Corrigendum: Bioassay-Guided Interpretation of Antimicrobial Compounds in Kumu, a TCM Preparation From Picrasma quassioides’ Stem via UHPLC-Orbitrap-Ion Trap Mass Spectrometer Combined With Fragmentation and Retention Time Calculation
- 1Department of Biology, Animal Physiology and Neurobiology Section, KU Leuven, Leuven, Belgium
- 2National Engineering Research Center for Modernization of Traditional Chinese Medicine - Hakka Medical Resources Branch, School of Pharmacy, Gannan Medical University, Ganzhou, China
- 3Laboratory for Functional Foods and Human Health, Center for Excellence in Postharvest Technologies, North Carolina Agricultural and Technical State University, North Carolina Research Campus, Kannapolis, NC, United States
- 4College of Life Sciences, NanKai University, Tianjin, China
The stem of Picrasma quassioides (PQ) was recorded as a prominent traditional Chinese medicine, Kumu, which was effective for microbial infection, inflammation, fever, and dysentery, etc. At present, Kumu is widely used in China to develop different medicines, even as injection (Kumu zhusheye), for combating infections. However, the chemical basis of its antimicrobial activity has still not been elucidated. To examine the active chemicals, its stem was extracted to perform bioassay-guided purification against Staphylococcus aureus and Escherichia coli. In this study, two types of columns (normal and reverse-phase) were used for speedy bioassay-guided isolation from Kumu, and the active peaks were collected and identified via an UHPLC-Orbitrap-Ion Trap Mass Spectrometer, combined with MS Fragmenter and ChromGenius. For identification, the COCONUT Database (largest database of natural products) and a manually built PQ database were used, in combination with prediction and calculation of mass fragmentation and retention time to better infer their structures, especially for isomers. Moreover, three standards were analyzed under different conditions for developing and validating the MS method. A total of 25 active compounds were identified, including 24 alkaloids and 1 triterpenoid against S. aureus, whereas only β-carboline-1-carboxylic acid and picrasidine S were active against E. coli. Here, the good antimicrobial activity of 18 chemicals was reported for the first time. Furthermore, the spectrum of three abundant β-carbolines was assessed via their IC50 and MBC against various human pathogens. All of them exhibited strong antimicrobial activities with good potential to be developed as antibiotics. This study clearly showed the antimicrobial chemical basis of Kumu, and the results demonstrated that HRMS coupled with MS Fragmenter and ChromGenius was a powerful tool for compound analysis, which can be used for other complex samples. Beta-carbolines reported here are important lead compounds in antibiotic discovery.
Introduction
Picrasma quassioides (D. Don) Benn, a prominent medicinal herb from southern and eastern Asia, named “Kumu,” “Kudanmu,” “Shanxiongdan,” and “Kupizi” in China due to its extremely bitter and lasting taste, is a deciduous tree of the Simaroubaceae family. It was described with a scientific name in 1884 (Flora of China Editorial Committee, 1997; Flora of China Editorial Committee, 2008), but as a medicine, its stem in slices or powder was first recorded as “Shanxiongdan” in Xin Yi Xue (AD 1972) for the treatment of dysentery, infections of the biliary tract, and burns and wounds (Medical Team of, 1972; Chinese Materia Medica Editoral Committee of National Administration of TCM, 1999). Although PQ does not have as long a documented history as other TCMs (traditional Chinese medicines), it has received much attention due to its effectiveness in inflammation, infection, and cancer. By now, PQ is widely used in China in different medicinal products, such as Kumu Xiaosanpian (tablet), Fufang Kumu Xiaoyanjiaonang (capsule), Kumu Zhusheye (injection) for treating influenza, upper respiratory tract infections, acute tonsillitis, enteritis, and bacterial dysentery. According to the Chinese Pharmacopeia, its dried stem and leaf are used as Kumu for anti-inflammation, microbial infection, fever, dysentery, and snake or insect bites. However, PQ’s thick stems are commonly used in TCM markets. Also, the Tamang people exploit its wood for fever and joint pain in Nepal (Ambu et al., 2020). In Korea and Japan, the heartwood of PQ, named ‘Picrasma wood’, is used as a herbal drug, consisting of chips, slices, or short pieces of wood (Ministry of Food and Drug Safety, 2012; The ministry of health and labour and welfare, 2016).
Phytochemical studies showed that alkaloids (β-carbolines, canthinones, and cinnamamides), triterpenoids (quassinoids, apotirucallanes, tirucallanes, and apotirucallanes), neolignans, and flavonoids are the major compounds in PQ (Jiao et al., 2010a; Xu et al., 2016; Bai et al., 2020; Mohd Jamil et al., 2020; Ren et al., 2020). They exhibit antitumor (Xie et al., 2020), anti-inflammatory (Jiao et al., 2011), antimicrobial (Chen et al., 2009b; Zhang J. et al., 2019), antiparasitic (Curcino Vieira and Braz-Felho, 2006), anti-dyspepsia (Babu Rao et al., 2013), antihypertensive (Zhao et al., 2013), anti-asthma (Shin et al., 2014), antioxidant (Jung et al., 2012), anti-osteoporosis (Kong et al., 2017), neuroprotective (Zhao et al., 2019c; Guo et al., 2019), and other biological activities. PQ also showed some toxicity and adverse effects in zebrafish embryos (Gong et al., 2018), cultured cells (Lou et al., 2018), and in clinical studies (Weiqi et al., 2019). However, comprehensive studies investigating PQ’s antimicrobial chemicals are still scarce, and the NMR chemical identification usually required milligrams of pure compound, which is time-consuming to purify. In recent years, the advanced Orbitrap Elite Hybrid Mass Spectrometer, which combines a novel high-field orbitrap mass analyzer with a premier dual-pressure linear ion trap mass spectrometer, greatly improved MS and MSn performance and versatility, and it has become a reliable technique for a targeted and a non-targeted analysis of the chemical structure, requiring only picogram of the sample (Denisov et al., 2012; Jiang et al., 2020; Zhao et al., 2020). LC-HRMS/MS (high-resolution mass spectrometry) and MSn provide retention time, extract m/z and its fragmentation, from which the structure normally can be inferred based on comparison with standards, or published spectra of compound databases, MS/MS libraries, and molecular networking (Allen et al., 2014; Schymanski et al., 2014; Fisher et al., 2021). But, it is still a challenge to distinguish isomers with similar structures. Recently, several computer-assisted structure elucidation software packages for MS were developed offering new ways for isomer interpretation, such as MS fragmentation prediction (MFP) (Tyrkkö et al., 2010; Hu et al., 2021; Krettler and Thallinger, 2021), retention time calculation (RTC) (Noreldeen et al., 2018; Aalizadeh et al., 2019), and ion mobility spectrometry tools (IMS)(Campuzano et al., 2012; Dodds et al., 2017). With contrast to MFP and RTC, IMS measures the collisional cross section of compounds based on their time-of-flight via a buffer gas in a drift tube, which is not available for all instruments, including orbitraps. Hence, in this study, MFP and RTC (MS Fragmenter and ChromGenius from ACD/Structure Elucidator Suite) were used to perform fragmentation and retention time calculations, aiding for the MS elucidation.
To figure out the active compounds in Kumu, two types of columns (normal and reverse phases) were utilized for fast separation of the more widely used PQ stem. Based on the bioassay test, the active peaks were collected and detected by LC-HRMS/MS for MSn. Meanwhile, a compound database of PQ was manually built for MS interpretation, and chemical structures were elucidated based on MSn ion fragments and retention time, comparing with several standards and the database. Notably, for “unknown” compounds (lack of authentic standards and scarce information, such as spectra or fragmentation), the fragmentation prediction program (MS Fragmenter) and the retention time calculation program (ChromGenius) were used to evaluate the chemical structures via predicting mass spectral fragments based on the cleavage rules, and calculating their retention time based on chemical similarity searching. Then all the active compounds were identified. This study demonstrates a rapid, reliable method to isolate and identify natural products via columns, LC-HRMS/MS, and computer-assisted interpretation softwares. The results will provide a basis for further development and utilization of Kumu.
Materials and Methods
Preparation of Reagents and Materials
For MS samples, three standard compounds were used in this study: methylnigakinone, nigakinone, and β-carboline-1-carboxylic acid. The first two were previously isolated and identified in our lab via MS and NMR comparison (Gong et al., 2016), and β-carboline-1-carboxylic acid (LOT: 2015-0005238) was purchased from Enamine Ltd (Kyiv, Ukraine). The purity of all chemicals was over 95%. Water was generated by a Milli-Q system (Millipore, Bedford, MA, United States), and formic acid, acetonitrile, methanol, and ammonium acetate (LC/MS grade) were from Thermo Fisher Scientific (Fair Lawn, NJ, United States). The stock solutions of three standards in methanol were prepared at 4 μg/ml and an additional mixed solution for MS detection, stored at 4°C until use.
For samples of antimicrobial tests, the plant materials were collected from Jinpen Mountain, Xinfeng County, Ganzhou, China (Figure 1). According to Flora of China, the samples were identified as Picrasma quassioides (D. Don) Benn. by Professor Haibo Hu. The voucher specimen was kept in our lab at Gannan Medical University (No. PQ-XF1701). PQ stems (Kumu) were dried at 40°C, and then grounded into a fine powder. One gram of each powder was extracted in 10 ml of H2O, MeOH, EtOAC, and hexane with the help of sonication (4 × 15 min with 4–6 h gap). Then after 5 min centrifuging at 3500 rpm, a 1-ml aliquot of each supernatant was sampled and evaporated in a fume hood (hexane and ethyl acetate), or dried (methanol and water) in a SpeedVac concentrator (SVC 200 H, Stratech Scientific, London, United Kingdom). The dried extracts and three standards were redissolved in DMSO as a 20 mg/ml for antimicrobial assessment. All the samples were stored at 4°C for further testing.
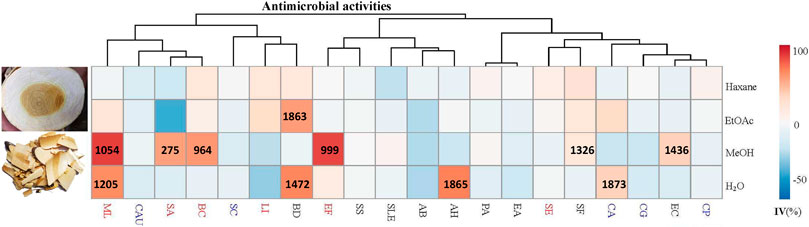
FIGURE 1. Antimicrobial activities (IC50) (μg/ml) of different solvent extracts of Picrasma quassioides stem; IV: inhibition value; The abbreviations are the initials of the scientific names of the tested microorganism, including five fungi [Candida albicans (CA), Candida parapsilosis (CP), Candida auris (CAU), Candida glabrata (CG), and Saccharomyces cerevisiae (SC)], nine G– bacteria [Escherichia coli (EC), Pseudomonas aeruginosa (PA), Shigella sonnei (SS), Acinetobacter baumannii (AB), Enterobacter aerogenes (EA), Brevundimonas diminuta (BD), Shigella flexneri (SF), Salmonella enterica subsp. enterica (SLE), and Aeromonas hydrophila (AH)], and six G+ bacteria [Staphylococcus aureus (SA), Staphylococcus epidermidis (SE), Micrococcus luteus (ML), Listeria innocua (LI), Enterococcus faecalis (EF), and Bacillus cereus (BC)], marked in blue, black, and red, respectively.
Antimicrobial Assay
The antibacterial activity of crude extracts was tested against 20 human pathogenic strains, including five fungi [Candida albicans (SC 5314), Candida parapsilosis (ATCC 22019), Candida auris (OS299), Candida glabrata (ATCC 2001), and Saccharomyces cerevisiae (ATCC 7754)], six Gram-positive bacteria [Staphylococcus aureus (ATCC6538, Rosenbach), Staphylococcus epidermidis (ATCC 1457), Micrococcus luteus (DPMB 3), Listeria innocua (LMG 11387), Enterococcus faecalis (HC-1909-5), and Bacillus cereus (LMG9610)], and nine Gram-negative bacteria [Escherichia coli (ATCC 47076), Pseudomonas aeruginosa (PAO1), Shigella sonnei (LMG 10473), Acinetobacter baumannii (RUH134), Enterobacter aerogenes (or Klebsiella aerogenes, ATCC 13048), Brevundimonas diminuta (a gift from Prof. Rob Lavigne at KU Leuven), Shigella flexneri (LMG10472), Salmonella enterica subsp. enterica (ATCC 13076), and Aeromonas hydrophila (ATCC 7966)]. The antimicrobial tests were performed by a 96-well microdilution method. In brief, for bacteria and fungi, 10 and 5 μl, respectively, of a two-fold dilution series of each extract (20 mg/ml stock), blank controls (DMSO) and positive controls (ciprofloxacin, 100 μg/ml stock and miconazole, 200 μg/ml stock) were transferred into 96-well plates with the test microogranism (OD: 0.003) and cultured for 20 h. Then the inhibition value (%, IV) was obtained by dividing the sample’s OD value minus that of an extract control without inoculation by the average OD of the blank controls (solvent) and multiplying by 100.
All the tests were repeated for confirmation, and IC50 (minimum inhibitory concentration to inhibit the growth of 50%) was calculated by dose-response fitting using non-linear least-squares sigmoid regression (GraphPad Prism 7.0, San Diego, CA). The data were clustered using a webtool (https://biit.cs.ut.ee/clustvis/) to obtain clustered heatmaps. MBC (minimum bactericidal concentration) was obtained directly from the agar tests of serial chemical dilution.
Fractionation and Isolation of Constituents
100 g of PQ stem was extracted three times aided by sonication (4 × 30 min with 4–6 h gap) with the selected solvent (methanol) according to the antimicrobial activities, and then dried in a Rotavapor (R-100, BÜCHI, Flawil, Switzerland). Then, 5.6 g of extract was finally obtained and mixed with 25 g silica gel (60 Å, LOT#MKCK 1888; Sigma-Aldrich, Germany). Afterward, a large-scale separation was performed on a silica gel column (ECOPLUS 35 × 500 mm, YMC, Kyoto, Japan) in a preparative liquid chromatography setup (Waters Delta 600 multi-solvent quaternary pump, Waters detector 2487, Waters 600 controller, Massachusetts, United States) using a mobile phase consisting of hexane (A), ethyl acetate (B), methanol (C), and 25% acetic acid in methanol (D), performed a flow rate of 40 ml/min, with a step gradient from 95% A and 5% B to 100% D (5–20% step every 10 min). Finally, 218 fractions were collected (1 fraction per minute) and 1 ml of each fraction was sampled and dried; then 100 μl DMSO was added per sample to prepare stocks for antimicrobial testing.
Based on the bioactivity results of 218 fractions, seven active fractions (F47, F60, F88, F108, F118, F125, and F181) were selected for further separation via HPLC (LC-20AT pumps combined with a SPD-M20A detector and DGU-20A3R degasser, SHIMADZU, Kyoto, Japan). Separation conditions of each fraction were optimized on several analytical columns (250 × 4.6 mm, 5 μm, C18, Symmetry, Waters; 150 × 4.6 mm, 4 μm, Polar-RP, Synergi, Phenomenex; 250 × 4.6 mm, 4 μm, Hydro-RP, Synergi, Phenomenex), and then were geometrically transferred to semi-preparative columns to ensure the same selectivity for peak collections. F47 and F60 were separated on a semi-preparative C18 column (250 × 10 mm, 5 μm, C18, Sunfire, Waters) and the rest by a Polar-RP column (250 × 10 mm, 4 μm, Polar-RP, Synergi, Phenomenex) using a mobile phase consisting of water (A) and acetonitrile (B), both containing 0.1% formic acid. For each fraction, the injected amount was kept around 2 mg, and all the subfraction (1 per minute) were collected via gradient elution with a 4 ml/min flow rate using the conditions listed in Table 1. After drying each fraction in a SpeedVac, 15 μl DMSO were added to each tube for antimicrobial testing. To eliminate possible errors during a single run, all subfractions were collected and tested twice. This was also done for all the active peaks collected under the same condition to confirm the activities. Then the confirmed peaks were collected, dried, and kept in 4°C for the MS analysis.
UHPLC and MS/MS Conditions
The UHPLC-MS/MS analysis was performed with an Ultimate 3000 UHPLC system (Dionex Thermo Scientific) equipped with an EasySpray C18 column (2 μm, 100 Å, 50 μm × 15 cm, Thermo Scientific), and Orbitrap Elite High-Field Orbitrap Hybrid Mass Spectrometer (Thermo Scientific, United States), which consisted of orbitrap high-resolution selection and ion trap scanning, was used for the chemical analysis of natural small-molecular compounds. With a 0.3 μl·min−1 flow rate, the C18 column was set at 35°C and the injection volume was 5 μl. The mobile phases were composed of water (eluent A) and 80% acetonitrile with 20% water (eluent B), both containing 0.1% formic acid (for negative mode detection, 0.1% ammonium acetate was used instead of formic acid). The gradients were set as follows: 0–3 min, 5% B; 3–20 min, 5%–100% B; 20–23 min, 100% B; 23–24 min, 100%–5% B; and 24–34 min, 5% B to re-equilibrate the system.
For MS detection, the Orbitrap-Ion Trap mass spectrometer was fitted with a heated electrospray ionization (ESI) ion source, and both negative and positive ionization modes were performed at a full scan mode ranging from m/z 100–2000. To aid the structural identification of the components, top 20 ions’ MS/MS fragmentation (dd-MS2-TOP 20) was operated with an m/z range of 50–2000. The MS parameters were optimized as reported previously (Hu et al., 2021) with the followings: spray voltage −2.1 Kv/+2.1 Kv; capillary temperature, 275°C; multipole RF amplifier, 800 Vp-p; reagent ion source temperature, 160°C. The stepped ITMS (Ion trap mass spectrometry) was set to 35 eV for MS/MS acquisition of the most intense ions from FTMS (Fourier transform mass spectrometry).
Data Processing and Analysis
All the MS data acquisition and analyses were done by Xcalibur 4.2 (Thermo Scientific, United States), ACD/MS Workbook Suite 2020, combined with MS Fragmenter, and ChromGenius (ACD/labs, Canada). The original UHPLC-MS data of each sample were exported, and their background was subtracted based on MS data of the blank solvent for both positive and negative ions. Then the processed data were imported into ACD/labs to extract peaks and align chromatograms to make the comparison more accurate than manually, including ITA (itellitarget) and IX 2.0 (IntelliXtract), which are algorithms to analyze LC(GC)/MS datasets for targeted and untargeted component detection and deconvolution, respectively. The assignment accuracy was set to 5 ppm for both.
Moreover, the published components of PQ were collected by checking SciFinder, Web of Science, the Dictionary of Natural Products, China National Knowledge Infrastructure (CNKI), and other databases, and then Spectrus DB (ACD/labs) was used to create a manually built compound library, including chemical names, molecular formulas, exact molecular mass, and structures. The MS1 and MS2 spectra of active peaks were analyzed by comparing the detected formulas and MS fragments with the component library we built and the COCONUT database (Sorokina et al., 2021) (the largest publicly available natural product database), and all the active compounds were targeted and identified. In detail, the spectrum of standards can figure out themselves easily, and those compounds without standards can be elucidated by their ion fragments and retention time (tR) according to fragmentation prediction by MS Fragmenter and retention time calculation by ChromGenius, from which all the fragmentation patterns of each compound in this study were summarized and further verified.
Results and Discussion
Antimicrobial Activities of Kumu
Thus far, antimicrobial tests mainly focused on PQ compounds, especially for its alkaloids. A previous study showed that the extract of total alkaloids exhibited antimicrobial activities against Streptococcus hemolytic-β, Staphylococcus aureus, Shigella castellani, Bacillus subtilis subsp, and Sporosarcina (Meng, 2007). Total alkaloids were reported to exhibit a wider spectrum and better antimicrobial activity than single alkaloids, possibly due to synergistic effects (Guihua, 2011). Notably, the fat-soluble alkaloids provided good inhibition against both wild type and two highly virulent strains of E. coli, even higher than berberine sulfate, mequindox, and ofloxacin, while water-soluble alkaloids hardly had bacteriostatic effects on these strains (He et al., 2008). Also, the essential oil of PQ inhibited both bacterial (e.g. Bacillus subtilis) and fungal (e.g. Ganoderma lucidum) species (Hanif et al., 2010). However, as a TCM, Kumu was always used as a whole extract. Comprehensive studies investigating the bioactivities of its total extracts are still scarce. Hence, in this study, four different solvents (hexane, ethyl acetate, methanol, and water) were used to extract the stem of PQ, and test these extracts against 20 different human pathogens. The results show that Kumu extracts can inhibit five G+, four G– bacteria, and one fungus (Figure 1), in which Kumu exhibited strongest activity against Staphylococcus aureus (IC50 = 275 μg/ml). These results support Kumu’s medicinal application as an anti-infection agent. Moreover, the results show that methanol is the best solvent to extract Kumu for its antimicrobial activity, and it was used to perform the large-scale extracting for bioassay guided purification work.
Bioassay-Guided Isolation
Based on the above antimicrobial activities, it is worth performing bioassay-guided purification for identifying the antimicrobial compounds of Kumu. By now, bioassay-guided isolation of natural products have been widely used for different activities, such as anti-inflammatory (Erdemoglu et al., 2008), antioxidant (Yang et al., 2020), anti-seizure (Brillatz et al., 2020b) (Brillatz et al., 2020a), vasorelaxant (Othman et al., 2006), aldose reductase inhibitory (Logendra et al., 2006), anti-plasmodial (Baldé et al., 2021), antiviral (Parvez et al., 2020), antibacterial (He et al., 2017), and antifungal (Yan et al., 2020). But most of these studies were performed with multiple purification steps, which consumed long time and large amounts of solvents. With the rapid development of chromatographic techniques, many different columns have been already used for fast collection of small-amount compounds. Hence, in this study, two types of columns were selected for the separation: a large normal-phase silica gel column and semi-preprative reverse-phase columns. The aim was to rapidly determine and collect the active fractions/peaks to increase the purification efficiency.
For the bioassays, Staphylococcus aureus (Gram-positive) and Escherichia coli (Gram-negative), which belong to top dangerous superbugs worldwide (World Health Organization, 2017), were selected for the antimicrobial activities of Kumu. After separation on a preparative silica gel column, 218 fractions (F1-218) were obtained for antimicrobial testing, and seven active groups were found including F46-52, F54-60, F88-96, F105-112, F113-119, F122-129, and F177-187 (Figure 2). From each, a representative fraction was selected for further purification based on their inhibition values: F47, F60, F88, F108, F118, F125, and F181. By comparing different columns and separation conditions, F47 and F60 were further separated on a semi-preparative C18 column, and the remaining fractions by the Polar-RP column. In either case, we used a mobile phase consisting of water (A) and acetonitrile (B), both containing 0.1% formic acid. Afterward, each HPLC-subfraction was tested against the two aforementioned bacteria, and their activities are shown as a heatmap (Figure 2). The subfractions showed much less activity against E. coli than S. aureus, presumably because G– bacteria have a multilayer outer membrane preventing many xenobiotics passing through the cell membrane, so it is more difficult to find activity against G– (Randall et al., 2013). Finally, 3, 5, 1, 10, 10, 2, and 5 active peaks were obtained from the seven fractions, respectively, and all these 36 peaks were collected for chemical identification via LC-HRMS/MS (Figure 3).
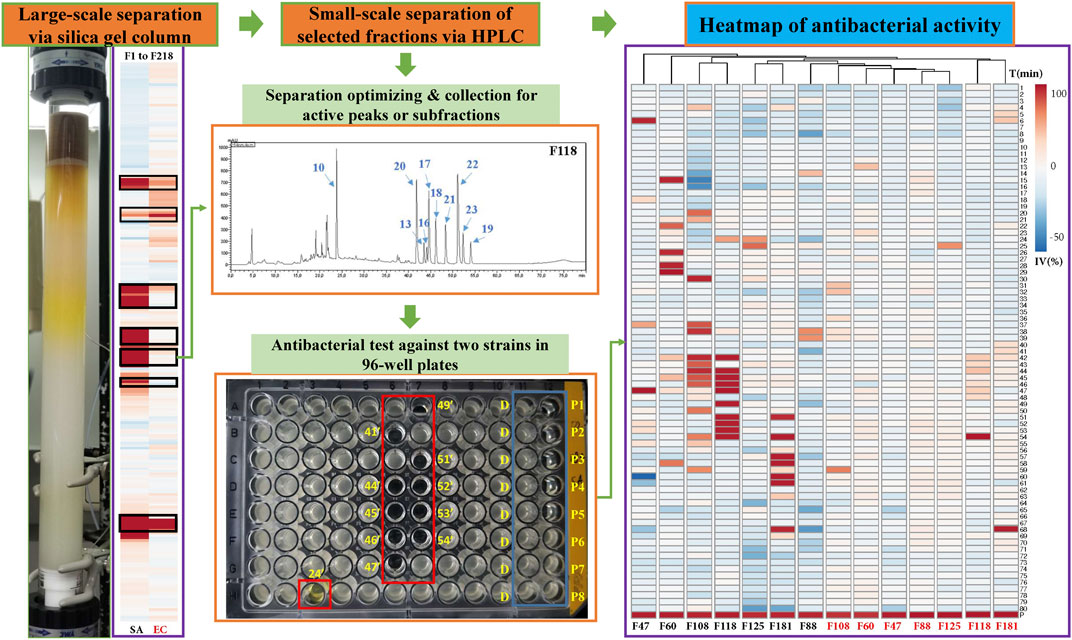
FIGURE 2. Separation of antibacterial compounds from Kumu based on successive chromatographic columns (normal and reversed-phase chromatography) and the heatmap of the bioactivity of their factions against a representative Gram-positive strain (SA: S. aureus) and Gram-negative strain (EC: E. coli). F: fractions of preparative silica gel column, D: negative control, DMSO, P1-8: positive control (ciprofloxacin) in different concentrations, IV: inhibition value, T: time for collection in minutes, such as 24’ and 41’.
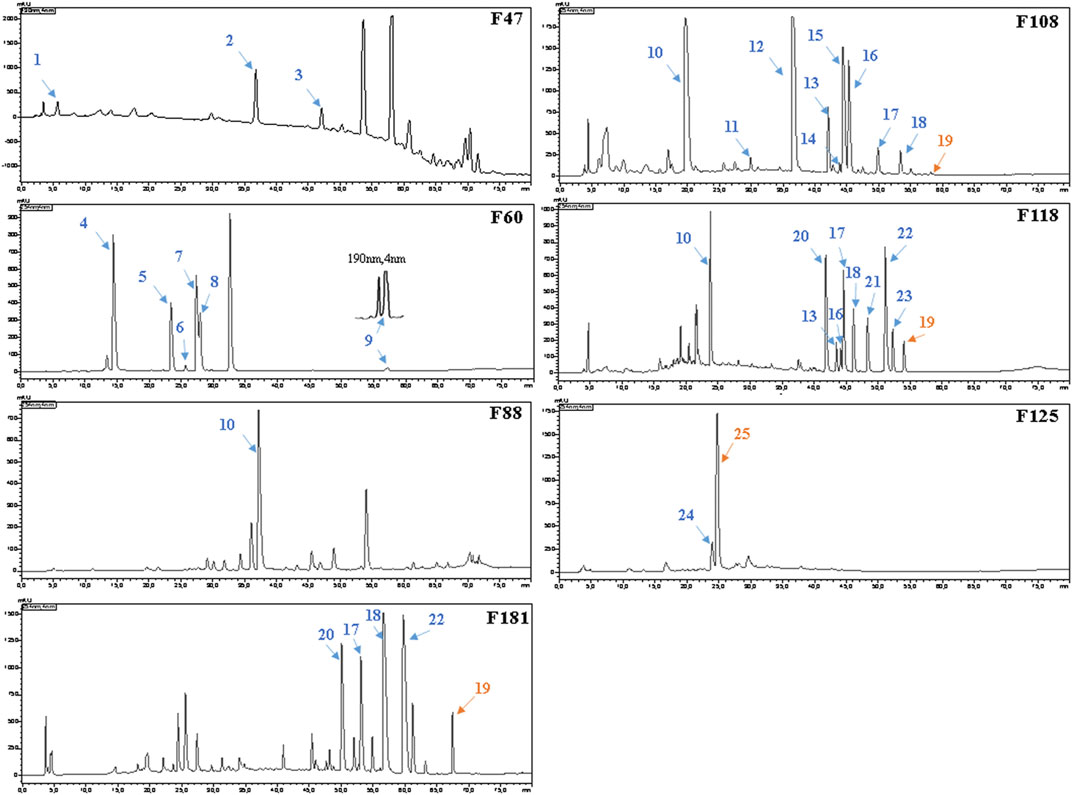
FIGURE 3. HPLC–UV chromatograms and active peaks of the selected fractions from the stem of Picrasma quassioides. The peaks active against S. aureus are marked in blue, while the ones active against both S. aureus and E. coli are numbered in orange.
Identification of Active Compounds in Selected Fractions via LC-HRMS/MS
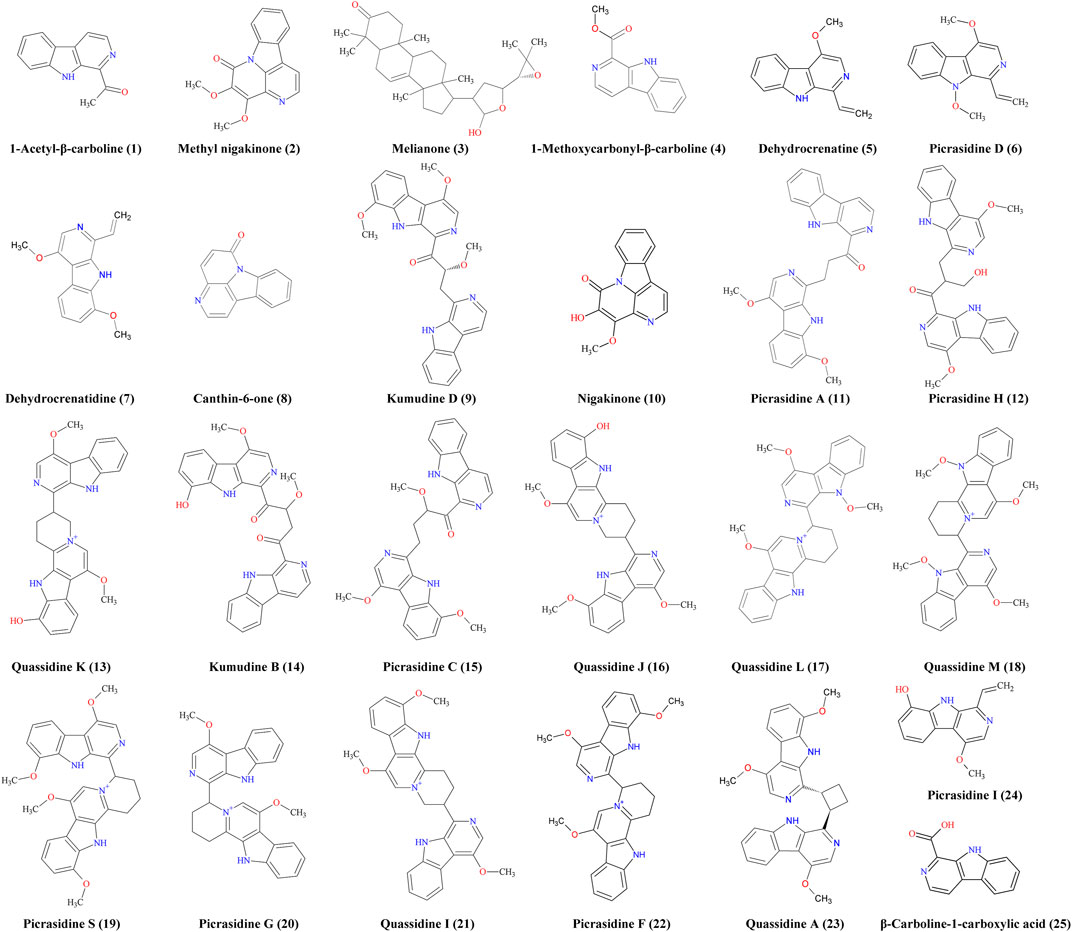
Before the MS analysis, all peaks were collected and tested to confirm their activities. The results showed that all 36 peaks were active against S. aureus, but only four peaks were also active against E. coli, including the subfractions at 59, 54, 26 , and 68 min of F108, 118, 125, and 181, respectively. Furthermore, mass responds of pure chemicals were easily affected by various noises of background in total ion chromatograms, due to the high sensitivity of mass detection. Hence, all 36 peaks and several blank samples were analyzed both in positive and negative ion mode by an UHPLC-Orbitrap-Ion-Trap Mass Spectrometer, based on the optimized experimental condition. Then all chemical analyses were confirmed by both positive and negative ions, except in a few cases where data were only available from one ion mode. By comparing with available standards, fragmentation of MS Fragmenter and retention time calculations, a total of 25 active compounds (some peaks were identified as same compounds, in Figure 3) were identified (Table 2), three of which were only detected in positive ion mode under our experiment conditions: 1-acetyl-β-carboline, methylnigakinone, and melianone. All the chemical structures are shown in Figure 4, in which compounds 1–2 and 4–25 are alkaloids, while compound 3 is a triterpenoid. Notably, only picrasidine S (19) and β-carboline-1-carboxylic acid (25) showed strong inhibition against both S. aureus and E. coli.
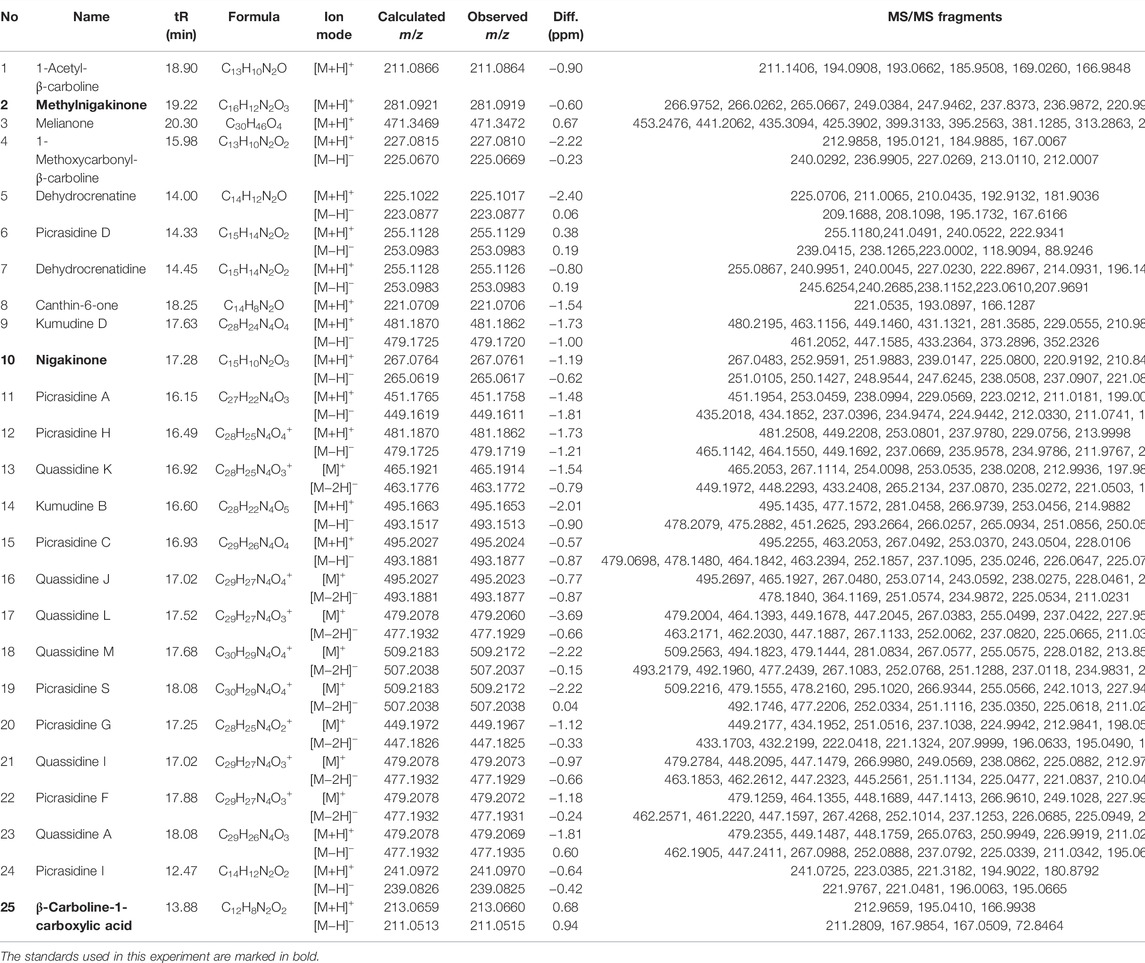
TABLE 2. Mass information of antimicrobial chemicals identified from the stem of Picrasma quassioides.
Structure Elucidation Based on Comparison With Published Spectra, Standards, and MS Fragmenter
For component identification, comparison of the MS1 and MS2 spectra with published ones was extensively used, but for most PQ components no matching published sources could be available. Thus, several standards were first analyzed under the same condition to assist the MS interpretation, including methylnigakinone, nigakinone, and β-carboline-1-carboxylic acid. As shown in Figure 5, the β-carboline-1-carboxylic acid standard (25) was detected in both negative and positive ion mode, with m/z 239.0826 and 241.0969, respectively, as precursor ions. The detected active peaks of F125 showed quite similar retention time (tR) and m/z compared to the standard both in MS1 and MS2, indicating that it was identical to the standard, compound 25. Meanwhile, another several active peaks of F47, 88, 108, and 118 (shown in Figure 3) were similarly identified as methylnigakinone (2) and nigakinone (10).
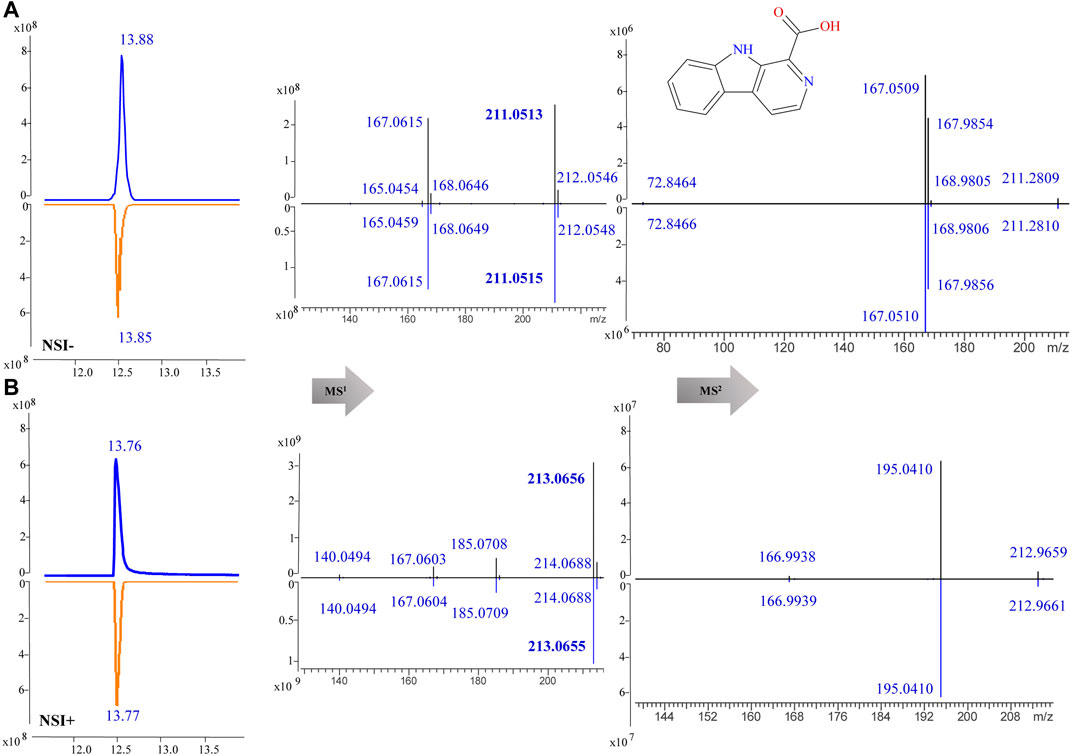
FIGURE 5. MS1 and MS2 spectra of β-carboline-1-carboxylic acid (25) in the negative (NSI-) (A) and the positive (NSI+) (B) ion mode (upper panel: sample, lower panel: standard).
For the other compounds where no standards or published spectra were available, the software, MS Fragmenter (Zhou et al., 2014; Zhang Y. et al., 2019) was used to predict and interpret their fragmentation in both positive and negative modes. Taking the fourth active peak in F108 as an example, it was detected in positive and negative ion mode with strong procedures, m/z 465.1914 [C28H25N4O3]+ and m/z 463.1772, [C28H23N4O3]-,respectively. After searching its structure via Mass Suite Book (ACD/labs) in the COCONUT database (Sorokina et al., 2021) (the biggest publicly available natural product database) and a manually built the PQ database (some components were not included in the COCONUT database), 15 compounds with chemical formula C28H25N4O3+ or C28H26N4O3 were retrieved. Then their mass fragmentation spectra were compared one by one via MS Fragmenter (Tyrkkö et al., 2010), and the best matched one, quassidine K (13), was identified as the target, also according to the comparisons of retention time and MS1 spectra in positive and negative ion mode. Its fragmentaion in positive ion mode, shown as an example in Figure 6, mainly originates from the cleavage of nitrogen-containing heterocycles. Via the fragmentation analysis, most active peaks of Kumu could be identified and are shown in Table 1.
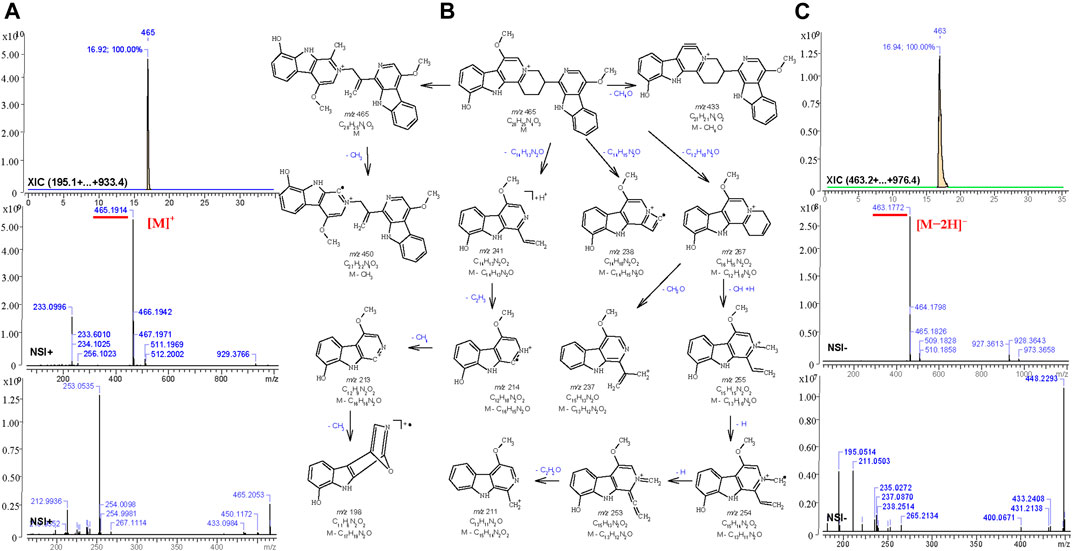
FIGURE 6. Total ion chromatography analysis (XIC) and MS1-2 spectra (A,C) of quassidine K (13) in the positive (m/z 465.1914, [C28H25N4O3]+) and the negative (m/z 463.1772, [C28H23N4O3]-) ion mode, and its main mass fragmentation pathway (B) in the positive ion mode generated by MS Fragmenter.
Isomers Distiguished by ChromGenius and MS Fragmenter
Although MS Fragmenter can distinguish different fragmentation pattern of compounds, it was still difficult to see the differences for some compounds with quite similar structures. Hence, retention time (tR) calculation can be utilized as a supplementary method to distinguish isomers. Notably, lots of isomers have been reported in PQ, and were also detected in this study. Fraction 60 and 108 showed active peaks with identical m/z 481.1862 in the positive ion mode, and m/z 479.1720 and 479.1719 in the negative ion mode. Hence, they were identified as the chemical formula, C28H24N4O4 or C28H25N4O4+. To distiguish their structure, three compounds: kumudine D, picrasidine H, and picrasidine T, were considered after MS Fragmenter analysis as mentioned in the above because they have the same calculated m/z (481.1870 in positive and 479.1725 in the negative ion mode).
Then ChromGenius was utilized for their retention time (tR) calculation, which was performed based on chemical similarity searching (Willett et al., 1998). The calculation was set as the UHPLC-MS/MS condition and showed good prediction according to the main parameters, correction equation (−0.03*tR2 + 1.936*tR −6.696) and method coefficient (0.9039). The calculated results are listed in Figure 7E, in which the retention factor k’ was calculated as: k' = (tR – t0)∕t0, where tR stands for retention time, t0 implied dead time, and the similarity coefficient (sim coeff) is calculated based on the hamming distance coefficient (the closer to 1, the better). The predicted tRs of these isomers are shown in Figure 7A, and 25 best-matched points for the calculation are regressed in the curves (Figures 7B–D). Finally, based on the predicted tR, the active peaks in F60 and 108 were identified as kumudine D (9) and picrasidine H (12), respectively.
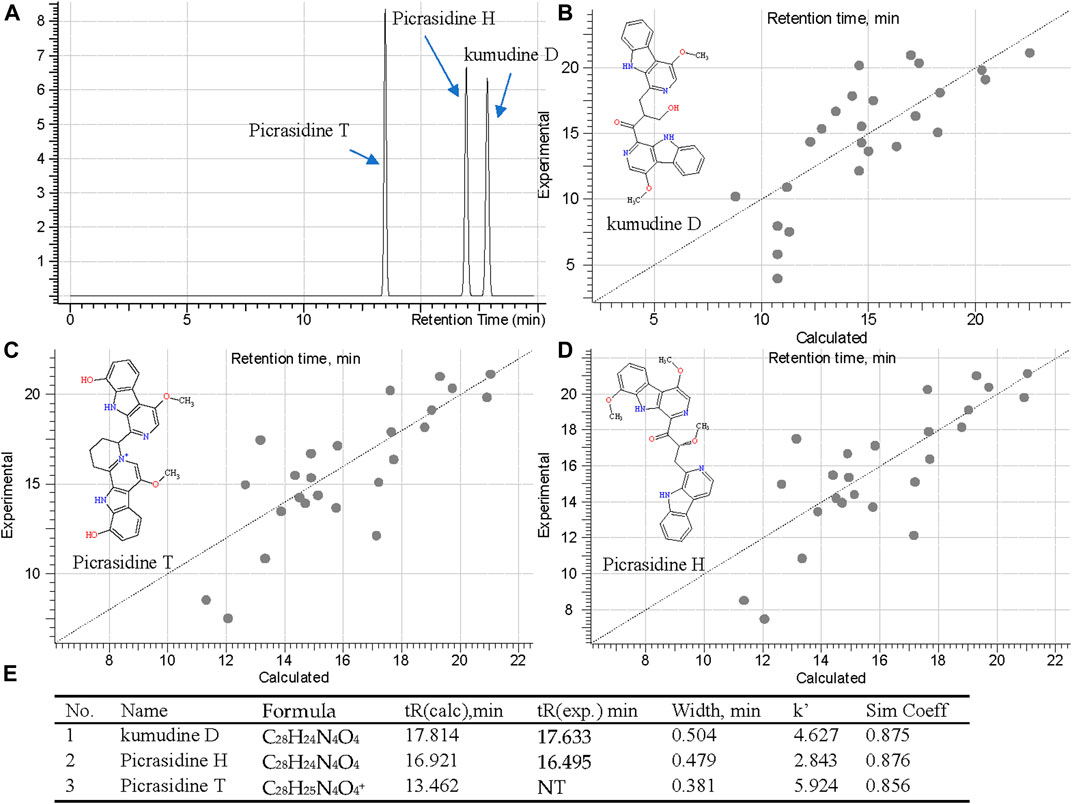
FIGURE 7. Retention time prediction in chromatography (A), regression curves of three isomers [kumudine D (B), picrasidine T (C), and picrasidine H (D)] via ChromGenius and the calculation parameters (E).
Four more sets of isomers were similarly analyzed based on their calculated tR: isomers of m/z 255.1128 (C15H14N2O2) in F60 were picrasidine D (6) and dehydrocrenatidine (7); isomers of m/z 479.2078 (C29H27N4O3+) in F108, 118, and 181 were identified as quassidine L (17), quassidine I (21), and picrasidine F (22), respectively; isomers of m/z 509.2183 (C30H29N4O4+) in F108, 118, and 181 were suggested to be quassidine M (18) and picrasidine S (19), while m/z 495.2027 (C29H26N4O4) in F108 and 118 were picrasidine C (15) and quassidine J (16), respectively.
To confirm the accuracy of fragmentation and tR calculation for identification, three standards were also subjected to the same calculated method. Taking β-carboline-1-carboxylic acid (25) as an example, the exact masses were 213.0659 ([M+H]+) and 211.0513 ([M−H]−), and its precursors were detected as 213.0656 and 211.0513 in MS1; thus, the chemical formula was calculated as C12H8N2O2. Searching in the COCONUT and PQ databases 14 compounds were retrieved. After assigning the MS2 fragmentation via MS Fragmenter, three compounds were considered as the best-matching ones, and then, tR calculations were performed to distinguish them. Finally, the compound was identified as β-carboline-1-carboxylic acid. Also, the other two standards could be identified by the same approach. Hence, this can be considered as a method validation for this study.
The Antimicrobial Spectrum of Three Representative β-carbolines
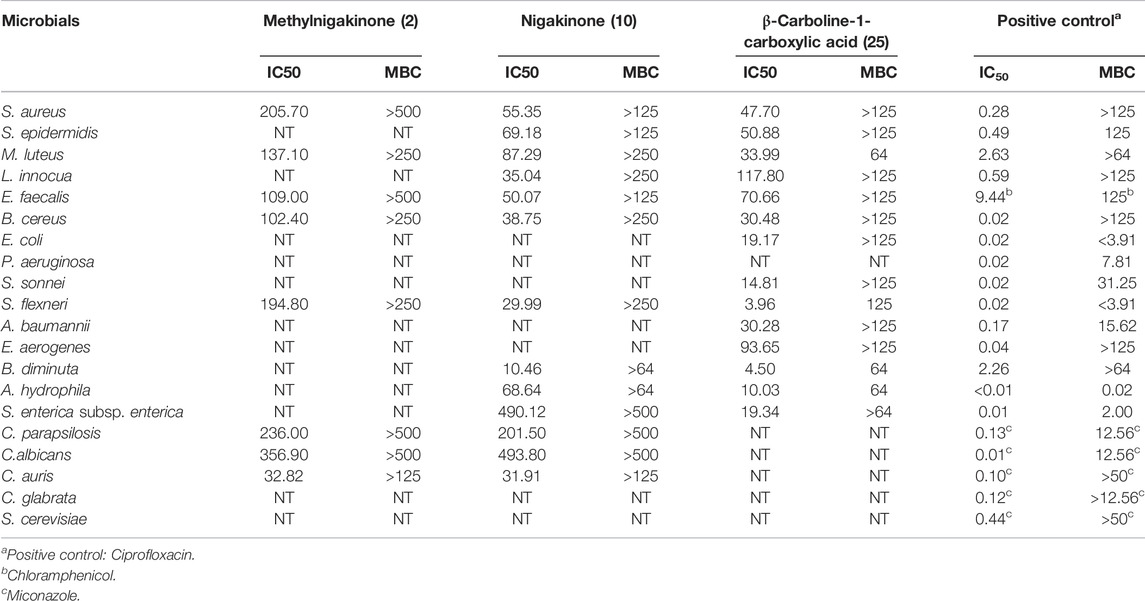
The standards of three β-carbolines were also assessed for their IC50 (minimum inhibitory concentration to inhibit the growth of 50%) and MBC (minimum bactericidal concentration): methylnigakinone (2), nigakinone (10), and β-carboline-1-carboxylic acid (25). Among them, only methylnigakinone’s antimicrobial activity was reported in a Chinese patent against Staphylococcus aureus (MIC = 0.76 mg/ml), Escherichia coli (3.09 mg/ml), Pseudomonas aeruginosa (3.12 mg/ml), Streptococcus hemolyticus (2.46 mg/ml), and Streptococcus pneumoniae (1.23 mg/ml) (Wu et al., 2007). As far as we know, our study is the first systematic evaluation of their antimicrobial activities. The tests were done by a 96-well microdilution method against 20 different microorganisms. All three β-carbalines strongly inhibit the growth of many microorganisms and even killed them. Also the GraphPad was used to calculate the IC50 while MBC was obtained directly from the agar tests of serial chemical dilution (Table 3, Figure 8).
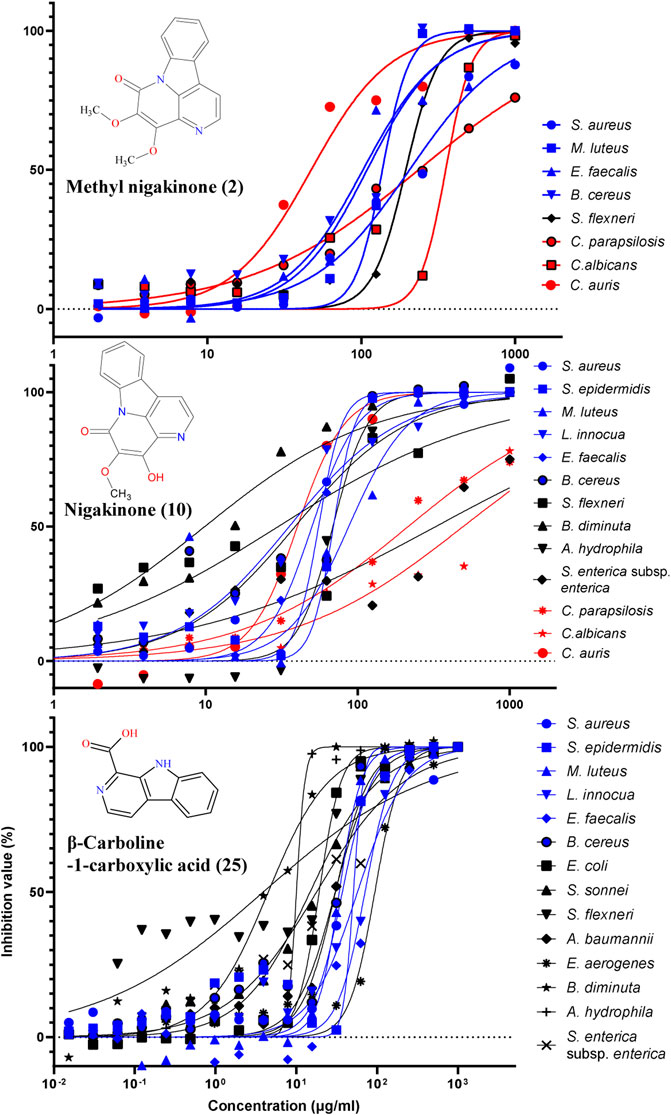
FIGURE 8. Inhibition of different microorganisms by three β-carbolines from the stem of Picrasma quassioides.
From the result of antimicrobial test, compound 2 (C2) can inhibit four Gram-positive, one Gram-negative bacteria, and three fungi; however, most of the IC50 values were above 100 μg/ml. C10 was active against more microorganisms, including six G+, four G– bacteria, and three fungi, and showed mostly somewhat higher activity than C2. C10 differs from C2 by a methylation at the 4’-hydroxy position. The methylation can reduce the chemical sensitivity, which was one of mechanism of antimicrobial resistance (Malone and Gordon, 2016; Warrier et al., 2016), while other study also indicated the methylation can also improve antimicrobial potency and broaden the spectrum (Benhamou et al., 2015), indicating that methylation worked differently for diverse compounds. According to the IC50 and MBC results, it was clear that the methylation reduced the antibacterial, but not the antifungal activity of C10, suggesting its different mechanism of action on bacteria vs. fungi.
An interesting result was obtained for Candida auris, an important emerging fungal pathogen, which was first described in 2009 and has spread across six continents as a cause of infections in hospitals (Meis and Chowdhary, 2018; Proctor et al., 2021). Both C2 and C10 were found to strongly suppress the growth of C. auris with a similar IC50 (around 30 μg/ml). Furthermore, C25 exhibited higher antibacterial activity and broader spectrum than both C2 and C10, including six G+ and eight G– bacteria, but it showed no antifungal activity. Notably, most G– bacteria, such as E. coli, S. sonnei, S. flexneri, B. diminuta, A. hydrophila, and S. enterica subsp. enterica were sensitive for C25 (with IC50 mostly below 20 μg/ml). Therefore, all three compounds were considered as having potential for antibiotic development.
Bioactivity Discussion of Kumu Compounds
To better comparing the bioactivities of the chemicals detected in this study, all the related literatures were checked, especially for their antimicrobial activity. 1-acetyl-β-carboline (1)(OHMOTO and KOIKE, 1982), methylnigakinone (2)(Jiang and Zhou, 2008), and melianone (3) were identified from fraction 47, where C1 was already reported as active against several laboratory and clinical strains (Savidge and Sorg, 2019)(Chen et al., 2018), such as S. aureus, with IC50 below 260 μg/ml (Shin et al., 2010). C2 has shown antiulcer effects (Omoja et al., 2014), significant cytotoxicity against CNE2 and HepG2 cancer cells (Jiang and Zhou, 2008; Mohd Jamil et al., 2020), and antiviral activity against Coxsackie virus B3 (with an IC50 as 7.4 μM (Zhang et al., 2013)), and its antimicrobial activity was documented and patented (Mitscher et al., 1975; Wu et al., 2007). Moreover, C3 (Zhao et al., 2019) was active against Salmonella ser. typhi with both MIC and MBC of 0.053 µM (Veni et al., 2020) against S. aureus with agar diffusion inhibition zone of 9 mm (Biavatti et al., 2001), and it was also documented for anticancer, antivirus, anti-gout, anti-depressant, and antifeeding activities (Su et al., 1990; Ishaq, 2016; Liu Y. et al., 2019; Zhao et al., 2019; Anita and Praveena, 2020; Sundar and Aarthy, 2020).
In fraction 60, six active chemicals were identified: 1-methoxycarbonyl-β-carboline (4), dehydrocrenatine (5), picrasidine D (6), dehydrocrenatidine (7), canthin-6-one (8), and kumudine D (9). Compound 4 can inhibit tobacco mosaic virus (TMV) replication with EC50 of 0.179 mM (Chen et al., 2009b) and had significant inhibitory activity on various plant pathogenic bacteria and fungi (Gao et al., 2020). C5 was reported for antimalarial, anti-inflammatory, antiparasitic, and antiprotozoal activities (Kita et al., 2004; Van Baelen et al., 2009; Liu P. et al., 2019; Saiin et al., 2019), but ours is the first report of its antibacterial activity to the best of our knowledge. C6 and C7 were reported for their cytotoxicity, anti-inflammatory, and antivirus activities (Ohmoto and Koike, 1988; Han et al., 2001; Chen et al., 2009b; Lee et al., 2009; Liu P. et al., 2019); their antibacterial activity is reported here for the first time. C8 showed anticancer, antimalarial, anti-inflammatory, antidiabetic, anti-plasmodium, antiprotozoal, antibacterial, antifungal, and antivirus activities (Lagoutte et al., 2008; Ross et al., 2008; Almeida et al., 2011; Arias et al., 2011; Cebrian-Torrejon et al., 2011; Agrawal et al., 2013; Rashed et al., 2013; Amanulla et al., 2017; Cho et al., 2018; Makong et al., 2019); it was active against S. aureus (of inhibition zone diameter of 6.1 mm), but no activity against E. coli was observed (Casciaro et al., 2019), which is the same as our result. C9 showed cytotoxicity against HepG2 and 3B cells, but there are no reports thus far about its antimicrobial activity.
Fraction 88 only showed one active peak, identified as nigakinone (10), which has documented cytotoxicity, antivirus, antimalarial, and anti-inflammatory activities (Kuo et al., 2003; Chen et al., 2009b; Liu P. et al., 2019). Both fraction 108 and 118 had 10 active peaks, as shown in Figure 3. The active peaks in F108 were identified as nigakinone (10), picrasidine A (11), picrasidine H (12), (±)-quassidine K (13), kumudine B (14), picrasidine C (15), quassidine J (16), quassidine L (17), quassidine M (18), and picrasidine S (19). Many similar compounds were found in F118, including C10, 13, 16, 17, 18, and 19, in addition to four other compounds: picrasidine G (20), quassidine I (21), picrasidine F (22), and quassidine A (23). It can be expected that the composition of compounds eluting from successive steps in a gradient may overlap, especially if the steps are small. For their bioactivities, C11 was reported as a TMV inhibitor (Chen et al., 2009a) and C12 (KOIKE and OHMOTO, 1986) showed none of anti-inflammatory effects and cytotoxicity against RAW 264.7 cells (Liu P. et al., 2019), while C13 had good cytotoxicity against the human cervical HeLa cell line, with an IC50 value around 20 μM (Guo et al., 2020). C14 has anti-hepatoma potential l (Zhao et al., 2019d), and C15 exhibited anti-inflammatory activity (Liu P. et al., 2019) and cytotoxicity against RAW 264.7 (Liu P. et al., 2019), esophageal squamous carcinoma cells (Shi et al., 2018), and also beneficial effects in cerebral protection (Sasaki et al., 2016). From the literatures, C16, C19, C20, and C21 had cytotoxicity against human cervical HeLa, gastric MKN-28 cancer, and mouse melanoma B-16 cancer cells (Jiao et al., 2015), and C21 also strongly inhibited osteoclast formation (Kong et al., 2017). C17 exerted beneficial effects in the cerebral protection (Sasaki et al., 2016), and the trifluoroacetate of both C17 and C18 showed potential anti-Alzheimer’s disease activity (Zhang et al., 2020). C20 had antiprotozoal activity (Kita et al., 2004) and was also active against RAW 264.7 (Liu P. et al., 2019) and triple-negative breast cancer cells (MDA-MB 468) (Yamashita et al., 2017). C22 was toxic against HeLa cells and active against several methicillin-sensitive and -resistant strains of S. aureus (MRSA), while C19 and C20 were also reported for their anti-MRSA activity (Guo-hua et al., 2015). For C23, only its anti-inflammatory activity was studied (Jiao et al., 2010b).
Only two active compounds were found in F125: picrasidine I (24) and β-carboline-1-carboxylic acid (25). C24 was known to have a strong antiosteoclastogenic effect (Kong et al., 2017) but no other effects were reported thus far. For C25, the previous bioactivity studies implied it can inhibit the growth of K-562 and SGC-7901 cell lines (Lai et al., 2014) and had potential for attenuating bleomycin-induced pulmonary fibrosis (Cui et al., 2019), while none anti-inflammatory activity (Liu P. et al., 2019) and none photocytotoxicity against lung (A549) and colon (Colon-26) cancer cells (Reddy et al., 2019).
To summarize, 18 compounds identified in our study are first reported here for their activity against S. aureus or E. coli, including 1-methoxycarbonyl-β-carboline (4), dehydrocrenatine (5), picrasidine D (6), dehydrocrenatidine (7), kumudine D (9), nigakinone (10), picrasidine A (11), picrasidine H (12), (±)-quassidine K (13), kumudine B (14), picrasidine C (15), quassidine J (16), quassidine L (17), quassidine M (18), quassidine I (21), quassidine A (23), Picrasidine I (24), and β-carboline-1-carboxylic acid (25). Notably, 24 active compounds were β-carbolines or dimers thereof. This class is a promising scaffold for the design and development of antibacterial compounds. It is worth to find lots of related bioactive compounds in plant extracts, since the biosynthesis of secondary metabolites often generates significant amounts of intermediates of the biosynthetic pathway, some of which may share bioactivities. The number of bioactive compounds identified from a single plant is large in this study, although it cannot be sure that all the identified β-carbolines originate from a single biosynthetic pathway here, which could be helpful and pending for future studies.
Conclusion
In summary of this study, the interpretation method using two types of columns for chemical isolation and UHPLC-Orbitrap- Ion-Trap Mass Spectrometer for chemical identification was developed. The standard comparison, database searching, MS Fragmenter, and ChromGenius were introduced for the structure analysis. The results showed that high-resolution mass spectrometry combined with mass fragmentation and retention time calculation was a powerful tool to identify natural products, especially for distinguishing isomers with similar structures. A total of 25 active compounds were isolated and identified from Kumu against S. aureus or E. coli, including five types of isomers, in which 18 components’ antimicrobial activities were firstly reported in this study. Notably, 24 active chemicals belonged to β-carboline or its dimer, which class exhibited as a promising scaffold for the design and development of potent and selective antibacterial compounds. The antimicrobial components reported here provide a basis for the medicinal usage of themselves and Kumu, and are beneficial for the scientific development of quality standards and its therapeutic utilization, indicating that the method can be used to figure out active chemical basis for other complex materials.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
Conceptualization, HH, CH, and WL; methodology, HH, AK, and AG; software, HH, JP, and DS.; validation, HH.; formal analysis, HH; investigation, HH, JP, and CH; resources, HH, JP, and CH; data curation, HH; writing—original draft preparation, HH; writing—review and editing, HH and WL; visualization, HH; supervision, WL; project administration, WL and HH; funding acquisition, WL and HH. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant Nos. 81660639). WL largely funded himself.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aalizadeh, R., Nika, M. C., and Thomaidis, N. S. (2019). Development and Application of Retention Time Prediction Models in the Suspect and Non-target Screening of Emerging Contaminants. J. Hazard. Mater. 363, 277–285. doi:10.1016/j.jhazmat.2018.09.047
Agrawal, R., Sethiya, N. K., and Mishra, S. H. (2013). Antidiabetic Activity of Alkaloids of Aerva Lanata Roots on Streptozotocin-Nicotinamide Induced Type-II Diabetes in Rats. Pharm. Biol. 51, 635–642. doi:10.3109/13880209.2012.761244
Allen, F., Greiner, R., and Wishart, D. (2014). Competitive Fragmentation Modeling of ESI-MS/MS Spectra for Putative Metabolite Identification. Metabolomics 11, 98–110. doi:10.1007/s11306-014-0676-4
Amanulla, S. S. D., Veerakumar, S., and Ramanathan, K. (2017). Screening of Potential Plant Compounds as Survivin Inhibitors and its Anti-Cancer Efficacy by Molecular Docking. Cei 13, 41–48. doi:10.2174/1573408012666160727165940
Ambu, G., Chaudhary, R. P., Mariotti, M., and Cornara, L. (2020). Traditional Uses of Medicinal Plants by Ethnic People in the Kavrepalanchok District, Central Nepal. Plants (Basel) 9, 759. doi:10.3390/plants9060759
A. Mitscher, L., Shipchandler, M., D. Hollis Showalter, H., and S. Bathala, M. (1975). Antimicrobial Agents from Higher Plants. Synthesis in the Canthin-6-One (6H-Indolo[3,2,1-De][1,5]naphthyridin-6-One) Series. Heterocycles 3, 7–14. doi:10.3987/r-1975-01-0007
Anita, B. R., and Praveena, B. (2020). A Review on Anti-cancer Potential of Phytochemicals Isolated from Simarouba Glauca. Indo Am. J. Pharm. Sci. 7, 281–286. doi:10.5281/zenodo.3746442
Arias, J. L., Cebrián-Torrejón, G., Poupon, E., Fournet, A., and Couvreur, P. (2011). Biodegradable Polymeric Nanoformulation Based on the Antiprotozoal Canthin-6-One. J. Nanopart Res. 13, 6737–6746. doi:10.1007/s11051-011-0580-z
Bai, M., Zhao, W.-Y., Xu, W., Zhang, Y.-Y., Huang, X.-X., and Song, S.-J. (2020). Triterpenoids from Picrasma Quassioides with Their Cytotoxic Activities. Phytochemistry Lett. 39, 128–131. doi:10.1016/j.phytol.2020.07.014
Baldé, M. A., Tuenter, E., Matheeussen, A., Traoré, M. S., Cos, P., Maes, L., et al. (2021). Bioassay-guided Isolation of Antiplasmodial and Antimicrobial Constituents from the Roots of Terminalia Albida. J. Ethnopharmacol. 267, 113624. doi:10.1016/j.jep.2020.113624
Benhamou, R. I., Shaul, P., Herzog, I. M., and Fridman, M. (2015). Di-N-Methylation of Anti-Gram-Positive Aminoglycoside-Derived Membrane Disruptors Improves Antimicrobial Potency and Broadens Spectrum to Gram-Negative Bacteria. Angew. Chem. Int. Ed. Engl. 54, 13617–13621. doi:10.1002/ange.20150681410.1002/anie.201506814
Biavatti, M. W., Vieira, P. C., da Silva Mfg, M. F. G. F., Fernandes, J. B., Albuquerque, S., Magalhães, C. M., et al. (2001). Chemistry and Bioactivity of Raulinoa Echinata Cowan, an Endemic Brazilian Rutaceae Species. Phytomedicine 8, 121–124. doi:10.1078/0944-7113-00016
Brillatz, T., Jacmin, M., Queiroz, E. F., Marcourt, L., Slacanin, I., Petit, C., et al. (2020a). Zebrafish Bioassay-Guided Isolation of Antiseizure Compounds from the Cameroonian Medicinal Plant Cyperus Articulatus L. Phytomedicine 70, 153175. doi:10.1016/j.phymed.2020.153175
Brillatz, T., Kubo, M., Takahashi, S., Jozukuri, N., Takechi, K., Queiroz, E. F., et al. (2020b). Metabolite Profiling of Javanese Ginger Zingiber Purpureum and Identification of Antiseizure Metabolites via a Low-Cost Open-Source Zebrafish Bioassay-Guided Isolation. J. Agric. Food Chem. 68, 7904–7915. doi:10.1021/acs.jafc.0c02641
Campuzano, I., Bush, M. F., Robinson, C. V., Beaumont, C., Richardson, K., Kim, H., et al. (2012). Structural Characterization of Drug-like Compounds by Ion Mobility Mass Spectrometry: Comparison of Theoretical and Experimentally Derived Nitrogen Collision Cross Sections. Anal. Chem. 84, 1026–1033. doi:10.1021/ac202625t
Casciaro, B., Calcaterra, A., Cappiello, F., Mori, M., Loffredo, M. R., Ghirga, F., et al. (2019). Nigritanine as a New Potential Antimicrobial Alkaloid for the Treatment of Staphylococcus Aureus-Induced Infections. Toxins (Basel) 11, 511. doi:10.3390/toxins11090511
Cebrian-Torrejon, G., Spelman, K., Leblanc, K., Munoz-Durango, K., Gutierrez, S. T., Ferreira, M. E., et al. (2011). The Antiplasmodium Effects of a Traditional South American Remedy: Zanthoxylum Chiloperone Var. Angustifolium against Chloroquine Resistant and Chloroquine Sensitive Strains of Plasmodium Falciparum. Rev. Bras. Farmacogn. 21, 652–661. doi:10.1590/S0102-695X2011005000104
Chen, J., Yan, X. H., Dong, J. H., Sang, P., Fang, X., Di, Y. T., et al. (2009a). Tobacco Mosaic Virus (TMV) Inhibitors from Picrasma Quassioides Benn. J. Agric. Food Chem. 57, 6590–6595. doi:10.1021/jf901632j
Chen, Q., Zhang, S., and Xie, Y. (2018). Characterization of a New Microbial Pictet-Spenglerase NscbB Affording the β-carboline Skeletons from Nocardiopsis Synnemataformans DSM 44143. J. Biotechnol. 281, 137–143. doi:10.1016/j.jbiotec.2018.07.007
Chinese Materia Medica Editorial Committee of National Administration of TCM (1999). Chinese Materia Medica (Volumn 5). Shanghai: Shanghai Science and Technology Press, 13–14.
Cho, S. K., Jeong, M., Jang, D. S., and Choi, J. H. (2018). Anti-inflammatory Effects of Canthin-6-One Alkaloids from Ailanthus Altissima. Planta Med. 84, 527–535. doi:10.1055/s-0043-123349
Cui, Y., Jiang, L., Yu, R., Shao, Y., Mei, L., and Tao, Y. (2019). β-Carboline Alkaloids Attenuate Bleomycin Induced Pulmonary Fibrosis in Mice through Inhibiting NF-Kb/p65 Phosphorylation and Epithelial-Mesenchymal Transition. J. Ethnopharmacol. 243, 112096. doi:10.1016/j.jep.2019.112096
Curcino Vieira, I. J., and Braz-Filho, R. (2006). “Quassinoids: Structural Diversity, Biological Activity and Synthetic Studies,” In Studies In Natural Products Chemistry 33, 433–492. doi:10.1016/S1572-5995(06)80032-3
de Souza Almeida, E. S., Filho, V. C., Niero, R., Clasen, B. K., Balogun, S. O., and de Oliveira Martins, D. T. (2011). Pharmacological Mechanisms Underlying the Anti-ulcer Activity of Methanol Extract and Canthin-6-One of Simaba Ferruginea A. St-Hil. In Animal Models. J. Ethnopharmacol. 134, 630–636. doi:10.1016/j.jep.2011.01.009
Denisov, E., Damoc, E., Lange, O., and Makarov, A. (2012). Orbitrap Mass Spectrometry with Resolving powers above 1,000,000. Int. J. Mass Spectrom. 325-327, 80–85. doi:10.1016/j.ijms.2012.06.009
Dodds, J. N., May, J. C., and McLean, J. A. (2017). Investigation of the Complete Suite of the Leucine and Isoleucine Isomers: Toward Prediction of Ion Mobility Separation Capabilities. Anal. Chem. 89, 952–959. doi:10.1021/acs.analchem.6b04171
Erdemoglu, N., Akkol, E. K., Yesilada, E., and Caliş, I. (2008). Bioassay-guided Isolation of Anti-inflammatory and Antinociceptive Principles from a Folk Remedy, Rhododendron Ponticum L. Leaves. J. Ethnopharmacol. 119, 172–178. doi:10.1016/j.jep.2008.06.021
Fisher, C. M., Croley, T. R., and Knolhoff, A. M. (2021). Data Processing Strategies for Non-targeted Analysis of Foods Using Liquid Chromatography/high-Resolution Mass Spectrometry. Trac Trends Anal. Chem. 136, 116188. doi:10.1016/j.trac.2021.116188
Flora of China Editorial Committee (1997). flora of china (Chinese Version, Volumn 43). Beijing: Science Press, 7–8.
Flora of China Editorial Committee (2008). Flora of China (English Version,volumn 11). St. Louis: Missouri Botanical Garden Press, 102.
Gao, X., Sheng, T., Wu, H., and Gu, Q. (2020). Inventor; Nanjing Agricultural University, Assignee. Preparation of 1-substituted β-carboline Derivatives for Antibacterial Agents. Chinese patent CN 111303148.
Gong, G., Jiang, L., Lin, Q., Liu, W., He, M. F., Zhang, J., et al. (2018). In Vivo toxic Effects of 4-Methoxy-5-Hydroxy-Canthin-6-One in Zebrafish Embryos via Copper Dyshomeostasis and Oxidative Stress. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 204, 79–87. doi:10.1016/j.cbpc.2017.11.014
Gong, G., Lin, Q., Xu, J., Ye, F., Jiang, L., Liu, W., et al. (2016). In Vivo SAR and STR Analyses of Alkaloids from Picrasma Quassioides Identify 1-Hydroxymethyl-8-Hydroxy-β-Carboline as a Novel Natural Angiogenesis Inhibitor. RSC Adv. 6, 9484–9494. doi:10.1039/C5RA22391A
Guihua, D. (2011). Study on chemical constituents and quality evaluation of Ramulus et Folium Picrasmae. Master’s Thesis. Guangzhou (GD): Guangzhou University of Chinese Medicine.
Guo, E., Hu, Y., Du, T., Zhu, H., Chen, L., Qu, W., et al. (2019). Effects of Picrasma Quassioides and its Active Constituents on Alzheimer's Disease In Vitro and In Vivo. Bioorg. Chem. 92, 103258. doi:10.1016/j.bioorg.2019.103258
Guo, X. M., Li, F., Zheng, F. F., Gong, N. N., Li, Y., Feng, W. Z., et al. (2020). (±)-Quassidine K, a Pair of Cytotoxic Bis-β-Carboline Alkaloid Enantiomers from Picrasma Quassioides. Nat. Prod. Res. 34, 489–493. doi:10.1080/14786419.2018.1489388
Guo-hua, S., Wei-hua, J., Fan, Y., and Hou-wen, L. (2015). Three Bis-β-Carboline Alkaloids from Picrasma Quassioides and Their Bioactivities. Chin. Tradit. Herb. Drugs 6, 803–807. doi:10.7501/j.issn.0253-2670.2015.06.004
Han, J. H., Hong, S. R., Ji, O. P., Kwon, H. C., Lee, B. G., Lee, G. N., et al. (2001). Inventor. Antiinflammatory Composition Containing -carboline Derivatives. Korean patent KR, 2001037653.
Hanif, M. A., Bhatti, H. N., Jamil, M. S., Anjum, R. S., Jamil, A., and Khan, M. M. (2010). Antibacterial and Antifungal Activities of Essential Oils Extracted from Medicinal Plants Using CO2 Supercritical Fluid Extraction Technology. Asian J. Chem. 22, 7787–7798.
He, Y., Hu, Z., Li, Q., Huang, J., Li, X. N., Zhu, H., et al. (2017). Bioassay-Guided Isolation of Antibacterial Metabolites from Emericella Sp. TJ29. J. Nat. Prod. 80, 2399–2405. doi:10.1021/acs.jnatprod.7b00077
He, Y., Liu, W., Chen, Z., Zhao, B., Zhong, Z., and Wei, Y. (2008). Research on the In Vitro Bacteriostatic Effect of Quassiawood Alkaloid on Escherichia coli. J. Anhui Agri.Sci. 36, 2777–2778.
Hu, H., Lee-Fong, Y., Peng, J., Hu, B., Li, J., Li, Y., et al. (2021). Comparative Research of Chemical Profiling in Different Parts of Fissistigma Oldhamii by Ultra-High-Performance Liquid Chromatography Coupled with Hybrid Quadrupole-Orbitrap Mass Spectrometry. Molecules 26, 960. doi:10.3390/molecules26040960
Hwisa, N. T., Gindi, S., Rao, C. B., Katakam, P., Rao Chandu, B., and Info, A. (2013). Evaluation of Antiulcer Activity of Picrasma Quassioides Bennett Aqueous Extract in Rodents. VRI-PM 1, 27. doi:10.14259/pm.v1i1.35
Ishaq, H. (2016). Anxiolytic and Antidepressant Activity of Different Methanolic Extracts of Melia Azedarach Linn. Pak. J. Pharm. Sci. 29, 1649–1655.
Jiang, C., Arthur, C. J., and Gates, P. J. (2020). A Computational and Experimental Study of the Fragmentation of L-Leucine, L-Isoleucine and L-Allo-Isoleucine under Collision-Induced Dissociation Tandem Mass Spectrometry. Analyst 145, 6632–6638. doi:10.1039/d0an00778a
Jiang, M. X., and Zhou, Y. J. (2008). Canthin-6-one Alkaloids from Picrasma Quassioides and Their Cytotoxic Activity. J. Asian Nat. Prod. Res. 10, 1009–1012. doi:10.1080/10286020802277956
Jiao, W. H., Chen, G. D., Gao, H., Li, J., Gu, B. B., Xu, T. T., et al. (2015). (±)-Quassidines I and J, Two Pairs of Cytotoxic Bis-β-Carboline Alkaloid Enantiomers from Picrasma Quassioides. J. Nat. Prod. 78, 125–130. doi:10.1021/np500801s
Jiao, W. H., Gao, H., Li, C. Y., Zhao, F., Jiang, R. W., Wang, Y., et al. (2010b). Quassidines A-D, Bis-Beta-Carboline Alkaloids from the Stems of Picrasma Quassioides. J. Nat. Prod. 73, 167–171. doi:10.1021/np900538r
Jiao, W. H., Gao, H., Li, C. Y., Zhou, G. X., Kitanaka, S., Ohmura, A., et al. (2010a). Beta-carboline Alkaloids from the Stems of Picrasma Quassioides. Magn. Reson. Chem. 48, 490–495. doi:10.1002/mrc.2602
Jiao, W. H., Gao, H., Zhao, F., Lin, H. W., Pan, Y. M., Zhou, G. X., et al. (2011). Anti-inflammatory Alkaloids from the Stems of Picrasma Quassioides BENNET. Chem. Pharm. Bull. (Tokyo) 59, 359–364. doi:10.1248/cpb.59.359
Jung, Y.-T., Lee, I.-S., Whang, K., and Yu, M.-H. (2012). Antioxidant Effects of Picrasma quassioides and Chamaecyparis obtusa (S. et Z.) ENDL Extracts. J. Life Sci. 22, 354–359. doi:10.5352/jls.2012.22.3.354
Kita, K., Yabu, Y., Nagai, K., Minagawa, N., and Hosokawa, K. , Inventor; Arigen Pharmaceuticals Inc., Can. , Assignee. Indole Alkaloids as Enhancers for Antiprotozoal Activity of Ascofuranone, Their Compositions and Kits, and Treatment of Protozoan Diseases with Them. Japanese patent JP 2004231601 (2004).
Koike, K., and Ohmoto, T. (1986). Studies on the Alkaloids from Picrasma Quassioides Bennet. VII. Structures of .BETA.-carboline Dimer Alkaloids, Picrasidines-H and -R. Chem. Pharm. Bull. 34, 2090–2093. doi:10.1248/cpb.34.2090
Kong, L., Wang, B., Yang, X., Guo, H., Zhang, K., Zhu, Z., et al. (2017). Picrasidine I from Picrasma Quassioides Suppresses Osteoclastogenesis via Inhibition of RANKL Induced Signaling Pathways and Attenuation of ROS Production. Cell. Physiol. Biochem. 43, 1425–1435. doi:10.1159/000481874
Krettler, C. A., and Thallinger, G. G. (2021). A Map of Mass Spectrometry-Based In Silico Fragmentation Prediction and Compound Identification in Metabolomics. Brief. Bioinform 22. doi:10.1093/bib/bbab073
Kuo, P. C., Shi, L. S., Damu, A. G., Su, C. R., Huang, C. H., Ke, C. H., et al. (2003). Cytotoxic and Antimalarial Beta-Carboline Alkaloids from the Roots of Eurycoma Longifolia. J. Nat. Prod. 66, 1324–1327. doi:10.1021/np030277n
Lagoutte, D., Nicolas, V., Poupon, E., Fournet, A., Hocquemiller, R., Libong, D., et al. (2008). Antifungal Canthin-6-One Series Accumulate in Lipid Droplets and Affect Fatty Acid Metabolism in Saccharomyces cerevisiae. Biomed. Pharmacother. 62, 99–103. doi:10.1016/j.biopha.2007.07.014
Lai, Z.-Q., Liu, W.-H., Ip, S.-P., Liao, H.-J., Yi, Y.-Y., Qin, Z., et al. (2014). Seven Alkaloids from Picrasma Quassioides and Their Cytotoxic Activities. Chem. Nat. Compd. 50, 884–888. doi:10.1007/s10600-014-1106-6
Lee, J. J., Oh, C.-H., Yang, J. H., Baek, N.-I., Kim, S.-H., Cho, C. H., et al. (2009). Cytotoxic Alkaloids from the wood of Picrasma Quassioides. J. Korean Soc. Appl. Biol. Chem. 52, 663–667. doi:10.3839/jksabc.2009.110
Liu, P., Li, H., Luan, R., Huang, G., Liu, Y., Wang, M., et al. (2019a). Identification of β-carboline and Canthinone Alkaloids as Anti-inflammatory Agents but with Different Inhibitory Profile on the Expression of iNOS and COX-2 in Lipopolysaccharide-Activated RAW 264.7 Macrophages. J. Nat. Med. 73, 124–130. doi:10.1007/s11418-018-1251-5
Liu, Y., Lai, L., Ju, Y., Liu, C., and Meng, D. (2019b). Chemical Constituents and Synergistic Anti-gout Studies on Eurycoma Longifolia and Potential Mechanisms Evaluation Based on Systemic Analysis Approach. Bioorg. Chem. 92, 103302. doi:10.1016/j.bioorg.2019.103302
Logendra, S., Ribnicky, D. M., Yang, H., Poulev, A., Ma, J., Kennelly, E. J., et al. (2006). Bioassay-guided Isolation of Aldose Reductase Inhibitors from Artemisia Dracunculus. Phytochemistry 67, 1539–1546. doi:10.1016/j.phytochem.2006.05.015
Lou, L. L., Yao, G. D., Wang, J., Zhao, W. Y., Wang, X. B., Huang, X. X., et al. (2018). Enantiomeric Neolignans from Picrasma Quassioides Exhibit Distinctive Cytotoxicity on Hepatic Carcinoma Cells through ROS Generation and Apoptosis Induction. Bioorg. Med. Chem. Lett. 28, 1263–1268. doi:10.1016/j.bmcl.2018.03.043
Makong, Y. S., Mouthé Happi, G., Djouaka Bavoua, J. L., Wansi, J. D., Nahar, L., Kamdem Waffo, A. F., et al. (2019). Cytotoxic Stilbenes and Canthinone Alkaloids from Brucea Antidysenterica (Simaroubaceae). Molecules 24, 4412. doi:10.3390/molecules24234412
Malone, K. M., and Gordon, S. V. (2016). Antibiotic Methylation: A New Mechanism of Antimicrobial Resistance. Trends Microbiol. 24, 771–772. doi:10.1016/j.tim.2016.08.003
Medical team of, C. P. L. A. (1972). Clinical Application of Chinese Herbal Medicine. Shanxiongdan. J. New Med. 26, 0956.
Meis, J. F., and Chowdhary, A. (2018). Candida Auris: a Global Fungal Public Health Threat. Lancet Infect. Dis. 18, 1298–1299. doi:10.1016/S1473-3099(18)30609-1
Meng, C. (2007).Alkaloids from Picrasma Quassioies (D.Dom)Benn. And Their Anti-inflammatory and Antibacterial Activities. Master’s Thesis. Yantai: Yantai University.
Ministry of Food and Drug Safety (2012). Korean Pharmacopeia (Tenth Edition,Monographs,Part II). Cheongju, Korea: Ministry of Food and Drug Safety, 1344.
Mohd Jamil, M. D. H., Taher, M., Susanti, D., Rahman, M. A., and Zakaria, Z. A. (2020). Phytochemistry, Traditional Use and Pharmacological Activity of Picrasma Quassioides: A Critical Reviews. Nutrients 12, 2584. doi:10.3390/nu12092584
Noreldeen, H. A. A., Liu, X., Wang, X., Fu, Y., Li, Z., Lu, X., et al. (2018). Quantitative Structure-Retention Relationships Model for Retention Time Prediction of Veterinary Drugs in Food Matrixes. Int. J. Mass Spectrom. 434, 172–178. doi:10.1016/j.ijms.2018.09.022
Ohmoto, T., and Koike, K. (1988). Antiherpes Activity of Simaroubaceae Alkaloids In Vitro. Shoyakugaku Zasshi 42, 160–162.
Ohmoto, T., and Koike, K. (1982). Studies on the Constituents of Picrasma Quassioides Bennet. I. On the Alkaloidal Constituents. Chem. Pharm. Bull. 30, 1204–1209. doi:10.1248/cpb.30.1204
Omoja, V., Ihedioha, T., Eke, G., PeterAjuzie, I., and Okezie, S. (2014). Evaluation of the Acute Toxicity, Phytochemical Constituents and Anti - Ulcer Properties of Methanolic Leaf Extract of Annona Muricata in Mice. J. Intercult Ethnopharmacol 3, 37–43. doi:10.5455/jice.20140111103203
Othman, R., Ibrahim, H., Mohd, M. A., Mustafa, M. R., and Awang, K. (2006). Bioassay-guided Isolation of a Vasorelaxant Active Compound from Kaempferia Galanga L. Phytomedicine 13, 61–66. doi:10.1016/j.phymed.2004.07.004
Parvez, M. K., Al-Dosari, M. S., Arbab, A. H., Al-Rehaily, A. J., and Abdelwahid, M. A. S. (2020). Bioassay-guided Isolation of Anti-hepatitis B Virus Flavonoid Myricetin-3-O-Rhamnoside along with Quercetin from Guiera Senegalensis Leaves. Saudi Pharm. J. 28, 550–559. doi:10.1016/j.jsps.2020.03.006
Proctor, D. M., Dangana, T., Sexton, D. J., Fukuda, C., Yelin, R. D., Stanley, M., et al. (2021). Integrated Genomic, Epidemiologic Investigation of Candida Auris Skin Colonization in a Skilled Nursing Facility. Nat. Med. 27, 1401–1409. doi:10.1038/s41591-021-01383-w
Randall, C. P., Mariner, K. R., Chopra, I., and O'Neill, A. J. (2013). The Target of Daptomycin Is Absent from Escherichia coli and Other Gram-Negative Pathogens. Antimicrob. Agents Chemother. 57, 637–639. doi:10.1128/AAC.02005-12
Rashed, K., Said, A., and Ahmed, M. (2013). Antiviral Activity and Phytochemical Analysis of Ailanthus Excelsa Roxb Bark. J. For. Prod. Ind. 2, 30–33.
Reddy, P. O. V., Shekar, K. P. C., Khandagale, S. B., Hara, D., Son, A., Ito, T., et al. (2019). Easy Access to Water-Soluble Cationic Porphyrin-β -Carboline Conjugates as Potent Photocytotoxic and DNA Cleaving Agents. Asian J. Org. Chem. 8, 269–274. doi:10.1002/ajoc.201800649
Ren, J.-X., Bai, M., Zhao, W.-Y., Huang, X.-X., and Song, S.-J. (2020). Chemical Constituents from Picrasma Quassioides (D.Don) Benn. And Their Network Analysis of Chemotaxonomic Significance. Biochem. Syst. Ecol. 93, 104160. doi:10.1016/j.bse.2020.104160
Ross, S. A., Krishnaven, K., Radwan, M. M., Takamatsu, S., and Burandt, C. L. (2008). Constituents of Zanthoxylum Flavum and Their Antioxidant and Antimalarial Activities. Nat. Product. Commun. 3, 1934578X0800300–794. doi:10.1177/1934578x0800300521
Saiin, C., Junwised, J., Junwised, J., Nuengchamnong, N., and Ingkaninan, K. (2019). Determination of β-carbolines in Thai Picrasma Javanica Bl.; the Source of Potential Antimalarial Agents. Adv. Med. Plant Res. 7, 61–67. doi:10.30918/ampr.72.19.025
Sasaki, T., Li, W., Ohmoto, T., and Koike, K. (2016). Evaluation of Canthinone Alkaloids as Cerebral Protective Agents. Bioorg. Med. Chem. Lett. 26, 4992–4995. doi:10.1016/j.bmcl.2016.09.006
Savidge, T., and Sorg, J. A. (2019). Role of Bile in Infectious Disease: the Gall of 7α-Dehydroxylating Gut Bacteria. Cell Chem. Biol. 26, 1–3. doi:10.1016/j.chembiol.2018.12.010
Schymanski, E. L., Jeon, J., Gulde, R., Fenner, K., Ruff, M., Singer, H. P., et al. (2014). Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 48, 2097–2098. doi:10.1021/es5002105
Shi, Y., Liu, X., Fredimoses, M., Song, M., Chen, H., Liu, K., et al. (2018). FGFR2 Regulation by Picrasidine Q Inhibits the Cell Growth and Induces Apoptosis in Esophageal Squamous Cell Carcinoma. J. Cel. Biochem. 119, 2231–2239. doi:10.1002/jcb.26385
Shin, H. J., Lee, H. S., and Lee, D. S. (2010). The Synergistic Antibacterial Activity of 1-Acetyl-Beta-Carboline and Beta-Lactams against Methicillin-Resistant Staphylococcus aureus (MRSA). J. Microbiol. Biotechnol. 20, 501–505. doi:10.4014/jmb.0910.10019
Shin, N. R., Shin, I. S., Jeon, C. M., Hong, J. M., Oh, S. R., Hahn, K. W., et al. (2014). Inhibitory Effects of Picrasma Quassioides (D.Don) Benn. On Airway Inflammation in a Murine Model of Allergic Asthma. Mol. Med. Rep. 10, 1495–1500. doi:10.3892/mmr.2014.2322
Sorokina, M., Merseburger, P., Rajan, K., Yirik, M. A., and Steinbeck, C. (2021). COCONUT Online: Collection of Open Natural Products Database. J. Cheminform. 13, 2. doi:10.1186/s13321-020-00478-9
Su, R. H., Kim, M., Yamamoto, T., and Takahashi, S. (1990). Antifeeding Constituents of Phellodendron Chinense Fruit against Reticulitermes Speratus. J. Pestic. Sci. 15, 567–572. doi:10.1584/jpestics.15.567
Sundar, J. V., and Aarthy, T. (2020). Structure-based Screetning of Potential Triterpenoids for Anti-viral Activity against SARS-CoV-2 and Related Coronaviruses [Preprint]. Available at: https://chemrxiv.org/engage/chemrxiv/article-details/60c74e64567dfe3d57ec54ef (Accessed September 15, 2021).
The Ministry of Health and Labour and Welfare (2016). The Japanese Pharmacopoeia. Tokyo: Pharmaceuticals and Medical Devices Agency. 17 edition.
Tyrkkö, E., Pelander, A., and Ojanperä, I. (2010). Differentiation of Structural Isomers in a Target Drug Database by LC/Q-TOFMS Using Fragmentation Prediction. Drug Test. Anal. 2, 259–270. doi:10.1002/dta.134
Van Baelen, G., Hostyn, S., Dhooghe, L., Tapolcsányi, P., Mátyus, P., Lemière, G., et al. (2009). Structure-activity Relationship of Antiparasitic and Cytotoxic Indoloquinoline Alkaloids, and Their Tricyclic and Bicyclic Analogues. Bioorg. Med. Chem. 17, 7209–7217. doi:10.1016/j.bmc.2009.08.057
Veni, A., Lokeswari, T. S., Krishna Kumari, G. N., Gayathri, D., and Sudandiradoss, C. (2020). Bioactivity of Melianone against Salmonella and In Silico Prediction of a Membrane Protein Target. 3 Biotech. 10, 460. doi:10.1007/s13205-020-02441-9
Warrier, T., Kapilashrami, K., Argyrou, A., Ioerger, T. R., Little, D., Murphy, K. C., et al. (2016). N-methylation of a Bactericidal Compound as a Resistance Mechanism in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A. 113, E4523–E4530. doi:10.1073/PNAS.1606590113
Weiqi, F., Jinhua, W., Wan, L., Lin, W., and Guanhua, D. (2019). The historical cognition and evaluation of the toxicity of picrasmaeramulus et folium. Pharmacol. Clin. Chin. Mater. Med. 35, 154–156. doi:10.13412/j.cnki.zyyl.2019.02.035
Willett, P., Barnard, J. M., and Downs, G. M. (1998). Chemical Similarity Searching. J. Chem. Inf. Comput. Sci. 38, 983–996. doi:10.1021/ci9800211
World Health Organization (2017). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (Accessed September 15, 2021).
Wu, W., Shao, M., Zhai, D., Liu, J., and Teng, H. (2007). Inventor; Shengke Natural Drug Academy Co., Ltd. Hainan, Assignee. Medical Composition Having Anti-inflammatory and Anti-infectious Function. Beijing: China National Intellectual Property Administration. CN 101036647A.
Xie, D. P., Gong, Y. X., Jin, Y. H., Ren, C. X., Liu, Y., Han, Y. H., et al. (2020). Anti-tumor Properties of Picrasma Quassioides Extracts in H-RasG12V Liver Cancer Are Mediated Through ROS-dependent Mitochondrial Dysfunction. Anticancer Res. 40, 3819–3830. doi:10.21873/ANTICANRES.14371
Xu, J., Xiao, D., Lin, Q. H., He, J. F., Liu, W. Y., Xie, N., et al. (2016). Cytotoxic Tirucallane and Apotirucallane Triterpenoids from the Stems of Picrasma Quassioides. J. Nat. Prod. 79, 1899–1910. doi:10.1021/acs.jnatprod.5b01137
Yamashita, N., Kondo, M., Zhao, S., Li, W., Koike, K., Nemoto, K., et al. (2017). Picrasidine G Decreases Viability of MDA-MB 468 EGFR-Overexpressing Triple-Negative Breast Cancer Cells through Inhibition of EGFR/STAT3 Signaling Pathway. Bioorg. Med. Chem. Lett. 27, 2608–2612. doi:10.1016/j.bmcl.2017.03.061
Yan, Y. F., Yang, C. J., Shang, X. F., Zhao, Z. M., Liu, Y. Q., Zhou, R., et al. (2020). Bioassay-guided Isolation of Two Antifungal Compounds from Magnolia Officinalis, and the Mechanism of Action of Honokiol. Pestic. Biochem. Physiol. 170, 104705. doi:10.1016/j.pestbp.2020.104705
Yang, L., Liu, S., Liu, R., and He, J. (2020). Bioassay-guided Isolation of Cyclooxygenase-2 Inhibitory and Antioxidant Phenylpropanoid Derivatives from the Roots of Dendropanax Dentiger. Bioorg. Chem. 104, 104211. doi:10.1016/j.bioorg.2020.104211
Zhang, J., Wang, C. X., Song, X. J., Li, S., Fan, C. L., Chen, G. D., et al. (2019a). A New Cinnamamide Derivative and Two New β-carboline Alkaloids from the Stems of Picrasma Quassioides. Fitoterapia 139, 104375. doi:10.1016/j.fitote.2019.104375
Zhang, J., Zhao, S. S., Xie, J., Yang, J., Chen, G. D., Hu, D., et al. (2020). N-methoxy-β-carboline Alkaloids with Inhibitory Activities against Aβ42 Aggregation and Acetylcholinesterase from the Stems of Picrasma Quassioides. Bioorg. Chem. 101, 104043. doi:10.1016/j.bioorg.2020.104043
Zhang, Y., Liu, Y. B., Li, Y., Ma, S. G., Li, L., Qu, J., et al. (2013). Sesquiterpenes and Alkaloids from the Roots of Alangium Chinense. J. Nat. Prod. 76, 1058–1063. doi:10.1021/np4000747
Zhang, Y., Pan, Z., Rong, C., Shao, Y., Wang, Y., and Yu, K. (2019b). Transformation of Antibacterial Agent Roxithromycin in Sodium Hypochlorite Disinfection Process of Different Water Matrices. Separat. Purif. Tech. 212, 528–535. doi:10.1016/j.seppur.2018.11.061
Zhao, W., Yu, J., Su, Q., Liang, J., Zhao, L., Zhang, Y., et al. (2013). Antihypertensive Effects of Extract from Picrasma Quassiodes (D. Don) Benn. In Spontaneously Hypertensive Rats. J. Ethnopharmacol. 145, 187–192. doi:10.1016/j.jep.2012.10.049
Zhao, W. Y., Chen, J. J., Zou, C. X., Zhang, Y. Y., Yao, G. D., Wang, X. B., et al. (2019). New Tirucallane Triterpenoids from Picrasma Quassioides with Their Potential Antiproliferative Activities on Hepatoma Cells. Bioorg. Chem. 84, 309–318. doi:10.1016/j.bioorg.2018.11.049
Zhao, W. Y., Song, X. Y., Zhao, L., Zou, C. X., Zhou, W. Y., Lin, B., et al. (2019c). Quassinoids from Picrasma Quassioides and Their Neuroprotective Effects. J. Nat. Prod. 82, 714–723. doi:10.1021/acs.jnatprod.8b00470
Zhao, W. Y., Zhou, W. Y., Chen, J. J., Yao, G. D., Lin, B., Wang, X. B., et al. (2019d). Enantiomeric β-carboline Dimers from Picrasma Quassioides and Their Anti-hepatoma Potential. Phytochemistry 159, 39–45. doi:10.1016/j.phytochem.2018.12.002
Zhao, Y., Zhao, Q., and Lu, Q. (2020). Purification, Structural Analysis, and Stability of Antioxidant Peptides from Purple Wheat Bran. BMC Chem. 14, 58–12. doi:10.1186/s13065-020-00708-z
Keywords: Picrasma quassioides, kumu, beta-carboline, orbitrap elite, MS Fragmenter, fragmentation prediction
Citation: Hu H, Hu C, Peng J, Ghosh AK, Khan A, Sun D and Luyten W (2021) Bioassay-Guided Interpretation of Antimicrobial Compounds in Kumu, a TCM Preparation From Picrasma quassioides’ Stem via UHPLC-Orbitrap-Ion Trap Mass Spectrometry Combined With Fragmentation and Retention Time Calculation. Front. Pharmacol. 12:761751. doi: 10.3389/fphar.2021.761751
Received: 20 August 2021; Accepted: 16 September 2021;
Published: 27 October 2021.
Edited by:
Mukhlesur Rahman, University of East London, United KingdomReviewed by:
Qiang Ma, Chinese Academy of Inspection and Quarantine (CAIQ), ChinaTing Han, Second Military Medical University, China
Copyright © 2021 Hu, Hu, Peng, Ghosh, Khan, Sun and Luyten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haibo Hu, haibo.hu@kuleuven.be; Walter Luyten, walter.luyten@kuleuven.be
 Haibo Hu
Haibo Hu Changling Hu3
Changling Hu3 Walter Luyten
Walter Luyten