- Department of Infectious Diseases, Shunde Hospital, Southern Medical University, Shunde, China
An increasing body of evidence supports the association of immune genes with tumorigenesis and prognosis of breast cancer (BC). This research aims at exploring potential regulatory mechanisms and identifying immunogenic prognostic markers for BC, which were used to construct a prognostic signature for disease-free survival (DFS) of BC based on artificial intelligence algorithms. Differentially expressed immune genes were identified between normal tissues and tumor tissues. Univariate Cox regression identified potential prognostic immune genes. Thirty-four transcription factors and 34 immune genes were used to develop an immune regulatory network. The artificial intelligence survival prediction system was developed based on three artificial intelligence algorithms. Multivariate Cox analyses determined 17 immune genes (ADAMTS8, IFNG, XG, APOA5, SIAH2, C2CD2, STAR, CAMP, CDH19, NTSR1, PCDHA1, AMELX, FREM1, CLEC10A, CD1B, CD6, and LTA) as prognostic biomarkers for BC. A prognostic nomogram was constructed on these prognostic genes. Concordance indexes were 0.782, 0.734, and 0.735 for 1-, 3-, and 5- year DFS. The DFS in high-risk group was significantly worse than that in low-risk group. Artificial intelligence survival prediction system provided three individual mortality risk predictive curves based on three artificial intelligence algorithms. In conclusion, comprehensive bioinformatics identified 17 immune genes as potential prognostic biomarkers, which might be potential candidates of immunotherapy targets in BC patients. The current study depicted regulatory network between transcription factors and immune genes, which was helpful to deepen the understanding of immune regulatory mechanisms for BC cancer. Two artificial intelligence survival predictive systems are available at https://zhangzhiqiao7.shinyapps.io/Smart_Cancer_Survival_Predictive_System_16_BC_C1005/ and https://zhangzhiqiao8.shinyapps.io/Gene_Survival_Subgroup_Analysis_16_BC_C1005/. These novel artificial intelligence survival predictive systems will be helpful to improve individualized treatment decision-making.
Introduction
As the most common malignant tumor in women, breast cancer (BC) resulted in 2,088,849 new cases and 626,679 deaths in 2018 (1). Although advances in diagnosis and treatments improved the survival rate of patients with early BC, but the survival rate of patients with advanced BC was still poor (2). Early identification of BC patients with poor prognosis and timely individualized treatments were helpful to improve the prognosis of BC patients.
The tremendous progress of bioinformatics has provided tremendous support for exploring the intrinsic mechanism of tumorigenesis and prognosis. (3–6). Tumor-infiltrating immune cells were reported to be associated with tumorigenesis and prognosis (7, 8). It was reported that there was a significant correlation relationship between tumor-infiltrating immune and prognosis in BC patients (9). Immune-related genes could be used to calculate the immune scores and evaluate the tumor infiltration of immune cells to analyze the tumor immune characteristics (10). There were several prognostic models for prediction of prognosis in BC patients (11–13). However, these prognostic models could only provide mortality risk prediction for patients in different subgroups, but not individual mortality risk prediction for a specific patient at the individual level. From a specific patient's point of view, the patient's own mortality risk prediction was more important than that of patients in different subgroups. Therefore, a prognostic model that can provide individualized mortality risk prediction for a specific patient is helpful to optimize individualized treatment and improve clinical prognosis.
The current research aimed at exploring the relationship of immune-related genes with transcription factor, immune-infiltrating cells, and disease-free survival (DFS) of BC patients. Based on different artificial intelligence algorithms, the current study focused on developing artificial intelligence survival predictive systems for providing individual mortality risk prediction for BC patients.
Materials and Methods
Study Datasets
The original gene expression dataset from The Cancer Genome Atlas (TCGA) database contained 21,205 mRNAs from 1,109 tumor specimens to 113 normal specimens. After removal of patients with survival time <1 month and duplicate samples, 1,030 were included in further survival analyses. The original gene expression values have been log10 transformed for TCGA dataset. GSE31448 dataset (GPL570 platform) contained 246 patients and 23,319 mRNAs.
Gencode.v29 was used for converting probe IDs name to gene symbols.
Differentially Expressed Analyses
Differentially expressed analyses were conducted with cutoff values of log2 |fold change| >1 and P < 0.05 by “edgeR” (14). Data were normalized by Trimmed mean of M values method.
Immune Gene and Transcription Factor
Immune genes were identified through Immunology Database and Analysis Portal database (15). Transcription factors were defined through Cistrome Cancer database (16).
Tumor Immune Infiltration
Six tumor-infiltrating immune cell data were obtained from Tumor IMmune Estimation Resource database (16). Single sample gene set enrichment analysis was used to evaluate tumor immune infiltration scores for 28 immune categories (17, 18).
Statistical Analyses
Statistical analyses were conducted by SPSS Statistics 19.0 (SPSS Inc., Chicago, IL, USA). Artificial intelligence algorithms were performed by Python language 3.7.2 and R software 3.5.2. Artificial intelligence algorithms were carried out according to the original articles: Cox survival regression (19), multitask logistic regression (20, 21), and random survival forest (22, 23). Threshold for statistically significant difference was P < 0.05.
Results
Study Datasets
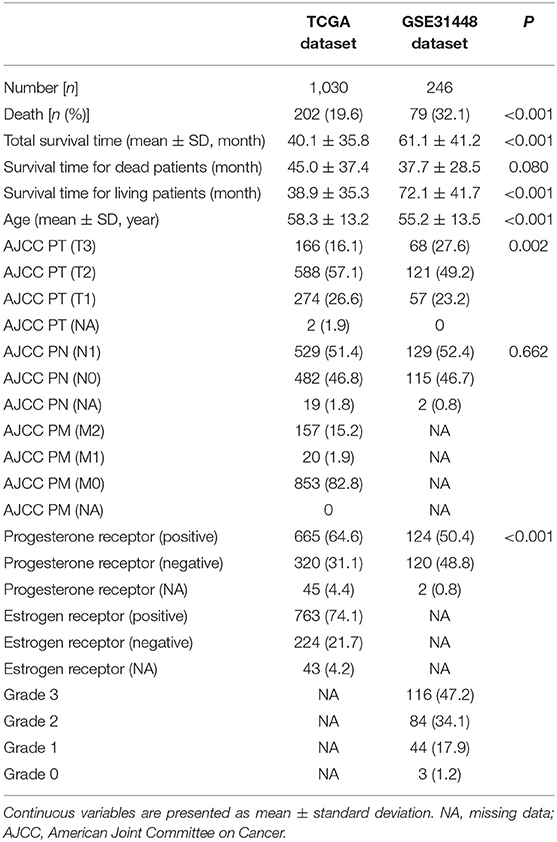
Details of research steps are displayed in Supplementary Figure 1. Table 1 displays the basic information of patients in the model dataset and validation dataset. The mortality rate in the validation dataset was 32.1% (79/246), which was significantly higher than 19.6% (202/1,030) in the model dataset.
Differentially Expressed Analyses
Volcano plots of 21,205 mRNAs and 3,627 immune genes are presented in Figures 1A,B. There were 265 up-regulated and 185 down-regulated immune genes in differentially expressed analyses.
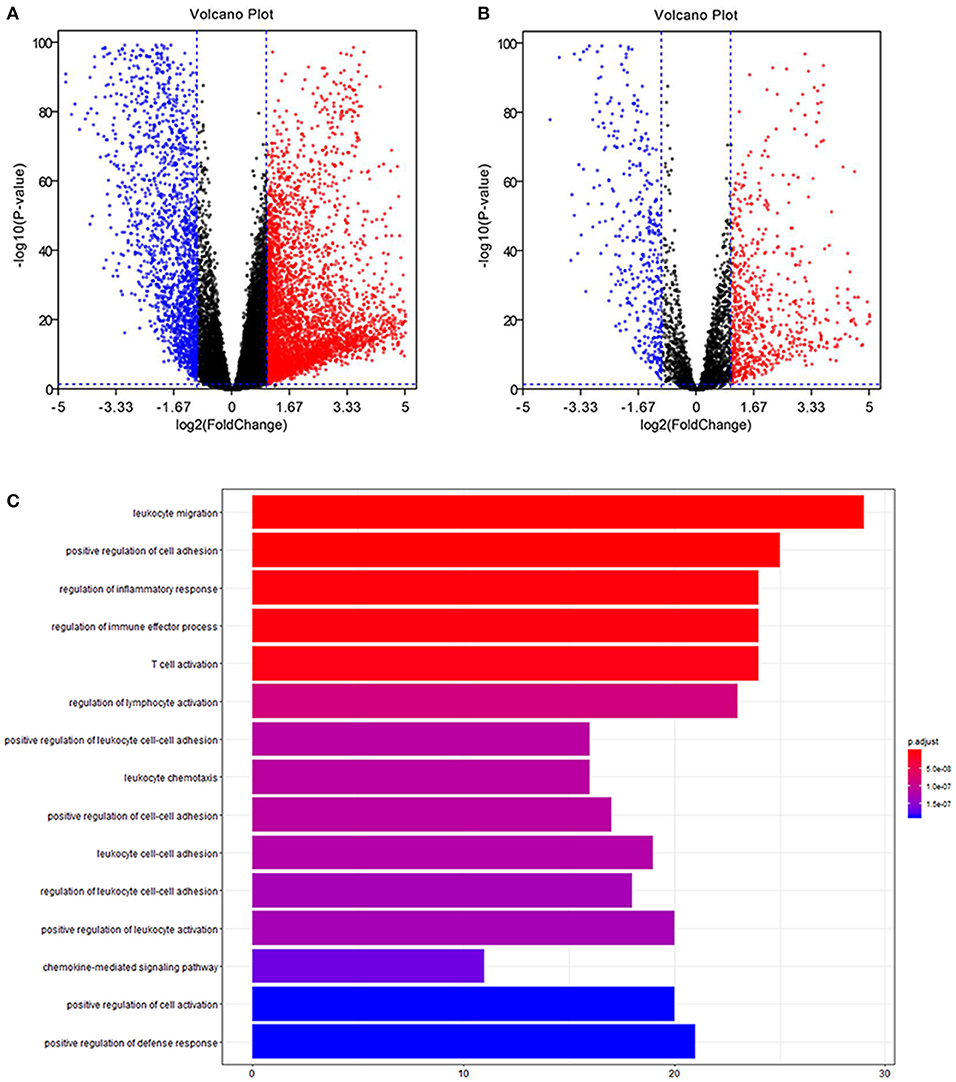
Figure 1. Differentially expression and functional enrichment. (A) Volcano chart of differentially expressed genes. (B) Volcano chart of immune differentially expressed genes. (C) Barplot chart for functional enrichment analysis.
Functional Enrichment Analyses
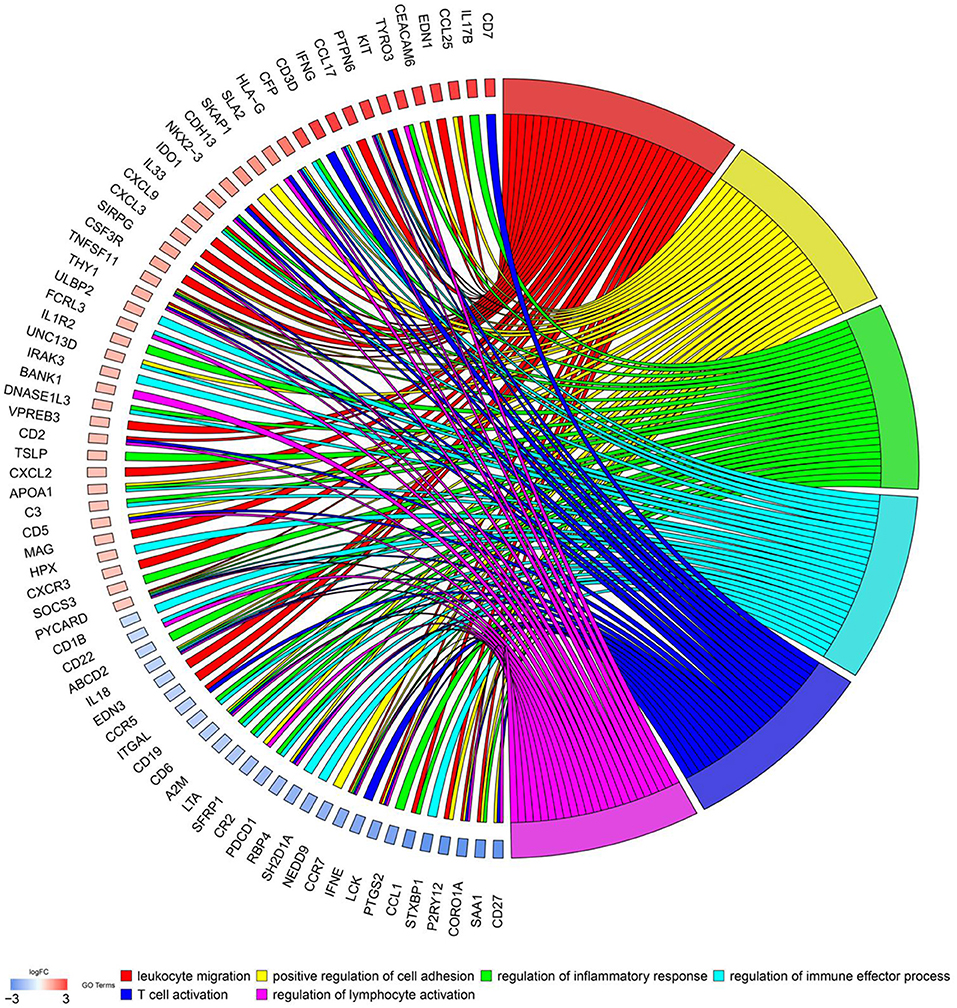
To explore biological functions of immune genes, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed. Bar plot (Figure 1C), GO chord chart (Figure 2), and KEGG chord plot (Supplementary Figure 2) showed that biological functions of immune genes were mainly enriched in leukocyte migration, positive regulation of cell adhesion, regulation of inflammatory response, regulation of immune effector process, T cell activation, regulation of lymphocyte activation, positive regulation of leukocyte cell–cell adhesion, leukocyte chemotaxis, positive regulation of cell–cell adhesion, and leukocyte cell–cell adhesion. The top five KEGG items were as follows: cytokine–cytokine receptor interaction, hematopoietic cell lineage, viral protein interaction with cytokine and cytokine receptor, human T cell leukemia virus 1 infection, and PI3K–Akt signaling pathway.
Immune Regulatory Network
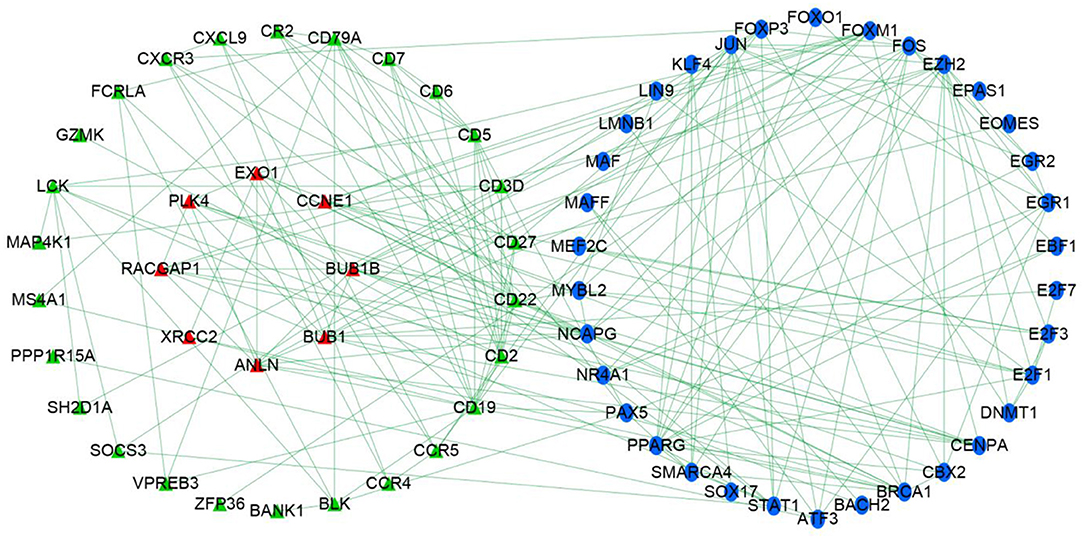
Univariate Cox regression determined 179 prognostic genes for DFS. The current research adopted three methods to explore the relationship between immune genes and transcription factors. First, with thresholds of correlation coefficient >0.5 and P < 0.01, the current study identified transcription factors that were highly correlated with prognostic immune genes. Second, prognostic immune genes and their highly correlated transcription factors were put in STRING database (medium confidence, 0.70) to explore relationships among prognostic immune genes and transcription factors. Finally, Cytoscape v3.6.1 was used to develop an immune regulatory network (Figure 3) on 34 immune genes and 34 transcription factors (24).
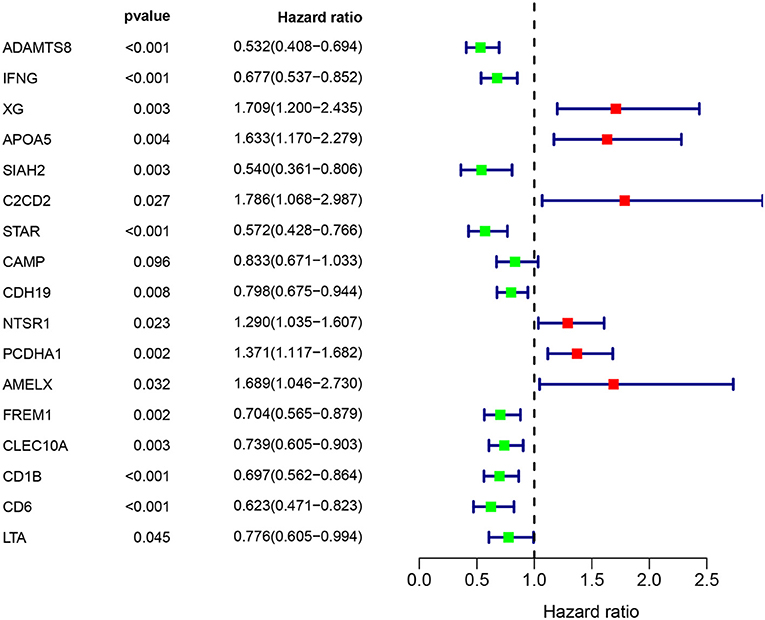
Construction of Prognostic Model
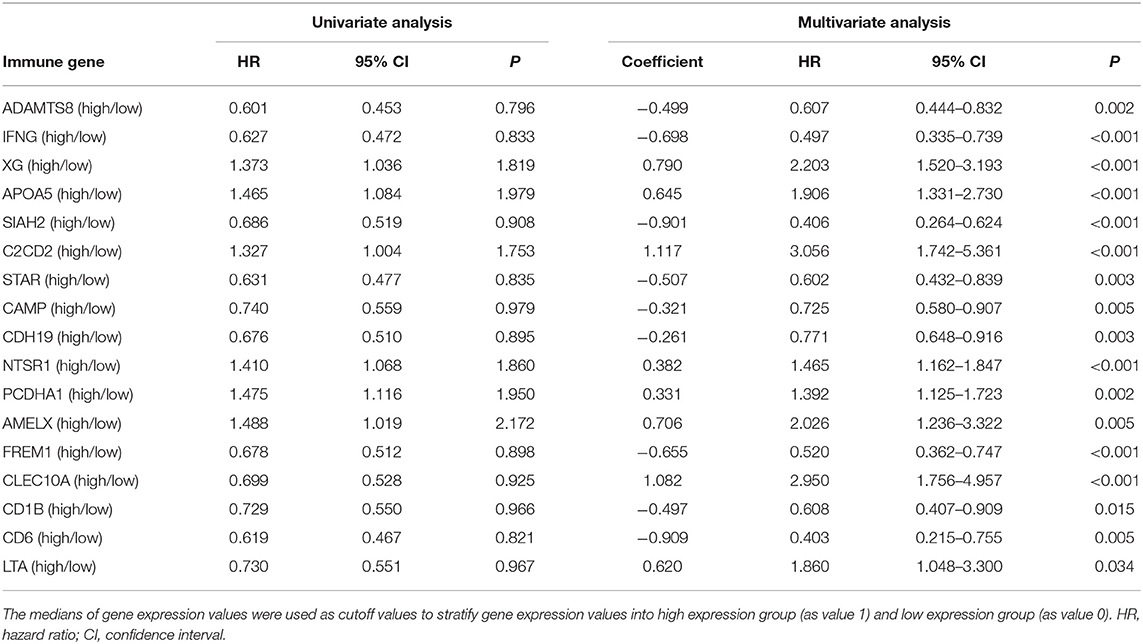
Multivariate Cox regression identified 17 genes as independent influence factors for BC (Table 2 and Figure 4). The formula of prognostic model was as follows:
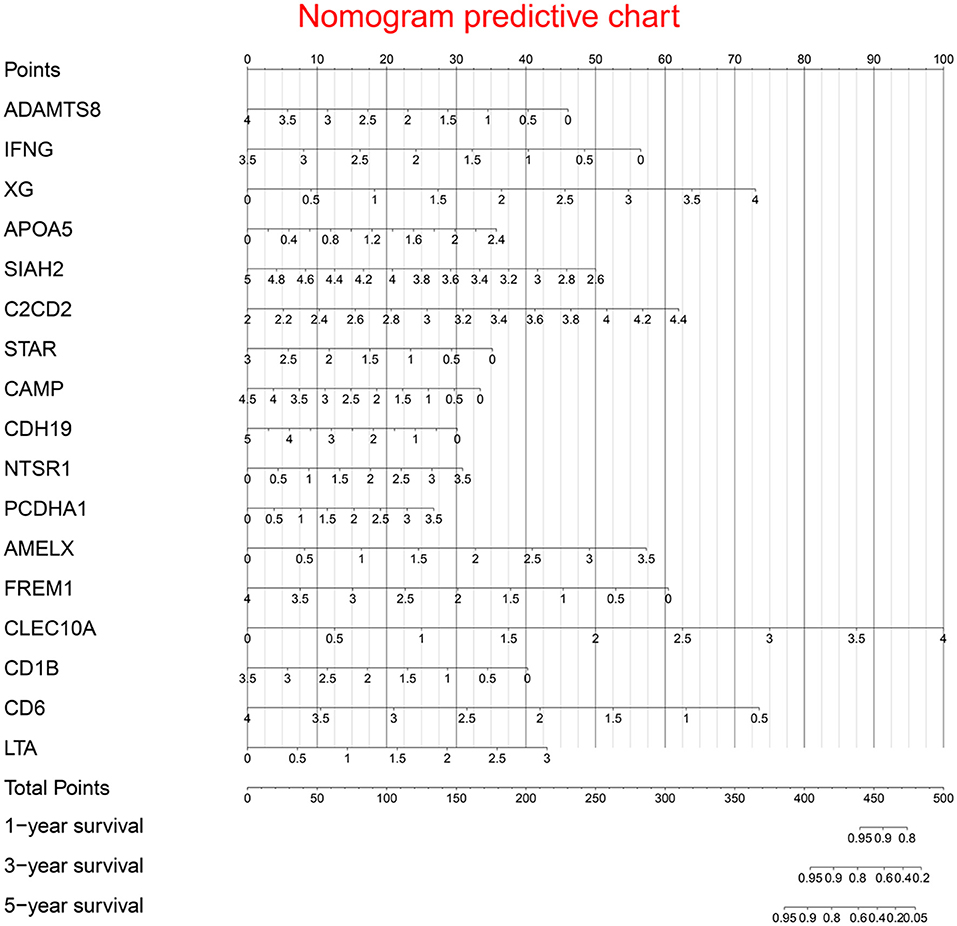
Prognostic nomogram is shown in Figure 5.
Survival curves of prognostic genes are shown in Supplementary Figure 3. Supplementary Figures 4, 5 are predictive value distribution chart and survival status scatterplot, respectively.
Clinical Performance of Model Cohort
Figure 6A displays survival curves in the high-risk group and low-risk group divided by the median of prognostic scores. Figure 6B demonstrates that concordance indexes were 0.782, 0.734, and 0.735 for 1-, 3-, and 5-year survival, respectively. Supplementary Figure 6 shows calibration curves of the model cohort.
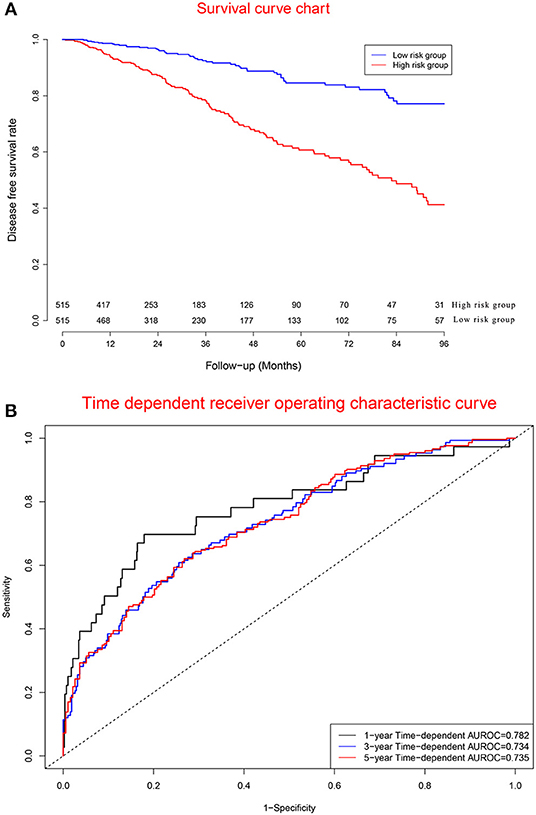
Figure 6. Clinical performance in model cohort. (A) Survival curves for high risk group and low risk group. (B) Time-dependent receiver operating characteristic curves.
Clinical Performance of Validation Cohort
Figure 7A displays survival curves in the high-risk group and low-risk group. Figure 7B demonstrates that concordance indexes were 0.778, 0.738, and 0.792 for 1-, 3-, and 5-year survival, respectively. Supplementary Figure 7 shows calibration curves of the validation cohort.
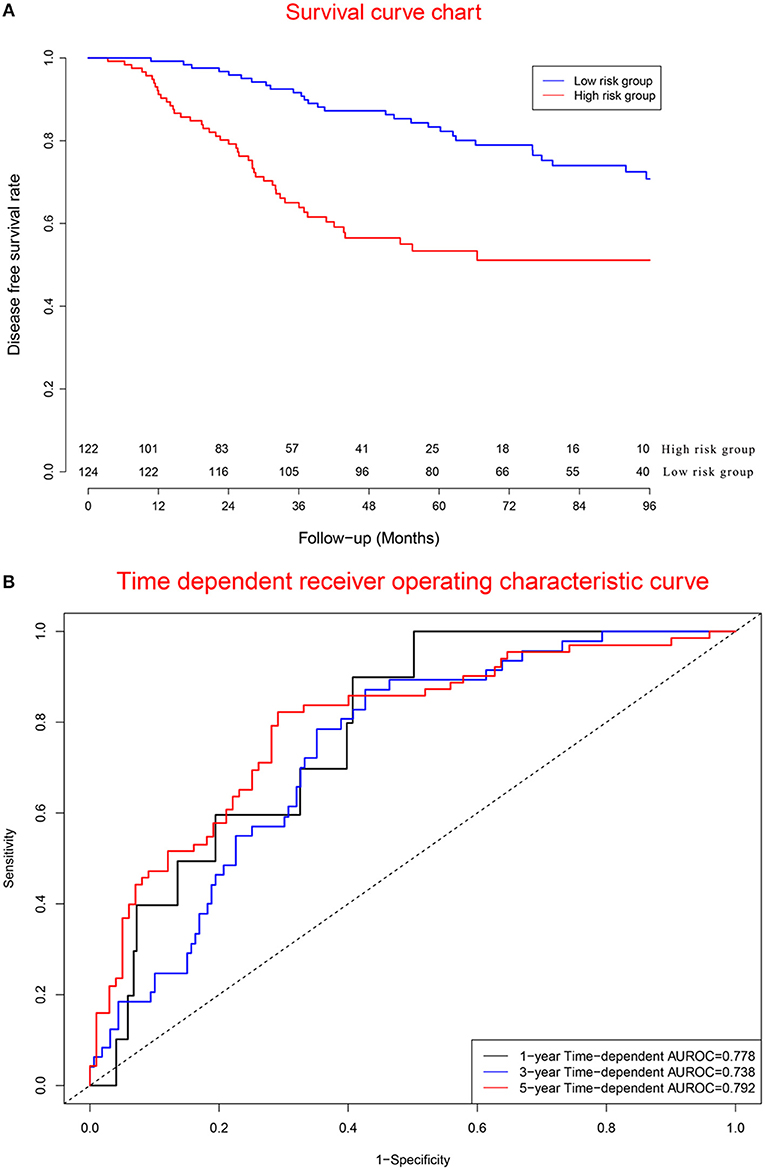
Figure 7. Clinical performance in validation cohort. (A) Survival curves for high risk group and low risk group. (B) Time-dependent receiver operating characteristic curves.
Artificial Intelligence Survival Prediction System
Artificial intelligence survival prediction system was constructed to provide individual mortality risk prediction for BC patients (Figure 8). This tool could provide three individual mortality risk predictive curves by using random survival forest algorithm (Figure 8A), multitask logistic regression algorithm (Figure 8B), and Cox survival regression algorithm (Figure 8C). Artificial intelligence survival prediction system is available at https://zhangzhiqiao7.shinyapps.io/Smart_Cancer_Survival_Predictive_System_16_BC_C1005/.
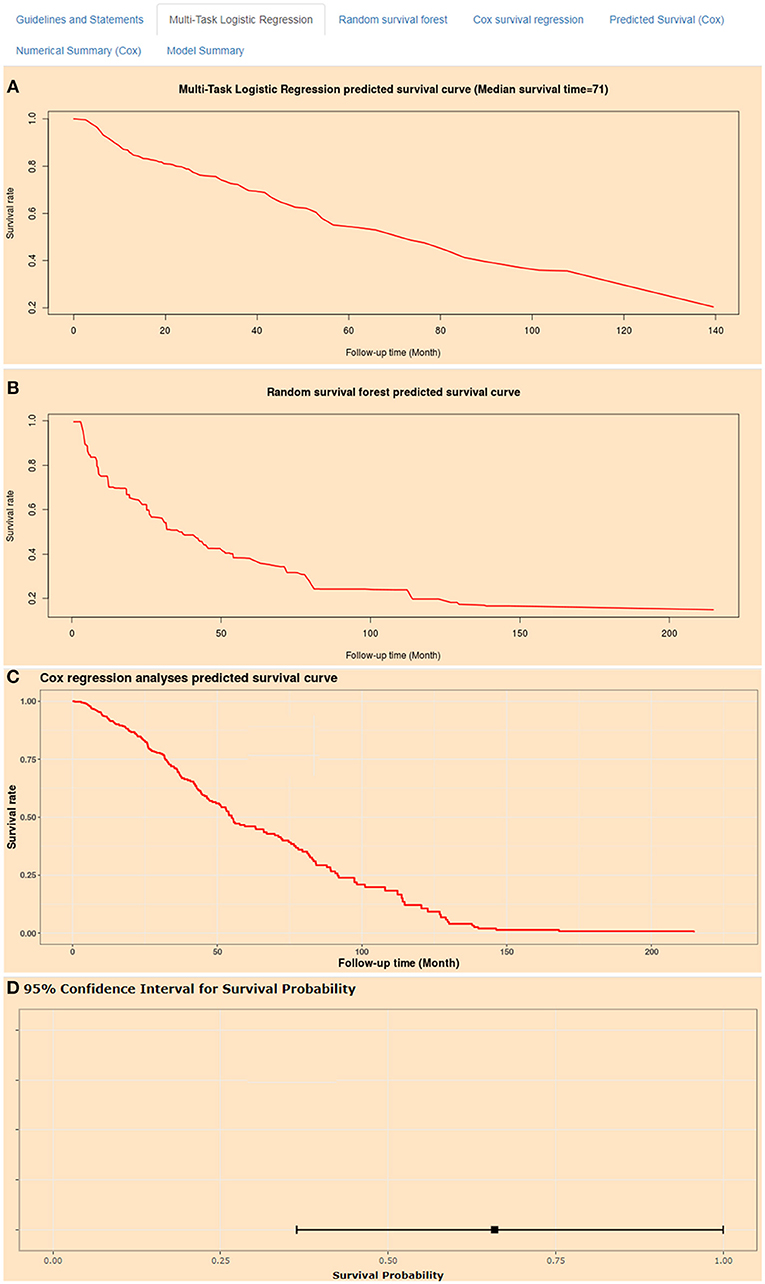
Figure 8. Home page of Smart Cancer Survival Predictive Predictive System. (A) Multi-Task logistic regression predicted survival curves. (B) Random survival forest predicted survival curves. (C) Cox survival regression predicted survival curves. (D) Cox survival regression predicted mortality percentage and 95% confidence interval.
Gene Survival Analysis Screen System
The Gene Survival Analysis Screen System was constructed for exploratory research of immune genes (Supplementary Figure 8). The Gene Survival Analysis Screen System is available at https://zhangzhiqiao8.shinyapps.io/Gene_Survival_Subgroup_Analysis_16_BC_C1005/.
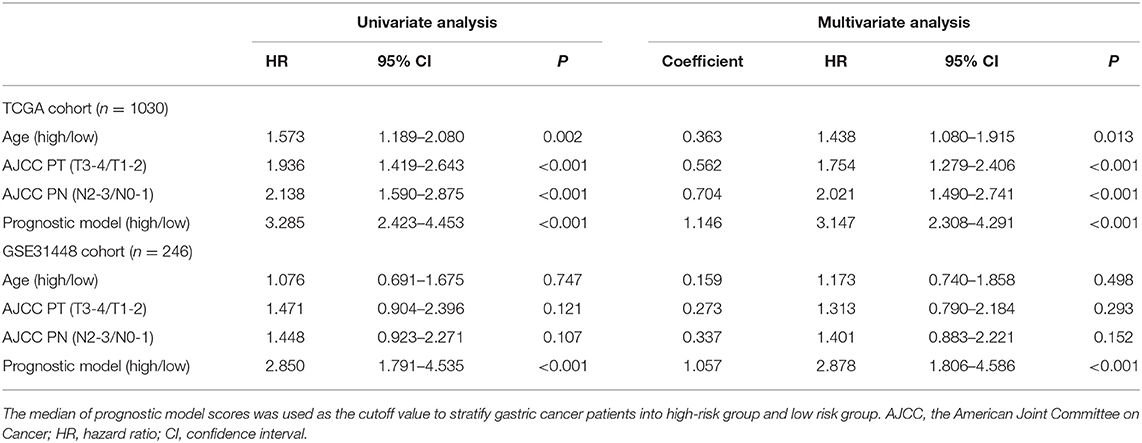
Independence Assessment
Prognostic signature, AJCC PT, and AJCC PN were independent risk factors for DFS in the model dataset (Table 3). In the validation dataset, prognostic signature was proven to be an independent risk factor for DFS.
Clinical Correlation Analyses
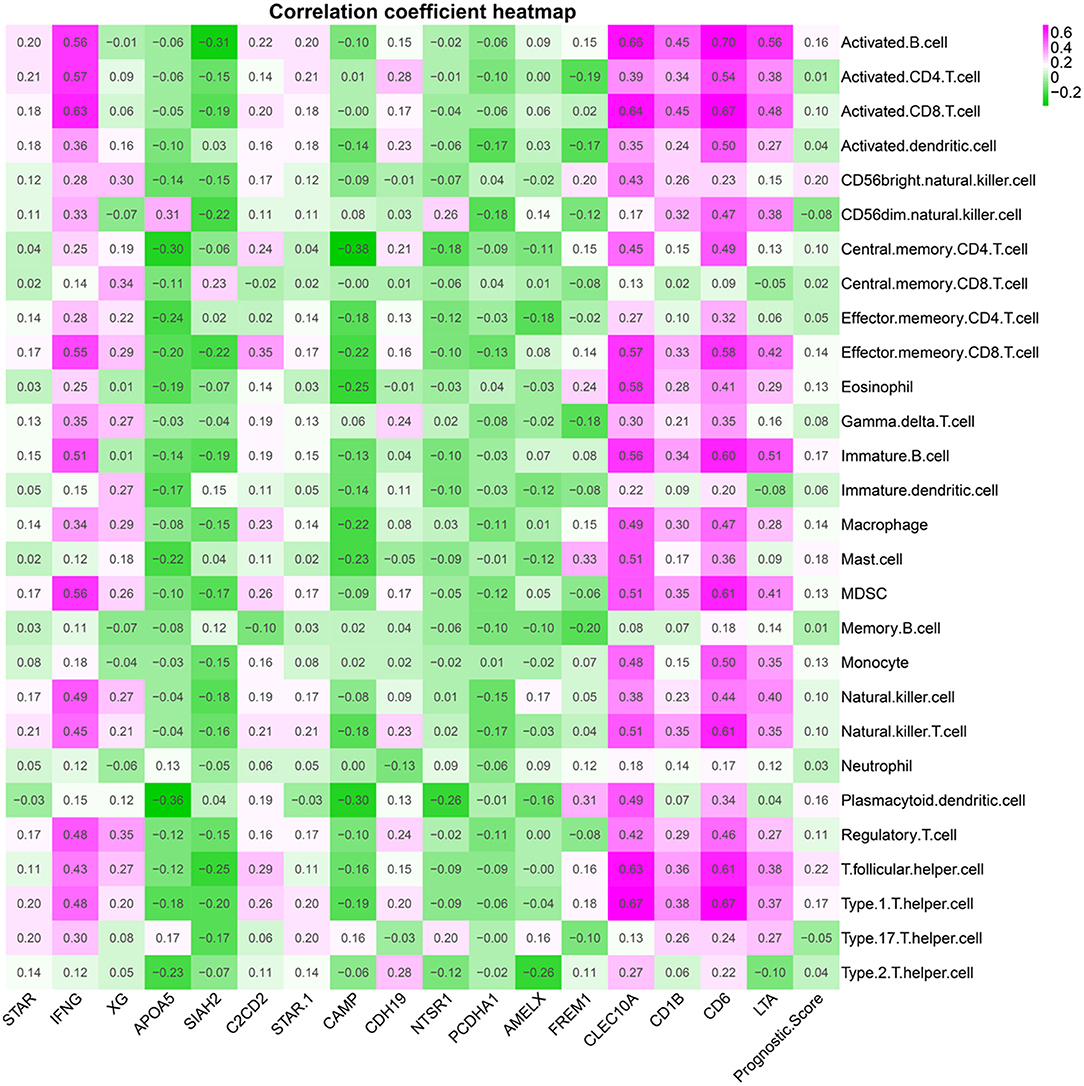
Figure 9 shows a correlation coefficient heatmap between prognostic genes and clinical variables. Supplementary Figure 9 presents correlation significance heatmap between prognostic genes and clinical variables.
Tumor Immune Infiltration Correlation Analyses
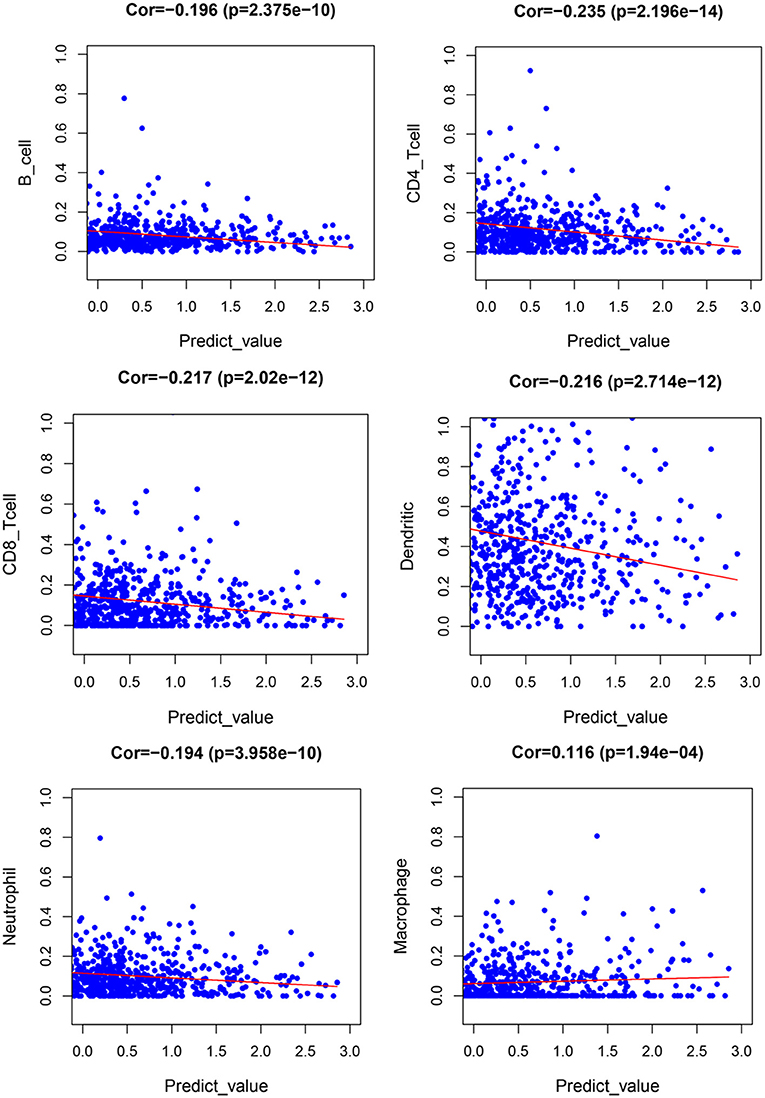
Figure 10 presents correlation coefficient heatmap between tumor immune infiltration and prognostic genes. Supplementary Figure 10 presents correlation significance heatmap between tumor immune infiltration and prognostic genes.
Tumor Immune Infiltration
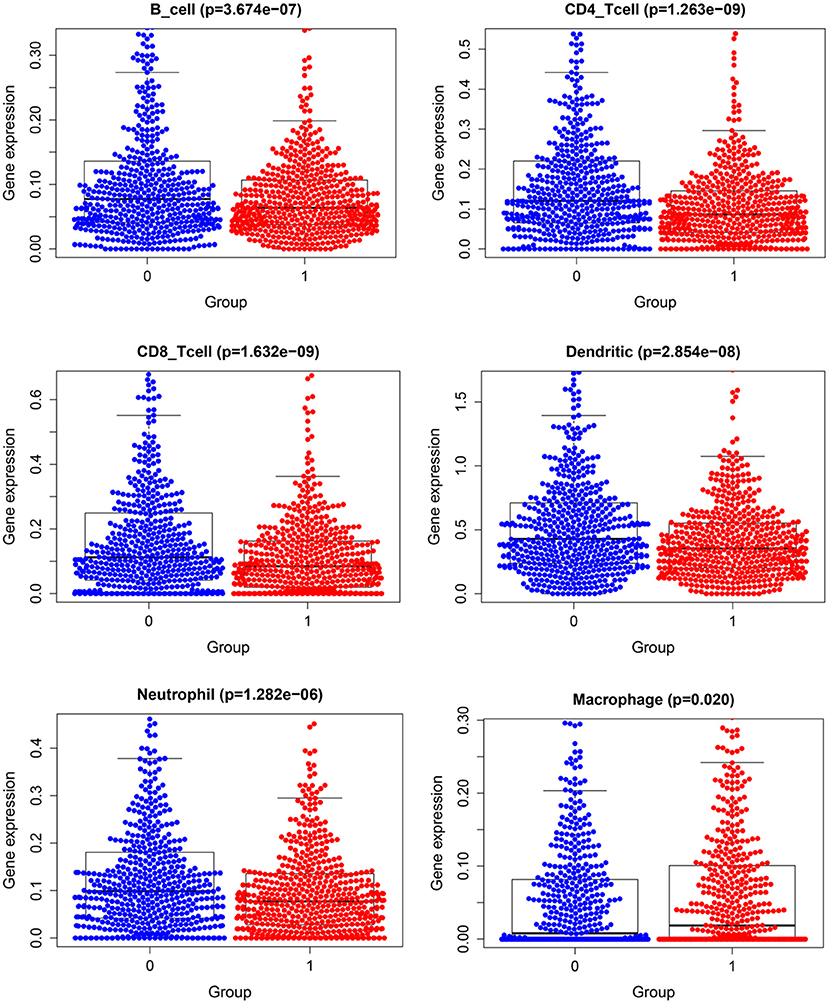
Figure 11 demonstrates expression levels of six tumor immune infiltration in the high-risk group and low-risk group. Figure 12 presents scatterplots between six tumor immune infiltrations and prognostic score.
Gene Differential Expression Between Normal Samples and Tumor Samples
To demonstrate the gene differential expression between normal samples and tumor samples at the molecular level, the current study performed group differential expression analyses between normal samples and tumor samples obtained from TCGA database. There were 1,109 tumor samples and 113 normal samples for group differential expression analyses. Supplementary Figure 11 presents the gene differential expression between normal samples and tumor samples at the molecular level.
Clinical Performance in Different Cancers
To explore the clinical performance of the current prognostic model for other cancers, four tumor datasets were obtained from TCGA database as external validation datasets. There were 348 patients in the hepatocellular carcinoma dataset, 265 patients in the colorectal cancer dataset, 494 patients in the lung cancer dataset, and 370 patients in the ovarian cancer dataset. The prognostic scores in external validation datasets were calculated according to the previous formula derived from the model dataset. Survival curve analyses indicated good diagnostic performance of the current prognostic model for hepatocellular carcinoma, colorectal cancer, lung cancer, and ovarian cancer (Supplementary Figure 12), suggesting that the current prognostic model might be useful for other malignant tumors.
External Validation of Accuracy and Clinical Validity
To validate the accuracy and clinical validity of the current prognostic model in other external validation dataset, hepatocellular carcinoma dataset, colorectal cancer dataset, lung cancer dataset, and ovarian cancer dataset were downloaded from TCGA database. These four malignant-tumor datasets were merged into one joint dataset as external validation dataset with 1,640 tumor patients. Supplementary Figure 13A demonstrates that concordance indexes were 0.832, 0.781, and 0.778 for 1-, 3-, and 5-year survival, respectively. Supplementary Figure 13B suggests that the current prognostic model could distinguish tumor patients with high mortality risk from those with low mortality risk. Calibration curves of external validation dataset showed good accordance between actual mortality percentage and predicted mortality percentage (Supplementary Figure 14).
Discussion
The current research determined 17 immune genes as prognostic biomarkers for DFS. Then the current research depicted regulatory relationships among transcription factors and immune genes through correlation analyses and STRING database. Based on these 17 immune genes, the current research created a prognostic nomogram to predict the DFS for BC patients. Based on the previous prognostic nomogram, the current research developed two artificial intelligence survival predictive systems for individual mortality risk prediction. These two artificial intelligence survival predictive systems were helpful to provide precise individual mortality risk prediction and improve individual treatment decision-making.
Several prognostic models have been built for predicting the prognosis in BC patients (11–13). The previous prognosis models could only provide the mortality curves for two groups of tumor patients with different characteristics, failing to provide the individual mortality curve for a special patient. The progress of artificial intelligence algorithms provides necessary basic conditions for the realization of individualized mortality risk prediction of cancer patients. Random survival forest algorithm (25–27), multitask logistic regression (28, 29), and Cox survival regression algorithm (30) have been proposed and used to improve the predictive performance of prognostic models. Based on three artificial intelligence algorithms above, we develop an artificial intelligence survival predictive system. Our artificial intelligence survival predictive system could display three individual mortality risk predictive curves by using random survival forest algorithm, multitask logistic regression algorithm, and Cox survival regression algorithm. At present, there are few prognostic models that can provide individual mortality risk prediction. The current study provides an interesting and feasible way for the transformation and application of artificial intelligence algorithm in the field of medicine.
Tumor immune infiltration acted an important role in oncogenesis and prognosis (7, 31). Immune genes could be used to predict the prognosis of BC patients (32, 33). Three prognostic models have been developed to predict the prognosis for BC patients (11–13). Compared with three previous prognostic models, the current prognostic model could provide individualized mortality risk prediction and online calculation function, which were of great significance for clinical application by patients and clinicians.
Biological processes of immune genes were explored via TISIDB databases (http://cis.hku.hk/TISIDB/index.php). Top biological processes of CD1b molecule (CD1B) were adaptive immune response, antigen processing and presentation, and antigen processing and presentation via major histocompatibility complex class Ib. Top biological processes of lymphotoxin α (LTA) were adaptive immune response, lymphocyte-mediated immunity, leukocyte-mediated immunity, and inflammatory response to antigenic stimulus. Top biological processes of CD6 molecule (CD6) were immunological synapse formation, cell recognition, acute inflammatory response, and inflammatory response to antigenic stimulus. Top biological processes of cathelicidin antimicrobial peptide (CAMP) were cell killing, antibacterial humoral response, innate immune response in mucosa, mucosal immune response, and organ- or tissue-specific immune response. Top biological processes of interferon γ9 (IFNG) were cell killing, neutrophil homeostasis, leukocyte homeostasis, and neutrophil apoptotic process. Top biological processes of ADAM metallopeptidase with thrombospondin type 1 motif 8 (ADAMTS8) were phosphate ion transport, anion transport, and inorganic anion transport. Top biological processes of apolipoprotein A-V (APOA5) were receptor-mediated endocytosis, tissue regeneration, and positive regulation of receptor-mediated endocytosis. Top biological processes of siah E3 ubiquitin protein ligase 2 (SIAH2) were proteasomal protein catabolic process, regulation of cysteine-type endopeptidase activity involved in apoptotic process, and negative regulation of cysteine-type endopeptidase activity involved in apoptotic process. Top biological processes of steroidogenic acute regulatory protein (STAR) were response to molecule of bacterial origin, response to oxidative stress, and response to reactive oxygen species. Top biological processes of cadherin 19, type 2 (CDH19), were homophilic cell adhesion via plasma membrane adhesion molecules and cell–cell adhesion via plasma-membrane adhesion molecules. Biological processes of C-type lectin domain family 10, member A (CLEC10A), were adaptive immune response.
ADAMTS8 was related with poor prognosis for breast invasive ductal carcinoma patients (34). ADAMTS8 could regulate invasion and apoptosis of hepatocellular carcinoma through ERK signaling pathway (35). The interaction between CD4+ T cells and lung cancer cells could up-regulate expression of DNMT and methylation of IFNG promoter (36). CpG methylation of IFNG gene could induce immunosuppression of tumor-infiltrating lymphocytes (37). High expression level of XG in Ewing sarcoma cell line could promote tumor migration and invasiveness (38). Highly expressed SIAH2 was associated with poor progression-free survival after tamoxifen treatment (39). SIAH2 participated in the regulation of EAF2 polyubiquitin in prostate cancer cells as E3 ligase of EAF2 polyubiquitination (40). Cathelicidin antimicrobial peptide directly activated exchange protein, which regulated migration and apoptosis of BC cells (41). Interleukin 24 enhanced apoptosis of BC cell via cAMP-dependent PKA pathway (42). Low expression of NTSR1 was associated with non-invasive growth of colorectal cancer (43). Interaction of CLEC10A with macrophages and dendritic cells might play an important role in tumor progression (44). As a functional ligand of CLEC10A, sv6D could induce the maturation of immune cells (45). Variation of STXBP6 might affect the response of TNF-α inhibitors in rheumatoid arthritis patients (46). There was a significant correlation between LTA RS909253GA genotype and the development of Asian gastric cancer (47). These previous studies revealed possible immune regulatory mechanisms and biological roles of previous 17 immune genes in tumorigenesis and progression.
Toll-like receptor–activated plasma-like dendritic cells inhibited growth of BC cells (48). CD56 enhanced formation of cytotoxic immune synapses and strengthened sensitivity of cytotoxicity mediated by natural killer cells (49). CD4 and CD8 T cell tumor infiltration driven by HER2-dendritic cells improved survival of BC mice (50). CD4+ T cells inhibited CD8+ T cell failure at the initiation stage of immune response in BC (51). V delta 2+ gamma delta T lymphocyte had cytotoxicity to MCF 7 BC cells (52). High expression of SEMA4C was correlated with the proliferation of tumor cells and the aggregation of macrophages in BC (53). Interleukin 32θ suppressed the growth of BC by regulating CCL18 secreted by macrophages (54). Macrophage adhesion regulated by integrin induces lymphovascular dissemination in BC (55). CCL5 induced recurrence of BC via aggregation of macrophages in residual tumors (56). High expression of mast cells induced the tumor size and the incidence of spontaneous metastasis in BC mice (57). Tumor-infiltrating myeloid-derived suppressor cells (MDSCs) was related with therapeutic effect and prognosis of neoadjuvant chemotherapy of BC (58). Neutrophil–lymphocyte ratio could predict prognosis of triple-negative BC patients (59). Interleukin 10 and interleukin 2 promoted proliferation and cytotoxicity of CD8+T cells (60).
Advantages of current research: First, two artificial intelligence survival predictive systems were developed based on immune genes for BC patients. These two tools could provide online individual mortality risk prediction and provide valuable prognostic information for optimizing individual treatment decision. Second, artificial intelligence survival prediction system provided three individual mortality risk predictive curves based on different artificial intelligence algorithms, providing different valuable individual mortality curves as the references of individual medical decision-making. Third, artificial intelligence survival prediction system provided predicted median survival time and 95% confidence interval of predicted mortality, which were of clinical practical values for optimizing individual medical decision-making.
Shortcomings of current research: First, the current research explored potential biological functions and regulatory mechanisms of immune genes in BC based on public databases, but the conclusion was not validated by confirmative experimental studies. Second, estrogen receptor, progesterone receptor, and erb-b2 receptor tyrosine kinase 2 are closely related to prognosis of BC patients. Subgroup studies based on these biomarkers are helpful to provide more accurate individual mortality risk prediction for BC patients in different subgroups. Third, the current studies did not include and analyze the impacts of several clinical factors, such as radiotherapy, chemotherapy, and targeted drug therapy, which should be taken into account for future studies. Fourth, the calculation process of artificial intelligence algorithms are too complex to perform and cannot be present through conventional formula. The operation process of artificial intelligence algorithm is opaque, just like a black box, which limits the clinical application and verification of artificial intelligence algorithm. Because it is difficult for artificial intelligence algorithm to perform repeated verification research, we provided three different artificial intelligence algorithms as references for each other. As the inherent deficiency of artificial intelligence, opaque computing process and lack of verification research need to be solved by future artificial intelligence algorithm research. Fifth, the current study identified 17 immune genes were correlated with prognosis of BC patients. However, the associations of these immune genes with tumor heterogeneity and tumor resistance were still unclear. Further basic immune research is helpful to clarify the associations of these immune genes with tumor heterogeneity and tumor resistance. Sixth, although the current study demonstrated the gene differential expression between normal samples and tumor samples, the current study was lack of external validation at the cell level and the animal model level. Further validation studies at the cell level and the animal model level were helpful to ascertain the differences of immune regulatory mechanism of BC patients compared with normal people.
In conclusion, comprehensive bioinformatics identified 17 immune genes as potential prognostic biomarkers, which might be potential candidates of immunotherapy targets in BC patients. The current study depicted regulatory network between transcription factors and immune genes, which was helpful to deepen the understanding of immune regulatory mechanisms for BC cancer. Two artificial intelligence survival predictive systems are available at https://zhangzhiqiao7.shinyapps.io/Smart_Cancer_Survival_Predictive_System_16_BC_C1005/ and https://zhangzhiqiao8.shinyapps.io/Gene_Survival_Subgroup_Analysis_16_BC_C1005/. These novel artificial intelligence survival predictive systems will be helpful to improve individualized treatment decision-making.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://zhangzhiqiao8.shinyapps.io/Gene_Survival_Subgroup_Analysis_16_BC_C1005/.
Ethics Statement
The studies in TCGA database and GEO database have received ethical approval from ethics committees of their respective research institutes. These studies obtained informed consent from patients before admission. The current study is a second study based on public datasets from TCGA database and GEO database. Details of all patients in public datasets have been anonymously processed and therefore the current research does not involve patients' privacy information. This study was performed according to public database policy and declaration of Helsinki. Ethical approval and informed consent were not applicable.
Author Contributions
ZZ, JD, JL, and TH: conceptualization, methodology and resources. ZZ, JD, JL, and TH: investigation, data curation, formal analysis, validation, software, project administration, and supervision. ZZ, and JD: writing and visualization. ZZ: funding acquisition.
Funding
The current research was funded by Medical Science and Technology Foundation of Guangdong Province (A2016450 and B2018237).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Gary S Collins (University of Oxford), Dr. Manali Rupji (Emory University), Mrs Qingmei Liu for help and support on development of precision medical tools.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00330/full#supplementary-material
Abbreviations
BC, breast cancer; TCGA, The Cancer Genome Atlas; GEO, the Gene Expression Omnibus; ROC, receiver operating characteristic; DFS, disease free survival; HR, hazard ratio; CI, confidence interval; AJCC, the American Joint Committee on Cancer; SD, standard deviation.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Foster TS, Miller JD, Boye ME, Blieden MB, Gidwani R, Russell MW. The economic burden of metastatic breast cancer: a systematic review of literature from developed cou ntries. Cancer Treat Rev. (2011) 37:405–15. doi: 10.1016/j.ctrv.2010.12.008
3. Zeng J, Cai X, Hao X, Huang F, He Z, Sun H, et al. LncRNA FUNDC2P4 down-regulation promotes epithelial-mesenchymal transition by reducing E-cadherin exp ression in residual hepatocellular carcinoma after insufficient radiofrequency ablation. Int J Hyperthermia. (2018) 34:802–11. doi: 10.1080/02656736.2017.1422030
4. Zhong X, Long Z, Wu S, Xiao M, Hu W. LncRNA-SNHG7 regulates proliferation, apoptosis and invasion of bladder cancer cells assurance guidel ines. J Buon. (2018) 23:776–781.
5. Shi X, Zhao Y, He R, Zhou M, Pan S, Yu S, et al. Three-lncRNA signature is a potential prognostic biomarker for pancreatic adenocarcinoma. Oncotarget. (2018) 9:24248–59. doi: 10.18632/oncotarget.24443
6. Huang Y, Xiang B, Liu Y, Wang Y, Kan H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. (2018) 437:56–66. doi: 10.1016/j.canlet.2018.08.024
7. Pagès F, Galon J, Dieu-Nosjean MC, Tartour E, Sautès-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. (2010) 29:1093–102. doi: 10.1038/onc.2009.416
8. Domingues P, González-Tablas M, Otero Á, Pascual D, Miranda D, Ruiz L, et al. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav Immun. (2016) 53:1–15. doi: 10.1016/j.bbi.2015.07.019
9. Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of immune Infiltration in breast cancer and their clinical implications: a gene-expression-B ased retrospective study. PLoS Med. (2016) 13:e1002194. doi: 10.1371/journal.pmed.1002194
10. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. (2013) 4:2612. doi: 10.1038/ncomms3612
11. Zhang Z, Ouyang Y, Huang Y, Wang P, Li J, He T, et al. Comprehensive bioinformatics analysis reveals potential lncRNA biomarkers for overall survival in pat ients with hepatocellular carcinoma: an on-line individual risk calculator based on TCGA cohort. Cancer Cell Int. (2019) 19:174. doi: 10.1186/s12935-019-0890-2
12. Cheng C, Wang Q, Zhu M, Liu K, Zhang Z. Integrated analysis reveals potential long non-coding RNA biomarkers and their potential biological functions for disease free survival in gastric cancer patients. Cancer Cell Int. (2019) 19:123. doi: 10.1186/s12935-019-0846-6
13. Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, et al. A signature of immune function genes associated with recurrence-free survival in breast cancer patien ts. Breast Cancer Res Treat. (2012) 131:871–80. doi: 10.1007/s10549-011-1470-x
14. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
15. Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. (2014) 58:234–9. doi: 10.1007/s12026-014-8516-1
16. Mei S, Meyer CA, Zheng R, Qin Q, Wu Q, Jiang P, et al. Cistrome cancer: a web resource for integrative gene regulation modeling in cancer. Cancer Res. (2017) 77:e19–22. doi: 10.1158/0008-5472.CAN-17-0327
17. Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. (2018) 9:5361. doi: 10.1038/s41467-018-07767-w
18. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of res ponse to checkpoint blockade. Cell Rep. (2017) 18:248–62. doi: 10.1016/j.celrep.2016.12.019
19. LD F, DY L. Time-dependent covariates in the Cox proportional-hazards regression model. Ann Rev Public Health. (1999) 20:145–57. doi: 10.1146/annurev.publhealth.20.1.145
20. Alaeddini A, Hong SH. A multi-way multi-task learning approach for multinomial logistic regression*. An application in joint prediction of appointment miss-opportunities across multiple clinics. Methods Inform Med. (2017) 56:294–307. doi: 10.3414/ME16-01-0112
21. Bisaso KR, Karungi SA, Kiragga A, Mukonzo JK, Castelnuovo B. A comparative study of logistic regression based machine learning techniques for prediction of early virological suppression in antiretroviral initiating HIV patients. BMC Med Inform Decis Mak. (2018) 18:77. doi: 10.1186/s12911-018-0659-x
22. Xu H, Gu X, Tadesse MG, Balasubramanian R. A modified random survival forests algorithm for high dimensional predictors and self-reported outcomes. J Comput Graph Stat. (2018) 27:763–72. doi: 10.1080/10618600.2018.1474115
23. Nasejje JB, Mwambi H. Application of random survival forests in understanding the determinants of under-five child mortality in Uganda in the presence of covariates that satisfy the proportional and non-proportional hazards assumption. BMC Res Notes. (2017) 10:459. doi: 10.1186/s13104-017-2775-6
24. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
25. Wang H, Liu D, Yang J. Prognostic risk model construction and molecular marker identification in glioblastoma multiforme based on mRNA/microRNA/long non-coding RNA analysis using random survival forest method. Neoplasma. (2019) 66:459–69. doi: 10.4149/neo_2018_181008N746
26. Shi M, Xu G. Development and validation of GMI signature based random survival forest prognosis model to predict clinical outcome in acute myeloid leukemia. BMC Med Genomics. (2019) 12:90. doi: 10.1186/s12920-019-0540-5
27. Wang H, Shen L, Geng J, Wu Y, Xiao H, Zhang F, et al. Prognostic value of cancer antigen−125 for lung adenocarcinoma patients with brain metastasis: a random survival forest prognostic model. Sci Rep. (2018) 8:5670. doi: 10.1038/s41598-018-23946-7
28. Lan Q, Sun H, Robertson J, Deng X, Jin R. Non-invasive assessment of liver quality in transplantation based on thermal imaging analysis. Comput Meth Prog Biomed. (2018) 164:31–47. doi: 10.1016/j.cmpb.2018.06.003
29. Halme HL, Parkkonen L. Across-subject offline decoding of motor imagery from MEGEEG. Sci Rep. (2018) 8:10087. doi: 10.1038/s41598-018-30241-y
30. Kim DW, Lee S, Kwon S, Nam W, Cha IH, Kim HJ. Deep learning-based survival prediction of oral cancer patients. Sci Rep. (2019) 9:6994. doi: 10.1038/s41598-019-43372-7
31. Gough MJ, Crittenden MR. Immune system plays an important role in the success and failure of conventional cancer therapy. Immunotherapy. (2012) 4:125–8. doi: 10.2217/imt.11.157
32. Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor ne gative breast cancer. Genome Biol. (2007) 8:R157. doi: 10.1186/gb-2007-8-8-r157
33. Yang B, Chou J, Tao Y, Wu D, Wu X, Li X, et al. An assessment of prognostic immunity markers in breast cancer. NPJ Breast Cancer. (2018) 4:35. doi: 10.1038/s41523-018-0088-0
34. Guo X, Li J, Zhang H, Liu H, Liu Z, Wei X. Relationship between ADAMTS8, ADAMTS18, and ADAMTS20 (A Disintegrin and metalloproteinase with thromb ospondin motifs) expressions and tumor molecular classification, clinical pathological parameters, a nd prognosis in breast invasive ductal carcinoma. Med Sci Monit. (2018) 24:3726–35. doi: 10.12659/MSM.907310
35. Zhao X, Yang C, Wu J, Nan Y. ADAMTS8 targets ERK to suppress cell proliferation, invasion, and metastasis of hepatocellular carcinoma. OncoTargets Ther. (2018) 11:7569–78. doi: 10.2147/OTT.S173360
36. Wang F, Xu J, Zhu Q, Qin X, Cao Y, Lou J, et al. Downregulation of IFNG in CD4(+) T cells in lung cancer through hypermethylation: a possible mechanis m of tumor-induced immunosuppression. PLoS ONE. (2013) 8:e79064. doi: 10.1371/journal.pone.0079064
37. Janson PC, Marits P, Thörn M, Ohlsson R, Winqvist O. CpG methylation of the IFNG gene as a mechanism to induce immunosuppression [correction of immunosupr ession] in tumor-infiltrating lymphocytes. J Immunol. (2008) 181:2878–86. doi: 10.4049/jimmunol.181.4.2878
38. Meynet O, Scotlandi K, Pradelli E, Manara MC, Colombo MP, Schmid-Antomarchi H, et al. Xg expression in Ewing's sarcoma is of prognostic value and contributes to tumor invasiveness. Cancer Res. (2010) 70:3730–8. doi: 10.1158/0008-5472.CAN-09-2837
39. van der Willik KD, Timmermans MM, van Deurzen CH, Look MP, Reijm EA, van Zundert WJ, et al. SIAH2 protein expression in breast cancer is inversely related with ER status and outcome to tamoxife n therapy. Am J Cancer Res. (2016) 6:270–84. doi: 10.1158/1538-7445.SABCS15-P5-08-51
40. Yu X, Ai J, Cai L, Jing Y, Wang D, Dong J, et al. Regulation of tumor suppressor EAF2 polyubiquitination by ELL1 and SIAH2 in prostate cancer cells. Oncotarget. (2016) 7:29245–54. doi: 10.18632/oncotarget.8588
41. Kumar N, Gupta S, Dabral S, Singh S, Sehrawat S. Role of exchange protein directly activated by cAMP (EPAC1) in breast cancer cell migration and apopt osis. Mol Cell Biochem. (2017) 430:115–25. doi: 10.1007/s11010-017-2959-3
42. Persaud L, Mighty J, Zhong X, Francis A, Mendez M, Muharam H, et al. IL-24 promotes apoptosis through cAMP-dependent PKA pathways in human breast cancer cells. Int J Mol Sci. (2018) 19:3561. doi: 10.3390/ijms19113561
43. Kamimae S, Yamamoto E, Kai M, Niinuma T, Yamano HO, Nojima M, et al. Epigenetic silencing of NTSR1 is associated with lateral and noninvasive growth of colorectal tumors. Oncotarget. (2015) 6:29975–90. doi: 10.18632/oncotarget.5034
44. Mortezai N, Behnken HN, Kurze AK, Ludewig P, Buck F, Meyer B, et al. Tumor-associated Neu5Ac-Tn and Neu5Gc-Tn antigens bind to C-type lectin CLEC10A (CD301, MGL). Glycobiology. (2013) 23:844–52. doi: 10.1093/glycob/cwt021
45. Eggink LL, Roby KF, Cote R, Kenneth Hoober J. An innovative immunotherapeutic strategy for ovarian cancer: CLEC10A and glycomimetic peptides. J Immunother Cancer. (2018) 6:28. doi: 10.1186/s40425-018-0339-5
46. Krintel SB, Essioux L, Wool A, Johansen JS, Schreiber E, Zekharya T, et al. CD6 and syntaxin binding protein 6 variants and response to tumor necrosis factor alpha inhibitors in Danish patients with rheumatoid arthritis. PLoS ONE. (2012) 7:e38539. doi: 10.1371/journal.pone.0038539
47. Lu R, Dou X, Gao X, Zhang J, Ni J, Guo L. A functional polymorphism of lymphotoxin-alpha (LTA) gene rs909253 is associated with gastric cancer risk in an Asian population. Cancer Epidemiol. (2012) 36:e380–6. doi: 10.1016/j.canep.2012.05.014
48. Wu J, Li S, Yang Y, Zhu S, Zhang M, Qiao Y, et al. TLR-activated plasmacytoid dendritic cells inhibit breast cancer cell growth in vitro and in vivo. Oncotarget. (2017) 8:11708–18. doi: 10.18632/oncotarget.14315
49. Taouk G, Hussein O, Zekak M, Abouelghar A, Al-Sarraj Y, Abdelalim EM, et al. CD56 expression in breast cancer induces sensitivity to natural killer-mediated cytotoxicity by enhan cing the formation of cytotoxic immunological synapse. Sci Rep. (2019) 9:8756. doi: 10.1038/s41598-019-45377-8
50. Kodumudi KN, Ramamoorthi G, Snyder C, Basu A, Jia Y, Awshah S, et al. Sequential Anti-PD1 therapy following dendritic cell vaccination improves survival in a HER2 mammary carcinoma model and identifies a critical role for CD4 T cells in mediating the response. Front Immunol. (2019) 10:1939. doi: 10.3389/fimmu.2019.01939
51. Kmieciak M, Worschech A, Nikizad H, Gowda M, Habibi M, Depcrynski A, et al. CD4+ T cells inhibit the neu-specific CD8+ T-cell exhaustion during the priming phase of immune respo nses against breast cancer. Breast Cancer Res Treat. (2011) 126:385–94. doi: 10.1007/s10549-010-0942-8
52. Bank I, Book M, Huszar M, Baram Y, Schnirer I, Brenner H. V delta 2+ gamma delta T lymphocytes are cytotoxic to the MCF 7 breast carcinoma cell line and can be detected among the T cells that infiltrate breast tumors. Clin Immunol Immunopathol. (1993) 67:17–24. doi: 10.1006/clin.1993.1040
53. Yang J, Zeng Z, Qiao L, Jiang X, Ma J, Wang J, et al. Semaphorin 4C promotes macrophage recruitment and angiogenesis in breast cancer. Mol Cancer Res. (2019) 17:2015–28. doi: 10.1158/1541-7786.MCR-18-0933
54. Pham TH, Bak Y, Kwon T, Kwon SB, Oh JW, Park JH, et al. Interleukin-32θ inhibits tumor-promoting effects of macrophage-secreted CCL18 in breast cancer. Cell Commun Signal. (2019) 17:53. doi: 10.1186/s12964-019-0374-y
55. Evans R, Flores-Borja F, Nassiri S, Miranda E, Lawler K, Grigoriadis A, et al. Integrin-Mediated macrophage adhesion promotes lymphovascular dissemination in breast cancer. Cell Rep. (2019) 27:1967–78.e1964. doi: 10.1016/j.celrep.2019.04.076
56. Walens A, DiMarco AV, Lupo R, Kroger BR, Damrauer JS, Alvarez JV. CCL5 promotes breast cancer recurrence through macrophage recruitment in residual tumors. Elife. (2019) 8:e43653. doi: 10.7554/eLife.43653
57. Aponte-López A, Fuentes-Pananá EM, Cortes-Muñoz D, Muñoz-Cruz S. Mast cell, the neglected member of the tumor microenvironment: role in breast cancer. J Immunol Res. (2018) 2018:2584243. doi: 10.1155/2018/2584243
58. Li F, Zhao Y, Wei L, Li S, Liu J. Tumor-infiltrating Treg, MDSC, and IDO expression associated with outcomes of neoadjuvant chemotherap y of breast cancer. Cancer Biol Ther. (2018) 19:695–705. doi: 10.1080/15384047.2018.1450116
59. Patel DA, Xi J, Luo J, Hassan B, Thomas S, Ma CX, et al. Neutrophil-to-lymphocyte ratio as a predictor of survival in patients with triple-negative breast cancer. Breast Cancer Res Treat. (2019) 174:443–52. doi: 10.1007/s10549-018-05106-7
Keywords: breast cancer, disease-free survival, immune gene, transcription factor, prognostic signature
Citation: Zhang Z, Li J, He T and Ding J (2020) Bioinformatics Identified 17 Immune Genes as Prognostic Biomarkers for Breast Cancer: Application Study Based on Artificial Intelligence Algorithms. Front. Oncol. 10:330. doi: 10.3389/fonc.2020.00330
Received: 15 November 2019; Accepted: 25 February 2020;
Published: 31 March 2020.
Edited by:
Shafiq Khan, Clark Atlanta University, United StatesReviewed by:
Ahmed Abdalla Agab Eldour, Kordofan University, South SudanSatyanarayana Alleboina, University of Tennessee Health Science Center (UTHSC), United States
Copyright © 2020 Zhang, Li, He and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianqiang Ding, jding18@foxmail.com
†These authors share first authorship
‡ORCID: Zhiqiao Zhang orcid.org/0000-0003-4631-8818
 Zhiqiao Zhang
Zhiqiao Zhang Jing Li†
Jing Li†