- Neural Circuit Research Group, Korea Brain Research Institute, Daegu, Republic of Korea
Background: Microglia, brain resident macrophages, play multiple roles in maintaining homeostasis, including immunity, surveillance, and protecting the central nervous system through their distinct activation processes. Identifying all types of microglia-driven populations is crucial due to the presence of various phenotypes that differ based on developmental stages or activation states. During embryonic development, the E8.5 yolk sac contains erythromyeloid progenitors that go through different growth phases, eventually resulting in the formation of microglia. In addition, microglia are present in neurological diseases as a diverse population. So far, no individual biomarker for microglia has been discovered that can accurately identify and monitor their development and attributes.
Summary: Here, we highlight the newly defined biomarker of mouse microglia, UGT1A7C, which exhibits superior stability in expression during microglia development and activation compared to other known microglia biomarkers. The UGT1A7C sensing chemical probe labels all microglia in the 3xTG AD mouse model. The expression of Ugt1a7c is stable during development, with only a 4-fold variation, while other microglia biomarkers, such as Csf1r and Cx3cr1, exhibit at least a 10-fold difference. The UGT1A7C expression remains constant throughout its lifespan. In addition, the expression and activity of UGT1A7C are the same in response to different types of inflammatory activators’ treatment in vitro.
Conclusion: We propose employing UGT1A7C as the representative biomarker for microglia, irrespective of their developmental state, age, or activation status. Using UGT1A7C can reduce the requirement for using multiple biomarkers, enhance the precision of microglia analysis, and even be utilized as a standard for gene/protein expression.
Introduction
Microglia are involved in immune responses as tissue-resident macrophages of the central nervous system (CNS) (1–3). In addition to the immune role, their cellular activities are involved in the neuronal array regarding the refinement of synaptic connections and the elaboration of neuromodulatory factors for cognitive ability (4–7). They take up approximately 5 ~ 12% of the total cells in the mouse brain, with a diversity of transcriptional module combinations and levels of crowd across the brain region (8–12). Given their functional role and prevalence in the brain, microglial regulation has a high potential to develop brain disease therapy (13–15). Indeed, recent studies have linked microglia to neurodevelopmental and psychiatric diseases and neurodegenerative diseases (16–19). Accumulated mouse in vivo lineage tracing results indicate that microglia at different developmental stages are characterized by unique molecular features (20–23). Erythromyeloid progenitors (CD45-c-Kit+) arise before the end of embryonic day (E) 8 during the first wave of hematopoiesis in the yolk sac (24). Erythromyeloid progenitors - derived primitive macrophage progenitors (CD45+c-KitloCX3CR1-) colonize the developing brain at E9.5 and further differentiate into microglia in a Myb-independent manner via the PU.1- and IRF8-dependent pathway (22, 25). Regardless of distinctive ontogeny, microglia also express general macrophage markers such as CD11b, CSF1-1 receptor CD115, surface glycoprotein F4/80, and fractalkine receptor CX3 chemokine receptor CX3CR1 (24, 26–28). Although the expression level is not very high, microglia even express the hematopoietic marker CD45 (24, 29). However, most of the general markers does not satisfy the requirements for covering all stages of microglia. F4/80 is present from E9.5, but its expression is undetectable at E8.5 in the brain according to fate-mapping analysis of CSF1R (CD115)-expressing cells (30). CX3CR1 is expressed in the gut region at E8.5 and is sparsely visualized throughout the embryo. At stage E8.5 to 9.0, it was detectable in neural tissue within telencephalic vesicle. At E9.5, CX3CR1 microglial precursor cells were detected in the surface ectoderm [(31), Figure 1A]. As a result of these restricted generalities, it is common for researchers to utilize at least two biomarkers in lineage tracing experiments [(32), Table 1]. However, this approach often results fragmented data and unclear interpretation during specific time periods.
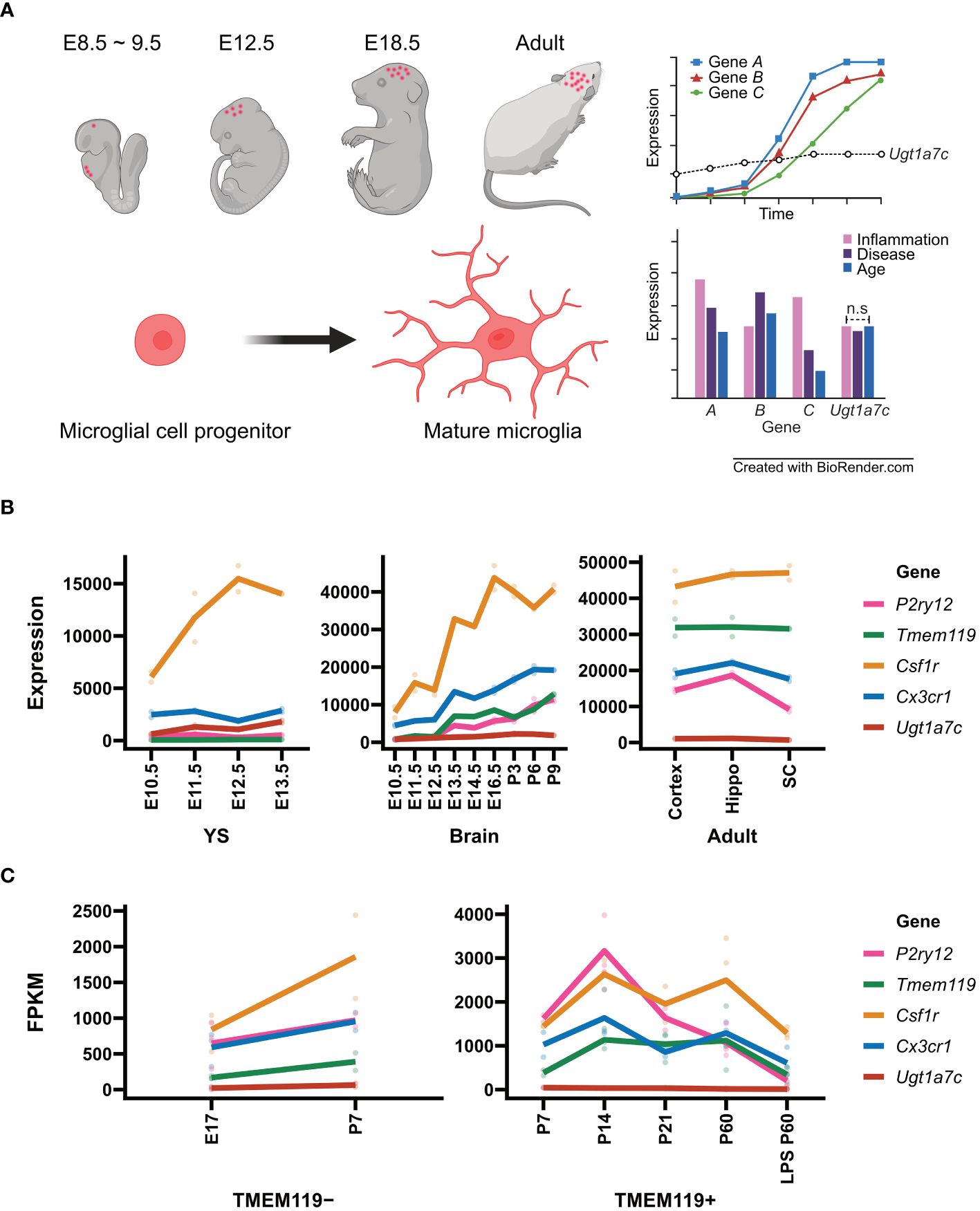
Figure 1 Microglial marker genes and Ugt1a7c. (A) Schematic diagram of microglial housekeeping gene. (B) Line plot of expression of microglial marker genes and Ugt1a7c across the microglial development including yolk sac (YS), pre-mature brain (Brain), and mature brain (Adult). From GSE79812, expression profiling data of microglial marker genes and Ugt1a7c were acquired. Units on the y axes are arbitrary. (C) Line plot of expression of microglial marker genes depending on TMEM119 protein expression. Data were acquired from NCBI BioProject (Accession PRJNA307271). For microglial marker genes, Csf1r, Cx3cr1, P2ry12, and Tmem119 were selected. A line represents the mean value of replications (colored spots). E, embryonic day; P, postnatal day; Hippo, hippocampus; SC, spinal cord; LPS, lipopolysaccharide. Created with BioRender.com.
Since the publication of the fate mapping study in 2010, there has been a significant increase in information regarding microglia’s ontogeny, maintenance, neuroimmune activities, and interactions (20). Microglia express heterogeneous profiles with different shapes, gene expression patterns, and even function (84, 85). The integration of several microglia biomarkers for translational/transcriptomic analysis is an informative feature based on information about microglia (84, 86–90). A single microglia biomarker was not sufficient to identify them and track their ontogeny and characteristics, especially when there was an interaction with other types of glia/neurons and a transition of status (13, 91–93). For example, P2RY12 downregulation occurs upon microglia activation (65). TMEM119 appears to be influenced by inflammatory responses and environmental factors including TGF-β and LPS (65, 81, 94). Sall1 expression is highly correlated with TGF-β1 signaling and varies between microglial cell lines (71, 72). The activation state of microglia also strongly impacts the expression of CD11b (59–61), CD115 (48, 49) and F4/80 (12, 33–35). Identifying a novel biomarker for monitoring microglia remains a major challenge in microglial biology.
Many aspects of microglial development and origin have been clarified through fate mapping studies in mice (20, 95). Depletion–repopulation experiments have demonstrated that microglia rely solely on self-renewal and are not influenced by other organs (96). Microglia were once considered to be uniform cells that respond to their environment because of their distinctive feature: “single origin” and “self-renewal.” However, recent research indicates that microglia exhibit high diversity in terms of morphology, function, and gene expression (27, 57, 71, 97–100). This heterogeneity in microglia is due to various factors, including intrinsic factors such as species, gender, and genetic background, and extrinsic factors such as pathogens, nutrition, and microbiota (10, 80, 101–103). The conventional in vitro-based classification distinguished “M1” and “M2” microglia, with M1 indicating neurotoxic and proinflammatory microglia and M2 representing neuroprotective and anti-inflammatory microglia (104–107). Nonetheless, the dichotomous classification has been replaced by multiple subclass-cluster classification based on transcript combinations and surface protein combinations with the advent of technologies such as single cell RNA seq and single cell mass spectrometry (CyTOF) (98, 108–115).
Low gene expression is often disregarded, as it is anticipated to have a negligible impact on cells. A recent study discovered a new microglial biomarker, UDP-glucuronosyltransferase 1a7c (UGT1A7C). The discovery was made during the analysis of the target gene to a microglia-specific BODIPY-based fluorescent dye called CDr20 (83). Thanks to the low yet sufficient level of the enriched UGT1A7C enzyme, the microglia population efficiently converts the small chemical into a fluorescent active form (57). The development of high-performance fluorogenic chemical probes enables the visualization of microglia with the biomarker and allows UGT1A7C to enter the microglia research area with its unique approaching capacity that can act both in vitro and in vivo (116). From this perspective, we present a concise overview of microglia and UGT1A7C, while also shedding light on areas that faced technical limitations and did not receive adequate attention due to previous doubts about their existence in the brain.
Discussion
Microglia originate from the yolk sac and go through different phases, such as erythromyeloid and macrophage precursors. Once they enter the brain, microglia take on the role of tissue-resident macrophages and become involved in neuromodulation, which explains the diverse protein expression patterns observed in different developmental stages and microglia functions. To track microglia, different markers that match their specific property of interest are utilized. As technology advances, the list of attributes specific to each stage of microglia, including the genes they express, is continuously growing. Consequently, microglia are undergoing additional subclassification according to their recognized features.
Microglia heterogeneity
Microglia exhibit variations across distinct brain regions. Microglia in the prefrontal cortex (PFC) express high levels of Cx3cr1, P2ry12, and Tmem119 and low levels of Apoe, Lyz2, and Spp1, which are involved in synaptic modulation and plasticity for learning and memory and in inflammation and immune response, respectively (117). Microglia in the striatum express significant amounts of Map1b, Map2, and Tubb2a and low levels of Cd68, Lgals3, and Mrc1, which are involved in cell movement and shape and for the suppressed activity for phagocytosis and lysosomal function against pathogens and injuries, respectively (117). Microglia in the midbrain express high levels of Ccl2, Ccl5, and Il1b and low levels of Cx3cr1, P2ry12, and Tmem119, caused by their active inflammation and immune response and low synaptic modulation and plasticity activities (117). Microglia in the cerebellum express high levels of Gpx1, Gpx4, and Sod2 and low levels of Cx3cr1, P2ry12, Tmem119, and Aif1, indicating that they are sensitive to oxidative stress and metabolism, but show less function related to synaptic modulation and identity (118).
In addition to regional differences, the specific microenvironment is also linked to the molecular signature of microglia. For example, microglia actively express the fractalkine receptor (CX3CR1) when in contact with neurons that express its ligand, called fractalkine (CX3CL1) (26, 119–121). The contacted microglia are then actively involved in neural plasticity by pruning excess and/or weak synapses through the receptor-specific signaling pathway (122, 123). Another example is the interaction between the microglial TREM2 receptor and the neuronal ApoE ligand (124–126). The interaction plays a role in regulating microglial phagocytosis and inflammation as well as neuronal lipid metabolism and function (127–130). Lastly, Brain-derived neurotrophic factor (BDNF) secreted from neurons binds to TrkB receptors in microglia, enhancing neuronal survival and function (131–133). These examples collectively indicate that the microglial signature genes are largely controlled by the activity of neurons and other glia such as astrocytes, meaning that most of the signature genes are highly and temporally regulated by the microenvironment status of the brain (18, 77, 84, 134).
Using a single microglial biomarker in diversity studies is insufficient for classifying microglia (135, 136). A new trend has arisen where functional studies are using multiple microglial biomarkers to define different “subtypes” of microglia (136). However, it is frustrating to use more than two or three biomarkers to distinguish a microglial population from other glia/neurons. The development of a tool capable of continuous detection of all types of microglial population in any of the brain regions is necessary for future microglial studies regarding its cellular function in any “phenotype” of microglia appearing in a region and a condition impacted by external factors (117, 118). Therefore, it is crucial to ascertain the count of microglial subtypes, comprehend their localization, and identify the adjacent cells in every brain region while determining these biomarkers. Brain region-specific cellular relationships are currently characterized across the human brain as well as the mouse’s through single cell RNA sequencing with anatomical dissections (111, 137–139). Siletti and colleagues comprehensively analyzed 105 anatomical dissections of 4 human brains across 10 brain regions, including the cerebral cortex, hippocampus, cerebral nuclei, hypothalamus, thalamus, midbrain, pons, cerebellum, medulla, and spinal cord. Each single cell was classified into 31 superclusters and 461 clusters using a hierarchical classification system. According to this dataset, microglia were classified as a single supercluster and subsequently divided into 9 clusters (111). In contrast, other glial cell types such as astrocytes were classified as superclusters, comprising 13 clusters. Compared with glia, neurons showed significant variation across different brain regions. For example, medium spiny neurons (MSNs) were classified into 32 clusters, including eccentric MSN clusters (111). This study compares cell clustering combinations in adjacent anatomical dissections and offers new insights into the impact of cell type diversity on regional-specific variations across the human brain through single cell-level transcriptome analysis of regions. Comparative analysis of neighboring anatomical dissections reveals that when a new neural circuit is established for a particular function, gradual changes occur in the neuron’s surroundings, including microglia and astrocytes, instead of creating a “primary function neuron” all at once. Subsequently, the existing “primary function neuron” undergoes fine-tuning to transform into a new “primary function neuron” through interactions with its environment (111, 137). This means that not only individual cell function, but also the function of individual brain regions is strongly influenced by interactions between cells. As a result, identifying genes that are expressed regardless of regional differences and cell-cell interactions becomes a priority.
The gene-based heterogeneity of microglia, however, is limited because current knowledge is provided by information only about a specific snapshot in time. The diversity of microglia due to environmental factors, such as cell–cell interactions and disease states, is more precisely delineated when observed over a broad period. To track environmentally responsive microglial phenotypes and associated protein functions over time, reliable stable markers that are constitutively expressed and independent of the environment are essential. We serendipitously discovered UGT1A7C as a new type of microglia biomarker during the development of a fluorogenic microglia probe called CDr20, an enzyme that marks microglia regardless of environmental differences.
UGT1A7C as a house-keeping biomarker of microglia
According to Matcovitch-Natan et al. RNA-seq data, the development of microglia was identified in three stages: early (until embryonic day 14), pre- (within a few weeks after birth), and adult microglia. They revealed a stepwise developmental program of microglia that is synchronized with the development of the brain. Early microglia were initially formatted with genes related to cell cycling and differentiation, such as Mcm5 and Dab2. Thereafter, the expression of genes related to neurodevelopment, such as Csf1 and Cxcr2, increased and reached its peak a few days before birth. Cd14 and Pmepa1, which are representative genes for mature microglia, were found to be expressed primarily in adult microglia (62). In this dataset, we examined the expression patterns of well-known microglial markers including P2ry12, Tmem119, Csf1r and Cx3cr1 compared to Ugt1a7c across different developmental stages (Figures 1B, C).
The gene P2ry12, which encodes P2RY12, is expressed approximately 82 times more in the adult stage than in the yolk sac stage [(62), Figure 1B]. P2RY12, which was initially identified on platelets as a mediator of platelet activation and blood clotting, is a Gi/o-coupled purinergic receptor expressed in the CNS specifically by homeostatic microglia (140–144). Activation of P2RY12 through ATP/ADP induces rapid microglial chemotaxis and directional branching of microglial processes (65, 145). Additionally, it is involved in activities such as substrate-dependent cell migration and extension in vitro and ex vivo, as well as the regulation of microglial migration (65, 146, 147). P2RY12 is highly expressed along the microglial membrane under normal circumstances (57).. However, after an injury, P2RY12 downregulation occurs (65, 145). This means that microglia may promptly identify alterations in brain homeostasis and react appropriately by expressing P2RY12 (57, 65, 148, 149). The gene encoding transmembrane protein 119 (TMEM119) was expressed 803 times higher in the adult stage than in the yolk sac stage (62). Tmem119, also referred to as Obif (Osteoblast induction factor), is expressed exclusively in microglia in the brains of mice and humans, allowing for distinction from infiltrating blood-derived macrophages (81, 89). Its function in microglia remains unclear (81, 82, 89, 94). TMEM119 is located in the endoplasmic reticulum and plasma membrane (150–152). It is also expressed in several organs like the alimentary system, genitourinary system, limb, and skeleton as well as the brain (153, 154). A previous study found that frozen tissue samples of the frontal cortex from patients with Alzheimer’s had increased levels of TMEM119 mRNA (81, 155). Transforming growth factor beta (TGF-β) induced an upregulation of Tmem119 gene expression in cultured mouse microglia (156). In contrast, lipopolysaccharide (LPS), interleukin 4 (IL–4), or interferon-gamma (IFN-γ) induced downregulation of TMEM119 gene expression in cultured human microglia (81, 89). The diverse expression patterns of TMEM119 appear to be influenced by inflammatory responses and environmental factors (82).
In contrast to the wide variation observed in the expression of P2ry12 and Tmem119, the expression of Csf1r and Cx3cr1 remains relatively stable across microglial developmental stages, with only 9-fold and 13-fold changes, respectively [(62), Figure 1B]. Csf1r encodes the colony-stimulating factor 1 receptor (CSF1R), a member of the tyrosine kinase receptor family (157). Upon stimulation by its ligands, including CSF1 and interleukin 34, CSF1R undergoes autophosphorylation of tyrosine residues in the intracellular domain, followed by activation of downstream signaling pathways (158). CSF1R is primarily expressed in the microglia of the brain and is crucial for their survival, proliferation, and differentiation (159–162). The gene Cx3cr1 encodes the receptor for the C-X3-C chemokine fractalkine (CX3CL1), which is found in numerous leukocyte cells during early development (163–165). Signaling through CX3CR1-CX3CL1 exerts distinct functions in various tissue compartments, including immune response, inflammation, cell adhesion, and chemotaxis (163, 166–169). It controls the inflammation process that triggers atherogenesis, by facilitating the recruitment of macrophages and monocytes to inflamed atherosclerotic plaques, thus promoting cell survival (170, 171). CX3CR1 plays a crucial role in regulating the inflammatory response and synapse maturation in CNS microglia (122, 172, 173). During postnatal brain development, the brain participates in synaptic pruning, a natural process in which brain microglia eliminate extra synapses (122, 123, 133, 174, 175). Interestingly, despite the relative stability of both Csf1r and Cx3cr1 expression, the fluctuation of their expression levels around 10-fold makes the two genes more suitable for distinguishing microglial subtypes rather than serving as housekeeping biomarkers for microglia (62).
Surprisingly, Ugt1a7c expression remains remarkably stable throughout the yolk sac, pre-microglia, and adult microglia stage, exhibiting only a 4-fold difference between maximum and minimum expression [(62), Figure 1B]. The peak expression period occurs between 3 and 6 days after birth, with expression levels remaining moderate thereafter as the microglia reach full maturity (62). The Ugt1a7c gene belongs to the UGT (UDP-Glycosyltransferase) gene family and is the only member enriched in microglia (176–179). The UGT gene families found in animals, plants, fungi, and bacteria facilitate phase II biotransformation reactions (180–184). They conjugate lipophilic substrates with glucuronic acid, from the UDP-glucuronic acid to the functional hydroxyl group of substrates, resulting in increasing hydrophilicity and facilitating excretion through bile and urine in the systemic organs eventually (176, 184–188). The role of Ugt1a7c in the brain, however, has not been extensively studied because its expression is not high compared with that of microglia-specific genes such as P2ry12 and Tmem119, although its expression remains constant throughout development (Figures 1B, C).
To assess the stability of Ugt1a7c expression in mouse brains across different ages, we employed the UGT1A7C-specific substrate, CDr20, and conducted further evaluation. Interestingly, the fluorescence intensity of CDr20 remained unchanged across all tested ages, ranging from 3 months to 18 months (Figure 2A). This indicates that Ugt1a7c activity persists throughout the lifespan of the mouse. In addition to physiological condition, treatment with various activators, such as LPS, IFNγ, LPS/IFNγ, ATP, IL-13, or Aβ, did not alter the activity or expression of Ugt1a7c in mouse microglia (Figures 2B, C). This indicates that the stability of Ugt1a7c expression is extensive, even when strong environmental factors or activators are present. The stability of Ugt1a7c expression was also confirmed, as all Iba1-positive microglia in an aged AD animal could be labeled with CDr20, irrespective of their localization to Aβ aggregates (Figure 2D).
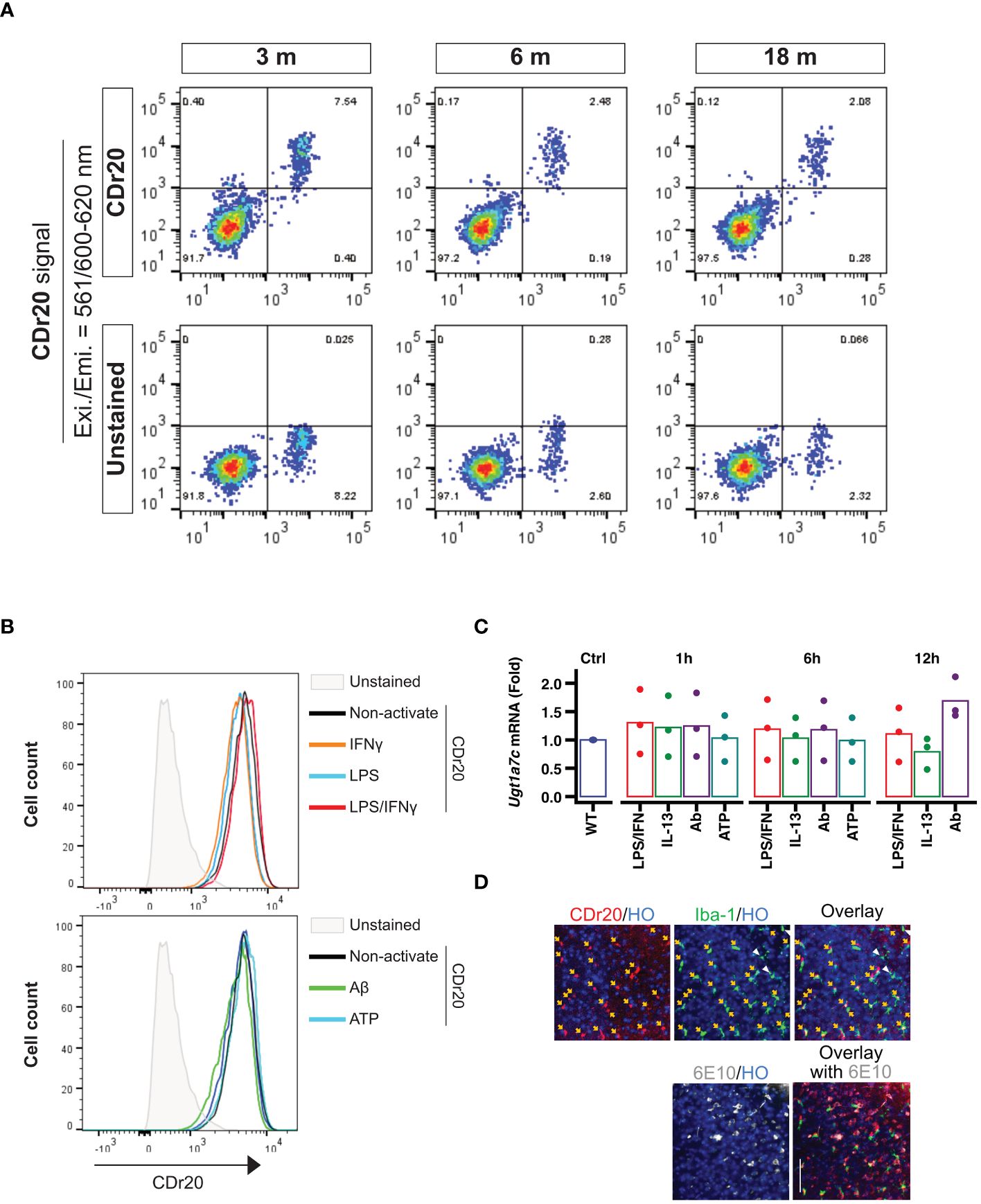
Figure 2 Environmental factor independent UGT1A7C. (A) Age-independent Ugt1a7c. The intensities from CDr20 (UGT1A7C-specific fluorescence substrate)-derived fluorescence were analyzed by treatment of live single cells dissociated from each age of the whole brain of a mouse. (B) Activation-independent Ugt1a7c. CDr20 fluorescence intensity was analyzed by each activation stimulation. (C) Activation-independent Ugt1a7c. Ugt1a7c mRNA expression levels were tracked at each time point. (D) AD-independent Ugt1a7c. Fluorescence image of the CDr20 (upper left, UGT1A7C-positive cells) in a live cortical brain slice after 30 minutes of treatment and of the immunostaining of Iba-1 (upper middle, microglia) and 6E10 (lower left, Aβ aggregates) after fixation of the tissue. Live and immunostaining images were superimposed (upper/lower right). Created with BioRender.com.
In summary, the expression of particular genes in microglia changes according to their developmental stage, the brain region they reside in, and the surroundings in which they communicate. Microglia express a variety of genes, allowing them to precisely adjust neural circuits and control inflammatory responses. Even though they are only expressed in microglia, there are still genes whose functions and sequences remain unknown. The UGT1A7C is essential for phase II biotransformation reactions, aiding in the elimination of lipophilic substances from the body, and is exclusive to microglia. While the role of Ugt1a7c in the brain is still uncertain, its recognition as a microglia marker has been achieved through the development of a fluorescent substrate, despite its low expression level. Ugt1a7c remains consistently expressed, regardless of microglial activity, developmental stage, or disease state, making it distinct from other markers (Figures 1, 2).
In recent years, fluorescent probes for functional enzymes have attracted considerable attention because of their inherent advantages, such as high sensitivity, cost-effectiveness, and applicability to high-throughput screening (HTS). However, developing a practical fluorescent probe for a given UGT enzyme remains challenging for the following two reasons. First, UGTs within a subfamily share high amino acid sequence homology (>65%) and usually exhibit broad and overlapping substrate specificity (184). Second, the fluorescence properties of many fluorophores are often “turned off” following O-glucuronidation at the hydroxyl group (116, 189–191). Interestingly, the novel fluorogenic microglia probe, CDr20, identified by unbiased high-content imaging screening with over a thousand of small fluorescent molecules, was a specific exogenous substrate of UGT1A7C after genome-wide CRISPR/Cas9 knockout screening in BV2 microglia. CDr20 was able to label only microglia with high specificity and sensitivity in the mixture of primary glia culture and even in the brain in vivo (83). This means that the low expression levels of UGT1A7C in microglia are functional enough for the specific labeling of microglia with its fluorescence substrate.
Conclusion
Microglial detection with CDr20 is not affected by developmental stages, disease, or environmental factors. This indicates that Ugt1a7c is a very stable gene in microglia like housekeeping genes and performs its enzymatic function in both silent and active states of microglia constantly. Although the function of this protein in microglia is not yet fully understood, similar to that of TMEM119, we speculate that this new biomarker is very interesting to other microglia biomarkers because of its unique property as the microglia’s housekeeping gene in the brain and the existence of its specific fluorogenic substrate.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) of Korea Brain Research Institute (KBRI). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
WK: Conceptualization, Data curation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. MK: Writing – review & editing. BK: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by KBRI basic research program through Korea Brain Research Institute, funded by Ministry of Science and ICT (24-BR-01-03, 24-BR-03-04, and 24-BR-04-04), and by the Basic Science Research Program (NRF-2021R1A2C1013975) through the National Research Foundation of Korea to BK. The funders had no role in study design, data collection, analysis, publication decision, or manuscript preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol. (2018) 18:225–42. doi: 10.1038/nri.2017.125
2. Aguzzi A, Barres BA, Bennett ML. Microglia: scapegoat, saboteur, or something else? Science. (2013) 339:156–61. doi: 10.1126/science.1227901
3. Prinz M, Erny D, Hagemeyer N. Ontogeny and homeostasis of CNS myeloid cells. Nat Immunol. (2017) 18:385–92. doi: 10.1038/ni.3703
4. Frost JL, Schafer DP. Microglia: architects of the developing nervous system. Trends Cell Biol. (2016) 26:587–97. doi: 10.1016/j.tcb.2016.02.006
5. Schafer DP, Stevens B. Microglia function in central nervous system development and plasticity. Cold Spring Harbor Perspect Biol. (2015) 7:a020545. doi: 10.1101/cshperspect.a020545
6. Cserép C, Pósfai B, Lénárt N, Fekete R, László ZI, Lele Z, et al. Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science. (2020) 367:528–37. doi: 10.1126/science.aax6752
7. Kierdorf K, Prinz M. Microglia in steady state. J Clin Investig. (2017) 127:3201–9. doi: 10.1172/JCI90602
8. Thion MS, Ginhoux F, Garel S. Microglia and early brain development: An intimate journey. Science. (2018) 362:185–9. doi: 10.1126/science.aat0474
9. Tan Y-L, Yuan Y, Tian L. Microglial regional heterogeneity and its role in the brain. Mol Psychiatry. (2020) 25:351–67. doi: 10.1038/s41380-019-0609-8
10. Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, et al. Microglial brain region–dependent diversity and selective regional sensitivities to aging. Nat Neurosci. (2016) 19:504–16. doi: 10.1038/nn.4222
11. Li Y, Li Z, Yang M, Wang F, Zhang Y, Li R, et al. Decoding the temporal and regional specification of microglia in the developing human brain. Cell Stem Cell. (2022) 29:620–34.e6. doi: 10.1016/j.stem.2022.02.004
12. Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. (1990) 39:151–70. doi: 10.1016/0306-4522(90)90229-W
13. Priller J, Prinz M. Targeting microglia in brain disorders. Science. (2019) 365:32–3. doi: 10.1126/science.aau9100
14. Oosterhof N, Chang IJ, Karimiani EG, Kuil LE, Jensen DM, Daza R, et al. Homozygous mutations in CSF1R cause a pediatric-onset leukoencephalopathy and can result in congenital absence of microglia. Am J Hum Genet. (2019) 104:936–47. doi: 10.1016/j.ajhg.2019.03.010
15. Mass E, Jacome-Galarza CE, Blank T, Lazarov T, Durham BH, Ozkaya N, et al. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature. (2017) 549:389–93. doi: 10.1038/nature23672
16. Chung H-Y, Wickel J, Hahn N, Mein N, Schwarzbrunn M, Koch P, et al. Microglia mediate neurocognitive deficits by eliminating C1q-tagged synapses in sepsis-associated encephalopathy. Sci Adv. (2023) 9:eabq7806. doi: 10.1126/sciadv.abq7806
17. Langston RG, Beilina A, Reed X, Kaganovich A, Singleton AB, Blauwendraat C, et al. Association of a common genetic variant with Parkinson’s disease is mediated by microglia. Sci Transl Med. (2022) 14:eabp8869. doi: 10.1126/scitranslmed.abp8869
18. Crotti A, Benner C, Kerman BE, Gosselin D, Lagier-Tourenne C, Zuccato C, et al. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci. (2014) 17:513–21. doi: 10.1038/nn.3668
19. Kim B, Suh YH, Joe E. LRRK2 decreases microglial actin dynamics by filamentous actin depolymerization and Rac1 inhibition. Anim Cells Syst (Seoul). (2022) 26:380–7. doi: 10.1080/19768354.2022.2158219
20. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. (2010) 330:841–5. doi: 10.1126/science.1194637
21. Schulz C, Perdiguero EG, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science. (2012) 336:86–90. doi: 10.1126/science.1219179
22. Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. (2013) 16:273–80. doi: 10.1038/nn.3318
23. Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Dev Brain Res. (1999) 117:145–52. doi: 10.1016/S0165-3806(99)00113-3
24. Cuadros MA, Sepulveda MR, Martin-Oliva D, Marín-Teva JL, Neubrand VE. Microglia and microglia-like cells: similar but different. Front Cell Neurosci. (2022) 16:816439. doi: 10.3389/fncel.2022.816439
25. Zhang W, Jiang J, Xu Z, Yan H, Tang B, Liu C, et al. Microglia-containing human brain organoids for the study of brain development and pathology. Mol Psychiatry. (2023) 28:96–107. doi: 10.1038/s41380-022-01892-1
26. Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. (2000) 20:4106–14. doi: 10.1128/MCB.20.11.4106-4114.2000
27. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. (2012) 13:1118–28. doi: 10.1038/ni.2419
28. Low D, Ginhoux F. Recent advances in the understanding of microglial development and homeostasis. Cell Immunol. (2018) 330:68–78. doi: 10.1016/j.cellimm.2018.01.004
29. Dijkstra CD, Döpp EA, Joling P, Kraal G. The heterogeneity of mononuclear phagocytes in lymphoid organs: distinct macrophage subpopulations in the rat recognized by monoclonal antibodies ED1, ED2 and ED3. Immunology. (1985) 54:589–99.
30. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. (2015) 518:547–51. doi: 10.1038/nature13989
31. Mizutani M, Pino PA, Saederup N, Charo IF, Ransohoff RM, Cardona AE. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J Immunol. (2012) 188:29–36. doi: 10.4049/jimmunol.1100421
32. Jurga AM, Paleczna M, Kuter KZ. Overview of general and discriminating markers of differential microglia phenotypes. Front Cell Neurosci. (2020) 14:198. doi: 10.3389/fncel.2020.00198
33. Rumianek AN, Davies B, Channon KM, Greaves DR, Purvis GSD. A human CD68 promoter-driven inducible cre-recombinase mouse line allows specific targeting of tissue resident macrophages. Front Immunol. (2022) 13:918636. doi: 10.3389/fimmu.2022.918636
34. Lin HH, Faunce DE, Stacey M, Terajewicz A, Nakamura T, Zhang-Hoover J, et al. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J Exp Med. (2005) 201:1615–25. doi: 10.1084/jem.20042307
35. Roesch S, Rapp C, Dettling S, Herold-Mende C. When immune cells turn bad-tumor-associated microglia/macrophages in glioma. Int J Mol Sci. (2018) 19. doi: 10.3390/ijms19020436
36. Xiao C, Liu N, Jacobson KA, Gavrilova O, Reitman ML. Physiology and effects of nucleosides in mice lacking all four adenosine receptors. PloS Biol. (2019) 17:e3000161. doi: 10.1371/journal.pbio.3000161
37. Bi J, Zheng C, Zheng X. Increased expression of adenosine A3 receptor in tumor-infiltrating natural killer cells. Cell Mol Immunol. (2021) 18:496–7. doi: 10.1038/s41423-020-00632-1
38. Coppi E, Cherchi F, Lucarini E, Ghelardini C, Pedata F, Jacobson KA, et al. Uncovering the mechanisms of adenosine receptor-mediated pain control: focus on the A(3) receptor subtype. Int J Mol Sci. (2021) 22. doi: 10.3390/ijms22157952
39. Thrupp N, Sala Frigerio C, Wolfs L, Skene NG, Fattorelli N, Poovathingal S, et al. Single-nucleus RNA-seq is not suitable for detection of microglial activation genes in humans. Cell Rep. (2020) 32:108189. doi: 10.1016/j.celrep.2020.108189
40. Eudy BJ, da Silva RP. Systematic deletion of adenosine receptors reveals novel roles in inflammation and pyroptosis in THP-1 macrophages. Mol Immunol. (2021) 132:1–7. doi: 10.1016/j.molimm.2021.01.018
41. Sasaki Y, Ohsawa K, Kanazawa H, Kohsaka S, Imai Y. Iba1 is an actin-cross-linking protein in macrophages/microglia. Biochem Biophys Res Commun. (2001) 286:292–7. doi: 10.1006/bbrc.2001.5388
42. Moore KJ, Andersson LP, Ingalls RR, Monks BG, Li R, Arnaout MA, et al. Divergent response to LPS and bacteria in CD14-deficient murine macrophages. J Immunol. (2000) 165:4272–80. doi: 10.4049/jimmunol.165.8.4272
43. Baumann CL, Aspalter IM, Sharif O, Pichlmair A, Bluml S, Grebien F, et al. CD14 is a coreceptor of Toll-like receptors 7 and 9. J Exp Med. (2010) 207:2689–701. doi: 10.1084/jem.20101111
44. Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, et al. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. (2011) 147:868–80. doi: 10.1016/j.cell.2011.09.051
45. Watanabe M, Fujihara C, Radtke AJ, Chiang YJ, Bhatia S, Germain RN, et al. Co-stimulatory function in primary germinal center responses: CD40 and B7 are required on distinct antigen-presenting cells. J Exp Med. (2017) 214:2795–810. doi: 10.1084/jem.20161955
46. Park CG, Thiex NW, Lee KM, Szot GL, Bluestone JA, Lee KD. Targeting and blocking B7 costimulatory molecules on antigen-presenting cells using CTLA4Ig-conjugated liposomes: in vitro characterization and in vivo factors affecting biodistribution. Pharm Res. (2003) 20:1239–48. doi: 10.1023/A:1025057216492
47. Lebedeva T, Dustin ML, Sykulev Y. ICAM-1 co-stimulates target cells to facilitate antigen presentation. Curr Opin Immunol. (2005) 17:251–8. doi: 10.1016/j.coi.2005.04.008
48. Li J, Chen K, Zhu L, Pollard JW. Conditional deletion of the colony stimulating factor-1 receptor (c-fms proto-oncogene) in mice. Genesis. (2006) 44:328–35. doi: 10.1002/(ISSN)1526-968X
49. Jenkins SJ, Ruckerl D, Thomas GD, Hewitson JP, Duncan S, Brombacher F, et al. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J Exp Med. (2013) 210:2477–91. doi: 10.1084/jem.20121999
50. Wada Y, Nagai A, Sheikh AM, Onoda K, Terashima M, Shiota Y, et al. Co-localization of cystatin C and prosaposin in cultured neurons and in anterior horn neurons with amyotrophic lateral sclerosis. J Neurol Sci. (2018) 384:67–74. doi: 10.1016/j.jns.2017.11.023
51. Jones BA, Beamer M, Ahmed S. Fractalkine/CX3CL1: a potential new target for inflammatory diseases. Mol Interv. (2010) 10:263–70. doi: 10.1124/mi.10.5.3
52. Ikeda K, Nakano R, Uraoka M, Nakagawa Y, Koide M, Katsume A, et al. Identification of ARIA regulating endothelial apoptosis and angiogenesis by modulating proteasomal degradation of cIAP-1 and cIAP-2. Proc Natl Acad Sci U S A. (2009) 106:8227–32. doi: 10.1073/pnas.0806780106
53. Hu Y, Yao Y, Qi H, Yang J, Zhang C, Zhang A, et al. Microglia sense and suppress epileptic neuronal hyperexcitability. Pharmacol Res. (2023) 195:106881. doi: 10.1016/j.phrs.2023.106881
54. Baker K, Rath T, Pyzik M, Blumberg RS. The role of fcRn in antigen presentation. Front Immunol. (2014) 5:408. doi: 10.3389/fimmu.2014.00408
55. Lorenz V, Stegner D, Stritt S, Vogtle T, Kiefer F, Witke W, et al. Targeted downregulation of platelet CLEC-2 occurs through Syk-independent internalization. Blood. (2015) 125:4069–77. doi: 10.1182/blood-2014-11-611905
56. Mukherjee S, Klaus C, Pricop-Jeckstadt M, Miller JA, Struebing FL. A microglial signature directing human aging and neurodegeneration-related gene networks. Front Neurosci. (2019) 13:2. doi: 10.3389/fnins.2019.00002
57. Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat Neurosci. (2014) 17:131–43. doi: 10.1038/nn.3599
58. Ochoa-Dragos Z, Imai DM, Reinholdt L, Schile A. Lysosomal Storage Disease Caused by a Spontaneous Hexb Gene Mutation in Immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ Mice. J Am Assoc Lab Anim Sci. (2019) 58:607–726.
59. Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. (2005) 115:56–65. doi: 10.1172/JCI22675
60. Jeong HK, Ji K, Min K, Joe EH. Brain inflammation and microglia: facts and misconceptions. Exp Neurobiol. (2013) 22:59–67. doi: 10.5607/en.2013.22.2.59
61. Chakrabarty P, Jansen-West K, Beccard A, Ceballos-Diaz C, Levites Y, Verbeeck C, et al. Massive gliosis induced by interleukin-6 suppresses Abeta deposition in vivo: evidence against inflammation as a driving force for amyloid deposition. FASEB J. (2010) 24:548–59. doi: 10.1096/fj.09-141754
62. Matcovitch-Natan O, Winter DR, Giladi A, Aguilar SV, Spinrad A, Sarrazin S, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. (2016) 353:aad8670. doi: 10.1126/science.aad8670
63. Xu J, Gong NL, Bodi I, Aronow BJ, Backx PH, Molkentin JD. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J Biol Chem. (2006) 281:9152–62. doi: 10.1074/jbc.M510217200
64. Li H, Wang F, Guo X, Jiang Y. Decreased MEF2A expression regulated by its enhancer methylation inhibits autophagy and may play an important role in the progression of alzheimer’s disease. Front Neurosci. (2021) 15:682247. doi: 10.3389/fnins.2021.682247
65. Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan W-B, et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci. (2006) 9:1512–9. doi: 10.1038/nn1805
66. McKinsey GL, Lizama CO, Keown-Lang AE, Niu A, Santander N, Larpthaveesarp A, et al. A new genetic strategy for targeting microglia in development and disease. Elife. (2020) 9. doi: 10.7554/eLife.54590
67. Amadio S, Parisi C, Montilli C, Carrubba AS, Apolloni S, Volonte C. P2Y(12) receptor on the verge of a neuroinflammatory breakdown. Mediators Inflamm. (2014) 2014:975849. doi: 10.1155/2014/975849
68. Kyrargyri V, Madry C, Rifat A, Arancibia-Carcamo IL, Jones SP, Chan VTT, et al. P2Y(13) receptors regulate microglial morphology, surveillance, and resting levels of interleukin 1beta release. Glia. (2020) 68:328–44. doi: 10.1002/glia.23719
69. Schluns KS, Williams K, Ma A, Zheng XX, Lefrancois L. Cutting edge: requirement for IL-15 in the generation of primary and memory antigen-specific CD8 T cells. J Immunol. (2002) 168:4827–31. doi: 10.4049/jimmunol.168.10.4827
70. Rice RA, Pham J, Lee RJ, Najafi AR, West BL, Green KN. Microglial repopulation resolves inflammation and promotes brain recovery after injury. Glia. (2017) 65:931–44. doi: 10.1002/glia.23135
71. Butovsky O, Weiner HL. Microglial signatures and their role in health and disease. Nat Rev Neurosci. (2018) 19:622–35. doi: 10.1038/s41583-018-0057-5
72. Kim JS, Kolesnikov M, Peled-Hajaj S, Scheyltjens I, Xia Y, Trzebanski S, et al. A binary cre transgenic approach dissects microglia and CNS border-associated macrophages. Immunity. (2021) 54:176–90 e7. doi: 10.1016/j.immuni.2020.11.007
73. Fixsen BR, Han CZ, Zhou Y, Spann NJ, Saisan P, Shen Z, et al. SALL1 enforces microglia-specific DNA binding and function of SMADs to establish microglia identity. Nat Immunol. (2023) 24:1188–99. doi: 10.1038/s41590-023-01528-8
74. Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol. (2016) 17:1397–406. doi: 10.1038/ni.3585
75. Norose K, Clark JI, Syed NA, Basu A, Heber-Katz E, Sage EH, et al. SPARC deficiency leads to early-onset cataractogenesis. Invest Ophthalmol Vis Sci. (1998) 39:2674–80.
76. Lloyd-Burton SM, York EM, Anwar MA, Vincent AJ, Roskams AJ. SPARC regulates microgliosis and functional recovery following cortical ischemia. J Neurosci. (2013) 33:4468–81. doi: 10.1523/JNEUROSCI.3585-12.2013
77. Gosselin D, Link VM, Romanoski Casey E, Fonseca Gregory J, Eichenfield Dawn Z, Spann Nathanael J, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. (2014) 159:1327–40. doi: 10.1016/j.cell.2014.11.023
78. Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H, et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol. (2005) 25:2832–45. doi: 10.1128/MCB.25.7.2832-2845.2005
80. Rothhammer V, Borucki DM, Tjon EC, Takenaka MC, Chao C-C, Ardura-Fabregat A, et al. Microglial control of astrocytes in response to microbial metabolites. Nature. (2018) 557:724–8. doi: 10.1038/s41586-018-0119-x
81. Satoh Ji, Kino Y, Asahina N, Takitani M, Miyoshi J, Ishida T, et al. TMEM119 marks a subset of microglia in the human brain. Neuropathology. (2016) 36:39–49. doi: 10.1111/neup.12235
82. Kaiser T, Feng G. Tmem119-EGFP and tmem119-creERT2 transgenic mice for labeling and manipulating microglia. eNeuro. (2019) 6:ENEURO.0448–18.2019. doi: 10.1523/eneuro.0448-18.2019
83. Kim B, Fukuda M, Lee JY, Su D, Sanu S, Silvin A, et al. Visualizing microglia with a fluorescence turn-on ugt1a7c substrate. Angew Chem. (2019) 131:8056–60. doi: 10.1002/ange.201903058
84. Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. (2017) 356. doi: 10.1126/science.aal3222
85. Goldmann T, Wieghofer P, Jordão MJC, Prutek F, Hagemeyer N, Frenzel K, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol. (2016) 17:797–805. doi: 10.1038/ni.3423
86. Macosko Evan Z, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. (2015) 161:1202–14. doi: 10.1016/j.cell.2015.05.002
87. Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell. (2018) 172:500–16.e16. doi: 10.1016/j.cell.2017.11.042
88. Johnson ECB, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med. (2020) 26:769–80. doi: 10.1038/s41591-020-0815-6
89. Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci. (2016) 113:E1738–E46. doi: 10.1073/pnas.1525528113
90. Eme-Scolan E, Dando SJ. Tools and approaches for studying microglia in vivo. Front Immunol. (2020) 11:583647. doi: 10.3389/fimmu.2020.583647
91. Wright-Jin EC, Gutmann DH. Microglia as dynamic cellular mediators of brain function. Trends Mol Med. (2019) 25:967–79. doi: 10.1016/j.molmed.2019.08.013
92. Hanamsagar R, Bilbo SD. Environment matters: microglia function and dysfunction in a changing world. Curr Opin Neurobiol. (2017) 47:146–55. doi: 10.1016/j.conb.2017.10.007
93. Lier J, Streit WJ, Bechmann I. Beyond activation: characterizing microglial functional phenotypes. Cells. (2021) 10:2236. doi: 10.3390/cells10092236
94. Ruan C, Elyaman W. A new understanding of TMEM119 as a marker of microglia. Front Cell Neurosci. (2022) 16:902372. doi: 10.3389/fncel.2022.902372
95. Utz SG, See P, Mildenberger W, Thion MS, Silvin A, Lutz M, et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell. (2020) 181:557–73 e18. doi: 10.1016/j.cell.2020.03.021
96. Huang Y, Xu Z, Xiong S, Sun F, Qin G, Hu G, et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat Neurosci. (2018) 21:530–40. doi: 10.1038/s41593-018-0090-8
97. Vidal-Itriago A, Radford RAW, Aramideh JA, Maurel C, Scherer NM, Don EK, et al. Microglia morphophysiological diversity and its implications for the CNS. Front Immunol. (2022) 13:997786. doi: 10.3389/fimmu.2022.997786
98. Masuda T, Sankowski R, Staszewski O, Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. (2020) 30:1271–81. doi: 10.1016/j.celrep.2020.01.010
99. Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, L-c W, TK M, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. (2013) 16:1896–905. doi: 10.1038/nn.3554
100. Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. (2014) 34:11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014
101. Soreq L, Bird H, Mohamed W, Hardy J. Single-cell RNA sequencing analysis of human Alzheimer’s disease brain samples reveals neuronal and glial specific cells differential expression. PloS One. (2023) 18:e0277630. doi: 10.1371/journal.pone.0277630
102. Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, et al. Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell. (2019) 179:1609–22.e16. doi: 10.1016/j.cell.2019.11.010
103. Hellwig S, Heinrich A, Biber K. The brain’s best friend: microglial neurotoxicity revisited. Front Cell Neurosci. (2013) 7:71. doi: 10.3389/fncel.2013.00071
104. Ransohoff RM. A polarizing question: do M1 and M2 microglia exist? Nat Neurosci. (2016) 19:987–91. doi: 10.1038/nn.4338
105. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. (2014) 6:13. doi: 10.12703/P
106. Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. (2005) 8:752–8. doi: 10.1038/nn1472
107. Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. (2005) 308:1314–8. doi: 10.1126/science.1110647
108. Fernández-Zapata C, Leman JKH, Priller J, Böttcher C. The use and limitations of single-cell mass cytometry for studying human microglia function. Brain Pathol. (2020) 30:1178–91. doi: 10.1111/bpa.12909
109. Gao C, Jiang J, Tan Y, Chen S. Microglia in neurodegenerative diseases: mechanism and potential therapeutic targets. Signal Transduct Target Ther. (2023) 8:359. doi: 10.1038/s41392-023-01588-0
110. Zhang M, Pan X, Jung W, Halpern AR, Eichhorn SW, Lei Z, et al. Molecularly defined and spatially resolved cell atlas of the whole mouse brain. Nature. (2023) 624:343–54. doi: 10.1038/s41586-023-06808-9
111. Siletti K, Hodge R, Albiach AM, Lee KW, Ding S-L, Hu L, et al. Transcriptomic diversity of cell types across the adult human brain. Science. (2023) 382:eadd7046. doi: 10.1126/science.add7046
112. Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. (2019) 101:207–23.e10. doi: 10.1016/j.neuron.2018.12.006
113. Psy NBB, Böttcher C, Schlickeiser S, Sneeboer MAM, Kunkel D, Knop A, et al. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat Neurosci. (2019) 22:78–90. doi: 10.1038/s41593-018-0290-2
114. Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar, Sindram E, et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat Neurosci. (2019) 22:2098–110. doi: 10.1038/s41593-019-0532-y
115. Mrdjen D, Pavlovic A, Hartmann FJ, Schreiner B, Utz SG, Leung BP, et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity. (2018) 48:380–95.e6. doi: 10.1016/j.immuni.2018.01.011
116. Feng L, Ning J, Tian X, Wang C, Zhang L, Ma X, et al. Fluorescent probes for bioactive detection and imaging of phase II metabolic enzymes. Coord Chem Rev. (2019) 399:213026. doi: 10.1016/j.ccr.2019.213026
117. Barko K, Shelton M, Xue X, Afriyie-Agyemang Y, Puig S, Freyberg Z, et al. Brain region- and sex-specific transcriptional profiles of microglia. Front Psychiatry. (2022) 13:945548. doi: 10.3389/fpsyt.2022.945548
118. Soares NL, Vieira HLA. Microglia at the centre of brain research: accomplishments and challenges for the future. Neurochem Res. (2022) 47:218–33. doi: 10.1007/s11064-021-03456-1
119. Camacho-Hernández NP, Peña-Ortega F. Fractalkine/CX3CR1-dependent modulation of synaptic and network plasticity in health and disease. Neural Plast. (2023) 2023:4637073. doi: 10.1155/2023/4637073
120. Imai T, Hieshima K, Haskell C, Baba M, Nagira M, Nishimura M, et al. Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell. (1997) 91:521–30. doi: 10.1016/S0092-8674(00)80438-9
121. Combadiere C, Salzwedel K, Smith ED, Tiffany HL, Berger EA, Murphy PM. Identification of CX 3CR1 A CHEMOTACTIC RECEPTOR FOR THE HUMAN CX 3C CHEMOKINE FRACTALKINE AND A FUSION CORECEPTOR FOR HIV-1*. J Biol Chem. (1998) 273:23799–804. doi: 10.1074/jbc.273.37.23799
122. Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. (2011) 333:1456–8. doi: 10.1126/science.1202529
123. Cornell J, Salinas S, Huang H-Y, Zhou M. Microglia regulation of synaptic plasticity and learning and memory. Neural Regener Res. (2021) 17:705–16. doi: 10.4103/1673-5374.322423
124. Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, Fatimy RE, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. (2017) 47:566–81.e9. doi: 10.1016/j.immuni.2017.08.008
125. McQuade A, Kang YJ, Hasselmann J, Jairaman A, Sotelo A, Coburn M, et al. Gene expression and functional deficits underlie TREM2-knockout microglia responses in human models of Alzheimer’s disease. Nat Commun. (2020) 11:5370. doi: 10.1038/s41467-020-19227-5
126. Chintamen S, Gaur P, Vo N, Bradshaw EM, Menon V, Kernie SG. Distinct microglial transcriptomic signatures within the hippocampus. PloS One. (2024) 19:e0296280. doi: 10.1371/journal.pone.0296280
127. McNamara NB, Munro DAD, Bestard-Cuche N, Uyeda A, Bogie JFJ, Hoffmann A, et al. Microglia regulate central nervous system myelin growth and integrity. Nature. (2023) 613:120–9. doi: 10.1038/s41586-022-05534-y
128. Safaiyan S, Besson-Girard S, Kaya T, Cantuti-Castelvetri L, Liu L, Ji H, et al. White matter aging drives microglial diversity. Neuron. (2021) 109:1100–17.e10. doi: 10.1016/j.neuron.2021.01.027
129. Sen MK, Mahns DA, Coorssen JR, Shortland PJ. The roles of microglia and astrocytes in phagocytosis and myelination: Insights from the cuprizone model of multiple sclerosis. Glia. (2022) 70:1215–50. doi: 10.1002/glia.24148
130. Fu R, Shen Q, Xu P, Luo JJ, Tang Y. Phagocytosis of microglia in the central nervous system diseases. Mol Neurobiol. (2014) 49:1422–34. doi: 10.1007/s12035-013-8620-6
131. Cramer T, Gill R, Thirouin ZS, Vaas M, Sampath S, Martineau F, et al. Cross-talk between GABAergic postsynapse and microglia regulate synapse loss after brain ischemia. Sci Adv. (2022) 8:eabj0112. doi: 10.1126/sciadv.abj0112
132. Ferrini F, Koninck YD. Microglia control neuronal network excitability via BDNF signalling. Neural Plast. (2013) 2013:429815. doi: 10.1155/2013/429815
133. Parkhurst Christopher N, Yang G, Ninan I, Savas Jeffrey N, Yates John R, Lafaille Juan J, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. (2013) 155:1596–609. doi: 10.1016/j.cell.2013.11.030
134. Ochocka N, Segit P, Walentynowicz KA, Wojnicki K, Cyranowski S, Swatler J, et al. Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages. Nat Commun. (2021) 12:1151. doi: 10.1038/s41467-021-21407-w
135. Clevers H, Rafelski S, Elowitz M, Klein A, Shendure J, Trapnell C, et al. What is your conceptual definition of “Cell type” in the context of a mature organism? Cell Syst. (2017) 4:255–9. doi: 10.1016/j.cels.2017.03.006
136. Stratoulias V, Venero JL, Tremblay MÈ, Joseph B. Microglial subtypes: diversity within the microglial community. EMBO J. (2019) 38:e101997. doi: 10.15252/embj.2019101997
137. Weninger A, Arlotta P. A family portrait of human brain cells. Science. (2023) 382:168–9. doi: 10.1126/science.adk4857
138. Jorstad NL, Close J, Johansen N, Yanny AM, Barkan ER, Travaglini KJ, et al. Transcriptomic cytoarchitecture reveals principles of human neocortex organization. Science. (2023) 382:eadf6812. doi: 10.1126/science.adf6812
139. Langlieb J, Sachdev NS, Balderrama KS, Nadaf NM, Raj M, Murray E, et al. The molecular cytoarchitecture of the adult mouse brain. Nature. (2023) 624:333–42. doi: 10.1038/s41586-023-06818-7
140. Lowery RL, Mendes MS, Sanders BT, Murphy AJ, Whitelaw BS, Lamantia CE, et al. Loss of P2Y12 has behavioral effects in the adult mouse. Int J Mol Sci. (2021) 22:1868. doi: 10.3390/ijms22041868
141. Sasaki Y, Hoshi M, Akazawa C, Nakamura Y, Tsuzuki H, Inoue K, et al. Selective expression of Gi/o-coupled ATP receptor P2Y12 in microglia in rat brain. Glia. (2003) 44:242–50. doi: 10.1002/glia.10293
142. Hollopeter G, Jantzen H-M, Vincent D, Li G, England L, Ramakrishnan V, et al. Identification of the platelet ADP receptor targeted by antithrombotic drugs. Nature. (2001) 409:202–7. doi: 10.1038/35051599
143. Kettenmann H, Hanisch U-K, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. (2011) 91:461–553. doi: 10.1152/physrev.00011.2010
144. Fontainhas AM, Wang M, Liang KJ, Chen S, Mettu P, Damani M, et al. Microglial morphology and dynamic behavior is regulated by ionotropic glutamatergic and GABAergic neurotransmission. PloS One. (2011) 6:e15973. doi: 10.1371/journal.pone.0015973
145. Honda S, Sasaki Y, Ohsawa K, Imai Y, Nakamura Y, Inoue K, et al. Extracellular ATP or ADP induce chemotaxis of cultured microglia through gi/o-coupled P2Y receptors. J Neurosci. (2001) 21:1975–82. doi: 10.1523/JNEUROSCI.21-06-01975.2001
146. Bisht K, Okojie KA, Sharma K, Lentferink DH, Sun Y-Y, Chen H-R, et al. Capillary-associated microglia regulate vascular structure and function through PANX1-P2RY12 coupling in mice. Nat Commun. (2021) 12:5289. doi: 10.1038/s41467-021-25590-8
147. Darabid H, St-Pierre-See A, Robitaille R. Purinergic-dependent glial regulation of synaptic plasticity of competing terminals and synapse elimination at the neuromuscular junction. Cell Rep. (2018) 25:2070–82.e6. doi: 10.1016/j.celrep.2018.10.075
148. Sipe GO, Lowery RL, Tremblay MÈ, Kelly EA, Lamantia CE, Majewska AK. Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat Commun. (2016) 7:10905. doi: 10.1038/ncomms10905
149. Burnstock G. Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev. (2007) 87:659–797. doi: 10.1152/physrev.00043.2006
150. Mizuhashi K, Chaya T, Kanamoto T, Omori Y, Furukawa T. Obif, a transmembrane protein, is required for bone mineralization and spermatogenesis in mice. PloS One. (2015) 10:e0133704. doi: 10.1371/journal.pone.0133704
151. Hisa I, Inoue Y, Hendy GN, Canaff L, Kitazawa R, Kitazawa S, et al. Parathyroid hormone-responsive smad3-related factor, tmem119, promotes osteoblast differentiation and interacts with the bone morphogenetic protein-runx2 pathway*. J Biol Chem. (2011) 286:9787–96. doi: 10.1074/jbc.M110.179127
152. Kanamoto T, Mizuhashi K, Terada K, Minami T, Yoshikawa H, Furukawa T. Isolation and characterization of a novel plasma membrane protein, osteoblast induction factor (obif), associated with osteoblast differentiation. BMC Dev Biol. (2009) 9:70. doi: 10.1186/1471-213X-9-70
153. Mizuhashi K, Kanamoto T, Ito M, Moriishi T, Muranishi Y, Omori Y, et al. OBIF, an osteoblast induction factor, plays an essential role in bone formation in association with osteoblastogenesis. Dev Growth Differ. (2012) 54:474–80. doi: 10.1111/j.1440-169X.2012.01333.x
154. Tanaka K-i, Inoue Y, Hendy GN, Canaff L, Katagiri T, Kitazawa R, et al. Interaction of Tmem119 and the bone morphogenetic protein pathway in the commitment of myoblastic into osteoblastic cells. Bone. (2012) 51:158–67. doi: 10.1016/j.bone.2012.04.017
155. Kenkhuis B, Somarakis A, Kleindouwel LRT, Roon-Mom W, Höllt T, Weerd L. Co-expression patterns of microglia markers Iba1, TMEM119 and P2RY12 in Alzheimer’s disease. Neurobiol Dis. (2022) 167:105684. doi: 10.1016/j.nbd.2022.105684
156. Attaai A, Neidert N, Ehr Av, Potru PS, Zöller T, Spittau B. Postnatal maturation of microglia is associated with alternative activation and activated TGFβ signaling. Glia. (2018) 66:1695–708. doi: 10.1002/glia.23332
157. Yeung YG, Jubinsky PT, Sengupta A, Yeung DC, Stanley ER. Purification of the colony-stimulating factor 1 receptor and demonstration of its tyrosine kinase activity. Proc Natl Acad Sci. (1987) 84:1268–71. doi: 10.1073/pnas.84.5.1268
158. Stanley ER, Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harbor Perspect Biol. (2014) 6:a021857. doi: 10.1101/cshperspect.a021857
159. Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet. (2012) 44:200–5. doi: 10.1038/ng.1027
160. Nandi S, Gokhan S, Dai X-M, Wei S, Enikolopov G, Lin H, et al. The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev Biol. (2012) 367:100–13. doi: 10.1016/j.ydbio.2012.03.026
161. Marzan DE, Brügger-Verdon V, West BL, Liddelow S, Samanta J, Salzer JL. Activated microglia drive demyelination via CSF1R signaling. Glia. (2021) 69:1583–604. doi: 10.1002/glia.23980
162. Green KN, Crapser JD, Hohsfield LA. To kill a microglia: A case for CSF1R inhibitors. Trends Immunol. (2020) 41:771–84. doi: 10.1016/j.it.2020.07.001
163. Haskell CA, Cleary MD, Charo IF. Molecular uncoupling of fractalkine-mediated cell adhesion and signal transduction RAPID FLOW ARREST OF CX3CR1-EXPRESSING CELLS IS INDEPENDENT OF G-PROTEIN ACTIVATION*. J Biol Chem. (1999) 274:10053–8. doi: 10.1074/jbc.274.15.10053
164. Pan Y, Lloyd C, Zhou H, Dolich S, Deeds J, Gonzalo J-A, et al. Neurotactin, a membrane-anchored chemokine upregulated in brain inflammation. Nature. (1997) 387:611–7. doi: 10.1038/42491
165. Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, et al. A new class of membrane-bound chemokine with a CX3C motif. Nature. (1997) 385:640–4. doi: 10.1038/385640a0
166. Combadiere C, Gao J, Tiffany HL, Murphy PM. Gene cloning, RNA distribution, and functional expression of mCX3CR1,a mouse chemotactic receptor for the CX3C chemokine fractalkine. Biochem Biophys Res Commun. (1998) 253:728–32. doi: 10.1006/bbrc.1998.9849
167. Loh SX, Ekinci Y, Spray L, Jeyalan V, Olin T, Richardson G, et al. Fractalkine signalling (CX3CL1/CX3CR1 axis) as an emerging target in coronary artery disease. J Clin Med. (2023) 12:4821. doi: 10.3390/jcm12144821
168. Harrison JK, Fong A, Swain PW, Chen S, Yu Y-R, Salafranca MN, et al. Mutational analysis of the fractalkine chemokine domain BASIC AMINO ACID RESIDUES DIFFERENTIALLY CONTRIBUTE TO CX3CR1 BINDING, SIGNALING, AND CELL ADHESION*. J Biol Chem. (2001) 276:21632–41. doi: 10.1074/jbc.M010261200
169. DiNatale A, Kaur R, Qian C, Zhang J, Marchioli M, Ipe D, et al. Subsets of cancer cells expressing CX3CR1 are endowed with metastasis-initiating properties and resistance to chemotherapy. Oncogene. (2022) 41:1337–51. doi: 10.1038/s41388-021-02174-w
170. Lesnik P, Haskell CA, Charo IF. Decreased atherosclerosis in CX3CR1–/– mice reveals a role for fractalkine in atherogenesis. J Clin Investig. (2003) 111:333–40. doi: 10.1172/JCI200315555
171. Landsman L, Bar-On L, Zernecke A, Kim K-W, Krauthgamer R, Shagdarsuren E, et al. CX3CR1 is required for monocyte homeostasis and atherogenesis by promoting cell survival. Blood. (2009) 113:963–72. doi: 10.1182/blood-2008-07-170787
172. Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. (2006) 9:917–24. doi: 10.1038/nn1715
173. Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci. (2014) 17:400–6. doi: 10.1038/nn.3641
174. Wang C, Lu J, Sha X, Qiu Y, Chen H, Yu Z. TRPV1 regulates ApoE4-disrupted intracellular lipid homeostasis and decreases synaptic phagocytosis by microglia. Exp Mol Med. (2023) 55:347–63. doi: 10.1038/s12276-023-00935-z
175. Basilico B, Ferrucci L, Ratano P, Golia MT, Grimaldi A, Rosito M, et al. Microglia control glutamatergic synapses in the adult mouse hippocampus. Glia. (2022) 70:173–95. doi: 10.1002/glia.24101
176. Buckley DB, Klaassen CD. Tissue- and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab Dispos. (2007) 35:121–7. doi: 10.1124/dmd.106.012070
177. Chik MW, Hazalin NAMN, Singh GKS. Regulation of phase I and phase II neurosteroid enzymes in the hippocampus of an Alzheimer’s disease rat model: A focus on sulphotransferases and UDP-glucuronosyltransferases. Steroids. (2022) 184:109035. doi: 10.1016/j.steroids.2022.109035
178. Yang N, Li S, Yan C, Sun R, He J, Xie Y, et al. Inhibitory effects of endogenous linoleic acid and glutaric acid on the renal glucuronidation of berberrubine in mice and on recombinant human UGT1A7, 1AB, and 1A9. Mol Pharmacol. (2018) 93:mol.117.110668. doi: 10.1124/mol.117.110668
179. Rowland A, Miners JO, Mackenzie PI. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int J Biochem Cell Biol. (2013) 45:1121–32. doi: 10.1016/j.biocel.2013.02.019
180. Yuan Q, Zhang J, Wang S, Zhang M, Nie J. Identification, evolution and expression analysis of the UDP-glycosyltransferase gene family in cucumber (Cucumis sativus L.). Sci Hortic. (2024) 324:112615. doi: 10.1016/j.scienta.2023.112615
181. Chen ML, Zhang SX, Guo PY, Qin QS, Meng LW, Yuan GR, et al. Identification and characterization of UDP-glycosyltransferase genes and the potential role in response to insecticides exposure in Bactrocera dorsalis. Pest Manag Sci. (2023) 79:666–77. doi: 10.1002/ps.7234
182. Wu Y, Liu J, Jiao B, Wang T, Sun S, Huang B. Genome-wide analysis of family-1 UDP-glycosyltransferases in potato (Solanum tuberosum L.): identification, phylogenetic analysis and determination of response to osmotic stress. Genes. (2023) 14:2144. doi: 10.3390/genes14122144
183. Gan Y, Yu B, Liu R, Shu B, Liang Y, Zhao Y, et al. Systematic analysis of the UDP-glucosyltransferase family: discovery of a member involved in rutin biosynthesis in Solanum melongena. Front Plant Sci. (2023) 14:1310080. doi: 10.3389/fpls.2023.1310080
184. Meech R, Hu DG, McKinnon RA, Mubarokah SN, Haines AZ, Nair PC, et al. The UDP-glycosyltransferase (UGT) superfamily: new members, new functions, and novel paradigms. Physiol Rev. (2019) 99:1153–222. doi: 10.1152/physrev.00058.2017
185. Rouleau M, Long FNV, Turcotte V, Caron P, Lacombe L, Aprikian A, et al. Extensive metabolic consequences of human glycosyltransferase gene knockouts in prostate cancer. Br J Cancer. (2023) 128:285–96. doi: 10.1038/s41416-022-02040-w
186. Tiwari P, Sangwan RS, Sangwan NS. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol Adv. (2016) 34:714–39. doi: 10.1016/j.biotechadv.2016.03.006
187. Bowles D, Isayenkova J, Lim E-K, Poppenberger B. Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol. (2005) 8:254–63. doi: 10.1016/j.pbi.2005.03.007
188. Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. (2011) 66:182–93. doi: 10.1111/j.1365-313x.2011.04493.x
189. Lv X, Ge G-B, Feng L, Troberg J, Hu L-H, Hou J, et al. An optimized ratiometric fluorescent probe for sensing human UDP-glucuronosyltransferase 1A1 and its biological applications. Biosens Bioelectron. (2015) 72:261–7. doi: 10.1016/j.bios.2015.05.003
190. Jin Q, Wu J, Wu Y, Li H, Finel M, Wang D, et al. Optical substrates for drug-metabolizing enzymes: Recent advances and future perspectives. Acta Pharm Sin B. (2022) 12:1068–99. doi: 10.1016/j.apsb.2022.01.009
Keywords: housekeeping gene, microglial cell, biomarker, UGT1A7C protein, mouse, embryo development, transcriptome analysis, neurologic disorders
Citation: Kim W, Kim M and Kim B (2024) Unraveling the enigma: housekeeping gene Ugt1a7c as a universal biomarker for microglia. Front. Psychiatry 15:1364201. doi: 10.3389/fpsyt.2024.1364201
Received: 01 January 2024; Accepted: 26 March 2024;
Published: 11 April 2024.
Edited by:
Kyoungho Suk, Kyungpook National University, Republic of KoreaReviewed by:
Joana Gonçalves, University of Coimbra, PortugalCopyright © 2024 Kim, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beomsue Kim, kimbs@kbri.re.kr
 Wonju Kim
Wonju Kim Minji Kim
Minji Kim Beomsue Kim
Beomsue Kim