- Division of Clinical Pharmacology and Toxicology, Department of Biomedicine and Department of Clinical Research, University Hospital Basel, Basel, Switzerland
3,4-Methylenedioxymethamphetamine (MDMA, ecstasy) is a recreational substance also investigated as medication for posttraumatic stress disorder. Dopamine (DA) system stimulation likely contributes to the acute mood effects of amphetamines, including MDMA. Genetic variants, such as single-nucleotide polymorphisms (SNPs), and polymorphic regions of the DA system genes may in part explain interindividual differences in the acute responses to MDMA in humans. We characterized the effects of common genetic variants within genes coding for key players in the DA system including the dopamine D2 receptor (DRD2/ANKK1 rs1800497, DRD2 rs6277, and rs107959), the dopamine transporter (DAT1 rs28363170, rs3836790, rs6347, rs11133767, rs11564774, rs460000, and rs463379), and dopamine D4 receptor [DRD4, variable-number tandem repeat (VNTR)] on the subjective and autonomic response to MDMA (125 mg) in pooled data from randomized, placebo-controlled, crossover studies in a total of 149 healthy subjects. Plasma concentrations of MDMA were used as covariate in the analysis to control for individual pharmacokinetic (metabolic and weight) differences. None of the tested genetic polymorphisms within the DA system altered effects of MDMA when adjusting for multiple comparisons. Genetic variations in genes coding for players of the DA system are unlikely to explain interindividual variations in the acute effects of MDMA in humans.
Introduction
3,4-Methylenedioxymethamphetamine (MDMA; ecstasy) is widely used recreationally for its euphoric effects. Additionally, recent investigations are looking into MDMA as a medication to assist psychotherapy in patients with posttraumatic stress disorder (PTSD) (1–3). MDMA acts mainly as a releaser and reuptake inhibitor of serotonin (5-HT), norepinephrine (NE), and dopamine (DA) via an interaction with the respective transporter (4–7). The subjective effects of MDMA have been shown to mainly depend on transporter-mediated release of 5-HT and NE (8). In animals, however, the possibility was raised that the importance of interaction with the DA system would increase with the amount of drug taken (9). To what extent DA is mediating the acute effects of MDMA in humans is unclear. For example, the positive effects of MDMA were diminished after pharmacological inhibition of DA receptors with haloperidol (10). In addition, MDMA-induced hyperactivity was reduced in knockout mice without the DA receptor D2 gene (DRD2) (11). However, in contrast to the strongly diminished effect of MDMA in subjects with a blocked serotonin transporter, preventing the interaction of MDMA with the DA transporter (DAT) by pretreatment with bupropion or methylphenidate had no effect on the acute mood effects of MDMA in humans (12–15). Studies on the influence of genetic polymorphisms in the DA system could add adjuvant information to this matter and may also explore the role of the DA system in the interindividual differences in the response to MDMA. So far, only genetic variations of the enzymes that are involved in MDMA metabolism (mainly CYP2D6) displayed a robust influence on MDMA plasma levels in several clinical studies (16–18) and also showed a concomitant modulation of the pharmacodynamic effects of MDMA. However, genetic variants of pharmacological targets of MDMA may also alter its pharmacodynamic effects. A few studies explored the role of genetic polymorphisms of the 5-HT, NE, and oxytocin systems and found only minimal influences on acute effects of MDMA (19–23).
The DAT is a key target for many stimulant-type drugs, including cocaine, amphetamine, methylphenidate, and MDMA (6, 24). Additionally, the transporter is involved in various psychiatric disorders and treatment approaches (25–27). Subsequently, genetic polymorphisms within the single copy gene coding for the DAT (DAT1, SLC6A3) were investigated in relation to cocaine dependence and abuse, methamphetamine psychosis, attention-deficit/hyperactivity disorder (ADHD) and treatment, and bipolar disorder (28–36). Two common variable-number tandem repeat (VNTR) polymorphisms were most extensively studied. One, the rs28363170, is located in the 3′ untranslated region (3′UTR) of the DAT1 gene and exhibits 9 or 10 repeats as most common forms (37). Homozygous carriers of the 9-repeat allele were found to be at a higher risk for persistent ADHD, and the 10/10 genotype was associated with ADHD in children (38). Subjects with the 9/9 genotype were less susceptible to the subjective effects of amphetamine (39). However, carriers of at least one 9-repeat allele showed higher ratings of “high,” “any drug effect,” “anxious,” and “stimulated” after cocaine (40). Conversely, homozygous 10-repeat carriers in combination with a 5-repeat allele of the other extensively studied VNTR in the DAT1, the rs3836790, displayed a lower response to “good drug effects,” “bad drug effects,” “depressed,” and “anxious” (40). The rs3836790 VNTR is located in intron 8 of the human DAT1 gene. The most common forms of this VNTR are 5 or 6 repeats (30). A study in a Brazilian sample found a positive association of the 6-repeat allele and cocaine addiction (28). In contrast, another yet smaller case-control study in a Spain sample showed an overrepresentation of the 5/5 genotype in cocaine abusers (33).
MDMA also directly and indirectly interacts with DA receptors (4). Especially the inhibition of the D2 with haloperidol showed a significant reduction in MDMA positive effects (10). MDMA-unrelated pharmacogenetic studies showed a positive association of the minor allele of the DRD2 single-nucleotide polymorphisms (SNPs) rs1079597 and rs1800497 with heroin dependence (41), rs6277 and rs1800497 with nicotine dependence (42), and rs6277 with alcohol dependence in males (43). The VNTR polymorphism within the gene coding for the subtype 4 of the DA receptors (DRD4) is also frequently studied in relation to psychiatric disorders and personality traits (44–47). DRD4 VNTR variations range from 2 repeats to 10 repeats, with 4 and 7 repeats as the most frequent forms (48). The presence of a 7-repeat allele has been linked with personal traits like high novelty seeking, risky decision making, and broad sexual interest (44, 47). Moreover, children and adolescents suffering from ADHD and carrying the 7-repeat allele had to take higher doses of methylphenidate to reach sufficient efficacy (49). This finding is in line with earlier results from an in vitro study showing a reduced sensitivity of the 7-repeat allele toward DA compared with the 2- and 4-repeat allele (50).
The present study is the first to explore the influence of variants within genes coding for the DA system on the acute effects of MDMA in humans. We analyzed DRD2/ANKK1 rs1800497, DRD2 rs6277, and rs107959, DAT1 rs28363170, rs3836790, rs6347, rs11133767, rs11564774, rs460000, and rs463379, DRD4 VNTR and their influence on acute subjective and autonomic effects of MDMA. Given the partially inconclusive pharmacogenetic studies in addition to the unclear degree to which MDMA effects are driven by the interaction with the DA system, we hypothesized that genetic mutations of the DA system would not influence cardiostimulant effects and have only minimal influence on the mood effects of MDMA.
Methods
Study Design
This was a pooled analysis of nine double-blind, placebo-controlled, crossover studies that used similar methods and were conducted in healthy subjects and in the same laboratory (14, 15, 51–55). The studies included a total of 164 healthy subjects. Seven studies included 16 subjects each, for a total of 112 subjects, who received 125 mg MDMA twice, once alone and once after pretreatment with a medication (14, 15, 51–54). Two additional studies included 24 and 28 subjects who received 125 mg MDMA alone, placebo, or other treatments (55; Holze et al., unpublished). In the present analysis, only data from the MDMA-alone and placebo sessions were used. In all of the studies, the washout periods between single-dose administrations of MDMA were at least 7 days to exclude possible carryover effects. The studies were all registered at ClinicalTrials.gov (NCT00886886, NCT00990067, NCT01136278, NCT01270672, NCT01386177, NCT01465685, NCT01771874, NCT01951508, and NCT03019822). All of the studies were approved by the local ethics committee and, if necessary, Swiss Agency for Therapeutic Products (Swissmedic). The studies were conducted in accordance with the Declaration of Helsinki. MDMA administration in healthy subjects was authorized by the Swiss Federal Office for Public Health (BAG), Bern, Switzerland. Written informed consent was obtained from all of the participants. All of the subjects were paid for their participation. Detailed pharmacokinetic and safety data from these studies have been reported elsewhere (17, 18, 56). Test sessions were conducted in a quiet hospital research ward with no more than two research subjects present per session. The participants were comfortably lying in hospital beds and were mostly listening to music and not engaging in physical activities. MDMA was given without food in the fasting state in the morning at 8:00–9:00 AM. A small standardized lunch was served at 12:00–1:00 PM.
Subjects
A total of 164 healthy subjects of European descent, 18–45 years old (mean ± SD = 25.3 ± 4 years), were recruited from the University of Basel campus and participated in the study. One genotyping sample was missing, three participants did not give consent for genotyping, and 11 subjects participated twice (only one participation that included all outcome measures was used), resulting in a final data set of 149 subjects (76 women). The mean ± SD body weight was 69 ± 11 kg (range: 46–97 kg).
The exclusion criteria included a history of psychiatric disorders, physical illness, a lifetime history of illicit drug use more than 10 times (with the exception of past cannabis use), illicit drug use within the past 2 months, and illicit drug use during the study, as determined by urine tests that were conducted before the test sessions, as reported in detail elsewhere (52–54). Fifty-five subjects had prior illicit drug experiences (1–8 times), of which 27 subjects had previously used MDMA (1–5 times), 14 subjects had previously used amphetamine or methamphetamine (1–2 times), 11 subjects had previously used cocaine (1–4 times), eight subjects had previously used lysergic acid diethylamide (1–2 times), and 11 subjects had previously used psilocybin (1–4 times).
Study Drug
(±)MDMA hydrochloride (Lipomed AG, Arlesheim, Switzerland) was administered orally in a single dose of 125 mg prepared as gelatin capsules (25 and 100 mg). Similar amounts of MDMA are found in ecstasy pills (57) and have been used in clinical trials in patients (1, 2). The doses were not adjusted for body weight or sex. The dose per body weight (mean ± SD) was 1.9 ± 0.3 mg/kg (range: 1.3–2.7 mg/kg).
Physiological Effects
Blood pressure, heart rate, and body temperature were assessed repeatedly before and 0, 0.33, 0.67, 1, 1.5, 2, 2.5, 3, 4, 5, and 6 h after MDMA or placebo administration. Systolic and diastolic blood pressures and heart rate were measured using an automatic oscillometric device (OMRON Healthcare Europe NA, Hoofddorp, Netherlands). The measurements were performed in duplicate at an interval of 1 min and after a resting time of at least 10 min. The averages were calculated for the analysis. Mean arterial pressure (MAP) was calculated as diastolic blood pressure + (systolic blood pressure – diastolic blood pressure)/3. The rate pressure product (RPP) was calculated as systolic blood pressure × heart rate. Core (tympanic) temperature was measured using a Genius 2 ear thermometer (Tyco Healthcare Group LP, Watertown, NY, USA). In two studies (N = 46), the 2-h time point was not used.
Pharmacodynamic Measures
Visual Analog Scales (VASs) were repeatedly used to assess subjective effects over time (58). The VASs included for instance “any drug effect,” “good drug effect,” and “stimulated.” The VASs were presented as 100 mm horizontal lines (0–100%), marked from “not at all” on the left to “extremely” on the right. Subjective effects like “concentration,” “appetite,” “tired,” “want to be hugged,” “want to hug,” and “talkative” were bidirectional (±50 mm). Not all VAS components were presented in all studies. Exact numbers of subjects per genotype group are reported in Tables 1–3. The VASs were applied before and 0, 0.33, 0.67, 1, 1.5, 2, 2.5, 3, 4, 5, and 6 h after MDMA or placebo administration. In two studies (N = 46), the 2-h time point is missing; additionally, in one study (N = 21), the 2.5-h time point is also missing.
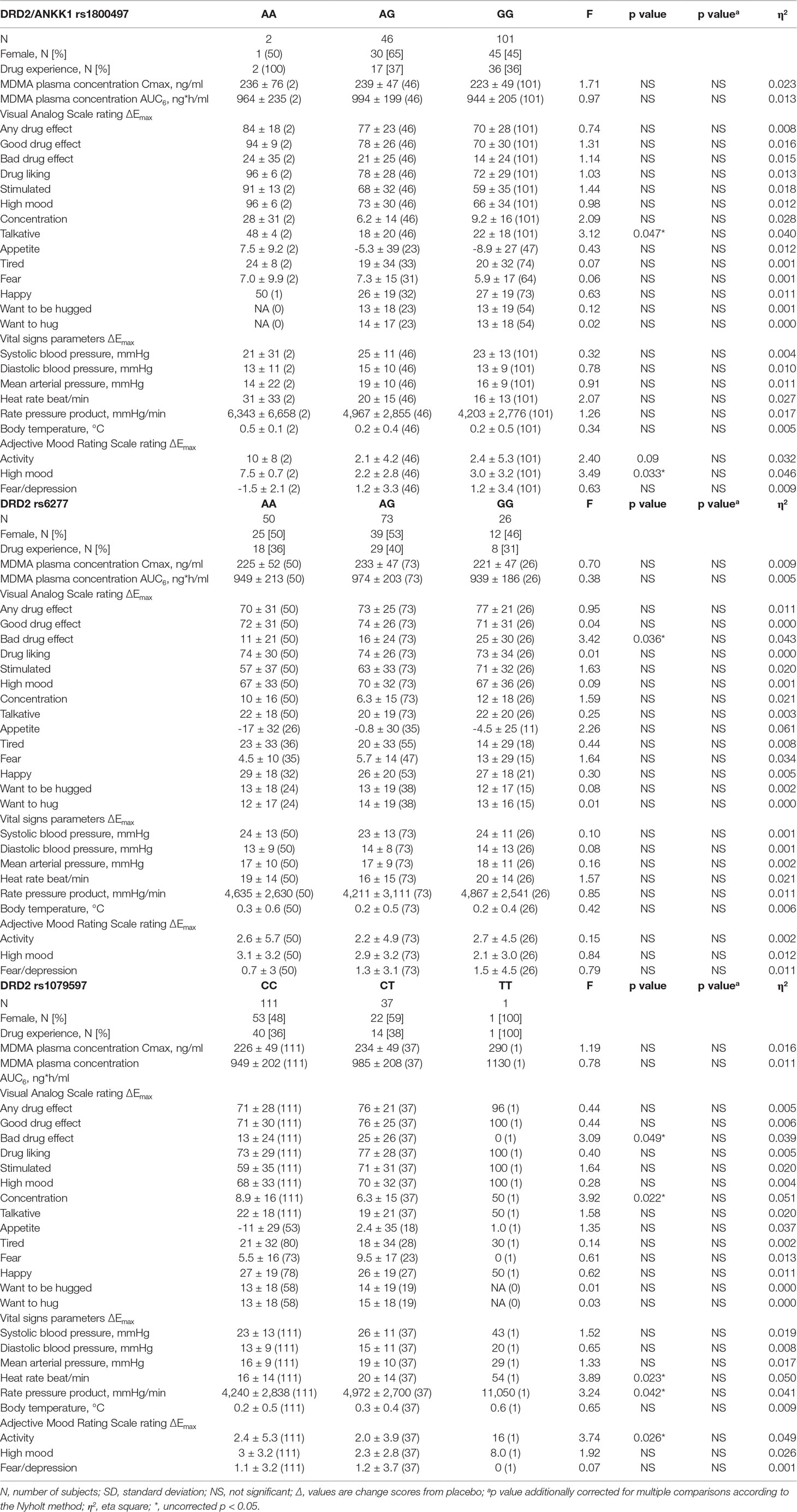
Table 1 Effects of polymorphisms in the dopamine receptor D2 gene on the maximal response to 125 mg MDMA (mean ± SD (N) and statistics) corrected with MDMA AUC6 (exclusive plasma concentrations).
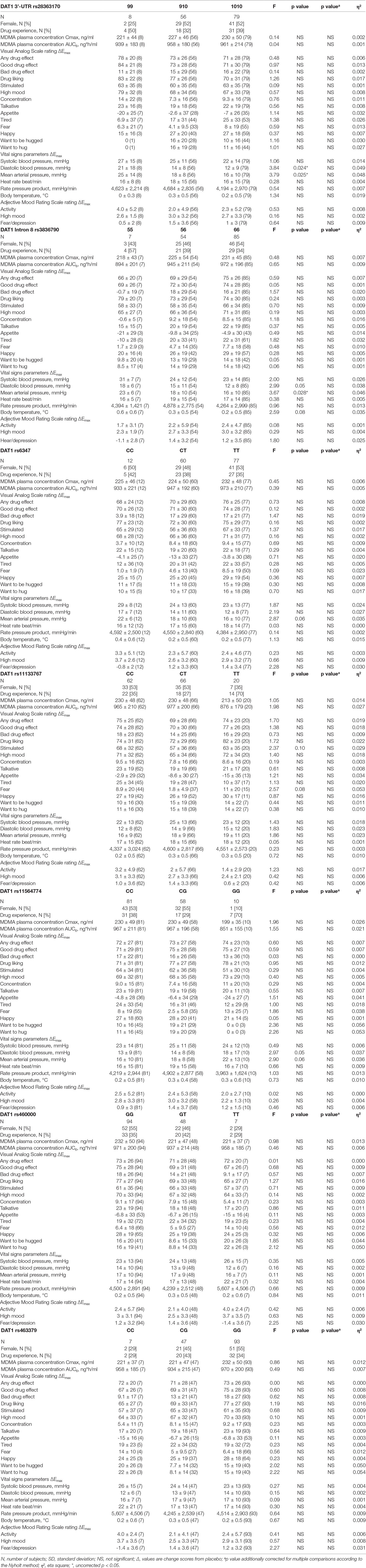
Table 2 Effects of polymorphisms in the dopamine transporter 1 gene on the maximal response to 125 mg MDMA (mean ± SD (N) and statistics) corrected with MDMA AUC6 (exclusive plasma concentrations).
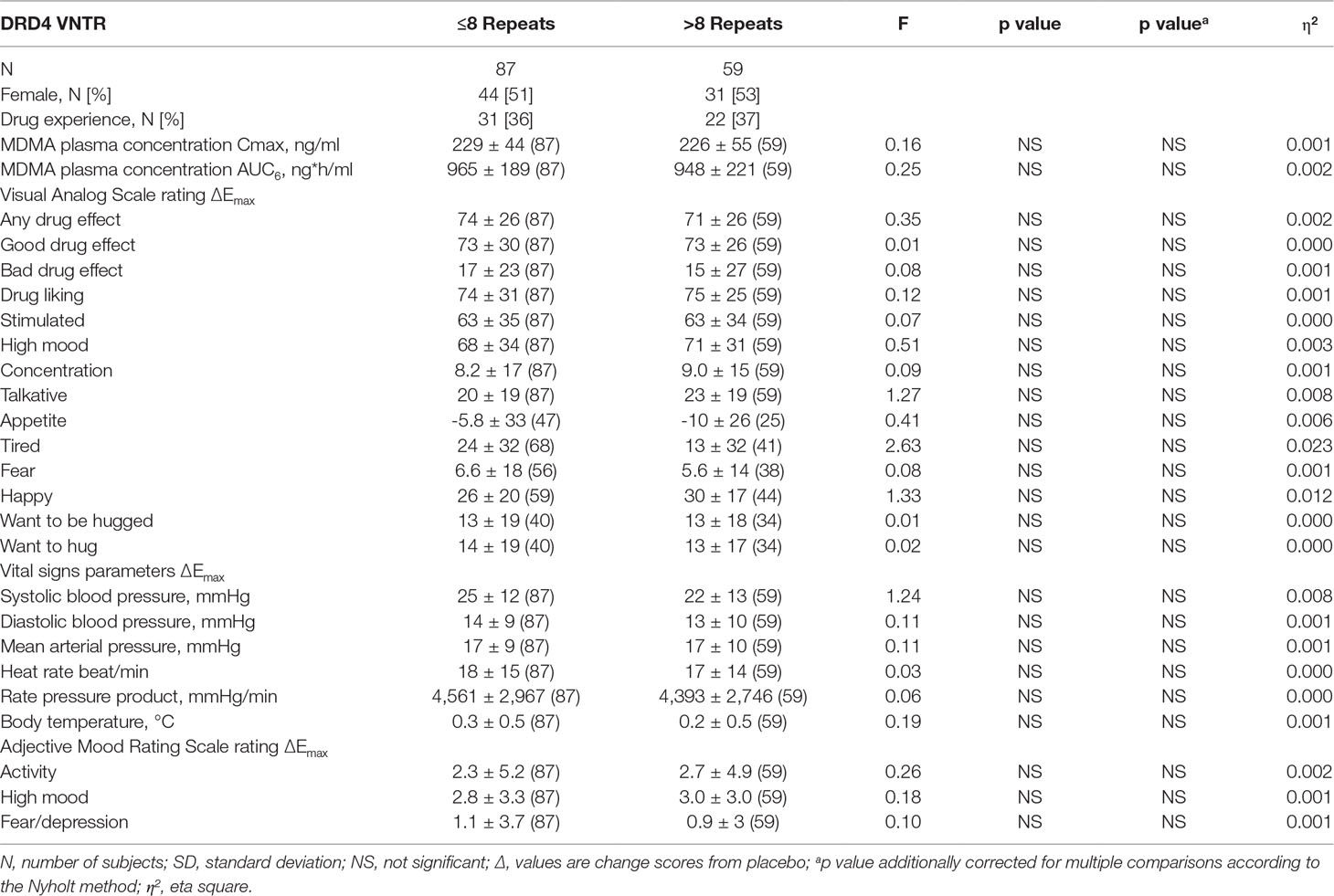
Table 3 Effects of the variable-number tandem repeat polymorphism in the dopamine receptor D4 gene on the maximal response to 125 mg MDMA (mean ± SD (N) and statistics) corrected with MDMA AUC6 (exclusive plasma concentrations).
The 60-item Likert-type scale of the short version of the Adjective Mood Rating Scale (AMRS) (59) was administered before and 1.25, 2, and 5 h after MDMA or placebo administration. The AMRS contains subscales for activity, well-being, and anxiety–depression.
Genotyping
Genomic DNA was extracted from whole blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hombrechtikon, Switzerland) and automated QIAcube system. SNP genotyping was performed using commercial TaqMan SNP genotyping assays (LuBio Science, Lucerne, Switzerland). We assayed the following SNPs: DRD2/ankyrin repeat and kinase domain containing 1 (ANKK1) SNPs rs1800497 (assay: C___7486676_10), DRD2 rs6277 (assay: C__11339240_10), and rs1079597 (assay: C___2278884_10), and DAT1 SNPs rs6347 (assay: C___8769902_10) and rs11133767 (assay: C___3024834_10) and rs11564774 (assay: C__25761679_10) and rs460000 (assay: C___3284837_10) and rs463379 (assay: C___3284827_10). We also used the following method to genotype the polymorphisms in DRD4 exon III VNTR, DAT1 3’UTR VNTR rs28363170, and DAT1 Intron 8 (5/6) VNTR rs3836790. Genotypes were determined by polymerase chain reaction (PCR) using 2.5, 1.25, and 1.25 units of HotStarTaq DNA polymerase (QIAGEN Instruments AG, Hombrechtikon, Switzerland), respectively; 1.5 µl PCR Buffer 10x each (15 mM Mg2+; QIAGEN Instruments AG, Hombrechtikon, Switzerland); 0.25, 1, and 1 µl dNTP Mix (40 mM), respectively; and primer set 5’-GCGACTACGTGGTCTACTCG and 5’-AGGACCCTCATGGCCTTG, 5’-TGTGGTGTAGGGAACGGCCTGAG and 5’-CTTCCTGGAGGTCACGGCTCAAGG, and 5’-GCATGTGGATGTGTTCTTGCA and 5’-TCATCCCAGGGACATCTGCTA (both 1 µl, both 0.5 µl, and both 0.5 µl, respectively) in a total reaction volume of 15 µl each. The following temperature profile was applied in a T100 thermal cycler (Bio-Rad, Cressier, Switzerland): for DRD4 (Exon III VNTR): initial activation step of 95°C (15 min) and 30 cycles of 98°C (60 s), 67.5°C (60 s), and 72°C (60 s), with final extension at 72°C (5 min); for DAT1 (3’UTR VNTR) rs28363170 and (intron8 5/6 VNTR) rs3836790: initial activation step of 95°C (15 min) and 30 cycles of 98°C (25 s), 95°C (35 s), and 72°C (45 s), with final extension at 72°C (5 min). The sizes of the resulting PCR products were assessed by 3.5% (for DRD4 exon III VNTR) and 2.5% (for DAT1 3’UTR VNTR rs28363170 and Intron 8 VNTR rs3836790) agarose gel electrophoresis. Amplicons of the DRD4 (Exon III VNTR in chromosome 11) of 379 bp were designated as 2 repeats (2R), and amplicons of every additional 48 bp were designated as 2+x times 48 bp variants [up to 9R (with 379 bp + 7 × 48 bp = 715 bp)]. Four and 7-repeat amplicons were the most common forms. Complete genotype and allele distributions are depicted in Supplementary Table S1. For the analysis, groups were made with cumulative ≤8 repeats or cumulative >8 repeats in both alleles. Amplicons of the DAT1 (3’UTR VNTR) rs28363170 of 448 bp were designated as 9 repeats (9R), and amplicons of 488 bp were designated as 10R. Individuals possessing other repeats were excluded from the analysis. Amplicons of the DAT1 (intron 8 5/6 VNTR) rs3836790 of 295 bp were designated as 5 repeats (5R), and amplicons of 325 bp were designated as 6 repeats (6R). The pairwise linkage disequilibrium (LD) and relative physical location of the determined SNPs on chromosome 11 (DRD2) and 5 (DAT1) are shown in Supplementary Figure 1. The tested genetic variants were consistent with Hardy–Weinberg equilibrium (p > 0.05).
Statistical Analysis
The statistical analyses were performed using Statistica 12 software (StatSoft, Tulsa, OK, USA). For repeatedly measured data, peak effects (Emax) and areas under the effect-time curve (AUEC) from 0- to 6-h values were determined for MDMA and placebo. Differences in Emax (Δ; MDMA-placebo) were then analyzed using one-way analysis of variance (ANOVA), with genotype as the between-group factor. The level of significance was set at p < 0.05. The Nyholt correction method was used to account for multiple comparisons and displayed separately in all tables (60). We thereby corrected for 17 subjective effect ratings (VAS+AMRS), and six vital parameters. In addition, this was then corrected for each of the 11 polymorphisms tested, resulting in (17 + 6) × 11 = 253 variables and an effective number of independent variables (Veff) of 183.6 according to Nyholt. Consequently, this led to a corrected significance threshold of p < 0.00027 to keep Type I error rate at 5%. To account for differences in plasma concentrations of MDMA that were caused by differences in body weight, dosing, or metabolizing enzymes (17, 18), the area under the MDMA plasma concentration–time curve from 0 to 6 h (AUC) was included as a covariate in the ANOVAs, and we report the corrected statistics. Additionally, modulatory effects of sex were explored by adding sex as a between-subjects factor in the ANOVAs (sex × genotype). Emax values were obtained directly from the observed data. AUC and AUEC values were calculated using the linear-log trapezoidal method in Phoenix WinNonlin 6.4 (Certara, Princeton, NJ, USA). The primary analysis was performed using an additive genotype model approach for SNPs. Recessive or dominant model analysis was also performed, the results of which are reported only when the additive model was initially significant.
Results
MDMA significantly altered all tested VAS and AMRS Emax values. Subjects did not significantly differ in MDMA plasma concentration or previous drug experience across genotype groups, with the exception of DAT1 rs11133767. Participants carrying two T-alleles showed disproportionately more illicit drug experiences than carriers of the C-allele (70% vs. 31%, respectively; χ2 = 11.2, p < 0.001).
The influence of polymorphisms within genes coding for the DRD2, DAT1, and DRD4 on the maximal acute subjective and autonomic effects of MDMA is shown in Tables 1–3, respectively. Supplementary Table S2 shows the data for the total response to MDMA over time (AUEC). Supplementary Tables S3 and S4 show the uncorrected statistics for Emax and AUEC, respectively. Homozygous A-allele carriers of the DRD2 rs1800497 showed a higher score in VASs “talkative” (F1,147 = 4.23, p < 0.05) and in AMRSs “activity” and “high mood” (F1,147 = 4.62, p < 0.05 and F1,147 = 4.50, p < 0.05, respectively) compared to carriers of the G-allele. Subjects with two 9R-alleles of the DAT1 rs28363170 had a higher MDMA-induced increase in diastolic blood pressure and MAP compared to subjects with a 10R-allele (F1,141 = 7.12, p < 0.01 and F1,141 = 6.56, p < 0.05, respectively). Regarding the DAT1 rs3836790, MDMA produced a higher increase in MAP in individuals homozygous for the 5R-allele compared to 6R-allele carriers (F1,144 = 4.31, p < 0.05).
Nyholt correction for multiple comparisons yielded statistics indicating that the genetic polymorphisms had no significant effect on the subjective and autonomic parameters. Sex did not significantly modulate the results.
Discussion
The current study expands previous research on whether the acute effects of MDMA are modulated by common genetic polymorphisms in pharmacological targets of MDMA. So far, the focus lied on the role of the NE and 5-HT system genetics in the acute effects of MDMA (22, 23). This is the first study to concentrate on a selection of genetic polymorphisms within the human DA system (namely, D2, D4, and DAT).
Action on the DA system is thought to be crucial for the effects of most psychostimulant substances (6, 24, 61), and pharmacogenetic studies demonstrated that different phenotypes are affected by various DA genotypes. As for MDMA, however, none of the herein investigated genetic polymorphisms significantly altered the acute effects after consideration of Type I error correction.
Nevertheless, this missing link between DA genetic variations and MDMA-related phenotypes might not solely be caused by a lack of genetic influence on the MDMA effects but rather the potentially minor role of DA in MDMA effects. Although MDMA is an amphetamine, it acts mainly on the 5-HT system and therefore leads to its classification as an entactogen (7, 62).
The present study has limitations. Although this analysis was done using the largest sample of healthy human subjects who received MDMA in placebo-controlled studies, the sample size is still relatively small when considering the partially small rare allele groups and mostly weak effect sizes for the influence of genetic variants on the MDMA response. This is especially influencing spurious, uncorrected effects (i.e., the AA carrier group for the SNP DRD2/ANKK1 rs1800497 with N = 2). Larger cohorts might show a more balanced sample distribution, which might lead to different results. Additionally, the study was conducted in healthy volunteers with a single dose of 125 mg MDMA. Therefore, the findings may not be applied to other populations and situations, such as psychiatric patients and the use of higher doses of MDMA. Furthermore, SNPs in genes of other targets of MDMA may also be involved. However, we corrected for the modulatory effects of known genetic variants that influence the metabolism of MDMA (17, 18) by taking interindividual differences in plasma MDMA concentrations into account. We also might have missed some relevant genetic polymorphisms. A novel potentially functional SNP within the DAT1 has been described in recent research. However, the SNP showed no significant alteration in the inhibition of DA uptake by MDMA in human embryonic kidney 293 cells (63). We have also not tested for rare haplotypes because a haplotype approach may lead to very small groups and more potential statistical artifacts. However, a haplotype suggested by Brewer et al., which consists of rs28363170 10/10 genotype and at least one rs3836790 5R-allele carriers, showed a reduced subjective response to cocaine compared to others (40). The same haplotype showed no effect in the present study. In fact, uncorrected results even implied opposite and incoherent effects, with 10R carriers showing lower MDMA-induced MAP changes and 5/5 carriers showing higher MAP changes than subjects with the 9/9 genotype or a 6R-allele, respectively. This incoherency may be attributable to the different substances used (cocaine vs. MDMA) and different cohorts (80% males of African descent vs. the sex-balanced sample of European descent) (40). Additionally, MDMA may interact with a different binding site on the DAT compared to other stimulants like cocaine (64). Finally, previous drug experiences were not equally distributed among DAT1 rs11133767 genotype groups, and effects might slightly depend on previous substance use experiences. Because of the involvement of DA in addiction, subjects carrying a TT genotype may be more prone to illicit substance use (65). Apart from this finding, given that our cohort included mostly drug-naive subjects with limited drug use experience, some alleles associated with increased drug use might even be underrepresented. However, the tested variants were consistent with the Hardy–Weinberg equilibrium and comparable with frequencies found in European genome databases.
We conclude that the present findings align with previous studies in that variations in genes coding for players of the monoaminergic systems are unlikely to explain interindividual variations in the acute effects of MDMA in humans.
Data Availability Statement
The datasets for this manuscript are not publicly available because the individual genotyping consent did not include storing in public repository. Requests to access the datasets should be directed to Matthias Liechti, Matthias.liechti@usb.ch.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethikkommission Nordwest- und Zentralschweiz (EKNZ). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
PV analyzed the data and wrote the manuscript. ML conceived the study, obtained funding, and wrote the manuscript.
Funding
This study was supported by the Swiss National Science Foundation (grant no. 320030_170249).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the assistance of C.M. Hysek, A. Rickli, Y. Schmid, P.C. Dolder, and F. Holze in conducting the clinical studies, H. Meyer zu Schwabedissen, K. Prestin, and Janine J. Hussner for assistance with genotyping, and M. Arends for text editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2019.00755/full#supplementary-material
References
1. Mithoefer MC, Wagner MT, Mithoefer AT, Jerome I, Doblin R. The safety and efficacy of ±3,4-methylenedioxymethamphetamine-assisted psychotherapy in subjects with chronic, treatment-resistant posttraumatic stress disorder: the first randomized controlled pilot study. J Psychopharmacol (2010) 25:439–52. doi: 10.1177/0269881110378371
2. Oehen P, Traber R, Widmer V, Schnyder U. A randomized, controlled pilot study of MDMA (± 3,4-methylenedioxymethamphetamine)-assisted psychotherapy for treatment of resistant, chronic post-traumatic stress disorder (PTSD). J Psychopharmacol (2013) 27(1):40–52. doi: 10.1177/0269881112464827
3. Mithoefer MC, Mithoefer AT, Feduccia AA, Jerome L, Wagner M, Wymer J, et al. 3,4-methylenedioxymethamphetamine (MDMA)-assisted psychotherapy for post-traumatic stress disorder in military veterans, firefighters, and police officers: a randomised, double-blind, dose-response, phase 2 clinical trial. Lancet Psychiatry (2018) 5(6):486–97. doi: 10.1016/S2215-0366(18)30135-4
4. Battaglia G, Brooks BP, Kulsakdinun C, De Souza EB. Pharmacologic profile of MDMA (3,4-methylenedioxymethamphetamine) at various brain recognition sites. Eur J Pharmacol (1988) 149(1–2):159–63. doi: 10.1016/0014-2999(88)90056-8
5. Verrico CD, Miller GM, Madras BK. MDMA (ecstasy) and human dopamine, norepinephrine, and serotonin transporters: implications for MDMA-induced neurotoxicity and treatment. Psychopharmacology (2007) 189(4):489–503. doi: 10.1007/s00213-005-0174-5
6. Luethi D, Liechti ME. Monoamine transporter and receptor interaction Profiles in vitro predict reported human doses of novel psychoactive stimulants and psychedelics. Int J Neuropsychopharmacol (2018) 21(10):926–31. doi: 10.1093/ijnp/pyy047
7. Simmler LD, Liechti ME. Pharmacology of MDMA- and amphetamine-like new psychoactive substances. Handb Exp Pharmacol. (2018) 252:143–164. doi: 10.1007/164_2018_113
8. Hysek CM, Simmler LD, Nicola V, Vischer N, Donzelli M, Krähenbühl S, et al. Duloxetine inhibits effects of MDMA (“ecstasy”) in vitro and in humans in a randomized placebo-controlled laboratory study. PLoS ONE (2012c) 7:e36476. doi: 10.1371/journal.pone.0036476
9. Webster JI, Harper DN, Schenk S. Generalization of serotonin and dopamine ligands to the discriminative stimulus effects of different doses of +/-3,4-methylenedioxymethamphetamine. Behav Pharmacol (2017) 28(4):245–54. doi: 10.1097/FBP.0000000000000282
10. Liechti ME, Vollenweider FX. Acute psychological and physiological effects of MDMA (“Ecstasy”) after haloperidol pretreatment in healthy humans. Eur Neuropsychopharmacol (2000a) 10(4):289–95. doi: 10.1016/S0924-977X(00)00086-9
11. Risbrough VB, Masten VL, Caldwell S, Paulus MP, Low MJ, Geyer MA. Differential contributions of dopamine D1, D2, and D3 receptors to MDMA-induced effects on locomotor behavior patterns in mice. Neuropsychopharmacology (2006) 31(11):2349–58. doi: 10.1038/sj.npp.1301161
12. Liechti ME, Baumann C, Gamma A, Vollenweider FX. Acute psychological effects of 3,4-methylenedioxymethamphetamine (MDMA, “Ecstasy”) are attenuated by the serotonin uptake inhibitor citalopram. Neuropsychopharmacology (2000) 22(5):513–21. doi: 10.1016/S0893-133X(99)00148-7
13. Tancer M, Johanson CE. The effects of fluoxetine on the subjective and physiological effects of 3,4-methylenedioxymethamphetamine (MDMA) in humans. Psychopharmacology (2007) 189(4):565–73. doi: 10.1007/s00213-006-0576-z
14. Hysek CM, Simmler LD, Schillinger N, Meyer N, Schmid Y, Donzelli M, et al. Pharmacokinetic and pharmacodynamic effects of methylphenidate and MDMA administered alone and in combination. Int J Neuropsychopharmacol (2014b) 17:371–81. doi: 10.1017/S1461145713001132
15. Schmid Y, Rickli A, Schaffner A, Duthaler U, Grouzmann E, Hysek CM, et al. Interactions between bupropion and 3,4-methylenedioxymethamphetamine in healthy subjects. J Pharmacol Exp Ther (2015) 353(1):102–11. doi: 10.1124/jpet.114.222356
16. de la Torre R, Yubero-Lahoz S, Pardo-Lozano R, Farre M. MDMA, methamphetamine, and CYP2D6 pharmacogenetics: what is clinically relvant? Front Genet (2012) 3:235. doi: 10.3389/fgene.2012.00235
17. Schmid Y, Vizeli P, Hysek CM, Prestin K, Meyer zu Schwabedissen HE, Liechti ME. CYP2D6 function moderates the pharmacokinetics and pharmacodynamics of MDMA in a controlled study in healthy subjects. Pharmacogenet Genom (2016) 26:397–401. doi: 10.1097/FPC.0000000000000231
18. Vizeli P, Schmid Y, Prestin K, Meyer zu Schwabedissen HE, Liechti ME. Pharmacogenetics of ecstasy: CYP1A2, CYP2C19, and CYP2B6 polymorphisms moderate pharmacokinetics of MDMA in healthy subjects. Eur Neuropsychopharmacol (2017) 27:232–8. doi: 10.1016/j.euroneuro.2017.01.008
19. Bershad AK, Weafer JJ, Kirkpatrick MG, Wardle MC, Miller MA, de Wit H. Oxytocin receptor gene variation predicts subjective responses to MDMA. Soc Neurosci (2016) 11(6):592–9. doi: 10.1080/17470919.2016.1143026
20. Kuypers KPC, de la Torre R, Farre M, Xicota L, de Sousa Fernandes Perna EB, Theunissen EL, et al. Depressive mood ratings are reduced by MDMA in female polydrug ecstasy users homozygous for the l-allele of the serotonin transporter. Sci Rep (2018) 8(1):1061. doi: 10.1038/s41598-018-19618-1
21. Vizeli P, Liechti ME. Oxytocin receptor gene variations and socio-emotional effects of MDMA: a pooled analysis of controlled studies in healthy subjects. PLoS ONE (2018) 13(6):e0199384. doi: 10.1371/journal.pone.0199384
22. Vizeli P, Meyer Zu Schwabedissen HE, Liechti ME. No major role of norepinephrine transporter gene variations in the cardiostimulant effects of MDMA. Eur J Clin Pharmacol (2018a) 74(3):275–83. doi: 10.1007/s00228-017-2392-2
23. Vizeli P, Meyer Zu Schwabedissen HE, Liechti ME. Role of serotonin transporter and receptor gene variations in the acute effects of MDMA in healthy subjects. ACS Chem Neurosci. (2018b) 10(7):3120–3131. doi: 10.1021/acschemneuro.8b00590
24. Hondebrink L, Zwartsen A, Westerink RHS. Effect fingerprinting of new psychoactive substances (NPS): what can we learn from in vitro data? Pharmacol Ther (2018) 182:193–224. doi: 10.1016/j.pharmthera.2017.10.022
25. Hahn MK, Blakely RD. Monoamine transporter gene structure and polymorphisms in relation to psychiatric and other complex disorders. Pharmacogenomics J (2002) 2(4):217–35. doi: 10.1038/sj.tpj.6500106
26. Madras BK, Miller GM, Fischman AJ. The dopamine transporter: relevance to attention deficit hyperactivity disorder (ADHD). Behav Brain Res (2002) 130(1–2):57–63. doi: 10.1016/S0166-4328(01)00439-9
27. Haddley K, Vasiliou AS, Ali FR, Paredes UM, Bubb VJ, Quinn JP. Molecular genetics of monoamine transporters: relevance to brain disorders. Neurochem Res (2008) 33(4):652–67. doi: 10.1007/s11064-007-9521-8
28. Barr CL, Xu C, Kroft J, Feng Y, Wigg K, Zai G, et al. Haplotype study of three polymorphisms at the dopamine transporter locus confirm linkage to attention-deficit/hyperactivity disorder. Biol Psychiatry (2001) 49(4):333–9. doi: 10.1016/S0006-3223(00)01053-2
29. Ujike H, Harano M, Inada T, Yamada M, Komiyama T, Sekine Y, et al. Nine- or fewer repeat alleles in VNTR polymorphism of the dopamine transporter gene is a strong risk factor for prolonged methamphetamine psychosis. Pharmacogenomics J (2003) 3(4):242–7. doi: 10.1038/sj.tpj.6500189
30. Guindalini C, Howard M, Haddley K, Laranjeira R, Collier D, Ammar N, et al. A dopamine transporter gene functional variant associated with cocaine abuse in a Brazilian sample. Proc Natl Acad Sci U S A (2006) 103(12):4552–7. doi: 10.1073/pnas.0504789103
31. Asherson P, Brookes K, Franke B, Chen W, Gill M, Ebstein RP, et al. Confirmation that a specific haplotype of the dopamine transporter gene is associated with combined-type ADHD. Am J Psychiatry (2007) 164(4):674–7. doi: 10.1176/ajp.2007.164.4.674
32. Friedel S, Saar K, Sauer S, Dempfle A, Walitza S, Renner T, et al. Association and linkage of allelic variants of the dopamine transporter gene in ADHD. Mol Psychiatry (2007) 12(10):923–33. doi: 10.1038/sj.mp.4001986
33. Fernandez-Castillo N, Ribases M, Roncero C, Casas M, Gonzalvo B, Cormand B. Association study between the DAT1, DBH and DRD2 genes and cocaine dependence in a Spanish sample. Psychiatr Genet (2010) 20(6):317–20. doi: 10.1097/YPG.0b013e32833b6320
34. Pinsonneault JK, Han DD, Burdick KE, Kataki M, Bertolino A, Malhotra AK, et al. Dopamine transporter gene variant affecting expression in human brain is associated with bipolar disorder. Neuropsychopharmacology (2011) 36(8):1644–55. doi: 10.1038/npp.2011.45
35. Sullivan D, Pinsonneault JK, Papp AC, Zhu H, Lemeshow S, Mash DC, et al. Dopamine transporter DAT and receptor DRD2 variants affect risk of lethal cocaine abuse: a gene-gene-environment interaction. Transl Psychiatry (2013) 3:e222. doi: 10.1038/tp.2012.146
36. Pagerols M, Richarte V, Sanchez-Mora C, Garcia-Martinez I, Corrales M, Corominas M, et al. Pharmacogenetics of methylphenidate response and tolerability in attention-deficit/hyperactivity disorder. Pharmacogenomics J (2017) 17(1):98–104. doi: 10.1038/tpj.2015.89
37. Vandenbergh DJ, Persico AM, Hawkins AL, Griffin CA, Li X, Jabs EW, et al. Human dopamine transporter gene (DAT1) maps to chromosome 5p15.3 and displays a VNTR. Genomics (1992) 14(4):1104–6. doi: 10.1016/S0888-7543(05)80138-7
38. Franke B, Vasquez AA, Johansson S, Hoogman M, Romanos J, Boreatti-Hummer A, et al. Multicenter analysis of the SLC6A3/DAT1 VNTR haplotype in persistent ADHD suggests differential involvement of the gene in childhood and persistent ADHD. Neuropsychopharmacology (2010) 35(3):656–64. doi: 10.1038/npp.2009.170
39. Lott DC, Kim SJ, Cook EH Jr., de Wit H. Dopamine transporter gene associated with diminished subjective response to amphetamine. Neuropsychopharmacology (2005) 30(3):602–9. doi: 10.1038/sj.npp.1300637
40. Brewer AJ 3rd, Nielsen DA, Spellicy CJ, Hamon SC, Gingrich J, Thompson-Lake DG, et al. Genetic variation of the dopamine transporter (DAT1) influences the acute subjective responses to cocaine in volunteers with cocaine use disorders. Pharmacogenet Genomics (2015) 25(6):296–304. doi: 10.1097/FPC.0000000000000137
41. Vereczkei A, Demetrovics Z, Szekely A, Sarkozy P, Antal P, Szilagyi A, et al. Multivariate analysis of dopaminergic gene variants as risk factors of heroin dependence. PLoS ONE (2013) 8(6):e66592. doi: 10.1371/journal.pone.0066592
42. Voisey J, Swagell CD, Hughes IP, van Daal A, Noble EP, Lawford BR, et al. A DRD2 and ANKK1 haplotype is associated with nicotine dependence. Psychiatry Res (2012) 196(2–3):285–9. doi: 10.1016/j.psychres.2011.09.024
43. Swagell CD, Lawford BR, Hughes IP, Voisey J, Feeney GF, van Daal A, et al. DRD2 C957T and TaqIA genotyping reveals gender effects and unique low-risk and high-risk genotypes in alcohol dependence. Alcohol Alcohol (2012) 47(4):397–403. doi: 10.1093/alcalc/ags047
44. Roussos P, Giakoumaki SG, Bitsios P. Cognitive and emotional processing in high novelty seeking associated with the L-DRD4 genotype. Neuropsychologia (2009) 47(7):1654–9. doi: 10.1016/j.neuropsychologia.2009.02.005
45. Ptacek R, Kuzelova H, Stefano GB. Dopamine D4 receptor gene DRD4 and its association with psychiatric disorders. Med Sci Monit (2011) 17(9):RA215–220. doi: 10.12659/MSM.881925
46. Goncalves VF, Tiwari AK, de Luca V, Kong SL, Zai C, Tampakeras M, et al. DRD4 VNTR polymorphism and age at onset of severe mental illnesses. Neurosci Lett (2012) 519(1):9–13. doi: 10.1016/j.neulet.2012.04.027
47. Halley AC, Boretsky M, Puts DA, Shriver M. Self-Reported Sexual Behavioral Interests and Polymorphisms in the Dopamine Receptor D4 (DRD4) Exon III VNTR in Heterosexual Young Adults. Arch Sex Behav (2016) 45(8):2091–100. doi: 10.1007/s10508-015-0646-6
48. Chang FM, Kidd JR, Livak KJ, Pakstis AJ, Kidd KK. The world-wide distribution of allele frequencies at the human dopamine D4 receptor locus. Hum Genet (1996) 98(1):91–101. doi: 10.1007/s004390050166
49. Hamarman S, Fossella J, Ulger C, Brimacombe M, Dermody J. Dopamine receptor 4 (DRD4) 7-repeat allele predicts methylphenidate dose response in children with attention deficit hyperactivity disorder: a pharmacogenetic study. J Child Adolesc Psychopharmacol (2004) 14(4):564–74. doi: 10.1089/cap.2004.14.564
50. Asghari V, Sanyal S, Buchwaldt S, Paterson A, Jovanovic V, Van Tol HH. Modulation of intracellular cyclic AMP levels by different human dopamine D4 receptor variants. J Neurochem (1995) 65(3):1157–65. doi: 10.1046/j.1471-4159.1995.65031157.x
51. Hysek CM, Simmler LD, Ineichen M, Grouzmann E, Hoener MC, Brenneisen R, et al. The norepinephrine transporter inhibitor reboxetine reduces stimulant effects of MDMA (“ecstasy”) in humans. Clin Pharmacol Ther (2011) 90(2):246–55. doi: 10.1038/clpt.2011.78
52. Hysek CM, Brugger R, Simmler LD, Bruggisser M, Donzelli M, Grouzmann E, et al. Effects of the α2-adrenergic agonist clonidine on the pharmacodynamics and pharmacokinetics of 3,4-methylenedioxymethamphetamine in healthy volunteers. J Pharmacol Exp Ther (2012a) 340:286–94. doi: 10.1124/jpet.111.188425
53. Hysek CM, Schmid Y, Rickli A, Simmler LD, Donzelli M, Grouzmann E, et al. Carvedilol inhibits the cardiostimulant and thermogenic effects of MDMA in humans. Br J Pharmacol (2012b) 166:2277–88. doi: 10.1111/j.1476-5381.2012.01936.x
54. Hysek CM, Liechti ME. Effects of MDMA alone and after pretreatement with reboxetine, duloxetine, clonidine, carvedilol, and doxazosin on pupillary light reflex. Psychopharmacology (Berl) (2012) 224:363–76. doi: 10.1007/s00213-012-2761-6
55. Dolder PC, Muller F, Schmid Y, Borgwardt SJ, Liechti ME. Direct comparison of the acute subjective, emotional, autonomic, and endocrine effects of MDMA, methylphenidate, and modafinil in healthy subjects. Psychopharmacology (Berl) (2018) 235(2):467–79. doi: 10.1007/s00213-017-4650-5
56. Vizeli P, Liechti ME. Safety pharmacology of acute MDMA administration in healthy subjects. J Psychopharmacol (2017) 31(5):576–88. doi: 10.1177/0269881117691569
57. Brunt TM, Koeter MW, Niesink RJ, van den Brink W. Linking the pharmacological content of ecstasy tablets to the subjective experiences of drug users. Psychopharmacology (Berl) (2012) 220(4):751–62. doi: 10.1007/s00213-011-2529-4
58. Hysek CM, Schmid Y, Simmler LD, Domes G, Heinrichs M, Eisenegger C, et al. MDMA enhances emotional empathy and prosocial behavior. Soc Cogn Affect Neurosci (2014a) 9:1645–52. doi: 10.1093/scan/nst161
60. Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet (2004) 74(4):765–9. doi: 10.1086/383251
61. Wise RA. Dopamine and reward: the anhedonia hypothesis 30 years on. Neurotox Res (2008) 14(2–3):169–83. doi: 10.1007/BF03033808
62. Nichols DE. Differences between the mechanism of action of MDMA, MBDB, and the classic hallucinogens. Identification of a new therapeutic class: entactogens. J Psychoactive Drugs (1986) 18(4):305–13. doi: 10.1080/02791072.1986.10472362
63. Zwartsen A, Litjens CHC, Hondebrink L, van den Heuvel J, Greupink R, Russel FGM, et al. Differential effects of psychoactive substances on human wildtype and polymorphic T356M dopamine transporters (DAT). Toxicology (2019) 422:69–75. doi: 10.1016/j.tox.2019.04.012
64. Heal DJ, Gosden J, Smith SL. Dopamine reuptake transporter (DAT) “inverse agonism” - A novel hypothesis to explain the enigmatic pharmacology of cocaine. Neuropharmacology (2014) 87C:19–40. doi: 10.1016/j.neuropharm.2014.06.012
Keywords: dopamine, SCL6A3, DAT1, DRD2, DRD4, MDMA
Citation: Vizeli P and Liechti ME (2019) No Influence of Dopamine System Gene Variations on Acute Effects of MDMA. Front. Psychiatry 10:755. doi: 10.3389/fpsyt.2019.00755
Received: 25 June 2019; Accepted: 19 September 2019;
Published: 24 October 2019.
Edited by:
Matthew W. Johnson, Johns Hopkins University, United StatesReviewed by:
Sean Dolan, Johns Hopkins University, United StatesZach Walsh, University of British Columbia Okanagan, Canada
Copyright © 2019 Vizeli and Liechti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthias E. Liechti, matthias.liechti@usb.ch
 Patrick Vizeli
Patrick Vizeli Matthias E. Liechti
Matthias E. Liechti