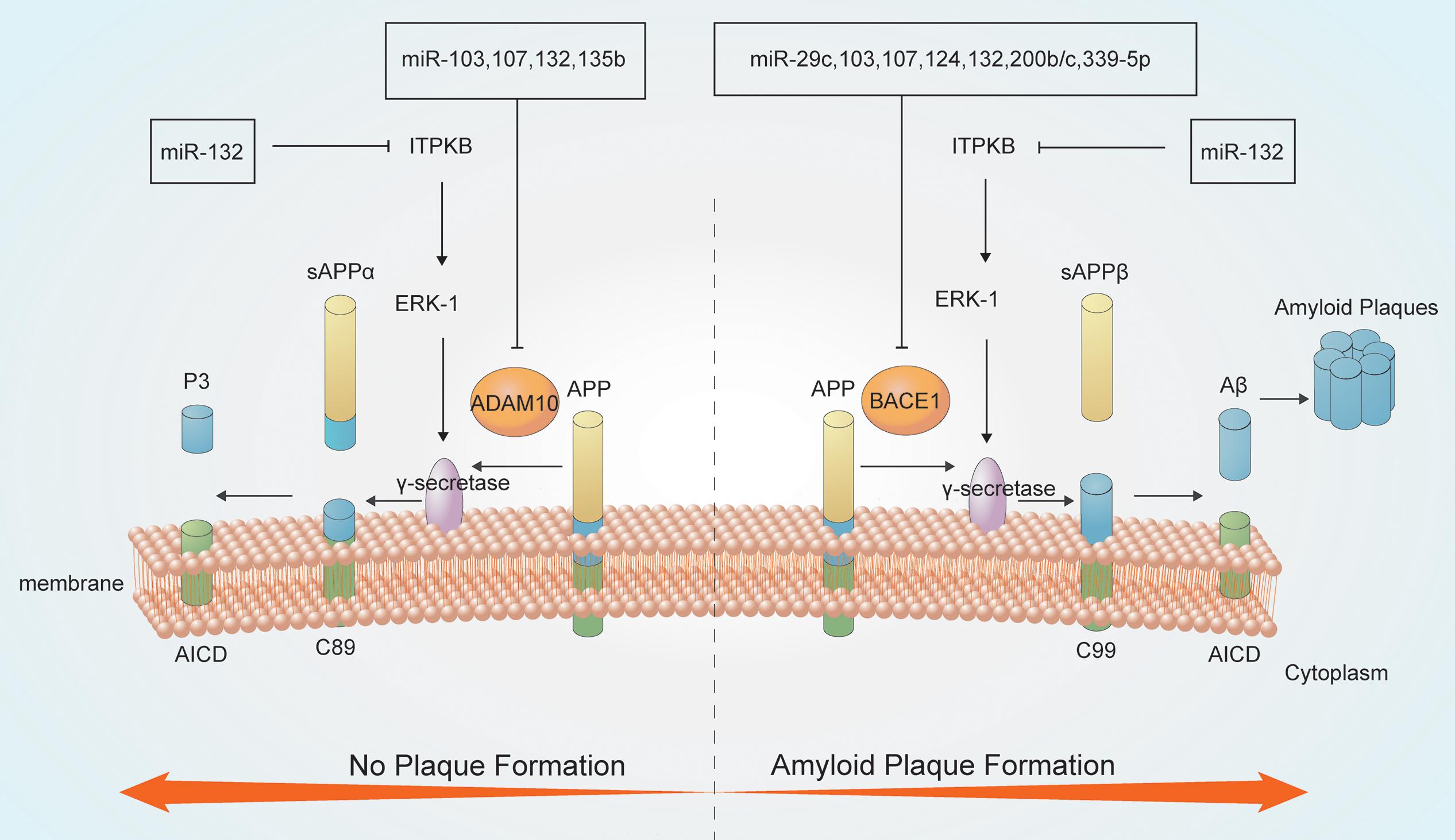
Figure 1. MicroRNAs (miRNAs) are involved in Aβ metabolism. Amyloid precursor protein (APP) is a type I integral inner membrane-localized protein. Under normal conditions, APP is hydrolyzed by α-secretase to produce the neuroprotective soluble external functional fragments (sAPP), P3 and the APP intracellular domain (AICD) (no plaque formation); in contrast, β-secretase-mediated APP hydrolysis generates plaque-forming Aβ, which is neurotoxic. The γ-secretase enzyme is crucial for both secretase pathways.
Basic Structure and Functions of MiRNAs
MicroRNAs are small, non-coding, single-stranded RNAs approximately 22 nucleotides long. Canonical miRNA biogenesis begins with the transcription of primary miRNAs (pri-miRNAs) by RNA polymerase II. These pri-miRNAs are processed into precursor miRNAs (pre-miRNAs) by Drosha in complex with Pasha/DGCR8, and then transported from nucleus to cytoplasm. Pre-miRNAs have a hairpin loop structure recognized for cleavage by dicer, leading to the production of mature miRNAs. One strand of the mature duplex is loaded onto a member of the Argonaute (Ago) family of proteins, forming RNA-induced silencing complexes (RISCs) that mediate gene silencing by recognizing the 3′ untranslated region (3′ UTR) of target mRNAs (Schwarz and Zamore, 2002; Friedman et al., 2009; Jinek and Doudna, 2009; Ipsaro and Joshua-Tor, 2015). Of specifically note, miRNA binding with AGO can promote gene expression. For example, miR-346 recruiting AGO2 targets the 5′ UTR of amyloid precursor protein (APP) mRNA, which competes with the translational suppressor, the iron response protein1, thus inducing stimulative translation of APP mRNA (Long et al., 2019).
Besides inhibiting the expression of target genes by binding to the 3′ UTR of mRNAs, increasing evidence has indicated that miRNAs also act in a non-canonical manner by changing binding partner. MiR-181c can target mitochondrial transcription (Das et al., 2012). MiRNAs can also interact with non-Ago proteins. Research reports that miR-let-7b activates Toll-like receptors (TLRs) as a signaling molecule (Lehmann et al., 2012). Apart from binding with protein, miRNAs can synergistically interact with non-coding RNAs, including long non-coding RNAs (lncRNAs) and circular RNAs (cirRNAs). Intriguingly, miRNAs can promote gene expression, such as miR-589, binding to the promoter RNA of COX-2 transcript, thus inducing transcriptional upregulation (Matsui et al., 2013). Another example is that miR-369 activates TNFα translation by recruiting AGO- FXR1 complex to the AU-rich elements of mRNA during G1/G0 phase. Finally, some pri-miRNAs are found to act as peptide encoding RNAs [miRNA-encoded peptides (miPEPs)], such as pri-miR-165a and pri-miR-171b (Lauressergues et al., 2015; Dragomir et al., 2018).
MiRNAs Are Crucial for the Onset and Pathological Progression of AD
miRNAs Are Involved in Aβ Metabolism
In 1984, George Glenner and Caine Wong found that the main component of senile plaques was a peptide of 39–43 amino acid residues, known as Aβ, thereby laying the foundation for the study of AD (Glenner and Wong, 1984). The Aβ hypothesis holds that Aβ aggregation is the causative factor in AD, leading to synaptic damage, tau protein phosphorylation, inflammation, oxidative stress, apoptosis, and eventually nerve cell damage and death (Sanabria-Castro et al., 2017). Mutations in the APP, presenilin 1 (PS1), PS2, and apolipoprotein E (APOE) genes all lead to abnormal APP processing and Aβ metabolism, resulting in Aβ deposition and subsequent neurocytotoxicity (Hardy and Higgins, 1992; Hardy and Selkoe, 2002). Other susceptibility genes for AD, such as CLU, CD2AP, PICALM, and ABCA7, affect Aβ generation and elimination (Gibson, 2010).
The APP gene is located in the middle segment of the long arm of chromosome 21 and contains 18 exons. APP is widely present in tissue cells throughout the body, but most abundantly expressing in neurons. APP is a type I integral inner membrane protein, comprising an intracellular domain (AICD) and an extracellular domain, and the aberrant function of APP can lead to an increase in Aβ production in AD patients (Goate et al., 1991; Rovelet-Lecrux et al., 2006). The PS1 and PS2 genes are located on chromosome 14 and chromosome 1, respectively, and their protein products have similar structure and function. They both contain 13 exons, and form PS proteins after transcription. PS1 possesses γ-secretase activity and participates in APP proteolysis. Mutations in PS1 or PS2 can affect the degradation and transport of APP, increase Aβ42 production and the Aβ42/Aβ40 ratio, and affect the interaction between tau protein and other cytoskeletal proteins, thereby attributing to the pathogenesis of AD (Kumar-Singh et al., 2006; Campion et al., 2016; Eggert et al., 2018).
APOE has the greatest correlation with late-onset AD (LOAD; age at onset ≥ 65 years). APOE has three alleles (ε2, ε3, and ε4) encoding APOE2, APOE3, and APOE4, respectively. APOE is a secreted glycoprotein consisting of 299 amino acids produced by astrocytes in the CNS and is associated with cholesterol transport. APOE2 and APOE3 can bind to Aβ and promote its clearance across the blood–brain barrier (BBB), while APOE4 has a relatively weak binding affinity for Aβ. The three APOE isoforms accelerate the deposition of Aβ, regulate the activity of tau-related kinases such as p35 and CDK5 through binding to receptors, and further regulate tau protein phosphorylation (Goate et al., 1991; Corder et al., 1993; Bu, 2009; Zhao et al., 2018).
MiRNAs have roles in APP degradation and Aβ metabolism through regulating the expression of related genes and associated pathways (Millan, 2017). APP degradation occurs mainly through the secretase pathway. APP is hydrolyzed by α-secretase to produce the neuroprotective soluble external functional fragments (sAPP), P3 and the AICD; in contrast, β-secretase hydrolyzes APP to produce the Aβ40 and Aβ42 forms. Aβ42 accumulates at a higher rate than Aβ40, thereby forming plaques and exerting neurotoxic effects. The γ-secretase enzyme is a key determinant of the Aβ40/Aβ42 ratio (Kaether et al., 2006; Cirrito et al., 2008; Haass et al., 2012; Roher et al., 2017).
MiRNAs can regulate the activities of key enzymes involved in APP lysis. Several miRNAs, including miR-339-5p, miR-29c, miR-15b, miR-195, and miR-124, participates in Aβ metabolism by modulating the activity of β-secretases such as BACE1 (Das et al., 2016; Selkoe and Hardy, 2016). Downregulation of miR-339-5p results in increased expression of BACE1, thus promoting Aβ deposition (Long et al., 2014). Moreover, both miR-29c and miR-135b negatively regulate BACE1 expression and show neuroprotective effects (Zhang et al., 2016b). Overexpression of hippocampal miR-188-3p reduces BACE1, Aβ, and neuroinflammation levels in APP transgenic mice (Zhang et al., 2014). AD-related ADAM metallopeptidase domain 10 (ADAM10), a member of the ADAM family of α-secretases, hydrolyzes APP to produce non-pathogenic Aβ. MiR-221 is downregulated in AD, which increases ADAM10 content (Manzine et al., 2018). MiR-140-5p is a negative regulator of ADAM10 and its transcription factor SOX2, is activated by Aβ (Akhter et al., 2018). MiRNAs have complicated interactions. MiR-107 targets BACE1 and ADAM10 also regulate APP metabolism, which suggested that single miRNA can target different genes or pathways producing additive effects (Nelson and Wang, 2010; Augustin et al., 2012; Goodall et al., 2013). BACE1 which is regulated by at least 10 more species of miRNA like miR-29c, miR-107, and miR-339-5p mentioned above. These miRNAs are both downregulated in AD showing negative correlation with BACE1. In addition, miR-221 and miR-140-5p can regulate ADAM10 negatively. However, miR-221 are downregulated in AD while miR-140-5p are upregulated, thus playing different roles. PS1 is an important component of the γ-secretory proteolytic system, and the PS1/γ-secretase system protects neurons by regulating miR-212 and PEA15 (Huang et al., 2018). Aph-1 homolog A (APH1A), a major mammalian APH1 subtype and a subunit of the γ-secretase complex. Overexpression of APH1A increases γ-secretase complex activity and consequently the levels of Aβ. MiR-151 involves in the formation of long-term episodic memory in the hippocampus by reducing the protein level of its target, APH1A (Xu et al., 2019). Increased β-secretase levels and activity elevate the levels of the AICD, which then stimulates the expression of APP and BACE1, thereby providing more substrate and enzyme for the amyloidogenic pathway. The AICD generated from the amyloidogenic pathway can translocate to the nucleus and function as a transcriptional regulator. AICD/miR-663 directly downregulates the expression of FBXL18 and CDK6, which affects the growth and differentiation of neuronal cells (Konietzko, 2012; Shu et al., 2015).
MiRNAs also involves in Aβ metabolism. MiR-15/107 family members, including miR-103 and miR-107, are downregulated in AD hippocampi, and enhance the generation of Aβ and phosphorylation of APP. This increases the levels of CDK5R1/p35 and, consequently, activates cyclin-dependent kinase 5 (CDK5) and finally leads to deterioration (Jiao et al., 2016; Moncini et al., 2017). MiR-132 is significantly downregulated in the middle and late stages of AD, leading to the upregulation of inositol 1,4,5-trisphosphate 3-kinase B (ITPKB) and increased ERK1/2 and BACE1 activity in AD patients (Salta et al., 2016; Zhu Q. B. et al., 2016). MiR-330 exerts a negative regulatory effect on vav guanine nucleotide exchange factor 1 (VAV1) via the MAPK signaling pathway, which promotes Aβ generation in the AD brain (Zhou et al., 2018). Upregulation of miR-33 in AD reduces ATP-binding cassette transporter A1 (ABCA1) levels, which can regulate APOE lipidation and Aβ metabolism, thus enhancing Aβ levels (Kim et al., 2015). MiR-128 targets peroxisome proliferator-activated receptor gamma (PPARG), which promotes Aβ pathology (Muller et al., 2014; Geng et al., 2018). Some miRNAs play neuroprotective roles by reducing the secretion and toxicity of Aβ. MiR-153 reduces the expression of APP (Long et al., 2012), while miR-200b/c and miR-302 inhibit PTEN to activate Akt via the PI3K/mTOR pathway and downstream Nanog signaling, thus alleviating Aβ-induced neurotoxicity (Li et al., 2016; Wu et al., 2016; Higaki et al., 2018).
In turn, aberrantly high levels of Aβ can affect the expression of miRNAs. Overexpression of APP inhibits miR-107 (Moncini et al., 2017), and Aβ42 oligomerization can reduce the expression of miR-188-5p in hippocampal neurons (Lee et al., 2016). When cortical neurons of APP/PS1 mice are exposed to Aβ, the levels of miR-34a initially increases, and then subsequently decreases (after 48 h) (Modi et al., 2016).
miRNAs Contribute to Abnormal Tau Protein Function
Tau is the most abundantly expressed microtubule-associated protein in neurons of the cerebral cortex, hippocampus, and axons of peripheral nerves in the human brain. There are six tau isomers in the human brain and are derived from exons 2, 3, and 10 by selective splicing (Goedert et al., 1992). Tau phosphorylation is important for its normal physiological functions, such as stabilizing the cytoskeleton, maintaining cell morphology, and ensuring intracellular transport; it also plays an important role in maintaining the protein composition of the PSD in dendritic spines in healthy neurons (Crimins et al., 2013; Iqbal et al., 2016). Aberrantly phosphorylated tau shows reduced binding affinity for microtubules, leading to tau aggregation and neurofibrillary tangle (NTF) formation, and also competitively binds to other normal microtubule-related proteins; this leads to the loss of the dynamic balance between microtubule assembly and disassembly, affects axonal transport and cell function, and results in neuronal degeneration (Sanabria-Castro et al., 2017). A variety of mechanisms, including gene mutation and an imbalance in tau protein-mediated regulation of enzyme function, lead to abnormal tau protein phosphorylation (Alonso et al., 2018; Davila-Bouziguet et al., 2019; Penke et al., 2019).
MiRNAs can not only directly affect tau protein synthesis, such as miR-219 directly targeting MAPT (Santa-Maria et al., 2015), but can also affect tau phosphorylation by regulating the activity of the relevant enzymes. GSK3, PKA, and CDK5 protein kinases can reveal or hide phosphorylation sites to synergistically adjust tau protein phosphorylation, primarily on serine and threonine residues, while phosphatases such as PP2A, PP2B, and PP1 dephosphorylate tau protein at multiple sites to varying degrees (Lee et al., 2011). CDK5, a proline-directed serine/threonine kinase, can regulate tau phosphorylation. P35/P25 are activators of CDK5, and calpain (CAPN)-induced cleavage of p35, which generates p25, gives rise to the aberrant activation of CDK5 and promotes tau hyperphosphorylation (Lopes and Agostinho, 2011). MiR-124-3p, which reduces in AD, inhibits the translation of CAPN1 mRNA, prevents the conversion of p35 to p25 and the subsequent formation of the p25/CDK5 complex, and reduces abnormal tau phosphorylation (Zhou et al., 2019). Overexpression of miR-125b leads to the upregulation of the p35, CDK5, and p44/42 MAPK (Erk1/2) signaling pathways, while the phosphatases DUSP6 and PPP1CA and the antiapoptotic factor Bcl-W are downregulated as direct targets of miR-125b, which promotes tau hyperphosphorylation (Banzhaf-Strathmann et al., 2014; Ma et al., 2017). GSK3 is also an important kinase for tau protein phosphorylation, and miR-219-5p downregulates GSK3 to inhibit tau phosphorylation in AD (Li et al., 2019).
Additional mechanisms are reported via which miRNAs can affect tau phosphorylation. MiR-132/212 plays an important role in memory formation and maintenance (Hernandez-Rapp et al., 2015). Its downregulation affects the balance of S-nitrosylation and induces tau phosphorylation and aggregation in a NOS1-dependent manner in vivo (Pichler et al., 2017; Salta and De Strooper, 2017; Wang et al., 2017). MiR-322, a rodent homolog of human miR-424, promotes tau phosphorylation by negatively regulating brain-derived neurotrophic factor (BDNF)-TrkB receptor activation (Zhang et al., 2018). MiR-146a upregulation results in tau hyperphosphorylation in neurons through modulation of the ROCK1/PTEN signaling pathway (Wang et al., 2016). MiR-138 overexpression induces tau hyperphosphorylation by targeting the RARA/GSK3β pathway, increasing tau phosphorylation at Thr231, Ser396, and Ser404 (Wang X. et al., 2015). MiR-106b inhibits Aβ42-induced tau phosphorylation at Tyr18 by targeting Fyn (Liu et al., 2016), while miR-137 exerts inhibitory actions on tau phosphorylation by suppressing CACNA1C expression (Jiang et al., 2018). Changes in tau protein levels also affect miRNA expression. Tau accumulation increases miR-92a levels in AD, thereby inducing anxiety through the miR-92a/vGAT/GABA signal in the mouse (Li et al., 2017).
miRNAs Regulate Synaptic Plasticity
Synapse formation is the basis of neural signal transduction, while synaptic plasticity is the basis of learning and memory. Memory impairment in AD patients is recognized to be a result of abnormal synaptic plasticity. Overexpression of miR-34c in hippocampal neurons influences AD pathogenesis by negatively regulating dendritic length and spine density (Kao et al., 2018). An increase in the level of soluble Aβ enhances glutamate release and excitatory toxicity (Edwards, 2019). Synapses are vulnerable to Aβ-induced neurotoxicity, and miRNAs regulates Aβ-mediated synaptic toxicity and plasticity. The CAMKK2/AMPK/Tau pathway is a key mediator of Aβ42 oligomer-mediated synaptic toxicity (Mairet-Coello et al., 2013). MiR-431 protects synapses and neurites from Aβ-induced toxic effects via the Wnt/β-catenin signaling pathway (Ross et al., 2018). MiR-188-5p can also alleviate Aβ42-mediated synaptic damage and dysfunction (Lee et al., 2016). The N-methyl-D-aspartate receptor (NMDAR) is an ion channel protein localized in the postsynaptic membrane and an important “molecular switch” for learning and memory. The excitation of NMDAR leads to a continuous increase in Ca2+ concentration, impairment of long-term potentiation (LTP), toxic damage, and loss of synapses. NMDA receptors drive glutamate-induced neuroexcitotoxicity, and a variety of factors, including p38 kinase, contribute to Aβ-mediated neurotoxicity (Hardingham and Bading, 2010). BDNF also contributes to the regulation of synaptic function. Inhibiting miR-132 reduces the increase in BDNF-dependent postsynaptic protein expression. MiR-132/212 family members play important roles in neural function and synaptic plasticity, and are continuously downregulated in early AD (Kawashima et al., 2010; Pichler et al., 2017). A potential target of miR-132 in cholinergic neurons may be a regulator of cholinergic transmission and synaptic plasticity, which may indirectly promote Aβ42 production and lead to cholinergic neurodegeneration (Zhu L. et al., 2016). MiR-200c is also downregulated in the frontal and temporal lobes of AD brains, exerting a protective effect against endoplasmic reticulum stress (ERS)-induced loss of cholinergic neurons (Wu et al., 2016).
miRNAs Participate in Neuronal Growth, Differentiation, and Apoptosis
Neural stem cells (NSCs) are characterized by their capacity to proliferate and differentiate into multiple neuronal cell types, including neurons, astrocytes, and oligodendrocytes. How to promote the differentiation of NSCs into neurons as a means of replacement therapy in AD is currently the subject of intensive research efforts. MiRNAs modulate the growth, development, maturation, and differentiation of neurons to varying degrees. MiR-142a-5p, miR-146a-5p, miR-155-5p, and miR-455-5p are upregulated in the AD brain and regulate neuronal function, and may also have a role in brain development and neurodegeneration (Arena et al., 2017; Sierksma et al., 2018). MiR-135a targets the thrombospondin 1 (THBS1) 3′ UTR, thereby promoting angiogenesis (Ko et al., 2015). MiR-511 increases neuronal differentiation and development as a functional regulator of FKBP5 in primary neurons (Zheng et al., 2016). MiR-302/367 induces the reprogramming of reactive astrocytes to neurons, contributing to neural repair, and may represent a potential therapeutic strategy to restore learning and memory (Ghasemi-Kasman et al., 2018).
In the AD brain, miRNAs play a two-way regulatory role in apoptosis, one of the main causes of neuronal loss in AD patients. On the one hand, miRNAs can promote apoptosis. For example, the downregulation of miR-512 in AD leads to an imbalance in proapoptotic/antiapoptotic factors, thereby promoting apoptosis and further deterioration (Mezache et al., 2015). Aβ also induces the downregulation of miR-34a, enhances the expression of cyclin-D1, and promotes the neuronal cell cycle through the MEK/ERK signaling pathway, which leads to apoptosis (Modi et al., 2016). MiR-146a inhibits LRP2 translation, which also leads to cell apoptosis (Zhang B. et al., 2016). On the other hand, miRNAs can also inhibit apoptosis. MiR-19 is a key component of the miR-17-92 cluster and inhibits aluminum-induced neuronal apoptosis (Zhu M. et al., 2016). MiR-214-3p negatively regulates the expression of ATG12 by targeting its 3′ UTR, inhibits autophagy, and reduces the levels of apoptosis in hippocampal neurons (Zhang et al., 2016a). Increased expression of miR-4487 decreases Aβ-induced apoptosis in neurons (Hu et al., 2018). Furthermore, miR-98 reduces Aβ production, inhibits the Notch signaling pathway, and suppresses the apoptosis of hippocampal neurons, thereby promoting their survival (Chen et al., 2019). MiR-124-3p attenuates tau phosphorylation-induced neuronal apoptosis by targeting the caveolin-1/PI3K/Akt/Gsk3β pathway (Kang et al., 2017). MiR-125b regulates inflammatory factors and oxidative stress through SphK1, thereby mediating the growth and apoptosis of neuronal cells (Jin et al., 2018). MiR-603 is an intronic miRNA of KIAA1217, a gene that is highly expressed in the human brain and elicits protective effects on neuroanl cells (Zhang C. et al., 2016).
miRNAs Mediate Immuno-Inflammatory Responses in AD
Immuno-inflammation is one of the pathological hallmarks of AD. Microglia and astrocytes both participate in the physiological function of central neuritis. Microglia, an immune effector cell in the brain, protects neurons from neuronal loss by eliminating harmful substances, but also exerts toxic effects on neurons through the activity of proinflammatory factors. Microglia neuroinflammation may act as an early trigger or as a sustained vulnerability factor that aggravates pathophysiological processes driving AD, leading to neurol loss. Aβ and oxidative stress can activate microglia and astrocytes, leading to Ca2+ influx and mitochondrial damage in synapses, followed by neurodegeneration. However, microglia effectively clears damaged synapses to prevent further extensive axonal damage (Kuchibhotla et al., 2008), and also releases cytokines and chemokines through a process known as “synaptic pruning,” assists and guides the process of neuronal differentiation, and mitigates Aβ-mediated toxic damage (Nayak et al., 2014; Salter and Beggs, 2014).
Numerous SNPs and rare coding variants in immune-related genes thought to be involved in microglial function have been identified as risk factors for AD in whole-genome sequencing and GWAS analyses, including TREM2, CR1, SHIP1, BIN1, CD33, PICALM, CLU, and the MS4A gene cluster (Gibson, 2010; Rosenberg et al., 2016; Bis et al., 2018). TLR-associated gene polymorphisms have been linked with susceptibility to LOAD (Sohrabifar et al., 2015). MiRNAs may activate TLRs and play a role in neuroinflammation under certain conditions (Bryniarski et al., 2015). TREM2 is an immunoglobulin superfamily receptor found in microglia, and mutations in the TREM2 gene increase the risk of LOAD (Cheng-Hathaway et al., 2018; Parhizkar et al., 2019). MiR-34a targets 299 nucleotides of the 3′ UTR of the TREM2 mRNA, resulting in the downregulation of TREM2 and microglia phagocytosis (Bhattacharjee et al., 2016). Knocking out TREM2 reduces neuroinflammation in AD mice (Leyns et al., 2017).
MiRNAs can play both protective and pathogenic roles by influencing neuroinflammatory responses through inflammation-associated cytokines. MiR-9, miR-34a, and miR-155 exhibit an anti-inflammatory effect through the modulation of downstream targets of proinflammatory mediators in the brain, including TNF receptor-associated factor 6 (TRAF6) and interleukin 1 receptor-associated kinase 1 (IL1R-AK1). Complement factor H (CFH) is a suppressor of the inflammatory response, and miR-125, miR-146a, and miR-155 enhance harmful CFH-induced proinflammatory events in AD, which may be associated with oligomeric Aβ induced inflammatory responses (Millan, 2017). MiR-139 has a negative regulatory effect on responses to proinflammatory stimuli, and prevent AD progression through the regulation of cannabinoid receptor type 2 (CB2)-mediated neuroinflammation (Tang et al., 2017). Prostaglandin E2 (PGE2) is a key mediator of the inflammatory response. PGE2 regulates CCAAT/enhancer-binding protein delta (CEBPD) in astrocytes through the EP4 receptor and protein kinase A, and CEBPD activation is associated with AD. Following PGE2 treatment, CEBPD induces miR-135a activation in astrocytes to inhibit THBS, suggesting that the CEBPD/miR135a/THBS1 axis may be a therapeutic target for the treatment of AD (Ko et al., 2015).
Spatial and Temporal Expression of MiRNAs in AD
As outlined above, miRNAs participate in the onset and pathological progression of AD. The expression levels of miRNAs show spatial and temporal differences in AD patients. MiRNAs like miR-9, miR-124, miR-125b, and miR-132, are expressed specifically in the CNS (Millan, 2017), and their dysregulation is associated with neurodegenerative diseases, such as AD.
MiRNA expression differs between gray matter and white matter in AD, although relatively few miRNAs are specifically altered in the white matter of AD brains. For example, the levels of miR-132 and miR-212 in AD brain reduce to different degrees in gray matter and white matter and more prominent in gray matter compared with healthy subjects (Pichler et al., 2017). Different miRNA species have different physiological functions, and their expression and distribution in AD also differ. The level of miR-107 in the hippocampus and temporal lobe decreases, while that of miR-146a increases during AD (Millan, 2017). Moreover, even the same miRNA shows different expression patterns between cerebral regions. MiR-29c is upregulated in the hippocampus of mice in the early stages of AD, but is significantly downregulated in the cortex (Zong et al., 2015). The difference can be partly explained by different cell composition and functions, as well as the properties of the different types of miRNA and differences in research methods (Smith et al., 2015; Salta et al., 2016). Gray matter and white matter are important components of the CNS. Gray matter comprises mostly neurons, astrocytes, endothelial cells, microglia, and relatively few oligodendrocytes, while white matter functions mainly in conduction (Wang et al., 2010). The cerebral cortex, closely related to learning and memory, is composed of gray matter. Moreover, the neural-specific pathological changes in AD, including amyloid plaques and NFTs, are primarily found in gray matter (Cech and Steitz, 2014). The pathological changes occurring in AD show temporal continuity, beginning in the entorhinal cortex at the base of hippocampus, and subsequently spreading to frontal lobe, temporal lobe, and occipital cortex with continued disease development, leading to impaired learning and memory function, as well as personality changes. This may also underlie the spatial and temporal changes of miRNA profiles. One example is that hippocampal MiR-128 elevates in the middle stage of AD, whereas decreases in the late stage (Muller et al., 2014; Geng et al., 2018). The expression of miR-132 in the nucleus basalis of Meynert is fairly stable in the early stage of AD, but is significantly downregulated in late stage (Zhu Q. B. et al., 2016). MiR-212 expression is similar to that of miR-132 (Pichler et al., 2017).
miRNAs Help With the Diagnosis of AD
AD is a pathophysiological continuum and can be divided into three stages according to clinical and pathological changes: the early preclinical stage, mild cognitive impairment (MCI), and subsequent dementia. Two commonly applied diagnostic criteria, NIA-AA and IWG-2, both recommend the application of a variety of biomarkers and methods for the stratification, classification, and differential diagnosis of AD. Most biomarkers and methods focus on the late stage of the disease, and can be summarized as follows: (1) Neuropsychological tests: Cognitive assessments such as the Mini-Mental State Examination (MMSE) can be used for early diagnosis to quantitatively assess the severity of cognitive impairment and record cognitive changes over time; however, this method depends on factors such as the patient’s education level and familiarity with the test, which limits its specificity and sensitivity. (2) Neuroimaging examination: Magnetic resonance imaging (MRI) and fluorodeoxyglucose (FDG)–positron emission tomography (PET) can be used to observe the pathological changes and functional abnormalities that can occur without obvious cognitive impairment, including medial temporal lobe atrophy and metabolic abnormalities. Although this method can be practical, it has important limitations in terms of time and cost. (3) Other biomarkers: Aβ1–40, Aβ1–42, total tau (t-tau), and phosphorylated tau (p-tau) proteins in the cerebrospinal fluid (CSF) are currently the best biomarkers for clinical research and the monitoring of AD. However, CSF acquisition requires a lumbar puncture, which is invasive and not easily accepted by the patients. Additionally, the detection of disease-causing genes also can be used. AD diagnosis still lacks efficient, simple, and inexpensive biomarkers, especially for the early stages of the disease.
In this respect, miRNAs have several advantages over classical biomarkers. Several studies have shown that specific species of miRNAs detected in the biofluid of AD patients are consistent with the observed pathological changes (Keller et al., 2016; Swarbrick et al., 2019; Takousis et al., 2019; Wiedrick et al., 2019). MiRNAs in serum, plasma, or CSF show great stability when they are enwrapped in liposomes or bound to lipoproteins, which prevents their degradation and allows them to withstand severe environmental conditions (van den Berg et al., 2020). Moreover, miRNAs can be easily obtained and quantified using real-time PCR, next-generation sequencing (NGS), or microarray. Some of the findings are summarized in Table 2.
The main potential applications are as follows: (1) A biomarker for the diagnosis of AD. A systematic review and meta-analysis of 10 studies comprising 770 AD patients and 664 normal controls indicated that miRNAs display excellent diagnostic performance, showing an overall sensitivity of 0.80 (95% CI: 0.75–0.83), a specificity of 0.83 (95% CI: 0.78–0.87), and a diagnostic odds ratio of 14 (95% CI: 11–19) (Zhang et al., 2019). Serum miRNA biomarkers related to AD prognosis show consistency with neuropsychological and neuroimaging assessments, and plasma levels of miR-34a-5p and miR-545-3p have potential as biomarkers in early AD; however, further large-scale research is still needed to confirm this (Cheng et al., 2015; Cosin-Tomas et al., 2017). Serum miR-133b levels in AD patients are positively correlated with the simple intelligence status test score. The area under the ROC curve of miR-133b in the diagnosis of AD was 0.907, with 1.7 as the critical value, with a sensitivity of 90.8% and a specificity of 74.3% (Yang et al., 2019). There was a significant positive correlation between the serum level of miR-193a-3p and the MMSE score in AD patients (Cao et al., 2020). Although some studies showed connection between miRNAs and cognition tests, there are inadequate studies directly focusing the combination of miRNAs and MMSE, as well as other clinical diagnosis including CSF Aβ and tau, PET imaging. MiR-193a-3p has potential for use as a new biomarker to distinguish AD patients from healthy people. The serum concentrations of miR-222, miR-29c-3p, and miR-19b-3p also have potential as biomarkers for AD (Wu et al., 2017; Zeng et al., 2017), as do miR-455-3p, miR-29a, miR-107, miR-106a-5p, and miR-324-3p (Wang T. et al., 2015; Muller et al., 2016; Yilmaz et al., 2016; Cai et al., 2018; Kumar and Reddy, 2018). Combining between two and four miRNAs can distinguish AD from controls with an accuracy of 75–82% (Lusardi et al., 2017). Discrimination analysis using a combination of miR-100, miR-103, and miR-375 could detect AD in CSF by positively classifying controls and AD cases with 96.4 and 95.5% accuracy, respectively (Denk et al., 2015). The combination of serum miR-223 and miR-125b levels provided improved sensitivity/specificity for AD prediction than either miRNA alone (Jia and Liu, 2016). A 54 months study found that an AD-specific 16-miRNA signature can predict AD with a sensitivity and specificity of 87 and 77%, respectively. Each participant were assessed by cognitive assessments and Aβ neuroimaging during this study, and those AD participants with normal clinical manifestations diagnosed by Aβ neuroimaging suggested a higher risk of progression toward AD (Cheng et al., 2015). Changes in plasma APOE, miR-107, and miR-650 levels may be a marker of neurodegeneration during AD associated with amyloid metabolism and cell cycle disorders (Prendecki et al., 2019). (2) A predictor of the conversion from mild cognitive impairment (MCI) to AD. Approximately 10–15% of MCI patients enter the dementia stage each year, and amnestic MCI (aMCI) patients may have a higher risk of developing AD (Giau et al., 2019). A 5-year follow-up study showed that an increased serum level of miR-206 may be a potential predictor of aMCI-to-AD conversion. There was a positive correlation between serum miR-206 levels and the rate of progression from aMCI to AD (Kenny et al., 2019). However, these results need to be confirmed in more studies. Bioinformatic analysis indicated that the serum levels of miR-519d-3p could be the bridge regulator between MCI and AD; however, this requires further verification (Tao et al., 2020). Plasma miR-92a-3P, miR-181c-5p, and miR-210-3p levels are significantly upregulated in MCI and AD patients. Patients with MCI progressing to AD had higher plasma levels of these miRNAs (Siedlecki-Wullich et al., 2019). MiR-135a and miR-384 levels were increased and miR-193b levels were decreased in patients with AD and MCI, while the combination of these three miRNAs could predict the risk of MCI onset and conversion to AD (Giau et al., 2019). However, no study has evaluated the diagnostic performance of differentially expressed miRNAs between MCI/AD patients and healthy controls (Pena-Bautista et al., 2019). Indeed, the lack of recognized and reliable reference genes in the analysis of miRNAs in patients with MCI seriously hinders the analysis and limits research on circulating miRNAs (Piscopo et al., 2019). (3) A tool to differentiate AD from other neurodegenerative diseases. The expression levels of exosomal miRNA-384 in the serum of AD and non-AD patients differ significantly. In addition, the serum level of exosomal miR-384 has potent differential diagnostic ability for AD and Parkinson’s disease dementia (PDD), as well as for AD and vascular dementia (VaD), with sensitivity/specificity indices of 97.2%/100% and 99.1%/100%, respectively (Yang et al., 2018). Using this 12-miRNA signature, the differentiation of AD from other neurological diseases is possible with accuracies of between 74 and 78%. The differentiation of the other CNS disorders from controls yields even higher accuracies (Leidinger et al., 2013). Gender differences have been found in the analysis of plasma miRNA in patients with frontotemporal dementia (FTD) (Grasso et al., 2019), but not in those with AD. Indeed, miRNA biomarkers show considerable inconsistencies between studies, and results are hard to reproduce. Studies with the same sample source and subjects of similar demographic background can generate different or even contradictory results (Jain et al., 2019; Pena-Bautista et al., 2019). This can be explained by the different procedures and methods of sample collection, miRNA detection, and data analyses. Differences in miRNA isolation procedures, cell contamination and hemolysis, quantitative methods, reference genes, and sample quality control can all affect the final results (Piscopo et al., 2019). For example, platelets are rich in miRNAs and release large amounts of miRNAs into the circulation during the coagulation process, leading to differences between serum and plasma miRNA concentrations (Wang et al., 2012). Additionally, miRNAs are more stable in platelets than in the corresponding plasma and serum samples, as indicated by the higher miRNA concentration in platelets (Lee et al., 2020). Similarly, CSF contamination with blood cells is a major confounding factor when analyzing CSF-derived miRNAs. In such a scenario, the analysis of cell-free CSF-derived exosomes could be superior to total CSF analysis and may also explain some of the discrepancies among the results (Müller et al., 2016). In addition, miRNA profiles vary with the biofluid or the exosome. A major reason for this variation is the selective transportation of miRNAs. Specifically, mature miRNAs can be actively sorted into microvesicles (MVs) by specific proteins (membrane-localized proteins, RNA-binding proteins) and then released into the biofluid (Li et al., 2018; Groot and Lee, 2020). Neuron-derived exosomes can cross the blood–brain barrier (BBB) and transfer their cargo to the CSF (Barbagallo et al., 2020). MiRNAs are also passively released from apoptotic bodies or platelets during coagulation. Biomarkers in exosomes are suggested to have higher diagnostic efficiency and be of better quality than those in biofluid (Nie et al., 2020). Notably, different approaches used for exosome purification may also lead to differences in results (Lee et al., 2019).
Summary and Outlook
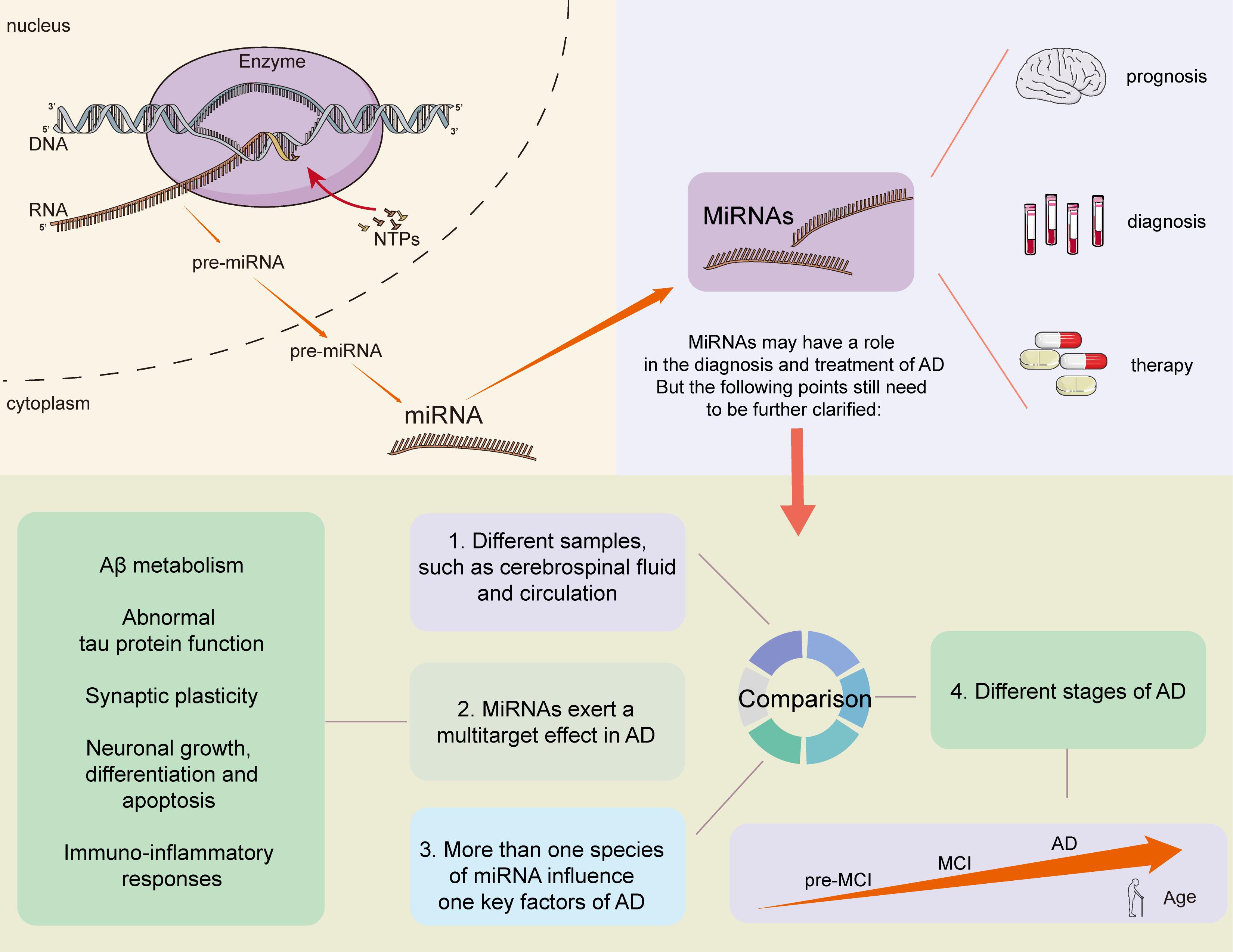
MiRNAs display wide distribution patterns throughout the CNS. They mainly interact with non-coding sequences of target messenger RNAs, and play important regulatory roles in the development, maturation, differentiation, and gene expression of neuronal cells. MiRNAs with different physiological functions are differentially expressed between brain regions, and influence various aspects of AD pathogenesis through different pathways. MiRNAs can collectively exert more pronounced effects. Several studies have noted that one miRNA acts on hundreds of targets, while multiple miRNAs also coordinate to act on the same mRNA sequence, resulting in an intricate network (Ameres and Zamore, 2013; Barry, 2014). These data suggest that, although the expression of individual miRNAs may have specific effects, the overall effect of miRNAs will not be fully understood until the global miRNA expression patterns in the brain have been elucidated. How to identify simple and effective pathways in this complex network, and then guide the theory and practice, is a future subject requiring intensive investigation. Although the mechanisms underlying the effects of miRNA dysregulation in AD are increasingly identified, research is still in the early stages. Most studies are relatively scattered. The breadth and depth of related studies need to be expanded to further screen the key mechanisms involving in the interactions between miRNAs and AD to provide new insights for the study of pathogenesis, to identify effective indicators and targets for diagnosis, and to administrator as a cognition-improving treatment.
In this review, we summarized existing evidence about miRNAs serving as diagnostic biomarkers in AD. The use of miRNAs as AD biomarkers still faces many challenges, even though a substantial number of miRNAs have been identified that have relatively high efficiency, specificity, and sensitivity for diagnosing AD. The following points need to be further clarified: (1) whether the changes in miRNA content in different brain regions, cerebrospinal fluid, and serum are related and whether the changes are AD-specific; (2) some miRNAs may show opposing trends during different stages of AD, and how to effectively divide the boundaries remains a major challenge; (3) because miRNAs exert a multitarget effect, and many key factors of AD are influenced by more than one species of miRNA: this is exemplified by BACE1 which is regulated by at least 10 more species of miRNA. Hence, it is important to effectively identify the individual roles of specific miRNAs, as well as the collective role of multiple miRNAs in AD. In addition, procedures for sample collection, miRNA detection, and data handling need to be standardized to increase the repeatability of results. Finally, the use of a combination of multiple miRNAs as markers, or combining miRNAs with other biofluid biomarkers, may perform better in the diagnosis, differentiation, and prediction of AD. Large sample trials are required to reach a robust conclusion (Figure 2).
Author Contributions
WW wrote the manuscript. Z-YW, L-NM, and T-TZ assisted in the manuscript writing. YC and HL assisted in ideas and modification of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Science and Technology Major Project for “Essential new drug research and development” (No. 2019ZX09301114) and National Projects for Leading Professionals in the Traditional Chinese Medicine (Qi Huang Scholar, No. 02045003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National Science and Technology Major Project and National Projects for Leading Professionals in Traditional Chinese Medicine for their financial support.
References
Akhter, R., Shao, Y., Shaw, M., Formica, S., Khrestian, M., Leverenz, J. B., et al. (2018). Regulation of ADAM10 by miR-140-5p and potential relevance for Alzheimer’s disease. Neurobiol. Aging 63, 110–119. doi: 10.1016/j.neurobiolaging.2017.11.007
Alonso, A. D., Cohen, L. S., Corbo, C., Morozova, V., ElIdrissi, A., Phillips, G., et al. (2018). Hyperphosphorylation of tau associates with changes in its function beyond microtubule stability. Front. Cell. Neurosci. 12:338. doi: 10.3389/fncel.2018.00338
Ameres, S. L., and Zamore, P. D. (2013). Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488. doi: 10.1038/nrm3611
Arena, A., Iyer, A. M., Milenkovic, I., Kovacs, G. G., Ferrer, I., Perluigi, M., et al. (2017). Developmental expression and dysregulation of miR-146a and miR-155 in down’s syndrome and mouse models of down’s syndrome and Alzheimer’s disease. Curr. Alzheimer Res. 14, 1305–1317. doi: 10.2174/1567205014666170706112701
Augustin, R., Endres, K., Reinhardt, S., Kuhn, P.-H., Lichtenthaler, S. F., Hansen, J., et al. (2012). Computational identification and experimental validation of microRNAs binding to the Alzheimer-related gene ADAM10. BMC Med. Genet. 13:35. doi: 10.1186/1471-2350-13-35
Banzhaf-Strathmann, J., Benito, E., May, S., Arzberger, T., Tahirovic, S., Kretzschmar, H., et al. (2014). MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 33, 1667–1680. doi: 10.15252/embj.201387576
Barbagallo, C., Mostile, G., Baglieri, G., Giunta, F., Luca, A., Raciti, L., et al. (2020). Specific signatures of serum miRNAs as potential biomarkers to discriminate clinically similar neurodegenerative and vascular-related diseases. Cell. Mol. Neurobiol. 40, 531–546. doi: 10.1007/s10571-019-00751-y
Barry, G. (2014). Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatry 19, 410–416. doi: 10.1038/mp.2013.196
Bhatnagar, S., Chertkow, H., Schipper, H. M., Yuan, Z., Shetty, V., Jenkins, S., et al. (2014). Increased microRNA-34c abundance in Alzheimer’s disease circulating blood plasma. Front. Mol. Neurosci. 7:2. doi: 10.3389/fnmol.2014.00002
Bhattacharjee, S., Zhao, Y., Dua, P., Rogaev, E. I., and Lukiw, W. J. (2016). microRNA-34a-mediated down-regulation of the microglial-enriched triggering receptor and phagocytosis-sensor TREM2 in age-related macular degeneration. PLoS One 11:e0150211. doi: 10.1371/journal.pone.0150211
Bis, J. C., Jian, X., Kunkle, B. W., Chen, Y., Hamilton-Nelson, K. L., Bush, W. S., et al. (2018). Whole exome sequencing study identifies novel rare and common Alzheimer’s-Associated variants involved in immune response and transcriptional regulation. Mol. Psychiatry 25, 1859–1875. doi: 10.1038/s41380-018-0112-7
Bryniarski, K., Ptak, W., Martin, E., Nazimek, K., Szczepanik, M., Sanak, M., et al. (2015). Free extracellular miRNA functionally targets cells by transfecting exosomes from their companion cells. PLoS One 10:e0122991. doi: 10.1371/journal.pone.0122991
Bu, G. (2009). Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 10, 333–344. doi: 10.1038/nrn2620
Cai, M., Wang, Y. W., Xu, S. H., Qiao, S., Shu, Q. F., Du, J. Z., et al. (2018). Regulatory effects of the long noncoding RNA RP11543N12.1 and microRNA3243p axis on the neuronal apoptosis induced by the inflammatory reactions of microglia. Int. J. Mol. Med. 42, 1741–1755. doi: 10.3892/ijmm.2018.3736
Campion, D., Pottier, C., Nicolas, G., Le Guennec, K., and Rovelet-Lecrux, A. (2016). Alzheimer disease: modeling an Abeta-centered biological network. Mol. Psychiatry 21, 861–871. doi: 10.1038/mp.2016.38
Cao, F., Liu, Z., and Sun, G. (2020). Diagnostic value of miR-193a-3p in Alzheimer’s disease and miR-193a-3p attenuates amyloid-beta induced neurotoxicity by targeting PTEN. Exp. Gerontol. 130:110814. doi: 10.1016/j.exger.2019.110814
Cech, T. R., and Steitz, J. A. (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94. doi: 10.1016/j.cell.2014.03.008
Cha, D. J., Mengel, D., Mustapic, M., Liu, W., Selkoe, D. J., Kapogiannis, D., et al. (2019). miR-212 and miR-132 Are Downregulated In Neurally Derived Plasma Exosomes Of Alzheimer’s patients. Front. Neurosci. 13:1208. doi: 10.3389/fnins.2019.01208
Chen, F. Z., Zhao, Y., and Chen, H. Z. (2019). MicroRNA-98 reduces amyloid beta-protein production and improves oxidative stress and mitochondrial dysfunction through the Notch signaling pathway via HEY2 in Alzheimer’s disease mice. Int. J. Mol. Med. 43, 91–102. doi: 10.3892/ijmm.2018.3957
Cheng, L., Doecke, J. D., Sharples, R. A., Villemagne, V. L., Fowler, C. J., Rembach, A., et al. (2015). Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 20, 1188–1196. doi: 10.1038/mp.2014.127
Cheng-Hathaway, P. J., Reed-Geaghan, E. G., Jay, T. R., Casali, B. T., Bemiller, S. M., Puntambekar, S. S., et al. (2018). The Trem2 R47H variant confers loss-of-function-like phenotypes in Alzheimer’s disease. Mol. Neurodegener. 13:29. doi: 10.1186/s13024-018-0262-8
Cirrito, J. R., Kang, J. E., Lee, J., Stewart, F. R., Verges, D. K., Silverio, L. M., et al. (2008). Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron 58, 42–51. doi: 10.1016/j.neuron.2008.02.003
Corder, E. H., Saunders, A. M., Strittmatter, W. J., Schmechel, D. E., Gaskell, P. C., Small, G. W., et al. (1993). Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923. doi: 10.1126/science.8346443
Cosin-Tomas, M., Antonell, A., Llado, A., Alcolea, D., Fortea, J., Ezquerra, M., et al. (2017). Plasma miR-34a-5p and miR-545-3p as early biomarkers of Alzheimer’s disease: potential and limitations. Mol. Neurobiol. 54, 5550–5562. doi: 10.1007/s12035-016-0088-8
Crimins, J. L., Pooler, A., Polydoro, M., Luebke, J. I., and Spires-Jones, T. L. (2013). The intersection of amyloid beta and tau in glutamatergic synaptic dysfunction and collapse in Alzheimer’s disease. Ageing Res. Rev. 12, 757–763. doi: 10.1016/j.arr.2013.03.002
Dangla-Valls, A., Molinuevo, J. L., Altirriba, J., Sanchez-Valle, R., Alcolea, D., Fortea, J., et al. (2017). CSF microRNA profiling in Alzheimer’s disease: a screening and validation study. Mol. Neurobiol. 54, 6647–6654. doi: 10.1007/s12035-016-0106-x
Das, S., Ferlito, M., Kent, O. A., Fox-Talbot, K., Wang, R., Liu, D., et al. (2012). Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 110, 1596–1603. doi: 10.1161/circresaha.112.267732
Das, U., Wang, L., Ganguly, A., Saikia, J. M., Wagner, S. L., Koo, E. H., et al. (2016). Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat. Neurosci. 19, 55–64. doi: 10.1038/nn.4188
Davila-Bouziguet, E., Targa-Fabra, G., Avila, J., Soriano, E., and Pascual, M. (2019). Differential accumulation of Tau phosphorylated at residues Thr231, Ser262 and Thr205 in hippocampal interneurons and its modulation by Tau mutations (VLW) and amyloid-beta peptide. Neurobiol. Dis. 125, 232–244. doi: 10.1016/j.nbd.2018.12.006
Denk, J., Boelmans, K., Siegismund, C., Lassner, D., Arlt, S., and Jahn, H. (2015). MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer‘s disease. PLoS One 10:e0126423. doi: 10.1371/journal.pone.0126423
Dragomir, M. P., Knutsen, E., and Calin, G. A. (2018). SnapShot: unconventional miRNA functions. Cell 174, 1038.e1–1038.e1. doi: 10.1016/j.cell.2018.07.040
Edwards, F. A. (2019). A Unifying Hypothesis for Alzheimer’s disease: from plaques to neurodegeneration. Trends Neurosci. 42, 310–322. doi: 10.1016/j.tins.2019.03.003
Eggert, S., Thomas, C., Kins, S., and Hermey, G. (2018). Trafficking in Alzheimer’s disease: modulation of APP transport and processing by the transmembrane proteins LRP1, SorLA, SorCS1c, sortilin, and calsyntenin. Mol. Neurobiol. 55, 5809–5829. doi: 10.1007/s12035-017-0806-x
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Geng, L., Zhang, T., Liu, W., and Chen, Y. (2018). Inhibition of miR-128 abates abeta-mediated cytotoxicity by targeting PPAR-gamma via NF-kappaB inactivation in primary mouse cortical neurons and Neuro2a cells. Yonsei Med. J. 59, 1096–1106. doi: 10.3349/ymj.2018.59.9.1096
Ghasemi-Kasman, M., Shojaei, A., Gol, M., Moghadamnia, A. A., Baharvand, H., and Javan, M. (2018). miR-302/367-induced neurons reduce behavioral impairment in an experimental model of Alzheimer’s disease. Mol. Cell. Neurosci. 86, 50–57. doi: 10.1016/j.mcn.2017.11.012
Giau, V. V., Bagyinszky, E., and An, S. S. A. (2019). Potential fluid biomarkers for the diagnosis of mild cognitive impairment. Int. J. Mol. Sci. 20:4149. doi: 10.3390/ijms20174149
Gibson, G. (2010). Hints of hidden heritability in GWAS. Nat. Genet. 42, 558–560. doi: 10.1038/ng0710-558
Glenner, G. G., and Wong, C. W. (1984). Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890. doi: 10.1016/s0006-291x(84)80190-4
Goate, A., Chartier-Harlin, M. C., Mullan, M., Brown, J., Crawford, F., Fidani, L., et al. (1991). Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349, 704–706. doi: 10.1038/349704a0
Goedert, M., Spillantini, M. G., Cairns, N. J., and Crowther, R. A. (1992). Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron 8, 159–168. doi: 10.1016/0896-6273(92)90117-v
Goodall, E. F., Heath, P. R., Bandmann, O., Kirby, J., and Shaw, P. J. (2013). Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front. Cell. Neurosci. 7:178. doi: 10.3389/fncel.2013.00178
Grasso, M., Piscopo, P., Talarico, G., Ricci, L., Crestini, A., Tosto, G., et al. (2019). Plasma microRNA profiling distinguishes patients with frontotemporal dementia from healthy subjects. Neurobiol. Aging 84, 240.e1–240.e12. doi: 10.1016/j.neurobiolaging.2019.01.024
Groot, M., and Lee, H. (2020). Sorting mechanisms for MicroRNAs into extracellular vesicles and their associated diseases. Cells 9:1044. doi: 10.3390/cells9041044
Guo, R., Fan, G., Zhang, J., Wu, C., Du, Y., Ye, H., et al. (2017). A 9-microRNA signature in serum serves as a noninvasive biomarker in early diagnosis of Alzheimer’s disease. J. Alzheimers Dis. 60, 1365–1377. doi: 10.3233/JAD-170343
Haass, C., Kaether, C., Thinakaran, G., and Sisodia, S. (2012). Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2:a006270. doi: 10.1101/cshperspect.a006270
Hajjri, S. N., Sadigh-Eteghad, S., Mehrpour, M., Moradi, F., Shanehbandi, D., and Mehdizadeh, M. (2020). Beta-amyloid-dependent mirnas as circulating biomarkers in Alzheimer’s disease: a preliminary report. J. Mol. Neurosci. 70, 871–877. doi: 10.1007/s12031-020-01511-0
Hara, N., Kikuchi, M., Miyashita, A., Hatsuta, H., Saito, Y., Kasuga, K., et al. (2017). Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 5:10. doi: 10.1186/s40478-017-0414-z
Hardingham, G. E., and Bading, H. (2010). Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat. Rev. Neurosci. 11, 682–696. doi: 10.1038/nrn2911
Hardy, J., and Selkoe, D. J. (2002). The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356. doi: 10.1126/science.1072994
Hardy, J. A., and Higgins, G. A. (1992). Alzheimer’s disease: the amyloid cascade hypothesis. Science 256, 184–185. doi: 10.1126/science.1566067
Hernandez-Rapp, J., Smith, P. Y., Filali, M., Goupil, C., Planel, E., Magill, S. T., et al. (2015). Memory formation and retention are affected in adult miR-132/212 knockout mice. Behav. Brain Res. 287, 15–26. doi: 10.1016/j.bbr.2015.03.032
Higaki, S., Muramatsu, M., Matsuda, A., Matsumoto, K., Satoh, J. I., Michikawa, M., et al. (2018). Defensive effect of microRNA-200b/c against amyloid-beta peptide-induced toxicity in Alzheimer’s disease models. PLoS One 13:e0196929. doi: 10.1371/journal.pone.0196929
Hu, L., Zhang, R., Yuan, Q., Gao, Y., Yang, M. Q., Zhang, C., et al. (2018). The emerging role of microRNA-4487/6845-3p in Alzheimer’s disease pathologies is induced by Abeta25-35 triggered in SH-SY5Y cell. BMC Syst. Biol. 12:119. doi: 10.1186/s12918-018-0633-3
Huang, Q., Voloudakis, G., Ren, Y., Yoon, Y., Zhang, E., Kajiwara, Y., et al. (2018). Presenilin1/gamma-secretase protects neurons from glucose deprivation-induced death by regulating miR-212 and PEA15. FASEB J. 32, 243–253. doi: 10.1096/fj.201700447RR
Ipsaro, J. J., and Joshua-Tor, L. (2015). From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 22, 20–28. doi: 10.1038/nsmb.2931
Iqbal, K., Liu, F., and Gong, C. X. (2016). Tau and neurodegenerative disease: the story so far. Nat. Rev. Neurol. 12, 15–27. doi: 10.1038/nrneurol.2015.225
Jain, G., Stuendl, A., Rao, P., Berulava, T., Pena Centeno, T., Kaurani, L., et al. (2019). A combined miRNA-piRNA signature to detect Alzheimer’s disease. Transl. Psychiatry 9:250. doi: 10.1038/s41398-019-0579-2
Jia, L. H., and Liu, Y. N. (2016). Downregulated serum miR-223 servers as biomarker in Alzheimer’s disease. Cell Biochem. Funct. 34, 233–237. doi: 10.1002/cbf.3184
Jiang, Y., Xu, B., Chen, J., Sui, Y., Ren, L., Li, J., et al. (2018). Micro-RNA-137 inhibits tau hyperphosphorylation in Alzheimer’s disease and targets the CACNA1C gene in transgenic mice and human neuroblastoma SH-SY5Y cells. Med. Sci. Monit. 24, 5635–5644. doi: 10.12659/msm.908765
Jiao, Y., Kong, L., Yao, Y., Li, S., Tao, Z., Yan, Y., et al. (2016). Osthole decreases beta amyloid levels through up-regulation of miR-107 in Alzheimer’s disease. Neuropharmacology 108, 332–344. doi: 10.1016/j.neuropharm.2016.04.046
Jin, Y., Tu, Q., and Liu, M. (2018). MicroRNA125b regulates Alzheimer’s disease through SphK1 regulation. Mol. Med. Rep. 18, 2373–2380. doi: 10.3892/mmr.2018.9156
Jinek, M., and Doudna, J. A. (2009). A three-dimensional view of the molecular machinery of RNA interference. Nature 457, 405–412. doi: 10.1038/nature07755
Kaether, C., Schmitt, S., Willem, M., and Haass, C. (2006). Amyloid precursor protein and Notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic 7, 408–415. doi: 10.1111/j.1600-0854.2006.00396.x
Kang, Q., Xiang, Y., Li, D., Liang, J., Zhang, X., Zhou, F., et al. (2017). MiR-124-3p attenuates hyperphosphorylation of Tau protein-induced apoptosis via caveolin-1-PI3K/Akt/GSK3beta pathway in N2a/APP695swe cells. Oncotarget 8, 24314–24326. doi: 10.18632/oncotarget.15149
Kao, Y. C., Wang, I. F., and Tsai, K. J. (2018). miRNA-34c overexpression causes dendritic loss and memory decline. Int. J. Mol. Sci. 19:2323. doi: 10.3390/ijms19082323
Kawashima, H., Numakawa, T., Kumamaru, E., Adachi, N., Mizuno, H., Ninomiya, M., et al. (2010). Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience 165, 1301–1311. doi: 10.1016/j.neuroscience.2009.11.057
Keller, A., Backes, C., Haas, J., Leidinger, P., Maetzler, W., Deuschle, C., et al. (2016). Validating Alzheimer’s disease micro RNAs using next-generation sequencing. Alzheimers Dement. 12, 565–576. doi: 10.1016/j.jalz.2015.12.012
Kenny, A., McArdle, H., Calero, M., Rabano, A., Madden, S. F., Adamson, K., et al. (2019). Elevated plasma microRNA-206 levels predict cognitive decline and progression to dementia from mild cognitive impairment. Biomolecules 9:734. doi: 10.3390/biom9110734
Kiko, T., Nakagawa, K., Tsuduki, T., Furukawa, K., Arai, H., and Miyazawa, T. (2014). MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J. Alzheimers Dis. 39, 253–259. doi: 10.3233/JAD-130932
Kim, J., Yoon, H., Horie, T., Burchett, J. M., Restivo, J. L., Rotllan, N., et al. (2015). microRNA-33 regulates ApoE lipidation and amyloid-beta metabolism in the brain. J. Neurosci. 35, 14717–14726. doi: 10.1523/jneurosci.2053-15.2015
Ko, C. Y., Chu, Y. Y., Narumiya, S., Chi, J. Y., Furuyashiki, T., Aoki, T., et al. (2015). CCAAT/enhancer-binding protein delta/miR135a/thrombospondin 1 axis mediates PGE2-induced angiogenesis in Alzheimer’s disease. Neurobiol. Aging 36, 1356–1368. doi: 10.1016/j.neurobiolaging.2014.11.020
Konietzko, U. (2012). AICD nuclear signaling and its possible contribution to Alzheimer’s disease. Curr. Alzheimer Res. 9, 200–216. doi: 10.2174/156720512799361673
Kuchibhotla, K. V., Goldman, S. T., Lattarulo, C. R., Wu, H. Y., Hyman, B. T., and Bacskai, B. J. (2008). Abeta plaques lead to aberrant regulation of calcium homeostasis in vivo resulting in structural and functional disruption of neuronal networks. Neuron 59, 214–225. doi: 10.1016/j.neuron.2008.06.008
Kumar, S., and Reddy, P. H. (2018). MicroRNA-455-3p as a potential biomarker for Alzheimer’s disease: an update. Front. Aging Neurosci. 10:41. doi: 10.3389/fnagi.2018.00041
Kumar, S., and Reddy, P. H. (2019). A new discovery of MicroRNA-455-3p in Alzheimer’s disease. J. Alzheimers Dis. 72, S117–S130. doi: 10.3233/JAD-190583
Kumar, S., Vijayan, M., and Reddy, P. H. (2017). MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum. Mol. Genet. 26, 3808–3822. doi: 10.1093/hmg/ddx267
Kumar-Singh, S., Theuns, J., Van Broeck, B., Pirici, D., Vennekens, K., Corsmit, E., et al. (2006). Mean age-of-onset of familial alzheimer disease caused by presenilin mutations correlates with both increased Abeta42 and decreased Abeta40. Hum. Mutat. 27, 686–695. doi: 10.1002/humu.20336
Lane, C. A., Hardy, J., and Schott, J. M. (2018). Alzheimer’s disease. Eur. J. Neurol. 25, 59–70. doi: 10.1111/ene.13439
Lauressergues, D., Couzigou, J. M., Clemente, H. S., Martinez, Y., Dunand, C., Bécard, G., et al. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature 520, 90–93. doi: 10.1038/nature14346
Lee, B. K., Kim, M. H., Lee, S. Y., Son, S. J., Hong, C. H., and Jung, Y. S. (2020). Downregulated platelet miR-1233-5p in patients with Alzheimer’s pathologic change with mild cognitive impairment is associated with abeta-induced platelet activation via P-Selectin. J. Clin. Med. 9:1624. doi: 10.3390/jcm9061642
Lee, K., Kim, H., An, K., Kwon, O. B., Park, S., Cha, J. H., et al. (2016). Replenishment of microRNA-188-5p restores the synaptic and cognitive deficits in 5XFAD mouse model of Alzheimer’s disease. Sci. Rep. 6, 34433. doi: 10.1038/srep34433
Lee, S., Hall, G. F., and Shea, T. B. (2011). Potentiation of tau aggregation by cdk5 and GSK3beta. J. Alzheimers Dis. 26, 355–364. doi: 10.3233/jad-2011-102016
Lee, S., Mankhong, S., and Kang, J. H. (2019). Extracellular vesicle as a source of Alzheimer’s biomarkers: opportunities and challenges. Int. J. Mol. Sci. 20:1728. doi: 10.3390/ijms20071728
Lehmann, S. M., Kruger, C., Park, B., Derkow, K., Rosenberger, K., Baumgart, J., et al. (2012). An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835. doi: 10.1038/nn.3113
Leidinger, P., Backes, C., Deutscher, S., Schmitt, K., Mueller, S. C., Frese, K., et al. (2013). A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 14:R78. doi: 10.1186/gb-2013-14-7-r78
Leyns, C. E. G., Ulrich, J. D., Finn, M. B., Stewart, F. R., Koscal, L. J., Remolina Serrano, J., et al. (2017). TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc. Natl. Acad. Sci. U.S.A. 114, 11524–11529. doi: 10.1073/pnas.1710311114
Li, D., Li, Y. P., Li, Y. X., Zhu, X. H., Du, X. G., Zhou, M., et al. (2018). Effect of regulatory network of exosomes and microRNAs on neurodegenerative diseases. Chin. Med. J. 131, 2216–2225. doi: 10.4103/0366-6999.240817
Li, H. H., Lin, S. L., Huang, C. N., Lu, F. J., Chiu, P. Y., Huang, W. N., et al. (2016). miR-302 attenuates amyloid-beta-induced neurotoxicity through activation of Akt signaling. J. Alzheimers Dis. 50, 1083–1098. doi: 10.3233/jad-150741
Li, J., Chen, W., Yi, Y., and Tong, Q. (2019). miR-219-5p inhibits tau phosphorylation by targeting TTBK1 and GSK-3beta in Alzheimer’s disease. J. Cell. Biochem. 120, 9936–9946. doi: 10.1002/jcb.28276
Li, X., Wang, Z., Tan, L., Wang, Y., Lu, C., Chen, R., et al. (2017). Correcting miR92a-vGAT-mediated GABAergic dysfunctions rescues human tau-induced anxiety in mice. Mol. Ther. 25, 140–152. doi: 10.1016/j.ymthe.2016.10.010
Liu, W., Zhao, J., and Lu, G. (2016). miR-106b inhibits tau phosphorylation at Tyr18 by targeting Fyn in a model of Alzheimer’s disease. Biochem. Biophys. Res. Commun. 478, 852–857. doi: 10.1016/j.bbrc.2016.08.037
Liu, Y., He, X., Li, Y., and Wang, T. (2018). Cerebrospinal fluid CD4+ T lymphocyte-derived miRNA-let-7b can enhances the diagnostic performance of Alzheimer’s disease biomarkers. Biochem. Biophys. Res. Commun. 495, 1144–1150. doi: 10.1016/j.bbrc.2017.11.122
Long, J. M., and Lahiri, D. K. (2011). MicroRNA-101 downregulates Alzheimer’s amyloid-beta precursor protein levels in human cell cultures and is differentially expressed. Biochem. Biophys. Res. Commun. 404, 889–895. doi: 10.1016/j.bbrc.2010.12.053
Long, J. M., Maloney, B., Rogers, J. T., and Lahiri, D. K. (2019). Novel upregulation of amyloid-beta precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5′-untranslated region: implications in Alzheimer’s disease. Mol. Psychiatry 24, 345–363. doi: 10.1038/s41380-018-0266-3
Long, J. M., Ray, B., and Lahiri, D. K. (2012). MicroRNA-153 physiologically inhibits expression of amyloid-beta precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 287, 31298–31310. doi: 10.1074/jbc.M112.366336
Long, J. M., Ray, B., and Lahiri, D. K. (2014). MicroRNA-339-5p down-regulates protein expression of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J. Biol. Chem. 289, 5184–5198. doi: 10.1074/jbc.M113.518241
Lopes, J. P., and Agostinho, P. (2011). Cdk5: multitasking between physiological and pathological conditions. Prog. Neurobiol. 94, 49–63. doi: 10.1016/j.pneurobio.2011.03.006
Lugli, G., Cohen, A. M., Bennett, D. A., Shah, R. C., Fields, C. J., Hernandez, A. G., et al. (2015). Plasma Exosomal miRNAs in Persons with and without Alzheimer disease: altered Expression and Prospects for Biomarkers. PLoS One 10:e0139233. doi: 10.1371/journal.pone.0139233
Lusardi, T. A., Phillips, J. I., Wiedrick, J. T., Harrington, C. A., Lind, B., Lapidus, J. A., et al. (2017). MicroRNAs in human cerebrospinal fluid as biomarkers for Alzheimer’s disease. J. Alzheimers Dis. 55, 1223–1233. doi: 10.3233/JAD-160835
Ma, X., Liu, L., and Meng, J. (2017). MicroRNA-125b promotes neurons cell apoptosis and Tau phosphorylation in Alzheimer’s disease. Neurosci. Lett. 661, 57–62. doi: 10.1016/j.neulet.2017.09.043
Madadi, S., Saidijam, M., Yavari, B., and Soleimani, M. (2020). Downregulation of serum miR-106b: a potential biomarker for Alzheimer disease. Arch. Physiol. Biochem. [Epub ahead of print]. doi: 10.1080/13813455.2020.1734842
Mairet-Coello, G., Courchet, J., Pieraut, S., Courchet, V., Maximov, A., and Polleux, F. (2013). The CAMKK2-AMPK kinase pathway mediates the synaptotoxic effects of Abeta oligomers through Tau phosphorylation. Neuron 78, 94–108. doi: 10.1016/j.neuron.2013.02.003
Manzine, P. R., Pelucchi, S., Horst, M. A., Vale, F. A. C., Pavarini, S. C. I., Audano, M., et al. (2018). microRNA 221 Targets ADAM10 mRNA and is Downregulated in Alzheimer’s Disease. J. Alzheimers Dis. 61, 113–123. doi: 10.3233/jad-170592
Marchegiani, F., Matacchione, G., Ramini, D., Marcheselli, F., Recchioni, R., Casoli, T., et al. (2019). Diagnostic performance of new and classic CSF biomarkers in agerelated dementias. Aging 11, 2420–2429. doi: 10.18632/aging.101925
Matsui, M., Chu, Y., Zhang, H., Gagnon, K. T., Shaikh, S., Kuchimanchi, S., et al. (2013). Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 41, 10086–10109. doi: 10.1093/nar/gkt777
McKeever, P. M., Schneider, R., Taghdiri, F., Weichert, A., Multani, N., Brown, R. A., et al. (2018). MicroRNA expression levels are altered in the cerebrospinal fluid of patients with young-onset Alzheimer’s disease. Mol. Neurobiol. 55, 8826–8841. doi: 10.1007/s12035-018-1032-x
Mezache, L., Mikhail, M., Garofalo, M., and Nuovo, G. J. (2015). Reduced miR-512 and the elevated expression of its targets cFLIP and MCL1 localize to neurons with hyperphosphorylated tau protein in Alzheimer disease. Appl. Immunohistochem. Mol. Morphol. 23, 615–623. doi: 10.1097/pai.0000000000000147
Millan, M. J. (2017). Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: an integrative review. Prog. Neurobiol. 156, 1–68. doi: 10.1016/j.pneurobio.2017.03.004
Modi, P. K., Jaiswal, S., and Sharma, P. (2016). Regulation of Neuronal Cell Cycle and Apoptosis by MicroRNA 34a. Mol. Cell. Biol. 36, 84–94. doi: 10.1128/mcb.00589-15
Moncini, S., Lunghi, M., Valmadre, A., Grasso, M., Del Vescovo, V., Riva, P., et al. (2017). The miR-15/107 family of microRNA genes regulates CDK5R1/p35 with implications for alzheimer’s disease pathogenesis. Mol. Neurobiol. 54, 4329–4342. doi: 10.1007/s12035-016-0002-4
Muller, M., Jakel, L., Bruinsma, I. B., Claassen, J. A., Kuiperij, H. B., and Verbeek, M. M. (2016). MicroRNA-29a is a candidate biomarker for Alzheimer’s disease in cell-free cerebrospinal fluid. Mol. Neurobiol. 53, 2894–2899. doi: 10.1007/s12035-015-9156-8
Müller, M., Jäkel, L., Bruinsma, I. B., Claassen, J. A., Kuiperij, H. B., and Verbeek, M. M. (2016). MicroRNA-29a is a candidate biomarker for alzheimer’s disease in cell-free cerebrospinal fluid. Mol. Neurobiol. 53, 2894–2899.
Muller, M., Kuiperij, H. B., Claassen, J. A., Kusters, B., and Verbeek, M. M. (2014). MicroRNAs in Alzheimer’s disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol. Aging 35, 152–158. doi: 10.1016/j.neurobiolaging.2013.07.005
Nayak, D., Roth, T. L., and McGavern, D. B. (2014). Microglia development and function. Annu. Rev. Immunol. 32, 367–402. doi: 10.1146/annurev-immunol-032713-120240
Nelson, P. T., and Wang, W. X. (2010). MiR-107 is reduced in Alzheimer’s disease brain neocortex: validation study. J. Alzheimers Dis. 21, 75–79. doi: 10.3233/jad-2010-091603
Nie, C., Sun, Y., Zhen, H., Guo, M., Ye, J., Liu, Z., et al. (2020). Differential expression of plasma Exo-miRNA in neurodegenerative diseases by next-generation sequencing. Front. Neurosci. 14:438. doi: 10.3389/fnins.2020.00438
Parhizkar, S., Arzberger, T., Brendel, M., Kleinberger, G., Deussing, M., Focke, C., et al. (2019). Loss of TREM2 function increases amyloid seeding but reduces plaque-associated ApoE. Nat. Neurosci. 22, 191–204. doi: 10.1038/s41593-018-0296-9
Pena-Bautista, C., Baquero, M., Vento, M., and Chafer-Pericas, C. (2019). Omics-based biomarkers for the early alzheimer disease diagnosis and reliable therapeutic targets development. Curr. Neuropharmacol. 17, 630–647. doi: 10.2174/1570159X16666180926123722
Penke, B., Bogar, F., Paragi, G., Gera, J., and Fulop, L. (2019). Key peptides and proteins in Alzheimer’s disease. Curr. Protein Pept. Sci. 20, 577–599. doi: 10.2174/1389203720666190103123434
Pichler, S., Gu, W., Hartl, D., Gasparoni, G., Leidinger, P., Keller, A., et al. (2017). The miRNome of Alzheimer’s disease: consistent downregulation of the miR-132/212 cluster. Neurobiol. Aging 50, 167.e1–167.e10. doi: 10.1016/j.neurobiolaging.2016.09.019
Piscopo, P., Grasso, M., Puopolo, M., D’Acunto, E., Talarico, G., Crestini, A., et al. (2018). Circulating miR-127-3p as a potential biomarker for differential diagnosis in frontotemporal dementia. J. Alzheimers Dis. 65, 455–464. doi: 10.3233/JAD-180364
Piscopo, P., Lacorte, E., Feligioni, M., Mayer, F., Crestini, A., Piccolo, L., et al. (2019). MicroRNAs and mild cognitive impairment: a systematic review. Ageing Res. Rev. 50, 131–141. doi: 10.1016/j.arr.2018.11.005
Prendecki, M., Florczak-Wyspianska, J., Kowalska, M., Ilkowski, J., Grzelak, T., Bialas, K., et al. (2019). APOE genetic variants and apoE, miR-107 and miR-650 levels in Alzheimer’s disease. Folia Neuropathol. 57, 106–116. doi: 10.5114/fn.2019.84828
Ren, R. J., Zhang, Y. F., Dammer, E. B., Zhou, Y., Wang, L. L., Liu, X. H., et al. (2016). Peripheral blood MicroRNA expression profiles in Alzheimer’s disease: screening, validation, association with clinical phenotype and implications for molecular mechanism. Mol. Neurobiol. 53, 5772–5781. doi: 10.1007/s12035-015-9484-8
Riancho, J., Vazquez-Higuera, J. L., Pozueta, A., Lage, C., Kazimierczak, M., Bravo, M., et al. (2017). MicroRNA profile in patients with Alzheimer’s disease: analysis of miR-9-5p and miR-598 in raw and exosome enriched cerebrospinal fluid samples. J. Alzheimers Dis. 57, 483–491. doi: 10.3233/JAD-161179
Roher, A. E., Kokjohn, T. A., Clarke, S. G., Sierks, M. R., Maarouf, C. L., Serrano, G. E., et al. (2017). APP/Abeta structural diversity and Alzheimer’s disease pathogenesis. Neurochem. Int. 110, 1–13. doi: 10.1016/j.neuint.2017.08.007
Rosenberg, R. N., Lambracht-Washington, D., Yu, G., and Xia, W. (2016). Genomics of Alzheimer disease: a review. JAMA Neurol. 73, 867–874. doi: 10.1001/jamaneurol.2016.0301
Ross, S. P., Baker, K. E., Fisher, A., Hoff, L., Pak, E. S., and Murashov, A. K. (2018). miRNA-431 prevents amyloid-beta-induced synapse loss in neuronal cell culture model of Alzheimer’s disease by silencing kremen1. Front. Cell. Neurosci. 12:87. doi: 10.3389/fncel.2018.00087
Rovelet-Lecrux, A., Hannequin, D., Raux, G., Le Meur, N., Laquerriere, A., Vital, A., et al. (2006). APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 38, 24–26. doi: 10.1038/ng1718
Sala Frigerio, C., Lau, P., Salta, E., Tournoy, J., Bossers, K., Vandenberghe, R., et al. (2013). Reduced expression of hsa-miR-27a-3p in CSF of patients with Alzheimer disease. Neurology 81, 2103–2106. doi: 10.1212/01.wnl.0000437306.37850.22
Salta, E., and De Strooper, B. (2017). microRNA-132: a key noncoding RNA operating in the cellular phase of Alzheimer’s disease. FASEB J. 31, 424–433. doi: 10.1096/fj.201601308
Salta, E., Sierksma, A., Vanden Eynden, E., and De Strooper, B. (2016). miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer’s brain. EMBO Mol. Med. 8, 1005–1018. doi: 10.15252/emmm.201606520
Salter, M. W., and Beggs, S. (2014). Sublime microglia: expanding roles for the guardians of the CNS. Cell 158, 15–24. doi: 10.1016/j.cell.2014.06.008
Sanabria-Castro, A., Alvarado-Echeverria, I., and Monge-Bonilla, C. (2017). Molecular pathogenesis of Alzheimer’s disease: an update. Ann. Neurosci. 24, 46–54. doi: 10.1159/000464422
Santa-Maria, I., Alaniz, M. E., Renwick, N., Cela, C., Fulga, T. A., Van Vactor, D., et al. (2015). Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J. Clin. Invest. 125, 681–686. doi: 10.1172/jci78421
Schwarz, D. S., and Zamore, P. D. (2002). Why do miRNAs live in the miRNP? Genes Dev. 16, 1025–1031. doi: 10.1101/gad.992502
Selkoe, D. J., and Hardy, J. (2016). The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608. doi: 10.15252/emmm.201606210
Shi, Z., Zhang, K., Zhou, H., Jiang, L., Xie, B., Wang, R., et al. (2020). Increased miR-34c mediates synaptic deficits by targeting synaptotagmin 1 through ROS-JNK-p53 pathway in Alzheimer’s disease. Aging Cell 19:e13125. doi: 10.1111/acel.13125
Shu, R., Wong, W., Ma, Q. H., Yang, Z. Z., Zhu, H., Liu, F. J., et al. (2015). APP intracellular domain acts as a transcriptional regulator of miR-663 suppressing neuronal differentiation. Cell Death Dis. 6:e1651. doi: 10.1038/cddis.2015.10
Siedlecki-Wullich, D., Catala-Solsona, J., Fabregas, C., Hernandez, I., Clarimon, J., Lleo, A., et al. (2019). Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer’s disease. Alzheimers Res. Ther. 11:46. doi: 10.1186/s13195-019-0501-4
Sierksma, A., Lu, A., Salta, E., Vanden Eynden, E., Callaerts-Vegh, Z., D’Hooge, R., et al. (2018). Deregulation of neuronal miRNAs induced by amyloid-beta or TAU pathology. Mol. Neurodegener. 13:54. doi: 10.1186/s13024-018-0285-1
Smith, P. Y., Hernandez-Rapp, J., Jolivette, F., Lecours, C., Bisht, K., Goupil, C., et al. (2015). miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 24, 6721–6735. doi: 10.1093/hmg/ddv377
Sohrabifar, N., Gharesouran, J., Talebi, M., Ghojazadeh, M., and Mohaddes Ardebili, S. M. (2015). Association of CLU and TLR2 gene polymorphisms with late-onset Alzheimer disease in a northwestern Iranian population. Turk. J. Med. Sci. 45, 1082–1086. doi: 10.3906/sag-1406-64
Souza, V. C., Morais, G. S. Jr., Henriques, A. D., Machado-Silva, W., Perez, D. I. V., Brito, C. J., et al. (2020). Whole-blood levels of MicroRNA-9 are decreased in patients with late-onset Alzheimer disease. Am. J. Alzheimers Dis. Other Demen. 35:1533317520911573. doi: 10.1177/1533317520911573
Swarbrick, S., Wragg, N., Ghosh, S., and Stolzing, A. (2019). Systematic review of miRNA as biomarkers in Alzheimer’s disease. Mol. Neurobiol. 56, 6156–6167. doi: 10.1007/s12035-019-1500-y
Takousis, P., Sadlon, A., Schulz, J., Wohlers, I., Dobricic, V., Middleton, L., et al. (2019). Differential expression of microRNAs in Alzheimer’s disease brain, blood, and cerebrospinal fluid. Alzheimers Dement. 15, 1468–1477. doi: 10.1016/j.jalz.2019.06.4952
Tan, L., Yu, J. T., Liu, Q. Y., Tan, M. S., Zhang, W., Hu, N., et al. (2014). Circulating miR-125b as a biomarker of Alzheimer’s disease. J. Neurol. Sci. 336, 52–56. doi: 10.1016/j.jns.2013.10.002
Tang, Y., Bao, J. S., Su, J. H., and Huang, W. (2017). MicroRNA-139 modulates Alzheimer’s-associated pathogenesis in SAMP8 mice by targeting cannabinoid receptor type 2. Genet. Mol. Res. 16:gmr16019166. doi: 10.4238/gmr16019166
Tao, Y., Han, Y., Yu, L., Wang, Q., Leng, S. X., and Zhang, H. (2020). The predicted key molecules, functions, and pathways that bridge mild cognitive impairment (MCI) and Alzheimer’s disease (AD). Front. Neurol. 11:233. doi: 10.3389/fneur.2020.00233
van den Berg, M. M. J., Krauskopf, J., Ramaekers, J. G., Kleinjans, J. C. S., Prickaerts, J., and Briede, J. J. (2020). Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog. Neurobiol. 185:101732. doi: 10.1016/j.pneurobio.2019.101732
Villa, C., Ridolfi, E., Fenoglio, C., Ghezzi, L., Vimercati, R., Clerici, F., et al. (2013). Expression of the transcription factor Sp1 and its regulatory hsa-miR-29b in peripheral blood mononuclear cells from patients with Alzheimer’s disease. J. Alzheimers Dis. 35, 487–494. doi: 10.3233/JAD-122263
Wang, G., Huang, Y., Wang, L. L., Zhang, Y. F., Xu, J., Zhou, Y., et al. (2016). MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci. Rep. 6:26697. doi: 10.1038/srep26697
Wang, K., Yuan, Y., Cho, J. H., McClarty, S., Baxter, D., and Galas, D. J. (2012). Comparing the MicroRNA spectrum between serum and plasma. PLoS One 7:e41561. doi: 10.1371/journal.pone.0041561
Wang, L., Liu, J., Wang, Q., Jiang, H., Zeng, L., Li, Z., et al. (2019). MicroRNA-200a-3p mediates neuroprotection in alzheimer-related deficits and attenuates amyloid-beta overproduction and tau hyperphosphorylation via coregulating BACE1 and PRKACB. Front. Pharmacol. 10:806. doi: 10.3389/fphar.2019.00806
Wang, T., Chen, K., Li, H., Dong, S., Su, N., Liu, Y., et al. (2015). The feasibility of utilizing plasma MiRNA107 and BACE1 messenger RNA gene expression for clinical diagnosis of amnestic mild cognitive impairment. J. Clin. Psychiatry 76, 135–141. doi: 10.4088/JCP.13m08812
Wang, W.-X., Huang, Q., Hu, Y., Stromberg, A. J., and Nelson, P. T. (2010). Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 121, 193–205. doi: 10.1007/s00401-010-0756-0
Wang, X., Tan, L., Lu, Y., Peng, J., Zhu, Y., Zhang, Y., et al. (2015). MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 589, 726–729. doi: 10.1016/j.febslet.2015.02.001
Wang, Y., Veremeyko, T., Wong, A. H., El Fatimy, R., Wei, Z., Cai, W., et al. (2017). Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer’s disease. Neurobiol. Aging 51, 156–166. doi: 10.1016/j.neurobiolaging.2016.12.015
Wei, H., Xu, Y., Xu, W., Zhou, Q., Chen, Q., Yang, M., et al. (2018). Serum exosomal miR-223 serves as a potential diagnostic and prognostic biomarker for dementia. Neuroscience 379, 167–176. doi: 10.1016/j.neuroscience.2018.03.016
Wiedrick, J. T., Phillips, J. I., Lusardi, T. A., McFarland, T. J., Lind, B., Sandau, U. S., et al. (2019). Validation of MicroRNA biomarkers for Alzheimer’s disease in human cerebrospinal fluid. J. Alzheimers Dis. 67, 875–891. doi: 10.3233/jad-180539
Wu, D. M., Wen, X., Wang, Y. J., Han, X. R., Wang, S., Shen, M., et al. (2018). Effect of microRNA-186 on oxidative stress injury of neuron by targeting interleukin 2 through the janus kinase-signal transducer and activator of transcription pathway in a rat model of Alzheimer’s disease. J. Cell. Physiol. 233, 9488–9502. doi: 10.1002/jcp.26843
Wu, H. Z. Y., Thalamuthu, A., Cheng, L., Fowler, C., Masters, C. L., Sachdev, P., et al. (2020). Differential blood miRNA expression in brain amyloid imaging-defined Alzheimer’s disease and controls. Alzheimers Res. Ther. 12:59. doi: 10.1186/s13195-020-00627-0
Wu, Q., Ye, X., Xiong, Y., Zhu, H., Miao, J., Zhang, W., et al. (2016). The protective role of microRNA-200c in Alzheimer’s disease pathologies is induced by beta amyloid-triggered endoplasmic reticulum stress. Front. Mol. Neurosci. 9:140. doi: 10.3389/fnmol.2016.00140
Wu, Y., Xu, J., Xu, J., Cheng, J., Jiao, D., Zhou, C., et al. (2017). Lower serum levels of miR-29c-3p and miR-19b-3p as biomarkers for Alzheimer’s disease. Tohoku J. Exp. Med. 242, 129–136. doi: 10.1620/tjem.242.129
Xie, B., Zhou, H., Zhang, R., Song, M., Yu, L., Wang, L., et al. (2015). Serum miR-206 and miR-132 as potential circulating biomarkers for mild cognitive impairment. J. Alzheimers Dis. 45, 721–731. doi: 10.3233/JAD-142847
Xu, X. F., Wang, Y. C., Zong, L., and Wang, X. L. (2019). miR-151-5p modulates APH1a expression to participate in contextual fear memory formation. RNA Biol. 16, 282–294. doi: 10.1080/15476286.2019.1572435
Yang, G., Song, Y., Zhou, X., Deng, Y., Liu, T., Weng, G., et al. (2015). MicroRNA-29c targets beta-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol. Med. Rep. 12, 3081–3088. doi: 10.3892/mmr.2015.3728
Yang, Q., Zhao, Q., and Yin, Y. (2019). miR-133b is a potential diagnostic biomarker for Alzheimer’s disease and has a neuroprotective role. Exp. Ther. Med. 18, 2711–2718. doi: 10.3892/etm.2019.7855
Yang, T. T., Liu, C. G., Gao, S. C., Zhang, Y., and Wang, P. C. (2018). The serum exosome derived MicroRNA-135a, -193b, and -384 were potential Alzheimer’s disease biomarkers. Biomed. Environ. Sci. 31, 87–96. doi: 10.3967/bes2018.011
Yilmaz, S. G., Erdal, M. E., Ozge, A. A., and Sungur, M. A. (2016). Can peripheral MicroRNA expression data serve as epigenomic (upstream) biomarkers of Alzheimer’s disease? OMICS 20, 456–461. doi: 10.1089/omi.2016.0099
Zeng, Q., Zou, L., Qian, L., Zhou, F., Nie, H., Yu, S., et al. (2017). Expression of microRNA222 in serum of patients with Alzheimer’s disease. Mol. Med. Rep. 16, 5575–5579. doi: 10.3892/mmr.2017.7301
Zhang, B., Wang, L. L., Ren, R. J., Dammer, E. B., Zhang, Y. F., Huang, Y., et al. (2016). MicroRNA-146a represses LRP2 translation and leads to cell apoptosis in Alzheimer’s disease. FEBS Lett. 590, 2190–2200. doi: 10.1002/1873-3468.12229
Zhang, C., Lu, J., Liu, B., Cui, Q., and Wang, Y. (2016). Primate-specific miR-603 is implicated in the risk and pathogenesis of Alzheimer’s disease. Aging 8, 272–290. doi: 10.18632/aging.100887
Zhang, J., Hu, M., Teng, Z., Tang, Y. P., and Chen, C. (2014). Synaptic and cognitive improvements by inhibition of 2-AG metabolism are through upregulation of microRNA-188-3p in a mouse model of Alzheimer’s disease. J. Neurosci. 34, 14919–14933. doi: 10.1523/jneurosci.1165-14.2014
Zhang, J., Liu, Z., Pei, Y., Yang, W., Xie, C., and Long, S. (2018). MicroRNA-322 cluster promotes tau phosphorylation via targeting brain-derived neurotrophic factor. Neurochem. Res. 43, 736–744. doi: 10.1007/s11064-018-2475-1
Zhang, Y., Li, Q., Liu, C., Gao, S., Ping, H., Wang, J., et al. (2016a). MiR-214-3p attenuates cognition defects via the inhibition of autophagy in SAMP8 mouse model of sporadic Alzheimer’s disease. Neurotoxicology 56, 139–149. doi: 10.1016/j.neuro.2016.07.004
Zhang, Y., Xing, H., Guo, S., Zheng, Z., Wang, H., and Xu, D. (2016b). MicroRNA-135b has a neuroprotective role via targeting of beta-site APP-cleaving enzyme 1. Exp. Ther. Med. 12, 809–814. doi: 10.3892/etm.2016.3366
Zhang, Y. H., Bai, S.-F., and Yan, J. Q. (2019). Blood circulating miRNAs as biomarkers of Alzheimer’s disease: a systematic review and meta-analysis. Biomark. Med. 13, 1045–1054. doi: 10.2217/bmm-2018-0341
Zhao, N., Liu, C. C., Qiao, W., and Bu, G. (2018). Apolipoprotein E, receptors, and modulation of Alzheimer’s disease. Biol. Psychiatry 83, 347–357. doi: 10.1016/j.biopsych.2017.03.003
Zheng, D., Sabbagh, J. J., Blair, L. J., Darling, A. L., Wen, X., and Dickey, C. A. (2016). MicroRNA-511 binds to FKBP5 mRNA, which encodes a chaperone protein, and regulates neuronal differentiation. J. Biol. Chem. 291, 17897–17906. doi: 10.1074/jbc.M116.727941
Zhou, Y., Deng, J., Chu, X., Zhao, Y., and Guo, Y. (2019). Role of post-transcriptional control of calpain by miR-124-3p in the development of Alzheimer’s disease. J. Alzheimers Dis. 67, 571–581. doi: 10.3233/jad-181053
Zhou, Y., Wang, Z. F., Li, W., Hong, H., Chen, J., Tian, Y., et al. (2018). Protective effects of microRNA-330 on amyloid beta-protein production, oxidative stress, and mitochondrial dysfunction in Alzheimer’s disease by targeting VAV1 via the MAPK signaling pathway. J. Cell. Biochem. 119, 5437–5448. doi: 10.1002/jcb.26700
Zhu, L., Li, J., Dong, N., Guan, F., Liu, Y., Ma, D., et al. (2016). mRNA changes in nucleus accumbens related to methamphetamine addiction in mice. Sci. Rep. 6:36993. doi: 10.1038/srep36993
Zhu, M., Huang, C., Ma, X., Wu, R., Zhu, W., Li, X., et al. (2016). Modulation of miR-19 in aluminum-induced neural cell apoptosis. J. Alzheimers Dis. 50, 1149–1162. doi: 10.3233/jad-150763
Zhu, Q. B., Unmehopa, U., Bossers, K., Hu, Y. T., Verwer, R., Balesar, R., et al. (2016). MicroRNA-132 and early growth response-1 in nucleus basalis of Meynert during the course of Alzheimer’s disease. Brain 139, 908–921. doi: 10.1093/brain/awv383
Keywords: microRNAs, Alzheimer’s disease, pathogenesis, diagnosis, biomarker
Citation: Wei W, Wang Z-Y, Ma L-N, Zhang T-T, Cao Y and Li H (2020) MicroRNAs in Alzheimer’s Disease: Function and Potential Applications as Diagnostic Biomarkers. Front. Mol. Neurosci. 13:160. doi: 10.3389/fnmol.2020.00160
Received: 11 May 2020; Accepted: 05 August 2020;
Published: 21 August 2020.
Edited by:
Albert I. Chen, Nanyang Technological University, SingaporeReviewed by:
Hyang-Sook Hoe, Korea Brain Research Institute, South KoreaLi Zeng, National Neuroscience Institute (NNI), Singapore
Copyright © 2020 Wei, Wang, Ma, Zhang, Cao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Cao, eXUuY2FvLXhpeXVhbkBvdXRsb29rLmNvbQ==; Hao Li, eHlocGxpaGFvMTk2NUAxMjYuY29t
 Wei Wei
Wei Wei Zhi-Yong Wang2
Zhi-Yong Wang2 Li-Na Ma
Li-Na Ma Yu Cao
Yu Cao Hao Li
Hao Li