- 1Division of Urology, The Hospital for Sick Children, Toronto, ON, Canada
- 2Temerty Faculty of Medicine, University of Toronto, Toronto, ON, Canada
- 3Division of Urology, Department of Surgery, University of Toronto, Toronto, ON, Canada
- 4Temerty Centre for AI Research and Education in Medicine, University of Toronto, Toronto, ON, Canada
- 5Department of Computer Science, University of Toronto. Toronto, ON, Canada
- 6Vector Institute, Toronto, ON, Canada
- 7Center for Computational Medicine, Hospital for Sick Children, Toronto, ON, Canada
- 8Department of Surgery, IWK Hospital, Halifax, NS, Canada
- 9Division of Urology, Seattle Children’s Hospital, University of Washington, Seattle, WA, United States
- 10Division of Urology, Children’s Hospital of Philadelphia, Philadelphia, PA, United States
- 11Department of Urology, Boston Children’s Hospital, Boston, MA, United States
Review Context and Objective: Artificial intelligence (AI) and machine learning (ML) offer new tools to advance care in pediatric urology. While there has been interest in developing ML models in the field, there has not been a synthesis of the literature. Here, we aim to highlight the important work being done in bringing these advanced tools into pediatric urology and review their objectives, model performance, and usability.
Evidence Acquisition: We performed a comprehensive, non-systematic search on MEDLINE and EMBASE and combined these with hand-searches of publications which utilize ML to predict outcomes in pediatric urology. Each article was extracted for objectives, AI approach, data sources, model inputs and outputs, model performance, and usability. This information was qualitatively synthesized.
Evidence Synthesis: A total of 27 unique ML models were found in the literature. Vesicoureteral reflux, hydronephrosis, pyeloplasty, and posterior urethral valves were the primary topics. Most models highlight strong performance within institutional datasets and accurately predicted clinically relevant outcomes. Model validity was often limited without external validation, and usability was hampered by model deployment and interpretability.
Discussion: Current ML models in pediatric urology are promising and have been applied to many major pediatric urology problems. These models still warrant further validation. However, with thoughtful implementation, they may be able to influence clinical practice in the near future.
Introduction
Artificial intelligence (AI) has been gaining popularity over the last decade, and with advances in computing, big data analysis, and expertise, the pace will continue to accelerate (1, 2). Powered by machine learning (ML), new clinical models are being deployed in urology to diagnose urological diseases, detect abnormal imaging findings, and predict a patient’s clinical trajectory (3). Although there has been great momentum and promise, ML-based models require rigorous validation, governance, and thoughtful deployment to achieve meaningful real-world utility (4, 5). Within urology, ML models have been applied to predict oncological outcomes, determine ideal surgical candidates in functional urology, and predict stone clearance in endourology, among many others (6–8).
How do machine learning models work?
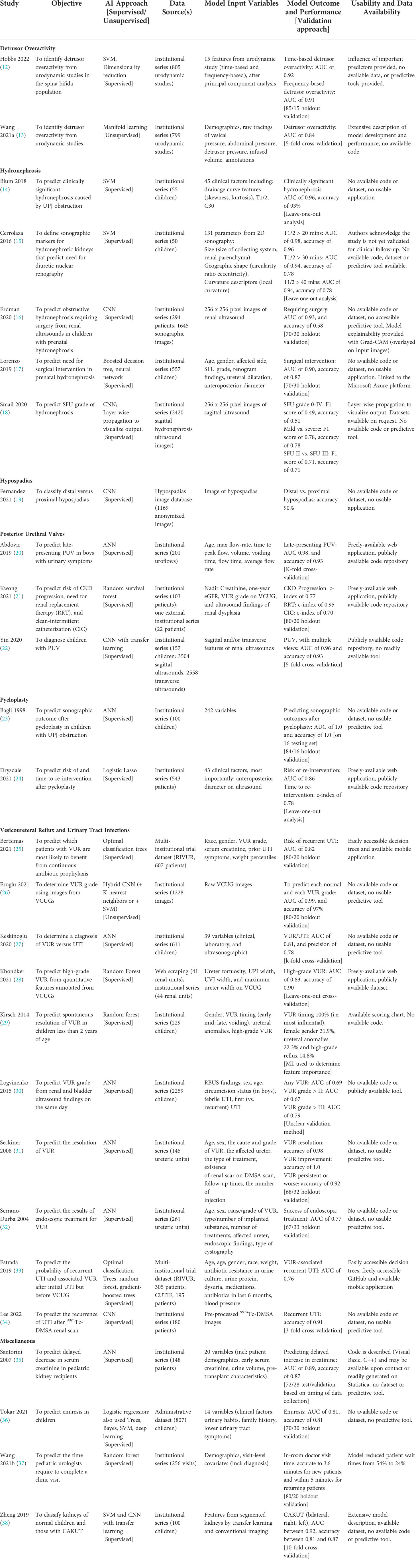
ML approaches differ from regression models by employing complex nonlinear mathematical models and automatically accounting for interactions between variables. In other words, combining clinical factors, or inputs, offers greater utility than each part alone in determining the output. ML models differ in their approach to combining input information when determining an output. The most common models harnessing ML generally fall into one of three categories: decision trees, support vector machines (SVM), or deep learning (i.e. neural networks) (Figure 1).
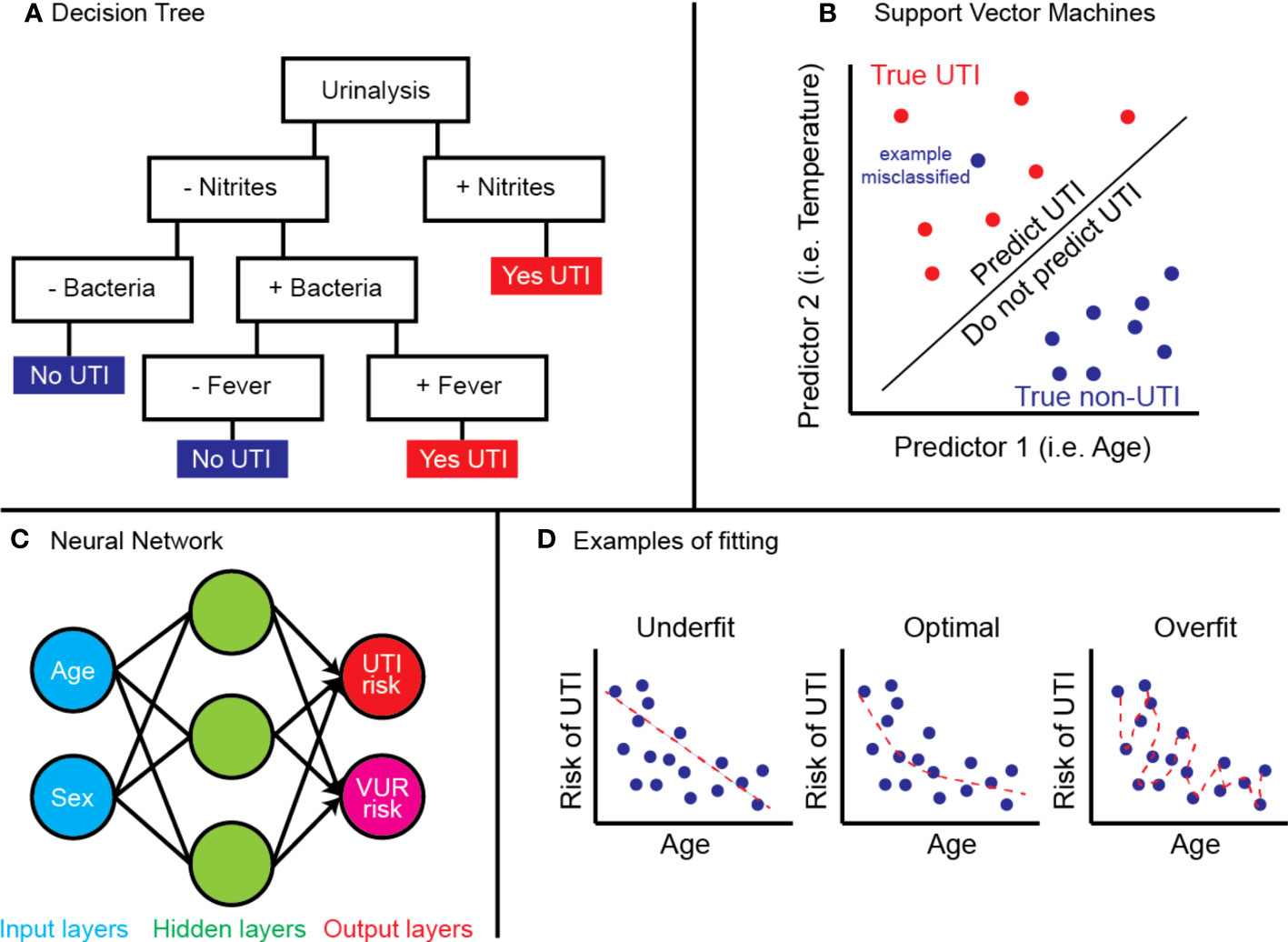
Figure 1 Examples of (A) decision tree predicting UTI by urinalysis and fever, (B) SVM model predicting true UTI by age and temperature, (C) neural network using age and sex to determine UTI and VUR risk, and (D) concept of model fitting.
Decision trees are best thought of as physical trees (Figure 1A) (9). The models are arranged with a starting root and flow through a series of branches, and each branch represents a decision. A leaf, or a final output, is at the end of a series of branches. The inputs will define which branches are selected and decide the final leaf. Decision trees can also be collated or “ensembled” such that the final output is based on a majority vote of the final leaf for each decision tree. Common types include random forest algorithms, optimizable trees, and gradient-boosted or bagged tree models.
SVMs are best thought of on a two-dimensional grid (Figure 1B) (10). Input values from multiple groups are organized along the grid, and the model is trained to create a decision boundary which separates the two groups. Common types include linear or non-linear SVM or kernel-based functions.
Neural networks work like biological neurons, where multiple inputs feed into a given node, activated once a certain threshold is reached (11). The node will propagate a signal which serves as the input for the next layer of nodes. As the model is trained, each node is given a certain weight in the network, and the combinations from the initial input via the weights inform an output. Common types include convolutional neural networks (CNN) or artificial neural networks (ANN) and are often applied to computer vision problems.
An important distinction should be made between supervised and unsupervised learning, which applies to all ML models. In supervised learning, the training datasets include both the inputs and labelled output so that the model can iteratively improve its accuracy. Classically, supervised learning is used for classification or regression. In contrast, unsupervised learning does not include labels and can determine “hidden” patterns in datasets. This is often applied to determine clusters of data, associations, or dimensionality reduction.
Lastly, it is important to understand how the performance of ML models are assessed. The most widely used metric is area-under-the-receiver-operator-curve (AUC), a number between 0 and 1 where 0.5 denotes random classification, and 1.0 denotes perfect classification. The AUC curve is intrinsic to a developed model – describing its ratio of true positive to false positive as the model’s classification criteria is tuned. Thus, AUC offers an aggregate measure of performance across all possible classification thresholds. Other performance metrics should be carefully considered depending on the clinical context. For example, a model that achieves high sensitivity at the cost of more false positives may be more clinically relevant if the disease is serious with life-threatening sequelae if missed.
Methods and evidence synthesis
In this state-of-the-art review, articles regarding ML applications and models applied to pediatric urology were considered. We decided against performing a systematic review a priori to ensure that we are able to provide a broad scope of a developing field while providing commentary on current progress.
Articles were retrieved from the MEDLINE and EMBASE databases without language restrictions and included hand-searched references in August 2022. A pediatrics filter was used with the following keywords: hydronephrosis, VUR, urethral obstruction, spina bifida, urodynamics, Wilms tumor, urolithiasis, hypospadias, artificial intelligence, and machine learning. These keywords were meant to represent common conditions that are managed by pediatric urologists. A sample search is provided in Supplementary Table 1.
Only full published articles were included. Editorials, conference abstracts, comments, reviews, and book chapters were excluded but were searched for relevant full articles. In cases where multiple publications about the same model were found, the article with the largest model training size was collated in the synthesis. Given the nature of this review and subject matter, we did not perform a critical appraisal of study quality.
Articles were included if they involved using ML models (i.e. beyond logistic or multivariable regression) to determine a specific outcome listed in their study objective. Articles including decision trees, SVMs, or deep learning were automatically included for this study. Articles were excluded if the performance metrics of an ML-trained model were not provided, such as accuracy or AUC. Articles were also excluded if the use of ML was for primarily basic science study (e.g. genetic risk scores, image segmentation) without addressing clinical outcomes.
Application of artificial intelligence in pediatric urology
In total, 453 records were retrieved. After confirming the titles and abstracts by two independent reviewers from both literature search and hand-search, 27 unique ML models were available from 31 individual publications (Table 1). There has been a surge in interest in ML-powered applications within pediatric urology, which now encompasses VUR, urinary tract infections (UTIs), hydronephrosis, pyeloplasty, posterior urethral valves, detrusor activity, hypospadias, and others (Figure 2). Model approaches included deep learning (16 models), tree-based classifiers (8 models), SVM (5 models), logistic lasso (1 model), manifold learning (1 model), or combinations thereof. We will discuss advances toward each clinical problem in the following sections.
Figure 2 (A) Number of ML models published per year, (B) distribution of research topic studied by included ML models.
Detrusor overactivity
Two models have been developed by Hobbs and Wang to predict detrusor overactivity from urodynamic studies with the goal of automating readings (12, 13). Both models use similar input variables, include > 800 samples each, and were thoughtfully developed by using feature extraction, data windowing, and describing their pre-processing workflow. Hobbs created a time-based and frequency-based model, achieving similar performance with AUC above 0.90. Meanwhile, Wang used manifold learning to characterize waveforms suggestive of detrusor overactivity on urodynamic studies and created a library to define the outcome of interest. The use of unsupervised learning to learn from tracings and output a binary outcome of detrusor overactivity is especially interesting to inform which patterns that providers should be monitoring for. This offers a new method to define detrusor overactivity, which may supersede current guidelines with further training and standardization.
Hydronephrosis
ML has been used to evaluate hydronephrosis severity, predict long-term renogram findings, and obstructive hydronephrosis using images from renal ultrasounds. Smail used deep learning, which inputs from sagittal images of renal ultrasounds and generates a Society of Fetal Urology (SFU) grade from I-IV (18). The model is believed to be comparable to physicians in predicting SFU grade II and III, where there is the most variability, and performed with an accuracy of 71%. Similarly, Erdman used sagittal and transverse images from renal ultrasounds to determine whether the hydronephrosis was obstructive or physiologic, with an accuracy of 58% and an AUC of 0.93 (16). Both models provided good explainability for deep learning models by identifying regions of interest to determine the corresponding outcome. These studies show promising results and the strength of deep learning models. It is unclear if the former model outperforms the reliability between clinicians performing SFU grading while the latter has not yet been clinically validated.
Cerrolaza similarly used renal ultrasounds; however, the authors manually pre-processed images and segmented the kidney (15). This resulted in 131 variables corresponding to size, geometric shape, and curvature. The variables were then used to predict T1/2 thresholds, which was superior to using SFU grade. This promising model could decrease the number of diuretic renograms in 62% of children safely. Furthermore, Blum predicted clinically significant obstruction using 43 variables directly from drainage curves (14). They used an SVM model, which primarily includes numeric inputs, and showed that tracer clearance 30 minutes after furosemide was the strongest predictor of the outcome. An important strength of this model is that no interpretation or manipulation is required; instead, the user provides recorded values from standardized renogram drainage curves. With both these models, clinical and external validation is warranted before translating into practice.
Alternatively, Lorenzo used patient variables and clinical factors without raw images to predict the need for surgery in prenatal hydronephrosis (17). The model performed well with an accuracy of 0.87 on a publicly available platform; however, this work did not include a usable model, had a limited description of the ML model development resulting in a “black box” phenomenon, and used the arguably subjective decision of performing surgery as the endpoint. Nevertheless, this work shows a potential application of ML from any readily available database at other institutions and describes methods by which researchers can accelerate the use of ML in the field.
Hypospadias
There is significant subjectivity in classifying hypospadias, which limits comparisons between surgeons and outcomes. Fernandez trained an image-based ML model on a large dataset of penile images to predict which patients had proximal vs. distal hypospadias (39). The model was 90% accurate in determining the correct classification, while urology practitioners were 97% accurate based on clinical assessment. Although the model falls short of urologist’s accuracy, the model had stronger reliability than urologists. Hypospadias classification is inherently subjective, and the authors should be commended for performing significant assessment of the reliability in the validity of the hypospadias outcome before implementing ML methods.
Posterior urethral valves
Two studies aimed to diagnose boys with PUV with very different approaches. Yin utilized computer vision models and features from the first postnatal renal ultrasounds to predict PUV in images showing hydronephrosis and achieved high accuracy and reliability. Importantly, their most accurate model used both transverse and sagittal views, for which the authors publicly shared their code (22). Alternatively, Abdovic used age and uroflowmetry findings to predict which boys from 3 to 17 years old with lower urinary tract symptoms have underlying PUV (20). They also employed a neural network model, achieving high accuracy and reliability. Notably, the authors released a publicly available user-friendly application for clinicians to use in practice.
Kwong attempted to predict which boys with PUV will have chronic kidney disease (CKD) progression, need renal replacement therapy, or require clean-intermittent catheterization by using nadir creatinine, VUR presence, and renal dysplasia findings (21). The model used random survival forests and Cox regression, representing the first instance of individualized survival curve prediction in pediatric urology. The authors shared their code and usable clinical tools publicly.
Pyeloplasty
The first documented ML model in pediatric urology was developed by Bagli, who used a neural network to predict the outcome of interest (improvement, same, worse) after pyeloplasty (23). In a sample of 100 children, the authors demonstrated that neural networks had superior sensitivity and specificity than simple linear regression and demonstrated the potential of ML. Since then, Drysdale has used a logistic Lasso model to predict recurrent obstruction risk and time to re-operation after failed pyeloplasty (24). They reported that the post-operative anteroposterior diameter was a significant predictor of this event. This was the first instance of post-selection inference techniques being deployed in pediatric urology ML models, which allows for accurate determination of which predictors, among many, are the most significant.
Vesicoureteral reflux and urinary tract infection
VUR and UTIs are among the most common conditions managed by pediatric urologists and have garnered significant interest in developing ML solutions. To predict the diagnosis of VUR in patients presenting with an uncomplicated UTI before voiding cystourethrogram (VCUG), Keskinoglu developed a neural network from clinical, laboratory, and ultrasound features (27). The model is limited by poor specificity, likely caused by the variable presentation of VUR on the model inputs used. Similarly, Logvinenko created a neural network to predict VUR grades in patients (30). The developers showed that predictors, such as circumcision status, UTI history, or ultrasound findings, can be significant in simple regression models yet futile in predictive ML models.
Two models have attempted to determine VUR grade from VCUGs (26, 28). The model by Eroglu inputs images of raw VCUGs, while the model in Khondker used four annotated features from each VCUG (ureter tortuosity and ureter diameter at proximal, distal and point of maximum dilatation). The former achieved reliability with an AUC of 1.0 in determining VUR grade with a hybrid deep learning approach. However, the validation of this model and inclusion of VCUGs without any VUR limit the model’s usability and uptake. The latter model in Khondker achieved a poorer AUC of 0.83 in predicting grade IV or greater VUR. However, the model was the first in pediatric urology to use explainable AI [set of frameworks to improve understanding of how ML models work to predict the specified outcome (40)] to show which VUR features directly with the grade (41), validated on two datasets, and the authors publicly released their code and data. These two distinct approaches have risks and benefits, but a direct comparison has not been conducted.
Three models by Kirsch, Seckiner, and Serrano-Durba were developed to predict the resolution of VUR (29, 31, 32). VURx, a score-based tool to predict the resolution of primary VUR, was developed by Kirsch. After traditional multivariate analysis to determine predictors for the outcome, the authors used random forest models to ascertain which predictors to use in the final scoring chart. This is a unique combination of the power of ML with score-based clinical assessments that are common in medicine to facilitate translation to practice. Seckiner similarly created a model to predict the resolution of VUR based on age, sex, and VUR grade and included a treatment approach into their model. Lastly, Serrano-Durba predicted the outcome of endoscopic treatment of VUR and showed that their ML model was superior to a simple regression model. They emphasize that the interaction between multiple predictors (VUR grade, age, sex, primary vs. secondary VUR) added value to prediction, while regression only determined VUR grade to be a predictor of resolution.
From large multi-center trial data, two models can predict which patients with VUR are at increased risk for recurrent UTI and who will benefit from continuous antibiotic prophylaxis. Estrada developed easily-accessible decision trees using simple clinical and laboratory measures to predict the risk of recurrent UTI in VUR with moderate reliability (33). Bertsimas built on this idea to determine which patients would best benefit from continuous prophylaxis by using the model to predict which children had a 10% recurrent UTI risk and offering treatment (25). Their retrospective analysis would significantly reduce the incidence of recurrent UTIs from 19.4% to 7.5%. Although prospective validation is warranted, this model has identified which children are in the highest risk groups by age, sex, VUR grade, symptoms, and weight percentiles.
Lastly, Lee used the presence of VUR, cortical defect, or split renal function from nuclear medicine scans to predict which patients would have recurrent UTIs (34). They showed that their ML model combining these features had superior accuracy and specificity than the features alone but similar sensitivity.
Current challenges with artificial intelligence in pediatric urology
Here, we demonstrate the immense potential of AI in pediatric urology and highlight models informing practice. We decided a priori not to critically appraise each model, as ML applications in pediatric urology are the early stages of development, and most were published prior to the establishment of current ML reporting standards (5). Instead, we identified four key areas for future development that apply to both the novice and seasoned developer and clinician: validity, outcome choice, transparency, and thoughtful implementation. Importantly, these future pursuits in AI need to foster multicenter collaboration to make significant impact.
First, validity is inherently difficult in pediatric research given smaller sample sizes and the lack of multi-institutional datasets relative to the adult population (42). For example, a ML model is ideally trained on data from a large number of individuals and externally validated across multiple institutions, which may be particularly challenging in pediatric urology. Most published articles used variations of holdout validation, where a single randomly selected subset of data is held from model training to act as a validation set. Holdout validation often results in inflated model performance metrics, which may not translate into real-world clinical use. Instead, a small external database from an outside institution or cross-validation is a preferred approach to improve the generalizability of ML models from training to practice.
Second, ML models which classify subjective outcomes are more susceptible to bias, such as hydronephrosis severity, VUR grade, or hypospadias classification. For example, ML models on hydronephrosis use pyeloplasty as the primary outcome (16, 24). This can be a subjective clinical decision, and its validity as a primary outcome is debated. ML can play a role in standardizing these outcomes in the long-term; however, at this time, authors should tie subjective outcomes to objective measures. In this case, follow-up nuclear imaging or kidney function measurements offer an additional secondary outcome which can validate the primary outcome. In conjunction, authors can determine the reliability in the subjective outcome itself and use this to perform bias assessment of their models (28, 39).
Next, a core principle in AI is transparency in model development and data sharing (43, 44). This is inherently difficult in medicine because of patient confidentiality and intellectual property conflicts. Regardless, most reviewed publications were clear in their ML approach description but unclear regarding their variable selection. For example, more than one-third of models included >10 variables to predict the respective outcome without justification for why each variable was included. To avoid creating a “black box” model, where users cannot understand a model’s complexity or “logic,” developers should be cautioned from a shotgun approach where all available data is used to train a model without thoughtful consideration. Authors can consider publicly depositing their model code with directions for use with the user’s institutional data. Guidelines have been developed to improve transparency and reproducibility to help standardize reporting of ML studies in urology. Before embarking on model development, developers should be aware of STREAM-URO and DECIDE-AI frameworks (4, 5). Recent advances have also helped reduce training time, improve data quality, and model performance when working with datasets with many variables. These dimensionality-reduction techniques, such as principal component analysis and discriminant analysis, retain the information from the data while removing noisy or irrelevant features to improve computational efficiency (45).
Lastly, most ML models in pediatric urology are difficult to interpret and poorly deployed (3, 17, 46). Most publications include some description of the model training, approach, and validation; however, only one-third of publications provided usable clinical web applications or clinically useful decision trees. The former is especially helpful for the “busy” clinician who is less interested in the technical aspects of ML and more interested in the direct clinical outcomes that each model predicts. Most ML models can be translated into data science web applications with open-source tools such as R Shiny and Streamlit (47, 48). Although application development takes time and learning, easy-to-use ML models facilitate improved model understanding and increase the likelihood of clinical uptake. At the very least, ML model developers should deposit training and validation code within the supplementary information or a publicly hosted platform (i.e. GitHub). Understandably, certain models and datasets will not be shared widely to protect private or commercial interests. This lack of transparency has created a “reproducibility crisis” that has been felt across all domains of AI research (49). Efforts should be made by researchers and journals to encourage data and model sharing; or at minimum, clearly outline their specific data sharing policy.
An outlook on artificial intelligence
The number of publications utilizing AI in medicine is increasing exponentially, and ML is becoming increasingly accessible to clinical research. Without a doubt, the future of personalized medicine will be driven by data, and AI will support clinical decision-making and define new areas of study. Future studies should explore the impact of these models on clinical practice. However, we must endorse caution with both developing and using ML models in pediatric urology. For developers, it is important to recognize that applying ML to a dataset does not justify its use without thoughtful consideration of included variables, intended utility, and clinical context. ML and its application to clinical problems are exciting and novel, but it is not necessarily superior to less complex models or clinical suspicion. Clinicians should recognize that AI in pediatric urology is still in its infancy compared to other medical fields, and further clinical impact assessment is required.
Author contributions
AK, JK, AL, MR contributed to the conception and design of the review and wrote the first draft of the manuscript. AK, JK, SM collected study data. All authors contributed to manuscript revision, read, and approved the submitted version. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fruro.2022.1024662/full#supplementary-material
References
1. Szolovits P. Artificial intelligence and medicine. In: Artificial intelligence in medicine. (UK: Taylor and Francis) (2019). p. 1–19.
2. Bägli DJ, Fernandez N. Artificial intelligence. how artificial is urology practice becoming? Rev Urol Colomb Urol J (2020) 29(01):5–6. doi: 10.1055/s-0040-1709124
3. Chen J, Remulla D, Nguyen JH, Aastha D, Liu Y, Dasgupta P, et al. Current status of artificial intelligence applications in urology and their potential to influence clinical practice. BJU Int (2019) 124(4):567–77. doi: 10.1111/bju.14852
4. Watkinson P, Clifton D, Collins G, McCulloch P, Morgan L, Group D-AS. DECIDE-AI: new reporting guidelines to bridge the development-to-implementation gap in clinical artificial intelligence. Nat Med (2021) 27(6):186–7. doi: 10.1038/s41591-021-01229-5
5. Kwong JCC, McLoughlin LC, Haider M, Goldenberg MG, Erdman L, Rickard M, et al. Standardized reporting of machine learning applications in urology: The STREAM-URO framework. Eur Urol Focus (2021) 7(4):672–82. doi: 10.1016/j.euf.2021.07.004
6. Pai RK, Van Booven DJ, Parmar M, Lokeshwar SD, Shah K, Ramasamy R, et al. A review of current advancements and limitations of artificial intelligence in genitourinary cancers. Am J Clin Exp Urol (2020) 8(5):152.
7. Bentellis I, Guérin S, Khene Z-E, Khavari R, Peyronnet B. Artificial intelligence in functional urology: how it may shape the future. Curr Opin Urol (2021) 31(4):385–90. doi: 10.1097/MOU.0000000000000888
8. Heller N, Weight C. “The algorithm will see you now”: The role of artificial (and real) intelligence in the future of urology. Eur Urol Focus (2021) 7(4):669–71. doi: 10.1016/j.euf.2021.07.010
9. Yiu T. Understanding random forest-towards data science. underst random for how algorithm work why it is so eff. (2019). Available at: https://towardsdatascience.com/understanding-random-forest-58381e0602d2. (Accessed: August 01, 2022)
10. Gandhi R, FR-C R-CNN, Faster R, Algorithms YD. Towards data science. support vector Mach to Mach learn algorithms (2018) (Accessed 26 December 2021).
11. Yiu T. Understanding neural networks-towards data science. Towar Data Sci (2019). Available at: https://towardsdatascience.com/understanding-neural-networks-19020b758230 (Accessed: August 01, 2022).
12. Hobbs KT, Choe N, Aksenov LI, Reyes L, Aquino W, Routh JC, et al. Machine learning for urodynamic detection of detrusor overactivity. Urology (2022) 159:247–54. doi: 10.1016/j.urology.2021.09.027
13. Wang HS, Cahill D, Panagides J, Nelson CP, Wu H, Estrada C. Pattern recognition algorithm to identify detrusor overactivity on urodynamics. Neurourol Urodyn (2021) 40(1):428–34. doi: 10.1002/nau.24578
14. Blum ES, Porras AR, Biggs E, Tabrizi PR, Sussman RD, Sprague BM, et al. Early detection of ureteropelvic junction obstruction using signal analysis and machine learning: A dynamic solution to a dynamic problem. J Urol (2018) 199(3):847–52. doi: 10.1016/j.juro.2017.09.147
15. Cerrolaza JJ, Peters CA, Martin AD, Myers E, Safdar N, Linguraru MG. Quantitative ultrasound for measuring obstructive severity in children with hydronephrosis. J Urol (2016) 195(4 Part 1):1093–9. doi: 10.1016/j.juro.2015.10.173
16. Erdman L, Skreta M, Rickard M, McLean C, Mezlini A, Keefe DT, et al. Predicting obstructive hydronephrosis based on ultrasound alone. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) - medical image computing and computer assisted intervention – MICCAI 2020. Springer Science and Business Media Deutschland GmbH (2020). p. 493–503.
17. Lorenzo AJ, Rickard M, Braga LH, Guo Y, Oliveria JP. Predictive analytics and modeling employing machine learning technology: The next step in data sharing, analysis, and individualized counseling explored with a Large, prospective prenatal hydronephrosis database. Urology (2019) 123:204–9. doi: 10.1016/j.urology.2018.05.041
18. Smail LC, Dhindsa K, Braga LH, Becker S, Sonnadara RR. Using deep learning algorithms to grade hydronephrosis severity: Toward a clinical adjunct. Front Pediatr (2020) 8(1). doi: 10.3389/fped.2020.00001
19. Fernandez N, Lorenzo AJ, Rickard M, Chua M, Pippi-Salle JL, Perez J, et al. Digital pattern recognition for the identification and classification of hypospadias using artificial intelligence vs experienced pediatric urologist. Urology (2021) 147:264–9. doi: 10.1016/j.urology.2020.09.019
20. Abdovic S, Cuk M, Cekada N, Milosevic M, Geljic A, Fusic S, et al. Predicting posterior urethral obstruction in boys with lower urinary tract symptoms using deep artificial neural network. World J Urol (2019) 37(9):1973–9. doi: 10.1007/s00345-018-2588-9
21. Kwong JCC, Khondker A, Kim JK, Chua M, Keefe DT, Dos Santos J, et al. Posterior urethral valves outcomes prediction (PUVOP): a machine learning tool to predict clinically relevant outcomes in boys with posterior urethral valves. Pediatr Nephrol (2022) 37(5):1067–74. doi: 10.1007/s00467-021-05321-3
22. Yin S, Peng Q, Li H, Zhang Z, You X, Fischer K, et al. Multi-instance deep learning of ultrasound imaging data for pattern classification of congenital abnormalities of the kidney and urinary tract in children. Urology (2020) 142:183–9. doi: 10.1016/j.urology.2020.05.019
23. Bagli DJ, Agarwal SK, Venkateswaran S, Shuckett B, Khoury AE, Merguerian PA, et al. Artificial neural networks in pediatric urology: prediction of sonographic outcome following pyeloplasty. J Urol (1998) 160(3 Part 2):980–3. doi: 10.1097/00005392-199809020-00003
24. Drysdale E, Khondker A, Kim JK, Kwong JCC, Erdman L, Chua M, et al. Personalized application of machine learning algorithms to identify pediatric patients at risk for recurrent ureteropelvic junction obstruction after dismembered pyeloplasty. World J Urol (2022) 40(2):593–9. doi: 10.1007/s00345-021-03879-z
25. Bertsimas D, Li M, Estrada C, Nelson C, Scott Wang H-H. Selecting children with vesicoureteral reflux who are most likely to benefit from antibiotic prophylaxis: Application of machine learning to RIVUR. J Urol (2021) 205(4):1170–9. doi: 10.1097/JU.0000000000001445
26. Eroglu Y, Yildirim K, Çinar A, Yildirim M. Diagnosis and grading of vesicoureteral reflux on voiding cystourethrography images in children using a deep hybrid model. Comput Methods Programs Biomed (2021) 210:106369. doi: 10.1016/j.cmpb.2021.106369
27. Keskinoglu A, Ozgur S. The use of artificial neural networks for differential diagnosis between vesicoureteral reflux and urinary tract infection in children. J Pediat Res (2020) 7(3):230–6. doi: 10.4274/jpr.galenos.2019.24650
28. Khondker A, Kwong JC, Rickard M, Skreta M, Keefe DT, Lorenzo AJ, et al. A machine learning-based approach for quantitative grading of vesicoureteral reflux from voiding cystourethrograms: Methods and proof of concept. J Pediatr Urol (2021) 18(1):78–e1. doi: 10.1016/j.jpurol.2021.10.009
29. Kirsch AJ, Arlen AM, Leong T, Merriman LS, Herrel LA, Scherz HC, et al. Vesicoureteral reflux index (VURx): a novel tool to predict primary reflux improvement and resolution in children less than 2 years of age. J Pediatr Urol (2014) 10(6):1249–54. doi: 10.1016/j.jpurol.2014.06.019
30. Logvinenko T, Chow JS, Nelson CP. Predictive value of specific ultrasound findings when used as a screening test for abnormalities on VCUG. J Pediatr Urol (2015) 11(4):176–e1. doi: 10.1016/j.jpurol.2015.03.006
31. Seckiner I, Seckiner SU, Erturhan S, Erbagci A, Solakhan M, Yagci F. The use of artificial neural networks in decision support in vesicoureteral reflux treatment. Urol Int (2008) 80(3):283–6. doi: 10.1159/000127342
32. Serrano-Durbá A, Serrano AJ, Magdalena JR, Martín JD, Soria E, Domínguez C, et al. The use of neural networks for predicting the result of endoscopic treatment for vesico-ureteric reflux. BJU Int (2004) 94(1):120–2. doi: 10.1111/j.1464-410X.2004.04912.x
33. Estrada CR, Nelson CP, Wang HH, Bertsimas D, Dunn J, Li M, et al. Targeted workup after initial febrile urinary tract infection: Using a novel machine learning model to identify children most likely to benefit from voiding cystourethrogram. J Urol (2019) 202(1):144–52. doi: 10.1097/JU.0000000000000186
34. Lee H, Yoo B, Baek M, Choi JY. Prediction of recurrent urinary tract infection in paediatric patients by deep learning analysis of 99mTc-DMSA renal scan. Diagnostics (2022) 12(2):424. doi: 10.3390/diagnostics12020424
35. Santori G, Fontana I, Valente U. Application of an artificial neural network model to predict delayed decrease of serum creatinine in pediatric patients after kidney transplantation. In: Transplantation proceedings. Elsevier (2007). p. 1813–9.
36. Tokar B, Baskaya M, Celik O, Cemrek F, Acikgoz A. Application of machine learning techniques for enuresis prediction in children. Eur J Pediatr Surg (2021) 31(5):414–9. doi: 10.1055/s-0040-1715655
37. Wang H-HS, Cahill D, Panagides J, Yang T-YT, Finkelstein J, Campbell J, et al. A machine learning model to maximize efficiency and face time in ambulatory clinics. Urol Pract (2021) 8(2):176–82. doi: 10.1097/UPJ.0000000000000202
38. Zheng Q, Furth SL, Tasian GE, Fan Y. Computer-aided diagnosis of congenital abnormalities of the kidney and urinary tract in children based on ultrasound imaging data by integrating texture image features and deep transfer learning image features. J Pediatr Urol (2019) 15(1):75–e1. doi: 10.1016/j.jpurol.2018.10.020
39. Fernandez N, Lorenzo AJ, Rickard M, Chua M, Pippi-Salle JL, Perez J, et al. Digital pattern recognition for the identification and classification of hypospadias using artificial intelligence vs experienced pediatric urologist. Urology (2021) 147:264–9. doi: 10.1016/j.urology.2020.09.019
40. Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell (2020) 2(1):56–67. doi: 10.1038/s42256-019-0138-9
41. Lundberg SM, Allen PG, Lee S-I. A unified approach to interpreting model predictions. Advan Neu Inform Proc Sys (2017) 30.
42. Holmbeck GN, Li ST, Schurman JV, Friedman D, Coakley RM. Collecting and managing multisource and multimethod data in studies of pediatric populations. J Pediatr Psychol (2002) 27(1):5–18. doi: 10.1093/jpepsy/27.1.5
43. Haibe-Kains B, Adam GA, Hosny A, Khodakarami F, Waldron L, Wang B, et al. Transparency and reproducibility in artificial intelligence. Nature (2020) 586(7829):E14–6. doi: 10.1038/s41586-020-2766-y
44. Hind M, Houde S, Martino J, Mojsilovic A, Piorkowski D, Richards J, et al. (2020). Experiences with improving the transparency of AI models and services, in: Extended Abstracts of the 2020 CHI Conference on Human Factors in Computing Systems. USA pp. 1–8.
45. Velliangiri S, Alagumuthukrishnan S. A review of dimensionality reduction techniques for efficient computation. Proc Comput Sci (2019) 165:104–11. doi: 10.1016/j.procs.2020.01.079
46. Baier L, Jöhren F, Seebacher S. Challenges in the deployment and operation of machine learning in practice. ECIS (2019).
47. Singh P. Machine learning deployment as a web service. In: Deploy machine learning models to production. (NY, USA:Springer) (2021). p. 67–90.
48. Sievert C. Interactive web-based data visualization with r, plotly, and shiny. (Canada:CRC Press) (2020).
Keywords: artificial intelligence, machine learning, pediatric urology, big data, personalized medicine
Citation: Khondker A, Kwong JCC, Malik S, Erdman L, Keefe DT, Fernandez N, Tasian GE, Wang H-HS, Estrada CR Jr, Nelson CP, Lorenzo AJ and Rickard M (2022) The state of artificial intelligence in pediatric urology. Front. Urol. 2:1024662. doi: 10.3389/fruro.2022.1024662
Received: 22 August 2022; Accepted: 28 September 2022;
Published: 13 October 2022.
Edited by:
Kiarash Taghavi, Monash Children’s Hospital, AustraliaReviewed by:
Marie-Klaire Farrugia, Chelsea and Westminster Hospital NHS Foundation Trust, United KingdomTariq O. Abbas, Weill Cornell Medicine, Qatar
Israel Franco, Yale University, United States
Anne Dudley, Connecticut Children’s Medical Center, United States
Neil Price, The University of Auckland, New Zealand
Copyright © 2022 Khondker, Kwong, Malik, Erdman, Keefe, Fernandez, Tasian, Wang, Estrada, Nelson, Lorenzo and Rickard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mandy Rickard, bWFuZHkucmlja2FyZEBzaWNra2lkcy5jYQ==
 Adree Khondker
Adree Khondker Jethro CC. Kwong
Jethro CC. Kwong Shamir Malik2
Shamir Malik2 Lauren Erdman
Lauren Erdman Daniel T. Keefe
Daniel T. Keefe Nicolas Fernandez
Nicolas Fernandez Carlos R. Estrada Jr.
Carlos R. Estrada Jr. Armando J. Lorenzo
Armando J. Lorenzo Mandy Rickard
Mandy Rickard