- 1International Joint Research Center for Animal Health Breeding, College of Animal Science and Technology, Shihezi University, Shihezi, China
- 2Collaborative Innovation Center for Prevention and Control of High Incidence Zoonotic Infectious Diseases in Western China, College of Animal Science and Technology, Shihezi University, Shihezi, China
- 3College of Life Sciences, Shihezi University, Shihezi, China
Dairy cow mastitis is a serious disease that is mainly caused by intramammary infection with Staphylococcus aureus and Streptococcus agalactiae [group B streptococcus (GBS)]. FnBP and ClfA are the virulence factors of S. aureus, while GapC is the respective factor for S. agalactiae. Sip is a highly immunogenic protein, and it is conserved in all GBS serotypes. In this study, we analyzed the abovementioned four genes prepared a FnBP+ClfA chimeric protein (FC), a GapC+Sip chimeric protein (GS), and a FnBP+ClfA+GapC+Sip chimeric protein (FCGS) based on the antigenic sites to evaluate their use in vaccine development. After expression and purification of the recombinant proteins in Escherichia coli, BALB/c mice were immunized with them to examine resistance effects. The total lethal and half lethal doses of S. aureus and S. agalactiae were then measured, and the immunoprotective effects of the fusion proteins were evaluated. The FC and FCGS chimeric proteins could induce mice to produce high levels of antibodies, and bacterial loads were significantly reduced in the spleens and livers after challenge. After immunization with FCGS, the recipients resisted the attacks of both S. aureus and S. agalactiae, indicating the potential of the fusion protein as a mastitis vaccine.
Introduction
Mastitis is a disease of dairy cows that causes significant economic losses in the dairy farming industry (1). Pathogenic microbial infection as well as physical and chemical damage can lead to dairy cow mastitis, although infection by pathogenic microorganisms is the main cause (2). Pathogens associated with mastitis include Staphylococcus aureus and Streptococcus agalactiae that both cause intramammary infections (IMIs) and thereby increase the difficulty of treating cow mastitis (3, 4).
Many studies have shown that S. aureus and S. agalactiae are the most common pathogens causing chronic, infectious, and refractory bovine mastitis (5, 6). Further, the infection prevalence of S. aureus in Chinese dairy herds is 29% (7), which is significantly higher than in western country herds (8). There is currently no effective vaccine for dairy cow mastitis, and the treatment of dairy cow mastitis is becoming increasingly more difficult with the emergence of bacterial resistance to treatment (9).
Bacterial adhesins including fibronectin-binding protein (FnBP) and aggregation factor A (ClfA) play key roles in pathogenic invasion (10). FnBP and ClfA are the two surface adhesins of S. aureus. ClfA binds to complement factor I and is an important factor that promotes evasion of neutrophil-related death (11). FnBP and elastin (12) mainly bind to plasminogen (13) and promote biofilm formation. Previous studies have shown that antibodies induced by FnBPs and ClfA can partially block the adhesion of S. aureus to breast tissue (14, 15). In addition, the mixed bivalent nucleic acid vaccine for S. aureus that targets FnBPs and ClfA (16) can effectively stimulate specific immune responses in dairy cows and the immunoprotective effects of an FnBP+fusion protein has been confirmed in mice (17).
Surface immunogenic protein (Sip) is an important adhesion factor on the surface of S. agalactiae. It is a highly conserved protein that is expressed in all S. agalactiae serotypes, and the protein can induce cross-immunity protection (18). In addition, GapC protein is a streptococcal surface dehydrogenase (SDH) with glyceraldehyde 3-phosphate dehydrogenase (DAPDH) activity (19). GapC exhibits considerable homology at the DNA and amino acid levels to homologs among different S. agalactiae strains (20). The protein is known to play an important role in the pathogenesis of dairy cow mastitis, and the use of GapC protein to immunize dairy cows yields significant immune protection (21). Consequently, GapC protein is an important target for cow mastitis vaccines (21, 22). However, the joint use of FnBP and ClfA from S. aureus in addition to GapC and Sip of S. agalactiae has rarely been studied, and no safe and effective mastitis vaccine has been reported.
Consequently, the goal of this study was to combine FnBP and ClfA of S. aureus with GapC and Sip of S. agalactiae to develop candidate recombinant proteins with activity against S. aureus and S. agalactiae infection. The recombinant proteins could then be used to further develop a cow mastitis vaccine. Specifically, the fusion proteins FnBP+ClfA (FC), GapC+Sip (GS), and FnBP+ClfA+GapC+Sip (FCGS) were constructed; and the effects of the fusion proteins on S. aureus and S. agalactiae infections were analyzed in mice. The results demonstrated that FC, GS, and FCGS can be used as potential vaccine proteins and are important targets for further research.
Materials and Methods
Ethical Approval
All animals were treated humanely and in accordance with institutional animal care guidelines. This study was approved by the Animal Care and Use Committee of Shihezi University.
Strains and Animals
Standard S. aureus [American Type Culture Collection (ATCC) 25923] and S. agalactiae (ATCC 13813) strains were provided by the ATCC and cultured in brain heart infusion (BHI) broth/agar (Hopebio, China) at 37°C. In addition, Escherichia coli strains DH5α (Sigma-Aldrich Corp., St. Louis, MO, USA) and C43 (DE3, Sigma, USA) were cultured in Luria–Bertani medium (Difco, Becton Dickinson, Franklin Lakes, New Jersey, USA). Five-week-old female BALB/c mice were purchased from the Experimental Animal Center of the Academy of Military Medical Science (Beijing, China). All experimental procedures and animal care protocols were performed in compliance with institutional animal care regulations.
Acquisition of FC, GS, and FCGS Gene Sequences and Bioinformatics Analysis
FnBP (gene ID: DQ447162) and ClfA (gene ID: EF207779) gene sequences from S. aureus in addition to GapC (gene ID: af421899) and Sip (gene ID: fj808732) gene sequences of S. agalactiae were retrieved from the GenBank database. The Kolaskar and Tongaonkar methods were used to predict and analyze the B cell epitopes of FnBP, ClfA, GapC, and Sip gene sequences (http://imed.med.ucm.es/Tools/antigenic.pl) (23). The SignalP 5.0 Server (http://www.cbs.dtu.dk/services/SignalP/) (24, 25) was used to predict the presence of signal peptides in the proteins; and fragments with excellent immunogenicity were selected and divided into FC, GS, and FCGS combination groups by adding a linker sequence (-GGGGSGGGGSGGGGS-) to tightly combine FC, GS, and FCGS. After the target genes were optimized, we commissioned GENERAL BIOL (Anhui, China) to synthesize the sequences and constructed each tandem sequence (FC, GS, and FCGS) by ligation into the vector pET28a/pET32a (EMD Biosciences, Novagen, San Diego, CA, USA). The Phyre2 software program (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) was used to predict the tertiary structures of the recombinant fusion proteins (26).
Expression and Identification of FC, GS, and FCGS Proteins
Isopropyl β-D-1-thiogalactopyranoside (IPTG; Solarbio, Beijing, China) was used to induce recombinant expression in the strains pET32a-FC-DE3, pET28a-GS-DE3, and pET32a-FCGS-DE3. After cells were collected, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was used to analyze the soluble expression of the target proteins. Target protein purification was performed with a His-Tagged Protein Purification Kit using an inclusion body protein (CWBIO, Beijing, China).
Target protein concentrations were quantified with a Micro BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), and Western blotting analysis was used to verify the reactogenicity of the purified proteins. The primary antibody used positive S. aureus/S. agalactiae antibody bovine serum (1:100) that was provided by International Joint Research Center for Animal Health Breeding (Shihezi University) (unpublished). The secondary antibody comprised rabbit anti-bovine IgG/horseradish peroxidase (HRP) (1:2,000; Solarbio, China). A SuperSignal West Femto Trial kit (Thermo, USA) was then used to develop color.
Determination of Lethal Bacterial Dosages in BALB/c Mice
The total lethal levels of S. aureus and S. agalactiae toward BALB/c mice were first determined. S. aureus was cultivated to the order of 2.1 × 1010 CFU/ml. BALB/c mice were divided into six groups that were intraperitoneally inoculated with 5 μl (1.05 × 108 CFU/mouse), 10 μl (2.1 × 108 CFU/mouse), 15 μl (3.15 × 108 CFU/mouse), 20 μl (4.2 × 108 CFU/mouse), 200 μl (4.2 × 109 CFU/mouse), and 400 μl (8.4 × 109 CFU/mouse) of culture. S. agalactiae was also cultivated to 3.3 × 1010 CFU/ml, and BALB/c mice were divided into six groups that were intraperitoneally injected with 50 μl (1.65 × 109 CFU/mouse), 100 μl (3.3 × 109 CFU/mouse), 150 μl (4.95 × 109 CFU/mouse), 200 μl (6.6 × 109 CFU/mouse), and 400 μl (1.32 × 1010 CFU/mouse) of S. agalactiae culture. Six mice were immunized with each dose, and the clinical manifestations and time of death for mice were observed and recorded for seven consecutive days.
The median lethal doses of S. aureus and S. agalactiae toward BALB/c mice were then determined. In these experiments, BALB/c mice were divided into six groups, and each group comprised six mice that were intraperitoneally injected with 16 μl (3.36 × 108 CFU/mouse), 18 μl (3.9 × 108 CFU/mouse), and 20 μl (4.2 × 108 CFU/mouse) of S. aureus culture. In addition, BALB/c mice were divided into six groups, and six mice in each group were intraperitoneally injected with 160 μl (5.28 × 109 CFU/mouse), 180 μl (6.0 × 109 CFU/mouse), and 200 μl (6.6 × 109 CFU/mouse) of S. agalactiae culture. The clinical manifestations and time of death for mice within each group were observed and recorded over 1 week.
Immunization Program of BALB/c Mice
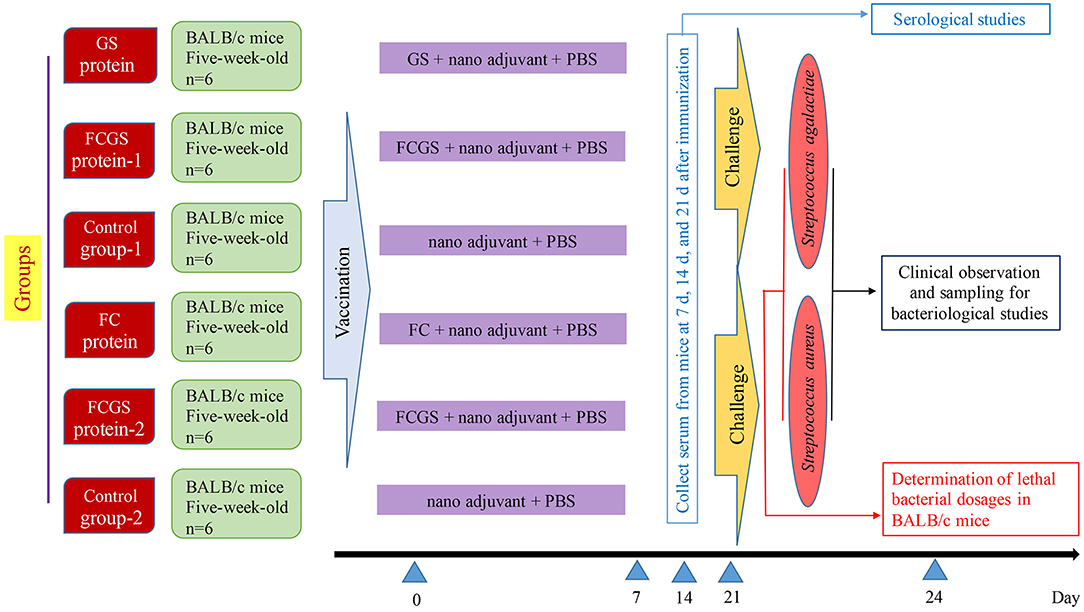
The final concentrations of FC, GS, and FCGS proteins were 4.5, 0.13, and 0.3 mg/ml. The proteins were diluted in PBS and thoroughly mixed with a nano-adjuvant in a 1:1 ratio to obtain FC, GS, and FCGS nano-adjuvant vaccines. We previously screened nano-adjuvants to identify immunological effects of the nano-adjuvants compared with ordinary adjuvants (27). BALB/c mice were randomly divided into six groups, with six mice in each group. The FC, GS, and FCGS nano-adjuvant vaccines were subcutaneously injected (300-μl injections with 40 μg of immunogen per mouse) to immunize mice. PBS and an equal volume of nano-adjuvant were emulsified and injected into mice as a control (Figure 1).
Indirect ELISA to Detect Antibody Levels
Orbital blood sampling was used to collect serum from mice at 7, 14, and 21 days after immunization. Indirect ELISA was then used to detect IgG antibody levels in mouse sera. ELISA coating solution (Solarbio, China) was used to dilute the antigen to a working concentration; and FC (4.5 ng/ml), GS (2.6 ng/ml), and FCGS (3.0 ng/ml) were added to 96-well plates and kept overnight at 4°C. Each well was washed three times with PBST, 5% skim milk was added, and the plates incubated at 37°C for 2 h. The liquid in each well was then discarded, wells were washed with PBST, and mouse serum (1:2,000) was added and again incubated at 37°C for 1 h. After being washed with PBST, rabbit anti-mouse IgG H&L (HRP) (1:5,000) (Abcam, Cambridge, UK) was added to each well and incubated at 37°C for 1 h. After being washed with PBST, a one-component TMB substrate color developing solution (Solarbio, China) was added, and plates were maintained at room temperature in the dark for 15 min. An ELISA stop solution (Solarbio, China) was then added to each well, and the absorbance at 450 nm (OD value) was measured with a microplate reader within 5 min.
Challenge Protection Test and Determination of Organ Load
At 21-day post-immunization, S. aureus and S. agalactiae were cultured to appropriate concentrations, and the mice were intraperitoneally injected with the half-lethal dose. After the challenge, the appearance and mental condition of mice in each group were evaluated, and mouse deaths were recorded.
At 72 h post-bacterial challenge, mice were sacrificed using CO2, immersed in 75% ethanol, and dissected under aseptic conditions. Livers and spleens were collected, and the organs were homogenized after adding PBS. Tissue homogenates were diluted to 10−1, 10−2, and 10−3. The homogenates were then spread on BHI Agar (Hopebio, China) and incubated at 37°C for 24–48 h, followed by colony enumeration. Tissue loads were calculated as CFU/g = average number of CFU in the plate × 5 × volume of homogenate (ml) × dilution factor/tissue weight (g). The design and schedule of the study are shown in Figure 1.
Data Analysis
Statistical significance in differences and correlation coefficients were calculated with SPSS Statistics 23 program. Student's t-tests, Student–Newman–Keuls (SNK) tests, and one-way ANOVAs were used to compare measurements among groups. All data are presented as means ± SEM, and data represent the results of three independent experiments. The GraphPad Prism software was used to construct figures.
Results
Sequence Selection and Bioinformatics Analysis
Epitope prediction was used as the basis for evaluating protein immunogenicity; and the epitopes of FnBP, ClfA, Sip, and GapC proteins served as the foundation for constructing and expression fusion proteins. The FnBP, ClfA, and GapC did not have signal peptide, while Sip did (20–40 amino acids in length) (Supplementary Figure 1). FnBP had three potential epitopes (propensity index = 0.9725), ClfA had 19 potential epitopes (propensity index = 0.9986), Sip had 19 potential epitopes (propensity index = 1.0315), and GapC had 17 potential epitopes (propensity index = 1.0260) (Supplementary Figure 2). Thus, the four proteins exhibited good immunogenicity. The tertiary structures of recombinant FC, GS, and FCGS were also predicting using Phyre2 (Supplementary Figure 3).
FC, GS, and FCGS Protein Acquisition
The fusion protein of S. aureus comprised residues 23–115 of FnBP and 159–282 of ClfA (Figure 2A), while the fusion protein of S. agalactiae comprised residues 183–312 of GapC and 162–338 of Sip (Figure 2B). The fusion protein of S. aureus and S. agalactiae comprised residues 23–115 of FnBP, 356–472 of ClfA, 88–162 of GapC, and 4–97 of Sip (Figure 2C). The amino acid sequences of FC, GS, and FCGS are shown in Supplementary Figure 4.
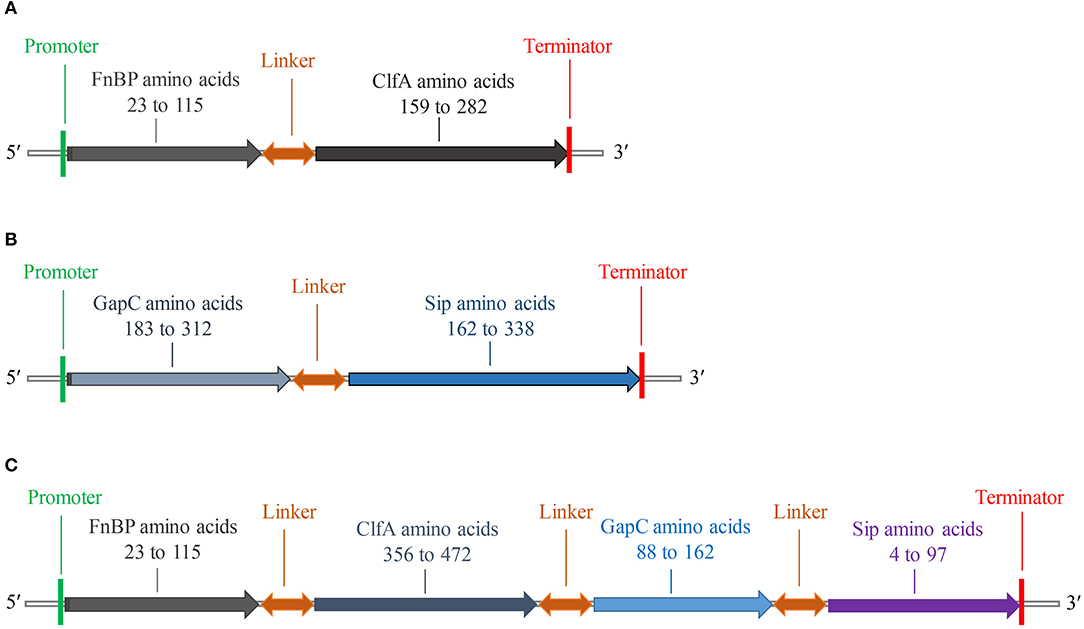
Figure 2. Schematic diagram of FC, GS, and FCGS recombinant proteins. (A) Staphylococcus aureus FnBP and ClfA (FC) recombinant protein structures. (B) Streptococcus agalactiae GapC and Sip (GS) recombinant protein structures. (C) S. aureus FnBP and ClfA, and S. agalactiae GapC and Sip (FCGS) recombinant protein structures. Linker sequences (-GGGGSGGGGSGGGGS-) were used as adapters.
The recombinant plasmids of each expression vector were transferred into E. coli DE3 competent cells, resulting in the expression of the FC (pET32a-FC) (Figure 3A), GS (pET28a-GS) (Figure 3B), and FCGS (pET32a-FCGS) fusion proteins (Figure 3C). SDS-PAGE analysis indicated that the FCGS, FC, and GS recombinant proteins were all expressed in bacterial inclusion bodies (Figures 3A–C). Post-purification, the purity of FC, GS, and FCGS was >90%, >85%, and >85%, respectively (Supplementary Figure 5). Western blotting analysis confirmed that the target proteins reacted with the positive sera of S. aureus and S. agalactiae and had good reactogenicity (Figure 3D). A semi-quantitative analysis of Western blotting bands was also conducted (Figure 3E).
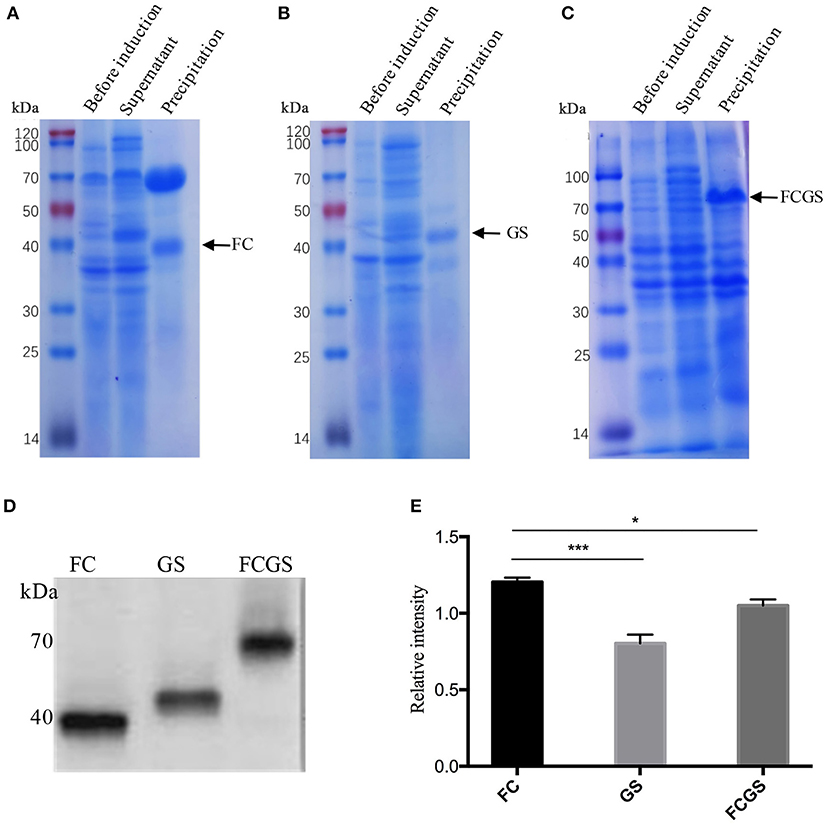
Figure 3. FC, GS, and FCGS recombinant protein identification and characterization. (A) Expression of FC recombinant fusion protein in Escherichia coli. (B) Expression of GS recombinant fusion protein in E. coli. (C) Expression of FCGS recombinant fusion protein in E. coli. IPTG was used to induce recombinant expression of pET32a-FC-DE3, pET28a-GS-DE3, and pET32a-FCGS-DE3 for 6 h; cells were collected by centrifugation and then subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis. (D) Western blotting identification of purified FC, GS, and FCGS recombinant proteins. (E) Relative intensities of FC, GS, and FCGS were semi-quantified using ImageJ software program. Data show means ± SEM, *p < 0.05 and ***p < 0.001. Results were obtained from three independent replicate experiments.
Determination of Lethal Doses in BALB/c Mice
BALB/c mice displayed clinical symptoms within 12 h after intraperitoneal injection of S. aureus and S. agalactiae. Initial manifestations included a lack of energy, slower movements, reduced eating, reduced drinking, curling up in a corner, and rapid breathing. The mice did not die after 72 h (Figure 4). The total lethal doses were 4.2 × 108 CFU/mouse for S. aureus and 6.6 × 109 CFU/mouse for S. agalactiae (Figures 4A–D). For the LD50 tests, mice showed clinical manifestations similar to total lethal doses, but the symptoms were alleviated, and they were able to crawl slightly. After 72 h, LD50 values were 3.9 × 108 CFU for S. aureus and 6.0 × 109 CFU for S. agalactiae (Figures 4E,F).
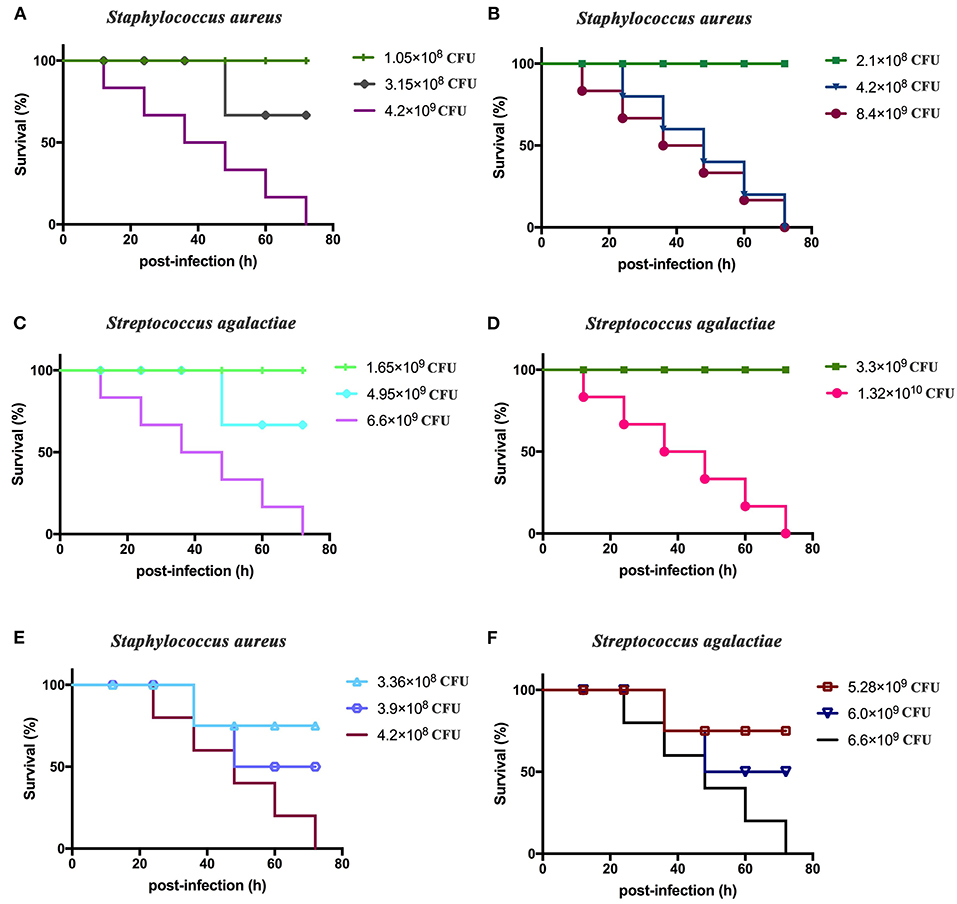
Figure 4. Staphylococcus aureus and Streptococcus agalactiae total and half lethal doses in BALB/c mouse challenges. After S. aureus and S. agalactiae cells were collected by centrifugation, BALB/c mice were immunized based on CFU/mouse dosages in the analyses; and mouse survival was recorded over 72 h. (A,B) The total lethal dose of S. aureus to BALB/c mice. (C,D) The total lethal dose of S. agalactiae to BALB/c mice. (E) The half lethal dose of S. aureus to BALBC/c mice; (F) the half lethal dose of S. agalactiae to BALB/c mice. The results represent three independent replicate experiments.
Levels of Antibodies Induced by FC, GS, and FCGS Recombinant Proteins
The procedure shown in Figure 1 was followed for the sequential study. FC, GS, and FCGS fusion proteins were used to coat the ELISA plates to detect the production of antibodies in mice immunized with the three proteins. On the 14th and 21st days post-immunization, antibody levels in mice induced by the FC recombinant protein were significantly higher than in the control group (p < 0.001) (Figure 5A). It is worth noting that the FC fusion protein rapidly induced antibody production in mice on the seventh day (p < 0.05) (Figure 5A). The FCGS fusion protein-1 group produced significantly higher antibody levels, higher than those of the control group on the 14th day after immunization (p < 0.01), and the difference was extremely significant on the 21st day (Figure 5B). The GS fusion protein induced the mice to produce higher antibody levels than the control group on the 21st day after immunization, but the difference was not significant (Figure 5C). In addition, compared with the control group on the 14th and 21st days after immunization, the FCGS protein-2 group had significant difference (p < 0.01) (Figure 5D). Thus, the antibody levels induced by the FCGS fusion protein were higher than those due to the FC and GS recombinant proteins.
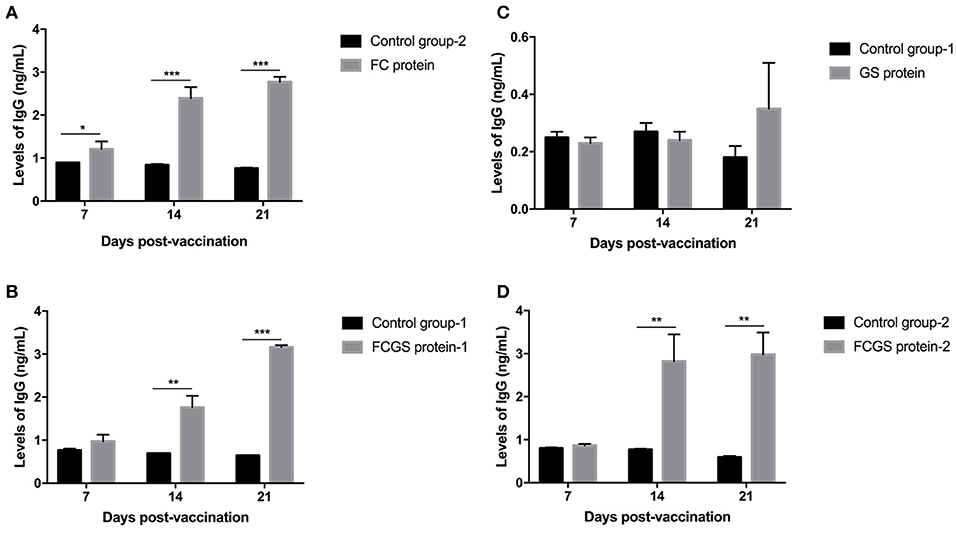
Figure 5. Levels of IgG antibodies induced by FC, GS, and FCGS recombinant proteins in BALB/c mice. Mice were immunized with protein and adjuvants, followed by collection of mouse sera at 7, 14, and 21 days post-immunization to detect IgG content with ELISA. (A) The specific IgG levels in the serum of mice at different time periods after FC immunization. (B,D) The specific IgG levels in the serum of mice at different time periods after FCGS immunization. (C) The specific IgG levels in the serum of mice at different time periods after GS immunization. Data are means ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001). The results were obtained from three independent replicate experiments.
Analysis of the Protective Effects of FC, GS, and FCGS Recombinant Proteins
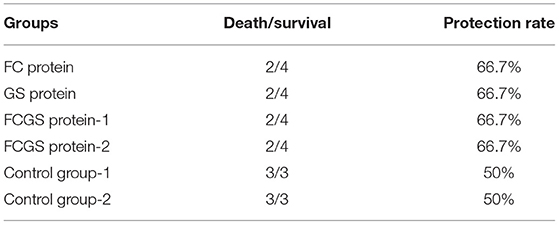
Standard S. aureus and S. agalactiae strains were used to challenge mice 21 days after immunization. The FC protein conferred good resistance to S. aureus, and the GS protein was effective against S. agalactiae. Significant resistance was also conferred by FCGS protein to S. agalactiae and S. aureus (Table 1).
At 72 h post-bacterial challenge, the bacterial loads in the spleens and livers were enumerated. Mice immunized with the FCGS and GS recombinant proteins withstood challenges with S. agalactiae, as indicated by fewer bacteria isolates compared with the control group (Figures 6A,B). The FCGS and FC recombinant proteins also conferred resistance to S. aureus after immunization compared with the control group, as evinced by significantly lower numbers of isolated colonies (Figures 6C,D). These results were consistent with FC, GS, and FCGS recombinant protein antibody levels (Figure 5) and challenge protection efficiency (Table 1), indicating that the FC, GS, and FCGS recombinant proteins can cause specific immune responses in mice. Thus, FC immunization was effective against S. aureus, and GS immunization was effective against S. agalactiae infection, while FCGS immunization was effective toward challenges of both S. aureus and S. agalactiae.
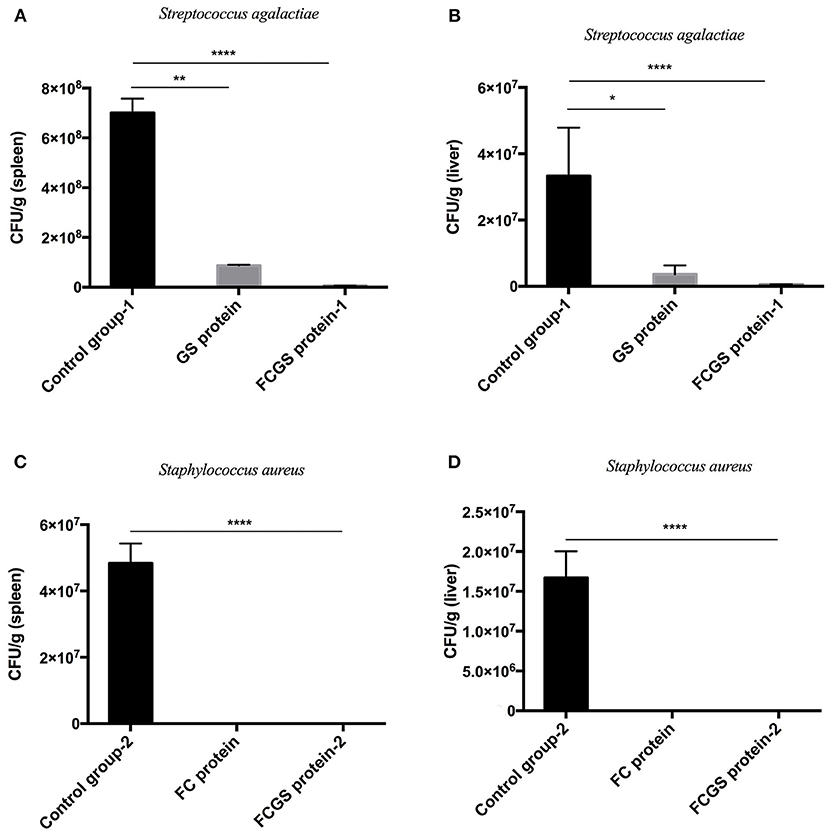
Figure 6. Bacterial loads in the spleens and livers of BALB/c mice after Staphylococcus aureus and Streptococcus agalactiae challenges. At 21 days after immunization with FC, GS, and FCGS proteins, mice were challenged with S. aureus and S. agalactiae. The spleens and livers of the mice were aseptically collected, homogenized, and spread on plates to count colonies. (A,B) After the challenge of S. agalactiae, the bacterial loads in the organs of the mice in the GS and FCGS groups are shown. (C,D) After the challenge of S. aureus, the bacterial loads in the organs of the mice in the FC and FCGS groups are shown. Data are shown as means ± SEM (*p < 0.05, **p < 0.01, ****p < 0.0001). Results represent three independent replicate experiments.
Discussion
Bioinformatics analysis, comparison of protein-associated information, and predictions of epitopes have become important methods in immunological research (28). Here, a signal peptide was observed at the N-terminus of the Sip protein, indicating that it is exported outside of the cell and may be related to bacterial cell wall function. Nevertheless, the mechanism mediating the association with cell walls has not been determined. FnBP, ClfA, GapC, and Sip proteins also were observed to exhibit multiple B cell epitopes, indicating that they can act as antigens to initiate immune response. Predictive analysis was used here to fuse the four genes of S. aureus FnBP and ClfA in addition to S. agalactiae GapC and Sip, thereby overcoming the low level of specific immunity induced by a single gene. Indeed, the recombinant fusion proteins FC, GS, and FCGS could stimulate the production of an immune response in mice. Further, immunizing mice with these recombinant proteins generated resistance to challenges with S. aureus and S. agalactiae. Thus, the FC, GS, and FCGS recombinant proteins identified here have potential value in mastitis vaccine development.
Adhesion is the first step during biofilm formation by pathogens or the invasion of host cells. Adhesion also protects bacteria from the host's immune system and promotes chronic infection. The primary mechanism by which S. aureus adheres to cells and invades non-specific phagocytes (FnBP-Fn-α5β1 integrin) is critical to target in the effective treatment of chronic staphylococcal infections (29). Castagliuolo et al. (30) used a mixture of pDNA encoding four adhesion hormones (ClfA, FnBPA, EfB, and Can) to intranasally immunize mice, with the treatment effectively reducing IMI caused by S. aureus. Delfani et al. (31) used a ClfA-ISDB-HLG recombinant protein to immunize BALB/C mice, wherein antibodies induced by the protein could promote enhanced phagocytosis of S. aureus by macrophages. Hall et al. (32) passively immunized mice with ClfA monoclonal antibodies, observing that this approach could prevent mastitis caused by S. aureus in mice. In addition, Song et al. (33) intramuscularly injected mice with XL1-Blue/LOG76 or XL1-Blue/LO11 strains in addition to recombinant GapC1−150. The immunized mice then resisted challenge with Streptococcus strains. Thus, the use of GapC1−150 on the surface of E. coli is a feasible vaccine approach against Streptococcus infection. El-Din et al. (34) immunized dairy cows with a DNA vaccine expressing ClfA plasmids, and the cows produced a strong specific antibody response against ClfA. Other studies have indicated that the Fc-Sip+Fc-FnBPB-ClfA dual subunit vaccine exhibits better therapeutic and preventive effects against S. agalactiae and S. aureus mastitis in dairy cows (35). Consequently, new anti-staphylococcal drugs are suitable vaccine agents and have been proposed based on structural models of ClfA–Fg interactions (36).
GapC chimeric proteins constructed from the non-conservative peptide regions of GapC from S. agalactiae and Shigella dysenteriae have been shown to retain the characteristics of the wild-type GapC protein of Streptococcus uberis (20). Further, other studies have reported that E. coli expression of recombinant Sips of Streptococcus can improve humoral immunity, while purified rSip can better induce the IgG anti-Sip immune responses and play an active role in reducing group B streptococcus (GBS) vaginal colonization (37). Chimeric CAMP (CAMP-3) of S. uberis and S. agalactiae can produce immune cross-reactive vaccine antigens that are more effective than any CAMP factor alone. In addition, inoculation of S. uberis several days after challenge can significantly reduce inflammation and provide protection (21). Herein, FC, GS, and FCGS recombinant proteins were obtained by expression and purification in bacteria, and their effective reactogenicity was confirmed by Western blotting analysis. Mice immunized with recombinant proteins were protected from bacterial irritation. Likewise, purified CP+Sip-FbsA conjugate mixed with aluminum salt adjuvant exhibited a stronger immunoprotective effect on Wistar rats infected with S. agalactiae during lactation (38). Importantly, the Sip subunit vaccine of S. agalactiae promotes the occurrence of humoral and local immune response in dairy cows and can reduce the number of somatic cells in milk (39).
The screening of the protective antigens from S. aureus including FnBP and ClfA in addition to S. agalactiae GapC and Sip provides a framework for the combined use of antigens with other virulence factors in the development of S. aureus and S. agalactiae vaccines. In particular, the FC, GS, and FCGS fusion proteins developed here have the potential to be used as vaccines against S. aureus or S. agalactiae infections. Among these, the FCGS fusion protein has the ability to simultaneously resist S. aureus and S. agalactiae infections, indicating that it is an important target for future studies. Both the FC and FCGS fusion proteins represent the Fnbp and ClfA genes, although the selected target fragments differ, implying that the same gene comprises different specific antigen fragments, and different combinations may alter the immune protection of recombinant proteins. It should also be noted that the results of this study are based on a limited number of mice, and although representative data were generated, a larger experimental population or an experiment with dairy cows is needed to verify the effectiveness of vaccines identified here.
Conclusion
FC, GS, and FCGS recombinant fusion proteins induced BALB/c mice to produce specific antibodies and provided effective protection against S. aureus and S. agalactiae infections. In particular, FCGS proteins conferred resistance to S. aureus and S. agalactiae and should be a key vaccine focus for future studies to mitigate mastitis.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Animal Care and Use Committee of Shihezi University.
Author Contributions
ZM, XY, and PW designed the study. ZM and PW were responsible for the development, integration, and writing of the manuscript. PW and CC reviewed the article. RH, YW, JY, and ZW provided help during the experiments. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Collaborative Innovation Project for Prevention and Control of High-incidence Zoonotic Infectious Diseases in Western China (2011 Plan).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.666098/full#supplementary-material
Supplementary Figure 1. FnBP, ClfA, GapC, and Sip protein signal peptide predictions. (A) FnBP protein signal peptide prediction. (B) ClfA protein signal peptide prediction. (C) Sip protein signal peptide prediction. (D) GapC protein signal peptide prediction. The predictions were generated with the SignalP 5.0 server (http://www.cbs.dtu.dk/services/SignalP/).
Supplementary Figure 2. FnBP, ClfA, GapC, and Sip protein B cell epitope predictions. (A) FnBP protein B cell epitope prediction. (B) ClfA protein B cell epitope prediction. (C) Sip protein B cell epitope prediction. (D) GapC protein B cell epitope prediction. The predictions were generated from http://imed.med.ucm.es/Tools/antigenic.pl.
Supplementary Figure 3. Three-dimensional structure prediction for FC, GS, and FCGS proteins. (A) FC protein structure prediction. (B) GS protein structure prediction. (C) FCGS protein structure prediction. The structures were generated with Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).
Supplementary Figure 4. Amino acid sequences of FC, GS, and FCGS proteins. (A) FC protein amino acid sequences. (B) GS protein amino acid sequences. (C) FCGS protein amino acid sequences.
Supplementary Figure 5. Purity identification of FC, GS, and FCGS proteins. The purified proteins was analyzed by SDS-PAGE to observe its purity. The pictures represent the results of three independent trials.
References
1. Halasa T, Huijps K, Østerås O, Hogeveen H. Economic effects of bovine mastitis and mastitis management: a review. Vet Q. (2007) 29:18–31. doi: 10.1080/01652176.2007.9695224
2. Zecconi A, Cesaris L, Liandris E, Dapr à V, Piccinini R. Role of several Staphylococcus aureus virulence factors on the inflammatory response in bovine mammary gland. Microb Pathogen. (2006) 40:177–83. doi: 10.1016/j.micpath.2006.01.001
3. Almeida A, Alves-Barroco C, Sauvage E, Bexiga R, Albuquerque P, Tavares F, et al. Persistence of a dominant bovine lineage of group B Streptococcus reveals genomic signatures of host adaptation. Environ Microbiol. (2016) 18:4216–29. doi: 10.1111/1462-2920.13550
4. Wang D, Liu L, Augustino S, Duan T, Hall T, MacHugh D, et al. Staphylococcus aureusIdentification of novel molecular markers of mastitis caused by using gene expression profiling in two consecutive generations of Chinese Holstein dairy cattle. J Anim Sci Biotechnol. (2020) 11:98. doi: 10.1186/s40104-020-00494-7
5. Hummerjohann J, Naskova J, Baumgartner A, Graber H. Enterotoxin-producing Staphylococcus aureus genotype B as a major contaminant in Swiss raw milk cheese. J Dairy Sci. (2014) 97:1305–12. doi: 10.3168/jds.2013-7643
6. Jørgensen H, Nordstoga A, Sviland S, Zadoks R, Sølverød L, Kvitle B, et al. Streptococcus agalactiae in the environment of bovine dairy herds–rewriting the textbooks? Vet Microbiol. (2016) 184:64–72. doi: 10.1016/j.vetmic.2015.12.014
7. Zhang L, Li Y, Bao H, Wei R, Zhou Y, Zhang H, et al. Population structure and antimicrobial profile of Staphylococcus aureus strains associated with bovine mastitis in China. Microb Pathogen. (2016) 97:103–9. doi: 10.1016/j.micpath.2016.06.005
8. Stalder U, Stephan R, Corti S, Bludau M, Maeschli A, Klocke P, et al. Short communication: Staphylococcus aureus isolated from colostrum of dairy heifers represent a closely related group exhibiting highly homogeneous genomic and antimicrobial resistance features. J Dairy Sci. (2014) 97:4997–5000. doi: 10.3168/jds.2013-7721
9. Shi D, Hao Y, Zhang A, Wulan B, Fan X. Antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis in China. Transboundary Emerging Dis. (2010) 57:221–4. doi: 10.1111/j.1865-1682.2010.01139.x
10. Brouillette E, Lacasse P, Shkreta L, Bélanger J, Talbot BG. DNA immunization against the clumping factor A (ClfA) of Staphylococcus aureus. Vaccine. (2002) 20:2348–57. doi: 10.1016/S0264-410X(02)00100-7
11. Hair P, Echague C, Sholl A, Watkins J, Geoghegan J, Foster T, et al. Clumping factor A interaction with complement factor I increases C3b cleavage on the bacterial surface of Staphylococcus aureus and decreases complement-mediated phagocytosis. Infect Immunity. (2010) 78:1717–27. doi: 10.1128/IAI.01065-09
12. Roche F, Downer R, Keane F, Speziale P, Park P, Foster T. The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J Biol Chem. (2004) 279:38433–40. doi: 10.1074/jbc.M402122200
13. Pietrocola G, Nobile G, Gianotti V, Zapotoczna M, Foster T, Geoghegan J, et al. Molecular interactions of human plasminogen with fibronectin-binding protein B (FnBPB), a fibrinogen/fibronectin-binding protein from Staphylococcus aureus. J Biol Chem. (2016) 291:18148–62. doi: 10.1074/jbc.M116.731125
14. Xiao-Juan J, Yong-Qing H, Ai-Rong Z, Xin F, Xiao-Jie C. Cloning and expression of the region A of clumping factor A gene of Staphylococcus aureus and its immunological characteristics. Chin Vet Sci. (2009) 7:65–70. doi: 10.16656/j.issn.1673-4696.2009.07.013
15. Dong-Yan S, Yong-Qing H, Ai-Rong Z. Expression of fibronectin-binding protein B (FnbpB) region D of Staphylococcus aureus and antiserum activity. Chin J Prevent Vet Med. (2010) 32:356–9. doi: 10.3724/SP.J.1011.2010.01385
16. Shkreta L, Talbot B, Diarra M, Lacasse P. Immune responses to a DNA/protein vaccination strategy against Staphylococcus aureus induced mastitis in dairy cows. Vaccine. (2004) 23:114–26. doi: 10.1016/j.vaccine.2004.05.002
17. Liu X, Hao Y, Wang S, Zhang A, Xu C. Assessment of immunogenicity of CP8, CLFA-FnBPB and CP8-CLFA-FnBPB antigens. Am J Anim Vet Sci. (2013) 8:134–41. doi: 10.3844/ajavsp.2013.134.141
18. Brodeur B, Boyer M, Charlebois I, Hamel J, Couture F, Rioux C, et al. Identification of group B streptococcal Sip protein, which elicits cross-protective immunity. Infect Immunity. (2000) 68:5610–8. doi: 10.1128/IAI.68.10.5610-5618.2000
19. Zhang L, Zhang H, Fan Z, Zhou X, Yu L, Sun H, et al. Identification of a conserved linear B-cell epitope of Streptococcus dysgalactiae GapC protein by screening phage-displayed random peptide library. PloS ONE. (2015) 10:e0131221. doi: 10.1371/journal.pone.0131221
20. Perez-Casal J, Prysliak T, Potter A. A GapC chimera retains the properties of the Streptococcus uberis wild-type GapC protein. Protein Exp Purific. (2004) 33:288–96. doi: 10.1016/j.pep.2003.09.011
21. Fontaine CM, Perez-Casal J, Song XM, Shelford J, Willson PJ, Potter AA. Immunisation of dairy cattle with recombinant Streptococcus uberis GapC or a chimeric CAMP antigen confers protection against heterologous bacterial challenge. Vaccine. (2002) 20:2278–86. doi: 10.1016/S0264-410X(02)00114-7
22. Kerro-Dego O, Prysliak T, Perez-Casal J, Potter A. Role of GapC in the pathogenesis of Staphylococcus aureus. Vet Microbiol. (2012) 156:443–7. doi: 10.1016/j.vetmic.2011.11.018
23. Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. (1990) 276:172–4. doi: 10.1016/0014-5793(90)80535-Q
24. Almagro Armenteros J, Tsirigos K, Sønderby C, Petersen T, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. (2019) 37:420–3. doi: 10.1038/s41587-019-0036-z
25. Nielsen H, Tsirigos K, Brunak S, von Heijne G. A brief history of protein sorting prediction. Protein J. (2019) 38:200–16. doi: 10.1007/s10930-019-09838-3
26. Ma Z, Li R, Hu R, Deng X, Xu Y, Zheng W, et al. Brucella abortus BspJ is a nucleomodulin that inhibits macrophage apoptosis and promotes intracellular survival of Brucella. Front Microbiol. (2020) 11:599205. doi: 10.3389/fmicb.2020.599205
27. Wu P, Zhang Y, Yin X, He Y, Zhang Q, Chen C. Layered double hydroxide nanoparticles as an adjuvant for inactivated foot-and-mouth disease vaccine in pigs. BMC Vet Res. (2020) 16:474. doi: 10.1186/s12917-020-02689-6
28. Blythe M, Flower D. Benchmarking B cell epitope prediction: underperformance of existing methods. Protein Sci. (2005) 14:246–8. doi: 10.1110/ps.041059505
29. Josse J, Laurent F, Diot A. Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Front Microbiol. (2017) 8:2433. doi: 10.3389/fmicb.2017.02433
30. Castagliuolo I, Piccinini R, Beggiao E, Pal ù G, Mengoli C, Ditadi F, et al. Mucosal genetic immunization against four adhesins protects against Staphylococcus aureus-induced mastitis in mice. Vaccine. (2006) 24:4393–402. doi: 10.1016/j.vaccine.2006.02.055
31. Delfani S, Imani Fooladi A, Mobarez A, Emaneini M, Amani J, Sedighian H. In silico analysis for identifying potential vaccine candidates against Staphylococcus aureus. Clin Exp Vaccine Res. (2015) 4:99–106. doi: 10.7774/cevr.2015.4.1.99
32. Hall A, Domanski P, Patel P, Vernachio J, Syribeys P, Gorovits E, et al. Characterization of a protective monoclonal antibody recognizing Staphylococcus aureus MSCRAMM protein clumping factor A. Infect Immun. (2003) 71:6864–70. doi: 10.1128/IAI.71.12.6864-6870.2003
33. Song B, Yang X, Sun H, Yu L, Ma J, Wu Z, et al. Immunogenicity of amino acids 1-150 of Streptococcus GapC displayed on the surface of Escherichia coli. Microb Pathogen. (2017) 105:288–97. doi: 10.1016/j.micpath.2017.02.003
34. El-Din ANN, Shkreta L, Talbot BG, Diarra MS, Lacasse P. DNA immunization of dairy cows with the clumping factor A of Staphylococcus aureus. Vaccine. (2006) 24:1997–2006. doi: 10.1016/j.vaccine.2005.11.033
35. Li S, Zhou Y, Lv T, Du L, Guo T, Hao Y. Immunological study of Streptococcus agalactia Fc-Sip and Staphylococcus aureus Fc-FnBPB-ClfA subunit vaccine against dairy cow mastitis. Chin J Prevent Vet Med. (2020) 42:70–6. doi: 10.3969/j.issn.1008-0589.201807004
36. Ganesh V, Rivera J, Smeds E, Ko Y, Bowden M, Wann E, et al. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathogens. (2008) 4:e1000226. doi: 10.1371/journal.ppat.1000226
37. Díaz-Dinamarca D, Jerias J, Soto D, Soto J, Díaz N, Leyton Y, et al. The optimisation of the expression of recombinant surface immunogenic protein of group B Streptococcus in Escherichia coli by Response surface methodology improves humoral immunity. Mol Biotechnol. (2018) 60:215–25. doi: 10.1007/s12033-018-0065-8
38. Du L, Lv T, Zhao H, Li S, Zhou X, Cui J, et al. The immune effect of Streptococcus agalactiae recombinant CP+Sip-FbsA fusion protein on S. agalactiae induced mouse mastitis. Chin J Prevent Vet Med. (2016) 38:976–80. doi: 10.3969/j.issn.1008-0425.2016.12.12
Keywords: Staphylococcus aureus, Streptococcus agalactiae, FnBP+ClfA+GapC+Sip gene, recombinant fusion expression, challenge protection experiment
Citation: Ma Z, Yin X, Wu P, Hu R, Wang Y, Yi J, Wang Z and Chen C (2021) The Recombinant Expression Proteins FnBP and ClfA From Staphylococcus aureus in Addition to GapC and Sip From Streptococcus agalactiae Can Protect BALB/c Mice From Bacterial Infection. Front. Vet. Sci. 8:666098. doi: 10.3389/fvets.2021.666098
Received: 09 February 2021; Accepted: 18 May 2021;
Published: 24 June 2021.
Edited by:
Shao-Lun Zhai, Guangdong Academy of Agricultural Sciences, ChinaReviewed by:
Mohanned Naif Alhussien, Technical University of Munich, GermanyDiego A. Díaz-Dinamarca, Instituto de Salud Pública de Chile, Chile
Alexander Suvorov, Institute of Experimental Medicine (RAS), Russia
Copyright © 2021 Ma, Yin, Wu, Hu, Wang, Yi, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Wu, 995176509@qq.com; Chuangfu Chen, chuangfu_chen@163.com
†These authors have contributed equally to this work
 Zhongchen Ma1,2,3†
Zhongchen Ma1,2,3† Chuangfu Chen
Chuangfu Chen