The pharmacogenomic landscape of an Indigenous Australian population
- 1Hunter Medical Research Institute, The University of Newcastle, New Lambton, NSW, Australia
- 2The University of Newcastle, Callaghan, NSW, Australia
- 3Maitland Hospital, Metford, NSW, Australia
- 4Hunter Medical Research Institute, New Lambton, NSW, Australia
- 5Lake Macquarie Private Hospital, Gateshead, NSW, Australia
A Commentary on
The pharmacogenomic landscape of an Indigenous Australian population
by Samarasinghe SR, Hoy W, Jadhao S, McMorran BJ, Guchelaar H-J and Nagaraj SH (2023). Front. Pharmacol. 14:1180640. doi: 10.3389/fphar.2023.1180640
We write to commend Samarasinghe and colleagues on their collaborative and progressive research in Indigenous Australian health and to acknowledge the important clinical implications of some of their findings. We address the significance of this article as it pertains to the safe prescription of chemotherapies for solid organ cancers.
Samarasinghe et al. conducted an analysis utilising whole genome sequencing on blood samples from 473 Tiwi Indigenous people looking specifically at the allele frequency of Very Important Pharmacogenes (VIP), as determined by PharmKGB (pharmkgb.org) (Samarasinghe et al., 2023). Our specific interest relates to the dihydropyrimidine dehydrogenase (DPYD) and uridine diphosphate-glucuronosyltransferase isoform 1A1 (UGT1A1) gene data.
Typical DPYD variants with clinical significance described within Caucasian populations include 4 main variants; c.1905 + 1G>A, (*2A, rs3918290), c.1679T>G (*13, rs55886062), c.2846A>T (rs67376798) and c.1236G>A/HapB3 (rs56038477) (Amstutz et al., 2018). Allele frequencies for these variants including Tiwi data are summarised in Table 1. Caucasian individuals who carry these variants typically have a deficiency in the dihydropyrimidine dehydrogenase (DPD) enzyme, the critical enzyme involved in the metabolism of fluoropyrimidine chemotherapies 5-fluorouracil and capecitabine, and can develop significant and life-threatening toxicity on exposure to these medications (Froehlich et al., 2015). Interestingly, of the 160+ known DPYD variants only seven distinct DPYD variants were identified within this Tiwi cohort; six variants deemed ‘normal metabolisers’ and c.1236G>A which is known to be clinically significant (Amstutz et al., 2018; Varughese et al., 2020; Samarasinghe et al., 2023). It is very significant that none of the 473 individuals screened carried c.1905 + 1G>A, c.1679T>G, c.2846A>T variants. Only one person was identified to carry c.1236G>A (HapB) in compound heterozygosity with ‘normal metaboliser’ variant c.1627A>G (*5, rs1801159). Fifty-seven individuals were found to be heterozygote for c.1627A>G with an additional 3 homozygote carriers. Nine individuals were found to carry c.2194G>A (*6, rs1801160). Both DPYD c.1627A>G and c.2194G>A are described as ‘non-significant’ variants in international guidelines, though there is conflicting data from non-Caucasian populations describing DPD deficiency and FP toxicity in carriers (Zhang et al., 2007; Offer et al., 2013; Patil et al., 2016; Amstutz et al., 2018; Naushad et al., 2020; White et al., 2021). Both these variants are found in higher frequency in the Tiwi population than other ethnic groups (Table 1) and so genotype/phenotype correlation should be performed to establish whether Tiwi carriers are truly ‘normal metabolisers’. We acknowledge that the small population size likely contributes to the high allele frequencies. We also note that a small sample size may account for the absence of rare variants such as c.1679T>G.
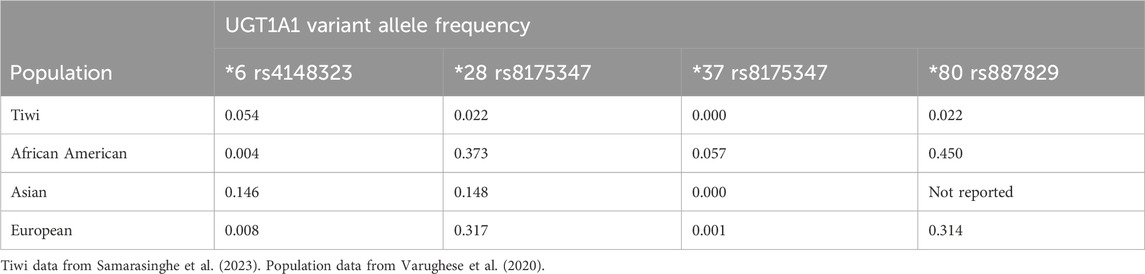
UGT1A1 was also sequenced by Samarasinghe et al in relation to the metabolism of antiretroviral Atazanavir (Samarasinghe et al., 2023). This gene holds clinical significance in the administration of irinotecan chemotherapy (Iyer et al., 1998). UGT1A1 is the critical enzyme of irinotecan metabolism, and all heterozygote carriers within this cohort are ‘intermediate metabolisers’ according to pharmacogenetics guidelines for irinotecan prescribing (Hulshof et al., 2022). Twenty-five heterozygote carriers of *6 and 18 heterozygous carriers of *28 (in compound with *80) were identified (Varughese et al., 2020; Hulshof et al., 2022; Samarasinghe et al., 2023). One homozygote of 28* (+80*) was identified, inferring ‘poor metabolism’ of irinotecan. Another significant variant, *37, was not identified within this cohort, and is typically only identified in African ethnic communities. Allele frequencies for these variants, including Tiwi data are described in Table 2. The clinical significance of *80 alone without compound of *28 or *37 is uncertain and it is not included as a clinically significant variant in current prescribing guidelines (Hulshof et al., 2022).
These data highlight the importance of conducting genomic sequencing in ethnically diverse populations to better understand VIP allele frequencies and improve pharmacogenomic (PGx) prescribing universally, not just for Caucasian communities. Due to the potential for ethnicity-based differences in gene interactions, additional effort should be made to determine the phenotype of individuals found to harbour clinically significant variants, and phenotyping could extend to all DPYD variants identified within the Tiwi community, as ‘normal metaboliser’ variants may in fact show different expression in this and other Indigenous populations. Larger population sampling within ethnic minority populations would help to establish the frequency of very rare variants and allow more rigorous extrapolation of polymorphism frequencies.
Fluoropyrimidines and irinotecan chemotherapies are prescribed for over 16,000 Australians per year for the treatment of solid organ malignancies (Australian Institute of Health and Welfare A, 2019). In various countries across Europe patients are genotyped prior to receiving fluoropyrimidine chemotherapies as standard of care. Patients found to hold certain DPYD variants have chemotherapy doses adjusted prior to exposure to reduce the likelihood of severe toxicity (Deenen et al., 2016; Henricks et al., 2018). This is not currently mandated in Australia, despite recognition of its importance in both international and national prescribing guidelines (Argiles et al., 2020; White et al., 2022; NSW, 2023). There is increasing data to show that differing ethnic populations may not express the same phenotypically significant DPYD variants that are important in European Caucasian communities and may carry alternate variants that have clinical significance (White et al., 2021). It is extremely important that as Australia moves toward developing PGx screening that we consider our ethnically diverse community and invest effort in determining genomic differences that need to be included in our screening modalities, rather than modelling our approach directly from European data.
Not enough is known about the clinical significance of the variants identified in the Tiwi community and in fact in many other non-European ethnicities. Further research is required before dose-personalisation of potentially life-threatening drugs can be developed. Understanding the genotype/phenotype interplay for variants determined in non-Caucasian communities will help to guide dose-personalisation to allow for safe administration of necessary chemotherapeutic agents such as fluoropyrimidines and irinotecan while reducing the risk of severe chemotherapy induced toxicity and even treatment related death. PGx gene panels can be tailored to the reflect DPYD genetic expression within local jurisdictions for national screening efforts (Suarez-Kurtz et al., 2023). Indigenous communities throughout Australia are as diverse in geographical location as they are in cultural practice and collaborative effort to develop culturally sensitive and inclusive genetic screening to determine important variants should be sought in an effort to improve safe prescribing for all Australian cancer patients.
We support the continuation and expansion of collaborative research such as this, in conjunction with Indigenous communities, to explore and enrich our understanding of the genomic landscape of our diverse ethnic populations. This will help to tailor health interventions such as pharmacogenomic screening programmes like prospective DPYD/UGT1A1 genotyping that will be meaningful for all ethnicities.
Author contributions
CW: Writing–original draft, Writing–review and editing. CP: Writing–review and editing. RS: Writing–review and editing. SA: Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amstutz, U., Henricks, L. M., Offer, S. M., Barbarino, J., Schellens, J. H. M., Swen, J. J., et al. (2018). Clinical pharmacogenetics implementation consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin. Pharmacol. Ther. 103 (2), 210–216. doi:10.1002/cpt.911
Argiles, G., Tabernero, J., Labianca, R., Hochhauser, D., Salazar, R., Iveson, T., et al. (2020). Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 31 (10), 1291–1305. doi:10.1016/j.annonc.2020.06.022
Australian Institute of Health and Welfare A (2019) Cancer in Australia 2019. Canberra: Australian Institute of Health and Welfare.
Deenen, M. J., Meulendijks, D., Cats, A., Sechterberger, M. K., Severens, J. L., Boot, H., et al. (2016). Upfront genotyping of DPYD*2A to individualize fluoropyrimidine therapy: a safety and cost analysis. J. Clin. Oncol. 34 (3), 227–234. doi:10.1200/JCO.2015.63.1325
Froehlich, T. K., Amstutz, U., Aebi, S., Joerger, M., and Largiader, C. R. (2015). Clinical importance of risk variants in the dihydropyrimidine dehydrogenase gene for the prediction of early-onset fluoropyrimidine toxicity. Int. J. Cancer 136 (3), 730–739. doi:10.1002/ijc.29025
Henricks, L. M., Lunenburg, C., de Man, F. M., Meulendijks, D., Frederix, G. W. J., Kienhuis, E., et al. (2018). DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: a prospective safety analysis. Lancet Oncol. 19 (11), 1459–1467. doi:10.1016/S1470-2045(18)30686-7
Hulshof, E. C., Deenen, M. J., Nijenhuis, M., Soree, B., de Boer-Veger, N. J., Buunk, A. M., et al. (2022). Dutch pharmacogenetics working group (DPWG) guideline for the gene-drug interaction between UGT1A1 and irinotecan. Eur. J. Hum. Genet. 31, 982–987. doi:10.1038/s41431-022-01243-2
Iyer, L., King, C. D., Whitington, P. F., Green, M. D., Roy, S. K., Tephly, T. R., et al. (1998). Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J. Clin. Invest. 101 (4), 847–854. doi:10.1172/JCI915
Naushad, S. M., Hussain, T., Alrokayan, S. A., and Kutala, V. K. (2020). Pharmacogenetic profiling of dihydropyrimidine dehydrogenase (DPYD) variants in the Indian population. J. Gene Med. 23, e3289. doi:10.1002/jgm.3289
NSW (2023). Dihydropyrimidine dehydrogenase (DPD) enzyme deficiency. Available at: https://www.eviq.org.au/clinical-resources/side-effect-and-toxicity-management/prophylaxis-and-treatment/1744-dihydropyrimidine-dehydrogenase-dpd-enzyme (Accessed May 24, 2023).
Offer, S. M., Lee, A. M., Mattison, L. K., Fossum, C., Wegner, N. J., and Diasio, R. B. (2013). A DPYD variant (Y186C) in individuals of african ancestry is associated with reduced DPD enzyme activity. Clin. Pharmacol. Ther. 94 (1), 158–166. doi:10.1038/clpt.2013.69
Patil, V. M., Noronha, V., Joshi, A., Zanwar, S., Ramaswamy, A., Arya, S., et al. (2016). Dihydropyrimidine dehydrogenase mutation in neoadjuvant chemotherapy in head and neck cancers: myth or reality? South Asian. J. Cancer 5 (4), 182–185. doi:10.4103/2278-330X.195338
Samarasinghe, S. R., Hoy, W., Jadhao, S., McMorran, B. J., Guchelaar, H. J., and Nagaraj, S. H. (2023). The pharmacogenomic landscape of an Indigenous Australian population. Front. Pharmacol. 14, 1180640. doi:10.3389/fphar.2023.1180640
Suarez-Kurtz, G., Fernandes, V. C., and Elias, A. B. R. (2023). Implementation of DPYD genotyping in admixed American populations: Brazil as a model case. Clin. Pharmacol. Ther. 114 (1), 23–24. doi:10.1002/cpt.2921
Varughese, L. A., Lau-Min, K. S., Cambareri, C., Damjanov, N., Massa, R., Reddy, N., et al. (2020). DPYD and UGT1A1 pharmacogenetic testing in patients with gastrointestinal malignancies: an overview of the evidence and considerations for clinical implementation. Pharmacotherapy 40, 1108–1129. doi:10.1002/phar.2463
White, C., Scott, R., Paul, C. L., and Ackland, S. P. (2022). Pharmacogenomics in the era of personalised medicine. Med. J. Aust. 217, 510–513. doi:10.5694/mja2.51759
White, C., Scott, R. J., Paul, C., Ziolkowski, A., Mossman, D., and Ackland, S. (2021). Ethnic diversity of DPD activity and the DPYD gene: review of the literature. Pharmgenomics Pers. Med. 14, 1603–1617. doi:10.2147/PGPM.S337147
Keywords: fluoropyrimidine, DPYD gene, UGT1A1 gene, ethnicity, pharmacogenenomics and personalised medicine
Citation: White C, Paul C, Scott RJ and Ackland S (2024) Commentary: The pharmacogenomic landscape of an Indigenous Australian population. Front. Pharmacol. 15:1373056. doi: 10.3389/fphar.2024.1373056
Received: 19 January 2024; Accepted: 03 May 2024;
Published: 15 May 2024.
Edited by:
Joseph Ciccolini, Assistance Publique Hôpitaux de Marseille, FranceReviewed by:
Andrea Orellana-Manzano, Facultad de Ciencias de la Vida (FCV), EcuadorAntonin Schmitt, Centre Georges François Leclerc, France
Copyright © 2024 White, Paul, Scott and Ackland. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cassandra White, cassandra.white10@uon.edu.au
 Cassandra White
Cassandra White Christine Paul4,2
Christine Paul4,2 Rodney J. Scott
Rodney J. Scott Stephen Ackland
Stephen Ackland