- Soonchunhyang Institute of Medi-bio Science, Soonchunhyang University, Cheonan-si, South Korea
Myeloid malignancies, including myelodysplastic syndromes and acute myeloid leukemia, are clonal diseases arising in hematopoietic stem or progenitor cells. In recent years, microRNA (miRNA) expression profiling studies have revealed close associations of miRNAs with cytogenetic and molecular subtypes of myeloid malignancies, as well as outcome and prognosis of patients. However, the roles of miRNA deregulation in the pathogenesis of myeloid malignancies and how they cooperate with protein-coding gene variants in pathological mechanisms leading to the diseases have not yet been fully understood. In this review, we focus on recent insights into the role of miRNAs in the development and progression of myeloid malignant diseases and discuss the prospect that miRNAs may serve as a potential therapeutic target for leukemia.
Introduction
MicroRNAs (miRNAs) in Cancer
MicroRNAs are small non-coding RNAs with 19–22 nucleotides to control gene expression through binding to mRNA of their cognate target genes and thereby participate in numerous biological processes such as cell proliferation, differentiation, development, metabolism, apoptosis, survival, and hematopoiesis (1). miRNA is transcribed by RNA pol II/III to generate a primary miRNA followed by nuclear cleavage by the RNase III endonuclease Drosha and its binding to the double-stranded RNA-binding protein DGCR8 to form a precursor miRNA (pre-miRNA) (2, 3). Subsequently, the pre-miRNA is transported by Exportin-5/RanGTP to the cytoplasm to be further cleaved by the RNase III endonuclease Dicer, leaving an unstable miRNA duplex that unwinds. The 5′ guide strand containing the mature miRNA sequence is incorporated into a ribonucleotide silencing complex, while the 3′ passenger strand undergoes rapid degradation (4–6).
While miRNAs located within chromosomes deleted in cancer play roles as tumor suppressors, miRNAs located in genomic regions amplified in cancer function as oncogenes. Deregulated miRNAs found in both solid tumors and hematopoietic malignancies target the transcripts of essential protein-coding genes involved in tumorigenesis (7, 8). Fingerprints of miRNAs’ expression are linked to clinical and biological characteristics of tumors including tissue type, aggressiveness, and therapy response. Abnormal expression of pre-miRNA is also found in various types of human cancer. Because sequence abnormalities of miRNAs’ genes and transcripts are also observed in the germline (8), the inherited subtle variations in miRNAs may have a great effect on the expression profiles of protein-coding genes in cancer.
miRNAs in Hematological Malignancies
Hematological malignancies comprise a collection of heterogeneous diseases, all originating from cells of the bone marrow or lymphatic system. Hematological malignancies include leukemias, lymphomas, myelodysplastic syndromes (MDS), and myeloproliferative neoplasms (9). Myeloid malignancies are clonal disorders that are characterized by excessive proliferation, abnormal self-renewal, and/or differentiation blocks of hematopoietic stem cells (HSCs) and myeloid progenitor cells (10, 11). miRNA expression profiling in myeloid malignancies has revealed distinct signatures associated with diagnosis, stage classification, progression, prognosis, and response to treatment of leukemias (Table 1). miRNAs can be regulated by epigenetic modifiers including DNA methylation and histone modification in leukemias, suggesting that aberrant expression of miRNAs by epigenetic mechanisms may trigger hematopoietic cell transformation.
In this review, we focus on recent advances in understanding the roles of miRNA deregulation in the pathogenesis of myeloid malignancies and discuss the prospect that miRNAs may serve as potential therapeutic targets for leukemias.
miRNA Deregulation in MDS
Myelodysplastic syndromes are HSC disorders characterized by ineffective hematopoiesis and a high risk of progression to acute myeloid leukemia (AML) (50). More than 70% of all human miRNAs are located within regions of recurrent copy-number alterations in MDS and AML cell lines (51). The targeted ablation of Dicer1 in murine hematopoietic system leads to abnormal hematopoiesis and MDS, supporting the relevance of miRNA deregulation to the pathogenesis of MDS (52).
Several recent studies have addressed the role of miRNAs in MDS pathogenesis. Vasilatou et al. have shown that miR-17-5p and miR-20a, as members of the miR-17–92 cluster, repress the transcription factor E2F1, which is highly expressed in 67% of patients with MDS (44). Similarly, let-7a downregulates KRAS, which is aberrantly expressed in high-risk MDS (53, 54). A subset of miRNAs involved in stage-specific regulation of erythropoiesis are also deregulated in MDS (55). Overexpression of miR-181, miR-221, miR-376b, miR-125b, miR-155, or miR-130a inhibits erythroid cell growth (56), and this event might be responsible for disease-associated ineffective erythropoiesis. miR-155 targeting CEBPB and CSF1R is significantly upregulated in high-risk MDS (57). High expressions of miR-155, miR-126, and miR-130 in MDS restrain megakaryopoiesis and may account for higher frequency of thrombocytopenia observed during disease progression (46). However, recent evidence reveals that reduction of Rho family members by miR-155 contributes to impaired neutrophil migration in MDS (58). miR-21 expression has been found to be increased in MDS, and its interaction with SMAD7 mRNA leads to ineffective, MDS-like hematopoiesis via overactivating TGFβ signaling (42). In addition, serum miR-21 level appears to act as a potential non-invasive biomarker that predicts a response following treatment with hypo-methylating agents, such as azacytidine or decitabine in MDS patients (59). In contrast, decreased expression of the miR-144/451 members targeting the erythroid transcription factor GATA-1 is closely associated with high-risk MDS (12, 49). Overall, both the aberrant expression and the function of miRNAs are the important factors contributing to MDS pathogenesis and prognosis.
Despite significant amount of evidence demonstrating miRNA expression and role in tumorigenesis is available, a very few studies illustrate mechanisms of miRNA deregulation and related mechanisms underlying MDS. miR-22 has been found to be overexpressed in MDS patients with poorer survival outcome (60). Furthermore, transgenic mice expressing hematopoietic miR-22 exhibit decreased global 5-hydroxymethylcytosine levels and increased HSC self-renewal along with defective differentiation and develop MDS and myeloid leukemia over time. miR-22 directly targets the DNA demethylating enzyme ten-eleven-translocation 2 (TET2) and affects the epigenetic landscape in the hematopoietic compartment, while forced expression of TET2 suppresses the miR-22-induced malignant phenotypes. A significant inverse correlation between miR-22 and TET2 observed in MDS patients suggest the miR-22-TET2 regulatory network as a reliable factor for MDS pathogenesis (60, 61). A better understanding of miR-22 deregulation in MDS disease progression and AML transformation will provide insight into the mechanisms of MDS pathogenesis and provide new therapeutic strategies against leukemia transformation.
Emerging Roles of miRNA Deregulation in the Pathogenesis of AML
Acute myeloid leukemia is characterized by the accumulation of immature myeloid cells in the bone marrow and shows genetic abnormalities including mutations and chromosomal translocations (10). Distinctive miRNA expression profiles have been demonstrated for cytogenetic subtypes and mutations in CEBPA, FLT3, and NPM1 of AML (62–64). miRNA profiles are also associated with AML prognosis, underscoring the importance of miRNAs in AML (65). As such, miRNAs impact AML development and progression through targeting known oncogenes or tumor suppressors or collaborating with them to promote or suppress myeloid malignancy.
miR-9 has shown to be overexpressed in MLL-rearranged AML and play a critical oncogenic role in MLL fusion-mediated leukemogenesis (66). Ectopic expression of miR-9 blocks neutrophil development in myeloid cell lines and in murine primary lineage-negative bone marrow cells by inhibiting ETS-related gene (67). Also, miR-9 exerts its tumor-suppressive effects through the cooperation with let-7 to repress the oncogenic Lin28b/HMGA2 axis in AML (68).
miR-125b is upregulated in AML patients and blocks the differentiation of AML blast cells by directly targeting the cytoplasmic tyrosine-protein kinase FES that is expressed exclusively in myeloid cells (33). Overexpression of miR-125b also leads to a reduction in expression of the RNA binding protein Lin28A (69), which is known to play an important role in stem cell biology.
miR-181 and miR-128 target Lin28, leading to the progression of myeloid leukemia and differentiation blockage of hematopoietic cells to their lineage (15, 70, 71). The inhibition of miR-181 expression partially reverses the lack of myeloid differentiation in AML patients and in the mice implanted with CD34+ hematopoietic stem/progenitor cells (HSPCs) from AML patients (72).
The targeted miR-126 reduction in cell lines and primary AML samples results in decreased AML growth through inhibiting multiple components of the PI3K/AKT/mTOR pathway (47, 73, 74). The attenuated expression of miR-126 also leads to expand normal HSC (75), suggesting that miR-126 dictates opposing self-renewal outcomes in normal and leukemic HSC. Furthermore, both gain- and loss-of-function in vivo studies of miR-126 in murine models demonstrate that either overexpression or knockout of miR-126 promotes development of AML in mice (76). This result suggests that miR-126 plays a dual role in leukemogenesis and supports a new layer of miRNA regulation in AML.
Overexpressed miR-155 is associated with poor outcome in AML patients. miR-155 promotes FLT3-ITD-induced myeloproliferative disorder through inhibition of the interferon (IFN) response, inositol 5-phosphatase 1 (SHIP1), CEBPB, and PU.1, while it is upregulated in FLT3-ITD+ and MLL-rearranged AML (57, 77–80). These results suggest that miR-155 can collaborate with FLT3-ITD to promote myeloid cell expansion, and this involves a multi-target mechanism that includes repression of IFN signaling.
miR-22 is overexpressed in AML, and its aberrant expression correlates with silencing of TET2 in AML patients (60). Approximately 70% of miR-22 transgenic mice develop AML by 2 years of age. Also, miR-22 impairs the MLL-AF9-induced leukemogenesis through repressing CREB and Myc pathways and relieves the monocyte/macrophage differentiation and the growth of AML by targeting MECOM (81, 82). Therefore, in AML, miR-22 can be both oncogenic and tumor-suppressive, depending on the specific individual backgrounds (e.g., early HSCs versus the committed myeloid progenitors).
The accumulation of peroxiredoxin III caused by decreased miR-26a leads to a marked reduction in reactive oxygen species (ROS) in primary AML granulocyte samples (83). Growing evidence demonstrates that ROS plays a key role in regulating the balance between self-renewal and differentiation of HSCs (84). Thus, the reduced ROS levels might drive HSCs toward differentiation into myeloid lineage fates, providing a potential mechanism for miR-26a’s role as a tumor suppressor.
miR-29a appears to be significantly increased in peripheral blood mononuclear cells and bone marrow white blood cells from AML patients. Increased miR-29a promotes differentiation into granulocytes and monocytes, while reduction of miR-29a suppresses myeloid differentiation in leukemic cells (85). In myeloid leukemogenesis, c-Myc inhibits miR-29a expression, resulting in increased AKT2 and Cyclin D2 expressions in AML (86). Conversely, ectopic expression of miR-29a in murine HSPCs leads to acquisition of self-renewal capacity by myeloid progenitors, biased myeloid differentiation, and the development of a myeloproliferative disorder that progresses to AML (87).
miR-34a is downregulated in AML and induces apoptosis via inhibition of autophagy by targeting HMGB1 in leukemic cells (21). miR-34b plays a critical role in AML pathogenesis by targeting CREB, and its expression is repressed due to its promoter hypermethylation in AML patients (22). The methylation of the miR-124a family, including miR-124a-1 and miR-124a-3, is also observed in AML patients independently of their cytogenetic subtypes (88). It is also noted that epigenetic silencing of miR-124a is associated with the expression of EVI1 in AML (89, 90).
While studies of miR-125b suggested that it has oncogenic role in AML, miR-125a is considered as a tumor-suppressive miRNA. miR-125a expression in cytogenetically normal AML appear to be most decreased in favorable and intermediate prognostic populations and associated with decreased survival (91).
In the context of AML caused by toxic DNA interstrand crosslinks (ICLs), miR-139 and miR-199a have opposite roles in hematopoietic cell expansion and leukemogenesis (92). The levels of miR-139 and miR-199a are elevated with age in myeloid progenitors from the nucleotide excision repair gene (Ercc1)-deficient mice. Ectopic expression of miR-139 inhibits proliferation of myeloid progenitors, whereas increased miR-199a enhances proliferation of progenitors and augments the AML phenotype. This study supports the oncogenic role of miR-199a and also indicates that the elevated miR-139 as a tumor suppressor is involved in the defective hematopoietic function in ICL-induced AML.
miR-223 decreases cell proliferation and enhances cell apoptosis in AML via targeting FBW7 (93, 94). miR-223 was originally identified as a critical regulator in granulopoiesis and transactivated by NFI-A and C/EBPα in acute promyelocytic leukemia (27). AML1/ETO oncoprotein induces epigenetic silencing of miR-223 through directly binding to the pre-miR-223 gene in AML (28). miR-223 targets E2F1 to inhibit cell cycle progression, thereby resulting in myeloid differentiation, and in turn, E2F1 represses miR-223 transcription, forming a negative feedback loop in AML (95–97). In summary, scientific evidence supporting the role of miRNAs in the pathogenesis of AML with proven tumor suppressors or oncogenic activities is becoming increasingly clear.
miRNA-Based Therapeutics in Myeloid Malignancies
As the understanding of miRNA expression and action in myeloid malignancies continues to evolve, miRNAs have a great potential to serve as both the non-invasive biomarkers and a potential therapeutic target for leukemia. For example, miRNA expression signatures classify leukemias of uncertain lineage as either AML or acute lymphoblastic leukemia (98). miRNA expression profiles can also predict progression of MDS to AML (99) and survival outcome of AML patients (100, 101). Furthermore, circulating miRNAs have been recently demonstrated as an economical, non-invasive, and sensitive tool to monitor for minimal residual disease, which refers to the persistence of a small number of leukemic blasts in the bone marrow after chemotherapy and can ultimately cause disease relapse. Indeed, AML patients have a distinctive circulating miRNA expression profiles compared to healthy controls (102, 103), and an altered expression signature of serum miRNAs is observed after standard chemotherapy (104).
Yet, cancer therapy is based on a therapy targeting a single gene or pathway: “one target, one drug” model. A treatment effectively targeting multiple genes and pathways of cancer concomitantly may be an important innovation. Such an approach would not only more effectively suppress cancer cell growth but also would inhibit the common emergence of resistance in a single gene or pathway. miRNAs form a complex network where each miRNA can regulate multiple genes and pathways and each gene or pathway can be regulated by multiple miRNAs. Thus, miRNAs hold promising potential for “multi-targeted therapy” in cancer patients. To date, miRNA replacement therapy has largely made use of synthetic miRNA mimics to restore lost tumor suppressor expression (105). Restoration of lost tumor suppressor miRNAs using synthetic double-stranded RNAs (with a delivery agent) has been successful in preclinical models of leukemia. For example, the targeted delivery of miR-29b mimics by transferrin-conjugated lipid nanoparticles in mice engrafted with human AML cells shows a significantly longer survival compared to control nanoparticles or free miR-29b (106).
Conversely, targeting overexpressed oncomiRs can be conducted mainly by three approaches: anti-miRNA oligonucleotides (AMOs; antagomiRs), miRNA masking, and miRNA sponges (Figure 1). AMOs that are chemically modified with the locked nucleic acid (LNA) can be systemically delivered to affect cancer-related pathway via the binding and inhibiting oncomiRs in leukemia. For instance, targeting nanoparticles containing miR-126 antagonists (antagomiR-126) results in an in vivo reduction of leukemic stem cells by depletion of the quiescent cell sub-population (74). miR-21- and miR-196b-specific antagomiRs inhibit in vitro leukemic colony-forming activity and in vivo leukemia-initiating cell activity of HOX-based leukemias, which have led to improved survival and delayed disease onset in murine AML models (107). miRNA-masking antisense oligonucleotides (miR-mask) can be used to achieve a gene-specific anti-miRNA therapy that masks the specific target mRNA from endogenous miRNA, and thus prevent the inhibitory action of miRNA. miRNA sponges are another approach to silence miRNAs with potentially important clinical utility, and have complementary binding sites to seed sequences of target miRNAs. This advantage gives them the ability to inhibit multiple miRNAs that have the same sequence in their seed region. It has been shown that using miR-22 sponges, both the leukemic cell proliferation and the activity of miR-22 are markedly impaired (60).
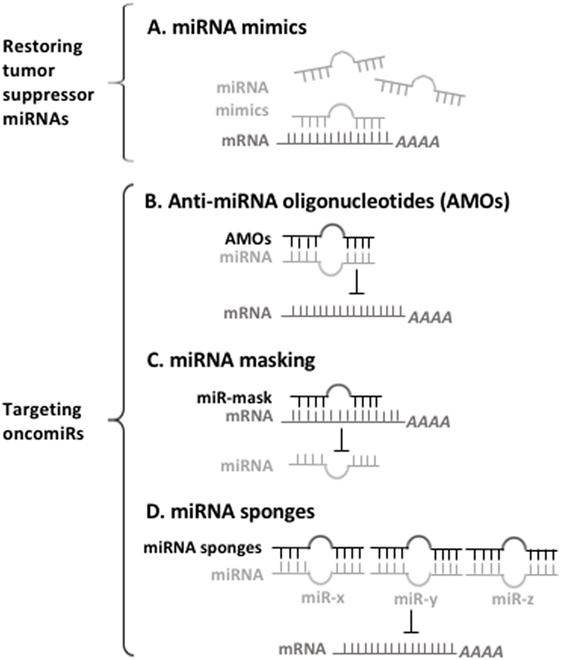
Figure 1. A prospect of miRNA-based therapy for myeloid malignancies. (A) Synthetic oligonucleotides used for restoring the depleted microRNAs (miRNAs) bind to their target mRNAs for inhibiting mRNAs of oncogenes. (B) Anti-miRNA oligonucleotides (AMOs) interact with oncomiRs, thus preventing them from interacting with their target mRNA. (C) miRNA-mask is designed to bind to 3′UTR of mRNAs, thus preventing oncomiRs recognition their target mRNAs. (D) miRNA sponges have multiple complementary sites against targeted miRNA, thereby inhibit the functions of oncomiRs.
In addition to these encouraging outcomes with the use of miRNA-based therapy in preclinical, animal models, the first therapies targeting miRNAs have now entered clinical trials (108). Treatment of an LNA inhibitor of miR-122 (known as Miravirsen) in patients with hepatitis C virus infection holds great promise of miRNA-based therapeutics (https://ClinicalTrials.gov, NCT01200420) (109). Furthermore, the first cancer-targeted miRNA drug—MRX34, a liposome-based miR-34 mimic—has entered clinical trials in patients with advanced or metastatic liver cancer (https://ClinicalTrials.gov, NCT01829971) (110). These studies provide a proof of principle that should encourage future endeavors of miRNA-directed therapy for leukemia.
Conclusion and Perspectives
There are many more miRNAs, shown in publication, that are involved in the pathogenesis of myeloid malignancies, suggesting intense enthusiasm for research in this area in recent years. However, the regulatory changes in miRNA levels are often small and might get lost in the biological noise when using a small number of samples. Using in vitro systems to study the miRNA phenotypes might be different from what happens in vivo. Also, the efficacy of overexpression or antagomiR tools should be validated using downstream target readout to convince the endogenous interaction between the miRNA and the targets.
MicroRNAs have emerged as the potential targets for therapeutic applications. Circulating miRNAs in exosomes/extracellular vesicles from serum or plasma represent a new source of promising biomarkers that may be applied to clinical settings. A specific MDS/AML-associated serum miRNA profiles could not only provide an exciting screening tool for early detection of leukemia in the clinic but could also be used to track leukemic blasts relapsed after chemotherapy. However, exosomal miRNAs loaded from leukemic cells can be transferred to stromal or normal HSC recipient cells and alter their functions, thereby promoting leukemic phenotypes. A further investigation of the relevance of exosomal miRNAs to the pathogenesis of myeloid malignancies is clearly warranted.
Although the case of miRNA-based therapeutics entering clinical trials continues to grow, no miRNA-based therapy has yet made its way to clinical trials particularly for the treatment of AML. A main obstacle of applying miRNA-based therapeutics for clinical use is the limitation of more efficient and specific delivery methods. Thus, many new approaches are currently being explored for improved delivery of miRNA-based therapies, including liposomes, nanoparticles, LNAs with increased stability, and peptide-based inhibitors. Further, how to precisely deliver miRNA mimics or antagomiRs into the targeted cells in vivo has also become another major barrier preventing the establishment of miRNA-directed strategies. But nevertheless, miRNA-based therapies may be available soon for the treatment of myeloid malignancies, and miRNA-based therapeutics may be efficacious when used in a combination with current chemotherapy regimen for leukemia.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
We thank Song’s lab members for critical discussion and editing of the manuscript. This work was supported by a grant of the National Research Foundation (NRF) of Korea funded by the Ministry of Science, ICT, and Future Planning (NRF-2016K1A4A3914725) to HK, and a grant of the National Research Foundation (NRF) of Korea funded by the Ministry of Science, ICT, and Future Planning (NRF-2016K1A4A3914725), and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health and Welfare, Republic of Korea (Grant number: HI15C2679) to SS.
References
1. Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol (2009) 10(2):116–25. doi:10.1038/nrm2621
2. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature (2004) 432(7014):231–5. doi:10.1038/nature03049
3. Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics (2010) 11(7):537–61. doi:10.2174/138920210793175895
4. Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science (2001) 293(5531):834–8. doi:10.1126/science.1062961
5. Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J (2002) 21(21):5875–85. doi:10.1093/emboj/cdf582
6. Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA (2004) 10(2):185–91. doi:10.1261/rna.5167604
7. Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med (2006) 12(12):580–7. doi:10.1016/j.molmed.2006.10.006
8. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer (2006) 6(11):857–66. doi:10.1038/nrc1997
9. Downing JR, Shannon KM. Acute leukemia: a pediatric perspective. Cancer Cell (2002) 2(6):437–45. doi:10.1016/S1535-6108(02)00211-8
10. Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med (2015) 373(12):1136–52. doi:10.1056/NEJMra1406184
11. Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer (2016) 16(3):145–62. doi:10.1038/nrc.2016.8
12. Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci U S A (2008) 105(9):3333–8. doi:10.1073/pnas.0712312105
13. Yao C, Zhang G, Walker A, Zhao KY, Li Y, Lyu L, et al. Potent induction of apoptosis by givinostat in BCR-ABL1-positive and BCR-ABL1-negative precursor B-cell acute lymphoblastic leukemia cell lines. Leuk Res (2017) 60:129–34. doi:10.1016/j.leukres.2017.08.003
14. Khalaj M, Woolthuis CM, Hu W, Durham BH, Chu SH, Qamar S, et al. miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J Exp Med (2017) 214(8):2453–70. doi:10.1084/jem.20161595
15. De Luca L, Trino S, Laurenzana I, Tagliaferri D, Falco G, Grieco V, et al. Knockdown of miR-128a induces Lin28a expression and reverts myeloid differentiation blockage in acute myeloid leukemia. Cell Death Dis (2017) 8(6):e2849. doi:10.1038/cddis.2017.253
16. Sadras T, Kok CH, Perugini M, Ramshaw HS, D’Andrea RJ. miR-155 as a potential target of IL-3 signaling in primary AML cells. Leuk Res (2017) 57:57–9. doi:10.1016/j.leukres.2017.02.010
17. Wurm AA, Zjablovskaja P, Kardosova M, Gerloff D, Brauer-Hartmann D, Katzerke C, et al. Disruption of the C/EBPalpha-miR-182 balance impairs granulocytic differentiation. Nat Commun (2017) 8(1):46. doi:10.1038/s41467-017-00032-6
18. Zhao H, Wang D, Du W, Gu D, Yang R. MicroRNA and leukemia: tiny molecule, great function. Crit Rev Oncol Hematol (2010) 74(3):149–55. doi:10.1016/j.critrevonc.2009.05.001
19. Han GT, Sun ZL. Up-regulation of serum miR-4262 predicts clinical outcome of patients with acute myeloid leukemia. Eur Rev Med Pharmacol Sci (2017) 21(9):2172–6.
20. Amodio N, Rossi M, Raimondi L, Pitari MR, Botta C, Tagliaferri P, et al. miR-29s: a family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget (2015) 6(15):12837–61. doi:10.18632/oncotarget.3805
21. Liu L, Ren W, Chen K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell Physiol Biochem (2017) 41(5):1981–92. doi:10.1159/000475277
22. Pigazzi M, Manara E, Baron E, Basso G. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res (2009) 69(6):2471–8. doi:10.1158/0008-5472.CAN-08-3404
23. Hu Y, Dong X, Chu G, Lai G, Zhang B, Wang L, et al. miR-137 downregulates c-kit expression in acute myeloid leukemia. Leuk Res (2017) 57:72–7. doi:10.1016/j.leukres.2017.01.028
24. Zhang Y, Liu Y, Xu X. Upregulation of miR-142-3p improves drug sensitivity of acute myelogenous leukemia through reducing P-glycoprotein and repressing autophagy by targeting HMGB1. Transl Oncol (2017) 10(3):410–8. doi:10.1016/j.tranon.2017.03.003
25. Dell’Aversana C, Giorgio C, D’Amato L, Lania G, Matarese F, Saeed S, et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia (2017) 31(11):2315–25. doi:10.1038/leu.2017.64
26. Xiao Y, Deng T, Su C, Shang Z. MicroRNA 217 inhibits cell proliferation and enhances chemosensitivity to doxorubicin in acute myeloid leukemia by targeting KRAS. Oncol Lett (2017) 13(6):4986–94. doi:10.3892/ol.2017.6076
27. Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell (2005) 123(5):819–31. doi:10.1016/j.cell.2005.09.023
28. Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell (2007) 12(5):457–66. doi:10.1016/j.ccr.2007.09.020
29. Zhang Y, Lin J, Huang W, Cao Y, Liu Y, Wang T, et al. The effect of circulating miR-223 on surveillance of different cancers: a meta-analysis. Onco Targets Ther (2017) 10:3193–201. doi:10.2147/ott.s137837
30. Liu X, Heng C, Li Y, Yu L. MiR-302a sensitizes leukemia cells to etoposide by targeting Rad52. Oncotarget (2017) 8(43):73884–91. doi:10.18632/oncotarget.17878
31. Su R, Gong JN, Chen MT, Song L, Shen C, Zhang XH, et al. c-Myc suppresses miR-451 dash, verticalYWTAZ/AKT axis via recruiting HDAC3 in acute myeloid leukemia. Oncotarget (2016) 7(47):77430–43. doi:10.18632/oncotarget.12679
32. Yuan C, Xu L, Du P, Pang J. miRNA-650 exerts anti-leukemia activity by inhibiting cell proliferation through Gfi1 targeting. Tumori (2017). doi:10.5301/tj.5000643
33. Hu J, Zheng L, Shen X, Zhang Y, Li C, Xi T. MicroRNA-125b inhibits AML cells differentiation by directly targeting Fes. Gene (2017) 620:1–9. doi:10.1016/j.gene.2017.04.002
34. Schotte D, Chau JC, Sylvester G, Liu G, Chen C, van der Velden VH, et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia (2009) 23(2):313–22. doi:10.1038/leu.2008.286
35. Ke S, Li RC, Lu J, Meng FK, Feng YK, Fang MH. MicroRNA-192 regulates cell proliferation and cell cycle transition in acute myeloid leukemia via interaction with CCNT2. Int J Hematol (2017) 106(2):258–65. doi:10.1007/s12185-017-2232-2
36. Sayadi M, Ajdary S, Nadali F, Rostami S, Edalati Fahtabad M. Tumor suppressive function of microRNA-192 in acute lymphoblastic leukemia. Bosn J Basic Med Sci (2017) 17(3):248–54. doi:10.17305/bjbms.2017.1921
37. Gao XN, Lin J, Li YH, Gao L, Wang XR, Wang W, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene (2011) 30(31):3416–28. doi:10.1038/onc.2011.62
38. Agirre X, Martinez-Climent JA, Odero MD, Prosper F. Epigenetic regulation of miRNA genes in acute leukemia. Leukemia (2012) 26(3):395–403. doi:10.1038/leu.2011.344
39. Liang YN, Tang YL, Ke ZY, Chen YQ, Luo XQ, Zhang H, et al. MiR-124 contributes to glucocorticoid resistance in acute lymphoblastic leukemia by promoting proliferation, inhibiting apoptosis and targeting the glucocorticoid receptor. J Steroid Biochem Mol Biol (2017) 172:62–8. doi:10.1016/j.jsbmb.2017.05.014
40. Nabhan M, Louka ML, Khairy E, Tash F, Ali-Labib R, El-Habashy S. MicroRNA-181a and its target Smad 7 as potential biomarkers for tracking child acute lymphoblastic leukemia. Gene (2017) 628:253–8. doi:10.1016/j.gene.2017.07.052
41. Xu L, Guo Y, Yan W, Cen J, Niu Y, Yan Q, et al. High level of miR-196b at newly diagnosed pediatric acute myeloid leukemia predicts a poor outcome. EXCLI J (2017) 16:197–209. doi:10.17179/excli2016-707
42. Bhagat TD, Zhou L, Sokol L, Kessel R, Caceres G, Gundabolu K, et al. miR-21 mediates hematopoietic suppression in MDS by activating TGF-beta signaling. Blood (2013) 121(15):2875–81. doi:10.1182/blood-2011-12-397067
43. Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood (2007) 109(11):4944–51. doi:10.1182/blood-2006-12-062398
44. Vasilatou D, Papageorgiou SG, Kontsioti F, Kontos CK, Tsiotra P, Mpakou V, et al. Expression analysis of mir-17-5p, mir-20a and let-7a microRNAs and their target proteins in CD34+ bone marrow cells of patients with myelodysplastic syndromes. Leuk Res (2013) 37(3):251–8. doi:10.1016/j.leukres.2012.11.011
45. Jia Q, Sun H, Xiao F, Sai Y, Li Q, Zhang X, et al. miR-17-92 promotes leukemogenesis in chronic myeloid leukemia via targeting A20 and activation of NF-kappaB signaling. Biochem Biophys Res Commun (2017) 487(4):868–74. doi:10.1016/j.bbrc.2017.04.144
46. Sokol L, Caceres G, Volinia S, Alder H, Nuovo GJ, Liu CG, et al. Identification of a risk dependent microRNA expression signature in myelodysplastic syndromes. Br J Haematol (2011) 153(1):24–32. doi:10.1111/j.1365-2141.2011.08581.x
47. Lechman ER, Gentner B, Ng SW, Schoof EM, van Galen P, Kennedy JA, et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell (2016) 29(2):214–28. doi:10.1016/j.ccell.2015.12.011
48. Guinn D, Lehman A, Fabian C, Yu L, Maddocks K, Andritsos LA, et al. The regulation of tumor-suppressive microRNA, miR-126, in chronic lymphocytic leukemia. Cancer Med (2017) 6(4):778–87. doi:10.1002/cam4.996
49. Zuo Z, Maiti S, Hu S, Loghavi S, Calin GA, Garcia-Manero G, et al. Plasma circulating-microRNA profiles are useful for assessing prognosis in patients with cytogenetically normal myelodysplastic syndromes. Mod Pathol (2015) 28(3):373–82. doi:10.1038/modpathol.2014.108
50. Nimer SD. Myelodysplastic syndromes. Blood (2008) 111(10):4841–51. doi:10.1182/blood-2007-08-078139
51. Starczynowski DT, Morin R, McPherson A, Lam J, Chari R, Wegrzyn J, et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood (2011) 117(2):595–607. doi:10.1182/blood-2010-03-277012
52. Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature (2010) 464(7290):852–7. doi:10.1038/nature08851
53. Fang J, Varney M, Starczynowski DT. Implication of microRNAs in the pathogenesis of MDS. Curr Pharm Des (2012) 18(22):3170–9. doi:10.2174/1381612811209023170
54. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell (2005) 120(5):635–47. doi:10.1016/j.cell.2005.01.014
55. Song SJ, Pandolfi PP. MicroRNAs in the pathogenesis of myelodysplastic syndromes and myeloid leukaemia. Curr Opin Hematol (2014) 21(4):276–82. doi:10.1097/MOH.0000000000000054
56. Rhyasen GW, Starczynowski DT. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia (2012) 26(1):13–22. doi:10.1038/leu.2011.221
57. O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med (2008) 205(3):585–94. doi:10.1084/jem.20072108
58. Cao M, Shikama Y, Kimura H, Noji H, Ikeda K, Ono T, et al. Mechanisms of impaired neutrophil migration by microRNAs in myelodysplastic syndromes. J Immunol (2017) 198(5):1887–99. doi:10.4049/jimmunol.1600622
59. Kim Y, Cheong JW, Kim YK, Eom JI, Jeung HK, Kim SJ, et al. Serum microRNA-21 as a potential biomarker for response to hypomethylating agents in myelodysplastic syndromes. PLoS One (2014) 9(2):e86933. doi:10.1371/journal.pone.0086933
60. Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell (2013) 13(1):87–101. doi:10.1016/j.stem.2013.06.003
61. Coutinho DF, Monte-Mor BC, Vianna DT, Rouxinol ST, Batalha AB, Bueno AP, et al. TET2 expression level and 5-hydroxymethylcytosine are decreased in refractory cytopenia of childhood. Leuk Res (2015) 39(10):1103–8. doi:10.1016/j.leukres.2015.07.005
62. Cancer Genome Atlas Research NetworkLey TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med (2013) 368(22):2059–74. doi:10.1056/NEJMoa1301689
63. Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood (2008) 111(10):5078–85. doi:10.1182/blood-2008-01-133355
64. Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med (2008) 358(18):1919–28. doi:10.1056/NEJMoa074256
65. Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood (2008) 111(6):3183–9. doi:10.1182/blood-2007-07-098749
66. Chen P, Price C, Li Z, Li Y, Cao D, Wiley A, et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci U S A (2013) 110(28):11511–6. doi:10.1073/pnas.1310144110
67. Nowek K, Sun SM, Bullinger L, Bindels EM, Exalto C, Dijkstra MK, et al. Aberrant expression of miR-9/9* in myeloid progenitors inhibits neutrophil differentiation by post-transcriptional regulation of ERG. Leukemia (2016) 30(1):229–37. doi:10.1038/leu.2015.183
68. Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F, et al. miR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia (2014) 28(5):1022–32. doi:10.1038/leu.2013.357
69. Wang J, Cao N, Yuan M, Cui H, Tang Y, Qin L, et al. MicroRNA-125b/Lin28 pathway contributes to the mesendodermal fate decision of embryonic stem cells. Stem Cells Dev (2012) 21(9):1524–37. doi:10.1089/scd.2011.0350
70. Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci U S A (2010) 107(50):21558–63. doi:10.1073/pnas.1016611107
71. Li X, Zhang J, Gao L, McClellan S, Finan MA, Butler TW, et al. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ (2012) 19(3):378–86. doi:10.1038/cdd.2011.127
72. Su R, Lin HS, Zhang XH, Yin XL, Ning HM, Liu B, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene (2015) 34(25):3226–39. doi:10.1038/onc.2014.274
73. de Leeuw DC, Denkers F, Olthof MC, Rutten AP, Pouwels W, Schuurhuis GJ, et al. Attenuation of microRNA-126 expression that drives CD34+38- stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res (2014) 74(7):2094–105. doi:10.1158/0008-5472.CAN-13-1733
74. Dorrance AM, Neviani P, Ferenchak GJ, Huang X, Nicolet D, Maharry KS, et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia (2015) 29(11):2143–53. doi:10.1038/leu.2015.139
75. Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell (2012) 11(6):799–811. doi:10.1016/j.stem.2012.09.001
76. Li Z, Chen P, Su R, Li Y, Hu C, Wang Y, et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood (2015) 126(17):2005–15. doi:10.1182/blood-2015-04-639062
77. Gerloff D, Grundler R, Wurm AA, Brauer-Hartmann D, Katzerke C, Hartmann JU, et al. NF-kappaB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia (2015) 29(3):535–47. doi:10.1038/leu.2014.231
78. Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrozek K, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol (2013) 31(17):2086–93. doi:10.1200/JCO.2012.45.6228
79. Schneider E, Staffas A, Rohner L, Krowiorz K, Heuser M, Dohner K, et al. MicroRNA-155 is upregulated in MLL-rearranged AML but its absence does not affect leukemia development. Exp Hematol (2016) 44(12):1166–71. doi:10.1016/j.exphem.2016.08.012
80. Wallace JA, Kagele DA, Eiring AM, Kim CN, Hu R, Runtsch MC, et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood (2017) 129(23):3074–86. doi:10.1182/blood-2016-09-740209
81. Jiang X, Hu C, Arnovitz S, Bugno J, Yu M, Zuo Z, et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat Commun (2016) 7:11452. doi:10.1038/ncomms11452
82. Shen C, Chen MT, Zhang XH, Yin XL, Ning HM, Su R, et al. The PU.1-modulated microRNA-22 is a regulator of monocyte/macrophage differentiation and acute myeloid leukemia. PLoS Genet (2016) 12(9):e1006259. doi:10.1371/journal.pgen.1006259
83. Jiang W, Min J, Sui X, Qian Y, Liu Y, Liu Z, et al. MicroRNA-26a-5p and microRNA-23b-3p up-regulate peroxiredoxin III in acute myeloid leukemia. Leuk Lymphoma (2015) 56(2):460–71. doi:10.3109/10428194.2014.924115
84. Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol (2014) 15(4):243–56. doi:10.1038/nrm3772
85. Wang XS, Gong JN, Yu J, Wang F, Zhang XH, Yin XL, et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood (2012) 119(21):4992–5004. doi:10.1182/blood-2011-10-385716
86. Gong JN, Yu J, Lin HS, Zhang XH, Yin XL, Xiao Z, et al. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ (2014) 21(1):100–12. doi:10.1038/cdd.2013.133
87. Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med (2010) 207(3):475–89. doi:10.1084/jem.20090831
88. Wong KY, So CC, Loong F, Chung LP, Lam WW, Liang R, et al. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS One (2011) 6(4):e19027. doi:10.1371/journal.pone.0019027
89. Hinai AA, Valk PJ. Review: aberrant EVI1 expression in acute myeloid leukaemia. Br J Haematol (2016) 172(6):870–8. doi:10.1111/bjh.13898
90. Vazquez I, Maicas M, Marcotegui N, Conchillo A, Guruceaga E, Roman-Gomez J, et al. Silencing of hsa-miR-124 by EVI1 in cell lines and patients with acute myeloid leukemia. Proc Natl Acad Sci U S A (2010) 107(44):E167–8; author reply E169–70. doi:10.1073/pnas.1011540107
91. Ufkin ML, Peterson S, Yang X, Driscoll H, Duarte C, Sathyanarayana P. miR-125a regulates cell cycle, proliferation, and apoptosis by targeting the ErbB pathway in acute myeloid leukemia. Leuk Res (2014) 38(3):402–10. doi:10.1016/j.leukres.2013.12.021
92. Alemdehy MF, Haanstra JR, de Looper HW, van Strien PM, Verhagen-Oldenampsen J, Caljouw Y, et al. ICL-induced miR139-3p and miR199a-3p have opposite roles in hematopoietic cell expansion and leukemic transformation. Blood (2015) 125(25):3937–48. doi:10.1182/blood-2014-11-612507
93. Gentner B, Pochert N, Rouhi A, Boccalatte F, Plati T, Berg T, et al. MicroRNA-223 dose levels fine tune proliferation and differentiation in human cord blood progenitors and acute myeloid leukemia. Exp Hematol (2015) 43(10):858–68.e7. doi:10.1016/j.exphem.2015.05.018
94. Xiao Y, Su C, Deng T. miR-223 decreases cell proliferation and enhances cell apoptosis in acute myeloid leukemia via targeting FBXW7. Oncol Lett (2016) 12(5):3531–6. doi:10.3892/ol.2016.5115
95. Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol (2009) 27(4):619–28. doi:10.1200/jco.2008.17.9812
96. Mueller BU, Pabst T. C/EBPalpha and the pathophysiology of acute myeloid leukemia. Curr Opin Hematol (2006) 13(1):7–14. doi:10.1097/01.moh.0000190110.08156.96
97. Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Muller-Tidow C, Bohlander SK, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood (2010) 115(9):1768–78. doi:10.1182/blood-2009-08-240101
98. de Leeuw DC, van den Ancker W, Denkers F, de Menezes RX, Westers TM, Ossenkoppele GJ, et al. MicroRNA profiling can classify acute leukemias of ambiguous lineage as either acute myeloid leukemia or acute lymphoid leukemia. Clin Cancer Res (2013) 19(8):2187–96. doi:10.1158/1078-0432.CCR-12-3657
99. Guo Y, Strickland SA, Mohan S, Li S, Bosompem A, Vickers KC, et al. MicroRNAs and tRNA-derived fragments predict the transformation of myelodysplastic syndromes to acute myeloid leukemia. Leuk Lymphoma (2017) 58(9):1–15. doi:10.1080/10428194.2016.1272680
100. Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood (2011) 117(4):1121–9. doi:10.1182/blood-2010-09-191312
101. Sun SM, Rockova V, Bullinger L, Dijkstra MK, Dohner H, Lowenberg B, et al. The prognostic relevance of miR-212 expression with survival in cytogenetically and molecularly heterogeneous AML. Leukemia (2013) 27(1):100–6. doi:10.1038/leu.2012.158
102. Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, et al. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci Rep (2015) 5:11295. doi:10.1038/srep11295
103. Zhi F, Cao X, Xie X, Wang B, Dong W, Gu W, et al. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS One (2013) 8(2):e56718. doi:10.1371/journal.pone.0056718
104. Koutova L, Sterbova M, Pazourkova E, Pospisilova S, Svobodova I, Horinek A, et al. The impact of standard chemotherapy on miRNA signature in plasma in AML patients. Leuk Res (2015) 39(12):1389–95. doi:10.1016/j.leukres.2015.08.009
105. Henry JC, Azevedo-Pouly AC, Schmittgen TD. MicroRNA replacement therapy for cancer. Pharm Res (2011) 28(12):3030–42. doi:10.1007/s11095-011-0548-9
106. Huang X, Schwind S, Yu B, Santhanam R, Wang H, Hoellerbauer P, et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin Cancer Res (2013) 19(9):2355–67. doi:10.1158/1078-0432.CCR-12-3191
107. Velu CS, Chaubey A, Phelan JD, Horman SR, Wunderlich M, Guzman ML, et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J Clin Invest (2014) 124(1):222–36. doi:10.1172/JCI66005
108. Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov (2014) 13(8):622–38. doi:10.1038/nrd4359
109. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med (2013) 368(18):1685–94. doi:10.1056/NEJMoa1209026
Keywords: acute myeloid leukemia, microRNAs, microRNA-based therapeutics, myelodysplastic syndrome, myeloid malignancies
Citation: Setijono SR, Kwon HY and Song SJ (2018) MicroRNA, an Antisense RNA, in Sensing Myeloid Malignancies. Front. Oncol. 7:331. doi: 10.3389/fonc.2017.00331
Received: 19 September 2017; Accepted: 26 December 2017;
Published: 30 January 2018
Edited by:
Keisuke Ito, Albert Einstein College of Medicine, United StatesReviewed by:
Linda Smit, VU University Medical Center, NetherlandsHideo Watanabe, Icahn School of Medicine at Mount Sinai, United States
Copyright: © 2018 Setijono, Kwon and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyog Young Kwon, hykwon@sch.ac.kr;
Su Jung Song, ssong1@sch.ac.kr
 Stephanie Rebecca Setijono
Stephanie Rebecca Setijono Hyog Young Kwon
Hyog Young Kwon Su Jung Song
Su Jung Song