- 1Department of Hematology/Oncology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, China
- 2Research Institute of Tsinghua University in Shenzhen, Shenzhen, China
- 3Department of Pathology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, China
- 4School of Pharmaceutical Sciences, Jilin University, Changchun, China
- 5Guangzhou Institute of Pediatrics, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangdong Provincial Clinical Research Center for Child Health, Guangzhou, China
Background: Rhabdomyosarcoma(RMS) is the most common soft tissue sarcoma in children and single nucleotide polymorphisms(SNPs) in certain genes influence risk of RMS. Although FOXO3 had been reported in multiple cancers including RMS, the role of FOXO3 polymorphisms in RMS remains unclear. In this case-control study, we evaluated the association of FOXO3 SNPs with RMS risk and prognosis in children.
Methods: Four FOXO3 SNPs(rs17069665 A>G, rs4946936 T>C, rs4945816 C>T and rs9400241 C>A) were genotyped in 110 RMS cases and 359 controls. The associations between FOXO3 polymorphisms and RMS risk were determined by odds ratios(ORs) with 95% confidence intervals(CIs). The associations of rs17069665 and rs4946936 with overall survival in RMS children were estimated using the Kaplan-Meier method and log-rank test. Functional analysis in silico was performed to estimate the probability that rs17069665 and rs4946936 might influence the regulation of FOXO3.
Results: We found that rs17069665 (GG vs. AA+AG, adjusted OR=2.96; 95%CI [1.10-3.32]; P=0.010) and rs4946936 (TC+CC vs. TT, adjusted OR=0.48; 95%CI [0.25-0.90]; P=0.023) were related to the increased and decreased RMS risk, respectively. Besides, rs17069665(P<0.001) and rs4946936(P<0.001) were associated with decreased and increased overall survival in RMS patients, respectively. Functional analysis showed that rs17069665 and rs4946936 might influence the transcription and expression of FOXO3 via altering the bindings to MYC, CTCF, and/or RELA.
Conclusions: This study revealed that FOXO3 polymorphisms influence the RMS susceptibility and prognosis in children, and might altered the expression of FOXO3. FOXO3 polymorphism was suggested as a biomarker for RMS susceptibility and prognosis.
Introduction
Rhabdomyosarcoma (RMS) is a myogenic tumor that arises from cells committed to skeletal muscle differentiation (1, 2). Although relatively rare, RMS is the most common soft tissue sarcoma and the third most prevalent extracranial solid tumor in children (3, 4). Despite of developments in the therapy and management of RMS patients in the past decades, the etiology of RMS remains unclear and the prognosis of RMS is also still poor (3, 5). Cancer occurs due to environmental exposure and/or genetic factors. In RMS initiation and development, genetic change has been suggested to play a vital role (3). One prevalent type of genetic variation in human disease is single nucleotide polymorphisms (SNPs). SNPs of certain genes have been validated to influence the incidence and outcome of multiple cancers including RMS (6, 7). However, many SNPs in genes associated with RMS are still not discovered and need to be explored.
Forkhead Box O3(FOXO3), also called Forkhead in Rhabdomyosarcoma-Like 1 (FKHRL1), belongs to the forkhead box O (FoxO) family of transcription factors which regulate the transcription of genes involved in multiple cellular processes (proliferation, apoptosis, autophagy, etc.) (8–10). Evidence supported that the aberrant expression or dysfunction of FOXO3 was associated with various types of cancers including RMS (11–14). In addition, gene polymorphism of FOXO3 has been validated to influence various human disorders including cancer, such as pancreatic cancer, colorectal cancer, active tuberculosis, polycystic ovary syndrome, and acute lymphoblastic leukemia (ALL) (15–20). However, the association of FOXO3 polymorphisms with childhood RMS risk and outcome has never been reported.
In this case-control study, we conducted genotyping assay for FOXO3 SNPs (rs17069665, rs4946936, rs4945816, and rs9400241) and estimated the association of FOXO3 polymorphisms with RMS risk and prognosis in children. Besides, the underlying mechanisms involved in FOXO3 regulation by these polymorphisms were explored via functional analysis in silico.
Materials and methods
Study subjects
In this current study, a total of 110 RMS cases and 359 non-cancer healthy controls were recruited from Guangzhou Women and Children’s Medical Center (GWCMC). Briefly, patients under 18 years old with RMS were identified with histological confirmation and enrolled in this study by pediatric clinicians. Each case was newly diagnosed and provided a detailed medical record. Patients with other malignancies, secondary disorders, or chemotherapy/radiotherapy records were excluded. Thus, 110 children with RMS were recruited during June 2012 to June 2019, and 359 age- and sex-matched healthy volunteers were collected as controls. The controls were randomly selected from children receiving routine physical examinations. All individuals included in this study were ethnic Han Chinese. Before participation, a written informed consent for each case was obtained in advance. This study was conducted under the guideline of the Institutional Review Board of GWCMC.
SNP Selection and genotyping
In this study, four SNPs were selected using data from SNPinfo and NCBI dbSNP databases as described previously (21–23). Briefly, the selection was based on three criteria as the following: (1) SNPs with the minor allele frequency (MAF) >5% in Chinese population; (2) located in or near the FOXO3 gene (i.e., 3’-UTR or 5’-UTR); (3) affecting splicing activity, transcription factor binding sites (TFBS) or miRNA-binding sites activity. At last, rs17069665 A>G, rs4946936 T>C, rs4945816 C>T, and rs9400241 C>A were retrieved for further analyses.
For genotyping, DNA was extracted from peripheral blood sample of each participant using the TIANamp DNAKit (TianGen, Beijing, China). In the genotyping assays, the corresponding SNP probes and primers (ABI, Massachusetts, USA) were used to genotype these four SNPs as previously described (21). For quality control, 10% samples were selected randomly for direct sequencing (24, 25), and a concordance rate of 100% was obtained.
Functional analysis in silico
The associations of rs17069665 A>G and rs4946936 T>C with FOXO3 expression were determined using the expression quantitative trait loci (eQTL) analysis in GTEx portal (https://www.gtexportal.org/home/) (26). The probability that these two polymorphisms might influence the regulation of FOXO3 was estimated by using the Roadmap Epigenome Browser (27, 28), the ENCODE Project (29, 30), and TFBIND software (31). In brief, promoter and enhancer were predicted via DNase hypersensitivity (DHS) and histone modification of muscle cells in Roadmap Epigenomics data. TFBIND was used to evaluate whether FOXO3 polymorphism rs17069665 A>G or rs4946936 T>C altered any TFBS, and then ENCODE ChIP-seq experiments of MYC, CTCF, and RELA (Experiment Series: ENCSR000DLR, ENCSR000ANS, ENCSR000EBD) were used to assess the binding signals and motifs overlapping rs17069665 or rs4946936.
Statistical analyses
The demographic variables and genotype distribution of each SNP between the RMS case and control groups were compared using a 2-sided χ 2 test. For each SNP in the control group, Hardy-Weinberg equilibrium (HWE) was assessed using goodness-of-fit chi-square test. The strength of the association between FOXO3 polymorphism and RMS susceptibility was estimated using logistic regression analysis with odds ratios (ORs) and 95% confidence intervals (CIs), adjusting for age and sex. False-positive report probability (FPRP) was conducted for significant findings, as described previously (32). We adopted OR of 2.00 and 0.50 for risk and protective effects, respectively. FPRP values < 0.2 were considered noteworthy. Overall survival was estimated with Kaplan-Meier method, and then differences between subgroups were compared using the log-rank test. All statistical analyses were performed with SAS software and GraphPad Prism software; a two-sided P < 0.05 was considered to be statistically significant.
Results
Characteristics of subject
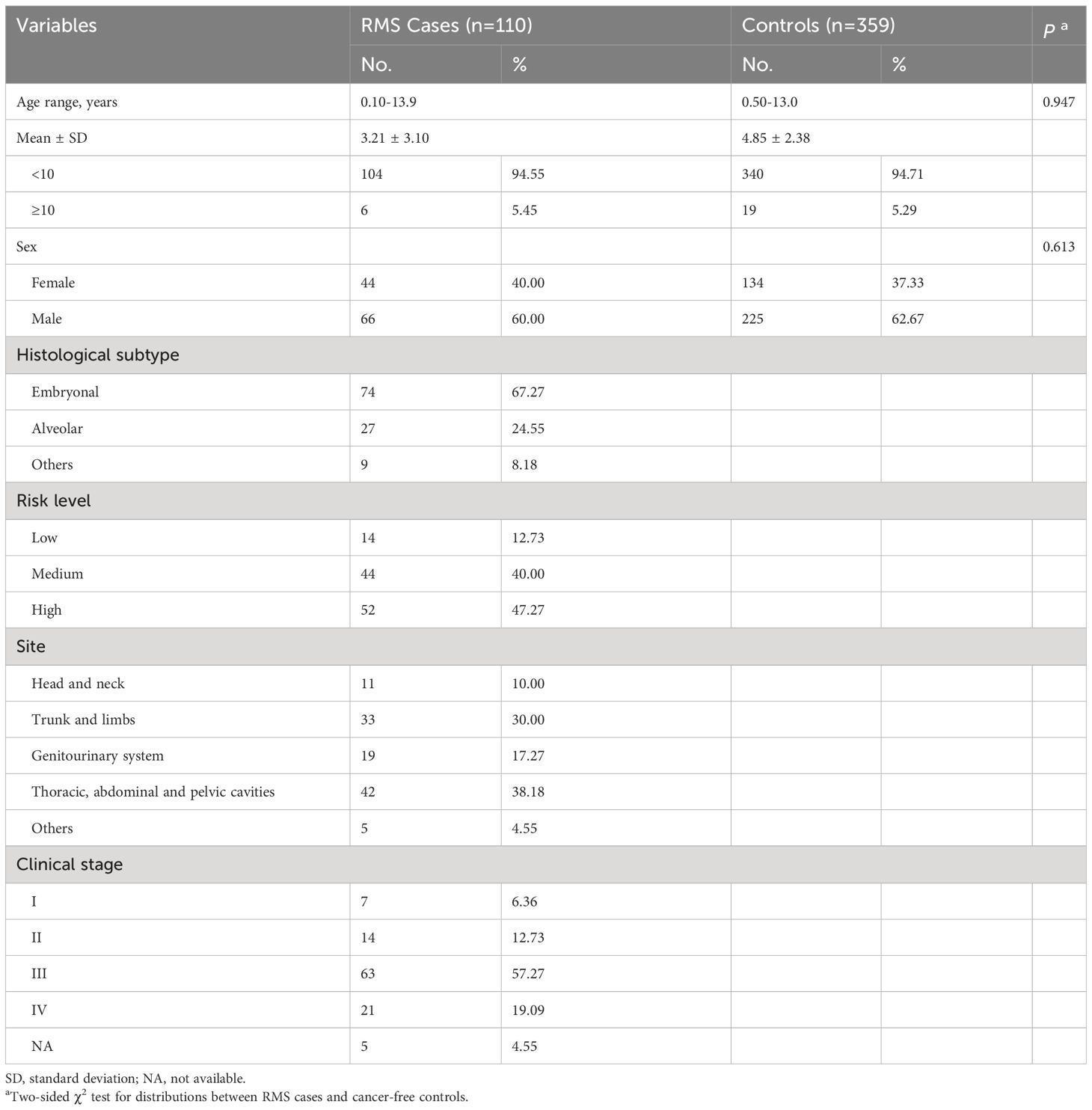
Totally, 110 RMS cases and 359 healthy controls were finally enrolled in this study. The detailed demographic characteristics are all summarized in Table 1. Briefly, no significant difference in the age (P = 0.947) or sex (P = 0.613) distribution was found between cases and controls. According to histological classification, 74 (67.27%) cases were diagnosed as embryonal RMS (ERMS), 27 (24.55%) cases as alveolar RMS (ARMS), and 9 (8.18%) cases as other subtypes including anaplastic and mixed RMS. In addition, other information including risk level, clinical stage, and site of origin were also included in Table 1.
Associations between FOXO3 gene polymorphisms and RMS susceptibility
According to the SNP selection strategy, four SNPs (rs17069665 A>G, rs4946936 T>C, rs4945816 C>T, and rs9400241 C>A) that overlap with TFBS or miRNA-binding sites were selected (Supplementary Table S1). The genotype frequencies of FOXO3 SNPs in all 110 cases and 359 controls and their association with RMS risk were displayed in Table 2. All these four SNPs were in HWE (PHWE > 0.05) among the control. Significant differences between RMS cases and controls were observed for rs17069665 A>G (P = 0.011) and rs4946936 T>C polymorphism (P = 0.025) in a recessive (GG vs. AA+AG) and a dominant (TC + CC vs. TT) model, respectively. After adjustment with age and sex, all these 2 polymorphisms, namely rs17069665 G allele (GG vs. AA+AG: adjusted OR = 2.96; 95% CI [1.10-3.32]; P = 0.010) and rs4946936 C allele (TC+CC vs. TT: adjusted OR=0.48; 95%CI [0.25-0.90]; P=0.023), were significantly related to the increased and decreased RMS risk, respectively. The remaining 2 polymorphisms (rs4945816 C>T and rs9400241 C>A), however, were not found to be in association with RMS risk.
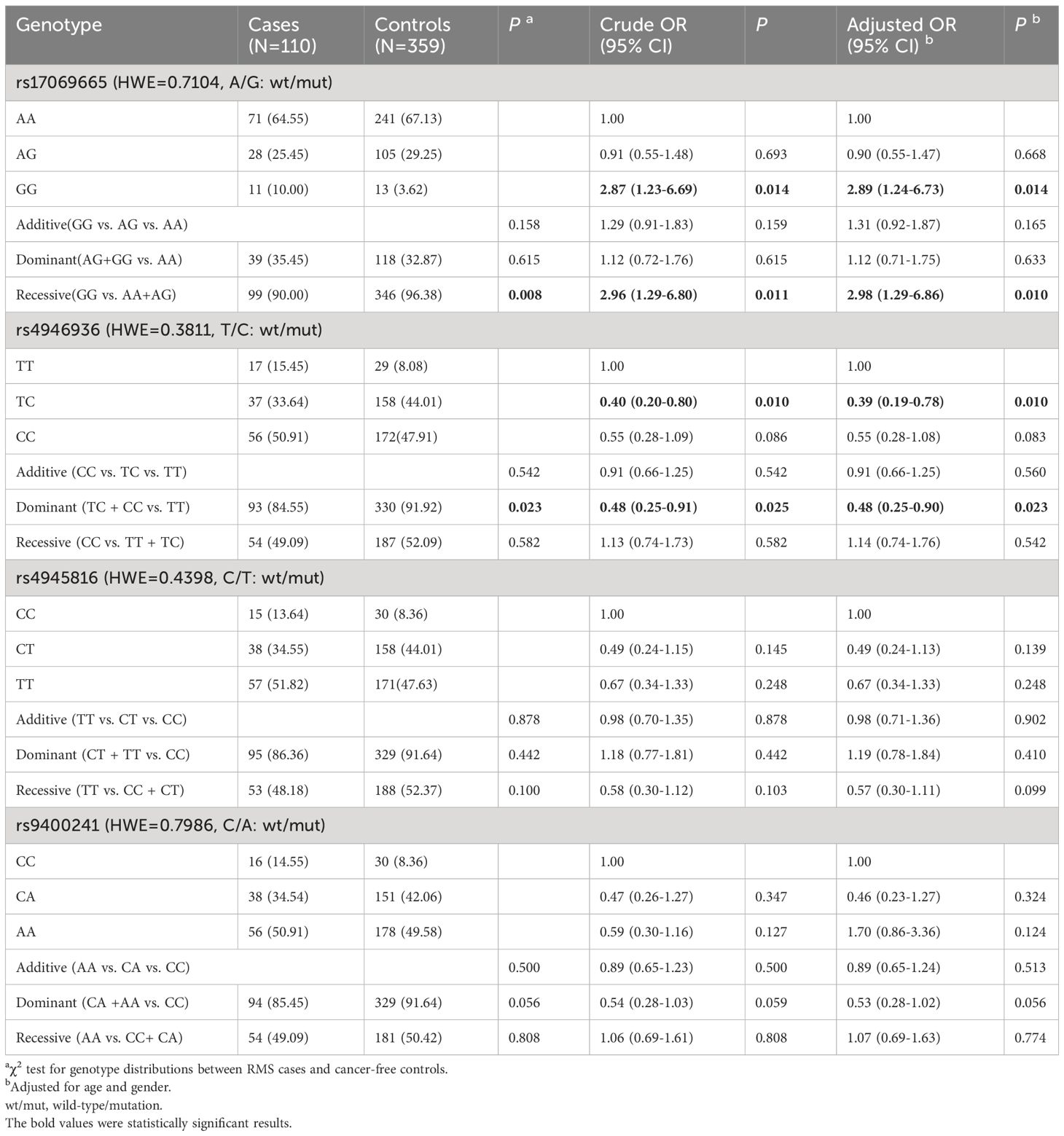
Table 2 Logistic regression analysis of associations between FOXO3 polymorphisms and RMS susceptibility.
Subgroup and stratification analyses
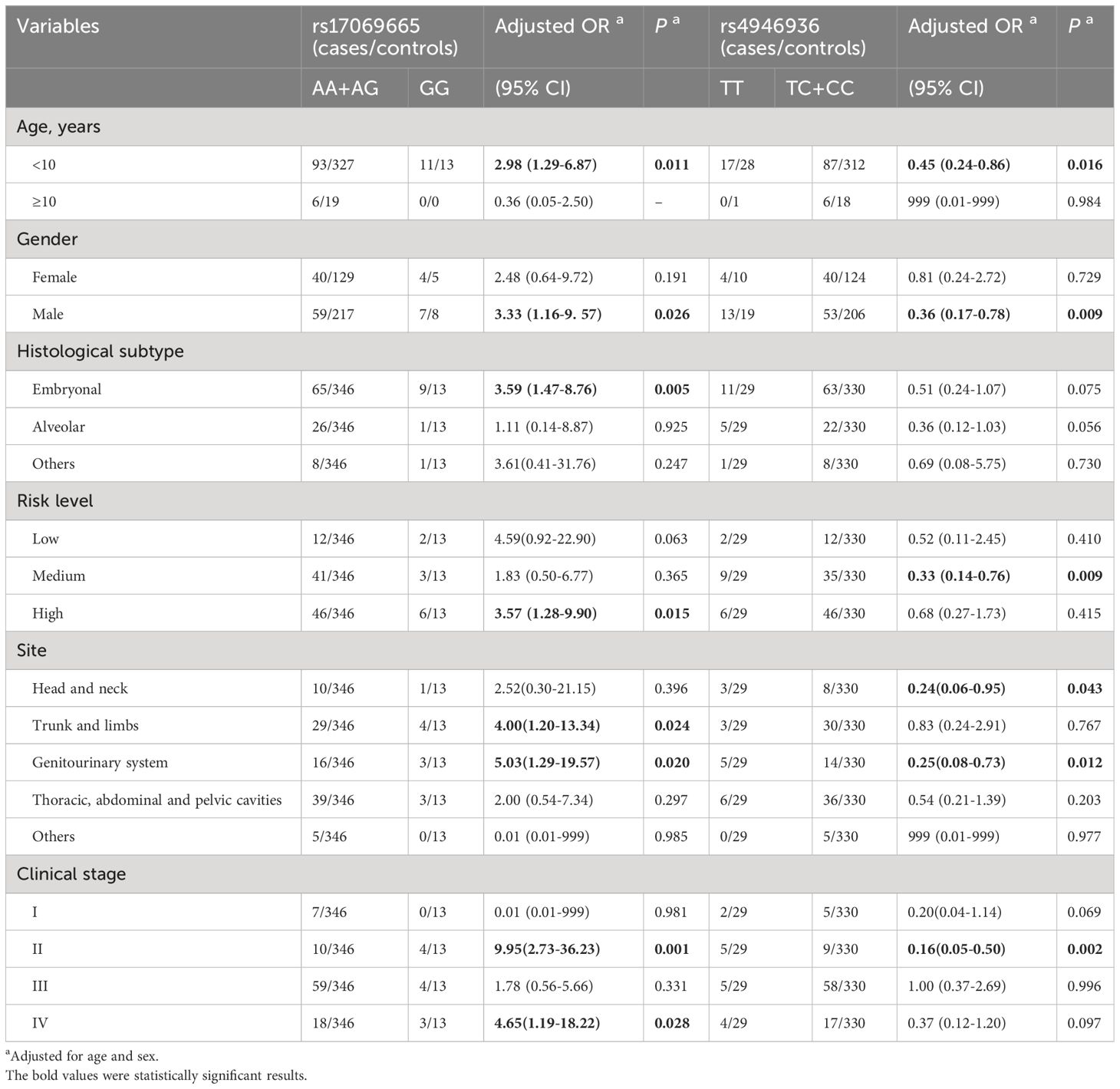
To further explore the association of the FOXO3 gene rs17069665 A>G and rs4946936 T>C polymorphisms with RMS susceptibility, we performed subgroup and stratification analyses in terms of age, sex, histological classification, risk level, clinical stage, and site of origin. As shown in Table 3, rs17069665 (GG vs. AA+AG) and rs4946936 (TC+CC vs. TT) were both associated with RMS in male (adjusted OR=3.33, 95% CI [1.16-9. 57], P =0.026; adjusted OR=0.36; 95% CI [0.17-0.78], P=0.009; respectively), in children aged < 10 years (adjusted OR=2.98, 95% CI [1.29-6.87], P =0.011; adjusted OR=0.45; 95% CI [0.24-0.86], P=0.016; respectively), in children with tumor of stage II (adjusted OR=9.95, 95% CI [2.73-36.23], P =0.001; adjusted OR=0.16; 95% CI [0.05-0.50], P=0.002; respectively), and in children with tumor in genitourinary system (adjusted OR=5.03, 95% CI [1.29-19.57], P =0.020; adjusted OR=0.25; 95% CI [0.08-0.73], P=0.012; respectively). In addition, rs17069665 (GG vs. AA+AG) was related to RMS in children with ERMS (adjusted OR=3.59, 95% CI [1.47-8.76], P =0.005), in children with tumor in high risk level (adjusted OR=3.57, 95% CI [1.28-9.90], P =0.015), in children with tumor of stage IV (adjusted OR=4.65, 95% CI [1.19-18.22], P =0.028), and in children with tumor in trunk and limbs (adjusted OR=4.00, 95% CI [1.20-13.34], P =0.024). Rs4946936 (TC+CC vs. TT) was related to RMS in children with tumor in medium risk level (adjusted OR=0.33, 95% CI [0.14-0.76], P =0.009) and in children with tumor in head and neck (adjusted OR=0.24, 95% CI [0.06-0.95], P =0.043). In the FPRP analysis (Supplementary Table S2), most of the above significant findings remained noteworthy at the prior probability level of 0.25(FPRP<0.200), which further strengthen the significant associations of rs17069665 and rs4946936 with RMS.
Overall survival analyses
The above results indicated that FOXO3 polymorphisms, rs17069665 and rs4946936, influenced RMS risk in children. However, it’s not clear whether they influence the prognosis of RMS. Therefore, we evaluated the associations of rs17069665 and rs4946936 with overall survival in RMS patients. The results were described in Figure 1. For rs17069665, RMS patients with GG allele had a poor overall survival than those with AA/AG (Figure 1A, P < 0.001). Compared with rs4946936 TT, TC/CC allele significantly contributed to the increased overall survival in RMS patients (Figure 1B, P < 0.001). The results suggested that FOXO3 polymorphisms were associated with the prognosis in RMS children.
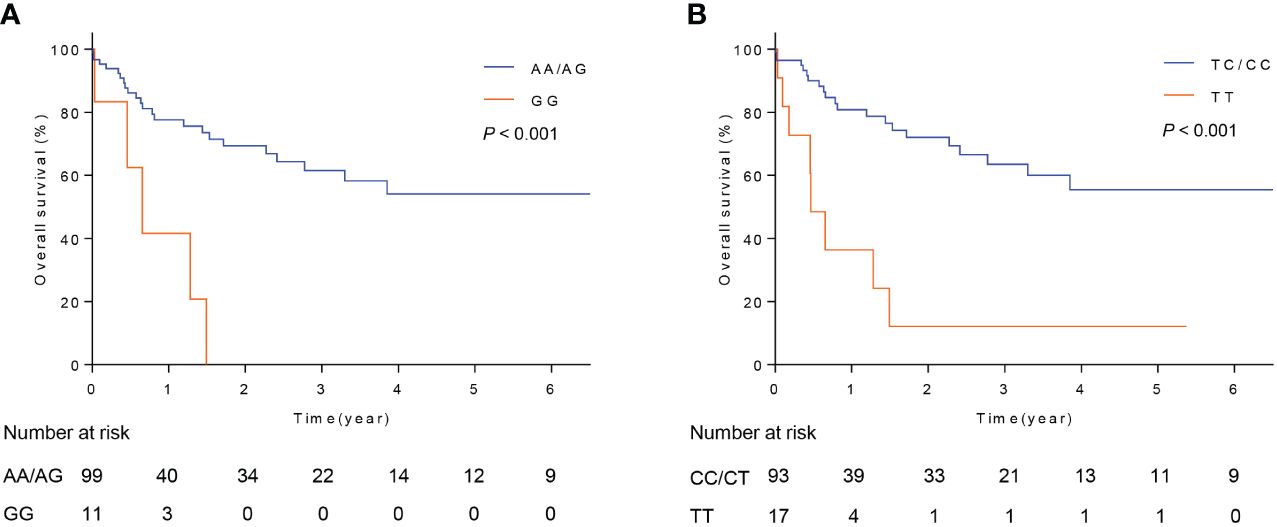
Figure 1 Overall survival of patients with RMS. (A) Survival curve of RMS patients with rs17069665 allele AA/AG or GG; (B) Survival curve of RMS patients with rs4946936 allele TC/CC or TT.
Functional analysis
To explore the possible mechanisms by which rs17069665 and rs4946936 influence the risk and prognosis of RMS, we evaluated the probability of rs17069665 and rs4946936 polymorphism altering expression regulation of FOXO3. In the eQTL analysis, rs17069665 G allele was associated with lower expression of FOXO3 gene (P = 8.52×10-4) (Supplementary Figure S1A). Rs4946936 C allele displayed a trend of high expression of FOXO3, although no significance was achieved (P = 3.32×10-1) (Supplementary Figure S1B). The Roadmap Epigenomics data showed that rs17069665 (Supplementary Figure S2A) and rs4946936 (Supplementary Figure S2B) both overlapped DHS marks and histone modifications related to the enhancer and promoter in multiple tissue types. These were further supported by H3K9ac, H3K27ac, H3K4me1, H3K4me3, and DHS ChIP data in muscle cells (Figures 2A, B). TFBIND analysis showed that rs17069665 altered the binding affinity to transcription factors including ARNT, BHLHE40, MYC, MYOD, NKX2-5 and USF. Rs4946936 altered the binding affinity to CAP1, CTCF, MYB, RELA and SETDB1. However, ENCODE ChIP-seq analysis showed that only MYC binds to DNA motif overlapping rs17069665 (Figure 3A). MYC has a higher preference for the non-risk allele A than risk allele G (Figure 3B). As for rs4946936, only CTCF and RELA bind to DNA motif overlapping it (Figure 3C). CTCF and RELA have higher preferences for the non-risk allele C than risk allele T (Figure 3D). The results revealed that rs17069665 and rs4946936 might influence the transcription of FOXO3 via altering the bindings to MYC, CTCF and/or RELA.
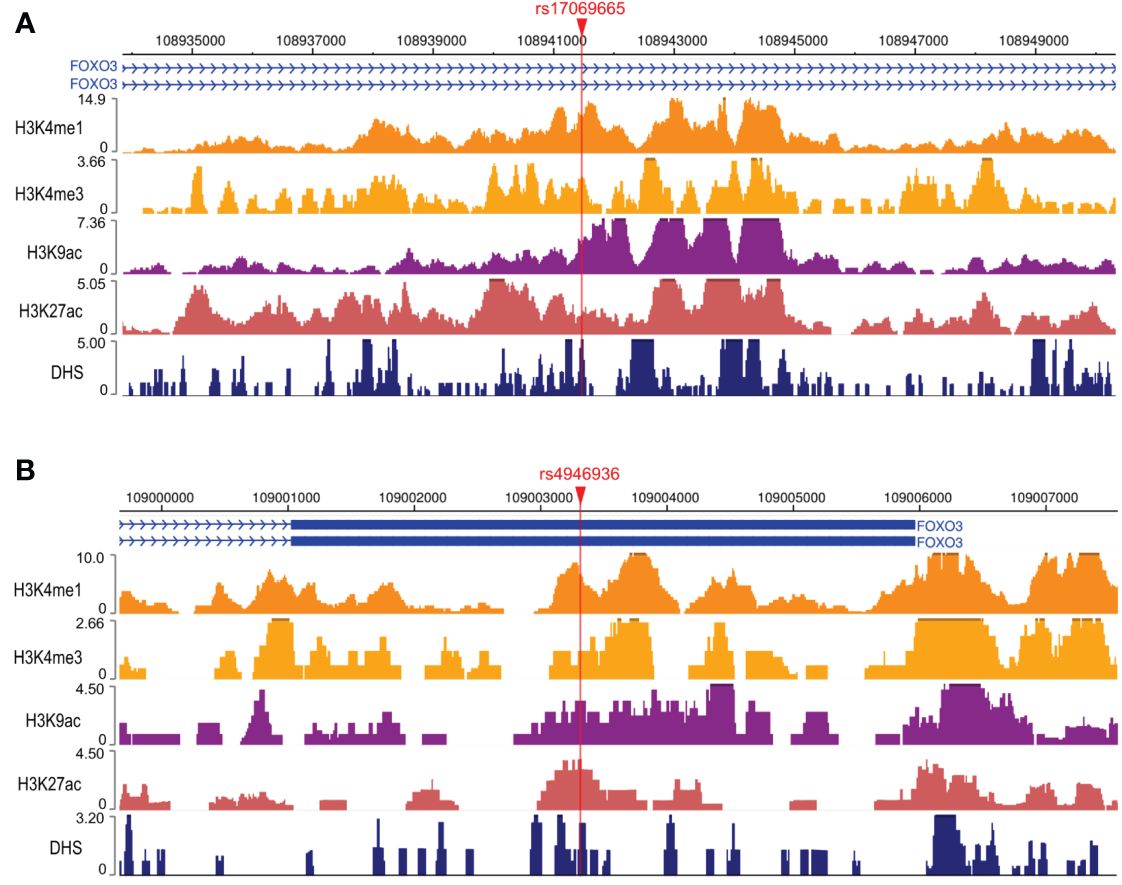
Figure 2 Rs17069665 and rs4946936 both overlap promoter and enhancer of FOXO3 gene. H3K4me1, H3K4me3, H3K9ac, H3K27ac, and DHS ChIP-seq signals at rs17069665 (A) and rs4946936 (B) loci.
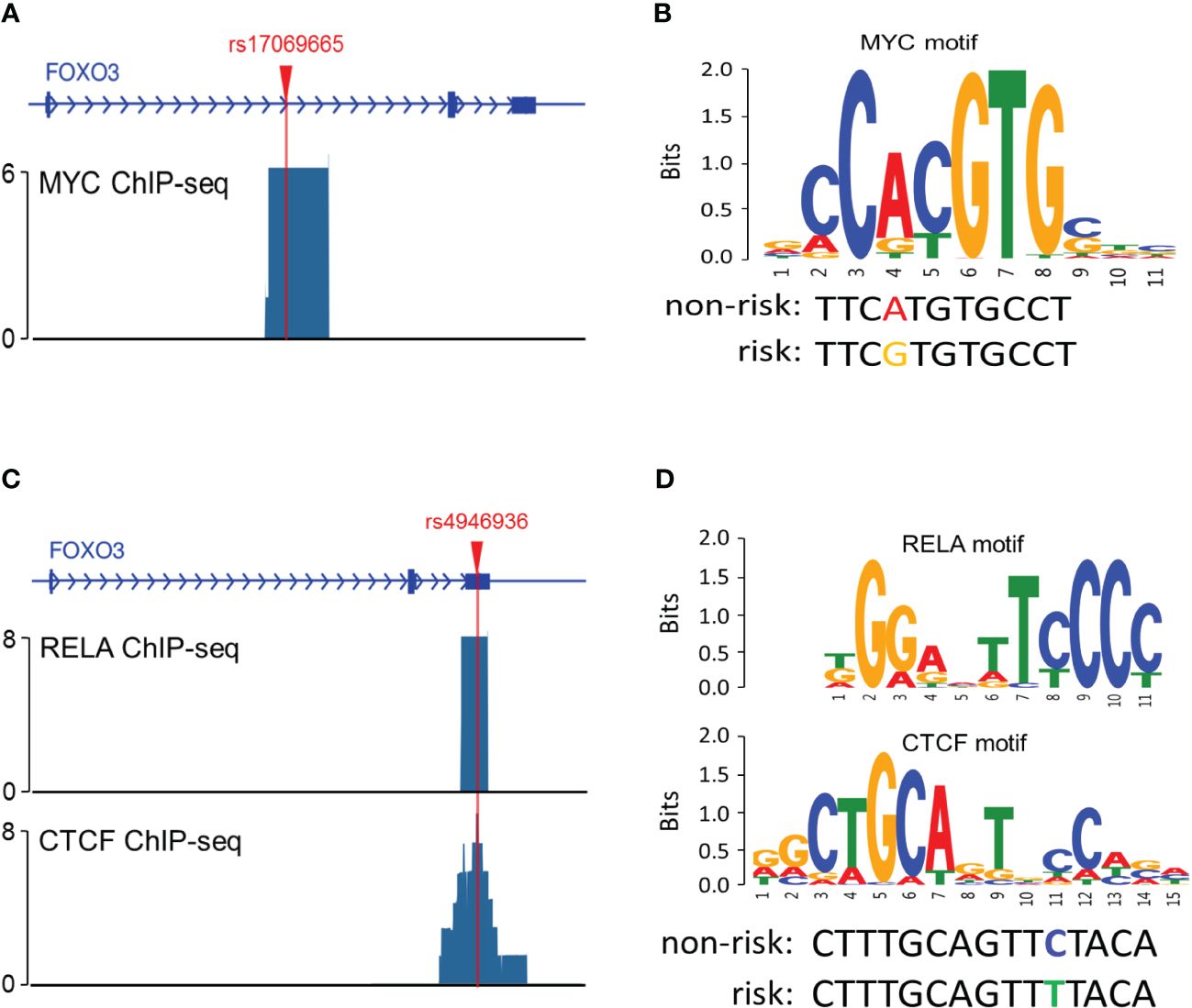
Figure 3 Rs17069665 and rs4946936 modulated the binding to MYC, RELA and/or CTCF. (A) MYC ChIP-seq signal at the rs17069665 locus; (B) Predicted preferential binding of MYC to the non-risk allele A of rs17069665; (C) RELA and CTCF ChIP-seq signals at the rs4946936 locus; (D) Predicted preferential bindings of RELA and CTCF to the non-risk allele C of rs4946936.
Discussion
In this case-control study with 110 RMS cases and 359 healthy controls, we evaluated the potential association of FOXO3 gene polymorphisms with RMS risk and prognosis in Chinese children. Among these four SNPs of FOXO3 in this study, we found that rs17069665 (GG vs. AA+AG) was significantly associated with increased RMS risk and poor prognosis, rs4946936 (TC+CC vs. TT) was significantly associated with decreased RMS risk and good prognosis. To our knowledge, the current study is the first one exploring the association between FOXO3 polymorphisms and RMS.
As a member of FOXO transcription factor family, FOXO3, as well as FOXO1, -4 and -6, controls transcription of target genes by binding to the Forkhead Response Element (33–35). Compared with the other three FOXO members, FOXO3 seemed to be more predominant in controlling cancer progression, although they potentially regulate the same target genes (36, 37). FOXO3 has been demonstrated to regulate various genes playing key roles in multiple cellular process including proliferation, apoptosis, drug resistance and stem cell properties (38–42). For instance, FOXO3 impaired the cancer stem cell phenotype of squamous cell carcinoma by controlling the transcriptional activity of SOX2 (11). The activation of FOXO3 sensitized tumors to anti-PD-1 therapy by inhibiting c-Myc and STAT3 (43). FOXO3 inhibited cell proliferation and induced apoptosis in colorectal cancer by regulating BIM expression (44). Certainly, FOXO3 is also regulated at multiple levels including genetic regulation and epigenetic modification (transcription, post-transcription, translation and post-translation) (40). FOXO3 protein was phosphorylated by proteins including ERK and AKT, which resulted in its inactivation and degradation (37, 45). FOXO3 gene polymorphisms were also investigated in multiple cancers, such as leukemia, pancreatic cancer, and hepatocellular carcinoma (16, 17, 46). However, FOXO3 polymorphisms have never been reported in RMS. It is urgent to conduct relevant investigations.
In the present study, we explored the association of four FOXO3 SNP sites (rs17069665, rs4946936, rs4945816 and rs9400241) with RMS. Rs17069665 was previously reported to increase ALL susceptibility (17), but had not been reported in other cancers. Rs4946936 was found to be associated with ALL (47), thyroid cancer (48), and head and neck cancer (49), but not with hepatocellular carcinoma (46). In this study, we found that rs17069665 and rs4946936 polymorphisms were associated with the increased and decreased RMS risk, respectively, and influence prognosis, for the first time. Rs17069665 is located in the intron one of FOXO3 gene, which overlaps with the FOXO3 promoter and enhancer. In the analyses of transcription factor binding, rs17069665 was found to disrupt the binding to MYC showing preferential binding of the non-risk allele A. Rs4946936, which is located in the 3’-UTR of FOXO3, was also found to overlap with the promoter and enhancer regions and to alter the binding to CTCF and RELA showing preferential binding of the non-risk allele C. SNP-gene expression analysis indicated that these two polymorphisms influenced the expression of FOXO3. Considering the tumor suppressing function of FOXO3, rs17069665 and rs4946936 might influence RMS risk and prognosis via regulating the expression of FOXO3 by altering the bindings to MYC, CTCF and/or RELA. The above potential mechanisms need to be validated in future studies.
As for the rest two SNPs (rs4945816 and rs9400241), both of which are located in the 3’UTR of FOXO3, no association between them and RMS risk was found. Several previous studies investigated the relationships between them and cancer risks. Rs4945816 was reported in studies on ALL and thyroid cancer, but no association was found (17, 48). Rs9400241 was associated with ALL and melanoma risks (17, 50). Along with the previous studies, our study indicates that the genetic variation of FOXO3 is complex, depending on cancer types. Besides, the variety in ethnicity and sample composition need to be taken into consideration.
Although this study is the first to investigate the association of FOXO3 polymorphisms with RMS risk and prognosis, several limitations should be considered. First, all participates were recruited from one hospital in south China, which may cause selection bias. Second, only 4 SNPs were genotyped in the present study and more potentially functional SNPs should be done in the future. Third, the sample size in this study was still not large enough because of the low incidence of RMS in China. Therefore, larger multicenter studies are warranted to further confirm the roles of FOXO3 in RMS. Finally, other factors including environment exposure and dietary intake were not available in this study. The functions of FOXO3 polymorphisms in the progression of RMS also need to be further explored.
Conclusion
In conclusion, the current study explored the association of FOXO3 polymorphisms (rs17069665, rs4946936, rs4945816 and rs9400241) with RMS in Chinese children and firstly demonstrated that rs17069665 was associated with increased RMS susceptibility and poor prognosis, while rs4946936 was associated with decreased RMS susceptibility and good prognosis, and that the two polymorphisms might influence the transcription of FOXO3 via altering the binding to MYC, CTCF and/or RELA. This study indicated that FOXO3 polymorphism might serve as a biomarker for RMS susceptibility and prognosis. Certainly, larger multicenter studies, as well as functional experiments, are encouraged to further elucidate the role of FOXO3 polymorphism and the underlying mechanisms in RMS.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Institutional Review Boards of Guangzhou Women and Children’s Medical Center. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
XZ: Conceptualization, Writing – original draft, Writing – review & editing, Methodology, Data curation, Formal analysis, Validation. YS: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Methodology, Funding acquisition. HN: Writing – original draft, Writing – review & editing, Investigation. PT: Writing – original draft, Writing – review & editing, Investigation. SL: Investigation, Writing – original draft, Writing – review & editing. XPL: Investigation, Writing – original draft, Writing – review & editing. XDL: Investigation, Writing – original draft, Writing – review & editing. AL: Investigation, Writing – original draft, Writing – review & editing. MC: Investigation, Writing – original draft, Writing – review & editing. YY: Investigation, Writing – original draft, Writing – review & editing. LX: Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Supervision. XY: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing, Methodology, Project administration.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from National Natural Science Foundation of China (82101959, 81870115, and 32001043), Guangdong Basic and Applied Basic Research Foundation (2020A1515110886), and Guangzhou Municipal Science and Technology Project (202102020344).
Acknowledgments
The authors thank all the patients and parents for their contribution and acknowledge the ENCODE Consortium and the ENCODE production laboratories generating the particular datasets.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1387735/full#supplementary-material
Supplementary Figure 1 | Functional relevance of rs3738067 A>G (A) and rs4946936 T > C (B) to FOXO3 expression in GTEx database.
Supplementary Figure 2 | Schematic of rs17069665 (A) and rs4946936 (B) regions with histone and DHS mark annotations in different tissue types in the Roadmap epigenomics data.
References
1. Gasparini P, Ferrari A, Casanova M, Limido F, Massimino M, Sozzi G, et al. MiRNAs as players in rhabdomyosarcoma development. Int J Mol Sci. (2019) 20:5818. doi: 10.3390/ijms20225818
2. Kashi VP, Hatley ME, Galindo RL. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nat Rev Cancer. (2015) 15:426–39. doi: 10.1038/nrc3961
3. Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975-2005. Cancer-Am Cancer Soc. (2009) 115:4218–26. doi: 10.1002/cncr.24465
4. Xu N, Hua Z, Ba G, Zhang S, Liu Z, Thiele CJ, et al. The anti-tumor growth effect of a novel agent DMAMCL in rhabdomyosarcoma in vitro and in vivo. J Exp Clin Canc Res. (2019) 38:118. doi: 10.1186/s13046-019-1107-1
5. McCarthy N, Dolgikh N, Logue S, Patterson JB, Zeng Q, Gorman AM, et al. The IRE1 and PERK arms of the unfolded protein response promote survival of rhabdomyosarcoma cells. Cancer Lett. (2020) 490:76–88. doi: 10.1016/j.canlet.2020.07.009
6. Labib RM A, Abdelrahim ME, Elnadi E, RM H, Yassin D. CYP2B6rs2279343 is associated with improved survival of pediatric rhabdomyosarcoma treated with cyclophosphamide. PloS One. (2016) 11:e158890. doi: 10.1371/journal.pone.0158890
7. Nishimura R, Takita J, Sato Otsubo A, Kato M, Koh K, Hanada R, et al. Characterization of genetic lesions in rhabdomyosarcoma using a high-density single nucleotide polymorphism array. Cancer Sci. (2013) 104:856–64. doi: 10.1111/cas.12173
8. Dong Z, Zhong X, Lei Q, Chen F, Cui H. Transcriptional activation of SIRT6 via FKHRL1/FOXO3a inhibits the Warburg effect in glioblastoma cells. Cell Signal. (2019) 60:100–13. doi: 10.1016/j.cellsig.2019.04.009
9. Guo X, Li Z, Zhu X, Zhan M, Wu C, Ding X, et al. A coherent FOXO3-SNAI2 feed-forward loop in autophagy. Proc Natl Acad Sci. (2022) 119:e2118285119. doi: 10.1073/pnas.2118285119
10. Kim CG, Lee H, Gupta N, Ramachandran S, Kaushik I, Srivastava S, et al. Role of Forkhead Box Class O proteins in cancer progression and metastasis. Semin Cancer Biol. (2018) 50:142–51. doi: 10.1016/j.semcancer.2017.07.007
11. Chen Y, Zhao H, Liang W, Jiang E, Zhou X, Shao Z, et al. Autophagy regulates the cancer stem cell phenotype of head and neck squamous cell carcinoma through the noncanonical FOXO3/SOX2 axis. Oncogene. (2022) 41:634–46. doi: 10.1038/s41388-021-02115-7
12. Coomans De Brachène A, Demoulin J. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. (2016) 73:1159–72. doi: 10.1007/s00018-015-2112-y
13. Panagopoulos I, Gorunova L, Andersen K, Lund-Iversen M, Tafjord S, Micci F, et al. Fusion of the paired box 3 (PAX3) and myocardin (MYOCD) genes in pediatric rhabdomyosarcoma. Cancer Genomics - Proteomics. (2021) 18:723–34. doi: 10.21873/cgp.20293
14. Yao J, Wang J, Xu Y, Guo Q, Sun Y, Liu J, et al. CDK9 inhibition blocks the initiation of PINK1-PRKN-mediated mitophagy by regulating the SIRT1-FOXO3-BNIP3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma. Autophagy. (2021) 18:1879–97. doi: 10.1080/15548627.2021.2007027
15. Miyamoto Y, Schirripa M, Suenaga M, Cao S, Zhang W, Okazaki S, et al. A polymorphism in the cachexia-associated gene INHBA predicts efficacy of regorafenib in patients with refractory metastatic colorectal cancer. PloS One. (2020) 15:e239439. doi: 10.1371/journal.pone.0239439
16. Yan H, Li Q, Wu J, Hu W, Jiang J, Shi L, et al. MiR-629 promotes human pancreatic cancer progression by targeting FOXO3. Cell Death Dis. (2017) 8:e3154. doi: 10.1038/cddis.2017.525
17. Yang X, Wu X, Fang N, Liu X, Liu X, Yang L, et al. FOXO3 gene polymorphisms influence the risk of acute lymphoblastic leukemia in Chinese children. J Cell Biochem. (2020) 121:2019–26. doi: 10.1002/jcb.29436
18. Uroog L, Rahmani AH, Alsahli MA, Almatroodi SA, Wani RA, Rizvi M. Genetic profile of FOXO3 Single Nucleotide Polymorphism in colorectal cancer patients. Oncology. (2023) 102:299–309. doi: 10.1159/000533729
19. Lu Y, Zhu Y, Wang X, Wang F, Peng J, Hou H, et al. FOXO3 rs12212067: T > G association with active tuberculosis in han chinese population. Inflammation. (2016) 39:10–5. doi: 10.1007/s10753-015-0217-y
20. Rakhshani Nejad A, Sargazi S, Ghasemi M, Samareh Moosavi S, Heidari Nia M, Saravani R. Association study to evaluate Foxo1 and Foxo3 gene polymorphisms in polycystic ovary syndrome: a preliminary case-control study and in silico analysis. Mol Biol Rep. (2023) 50:3569–80. doi: 10.1007/s11033-023-08292-w
21. Guo H, Li N, Sun Y, Wu C, Deng H, Xu L, et al. MYBL2 gene polymorphism is associated with acute lymphoblastic leukemia susceptibility in children. Front Oncol. (2021) 11:734588. doi: 10.3389/fonc.2021.734588
22. He J, Qiu L, Wang M, Hua R, Zhang R, Yu H, et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. (2012) 131:1235–44. doi: 10.1007/s00439-012-1152-8
23. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. (2009) 37:W600–5. doi: 10.1093/nar/gkp290
24. Lou J, Gong J, Ke J, Tian J, Zhang Y, Li J, et al. A functional polymorphism located at transcription factor binding sites, rs6695837 near LAMC1 gene, confers risk of colorectal cancer in Chinese populations. Carcinogenesis. (2017) 38:w204. doi: 10.1093/carcin/bgw204
25. Yang X, He J, Chang Y, Luo A, Luo A, Zhang J, et al. HOTAIR gene polymorphisms contribute to increased neuroblastoma susceptibility in Chinese children. Cancer-Am Cancer Soc. (2018) 124:2599–606. doi: 10.1002/cncr.31353
26. Consortium TG. The genotype-tissue expression (GTEx) project. Nat Genet. (2013) 45:580–5. doi: 10.1038/ng.2653
27. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. (2015) 518:317–30. doi: 10.1038/nature14248
28. Zhou X, Li D, Zhang B, Lowdon RF, Rockweiler NB, Sears RL, et al. Epigenomic annotation of genetic variants using the Roadmap Epigenome Browser. Nat Biotechnol. (2015) 33:345–6. doi: 10.1038/nbt.3158
29. Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. (2012) 489:57–74. doi: 10.1038/nature11247
30. Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. (2018) 46:D794–801. doi: 10.1093/nar/gkx1081
31. Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. (1999) 15:622–30. doi: 10.1093/bioinformatics/15.7.622
32. Wacholder S, Chanock S, Garcia-Closas M, El GL, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. (2004) 96:434–42. doi: 10.1093/jnci/djh075
33. Chiacchiera F, Simone C. The AMPK-FoxO3A axis as a target for cancer treatment. Cell Cycle. (2010) 9:1091–6. doi: 10.4161/cc.9.6.11035
34. Jiramongkol Y, Lam EW. FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev. (2020) 39:681–709. doi: 10.1007/s10555-020-09883-w
35. Yusuf D, Butland SL, Swanson MI, Bolotin E, Ticoll A, Cheung WA, et al. The transcription factor encyclopedia. Genome Biol. (2012) 13:R24. doi: 10.1186/gb-2012-13-3-r24
36. Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. (2013) 13:482–95. doi: 10.1038/nrc3539
37. Wu W, Lin J, Pan C, Chan T, Liu C, Wu W, et al. Amplification-driven BCL6-suppressed cytostasis is mediated by transrepression of FOXO3 and post-translational modifications of FOXO3 in urinary bladder urothelial carcinoma. Theranostics. (2020) 10:707–24. doi: 10.7150/thno.39018
38. Lee HK, Cha HS, Nam MJ, Park K, Yang Y, Lee J, et al. Broussochalcone A induces apoptosis in human renal cancer cells via ROS level elevation and activation of FOXO3 signaling pathway. Oxid Med Cell Longev. (2021) 2021:1–17. doi: 10.1155/2021/2800706
39. Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y, et al. Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differentiat. (2020) 27:966–83. doi: 10.1038/s41418-019-0389-3
40. Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X, et al. Critical role of FOXO3a in carcinogenesis. Mol Cancer. (2018) 17:104. doi: 10.1186/s12943-018-0856-3
41. Luo L, Zhang Z, Qiu N, Ling L, Jia X, Song Y, et al. Disruption of FOXO3a-miRNA feedback inhibition of IGF2/IGF-1R/IRS1 signaling confers Herceptin resistance in HER2-positive breast cancer. Nat Commun. (2021) 12:2699. doi: 10.1038/s41467-021-23052-9
42. Qin Y, Sun W, Wang Z, Dong W, He L, Zhang T, et al. RBM47/SNHG5/FOXO3 axis activates autophagy and inhibits cell proliferation in papillary thyroid carcinoma. Cell Death Dis. (2022) 13:270. doi: 10.1038/s41419-022-04728-6
43. Chung YM, Khan PP, Wang H, Tsai W, Qiao Y, Yu B, et al. Sensitizing tumors to anti-PD-1 therapy by promoting NK and CD8+ T cells via pharmacological activation of FOXO3. J Immunother Cancer. (2021) 9:e2772. doi: 10.1136/jitc-2021-002772
44. Ju H, Tan JY, Cao B, Song MQ, Tian ZB. Effects of miR-223 on colorectal cancer cell proliferation and apoptosis through regulating FoxO3a/BIM. Eur Rev Med Pharmacol Sci. (2018) 22:3771–8. doi: 10.26355/eurrev_201806_15259
45. Yang J, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. (2008) 10:138–48. doi: 10.1038/ncb1676
46. Tan C, Liu S, Tan S, Zeng X, Yu H, Li A, et al. Polymorphisms in microRNA target sites of forkhead box O genes are associated with hepatocellular carcinoma. PloS One. (2015) 10:e119210. doi: 10.1371/journal.pone.0119210
47. Wang Y, Zhou L, Chen J, Li J, He L, Wu P, et al. Association of the 3’UTR FOXO3a polymorphism rs4946936 with an increased risk of childhood acute lymphoblastic leukemia in a Chinese population. Cell Physiol Biochem. (2014) 34:325–32. doi: 10.1159/000363002
48. Röhlen N, Döring C, Hansmann ML, Grünwald F, Vorländer C, Bechstein WO, et al. FOXO3a Polymorphism rs4946936 and its interaction with vitamin D in differentiated thyroid cancer. Exp Clin Endocrinol Diabetes. (2014) 122:P84. doi: 10.1055/s-0034-1372101
49. Ahmed MW, Mahjabeen I, Gul S, Khursheed A, Mehmood A, Kayani MA. Relationship of single nucleotide polymorphisms and haplotype interaction of mitochondrial unfolded protein response pathway genes with head and neck cancer. Future Oncol. (2019) 15:3819–29. doi: 10.2217/fon-2019-0365
Keywords: FOXO3, single nucleotide polymorphisms (SNPs), rhabdomyosarcoma (RMS), susceptibility, prognosis
Citation: Zhang X, Sun Y, Niu H, Tan P, Liu S, Liu X, Liu X, Luo A, Cai M, Yan Y, Xu L and Yang X (2024) FOXO3 polymorphisms influence the risk and prognosis of rhabdomyosarcoma in children. Front. Oncol. 14:1387735. doi: 10.3389/fonc.2024.1387735
Received: 18 February 2024; Accepted: 11 April 2024;
Published: 24 April 2024.
Edited by:
Hong Duan, Sichuan University, ChinaCopyright © 2024 Zhang, Sun, Niu, Tan, Liu, Liu, Liu, Luo, Cai, Yan, Xu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Yang, wwwyangxu@163.com; Ling Xu, luoxul64@126.com
†These authors have contributed equally to this work
 Xiaohong Zhang1†
Xiaohong Zhang1† Xiaoping Liu
Xiaoping Liu Ling Xu
Ling Xu Xu Yang
Xu Yang