- 1Department of Pathology and Laboratory Medicine, McGovern Medical School, The University of Texas Health Science Center at Houston, Houston, TX, United States
- 2Department of Pathology, Duke University Medical Center, Durham, NC, United States
Nuclear protein in testis (NUT) carcinoma is a rare, highly aggressive, poorly differentiated carcinoma occurring mostly in adolescents and young adults. This tumor usually arises from the midline structures of the thorax, head, and neck, and exhibits variable degrees of squamous differentiation. NUT carcinoma is defined by the presence of a NUTM1 (15q14) rearrangement with multiple other genes. In about 70-80% of the cases, NUTM1 is involved in a balanced translocation with the BRD4 gene (19p13.12), leading to a BRD4-NUTM1 fusion oncogene. Other variant rearrangements include BRD3-NUTM1 fusion (~15-20%) and NSD3-NUTM1 fusion (~6%), among others. The diagnosis of NUT carcinoma requires the detection of nuclear expression of the NUT protein by immunohistochemistry. Additional methods for diagnosis include the detection of a NUTM1 rearrangement by fluorescence in situ hybridization or by reverse transcriptase PCR. NUT carcinoma is usually underrecognized due to its rarity and lack of characteristic histological features. Therefore, the goal of this review is to provide relevant recent information regarding the clinicopathologic features of NUT carcinoma, the role of the multiple NUTM1 gene rearrangements in carcinogenesis, and the impact of understanding these underlying molecular mechanisms that may result in the development of possible novel targeted therapies.
Introduction
Nuclear protein in testis (NUT) carcinoma (NC), is a rare carcinoma characterized by a chromosomal rearrangement involving the NUT midline carcinoma family member 1 (NUTM1) gene, also known as NUT gene, located on chromosome 15q14 (1–5). This entity was first described in 1991 in two independent case reports of mediastinal carcinomas characterized by the t(15;19) translocation (6, 7). Since then, tumors harboring NUTM1 translocation have been increasingly recognized with numerous cases reported in the literature. Similarly, the mechanisms underlying the multiple NUTM1 gene rearrangements have opened the door to better understand tumor pathogenesis and the role of NUT and other proteins in the epigenetics of this rare neoplasm and multiple other cancers. Here, we review the clinicopathologic features, methods of diagnosis, and the molecular genetics and epigenetic alterations known to date in NC.
Epidemiology and Clinical Features
NC is a rare, poorly differentiated carcinoma characterized by an aggressive clinical behavior and advanced stage at diagnosis (8). Although the cell of origin is unknown, it has been speculated that NC may arise from primitive neural crest-derived cells (8). Despite lack of knowledge regarding a definitive anatomical site of origin, NC has been described to typically originate from midline structures of the thorax or from head and neck, hence the original term “NUT midline carcinoma”, and to predominantly affect young patients or adolescents. However, over the last decade, this entity has been identified in patients of all ages (0 - 81.7 years) with a median age varying from 16 to 24 years, observed in four meta-analysis studies (9–12), and affecting females and males almost equally (9, 10, 12). The most common location of primary NC has been found in the thorax (~50%) followed by the head and neck region (~40%) (12), but NC can also rarely arise outside of midline locations such as bladder (13), ocular globe (13), salivary glands (14–28), brain (29) kidney (29–31), stomach (29), adrenal gland (2, 20), pancreas (32), soft tissue (29), and bone (33).
Histopathologic Features
By histology, NC is a poorly differentiated malignant neoplasm that usually grows as nests and sheets of primitive cells without an overlying in situ component (34), most of them (~55%) without squamous differentiation (12), and often with areas of confluent necrosis (34–36) (Figure 1A). The cells may have little or moderate amount of eosinophilic or amphophilic cytoplasm with indistinct borders, imparting the appearance of a cellular syncytium (Figures 1A–C). Focal cytoplasmic clearing or vacuolization (Figure 1C) (28, 34) as well as lumen or pseudo-lumen formation have been described (34). The tumor cells can be widely infiltrative and demonstrate a high mitotic rate (34, 35). The nuclei are usually large and quite monotonous, with vesicular chromatin and distinct nucleoli, lacking the pleomorphism typically encountered in high-grade carcinomas (3, 34, 35). A peculiar feature described in the literature as a clue to the diagnosis is the presence of keratinization in the form of large cells with intercellular bridges, focal squamous “pearls” or frank abrupt keratinization (i.e., poorly differentiated cells immediately adjacent to well differentiated squamous cells) (Figure 1B). However, this morphologic feature is only observed in about 30% of the cases (12) and it can also be seen in HPV-associated basaloid squamous cell carcinomas (3, 35). The background stroma varies from edematous, slightly myxoid to fibrous with variable amounts of desmoplasia (14, 36). The presence of an intratumoral neutrophilic infiltrate is common and can be very prominent (Figure 1C), and occasionally an intraepithelial and stromal lymphocytic infiltrate may also be observed (14, 28, 37).
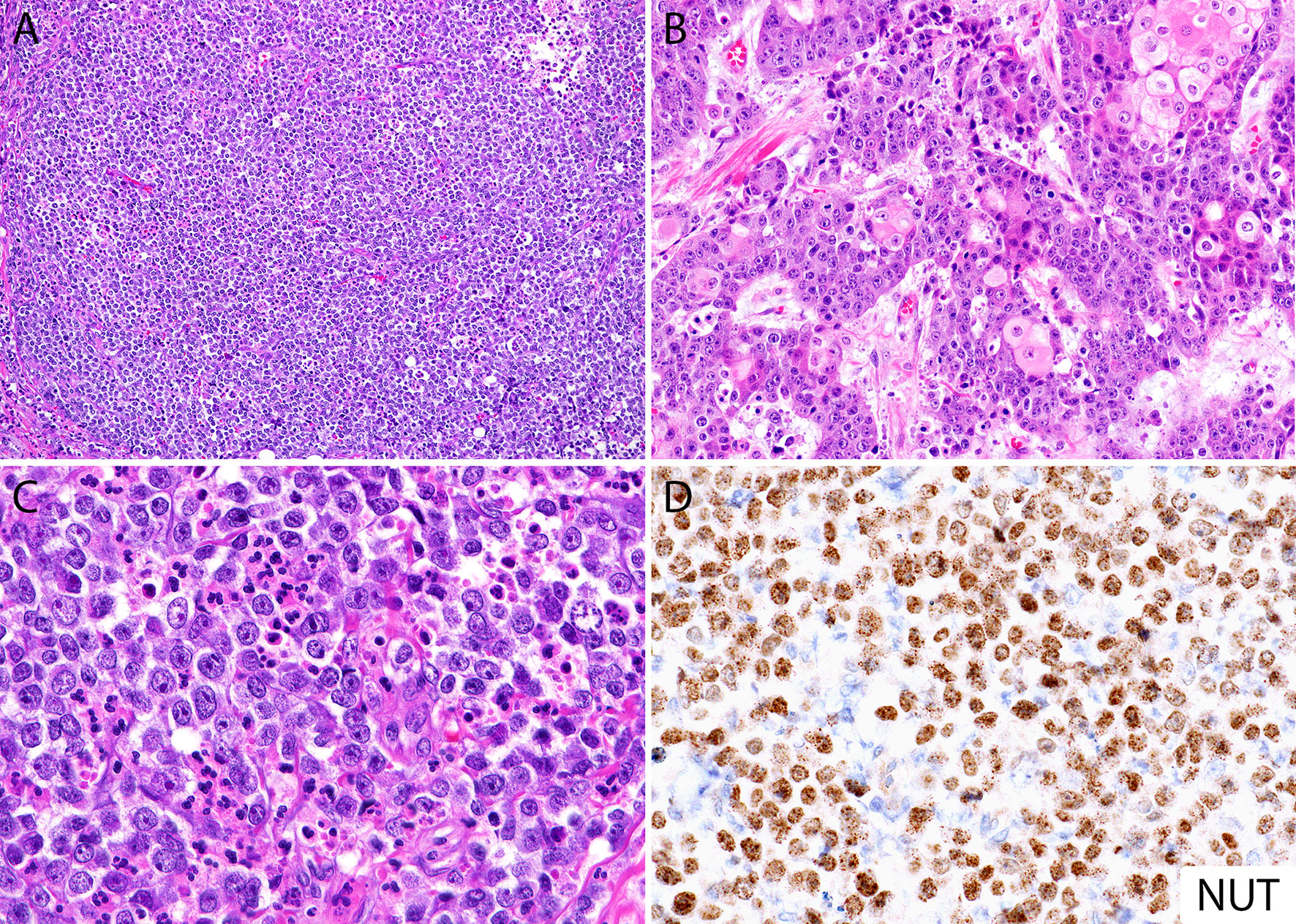
Figure 1 Histomorphology of NUT carcinoma and diagnosis by immunohistochemistry. (A) Diffuse sheets of poorly differentiated monotonous round cells with focal necrosis (top right). (B) Areas with abrupt keratinization in NC can be seen in up to 30% of cases. (C) Focal cytoplasmic clearing and intratumoral neutrophilic inflammatory infiltrate. (D) NUT immunohistochemical stain (monoclonal antibody, clone C52) shows diffuse nuclear labeling, often with a speckled pattern. Same case from panel (B).
Molecular Genetics and Epigenetics
The pathogenesis of NC is characterized by translocation-associated fusion oncoproteins that block cell differentiation and promote cellular growth (5). This distinct feature of single chromosomal translocation resembles those found in hematopoietic and mesenchymal malignancies and distinguishes NC from other epithelial tumors where multiple sequential mutations are required for tumorigenesis (38).
NC is defined by the rearrangement of NUTM1 gene on chromosome 15q14, which is frequently fused to the bromodomain containing protein 4 (BRD4) gene on chromosome 19p13.12, resulting in the most characteristic reciprocal translocation t(15;19) observed in 70-88% of the cases (9–13, 39). The predominant oncogenic variant involves the in-frame fusion of BRD4 exon 11 to the start of NUTM1 exon 2 (4, 6, 40, 41). However, variations of the specific exon fusions are known to occur including BRD4 exon 11 to NUTM1 exon 1b (41), BRD4 exon 15 to NUTM1 exon 2 (41), and BRD4 exon 15 to a partially deleted NUTM1 exon 2 (fusion at the last 124 nucleotides of NUTM1 exon 2) (42). The BRD4-NUTM1 fusion gene contains nearly the whole coding region for NUTM1 (exons 1b/2 to 7) and the three well characterized domains of BRD4 including the two bromodomains (BD 1 and BD2) and the extra-terminal (ET) domain, and a bipartite nuclear localization sequence (NLS) (4, 5). Its carboxyl-terminal domain (CTD), known to interact with the core positive transcription elongation factor b (P-TEFb), is absent (43) (Figure 2). The NUTM1 gene encodes an unstructured protein with two acidic transcriptional activation domains (AD1 and AD2), a NLS, and a nuclear export signal (NES) (5) (Figure 2). About 15-30% of NCs have a variant translocation involving exon 9 of the bromodomain containing protein 3 (BRD3) gene on chromosome 9q34.2 (5, 12). BRD3 gene encodes a protein similar to BRD4. The in-frame fusion involving BRD3 partner gene includes almost the entire NUTM1 structure (exons 2 to 7) along with the dual BDs, ET domain and the bipartite NLS of BRD3 (5) (Figure 2).
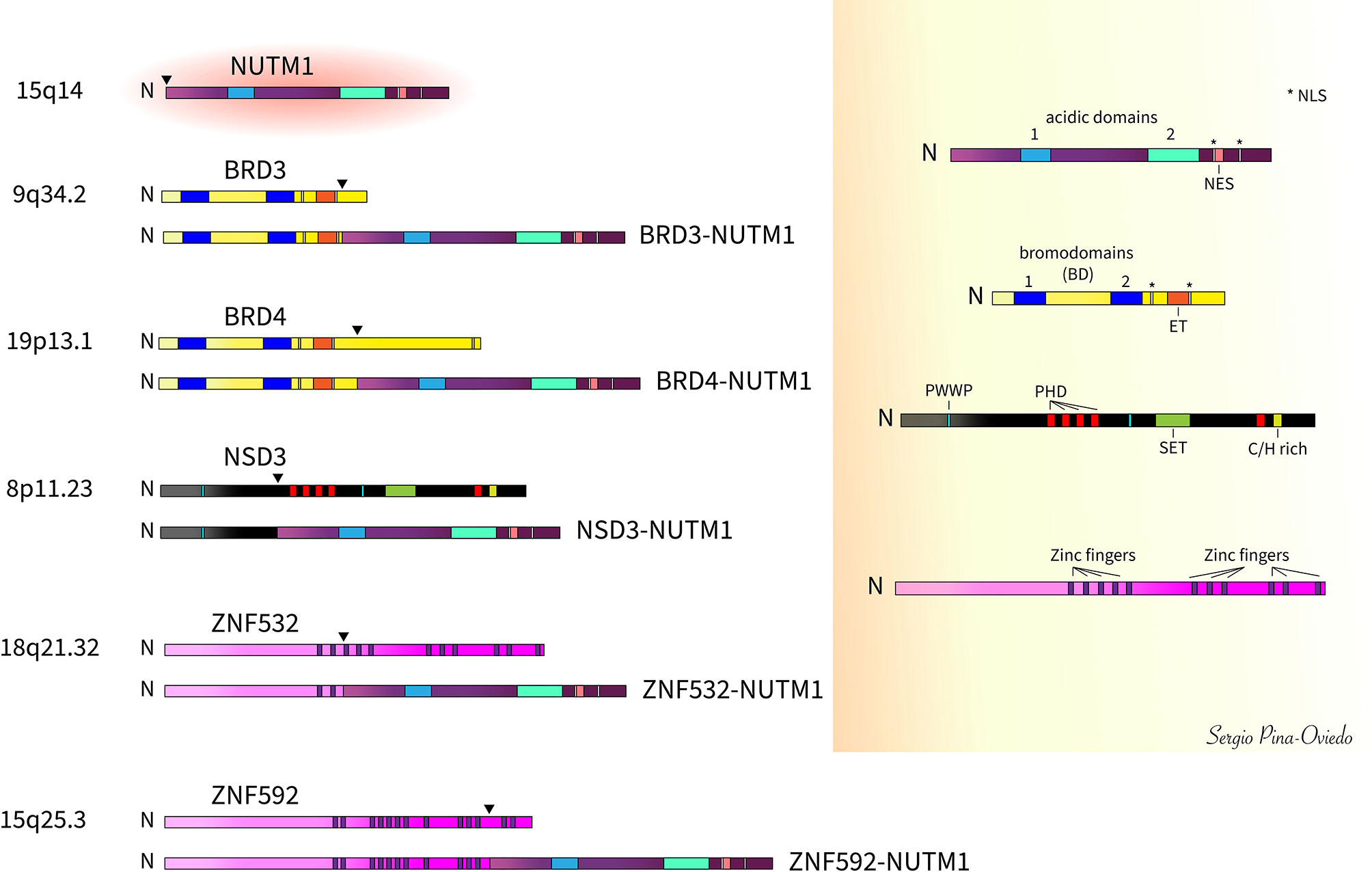
Figure 2 Schematic of NUTM1 fusions with BRD3, BRD4, NSD3, ZNF532, and ZNF592 and respective wild-type proteins (arrowheads denote fusion breakpoints). The most common of these fusions is BRD4-NUTM1 comprising about 70-80% of all cases. [N, amino- or N-terminal; NLS, nuclear localization signal; NES, nuclear export signal; BD, bromodomains (BD1 and BD2); ET, extra-terminal domain; PWWP, Proline-Tryptophan-Tryptophan-Proline domain; PHD, plant homeo-domain-type zinc-finger motifs; SET, Su(var)3-9, Enhancer-of-zeste and Trithorax (SET) domain; C/H rich, SET-associated Cys-His-rich (SAC) domain].
Other rarer NUTM1 fusion variants have been reported in about 6% of cases (12) including the nuclear receptor binding SET domain protein 3 (NSD3) gene (38), the zinc finger-containing protein encoding genes ZNF532 (44) and ZNF592 (45), and other yet unknown genes. The NSD3 gene is located on chromosome 8p11.23 and is considered a BET-binding protein. The NSD3 portion of the fusion (exons 1 to 7) lacks the Su(var)3-9, Enhancer-of-zeste and Trithorax (SET) domain and contains only one of the two Proline-Tryptophan-Tryptophan-Proline motif (PWWP) domains, whereas nearly all NUTM1 (exons 2 to 7) is included in the fusion (38) (Figure 2). The ZNF532-NUTM1 fusion gene encodes only the first 2 of 12 zinc finger domains from ZNF532 and almost the entire NUTM1 coding sequence (part of intron 1 of NUTM1 and its remaining exons 2 to 7) (44) (Figure 2). The first zinc finger included in the ZNF532-NUTM1 fusion gene encodes a putative zinc-ribbon domain that is predicted to bind nucleic acids directly (44). The ZNF592-NUTM1 resultant fusion protein contains the coding sequence of ZNF592 up to exon 10 fused with exons 2 to 10 of NUTM1. The ZNF592 moiety of the fusion protein retains the first 11 of 13 zinc finger domains (45) (Figure 2).
In a subset of malignant solid tumors from soft tissue and other organs, of uncertain relationship to NCs, NUTM1 has been reported to be fused with YAP1 (46, 47), MXD1 (29), MXD4 (39, 48, 49), CIC (50, 51), BCORL1 (29), ATXN1 (52), and MGA (39, 53, 54), in which most of them have been described to occur within the context of histologically defined high-grade sarcomas likely to be associated with a distinct pathogenetic pathway.
Bromodomain and Extra-Terminal (BET) Proteins
The BRD4 protein, encoded by the BRD4 gene, is the most extensively studied member of the bromodomain and extra-terminal (BET) protein family (i.e., BRD2, BRD3, BRD4, and BRDT). BET proteins bind to transcriptionally active chromatin through associations of one of their bromodomains (BD1 or BD2) to acetyl-lysin residues of histones (H3 and H4) affecting cell cycle progression and cellular proliferation (55). In cancer, BET BDs promote M to G1 cell cycle progression (56). These BDs are also responsible for tethering of the BRD4-NUTM1 to chromatin (5, 57). In addition, BET proteins activate transcription of target genes through recruitment of the multiprotein Mediator complex and P-TEFb, in which the Mediator complex is a transcriptional coactivator and the P-TEFb is a cyclin dependent kinase (CDK) containing the catalytic subunit CDK9 and one of several regulatory cyclin subunits (cyclin T1, T2, and K), thereby enhancing transcriptional elongation through phosphorylation of RNA Polymerase II on serine 2 of its CTD (58, 59). Although the CTD of the BRD4 is absent in the BRD4-NUTM1 fusion protein, the BD2 plays an important role binding P-TEFb and also interacting with wild-type BRD4 and other BRDs through the N-terminal portions of BRD4 representing, most likely, the required factors for the transcriptional activating function of BRD4-NUTM1 oncogene (43, 44). The ET domain of BET proteins is a protein-protein interaction module that binds NSD1-3, and other mediator components (43). BRD3/BRD4 also bind strongly to the regulatory regions of E2F1 transcriptional targets to enhance their activation; hence, BRD3/BRD4 promote specific cell cycle gene progression by activation of oncogenes (e.g., MYC, BCL6, and TP63) (60–64) and resistance to apoptosis by upregulating anti-apoptotic family member genes including BCL2 and the cyclin dependent kinase CDK6 (63). In addition, BET-family members appear to work co-operatively to control the release of pro-inflammatory cytokines from macrophages (65) and tumor cells (66), and BRD4 might act as a co-activator of transcription mediated by the pro-inflammatory molecule nuclear factor κB (NF-κB) (67, 68). Therefore, BET proteins may contribute with the role of immune cells within the tumor microenvironment in facilitating tumor growth and metastasis, through cytokine release.
Nuclear Protein in Testes (NUTM1)
Native NUTM1 protein expression is localized to the nucleus and it has been identified in germ cells of the testis (4, 69) and ovary (69), ciliary body (36), germ cell tumors (where protein expression is weak), and NUTM1-rearranged tumors (36, 69). While the normal function of the NUTM1 protein is related to spermatogenesis (70), overexpression of NUTM1 fusion genes leads to nuclear entrapment of the NUTM1 protein where it blocks cell differentiation and induces tumor growth (5, 14, 38, 57). The NES and NLS portions of NUTM1 allows the protein to shuttle between the nucleus and cytoplasm when transgenically expressed in cultured cells (5); hence, the tethering of NUTM1 to chromatin by BDs of BRD4 is critical to BRD4-NUTM1 oncoprotein function (5, 57). The AD1 of NUTM1 binds to and activates EP300, a histone acetyltransferase (44, 71), and this interaction plays a critical role in the oncogenic function of BRD-NUTM1 fusion protein (44, 57, 71, 72).
Nuclear Receptor Binding SET Domain Protein 3 (NSD3)
The NSD3 (also known as Wolf-Hirschhorn syndrome candidate 1-like 1, WHSC1L1) encodes a histone lysine methyltransferase that belongs to the mammalian NSD protein family of SET domain-containing methyltransferases (i.e., NSD1, NSD2, NSD3). Both NSD3 and NSD2 are known to bind the ET domain of BRD4. The NSD3 protein is considered an enzymatic protein (i.e., histone lysine methyltransferase or HMTases) involved in the methylation of histone lysine marks, regulating chromatin integrity and gene expression (73). Histone marks created by lysine HMTases are associated with either active transcription (e.g., H3K4me or H3K36me2) or repressed transcription (e.g., H3K27me or H2K9me) (74, 75). Only the N-terminus of NSD3, which binds to BRD4, is included in the genetic NSD3-NUTM1 fusion process, whereas its methyltransferase domain is absent (38, 76, 77). This interaction of BRD4 with NSD3 may be critical to BRD4-NUTM1 oncoprotein function. NSD3 may block differentiation in BRD4-NUTM1 expressing NC cells either through regulation of H3K36 methylation, leading to activation of specific gene expression (38, 43), or through interactions with the histone protein variant macroH2A1 where it represses or activates transcription of specific genes (78, 79). In addition, the NSD3-NUTM1 fusion oncogene encodes a protein that is necessary and sufficient for the blockade of differentiation in NC (38).
Zinc Finger Proteins
The ZNF532, ZNF592, ZNF687, and ZMYND8 have been collectively termed “Z4” protein factors. Although understanding of Z4 function is limited, some of them have been shown to play a role in cancer pathogenesis, either as oncogenes or tumor-suppressor genes (45). These zinc finger proteins have been shown to co-localize with BRD4/BRD4-NUTM1 chromatin complex proteins within megadomain regions (43–45). The ZNF532-NUTM1 and ZNF592-NUTM1 resultant fusion proteins form megadomains of hyperacetylated chromatin, similar to those formed by BRD4-NUTM1, suggesting that ZNF532 and ZNF592 genes are involved in a common feed-forward regulatory mechanism for megadomain formation that drive propagation of the oncogenic chromatin complex in BRD4-NUTM1 cells (44, 45). Moreover, BRD4-NUTM1-driven foci formation of Z4 factors suggests a mechanism of pathologic sequestration that may alter the normal function of Z4 proteins while enhancing those of BRD4-NUTM1 (45, 71, 80).
BRD-NUTM1 Protein Function
The tethering of NUTM1 to acetylated chromatin by BRD3/BRD4 leads to local chromatin acetylation by recruitment of EP300, resulting in a feed-forward expansion of acetylated chromatin and BRD-NUTM1 chimeric oncoprotein formation over massive genomic domains (megadomains), often filling entire topologically associating domains (5, 64, 71) (Figure 3). The number and magnitude of these “megadomains”, measuring from 100 kilobases up to 2 megabases in size, correlate with the characteristic nuclear foci seen in diagnostic patient tumor samples or in cultured NC cells stained with a NUT-specific antibody (5, 42, 64, 71). Additionally, the association of BRD4 to those regions has also been defined by the presence or co-localization of H3K27ac and H3K18ac, and the absence of H3K4m3 (60, 62, 81). In contrast, areas away from the megadomains become hypoacetylated, resulting in transcriptional repression of pro-differentiation genes (64, 72). As a result, these megadomain regions can drive targeted oncogene transcription. MYC has been shown to be a downstream oncogene target of BRD4-NUTM1 that blocks NC cellular differentiation and maintains a proliferative state (57). Moreover, there is evidence that BRD4-NUTM1 facilitates acetylation of TP53 through EP300, leading to its sequestration and inactivation within BRD4-NUTM1 foci (71). Also, it has been found that TP63, a TP53-related squamous cell-expressed gene, is regulated by BRD4-NUTM1 megadomains in all NC cells tested. Hence, the increased expression of TP63, a negative regulator of TP53, might represent another mechanism by which BRD4-NUTM1 NC cells evade gate keeper functions of TP53 (64). Another oncogenic target of BRD4-NUTM1 is the sex-determining region Y-box protein 2 (SOX2) which is a transcription factor essential for stem cell self-renewal and pluripotency (82). Although SOX2 expression is normally restricted to stem cells, aberrant overexpression has been linked to its ability to promote tumorigenicity and poorly differentiated morphology (83–86). BRD4-NUTM1 has been shown to drive overexpression of SOX2 in NC cells, which induces an aberrant stem cell-like growth feature (87). Furthermore, MED24, a Mediator subunit known to interact physically with BRD4 (43, 63), has been found to participate as both an oncogenic target gene and a cofactor of BRD4-NUTM1 complexes likely to provide another positive reinforcement loop for the establishment of megadomains and their transcriptional activity (64) (Figure 3). MED24 plays a role in transcriptional regulation during embryonic development (88), while its post-embryonic role appears to be tissue-specific coactivation of gene expression (89). These findings have been demonstrated in several in vitro studies when small interfering RNAs against NUTM1 or small-molecule BET inhibitors, such as JQ1, have been used to knockdown BRD3/4-NUTM1 and NSD3-NUTM1 patient-derived-tumor cells, leading to cellular differentiation and growth arrest (5, 38, 57, 64, 90). This indicates that NUTM1 fusion proteins act to maintain growth and block squamous-cell differentiation, in a mechanism dependent on the targeting of MYC, SOX2, MED24, and TP63 genes by BRD3/4-NUTM1 megadomains (10, 44, 57, 87).
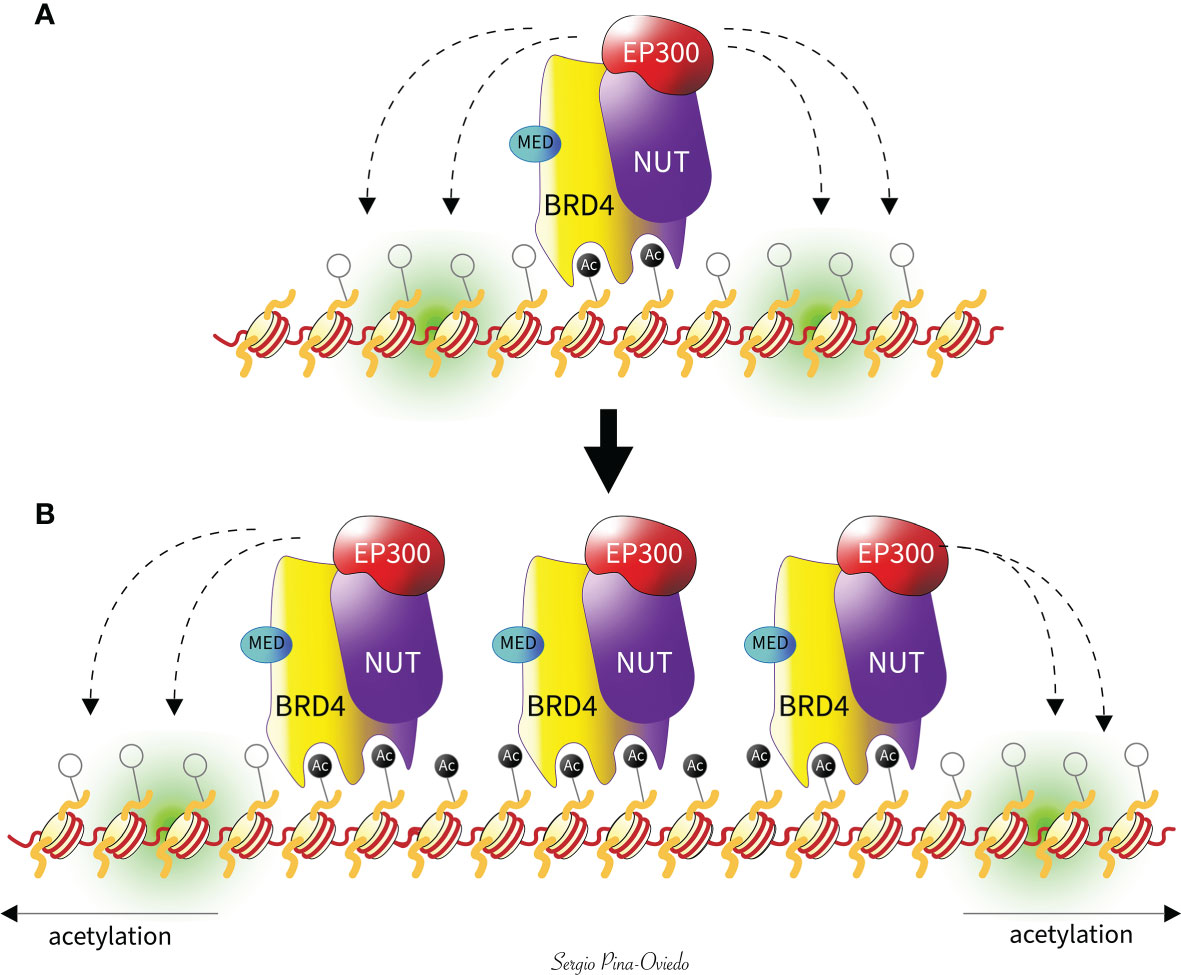
Figure 3 Feed-forward model of megadomain formation. (A) BRD4-NUTM1 is tethered to acetylated chromatin by BRD4 bromodomains. EP300 is recruited by the NUTM1 portion of the fusion, leading to increased local histone acetylation and, subsequently, (B) a self-perpetuating recruitment of BRD4-NUTM1 complexes. The resultant uncontrolled spreading of BRD4-NUTM1 within the chromatin leads to the formation of megadomains that are typically limited only by topologically associating domain boundaries (not shown). The Mediator subunit 24 (MED) interacts physically with BRD4 as a target and a cofactor of BRD4-NUTM1, to provide another positive reinforcement loop for the establishment of megadomains and their transcriptional activity.
Furthermore, wild-type BRD3, NSD3, and other Z4 protein factors (ZNF532, ZNF592, ZNF687 and ZMYND8) interact with BRD4, and its fusion with NUTM1 results in a powerful oncogenic complex. All these rare fusion partners of NUTM1 are functionally related to BRD4, indicating that the recruitment of NUTM1 to the chromatin through the BET family proteins is necessary in NC pathogenesis (38, 43–45).
Methods of Diagnosis
A definitive diagnosis of NC requires demonstrating the presence of NUTM1 gene rearrangement, which can be confirmed by immunohistochemistry with a NUT specific monoclonal antibody (clone C52) (Figure 1D). The immunohistochemical stain detects the presence of NUTM1 protein and has been reported as a relatively sensitive (87%) and highly specific (nearly 100%) tool for the diagnosis of NC (69). Diffuse (>50%) and strong nuclear positivity for NUTM1 is considered sufficient evidence for NUTM1 rearrangement, obviating the need of highly specialized genetic testing (1, 69). An alternative to NUT immunohistochemistry is molecular analysis to detect a NUTM1 gene rearrangement using fluorescence in situ hybridization, reverse-transcriptase polymerase chain reaction, cytogenetics, next generation sequencing, or whole-exome sequencing-based approaches. These methods should be considered if NUT immunohistochemistry is not available or if the result is negative or equivocal, and suspicion of NC is still high (29, 41, 51, 91–93).
Prognosis and Predictive Factors
The prognosis and outcomes of NC are very dismal with a median survival of 6.5 months (12) and poor response to conventional chemotherapeutic agents or radiotherapy. About 50% of patients present with lymph node involvement or distant metastatic disease (3, 9), frequently seen in lung and bones, and rarely in adrenal glands, brain, bone marrow, and liver (10, 14, 26, 36).
Although not required for diagnosis, molecular techniques can be used to determine the specific NUTM1 fusion partner which could be of potential prognostic and therapeutic significance. Recently, Chau et al. (12) proposed a prognostic risk classification model for NC survival outcomes based in the largest cohort of NC patients (n = 141) analyzed to date. In this study, they identified three distinct risk groups of patients based on anatomic site and NUTM1 fusion type, composed by the following: (1) Group A, patients with non-thoracic primary NC and presence of BRD3- or NSD3-NUTM1 fusion, (2) Group B, patients with non-thoracic primary NC and presence of BRD4-NUTM1 fusion, and (3) Group C, patients with thoracic primary NC regardless of the type of NUTM1 fusion. Interestingly, NSD3- or BRD3-NUTM1-positive tumors of non-thoracic origin are associated with significantly better overall survival, followed by the group of non-thoracic primary NC with BRD4-NUTM1 fusion. On the other hand, those patients with thoracic primary tumors, regardless of the NUTM1 fusion have worst prognosis than the other subgroups (12).
Therapeutics
At the current time, there is no standard treatment for this rare and aggressive form of cancer. However, a multimodal approach with aggressive initial surgical resection, systemic chemotherapy, and radiation therapy is currently adopted in clinical practice (11). A variety of chemoradiation therapy regimens have been used including intensive treatments commonly applied in other carcinomas, sarcomas, germ cell tumors, and other solid neoplasms. Some of the chemotherapeutic agents that have been used with some success include cisplatin, taxanes and alkylating agents (9, 11, 24, 33, 94). However, despite rapid response, tumors become treatment-refractory with early progression and poor overall outcome (9, 13).
Targeted therapy using small-molecule BET inhibitors, which are acetyl-lysine histone mimetic drugs, result in depletion of megadomains, proliferation arrest, and cellular differentiation (1, 90). BET inhibitors (e.g., Birabresib aka OTX015/MK-8628, Molibresib aka GSK525762, RO6870810, ODM-207, and NEO2734) have shown activity but no obvious survival benefit (3, 95–100), most likely due to toxicity effects (i.e., severe thrombocytopenia, gastrointestinal symptoms, anemia, and fatigue), limiting its use (3, 96, 98). However, by the time of the writing of this manuscript, there is an ongoing clinical trial where pediatric patients with solid tumors, brain tumors and lymphoma are being enrolled on a Phase I research study to evaluate the use of BET inhibitors (known as BMS-986158 and BMS-986378) in those patients described above and as possible treatments for NUT carcinoma in children. Both of these drugs are currently still being studied in adult patients (101). Other preclinical studies have shown that the BRD4-NUTM1 fusion gene is associated with global decreased histone acetylation and transcriptional repression of genes required for differentiation. Some in vitro and xenograft models have shown that this acetylation can be restored with histone deacetylase (HDAC) inhibitors such as Vorinostat, resulting in global increase in histone acetylation, squamous differentiation, and growth arrest (72, 102). However, the use of HDAC inhibitors has been limited due to the toxicity effects like those seen with BET inhibitors (72). Both of these novel targeted agents hold great promise, either alone or in combination with chemotherapy (37). In particular, preclinical studies have highlighted that BET inhibitors show synergism with immune checkpoint modulators (103–105).
Currently, patients can be enrolled into the International NUT Midline Carcinoma Registry (http://www.nmcregistry.org) which follows patient’s outcomes and may direct them to the institution running these trials (1, 35, 37). This international registry was originally established in 2010 and was created to raise awareness and disseminate the most updated information about NC, provide pathologic review to assist in the diagnosis of NC, and collect clinical data and response to treatment. This has allowed the creation of a repository of clinical specimens that will support future research (106).
Conclusions
In summary, this an overview of NUT carcinoma with a brief discussion of the main epidemiologic and clinicopathologic features, along with the main molecular genetics/epigenetics findings and therapeutics for this tumor. Although several NUTM1 translocations have been found to be associated with NUT carcinoma, to the best of our knowledge, up to this day there is no known specific etiology for the NUTM1 translocation. In addition, given the rarity and relatively recent description of this entity, studies about the complete mutational landscape of NUT carcinoma are not yet available. Therefore, additional mutations that could play a role in oncogenesis are not well studied or known at this point in time. The awareness of this deadly tumor and the understanding of the underlying molecular mechanisms, genetics, and epigenetics will be helpful in future research for the development of novel targeted therapies.
Author Contributions
Conceptualization: VM and SP-O. All authors VM, KS, and SP-O drafted, revised, and approved the submitted version of this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
2. French CA. Pathogenesis of NUT Midline Carcinoma. Annu Rev Pathol (2012) 7:247–65. doi: 10.1146/annurev-pathol-011811-132438
3. French CA. NUT Carcinoma: Clinicopathologic Features, Pathogenesis, and Treatment. Pathol Int (2018) 68(11):583–95. doi: 10.1111/pin.12727
4. French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT Fusion Oncogene: A Novel Mechanism in Aggressive Carcinoma. Cancer Res (2003) 63(2):304–7.
5. French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, et al. BRD-NUT Oncoproteins: A Family of Closely Related Nuclear Proteins That Block Epithelial Differentiation and Maintain the Growth of Carcinoma Cells. Oncogene (2008) 27(15):2237–42. doi: 10.1038/sj.onc.1210852
6. Kees UR, Mulcahy MT, Willoughby ML. Intrathoracic Carcinoma in an 11-Year-Old Girl Showing a Translocation T(15;19). Am J Pediatr Hematol Oncol (1991) 13(4):459–64. doi: 10.1097/00043426-199124000-00011
7. Kubonishi I, Takehara N, Iwata J, Sonobe H, Ohtsuki Y, Abe T, et al. Novel T(15;19)(Q15;P13) Chromosome Abnormality in a Thymic Carcinoma. Cancer Res (1991) 51(12):3327–8.
8. French CA. Demystified Molecular Pathology of NUT Midline Carcinomas. J Clin Pathol (2010) 63(6):492–6. doi: 10.1136/jcp.2007.052902
9. Bauer DE, Mitchell CM, Strait KM, Lathan CS, Stelow EB, Luer SC, et al. Clinicopathologic Features and Long-Term Outcomes of NUT Midline Carcinoma. Clin Cancer Res (2012) 18(20):5773–9. doi: 10.1158/1078-0432.CCR-12-1153
10. Giridhar P, Mallick S, Kashyap L, Rath GK. Patterns of Care and Impact of Prognostic Factors in the Outcome of NUT Midline Carcinoma: A Systematic Review and Individual Patient Data Analysis of 119 Cases. Eur Arch Otorhinolaryngol (2018) 275(3):815–21. doi: 10.1007/s00405-018-4882-y
11. Chau NG, Hurwitz S, Mitchell CM, Aserlind A, Grunfeld N, Kaplan L, et al. Intensive Treatment and Survival Outcomes in NUT Midline Carcinoma of the Head and Neck. Cancer (2016) 122(23):3632–40. doi: 10.1002/cncr.30242
12. Chau NG, Ma C, Danga K, Al-Sayegh H, Nardi V, Barrette R, et al. An Anatomical Site and Genetic-Based Prognostic Model for Patients With Nuclear Protein in Testis (NUT) Midline Carcinoma: Analysis of 124 Patients. JNCI Cancer Spectr (2020) 4(2):pkz094. doi: 10.1093/jncics/pkz094
13. French CA, Kutok JL, Faquin WC, Toretsky JA, Antonescu CR, Griffin CA, et al. Midline Carcinoma of Children and Young Adults With NUT Rearrangement. J Clin Oncol (2004) 22(20):4135–9. doi: 10.1200/JCO.2004.02.107
14. Agaimy A, Fonseca I, Martins C, Thway K, Barrette R, Harrington KJ, et al. NUT Carcinoma of the Salivary Glands: Clinicopathologic and Molecular Analysis of 3 Cases and a Survey of NUT Expression in Salivary Gland Carcinomas. Am J Surg Pathol (2018) 42(7):877–84. doi: 10.1097/PAS.0000000000001046
15. Andreasen S, French CA, Josiassen M, Hahn CH, Kiss K. NUT Carcinoma of the Sublingual Gland. Head Neck Pathol (2016) 10(3):362–6. doi: 10.1007/s12105-015-0672-7
16. Cho Y, Keam BS, Jung KC, Kim BH. A Case of Nuclear Protein in Testis Midline Carcinoma Arising From the Submandibular Gland Duct in a Pregnant Patient. J Oral Maxillofac Surg (2017) 75(9):2020–4. doi: 10.1016/j.joms.2017.02.002
17. den Bakker MA, Beverloo BH, van den Heuvel-Eibrink MM, Meeuwis CA, Tan LM, Johnson LA, et al. NUT Midline Carcinoma of the Parotid Gland With Mesenchymal Differentiation. Am J Surg Pathol (2009) 33(8):1253–8. doi: 10.1097/PAS.0b013e3181abe120
18. Esteves G, Ferreira J, Afonso R, Martins C, Zagalo C, Felix A. HDAC Overexpression in a NUT Midline Carcinoma of the Parotid Gland With Exceptional Survival: A Case Report. Head Neck Pathol (2020) 14(4):1117–22. doi: 10.1007/s12105-020-01130-6
19. Klijanienko J, Le Tourneau C, Rodriguez J, Caly M, Theocharis S. Cytological Features of NUT Midline Carcinoma Arising in Sino-Nasal Tract and Parotid Gland: Report of Two New Cases and Review of the Literature. Diagn Cytopathol (2016) 44(9):753–6. doi: 10.1002/dc.23506
20. Lemelle L, Pierron G, Freneaux P, Huybrechts S, Spiegel A, Plantaz D, et al. NUT Carcinoma in Children and Adults: A Multicenter Retrospective Study. Pediatr Blood Cancer (2017) 64(12):e26693. doi: 10.1002/pbc.26693
21. Park HS, Bae YS, Yoon SO, Lim BJ, Hong HJ, Ro JY, et al. Usefulness of Nuclear Protein in Testis (NUT) Immunohistochemistry in the Cytodiagnosis of NUT Midline Carcinoma: A Brief Case Report. Korean J Pathol (2014) 48(4):335–8. doi: 10.4132/KoreanJPathol.2014.48.4.335
22. Saik WN, Da Forno P, Thway K, Khurram SA. NUT Carcinoma Arising From the Parotid Gland: A Case Report and Review of the Literature. Head Neck Pathol (2021) 15(3):1064–8. doi: 10.1007/s12105-020-01254-9
23. Seim NB, Philips RHW, Schoenfield L, Teknos TN, Rocco JW, Agrawal A, et al. NUT Midline Carcinoma of the Sublingual Gland: Clinical Presentation and Review. Head Neck Pathol (2017) 11(4):460–8. doi: 10.1007/s12105-017-0809-y
24. Storck S, Kennedy AL, Marcus KJ, Teot L, Vaughn J, Gnekow AK, et al. Pediatric NUT-Midline Carcinoma: Therapeutic Success Employing a Sarcoma Based Multimodal Approach. Pediatr Hematol Oncol (2017) 34(4):231–7. doi: 10.1080/08880018.2017.1363839
25. Vulsteke C, Lurquin E, Debiec-Rychter M, Gheysens O, Nuyts S, Schoenaers J, et al. First Evidence of Treatment Efficacy in Metastatic Carcinoma of the Parotid Gland With BRD4/NUT Translocation. J Chemother (2016) 28(3):242–6. doi: 10.1179/1973947815Y.0000000046
26. Wang H, Weiss VL, Hoffman RD, Abel T, Ho RH, Borinstein SC, et al. Salivary Gland NUT Carcinoma With Prolonged Survival in Children: Case Illustration and Systematic Review of Literature. Head Neck Pathol (2021) 15(1):236–43. doi: 10.1007/s12105-020-01141-3
27. Ziai J, French CA, Zambrano E. NUT Gene Rearrangement in a Poorly-Differentiated Carcinoma of the Submandibular Gland. Head Neck Pathol (2010) 4(2):163–8. doi: 10.1007/s12105-010-0174-6
28. Moreno V, French CA, Guo T, Zhu H, Zhang S, Duncan D, et al. NUT Carcinoma of the Submandibular Gland: A Case at This Uncommon Site With Review of the Literature. Head Neck Pathol (2021). doi: 10.1007/s12105-021-01400-x
29. Dickson BC, Sung YS, Rosenblum MK, Reuter VE, Harb M, Wunder JS, et al. NUTM1 Gene Fusions Characterize a Subset of Undifferentiated Soft Tissue and Visceral Tumors. Am J Surg Pathol (2018) 42(5):636–45. doi: 10.1097/PAS.0000000000001021
30. Bishop JA, French CA, Ali SZ. Cytopathologic Features of NUT Midline Carcinoma: A Series of 26 Specimens From 13 Patients. Cancer Cytopathol (2016) 124(12):901–8. doi: 10.1002/cncy.21761
31. Yang F, Shen D, Shi J. Primary Renal NUT Carcinoma Identified by Next-Generation Sequencing: A Case Report and Literature Review. Int J Clin Exp Pathol (2021) 14(5):662–9.
32. Shehata BM, Steelman CK, Abramowsky CR, Olson TA, French CA, Saxe DF, et al. NUT Midline Carcinoma in a Newborn With Multiorgan Disseminated Tumor and a 2-Year-Old With a Pancreatic/Hepatic Primary. Pediatr Dev Pathol (2010) 13(6):481–5. doi: 10.2350/09-10-0727-CR.1
33. Mertens F, Wiebe T, Adlercreutz C, Mandahl N, French CA. Successful Treatment of a Child With T(15;19)-Positive Tumor. Pediatr Blood Cancer (2007) 49(7):1015–7. doi: 10.1002/pbc.20755
34. Stelow EB, Bellizzi AM, Taneja K, Mills SE, Legallo RD, Kutok JL, et al. NUT Rearrangement in Undifferentiated Carcinomas of the Upper Aerodigestive Tract. Am J Surg Pathol (2008) 32(6):828–34. doi: 10.1097/PAS.0b013e31815a3900
35. Bishop JA. Recently Described Neoplasms of the Sinonasal Tract. Semin Diagn Pathol (2016) 33(2):62–70. doi: 10.1053/j.semdp.2015.12.001
36. Vargas SO. Childhood Carcinoma. Surg Pathol Clin (2010) 3(3):689–710. doi: 10.1016/j.path.2010.06.008
37. Napolitano M, Venturelli M, Molinaro E, Toss A. NUT Midline Carcinoma of the Head and Neck: Current Perspectives. Onco Targets Ther (2019) 12:3235–44. doi: 10.2147/OTT.S173056
38. French CA, Rahman S, Walsh EM, Kuhnle S, Grayson AR, Lemieux ME, et al. NSD3-NUT Fusion Oncoprotein in NUT Midline Carcinoma: Implications for a Novel Oncogenic Mechanism. Cancer Discov (2014) 4(8):928–41. doi: 10.1158/2159-8290.CD-14-0014
39. Stevens TM, Morlote D, Xiu J, Swensen J, Brandwein-Weber M, Miettinen MM, et al. NUTM1-Rearranged Neoplasia: A Multi-Institution Experience Yields Novel Fusion Partners and Expands the Histologic Spectrum. Mod Pathol (2019) 32(6):764–73. doi: 10.1038/s41379-019-0206-z
40. Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Gonzalez A, Gazdar AF, et al. Cloned Fusion Product From a Rare T(15;19)(Q13.2;p13.1) inhibit S phase in vitro. J Med Genet (2005) 42(7):558–64. doi: 10.1136/jmg.2004.029686
41. Thompson-Wicking K, Francis RW, Stirnweiss A, Ferrari E, Welch MD, Baker E, et al. Novel BRD4-NUT Fusion Isoforms Increase the Pathogenic Complexity in NUT Midline Carcinoma. Oncogene (2013) 32(39):4664–74. doi: 10.1038/onc.2012.487
42. Stirnweiss A, McCarthy K, Oommen J, Crook ML, Hardy K, Kees UR, et al. A Novel BRD4-NUT Fusion in an Undifferentiated Sinonasal Tumor Highlights Alternative Splicing as a Contributing Oncogenic Factor in NUT Midline Carcinoma. Oncogenesis (2015) 4(11):e174. doi: 10.1038/oncsis.2015.33
43. Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 Extraterminal Domain Confers Transcription Activation Independent of pTEFb by Recruiting Multiple Proteins, Including NSD3. Mol Cell Biol (2011) 31(13):2641–52. doi: 10.1128/mcb.01341-10
44. Alekseyenko AA, Walsh EM, Zee BM, Pakozdi T, Hsi P, Lemieux ME, et al. Ectopic Protein Interactions Within BRD4-Chromatin Complexes Drive Oncogenic Megadomain Formation in NUT Midline Carcinoma. Proc Natl Acad Sci USA (2017) 114(21):E4184–E92. doi: 10.1073/pnas.1702086114
45. Shiota H, Elya JE, Alekseyenko AA, Chou PM, Gorman SA, Barbash O, et al. "Z4" Complex Member Fusions in NUT Carcinoma: Implications for a Novel Oncogenic Mechanism. Mol Cancer Res (2018) 16(12):1826–33. doi: 10.1158/1541-7786.MCR-18-0474
46. Agaimy A, Tögel L, Haller F, Zenk J, Hornung J, Märkl B. YAP1-NUTM1 Gene Fusion in Porocarcinoma of the External Auditory Canal. Head Neck Pathol (2020) 14(4):982–90. doi: 10.1007/s12105-020-01173-9
47. Sekine S, Kiyono T, Ryo E, Ogawa R, Wakai S, Ichikawa H, et al. Recurrent YAP1-MAML2 and YAP1-NUTM1 Fusions in Poroma and Porocarcinoma. J Clin Invest (2019) 129(9):3827–32. doi: 10.1172/jci126185
48. Tamura R, Nakaoka H, Yoshihara K, Mori Y, Yachida N, Nishikawa N, et al. Novel MXD4-NUTM1 Fusion Transcript Identified in Primary Ovarian Undifferentiated Small Round Cell Sarcoma. Genes Chromosomes Cancer (2018) 57(11):557–63. doi: 10.1002/gcc.22668
49. Van Treeck BJ, Thangaiah JJ, Torres-Mora J, Stevens TM, Rothermundt C, Fassan M, et al. NUTM1-Rearranged Colorectal Sarcoma: A Clinicopathologically and Genetically Distinctive Malignant Neoplasm With a Poor Prognosis. Mod Pathol (2021) 34(8):1547–57. doi: 10.1038/s41379-021-00792-z
50. Le Loarer F, Pissaloux D, Watson S, Godfraind C, Galmiche-Rolland L, Silva K, et al. Clinicopathologic Features of CIC-NUTM1 Sarcomas, a New Molecular Variant of the Family of CIC-Fused Sarcomas. Am J Surg Pathol (2019) 43(2):268–76. doi: 10.1097/PAS.0000000000001187
51. Schaefer IM, Dal Cin P, Landry LM, Fletcher CDM, Hanna GJ, French CA. CIC-NUTM1 Fusion: A Case Which Expands the Spectrum of NUT-Rearranged Epithelioid Malignancies. Genes Chromosomes Cancer (2018) 57(9):446–51. doi: 10.1002/gcc.3
52. Siegfried A, Masliah-Planchon J, Roux FE, Larrieu-Ciron D, Pierron G, Nicaise Y, et al. Brain Tumor With an ATXN1-NUTM1 Fusion Gene Expands the Histologic Spectrum of NUTM1-Rearranged Neoplasia. Acta Neuropathol Commun (2019) 7(1):220. doi: 10.1186/s40478-019-0870-8
53. Diolaiti D, Dela Cruz FS, Gundem G, Bouvier N, Boulad M, Zhang Y, et al. A Recurrent Novel MGA-NUTM1 Fusion Identifies a New Subtype of High-Grade Spindle Cell Sarcoma. Cold Spring Harb Mol Case Stud (2018) 4(6):a003194. doi: 10.1101/mcs.a003194
54. Mantilla JG, Ricciotti RW, Chen E, Hoch BL, Liu YJ. Detecting Disease-Defining Gene Fusions in Unclassified Round Cell Sarcomas Using Anchored Multiplex PCR/targeted RNA Next-Generation Sequencing-Molecular and Clinicopathological Characterization of 16 Cases. Genes Chromosomes Cancer (2019) 58(10):713–22. doi: 10.1002/gcc.22763
55. Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The Double Bromodomain Protein Brd4 Binds to Acetylated Chromatin During Interphase and Mitosis. Proc Natl Acad Sci USA (2003) 100(15):8758–63. doi: 10.1073/pnas.1433065100
56. Yang Z, He N, Zhou Q. Brd4 Recruits P-TEFb to Chromosomes at Late Mitosis to Promote G1 Gene Expression and Cell Cycle Progression. Mol Cell Biol (2008) 28(3):967–76. doi: 10.1128/mcb.01020-07
57. Grayson AR, Walsh EM, Cameron MJ, Godec J, Ashworth T, Ambrose JM, et al. MYC, a Downstream Target of BRD-NUT, is Necessary and Sufficient for the Blockade of Differentiation in NUT Midline Carcinoma. Oncogene (2014) 33(13):1736–42. doi: 10.1038/onc.2013.126
58. Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The Bromodomain Protein Brd4 is a Positive Regulatory Component of P-TEFb and Stimulates RNA Polymerase II-Dependent Transcription. Mol Cell (2005) 19(4):523–34. doi: 10.1016/j.molcel.2005.06.027
59. Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for Stimulation of Transcriptional Elongation by the Bromodomain Protein Brd4. Mol Cell (2005) 19(4):535–45. doi: 10.1016/j.molcel.2005.06.029
60. Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and Characterization of Super-Enhancer-Associated Dependencies in Diffuse Large B Cell Lymphoma. Cancer Cell (2013) 24(6):777–90. doi: 10.1016/j.ccr.2013.11.003
61. Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target C-Myc. Cell (2011) 146(6):904–17. doi: 10.1016/j.cell.2011.08.017
62. Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell (2013) 153(2):320–34. doi: 10.1016/j.cell.2013.03.036
63. Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET Recruitment to Chromatin as an Effective Treatment for MLL-Fusion Leukaemia. Nature (2011) 478(7370):529–33. doi: 10.1038/nature10509
64. Alekseyenko AA, Walsh EM, Wang X, Grayson AR, Hsi PT, Kharchenko PV, et al. The Oncogenic BRD4-NUT Chromatin Regulator Drives Aberrant Transcription Within Large Topological Domains. Genes Dev (2015) 29(14):1507–23. doi: 10.1101/gad.267583.115
65. Dey A, Yang W, Gegonne A, Nishiyama A, Pan R, Yagi R, et al. BRD4 Directs Hematopoietic Stem Cell Development and Modulates Macrophage Inflammatory Responses. EMBO J (2019) 38(7):e100293. doi: 10.15252/embj.2018100293
66. Gilan O, Rioja I, Knezevic K, Bell MJ, Yeung MM, Harker NR, et al. Selective Targeting of BD1 and BD2 of the BET Proteins in Cancer and Immunoinflammation. Science (2020) 368(6489):387–94. doi: 10.1126/science.aaz8455
67. Belkina AC, Nikolajczyk BS, Denis GV. BET Protein Function is Required for Inflammation: Brd2 Genetic Disruption and BET Inhibitor JQ1 Impair Mouse Macrophage Inflammatory Responses. J Immunol (2013) 190(7):3670–8. doi: 10.4049/jimmunol.1202838
68. Wienerroither S, Rauch I, Rosebrock F, Jamieson AM, Bradner J, Muhar M, et al. Regulation of NO Synthesis, Local Inflammation, and Innate Immunity to Pathogens by BET Family Proteins. Mol Cell Biol (2014) 34(3):415–27. doi: 10.1128/mcb.01353-13
69. Haack H, Johnson LA, Fry CJ, Crosby K, Polakiewicz RD, Stelow EB, et al. Diagnosis of NUT Midline Carcinoma Using a NUT-Specific Monoclonal Antibody. Am J Surg Pathol (2009) 33(7):984–91. doi: 10.1097/PAS.0b013e318198d666
70. Shiota H, Barral S, Buchou T, Tan M, Couté Y, Charbonnier G, et al. Nut Directs P300-Dependent, Genome-Wide H4 Hyperacetylation in Male Germ Cells. Cell Rep (2018) 24(13):3477–87.e6. doi: 10.1016/j.celrep.2018.08.069
71. Reynoird N, Schwartz BE, Delvecchio M, Sadoul K, Meyers D, Mukherjee C, et al. Oncogenesis by Sequestration of CBP/p300 in Transcriptionally Inactive Hyperacetylated Chromatin Domains. EMBO J (2010) 29(17):2943–52. doi: 10.1038/emboj.2010.176
72. Schwartz BE, Hofer MD, Lemieux ME, Bauer DE, Cameron MJ, West NH, et al. Differentiation of NUT Midline Carcinoma by Epigenomic Reprogramming. Cancer Res (2011) 71(7):2686–96. doi: 10.1158/0008-5472.Can-10-3513
73. Bennett RL, Swaroop A, Troche C, Licht JD. The Role of Nuclear Receptor-Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harb Perspect Med (2017) 7(6):a026708. doi: 10.1101/cshperspect.a026708
74. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell (2007) 129(4):823–37. doi: 10.1016/j.cell.2007.05.009
75. Lachner M, O'Sullivan RJ, Jenuwein T. An Epigenetic Road Map for Histone Lysine Methylation. J Cell Sci (2003) 116(Pt 11):2117–24. doi: 10.1242/jcs.00493
76. French CA. Small-Molecule Targeting of BET Proteins in Cancer. Adv Cancer Res (2016) 131:21–58. doi: 10.1016/bs.acr.2016.04.001
77. Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, et al. NSD3-Short Is an Adaptor Protein That Couples BRD4 to the CHD8 Chromatin Remodeler. Mol Cell (2015) 60(6):847–59. doi: 10.1016/j.molcel.2015.10.033
78. Buschbeck M, Uribesalgo I, Wibowo I, Rué P, Martin D, Gutierrez A, et al. The Histone Variant Macroh2a is an Epigenetic Regulator of Key Developmental Genes. Nat Struct Mol Biol (2009) 16(10):1074–9. doi: 10.1038/nsmb.1665
79. Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The Histone Variant Macroh2a1 Marks Repressed Autosomal Chromatin, But Protects a Subset of its Target Genes From Silencing. Genes Dev (2010) 24(1):21–32. doi: 10.1101/gad.1876110
80. Wang R, You J. Mechanistic Analysis of the Role of Bromodomain-Containing Protein 4 (BRD4) in BRD4-NUT Oncoprotein-Induced Transcriptional Activation. J Biol Chem (2015) 290(5):2744–58. doi: 10.1074/jbc.M114.600759
81. Zee BM, Dibona AB, Alekseyenko AA, French CA, Kuroda MI. The Oncoprotein BRD4-NUT Generates Aberrant Histone Modification Patterns. PloS One (2016) 11(10):e0163820. doi: 10.1371/journal.pone.0163820
82. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell (2005) 122(6):947–56. doi: 10.1016/j.cell.2005.08.020
83. Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, et al. SOX2 Silencing in Glioblastoma Tumor-Initiating Cells Causes Stop of Proliferation and Loss of Tumorigenicity. Stem Cells (2009) 27(1):40–8. doi: 10.1634/stemcells.2008-0493
84. Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, et al. Sox2 Expression in Breast Tumours and Activation in Breast Cancer Stem Cells. Oncogene (2012) 31(11):1354–65. doi: 10.1038/onc.2011.338
85. Sanada Y, Yoshida K, Ohara M, Oeda M, Konishi K, Tsutani Y. Histopathologic Evaluation of Stepwise Progression of Pancreatic Carcinoma With Immunohistochemical Analysis of Gastric Epithelial Transcription Factor SOX2: Comparison of Expression Patterns Between Invasive Components and Cancerous or Nonneoplastic Intraductal Components. Pancreas (2006) 32(2):164–70. doi: 10.1097/01.mpa.0000202947.80117.a0
86. Wu F, Zhang J, Wang P, Ye X, Jung K, Bone KM, et al. Identification of Two Novel Phenotypically Distinct Breast Cancer Cell Subsets Based on Sox2 Transcription Activity. Cell Signal (2012) 24(11):1989–98. doi: 10.1016/j.cellsig.2012.07.008
87. Wang R, Liu W, Helfer CM, Bradner JE, Hornick JL, Janicki SM, et al. Activation of SOX2 Expression by BRD4-NUT Oncogenic Fusion Drives Neoplastic Transformation in NUT Midline Carcinoma. Cancer Res (2014) 74(12):3332–43. doi: 10.1158/0008-5472.CAN-13-2658
88. Ito M, Okano HJ, Darnell RB, Roeder RG. The TRAP100 Component of the TRAP/Mediator Complex is Essential in Broad Transcriptional Events and Development. EMBO J (2002) 21(13):3464–75. doi: 10.1093/emboj/cdf348
89. Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, et al. A Novel Human SRB/MED-Containing Cofactor Complex, SMCC, Involved in Transcription Regulation. Mol Cell (1999) 3(1):97–108. doi: 10.1016/s1097-2765(00)80178-1
90. Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective Inhibition of BET Bromodomains. Nature (2010) 468(7327):1067–73. doi: 10.1038/nature09504
91. Mao N, Liao Z, Wu J, Liang K, Wang S, Qin S, et al. Diagnosis of NUT Carcinoma of Lung Origin by Next-Generation Sequencing: Case Report and Review of the Literature. Cancer Biol Ther (2019) 20(2):150–6. doi: 10.1080/15384047.2018.1523852
92. McLean-Holden AC, Moore SA, Gagan J, French CA, Sher D, Truelson JM, et al. NUT Carcinoma in a Patient With Unusually Long Survival and False Negative FISH Results. Head Neck Pathol (2021) 15(2):698–703. doi: 10.1007/s12105-020-01220-5
93. Paver E, O'Toole S, Cheng XM, Mahar A, Cooper WA. Updates in the Molecular Pathology of Non-Small Cell Lung Cancer. Semin Diagn Pathol (2021) 38(5):54–61. doi: 10.1053/j.semdp.2021.04.001
94. Elomaa I, Blomqvist CP, Saeter G, Akerman M, Stenwig E, Wiebe T, et al. Five-Year Results in Ewing's Sarcoma. The Scandinavian Sarcoma Group Experience With the SSG IX Protocol. Eur J Cancer (2000) 36(7):875–80. doi: 10.1016/s0959-8049(00)00028-9
95. Lewin J, Soria JC, Stathis A, Delord JP, Peters S, Awada A, et al. Phase Ib Trial With Birabresib, a Small-Molecule Inhibitor of Bromodomain and Extraterminal Proteins, in Patients With Selected Advanced Solid Tumors. J Clin Oncol (2018) 36(30):3007–14. doi: 10.1200/jco.2018.78.2292
96. Piha-Paul SA, Hann CL, French CA, Cousin S, Braña I, Cassier PA, et al. Phase 1 Study of Molibresib (GSK525762), a Bromodomain and Extra-Terminal Domain Protein Inhibitor, in NUT Carcinoma and Other Solid Tumors. JNCI Cancer Spectr (2020) 4(2):pkz093. doi: 10.1093/jncics/pkz093
97. Shapiro GI, LoRusso P, Dowlati A, TD K, Jacobson CA, Vaishampayan U, et al. A Phase 1 Study of RO6870810, a Novel Bromodomain and Extra-Terminal Protein Inhibitor, in Patients With NUT Carcinoma, Other Solid Tumours, or Diffuse Large B-Cell Lymphoma. Br J Cancer (2021) 124(4):744–53. doi: 10.1038/s41416-020-01180-1
98. Stathis A, Zucca E, Bekradda M, Gomez-Roca C, Delord JP, de la Motte Rouge T, et al. Clinical Response of Carcinomas Harboring the BRD4-NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer Discov (2016) 6(5):492–500. doi: 10.1158/2159-8290.Cd-15-1335
99. Ameratunga M, Brana I, Bono P, Postel-Vinay S, Plummer R, Aspegren J, et al. First-In-Human Phase 1 Open Label Study of the BET Inhibitor ODM-207 in Patients With Selected Solid Tumours. Br J Cancer (2020) 123(12):1730–6. doi: 10.1038/s41416-020-01077-z
100. Morrison-Smith CD, Knox TM, Filic I, Soroko KM, Eschle BK, Wilkens MK, et al. Combined Targeting of the BRD4-NUT-P300 Axis in NUT Midline Carcinoma by Dual Selective Bromodomain Inhibitor, Neo2734. Mol Cancer Ther (2020) 19(7):1406–14. doi: 10.1158/1535-7163.Mct-20-0087
101. ClinicalTrials.gov. Study of the Bromodomain (BRD) and Extra-Terminal Domain (BET) Inhibitors BMS-986158 and BMS-986378 in Pediatric Cancer (2019). Available at: https://clinicaltrials.gov/ct2/show/NCT03936465 (Accessed 01/12/2022).
102. Maher OM, Christensen AM, Yedururi S, Bell D, Tarek N. Histone Deacetylase Inhibitor for NUT Midline Carcinoma. Pediatr Blood Cancer (2015) 62(4):715–7. doi: 10.1002/pbc.25350
103. Hogg SJ, Vervoort SJ, Deswal S, Ott CJ, Li J, Cluse LA, et al. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep (2017) 18(9):2162–74. doi: 10.1016/j.celrep.2017.02.011
104. Kagoya Y, Nakatsugawa M, Yamashita Y, Ochi T, Guo T, Anczurowski M, et al. BET Bromodomain Inhibition Enhances T Cell Persistence and Function in Adoptive Immunotherapy Models. J Clin Invest (2016) 126(9):3479–94. doi: 10.1172/jci86437
105. Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, et al. BET Bromodomain Inhibition Promotes Anti-Tumor Immunity by Suppressing PD-L1 Expression. Cell Rep (2016) 16(11):2829–37. doi: 10.1016/j.celrep.2016.08.032
106. NUT Midline Carcinoma Registry. Our Goals are (2021). Available at: http://www.nmcregistry.org (Accessed 08/12/2021).
Keywords: NUT carcinoma, NUT midline carcinoma, BRD-NUTM1, NSD3, zinc finger proteins, BET inhibitors, HDAC inhibitors
Citation: Moreno V, Saluja K and Pina-Oviedo S (2022) NUT Carcinoma: Clinicopathologic Features, Molecular Genetics and Epigenetics. Front. Oncol. 12:860830. doi: 10.3389/fonc.2022.860830
Received: 23 January 2022; Accepted: 24 February 2022;
Published: 16 March 2022.
Edited by:
Luis Del Valle, Louisiana State University, United StatesReviewed by:
John Charles Rotondo, University of Ferrara, ItalyPeter C. Angeletti, University of Nebraska-Lincoln, United States
Copyright © 2022 Moreno, Saluja and Pina-Oviedo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergio Pina-Oviedo, sergio.pinaoviedo@duke.edu
 Vanessa Moreno
Vanessa Moreno Karan Saluja
Karan Saluja Sergio Pina-Oviedo
Sergio Pina-Oviedo