- Department of Natural Resource Sciences, Faculty of Agricultural and Environmental Sciences, McGill University, Sainte-Anne-de-Bellevue, QC, Canada
A natural inhabitant of freshwater microbial ecology, Legionella pneumophila is an opportunistically pathogenic bacteria that has found a niche in hot water distribution systems (HWDS) where it challenges hospitals with the spread of nosocomial infection. Superheat-and-flush is a high-temperature pasteurization which can be temporarily effective, but systems often test positive for contamination soon after pasteurization. Because of the centrality of pasteurization in HWDS sanitation schemes, an understanding of the L. pneumophila intrinsic heat shock response may help improve the strategies used to counter its proliferation and ability to trigger nosocomial outbreaks. We tested a group of strains to assess differences in heat shock tolerance between clinical, laboratory, and environmental strains of different origin. The transcriptome of a model Philadelphia-1 L. pneumophila strain in response to heat shock was determined using microarray as an exploratory analysis of the heat shock response: 401 genes were induced, including genes involved in determining protein fate and ribosome biosynthesis while 43 genes were downregulated. Mutants of 3 individual genes were tested to find their direct effects on heat shock tolerance. Deletions of dksA or rpoS, genes involved in the regulation of life cycle switching and important for surviving long-term nutrient deprivation in freshwater lowered heat shock tolerance, suggesting an overlap in the pathways required to tolerate these stressors. Surprisingly, the deletion of htpG, the 90-kilodalton heat shock protein, was found to increase the ability to survive under transient heat shock. Taken altogether, our results show that L. pneumophila exhibits most components of the conserved bacterial heat shock response. Based on this exploratory transcriptomic study, we have provided data that can act as a platform for the research of L. pneumophila's survival to pasteurization in hot water systems.
Introduction
Since the Philadelphia outbreak in 1976 that gave its name, Legionella pneumophila has steadily risen in prominence as a public health challenge in the ensuing years (McDade et al., 1977; Herwaldt and Marra, 2018). In a retrospective (Winn, 1988) on the unmasking of the “Philly Killer,” Winn details the CDC-led effort to trace the Legionnaires' Disease Bacterium, culminating in the isolation of the then-novel waterborne pathogen. L. pneumophila infection most often presents either as Legionnaires' Disease—a life-threatening pneumonia—or as Pontiac Fever—a mild respiratory illness (Cianciotto, 2001; Cunha et al., 2016), though cases of extrapulmonary legionellosis are also known (Padrnos et al., 2014; Chitasombat et al., 2018). A compilation of waterborne disease burden in 2014 in the United States found that Legionnaires' Disease had the second highest cost per hospital visit, costing an estimated total of 402 million 2014 dollars−12.1% of all direct healthcare costs attributed to waterborne disease. Cases were found to have a 98.1% hospitalization rate and 9.0% fatality rate, causing 15.0% of US domestic deaths due to waterborne disease (Collier et al., 2021). L. longbeachae, L. micdadei, and other members of the genus are also more rarely associated with human disease (Gomez-Valero et al., 2014; Chambers et al., 2021).
The aerosolization of contaminated water—commonly from cooling towers, showers, faucets, or some medical instruments—spreads the bacteria (Principe et al., 2017). Fortunately, L. pneumophila is exclusively transmitted directly from environmental reservoirs to humans [with one presumed instance of human-to-human transmission (Borges et al., 2016; Correia et al., 2016)] and therefore outbreaks are locally contained (Phin et al., 2014). Due to the dynamics of airborne transmission, a single source of L. pneumophila contaminated aerosols can seed infection in many exposed individuals (Addiss et al., 1989; David et al., 2017). Colonization of hot water distribution systems (HWDS) by L. pneumophila has been reported by hospitals worldwide, where they are associated with inpatient and staff infections (Darelid et al., 1994, 2002; Berthelot et al., 1998; Kool et al., 1999; Falkinham et al., 2015; Bédard et al., 2016a). As a result of comorbidities in hospital inpatients, nosocomial case fatality rates are higher than those for community-acquired infections (Soda et al., 2017).
Routine suppression of L. pneumophila populations in HWDS relies heavily on control of water temperatures within them (Springston and Yocavitch, 2017; Van Kenhove et al., 2019). Though operational guidelines vary across global health systems (reviewed in Van Kenhove et al., 2019), an example best practice can be drawn from the European Working Group for Legionella Infections (EWGLI). Recommendations include the setting of hot water heaters to above 60◦C, that water throughout the plumbing network hold a temperature >50◦C, and that hot water at terminal outlets be able to reach 70◦C if necessary (Van Kenhove et al., 2019). The superheat-and-flush process, similar to pasteurization, is commonly used to clear L. pneumophila populations from water distribution systems because of its low upfront costs (Whiley et al., 2017). L. pneumophila has proven to be able to re-populate pasteurized water systems, despite adherence to these guidelines (Allegra et al., 2008, 2011). This regrowth could be associated with structural issues in water system topology such as dead ends and stagnation zones where reduced fluid velocity allows biofilm harboring L. pneumophila to develop (Boppe et al., 2016). Incomplete pasteurization of the system can be caused by water use patterns and plumbing design flaws that mix recently heated water from recirculation loops and cooler water in stagnant pipes (Rhoads et al., 2015, 2016; Ji et al., 2017). Direct comparisons of culturability after heat shock between 4 strains of L. pneumophila (two commercial type strains and two environmental strains collected from a HWDS) and one type strain of L. longbeachae showed disimilarities in their abilities to tolerate heat shock (Cervero-Aragó et al., 2015). L. longbeachae was more sensitive to heat shock than any of the pneumophila strains, but no clear pattern emerged concerning the heat shock tolerance of the pneumophila strains exposed to temperatures between from 50 to 70◦C. Nutrient deprivation in HWDS may contribute to heat resistance, as water-suspended cells show an increased expression of heat shock protein genes, as well as other hallmarks of the canonical stringent response (Li et al., 2015; Trigui et al., 2015). Transition to the VBNC stage is associated with long-term nutrient deprivation, as well as with exposure to sanitation procedures like free chlorine or heat treatment (Dietersdorfer et al., 2018; Schrammel et al., 2018; Cervero-Aragó et al., 2019). These VBNC cells are under-counted in standard culture-based methods to assess contamination, but can be resuscitated by passage through the natural eukaryotic hosts of L. pneumophila, and are likely to contribute to survival and re-growth after superheat-and-flush (Casini et al., 2018; Dietersdorfer et al., 2018; Nisar et al., 2020).
The bacterial heat shock response is an integrated network of molecular pathways that works toward the maintenance of cellular homeostasis (Lema et al., 1988; Richter et al., 2010). Studies in E. coli have identified RpoH, or σ32, an alternative sigma factor which regulates transcription of the heat shock regulon as the high-level transcriptional regulator of the heat shock response (Cowing et al., 1985; Jenkins et al., 1991; Chakraborty et al., 2014). Many of the heat shock proteins of the heat shock response are highly-conserved, such as the 90, 70, and 40-kilodalton heat shock proteins—known as Hsp90, Hsp70, and Hsp40 in eukaryotes and as HtpG, DnaK, and DnaJ in bacteria, respectively (Spence et al., 1990; Feder and Hofmann, 1999; Sørensen et al., 2003). Though their precise mechanisms of action vary between species and are often unclear, their general functions are to refold misfolded polypeptides and resolve toxic protein aggregates (Bepperling et al., 2012; Choi et al., 2012; Johnson, 2012; Honoré et al., 2017).
Early radiolabelling studies in L. pneumophila found five high temperature-induced proteins (Lema et al., 1988). HtpB, a Legionella homolog of the E. coli GroEL chaperonin, has received particular attention due to its diverse but crucial roles in bacterial protein folding, aquatic survival, and eukaryotic host infection (reviewed in Garduño et al., 2011). HtpB is under transcriptional control by both the heat shock σ32 and housekeeping σ70 promoters (Garduño et al., 2011). Compared to its homolog in E. coli whose expression is low under steady-temperature conditions, the HtpB protein is abundant in L. pneumophila at all temperatures that support growth (Hoffman et al., 1989). This difference in regulation suggest a different and potentially more significant role of HtpB in L. pneumophila than in other bacteria (Hoffman et al., 1989, 1990).
Because of the importance of pasteurization in the control of L. pneumophila, we conducted a study into its response to transient heat shock in order to understand transcriptomic factors contributing to its survival in hospital hot water systems. Because epidemiological investigations have traced L. pneumophila contamination to a number of different source types, we looked to confirm prior studies that showed differences in the heat shock resistance of strains from different backgrounds by comparing a group of environmental isolates to the laboratory model strain, Philadelphia-1. Though prior research has shown a broadly-conserved bacterial heat shock response, there have been, to the best of our knowledge, no recent studies conducted using L. pneumophila. To address this gap, we performed an exploratory investigation into the transcriptomic response of L. pneumophila to a transient heat shock by microarray to provide insight into the ability of L. pneumophila to resist clearance from HWDS in the face of superheat-and-flush treatments. To validate the transcriptomic assessment of its heat shock response, we also directly characterized the importance of the heat shock and regulatory proteins encoded by htpG, dksA, and rpoS by direct genetic means.
Materials and Methods
Culture Conditions
All strains used in this study are listed in Table 1. All strains were stored at −80◦C. Cultures of E. coli were grown in LB broth or on LB agar (Li et al., 2015), supplemented where indicated with 25 μg/ml chloramphenicol, and incubated overnight at 37◦C. Cultures of L. pneumophila were grown on N-(2-Acetamido)2-aminoethanesulfonic acid (ACES)-buffered Charcoal Yeast Extract (BCYE) Agar plates or in ACES-buffered Yeast Extract (AYE) broth (Li et al., 2015). L. pneumophila was routinely grown at 37◦C for 72 h on BCYE plates or in liquid culture at 37◦C with shaking at 250 rpm to post-exponential phase, as determined by a dark-brown pigment in the culture tube. Where indicated, media were supplemented with 25 μg/ml kanamycin, 5 μg/ml chloramphenicol, and/or 1 mM IPTG.
Heat Shock Tolerance Assay
To assess the heterogeneity of the heat shock response, multiple strains of L. pneumophila were tested for differences in their abilities to survive heat shock. Strains, listed in Table 1, were chosen to represent different cross-section of L. pneumophila diversity. Philadelphia-1, ATCC 33152, represents the 1976 outbreak in Philadelphia (McDade et al., 1977)—though it is important to note that the strain available from the ATCC is slightly divergent from the canonical Philadelphia-1 strain from the CDC (i.e., the first wholly sequenced strain of L. pneumophila and the ancestor of laboratory domesticates JR32 and KS79) (Rao et al., 2013; Mercante et al., 2016). In addition, we tested strains collected from different environmental sources, including cooling towers, hot water plumbing, and clinical isolates (Lévesque et al., 2014; Bédard et al., 2021), as well as a control letS mutant known to be extremely sensitive to heat shock.
Survival of L. pneumophila was evaluated by CFU counts from heat shocked suspensions and from unheated suspensions through serial dilutions in Fraquil and spot plating on BCYE plates. A single colony of L. pneumophila was used to inoculate 10 mL AYE broth and grown overnight to post-exponential phase, as described above. Cells were pelleted by centrifugation for 5 min at 5,000 × g and washed 3 times in Fraquil, a defined artificial freshwater (Li et al., 2015). After washing, bacteria were suspended in Fraquil at a OD600nm of 1.0 and incubated at room temperature (20–25◦C) for 24 h to starve the cells in a low-nutrient environment. Suspensions were dispensed in 1 ml aliquots into 13 ml polypropylene tubes (Sarstedt) and were maintained at room temperature throughout or heat shocked by immersion in a 55◦C circulating water bath for 5 or 20 min and subsequently cooled passively to room temperature and used immediately afterwards to perform CFU counting. Significance analyses were conducted on log-transformed data using Student's two-tailed t-test with unequal variances using GraphPad Prism version 8.3.1.
Sample Preparation for Microarray
Single colonies of L. pneumophila were grown in AYE and suspended in 10 mL Fraquil aliquots at a OD600nm of 1.0, as described above, and incubated at room temperature for 1 h. To minimize the time elapsed before reaching the target temperature, 125 ml glass Erlenmeyer flasks were pre-heated to 55◦C in a circulating water bath. Ten milliliter cell suspensions were poured into the flasks and incubated at 55◦C for 5 min. At the end of heat shock periods, heat-treated cultures were removed from the water baths and 8 ml from each replicate suspension was pelleted by centrifugation at 2,000 × g for 5 min for RNA extraction. Control samples were prepared by pelleting 8 mL of suspensions which had been kept at room temperature.
Nucleic Acid Extraction
Pellets collected from samples treated as above were used for RNA extraction using TRIzol reagent (Invitrogen), following the manufacturer's protocol, and RNA was stored in nuclease-free water (Ambion). Purity and concentration of RNA samples were evaluated using a NanoDrop 1000 spectrophotometer. Genomic DNA (gDNA) was extracted from overnight cultures of Philadelphia-1 using the Wizard Genomic DNA Purification Kit (Promega), following the manufacturer's protocol, and stored in nuclease-free water.
Microarray Analysis
Transcriptomic analysis of the heat shock response was performed using a microarray following a two-color common reference design (Talaat et al., 2002; Faucher and Shuman, 2013). Fifteen micrograms of RNA were labeled with aminoallyl dUTP during reverse transcription into complementary DNA (cDNA) using random hexamer primers and ProtoScript II reverse transcriptase [New England Biolabs (NEB)]. Aminoallyl dUTP labeling was also performed on 2.5 μg of gDNA with random primers and an exonuclease-negative Klenow fragment. Labeled cDNA was coupled with AlexaFluor-647 and labeled gDNA was coupled with AlexaFluor-546, following the manufacturer's protocol, and purified into nuclease-free water via MinElute column (QIAgen).
Microarray slides (GPL19458) were designed with probes complementary to 2,994 open reading frames (ORFs) and 251 small RNA (sRNA) products in the L. pneumophila genome, as previously described (Trigui et al., 2015). Before DNA hybridization, slides were prepared by immersion in pre-hybridization buffer (5x saline sodium citrate, 0.1% SDS, 1 mg/mL bovine serum albumin), as described in Trigui et al. (2015). Microarray slides were hybridized with gDNA, as a reference channel, and cDNA before scanning fluorescence in an InnoScan microarray scanner.
Data analysis was performed in R 3.6.1 using package limma v3.41.2 (Ritchie et al., 2015; Phipson et al., 2016). Background fluorescence was corrected independently for each channel using normexp, an adaptive background correction method. Normalization within and between arrays was performed by variance-stabilizing normalization [implemented in limma as normalizeVSN()]. A linear model was fit to expression data, and an empirical Bayes method was used to compute significance statistics (Benjamini-Hochberg false discovery rate). Differentially regulated transcripts were considered to be those with a magnitude fold-change of ±2 or greater and an adjusted P-value of <0.05. Data deposited in Gene Expression Omnibus, accession number GSE196713.
RT-qPCR
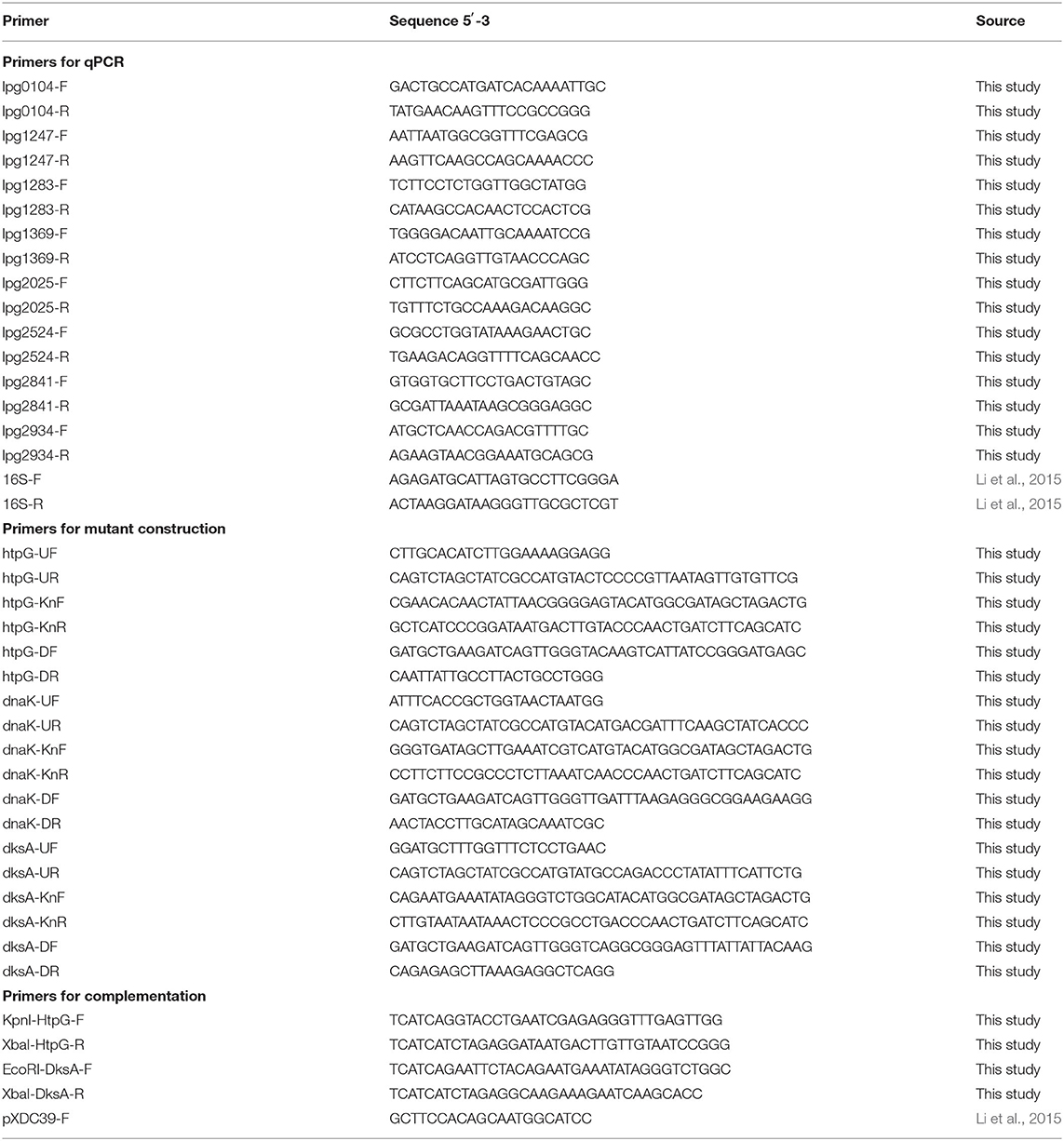
Microarray analysis was validated using quantitative reverse transcription PCR (RT-qPCR) targeted at eight genes identified as differentially regulated−5 upregulated and 3 downregulated—under heat shock. Primer pairs (Table 2) designed using NCBI Primer-BLAST had their amplification efficiencies confirmed using Philadelphia-1 gDNA dilutions between 10 and 0.001 ng/μl. All primers showed efficiencies of >90% (90.02–95.11%). RT-qPCR was conducted as described previously (Trigui et al., 2015). One microgram of RNA was reverse transcribed using Protoscript II reverse transcriptase (NEB). qPCR analysis was performed using iTaq Universal SYBR Green (Bio-Rad) and a CFX Connect Real-Time PCR Detection System (Bio-Rad). Analysis of RT-qPCR expression data was done using the ddCT method normalized to 16S RNA, as described previously (Li et al., 2015).
Mutant Construction
Deletion mutants of htpG (lpg1369) and dksA (lpg2338) were constructed through allelic exchange (Mendis et al., 2018). Efforts to construct a deletion mutant of dnak (lpg2025) were not successful. Flanking regions of each gene ~1 kbp in length were amplified from purified Philadelphia-1 gDNA using OneTaq polymerase (NEB) with their respective upstream region UF-UR primer pair and downstream region DF-DR primer pair (listed in Table 2). A kanamycin resistance cassette was amplified from pSF6 using oligonucleotide primers (gene-Kn-F and gene-Kn-R) that partially overlap with each gene's flanking sequence. Flanking region and kanamycin cassette PCR products were purified using the QIAquick gel extraction kit (QIAgen). DNA fragments were ligated with Phusion polymerase (NEB) using terminal UF-DR primer pairs. Purified ligation products were introduced into L. pneumophila KS79 via natural transformation and resulting transformants were selected on BCYE supplemented with kanamycin. Successful transformation was confirmed by fragment size analysis of PCR amplicons. KS79 was used as the wild-type control strain for phenotypic assays.
To generate hemi-diploid mutants with transcomplementation of the deleted alleles, we re-introduced plasmid-borne copies of each gene. The gene htpG was amplified with its promoter from the Philadelphia-1 genome using primers KpnI-HtpG-F and XbaI-HtpG-R. The amplicon and the plasmid pXDC39 were digested using KpnI and XbaI (NEB) restriction enzymes. Purified digests were ligated using T4 DNA ligase (NEB) and transformed into E. coli strain DH5α made chemically competent with CCMB, as per (Mendis et al., 2018). Plasmid-carrying bacteria were selected for on LB supplemented with chloramphenicol. The first attempt to clone dksA with its own promoter was unsuccessful. Therefore, we cloned it under the control of the Ptac promoter in pMMB207c (Charpentier et al., 2008). Primers EcoRI-DksA-F and XbaI-DksA-R were used to amplify dksA without its promoter. The amplicons and the plasmid were digested with EcoRI and XbaI (NEB) restriction enzymes, ligated and transformed into E. coli as described above. Each plasmid was checked for proper orientation of insert using the pXDC39-F primer and a gene-specific reverse primer. Plasmids were introduced into KS79 and into the deletion mutants through electroporation, as previously described (Mendis et al., 2018), and selected for on BCYE supplemented with chloramphenicol.
Results
Strain-Wise Differences in Heat Shock Tolerance
To support the use of Philadelphia-1 as a model strain of heat shock survival, we tested multiple strains of L. pneumophila to compare their heat resistances. Between-strain differences in ability to tolerate high temperatures were apparent after 20 min of exposure to 55◦C (Figure 1). At this time point, the model strain Philadelphia-1 suffered a 1.69 log10-fold drop in culturability. Our group has previously constructed a ΔletS mutant (Mendis et al., 2018), with an inactive LetA/S two-component signal transduction system causing metabolic dysregulations that render it more sensitive to heat and starvation stresses. Included as a control, this strain did show a significantly greater drop in culturability than Philadelphia-1 (4.99 log10-fold after 20 min). Three of the tested strains, 1427.B.F, 1427.W.HE, and 1427.B.HE, are closely related serotype 1427-like strains isolated from the hot water system of a hospital in Sherbrooke, Canada during the investigation of an outbreak in 2014 (Bédard et al., 2016b, 2021). 1427.W.HE, isolated from the water in a heat exchanger, experienced a loss of culturability comparable to Philadelphia-1 at 5 and 20 min of heat shock. Two biofilm-derived isolates, 1427.B.F and 1427.B.HE, isolated from a faucet and a heat exchanger, respectively, suffered greater losses of culturability−2.05 and 2.77 log10-fold drops, respectively. LSPQ120292 and LSPQ120329 were collected during an outbreak in 2012 in Québec City, Canada (Lévesque et al., 2014). They were isolated from the epidemic source cooling tower and from an unrelated cooling tower, respectively. Compared to Philadelphia-1, the surviving population of LSPQ120292 was not significantly different at either time point. In contrast, LSPQ120329 experienced 1.14 and 3.84 log10-fold drops in culturability after 5 and 20 min, both significantly greater than that of Philadelphia-1. These results show that Philadelphia-1, despite its origin as a cooling tower-derived strain and subsequent history as a laboratory strain, maintains a high level of heat tolerance compared to contemporary environmental isolates and is a usable model strain for our transcriptomic study.
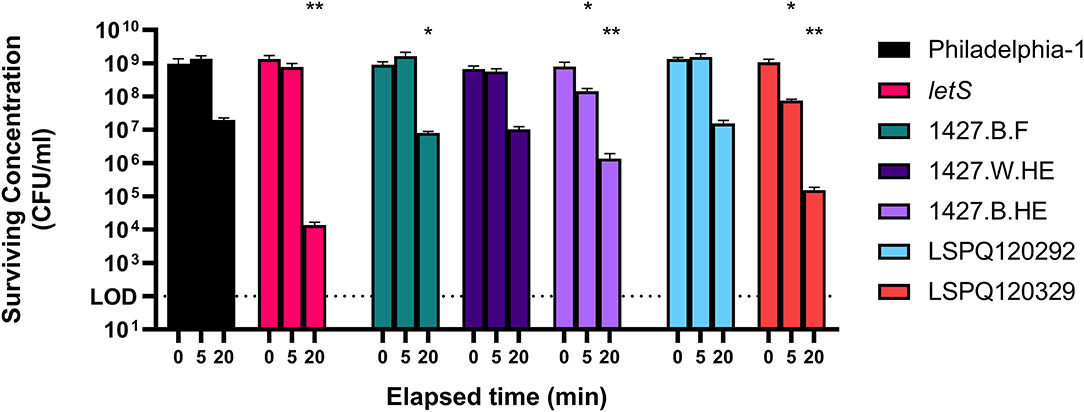
Figure 1. Survival of L. pneumophila strains at 55◦C. Isolates were washed in Fraquil and acclimated for 1 h and heat shocked at 55◦C for 5 or 20 min. Error bars indicate SD of three biological replicates. Significance is shown relative to population concentration of Philadelphia-1 after the same elapsed time and is calculated using a two tailed Student's t statistic with log-transformed data (ns = not significant, * < 0.05, ** < 0.01 with Bonferroni's correction for 7 comparisons).
The Early Heat Shock Regulon
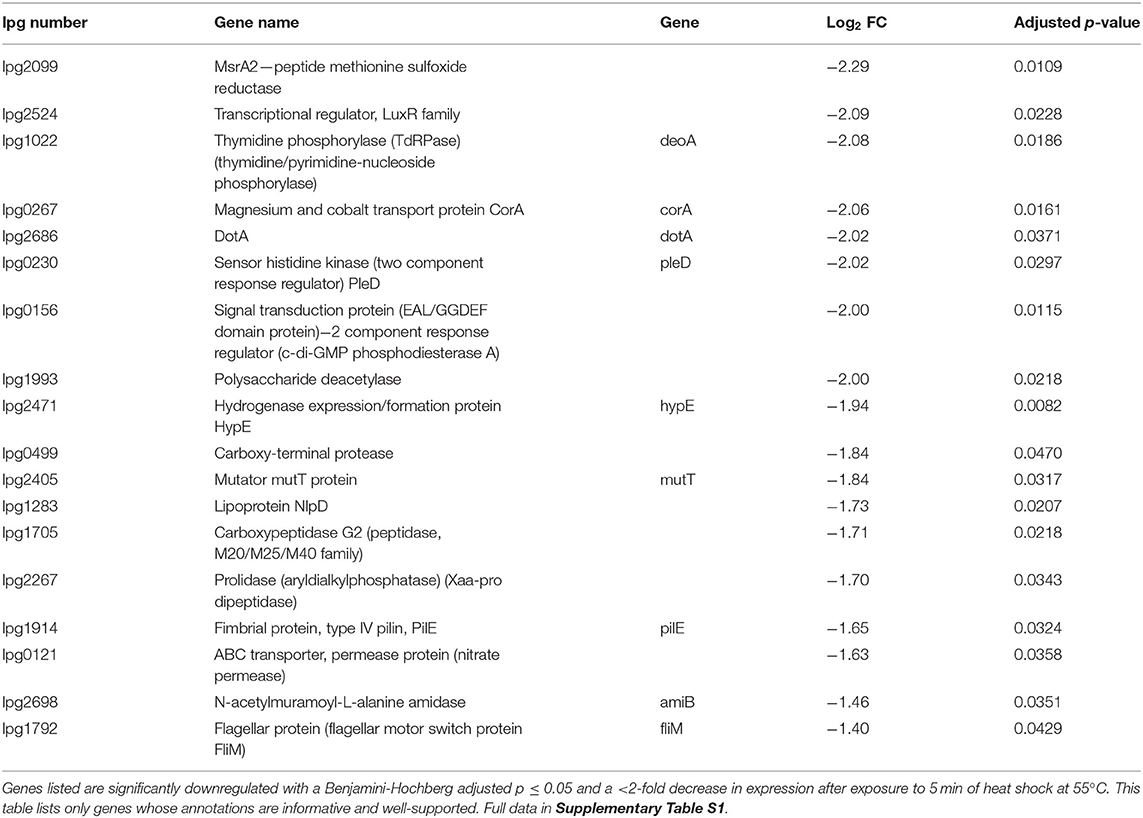
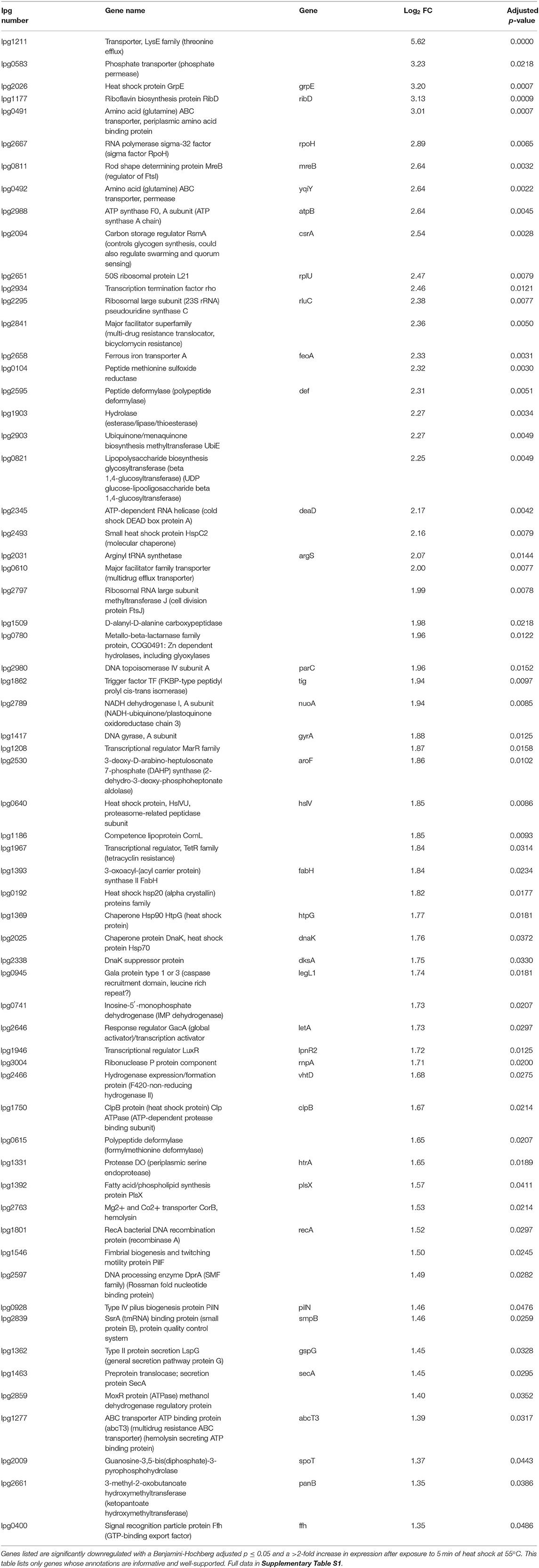
RNA was extracted from starved Philadelphia-1 suspensions before and after experiencing 55◦C temperatures for 5 min. Microarray analysis showed that most genes were not differentially regulated (absolute log2 fold-change ≥ 1 and Benjamini-Hochberg-adjusted p ≤ 0.05) in these conditions (Figure 2A, Supplementary Table S1). Of the 444 regulated genes, 43 were significantly downregulated (selected genes listed in Table 3) while 401 were upregulated (selected genes listed in Table 4). Global transcriptomic analysis by microarray was validated through RT-qPCR (Figure 2B) on a set of eight differentially regulated genes, selected to represent both upregulated and downregulated genes in multiple cellular pathways. Except for one gene (lpg2934), the direction of change was concordant overall between the two methods (Spearman's rho = 0.5476, p = 0.171).
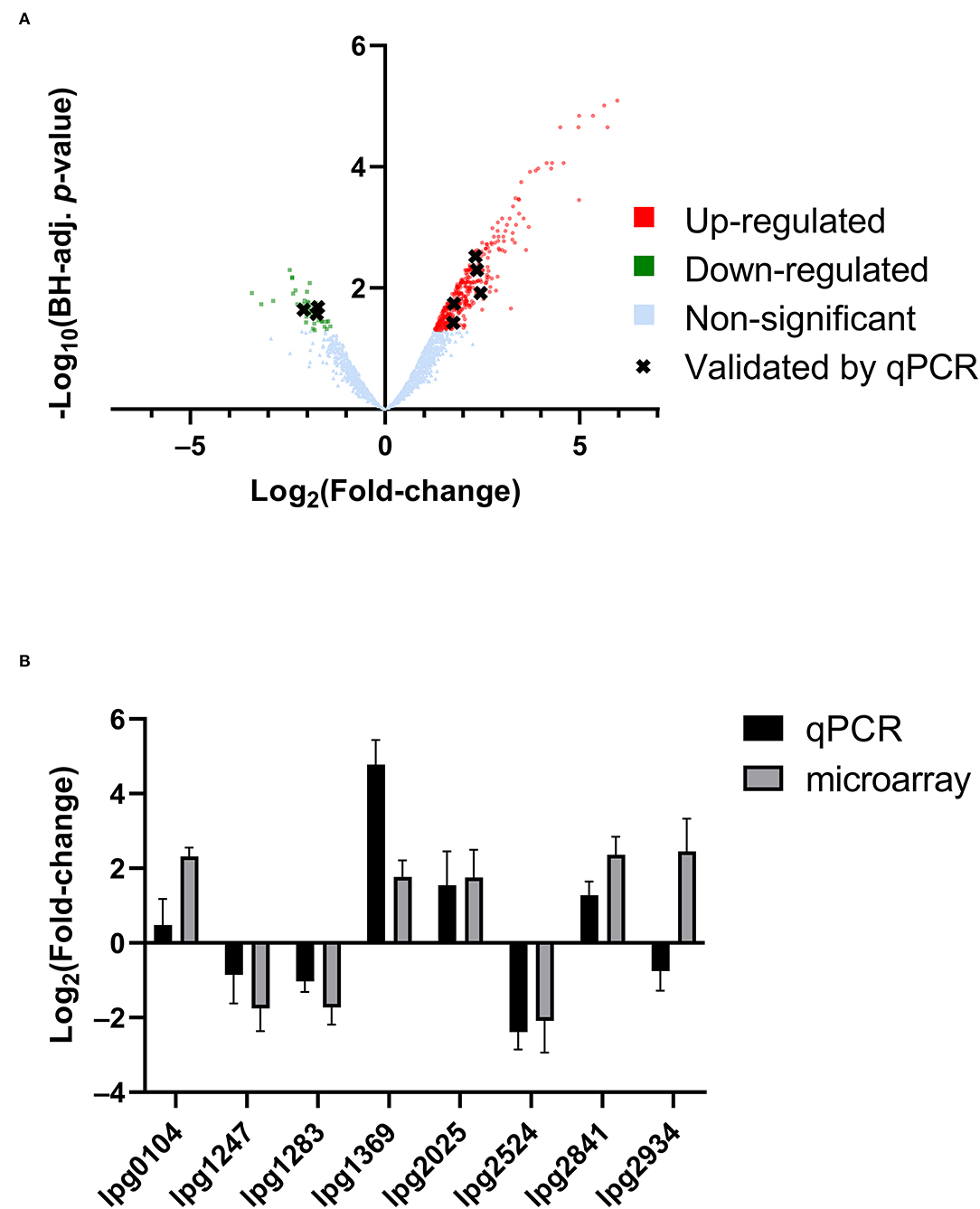
Figure 2. Transcriptomic response of L. pneumophila to heat shock. (A) Volcano plot of microarray data, showing significantly differentially regulated genes. (B) qPCR validation of microarray expression data, showing overall agreement of up-regulation and down-regulation profiles.
To discriminate patterns in gene expression, differentially regulated genes were classified by functional categories based on their annotated GenBank functions (Figure 3A). Like with the individual genes (Figure 2A), there was a global increase in expression of most functional categories (Figure 3A) based on their mean fold-changes. Only transcripts associated with biosynthesis of secondary metabolites, chemotaxis/motility/cell division, and rRNA were not significantly upregulated, with only rRNA transcripts showing an overall (non-significant) down-regulation. Genes involved in detoxification/adaptation and cell envelope synthesis had the second and fourth highest median fold changes in expression, respectively, as could be expected during the early repair of heat shock-induced damage to the cell (Guyot et al., 2010; Meyer and Baker, 2011; Li and Gänzle, 2016). Though patterns in the strict count of genes under regulation (Figure 3B) in each functional category is complicated by variations in the number of transcripts under each category and by the annotation of the L. pneumophila genome leaving many ORFs and proteins of unknown function, the category with the highest number of downregulated genes is transport and binding. Normalizing by percentage to account for variation in category size (Figure 3C), we find that tRNA transcripts are the most up-regulated (30.0%) and that protein fate/hydrolases/secretion transcripts are the most down-regulated (3.12%).
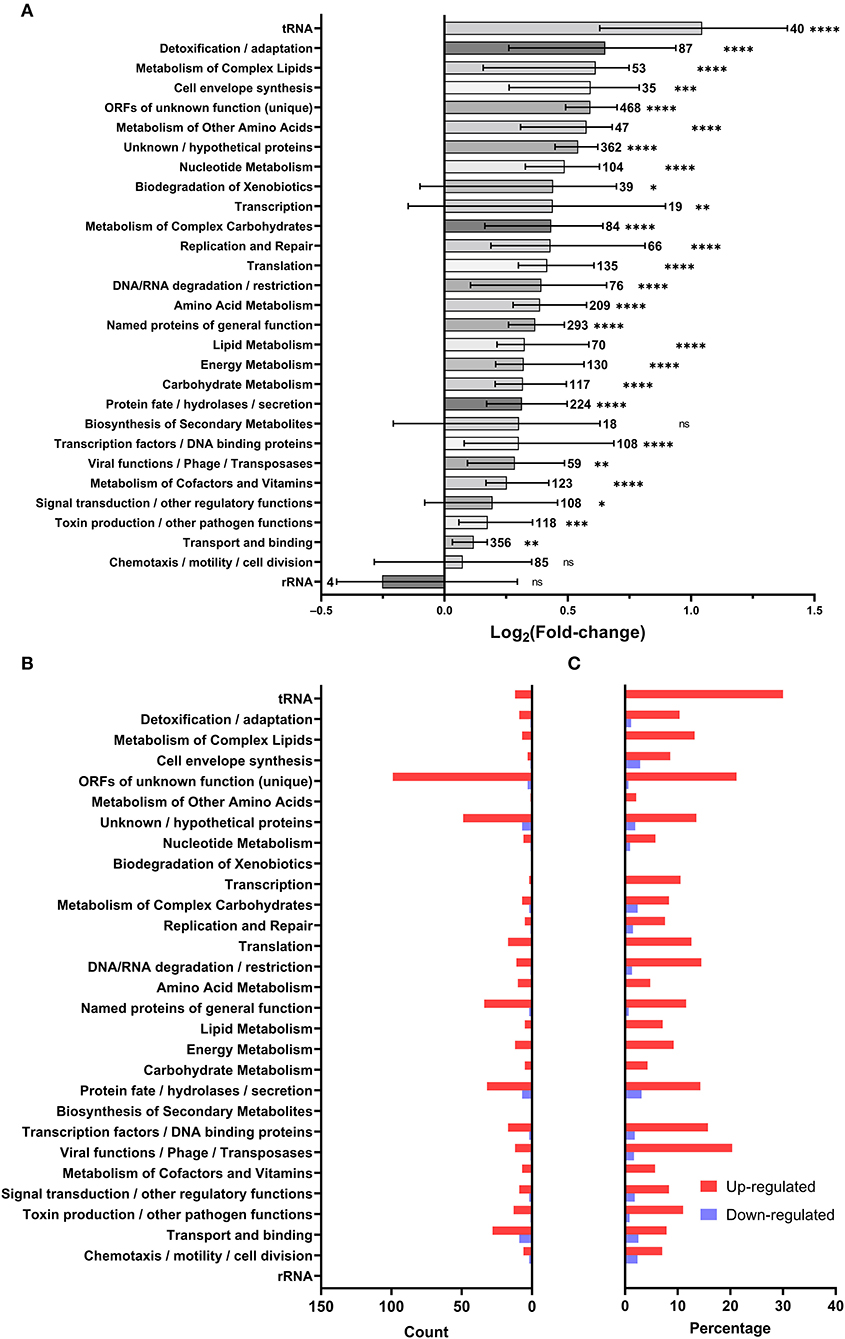
Figure 3. Microarray data summarized by GenBank annotated function. Each gene was assigned to the GenBank categories included in its annotated function: (A) the distribution of log2 fold-change of genes included in each category (bars and error bars show mean and 95% confidence interval; sample size adjacent to each bar; a one-sample two-tailed Wilcoxon signed rank test in Prism 9.3.1 was used to evaluate the significance of discrepancy between the observed log2 fold-changes and a hypothetical log2 fold-change of 0; ns = not significant, * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001), (B) count of differentially regulated genes in each Genbank category, (C) percentage of genes differentially regulated in each Genbank category.
Genes With the Same Expression Scheme Have Different Impacts on Heat Shock Survival
To further dissect the L. pneumophila response to heat shock, three upregulated genes were chosen for further investigation (Table 4). HtpG encodes a bacterial heat shock protein of the Hsp90 family. While homologs of HtpG are strongly conserved across the bacterial domain, its functions are diverse and difficult to characterize (Spence et al., 1990; Genest et al., 2011; Choi et al., 2012; Honoré et al., 2017; Grudniak et al., 2018). Lpg2025 encodes a heat shock protein homologous to the Hsp70-family DnaK (Genest et al., 2011). Finally, lpg2338 encodes DksA, a protein implicated in the regulation of the stringent response and life phase switching in L. pneumophila (Dalebroux et al., 2010). Using allelic exchange, single deletion mutants of KS79 were produced by replacing each gene in turn with a kanamycin resistance cassette. No deletion mutants of lpg2025 were successfully produced, even when the temperature of incubation during mutant construction was lowered from 37 to 25◦C to allow potentially thermo-sensitive mutants to grow.
The two successfully constructed mutant strains were tested for their heat shock tolerances at the same temperatures used in the microarray protocol. Though both strains were found to significantly differ from the wild type in their culturability after heat shock, the effects differed directionally. The ΔhtpG mutant, unexpectedly, maintained culturability to a significantly greater degree than its ancestor—a 1.29 log10-fold higher survivability at 20 min of heat shock (Figure 4A). In contrast, the ΔdksA mutant showed a lower heat shock tolerance, with the surviving population reaching the lower limit of detection after 20 min (Figure 4B). Both phenotypes were successfully complemented by expressing the deleted gene in trans (Figures 4A,B).
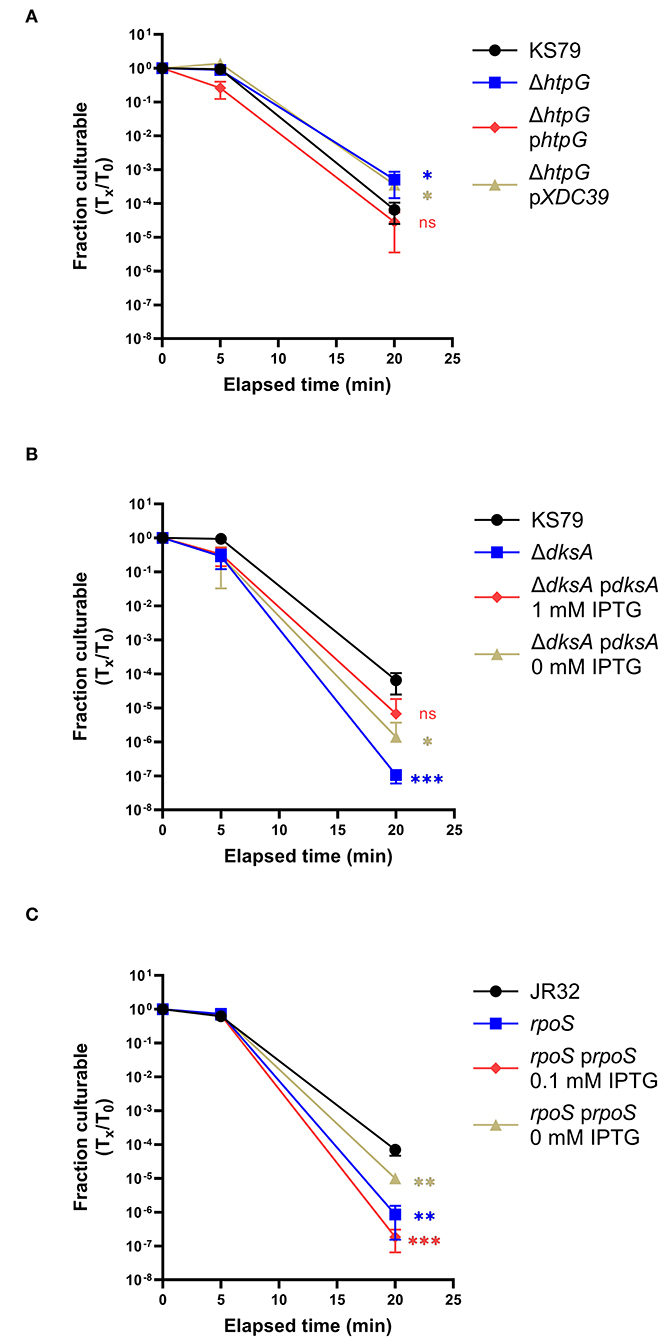
Figure 4. Effects of htpG, dksA, and rpoS on heat shock tolerance. Strains washed in and acclimated to Fraquil were heat shocked at 55◦C for 5 or 20 min. (A) ΔhtpG was compared with its ancestor and complemented with a plasmid-borne copy of htpG [phtpG or with an empty vector control (pXDC39)]. (B) ΔdksA was compared with its ancestor and complemented with a plasmid-borne copy induced with 1 mM IPTG. (C) An RpoS-null mutant was compared to its ancestor strain JR32 and complemented with a plasmid-borne copy induced with 0.1 mM IPTG. Error bars indicate SD, n = 3 biological replicates. Significance was measured compared to the ancestral strain at the same time point using a two-tailed Student's t statistic on log-transformed values (ns = not significant, * < 0.05, ** < 0.01, *** < 0.001 with Bonferroni's correction for 3 comparisons).
Because RpoS and DksA are known to collaborate in regulation of life cycle switching in L. pneumophila (Dalebroux et al., 2010), we extended prior research (Hales and Shuman, 1999; Trigui et al., 2015) on the stress resistance role of RpoS to our present study conditions. To measure the role of RpoS on heat shock survival, we used a insertion mutant strain constructed from a JR32 ancestor strain (Hales and Shuman, 1999). This mutant showed a diminished survivability in response to heat stress, consistent with the role of RpoS in E. coli and Pseudomonas aeruginosa (Hengge-Aronis et al., 1991; Suh et al., 1999). Compared to the JR32 strain, which had a 4.17 log10-fold drop in culturability after 20 min of exposure to 55◦C, the RpoS deletion mutant had a 6.20 log10-fold drop (Figure 4C). This phenotype was partially complemented by IPTG induction of plasmid-borne RpoS expression (Figure 4C).
Discussion
The Influence of Environmental Background on Heat Shock Tolerance
Legionella control regimes in hot water distribution systems are most often centered around baseline temperature set-points and remedial pasteurization processes (Whiley et al., 2017). We hypothesized that strains collected from hot water systems, which had presumably adapted to higher temperatures from exposure during hot water use or from failed pasteurizations, would show higher heat tolerances than clinical isolates or isolates collected from non-hot water systems, which are more likely to have adapted to other circumstances. We tested a panel of six isolates representing three outbreaks: the original 1976 Philadelphia outbreak from which L. pneumophila was identified (McDade et al., 1977; Mercante et al., 2016), a 2012 cooling tower-associated outbreak in Quebec City (Lévesque et al., 2014), and a 2014 HWDS-associated outbreak in Sherbrooke (Bédard et al., 2016b, 2021). After acclimatization to starvation in water, heat shock at 55◦C assay revealed differences in these strains' abilities to tolerate heat shock. Of the related 1427-like isolates collected from spatially separated locations in a hospital HWDS, the isolate collected from heat exchanger water (1427.W.HE) was more resistant to heat shock than related isolates collected from the biofilm in the heat exchanger or from an internal faucet surface. Biofilm isolates, collected from fouled metal surfaces, were found to have a higher resistance to copper attributed to local adaptation to the heavy-metal rich environment (Bédard et al., 2021); we speculate that the higher heat resistance in 1427.W.HE was due to similar in situ adaptation to transient heat shocks while in water, which is more closely mimicked in our experimental conditions. Surprisingly, the two clinically-isolated strains, Philadelphia-1 and LSPQ120292, were the most robust, while the remaining four environmental isolates suffered greater decreases in culturability at 55◦C. Though this contradicts our hypothesis that the heat exchanger strains with prior exposure to heat shock conditions would be the most heat resistant, it is consistent with the findings of Epalle et al., who found a higher retention of culturability and viability in a clinical strain of L. pneumophila compared both to either a laboratory or an environmental strain when exposed as saline suspended cells to temperatures between 50 and 70◦C (Epalle et al., 2015).
L. pneumophila Engages a Core Bacterial Heat Shock Response
Our microarray study of the L. pneumophila Philadelphia-1 transcriptional response to heat shock investigates an important aspect of its resistance to microbial control strategies. We determined that the L. pneumophila early heat shock response involves a co-ordinated up-regulation of genes necessary to cope with the immediate challenges of sudden stress (Figure 2A, Table 4), highlighted by an up-regulation in the transcription of rpoH, an alternative sigma factor canonically recruited by bacteria to direct transcription toward a heat shock response profile (Cowing et al., 1985; Gruber and Gross, 2003; Chakraborty et al., 2014). These include canonical heat shock proteins (such as HtpG and GrpE) and proteases (such as HtrA and ClpB) which help resolve protein damage either by promoting the re-folding of damaged polypeptides or by recruiting proteolytic machinery (Guisbert et al., 2008; Genest et al., 2011). Other upregulated proteins include FeoA (lpg2658), involved in ferrous iron transport, and two polypeptide deformylase genes lpg2595 and lpg0615. These proteins have been shown to play roles in re-establishing polypeptide structure and function (Cianciotto, 2001; Imlay, 2013). Genes related to DNA integrity were also identified in the heat shock up-regulon, including GyrA, RecA, and ParC, highlighting the stresses to the cell caused by heat shock (Piao et al., 2006; Almahmoud et al., 2009; Murata et al., 2011, 2018).
Can these transcriptomic results explain the differences in thermosensitivity observed between clinical and environmental strains of L. pneumophila or the known thermosensitivity of other Legionella species, such as L. longbeachae (Cervero-Aragó et al., 2015)? Though the Legionella genus pan-genome is large and highly variable (Gomez-Valero et al., 2019), the bacterial heat shock response and the heat shock proteins involved are highly conserved and it seems unlikely that the presence or absence of these genes could explain the differences seen in heat shock tolerance. Instead, we find it more plausible that more subtle regulatory changes are responsible for this variation. This is supported by the reduced heat shock resistance of mutants with disruptions in dksA, rpoS, or letS—genes important in the central regulation of virulence and life phase switching (Dalebroux et al., 2010; Trigui et al., 2015; Mendis et al., 2018). We propose therefore that mutations in regulators or promoter of heat shock proteins could most likely explain the variability in heat shock resistance seen between strains.
Transcriptional Involvement of Ribosome Biogenesis Genes
The molecular functions of several significantly upregulated genes (Table 4) implicate a possible role for ribosome biogenesis in the resolution of early heat shock including lpg2295 (rluC), lpg2345 (deaD/csdA), and lpg2797 (ftsJ/rrmJ/rlmE). In addition, the BipA ribosome-associated GTPase encoded at locus lpg2822 is also upregulated, though its corrected significance of 0.0529 falls just outside of a p < 0.05 statistical cut-off. Heat shock is known to independently cause the accumulation of immature ribosomal particle precursors, likely through titration of the DnaK/DnaJ/GrpE chaperone complex away from a putative thermosensitive ribosomal assembly factor (Alix and Guérin, 1993; René and Alix, 2011). One of the early irreversible denaturations during the thermal inactivation of E. coli is the disassembly of the 30S ribosomal subunit, with the remainder of the superstructure sequentially disassembling as temperature increases (Mackey et al., 1991).
BipA is a well-studied protein in E. coli whose deletion is associated with a number of phenotypic changes including virulence defects, hypermotility, and, interestingly, impaired growth at suboptimal temperatures (Farris et al., 1998; Pfennig and Flower, 2001; Krishnan and Flower, 2008; Choudhury and Flower, 2015; Choi and Hwang, 2018; Gibbs et al., 2020; Goh et al., 2021). The major function of BipA at low temperatures appears to be the assembly of the 50S subunit with deletion mutants accumulating 44S precursors that fail to integrate the L6 ribosomal protein (Choudhury and Flower, 2015; Choi and Hwang, 2018). The cold-sensitivity of ΔbipA mutants is complemented by the additional deletion of RluC, a pseudouridine synthase that modifies the peptidyl transferase center in the 50S subunit (Krishnan and Flower, 2008; Choudhury and Flower, 2015). CsdA, a DEAD-box RNA helicase, is another protein involved in the ribosomal biogenesis in E. coli through its effects in the maturation of the 50S subunit (Choudhury and Flower, 2015). Like with BipA, ΔCsdA mutants accumulate a 40S immature precursor of the 50S subunit at low temperatures (Charollais et al., 2004; Peil et al., 2008). Double mutants lacking both CsdA and BipA experience greater growth defects at low temperature and a more severe accumulation of immature ribosome subunit fractions, suggesting independent molecular mechanisms with overlapping effects (Choudhury and Flower, 2015). RrmJ is a methyltransferase whose role in ribosome biogenesis is also in the assembly of the 50S subunit (Caldas et al., 2000; Arai et al., 2015; Wang et al., 2020). Knockout of its methylation function causes delayed growth at lower temperatures.
Regulatory Protein Mutants Have Altered Heat Shock Tolerances
DksA, RpoS, and LetA/S are all involved in regulatory systems that control L. pneumophila's response to starvation and cellular stress, while influencing virulence and shifts into the transmissive or stationary phases (Dalebroux et al., 2010; Trigui et al., 2015; Mendis et al., 2018). Prior work has shown the importance of the LetA/S-RsmY/Z-CsrA regulatory cascade in suppressing growth-related gene transcription in water-suspended bacteria and, reciprocally, that a deletion of LetS causes pre-mature loss of culturability in water and increases susceptibility to heat shock (Mendis et al., 2018). It was also shown that the production of RsmY and RsmZ, downstream small regulatory RNA effectors of LetA/S, involves RpoS control (Hovel-Miner et al., 2009; Mendis et al., 2018). RpoS, an alternative sigma factor, is a major organizer of the bacterial general stress response and, in L. pneumophila, alters gene transcription under nutritional deprivation and allows the bacteria to differentiate from a replicative state to a more resistant transmissive form (Hughes et al., 2019). Proper function of RpoS is required for L. pneumophila to replicate in protozoan hosts (Hales and Shuman, 1999; Hovel-Miner et al., 2009) and to maintain long-term culturability in freshwater (Trigui et al., 2015). RpoS-null mutants of L. pneumophila retain a stationary-phase advantage in resistance to osmotic, oxidative, and acid stressors (Hales and Shuman, 1999) unlike in E. coli where RpoS is necessary to adopt this resistant form. While this appears to stand at odds with our observations of increased heat shock susceptibility in the tested RpoS-null mutants, we note that Hales and Shuman studied L. pneumophila cells immediately after harvest from post-exponential growth in broth while our results were found using cells that had been starved for 24 h in freshwater. We suggest that the pre-mature death seen when RpoS-null mutants are suspended in freshwater (Trigui et al., 2015) reflects a cell that is more sensitive to other stressors, including heat shock. Alternatively, it may be that heat resistance depends on pathways more deeply tied to RpoS function than osmotic, oxidative, or acid stresses, and so the loss of RpoS causes a greater phenotypic change.
DksA is a bacterial transcriptional regulator that binds to RNA polymerase and influences its affinity for different gene promoter sequences. DksA works with the alarmone ppGpp during starvation to allow the bacteria to transition from its replicative life stage to a motile transmissive form (Dalebroux et al., 2010), in co-ordination with RpoS. Both ppGpp and DksA are also involved in a negative feedback loop centered on CsrA, a post-transcriptional regulator that plays simultaneous roles in the control of iron homeostasis, thiamine biosynthesis, quorum sensing, flagellar biosynthesis, central carbon flux, virulence, and the general stress response (Sahr et al., 2017). L. pneumophila lacking DksA show a premature loss of viability in stationary phase compared to wild-type and also develop a filamentous cell shape (Dalebroux et al., 2010). Filamentation is also seen in sessile L. pneumophila cultured at higher temperatures, while planktonic cells and cells grown at 25◦C retain a rod-shaped morphology (Piao et al., 2006). The deletion mutant strain lacking DksA showed a greater sensitivity to heat shock conditions than its wild-type ancestor.
HtpG Deletion Causes Unanticipated Increase in Heat Shock Tolerance
HtpG is a homolog of the ubiquitous 90 kilodalton bacterial heat shock protein. In bacterial model systems, it responds to heat shock, and other forms of cell stress, through a coordinated chaperone response that disaggregates protein tangles and resolves misfolded polypeptides (Genest et al., 2011, 2015). In other bacterial species, HtpG is involved in numerous cell functions including virulence, hot and cold tolerance, and motility (Choi et al., 2012; Grudniak et al., 2018). Surprisingly, given its historically designated role as a heat shock protein, we observed that the deletion of the HtpG-coding locus in L. pneumophila improved heat-shock tolerance. A similar phenomenon was shown by Fauvet et al. in a proteomic study of E. coli, where the deletion of htpG improved growth at elevated temperatures in a temperature-sensitive ΔdnaKdnaJ double mutant (Fauvet et al., 2021). DnaK and DnaJ are, respectively, E. coli homologs of HSP70 and HSP40 which act cooperatively with HtpG to direct re-folding of misfolded or of nascent unfolded polypeptides (Amemura-Maekawa and Watanabe, 1997; Richter et al., 2010). When DnaK/J foldase activities are overwhelmed by massive disruptions of proteostasis during severe cellular stress or outright missing, as in a double mutant, the authors suggest that HtpG directs its substrates toward proteolysis which depletes the cellular inventory of proteins necessary for survival and growth under these conditions. A similar phenomenon may be occurring in our bacterial system, with an overzealous HtpG protein causing off-target proteolysis during transient heat shock.
Conclusion
As a natural member of the freshwater microbiome, L. pneumophila has been able to expand to similar niches in the human built environment (Kirschner, 2016; Lesnik et al., 2016). These ecosystems have repeatedly demonstrated the ability to seed L. pneumophila outbreaks of tremendous public health consequence (McDade et al., 1977; Lévesque et al., 2014). Despite the best efforts of premise plumbing managers, populations can often survive pasteurization (Allegra et al., 2011; Whiley et al., 2017) making it important to understand the heat shock response in L. pneumophila. The present study is, to our knowledge, the first transcriptomic study of L. pneumophila's response to heat shock under starvation conditions simulating a HWDS. As an exploratory platform for study, the microarray has narrowed the scope of possible genes involved in heat shock resistance for future experimental or comparative genomic studies to explain the differences in heat resistance seen in different strains and species of Legionella. We show the recruitment of canonical bacterial heat shock factors, as well as a possible role for ribosome biogenesis in resolving the disruption of protein homeostasis caused by a sudden change in temperature. The ability of the bacterium to accommodate thermal stress could be related to the overall organization of its life cycle, with genes such as letS, dksA, and rpoS that are involved in the life cycle control (Sahr et al., 2017) also affecting thermotolerance when genetically disrupted. Though this study gives a preliminary exploration of factors underlying the heat shock tolerance of L. pneumophila, there are further research paths that it leaves unaddressed. Though strain-wise differences are observed in heat shock tolerances, transcriptomic and genetic studies of a larger corpus of strains could give a deeper understanding of the pathways and genes involved in heat resistance that this study could not fully resolve. In conclusion, we find a complex regulatory network underlying the heat shock resistance of L. pneumophila that allows it to persist in engineered water systems following pasteurization.
Data Availability Statement
The microarray datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: GSE196713].
Author Contributions
JL and SF conceived the project, designed experiments, prepared manuscript, and edits. JL performed experiments and data analysis. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by a Natural Science and Engineering Research Council of Canada Discovery Grant (RGPIN/04499-2018) to SF. JL was the recipient of a Canada Graduate Scholarship—Master's award from Natural Science and Engineering Research Council of Canada.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2022.891477/full#supplementary-material
References
Addiss, D. G., Davis, J. P., Laventure, M., Wand, P. J., Hutchmson, M. A., and Mckinney, R. M. (1989). Community-acquired legionnaires' disease associated with a cooling tower: evidence for longer-distance transport of Legionella pneumophila. Am. J. Epidemiol. 130, 557–568. doi: 10.1093/oxfordjournals.aje.a115370
Alix, J. H., and Guérin, M. F. (1993). Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 9725–9729. doi: 10.1073/pnas.90.20.9725
Allegra, S., Berger, F., Berthelot, P., Grattard, F., Pozzetto, B., and Riffard, S. (2008). Use of flow cytometry to monitor Legionella viability. Appl. Environ. Microbiol. 74, 7813–7816. doi: 10.1128/AEM.01364-08
Allegra, S., Grattard, F., Girardot, F., Riffard, S., Pozzetto, B., and Berthelot, P. (2011). Longitudinal evaluation of the efficacy of heat treatment procedures against Legionella spp. in hospital water systems by using a flow cytometric assay. Appl. Environ. Microbiol. 77, 1268–1275. doi: 10.1128/AEM.02225-10
Almahmoud, I., Kay, E., Schneider, D., and Maurin, M. (2009). Mutational paths towards increased fluoroquinolone resistance in Legionella pneumophila. J. Antimicrob. Chemother. 64, 284–293. doi: 10.1093/jac/dkp173
Amemura-Maekawa, J., and Watanabe, H. (1997). Cloning and sequencing of the dnaK and grpE genes of Legionella pneumophila. Gene 197, 165–168. doi: 10.1016/S0378-1119(97)00257-6
Arai, T., Ishiguro, K., Kimura, S., Sakaguchi, Y., Suzuki, T. T. T., and Suzuki, T. T. T. (2015). Single methylation of 23S rRNA triggers late steps of 50S ribosomal subunit assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E4707–E4716. doi: 10.1073/pnas.1506749112
Bédard, E., Boppe, I., Kouamé, S., Martin, P., Pinsonneault, L., Valiquette, L., et al. (2016a). Combination of heat shock and enhanced thermal regime to control the growth of a persistent Legionella pneumophila strain. Pathogens 5, 1–3. doi: 10.3390/pathogens5020035
Bédard, E., Lévesque, S., Martin, P., Pinsonneault, L., Paranjape, K., Lalancette, C., et al. (2016b). Energy conservation and the promotion of Legionella pneumophila growth: the probable role of heat exchangers in a nosocomial outbreak. Infect. Control Hosp. Epidemiol. 37, 1475–1480. doi: 10.1017/ice.2016.205
Bédard, E., Trigui, H., Liang, J., Doberva, M., Paranjape, K., Lalancette, C., et al. (2021). Local Adaptation of Legionella pneumophila within a hospital hot water system increases tolerance to copper. Appl. Environ. Microbiol. 87, 1–13. doi: 10.1128/AEM.00242-21/SUPPL_FILE/AEM.00242-21-S0001.PDF
Bepperling, A., Alte, F., Kriehuber, T., Braun, N., Weinkauf, S., Groll, M., et al. (2012). Alternative bacterial two-component small heat shock protein systems. Proc. Natl. Acad. Sci. U.S.A. 109, 20407–20412. doi: 10.1073/pnas.1209565109
Berthelot, P., Grattard, F., Ros, A., Lucht, F., and Pozzetto, B. (1998). Nosocomial legionellosis outbreak over a three-year period: investigation and control. Clin. Microbiol. Infect. 4, 385–391. doi: 10.1111/j.1469-0691.1998.tb00082.x
Boppe, I., Bédard, E., Taillandier, C., Lecellier, D., Nantel-Gauvin, M. A., Villion, M., et al. (2016). Investigative approach to improve hot water system hydraulics through temperature monitoring to reduce building environmental quality hazard associated to Legionella. Build. Environ. 108, 230–239. doi: 10.1016/j.buildenv.2016.08.038
Borges, V., Nunes, A., Sampaio, D. A., Vieira, L., Machado, J., Simões, M. J., et al. (2016). Legionella pneumophila strain associated with the first evidence of person-to-person transmission of Legionnaireś disease: a unique mosaic genetic backbone. Sci. Rep. 6, 1–11. doi: 10.1038/srep26261
Caldas, T., Binet, E., Bouloc, P., Costa, A., Desgres, J., and Richarme, G. (2000). The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275, 16414–16419. doi: 10.1074/jbc.M001854200
Casini, B., Baggiani, A., Totaro, M., Mansi, A., Costa, A. L., Aquino, F., et al. (2018). Detection of viable but non-culturable legionella in hospital water network following monochloramine disinfection. J. Hosp. Infect. 98, 46–52. doi: 10.1016/j.jhin.2017.09.006
Cervero-Aragó, S., Rodríguez-Martínez, S., Puertas-Bennasar, A., and Araujo, R. M. (2015). Effect of common drinking water disinfectants, chlorine and heat, on free Legionella and amoebae-associated Legionella. PLoS ONE 10, e0134726. doi: 10.1371/journal.pone.0134726
Cervero-Aragó, S., Schrammel, B., Dietersdorfer, E., Sommer, R., Lück, C., Walochnik, J., et al. (2019). Viability and infectivity of viable but nonculturable Legionella pneumophila strains induced at high temperatures. Water Res. 158, 268–279. doi: 10.1016/j.watres.2019.04.009
Chakraborty, A., Mukherjee, S., Chattopadhyay, R., Roy, S., and Chakrabarti, S. (2014). Conformational adaptation in the E. coli sigma 32 protein in response to heat shock. J. Phys. Chem. B 118, 4793–4802. doi: 10.1021/jp501272n
Chambers, S. T., Slow, S., Scott-Thomas, A., and Murdoch, D. R. (2021). Legionellosis caused by non- Legionella pneumophila species, with a focus on Legionella longbeachae. Microorganisms 9, 291. doi: 10.3390/microorganisms9020291
Charollais, J., Dreyfus, M., and Iost, I. (2004). CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32, 2751–2759. doi: 10.1093/nar/gkh603
Charpentier, X., Faucher, S. P., Kalachikov, S., and Shuman, H. A. (2008). Loss of RNase R induces competence development in Legionella pneumophila. J. Bacteriol. 190, 8126–8136. doi: 10.1128/JB.01035-08
Chitasombat, M. N., Ratchatanawin, N., and Visessiri, Y. (2018). Disseminated extrapulmonary Legionella pneumophila infection presenting with panniculitis: case report and literature review. BMC Infect. Dis. 18, 467. doi: 10.1186/s12879-018-3378-0
Choi, E., and Hwang, J. (2018). The GTPase BipA expressed at low temperature in Escherichia coli assists ribosome assembly and has chaperone-like activity. J. Biol. Chem. 293, 18404–18419. doi: 10.1074/jbc.RA118.002295
Choi, S., Jang, K., Choi, S., Yun, H., and jee Kang, D. H. (2012). Identification of the Vibrio vulnificus htpG gene and its influence on cold shock recovery. J. Microbiol. 50, 707–711. doi: 10.1007/s12275-012-2294-z
Choudhury, P., and Flower, A. M. (2015). Efficient assembly of ribosomes is inhibited by deletion of bipA in Escherichia coli. J. Bacteriol. 197, 1819–1827. doi: 10.1128/JB.00023-15
Cianciotto, N. P (2001). Pathogenicity of Legionella pneumophila. Int. J. Med. Microbiol. 291, 331–343. doi: 10.1078/1438-4221-00139
Collier, S. A., Deng, L., Adam, E. A., Benedict, K. M., Beshearse, E. M., Blackstock, A. J., et al. (2021). Estimate of burden and direct healthcare cost of infectious waterborne disease in the United States. Emerg. Infect. Dis. 27, 140–149. doi: 10.3201/eid2701.190676
Correia, A. M., Ferreira, J. S., Borges, V., Nunes, A., Gomes, B., Capucho, R., et al. (2016). Probable person-to-person transmission of Legionnaires' disease. N. Engl. J. Med. 374, 497–498. doi: 10.1056/nejmc1505356
Cowing, D. W., Bardwell, J. C. A., Craig, E. A., Woolford, C., Hendrix, R. W., and Gross, C. A. (1985). Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci. U.S.A. 82, 2679–2683. doi: 10.1073/pnas.82.9.2679
Cunha, B. A., Burillo, A., and Bouza, E. (2016). Legionnaires' disease. Lancet 387, 376–385. doi: 10.1016/S0140-6736(15)60078-2
Dalebroux, Z. D., Yagi, B. F., Sahr, T., Buchrieser, C., and Swanson, M. S. (2010). Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol. Microbiol. 76, 200–219. doi: 10.1111/j.1365-2958.2010.07094.x
Darelid, J., Bengtsson, L., Gästrin, B., Hallander, H., Löfgren, S., Malmvall, B. E., et al. (1994). An outbreak of legionnaires' disease in a swedish hospital. Scand. J. Infect. Dis. 26, 417–425. doi: 10.3109/00365549409008615
Darelid, J., Löfgren, S., and Malmvall, B. E. (2002). Control of nosocomial Legionnaires' disease by keeping the circulating hot water temperature above 55◦C: experience from a 10-year surveillance programme in a district general hospital. J. Hosp. Infect. 50, 213–219. doi: 10.1053/jhin.2002.1185
David, S., Afshar, B., Mentasti, M., Ginevra, C., Podglajen, I., Harris, S. R., et al. (2017). Seeding and establishment of Legionella pneumophila in hospitals: implications for genomic investigations of nosocomial legionnaires' disease. Clin. Infect. Dis. 64, 1251–1259. doi: 10.1093/cid/cix153
De Felipe, K. S., Glover, R. T., Charpentier, X., Anderson, O. R., Reyes, M., Pericone, C. D., et al. (2008). Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4, e1000117. doi: 10.1371/journal.ppat.1000117
Dietersdorfer, E., Kirschner, A., Schrammel, B., Ohradanova-Repic, A., Stockinger, H., Sommer, R., et al. (2018). Starved viable but non-culturable (VBNC) Legionella strains can infect and replicate in amoebae and human macrophages. Water Res. 141, 428–438. doi: 10.1016/j.watres.2018.01.058
Epalle, T., Girardot, F., Allegra, S., Maurice-Blanc, C., Garraud, O., and Riffard, S. (2015). Viable but not culturable forms of Legionella pneumophila generated after heat shock treatment are infectious for macrophage-like and alveolar epithelial cells after resuscitation on Acanthamoeba polyphaga. Microb. Ecol. 69, 215–224. doi: 10.1007/s00248-014-0470-x
Falkinham, J. O., Pruden, A., and Edwards, M. (2015). Opportunistic premise plumbing pathogens: increasingly important pathogens in drinking water. Pathogens 4, 373–386. doi: 10.3390/pathogens4020373
Farris, M., Grant, A., Richardson, T. B., and O'Connor, C. D. (1998). BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol. Microbiol. 28, 265–279. doi: 10.1046/j.1365-2958.1998.00793.x
Faucher, S. P., Mueller, C. A., and Shuman, H. A. (2011). Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front. Microbiol. 2, 60. doi: 10.3389/fmicb.2011.00060
Faucher, S. P., and Shuman, H. A. (2013). Methods to study legionella transcriptome in vitro and in vivo. Methods Mol. Biol. 954, 567–582. doi: 10.1007/978-1-62703-161-5_35
Fauvet, B., Finka, A., Castanié-Cornet, M. P., Cirinesi, A. M., Genevaux, P., Quadroni, M., et al. (2021). Bacterial Hsp90 facilitates the degradation of aggregation-prone Hsp70–Hsp40 substrates. Front. Mol. Biosci. 8, 225. doi: 10.3389/fmolb.2021.653073
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282. doi: 10.1146/annurev.physiol.61.1.243
Garduño, R. A., Chong, A., Nasrallah, G. K., and Allan, D. S. (2011). The Legionella pneumophila chaperonin - an unusual multifunctional protein in unusual locations. Front. Microbiol. 2, 122. doi: 10.3389/fmicb.2011.00122
Genest, O., Hoskins, J. R., Camberg, J. L., Doyle, S. M., and Wickner, S. (2011). Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc. Natl. Acad. Sci. U.S.A. 108, 8206–8211. doi: 10.1073/pnas.1104703108
Genest, O., Hoskins, J. R., Kravats, A. N., Doyle, S. M., and Wickner, S. (2015). Hsp70 and Hsp90 of E. coli directly interact for collaboration in protein remodeling. J. Mol. Biol. 427, 3877–3889. doi: 10.1016/j.jmb.2015.10.010
Gibbs, M. R., Moon, K. M., Warner, B. R., Chen, M., Bundschuh, R., Foster, L. J., et al. (2020). Functional analysis of BipA in E. coli reveals the natural plasticity of 50s subunit assembly. J. Mol. Biol. 432, 5259–5272. doi: 10.1016/j.jmb.2020.07.013
Goh, K. J., Ero, R., Yan, X. F., Park, J. E., Kundukad, B., Zheng, J., et al. (2021). Translational GTPase BipA is involved in the maturation of a large subunit of bacterial ribosome at suboptimal temperature. Front. Microbiol. 12, 686049. doi: 10.3389/fmicb.2021.686049
Gomez-Valero, L., Rusniok, C., Carson, D., Mondino, S., Pérez-Cobas, A. E., Rolando, M., et al. (2019). More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc. Natl. Acad. Sci. U.S.A. 116, 2265–2273. doi: 10.1073/pnas.1808016116
Gomez-Valero, L., Rusniok, C., Rolando, M., Neou, M., Dervins-Ravault, D., Demirtas, J., et al. (2014). Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires' disease. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0505-0
Gruber, T. M., and Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466. doi: 10.1146/annurev.micro.57.030502.090913
Grudniak, A. M., Klecha, B., and Wolska, K. I. (2018). Effects of null mutation of the heat-shock gene htpG on the production of virulence factors by Pseudomonas aeruginosa. Future Microbiol. 13, 69–80. doi: 10.2217/fmb-2017-0111
Guisbert, E., Yura, T., Rhodius, V. A., and Gross, C. A. (2008). Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol. Mol. Biol. Rev. 72, 545–554. doi: 10.1128/mmbr.00007-08
Guyot, S., Pottier, L., Ferret, E., Gal, L., and Gervais, P. (2010). Physiological responses of Escherichia coli exposed to different heat-stress kinetics. Arch. Microbiol. 192, 651–661. doi: 10.1007/s00203-010-0597-1
Hales, L. M., and Shuman, H. A. (1999). The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181, 4879–4889. doi: 10.1128/jb.181.16.4879-4889.1999
Hengge-Aronis, R., Klein, W., Lange, R., Rimmele, M., and Boos, W. (1991). Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173, 7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991
Herwaldt, L. A., and Marra, A. R. (2018). Legionella: a reemerging pathogen. Curr. Opin. Infect. Dis. 31, 325–333. doi: 10.1097/QCO.0000000000000468
Hoffman, P. S., Butler, C. A., and Quinn, F. D. (1989). Cloning and temperature-dependent expression in Escherichia coli of a Legionella pneumophila gene coding for a genus-common 60-kilodalton antigen. Infect. Immun. 57, 1731–1739. doi: 10.1128/iai.57.6.1731-1739.1989
Hoffman, P. S., Houston, L., Butler, C. A., and Butler,', C. A. (1990). Legionella pneumophila htpAB heat shock operon: nucleotide sequence and expression of the 60-kilodalton antigen in L. pneumophila-infected HeLa cells. Infect. Immun. 58, 3380–3387. doi: 10.1128/iai.58.10.3380-3387.1990
Honoré, F. A., Méjean, V., and Genest, O. (2017). Hsp90 is essential under heat stress in the bacterium Shewanella oneidensis. Cell Rep. 19, 680–687. doi: 10.1016/j.celrep.2017.03.082
Hovel-Miner, G., Pampou, S., Faucher, S. P., Clarke, M., Morozova, I., Morozov, P., et al. (2009). σ S controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J. Bacteriol. 191, 2461–2473. doi: 10.1128/JB.01578-08
Hughes, E. D., Byrne, B. G., and Swanson, M. S. (2019). A two-component system that modulates cyclic di-gmp metabolism promotes Legionella pneumophila differentiation and viability in low-nutrient conditions. J. Bacteriol. 201, 1–19. doi: 10.1128/JB.00253-19
Imlay, J. A (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454. doi: 10.1038/nrmicro3032
Jenkins, D. E., Auger, E. A., and Matin, A. (1991). Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival. J. Bacteriol. 173, 1992–1996. doi: 10.1128/jb.173.6.1992-1996.1991
Ji, P., Rhoads, W. J., Edwards, M. A., and Pruden, A. (2017). Impact of water heater temperature setting and water use frequency on the building plumbing microbiome. ISME J. 11, 1318–1330. doi: 10.1038/ismej.2017.14
Johnson, J. L (2012). Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta Mol. Cell Res. 1823, 607–613. doi: 10.1016/j.bbamcr.2011.09.020
Kirschner, A. K. T (2016). Determination of viable legionellae in engineered water systems: do we find what we are looking for? Water Res. 93, 276–288. doi: 10.1016/j.watres.2016.02.016
Kool, J. L., Bergmire-Sweat, D., Butler, J. C., Brown, E. W., Peabody, D. J., Massi, D. S., et al. (1999). Hospital characteristics associated with colonization of water systems by Legionella and risk of nosocomial Legionnaires' disease: a cohort study of 15 hospitals. Infect. Control Hosp. Epidemiol. 20, 798–805. doi: 10.1086/501587
Krishnan, K., and Flower, A. M. (2008). Suppression of ΔbipA phenotypes in Escherichia coli by abolishment of pseudouridylation at specific sites on the 23S rRNA. J. Bacteriol. 190, 7675–7683. doi: 10.1128/JB.00835-08
Lema, M. W., Brown, A., Butler, C. A., and Hoffman, P. S. (1988). Heat-shock response in Legionella pneumophila. Can. J. Microbiol. 34, 1148–1153. doi: 10.1139/m88-202
Lesnik, R., Brettar, I., and Höfle, M. G. (2016). Legionella species diversity and dynamics from surface reservoir to tap water: from cold adaptation to thermophily. ISME J. 10, 1064–1080. doi: 10.1038/ismej.2015.199
Lévesque, S., Plante, P.-L., Mendis, N., Cantin, P., Marchand, G., Charest, H., et al. (2014). Genomic characterization of a large outbreak of Legionella pneumophila serogroup 1 strains in Quebec City, 2012. PLoS ONE 9, e103852. doi: 10.1371/journal.pone.0103852
Li, H., and Gänzle, M. (2016). Some like it hot: heat resistance of Escherichia coli in food. Front. Microbiol. 7, 1763. doi: 10.3389/fmicb.2016.01763
Li, L., Mendis, N., Trigui, H., and Faucher, S. P. (2015). Transcriptomic changes of Legionella pneumophila in water. BMC Genom. 16, 637. doi: 10.1186/s12864-015-1869-6
Mackey, B. M., Miles, C. A., Parsons, S. E., and Seymour, D. A. (1991). Thermal denaturation of whole cells and cell components of Escherichia coli examined by differential scanning calorimetry. J. Gen. Microbiol. 137, 2361–2374. doi: 10.1099/00221287-137-10-2361/CITE/REFWORKS
McDade, J. E., Shepard, C. C., Fraser, D. W., Tsai, T. R., Redus, M. A., and Dowdle, W. R. (1977). Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297, 1197–1203. doi: 10.1056/NEJM197712012972202
Mendis, N., McBride, P., Saoud, J., Mani, T., and Faucher, S. P. (2018). The LetA/S two-component system regulates transcriptomic changes that are essential for the culturability of Legionella pneumophila in water. Sci. Rep. 8, 6764. doi: 10.1038/s41598-018-24263-9
Mercante, J. W., Morrison, S. S., Desai, H. P., Raphael, B. H., and Winchell, J. M. (2016). Genomic analysis reveals novel diversity among the 1976 Philadelphia Legionnaires' disease outbreak isolates and additional ST36 strains. PLoS ONE 11, e164074. doi: 10.1371/journal.pone.0164074
Meyer, A. S., and Baker, T. A. (2011). Proteolysis in the Escherichia coli heat shock response: a player at many levels. Curr. Opin. Microbiol. 14, 194–199. doi: 10.1016/j.mib.2011.02.001
Murata, M., Fujimoto, H., Nishimura, K., Charoensuk, K., Nagamitsu, H., Raina, S., et al. (2011). Molecular strategy for survival at a critical high temperature in eschierichia coli. PLoS ONE 6, e20063. doi: 10.1371/journal.pone.0020063
Murata, M., Ishii, A., Fujimoto, H., Nishimura, K., Kosaka, T., Mori, H., et al. (2018). Update of thermotolerant genes essential for survival at a critical high temperature in Escherichia coli. PLoS ONE 13, e0189487. doi: 10.1371/journal.pone.0189487
Nisar, M. A., Ross, K. E., Brown, M. H., Bentham, R., and Whiley, H. (2020). Legionella pneumophila and protozoan hosts: implications for the control of hospital and potable water systems. Pathogens 9, 286. doi: 10.3390/pathogens9040286
Padrnos, L. J., Blair, J. E., Kusne, S., Dicaudo, D. J., and Mikhael, J. R. (2014). Cutaneous legionellosis: case report and review of the medical literature. Transpl. Infect. Dis. 16, 307–314. doi: 10.1111/tid.12201
Peil, L., Virumäe, K., and Remme, J. (2008). Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 275, 3772–3782. doi: 10.1111/j.1742-4658.2008.06523.x
Pfennig, P. L., and Flower, A. M. (2001). BipA is required for growth of Escherichia coli k12 at low temperature. Mol. Genet. Genomics 266, 313–317. doi: 10.1007/s004380100559
Phin, N., Parry-Ford, F., Harrison, T., Stagg, H. R., Zhang, N., Kumar, K., et al. (2014). Epidemiology and clinical management of Legionnaires' disease. Lancet Infect. Dis. 14, 1011–1021. doi: 10.1016/S1473-3099(14)70713-3
Phipson, B., Lee, S., Majewski, I. J., Alexander, W. S., and Smyth, G. K. (2016). Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 10, 946–963. doi: 10.1214/16-AOAS920
Piao, Z., Sze, C. C., Barysheva, O., Iida, K. I., and Yoshida, S. I. (2006). Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila. Appl. Environ. Microbiol. 72, 1613–1622. doi: 10.1128/AEM.72.2.1613-1622.2006
Principe, L., Tomao, P., and Visca, P. (2017). Legionellosis in the occupational setting. Environ. Res. 152, 485–495. doi: 10.1016/j.envres.2016.09.018
Rao, C., Benhabib, H., and Ensminger, A. W. (2013). Phylogenetic reconstruction of the Legionella pneumophila Philadelphia-1 laboratory strains through comparative genomics. PLoS ONE 8, e64129. doi: 10.1371/journal.pone.0064129
René, O., and Alix, J. H. (2011). Late steps of ribosome assembly in E. coli are sensitive to a severe heat stress but are assisted by the HSP70 chaperone machine. Nucleic Acids Res. 39, 1855–1867. doi: 10.1093/nar/gkq1049
Rhoads, W. J., Ji, P., Pruden, A., and Edwards, M. A. (2015). Water heater temperature set point and water use patterns influence Legionella pneumophila and associated microorganisms at the tap. Microbiome 3, 67. doi: 10.1186/s40168-015-0134-1
Rhoads, W. J., Pruden, A., and Edwards, M. A. (2016). Convective mixing in distal pipes exacerbates Legionella pneumophila growth in hot water plumbing. Pathogens 5, 29. doi: 10.3390/pathogens5010029
Richter, K., Haslbeck, M., and Buchner, J. (2010). The heat shock response: life on the verge of death. Mol. Cell 40, 253–266. doi: 10.1016/j.molcel.2010.10.006
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. doi: 10.1093/nar/gkv007
Sadosky, A. B., Wiater, L. A., and Shuman, H. A. (1993). Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61, 5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993
Sahr, T., Rusniok, C., Impens, F., Oliva, G., Sismeiro, O., Coppée, J. Y., et al. (2017). The Legionella pneumophila genome evolved to accommodate multiple regulatory mechanisms controlled by the CsrA-system. PLoS Genet. 13, 1–38. doi: 10.1371/journal.pgen.1006629
Schrammel, B., Cervero-Aragó, S., Dietersdorfer, E., Walochnik, J., Lück, C., Sommer, R., et al. (2018). Differential development of Legionella sub-populations during short- and long-term starvation. Water Res. 141, 417–427. doi: 10.1016/j.watres.2018.04.027
Soda, E. A., Barskey, A. E., Shah, P. P., Schrag, S., Whitney, C. G., Arduino, M. J., et al. (2017). Vital signs: health care–associated Legionnaires' disease surveillance data from 20 states and a large metropolitan area—United States, 2015. Am. J. Transplant. 17, 2215–2220. doi: 10.1111/ajt.14407
Sørensen, J. G., Kristensen, T. N., and Loeschcke, V. (2003). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6, 1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x
Spence, J., Cegielska, A., and Georgopoulos, C. (1990). Role of Escherichia coli heat shock proteins DnaK and HtpG (C62.5) in response to nutritional deprivation. J. Bacteriol. 172, 7157–7166. doi: 10.1128/jb.172.12.7157-7166.1990
Springston, J. P., and Yocavitch, L. (2017). Existence and control of Legionella bacteria in building water systems: a review. J. Occup. Environ. Hyg. 14, 124–134. doi: 10.1080/15459624.2016.1229481
Suh, S. J., Silo-Suh, L., Woods, D. E., Hassett, D. J., West, S. E. H., and Ohman, D. E. (1999). Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181, 3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999
Talaat, A. M., Howard, S. T., Hale, W., Lyons, R., Garner, H., and Johnston, S. A. (2002). Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30, e104. doi: 10.1093/nar/gnf103
Trigui, H., Dudyk, P., Oh, J., Hong, J. I., and Faucher, S. P. (2015). A regulatory feedback loop between RpoS and SpoT supports the survival of Legionella pneumophila in water. Appl. Environ. Microbiol. 81, 918–928. doi: 10.1128/AEM.03132-14
Van Kenhove, E., Dinne, K., Janssens, A., and Laverge, J. (2019). Overview and comparison of Legionella regulations worldwide. Am. J. Infect. Control 47, 968–978. doi: 10.1016/j.ajic.2018.10.006
Wang, W., Li, W., Ge, X., Yan, K., Mandava, C. S., Sanyal, S., et al. (2020). Loss of a single methylation in 23s rrna delays 50s assembly at multiple late stages and impairs translation initiation and elongation. Proc. Natl. Acad. Sci. U.S.A. 117, 15609–15619. doi: 10.1073/pnas.1914323117
Whiley, H., Bentham, R., and Brown, M. H. (2017). Legionella persistence in manufactured water systems: pasteurization potentially selecting for thermal tolerance. Front. Microbiol. 8, 1330. doi: 10.3389/fmicb.2017.01330
Keywords: Legionella pneumophila, heat shock response, transcriptome, water distribution systems, pasteurization, water-borne infection, disinfection
Citation: Liang J and Faucher SP (2022) Transcriptomic Adaptation of Legionella pneumophila to Transient Heat Shock. Front. Water 4:891477. doi: 10.3389/frwa.2022.891477
Received: 07 March 2022; Accepted: 09 May 2022;
Published: 31 May 2022.
Edited by:
William Rhoads, Swiss Federal Institute of Aquatic Science and Technology, SwitzerlandReviewed by:
Connor Brown, Virginia Tech, United StatesNicholas Cianciotto, Northwestern University, United States
Michael Waak, SINTEF, Norway
Copyright © 2022 Liang and Faucher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastien P. Faucher, c2ViYXN0aWVuLmZhdWNoZXIyQG1jZ2lsbC5jYQ==
 Jeffrey Liang
Jeffrey Liang Sebastien P. Faucher
Sebastien P. Faucher