- 1Department for Community Health, Faculty of Science and Medicine, University of Fribourg, Fribourg, Switzerland
- 2Department of Paediatrics, Fribourg Hospital, Fribourg, Switzerland
- 3Istanbul Faculty of Medicine, Istanbul University, Istanbul, Türkiye
- 4Department of Paediatrics, The University of Melbourne, Parkville, VIC, Australia
- 5Infectious Diseases Research Group, Murdoch Children’s Research Institute, Parkville, VIC, Australia
- 6Pediatric Infectious Diseases Unit, Geneva University Hospitals and Faculty of Medicine, Geneva, Switzerland
- 7Microbiology Laboratory, Fribourg Hospital, Fribourg, Switzerland
- 8Department of Biology, University of Fribourg and Swiss Institute of Bioinformatics, Fribourg, Switzerland
- 9Infectious Diseases Unit, The Royal Children’s Hospital Melbourne, Parkville, VIC, Australia
Introduction: The early-life intestinal microbiome plays a crucial role in the development and regulation of the immune system. Perturbations in its composition during this critical period have been linked to the development of allergic diseases.
Objective: This study aimed to investigate the association between the composition of the early-life intestinal microbiome and the presence of eczema in the first year of life using shotgun metagenomic sequencing and functional analyses (metabolic pathways).
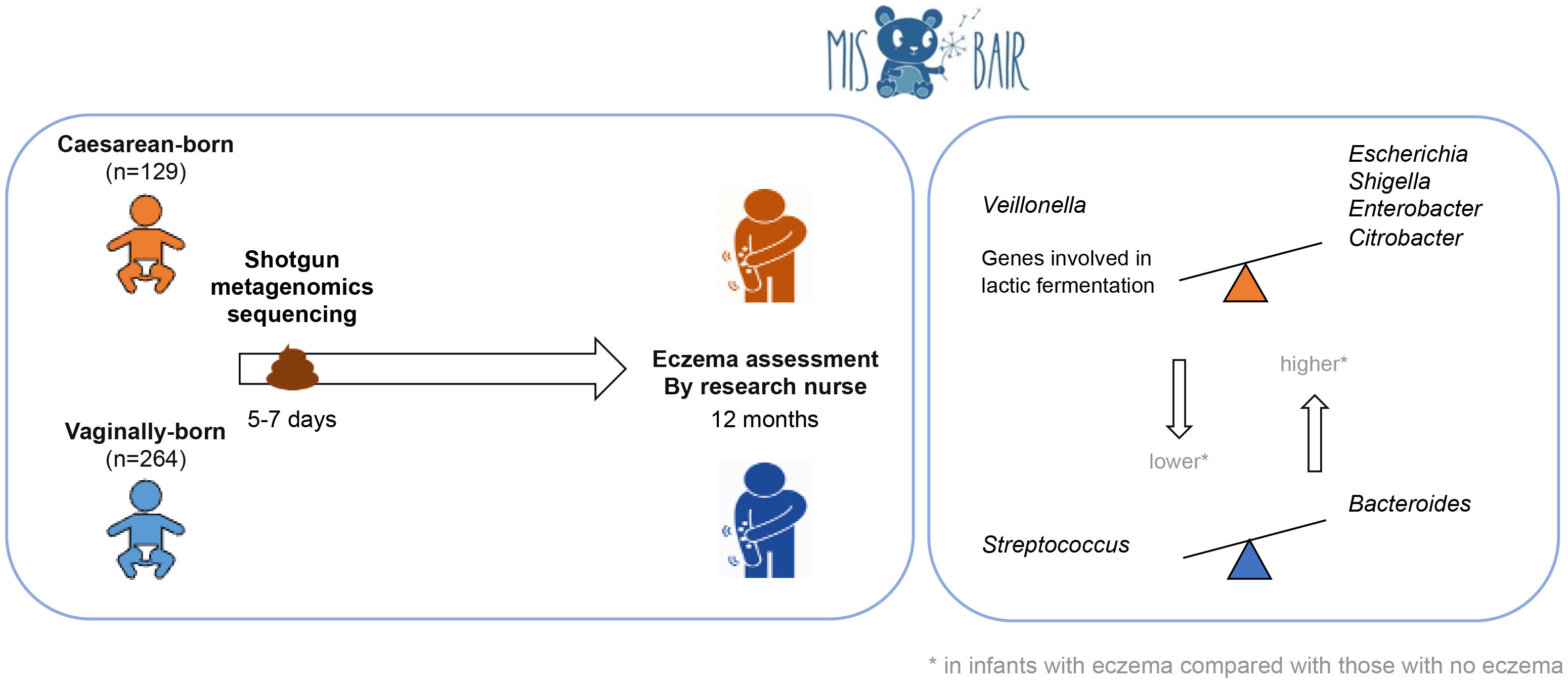
Methods: Stool samples from 393 healthy term infants collected at 1 week of age were analyzed with shotgun metagenomic sequencing. Environmental and clinical data were prospectively collected using 3-monthly validated questionnaires. Participants were clinically assessed during study visits at 12 months of age. Eczema was diagnosed by the UK diagnostic tool and by a research nurse. Data analysis was stratified by delivery mode.
Results: Eczema was diagnosed in 16.4% (60/366) of participants by nurse diagnosis. Infants born by cesarean section (CS) with nurse-diagnosed eczema had a higher relative abundance of Escherichia, Shigella, Enterobacter, and Citrobacter and a lower relative abundance of Veillonella than CS-born infants without eczema. In addition, CS-born infants without eczema had a higher abundance of genes involved in lactic fermentation. Vaginally born infants with eczema had a higher relative abundance of Bacteroides and a lower abundance of Streptococcus.
Conclusion: There is an association between the bacterial composition of the intestinal microbiome at 1 week of age and the presence of eczema in the first 12 months of life.
Introduction
Atopic eczema, also called atopic dermatitis, is an inflammatory disease of the skin characterized by a variety of symptoms, including skin dryness, itchiness, reddish patches, and xerosis. Eczema is considered the first manifestation of the atopic march, and infants affected by eczema are more likely to suffer from other allergic diseases later in life (van der Hulst et al., 2007; Hill and Spergel, 2018; Sroka-Tomaszewska and Trzeciak, 2021). Furthermore, severe eczema is associated with the presence of other diseases, such as diabetes, cardiovascular and autoimmune diseases, and mental health disorders (Sroka-Tomaszewska and Trzeciak, 2021).
Infants are the age group most frequently affected by eczema (de Lusignan et al., 2021). Risk factors for eczema include being first born, being of Asian or black ethnicity, having a higher socioeconomic status, and living in an urban area (Penders et al., 2013; de Lusignan et al., 2021). The intestinal microbiome also plays a crucial role in the development and regulation of immune responses. Consistent with this, there is evidence that allergic diseases, such as eczema, are influenced by the composition of the intestinal microbiome (Zimmermann et al., 2019). As a critical step in developing immunotolerance, the immune system learns to recognize commensal microbiota. It has been hypothesized that reduced exposure to microbial immune stimulation, particularly via reduced microbiome diversity, impairs the healthy development of the immune system and increases the risk of allergic disease (Rook and Stanford, 1998). However, a recent systematic review suggests that particular microbes might be more important than bacterial diversity (Zimmermann et al., 2019). Children who develop eczema, allergic sensitization, or asthma have been reported to have a greater abundance of Clostridiaceae and Enterobacteriaceae and a lower abundance of Lactobacillaceae (Zimmermann et al., 2019). The findings reported from studies in neonates are more consistent than those reported from studies in older children and suggest that the early-life microbiome is crucial.
This is the first study to use shotgun metagenomic sequencing and functional analyses to investigate whether the composition of the early-life intestinal microbiome is associated with the prevalence and severity of eczema in the first year of life in a large cohort of infants. Understanding this relationship might help with the early detection of infants at risk of developing eczema and provide a basis for evidence-based interventions to prevent or correct dysbiosis of the microbiome.
Methods
Study design and participants
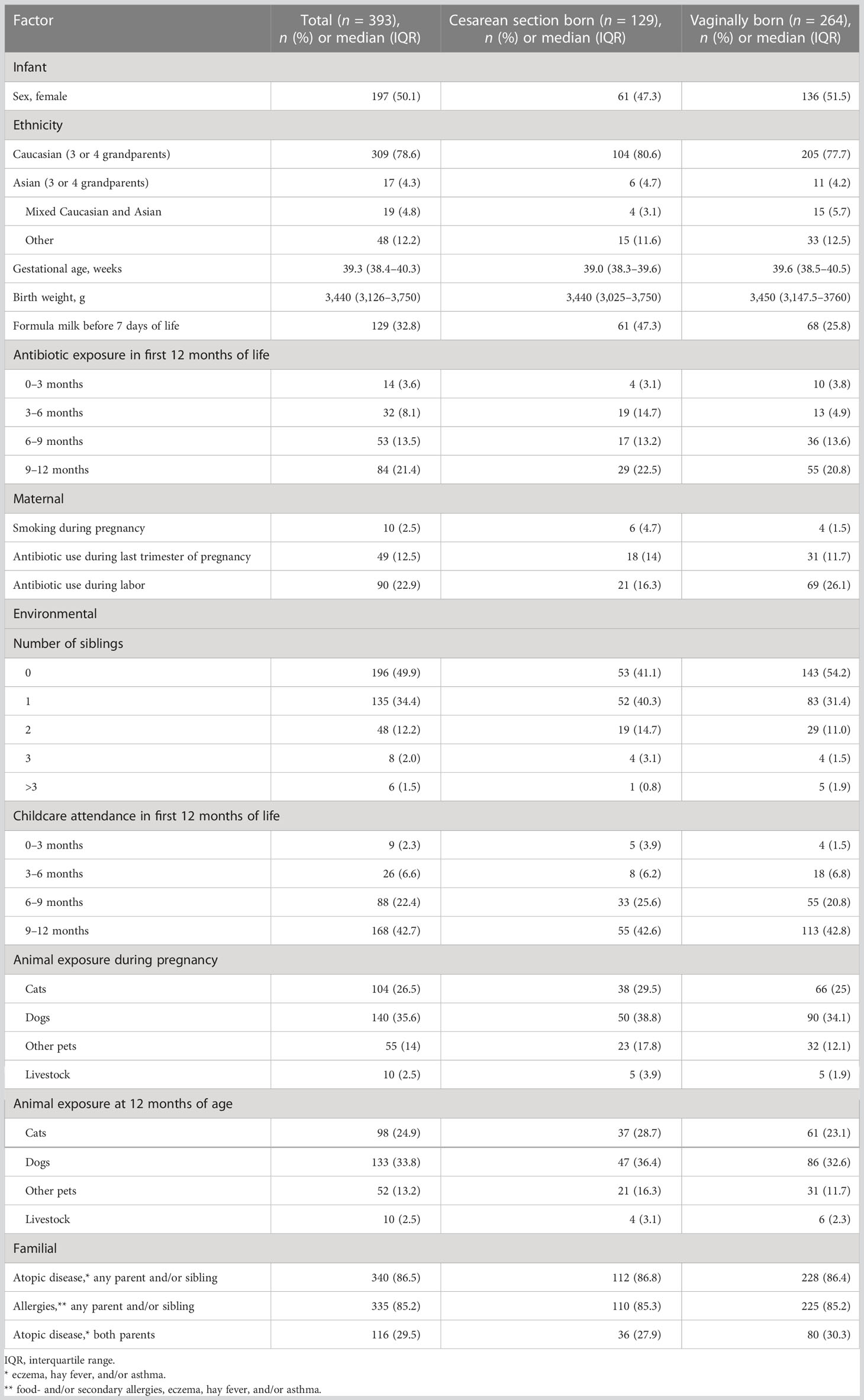
The participants were a subset of infants from the Melbourne Infant Study: BCG for Allergy and Infection Reduction (MIS BAIR) (Messina et al., 2019). In this randomized trial, 1,272 healthy infants were recruited antenatally to investigate whether Bacille Calmette–Guérin (BCG) immunization given in the first 10 days of life protects against childhood infection, allergy, and asthma. Inclusion criteria for infants were as follows: born after at least 32 weeks of gestation; birth weight greater than 1,500 g; absence of symptoms or signs of illness, including skin diseases; and no contraindication to BCG vaccination. Detailed data on demographics, pregnancy, and the perinatal period were collected at baseline. Three-monthly validated parent questionnaires were used to prospectively collect data on eczema-related symptoms, severity, and management. A visit by a study nurse took place when children were 12 months of age to clinically assess the incidence and severity of eczema.
Eczema diagnostic tools
The presence of eczema in the first 12 months of life was evaluated with two different complementary methods: (1) the cumulative incidence of any eczema in the first 12 months of life assessed by the UK diagnostic tool using data from the 3-monthly questionnaires (UK diagnostic tool) (Williams et al., 1994a; Williams et al., 1994b); and (2) the point prevalence of research nurse-diagnosed eczema at 12 months of age assessed by clinical assessment using the SCORing Atopic Dermatitis (SCORAD) system (nurse diagnosis).
The UK diagnostic tool was adapted in accordance with the participants’ ages, as well as for online data collection (Pittet et al., 2022a). Eczema was defined as an itchy skin condition (major criterion) accompanied by at least three of the following minor criteria: (1) generally dry skin; (2) lesions in skin creases or cheeks; (3) atopic disease in a first-degree relative; and (4) visible eczema involving the flexures, head, or limbs. Previous data show that parents are able to accurately report eczema in their infants (Fleming et al., 2001).
In the absence of an agreed tool to reliably diagnose eczema in infancy (Pittet et al., 2022b), the use of two complementary tools improves the reliability of the results. Although the point prevalence estimate by a trained professional (nurse diagnosis outcome) is the most robust assessment, it captures lesions present on the day of visit only, depends on the management of eczema (i.e., effective treatment will mean lesions are absent), may miss seasonal fluctuation of eczema, and could capture lesions that are not necessarily atopic dermatitis (Pittet et al., 2022b). Being widely used, the UK diagnostic tool enables comparison with other studies; however, it is less relevant for infant eczema, with one of the major limitations being that the score requires the subject to experience itching (which excludes infants who are not yet developmentally capable of scratching themselves). In addition, the minor criteria are primarily based on flexural lesions, and not on the typical distribution of eczema in infancy (i.e., trunk and face, with only the latter included in the modified version) (Pittet et al., 2022b).
Age at onset of eczema, use of topical steroids, and eczema severity were also evaluated. Eczema severity was assessed using the Patient-Oriented Eczema Measure (POEM) score (Charman et al., 2004) included in each 3-monthly questionnaire and the SCORAD score no author, (no author, 1993) assessed by the research nurse at the study visit at 12 months.
Stool collection
Parents were asked to collect stool samples from their infants in sterile tubes on each of the first 10 days of life. To minimize variation, parents were asked to collect from the first bowel movement of the day (with date and time recorded) and immediately freeze the stool in their domestic freezers (–18°C or below). Samples were collected from parents' homes by a study team member and kept frozen during transportation to the laboratory, where they were aliquoted for storage at –80°C. One stool sample collected between days 5 and 7 of life was analyzed for each infant.
DNA extraction and shotgun metagenomic sequencing
Samples were thawed at room temperature and 100 mg of stool was mixed with 1x phosphate-buffered saline (PBS). DNA was extracted using the FastDNA™ SPIN Kit for Soil (MP Biomedicals, Illkirch-Graffenstaden, France). Of this stool suspension, 300 µl was mixed with 978 µL of PBS (provided by the kit) and 122 µL of MT buffer and transferred into a lysing matrix E (LME) tube. Sample homogenization was performed with the FastPrep96 homogenizer using the program for human stool samples (settings: 1,600 rpm, 40 s). The LME tubes were centrifuged at 14,000 g for 10 minutes and the supernatant was mixed with 250 µL of protein precipitation solution in a 2-mL microcentrifuge tube. The tubes were inverted by hand 10 times and centrifuged at 14,000 g for a further 5 minutes. Subsequently, 1 mL of binding matrix solution was then added to the supernatant. The tubes were inverted for 2 minutes, then rested for 5 minutes, and 800 µL of the supernatant was removed. The remaining supernatant was resuspended in the binding matrix. At this point, 700 µL was transferred into a new spin filter tube and centrifuged for 1 minute at 14,000 g. This step was repeated twice. After this, 500 µL of ethanol-based washing solution (SEWS-M wash buffer) was mixed with the binding matrix. Samples were centrifuged three times for 1 minute at 14,000 g and allowed to dry at room temperature for 5 minutes. DNA was eluted in 50 µL of DNase- and pyrogen-free water.
The DNA concentrations of samples and a negative control (i.e., no sample DNA) were measured using Qubit™ dsDNA High Sensitivity Assay kits (Life Technologies, CA, USA). The mean DNA concentration in samples was 367 ng; the negative control was below the detection limit (i.e., < 0.01 ng/µL) and was therefore not sequenced. Sequencing libraries were prepared with the Nextera DNA Flex library preparation kits (Illumina, San Francisco, CA, USA) to obtain insert sizes of approximately 600 bp. Illumina PhiX DNA was added to the libraries, which were then sequenced (2x149) on a NextSeq 550 system (Illumina) with high-output flow cells. Bacterial and fungal evenly distributed mock communities (Gut Microbiome Whole Cell Mix MSA-2006™ and Mycobiome Whole Cell MixTM, respectively; ATCC, Manassas, VA, USA) were sequenced as positive controls.
Pre-processing, taxonomic assignment, and relative abundance computation
The overall quality of the sequencing run was inspected with the Illumina Sequencing Analysis Viewer v. 2.4.7. The quality of the raw reads was assessed by FastQC v. 0.11.7 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and scanned with Trimmomatic v. 0.39 by using a sliding window of 20 nt (Bolger et al., 2014). Reads with an average quality below a Phred score of 28 and with a length of less than 100 nt (before or after trimming) were filtered out. Quality-filtered reads were then analyzed with Kraken v. 2.0.9-beta (Wood et al., 2019) and sequences mapping to the human (vGRCh38.p11) genome with a confidence score of more than 0.1 were discarded. The remaining reads were assigned to bacterial, archaeal, viral, and fungal genomes with Kraken2 (with a confidence score of more than 0.1) and with MetaPhlAn v. 3.0.7-1 (Segata et al., 2012). Compared with Kraken2, MetaPhlAn 3 uses a smaller reference sequences database, made up of clade-specific gene markers. Data obtained by MetaPhlAn 3 were used to confirm the results from the Kraken2/Bracken pipeline.
Read counts obtained with Kraken2 were corrected with Bracken v. 2.6.2 (Wood et al., 2019; Lu and Salzberg, 2020) at phylum, family, genus, and species levels. All the reference genome sequences were collected and analyzed with kmer2read_distr [settings: k-mer length (-k) of 35 and read length (–l) of 100] to finally generate a k-mer distribution file with generate_kmer_distribution.py. Eventually, the read abundance of each phylum, family, genus, and species in each sample was estimated with est_abundance.py (settings: minimum number of reads necessary for taxonomic classification (–t) = 10).
For each taxonomic level, the relative abundance was computed as a percentage by dividing the number of Bracken-corrected read counts in a given taxon by the total number of Bracken-corrected reads assigned to that taxonomic level in any given sample. MetaPhlAn 3 provided taxa and relative abundance already expressed as percentages.
Phages
The phage content was investigated by selecting reads assigned to viruses from the Caudovirales order. Caudovirales includes double-stranded DNA phages from the families Siphoviridae, Myoviridae, and Podoviridae, which are the most abundant bacteriophages present in the human intestine (Hoyles et al., 2014; Shkoporov et al., 2019). The relative abundance of phages was computed by dividing the number of reads mapped to phage genomes by the total number of quality-filtered no-human reads. The ratio was then multiplied by 1,000,000 and the relative abundance was expressed as counts per million (CPM).
Metabolic pathway analyses
Microbial metabolic pathways were identified by analyzing quality-filtered non-human reads with the HUMAnN v. 3.5 tool with default parameters (Beghini et al., 2021). Forward and reverse read files from each sample were concatenated in a unique FASTQ file. UniRef90 was used as a database. Normalized gene copies per million were then obtained from pathabundance output tables with human_renorm_table HUMAnN 3 code. The interpretation of metabolic pathways and their putative presence in each species were investigated by querying the MetaCyc database (https://biocyc.org/) (Caspi et al., 2006).
Reference databases
The human genome (v. GRCh38.p11) and 15,009 bacterial, 511 archaeal, 11,833 viral, and 420 fungal genomes were downloaded from NCBI RefSeq on 3 March 2022.
Statistical analysis
All statistical analyses were performed with the R software v. 4.2.0. The codes used for statistical analyses are reported in Supplementary File 1.
To compare the bacterial compositions of the stool microbiomes at the species and genus level, principal coordinate analyses (PCoAs) were performed. For the PCoAs, Bray-Curtis dissimilarity distance was computed on the genera or species’ relative abundance after square-root transformation with the vegdist function of the vegan R package v. 2.6-2. The resulting data were analyzed with the betadisper function (vegan). Differences in bacterial composition according to a given variable were assessed at the species, genus, family, and phylum taxonomic levels by permutational multivariate analysis of variance (PERMANOVA) tests. PERMANOVA was performed with the adonis2 function (vegan) using Bray–Curtis dissimilarity distance computed on square root-transformed data and using 9,999 permutations.
Ecological indices, richness, and Shannon diversity were computed after rarefying read counts analyzed with the Kraken2/Bracken pipeline to 35,000 with the rrarefy function (vegan). Shannon indexes were computed with the diversity function (vegan).
To identify differentially abundant species, DESeq2 (Love et al., 2014) v. 1.36.0 and linear discriminant analysis of effect size [LEfSe (Segata et al., 2011)] as implemented in the microbiomeMarker package v. 1.2.2 were performed with the following settings: wilcoxon_cutoff = 0.05, norm = “CPM”, kw_cutoff = 0.05, lda_cutoff = 2, and enrich_group = 0.
Prior to running differentially abundant analyses, we filtered out genera and species identified with the Kraken2/Bracken pipeline that had less than 10 reads in less than 100 (25.5%) of 393 samples. LEfSe was also used to identify differentially abundant pathways with the same settings as described above, except the lda_cutoff was set at 1.5. For all other statistical testing, the Wilcoxon rank-sum test was used. Results were considered significant when associated p-values (or false-discovery-rate-adjusted p-values for DESeq2 analyses) were less than 0.05. Plotting was performed with the ggplot2 package v. 3.3.6.
As delivery mode was expected to have a strong effect on the intestinal microbiome (Zimmermann and Curtis, 2018), data analysis was stratified according to delivery mode. Further factors investigated for their effect on the composition of the intestinal microbiome included infant (sex, ethnicity, gestational age, birth weight, feeding method, and antibiotic exposure), maternal (antibiotic use during the last trimester of pregnancy or during labor, and smoking), environmental (number of siblings, childcare attendance, number of smokers in the household, and animal exposure during pregnancy or at 12 months of age), and familial [family history of atopic disease (defined as eczema, hay fever, or asthma) or allergies (food and/or secondary allergies, eczema, hay fever, and/or asthma)] factors.
Ethics
Informed consent was obtained from participants’ parents or guardians. The study was approved by the Royal Children’s Hospital Human Research Ethics Committee (HREC, authorization 53543). MIS BAIR is registered with the Australia and New Zealand Clinical Trials Registry (1051228) and the US National Institutes of Health (NCT01906853).
Results
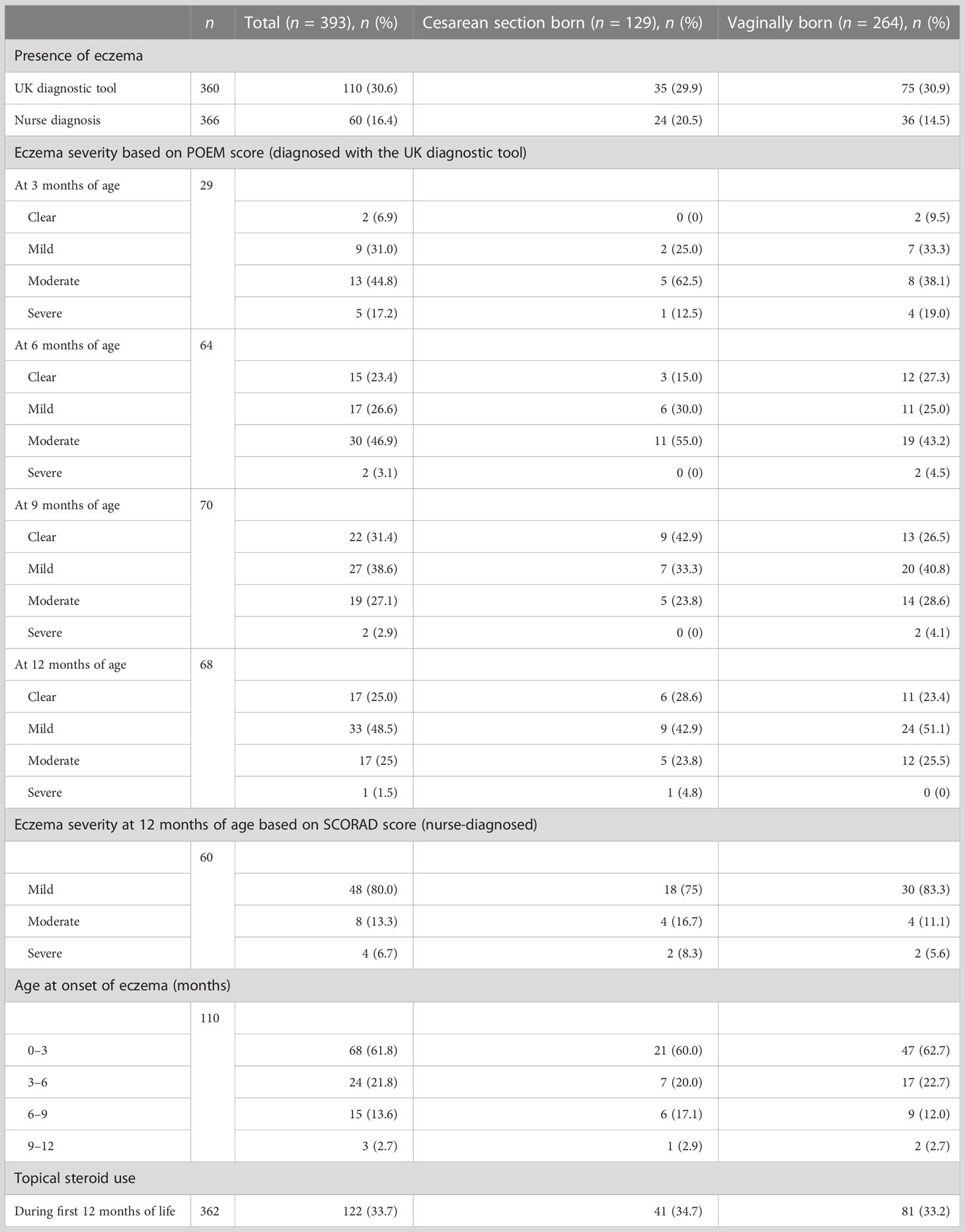
Of a total of 393 included term-born infants, 32.8% (129/393) were born by cesarean section (CS). Characteristics of the study cohort are summarized in Table 1. All four 3-monthly questionnaires were completed for 91.6% (360/393) of infants, and 93.1% (366/393) of infants attended the 12-month visit, with a median age of 13.3 months [interquartile range (IQR) 12.8–13.9 months]. Eczema was diagnosed in 30.6% (110/360) and 16.4% (60/366) of infants by the UK diagnostic tool and nurse diagnosis, respectively (Table 2).
When evaluated by the UK diagnostic tool, eczema symptoms in participants started in 18.9% (68/360) at 0 to 3 months of age, in 6.7% (24/360) at 3 to 6 months of age, in 4.2% (15/360) at 6 to 9 months of age, and in 0.8% (3/360) at 9 to 12 months of age. The proportions of infants with different degrees of eczema severity based on POEM score were as follows: 6.9% (2/29) clear, 31% (9/29) mild, 44.8% (13/29) moderate, and 17.2% (5/29) severe at 3 months of age; 23.4% (15/64) clear, 26.6% (17/64) mild, 46.9% (30/64) moderate, and 3.1% (2/64) severe at 6 months of age; 31.4% (22/70) clear, 38.6% (27/70) mild, 27.1% (19/70) moderate, and 2.9% (2/70) severe at 9 months of age; and 25% (17/68) clear, 48.5% (33/68) mild, 25% (17/68) moderate, and 1.5% (1/68) severe at 12 months of age. The eczema severity at 12 months of age based on the SCORAD score was mild in 80% (48/60) of infants, moderate in 13.3% (8/60) of infants, and severe in 6.7% (4/60) of infants. During the first 12 months of life, steroids were administered topically to 33.7% (122/362) of infants (Table 2).
There was no association between eczema prevalence and clinical variables such as delivery mode sex and family history (Supplementary File 2, Table 1).
Overall microbiome profile
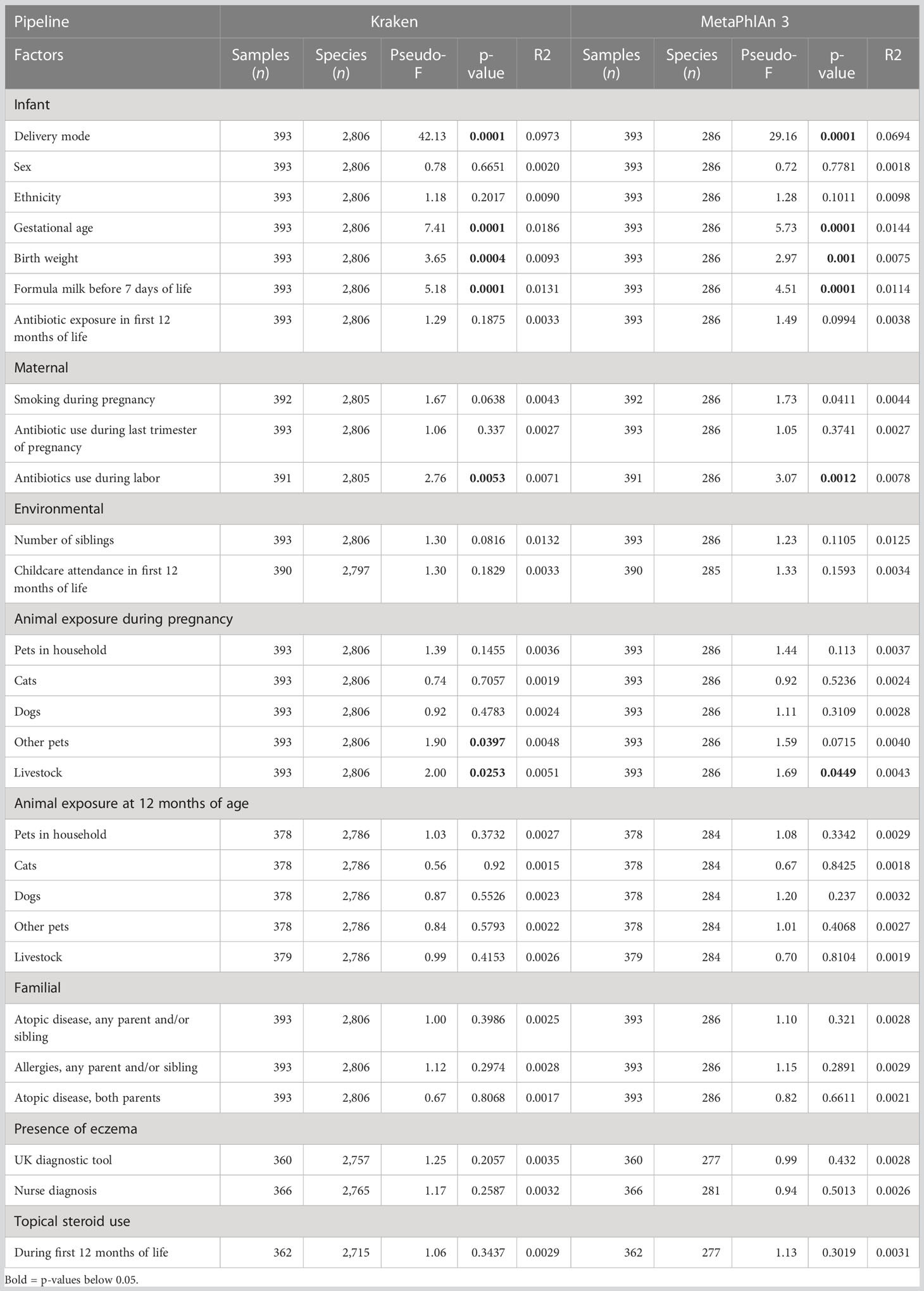
On average, 7.3 million read pairs (range 107,000–41,000,000) were obtained per sample. Overall, 19 and 4 bacterial phyla, 212 and 48 families, 882 and 97 genera, and 2,806 and 286 species were identified by Kraken2/Bracken and MetaPhlAn 3, respectively.
Delivery mode, gestational age, formula milk, antibiotics during labor, and number of siblings had a strong effect on the bacterial composition at all taxonomic levels (PERMANOVA test, Table 3; Supplementary File 2, Tables 2-4). Birth weight had an effect on the composition of bacterial families, genera, and species (Table 3; Supplementary File 2, Tables 3, 4). Concerning animal exposure, we did not find consistent results among the taxonomic levels: exposure to livestock at 12 months was significantly associated with bacterial composition at the phylum level only (Supplementary File 2, Table 2); exposure to pets (excluding cats and dogs) and livestock during pregnancy was associated with family, genus, and species composition (Table 3; Supplementary File 2, Tables 3, 4). Childcare attendance was significantly associated with phylum composition, as investigated by MetaPhlAn 3 (Supplementary File 2, Table 2). No associations were found between microbiome composition and sex, ethnicity, antibiotic exposure during the first year of life, smoking during pregnancy, antibiotic use during the last trimester of pregnancy, presence of smokers in the household at 12 months of age, cat and dog exposure during pregnancy or at 12 months of age, familial factors (presence of atopic disease or allergies in any parent and/or sibling, or presence of atopic disease in both parents), or use of topical steroids during the first 12 months of life.
No archaeal taxa were identified by MetaPhlAn 3. The only archaeal taxon detected by Kraken2\Bracken was the family of Methanobacteriaceae from the phylum Euryarchaeota. This family was detected in only two samples (read counts 12 and 28, respectively).
In total, 29 fungal species were identified by Kraken2\Bracken, belonging to 3 phylum, 21 families, and 21 genera. These species were identified in 68 (17.3%) samples and had read counts that did not allow further analyses (on average 0.03% of total non-human quality filtered reads). One fungal species (Candida albicans) was detected by MetaPhlAn 3 in three samples with a relative abundance between 0.02% and 0.29%. Because of the low abundance of archaea and fungi, the association between their composition in the intestinal microbiota and eczema outcomes could not be analyzed.
Intestinal bacterial microbiome composition and the presence of eczema
As expected, in PCoA plots, the stool microbiome profiles of CS-born infants clustered further apart from those of vaginally born infants than the stool microbiome profiles of infants with eczema clustered from those of infants without eczema (Figure 1; Supplementary File 2, Figures 1-3). There was no difference in bacterial species diversity or richness between CS-born or vaginally born infants with or without eczema (Supplementary File 2, Figure 4). However, in CS-born infants with nurse-diagnosed eczema, the microbiome composition was significantly different at all taxonomic levels to that of CS-born infants without eczema (PERMANOVA test, p-values 0.0297, 0.0162, 0.0176, and 0.0125 for phylum, family, genus, and species, respectively; Supplementary File 2, Table 5).
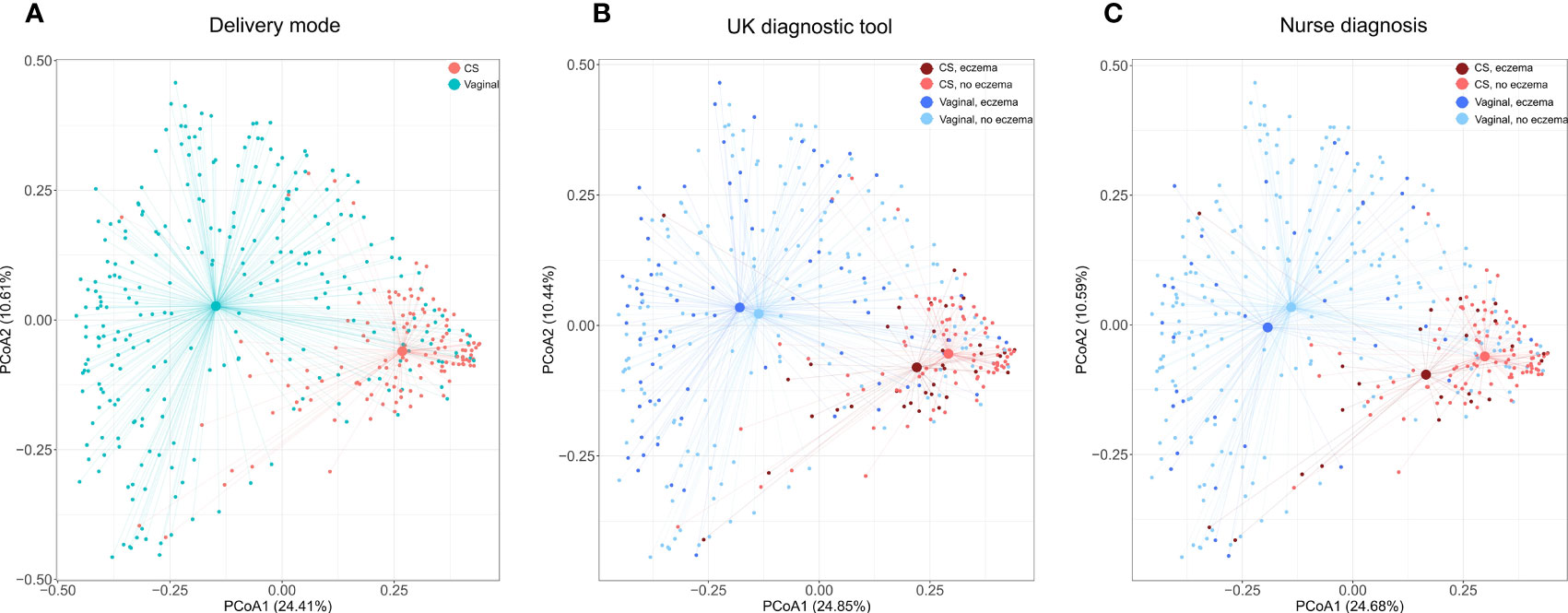
Figure 1 Species microbiome profiles according to delivery mode and eczema outcomes. Principal coordinate analyses (PCoAs) were performed on the relative abundance of 2,806 species detected in 393 infants with the Kraken2/Bracken pipeline. Axes indicate the percentage of total variance. Colors indicate delivery mode (A) and delivery mode and eczema outcomes (B, C). For eczema outcomes, participants with missing values were excluded from the analysis and plotting; thus, (B) was computed on 91.6% (360/393) of samples and 98.3% (2,757/2,806) of species, and (C) on 93.1% (366/393) of samples and 98.5% (2,765/2,806) of species. The centroid indicates average microbiome species composition. Lines connect centroids to corresponding data points.
Overall, the four most abundant phyla found in the intestinal microbiome were Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria. No significant differences in the relative abundance of these phyla were detected in relation to eczema outcomes in either CS- or vaginally born infants (Supplementary File 2, Figure 5).
The 10 most abundant families were Enterobacteriaceae, Bacteroidaceae, Bifidobacteriaceae, Veillonellaceae, Streptococcaceae, Staphylococcaceae, Pasteurellaceae, Tannerellaceae, Clostridiaceae, and Enterococcaceae. Compared with CS-born infants without eczema, CS-born infants with eczema had a higher relative abundance of Enterobacteriaceae (p-values of 0.06 and 0.15 for the UK diagnostic tool and nurse diagnosis, respectively) and a lower relative abundance of Veillonellaceae (Bacillota) (p-values of 0.48 and 0.006 for the UK diagnostic tool and nurse diagnosis, respectively) (Supplementary File 2, Figure 6). Vaginally born infants with eczema had a lower relative abundance of Streptococcaceae than vaginally born infants without eczema (p-values 0.01 and 0.36 for the UK diagnostic tool and nurse diagnosis, respectively) (Supplementary File 2, Figure 6).
The 10 most abundant genera were Bifidobacterium, Veillonella, Bacteroides, Escherichia, Streptococcus, Klebsiella, Shigella, Phocaeicola, Staphylococcus, and Parabacteroides, and the 10 most abundant species were Escherichia coli, Bifidobacterium longum, Veillonella parvula, Parabacteroides distasonis, Staphylococcus epidermidis, Haemophilus parainfluenzae, Streptococcus salivarius, Phocaeicola dorei, Bifidobacterium breve, and Shigella sonnei. Genera and species, which were identified by Kraken2/Bracken and had >10 reads in at least one-quarter of the sample cohort, were kept for further analyses. After this filtering step, 12.6% (111/882) of genera and 11.6% (326/2806) of species were retained. CS-born infants with eczema had a higher relative abundance of the genera Escherichia and Shigella than CS-born infants without eczema (p-values 0.049 and 0.008, respectively, for nurse diagnosis) and a lower relative abundance of Veillonella (p-value 0.004 for nurse diagnosis) (Supplementary File 2, Figure 7). Although not among the 10 most abundant genera, Enterobacter and Citrobacter were also more abundant in CS-born infants with eczema than CS-born infants without eczema. Vaginally born infants with eczema had a higher abundance of Bacteroides and a lower abundance of Streptococcus (p-values 0.045 and 0.011, respectively, for the UK diagnostic tool) (Figure 2; Supplementary File 2, Figures 7, 8).
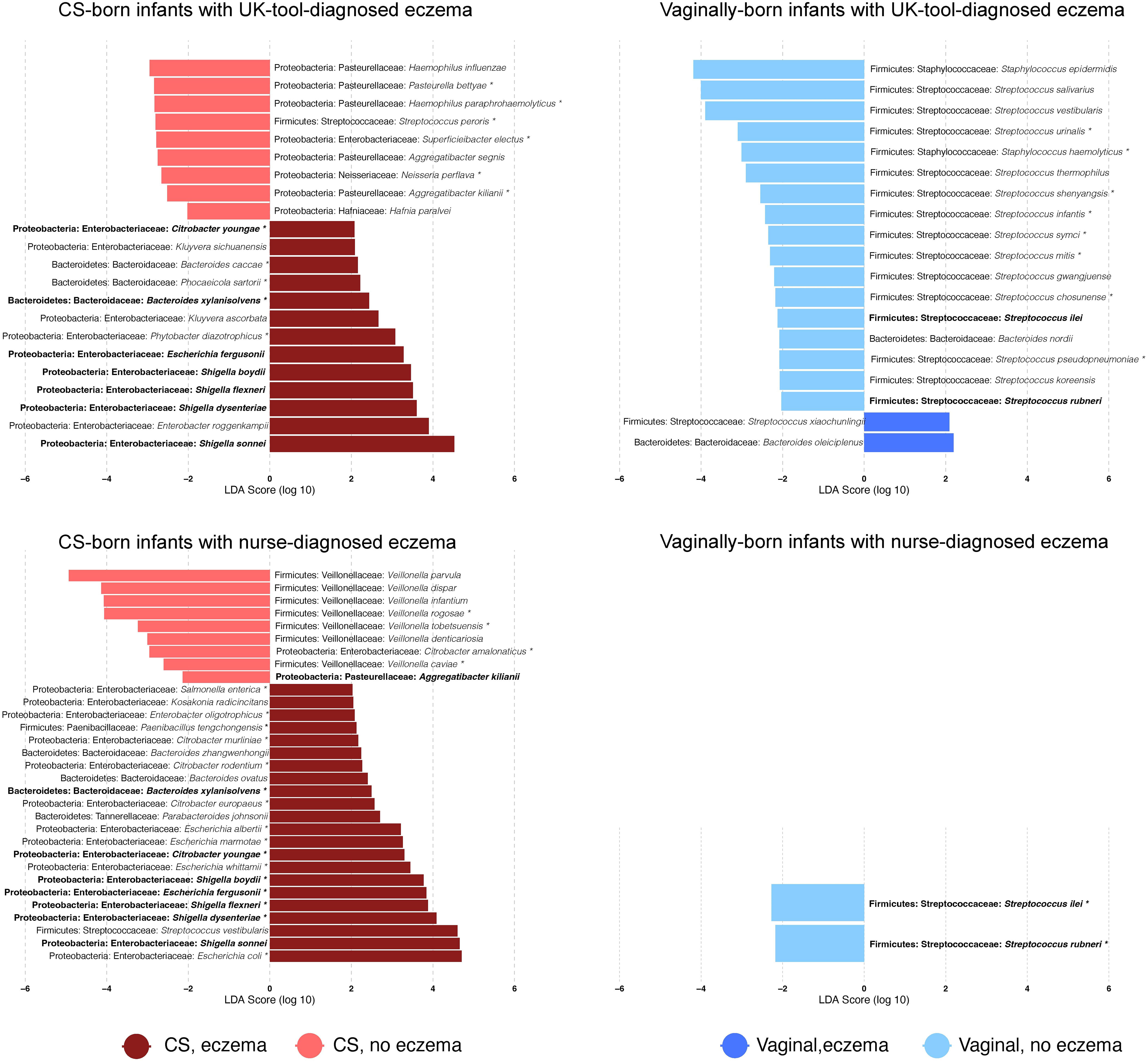
Figure 2 Differentially abundant bacterial species. Bar plots show the log10-transformed linear discriminant analysis (LDA) score for a given species obtained by LEfSe analysis according to the delivery mode and the presence of eczema diagnosed with a given tool. LDAs report which are the major species explaining the differences in microbiome species relative abundance between infants with and without eczema. Positive values of LDA correspond to species that are overrepresented in infants with eczema; negative values to those without eczema. * = species that were also detected as differentially abundant with DESeq2 (FDR-adjusted p-value < 0.05). Bold = species that were found to be differentially abundant by both eczema diagnosis tools in CS- or vaginally born infants. Values are also reported in Supplementary File 3.
The seven species Bacteroides xylanisolvens, Citrobacter youngae, Shigella boydii, Escherichia fergusonii, Shigella flexneri, Shigella dysenteriae, and Shigella sonnei were found to be discriminative for CS-born infants with eczema diagnosed by the UK diagnostic tool or nurse diagnosis (Figure 2), with a significantly higher abundance in infants with eczema. Seven Veillonella species, V. parvula, V. dispar, V. infantium, V. rogosae, V. tobetsuensis, V. denticariosia, and V. caviae, were found to be discriminative by LEfSe analyses, with a lower abundance in CS-born infants with nurse-diagnosed eczema (Figure 2; Supplementary File 3). Compared with vaginally born infants without eczema, Streptococcus rubneri and Streptococcus ilei were less abundant in vaginally born infants with both UK-diagnostic-tool- and nurse-diagnosed eczema.
Metabolic pathways differentially abundant in the intestinal microbiome and the presence of eczema
Eighteen pathways were significantly overrepresented in CS-born infants with both UK-diagnostic-tool- and nurse-diagnosed eczema. These were linked to:
(1) amine and polyamine biosynthesis (superpathway of polyamine biosynthesis I);
(2) amino acid synthesis (L-ornithine biosynthesis II and superpathway of arginine and polyamine biosynthesis, L-methionine biosynthesis, L-homoserine and L-methionine biosynthesis, and S-adenosyl-L-methionine biosynthesis);
(3) antibiotic resistance (polymyxin resistance)
(4) carbohydrate degradation (L-rhamnose degradation I, chitin deacetylation, starch degradation III, and sucrose degradation II)
(5) cofactor, carrier, and vitamin biosynthesis (superpathway of pyridoxal 5-phosphate biosynthesis and salvage, and pyridoxal 5-phosphate biosynthesis I)
(6) fatty acid degradation (fatty acid beta-oxidation I and superpathway of glyoxylate cycle and fatty acid degradation); and
(7) Tricarboxylic acid (TCA cycle I, TCA cycle V, and superpathway of glyoxylate bypass). Pathways from amine and polyamine biosynthesis; amino acid synthesis; cofactor, carrier, and vitamin biosynthesis; and fatty acid degradation are present in Escherichia coli species according to the MetaCyc database. Genes involved in homolactic fermentation and biotin biosynthesis II were significantly overrepresented in CS-born infants without eczema when compared with CS-born infants with eczema, irrespective of diagnostic tool (Supplementary File 2, Figure 9).
Functional analyses of the vaginally born cohort did not result in overlapping findings between the UK diagnostic tool and nurse diagnosis.
Intestinal phage composition and the presence of eczema
In total, viruses were detected in 379 infants (96.4%) and accounted for 0.2% (IQR 0.05%–0.48%) of quality-filtered non-human reads. To investigate phages, species belonging to the Caudovirales order were selected. These were the only detected viruses in 359 samples (91.3%). The most abundant detected phages were those infecting Bacteroides, Enterococcus, Escherichia, Enterobacteria, Streptococcus, and Staphylococcus. No significant differences were detected in the composition of phages according to eczema outcomes (Supplementary File 2, Figure 10).
Discussion
In this study, in the first week of life, CS-born infants who subsequently developed eczema had a significantly different microbiome composition at all taxonomic levels from CS-born infants without eczema. Compared with the UK diagnostic tool, nurse diagnosis at 1 year of age is a more robust assessment as it captures infants with more severe and persistent eczema. This explains the larger differences in the composition of the intestinal microbiome observed between infants with and without nurse-diagnosed eczema compared with the differences observed using the UK diagnostic tool, as the latter also captures infants who might have only transitorily suffered from mild eczema.
Consistent with our findings, previous studies have also reported that infants who develop eczema have a higher relative abundance of Enterobacteriaceae, especially Escherichia, Shigella, and Klebsiella, in their intestinal microbiome (Penders et al., 2006; Penders et al., 2007; Hong et al., 2010; Ta et al., 2020). Furthermore, a higher relative abundance of Enterobacteriaceae, especially Escherichia, Shigella, Enterobacter, and Citrobacter, have been found in the intestinal microbiome of children who develop food allergies and asthma (Gosalbes et al., 2013; Azad et al., 2015; Zheng et al., 2016; Tanaka et al., 2017).
Similar to our findings, a lower relative abundance of Veillonella in the intestinal microbiome has been found to be associated with the development of atopic disease and food allergies in children (Arrieta et al., 2015; Chen et al., 2016; Tanaka et al., 2017). In contrast, one small study investigating the intestinal microbiome and the prevalence of eczema simultaneously at 1 year of age found a positive association between the relative abundance of Veillonella and eczema (Zheng et al., 2016). This could be explained by the intestinal microbiome being analyzed at 1 year of age rather than 1 week of age.
Bacteroides, especially Bacteroides fragilis, have previously been reported to be more abundant in the intestine of infants who develop eczema (Vael et al., 2008; Vael et al., 2011; Wang et al., 2016). We found this association only in infants who were vaginally born. This was likely to be because of the low abundance of Bacteroides in the CS-born infants, which is a common observation in infants born by CS (Zimmermann and Curtis, 2018). Immunoglobulin (Ig) A is produced in mucosal tissue, including at high concentrations in the intestine. IgA promotes intestinal barrier function and is involved in the maintenance of host–commensal mutualism (Fagarasan et al., 2010). The binding of IgA to Bacteroides thetaiotaomicron inhibits the innate immune response (Peterson et al., 2007). Bacteroides are the main producer of the short-chain fatty acid propionate (Louis and Flint, 2017) and have been associated with both increased and decreased inflammation of the intestine (Hertli and Zimmermann, 2022). Propionate increases glucose-6-phosphatase activity in the jejunum, which induces gluconeogenesis (Kimura et al., 2011; De Vadder et al., 2014; Chenard et al., 2020). Increased glucose levels in stool samples have previously been found in infants who develop eczema (Ta et al., 2020). However, in the study that found increased glucose levels, infants who developed eczema were reported to have delayed colonization by B. fragilis. It has been hypothesized that the increased glucose levels result from a depletion of bacterial groups with the ability to derive butyrate and propionate through direct or indirect pathways, which results in reduced glycolysis (Ta et al., 2020).
A lower relative abundance of Streptococcus in the intestinal microbiome has previously been associated with the development of eczema in children (Zheng et al., 2016; Yu et al., 2021). In our study, we found a lower relative abundance of Streptococcus in both CS- and vaginally born infants with eczema; however, the difference was statistically significant only in the latter. This could be because a higher proportion of infants born by CS received formula milk, which has been associated with a lower abundance of Streptococcus (Zimmermann and Curtis, 2018).
Previous studies have reported an association between a lower relative abundance of Bifidobacterium and the development of eczema (Watanabe et al., 2003; Murray et al., 2005; Mah et al., 2006; Hong et al., 2010; Penders et al., 2010). Although Bifidobacterium was among the most abundant genera identified in our study, we did not observe such an association. Similarly, Clostridioides difficile has previously been found to be more abundant in the first month of life in infants who develop eczema (Penders et al., 2007; van Nimwegen et al., 2011), an association we also did not find in our study. In mice, Clostridia-related bacteria called segmented filamentous bacteria are reported to promote interleukin (IL)-6 and IL-26 production by dendritic cells, which leads to the induction of T helper (Th) 17 cell differentiation (Ivanov et al., 2009). Th17 cells produce pro-inflammatory cytokines such as IL-17A, IL-17F, and IL-22, which have been associated with the development of autoimmune disease (Littman and Rudensky, 2010). Clostridium spp. cluster IV and XIVa strains, as well as B. fragilis, have been associated with an increase in IL-10-producing T regulatory (Treg) cells (Atarashi et al., 2011; Round et al., 2011). Treg cells accumulate in the intestine and play a critical role in the maintenance of immune homeostasis and the development of autoimmunity and allergies (Turner et al., 2020).
A higher relative abundance of Lactobacillus paracasei in the early intestinal microbiome has been found to reduce the risk of developing eczema (Penders et al., 2010). Administered as a probiotic, Lactobacillus rhamnosus, which belongs to the same taxonomic group as L. paracasei, halves the risk of developing eczema (Wickens et al., 2008). In in vitro studies, exopolysaccharides from L. rhamnosus have been shown to stimulate TNF, IL-6, and IL-12 secretion from human peripheral blood mononuclear cells (Chabota et al., 2001).
In our study, a lower abundance of genes involved in lactic fermentation was found in CS-born infants with eczema. Veillonella metabolize lactate, the final product of lactic fermentation, into the short-chain fatty acids acetate and propionate (Ng and Hamilton, 1973). These metabolites have anti-inflammatory activities and can help to maintain the function of the intestinal barrier (Nurmi et al., 2005; Tedelind et al., 2007; Suzuki et al., 2008). It remains unclear whether lactate has pro- or anti-inflammatory properties (Manosalva et al., 2022). It is also not yet known whether a lower abundance of Veillonella is associated with higher levels of lactate and whether this is associated with an increase in intestinal inflammation. Other bacteria that can ferment lactate, such as Lactobacillus, Streptococcus, and Bifidobacterium, have also been found to be less abundant in infants who develop eczema (Watanabe et al., 2003; Murray et al., 2005; Mah et al., 2006; Hong et al., 2010; Penders et al., 2010; Zheng et al., 2016; Yu et al., 2021).
Although it has been reported that being born by CS and receiving formula milk increases the risk of developing eczema (Kull et al., 2005; Yu et al., 2015), the evidence for this is not robust. In fact, recent studies and meta-analyses report that infants born by CS do not have an increased risk for developing eczema (Renz-Polster et al., 2005; Bager et al., 2008; Polos and Fletcher, 2019). Similarly, a protective effect of breastfeeding against developing eczema is not certain (Elbert et al., 2017; Wang et al., 2017; Bigman, 2020). In line with this, there is only a partial overlap between the differences found in the intestinal microbiome after being born by CS or receiving formula milk and the differences in the composition of the intestinal microbiome associated with the development of eczema. Infants born by CS are known to have a higher relative abundance of Clostridioides and Enterobacteriaceae other than Escherichia (e.g., Citrobacter and Enterobacter) in their intestinal microbiome, while the relative abundance of Escherichia has been reported to be lower (Zimmermann and Curtis, 2018). Furthermore, infants born by CS have a lower relative abundance of Bacteroides and Bifidobacterium, while the findings on the abundance of Streptococcus remain contradictory (Zimmermann and Curtis, 2018). A higher relative abundance of Veillonella has been found in infants born by CS (Hesla et al., 2014). Similarly, breastfed infants have been reported to have a higher relative abundance of Bifidobacterium and a lower relative abundance of Bacteroides and Clostridioides, while the findings for Streptococcus and Veillonella are contradictory (Zimmermann and Curtis, 2018).
Although preterm-born infants are known to have a higher relative abundance of Enterobacteriaceae (especially Escherichia, Enterobacter, and Citrobacter) and Bacteroides, as well as a lower relative abundance of Bifidobacterium, in their intestinal microbiome (Zimmermann and Curtis, 2018), they have not been found to develop eczema more frequently than term-born infants (Gerner et al., 2022). However, in addition to the findings above, preterm-born infants have also been found to have a higher relative abundance of Streptococcus and Veillonella (Zimmermann and Curtis, 2018), which might protect them from eczema. Infants exposed to antibiotics have been reported to have a significantly higher risk of developing eczema and other allergic diseases (Duong et al., 2022). Although infants exposed to antibiotics have been reported to have a lower relative abundance of Bifidobacterium and a higher relative abundance of Enterobacteriaceae in their intestinal microbiome, they have also been found to have a lower relative abundance of Bacteroides, while findings for Clostridioides remain contradictory (Zimmermann and Curtis, 2018). However, the changes in the intestinal microbiome induced by antibiotics are influenced by many other factors, such as the condition leading to the antibiotic use, underlying disease, hospital admission, and, even more importantly, the dose, duration, administration route, and spectrum of the antibiotic (Zimmermann and Curtis, 2019). Exposure to pets has been associated with a higher relative abundance of Veillonella (Azad et al., 2013), which might partially explain why children exposed to pets are less prone to eczema (Epstein et al., 2011). However, again, the association between pet exposure and a decreased risk of eczema is not clear (Langan et al., 2007).
In conclusion, our study of a large cohort of infants, using shotgun metagenomic sequencing and independent bioinformatic and statistical pipelines, found that the bacterial composition of the intestinal microbiome at 1 week of age is associated with the subsequent presence of eczema in the first year of life. Our study also identified putative genera (Escherichia, Shigella, Enterobacter, Citrobacter, and Bacteroides) that might play a role in the development of eczema and others (Streptococcus and Veillonella) that seem to be protective.
The limitations of our study include the fact that the bioinformatics analyses identified taxa by mapping reads to known reference genomes without using de novo assembly. Furthermore, reads were mapped individually in each sample. In future, new approaches based on assembling metagenomes by pooling reads from all samples might improve the analyses of the composition of the microbiome, while also enabling the recovery of the genome of community-shared strains that are not yet discovered (Uritskiy et al., 2018; Churcheward et al., 2022; Krakau et al., 2022). The DNA extraction protocol used was not specifically designed to target viral particles, which means that the abundance of bacteriophages might have been underestimated. Also, many intestinal phages are yet to be characterized (Sutton and Hill, 2019).
Differences between the findings in our study and other studies investigating the association between the intestinal microbiome and the risk of developing eczema might be explained by (1) differences in the investigated time points, i.e., 1 week of age might be too early to detect significant differences in the abundance of Lactobacillus, Bifidobacterium, and Clostridioides, which are more prevalent later in infancy; (2) differences in the definition of eczema outcomes; (3) differences in the methodologies used to investigate the microbiome; and (4) smaller cohort sizes.
Future studies should include multiple sampling time points to investigate the trajectory of the intestinal microbiome that is associated with the development of eczema and atopic disease in general. Furthermore, the exact mechanistic roles of individual bacterial species and the role of the interaction between these species in atopic diseases should be addressed. Findings from these studies will form the basis for evidence-based interventions to modulate the intestinal microbiome in infants and children at risk of eczema and atopic disease, such as modifying breast milk and intestinal microbiota with directed prebiotics and probiotics, and the use of bacteriophages.
Data availability statement
FASTQ files containing forward and reverse quality-filtered non-human reads were deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under the project PRJEB55714. Codes used for the preprocessing of sequencing reads (quality-filtering, removal of human DNA, etc.) are deposited at GitHub: https://github.com/miraclelabunifr/Eczema.
Ethics statement
The studies involving human participants were reviewed and approved by the Royal Children’s Hospital Human Research Ethics Committee HREC, authorisation (53543). Written informed consent to participate in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
PZ conceptualized and supervised the study. NC is the chief principal investigator of the MIS BAIR trial. WJ performed the sequencing. SL, PZ, LP and NM helped with data cleaning. SL, OC, LP, PZ, and LF analyzed the data. SL and PZ wrote the initial draft of the manuscript. SL, OC, LP, LF, NC, and PZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Australian National Health and Medical Research Council (NHMRC) (grants GNT1051228 and GNT1099680). PZ is supported by grants from the Swiss National Foundation (PZPGP3_193140), the University of Fribourg, and the Fribourg Hospital, Switzerland. None of the funders had a role in designing the study or in the study conduct. NC is supported by a National Health and Medical Research Council (NHMRC) Investigator Grant (GNT1197117).
Acknowledgments
We thank Susie Germano, Rhian Bonnici, Tamblyn Thomason, and Samuel Forster for their help with data entry and/or the processing of samples, and Kaya Gardiner for project coordination.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frmbi.2023.1147082/full#supplementary-material
Supplementary File 1 | R code.
Supplementary File 2 | Supplementary figures and tables.
Supplementary File 3 | Excel tables containing results of LEfSe and DESeq2 analyses.
References
Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7 (307), 307ra152. doi: 10.1126/scitranslmed.aab2271
Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. (2011). Induction of colonic regulatory T cells by indigenous clostridium species. Science 331 (6015), 337–341. doi: 10.1126/science.1198469
Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Sears MR, et al. (2013). Infant gut microbiota and the hygiene hypothesis of allergic disease: Impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin. Immunol. 9 (1), 15. doi: 10.1186/1710-1492-9-15
Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. (2015). Infant gut microbiota and food sensitization: Associations in the first year of life. Clin. Exp. Allergy 45 (3), 632–643. doi: 10.1111/cea.12487
Bager P., Wohlfahrt J., Westergaard T. (2008). Caesarean delivery and risk of atopy and allergic disease: Meta-analyses. Clin. Exp. Allergy 38 (4), 634–642. doi: 10.1111/j.1365-2222.2008.02939.x
Beghini F, McIver LJ, Blanco-Miguez A, Dubois L, Asnicar F, Maharjan S, et al. (2021). Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10, e65088. doi: 10.7554/eLife.65088.sa2
Bigman G. (2020). The relationship of breastfeeding and infant eczema: The role of reverse causation. Breastfeed Med. 15 (2), 114–116. doi: 10.1089/bfm.2019.0269
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Caspi R, Foerster H, Fulcher CA, Hopkinson R, Ingraham J, Kaipa P, et al. (2006). MetaCyc: A multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 34 (Database issue), D511–D516. doi: 10.1093/nar/gkj128
Chabota S, Yub HL, De Léséleucb L, Cloutierb D, Van Calsterenc M, Lessardd M, et al. (2001). Exopolysaccharides from lactobacillus rhamnosus RW-9595M stimulate TNF, IL-6 and IL-12 in human and mouse cultured immunocompetent cells, and IFN-gamma in mouse splenocytes. Lait 81, 683–697. doi: 10.1051/lait:2001157
Charman C. R., Venn A. J., Williams H. C. (2004). The patient-oriented eczema measure: development and initial validation of a new tool for measuring atopic eczema severity from the patients' perspective. Arch. Dermatol. 140 (12), 1513–1519. doi: 10.1001/archderm.140.12.1513
Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. (2016). Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr. Allergy Immunol. 27 (3), 254–262. doi: 10.1111/pai.12522
Chenard T, Prevost K, Dube J, Masse E. (2020). Immune system modulations by products of the gut microbiota. Vaccines (Basel) 8 (3), 461. doi: 10.3390/vaccines8030461
Churcheward B, Millet M, Bihouée A, Fertin G, Chaffron S. (2022). MAGNETO: An automated workflow for genome-resolved metagenomics. mSystems 7 (4), e00432–e00422. doi: 10.1128/msystems.00432-22
de Lusignan S, Alexander H, Broderick C, Dennis J, McGovern A, Feeney C, et al. (2021). The epidemiology of eczema in children and adults in England: A population-based study using primary care data. Clin. Exp. Allergy 51 (3), 471–482. doi: 10.1111/cea.13784
De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. (2014). Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156 (1), 84–96. doi: 10.1016/j.cell.2013.12.016
Duong QA, Pittet LF, Curtis N, Zimmermann P. (2022). Antibiotic exposure and adverse long-term health outcomes in children: A systematic review and meta-analysis. J. Infect 85(3), 213–300. doi: 10.1016/j.jinf.2022.01.005
Elbert NJ, van Meel ER, den Dekker HT, de Jong NW, Nijsten TEC, Jaddoe VWV, et al. (2017). Duration and exclusiveness of breastfeeding and risk of childhood atopic diseases. Allergy 72 (12), 1936–1943. doi: 10.1111/all.13195
Epstein TG, Bernstein DI, Levin L, Khurana Hershey GK, Ryan PH, Reponen T, et al. (2011). Opposing effects of cat and dog ownership and allergic sensitization on eczema in an atopic birth cohort. J. Pediatr. 158 (2), 265–71.e1-5. doi: 10.1016/j.jpeds.2010.07.026
European Task Force on Atopic Dermatitis. (1993). Severity scoring of atopic dermatitis: the SCORAD index. consensus report of the European task force on atopic dermatitis. Dermatology 186 (1), 23–31. doi: 10.1159/000247298
Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. (2010). Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 28, 243–273. doi: 10.1146/annurev-immunol-030409-101314
Fleming S, Bodner C, Devereux G, Russell G, Campbell D, Godden D, et al. (2001). An application of the united kingdom working party diagnostic criteria for atopic dermatitis in Scottish infants. J. Invest. Dermatol. 117 (6), 1526–1530. doi: 10.1046/j.0022-202x.2001.01579.x
Gerner T, Rasmussen Rinnov M, Halling AS, Haarup Ravn N, Hjorslev Knudgaard M, Ewertsen C, et al. (2022). Differences in occurrence, risk factors and severity of early-onset atopic dermatitis among preterm and term children. Acta Derm Venereol 102, adv00737. doi: 10.2340/actadv.v102.575
Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, Francino MP. (2013). Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 43 (2), 198–211. doi: 10.1111/cea.12063
Hertli S., Zimmermann P. (2022). Molecular interactions between the intestinal microbiota and the host. Mol. Microbiol. 117 (6), 1297–1307. doi: 10.1111/mmi.14905
Hesla HM, Stenius F, Jaderlund L, Nelson R, Engstrand L, Alm J, et al. (2014). Impact of lifestyle on the gut microbiota of healthy infants and their mothers-the ALADDIN birth cohort. FEMS Microbiol. Ecol. 90 (3), 791–801. doi: 10.1111/1574-6941.12434
Hill D. A., Spergel J. M. (2018). The atopic march: Critical evidence and clinical relevance. Ann. Allergy Asthma Immunol. 120 (2), 131–137. doi: 10.1016/j.anai.2017.10.037
Hong P-Y, Lee BW, Aw M, Shek LPC, Yap GC, Chua KY, et al. (2010). Comparative analysis of fecal microbiota in infants with and without eczema. PloS One 5 (4), e9964. doi: 10.1371/journal.pone.0009964
Hoyles L, McCartney AL, Neve H, Gibson GR, Sanderson JD, Heller KJ, et al. (2014). Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 165 (10), 803–812. doi: 10.1016/j.resmic.2014.10.006
Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139 (3), 485–498. doi: 10.1016/j.cell.2009.09.033
Kimura I, Inoue D, Maeda T, Hara T, Ichimura A, Miyauchi S, et al. (2011). Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc. Natl. Acad. Sci. 108 (19), 8030–8035. doi: 10.1073/pnas.1016088108
Krakau S, Straub D, Gourlé H, Gabernet G, Nahnsen S. (2022). Nf-core/mag: a best-practice pipeline for metagenome hybrid assembly and binning. NAR Genomics Bioinf. 4 (1), lqac007. doi: 10.1093/nargab/lqac007
Kull I, Böhme M, Wahlgren CF, Nordvall L, Pershagen G, Wickman M. (2005). Breast-feeding reduces the risk for childhood eczema. J. Allergy Clin. Immunol. 116 (3), 657–661. doi: 10.1016/j.jaci.2005.04.028
Langan S. M., Flohr C., Williams H. C. (2007). The role of furry pets in eczema: a systematic review. Arch. Dermatol. 143 (12), 1570–1577. doi: 10.1001/archderm.143.12.1570
Littman D. R., Rudensky A. Y. (2010). Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140 (6), 845–858. doi: 10.1016/j.cell.2010.02.021
Louis P., Flint H. J. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19 (1), 29–41. doi: 10.1111/1462-2920.13589
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Lu J., Salzberg S. L. (2020). Ultrafast and accurate 16S rRNA microbial community analysis using kraken 2. Microbiome 8 (1), 124. doi: 10.1186/s40168-020-00900-2
Mah KW, Björkstén B, Lee BW, van Bever HP, Shek LP, Tan TN, et al. (2006). Distinct pattern of commensal gut microbiota in toddlers with eczema. Int. Arch. Allergy Immunol. 140 (2), 157–163. doi: 10.1159/000092555
Manosalva C, Quiroga J, Hidalgo AI, Alarcón P, Anseoleaga N, Hidalgo MA, et al. (2022). Role of lactate in inflammatory processes: Friend or foe. Front. Immunol. 12, 808799. doi: 10.3389/fimmu.2021.808799
Messina NL, Gardiner K, Donath S, Flanagan K, Ponsonby AL, Shann F, et al. (2019). Study protocol for the Melbourne infant study: BCG for allergy and infection reduction (MIS BAIR), a randomised controlled trial to determine the non-specific effects of neonatal BCG vaccination in a low-mortality setting. BMJ Open 9 (12), e032844. doi: 10.1136/bmjopen-2019-032844
Murray CS, Tannock GW, Simon MA, Harmsen HJ, Welling GW, Custovic A, et al. (2005). Fecal microbiota in sensitized wheezy and non-sensitized non-wheezy children: a nested case-control study. Clin. Exp. Allergy 35 (6), 741–745. doi: 10.1111/j.1365-2222.2005.02259.x
Ng S. K., Hamilton I. R. (1973). Carbon dioxide fixation by veillonella parvula m 4 and its relation to propionic acid formation. Can. J. Microbiol. 19 (6), 715–723. doi: 10.1139/m73-116
Nurmi J. T., Puolakkainen P. A., Rautonen N. E. (2005). Bifidobacterium lactis sp. 420 up-regulates cyclooxygenase (Cox)-1 and down-regulates cox-2 gene expression in a caco-2 cell culture model. Nutr. Cancer 51 (1), 83–92. doi: 10.1207/s15327914nc5101_12
Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, Van Ree R, et al. (2006). Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin. Exp. Allergy 36 (12), 1602–1608. doi: 10.1111/j.1365-2222.2006.02599.x
Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. (2007). Gut microbiota composition and development of atopic manifestations in infancy: The KOALA birth cohort study. Gut 56 (5), 661–667. doi: 10.1136/gut.2006.100164
Penders J, Thijs C, Mommers M, Stobberingh EE, Dompeling E, Reijmerink NE, et al. (2010). Intestinal lactobacilli and the DC-SIGN gene for their recognition by dendritic cells play a role in the aetiology of allergic manifestations. Microbiology 156 (Pt 11), 3298–3305. doi: 10.1099/mic.0.042069-0
Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. (2013). Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J. Allergy Clin. Immunol. 132 (3), 601–607.e8. doi: 10.1016/j.jaci.2013.05.043
Peterson DA, McNulty NP, Guruge JL, Gordon JI. (2007). IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2 (5), 328–339. doi: 10.1016/j.chom.2007.09.013
Pittet LF, Messina NL, Gardiner K, Freyne B, Abruzzo V, Francis KL, et al. (2022a). Prevention of infant eczema by neonatal bacillus calmette-guérin vaccination: The MIS BAIR randomized controlled trial. Allergy 77 (3), 956–965. doi: 10.1111/all.15022
Pittet LF, Messina NL, Gardiner K, Freyne B, Abruzzo V, Morrison C, et al. (2022b). Discordance between diagnosis tools for assessing eczema in infants: A challenge for intervention trials. Dermatitis 33 (3), 207–214. doi: 10.1097/DER.0000000000000842
Polos J., Fletcher J. (2019). Caesarean section and children's health: A quasi-experimental design. Popul Stud. (Camb) 73 (3), 353–368. doi: 10.1080/00324728.2019.1624810
Renz-Polster H, David MR, Buist AS, Vollmer WM, O'Connor EA, Frazier EA, et al. (2005). Caesarean section delivery and the risk of allergic disorders in childhood. Clin. Exp. Allergy 35 (11), 1466–1472. doi: 10.1111/j.1365-2222.2005.02356.x
Rook G. A., Stanford J. L. (1998). Give us this day our daily germs. Immunol. Today 19 (3), 113–116. doi: 10.1016/S0167-5699(98)80008-X
Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. (2011). The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332 (6032), 974–977. doi: 10.1126/science.1206095
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12 (6), R60. doi: 10.1186/gb-2011-12-6-r60
Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9 (8), 811–814. doi: 10.1038/nmeth.2066
Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, et al. (2019). The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26 (4), 527–541.e5. doi: 10.1016/j.chom.2019.09.009
Sroka-Tomaszewska J., Trzeciak M. (2021). Molecular mechanisms of atopic dermatitis pathogenesis. Int. J. Mol. Sci. 22 (8), 4130. doi: 10.3390/ijms22084130
Sutton T. D. S., Hill C. (2019). Gut bacteriophage: Current understanding and challenges. Front. Endocrinol. 10. doi: 10.3389/fendo.2019.00784
Suzuki T., Yoshida S., Hara H. (2008). Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br. J. Nutr. 100 (2), 297–305. doi: 10.1017/S0007114508888733
Ta LDH, Chan JCY, Yap GC, Purbojati RW, Drautz-Moses DI, Koh YM, et al. (2020). A compromised developmental trajectory of the infant gut microbiome and metabolome in atopic eczema. Gut Microbes 12 (1), 1801964. doi: 10.1080/19490976.2020.1801964
Tanaka M, Korenori Y, Washio M, Kobayashi T, Momoda R, Kiyohara C, et al. (2017). Signatures in the gut microbiota of Japanese infants who developed food allergies in early childhood. FEMS Microbiol. Ecol. 93 (8). doi: 10.1093/femsec/fix099
Tedelind S, Westberg F, Kjerrulf M, Vidal A. (2007). Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J. Gastroenterol. 13 (20), 2826–2832. doi: 10.3748/wjg.v13.i20.2826
Turner JA, Stephen-Victor E, Wang S, Rivas MN, Abdel-Gadir A, Harb H, et al. (2020). Regulatory T cell-derived TGF-β1 controls multiple checkpoints governing allergy and autoimmunity. Immunity 53 (6), 1202–1214.e6. doi: 10.1016/j.immuni.2020.10.002
Uritskiy G. V., DiRuggiero J., Taylor J. (2018). MetaWRAP–a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6 (1), 158. doi: 10.1186/s40168-018-0541-1
Vael C, Nelen V, Verhulst SL, Goossens H, Desager KN. (2008). Early intestinal bacteroides fragilis colonisation and development of asthma. BMC Pulm Med. 8, 19. doi: 10.1186/1471-2466-8-19
Vael C, Vanheirstraeten L, Desager KN, Goossens H. (2011). Denaturing gradient gel electrophoresis of neonatal intestinal microbiota in relation to the development of asthma. BMC Microbiol. 11, 68. doi: 10.1186/1471-2180-11-68
van der Hulst A. E., Klip H., Brand P. L. P. (2007). Risk of developing asthma in young children with atopic eczema: A systematic review. J. Allergy Clin. Immunol. 120 (3), 565–569. doi: 10.1016/j.jaci.2007.05.042
van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. (2011). Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 128 (5), 948–55.e1-3. doi: 10.1016/j.jaci.2011.07.027
Wang H, Li Y, Feng X, Li Y, Wang W, Qiu C, et al. (2016). Dysfunctional gut microbiota and relative co-abundance network in infantile eczema. Gut Pathog. 8, 36. doi: 10.1186/s13099-016-0118-0
Wang J, Ramette A, Jurca M, Goutaki M, Beardsmore CS, Kuehni CE. (2017). Association between breastfeeding and eczema during childhood and adolescence: A cohort study. PloS One 12 (9), e0185066. doi: 10.1371/journal.pone.0185066
Watanabe S, Narisawa Y, Arase S, Okamatsu H, Ikenaga T, Tajiri Y, et al. (2003). Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J. Allergy Clin. Immunol. 111 (3), 587–591. doi: 10.1067/mai.2003.105
Wickens K, Black PN, Stanley TV, Mitchell E, Fitzharris P, Tannock GW, et al. (2008). A differential effect of 2 probiotics in the prevention of eczema and atopy: A double-blind, randomized, placebo-controlled trial. J. Allergy Clin. Immunol. 122 (4), 788–794. doi: 10.1016/j.jaci.2008.07.011
Williams HC, Burney PG, Pembroke AC, Hay RJ. (1994a). The U.K. working party's diagnostic criteria for atopic dermatitis. III. independent hospital validation. Br. J. Dermatol. 131 (3), 406–416. doi: 10.1111/j.1365-2133.1994.tb08532.x
Williams HC, Burney PG, Hay RJ, Archer CB, Shipley MJ, Hunter JJ, et al (1994b). The U.K. working party's diagnostic criteria for atopic dermatitis . I. Derivation of a minimum set of discriminators for atopic dermatitis. Br. J. Dermatol. 131 (3), 383–396. doi: 10.1111/j.1365-2133.1994.tb08530.x
Wood D. E., Lu J., Langmead B. (2019). Improved metagenomic analysis with kraken 2. Genome Biol. 20 (1), 257. doi: 10.1186/s13059-019-1891-0
Yu M, Han K, Kim DH, Nam GE. (2015). Atopic dermatitis is associated with caesarean sections in Korean adolescents, but asthma is not. Acta Paediatr. 104 (12), 1253–1258. doi: 10.1111/apa.13212
Yu L, Deng Y-H, Huang Y-H, Ke H-J, Guo Y, Wu J-L. (2021). Comparison of gut microbiota between infants with atopic dermatitis and healthy controls in guangzhou, China. J. Asthma Allergy 14, 493–500. doi: 10.2147/JAA.S304685
Zheng H, Liang H, Wang Y, Miao M, Shi T, Yang F, et al. (2016). Altered gut microbiota composition associated with eczema in infants. PloS One 11 (11), e0166026. doi: 10.1371/journal.pone.0166026
Zimmermann P., Curtis N. (2018). Factors influencing the intestinal microbiome during the first year of life. Pediatr. Infect. Dis. J. 37 (12), e315–e335. doi: 10.1097/INF.0000000000002103
Zimmermann P., Curtis N. (2019). The effect of antibiotics on the composition of the intestinal microbiota - a systematic review. J. Infect. 79 (6), 471–489. doi: 10.1016/j.jinf.2019.10.008
Keywords: atopic dermatitis, allergies, microbiota, stool, intestine, shotgun metagenomic sequencing, infants, gut
Citation: Leo S, Cetiner OF, Pittet LF, Messina NL, Jakob W, Falquet L, Curtis N and Zimmermann P (2023) The association between the composition of the early-life intestinal microbiome and eczema in the first year of life. Front. Microbiomes 2:1147082. doi: 10.3389/frmbi.2023.1147082
Received: 18 January 2023; Accepted: 23 February 2023;
Published: 16 March 2023.
Edited by:
Jin Xu, Moffitt Cancer Center, United StatesReviewed by:
Ana Elena Pérez-Cobas, Ramón y Cajal Institute for Health Research, SpainYuanqiang Zou, Beijing Genomics Institute (BGI), China
Copyright © 2023 Leo, Cetiner, Pittet, Messina, Jakob, Falquet, Curtis and Zimmermann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Zimmermann, cGV0cmEuemltbWVybWFubkB1bmlmci5jaA==
 Stefano Leo
Stefano Leo Omer Faruk Cetiner
Omer Faruk Cetiner Laure F. Pittet4,5,6
Laure F. Pittet4,5,6 Nicole L. Messina
Nicole L. Messina Laurent Falquet
Laurent Falquet Nigel Curtis
Nigel Curtis Petra Zimmermann
Petra Zimmermann