- 1Guangdong and Shenzhen Key Laboratory of Reproductive Medicine and Genetics, Department of Urology, Peking University Shenzhen Hospital, Shenzhen, China
- 2Shantou University Medical College, Shantou, Guangdong, China
- 3Shenzhen University Medical College, Shenzhen, Guangdong, China
Background: Renal cell carcinoma (RCC) stands as the most prevalent form of urogenital cancer. However, there is currently no universally accepted method for predicting the prognosis of RCC. MiRNA holds great potential as a prognostic biomarker for RCC.
Methods: A total of 100 cases with complete paraffin specimens and over 5-year follow-up data meeting the requirements were collected. Utilizing the clinical information and follow-up data of the specimens, an information model was developed. The expression levels of eight microRNAs were identified using RT-qPCR. Finally, determine and analyze the clinical application value of these microRNAs as prognostic markers for RCC.
Results: Significant differences were observed in the expression of two types of miRNAs (miR-378a-5p, miR-23a-5p) in RCC tissue, and three types of miRNAs (miR-378a-5p, miR-642a-5p, miR-23a-5p) were found to be linked to the prognosis of RCC. Establish biomarker combinations of miR-378a-5p, miR-642a-5p, and miR-23a-5p to evaluate RCC prognosis.
Conclusion: The combination of three microRNA groups (miR-378a-5p, miR-642a-5p, and miR-23a-5p) identified in paraffin section specimens of RCC in this study holds significant potential as biomarkers for assessing RCC prognosis.
Introduction
In 2020, there were approximately 431288 newly diagnosed instances of kidney cancer (KC) worldwide, with the death toll reaching 179368 (1). Histologically, kidney cancer (KC) cases are overwhelmingly attributed to RCC, which accounts for 90% of all cases (2).. Arising from the nephron, renal cell carcinomas (RCC) form a diverse collection of malignant neoplasms with varying characteristics (3).. Early-stage cancer typically does not present with obvious symptoms, resulting in over 60% of patients being incidentally detected during routine ultrasound examinations (4, 5). Unfortunately, many patients are already in the advanced stage by the time they receive their initial diagnosis. Even after undergoing radical nephrectomy, about 30% of patients may still experience tumor recurrence (6). The survival rate is significantly influenced by the stage at which the diagnosis occurs. Localized disease, representing stage I, carries a 5-year relative survival rate of 93%. When the disease progresses to regional involvement (stage II/III), with local lymph nodes affected, the 5-year relative survival rate drops to 72.5%. However, when the disease reaches the metastatic stage (stage IV), the 5-year relative survival rate plummets to a mere 12% (7).. The earlier the prognosis of a patient is determined and treatment is initiated, the better the treatment outcome is expected to be. As a result, regular comprehensive CT scans are scheduled for patients to promptly detect tumor recurrence or metastasis to other areas following surgery, facilitating timely and effective treatment (8). However, frequent CT scans expose the patient’s body to long-term radiation imaging and impose burdens of cost and time loss (9, 10). Currently, there is no universally accepted standard in clinical practice for accurately evaluating the prognosis of renal cancer patients. Most evaluations rely on pathological staging and classification, which do not fully meet clinical needs (11, 12). Consequently, there is a requirement for a biomarker in clinical practice that can precisely forecast the prognosis of RCC. This would enable precise treatment tailored to individual prognoses and reduce the physical and financial burdens experienced by patients.
Ranging from 19 to 25 nucleotides (nt) in length, microRNAs (miRNAs) are noncoding RNA molecules with the ability to control gene expression at both transcriptional and translational levels (13, 14). They are powerful regulators of various cellular activities including cell growth, differentiation, development, and apoptosis (15). In cancer, miRNAs that are dysregulated are categorized into oncogenes (oncomiRs) or tumor suppressors. OncomiRs enhance and suppress their target tumor suppressor genes within cancer. Conversely, in malignant tumors, tumor suppressor miRNAs are reduced, leading to the increased expression of their target oncogenes. This suggests that inhibition or overexpression of miRNA may serve as a pathway for predicting tumor occurrence (16, 17). Compared to proteins and mRNA, miRNA exhibits stronger stability, ease of detection, reduced susceptibility to degradation, and cost-effectiveness (18). Furthermore, miRNA can be reliably extracted from cells, tissues, and body fluids (19). As a result, microRNAs in paraffin pathological sections have significant potential to serve as prognostic markers for renal cell carcinoma.
Materials and methods
Specimen collection
The 100 specimens were acquired from Peking University Shenzhen Hospital. The surgeries took place before 2015, and the follow-up data spans over 5 years. None of the participants underwent treatments such as chemotherapy or radiotherapy before their surgeries. All specimens were surgically removed and pathologically diagnosed as RCC, with a 2010 AJCC renal cancer TNM stage of ≥ II. Following resection, the specimens were promptly placed in RNA protective solution (Qiagen GmbH, Hilden, Germany), and then preserved in a refrigerator at -80°C. Permission for this subsequent information was secured from the Independent Ethics Committee of Peking University Shenzhen Hospital, and the supervision of the committee was adhered to during the subsequent procedures.
Clinicopathological parameters of RCC patients
By collecting the hospitalization history of patients, we were able to gather clinical information such as age, gender, date of birth, operation date, tumor size, tumor stage, and the treatment received. In addition, we categorized the histology and tumor stage of RCC according to medical and pathological perspectives.
Determination of miRNA expression
Adhering to the RNA extraction from paraffin protocol (miRNeasy FFPE Kit), the sample was deposited in a 1.5 ml centrifuge tube. Then, 160 μl of Deparaffinization Solution was added before the EP tube was incubated at 56°C in a water bath for 3 minutes. After reaching room temperature, 150 μl of Buffer PKD was introduced and blended via shaking, and then centrifuged at 4°C for 1 minute at 11,000g. Drawing from the lower segment of the EP tube with a pipette gun, 10 μl of Proteinase K was dispensed and the tube was then incubated at 56°C for 15 minutes before immediate transfer to an 80°C water bath for 15 minutes. Thereafter, the EP tube would be stratified and the colorless fluid from the bottom section transferred to a new EP tube, to be chilled on ice for 3 minutes and then centrifuged at 20,000 g for 15 minutes at 4°C. The newly retrieved supernatant was then relocated to a fresh microcentrifuge tube. 15 μl of DNase Booster Buffer and 10 μl of DNase I stock solution were subsequently added, followed by a short period of centrifugation and a 15-minute room temperature incubation. An additional 320 μl of Proteinase K was then added and after another brief round of centrifugation and room temperature incubation, 320 μl of Buffer RBC and 1120 μl of Anhydrous Ethanol (100%) were mixed in by pipetting. This was then transferred to a 2 ml collection tube without centrifugation and was centrifuged for 15 seconds at ≥8000 x g (≥10000 rpm). The precipitate in the collection tube was discarded and the process was repeated, ensuring the entire sample passed through the collection column. 500 μl of Buffer RPE was then added and the tube was agitated before a 15-second centrifugation at ≥8000 x g (≥10000 rpm). The residue was discarded. An RNeasy MinElute Spin Column was then placed in a new 2 ml collection tube and centrifuged at maximum speed for 5 minutes. The liquid was discarded and the RNeasy MinElute Spin Column placed in a new 1.5 ml collection tube. 14-30 μl of RNase-free water was then directly added to the spin column membrane which was then washed with a full-speed centrifuge for one minute.
In the following steps, a mixture was prepared by combining 1 μg of total RNA, 4 μl of 5× Hispec buffer, 2 μl of 10× Nuclecs Mix, and 2 μl of RT, with the volume adjusted to 20 μl using RNase-free water. This mixture was subsequently placed into a real-time quantitative PCR system (LightCycler® 480 Fluorescent Quantitative PCR System, Roche Diagnostics, Basel, Switzerland).The real-time quantitative PCR procedure commenced with an initial denaturation at 95°C for 15 minutes, succeeded by 40 rounds of 94°C for 15 s, 55°C for 30 s, and 72°C for 30 s. The 2−△△Cq method was employed to ascertain the relative expression levels of the target miRNAs. The primer information is shown in Table 1.
Statistical analysis
For this study, SPSS 20.0 was utilized to analyze the data. We determined the optimal cutoff value by using the maximum Jordan index method based on the expression levels of different miRNAs. 2−△△Cq >1 indicates high expression, while 2−△△Cq method<1 indicates low expression. Model construction was performed for the collected clinical information of the patients. Independent samples t-test was conducted for the measurement data, and rank sum test was performed for the count data. Kaplan-Meier survival analysis was utilized for one-way analysis, with p < 0.05 denoting statistical significance.
Result
Expression levels of miRNA and clinicopathologic characteristics of patients
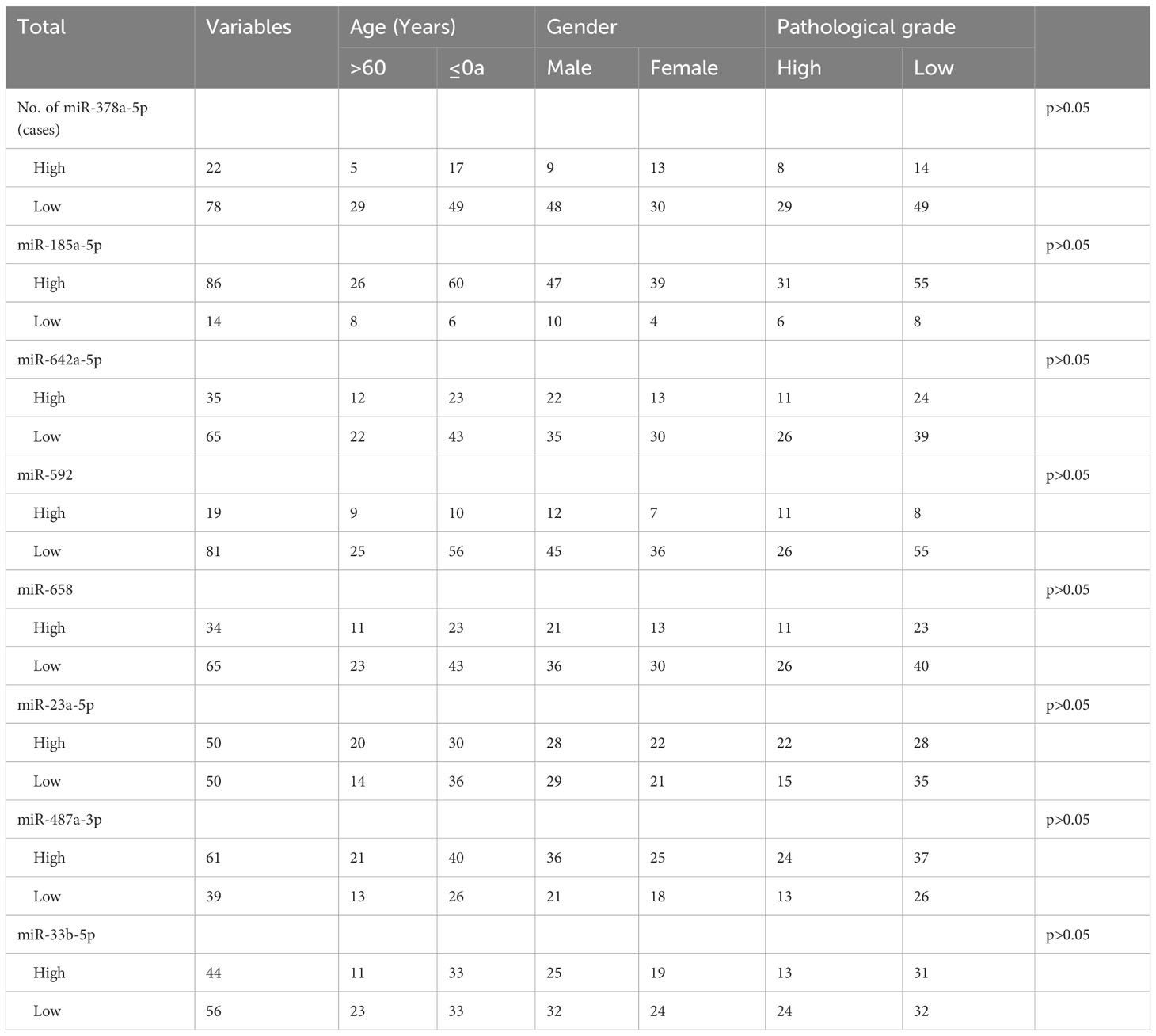
The relationship between the expression levels of different miRNAs and patients’ clinical characteristics was assessed using the chi-square test. As depicted in Table 2, the findings show no significant association between the miRNAs’ relative expression levels and the clinical features of the cases (p > 0.05).
Expression level of miRNA between survival and death
Based on the five-year follow-up survival status, 65 individuals were classified into the survival group, while 35 individuals were categorized into the death group. Figure 1 shows that in the survival group, the expression level of miR-378a-5p is markedly elevated (p<0.05), whereas the expression level of miR-23a-5p is significantly reduced (p<0.05).
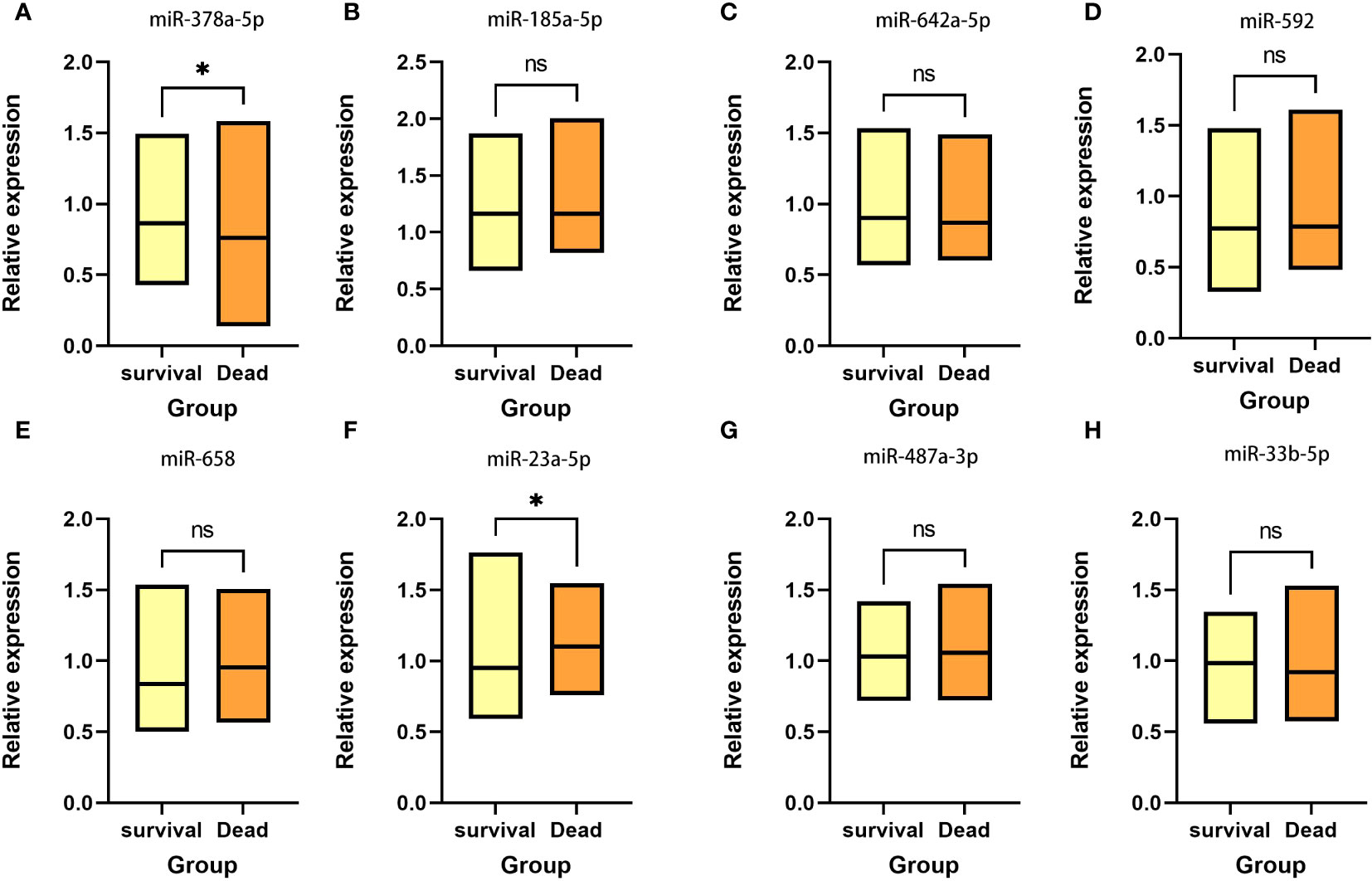
Figure 1 Expression between survival group and death group There is a significant elevation in the expression level of (A) miR-378a-5p within the survival group, whereas there is a considerable decrease in the expression level of (F) miR-23a-5p. No statistical significance was detected in (B) miR-185a-5p, (C) miR-642a-5p, (D) miR-592, (E) miR-658, (G) miR-487a-3p and (H) miR-33b-5p. *p < 0.05; ns, Non-significant.
Prognostic correlation analysis of miR-378a-5p, miR-642a-5p, and miR-23a-5p in RCC
Cox regression analysis indicated that higher expression levels of miR-378a-5p (Figure 2A, HR=3.063, 95% CI=1.076-8.721, p<0.05) and miR-642a-5p (Figure 2B, HR=0.341, 95% CI=0.142-0.822, p<0.05) correlate positively with the survival rates following surgery for RCC. In contrast, a negative correlation with postoperative survival was observed for the expression levels of miR-23a-5p (Figure 2C, HR=0.291, 95% CI=1.076-8.721, p<0.05). This suggests that increased levels of miR-378a-5p and miR-642a-5p are linked to longer overall survival, while higher expression levels of miR-23a-5p are correlated with shorter overall survival.
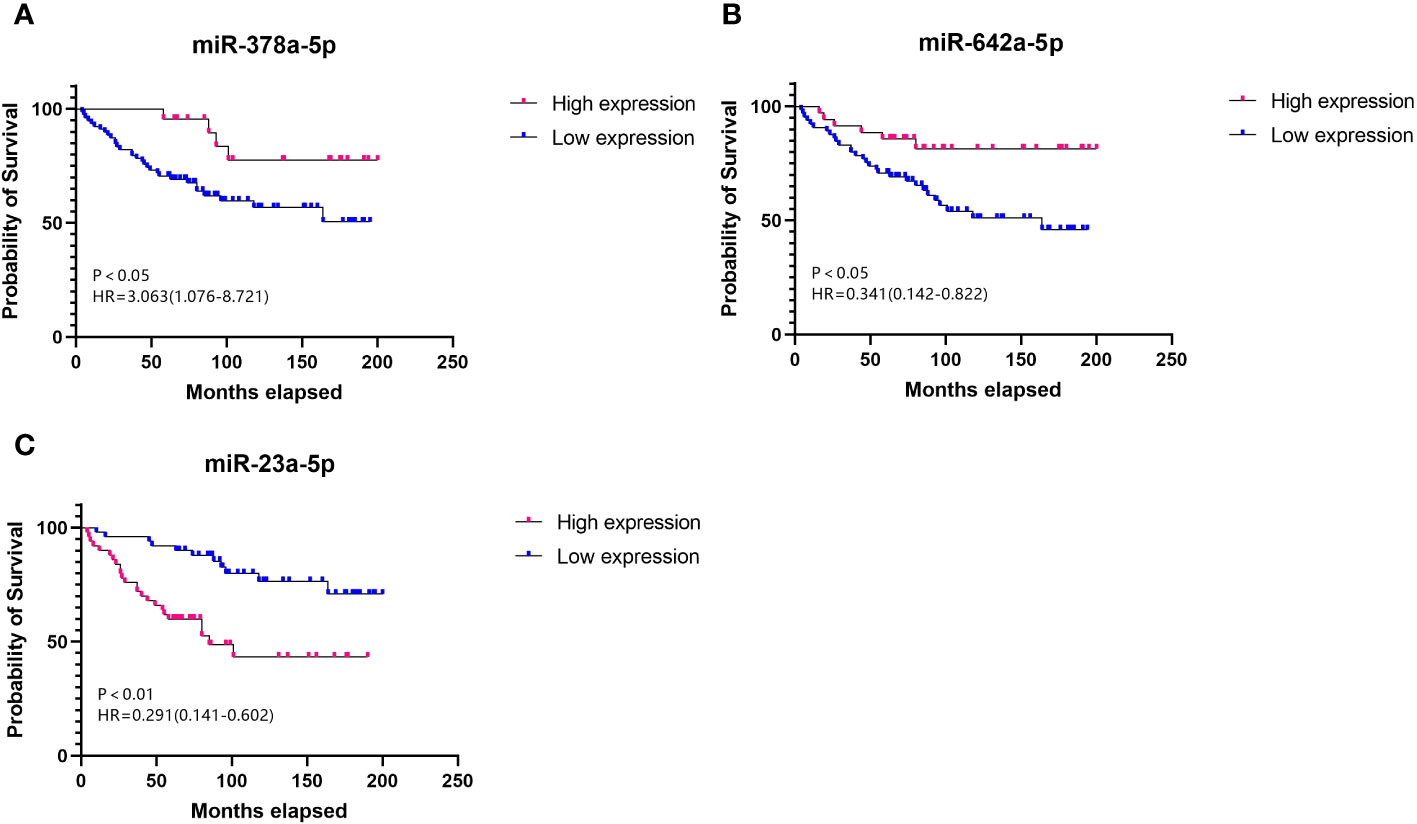
Figure 2 Survival curve Elevated levels of (A) miR-378a-5p and (B) miR-642a-5p, along with reduced levels of (C) miR-23a-5p, serve as prognostic protective factors for RCC.
A composite miRNA panel for the evaluation of RCC prognosis
We assessed the predictive potential of various miRNAs for renal cancer prognosis, recognizing that combining multiple miRNAs may yield superior predictive performance compared to using a single miRNA. The formula for the final logistic regression model established is as follows: logit (P) = 104.33 + 41.153 x miR-378a-5p + 43.598 x miR-642a-5p - 82.576 x miR-23a-5p. As depicted in Figure 3A, the established equation exhibits a high degree of fitting and is deemed reliable for usage. Utilizing a panel of miR-378a-5p, miR-642a-5p, and miR-23a-5p as biomarkers (p<0.05) may be an effective approach for predicting the prognosis of RCC. As depicted in Figure 3B, the AUC value of the combination of these three miRNAs is 0.712 (p<0.05), indicating moderate accuracy in prediction.
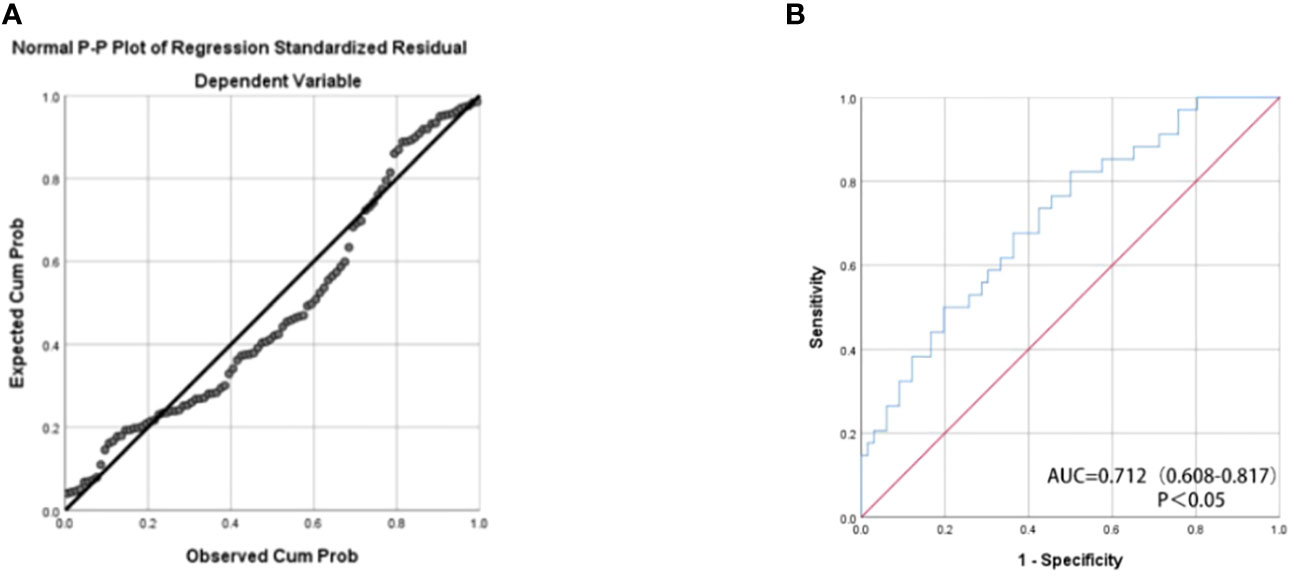
Figure 3 (A) Normal P-P Plot of Regression Standardized Residual The actual observed curve fits well with the predicted curve of the panel. (B) The receiver operating characteristic (ROC) curve analyses for the three miRNA panel (miR-378a-5p, miR-642a-5p, and miR-23a-5p).The combination of these three miRNAs has moderate predictive accuracy.
Discussion
Comprising 2-3% of all malignant tumors in adults, renal cell carcinoma (RCC) stands as the most common type of urogenital carcinoma. It is more frequently diagnosed in men than in women. Around 33% of RCC patients develop metastases, and the mortality rate can reach as high as 30-40% (7, 20, 21). This clearly demonstrates that RCC poses a significant risk to human life and well-being. The survival rate of RCC patients over a 5-year period varies significantly depend on their stages and grades. Accurately determining the prognosis is a crucial step in evaluating renal cell carcinoma patients. It can facilitate the initiation of new adjuvant therapies, such as the targeted drug sunitinib, and predict the progression of the disease (22). Given the limited efficacy of traditional chemotherapy or immunotherapy, a better understanding of prognosis can help patients select the optimal timing and methods of treatment, thereby maximizing treatment effectiveness and reducing the likelihood of recurrence or metastasis (23). This, in turn, can reduce the physical and economic burden caused by regular follow-up examinations. Regrettably, there is currently no gold standard in clinical practice that can accurately assess the prognosis of RCC (24). Consequently, there is an urgent need for effective biomarkers that can evaluate the prognosis of RCC in clinical practice. MiRNA in RCC issue has tremendous potential and is expected to become a biomarker that fulfills clinical requirements for predicting the prognosis of RCC.
The experimental findings suggest that miR-378a-5p, miR-642a-5p, and miR-23a-5p are correlated with the prognosis of renal cell carcinoma. Nevertheless, there is no noteworthy association between the case’s clinical characteristics and the relative expression levels (p>0.05). Furthermore, the combination of these miRNAs exhibits improved predictive performance and holds potential as a prognostic biomarker for RCC patients. MiRNA is a potent regulatory factor and has been demonstrated to drive carcinogenic pathways (25). There is a wealth of evidence indicating that miR-378a-5p is commonly downregulated in colorectal cancer, resulting in the suppression of cell proliferation and increased resistance to apoptosis. It also downregulates CDK1 levels, thereby inhibiting the development of colorectal cancer (26). In addition, targeting the VEGF pathway, miR-378a-5p has been discovered to enhance the prognosis and hinder the progression of hepatocellular carcinoma (27).. In prostate cancer, miR-642a-5p functions as a suppressor of tumor growth, effectively diminishing the proliferation of cells associated with prostate cancer (28).. Increased expression of miR-642a-5p has demonstrated a suppressive effect on the migratory and invasive capabilities of colon cancer cells. Conversely, decreased levels of miR-642a-5p in individuals with colon cancer are correlated with an unfavorable prognosis (29).. Additionally, miR-23a-5p has been identified to suppress the growth and invasion of pancreatic ductal adenocarcinoma cells through the inhibition of ECM1 expression (30). It is also involved in the growth inhibition of liver cancer induced by andrographolide (31). These investigations have demonstrated that a solitary miRNA possesses the ability to modulate numerous target genes, thereby exerting influence over a broad spectrum of biological functions (32).. Notably, these investigations have shed light on the tumor-suppressive impacts of miR-378a-5p, miR-642a-5p, and miR-23a-5p, highlighting their significant correlation with tumor prognosis. They also suggest the significant potential of miR-378a-5p, miR-642a-5p, and miR-23a-5p as biomarkers for evaluating the prognosis of renal cell carcinoma.
Conclusions
In conclusion, the panel of miR-378a-5p, miR-642a-5p, and miR-23a-5p holds promise as a prognostic biomarker for renal cell carcinoma, addressing the pressing clinical needs. Although numerous studies have verified the suppressive effects of miR-378a-5p, miR-642a-5p, and miR-23a-5p on tumor growth and invasion, additional investigation is required to clarify their precise functions in the molecular pathways that contribute to the initiation and progression of renal cell carcinoma. Subsequent studies in this area can enhance their clinical value as prognostic markers for renal cell carcinoma.
Data availability statement
Datasets are available on request: The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Independent Ethics Committee of Peking University Shenzhen Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
WC: Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. WW: Writing – original draft, Writing – review & editing. ZZ: Data curation, Writing – review & editing. ZW: Data curation, Writing – review & editing. YLi: Data curation, Writing – review & editing. ZG: Data curation, Writing – review & editing. YLa: Conceptualization, Writing – review & editing. LN: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by Shenzhen High-level Hospital Construction Fund, Clinical Research Project of Peking University Shenzhen Hospital (LCYJ2017001, LCYJ2020002, LCYJ2020015, LCYJ2020020), Science and Technology Development Fund Project of Shenzhen (no. JCYJ20180507183102747), the Scientific Research Projects of Medical and Health Institutions of Longhua District, Shenzhen (Grant No.2021055).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol. (2016) 70:93–105. doi: 10.1016/j.eururo.2016.02.029
3. Bahadoram S, Davoodi M, Hassanzadeh S, Bahadoram M, Barahman M, Mafakher L. Renal cell carcinoma: an overview of the epidemiology, diagnosis, and treatment. G Ital Nefrol. (2022) 39(3):2022.
4. Petejova N, Martinek A. Renal cell carcinoma: Review of etiology, pathophysiology and risk factors. BioMed Pap Med Fac Univ Palacky Olomouc Czech Repub. (2016) 160:183–94. doi: 10.5507/bp.2015.050
5. Khanna A, Crane A, Yerram N, Sun D, Ericson K, Lundy SD, et al. Contemporary management of advanced renal cell carcinoma. Clin Adv Hematol Oncol. (2018) 16:438–46.
6. Klatte T, Rossi SH, Stewart GD. Prognostic factors and prognostic models for renal cell carcinoma: a literature review. World J Urol. (2018) 36:1943–52. doi: 10.1007/s00345-018-2309-4
7. Padala SA, Barsouk A, Thandra KC, Saginala K, Mohammed A, Vakiti A, et al. Epidemiology of renal cell carcinoma. World J Oncol. (2020) 11:79–87. doi: 10.14740/wjon1279
8. Ljungberg B, Albiges L, Abu-Ghanem Y, Bedke J, Capitanio U, Dabestani S, et al. European association of urology guidelines on renal cell carcinoma: the 2022 update. Eur Urol. (2022) 82:399–410. doi: 10.1016/j.eururo.2022.03.006
9. Ferrero A, Takahashi N, Vrtiska TJ, Krambeck AE, Lieske JC, McCollough CH. Understanding, justifying, and optimizing radiation exposure for CT imaging in nephrourology. Nat Rev Urol. (2019) 16:231–44. doi: 10.1038/s41585-019-0148-8
10. Albert JM. Radiation risk from CT: implications for cancer screening. AJR Am J Roentgenol. (2013) 201:W81–7. doi: 10.2214/AJR.12.9226
11. Gray RE, Harris GT. Renal cell carcinoma: diagnosis and management. Am Fam Physician. (2019) 99:179–84.
12. Lane BR, Kattan MW. Prognostic models and algorithms in renal cell carcinoma. Urol Clin North Am. (2008) 35:613–25; vii. doi: 10.1016/j.ucl.2008.07.003
13. Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. (2008) 14:1–6. doi: 10.1097/PPO.0b013e318164145e
14. Hussen BM, Hidayat HJ, Salihi A, Sabir DK, Taheri M, Ghafouri-Fard S. MicroRNA: A signature for cancer progression. BioMed Pharmacother. (2021) 138:111528. doi: 10.1016/j.biopha.2021.111528
15. Hashemi A, Gorji-Bahri G. MicroRNA: promising roles in cancer therapy. Curr Pharm Biotechnol. (2020) 21:1186–203. doi: 10.2174/1389201021666200420101613
16. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. (2019) 234:5451–65. doi: 10.1002/jcp.27486
17. Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. (2016) 98:12–23. doi: 10.1016/j.critrevonc.2015.10.003
18. Hashiguchi Y, Nishida N, Mimori K, Sudo T, Tanaka F, Shibata K, et al. Down-regulation of miR-125a-3p in human gastric cancer and its clinicopathological significance. Int J Oncol. (2012) 40:1477–82. doi: 10.3892/ijo.2012.1363
19. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. (2018) 141:1202–7. doi: 10.1016/j.jaci.2017.08.034
20. Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. (2009) 373:1119–32. doi: 10.1016/S0140-6736(09)60229-4
21. Flanigan RC, Campbell SC, Clark JI, Picken MM. Metastatic renal cell carcinoma. Curr Treat Options Oncol. (2003) 4:385–90. doi: 10.1007/s11864-003-0039-2
22. Le Saux O, Freyer G, Negrier S. First-line treatments for poor-prognosis metastatic renal cell carcinoma: experts' Prescribing practices and systematic literature review. Clin Drug Investig. (2016) 36:389–99. doi: 10.1007/s40261-016-0384-0
23. Mejean A, Oudard S, Thiounn N. Prognostic factors of renal cell carcinoma. J Urol. (2003) 169:821–7. doi: 10.1097/01.ju.0000051378.14270.2a
24. Suarez C, Campayo M, Bastus R, Castillo S, Etxanitz O, Guix M, et al. Prognostic and predictive factors for renal cell carcinoma. Target Oncol. (2018) 13:309–31. doi: 10.1007/s11523-018-0557-2
25. Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. (2021) 14(4):dmm047662. doi: 10.1242/dmm.047662
26. Li K, Zhang J, Zhang M, Wu Y, Lu X, Zhu Y. miR-378a-5p inhibits the proliferation of colorectal cancer cells by downregulating CDK1. World J Surg Oncol. (2021) 19:54. doi: 10.1186/s12957-021-02166-w
27. Zou H, Yang L. miR-378a-5p improved the prognosis and suppressed the progression of hepatocellular carcinoma by targeting the VEGF pathway. Transl Cancer Res. (2020) 9:1558–66. doi: 10.21037/tcr
28. Beveridge DJ, Richardson KL, Epis MR, Brown RAM, Stuart LM, Woo AJ, et al. The tumor suppressor miR-642a-5p targets Wilms Tumor 1 gene and cell-cycle progression in prostate cancer. Sci Rep. (2021) 11:18003. doi: 10.1038/s41598-021-97190-x
29. Wang X, Song Z, Hu B, Chen Z, Chen F, Cao C. MicroRNA−642a−5p inhibits colon cancer cell migration and invasion by targeting collagen type I alpha1. Oncol Rep. (2021) 45:933–44. doi: 10.3892/or.2020.7905
30. Huang W, Huang Y, Gu J, Zhang J, Yang J, Liu S, et al. miR-23a-5p inhibits cell proliferation and invasion in pancreatic ductal adenocarcinoma by suppressing ECM1 expression. Amfi 2 J Transl Res. (2019) 11:2983–94.
31. Lu B, Sheng Y, Zhang J, Zheng Z, Ji L. The altered microRNA profile in andrographolide-induced inhibition of hepatoma tumor growth. Gene. (2016) 588:124–33. doi: 10.1016/j.gene.2016.05.012
Keywords: biomarker, renal cell carcinoma, kidney cancer, cancer diagnosis, miRNA, paraffin tissue
Citation: Chen W, Wang W, Zhao Z, Wen Z, Li Y, Ge Z, Lai Y and Ni L (2024) A three miRNAs panel in paraffin tissue serves as tool for predicting prognosis of renal cell carcinoma. Front. Oncol. 14:1391844. doi: 10.3389/fonc.2024.1391844
Received: 26 February 2024; Accepted: 04 April 2024;
Published: 24 April 2024.
Edited by:
Hua Tan, National Human Genome Research Institute (NIH), United StatesReviewed by:
Yibo Li, Southern Methodist University, United StatesWencao Zhao, University of Pennsylvania, United States
Copyright © 2024 Chen, Wang, Zhao, Wen, Li, Ge, Lai and Ni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangchao Ni, lncord@163.com; Yongqing Lai, yqlord@163.com
†These authors have contributed equally to this work and share first authorship
 Wenkang Chen1,2†
Wenkang Chen1,2† Yongqing Lai
Yongqing Lai