- 1Dix Laboratory, Department of Drug Discovery & Biomedical Sciences, Medical University of South Carolina, Charleston, SC, United States
- 2Norris Laboratory, Department of Regenerative Medicine and Cell Biology, Medical University of South Carolina, Charleston, SC, United States
- 3College of Medicine, Medical University of South Carolina, Charleston, SC, United States
- 4College of Pharmacy, The Ohio State University, Columbus, OH, United States
- 5Pritzker School of Medicine, The University of Chicago, Chicago, IL, United States
The emergence of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; COVID-19) in China, reported to the World Health Organization on December 31, 2019, has led to a large global pandemic and is a major public health issue. As a result, there are more than 200 clinical trials of COVID-19 treatments or vaccines that are either ongoing or recruiting patients. One potential therapy that has garnered international attention is hydroxychloroquine; a potent immunomodulatory agent FDA-approved for the treatment of numerous inflammatory and autoimmune conditions, including malaria, lupus, and rheumatoid arthritis. Hydroxychloroquine has demonstrated promise in vitro and is currently under investigation in clinical trials for the treatment of COVID-19. Despite an abundance of empirical data, the mechanism(s) involved in the immunomodulatory activity of hydroxychloroquine have not been characterized. Using the unbiased chemical similarity ensemble approach (SEA), we identified C-C chemokine receptor type 4 (CCR4) as an immunomodulatory target of hydroxychloroquine. The crystal structure of CCR4 was selected for molecular docking studies using the SwissDock modeling software. In silico, hydroxychloroquine interacts with Thr-189 within the CCR4 active site, presumably blocking endogenous ligand binding. However, the CCR4 antagonists compound 18a and K777 outperformed hydroxychloroquine in silico, demonstrating energetically favorable binding characteristics. Hydroxychloroquine may subject COVID-19 patients to QT-prolongation, increasing the risk of sudden cardiac death. The FDA-approved CCR4 antagonist mogalizumab is not known to increase the risk of QT prolongation and may serve as a viable alternative to hydroxychloroquine. Results from this report introduce additional FDA-approved drugs that warrant investigation for therapeutic use in the treatment of COVID-19.
Introduction
The novel coronavirus SARS-CoV-2 (SARS-CoV-2; COVID-19) has emerged as an international outbreak of acute respiratory illness. The first case of COVID-19 was reported to the World Health Organization on December 31, 2019, although news reports have described the outbreak initiating as early as November 17, 2019 (Ma, 2020; Scher, 2020; Walker, 2020). While a majority of COVID-19 infections present as a mild disease, at risk patients may experience life-threatening symptoms, including fever, coughing, breathing difficulties, fatigue, myalgia, multi-organ failure, and sepsis. As of July 19, there has been a total of 3,698,161 confirmed cases of COVID-19 in the United States, with 139,659 reported deaths (Centers for Disease Control and Prevention, 2020). As a result, there are currently 242 clinical trials of COVID-19 treatments or vaccines that are either ongoing or recruiting patients (Live Science, 2020; RECOVERY, 2020). One treatment that has garnered international attention is hydroxychloroquine, a potent immunomodulatory agent FDA-approved for the treatment of malaria, lupus, and rheumatoid arthritis. Hydroxychloroquine has demonstrated promise in vitro and is currently being investigated in clinical trials for use as pre-exposure or post-exposure prophylaxis of COVID-19 infection, as well as in the treatment of patients with an active COVID-19 infection (World Health Organization, 2020). Results from a clinical trial hosted at Renmin Hospital of Wuhan University indicated that hydroxychloroquine treatment led to a reduction in time to clinical recovery (TTCR); reduced the body temperature recovery time, and shortened the duration of pneumonia relative to control subjects. In a small study commissioned by the French government, all patients taking the combination therapy hydroxychloroquine/azithromycin were virologically cured within 6 days of treatment (Gautret et al., 2020). Despite promising data in Europe and China, ongoing clinical trials in the United States have not yet demonstrated that hydroxychloroquine is safe and effective in the treatment of patients with COVID-19 (Food and Drug Administration, 2020).
Hydroxychloroquine was identified as a potential candidate for COVID-19 treatment due to its ability to efficiently interfere with the replication cycle of various parasites, fungi, bacteria, and viruses (Raoult et al., 1990; Raoult et al., 1999; Boulos et al., 2004; Rolain et al., 2007; Devaux et al., 2020). Preliminary data suggests that hydroxychloroquine related compounds impede COVID-19 infection in multiple ways: by increasing the endosomal pH required for virus-cell fusion and by interfering with the glycosylation of cellular receptors involved in SARS-CoV-2 spike protein cleavage-induced cell membrane fusion (Simmons et al., 2011; Chen et al., 2020; Meng et al., 2020; Wang et al., 2020). Additionally, chloroquine and hydroxychloroquine interfere with the proteolytic processing of M-protein and modify virion assembly and budding (Vincent et al., 2005; Wang et al., 2020). Notwithstanding promising data suggesting altered viral uptake, replication, and processing, the greatest benefit of hydroxychloroquine treatment may due to its potent immunomodulatory effects, given a majority of patients with severe COVID-19 infections succumb to cytokine release syndrome as a result of septic shock (Rainsfor et al., 2015; Liang et al., 2018; Ankrum, 2020; Moore and June, 2020). In addition to inhibiting normal lysosomal activity, it has been hypothesized that hydroxychloroquine modulates signaling pathways and the transcriptional regulation of genes responsible for governing cytokine production and regulating the activity of co-stimulatory effectors (Schrezenmeier and Dorner, 2020). However, despite being used in the treatment of numerous inflammatory and autoimmune conditions, the precise mechanism of action responsible for hydroxychloroquine induced immunomodulation remains unknown. In this study, we used an unbiased chemical similarity ensemble approach (SEA) to identify potential immunomodulatory targets of hydroxychloroquine.
Materials and Methods
A step-wise in silico analysis was performed in order to uncover novel immunomodulatory targets of hydroxychloroquine (Figure 1).
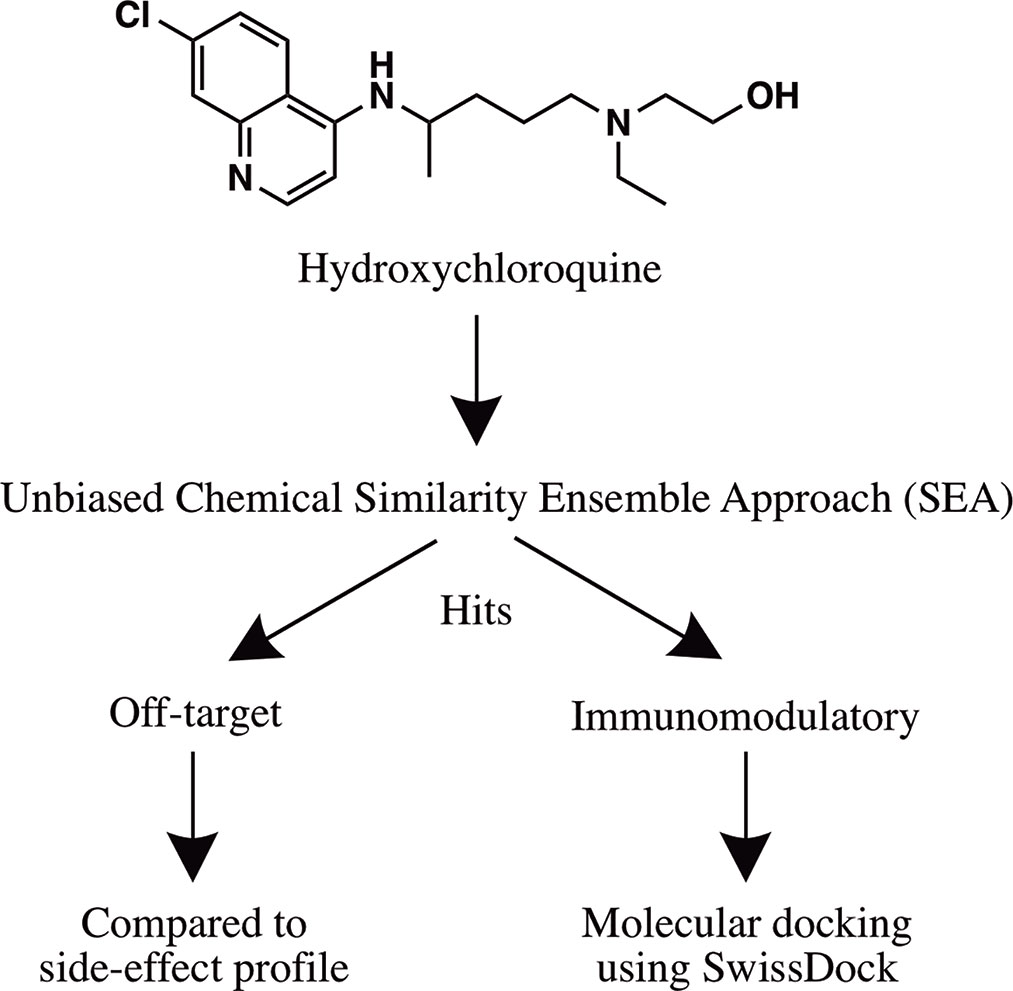
Figure 1 Flowchart summarizing the in silico approach employed to identify the immunomodulatory targets of hydroxychloroquine.
Similarity Ensemble Approach
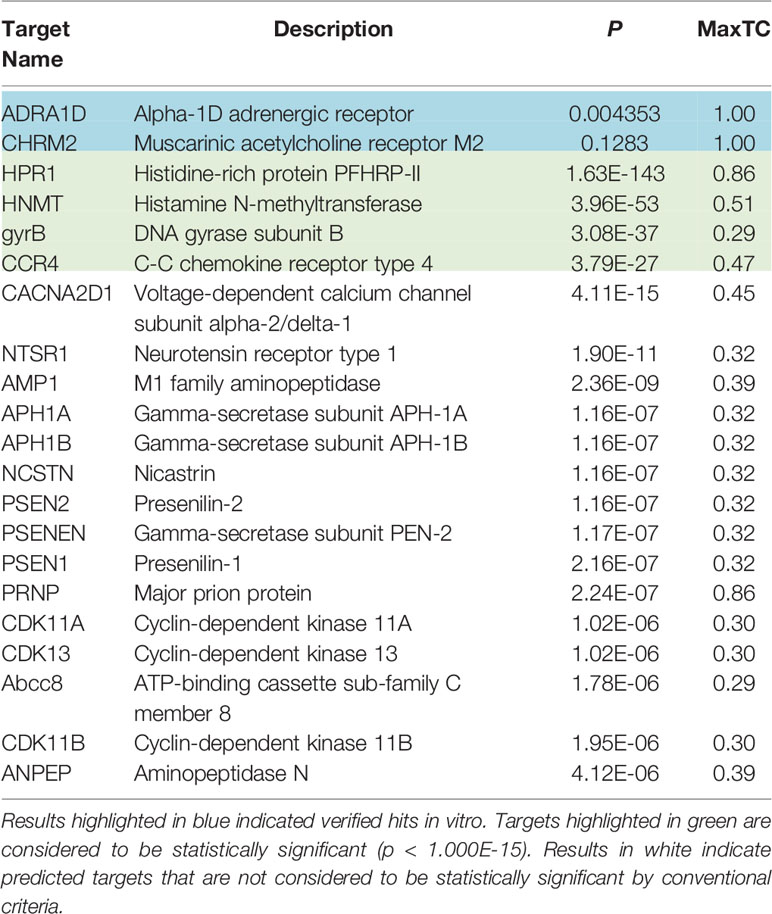
The average FDA-approved drug has eight targets, making in silico target prediction of compounds an integral component of the drug discovery process (Ma’ayan et al., 2008; Santos et al., 2016; Wang et al., 2016). The chemical similarity ensemble approach (SEA) relates protein targets based on the set-wise chemical similarity among their ligands (Keiser et al., 2007). Using the ZINC database of over 2.7 million compounds, SEA is capable of generating highly accurate and robust predictions useful in drug repurposing and mechanism-of-action target prediction. SEA utilizes an unbiased approach; a single uniform submission query is employed to generate a list of prospective drug targets. The SMILES code of hydroxychloroquine [CCN(CCCC(C)NC1=C2C=CC(=CC2=NC=C1)Cl)CCO] (Figure 1) was submitted to the SEA server for target prediction. Predicted targets with maximum Tanimoto coefficient (MaxTc) values and p-values were generated. The MaxTc is defined as the percent chemical similarity of the investigational ligand relative to the most similar ligand for the target of interest, with scores ranging from 0 to +1 (where +1 is the highest similarity). A p-value of less than 1.000e-15 was used as the cut-off for statistical significance as previously described (Keiser et al., 2007; Wang et al., 2016).
Molecular Docking
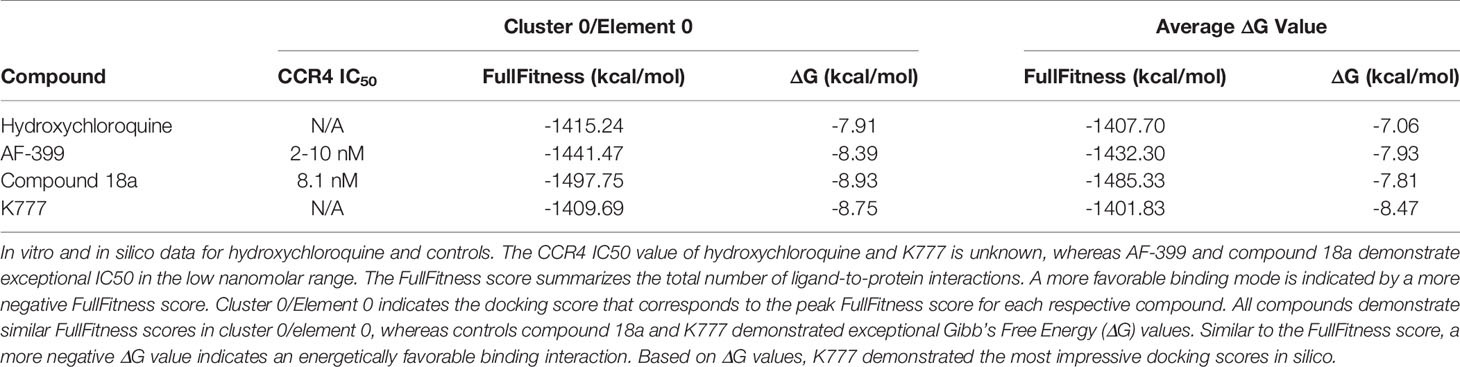
Molecular docking was performed using the SwissDock modeling software; a server dedicated to carrying out protein-ligand docking simulations hosted by the Swiss Bioinformatics Institute (Grosdidier et al., 2011; Bitencourt-Ferreira and Azevedo, 2019). The crystal structure of the chemokine receptor type 4 (CCR4) was selected for molecular docking assays (UniProt, 2020). Hydroxychloroquine was converted into a Mol.2 file using Marvin Sketch and submitted for molecular docking. Binding modes are scored using their FullFitness and clustered. Clusters are then ranked according to the average FullFitness of their elements with their respective Gibbs free energy (ΔG) values (Grosdidier et al., 2007). The ViewDock plug-in of UCSF Chimera was used to visualize predicted binding modes (Pettersen et al., 2004). Hydrogens were added in the ViewDock plug-in to optimize the predicted hydrogen bonding network and to allow UCSF Chimera to determine the protonation state (Lang, 2018). The known CCR4 antagonists AF-399, compound 18a, and K777 (Figure S1) were used as a positive control for comparison.
Results
Similarity Ensemble Approach
Results using the chemical Similarity Ensemble Approach (SEA) indicated six potential protein targets for hydroxychloroquine (Table 1). Two of the hits, including the alpha-1D adrenergic receptor (p = 0.004353) and muscarinic acetylcholine receptor M2 (p = 0.1283), did not possess significant p-values but have been verified in vitro. Four hits with significant p-values were discovered: Histidine-rich protein PFHRP-II (p = 1.63E-143), histamine N-methyltransferase (p=3.96E-53), DNA gyrase subunit B (3.08E-37), and C-C chemokine receptor type 4 (CCR4) (p = 3.79E-27). An additional 15 non-significant interactions were detected using SEA. Of the four hits with significant p-values, the target CCR4 presents as a target of interest due to its immunomodulatory function. CCR4 was selected for molecular docking assays using SwissDock and UCSF Chimera.
Molecular Docking
A total of 42, 35, 41, and 32 binding clusters were calculated for hydroxychloroquine, AF-399, compound 18a, K777, respectively. Clusters were then ranked according to the average FullFitness of their elements beginning with cluster 0/element 0 and their corresponding Gibbs free energy (ΔG) values. More negative ΔG values indicate favorable binding characteristics. At cluster 0/element 0, hydroxychloroquine demonstrated a ∆G of -7.91, whereas positive controls AF-399, compound 18a, and K777 had ∆G values of -8.39, -8.93, and -8.75, respectively (Table 2). The average ∆G of hydroxychloroquine was -7.06, whereas AF-399, compound 18a, and K777 had average ∆G values of -7.93, -7.81, and -8.47, respectively. UCSF Chimera was used to visualize the predicted binding conformations of hydroxychloroquine and controls. All drugs interact with the active site of CCR4 (Figure 2). All compounds formed a hydrogen bond with Thr-189. In cluster 3/element 6, hydroxychloroquine also interacted with Lys-35. At cluster 1/element 0, K777 interacted with both Thr-189 and Cys-110. Lastly, in clusters 4 and 5, compound 18a interacted with Thr-189, Pro-171, and Cys-187.
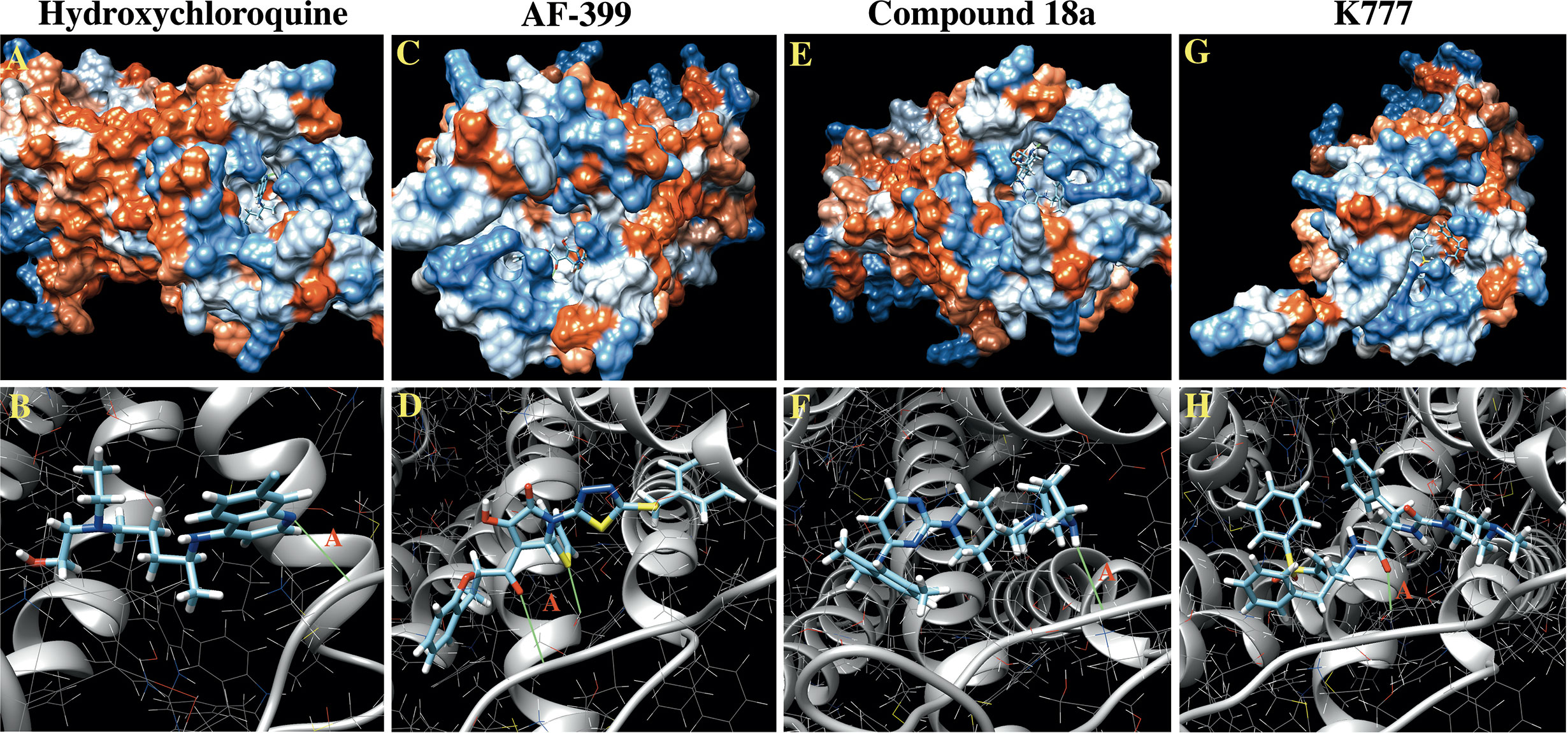
Figure 2 In Silico docking images for hydroxychloroquine and controls against CCR4. (A) Hydrophobicity plot demonstrating hydroxychloroquine interacting with the CCR4 active site. (B) Hydroxychloroquine forms a hydrogen bond with Thr-189 (A) in the CCR4 active site, as depicted in green. (C) Hydrophobicity plots demonstrating AF-399 interacting with the CCR4 active site. (D) AF-399 forms a hydrogen bond with Thr-189 (A) in the CCR4 active site. (E) Hydrophobicity plots demonstrating compound 18a interacting with the CCR4 active site. (F) Compound 18a forms a hydrogen bond with Thr-189 (A) in the CCR4 active site. (G) Hydrophobicity plots demonstrating K777 interacting with the CCR4 active site. (H) K777 forms a hydrogen bond with Thr-189 (A) in the CCR4 active site.
Discussion
Results from the present study indicate that the FDA-approved drug hydroxychloroquine interacts with the C-C chemokine receptor type 4 (CCR4) in silico. CCR4 is a seven transmembrane G protein-coupled receptor (GPCR) expressed throughout the human body with highest expression levels in the bone marrow and lymphoid tissue (CCR4 n.d.). Its endogenous ligands, the chemokines CCL17 and CCL22, are regulators of the immune system, contributing to lymphocyte trafficking and cytokine release. CCR4 plays both beneficial and deleterious effects in the immune response. Furthermore, CCR4 plays a beneficial role in recruiting immune cells to the site of infection, triggering a downstream immune response; however, CCR4 can also play deleterious roles, negatively impacting wound healing and contributing to sepsis (Barros et al., 2019; Traeger et al., 2008). Additionally, CCR4 induces Th2 effector T-cell polarization, suppressing Th1-dependent cellular immune responses that protect the body against pathogenic substances (Bayry et al., 2008; Szabo et al., 2003). In rheumatoid arthritis, CCR4 plays a critical role in CD4+ T cell migration to the synovium contributing to joint inflammation (Yang et al., 2004). In patients with rheumatoid arthritis and lupus, the percentage of circulating CD4+/CCR4+ T cells is significantly elevated relative to heathy controls (Yang et al., 2004). For patients with lupus in particular, an increase in CCR4-expressing Th2 cells are increased in patients with active disease and is thought to play a role in the development of autoantibodies contributing to disease pathogenesis (Hase et al., 2001). Additionally, significant numbers of CD8+/CCR4+ T lymphocytes were identified in skin biopsies of patients with cutaneous lupus erythematosus (CLE) and correlated with aggressive skin scarring (Wenzel et al., 2005). The putative ability of hydroxychloroquine to inhibit CCR4 signal transduction likely explains its efficacy in the treatment of rheumatoid arthritis and lupus. A recent study investigating the immune cell transcriptome profiles of COVID-19 patients with pneumonia identified CCR4 expression as an upregulated target relative to healthy controls (De Basi et al., 2020). CCR4 receptor antagonists have demonstrated efficacy in the prevention and treatment of septic shock (Power et al., 2000). In a lipopolysaccharide model of sepsis, transgenic CCR4-/- knockout mice showed improved survival rates due to a significant reduction in pro-inflammatory cytokine release and associated sepsis (Traeger et al., 2008). These data suggest a potential role for the use of CCR4 antagonists in the treatment of COVID-19 patients.
The chemical similarity ensemble approach (SEA) yielded interactions with several off-target proteins that likely contribute to the side-effect profile of hydroxychloroquine. Two of the hits, including the alpha-1D adrenergic receptor and muscarinic acetylcholine receptor M2, have been verified in vitro (Auerbach, 2020; Gordon et al., 2020). Inhibition of the alpha-1D adrenergic receptor may explain the rare, but reported, side-effects such as palpitations, headaches, and muscle weakness [Drugs.com, 2020j (Hydroxychloroquine)]. Side-effects such as blurred vision, constipation, dry mouth, and urinary retention are likely a result of the anti-cholinergic manifestations associated with muscarinic M2 receptor interaction. Also noteworthy, inhibition of several cyclin dependent kinases (CDKs), such as CDK11A, CDK11B, and CDK13 may explain the commonly reported gastrointestinal side-effects such as diarrhea, nausea, and vomiting.
Based on our molecular docking results, the control CCR4 inhibitors AF-399, compound 18a, and K777 demonstrate more pronounced thermodynamic binding characteristics to CCR4 relative to hydroxychloroquine. CCR4 antagonists are used in the United States for the treatment of mycosis fungoides and Sézary disease, and used globally for the treatment of adult T-cell leukemia/lymphoma (ATCLL) and CCR4+ cutaneous T cell lymphoma (CTCL) (Yu et al., 2007; FDA, 2018). AF-399 and compound 18a exhibit exceptional binding characteristics against CCR4 in vitro (IC50 = 2-10 and 8.1 nM, respectively) (Bayry et al., 2008; Klein et al., 2017; Shukla et al., 2016). Additionally, K777 is a broad-spectrum antiviral that prevents cathepsin-mediated cell entry (Jacobsen et al., 2000). K777 inhibits pseudovirus entry of SARS-CoV and EBOV with IC50 values of 0.68 nM and 0.87 nM, respectively (Jacobsen et al., 2000; Sato et al., 2013; Zhao, et al., 2015). Unlike hydroxychloroquine, none of these compounds, nor the FDA-approved CCR4 antagonist mogamulizumab, are known to increase the risk of QT prolongation (Mazzanti et al., 2020). Excessive QT prolongation can trigger life threatening arrhythmias such as torsades de pointes. It is important to note that hydroxychloroquine has been used for over half a century and has a well-established safety profile in the use for chronic conditions such as rheumatoid arthritis and lupus. Principally relevant to the current pandemic, a majority of patients hospitalized for COVID-19 infection have underlying comorbidities, such as cardiovascular disease, and are more likely to be elderly. Thus, confounding variables exist that may increase patient risk of adverse effects following hydroxychloroquine administration in the acute management of COVID-19 infection. Drugs that inhibit CCR4 in absence of QT prolongation, such as mogalizumab, may serve as viable alternatives to hydroxychloroquine in the treatment of patients with severe COVID-19 manifestations.
In addition to CCR4 inhibitors, the evaluation of various other classes of anti-inflammatory agents, particularly those drugs that limit cytokine release, may show promise in the treatment of COVID-19 infections. There are currently 242 clinical trials of COVID-19 treatments or vaccines (RECOVERY, 2020). Notable drugs include hydroxychloroquine, dexamethasone, and remdesivir (Table S1) (Stewart, 2020). Particularly noteworthy, the use of dexamethasone reduced death by up to one third in hospitalized patients with severe respiratory complications of COVID-19. A study in Wuhan, China demonstrated that plasma concentrations of interleukin (IL)-2, IL-7, IL-10, granulocyte-colony stimulating factor (GCSF), interferon gamma-induced protein 10 (IP-10 or CXCL10), monocyte chemoattractant protein-1 (MCP1), macrophage inflammatory protein 1-alpha (M1P1A), and tumor necrosis factor-alpha (TNF-α) were statistically elevated in ICU patients relative to non-ICU patients (Huang et al., 2020). While IL-2, CXCL10, and TNF-α contribute to the pathogenesis of sepsis, IL-7, IL-10, GCSF, MCP1, M1P1A are likely elevated as an endogenous attempt to combat sepsis (Snydman et al., 1990; Latifi et al., 2002; Takahashi et al., 2002; Schefold, 2011; Gaballa et al., 2012; Gomes et al., 2013; Herzig et al., 2014; Francois et al., 2018). IL-2 promotes the proliferation of T and B lymphocytes (Ross and Cantrell, 2018). While IL-2 plays a beneficial role in preventing autoimmune disease development, high levels of IL-2 lead to capillary leak syndrome and are associated with nosocomial sepsis (Snydman et al., 1990). FDA-approved IL-2 inhibitors that may warrant investigation in the treatment of patients with severe coronavirus disease include basiliximab and daclizumab [Drugs.com, 2020d (basiliximab); Drugs.com, 2020g (Daclizumab)] (Table S2). The chemokine CXCL10 (IP-10) plays a deleterious role in infection and inflammation by activating the chemokine receptor CXCR3, an important regulator of lymphocyte trafficking and activation (Herzig et al., 2014). The experimental compound ganodermycin is a potent inhibitor of the chemoattractant CXCL10 and may benefit patients with severe COVID-19 infection (Jung et al., 2011). Additionally, the FDA-approved 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase inhibitor atorvastatin inhibits CXCL10 and may be useful in the treatment of COVID-19 (Grip and Janciauskiene, 2009). Tumor necrosis factor alpha (TNF-α) is an endogenous pyrogen which mediates fever and inflammatory processes (Stefferl et al., 1996; Luheshi et al., 1997). TNF-α inhibitors have also proven beneficial in treating cytokine storm, a common cause of death in patients with severe coronavirus disease (Gaballa et al., 2012). TNF-α inhibitors, such as adalimumab, certolizumab, etanercept, golimumab, and infliximab may serve as prospective therapeutics in COVID-19 treatment [Drugs.com, 2020a (Adalimumab); Drugs.com, 2020f (Certolizumab); Drugs.com, 2020h (Etanercept); Drugs.com, 2020i (Golimumab); Drugs.com, 2020k (Infliximab)]. Additionally, inhibitors of pro-inflammatory targets such as interleukin (IL)-1 beta (IL-1β), IL-6, and Janus Kinase (JAK) 1/2 may be useful in reducing fever and preventing cytokine storm (Bird, 2018; Dinarello, 2015; Ekilsson et al., 2014; Gabay et al., 2010; Roskoski, 2016; Drugs.com, 2020b; Drugs.com, 2020c; Drugs.com, 2020e; Drugs.com, 2020l; Drugs.com, 2020m; Drugs.com, 2020n; Drugs.com, 2020o). (Table S2).
In summary, the use of hydroxychloroquine in the treatment of COVID-19 has received significant media attention over the past few months. Despite promising data from clinical trials hosted in China and France, hydroxychloroquine has not yet proven to be safe in preliminary U.S. clinical trials. QT prolongation in patients taking hydroxychloroquine along with azithromycin, a combination that has proven effective in European clinical trials, has raised concern regarding the use of these medicines in COVID-19 treatment (Mazzanti et al., 2020). This study has identified CCR4 as the immunomodulatory target for hydroxychloroquine. CCR4 antagonists that do not promote QT prolongation, such as the FDA-approved drug mogamulizumab or experimental compound K777, may warrant investigation in the treatment of severe coronavirus disease as either monotherapy or in combination with antivirals. The administration of anti-inflammatory agents may improve health outcomes in patients at risk of cytokine storm and warrant further investigation in clinical trials.
Limitations of the present study include a lack of in vitro verification of CCR4/hydroxychloroquine interaction. The use of the unbiased similarity ensemble approach (SEA) demonstrated that CCR4 is an immunomodulatory target for hydroxychloroquine. Results from this study may help identify FDA-approved drugs suitable for repurposing in the treatment of severe COVID-19 infection.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
All authors listed have made substantial, direct and intellectual contribution to the work and approved it for publication. TB and RN arranged the research, designed the experiments, examined and construed the data, wrote the manuscript, provided financial support, and guided all the experiments. KB, CH, RH, and TD examined and construed the data, and wrote and edited the manuscript.
Funding
This work was supported by an NIH training grant HL007260 to TB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to acknowledge Craig C. Beeson for introducing us to the similarity ensemble approach.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.01253/full#supplementary-material
References
Ankrum, J. (2020). Can cell therapies halt cytokine storm in severe COVID-19 patients? Sci. Tranl. Med. 12 (540), eabb5673. doi: 10.1126/scitranslmed.abb5673
Auerbach, P. (2020). DrugMatrix in vitro pharmacology data (National Toxicology Program). Available at: https://ntp.niehs.nih.gov/drugmatrix/index.html (Accessed on July 22, 2020).
Barros, J. F., Waclawiak, I., Pecli, C., Borges, P. A., Georgii, J. L., Ramos-Junior, E. S., et al. (2019). Role of Chemokine Receptor CCR4 and Regulatory T Cells in Wound Healing of Diabetic Mice. J Invest Dermatol. 139 (5), 1161–1170. doi: 10.1016/j.jid.2018.10.039
Bayry, J., Tchilian, E. Z., Davies, M. N., Forbes, E. K., Draper, S. J., Kaveri, S. V., et al. (2008). In silico identified CCR4 antagonists target regulatory T cells and exert adjuvant activity in vaccination. Proc. Natl. Acad. Sci. U. S. A. 105(29), 10221–10226. doi: 10.1073/pnas.0803453105
Bird, L. (2018). Calming the cytokine storm. Nat. Rev. Immunol. 18 (7), 417. doi: 10.1038/s41577-018-0030-6
Bitencourt-Ferreira, G., Azevedo, W. F. (2019). Docking with SwissDock. Methods Mol. Biol. 2053, 189–202. doi: 10.1007/978-1-4939-9752-7_12
Boulos, A., Rolain, J. M., Raoult, D. (2004). Antibiotic susceptibility of Tropheryma whipplei in MRC5 cells. Antimicrob. Agents Chemother. 48, 747–752. doi: 10.1128/AAC.48.3.747-752.2004
Centers for Disease Control and Prevention (2020). Cases of Coronavirus Disease (COVID-19) in the U.S. Available at: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/cases-in-us.html (Accessed July 19, 2020).
Chen, A., Hu, J., Zhang, Z., Jiang, S., Han, S., Yan, D., et al. (2020).Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial (Accessed April 15, 2020).
De Basi, S., Meschiari, M., Gibellini, L., Bellinazzi, C., Borella, R., Fidanza, L., et al. (2020). Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Comm. 11, 3434. doi: 10.1038/s41467-020-17292-4
Devaux, C. A., Rolain, J. M., Colson, P., Raoult, D. (2020). New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 55 (5), 105938. doi: 10.1016/j.ijantimicag.2020.105938
Dinarello, C. A. (2015). The History of Fever, Leukocytic Pyrogen and Interleukin-1. Temp. (Austin). 2 (1), 8–16. doi: 10.1080/23328940.2015.1017086
Drugs.com (2020a). Adalinumab. Available at: https://www.drugs.com/mtm/adalimumab.html (Accessed on April 26, 2020).
Drugs.com (2020b). Anakinra. Available at: https://www.drugs.com/cdi/anakinra.html (Accessed on April 27, 2020).
Drugs.com (2020c). Barcitinib. Available at: https://www.drugs.com/mtm/baricitinib.html (Accessed on April 26, 2020).
Drugs.com (2020d). Basiliximab. Available at: https://www.drugs.com/mtm/basiliximab.html (Accessed on April 28, 2020).
Drugs.com (2020e). Canakinumab. Available at: https://www.drugs.com/mtm/canakinumab.html (Accessed on April 26, 2020).
Drugs.com (2020f). Certolizumab. Available at: https://www.drugs.com/mtm/certolizumab.html (Accessed on August 8, 2020).
Drugs.com (2020g). Daclizumab. Available at: https://www.drugs.com/mtm/daclizumab.html (Accessed on April 28, 2020).
Drugs.com (2020h). Etanercept. Available at: https://www.drugs.com/cdi/etanercept-auto-injectors.html (Accessed on April 26, 2020).
Drugs.com (2020i). Golimumab. Available at: https://www.drugs.com/cdi/golimumab.html (Accessed on August 8, 2020).
Drugs.com (2020j). Hydroxychloroquine. Available at: https://www.drugs.com/sfx/hydroxychloroquine-side-effects.html (Accessed on July 20, 2020).
Drugs.com (2020k). Infliximab. Available at: https://www.drugs.com/mtm/infliximab.html (Accessed on April 26, 2020).
Drugs.com (2020l). Rilonacept. Available at: https://www.drugs.com/ppa/rilonacept.html (Accessed on April 26, 2020).
Drugs.com (2020m). Ruxolitinib. Available at: https://www.drugs.com/cdi/ruxolitinib.html (Accessed on April 26, 2020).
Drugs.com (2020n). Siltuximab. Available at: https://www.drugs.com/mtm/siltuximab.html (Accessed on April 27, 2020).
Drugs.com (2020o). Tocilizumab. Available at: https://www.drugs.com/mtm/tocilizumab.html (Accessed on April 26, 2020).
Ekilsson, A., Mirraskhian, E., Dufour, S., Schwaninger, M., Engbolm, D., Blomqvist, A. (2014). Immune-induced fever is mediated by IL-6 receptors on brain Endothelial cells coupled to STAT3-dependent induction of brain Endothelial Prostaglandin synthesis. J. Neurosci. 34 (48), 15957–15961. doi: 10.1523/JNEUROSCI.3520-14.2014
Food and Drug Administration (2018). FDA approves treatment for two rare types of non-Hodgkin lymphoma.https://www.fda.gov/news-events/press-announcements/fda-approves-treatment-two-rare-types-non-hodgkin-lymphoma (Accessed August 2, 2020).
Food and Drug Administration (2020). FDA cautions against use of hydroxychloroquine or chloroquine for COVID-19 outside of the hospital setting or a clinical trial due to risk of heart rhythm problems. Available at: https://www.fda.gov/drugs/drug-safety-and-availability/fda-cautions-against-use-hydroxychloroquine-or-chloroquine-covid-19-outside-hospital-setting-or (Accessed April 24, 2020).
Francois, B., Jeanett, R., Daix, T., Walton, A. H., Shotwell, M. S., Unsinger, J., et al. (2018). Interleukin-7 Restores Lymphocytes in Septic Shock: The IRIS-7 Randomized Clinical Trial. JCI Insight 3 (5), e98960. doi: 10.1172/jci.insight.98960
Gaballa, S., Besa, E. C., Alpdogan, O. (2012). The Use of Infliximab to Control Recurrent Cytokine Storms in EBV-Associated Angioimmunoblastic Lymphoma and Hemophagocytic Syndrome. Blood 120 (12), 4887. doi: 10.1182/blood.V120.21.4887.4887
Gabay, C., Lamacchia, C., Palmer, G. (2010). IL-1 Pathways in Inflammation and Human Diseases. Nat. Rev. Rheumatol. 6 (4), 232–241. doi: 10.1038/nrrheum.2010.4
Gautret, P., Lagier, J. C., Parola, P., Hoang, V. T., Meddeb, L., Mailhe, M., et al. (2020). Hydroxychloroquine and Azithromycin as a Treatment of COVID-19: Results of an Open-Label Non-Randomized Clinical Trial. Int. J. Antimicrob. Agents 56 (1), 105949. doi: 10.1016/j.ijantimicag.2020.105949
Gomes, R. N., Teixeira-Cunha, M. G. A., Figueiredo, R. T., Almeida, P. E., Alves, S. C., Bozza, P. T., et al. (2013). Bacterial clearance in septic mice is modulated by MCP-1/CCL2 and nitric oxide. Shock 39 (1), 63–69. doi: 10.1097/SHK.0b013e31827802b5
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. doi: 10.1038/s41586-020-2286-9
Grip, O., Janciauskiene, S. (2009). Atorvastatin reduces plasma levels of chemokine (CXCL10) in patients with Crohn’s disease. PloS One 4, e5263. doi: 10.1371/journal.pone.0005263
Grosdidier, A., Zoete, V., Michielin, O. (2007). EADock: docking of small molecules into protein active sites with a multiobjective evolutionary optimization. Proteins 67 (4), 1010–1025. doi: 10.1002/prot.21367
Grosdidier, A., Zoete, V., Michielin, O. (2011). SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277. doi: 10.1093/nar/gkr366
Hase, K., Tani, K., Simizu, T., Ohmoto, Y., Matsushima, K., Sone, S. (2001). Increased CCR4 expression in active systemic lupus erythematosus. J Leukoc Biol. 70 (5), 749–755.
Herzig, D. S., Luan, L., Toliver-kinsky, T. E., Guo, Y., Sherwood, E. R. (2014). The role of CXCL10 in the pathogenesis of experimental septic shock. Crit. Care 18 (3), R113. doi: 10.1186/cc13902
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Zhang, J., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi: 10.106/S0140-6736(20)30183-5
CCR4. (n.d.). The Human Protein Atlas. Available at: https://www.proteinatlas.org/ENSG00000183813-CCR4 (Accessed on August 14, 2020).
Jacobsen, W., Christians, U., Benet, L. Z. (2011). In vitro evaluation of the disposition of A novel cysteine protease inhibitor. Drug Metab Dispos. 28 (11), 1343–1351.
Jung, M., Liermann, J. C., Opatz, T., Erkel, G. (2011). Ganodermycin, a novel inhibitor of CXCL10 expression from Ganoderma applanatum. J. Antibiot. 64, 683–686. doi: 10.1038/ja.2011.64
Keiser, M. J., Roth, B. L., Armbruster, B. N., Ernsberger, P., Irwin, J. J., Shoichet, B. K., et al. (2007). Relating Protein Pharmacology by Ligand Chemistry. Nat. Biotechnol. 25 (2), 197–206. doi: 10.1038/nbt1284
Klein, A., Sagi-Assif, O., Meshel, T., Telerman, A., Izraely, S., Ben-Menachem, S., Byry, J., et al. (2017). CCR4 is a determinant of melanoma brain metastasis. Oncotarget 9 8 (19), 31079–31091. doi: 10.18632/oncotarget.16076
Kohler, C., Nittner, D., Rambow, F., Radaelli, E., Stanchi, F., Vandamme, N., et al. (2017). Mouse Cutaneous Melanoma Induced by Mutant BRaf Arises from Expansion.
Lang, P. T. (2018). Preparing Molecules for DOCKing. Available at: http://dock.compbio.ucsf.edu/DOCK_6/tutorials/struct_prep/prepping_molecules.htm (Accessed on July 22nd, 2020).
Latifi, S. Q., O’Riordan, M. A., Levine, A. D. (2002). Interleukin-10 Controls the Onset of Irreversible Septic Shock. Infect. Immun. 70 (8), 4441–4446. doi: 10.1128/IAI.70.8.4441-4446.2002
Liang, N., Zhong, Y., Zhou, J., Liu, B., Lu, R., Guan, Y., et al. (2018). Immunosuppressive effects of hydroxychloroquine and artemisinin combination therapy via the nuclear factor-κB signaling pathway in lupus nephritis mice. Exp. Ther. Med. 15 (3), 2436–2442. doi: 10.3892/etm.2018.5708
Live Science (2020). Treatments for COVID-19: Drugs being tested against the coronavirus. Available at: https://www.livescience.com/coronavirus-covid-19-treatments.html (Accessed on April 21, 2020).
Luheshi, G. N., Stefferl, A., Turnbull, A. V., Dascombe, M. J., Brouwer, S., Hopkins, S. J., et al. (1997). Febrile Response to Tissue Inflammation Involves Both Peripheral and Brain IL-1 and TNF-alpha in the Rat. Am. J. Physiol. 272 (3 Pt 2), R862–R868. doi: 10.1152/ajpregu.1997.272.3.R862
Ma, J. (2020). Coronavirus: China"s first confirmed Covid-19 case traced back to November 17 (South China Morning Post). Available at: https://www.scmp.com/news/china/society/article/3074991/coronavirus-chinas-first-confirmed-covid-19-case-traced-back (Accessed on April 27, 2020).
Ma’ayan, A., Jenkins, S. J., Goldfarb, J., Iyengar, R. (2008). Network Analysis of FDA Approved Drugs and their Targets. Mt. Sinai J. Med. 74 (1), 27–32. doi: 10.1002/msj.20002
Mazzanti, A., Briani, M., Kukavica, D., Bulian, F., Marelli, S., Trancuccio, A., et al. (2020). Association of Hydroxychloroquine with QTc Interval in Patients with COVID-19. Online ahead of print. Ciculation doi: 10.1161/CIRCULATIONAHA.120.048476
Meng, T., Coa, H., Zhang, H., Kang, Z., Xu, D., Gong, H., et al. (2020). The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS (Accessed April 15, 2020).
Moore, J. B., June, C. H. (2020). Cytokine release syndrome in severe COVID-19. Science 368, eabb8925. doi: 10.1126/science.abb8925
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 25 (13), 1605–1612. doi: 10.1002/jcc.20084
Power, C. A., Chvatchko, Y., Conquet, F. (2000). CCR4 Antagonists in Sepsis. WO2000067791A1 World Intellectual Property Organization, 16 (13).
Protein Atlas (n.d.). CCR4 https://www.proteinatlas.org/ENSG00000183813-CCR4/tissue. (Accessed on August 2, 2020)
Rainsfor, K. D., Parke, A. L., Clifford-Rashotte, M., Kean, W. F. (2015). Therapy and Pharmacological Properties of Hydroxychloroquine and Chloroquine in Treatment of Systemic Lupus Erythematosus, Rheumatoid Arthritis and Related Diseases. Inflammopharmacology 23 (5), 231–269. doi: 10.1007/s10787-015-0239-y
Raoult, D., Drancourt, M., Vestris, G. (1990). Bactericidal effect of doxycycline associated with lysosomotropic agents on Coxiella burnetii in P388D1 cells. Antimicrob. Agents Chemother. 34 (8), 1512–1514. doi: 10.1128/aac.34.8.1512
Raoult, D., Houpikian, P., Dupont, H. T., Riss, J. M., Arditi-Dijane, J., Brouqui, P. (1999). Comparison of 2 Regimens containing Doxycycline and Ofloxacin or Hydroxychloroquine. Arch. Intern. Med. 159 (2), 167–173. doi: 10.1001/archinte.159.2.167
RECOVERY Randomised Evaluation of COVID-19 Therapy (University of Oxford). Available at: https://www.recoverytrial.net/ (Accessed on July 22, 2020).
Rolain, J. M., Colson, P., Raoult, D. (2007). Recycling of chloroquine and its hydroxyl analogue to face bacterial, fungal and viral infection in the 21st century. Int. J. Antimicrob. Agents 30, 297–308. doi: 10.1016/j.ijantimicag.2007.05.015
Roskoski, R. (2016). Janus Kinase (JAK) inhibitors in the treatment of inflammatory and neoplastic diseases. Pharmacol. Res. 111, 784–803. doi: 10.1016/j.phrs.2016.07.038
Ross, S. H., Cantrell, D. A. (2018). Signaling and function of Interleukin-2 in T Lymphocytes. Ann. Rev. Immun. 36, 411–433. doi: 10.1146/annurev-immunol-042617-053352
Santos, R., Ursu, O., Gaulton, A., Bento, A. P., Donadi, R. S., Bologa, C. G., et al. (2016). A comprehensive map of molecular targets. Nat. Rev. Drug Discovery 16 (1), 19–34. doi: 10.1038/nrd.2016.230
Sato, T., Iwase, M., Miyama, M., Komai, M., Ohshima, E., Asai, A., et al. (2013). Internalization of CCR4 and inhibition of chemotaxis by K777, a potent and selective CCR4 antagonist. Pharmacology. 91 (5–6), 305–313. doi: 10.1159/000350390.
Schefold, J. C. (2011). Immunostimulation using granulocyte- and granulocyte-macrophage colony stimulating factor in patients with severe sepsis and septic shock. Crit. Care 15 (2), 136. doi: 10.1186/cc10092
Scher, I. (2020). The first COVID-19 case originated on November 17, according to Chinese officials searching for "patient zero" (Business Insider). Available at: https://www.businessinsider.com/coronavirus-patients-zero-contracted-case-november-2020-3 (Accessed on April 27, 2020).
Schrezenmeier, E., Dorner, T. (2020). Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 16 (3), 155–166. doi: 10.1038/s41584-020-0372-x
Shukla, L., Ajram, L. A., Begg, M., Evans, B., Graves, R. H., Hodgson, S. T., et al. (2016). 2,8-Diazaspiro[4.5]decan-8-yl)pyrimidin-4-amine potent CCR4 antagonists capable of inducing receptor endocytosis. Eur J Med Chem. 115, 14–25. doi: 10.1016/j.ejmech.2016.02.058
Simmons, G., Bertram, S., Glowacka, I., Steffen, I., Chaipan, C., Agudelo, J., et al. (2011). Different host cell proteases activate the SARS-coronavirus spike-protein for cell–cell and virus–cell fusion. Virology 413, 265–274. doi: 10.1016/j.virol.2011.02.020
Snydman, D. R., Sullivan, B., Gill, M., Gould, J. A., Parkinson, D. R., Atkins, M. B. (1990). Nosocomial Sepsis associated with Interleukin-2. Ann. Intern. Med. 112 (2), 102–107. doi: 10.7326/0003-4819-112-2-102
Stefferl, A., Hopkins, S. J., Rothwell, N. J., Luheshi, G. N. (1996). The role of TNF-alpha in fever: opposing actions of human and murine TNF-alpha and interactions with IL-beta in the rat. Br. J. Pharmacol. 118 (8), 1919–1924. doi: 10.1111/j.1476-5381.1996.tb15625.x
Stewart, J. (2020). COVID-19: Prevention & Investigational Treatments (Drugs.com). Available at: https://www.drugs.com/condition/covid-19.html (Accessed on April 15, 2020).
Szabo, S. J., Sullivan, B. M., Peng, S. L., Glimcher, L. H. (2003). Molecular mechanisms regulating TH1 immune responses. Annu Rev Immunol 21, 713–758.
Takahashi, H., Tashiro, T., Miyazaki, M., Kobayashi, M., Pollard, R. B., Suzuki, F. (2002). An essential role of macrophage inflammatory protein 1α/CCL3 on the expression of host’s innate immunities against infectious complications. J. Leukocyte Biol. 72, 1190–1197. doi: 10.1189/jlb.72.6.1190
Traeger, T., Kessler, W., Assfalg, V., Cziupka, K., Koerner, P., Dassow, C., et al. (2008). Detrimental Role of CC Chemokine Receptor 4 in Murine Polymicrobial Sepsis. Infect Immun. 76 (11), 5285–5293. doi: 10.1128/IAI.00310-08
UniProt (2020). UniProtKB – C-C Chemokine Receptor Type 4. https://www.uniprot.org/uniprot/P51679. (Accessed on August 1, 2020).
Vincent, M. J., Bergeron, E., Benjannet, S., Erikson, B. R., Rollien, P. E., Ksiazek, T. G., et al. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2, 69. doi: 10.1186/1743-422X-2-69
Walker, J. (2020). China traces coronavirus to first confirmed case, nearly identifying "patient zero" (Newsweek). Available at: https://www.newsweek.com/china-traces-coronavirus-back-first-confirmed-patient-zero-1492327 (Accessed on April 27, 2020).
Wang, Z., Liang, L., Yin, Z., Lin, J. (2016). Improving chemical similarity ensemble approach in target prediction. J. Cheminfo. 8, 20. doi: 10.1186/s13321-016-0130-x
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronaviru 2019-nCoV) in vitro. Cell Res. 30, 269–271. doi: 10.1038/s41422-020-0282-0
Wenzel, J., Henze, S., Worenkamper, E., Basner-Tschakarjan, E., Sokolowska-Wojdylo, M., Steitz, J., et al. (2005). Role of the Chemokine Receptor CCR4 and its Ligand Thymus- and Activation-Regulated Chemokine/CCL17 for Lymphocyte Recruitment in Cutaneous Lupus Erythematosus. J Invest Dermatol. 124 (6), 1241–1248. doi: 10.1111/j.0022-202X.2005.23755.x
World Health Organization (2020). Global research on coronavirus research. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov (Accessed on April 19, 2020).
Yang, P. T., Kaisai, H., Zhao, L. J., Xiao, W. G., Tanabe, F., Ito, M. (2004). Increased CCR4 expression on circulating CD4+ T cells in ankylosing spondylitis, rheumatoid arthritis and systemic lupus erythematosus. Clin Exp Immunol. 138 (2), 342–347. doi: 10.1111/j.1365-2249.2004.02617.x
Keywords: COVID-19, hydroxychloroquine, CCR4, SARS-CoV-2, immunomodulation
Citation: Beck TC, Beck KR, Holloway CB, Hemings RA Jr., Dix TA and Norris RA (2020) The C-C Chemokine Receptor Type 4 Is an Immunomodulatory Target of Hydroxychloroquine. Front. Pharmacol. 11:1253. doi: 10.3389/fphar.2020.01253
Received: 28 April 2020; Accepted: 30 July 2020;
Published: 28 August 2020.
Edited by:
Stefania Tacconelli, University of Studies G. d’Annunzio Chieti and Pescara, ItalyReviewed by:
Emanuela Marcantoni, New York University, United StatesMahmood Yaseen Hachim, Mohammed Bin Rashid University of Medicine and Health Sciences, United Arab Emirates
Isabella Zanella, University of Brescia, Italy
Copyright © 2020 Beck, Beck, Holloway, Hemings, Dix and Norris. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tyler C. Beck, beckt@musc.edu; Russell A. Norris, norrisra@musc.edu
 Tyler C. Beck
Tyler C. Beck Kyle R. Beck4
Kyle R. Beck4 Calvin B. Holloway
Calvin B. Holloway Richard A. Hemings Jr.
Richard A. Hemings Jr. Thomas A. Dix
Thomas A. Dix Russell A. Norris
Russell A. Norris